Abstract
During recombination-mediated repair of DNA double-strand breaks, strand transfer proteins must distinguish a homologous repair template from closely related genomic sequences. However, some tolerance by strand transfer proteins for sequence differences is also critical: too much stringency will prevent recombination between different alleles of the same gene, but too much tolerance will lead to illegitimate recombination. We characterized the heterology tolerance of Saccharomyces cerevisiae Rad51 by testing bypass of small heterologous inserts in either the single- or double-stranded substrate of an in vitro strand transfer reaction that models the early steps of homologous recombination. We found that the yeast protein is rather stringent, only tolerating heterologies up to 9 bases long. The efficiency of heterology bypass depends on whether the insert is in the single- or double-stranded substrate, as well as on the location of the insert relative to the end of the double-stranded linear substrate. Rad51 is distinct in that it can catalyze strand transfer in either the 3′→5′ or 5′→3′ direction. We found that bypass of heterology was independent of the polarity of strand transfer, suggesting that the mechanism of 5′→3′ transfer is the same as that of 3′→5′ transfer.
INTRODUCTION
During the repair of DNA double-strand breaks, strand transfer proteins, such as the Saccharomyces cerevisiae Rad51 protein, bind to single-stranded sequences generated at the break site and search the genome for homologous repair templates (1). If the stretch of DNA that contains the double-strand break has been recently replicated, the sister copy will provide a near perfect template. However, the more likely case in diploid organisms is that the repair template will be the allelic locus on the homologous chromosome. Because these chromosomes come from different parents, sequence differences will exist between the damaged DNA and the repair template. For repair to be successful, the ability of the strand transfer protein to tolerate sequence differences (‘heterology tolerance’) must be finely tuned. If the protein is too stringent, recombination will be blocked by the polymorphisms between parents. If the protein is too tolerant, an incorrect template may be chosen from the potentially large number of closely related sequences throughout the genome, leading to chromosomal rearrangement.
Strand transfer proteins such as Rad51 have been characterized using an in vitro strand transfer reaction that mimics the early steps in double-strand break repair (1,2). In this reaction (see Fig. 1A), a nucleoprotein filament consisting of a single-stranded circular (ssC) DNA coated with Rad51 reacts with a homologous double-stranded linear (dsL) DNA. A single-stranded DNA-binding protein, such as yeast RPA, is required for efficient strand transfer (3–5). During transfer, the complementary strand of the dsL substrate anneals to the ssC substrate to form a strand transfer intermediate with three DNA strands (2,6; Scandellari et al., submitted for publication). If the substrates are homologous, ATP-dependent unidirectional branch migration continues until nicked circular (NC) and single-stranded linear (ssL) products are released (3,7). The two substrates, intermediate joint molecules, and two product DNAs are easily separated by agarose gel electrophoresis.
Figure 1.
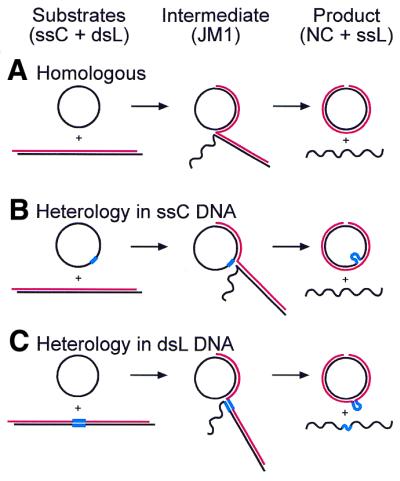
Strand transfer reactions. (A) Strand transfer reaction between completely homologous substrates. Two substrate DNAs, a ssC and a dsL, are shown on the left. DNA strands identical in sequence to the ssC are shown in black and the complementary strands in red. During the reaction, the complementary strand of the dsL is transferred to the ssC, yielding intermediate joint molecules (JM) that resolve into two products, a ssL and a double-stranded NC. (B) Strand transfer reaction with a heterologous insert (blue) in the ssC substrate. The JM in the center is shown paused at the insert. Note that before products can be formed, the single-stranded insert must be looped out of the double-stranded circle, as shown on the right. (C) Strand transfer reaction with a heterologous insert in the dsL substrate. The duplex insert must be both denatured and looped out before products can be formed, as shown on the right.
We studied the heterology tolerance of Rad51 using defined insertions in the ssC or dsL substrates. If the insert is in the ssC substrate, the heterologous sequence must be extruded from the nucleoprotein filament before the ssC and dsL DNA can be brought back into register to complete the reaction (Fig. 1B). If the insert is in the dsL substrate, the insert must be denatured and then looped out before strand transfer can continue (Fig. 1C). Because the base pairs broken when bypassing the duplex insert are not reformed in the NC product, bypass of double-stranded inserts has an energetic penalty that bypass of single-stranded inserts does not have (8).
The ability of the Escherichia coli RecA protein to bypass heterologies during strand transfer has been well defined (8–10). RecA easily bypasses inserts of 20 bp and can even bypass inserts of up to 200 bp. While non-hydrolyzable ATP analogs allow RecA to promote substrate pairing and product formation by random branch migration, bypass of heterologous inserts requires ATP hydrolysis (11). Interestingly, the efficiency of bypass increases as the heterology is positioned closer to the end of the dsL substrate at which strand transfer initiates (9,12). This position effect suggests that the nucleoprotein filament interacts with homologous DNA distal to the insert and that this interaction aids bypass, perhaps by generating topological strain (12,13). There is evidence for distal interactions with RecA (12,14), but the mechanism of bypass enhancement is not understood.
Whereas RecA is the paradigmatic prokaryotic strand transfer protein, Rad51 protein, particularly that of yeast, has been central to the understanding of eukaryotic general recombination (15). Rad51 is homologous to RecA over about half its length and is the key homolog in vivo (3,16). RecA and Rad51 form similar nucleoprotein filaments, though RecA polymerizes along single-stranded DNA in the 5′→3′ direction, whereas Rad51 polymerizes 3′→5′ (15,17). For RecA, the direction of strand transfer, defined relative to the ssC substrate, is the same as the direction of polymerization (15). Rad51, however, is unlike any other strand transfer protein studied in that it can catalyze strand transfer in either the 3′→5′ or 5′→3′ direction. Rad51 always begins strand transfer at the single-stranded overhang that is complementary to the ssC DNA (18). The 5′→3′ reaction was reported to yield more product (19). Both RecA and Rad51 require ATP hydrolysis for efficient strand transfer (3,20). Initial experiments characterizing the heterology bypass activity of Rad51 have been carried out (19).
We have described elsewhere the formation and structure of Rad51 strand transfer intermediates that accumulate when strand transfer is blocked by large heterologies (Scandellari et al., submitted for publication). The simplest intermediate, which we call joint molecule 1 (JM1), consists of a ssC substrate annealed to a dsL substrate (depicted in Fig. 1). A more complex intermediate, joint molecule 2 (JM2), is formed when a second dsL substrate reacts with a JM1 molecule. Both intermediate species can resolve to product.
Here we have studied the bypass of heterologies by Rad51 in a systematic fashion. We find that Rad51 is clearly more stringent than RecA, bypassing inserts no longer than 9 bp. Double-stranded inserts are bypassed less efficiently than single-stranded inserts, though the difference was much less than observed for RecA (9). The extent of bypass decreased as the insert was moved closer to the distal end of the dsL substrate. Interestingly, heterology bypass by Rad51 was identical in the 3′→5′ and 5′→3′ directions.
MATERIALS AND METHODS
Protein purification
Rad51 was purified as described (3) except that the Sephacryl S200 column was omitted. In addition, the final MonoQ column was used twice, first to purify and then to concentrate the protein. Small aliquots of purified Rad51 were frozen in liquid nitrogen and stored at –80°C. Replication protein A (RPA) was purified from yeast over-expressing all three subunits (21,22). It was stored in small, single-use aliquots at –80°C in 25 mM Tris–HCl pH 7.5, 0.35 M KCl, 0.5 mM dithiothreitol, 0.5 mM EDTA and 10% glycerol.
Generation of pBluescript derivatives
Deletion derivatives of the pBluescript II SK+ phagemid (pBS; Stratagene) were constructed as described (Scandellari et al., submitted for publication) by digesting the wild-type plasmid at unique sites in the multiple cloning sequence with appropriate restriction enzymes, filling in overhangs with Klenow polymerase if necessary and ligating with DNA ligase. The resulting plasmids were named pBSdX, where X is the number of nucleotides deleted. pBSd4 was generated by cutting at EcoRV and EcoRI sites and filling in the latter overhang before ligation. Two pBSd6 derivatives were made: one cut with XhoI and SalI and the other with SpeI and XbaI. While all data presented here used the XhoI–SalI derivative, results from the other plasmid were indistinguishable. pBSd9 was made by digesting with ClaI and AccI. pBSd18 was made by digesting with EcoRV and SmaI. All deletion derivatives were verified by direct sequencing.
Whereas a deletion in one strand transfer reaction substrate is equivalent to an insert in the other, we find it clearer to discuss all reactions in terms of heterologous inserts.
DNA preparation
Supercoiled plasmid DNA was purified from E.coli using two cesium chloride/ethidium bromide isopycnic bandings (23). Double-stranded linear DNA substrates were obtained by digesting supercoiled DNA with the restriction enzymes (NEB) indicated. Single-stranded circular phagemid DNA was rescued from XL1-Blue E.coli cells containing pBS derivatives as described (24).
Strand transfer reactions
The Rad51 strand transfer reaction was performed as described (25). An aliquot of 7 µg Rad51 was added to 0.12 µg ssC DNA in 7.5 µl of 40 mM potassium MOPS pH 7.2, 3 mM MgCl2, 1 mM dithiothreitol and 2.5 mM ATP. After 5 min at 37°C, 1.5 µg RPA was added and incubation continued for 10 min. Finally, 0.24 µg dsL DNA and spermidine to 4 mM were added for a total reaction volume of 13 µl. Incubation was continued for 120 min. The reaction was stopped by adding 1 vol of 20 mM Tris–HCl pH 7.5, 20 mM EDTA pH 8.0, 1% SDS and 1 mg/ml proteinase K, and incubating for another 30 min at 37°C. The DNA was resolved by electrophoresis through a 0.9% agarose gel in Tris–borate buffer (1.1–1.9 V/cm for 600–850 V·h) and then visualized by staining with ethidium bromide or by Southern blotting. Quantification of Southern blots was done with a Fuji phosphorimager.
The apparent low level of bypass of even very long inserts (Fig. 4) is an artifact due to trace NC DNA in either the dsL or the ssC DNA preparation. We therefore defined the baseline for true heterologous bypass as that in the reactions with 14 or 18 base inserts, which completely block bypass.
Figure 4.
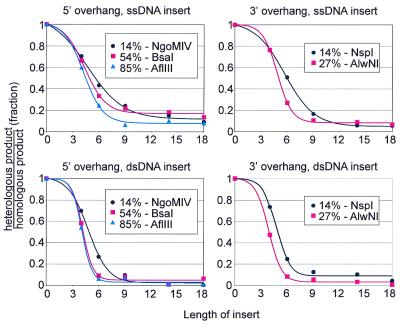
Rad51 heterology bypass is independent of strand transfer polarity. Quantification of Rad51 heterology bypass as a function of length, position, insert type and direction of strand transfer. The top two graphs show bypass of inserts in the ssC substrate; the bottom two show bypass of inserts in the dsL substrate. The left two graphs are for dsL substrates with 5′-overhangs and reactions in the 3′→5′ direction; the right two graphs are for 3′-overhangs and 5′→3′ reactions. Within each graph, data for dsL substrates linearized with different restriction enzymes are distinguished by symbols and colors. The distance of the restriction site from the initiating end for strand transfer is noted for each enzyme. Curve fits are standard Boltzmann sigmoids. Error bars are omitted for clarity.
RESULTS
Effect of the length of a heterologous insert
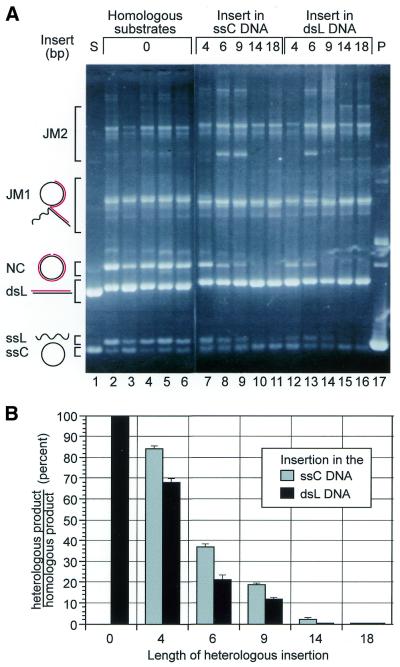
We measured the ability of Rad51 to bypass heterologies as a function of the length of single-stranded or double-stranded inserts, as diagrammed in Figure 1. Each experiment used dsL substrates linearized with the same restriction enzyme so that the insert position was constant and only the insert length varied. Homologous reactions were run for each experiment (Fig. 2A, lanes 2–6) to verify that all the DNAs were competent for strand transfer and to serve as standards for normalization. Each heterologous reaction was compared to the two homologous reactions that corresponded to its substrates. In reactions with single-stranded inserts we found that NC product formation decreased rapidly as insert length increased (Fig. 2A, lanes 7–11). With a 9 base insert, only 20% as much product was formed compared with the homologous control reactions (Fig. 2B). With inserts of 14 or 18 bases, no product was detected. We interpolated the insert length at which the heterology blocked 50% of strand transfer to be 4.9 bases. Double-stranded inserts caused a sharper fall-off of product formation with insert length (Fig. 2A, lanes 12–16, and B). NC product decreased to ∼20% of controls with only a 6 bp insert.
Figure 2.
Rad51 heterology bypass as a function of heterology length. (A) Rad51 strand transfer reactions with substrates containing single- or double-stranded inserts of varying lengths. Bands are labeled schematically as in Figure 1. The position of the more complex JM2 intermediate is indicated. Lane 1 shows the ssC and dsL substrates. Lanes 2–6 contain homologous reactions with ssC and dsL substrate pairs of identical sequence: pBS, pBSd4, pBSd6, pBSd9 and pBSd18, respectively (see Materials and Methods for plasmid descriptions). All dsL substrates were cut with BsaI to position the heterologies midway along the dsL. Lanes 7–16, from another gel, contain reactions between wild-type pBS and deletion derivatives. Lanes 10 and 15 contain reactions between the pBSd4 and pBSd18 substrates, whose deletions overlap to form a 14 base heterology. Lane 17 contains markers for the NC product band. The image is an ethidium bromide stained gel. (B) Quantification of the experiment in (A). Gray columns correspond to inserts in the ssC DNA. Black columns correspond to inserts in the dsL DNA. The amount of product in any reaction is defined as the amount of DNA in the NC product band divided by the sum of the DNA in the dsL, NC and JM bands. Error bars indicate one standard deviation; data are from three repetitions of the experiment.
Failure to form product was not due to an inability of the substrates to initiate strand transfer, as kinetic experiments showed that joint molecules were formed with identical efficiency in homologous and heterologous reactions (data not shown). As expected, joint molecules comprise a greater percentage of the reacted DNA in heterologous reactions than in homologous reactions, as they cannot resolve to product (Fig. 2A).
Effect of the position of a heterologous insert
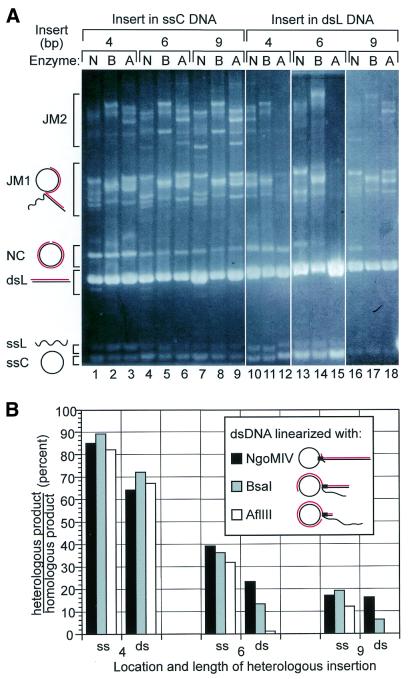
Having characterized the dependence of bypass on insert length, we determined whether the dependence on insert position seen for RecA (9) is conserved in the eukaryotic protein. We digested pBS derivatives with NgoMIV, BsaI or AflIII restriction endonuclease to generate circularly permuted dsL substrates such that the insert was positioned at the beginning (proximal), middle or end (distal), respectively, of strand transfer.
We found a dramatic position dependence for bypass of inserts in the dsL substrate that depended sharply on the insert length (Fig. 3A, lanes 10–18). While bypass of 4 bp inserts showed minimal position dependence, bypass of 6 bp inserts dropped 10-fold when the insert was moved from a proximal to a distal location (Fig. 3B). Bypass of 9 bp inserts at the most distal insert position was undetected.
Figure 3.
Rad51 heterology bypass as a function of heterology position. (A) Rad51 strand transfer reactions with 4–9 base inserts at different positions along the dsL substrate. Bands are labeled schematically as in Figure 2. Lanes 10–18, assembled from three gels, contain reactions where the insert is in the dsL substrate. Restriction enzyme digestion with NgoMIV (N), BsaI (B) and AflIII (A) positioned the insert at 14, 54 or 85%, respectively, along the dsL substrate. Lanes 1–9 contain reactions where the heterologous insert is in the same location in the ssC substrate. The homologous reactions are not shown. The image is assembled from ethidium bromide stained gels. (B) Quantification of the experiment in (A). Results are grouped by insert length and by whether the insert is in the single- (ss) or double-stranded (ds) substrate. The inset shows the position of the heterology as a black rectangle and the length of homology beyond the heterology as the double-stranded tail of a JM1 molecule.
In contrast, when the insert was in the same three positions in the ssC substrate, there was only a minimal position effect (Fig. 3A, lanes 1–9). Whereas no position effect was seen for the 4 base insert, the 6 and 9 base inserts showed slight decreases in bypass as the insert was moved closer to the position corresponding to the distal end of the dsL substrate (Fig. 3B).
The significant effect of heterology position with double-stranded inserts suggests that the need for interactions with distal homology is a conserved property between Rad51 and RecA. Interestingly, Rad51 shares with RecA a tempered position effect for single-stranded inserts (8,9).
Comparison of 5′→3′ and 3′→5′ heterology bypass
Rad51 is unique among known strand transfer proteins in that it can catalyze strand transfer in vitro in either the 3′→5′ or 5′→3′ direction. For RecA, it was believed that the unique polarity of nucleoprotein formation dictated the polarity of subsequent strand transfer (26). This cannot be the case for Rad51: either the polarity of strand transfer is independent of the polarity of polymerization or the 3′→5′ and 5′→3′ reactions use different mechanisms. Heterology bypass provides a sensitive assay to compare the 3′→5′ and 5′→3′ reactions. Because bypass is measured by comparing product formation from heterologous and homologous reactions, differences in the rate of product formation between the 3′→5′ and 5′→3′ reactions are controlled for.
The experiments in Figures 2 and 3 used substrates with 5′-overhangs and thus proceeded in the 3′→5′ direction. To test bypass 5′→3′, the transfer direction of RecA, DNAs were digested with NspI and AlwNI to generate substrates with 3′-overhangs and inserts positioned at the beginning or middle of the dsL, respectively. Both heterology length and position effects were monitored.
We found no appreciable difference between the 3′→5′ and 5′→3′ directions of strand transfer in either the bypass of single- and double-stranded inserts (Fig 4, compare top and bottom panels) or in the position effect (Fig 4, bottom panels, compare NspI and AlwNI to NgoMIV and BsaI). The similarity between insert length dependences can be seen readily by comparing the NgoMIV and NspI reactions that are matched for heterology position (Fig 4, black curves in the left and right panels). The interpolated insert length at which strand transfer was 50% blocked was ∼5 bases for these reactions, whether the insert was single- or double-stranded. We conclude that the mechanism of bypass is fundamentally the same in both directions of Rad51 strand transfer.
DISCUSSION
We found that Rad51 is proficient but stringent in strand transfer through heterologies. Inserts 4 bases or base pairs long are bypassed with between 50 and 70% efficiency. Bypass quickly drops to zero at insert lengths of >9 bases. Strand transfer is 50% blocked, depending on the type and position of insert, at insert lengths between 4 and 6 bases. Rad51 is an order of magnitude more stringent than RecA but shares the position dependence of bypass efficiency.
We were impressed by the sharp transition between efficient bypass of 4 base inserts and the relatively poor bypass of 6 base inserts. A sigmoidal dependence of bypass on insert length has also been seen in studies using RecA heterology bypass, where product formation dropped sharply between distal insert lengths of 16 and 26 bases (9). There appears to be a critical heterology limit intrinsic to the strand transfer mechanism below which bypass is very efficient. Indeed, Rad51 bypass of 4 base inserts is relatively immune to position effects.
We found the difference in bypass of single-stranded and double-stranded inserts to be surprisingly small. Averaged over the five substrates used, the ratio of single-stranded insert lengths to double-stranded insert lengths where bypass was 50% blocked was only ∼1 bp. Perhaps the energetic penalty of freeing single-stranded insert DNA from the nucleoprotein filament, either by extrusion or by filament reorganization, is on a par with the energetic cost of denaturing and looping out the duplex insert. Studies of RecA found a much larger difference, but the cost of denaturing much longer duplex inserts could have overwhelmed the costs of other steps (8,9).
Several models have been proposed for the mechanism of heterology bypass during strand transfer (8,13,14). These models differ in detail, but all attempt to explain the requirements for ATP hydrolysis and distal homology by suggesting that the nucleoprotein filament must denature the heterology through helicase-like activity or by generating and trapping negative supercoiling. Indeed, RecA has been shown to open the duplex dramatically during strand transfer (27). Rad51 shares with RecA the relevant mechanistic requirements (this work; 19) and can also be modeled in these ways. We imagine that the stringency of Rad51 is determined by the number of base pairs that can be denatured and thus is limited by the mechanical force that Rad51 generates. Yeast Rad51 is roughly 10-fold more stringent than RecA and has been shown to hydrolyze ATP ∼20 times more slowly (3).
Why has the eukaryotic protein shifted its heterology tolerance towards stringency? Eukaryotic genomes have increased their sequence redundancy through evolutionary genome duplication, divergence of related genes and multiplication of transposable elements (28,29). As a result, the danger of recombinational chromosome rearrangement is much greater for even the simple yeast genome than for the E.coli genome, which has scant repetitions (30). Strand transfer proteins may have adapted to minimize this danger, increasing the accuracy of recombinational repair even while sacrificing overall recombination efficiency.
Acknowledgments
ACKNOWLEDGEMENTS
V.F.H. is a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by grants to N.R.C. from the General Medical Institute of the NIH.
REFERENCES
- 1.Kowalczykowski S.C. (2000) Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci., 25, 156–165. [DOI] [PubMed] [Google Scholar]
- 2.Cox M.M. (1999) Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acid Res. Mol. Biol., 63, 311–366. [DOI] [PubMed] [Google Scholar]
- 3.Sung P. (1994) Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science, 265, 1241–1243. [DOI] [PubMed] [Google Scholar]
- 4.Cox M.M., Soltis,D.A., Livneh,Z. and Lehman,I.R. (1983) On the role of single-stranded DNA binding protein in recA protein-promoted DNA strand exchange. J. Biol. Chem., 258, 2577–2585. [PubMed] [Google Scholar]
- 5.West S.C., Cassuto,E. and Howard-Flanders,P. (1982) Role of SSB protein in RecA promoted branch migration reactions. Mol. Gen. Genet., 186, 333–338. [DOI] [PubMed] [Google Scholar]
- 6.Kowalczykowski S.C., Dixon,D.A., Eggleston,A.K., Lauder,S.D. and Rehrauer,W.M. (1994) Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev., 58, 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain S.K., Cox,M.M. and Inman,R.B. (1994) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. III. Unidirectional branch migration and extensive hybrid DNA formation. J. Biol. Chem., 269, 20653–20661. [PubMed] [Google Scholar]
- 8.Bianchi M.E. and Radding,C.M. (1983) Insertions, deletions and mismatches in heteroduplex DNA made by RecA protein. Cell, 35, 511–520. [DOI] [PubMed] [Google Scholar]
- 9.Morel P., Stasiak,A., Ehrlich,S.D. and Cassuto,E. (1994) Effect of length and location of heterologous sequences on RecA-mediated strand exchange. J. Biol. Chem., 269, 19830–19835. [PubMed] [Google Scholar]
- 10.Bucka A. and Stasiak,A. (2001) RecA-mediated strand exchange traverses substitutional heterologies more easily than deletions or insertions. Nucleic Acids Res., 29, 2464–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.I., Cox,M.M. and Inman,R.B. (1992) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I. Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem., 267, 16438–16443. [PubMed] [Google Scholar]
- 12.Jwang B. and Radding,C.M. (1992) Torsional stress generated by RecA protein during DNA strand exchange separates strands of a heterologous insert. Proc. Natl Acad. Sci. USA, 89, 7596–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Honigberg S.M. and Radding,C.M. (1988) The mechanics of winding and unwinding helices in recombination: torsional stress associated with strand transfer promoted by RecA protein. Cell, 54, 525–532. [DOI] [PubMed] [Google Scholar]
- 14.MacFarland K.J., Shan,Q., Inman,R.B. and Cox,M.M. (1997) RecA as a motor protein. Testing models for the role of ATP hydrolysis in DNA strand exchange. J. Biol. Chem., 272, 17675–17685. [DOI] [PubMed] [Google Scholar]
- 15.Bianco P.R., Tracy,R.B. and Kowalczykowski,S.C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci., 3, D570–D603. [DOI] [PubMed] [Google Scholar]
- 16.Ogawa T., Yu,X., Shinohara,A. and Egelman,E.H. (1993) Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science, 259, 1896–1899. [DOI] [PubMed] [Google Scholar]
- 17.Sung P. and Robberson,D.L. (1995) DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–561. [DOI] [PubMed] [Google Scholar]
- 18.Namsaraev E.A. and Berg,P. (1998) Branch migration during Rad51-promoted strand exchange proceeds in either direction. Proc. Natl Acad. Sci. USA, 95, 10477–10481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Namsaraev E.A. and Berg,P. (2000) Rad51 uses one mechanism to drive DNA strand exchange in both directions. J. Biol. Chem., 275, 3970–3976. [DOI] [PubMed] [Google Scholar]
- 20.Namsaraev E. and Berg,P. (1997) Characterization of strand exchange activity of yeast Rad51 protein. Mol. Cell. Biol., 17, 5359–5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alani E., Thresher,R., Griffith,J.D. and Kolodner,R.D. (1992) Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J. Mol. Biol., 227, 54–71. [DOI] [PubMed] [Google Scholar]
- 22.Brill S.J. and Stillman,B. (1989) Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature, 342, 92–95. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Yamamoto K.R., Alberts,B.M., Benzinger,R., Lawhorne,L. and Treiber,G. (1970) Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology, 40, 734–744. [DOI] [PubMed] [Google Scholar]
- 25.Sung P. and Stratton,S.A. (1996) Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J. Biol. Chem., 271, 27983–27986. [DOI] [PubMed] [Google Scholar]
- 26.Klapstein K. and Bruinsma,R. (2000) RecA force generation by hydrolysis waves. J. Biol. Chem., 275, 16073–16083. [DOI] [PubMed] [Google Scholar]
- 27.Voloshin O.N. and Camerini-Otero,R.D. (1997) The duplex DNA is very underwound in the three-stranded RecA protein-mediated synaptic complex. Genes Cells, 2, 303–314. [DOI] [PubMed] [Google Scholar]
- 28.Mewes H.W., Albermann,K., Bahr,M., Frishman,D., Gleissner,A., Hani,J., Heumann,K., Kleine,K., Maierl,A., Oliver,S.G., Pfeiffer,F. and Zollner,A. (1997) Overview of the yeast genome. Nature, 387 (suppl.), 7–65. [DOI] [PubMed] [Google Scholar]
- 29.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 30.Blattner F.R., Plunkett,G.,III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]