Abstract
Respiratory syncytial virus (RSV) and human metapneumovirus (HMPV) cause a high burden of disease, particularly in children and the elderly. With the aim to add knowledge on RSV and HMPV infections in Italy, a prospective, multicenter study was conducted by eight centers of the Working Group on Respiratory Virus Infections (GLIViRe), from December 2018–April 2019. Weekly distribution and patients’ demographic and clinical data were compared in 1300 RSV and 222 HMPV-positive cases. Phylogenetic analysis of the G-glycoprotein coding region was performed to characterize circulating strains. RSV positivity ranged from 6.4% in outpatients of all ages to 31.7% in hospitalized children; HMPV positivity was 4–1.2% with no age-association. RSV season peaked in February and ended in mid-April: HMPV circulation was higher when RSV decreased in early spring. RSV was more frequent in infants, whereas HMPV infected comparatively more elderly adults; despite, their clinical course was similar. RSV-B cases were two-thirds of the total and had similar clinical severity compared to RSV-A. Phylogenetic analysis showed the circulation of RSV-A ON1 variants and the predominance of RSV-B genotype BA10. HMPV genotype A2c was the prevalent one and presented insertions of different lengths in G. This first multicenter Italian report on seasonality, age-specific distribution, and clinical presentation of RSV and HMPV demonstrated their substantial disease burden in young patients but also in the elderly. These data may provide the basis for a national respiratory virus surveillance network.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-022-00973-3.
Keywords: Respiratory syncytial virus, Human metapneumovirus, Respiratory viruses, Molecular epidemiology, Bronchiolitis, Pneumonia
Introduction
Respiratory syncytial virus (RSV) infection is the main cause of hospitalization in children, causing 1.4 million hospitalizations, and 45,700 deaths in infants less than 6 months, estimated worldwide in 2019 [1]. Most children get infected with RSV in the first years of life, but only a fraction develops severe lower respiratory tract infection, with bronchiolitis being the most common severe clinical manifestation associated with RSV infection in infants under 1 year of age [1]. Risk factors associated with severe disease such as very young age, premature birth, and underlying chronic diseases have been well documented [2, 3]; nonetheless, it has been reported that most children hospitalized for RSV bronchiolitis were previously healthy [2, 3]. In older children, RSV may cause pneumonia, wheezing episodes and acute asthma exacerbations [1–3]. Reinfections occur frequently, causing less severe disease [2, 3], until late in life when RSV-related hospitalizations and death occur at rates similar to those of seasonal influenza A virus infection [4, 5]. Although the ever-increasing awareness on RSV disease burden, this viral infection is not routinely diagnosed nor can be differentiated on a clinical basis, particularly in adult patients.
Human metapneumovirus (HMPV) was identified in 2001 in the Netherlands [6] and is structurally similar to RSV; both were recently included in the novel family Pneumoviridae, in the genera Metapneumovirus and Orthopneumovirus, respectively [7]. With respect to RSV, HMPV has a smaller genome and a different gene order and lacks two non-structural proteins [6, 7]. HMPV causes a broad range of illnesses ranging from upper respiratory tract infection to severe lower respiratory tract disease, among children [8] and older adults [9, 10], with lower prevalence rates with respect to RSV [8–10]. Moreover, in comparative studies, HMPV has resulted less pathogenic than RSV, having lesser capacity to counteract the interferon-mediated antiviral response and to induce inflammatory cytokine [11].
Extensive data on these important respiratory pathogens are required to guide health authorities in priorities for disease control at national and international level. In Italy, several studies have previously characterized RSV and HMPV-related diseases, investigating on respiratory viral infections in children or in high-risk patients of any age [12–15], but data were not aggregated at the national level. With the aim to assess the impact of respiratory viruses’ infection, in different age and risk groups, a national network, named GLIViRe, was initiated in 2018, by eight research or diagnostic laboratories. The eight GLIViRe centers, during the first period of surveillance, from December 2018–April 2019, tested more than 11,500 respiratory samples, and in this study, data about RSV and HMPV infections are reported. The distribution of RSV and HMPV-positive cases was analyzed by week of occurrence and by age group; moreover, clinical data, available from around half of the RSV- and HMPV-positive samples, were compared. A phylogenetic analysis based on sequences of the external glycoprotein (G) was performed to characterize circulating strains of RSV and HMPV.
Methods
The network
GLIViRe (the Italian acronym for Working Group on Respiratory Viral Infections) was established in November 2018, during the 47th National Congress of the Italian Association of Clinical Microbiologists (AMCLI) affiliated to FEMS (Federation of European Microbiology Societies). GLIViRe is composed by eight centers (from North-West, North-East and Central Italy, Supplementary Fig. 1): 1. Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo, Pavia (PV); 2. Department of Biomedical Sciences for Health, University of Milan, Milan (UNI-MI); 3. Virology Unit, Fondazione IRCCS Ca' Granda Ospedale Maggiore Policlinico, Milan (POL-MI); 4. Laboratory of Microbiology and Virology, Azienda Sanitaria dell'Alto Adige, Bolzano (BZ); 5. Department of Clinical Pathology, AULSS 2, Marca Trevigiana, Treviso (TV); 6. Laboratory of Virology, Microbiology Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna (BO); 7. Department of Biomedical Sciences and Public Health, Virology Unit, University Politecnica Marche, Ancona (AN); and 8. Virology Laboratory, Department of Molecular Medicine, Sapienza University, Rome (RM).
GLIViRe included: three large hospitals accredited by the Italian Ministry of Health for biomedical research (the IRCCS centers PV, POL-MI and BO), two large territorial health structures (BZ and TV), and three University Hospital Virology Units (UNI-MI, AN and RM). The institutional review board and the Ethics Committee of Rome University Hospital approved the study (Prot. 107/12); informed consent was obtained from individual participants or their parents. Patients' demographic and clinical data were retrospectively extracted from healthcare records, and in line with confidentiality requirements, the database was anonymized.
Patients and samples
The study involved respiratory samples (n = 11,577) performed for diagnostic purpose by the eight centers from December 1, 2018, to April 30, 2019; most of these (11,008/11,577; 95.1%) were also tested for HMPV. In six out of eight centers, testing for respiratory viruses was performed on patients of any age, upon request of the attending physician, on the judgment that the illness was due to a respiratory infection. The University of Milan participates in the InfluNet national surveillance of influenza, tested RSV and other respiratory viruses, in influenza-like illness (ILI) cases, using the ECDC definition of ILI, i.e., fever (temperature ≥ 37.8 °C), with at least one systemic symptom (i.e., headache, asthenia, myalgia), and at least one respiratory symptom (i.e., cough, dyspnea, acute pharyngitis). In Rome, respiratory viruses were tested only in children up to 3 years of age (< 3 y), hospitalized in Pediatric Departments of Sapienza University Hospital, with a clinical diagnosis of bronchiolitis (< 1y) or acute respiratory infection.
Respiratory samples were mainly nose–throat swabs and nasopharyngeal aspirates; lower respiratory tract samples were about 5% of total samples. RSV, HMPV and other respiratory viruses were tested using different molecular methods (Table 1). PV, UNI-MI, and RM centers used home-made, qualitative or real-time PCRs previously validated and published [12, 14, 15] to detect respiratory viruses; the other centers detected respiratory viruses using multiplex real-time PCR assays [Anyplex™ II RV16 (Seegene), Allplex™ Respiratory Panel (Seegene), Biofire FilmArray™ (Biomerieux), FTD21plus™ (Siemens)] (Table 1). In order to subtype RSV-positive cases, PV, UNI-MI, BO, AN and RM used an in-house real-time PCR assay, targeting the matrix (M) gene, as described [16].
Table 1.
Respiratory virus diagnosis and RSV- and HMPV-positive rates in the GLIViRe centers
| Center | 1 PV | 2 UNI-MI | 3 POL-MI | 4 BZ | 5 TV | 6 BO | 7 AN | 8 RM |
|---|---|---|---|---|---|---|---|---|
| N. of samples tested | 1749 | 569 | 1849 | 1978 | 3267 | 921 | 1036 | 208 |
| Molecular method | In-house assay [16] | In-house assay [16] | Anyplex™ II RV16 | Allplex™ Respiratory Panel | Allplex™ Respiratory Panel | FilmArray™ | FTD21™ | In-house assay [12] |
| Inclusion criteria | Clinic (all ages) | ILI (all ages) | Clinic (all ages) | Clinic (all ages) | Clinic (all ages) | Clinic (all ages) | Clinic (all ages) | Clinic (0–3 y) |
| Outpatients tested | yes | yes | yes | yes | yes | no | yes | no |
| N. of RSV-positive samples (%) | 157 (9.0%) | 46 (8.1%) | 304 (16.4%) | 264 (13.3%) | 210 (6.4%) | 131 (14.2%) | 122 (11.8%) | 66 (31.7%) |
| N. of RSV-A-positive samples (%)a | 20 (43.5%) | 16 (42.1%) | 98 (41.7%) | 26 (9.9%) | 60 (28.8) | 22 (42.3%) | 33 (28.2%) | 8 (16.7%) |
| N. of RSV-B-positive samples (%)a | 26 (56.5%) | 22 (57.9%) | 137 (58.3%) | 236 (90.1%) | 148 (71.2%) | 30 (57.7%) | 84 (71.8) | 48 (83.3%) |
| RSV-positive patients’ median age years (range) | 44.7 (0.04–93.5) | 9.7 (0.41–86.1) | 0.93 (0.02–92.5) | 0.89 (0.03–98.2) | 64.3 (0.04–98.5) | 0.43 (0.04–14.8) | 0.85 (0.04–91.4) | 0.84 (0.05–1.77) |
| HMPV-positive samples | 21 (1.2%) | ND | 77 (4.2%) | 32 (1.6%) | 58 (1.8%) | 13 (1.4%) | 17 (1.6%) | 4 (1.9%) |
| HMPV positive patients’ median age days (range) | 17.8 (0.06–84.9) | ND | 3.3 (0.11–84.3) | 4.6 (0.15 –85.1) | 69.2 (0.11 –97.4) | 1.3 (0.12 –3.5) | 3.1 (0.16 –57.4) | 0.76 (0.26 –1.3) |
RSV respiratory syncytial virus, HMPV human metapneumovirus, ILI influenza-like illness, ND not done
athe subtype % is calculated on the RSV-positive samples in the which one subtype could be assigned
Phylogenetic analysis
RSV-A and -B positive samples were randomly selected for genomic characterization of around 25% of cases. Amplicons for RSV sequencing were obtained with subtype-specific forward primers (A-Forseq and B-Forseq) designed in our previous study [17], (position 481–498 of the RSV-A2 reference strain G gene), and the F1 reverse primer targeting both subtypes at the 5’ end of the fusion protein gene [18]. HMPV sequences were performed on amplicons of the whole G gene obtained with published primers [19].
Good quality sequences were aligned with reference sequences using Bioedit v7.1.3. The best-fit evolutionary model and parameters were selected using Model test on MEGA 11 software. For each dataset, the phylogenetic tree was constructed using the maximum likelihood method based on the Tamura–Nei model and a discrete Gamma distribution with five categories (+ G) to model evolutionary rate differences among sites, with bootstrap values of 1,000.
Statistical analysis
For categorical variables, Pearson's chi-square was used to test the difference between independent groups. For numerical variables, the Mann–Whitney U was used for comparing two groups when variables were non-normally distributed. Data were analyzed using SPSS version 27 (IBM Corp, New York, USA); a p value < 0.05 was considered as statistically significant.
Results
RSV and HMPV detection rates
In the eight GLIViRe centers, located in North-West (PV, UNI-MI, POL-MI), North-East (BZ, TV, BO), and central Italy, eastward (AN) and westward (RM) (Figure S1), a total of 11,577 respiratory samples were tested by molecular assays during the 2018/19 epidemic season (Table 1).
Cumulatively, RSV-positive samples were 1300/11,577 (11.2%). RSV positivity rates ranged from 6.4% (TV, a territorial health unit where most samples were from adult patients) to 31.7% (RM University Hospital, where only hospitalized children were tested) (Table 1). Overall, HMPV was detected in 2% of tested samples (222/11,008), with rates ranging from 1.2% to 4% with no relationship with patients’ characteristics (Table 1).
RSV and HMPV circulation by calendar week and geographical location
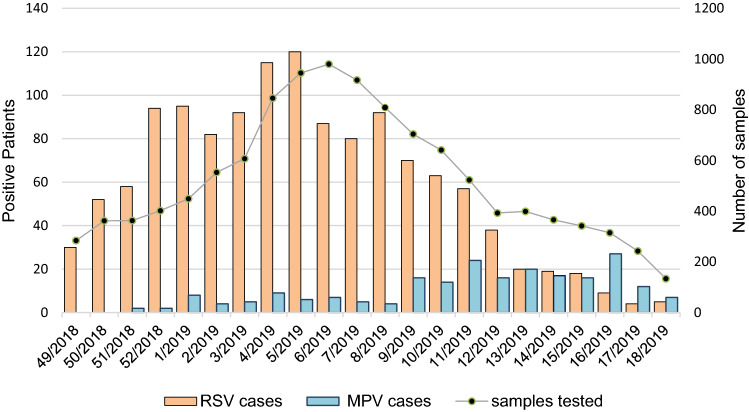
The total number of tested samples and the rate of RSV and HMPV positivity are shown in Fig. 1, by week of sample collection.
Fig. 1.
Weekly distribution of the total number of tested respiratory samples, the respiratory syncytial virus (RSV) and human metapneumovirus (HMPV)-positive cases. On the X-axis, the calendar week of the study period (December 2018–April 2019) is represented; on the left Y-axis, the number of RSV and HMPV-positive patients and on the right Y-axis, the total number of samples tested by the GLIViRe centers are reported
Using a definition of the epidemic season starts as the week when RSV cases exceed 1.2% of total RSV-positive specimens [20, 21], in the first week of December, RSV activity (30 detections out of 1300 = 2.3%) was already over the start of season and then rapidly increased. The seasonal peak of RSV circulation was recorded in week 05-2019; other two smaller peaks were recorded in week 52-2018 and in week 08-2019 (Fig. 1). After the cases’ decline in weeks 12- and 13-2019 (Fig. 1), week 15–2019 was the last one with RSV circulation above the epidemic threshold (18/1300 = 1.38%).
During RSV circulation, HMPV activity was very low in all centers; HMPV frequency of detection increased starting from week 09-2019 (16/222 = 7.2%), apparently reached a peak in week 16-2019 (27/222 = 12.2%) and suddenly declined (Fig. 1).
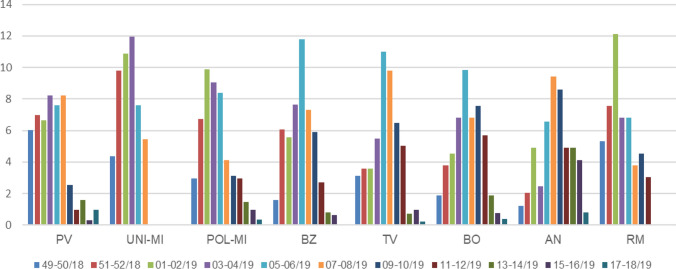
Circulation of RSV was further analyzed in the different locations to appreciate geographical differences. Therefore, the biweekly percentage of RSV detection, i.e., the ratio between positive cases in two weeks (indicated in the legend) and the total RSV-positive cases in that center, was calculated (Fig. 2). Interestingly, the centers that are located in north-western (PV, UNI-MI, and POL-MI) and central-western Italy (RM) had most positive cases in December and January, the epidemic peak in January and relatively few cases in March, whereas the centers located eastward (BZ, TV, BO and AN) had relatively lower rates of RSV positivity in December, the epidemic peak in mid-February and a sustained RSV circulation in March (Fig. 2). The percentage of RSV-positive cases from total numbers of tested samples at each center was also calculated biweekly and depicted in Supplementary Fig. 2.
Fig. 2.
Biweekly distribution of the RSV cases in the eight GLIViRe centers. The number of RSV cases per week with respect of the total RSV-positive cases in each center was calculated in percentage and values from two weeks aggregated, as shown in the color legend
The low number of weekly HMPV cases does not allow a separate analysis by geographical location.
Demographic and clinical characterization of RSV- and HMPV-positive patients
Demographic data (age and sex), the presence of viral co-infections, the hospitalization status, and the need for intensive care were reported by the GLIViRe centers for the majority of RSV- and HMPV-positive cases (Table 2).
Table 2.
Comparison of demographic and clinical data between RSV and HMPV-positive cases
| Categories | RSV-positivea (N = 1300) | HMPV-positivea (N = 222) | P value | |
|---|---|---|---|---|
| Sex (male) | 661 (50.8%) | 106 (47.7%) | ns | |
| Age group | 0–6 m | 481 (37.4%) | 49 (22.1%) | < 0.001 |
| 6 m–3 y | 278 (21.6%) | 46 (20.7%) | ||
| 3 y–16 y | 104 (8.1%) | 22 (9.9%) | ||
| 16 y–65 y | 148 (11.5%) | 37 (16.6%) | ||
| 65 y–80 y | 127 (9.9%) | 26 (11.7%) | ||
| > 80 y | 148 (11.5%) | 42 (18.9%) | ||
| Median age in years (range) | 1.25 (0.02–98.6) | 9.3 (0.06–97.4) | < 0.001 | |
| Pediatric cases (< 16 y) | 864/1286 (67.2%) | 117/222 (52.7%) | < 0.001 | |
| Older adults (> 65 y) | 275/1286 (21.4%) | 68/222 (30.6%) | 0.0026 | |
| Ct value b | 26.0 ± 6.6 (n = 868) | 27.9 ± 5.4 (n = 166) | 0.001 | |
| Co-infectionsc | 278/1114 (25.0%) | 47/211 (22.3%) | ns | |
| Hospitalized | 1124/1256 (89.5%) | 172/195 (88.2%) | ns | |
| Intensive care | 101/1027 (9.8%) | 17/150 (11.3%) | ns | |
| Diagnosis | Bronchiolitis | 347/742 (46.8%) | 27/113 (23.9%) | < 0.001 |
| Bronchopneumonia | 94/742 (12.7%) | 31/113 (27.4%) | ||
| Influenza-like illness | 85/742 (11.4%) | 3/113 (2.6%) | ||
| Other acute respiratory diagnoses | 137/742 (18.5%) | 40/113 (35.4%) | ||
| Other diagnoses | 79 /742 (10.6%) | 12/113 (10.6%) | ||
| Fever (T > 37.5 °C) | 404/660 (61.2%) | 77/124 (62.1%) | ns | |
| Cough | 443/612 (72.4%) | 92/115 (80.0%) | ns | |
| Productive cough | 206/443 (46.5%) | 55/92 (59.8%) | 0.009 | |
| SaO2 in room air | > 95% | 235/568 (41.4%) | 36/113 (31.9%) | 0.058 |
| 95–90% | 227/568 (40.0%) | 46/113 (40.7%) | ||
| < 90% | 106/568 (18.6%) | 31/112 (27.4%) | ||
| Dyspnea | 425/602 (70.6%) | 77/112 (68.7%) | ns | |
| Antibiotic therapy | 358/613 (58.4%) | 81/114 (71.1%) | 0.012 | |
RSV respiratory syncytial virus, HMPV human metapneumovirus
aThe number of cases in which information was available is indicated at the denominator
bDiagnostic test cycle threshold (Ct) value is reported as mean ± standard deviation
cCases of RSV and HMPV co-infections were not included
Around two-third (67.2%) of RSV-positive cases were in pediatric age (< 16 y), particularly in infants under six months of age (Table 2); it is also relevant that about 20% of the RSV-positive cases were elderly people (> 65 y), with a similar rate in the group including 65–80 y and in the over 80 y (Table 2). Cases positive for HMPV in pediatric age were 117/222 (52.7%) and were similarly distributed in the first two age classes; of note, 30% of the HMPV cases were older adults (> 65 y) (Table 2).
The age distribution of RSV and HMPV infections was significantly different (p < 0.001, Table 2); HMPV patients’ median age was higher (p < 0.001), and positivity rates were significantly lower in pediatric patients (p < 0.001), with respect to RSV (Table 2). Contrastingly, HMPV cases were relatively more frequent in older adults (> 65 y), compared to RSV (p = 0.0026, Table 2).
The diagnostic Ct value was significantly lower (i.e., higher viral load) in RSV than in HMPV cases (Table 2).
Most respiratory samples were tested also for the other respiratory viruses: viral co-infections were detected in 25% of the RSV-positive and 22.3% of the HMPV-positive cases (Table 2).
About 10% (101/1027) and 11.3% (17/150) of all hospitalized patients with an RSV and HMPV infections needed intensive care (Table 2): this rate raised to nearly 12% (64/537) when considering RSV-positive children under one year.
Clinical data were available for around half of the RSV- and HMPV-positive cases (Table 2). About half of the RSV cases were hospitalized for bronchiolitis (Table 2), and bronchiolitis represented 86% (344/399) of the total clinical diagnosis among the RSV-positive cases up to one year of age. Other frequent causes of hospitalization among RSV-positive cases were acute respiratory distress and bronchopneumonia (Table 2). HMPV-positive cases were more frequently hospitalized for acute respiratory distress, followed by bronchopneumonia (Table 2).
RSV- and HMPV-hospitalized cases did not differ in clinical signs and symptoms; they presented similar rates of fever, dyspnea and cough. Of note, productive cough was more frequent among HMPV-positive than in RSV-positive cases (Table 2). An antibiotic therapy was given more frequently to HMPV infected than to the RSV-positive patients (p = 0.012). Moreover, the antibiotic use was significantly different by age group (p < 0.0001); in the 0–6 months age group, the antibiotic use was 42%, whereas an antibiotic therapy was given at very high percentages in the other groups (74,4% at 6 m-3 y; 85.7% at 3 y–16 y; 63.1% at 16 y–65 y; 63.3% at 65–80 y; 79.3% in the over 80 y).
Demographic and clinical characterization of RSV-A and RSV–B cases
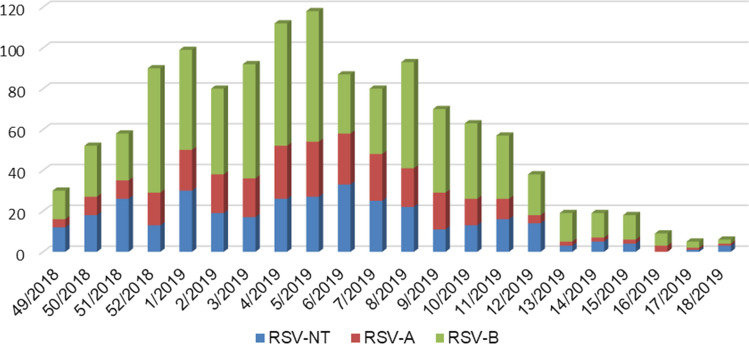
On most RSV-positive cases (975/1300: 75%) subtyping was performed: 273 cases were positive for RSV-A, 695 to RSV-B and 7 were positive for both RSV-A and RSV–B. At the national level and in every center, RSV-B was more abundant than RSV-A, but with different percentages (Table 1); the weekly distribution of subgroups was similar (Fig. 3).
Fig. 3.
RSV-A and -B cases weekly distribution. On the X-axis, the calendar week of the study period (December 2018–April 2019) is reported; on the Y-axis, the number of cases positive for RSV is represented. RSV-positive cases that were not typed (NT) are represented in blue; in red are those positive for RSV-A; in green, the positive for RSV–B
Demographic and clinical data were compared according to the RSV infecting subgroup (Table 3). RSV-B cases were significantly older (p = 0.021) and less frequently co-infected with other respiratory viruses (p < 0.001), with respect to RSV-A cases (Table 3). Besides, RSV-A and -B did not differ significantly as regards the overall clinical severity, nor in the clinical diagnosis; the only difference in clinical presentation was cough, more frequent (p = 0.021) in the RSV-B group (Table 3).
Table 3.
Comparison of demographic and clinical data between RSV-A and RSV-B positive cases
| Categories | RSV-Aa (N = 273) | RSV-Ba (N = 695) | P value | |
|---|---|---|---|---|
| Sex (male) | 138 (50.5%) | 352 (50.6%) | ns | |
| Age group | 0–6 m | 116/273 (42.5%) | 254/694 (36.6%) | 0.012 |
| 6 m–3 y | 72/273 (26.4%) | 141/694 (20.3%) | ||
| 3 y–16 y | 19/273 (7.0%) | 56/694 (8.1%) | ||
| 16 y–65 y | 15/273 (5.5%) | 77/694 (11.1%) | ||
| 65 y–80 y | 23/273 (8.4%) | 70/694 (10.1%) | ||
| > 80 y | 28/273 (10.2%) | 96/694 (13.8%) | ||
| Median age in years (range) | 0.77 (0.02–98.2) | 1.31 (0.03–98.6) | 0.021 | |
| Pediatric cases (< 16 y) | 207/273 (75.8%) | 451/694 (65.0%) | 0.0015 | |
| Older adults (> 65 y) | 51/273 (18.7%) | 166/694 (23.9%) | 0.078 | |
| Ct valueb | 25.3 ± 6.1 (n = 233) | 26.4 ± 7.1 (n = 462) | 0.054 | |
| Co-infections | 84/242 (34.7%) | 146/618 (23.6%) | < 0.001 | |
| Hospitalized | 255/273 (93.4%) | 652/687 (94.9%) | ns | |
| Intensive care | 25/216 (9.7%) | 56/567 (8.1%) | ns | |
| Diagnosis | Bronchiolitis | 89/165 (53.9%) | 158/327 (48.3%) | ns |
| Bronchopneumonia | 16/165 (9.7%) | 56/327 (17.1%) | ||
| Influenza-like illness | 9/165 (5.5%) | 24/327 (7.3%) | ||
| Other acute respiratory diagnoses | 36/165 (21.8%) | 72/327 (22.0%) | ||
| Other diagnoses | 15/165 (9.1%) | 17/327 (5.2%) | ||
| Fever (T > 37.5 °C) | 107/166 (64.5%) | 179/297 (60.3%) | ns | |
| Cough | 114/155 (73.5%) | 224/263 (85.2%) | 0.005 | |
| Productive cough | 43/114 (37.7%) | 102/224 (45.5%) | 0.013 | |
| SaO2 in room air | > 95% | 53/145 (36.6%) | 118/275 (42.9%) | ns |
| 95–90% | 65/145 (44.8%) | 104/275 (37.8%) | ||
| < 90% | 27/145 (18.6%) | 53/275 (19.3%) | ||
| Antibiotic therapy | 83/151(55.0%) | 165/268 (61.6%) | ns | |
RSV respiratory syncytial virus
aThe number of cases in which information was available is indicated at the denominator
bDiagnostic test cycle threshold (Ct) value is reported as mean ± standard deviation
Phylogenetic analysis of RSV and HMPV circulating strains
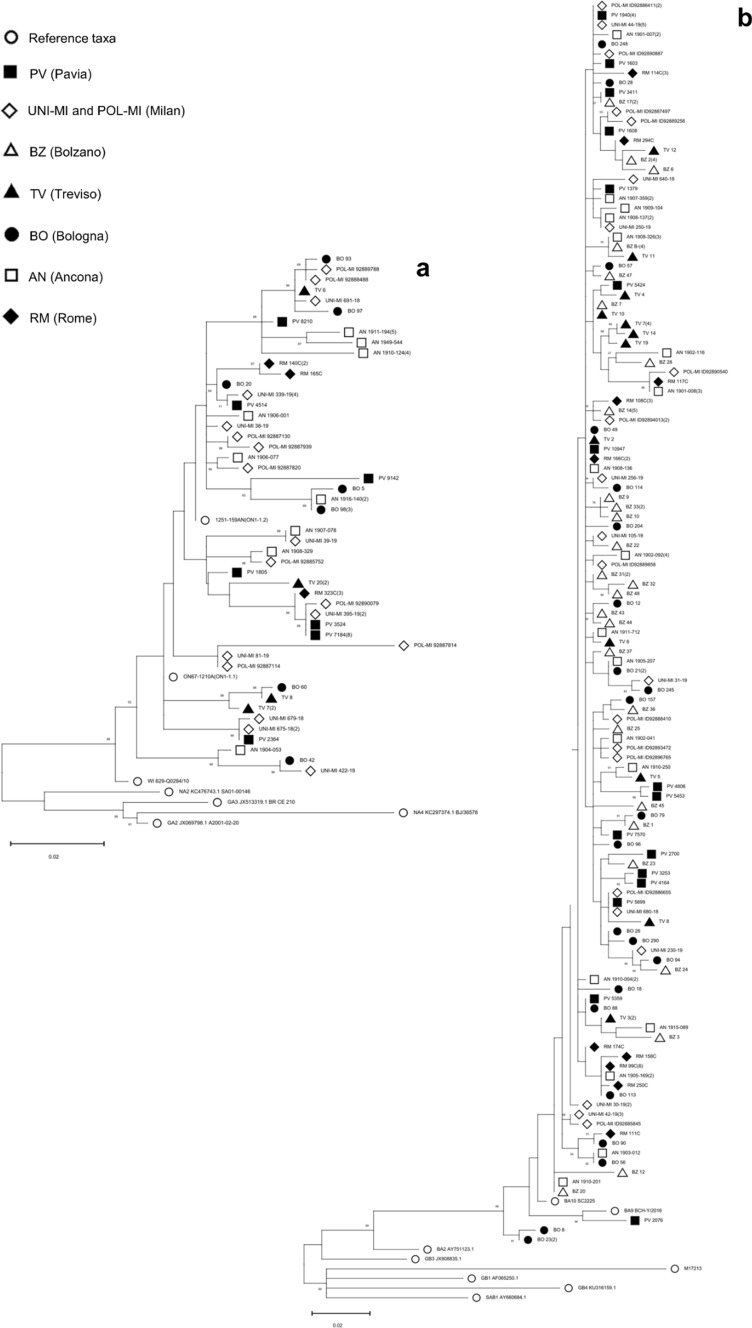
Approximately a quarter of RSV-A and -B positive samples from all centers were sequenced in the second half of the G-glycoprotein gene (G gene position 516–891); of these around 20% were not further analyzed because of poor chromatograms. The final datasets included 77 RSV-A strains (13 PV, 14 UNI-MI, 9 POL-MI, 7 TV, 9 BO, 17 AN, 8 RM) and 181 RSV-B (19 PV, 17 UNI-MI, 15 POL-MI, 41 BZ, 16 TV, 25 BO, 30 AN, 18 RM); identical RSV sequences from a center were grouped and their number of occurrences reported in the tree (Fig. 4 a and b).
Fig. 4.
Phylogenetic analysis of the 2nd half of the G gene of the RSV strains circulating in Italy (December 2018–April 2019). The phylogenetic tree of RSV-A (panel a) includes 77 GLIViRe sequences and 7 reference strains; the RSV-B tree (panel b) includes 181 GLIViRe sequences and 8 reference strains. The legend on the left indicates the symbols’ shape and filling for reference strains and the GLIViRe centers. The number of occurrences of identical RSV sequences from a center is reported in parenthesis. Numbers at nodes are bootstrap values for 1,000 iterations; only bootstrap values of > 50% are shown. Below the trees, scale bar shows the number of substitutions per site
All RSV-A sequences were identified as ON1 genotype [22], for the presence of the 72-nucleotide (nt) insertion, in the C-terminal region of the G-glycoprotein. Phylogenetic reconstruction (Fig. 4a) showed that numerous variants of the ON1 genotype circulated in Italy during 2018/19 season, clustering into at least two distinct clades containing sequences from several centers. The first clade of strains is derived from the ON1 reference strain [22], now named ON1-1.1 [23]; a second clade departs from the reference sequence of a sub-genotype, named ON1-1.2 [23], that was detected by two centers (AN and RM) [17] since the 2012/13 epidemic season (Fig. 4a). In addition, several sequences cluster together with high bootstrap values and may constitute new lineages with some regional clustering appearing in the phylogenetic tree (Fig. 4a).
All Italian RSV-B isolates have the 60-nt duplication in the 3’-terminal region of the G protein, typical of the BA genotype that first emerged in 1999 [24] and then differentiated in several other genotypes (BA1-BA14) [25]. Only one sequence (from PV) grouped with genotype BA9, whereas most study strains showed similarity with genotype BA10. The more relevant difference between BA9 and BA10 is the presence of an anticipated stop codon in BA10, which shortens the G protein of seven amino acids [25]. Several RSV-B study cases clustered by geographical location (Fig. 4b), for instance, three divergent sequences from BO center that do not cluster with either BA9 or BA10 (bottom of Fig. 4b).
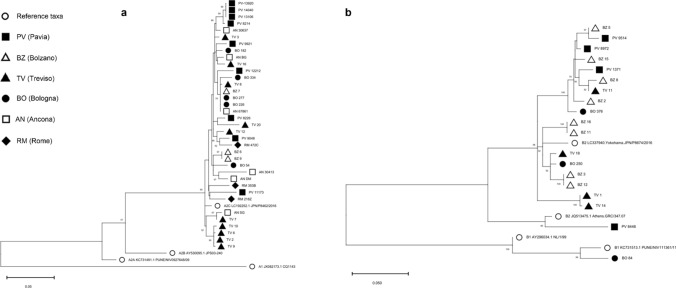
Around 30% of the HMPV-positive samples were sequenced by six centers; the phylogenetic analysis (Fig. 5, a and b) was conducted on a total of 55 good quality sequences of the entire G-glycoprotein gene: 36 (65.5%) were HMPV-A strains (9 PV, 3 BZ, 10 TV, 5 BO, 6 AN, 3 RM), and 19 (34.5%) were HMPV-B (4 PV, 8 BZ, 4 TV, 3 BO). All HMPV-A strains belonged to subgroup A2c and carried an insertion in the G gene, with the exception of one PV and one BZ strain: 9 strains (from TV, AN and RM) carried the 180 nt insertion identified about 10 years ago [26, 27] and 25 (from all centers) had the shorter 111 nt insertion that was detected more recently in Japan and Spain [28, 29]. HMPV-B isolates belonged to subgroup B2 except for one strain (from BO) that had high similarity with B1 subtype (Fig. 5 b).
Fig. 5.
Phylogenetic analysis of the G gene of the HMPV strains circulating in Italy (December 2018–April 2019). The phylogenetic tree of HMPV-A (panel a) includes 36 GLIViRe sequences and 4 reference strains; the HMPV-B tree (panel b) includes 19 GLIViRe sequences and 4 reference strains. The legend on the left indicates the symbols’ shape and filling for reference strains and the GLIViRe centers. The number of occurrences of identical HMPV sequences from a center is reported in parenthesis. Numbers at nodes are bootstrap values for 1,000 iterations; only bootstrap values of > 50% are shown. Below the trees, scale bar shows the number of substitutions per site
Discussion
The GLIViRe study group has been created in 2018 as a spontaneous, voluntary, laboratory-based surveillance network with the aim to monitor the circulation of respiratory viruses in Italy, utilizing results of the routine molecular tests. In the present study, we report epidemiological, clinical and laboratory data of RSV and HMPV cases prospectively assessed during the 2018/19 season, the last complete respiratory virus epidemic season before SARS-CoV-2 pandemic. The highest detected RSV positivity rate was in Rome (31.7%) where all tested cases were collected from hospitalized children < 3 years, in accordance with the greater burden of RSV-associated disease in infants [1–3]; rates were around 12–16% in the four centers that tested patients of all age (POL-MI, BZ, BO, AN) but with a median age below one year, whereas it was 6–9% in the other centers (PV, UNI-MI, TV) that tested patients with higher median age. Contrastingly, HMPV positivity rate was similar (1–2%) in all centers except for POL-MI, where it was two times higher (4%), but this difference was not apparently related to patients’ age. In recent Italian studies, HMPV median prevalence was 1.6% in bronchiolitis cases in Rome over ten years [13], and 3.2% in hospitalized children in northern Italy over five epidemic seasons with alternating pattern of incidence [30].
According to the GLIViRe data, the RSV season started probably in the first weeks of November, earlier than the influenza season and ended in mid-April, a length similar to the average of influenza season in Italy (18–20 weeks). Overall, the RSV epidemic season reported by GLIViRe is comparable to those reported in other European countries in 2018/19 [31] and very similar to that predicted for Italy, by modeling RSV historical epidemics on the base of 2018/19 temperature and relative humidity [31]. Moreover, the regional weekly occurrence of RSV cases had a longitudinal (west-to-east) distribution consistent with that reported in the recent European study [31]; differently, other papers detected both latitudinal and longitudinal gradients in the start of RSV seasonal epidemics in European or other temperate countries [21, 32].
In this study, HMPV circulation was detected mainly in spring with the epidemic peak registered 11 weeks later than that of RSV; the geographical distribution was not further analyzed due to the low number of HMPV weekly cases. Indeed, HMPV has been less studied with respect to RSV or other respiratory viruses. According to a global systematic analysis of respiratory viruses’ monthly activity [33], HMPV epidemics occur a median of 1.7 months later than RSV, in the temperate region. A recent US multicenter study found sustained HMPV circulation in winter and spring, over three years [10]. In several epidemic seasons, MPV circulation was higher from January to March in northern Europe [34], whereas in central Europe alternating HMPV epidemics in winter and spring–summer have been observed every two years [35, 36]. In a previous study spanning five epidemic seasons in Italy, HMPV incidence varied biannually with no clear seasonality in the annual distribution of cases [30].
The analysis of patients’ demographic and clinical data consolidated the notion that bronchiolitis is the more frequent and severe RSV-related clinical diagnosis, and nearly 60% of RSV cases were under three years of age. In comparison with RSV, HMPV-associated illnesses were more evenly distributed by age groups in pediatric age, consistent with a recent US multicenter study [10]. For the first time in Italy, this study reported that more than 20% of the RSV cases and 30% of the HMPV were in patients > 65 years, confirming the elevated burden of disease of these important pathogens in the older adults [4, 5, 9, 10]. Among GLIViRe cases, HMPV was relatively more frequent than RSV, in the eldest patients (> 80 y), differently from the US multicenter study [10]. Importantly, 10–12% of all-age patients infected with either RSV or HMPV, needed intensive care. Overall, an elevated antibiotic therapy was administered to hospitalized patients, with higher rates in the HMPV infected. Nevertheless, the difference between RSV and HMPV was likely driven by the high antibiotic prescription in all age groups (ranging from 63% in the young and middle-aged adults to around 80% in the eldest and in young children) with the exception of the infants 0–6 m (42%). Probably, the notion that bronchiolitis is nearly always caused by a viral infection, mostly RSV, avoided antibiotic use in several cases. The high numbers of RSV-positive cases allowed subtype comparisons. RSV-A positive cases were younger than those RSV–B infected; despite their higher median age, the overall clinical severity in RSV-B patients was comparable to that of the RSV-A cases. Characteristics that would differentiate subtypes were the higher co-infection rate in RSV-A patients and the more frequent presence of cough in the RSV-B positive cases. There is still debate on the possible different clinical presentation between RSV subtypes A and B; certainly, a different disease course may also reflect different RSV genotypes and variants [36–38].
Sequences of the highly variable G gene from around one-quarter of the RSV- and HMPV-positive cases were analyzed to characterize whether the co-circulation of multiple genotypes and strains during one epidemic season is similar at a national level or different patterns of local transmission occur. The RSV-A genotypes ON1-1.1 and ON1-1.2 continued to diverge, creating novel clades/sub-genotypes, circulating in several, but not all, Italian centers. ON1 strains circulating in previous epidemic seasons in northern [38] and central Italy [17, 37] were not detected in any of the samples sequenced in this study. In accordance with previous reports [23, 36], these results would suggest a fast appearance of RSV-A variant strains, circulating at a regional level, that will not be conserved among different epidemic seasons. Indeed, the 2018/19 season was characterized by a sustained RSV-B circulation; genotype BA10 was found nearly exclusively, differently from the previous study in northern Italy, that found a prevalence of BA9 and only a few strains of the BA10 genotype [38]. Also, other recent phylogenetic studies reported that BA9 was the most prevalent genotype, and somewhere the only circulating worldwide up to 2017/18 [39, 40]. It is tempting to speculate that the (re)introduction of the BA10 genotype may have contributed to the higher prevalence of RSV-B cases and to their clinical relevance in the 2018/19 season in Italy. More molecular epidemiology studies are needed to understand whether the alternating predominance of RSV-A and RSV-B may be dictated by the introduction of novel genotypes/variants and at which extent, RSV antigenic diversity may have an impact on the hospitalization rate. Sequences of the HMPV G gene were obtained from six GLIViRe centers; two-third of cases were categorized as HMPV-A, subgroup A2c. The A2c strains carrying the G gene insertion of 111-nt were found in all centers and were more numerous than those with the 180-nt insertion. This finding is consistent with the hypothesis that the 111-nt insertion would confer an evolutive advantage, greater than the longer insertion, that could lead to the predominance of these recently described HMPV strains [28, 29] and to the disappearance of the HMPV with no nt insertion in G, similarly to what happened with RSV-A and -B.
In conclusion, this study constitutes the first multicenter report on seasonality, age-specific distribution, and clinical presentation of RSV and HMPV cases, as well as their phylogenetic analysis, with a large geographic coverage of northern and central Italy. We recognize that the GLIViRe centers used different molecular methods (in-house assays, qualitative or real-time PCRs, or CE-IVD respiratory viral panels) that may have different analytical sensitivity. Notwithstanding, RSV positivity rates varied among centers coherently with the different populations tested, and their age distribution. Aggregated data are useful to define the annual RSV season threshold, to assess the performance of the case definition for RSV infection, and to estimate the age-specific annual RSV disease burden in Italy, in view of the up-coming immune-prophylactic treatment indicated for all newborns. Our data showed, for the first time in Italy, that RSV infections represent a substantial disease burden in the elderly, a target population of RSV vaccines in the near future. As regards to HMPV, this study provides the first epidemiological and clinical data on this important pathogen, at the national level; in comparison with RSV, the cumulative HMPV hospitalization rate was about 5 times lower, but relatively higher in older patients. Further studies are needed in more winter seasons to better characterize HMPV epidemics and its clinical impact in Italy, which may rise if RSV infections are more controlled by future vaccines.
After pandemic restrictions have been lifted, several respiratory viruses circulate together during the same period and may cause severe outbreaks; hence, it is particularly important to extend testing of respiratory infections beyond SARS-CoV-2. The GLIViRe collaborative network, that is being extended to southern Italian regions, will aid a rapid collection and analysis of diagnostic data and provide the foundation of a national integrated surveillance system that could monitor unusual peaks of severe respiratory infections, thus improving pandemic preparedness.
Supplementary Information
Below is the link to the electronic supplementary material.
Geographic location of the eight centers that constitute the GLIViRe network, Italy
Supplementary file1 (TIF 172 KB)
Biweekly distribution of the RSV cases in the eight GLIViRe centers. The number of RSV cases per week with respect of the total samples tested in two weeks in each center was calculated in percentage and values from two weeks aggregated, as shown in the color legend
Supplementary file2 (TIF 150 KB)
Acknowledgments
Giovanna Lunghi passed away in 2020. She strongly contributed to the planning of the research, design of the experiments and early discussion of the results. This work is dedicated to her memory, she was a mentor, a colleague, and a friend of all of us. We gratefully acknowledge nurses and physicians for collecting samples, technicians of the diagnostic laboratory, and the other Working Group on Respiratory Virus Infections (GLIViRe) components, not listed as authors.
Author contribution
AP, AP, GL and FB were involved in conceptualization; EP, EP, EV, TL, and SM contributed to methodology; SUR and CG were involved in formal analysis; GG, EG, VB, MLF, FN, GF, and GO contributed to investigation; SB, GP, LP, AL, and LP were involved in data acquisition and analysis; AP, AP, and FB contributed to writing, review and editing; GA was involved in supervision; AP contributed to funding acquisition. The manuscript has been read and approved by all authors and was not submitted, published, and accepted for publication elsewhere.
Funding
This study was supported by a grant “Medie attrezzature” from Sapienza University to Alessandra Pierangeli: n. MA120172B4A32AE4. The funding organization had no role in the design and conduct of the study and in writing of the paper.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This study was conducted in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration. The institutional review board and the Ethics Committee of Rome University Hospital approved this study (Prot. 107/12); informed consent was obtained from individual participants or their parents. Results of the diagnostic tests and clinical data were retrospectively extracted from patients' healthcare records, and in line with confidentiality requirements, the database was anonymized.
Footnotes
Giovanna Lunghi: Deceased.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Alessandra Pierangeli and Antonio Piralla contributed equally to this study as co-first authors.
Contributor Information
Alessandra Pierangeli, Email: alessandra.pierangeli@uniroma1.it.
Antonio Piralla, Email: a.piralla@smatteo.pv.it.
Sara Uceda Renteria, Email: sara.ucedarenteria@policlinico.mi.it.
Giovanni Giacomel, Email: giovanni.giacomel@policlinico.mi.it.
Elisabetta Pagani, Email: elisabetta.pagani@sabes.it.
Elisabetta Giacobazzi, Email: elisabetta.giacobazzi@sabes.it.
Elisa Vian, Email: elisa.vian@aulss2.veneto.it.
Valeria Biscaro, Email: valeria.biscaro@aulss2.veneto.it.
Giulia Piccirilli, Email: giulia.piccirilli@aosp.bo.it.
Tiziana Lazzarotto, Email: tiziana.lazzarotto@unibo.it.
Stefano Menzo, Email: menzo@univpm.it.
Monica Lucia Ferreri, Email: mluferreri@gmail.com.
Federica Novazzi, Email: federica.novazzi@asst-settelaghi.it.
Laura Petrarca, Email: laura.petrarca@uniroma1.it.
Amelia Licari, Email: amelia.licari@unipv.it.
Guglielmo Ferrari, Email: guglielmo.ferrari01@universitadipavia.it.
Giuseppe Oliveto, Email: giuseppe.oliveto@uniroma1.it.
Guido Antonelli, Email: guido.antonelli@uniroma1.it.
Sandro Binda, Email: sandro.binda@unimi.it.
Cristina Galli, Email: cristina.galli@unimi.it.
Laura Pellegrinelli, Email: laura.pellegrinelli@unimi.it.
Elena Pariani, Email: elena.pariani@unimi.it.
Fausto Baldanti, Email: f.baldanti@smatteo.pv.it.
References
- 1.Li Y, Wang X, Blau DM, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet. 2022;399:2047–2064. doi: 10.1016/S0140-6736(22)00478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holman RC, Shay DK, Curns AT, Lingappa JR, Anderson LJ. Risk factors for bronchiolitis-associated deaths among infants in the United States. Pediatr Infect Dis J. 2003;22:483–490. doi: 10.1097/01.inf.0000069765.43405.3b. [DOI] [PubMed] [Google Scholar]
- 3.Bont L, Checchia PA, Fauroux B, et al. Defining the epidemiology and burden of severe respiratory syncytial virus infection among infants and children in western countries. Infect Dis Ther. 2016;5:271–298. doi: 10.1007/s40121-016-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;28(352):1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 5.Shi T, Denouel A, Tietjen AK, et al. Global disease burden estimates of respiratory syncytial virus-associated acute respiratory infection in older adults in 2015: a systematic review and meta-analysis. J Infect Dis. 2020;222:S577–S583. doi: 10.1093/infdis/jiz059. [DOI] [PubMed] [Google Scholar]
- 6.van den Hoogen BG, de Jong JC, Groen J, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rima B, Collins P, Easton A, et al. ICTV virus taxonomy profile: pneumoviridae. J Gen Virol. 2017;98:2912–2913. doi: 10.1099/jgv.0.000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Li Y, Deloria-Knoll M, et al. Global burden of acute lower respiratory infection associated with human metapneumovirus in children under 5 years in 2018: a systematic review and modelling study. Lancet Glob Health. 2021;9:e33–e43. doi: 10.1016/S2214-109X(20)30393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi T, Arnott A, Semogas I, et al. The Etiological role of common respiratory viruses in acute respiratory infections in older adults: a systematic review and meta-analysis. J Infect Dis. 2020;222:S563–S569. doi: 10.1093/infdis/jiy662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard LM, Edwards KM, Zhu Y, et al. Clinical features of human metapneumovirus-associated community-acquired pneumonia hospitalizations. Clin Infect Dis. 2021;72:108–17. doi: 10.1093/cid/ciaa088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soto JA, Gálvez NMS, Benavente FM, et al. Human metapneumovirus: mechanisms and molecular targets used by the virus to avoid the immune system. Front Immunol. 2018;9:2466. doi: 10.3389/fimmu.2018.02466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierangeli A, Gentile M, Di Marco P, et al. Detection and typing by molecular techniques of respiratory viruses in children hospitalized for acute respiratory infection in Rome. Italy J Med Virol. 2007;79:463–468. doi: 10.1002/jmv.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cangiano G, Nenna R, Frassanito A, et al. Bronchiolitis: analysis of 10 consecutive epidemic seasons. Pediatr Pulmonol. 2016;51:1330–1335. doi: 10.1002/ppul.23476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piralla A, Mariani B, Rovida F, Baldanti F. Frequency of respiratory viruses among patients admitted to 26 intensive care units in seven consecutive winter-spring seasons (2009–2016) in Northern Italy. J Clin Virol. 2017;92:48–51. doi: 10.1016/j.jcv.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellegrinelli L, Galli C, Bubba L, et al. Respiratory syncytial virus in influenza-like illness cases: epidemiology and molecular analyses of four consecutive winter seasons (2014–2015/2017–2018) in Lombardy (Northern Italy) J Med Virol. 2020;92:2999–3006. doi: 10.1002/jmv.25917. [DOI] [PubMed] [Google Scholar]
- 16.Piralla A, Lunghi G, Percivalle E, et al. FilmArray® respiratory panel performance in respiratory samples from neonatal care units. Diagn Microbiol Infect Dis. 2014;79:183–186. doi: 10.1016/j.diagmicrobio.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pierangeli A, Trotta D, Scagnolari C, et al. Rapid spread of the novel respiratory syncytial virus A ON1 genotype, central Italy, 2011–2013. Euro Surveill. 2014;19:20843. doi: 10.2807/1560-7917.es2014.19.26.20843. [DOI] [PubMed] [Google Scholar]
- 18.Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79:2221–9. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- 19.Banerjee S, Sullender WM, Choudekar A, et al. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in India. J Med Virol. 2011;83:1799–810. doi: 10.1002/jmv.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bloom-Feshbach K, Alonso WJ, Charu V, et al. Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PLoS One. 2013;8:e54445. doi: 10.1371/journal.pone.0054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broberg EK, Waris M, Johansen K, Snacken R, Penttinen P. European influenza network seasonality and geographical spread of respiratory syncytial virus epidemics in 15 European countries 2010–2016. Euro Surveill. 2018;23:17–00284. doi: 10.2807/1560-7917.ES.2018.23.5.17-00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eshaghi A, Duvvuri VR, Lai R, et al. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS ONE. 2012;7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duvvuri VR, Granados A, Rosenfeld P, Bahl J, Eshaghi A, Gubbay JB. Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: global and local transmission dynamics. Sci Rep. 2015;5:14268. doi: 10.1038/srep14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trento A, Galiano M, Videla C, et al. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84:3115–3120. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- 25.Zlateva KT, Lemey P, Moës E, Vandamme AM, Van Ranst M. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J Virol. 2005;79:9157–67. doi: 10.1128/JVI.79.14.9157-9167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saikusa M, Kawakami C, Nao N, et al. 180-nucleotide duplication in the G gene of human metapneumovirus A2b subgroup strains circulating in Yokohama City, Japan, since 2014. Front Microbiol. 2017;8:402. doi: 10.3389/fmicb.2017.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piñana M, Vila J, Gimferrer L, et al. Novel human metapneumovirus with a 180-nucleotide duplication in the G gene. Future Microbiol. 2017;12:565–571. doi: 10.2217/fmb-2016-0211. [DOI] [PubMed] [Google Scholar]
- 28.Saikusa M, Nao N, Kawakami C, et al. Predominant detection of the subgroup A2b human metapneumovirus strain with a 111-nucleotide duplication in the G gene in Yokohama City, Japan in 2018. Jpn J Infect Dis. 2019;72:350–352. doi: 10.7883/yoken.JJID.2019.124. [DOI] [PubMed] [Google Scholar]
- 29.Piñana M, Vila J, Maldonado C, et al. Insights into immune evasion of human metapneumovirus: novel 180- and 111-nucleotide duplications within viral G gene throughout 2014–2017 seasons in Barcelona. Spain J Clin Virol. 2020;132:104590. doi: 10.1016/j.jcv.2020.104590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apostoli P, Zicari S, Lo Presti A, et al. Human metapneumovirus-associated hospital admissions over five consecutive epidemic seasons: evidence for alternating circulation of different genotypes. J Med Virol. 2012;84:511–516. doi: 10.1002/jmv.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Wang X, Broberg EK, Campbell H, Nair H. European RSV surveillance network. seasonality of respiratory syncytial virus and its association with meteorological factors in 13 European countries, week 40, 2010–week 39 2019. Euro Surveill. 2022;27:2100619. doi: 10.2807/1560-7917.ES.2022.27.16.2100619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Reeves RM, Wang X, et al. Global patterns in monthly activity of influenza virus, respiratory syncytial virus, parainfluenza virus, and metapneumovirus: a systematic analysis. Lancet Glob Health. 2019;7:e1031–e1045. doi: 10.1016/S2214-109X(19)30264-5. [DOI] [PubMed] [Google Scholar]
- 33.Moe N, Stenseng IH, Krokstad S, et al. The burden of human metapneumovirus and respiratory syncytial virus infections in hospitalized Norwegian children. J Infect Dis. 2017;216:110–116. doi: 10.1093/infdis/jix262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aberle JH, Aberle SW, Redlberger-Fritz M, Sandhofer MJ, Popow-Kraupp T. Human metapneumovirus subgroup changes and seasonality during epidemics. Pediatr Infect Dis J. 2010;29:1016–1018. doi: 10.1097/INF.0b013e3181e3331a. [DOI] [PubMed] [Google Scholar]
- 35.Heininger U, Kruker AT, Bonhoeffer J, Schaad UB. Human metapneumovirus infections–biannual epidemics and clinical findings in children in the region of Basel. Switzerland Eur J Pediatr. 2009;168:1455–1460. doi: 10.1007/s00431-009-0949-5. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Fernandez R, Tapia LI, Yang CF, et al. Respiratory syncytial virus genotypes, host immune profiles, and disease severity in young children hospitalized with bronchiolitis. J Infect Dis. 2017;217:24–34. doi: 10.1093/infdis/jix543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Midulla F, Di Mattia G, Nenna R, et al. Novel variants of respiratory syncytial virus A ON1 associated with increased clinical severity of bronchiolitis. J Infect Dis. 2020;222:102–110. doi: 10.1093/infdis/jiaa059. [DOI] [PubMed] [Google Scholar]
- 38.Esposito S, Piralla A, Zampiero A, et al. Characteristics and their clinical relevance of respiratory syncytial virus types and genotypes circulating in northern italy in five consecutive winter seasons. PLoS ONE. 2015;10:e0129369. doi: 10.1371/journal.pone.0129369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sáez-López E, Cristóvão P, Costa I, et al. Epidemiology and genetic variability of respiratory syncytial virus in Portugal, 2014–2018. J Clin Virol. 2019;121:104200. doi: 10.1016/j.jcv.2019.104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu JM, Fu YH, Peng XL, Zheng YP, He JS. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci Rep. 2021;11:12941. doi: 10.1038/s41598-021-92435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Geographic location of the eight centers that constitute the GLIViRe network, Italy
Supplementary file1 (TIF 172 KB)
Biweekly distribution of the RSV cases in the eight GLIViRe centers. The number of RSV cases per week with respect of the total samples tested in two weeks in each center was calculated in percentage and values from two weeks aggregated, as shown in the color legend
Supplementary file2 (TIF 150 KB)