Abstract
Attachment of oligodeoxynucleotides (ODNs) containing benzaldehyde (BAL) groups to semicarbazide-coated glass (SC-glass) slides is described. 5′-BAL-ODNs are prepared using automated DNA synthesis and an acetal-protected BAL phosphoramidite reagent. The hydrophobic protecting group simplifies purification of BAL-ODNs by reverse phase HPLC and is easily removed using standard acid treatment. The electrophilic BAL-ODNs are stable in solution, but react specifically with semicarbazide groups to give semicarbazone bonds. Glass slides were treated with a semicarbazide silane to give SC-glass. BAL-ODNs are coupled to the SC-glass surface by a simple one-step procedure that allows rapid, efficient and stable attachment. Hand-spotted arrays of BAL-ODNs were prepared to evaluate loading density and hybridization properties of immobilized probes. Hybridization to radiolabeled target strands shows that at least 30% of the coupled ODNs were available for hybridization at maximum immobilization density. The array was used to probe single nucleotide polymorphisms in synthetic DNA targets, and PCR products were correctly genotyped using the same macroarray. Application of this chemistry to manufacturing of DNA microarrays for sequence analysis is discussed.
INTRODUCTION
Hybridization of nucleic acids to microarrays prepared from DNA immobilized on solid supports has become a fundamental technique for detection, isolation, quantitation and analysis of nucleic acid sequences (1). Immobilized cDNA arrays are commonly used for gene expression analysis (2), but synthetic oligodeoxynucleotides (ODNs) are increasingly being used (3,4) since structure, quality and hybridization performance are more easily controlled. Attachment of cDNA by UV or thermal crosslinking can give variable results due to probe damage or conformational restriction of the DNA strands near the multiple attachment points. Some assay formats require specific end-point attachment that only synthetic ODNs can provide. In particular, techniques such as screening of genetic polymorphisms (5), DNA sequencing by nucleic acid hybridization (6) or minisequencing (7) rely upon simple and highly reproducible methods for manufacturing ODN microarrays. The success of DNA microarray technology depends on two major issues: feasibility of array fabrication and hybridization performance of arrayed ODNs.
There are two distinct approaches to manufacturing ODN arrays. In one approach (in situ synthesis) ODN probes are synthesized directly on the surface using photolithographic techniques and modified ODN synthesis chemistry (8), conventional ODN synthesis chemistry in conjunction with ink jet printing (3), or microfluidic channeling techniques (9). This approach provides wide flexibility in design of custom arrays without preliminary efforts in synthesis and maintenance of a vast library of modified ODNs. On the other hand in situ synthesis does not allow quality control and purification of generated features, and the less efficient phosphoramidite monomer coupling gives poor yields for longer ODN probes.
The other approach to array manufacturing (immobilization) uses pre-synthesized ODNs containing end group modifications that react with functional groups on the solid support. The ability to purify full-length products before arraying is a major advantage of immobilization. Several solid supports are currently used for preparation of DNA arrays, but glass microscope slides are the most popular substrate due to their availability, physical and chemical inertness, low intrinsic fluorescence and well-developed methods for glass modification (10). Immobilization chemistry must be specific, fast and provide stable chemical bonds. Large-scale, high-throughput manufacturing of ODN arrays also requires that the chemistry is simple and reproducible for both surface and modified ODN preparation. Shelf-life of the modified surface and modified ODNs is another important prerequisite. A comparison of immobilization chemistries for preparation of ODN microarrays was recently published (11).
Most immobilization chemistry for preparation of ODN microarrays involves reaction of electrophilic glass surfaces with nucleophilic ODNs. DNA synthesis reagents for preparation of alkylamine-modified ODNs are commercially available and provide a convenient nucleophilic linker. Electrophilic glass coatings such as epoxysilane (12) or phenylisothiocyanate (13) have been prepared by direct silylation, but chemical instability of the reactive molecular monolayer can give variation in performance. A commercial slide (3D-Link; Motorola) is available that has N-hydroxysuccinimide ester groups on the surface, but in our hands performance was variable. In general, glass surfaces with strongly electrophilic groups have poor shelf-life and are difficult to control. Glass supports coated with milder electrophiles such as aliphatic aldehydes or carboxylic acids have also been used for preparation of microarrays (14). Although these supports are more stable, reaction of alkylamine-modified ODNs with aldehyde-modified supports requires post-arraying reduction of the unstable Schiff base linkages with sodium borohydride. Use of carboxylic acid supports requires use of carbodiimide coupling agents. More commonly, amine-modified slides are activated with homo- or hetero-bifunctional coupling agents to provide an electrophilic surface just prior to arraying (5,15). Use of coupling agents is difficult for researchers since the conditions are difficult to reproduce and reagents are hazardous.
There have been few reports of ODNs with electrophilic groups being used for immobilization to nucleophilic solid supports, presumably because of the poor stability of the electrophilic ODNs. Cyanuric chloride-modified ODNs have been coupled to amine-modified nylon beads, but these electrophilic ODNs required a post-DNA synthesis coupling step and degraded on drying (16). ODNs bearing aliphatic aldehydes have been used for coupling to hydrazide-modified latex beads, but the electrophilic aldehyde groups were generated after DNA synthesis using periodate oxidation of a vicinal diol (17). In addition, aliphatic aldehydes undergo side reactions and a borohydride reduction step was required to obtain stable linkages.
In this paper, we present our studies on covalent attachment of benzaldehyde-modified ODNs (BAL-ODNs) to semicarbazide-modified glass (SC-glass) to generate semicarbazone linkages (Fig. 1). These bonds were stable to typical hybridization conditions without the need for borohydride reduction. The mildly electrophilic BAL-ODNs were easily prepared by automated DNA synthesis, were stable in solution, and were easily stored and handled. Derivatized triethoxysilane was used to introduce semicarbazide groups onto the glass surface via conventional silylation. The SC-glass slides were easily prepared and the slide surface did not suffer from shelf-life problems. 32P-labeled ODNs and hand-spotted ‘macroarrays’ were used to explore properties of the immobilized ODNs. Reaction kinetics and pH dependence of ODN immobilization were examined. The reactive glass surface was blocked after immobilization, and hybridization properties were studied. Short (14–15mer) BAL-ODN probes were used to discriminate single mismatches in 32P-labeled 36mer ODN targets. A macroarray of six BAL-ODN probes was used to successfully genotype a complex single-stranded PCR product.
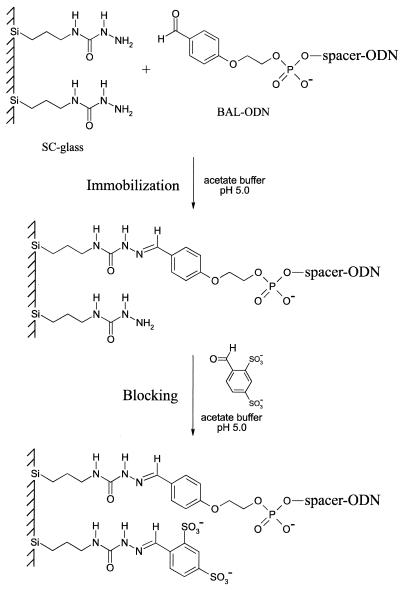
Figure 1.
Immobilization of BAL-ODNs to SC-glass. BAL-ODN is spotted on an SC-glass slide in an acidic buffer and incubated at 37°C for 3 h in a humid chamber. During blocking excess BAL-ODN is removed and unreacted semicarbazide residues are blocked with negatively charged groups. The blocking step prevents spots from smearing and reduces background during hybridization.
MATERIALS AND METHODS
Chemicals and reagents
Chemical reagents were obtained from commercial suppliers and used without further purification. Moisture-sensitive reactions were performed under an argon atmosphere. Analytical thin-layer chromatography (TLC) was conducted on silica gel 60 F-254 aluminum-backed plates (EM Science). For flash chromatography, silica gel 60 Geduran 35–75 µm (EM Science) was employed. NMR spectra were recorded at 300 MHz on a Varian Gemini-300 instrument. Chemical shift values are expressed in δ (parts per million) relative to tetramethylsilane as an internal standard. MALDI-TOF mass spectrometry of modified ODNs was performed by Midland Certified Reagent Company, Inc. (Midland, TX).
2-[4-(5,5-Diethyl-1,3-dioxan-2-yl)phenoxy]ethan-1-ol (1)
A solution of 4-(2-hydroxyethoxy)benzaldehyde (18) (4.0 g, 24 mmol) and 2,2-diethyl-1,3-propanediol (4.0 g, 30 mmol) in 30 ml of anhydrous methylene chloride was prepared. Boron trifluoride diethyl etherate (3.0 ml, 24 mmol) was added and the reaction was kept at room temperature for 15 min. TLC analysis (hexanes/EtOAc, 1:1) with 2,4-dinitrophenylhydrazine development showed ∼75% completion. Triethylamine (10 ml) was added to quench the reaction and the solution was concentrated under reduced pressure. The resultant oil was applied to a silica gel column (5 × 30 cm), eluting with hexanes/EtOAc (1:1). Concentration of the pure product fractions afforded 4.55 g (68%) of acetal 1 as an oil that slowly crystallized upon drying (m.p. 58–60°). 1H NMR (DMSO-d6): δ 7.31 (d, J = 8 Hz, 2H), 6.90 (d, J = 8 Hz, 2H), 5.34 (s, 1H), 4.85 (br s, 1H), 3.97 (t, J = 5 Hz, 2H), 3.83 (s, 1H), 3.80 (s, 1H), 3.70 (t, 2H), 3.57 (s, 1H), 3.51 (s, 1H), 1.71 (q, J = 7.5 Hz, 2H), 1.08 (q, J = 7.6 Hz, 2H), 0.82 (t, J = 7.6 Hz, 3H), 0.75 (t, J = 7.7 Hz, 3H). 13C NMR (DMSO-d6): 158.79, 131.14, 127.43, 113.79, 100.91, 73.73, 69.46, 59.51, 34.19, 23.68, 22.21, 7.39, 6.48.
BAL-phosphoramidite: 3-({2-[4-(5,5-diethyl(1,3-dioxan-2-yl))phenoxy]ethoxy}[bis(methylethyl)amino]phosphinooxy)propanenitrile (2)
A solution of 1 (3.1 g, 11.0 mmol) and 10 ml of ethyldiisopropylamine in 40 ml of anhydrous CH2Cl2 was prepared and 2-cyanoethyldiisopropylchlorophosphoramidite (2.7 g, 11.4 mmol) was added dropwise. After stirring at room temperature for 1 h the solution was concentrated under reduced pressure to an oily residue. The residue was purified by flash chromatography on neutralized silica gel (triethylamine/EtOAc) eluting with hexanes/EtOAc (3:1) to afford 4.1 g (77%) of the desired phosphoramidite 2 as a colorless oil. This material slowly solidified upon drying in vacuum followed by freezing at –20°C. 1H NMR (DMSO-d6): δ 7.31 (d, J = 8.7 Hz, 2H), 6.91 (d, J = 8.7 Hz, 2H), 5.34 (s, 1H), 4.11 (t, J = 5 Hz, 2H), 3.90 (m, 2H), 3.83 (s, 1H), 3.80 (s, 1H), 3.73 (m, 2H), 3.59 (m, 2H), 3.56 (s, 1H), 3.52 (s, 1H), 2.75 (t, J = 6 Hz, 2H), 1.71 (q, J = 7.5 Hz, 2H), 1.35 (dd, J1 = 6 Hz, J2 = 3 Hz, 6H), 1.07 (q, J = 7.6 Hz, 2H), 0.82 (t, J = 7.6 Hz, 3H), 0.75 (t, J = 7.7 Hz, 3H). 31P NMR (DMSO-d6): δ148.00 (s).
SC-silane: 3-(4-semicarbazido)propyltriethoxysilane (3)
Anhydrous hydrazine (32 ml, 1 mol) was dissolved in 30 ml of anhydrous acetonitrile. 3-Isocyanatopropyltriethoxysilane (25 ml, 0.1 mol) was added dropwise with vigorous stirring. The reaction mixture was stirred for 1 h at room temperature and the solvent was removed in vacuum (caution: distillation of anhydrous hydrazine in air can lead to an explosion). The oily residue was dissolved in anhydrous ethanol and the solution was evaporated under reduced pressure. The last step was repeated twice to remove unreacted hydrazine. The resulting viscous residue was dried in vacuum overnight to afford 27 g (96% yield) of the desired product as a colorless oil that was used without further purification. 1H NMR (DMSO-d6): δ 6.82 (s, 1H), 6.32 (t, J = 5.4 Hz, 1H), 4.05 (br s, 2H), 3.72 (q, J = 7 Hz, 6H), 2.96 (q, J = 6.6 Hz), 1.41 (m, 2H), 1.26 (t, J = 6.9 Hz, 9H), 0.49 (m, 2H).
Silylation of glass slides
Glass slides were silylated according to a standard procedure described below. Pre-cleaned microscope slides (Corning Life Sciences, Corning, NY) were treated with 1 N nitric acid for 1 h at room temperature and then rinsed with running deionized water followed by anhydrous ethanol wash. The slides were then immersed in a 1% solution of SC-silane 3 in 95% ethanol for 30 min. The slides were washed with 95% ethanol for 5 min, twice with acetonitrile, 5 min per wash, and finally with diethyl ether. The slides were then cured for 45 min at 110°C. The coated slides containing semicarbazidopropyl functional groups can be stored for at least a month on a benchtop in a dust-proof container without noticeable loss of activity.
DNA synthesis
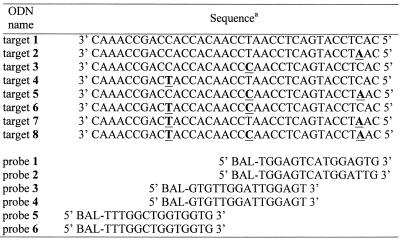
All ODNs were synthesized on an ABI 394 DNA synthesizer using standard phosphoramidite chemistry. A polyethylene glycol spacer (Spacer 18; Glen Research, Sterling, VA) was introduced between the BAL terminus and the 5′ end of all the probes. BAL-phosphoramidite 2 was added using standard coupling time and conditions. The hydrophobic acetal-protected BAL-ODNs were purified by reverse-phase HPLC using standard methods and dried in vacuo. The acetal protecting group was removed by treatment with 80% acetic acid at room temperature for 1 h. Concentrations were determined by UV spectrophotometry at 260 nm using extinction coefficients calculated using a nearest neighbor model (19). Sequences of all ODN probes and targets used in this study are listed in Table 1.
Table 1. Sequences of oligodeoxynucleotide targets and probes used in this study.
aPositions of nucleotide substitutions are underlined. Sequences of targets and probes have reversed orientation and are aligned relative to their binding sites.
Preparation and aqueous stability of BAL-ODN semicarbazone conjugates
A solution of 50 nmol of probe 1 in 50 µl of 100 mM sodium acetate (pH 5.0) was treated with 50 µl of a 1 M solution of 4-benzyl-semicarbazide (20) in DMSO. After incubation for 2 h at room temperature the reaction mixture was precipitated by adding 1 ml of 2% LiClO4 in acetone. The precipitate was separated by centrifugation, washed with 1.5 ml of acetone and dried in vacuum. The longer retention time semicarbazone conjugate was isolated by C18 HPLC. Usually >90% conversion was observed. Probe 1 and the corresponding semicarbazone conjugate were analyzed by MALDI-TOF mass spectrometry. Probe 1: calculated mass, 5260.3; observed mass, 5260.5. Semicarbazone conjugate of probe 1: calculated mass, 5407.4; observed mass, 5408.3.
A 0.1 mM solution of the conjugate in 100 mM HEPES (pH 7.5) was incubated at 65°C and aliquots were analyzed by C18 HPLC at 0, 4 and 24 h. HPLC used a 4.6 × 150 mm Microsorb C18 column (Varian, Walnut Creek, CA) and a gradient of 0–50% acetonitrile in 0.1 M TEAA over 20 min (flow rate = 1 ml/min).
3′-Radiolabeling of BAL-ODN probe 1
Probe 1 was radiolabeled at the 3′ terminus using [α-32P]ddATP (NEN, Boston, MA) and terminal deoxynucleotidyl transferase (Promega, Madison, WI). Briefly, 1.2 nmol of ODN and 100 µCi of radiolabeled triphosphate were reacted using conditions specified by the manufacturer. The labeled probe was purified using a NENSORB™ 20 cartridge (NEN, Boston, MA). The eluent containing labeled probe was dried in vacuo and dissolved in 100 µl of 100 mM sodium acetate buffer (pH 5.0) containing 9 nmol of unlabeled probe 1 to give 100 µM final concentration. This stock solution was used for the quantitative experiments described below. Immobilized probe was quantified by phosphorimaging using a Bio-Rad GS-250 Molecular Imager. The data from the phosphorimager were converted to fmol/spot by comparing with standard curves generated from a serial dilution of known amounts of the same labeled probes spotted on a microscope slide and dried without washing.
General procedure for immobilization of BAL-ODN probes
Probe 1 was dissolved in 100 mM sodium acetate buffer (pH 5.0) at the indicated concentration and spotted manually with a 2 µl Pipetteman on the derivatized slide as 0.5 µl droplets following a grid pattern on a wet filter paper template underneath the slide. Slides were incubated at 37°C in a covered 10 cm Petri dish located in a humid container for 1–5 h. To block unreacted semicarbazide groups on the glass surface the slides were treated with a 100 mM solution of 5-formyl-1,3-benzenedisulfonic acid disodium salt (Aldrich, Milwaukee, WI) in 100 mM sodium acetate buffer (pH 5.0) for 1 h at 37°C in a 50 ml polypropylene centrifuge tube. The slides were then rinsed with water, washed for 30 min at 37°C with 30% methanol in 0.5 M sodium phosphate buffer (pH 7.0), followed by a 30 min wash in 5× SSPE, 0.1% Triton X-100 at the same temperature. The slides were rinsed thoroughly with water, dried at room temperature, and were ready for use in hybridization experiments.
Effect of pH on BAL-ODN immobilization
Probe 1 (32P-labeled) was immobilized as described above using a standard time of 3 h. Three different buffers were used to study the effect of concentration or pH on immobilization efficiency. Buffer A: 100 mM sodium acetate, pH 5.0; buffer B: 100 mM sodium acetate, pH 6.0; buffer C: 100 mM HEPES, pH 8.0. After blocking and washing as usual, the amount of immobilized probe in each spot was measured by phosphorimaging.
Determination of BAL-ODN loading efficiency
Serial dilution of the 32P-labeled probe 1 stock solution was made using the pH 5 buffer with 2-fold decrements. Probe 1 solution (0.5 µl) at various concentrations was applied in quadruplicate to an SC-glass slide and allowed to react at 37°C for 3 h. The glass surface was blocked and washed as described above, and the amount of immobilized probe in each spot was measured by phosphorimaging. Specificity of reaction at the BAL residues was determined by immobilizing a 5′-32P-labeled C15 ODN (no BAL) as described above. Radioactive background was measured after standard blocking and washing.
Determination of hybridization efficiency
To determine hybridization efficiency of attached BAL-ODN probes, non-labeled probe 1 was immobilized using the general procedure described above. A 2.4 × 5.0 cm cover slip was positioned over the spotted area using 0.2-mm-thick spacers made of electrical tape. 36mer complementary target 1 was 32P-labeled at the 5′ end using T4 polynucleotide kinase, [γ-32P]ATP and standard conditions (21). Hybridization mixture (80–100 µl) (1 µM 32P-labeled target 1, 5× SSPE, 0.1% Triton X-100) was applied by capillary action between the slide and the cover slip. The slide was incubated overnight at 37°C in a closed Petri dish over wet Whatman 3MM paper in a humid container to prevent evaporation of the hybridization solution. Two 30 min washes were performed at room temperature on an orbital shaker using 25 ml of hybridization buffer per slide. The level of hybridization was quantified by phosphorimaging as described above.
PCR conditions and DNA strand separation
Female and male human genomic DNA samples were obtained from the Coriell Institute of Medical Research (NIGMS Human Genetic Mutant Cell Repository, Camden, NJ). The 235 bp amelogenin gene fragment corresponding to exon 3 was amplified by PCR using a set of primers, 5′-GCTGCACCACCAAATCATCCC-3′ and 5′-biotin-CTGGTGAGGCTGTGGCTGAAC-3′. The amplification reaction (100 µl) contained 50 mM KCl, 10 mM Tris–HCl (pH 8.3), 1.5 mM MgCl2, 0.001% gelatin, 100 ng DNA, 1 µM each primer, 200 µM each dATP, dCTP, dTTP and dGTP, and 2.5 U of JumpStart™ Taq DNA polymerase (Sigma, St Louis, MO). PCR was performed in a Stratagene RoboCycler Gradient 40 Temperature Cycler (Stratagene, La Jolla, CA) using 35 cycles (95°C for 1 min, 65°C for 1 min, 72°C for 1 min).
PCR products were purified by 4% non-denaturing PAGE and eluted from the gel. One DNA strand of PCR products derived from the non-biotinylated primer was 5′ end labeled using [γ-32P]ATP and T4 polynucleotide kinase. This labeled strand was separated using streptavidin-coupled magnetic beads (Dynabeads M-280; Dynal, Inc., Lake Success, NY) according to the manufacturer’s instructions. Labeling reaction mixture (50 µl) containing 1–2 µg of PCR product was diluted twice with 2× B&W buffer [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 2 M NaCl] and added to 1 mg of pre-washed Dynabeads. The suspension was incubated at room temperature for 15 min with occasional mixing. Beads were separated using a magnet, washed three times with 100 µl of B&W buffer and treated with 75 µl of 0.1 N NaOH to denature the DNA strands. The mixture was incubated at room temperature for 5 min, supernatant was collected, and alkaline treatment was repeated one more time. Combined supernatants were neutralized with an equal volume of 0.1 N HCl and ethanol precipitated. Specificity of amplification was confirmed by Maxam–Gilbert sequencing (22).
Hybridization of BAL-ODN arrays to short ODN or single-stranded PCR targets
Hybridization of ODN arrays consisting of BAL-ODN probes 1–6 spotted in triplicate with 5′-32P-labeled 36mer ODN targets 1–8 was accomplished as described above, except for the hybridization time, which was reduced to 3 h. After hybridization, slides were washed for 15 min at room temperature in hybridization buffer (5× SSPE, 0.1% Triton X-100). Slides were then washed twice for 15 min at 42°C in 0.5× SSPE, 0.1% Triton X-100. In some cases an additional 15 min wash was necessary to improve mismatch discrimination. Finally, the slides were air dried and analyzed by phosphorimaging. For hybridizion of single-stranded PCR products to the ODN array, the concentration of target DNA was 10–20 nM. Overnight hybridization at 37°C was followed by a 15 min wash in hybridization buffer, and two 15 min washes at 37°C in 0.5× SSPE, 0.1% Triton X-100.
RESULTS AND DISCUSSION
The reaction between semicarbazide-modified ligands and carbonyl groups on biomolecules is a common method for preparation of protein conjugates (23), but has been little used for nucleic acid conjugates. Aliphatic aldehydes have been generated in ODNs by periodate oxidation of a ribonucleotide residue and conjugated to poly-l-lysine for use as DNA delivery agents (24). Oxidation of other diol residues has also been described to generate aliphatic aldehydes in ODNs for reaction to hydrazide-coated beads (17). For these nucleic acid applications, reduction of the Schiff base by treatment with sodium borohydride was required to give a stable alkylamine bond. ODNs containing aliphatic aldehydes are also unstable and prone to side reactions and are therefore not commonly used. High-throughput manufacturing of ODN arrays using aliphatic aldehyde-modified ODNs would be costly and labor intensive. The immobilization strategy we chose uses BAL-modified ODNs and semicarbazide-coated glass as illustrated in Figure 1. BAL-ODNs are easily prepared using standard DNA synthesis methods and stable to standard ODN handling and storage conditions. Post-immobilization reduction of the stable semicarbazone is not required for hybridization assays. Many immobilization methods reported in the literature suffer from high background during the hybridization step due to the inherent affinity of DNA to glass (25). Here we used a disulfonated BAL blocking group to cap unreacted semicarbazide residues and decrease non-specific background (Fig. 1).
Reagents for ODN and glass surface modification
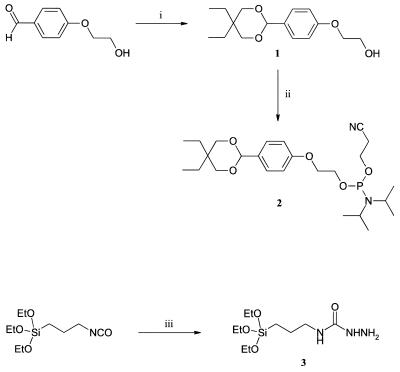
In order to implement this immobilization chemistry in ODN array production we developed new reagents for modification of both glass surfaces and ODNs. To introduce benzaldehyde groups into ODNs, we synthesized BAL phosphoramidite reagent 2 (Fig. 2) where the BAL moiety is protected by a cyclic acetal group. Use of a symmetrical diol to form the acetal gave intermediate 1 as a single stereoisomer. Other acetal structures were explored (data not shown) but this structure provided several advantages. First of all, the hydrophobic protecting group in 2 was optimized to allow ‘acetal-ON’ (very similar to trityl-ON) purification of the BAL-ODNs by reverse phase HPLC or disposable cartridges. Unsymmetrical diols gave diastereomer products after phosphitylation that complicated purification of both the phosphoramidite reagent and ODNs. Finally, the 1,3-dioxanyl protecting group was readily hydrolyzed under standard DNA synthesis detritylation conditions (80% acetic acid). BAL phosphoramidite 2 allows direct introduction of BAL groups during automated DNA synthesis. DNA coupling yields were not measured directly, but overall yield of synthetic probes was not compromised. HPLC purification and deprotection conditions use standard methods. To our knowledge this is the first report of a method for direct introduction of a protected aldehyde modification into ODNs during DNA synthesis. Unlike other electrophilic ODNs, BAL-ODNs can be handled like unmodified ODNs and do not suffer from degradation upon precipitation or drying. Solutions of BAL-ODNs are stable on the benchtop and can be stored frozen for months with no loss of reactivity.
Figure 2.
Preparation of BAL-phosphoramidite (2) and SC-silane (3). Reagents and conditions: (i) 2,2-diethyl-1,3-propanediol, Et2O·BF3, CH2Cl2, room temperature, 15 min; (ii) 2-cyanoethyldiisopropylchlorophosphoramidite, DIEA, CH2Cl2, room temperature, 1 h; (iii) hydrazine, CH3CN, room temperature, 1 h.
SC-glass slides were prepared using semicarbazidopropyltriethoxysilane (SC-silane 3). SC-silane was easily prepared by the reaction of commercially available isocyanopropyltriethoxysilane with hydrazine, and suitable purity was obtained by simply removing excess hydrazine in vacuo. The pre-washed glass slides were treated with a 1% solution of SC-silane in 95% EtOH for 30 min, washed, dried and cured at 110°C. Silylation in organic solution presumably gives the desired molecular monolayer surface whereas aqueous methods can give polymeric siloxanes (10). SC-glass slides were kept in a slide box at room temperature in a dust-free environment. No special precautions such as vacuum, dry or inert atmosphere were necessary for storing the slides.
Aqueous stability of BAL-ODN semicarbazone conjugates
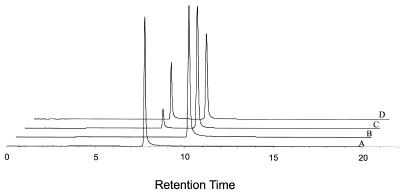
5′-BAL-ODN probe 1 (Table 1) was used for immobilization experiments described below. To explore the aqueous stability of the semicarbazone linkage, probe 1 was reacted with 4-benzyl-semicarbazide (20) to prepare a model conjugate. Probe 1 reacted at pH 5 to give the desired semicarbazone conjugate in >90% yield. This shows that the 5′-BAL was present in probe 1 and fully reactive. The molecular weight of probe 1 and the corresponding semicarbazone conjugate were confirmed by MALDI-TOF mass spectrometry. Stability of the semicarbazone conjugate was analyzed using C18 HPLC. Figure 3 shows that the half-life in neutral buffer at 65°C is >24 h. This is sufficiently stable for most hybridization applications including PCR on the slide. If necessary, the semicarbazone bond can be reduced by sodium borohydride to create an alkyl-substituted semicarbazide linkage that is even more stable.
Figure 3.
Aqueous stability of semicarbazone conjugates. C18 HPLC chromatograms of (A) BAL-ODN probe 1; (B) model semicarbazone conjugate of probe 1 and 4-benzyl-semicarbazide; (C) same semicarbazone conjugate after incubation in 100 mM HEPES, pH 7.5 for 4 h and (D) 24 h at 65°C.
Immobilization of ODNs to SC-glass surfaces
In order to optimize the immobilization chemistry, effects of pH, BAL-ODN concentration, reaction time and temperature on the density of the oligonucleotide attachment were studied. BAL-ODN probe 1 was 32P radiolabeled at the 3′-terminus to provide a method for quantitative analysis of probe immobilization. Use of radiolabeling avoids certain complications caused by self-quenching of fluorescently labeled ODNs at high concentrations (8). Hand-spotted ‘macroarrays’ were used for initial studies to avoid variables associated with microarraying small volumes of ODNs. The immobilization reaction was performed in a humid Petri dish that was placed in another humid chamber to ensure that the microdrops remained hydrated. After the immobilization reaction was complete, the slide with spotted probes was immediately immersed into the blocking solution to prevent unreacted ODNs from migrating to surrounding areas of a slide. This resulted in good spot morphology and generates a negatively charged slide surface that prevented non-specific adsorption of DNA during hybridization. Similar blocking procedures are used in preparation of cDNA arrays on poly-l-lysine where succinic anhydride is used to block residual amine groups (2). The highly anionic aromatic disulfonate blocking group may have advantages in other immobilization methods.
We performed a control immobilization to demonstrate that the BAL group was reacting specifically with semicarbazide groups on the glass surface. A dC15 oligo (no BAL) was 5′ labeled with 32P and spotted, blocked and washed as usual. We conjectured that semicarbazide residues may form adducts with dC since a similar reaction of hydrazine with the C6 position of dC forms the basis of Maxam–Gilbert sequencing (22). Less than 5% of this control oligo remained bound to the glass surface after blocking, indicating that the BAL group is required for immobilization and there is little non-specific binding of ODNs to the SC-glass.
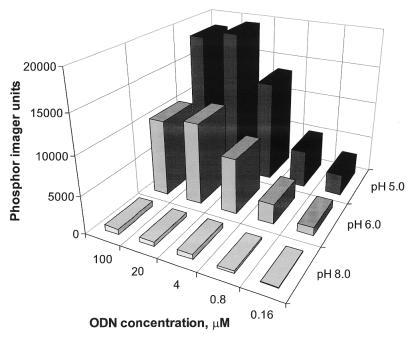
Effect of BAL-ODN concentration and pH on immobilization
The efficiency of BAL-ODN attachment to SC-glass slides was tested using reaction buffers with different pH (Fig. 4). The goal of these studies was to find immobilization conditions that were rapid and would provide high probe densities. Solutions of 3′-32P-labeled probe 1 in three buffers were prepared, and varying concentrations were spotted to SC-glass as usual. At 3 h, each slide was blocked, washed and phosphorimaged. The results clearly indicate that at lower pH, higher probe density is achieved. This is consistent with acid-catalyzed Schiff base formation and has been observed for preparation of protein semicarbazone conjugates (23). The pH 5 acetate buffer was chosen for further experiments. The probe density reaches maximum capacity at a concentration of ∼20 µM. Immobilization at pH 4 showed comparable performance (data not shown).
Figure 4.
pH dependence of BAL-ODN immobilization to SC-glass slide. Radiolabeled probe 1 was diluted in three buffers of different pH ranging from 8.0 to 5.0 and deposited on the slide as 0.5 µl droplets. After incubation, blocking and washing the immobilized probe was quantified by phosphorimaging. The average spot size had a diameter of 1.5 mm. Values are presented as phosphorimager units per spot (an average of four spots).
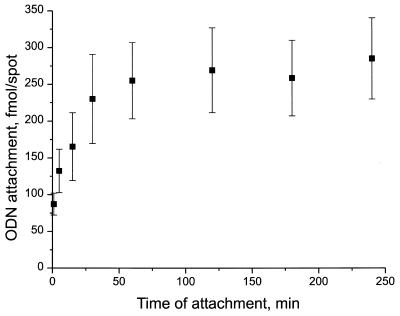
Time course of immobilization
The effect of reaction time on attachment of BAL-ODN to SC-glass was studied at pH 5. 3′-32P-labeled probe 1 was attached to SC-glass as usual. The reaction was performed using 50 µM probe 1. Aliquots (0.5 µl) were spotted at different times and incubated over different time periods ranging from 5 min to 4 h. Quantitative analysis was carried out using phosphorimaging and attached ODN was determined in fmol per spot by reference to calibration spots of known concentration (Fig. 5). The results indicate that immobilization goes almost to completion within the first hour of incubation with a maximum loading of ∼300 fmol/spot at 4 h.
Figure 5.
Kinetics of BAL-ODN immobilization. An SC-glass slide was incubated in a humidified chamber at 37°C. A 100 mM solution of 32P-labeled probe 1 in 100 mM NaOAc, pH 5.0 was applied to the slide at different time points during the 4 h incubation period. The slide was blocked, washed and the amount of bound probe was determined by phosphorimaging. The average spot size had a diameter of 1.5mm. Attachment (fmol/spot) is plotted versus time. Values presented are means ± SD from four spots.
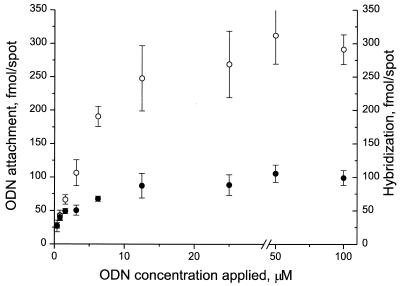
Immobilized probe density
To determine the maximum density of covalent attachment of BAL-ODNs to SC-glass, 3′-32P-labeled probe 1 with concentrations ranging from 0.3 to 100 µM were used. The hydrophobic nature of the surface prevented the droplets from spreading, and 0.5 µl ODN aliquots produced spots of ∼1.5 mm in diameter. Quantitative data (fmol/spot) were obtained using phosphorimaging and represent the averages of, minimally, four replicates. The results show that coupling efficiency increases with increasing concentration of applied ODN and reaches a plateau between 10 and 20 µM (Fig. 6, open circles). The maximum value of 300 fmol/spot attachment density corresponds to ∼1.8 × 105 molecules/µm2. This is the same order of magnitude as optimized probe densities for ODN arrays prepared by in situ synthesis (26).
Figure 6.
Concentration dependence of BAL-ODN probe density and effect on hybridization performance. A dilution series of 32P-labeled or unlabeled probe 1 in 100 mM NaOAc, pH 5.0 was immobilized on two separate SC-glass slides for 3 h at 37°C. Slides were blocked and washed and phosphorimaged. The slide containing labeled probe was quantified to determine immobilization efficiency (open circles). The second slide was hybridized overnight at 37°C with 1 µM 32P-labeled target 1 in 5× SSPE containing 0.1% Triton X-100. After extensive room temperature washing in hybridization buffer the slides were quantified to determine the amount of hybridized target ODN (closed circles). Values presented are means ± SD from four spots.
Hybridization properties
As a model system for hybridization studies we chose a fragment of the human amelogenin gene. The copy of the gene located in the X chromosome is significantly different in sequence from that located in the Y chromosome (27). The presence of multiple deletions and single nucleotide variations between two alleles makes this gene a convenient model for studies addressing problems of single mismatch discrimination by hybridization with short ODN probes.
Accessibility. First, we addressed the issue of efficiency of hybridization. One of the major concerns in ODN array technology is accessibility of the surface-bound ODN probes for hybridization (3). It has been reported (26) that having a spacer between the glass surface and ODN probes is critical for their hybridization performance. Therefore, we introduced an 18-atom-long polyethylene glycol spacer between the BAL-modification and ODN sequences using a commercially available phosphoramidite reagent. Hybridization efficiency is further increased with up to three additional spacers, but DNA synthesis yields can suffer (data not shown). Overloading the surface with probe molecules can lower the accessibility of the immobilized probes for hybridization with the target DNA. In order to answer this question, we immobilized the same concentration series (0.3–100 µM) of probe 1 on SC-glass as we did for the attachment efficiency study. But this time the BAL-ODN was not labeled. The slide was then hybridized to 5′-32P-labeled target 1, which is identical in sequence to a fragment of the X copy of amelogenin. The hybridized material was quantified and plotted on the same graph (Fig. 6, closed circles). The results indicate that at least 30% of immobilized probes are accessible for hybridization at the maximum immobilization density. At lower probe density increased hybridization efficiency was observed. This fact is consistent with a crowding effect taking place at high probe density.
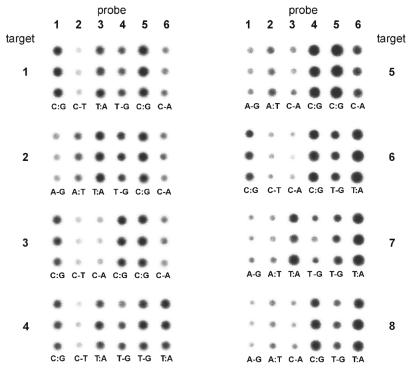
ODN targets. The second issue we addressed was specificity of hybridization and mismatch discrimination. The main goal of this study was to show that the immobilization chemistry is reliable and hybridization performance of arrayed ODN probes is predictable and consistent with thermodynamic properties of formed DNA duplexes in solution. An array of six BAL-ODN probes was prepared and hybridized to eight different 36mer target ODNs (see Table 1). The eight different targets started from a sequence identical to the X copy of the gene, and changed the nucleotide at each single nucleotide polymorphism (SNP) site to its variant in the Y copy. Three pairs of 14- or 15mer BAL-ODN probes span this 30-nt-long target sequence and each contain one of the SNPs. Each probe in the pair is complementary to either the X or the Y copy. With these six short probes arrayed on the glass slide, the hybridization to each ODN target generates three match/mismatch combinations. Hybridization results are presented in Figure 7. In all cases one can see a difference in hybridization signal between fully matched probe and the one containing single mismatch with a hybridized target. This difference varies depending on the type of mismatch. For example, T-G mismatch versus T:A match is well known as being tough to discriminate. The same statement is true for A-G versus A:T (see target 7). The array is not optimized in terms of hybridization properties, but performance was consistent with expected properties of DNA duplexes in solution.
Figure 7.
Results of eight parallel hybridizations of an array consisting of six BAL-ODN probes with eight different 32P-labeled 36mer ODN targets. Each probe is spotted in triplicate (spotted vertically) giving an array of 3 × 6 spots. Targets 1 and 8 correspond to X and Y copy of the amelogenin gene, respectively. All others contain nucleotide substitutions at the underlined positions (see Table 1 for the sequences). Match or mismatch base pairs formed between target and probe are indicated at the bottom of each triplicate.
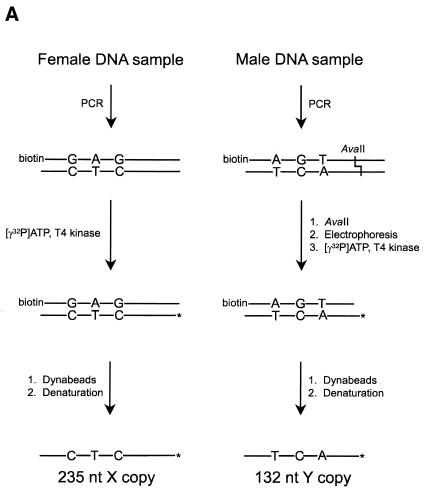
PCR product targets. Finally, a simple genotyping experiment was performed to demonstrate application of the ODN array chemistry to SNP analysis. We designed a pair of PCR primers to generate a 235 bp fragment of the presumed exon 3 of the amelogenin gene (27). This short PCR product contains more then a dozen SNPs. For example, the central segment of the amplicon (36 nt in length) contains three SNPs, and was chosen as a basis for the short oligonucleotide targets that were used for mismatch discrimination studies presented above. The same array of six ODN probes was used for hybridization with single-stranded PCR products.
In order to isolate single-stranded PCR products of the amelogenin gene, we used a combination of biotinylated primer for the strand complementary to the strand of interest and streptavidin-coated magnetic beads (Fig. 8A). Pure female (X) PCR products were easily isolated after labeling with 32P. Pure male (Y) copy of the amplicon cannot be obtained by simple PCR due to ‘heterozygosity’ of the male genome. The Y copy was cleaved with AvaII at one of the single nucleotide substitutions. The cleavage produced a shorter 132 bp fragment containing the target region and was easily isolated electrophoretically. This shorter fragment still contained the biotin residue and the same procedure was used to label and isolate the DNA strand of interest. The ‘heterozygous’ (XY) target DNA was generated by processing the 235 bp PCR fragment derived from the male genomic DNA sample without performing a restriction digest. The hybridization results of purified single-stranded PCR products to an array of six ODN probes are presented in Figure 8B. As expected, the hybridization pattern for ‘homozygous’ DNA targets was the same as for ODN targets 1 and 8. Again, it was hard to genotype the Y copy in the case of T:A versus T-G mismatch. Despite that fact, one could easily make a call for the other two SNPs. All three SNPs in the case of X copy of the gene were unmistakably discriminated. As expected, the ‘heterozygous’ target hybridized equally well to all six arrayed ODN probes.
Figure 8.
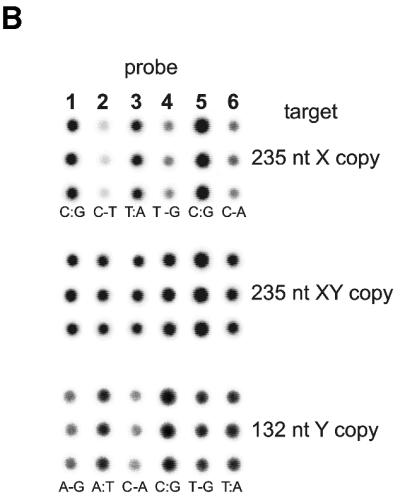
(A) Schematic representation of procedure used for 32P labeling and isolation of single-stranded PCR products. (B) Hybridization of the same array of six probes shown in Figure 7 to single-stranded 235 nt PCR products generated from female or male human genomic DNAs (X and XY, respectively) as well as to 132 nt product representing pure male copy of amelogenin gene fragment (Y). PCR product generated from the heterozygous male DNA sample shows an equimolar mixture of female and male copies of the gene fragment.
CONCLUSIONS
The semicarbazone immobilization chemistry presented here has a number of advantages. First of all, the starting materials are chemically stable. SC-glass surfaces should not be any more susceptible to degradation than aminopropyl-modified glass slides currently being used for DNA array preparation. Mildly electrophilic BAL-modified ODNs do not require any special handling or storage precautions. We have stored BAL-ODN samples at –20°C for several months without significant loss in activity. Ease of preparation of the arrays is also an advantage. No separate reduction steps or handling of toxic coupling reagents are required. The blocking step provides low background and the resulting arrays have good signals and predictable hybridization performance. In our opinion, the results presented above are encouraging in anticipation of the use of ODN arrays to detect point mutations in amplified gene fragments. The study was carried out on ‘macroarrays’ solely for ease of quantification and optimization of reaction conditions. We have successfully adopted the immobilization chemistry to a microarray format using automated spotters and fluorescence detection using commercially available array scanners, and intend to commercialize the BAL phosphoramidite reagent and SC-glass slides. BAL-ODNs can also be coupled to semicarbazide groups on other solid supports, and we are exploring these applications.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Vladimir Gorn for the synthesis of the oligonucleotides used in this study, Dr Michael Singer for suggestion of the amelogenin model, and Dr Richard Case for performing the mass spectrometry. Help from employees of Epoch Biosciences involved in this project as well as many useful discussions during preparation of this manuscript is appreciated.
REFERENCES
- 1.Southern E.M. (2001) DNA Microarrays: History and Overview. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 2.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 3.Hughes T.R., Mao,M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W., Lefkowitz,S.M., Ziman,M., Schelter,J.M., Meyer,M.R. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 4.Lockhart D.J., Dong,H., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittman,M., Wang,C., Kobayashi,M., Horton,H. et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 5.Guo Z., Guilfoyle,R.A., Thiel,A.J., Wang,R. and Smith,L.M. (1994) Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res., 22, 5456–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drmanac R., Drmanac,S., Stresoska,Z., Paunesku,T., Labat,I., Zeremski,M., Snoddy,V., Funkhouser,W.K., Koop,B., Hood,L. et al. (1993) DNA sequencing by hybridization: a strategy for efficient large-scale sequencing. Science, 260, 1649–1652. [DOI] [PubMed] [Google Scholar]
- 7.Pastinen T., Kurg,A., Metspalu,A., Peltonen,L. and Syvanen,A.-C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res., 7, 606–614. [DOI] [PubMed] [Google Scholar]
- 8.McGall G.H., Barone,A.D., Diggelmann,M., Fodor,S.P.A., Gentalen,E. and Ngo,N. (1997) The efficiency of light-directed synthesis of DNA arrays on glass substrates. J. Am. Chem. Soc., 119, 5081–5090. [Google Scholar]
- 9.Southern E.M., Case-Green,S.C., Elder,J.K., Johnson,M., Mir,K.U., Wang,L. and Williams,J.C. (1994) Arrays of complementary oligonucleotides for analysing the hybridisation behaviour of nucleic acids. Nucleic Acids Res., 22, 1368–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arkles B. (1977) Tailoring surfaces with silanes. Chemtech., 7, 766–778. [Google Scholar]
- 11.Lindroos K., Liljedahl,U., Raitio,M. and Syvanen,A.-C. (2001) Minisequencing on oligonucleotide microarrays: comparison of immobilisation chemistries. Nucleic Acids Res., 29, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamture J.B., Beattie,K.L., Burke,B.E., Eggers,M.D., Ehrlich,D.J., Fowler,R., Hollis,M.A., Kosicki,B.B., Reich,R.K., Smith,S.R. et al. (1994) Direct detection of nucleic acid hybridization on the surface of a charge coupled device. Nucleic Acids Res., 22, 2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen D., Yan,Z., Cole,D.L. and Srivatsa,G.S. (1999) Analysis of internal (n–1)mer deletion sequences in synthetic oligodeoxyribonucleotides by hybridization to an immobilized probe array. Nucleic Acids Res., 27, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zammatteo N., Jeanmart,L., Hamels,S., Courtois,S., Louette,P., Hevesi,L. and Remacle,J. (2000) Comparison between different strategies of covalent attachment of DNA to glass surfaces to build DNA microarrays. Anal. Biochem., 280, 143–150. [DOI] [PubMed] [Google Scholar]
- 15.Beier M. and Hoheisel,J.D. (1999) Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res., 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanNess J., Kalbfleisch,S., Petrie,C.R., Reed,M.W., Tabone,J.C. and Vermeulen,N.M.J. (1991) A versatile solid support system for oligodeoxynucleotide probe-based hybridization assays. Nucleic Acids Res., 19, 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kremsky J.N., Wooters,J.L., Dougherty,J.P., Meyers,R.E., Collins,M. and Brown,E.L. (1987) Immobilization of DNA via oligonucleotides containing an aldehyde or carboxylic acid group at the 5′ terminus. Nucleic Acids Res., 15, 2891–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein J., Yale,H.L., Losee,K., Holsing,M., Martins,J. and Lott,W.A. (1951) The chemotherapy of experimental tuberculosis. III. The synthesis of thiosemicarbazones and related compounds. J. Am. Chem. Soc., 73, 906–912. [Google Scholar]
- 19.Cantor C.R., Warshaw,M.M. and Shapiro,H. (1970) Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxynucleotides. Biopolymers, 9, 1059–1077. [DOI] [PubMed] [Google Scholar]
- 20.Klich M. and Teutsch,G. (1986) Synthesese de N-(tetrazole-5-yl)azetidin-2-ones. Tetrahedron, 42, 2677–2684. [Google Scholar]
- 21.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) John Wiley, New York, NY.
- 22.Maxam A.M. and Gilbert,W. (1977) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 23.Dent A.H. and Aslam,M. (1998) Other Categories of Protein Coupling. Macmillan, New York, NY.
- 24.Lemaitre M., Dayard,B. and Lebleu,B. (1987) Specific antiviral activity of a poly(l-lysine)-conjugated oligodeoxyribonucleotide sequence complementary to vesicular stomatitis virus N protein mRNA initiation site. Proc. Natl Acad. Sci. USA, 84, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Call D.R., Chandler,D.P. and Brockman,F. (2001) Fabrication of DNA microarrays using unmodified oligonucleotide probes. Biotechniques, 30, 368–379. [DOI] [PubMed] [Google Scholar]
- 26.Shchepinov M.S., Case-Green,S.C. and Southern,E.M. (1997) Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res., 25, 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakahori Y., Takenaka,O. and Nakagome,Y. (1991) A human X-Y homologous region encodes ‘amelogenin’. Genomics, 9, 264–269. [DOI] [PubMed] [Google Scholar]