Abstract
Bacteria are causative agents of periodontal diseases. Interactions between oral bacteria and gingival epithelial cells are essential aspects of periodontal infections. Using an in vitro tissue culture model, a selected group of gram-negative anaerobic bacteria frequently associated with periodontal diseases, including Bacteroides forsythus, Campylobacter curvus, Eikenella corrodens, Fusobacterium nucleatum, Porphyromonas gingivalis, and Prevotella intermedia, were examined for their ability to adhere to and invade primary cultures of human gingival epithelial cells (HGEC). The effects of these bacteria on the production of interleukin-8 (IL-8), a proinflammatory chemokine, were also measured. These studies provided an initial demonstration that F. nucleatum adhered to and invaded HGEC and that this was accompanied by high levels of IL-8 secretion from the epithelial cells. The attachment and invasion characteristics of F. nucleatum were also tested using KB cells, an oral epithelial cell line. The invasion was verified by transmission electron microscopy and with metabolic inhibitors. Invasion appeared to occur via a “zipping” mechanism and required the involvement of actins, microtubules, signal transduction, protein synthesis, and energy metabolism of the epithelial cell, as well as protein synthesis by F. nucleatum. A spontaneous mutant, lam, of F. nucleatum, isolated as defective in autoagglutination, was unable to attach to or invade HGEC or KB cells, further indicating the requirement of bacterial components in these processes. Sugar inhibition assays indicated that lectin-like interactions were involved in the attachment of F. nucleatum to KB cells. Investigation of these new virulence phenotypes should improve our understanding of the role of F. nucleatum in periodontal infections.
Gram-negative anaerobic bacteria are etiologic agents of periodontal diseases. Depending on the severity, the disease can be broadly classified into two stages: gingivitis, characterized by tissue inflammation, and periodontitis, associated with attachment loss, alveolar bone resorption, and tooth loss (6). Among more than 300 species identified in the oral cavity, a relatively small group of gram-negative organisms, including Actinobacillus actinomycetemcomitans, Bacteroides forsythus, Campylobacter spp., Capnocytophoga spp., Eikenella corrodens, Fusobacterium nucleatum, Porphyromonas gingivalis, and Prevotella intermedia, along with oral spirochetes, are the bacteria most frequently isolated from infected periodontal pockets and are thus recognized as potential periodontal pathogens (28, 37). Interactions between bacteria and their surrounding epithelium are critical factors in bacterial infections (11, 12, 27). Adherence to epithelial cells is important for colonization. Invasion allows the bacteria not only to evade the host immune surveillance but also to spread into deeper tissues. Histological studies of periodontal infections also indicated penetration of deeper tissues by cocci, rods, and fusi-spirochetal forms of bacteria in advanced periodontitis (1, 14, 25, 33). So far, analysis of tissue attachment and invasion by oral bacteria has been focused on A. actinomycetemcomitans and P. gingivalis (8, 10, 22, 23, 26, 29, 34). Both organisms invade by a “ruffling” mechanism; that is, they cause dramatic ruffling of host cell membranes at the site of entry, resulting in bacteria internalized in the form of spacious vacuoles (22, 26). This ruffling mechanism is one of the two major penetration mechanisms used by invasive bacteria (11). The other major entry mechanism, termed zipping, in which the invading bacteria remain in close contact with the host membrane during penetration, has not been reported for any oral bacteria.
Besides serving as a physical barrier, the epithelium also functions as a sensor for the presence of bacteria. The direct physical contact between bacteria and the mucosal surface triggers the expression of a variety of immune response mediators from epithelial cells (19, 30). One such modulator is interleukin-8 (IL-8), a low-molecular-weight, proinflammatory chemokine which attracts and activates neutrophils (2, 30). Expression of IL-8 has been suggested to represent an important regulatory mechanism leading to neutrophil migration into the gingival sulcus (2, 30). Recent studies revealed that the effects of periodontal pathogens on the production of IL-8 from epithelial cells also vary (6, 18).
Since periodontal diseases result from complex interactions of multiple microorganisms, it is essential to investigate the interactions between different periodontal bacteria and epithelial cells. Using primary cultures of human gingival epithelial cells (HGEC), we examined a selected group of putative periodontal pathogens for their tissue attachment and invasion properties and their roles in stimulating IL-8 production. Our findings revealed Fusobacterium nucleatum as a potentially important organism in periodontal infections. F. nucleatum is a filamentous human pathogen strongly associated with periodontal diseases (28) as well as infections and abscesses in other parts of the body (3, 5, 17). Several potential virulence mechanisms have been proposed for F. nucleatum (7, 13, 15, 21, 24, 31, 35, 36, 38). In this study, F. nucleatum 12230, a clinical transtracheal isolate used as the working strain in our laboratory, was found to adhere to and invade HGEC and to induce high levels of IL-8 secretion. The novel characteristics of tissue attachment and invasion were further verified using KB cells, and additional F. nucleatum strains were also tested.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. All bacteria were maintained either in brain heart infusion (BHI) broth (Difco, Detroit, Mich.) supplemented with hemin (5 μg/ml; Sigma, St. Louis, Mo.) and menadione (1 μg/ml; Sigma) or on BHI agar (Difco) plates, also supplemented with hemin and menadione, with or without 5% defibrinated sheep blood agar (Crane Laboratories, Syracuse, N.Y.). Cultures were incubated for 2 to 4 days at 37°C in an anaerobic chamber (Forma Scientific, Marietta, Ohio) containing 10% CO2, 5% H2, and 85% N2. F. nucleatum 12230 lam (for “less adhesive mutant”) was a spontaneous mutant, defective in autoaggregation, that was isolated by progressive enrichment of the nonaggregating fractions of the liquid cultures. Briefly, F. nucleatum 12230 was grown to stationary phase in 10 ml of BHI broth. Avoiding the precipitated aggregates at the bottom of the tube, 1 ml of the culture was transferred to a sterile Eppendorf tube and centrifuged at 5,000 rpm for 5 min. After centrifugation, 200 μl of the supernatant was removed and inoculated into fresh BHI broth, which was incubated at 37°C until the bacteria reached the stationary phase. The procedures were repeated six to eight times until a homogeneous culture without any precipitation was obtained. Single colonies were then purified from this culture, reinoculated into fresh BHI broth, and grown to stationary phase to determine their autoaggregation properties. One colony, when grown in broth, consistently yielded homogeneous cultures with no precipitation of autoaggregates. When the broth was examined under a light microscope, the organism remained singular, in contrast to the formation of large autoaggregates seen with the wild-type F. nucleatum 12230 (data not shown). Therefore, this isolate was saved and designated lam.
TABLE 1.
Bacterial strains used in this study
| Strain | Source |
|---|---|
| Bacteroides forsythus ATCC 43037 | American Type Culture Collection |
| Campylobacter curvus ATCC 35224 | American Type Culture Collection |
| Eikenella corrodens ATCC 23834 | American Type Culture Collection |
| Fusobacterium nucleatum ATCC 10953 | American Type Culture Collection |
| Fusobacterium nucleatum ATCC 25586 | American Type Culture Collection |
| Fusobacterium nucleatum PK 1594 | P. Kolenbrander |
| Fusobacterium nucleatum 12230 | S. Finegold |
| Fusobacterium nucleatum 12230 lam | This study |
| Porphyromonas gingivalis 381 | Forsyth Dental Center |
| Prevotella intermedia ATCC 49046 | American Type Culture Collection |
Epithelial cell cultures.
Primary cultures of HGEC were prepared as previously described (32). Gingival tissues were collected during oral surgery from dental patients with or without periodontal infections. The collected gum tissues were digested with dispase (2.4 mg/ml; Boehringer Mannheim, Indianapolis, Ind.) and collagenase (150 U/ml; Sigma). Following incubation, the surface epithelium was digested further in 0.05% trypsin solution (GIBCO, Buffalo, N.Y.) for 5 to 10 min. The cells were collected and resuspended in keratinocyte growth medium 2 (KGM-2; Clonetics, San Diego, Calif.). The cultures were incubated at 37°C under 5% CO2 until confluent. The cells were then suspended with trypsin and seeded into either 24-well trays or 6-mm-diameter dishes for tissue attachment and invasion assays or for electron microscopy studies, respectively. The KB cells were maintained in Dulbecco minimal essential medium (GIBCO) containing 10% fetal calf serum (GIBCO) and 50 μg of gentamicin (GIBCO) per ml and were processed as described above for the assays.
Tissue attachment, invasion assays, and detection of secreted IL-8.
Attachment and invasion assays were conducted as previously described (16) with modifications for oral bacteria. Epithelial cells were grown to near confluency (5 × 104 to 8 × 104 cells per well) for the assays. Bacteria were added at a multiplicity of infection of 50 to 100 and incubated under 5% CO2 at 37°C. For invasion assays, the incubation was continued for 3 to 4 h. Following incubation, the supernatant was collected and stored at −20°C if the amount of IL-8 was to be determined. The monolayers were washed twice with sterile phosphate-buffered saline (PBS), fresh medium containing gentamicin (300 μg/ml) and metronidazole (200 μg/ml; Sigma) was added, and the monolayers were incubated for an additional 1 h to kill extracellular bacteria. The monolayers were rinsed again with PBS before being lysed with sterile water. Internalized bacteria were enumerated on BHI agar plates. For attachment assays, the bacteria were incubated with the monolayers for 1 h and then washed four times with PBS. The monolayers were lysed with water, and total cell-associated bacteria were enumerated on BHI agar plates. Control experiments showed that bacterial viability was not affected by the water treatment during cell lysis. Multiplication of the bacteria in the tissue culture medium during the assay was minimal. Therefore, unless otherwise indicated, the levels of attachment and invasion were expressed as the percentage of bacteria retrieved following cell lysis relative to the total number of bacteria initially added. All experiments were performed in duplicate or triplicate and repeated at least three times. For the inhibitory attachment assays, various sugars were incubated with the monolayers at 37°C for 30 min at a final concentration of 33 mg/ml, prior to the addition of F. nucleatum. The inhibitory invasion assays were carried out as described by Deshpande et al. (8). The inhibitors did not affect the viability of either the epithelial cells or F. nucleatum at the concentrations used. To visually demonstrate attachment of wild-type and mutant F. nucleatum, gingival epithelial cells were seeded onto chambered, tissue culture-treated glass slides (Falcon, Franklin Lakes, N.J.). Following a 1-h infection, the chambers were rinsed with PBS and photographed using a Zeiss microscope camera with a 40× objective. To determine the amount of IL-8 secreted by HGEC, enzyme-linked immunosorbent assays were conducted by the procedures of Huang et al. (18).
Electron microscopy.
HGEC or KB cells were grown to near confluency before infected with bacteria. After a 3-h incubation, the monolayers were washed 6 to 10 times with PBS and then treated for 2 min with 0.25% trypsin–1 mM EDTA (GIBCO) (for HGEC) or for 5 min with cell dissociation buffer (GIBCO) (for KB cells). Detached cells were collected in microcentrifuge tubes, rinsed with PBS, and fixed in 4% buffered glutaraldehyde fixative. Samples were postfixed in osmium tetroxide, dehydrated in graded acetone, and embedded in Spurr low-viscosity embedding media. Thin sections of the specimens were viewed and photographed with a Simens Elmiskop 101 transmission electron microscope.
RESULTS
Attachment to and invasion of HGEC.
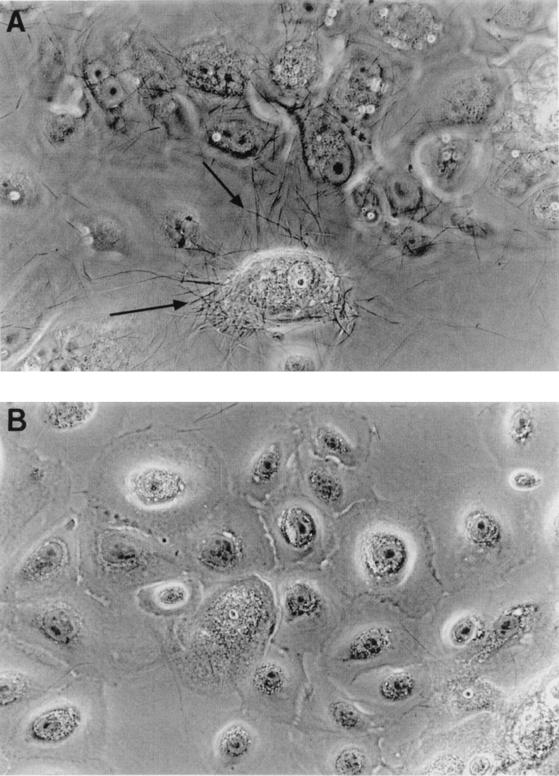
Attachment to and invasion of HGEC by oral bacteria were tested using primary cultures of gingival tissues obtained from dental patients with or without periodontal infections. P. gingivalis 381 was included as a positive control. All values obtained with other bacteria were compared with those obtained with P. gingivalis 381. For each individual assay, HGEC isolated from the same patient were used for all bacteria. Shown in Table 2 are the results of one representative assay. Except for C. curvus 35224, all strains tested adhered to HGEC. However, only F. nucleatum 12230 demonstrated an invasion activity comparable to that of P. gingivalis 381. The attachment and invasion levels of the lam mutant of F. nucleatum 12230 were about 15- to 20-fold lower than those of the wild-type F. nucleatum. Light micrographs (Fig. 1) also revealed differential attachment of wild-type and mutant F. nucleatum 12230 to HGEC. Prevotella intermedia ATCC 49046, B. forsythus ATCC 43037, C. curvus ATCC 35224, and E. corrodens ATCC 23834 appeared to be weakly invasive or noninvasive. When the assays were repeated using HGEC isolated from different patients, the attachment and invasion values remained consistent compared to those of P. gingivalis 381 (data not shown).
TABLE 2.
Bacterial attachment to and invasion of, and IL-8 production by, HGECa
| Strain | Attachment (%)b | Invasion (%)b | IL-8 production (pg/ml)c |
|---|---|---|---|
| B. forsythus ATCC 43037 | 1.90 ± 0.22 | 0.08 ± 0.04 | 146 ± 10 |
| C. curvus ATCC 35224 | 0.04 ± 0.01 | 0.03 ± 0.01 | 794 ± 163 |
| E. corrodens ATCC 23834 | 2.10 ± 0.30 | 0.01 ± 0.00 | 3,470 ± 41 |
| F. nucleatum 12230 | 8.16 ± 0.07 | 2.88 ± 0.22 | 5,029 ± 1,807 |
| F. nucleatum 12230 lam | 0.41 ± 0.14 | 0.16 ± 0.02 | NDd |
| P. gingivalis 381 | 8.57 ± 3.71 | 2.86 ± 0.14 | 312 ± 18 |
| P. intermedia ATCC 49046 | 4.78 ± 0.48 | 0.13 ± 0.01 | 1,154 ± 323 |
The assays were carried out as described in the text. Shown here are the mean results of one representative assay with standard deviations. Each experiment was carried out in duplicate.
The attachment and invasion levels were expressed as the percentage of bacteria retrieved following cell lysis relative to the total number of bacteria initially added. The background was <0.01% ± 0.00%.
The background was 68 ± 31 pg/ml.
ND, not determined.
FIG. 1.
Light micrographs of attachment of F. nucleatum 12230 to HGEC. (A) Wild-type bacteria; (B), the lam mutant. The images were taken using a 40× objective. The filaments indicated by the arrows are F. nucleatum.
Production of IL-8 upon infection by oral bacteria.
The levels of IL-8 secreted by HGEC at 4 h after infection by oral bacteria were also measured (Table 2). All strains induced IL-8 production by HGEC in this assay. F. nucleatum stimulated the greatest IL-8 production, while P. gingivalis induced the least. When HGEC from different individuals were tested, C. curvus, E. corrodens, F. nucleatum, and Prevotella intermedia consistently stimulated the production of IL-8, with F. nucleatum acting as one of the strongest stimulators (data not shown). B. forsythus induced very low levels of IL-8 secretion. The levels of IL-8 produced upon infection by P. gingivalis fluctuated from moderately increased to moderately decreased (data not shown).
Attachment to and invasion of KB cells.
F. nucleatum 12230 and additional strains of this species were tested for their ability to adhere to and invade KB cells (Table 3). F. nucleatum 12230 and its lam mutant exhibited similar levels of attachment to and invasion of KB cells as with HGEC. F. nucleatum ATCC 10953, ATCC 25586, and PK 1594 all adhered strongly to KB cells. F. nucleatum ATCC 10953 and ATCC 25586 were also highly invasive, whereas PK 1594 was only slightly invasive.
TABLE 3.
Attachment and invasion of KB cells by F. nucleatuma
| Strain | Attachment (%)b | Invasion (%)b |
|---|---|---|
| F. nucleatum ATCC 10953 | 19.2 ± 0.3 | 7.4 ± 1.1 |
| F. nucleatum ATCC 25586 | 16.8 ± 0.8 | 7.3 ± 2.0 |
| F. nucleatum PK 1594 | 14.4 ± 0.8 | 0.3 ± 0.0 |
| F. nucleatum 12230 | 17.7 ± 0.5 | 4.5 ± 1.9 |
| F. nucleatum 12230 lam | 1.5 ± 0.4 | 0.1 ± 0.0 |
The assays were carried out as described in the text. Shown here are representative results (mean ± standard deviation). The assay was carried out in duplicate and repeated at least three times.
The attachment and invasion levels were expressed as indicated in the text and Table 2, footnote b. The background was <0.01% ± 0.00%.
Inhibition of invasion of HGEC and KB cells.
To dissect the invasion process, a group of metabolic inhibitors were tested (Table 4). The extent of inhibition varied when HGEC from different individuals were used (data not shown). Nonetheless, the results indicated that for both HGEC and KB cells, invasion by F. nucleatum required multiple components of the host, including actin, microtubules, signal transduction, protein synthesis, and energy metabolism. Chloramphenicol inhibited protein synthesis without affecting the viability of F. nucleatum (data not shown). The results indicated the requirement for a bacterial protein component(s) in the process.
TABLE 4.
Effects of metabolic inhibitors on invasion by F. nucleatum 12230
| Inhibitora | Target | Invasion of HGEC (%)b | Invasion of KB (%)b |
|---|---|---|---|
| None | 100 ± 3.0 | 100 ± 17.7 | |
| Cytochalasin D (HC) | Actin | 4.7 ± 1.3 | 5.3 ± 0.2 |
| Nocodazole (HC) | Microtubule | 1.7 ± 0.9 | 42.5 ± 14.2 |
| Staurosporine (HC) | Protein kinase | 32.6 ± 7.7 | 0.9 ± 0.6 |
| Cycloheximide (HC) | Protein synthesis | 8.6 ± 2.6 | 21.2 ± 1.8 |
| Sodium azide (HC) | Energy metabolism | 22.7 ± 4.7 | 3.5 ± 0.9 |
| Chloramphenicol (Fn) | Protein synthesis | 17.6 ± 3.0 | 2.2 ± 1.4 |
The inhibitors were preincubated with either the host cells (HC) or F. nucleatum (Fn) as described in the text. The concentrations of the chemicals were as follows: cytochalasin D, 1 μg/ml; nocodazole, 10 μg/ml; staurosporine, 1 μM; cycloheximide, 100 μg/ml; sodium azide, 50 mM; and chloramphenicol, 5 μg/ml.
The level of invasion was expressed as a percentage of the control level without any inhibitors. Means and standard deviations are given.
Inhibition of attachment to KB cells.
To determine whether the attachment of F. nucleatum to epithelial cells involves a lectin-like adhesin(s), various sugars were tested (Table 5). Galactose-containing sugars (galactose, N-acetylgalactosamine, and lactose) strongly inhibited the adherence of F. nucleatum to KB cells, whereas non-galactose-containing sugars had little or no effect.
TABLE 5.
Effects of sugars on attachment of F. nucleatum to KB cells
| Sugara | Attachment (%)b |
|---|---|
| None | 100 ± 3.3 |
| Galactose | 5.2 ± 0.6 |
| N-Acetylgalactosamine | 2.7 ± 1.3 |
| Lactose | 5.4 ± 1.3 |
| Glucose | 63.1 ± 1.5 |
| N-Acetylglucosamine | 65.9 ± 2.7 |
| Mannose | 63.1 ± 1.5 |
| Fucose | 74.0 ± 3.1 |
| Fructose | 94.8 ± 7.4 |
| Sucrose | 78.7 ± 7.9 |
| N-Acetylneuraminic acid | 60.6 ± 3.1 |
The sugars were added to KB cells at a final concentration of 33 mg/ml prior to the addition of F. nucleatum, as described in the text.
The level of attachment was expressed as a percentage of the control level without any sugar added. Means ± standard deviations are given.
Electron microscopy.
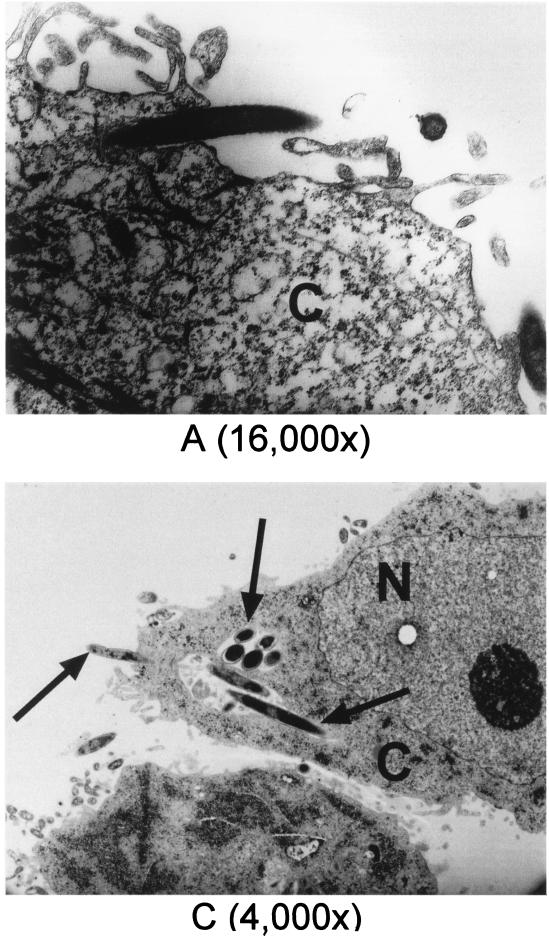
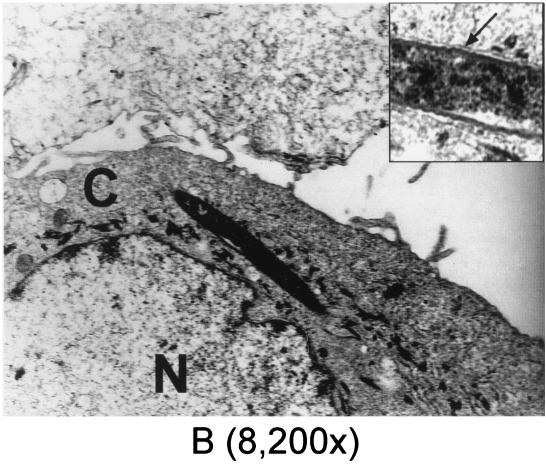
Transmission electron microscopy (TEM) was performed to further verify invasion of HGEC and KB cells by F. nucleatum 12230. Shown in Fig. 2 are transmission electron micrographs elucidating different invasion stages, ranging from the bacterium halfway inside the cell (Fig. 2A and C) to completely internalized (Fig. 2B and C). Once internalized, the bacterium resided in the cytoplasm and was confined to a membrane-bound vacuole (Fig. 2B and C, inset). This process has characteristics of a zipping mechanism (11). The frequencies of intracellular F. nucleatum 12230 observed under TEM correlated well with the invasion activities obtained in the quantitative assay (data not shown). Upon screening of samples prepared with HGEC infected with F. nucleatum 12230 lam, C. curvus ATCC 35224, or E. corrodens ATCC 23834, respectively, no intracellular bacteria were detected (data not shown); the same negative result was also observed with B. forsythus ATCC 43037 (S. Holt, personal communication). Only one internalized Prevotella intermedia ATCC 49046 was detected by TEM screening (data not shown).
FIG. 2.
Transmission electron micrographs of invasion of HGEC and KB by F. nucleatum 12230. (A) F. nucleatum entering HGEC; (B) F. nucleatum internalized in HGEC, with the membrane of the vacuole indicated by the arrow in the inset; (C) F. nucleatum invading and internalized in KB, as indicated by the arrows. C, cytoplasm; N, nucleus. Numbers in parentheses indicate original magnifications.
DISCUSSION
Periodontal diseases result from complex actions of a group of periodontal bacteria, mostly gram-negative anaerobes. However, so far only a few, such as A. actinomycetemcomitans and P. gingivalis, have been characterized as bona fide periodontal pathogens. Other gram-negative organisms, although often linked to various forms of periodontal diseases, are considered only putative pathogens, largely due to our limited understanding of their virulence potential. Therefore, we set out to analyze the virulence mechanisms of a selected group of putative periodontal pathogens, including B. forsythus, C. curvus, E. corrodens, F. nucleatum, and Prevotella intermedia, while P. gingivalis served as a positive control. The focus of our study was on interactions between these bacteria and oral epithelial cells. To mimic the in vivo situation as much as possible, primary cultures of HGEC were initially used in the in vitro model system. Three biological activities often associated with virulence were measured: bacterial attachment, invasion, and induction of IL-8 production. Our rationale for such a general screening was to identify potential virulence mechanisms of these oral bacteria to help elucidate their relevance to periodontal disease.
Adherence is a common characteristic shared by many pathogens since it is a crucial step for establishing an infection. All strains tested in this study adhered to HGEC except C. curvus ATCC 35224, suggesting that these bacteria have the potential to interact with the epithelium to establish colonization. Since only one strain from each genus was selected in our screening, it is possible that variations among species exist. For instance, other oral campylobacters may exhibit adherence properties despite the inability of C. curvus ATCC 35224 to attach to HGEC.
Invasion of epithelial cells by oral bacteria has attracted considerable attention (27). The majority of the putative pathogens screened in this study, however, exhibited limited invasiveness. Prevotella intermedia ATCC 49046 was weakly invasive, if at all, with its invasion level being about 1/20 that of P. gingivalis 381. Dorn et al. recently reported invasion of KB cells by a clinical isolate of this species, Prevotella intermedia 17, with its invasion level also being about 1/20 of that of P. gingivalis 381 (9). B. forsythus ATCC 43037, C. curvus ATCC 35224 and E. corrodens ATCC 23834 were noninvasive in our study. However, it cannot be ruled out that other invasive strains of these species may exist.
F. nucleatum 12230, on the other hand, was distinguished from the rest of the group in the invasion assays. This organism was demonstrated to be highly invasive, with its activity being comparable to that of P. gingivalis. To the best of our knowledge, this is the first documented study demonstrating such invasive properties for F. nucleatum. The fact that the lam mutant was defective in attachment and invasion of HGEC implied the requirement of a bacterial component(s) during these processes. Therefore, the possibility of an invasion artifact was unlikely. The spontaneous lam mutant was defective in aggregation with human lymphocytes and coaggregation with P. gingivalis as well (unpublished results). Several adherence modes and putative adhesins have been proposed for F. nucleatum (20, 24, 35, 36, 38). It is possible that one or more adhesins are involved in various adherence and invasion processes. Alternatively, different components may be required for different adherence functions and lam was simply pleiotropic. The nature of the mutation in this lam mutant is under investigation.
Once the invasion phenotype of F. nucleatum was established using HGEC, invasion of KB cells was examined. Both F. nucleatum 12230 and its lam mutant exhibited similar relative levels of invasion of KB cells, as with HGEC. Electron microscopy also revealed similar patterns of invasion of HGEC and KB cells by F. nucleatum 12230. Due to the distinct large size of this organism, different stages of penetration were easily visualized. It appeared that F. nucleatum 12230 invaded both cell types via a zipping mechanism, which is the first report of this mechanism for an oral bacterium. Upon internalization, the organism apparently resided in vacuoles. Furthermore, metabolic inhibition assays showed that invasion required the participation of host actin, microtubules, signal transduction, and energy metabolism. Invasion required protein synthesis by F. nucleatum as well. These observations suggest that invasion of HGEC and KB cells have significant similarities. Therefore, KB cells may be employed in certain studies in the future in place of HGEC for ease of manipulation. The adherence of F. nucleatum 12230 to KB cells involved a galactose-binding lectin(s), since the attachment was greatly inhibited by galactose-containing sugars. This property is currently being utilized to identify adhesins of this organism. It is possible that a non-lectin-like adhesin(s) may also be involved in the interactions between F. nucleatum and the host cells. Using KB cells, additional strains of F. nucleatum were tested for attachment and invasion. The results suggested that invasion was a rather common feature of the species, supporting early histological findings of penetration of deeper tissues by fusiform bacteria in periodontal patients (1, 14, 25, 33).
Besides bacterial penetration into deeper tissues, one other important characteristic of periodontal infection is inflammation, a result of neutrophil infiltration into infected sites (2, 30). Continuous production of proinflammatory cytokines, such as IL-8, appears to be important for the progression of periodontal infections and tissue destruction (30). With the exception of P. gingivalis 381, all the bacteria tested in this study consistently stimulated the production of IL-8 as early as 4 h postinfection, implicating their ability to induce inflammation. Previous studies revealed that when epithelial cells were infected at high doses, or if the infection persisted, P. gingivalis 381 suppressed the production of IL-8 (6). In the present study, the effect of P. gingivalis 381 on IL-8 production was minimal, varying from moderate suppression to moderate stimulation. This discrepancy could be attributed to two factors: a low infectious dose (multiplicity of infection, <100:1), and a relatively short period of infection (4 h). The initiation and progression of periodontal infections may be affected by the dynamic competition between suppression and stimulation of immune mediators by various periodontal bacteria. Combining evidence obtained in this study with the results of an earlier study (6), we found that F. nucleatum is likely to be a strong stimulator of IL-8 production throughout the course of infection. This conclusion is consistent with clinical studies demonstrating that F. nucleatum is highly prevalent during the early stages of inflammation associated with gingivitis (28).
In summary, we have demonstrated that several potential periodontal pathogens were able to adhere to oral epithelial cells. The majority of these organisms were also capable of inducing IL-8 production from these cells. In particular, F. nucleatum was highly invasive for both HGEC and KB cells and was a potent stimulator of IL-8 expression. With its length of 10 times that of Escherichia coli, F. nucleatum 12230 is by far the largest bacteria demonstrated to invade a mammalian cell. Thus, it could potentially be developed into an excellent model system to study the cellular processes of invasion. In addition, F. nucleatum may be an important contributor to periodontal disease either directly or by serving as a mediator of plaque colonization for other virulent anaerobes.
ACKNOWLEDGMENTS
We thank S. Finegold and P. Kolenbrander for kindly providing F. nucleatum 12230 and PK1594, respectively.
This work was supported in part by NIH grants AI09268, GM54666, DE08293, and DE08240.
REFERENCES
- 1.Allenspach-Petrzilka G E, Guggenheim B. Bacterial invasion of the periodontium; an important factor in the pathogenesis of periodontitis? J Clin Periodontol. 1983;10:609–617. doi: 10.1111/j.1600-051x.1983.tb01299.x. [DOI] [PubMed] [Google Scholar]
- 2.Bickel M. The role of interleukin-8 in inflammation and mechanisms of regulation. J Periodontol. 1993;64:456–460. [PubMed] [Google Scholar]
- 3.Brook I. Recovery of anaerobic bacteria from clinical specimens in 12 years at two military hospitals. J Clin Microbiol. 1988;26:1181–1188. doi: 10.1128/jcm.26.6.1181-1188.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown L J, Loe H. Prevalence, extent, severity and progression of periodontal disease. Perio 2000. 1993;2:57–71. doi: 10.1111/j.1600-0757.1993.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 5.Civen R, Jousimies-Somer H, Marina M, Borenstein L, Shah H, Finegold S M. A retrospective review of cases of anaerobic empyema and update of bacteriology. Clin Infect Dis. 1995;20:S224–S229. doi: 10.1093/clinids/20.supplement_2.s224. [DOI] [PubMed] [Google Scholar]
- 6.Darveau R P, Belton C M, Reife R A, Lamont R J. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demuth D R, Savary R, Golub E, Shenker B J. Identification and analysis of fipA, a Fusobacterium nucleatum immunosuppressive factor gene. Infect Immun. 1996;64:1335–1341. doi: 10.1128/iai.64.4.1335-1341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande R G, Khan M B, Genco C A. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun. 1998;66:5337–5343. doi: 10.1128/iai.66.11.5337-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn B R, Leung K-P, Progulske-Fox A. Invasion of human oral epithelial cells by Prevotella intermedia. Infect Immun. 1998;66:6054–6057. doi: 10.1128/iai.66.12.6054-6057.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan M J, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay B B, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 12.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillece-Castro B L, Prakobphol A, Bburlingame A L, Leffler H, Fisher S J. Structure and bacterial receptor activity of human salivary proline-rich glycoprotein. J Biol Chem. 1991;266:17358–17368. [PubMed] [Google Scholar]
- 14.Giuliana G, Ammatuna P, Pizzo G, Capone F, D'Angelo M. Occurrence of invading bacteria in radicular dentin of periodontally diseased teeth: microbiological findings. J Clin Periodontol. 1997;24:478–485. doi: 10.1111/j.1600-051x.1997.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 15.Grenier D, Michaud J. Demonstration of human immunoglobulin G Fc-binding activity in oral bacteria. Clin Diagn Lab Immunol. 1994;1:247–249. doi: 10.1128/cdli.1.2.247-249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han Y W, Miller V L. Reevaluation of the virulence phenotype of the inv yadA double mutants of Yersinia pseudotuberculosis. Infect Immun. 1997;65:327–330. doi: 10.1128/iai.65.1.327-330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill G B. Preterm birth: associations with genital and possibly oral microflora. Ann Periodontol. 1998;3:222–232. doi: 10.1902/annals.1998.3.1.222. [DOI] [PubMed] [Google Scholar]
- 18.Huang G T-J, Kinder Haake S, Kim J-W, Park N-H. Differential expression of interleukin-8 and intercellular adhesion molecule-1 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans or Porphyromonas gingivalis infection. Oral Microbiol Immunol. 1998;13:301–309. doi: 10.1111/j.1399-302x.1998.tb00711.x. [DOI] [PubMed] [Google Scholar]
- 19.Kagnoff M F, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Investig. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinder S, Holt S C. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J Bacteriol. 1993;175:840–850. doi: 10.1128/jb.175.3.840-850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander P E. Coaggregations among oral bacteria. Methods Enzymol. 1995;253:385–397. doi: 10.1016/s0076-6879(95)53033-9. [DOI] [PubMed] [Google Scholar]
- 22.Lamont R J, Chan A, Belton C M, Izutsu K T, Vasel D, Weinberg A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect Immun. 1995;63:3878–3885. doi: 10.1128/iai.63.10.3878-3885.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madianos P N, Papapanou P N, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–664. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangan D F, Novak M J, Vora S A, Mourad J, Kriger P S. Lectinlike interactions of Fusobacterium nucleatum with human neutrophils. Infect Immun. 1989;57:3601–3611. doi: 10.1128/iai.57.11.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manor A, Lebendiger M, Shiffer A, Tovel H. Bacterial invasion of periodontal tissues in advanced periodontitis in humans. J Periodontol. 1984;55:567–573. doi: 10.1902/jop.1984.55.10.567. [DOI] [PubMed] [Google Scholar]
- 26.Meyer D H, Lippmann J E, Fives-Taylor P M. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect Immun. 1996;64:2988–2997. doi: 10.1128/iai.64.8.2988-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer D H, Mintz K P, Fives-Taylor P M. Models of invasion of enteric and periodontal pathogens into epithelial cells: a comparative analysis. Crit Rev Oral Biol Med. 1997;8:389–409. doi: 10.1177/10454411970080040301. [DOI] [PubMed] [Google Scholar]
- 28.Moore W E C, Moore L V H. The bacteria of periodontal diseases. Perio 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 29.Njoroge T, Genco R J, Sojar H T, Hamada N, Genco C A. A role for fimbriae in invasion of oral epithelial cells. Infect Immun. 1997;65:1980–1984. doi: 10.1128/iai.65.5.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–266. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- 31.Ozaki M, Miyake Y, Shirakawa M, Takemoto T, Okamoto H, Suginaka H. Binding specificity of Fusobacterium nucleatum to human erythrocytes, polymorphonuclear leukocytes, fibroblasts, and HeLa cells. J Periodont Res. 1990;25:129–134. doi: 10.1111/j.1600-0765.1990.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 32.Park N H, Min B M, Li S L, Huang M Z, Cherick H M, Doniger J. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis. 1991;12:1627–1631. doi: 10.1093/carcin/12.9.1627. [DOI] [PubMed] [Google Scholar]
- 33.Saglie R, Newman M G, Carranza Jr F A, Pattison G L. Bacterial invasion of gingiva in advanced periodontitis in humans. J Periodontol. 1982;53:217–222. doi: 10.1902/jop.1982.53.4.217. [DOI] [PubMed] [Google Scholar]
- 34.Sandros J, Papapanou P N, Nannmark U, Dahlen G. Porphyromonas gingivalis invades human pocket epithelium in vitro. J Periodont Res. 1994;29:62–69. doi: 10.1111/j.1600-0765.1994.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 35.Shaniztki B, Ganeshkumar N, Weiss E I. Characterization of a novel N-acetylneuraminic acid-specific Fusobacterium nucleatum PK1594 adhesin. Oral Microbiol Immunol. 1998;13:47–50. doi: 10.1111/j.1399-302x.1998.tb00750.x. [DOI] [PubMed] [Google Scholar]
- 36.Shaniztki B, Hurwitz D, Smorodinsky N, Ganeshkumar N, Weiss E I. Identification of a Fusobacterium nucleatum PK1594 galactose-binding adhesin which mediates coaggregation with periopathogenic bacteria and hemagglutination. Infect Immun. 1997;65:5231–5237. doi: 10.1128/iai.65.12.5231-5237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Socransky S S, Haffajee A D. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 38.Tuttle R S, Strubel N A, Mourad J, Mangan D F. A non-lectin-like mechanism by which Fusobacterium nucleatum 10953 adheres to and activates human lymphocytes. Oral Microbiol Immunol. 1992;7:78–83. doi: 10.1111/j.1399-302x.1992.tb00513.x. [DOI] [PubMed] [Google Scholar]