Abstract
Objective
Lipid dysregulation and complement system (CS) activation are 2 important pathophysiology pathways for age-related macular degeneration (AMD). We hypothesized that the relationship between lipids and AMD may also differ according to CS genotype profile. Thus, the objective was to investigate the relationships between lipid-related metabolites and AMD according to CS genotypes.
Design
Population-based cross-sectional study.
Participants
A total of 6947 participants from Singapore Epidemiology of Eye Diseases study with complete relevant data were included.
Methods
We investigated a total of 32 blood lipid–related metabolites from nuclear magnetic resonance metabolomics data including lipoproteins and their subclasses, cholesterols, glycerides, and phospholipids, as well as 4 CS single nucleotide polymorphisms (SNPs): rs10922109 (complement factor H), rs10033900 (complement factor I), rs116503776 (C2-CFB-SKIV2L), and rs2230199 (C3). We first investigated the associations between AMD and the 32 lipid-related metabolites using multivariable logistic regression models. Then, to investigate whether the effect of lipid-related metabolites on AMD differ according to the CS SNPs, we tested the possible interactions between the CS SNPs and the lipid-related metabolites.
Main Outcome Measures
Age-related macular degeneration was defined using the Wisconsin grading system.
Results
Among the 6947 participants, the prevalence of AMD was 6.1%, and the mean age was 58.3 years. First, higher levels of cholesterol in high-density lipoprotein (HDL) and medium and large HDL particles were associated with an increased risk of AMD, and higher levels of serum total triglycerides (TG) and several very-low–density lipoprotein subclass particles were associated with a decreased risk of AMD. Second, these lipids had significant interaction effects on AMD with 2 CS SNPs: rs2230199 and rs116503776 (after correction for multiple testing). For rs2230199, in individuals without risk allele, higher total cholesterol in HDL2 was associated with an increased AMD risk (odds ratio [OR] per standard deviation increase, 1.20; 95% confidence interval (CI), 1.06–1.37; P = 0.005), whereas, in individuals with at least 1 risk allele, higher levels of these particles were associated with a decreased AMD risk (OR, 0.69; 95% CI, 0.45–1.05; P = 0.079). Conversely, for rs116503776, in individuals without risk allele, higher serum total TG were associated with a decreased AMD risk (OR, 0.84; 95% CI, 0.74–0.95; P = 0.005), whereas, in individuals with 2 risk alleles, higher levels of these particles were associated with an increased risk of AMD (OR, 2.3, 95% CI, 0.99–5.39, P = 0.054).
Conclusions
Lipid-related metabolites exhibit opposite directions of effects on AMD according to CS genotypes. This indicates that lipid metabolism and CS may have synergistic interplay in the AMD pathogenesis.
Keywords: Age-related macular degeneration, Complement system, Lipids, Metabolites
Abbreviations and Acronyms: AMD, age-related macular degeneration; CFH, complement factor H; CS, complement system; HDL, high-density lipoprotein; NMR, nuclear magnetic resonance; OR, odds ratio; RPE, retinal pigment epithelium; SNP, single nucleotide polymorphism; TG, triglycerides; VLDL, very-low–density lipoprotein
Age-related macular degeneration (AMD) is a leading cause of irreversible blindness in Asia and worldwide.1 It is known to be a multifactorial disease caused by a combination of lifestyle and genetic factors.2, 3, 4, 5 Several pathways are involved in the pathogenesis of AMD, such as complement system (CS) activation and lipid dysregulation.6, 7, 8, 9, 10 Deregulation of the alternative CS pathway leads to an increase in inflammation, which results in photoreceptor degeneration and the loss of central vision over time.11 In recent studies, the retinal pigment epithelium (RPE) and the choroid have been identified as being key sites of the CS pathway.12,13 Although progress is being made in the understanding of the CS pathway, how the dysregulation of lipids and lipoproteins contribute to AMD remains unclear. The latest epidemiologic studies have reported that increased levels of high-density lipoprotein (HDL) cholesterol are associated with a higher risk of AMD and triglycerides (TG) with lower risk.14, 15, 16, 17
However, there are known links between lipid metabolism and CS activity, which could potentially confound our understanding of the role of lipids in AMD pathophysiology. First, recent proteomic analyses revealed that several types of HDL particles contain CS components, such as C3, C4, or C9.18,19 Furthermore, a subset of HDL particles carry very specific complement components or regulators, such as C4a, C4b, C9, and vitronectin.20 Studies show that the complex interactions between CS components and lipids can lead to both changes in the lipid properties and regulation of the CS. For example, during an acute-phase response, the HDL could become dysfunctional and change from anti-inflammatory to proinflammatory.21 Therefore, to fully understand the role of lipids in the pathogenesis of AMD, the CS needs to be accounted for.
We hypothesized that the relationship between lipid-related metabolites and the risk of AMD may differ according to CS genotypes. Because the size and composition of lipoprotein particles may impact their functions, we considered measurements of lipoprotein particle subclasses using nuclear magnetic resonance (NMR) metabolomics. The objective of this study was to determine the relationships between NMR lipid–related metabolites and AMD according to CS genotype profiles. We used a data-driven approach using this large set of lipid-related metabolites along with the main CS genetic variants to identify which components of the lipid metabolism and CS activity interact with regard to the risk of AMD. This study will help generate more specific hypotheses for future studies.
Methods
Study Population
Participants were enrolled from the Singapore Epidemiology of Eye Diseases study (n = 10 033). Singapore Epidemiology of Eye Diseases is a multiethnic population-based study that has recruited subjects from 3 major ethnic groups (Chinese [n = 3353], Malay [n = 3280], and Indian [n = 3400]) to investigate risk factors for major age-related eye diseases. Participants underwent a standardized interview, clinical examination, and laboratory investigations. Singapore Epidemiology of Eye Diseases baseline examinations were conducted from 2004 to 2011. The detailed methodology of Singapore Epidemiology of Eye Diseases has been published previously.21, 22, 23 Briefly, an age-stratified random sampling was used to select subjects aged ≥40 years from each ethnic group living across southwestern Singapore (overall response rate of 75.6%). This study was approved by the SingHealth Centralized Institutional Review Board and was conducted according to the tenets of the Declaration of Helsinki after informed consent was obtained.
Clinical Examination
Age-related macular degeneration was graded from retinal photographs using the Wisconsin AMD classification.24 Several features were assessed to determine the grading: presence and extent of RPE degeneration, increased pigment, RPE detachment and serous detachment of the sensory retina, retinal hard exudate, subretinal and sub-RPE hemorrhage, subretinal and sub-RPE fibrous tissue, geographic atrophy, retinal edema (thickening), and retinal hemorrhages. Moreover, the following demographic and clinical information were collected: age, sex, ethnicity (Chinese, Indian, or Malay), hypertension (yes/no), diabetes (yes/no), smoking status (current or past smoker, never smoked), body mass index, and the use of lipid-lowering medication (statin, fibrate, or other cholesterol-lowering drugs) (yes/no).
Blood Lipid–Related Metabolites
A total of 228 blood metabolites were quantified from stored serum (Chinese and Indian) and plasma (Malay) samples using a high-throughput proton NMR metabolomics platform at Nightingale Health Ltd (Helsinki, Finland). This method provides simultaneous quantification of routine lipids, lipoprotein subclass profiling with lipid concentrations within 14 subclasses, fatty acid composition, and various low molecular metabolites, including amino acids, ketone bodies, and gluconeogenesis-related metabolites, in molar concentration units. Details of the methodology and applications of the NMR metabolomics platform have been described previously.25 This study focused on a subset of 32 lipid-related metabolites divided into 4 categories—lipoproteins subclasses, lipoprotein particle sizes, cholesterol, glycerides and phospholipids (Fig S1, available at www.ophthalmologyscience.org). Lipoprotein subfraction concentrations (e.g., the concentration of cholesteryl esters or free cholesterol within each lipoprotein subclass) were disregarded, and the overall concentration of each lipoprotein subclass (e.g., the concentration of large HDL) was kept. Because the lipid-related metabolites were available in 2 different units, we disregarded the ratio to total lipids and analyzed concentrations (as done in previous studies).26,27 Removing these metabolites mitigated multicollinearity issues caused by (very) high correlations.
Complement System Single Nucleotide Polymorphisms
We considered the lead single nucleotide polymorphisms (SNPs) identified to be associated with AMD in the latest large international genome-wide association studies conducted on 16 144 patients with AMD and 17 832 controls.3 They were located in the following loci: complement factor H (CFH), complement factor I, C9, C2-CFB-SKIV2L, and C3. However, the SNP located in the locus C9 (rs62358361) was not available in our genotype data. Therefore, we considered the following 4 SNPs: rs10922109 (CFH), rs10033900 (complement factor I), rs116503776 (C2-CFB-SKIV2L), and rs2230199 (C3). Figure S2 (available at www.ophthalmologyscience.org) shows the allele frequencies according to the ethnic groups.
Statistical Analyses
The lipid-related metabolites were first log-transformed, then centered and scaled (thus expressed as increase or decrease in standard deviation). Moreover, we removed the outliers, defined as having a value lower than −4 standard deviation or higher than +4 standard deviation. The SNPs were considered as continuous variables (0, homozygous reference allele; 1, heterozygous; and 2, homozygous risk allele), with the common underlying assumption of linearity.
First, we investigated the associations between AMD and the 32 lipid-related metabolites and the 4 CS SNPs using a multivariable logistic regression model. These models were adjusted for age, sex, ethnicity, lipid-lowering medication, hypertension, diabetes, body mass index, and smoking status. Second, to investigate whether the effect of lipid-related metabolites on AMD differs according to the CS SNPs, we tested the 128 possible interactions between the 4 CS SNPs and the 32 lipid-related metabolites. One separate model was run for each interaction using multivariable logistic regression models adjusted on age, sex, ethnicity, lipid-lowering medication, hypertension, diabetes, body mass index, and smoking status. Moreover, to correct for multiple testing and, thus, reduce the risk of false-positive results, we used the false discovery rate method.28,29 Third, for significant (after false discovery rate correction) interactions, we estimated the effect of the corresponding lipid-related metabolites on AMD in different subgroups on the basis of the CS SNP risk alleles. We used 3 categories—that is, 0, homozygous reference allele; 1, heterozygous; and 2, homozygous risk allele—if the sample size was large enough, especially for category 2. Alternatively, we combined categories 1 and 2 into 1 category, thus corresponding to at least 1 risk allele. For all the analyses, the metabolites were considered as continuous variables. The odds ratios (ORs) corresponding to the effects of lipid-related metabolites on AMD were calculated as the exponential of the logistic regression’s coefficients and were given with their 95% confidence intervals. All statistical analyses were performed with R, version 4.0.4. Finally, to account for the genetic structure of the population, we further adjusted the analyses on the first 5 principal components.
Results
The prevalence of AMD in our population was 6.1% (n = 423/6947). Of the 423 AMD cases, 393 (92.9%) were early cases. Those with AMD, compared with those without AMD, were more likely to be male (60.3% vs. 49.6%), to be older (65.8 vs. 57.8 years), to be on lipid-lowering medication (31.4% vs. 23.1%), and to have hypertension (80.1% vs. 61.6%) and diabetes (31.7% vs. 28.3%) (Table 1).
Table 1.
Characteristics of the Study Population According to the AMD Status
| Free of AMD (N = 6524) | AMD (N = 423) | P Value | |
|---|---|---|---|
| Age (yrs), mean (SD) | 57.84 (9.8) | 65.82 (10.1) | < 0.001 |
| Female, n (%) | 3287 (50.4) | 168 (39.7) | < 0.001 |
| Ethnicity, n (%) | 0.561 | ||
| Chinese | 2087 (32.0) | 145 (34.3) | |
| Indian | 2220 (34.0) | 143 (33.8) | |
| Malay | 2217 (34.0) | 135 (31.9) | |
| Hypertension, n (%) | 4019 (61.6) | 339 (80.1) | < 0.001 |
| Diabetes, n (%) | 1846 (28.3) | 134 (31.7) | 0.135 |
| BMI (kg/m2), mean (SD) | 25.5 (4.7) | 25.26 (5.0) | 0.301 |
| Lipid-lowering medication, n (%) | 1504 (23.1) | 133 (31.4) | < 0.001 |
| Smoking status, n (%) | < 0.001 | ||
| Never smoked | 4551 (69.8) | 279 (66.0) | |
| Current smoker | 1066 (16.3) | 56 (13.2) | |
| Past smoker | 907 (13.9) | 88 (20.8) |
AMD = age-related macular degeneration; BMI = body mass index; SD = standard deviation.
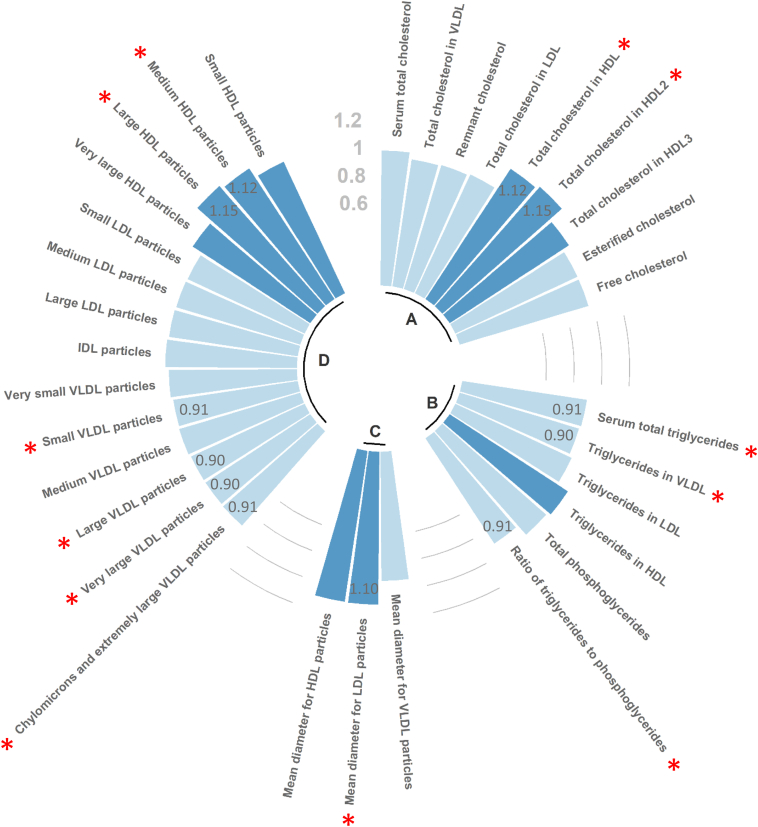
On one hand, as shown in Figure 1, of the 32 lipid-related metabolites, 5 were positively associated (all P < 0.05) with AMD (levels of cholesterol in HDL and HDL2, mean diameter for low-density lipoprotein particle, and concentrations of medium and large HDL particles). On the other hand, 7 were negatively associated (all P < 0.05) with AMD (total level of TG, level of TG in very-low–density lipoprotein [VLDL] particles, ratio of TG to phosphoglycerides, and concentrations of chylomicrons, extremely large, very large, and small VLDL particles). Regarding the associations between the 4 CS SNPs and AMD, we found the following ORs: 0.88 (0.77–1.01) (P = 0.06) for rs10922109, 1.07 (0.93–1.23) (P = 0.37) for rs10033900, 0.81 (0.65–1.00) (P = 0.05) for rs116503776, and 1.30 (0.93–1.81) (P = 0.12) for rs2230199.
Figure 1.
Associations between lipid-related metabolites and age-related macular degeneration, expressed as odds ratios (ORs). The metabolites were considered as continuous variables. They were log-transformed, then centered and scaled so that ORs corresponded to the variation of age-related macular degeneration risk for 1 standard deviation increase in metabolite levels. Odds ratios were calculated as the exponential of the logistic regression’s coefficients. Dark and light blue corresponded to an OR higher and lower than 1, respectively. The red stars correspond to significant associations (P < 0.05), and for these associations, the corresponding values of the ORs are reported in dark gray in the bars. A indicates cholesterols, B indicates glycerides and phospholipids, C indicates lipoprotein particle sizes, and D indicates lipoprotein subclasses. HDL = high-density lipoprotein; IDL = intermediate-density lipoprotein; LDL = low-density lipoprotein; VLDL = very-low–density lipoprotein.
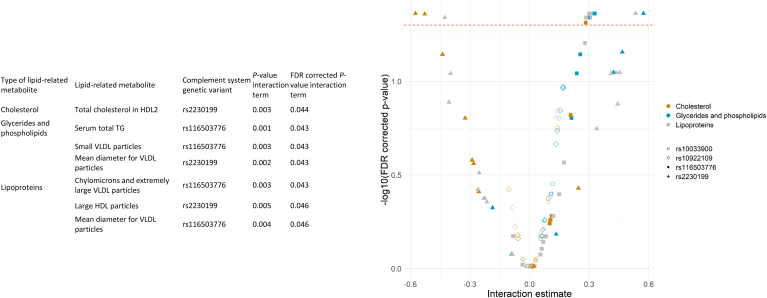
Of 128 interactions tested between lipid-related metabolites and CS SNPs with regard to AMD, 38 were significant at the P threshold of 0.05, without correction for multiple testing, involving 21 lipid-related metabolites and 3 SNPs. After false discovery rate correction, 13 interactions were considered significant, involving 11 lipid-related metabolites and 2 SNPs (Fig 2). We removed 6 interactions owing to the very high correlations between lipids. For example, 2 significant interactions were total cholesterol in HDL × rs2230199 and total cholesterol in HDL2 × rs2230199. The correlation between the 2 cholesterols was 0.99, implying that the 2 interactions represent similar information. We removed total cholesterol in HDL × rs2230199 because this metabolite had higher correlations with the other metabolites than total cholesterol in HDL2. By doing so, no metabolites involved in the remaining interactions had a correlation higher than 0.95 (Fig S3, available at www.ophthalmologyscience.org). Finally, a total of 7 interactions involving 6 lipid-related metabolites (1 cholesterol, 1 glyceride, and 4 lipoprotein particles) and 2 SNPs (rs2230199 and rs116503776) remained (Fig 2).
Figure 2.
Table and graph showing the interaction effects between lipid-related metabolites and complement system (CS) genetic variants on age-related macular degeneration. In the graph, each of the 128 points corresponds to 1 interaction between 1 metabolite and 1 CS genetic variant. Each interaction was tested in a separate model adjusted for the confounders mentioned in the statistical analyses section. The metabolites and CS genetic variants were considered continuous, so the models returned 1 coefficient per interaction between 1 metabolite and 1 genetic variant. The table shows significant interactions after correction for multiple testing using the false discovery rate (FDR) method (those above the red dotted line in the graph). The color legend indicates the type of lipid-related metabolite. The shape legend indicates the CS genetic variant. HDL = high-density lipoprotein; TG = triglycerides; VLDL= very-low–density lipoprotein.
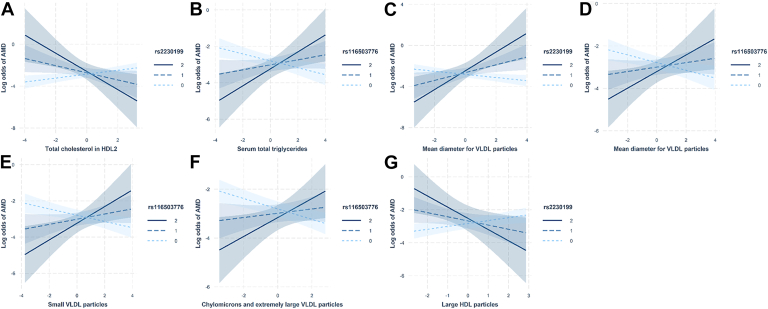
The effect of these 7 lipid-related metabolites (which had significant interaction with CS SNP) on AMD are presented in Figure 3 and Table 2. For total cholesterol in HDL2 and large HDL particles, the associations with AMD were positive (OR, 1.20 [1.06–1.37] and 1.19 [1.05–1.35]) in individuals with no risk allele (category 0) and negative (OR, 0.69 [0.45–1.05] and 0.75 [0.52–1.08]) in individuals with a single or 2 risk alleles (categories 1 or 2). For serum total TG and the different VLDL particles, the associations with AMD were negative in individuals with no risk allele and positive otherwise. For example, the level of chylomicrons and extremely large VLDL particles was significantly negatively associated with AMD in individuals with no risk allele for rs116503776 (OR, 0.82 [0.73–0.91]; P < 0.001) and significantly positively associated in individuals with 2 risk alleles (OR, 3.83 [1.04–14.18]; P = 0.04). Finally, the results were very similar after further adjusting on the 5 first principal components to account for the genetic structure of the population (Table S1, available at www.ophthalmologyscience.org).
Figure 3.
Effects of the lipid-related metabolites on AMD according to the complement system genetic variants. These interactions correspond to the significant interactions (see Figure 2) after correction for multiple testing. Each panel corresponds to the effect of one metabolite on AMD according to one complement genetic variant. The metabolites were log-transformed, then centered and scaled (x-axis corresponds to standard deviations). The gray areas around the solid lines corresponded to the 95% confidence intervals. AMD = age-related macular degeneration; HDL = high-density lipoprotein; VLDL = very-low–density lipoprotein.
Table 2.
Effects of Lipid-Related Metabolites According to the Complement System Genetic Variants
| Lipid-Related Metabolite | Complement System Genetic Variant | Allele | Sample Size | Odds Ratio (95% CI) | P Value |
|---|---|---|---|---|---|
| Total cholesterol in HDL2 | rs2230199 | 0 | 6481 | 1.20 (1.06–1.37) | 0.005 |
| 1/2∗ | 550 | 0.69 (0.45–1.05) | 0.079 | ||
| Serum total TG | rs116503776 | 0 | 5249 | 0.84 (0.74–0.95) | 0.005 |
| 1 | 1620 | 1.04 (0.83–1.3) | 0.730 | ||
| 2 | 178 | 2.30 (0.99–5.39) | 0.054 | ||
| Mean diameter for VLDL particles | rs2230199 | 0 | 6497 | 0.86 (0.77–0.96) | 0.010 |
| 1/2∗ | 552 | 1.46 (1.01–2.11) | 0.043 | ||
| Small VLDL particles | rs116503776 | 0 | 5247 | 0.85 (0.76–0.96) | 0.010 |
| 1 | 1620 | 1.06 (0.84–1.33) | 0.628 | ||
| 2 | 177 | 1.88 (0.81–4.36) | 0.140 | ||
| Chylomicrons and extremely large VLDL particles | rs116503776 | 0 | 5252 | 0.82 (0.73–0.91) | 0.000 |
| 1 | 1623 | 0.99 (0.81–1.21) | 0.925 | ||
| 2 | 178 | 3.83 (1.04–14.18) | 0.044 | ||
| Large HDL particles | rs2230199 | 0 | 6501 | 1.19 (1.05–1.35) | 0.005 |
| 1/2∗ | 552 | 0.75 (0.52–1.08) | 0.127 | ||
| Mean diameter for VLDL particles | rs116503776 | 0 | 5250 | 0.85 (0.75–0.96) | 0.009 |
| 1 | 1621 | 1.01 (0.8–1.26) | 0.945 | ||
| 2 | 179 | 2.40 (0.98–5.86) | 0.055 |
The metabolites were considered as continuous variables. They were log-transformed, then centered and scaled so that the odds ratios corresponded to the variation of AMD risk for 1 standard deviation increase in metabolites levels. The interactions presented in this table are the ones significant after correction for multiple testing using the false discovery rate method.
CI = confidence interval; HDL = high-density lipoprotein; TG = triglycerides; VLDL = very-low–density lipoprotein.
Because of the limited sample size (only 26 individuals with 2 risk alleles for this single nucleotide polymorphism), we combined categories 1 and 2.
Discussion
In this large population-based study using NMR metabolomics data, we first identified lipid-related metabolites that were associated with AMD. Some were positively associated, such as total cholesterol in HDL2 and medium and large HDL particles; others were negatively associated, such as serum total TG and several VLDL subclass particles. Next, we found that the effects of these metabolites exhibited significant interactions (after correction for multiple testing) with 2 CS genotypes (rs2230199 [C3] and rs116503776 [C2-CFB-SKIV2L]). Indeed, some metabolites had opposite directions of effects on AMD (significant in both directions), depending on the CS allele distribution. The key observations are as follows: in individuals without risk allele for C3 SNP, higher total cholesterol in HDL2 and large HDL particles were associated with an increased risk of AMD, whereas, in individuals with at least 1 risk allele, higher levels of these particles were associated with a decreased risk of AMD. In contrast, in individuals without risk allele for C2 SNP, higher serum total TG and VLDL particle subclasses were associated with a decreased risk of AMD, whereas, in individuals with 2 risk alleles, higher levels of these particles were associated with an increased risk of AMD. In other words, when C3 SNP has a risk allele, the properties of HDL particles and cholesterol subfractions become protective (e.g., via a shift from proinflammatory to anti-inflammatory properties), whereas, when C2 SNP has risk alleles, TG and VLDL particles subclasses become risk factors (e.g., via a shift from anti-inflammatory to proinflammatory properties).
Our findings regarding the overall effects of HDL cholesterol and TG on AMD were similar to those in the latest large studies14, 15, 16: HDL cholesterol was positively associated with AMD, whereas TG was negatively associated with AMD. However, more importantly, we found that these effects could invert in the opposite direction, depending on the CS genotype. Under certain circumstances, HDL properties can indeed change from anti-inflammatory to proinflammatory.20,30,31 For example, in individuals with chronic conditions that promote systemic oxidative stress and inflammation, such as diabetes, HDL may actually become proinflammatory.20 When converted into a dysfunctional proinflammatory particle, HDL cannot promote cholesterol efflux or prevent low-density lipoprotein oxidation.32, 33, 34 These modifications in HDL properties result from changes in the HDL composition, for example, decreased levels and activities of anti-inflammatory and antioxidant factors, such as apolipoprotein AI (apoA-I) and paraoxonase PON1.35, 36, 37 These changes in HDL inflammatory properties may also be associated with the immune innate response.20
There are intimate links between lipid metabolism and CS. On one hand, the CS can regulate lipid metabolism. For instance, changes in CFH concentration can modify the anti-inflammatory properties of large HDL particles.38 The enrichment of CFH in HDL allows it to prevent the organization of complement membrane attack complex and suppress inflammation.39 Moreover, C3adesarg, the degradation product of C3, can stimulate TG synthesis in adipocytes.40 Taken all together, the complex interactions between CS components and lipids may lead to modifications of the properties of the lipids, such as the inflammatory and atherogenic properties of HDL, which in turn modify their effects on the development of AMD. This seems to explain the interactions we found between HDL cholesterol and some CS genetic variants. More investigations need to be performed regarding the interactions between CS components and other types of lipids and lipid subfractions, such as TG and VLDL particles. On the other hand, lipids can also play a role in regulating the CS. High-density lipoprotein particles can, for example, inhibit the complement lytic mechanism.41 Moreover, chylomicrons carry transthyretin, which has been shown to stimulate C3 and C3adesarg, the degradation product of C3, in a dose-dependent manner.42 Furthermore, apolipoprotein J (an HDL component) was suggested to be a central player in the inhibition of the complement-mediated lysis via binding to C7, C8, and C9.43, 44, 45 Finally, apolipoprotein E (APOE) binds to CFH and subsequently regulates complement activation.46 More research is needed to better understand these complex interactions and how they play a role in the pathophysiology of AMD.
It is crucial to better understand the role of lipids in the development of AMD because they may be involved in the early stages of the disease pathologies. Colijn et al14 found that the magnitude of the effects of HDL cholesterol and TG were higher for early than for late AMD cases. In the present study, the great majority of AMD cases were early (93%), and there were unfortunately not enough late cases to allow stratification by severity. Specifically, understanding precisely how the lipid dysregulation and which lipids are involved in early AMD will help clarify whether statins can be useful for AMD therapeutics. Many studies have been conducted; however, so far, there is no clear answer.47, 48, 49, 50 Given our results, it is possible that statin medication could be beneficial for individuals with a risk allele on the C3 SNP. Indeed, statin increases the level of HDL, and for people with such a genotype, a higher level of HDL is protective for AMD. A specific study to investigate the effect of statin according to the CS genotypes is required to fully answer this question.
This population-based study has the following strengths. First, the large sample size allowed us to investigate the effect of lipids on AMD in the different subgroups corresponding to the CS genotypes, even though some allele frequencies were low. Second, we used an NMR approach to measure the lipid-related metabolites. That made it possible to quantify many lipoproteins subclasses, which inform better on their functionalities. For example, we could show some novel associations between AMD and the levels in TG in VLDL or between AMD and the mean diameter of low-density lipoprotein particles. More studies are needed to investigate the specific roles of these lipid subfractions in the development of AMD. However, this study has limitations. First, the NMR metabolite quantification has not been done in the same type of sample according to the ethnicity: serum for Chinese and Indians and plasma for Malay. However, the correlation between lipid-related metabolites concentrations in these 2 matrixes is high51 and should, thus, not have introduced major bias into our findings. Moreover, the patients were not fasting at the time of blood draw, which could confound the results. Second, we only investigated 4 lead CS genetic variants that were associated with AMD at the genome-wide significance level3; however, more genetic variants, that is, not just lead SNPs, at a less conservative threshold could be considered. That would allow determining whether other CS genetic variants are interacting with lipids. Finally, here we used the CS genotypes as a surrogate of the CS activity; the next step will thus be to use the quantification of the CS proteins to identify which are interacting with the lipids metabolism.
The contribution of lipid dysregulation to AMD pathophysiology is poorly understood. We show in this study that lipid-related metabolites can exhibit opposite directions of effects on AMD according to CS genotypes. Specific interactions between CS components and lipids may change lipid properties and, thus, modify the risk of AMD. These results suggest that lipid dysregulation and CS activation may have synergistic interplay in AMD pathogenesis.
Manuscript no. XOPS-D-22-00053R2.
Footnotes
Supplemental material available at www.ophthalmologyscience.org.
Disclosures:
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures: C.M.G.C: Consultant – Roche, Bayer, Novartis; Speaker – Roche, Bayer, Novartis.
T.Y.W.: Consultant – Bayer, Boehringer-Ingelheim, Eden Ophthalmic, Genentech, Iveric Bio, Merck, Novartis, Oxurion, Plano, Roche, Samsung, Shanghai Henlius, Zhaoke Pharmacecutical; Inventor, Patents, Co-founder – EyRIS and VisRE.
Supported by the National Medical Research Council (NMRC), Singapore (grant nos.: NMRC/CIRG/1488/2018 and NMRC/OFLCG/004a/2018).
HUMAN SUBJECTS: Human subjects were included in this study. This study was approved by the SingHealth Centralized Institutional Review Board and was conducted according to the tenets of the Declaration of Helsinki after informed consent was obtained.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Hui, Tham, Sabanayagam, Cheung, Wong, Cheng, Nusinovici.
Data collection: Hui, Betzler, Zhou, Wang, Cheng, Nusinovici.
Analysis and interpretation: Hui, Tham, Zhou, Wang, Sabanayagam, Cheung, Wong, Cheng, Nusinovici.
Obtained funding: N/A; Study was performed as part of regular employment duties at SERI/SNEC. No additional funding was provided.
Overall responsibility: Hui, Cheng, Nusinovici.
Supplementary Data
References
- 1.Wong W.L., Su X., Li X., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P., Liew G., Gopinath B., Wong T.Y. Age-related macular degeneration. Lancet. 2018;392:1147–1159. doi: 10.1016/S0140-6736(18)31550-2. [DOI] [PubMed] [Google Scholar]
- 3.Fritsche L.G., Igl W., Bailey J.N.C., et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saunier V., Merle B.M.J., Delyfer M.N., et al. Incidence of and risk factors associated with age-related macular degeneration: four-year follow-up from the ALIENOR Study. JAMA Ophthalmol. 2018;136:473–481. doi: 10.1001/jamaophthalmol.2018.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Bedell M., Zhang K. Age-related macular degeneration: genetic and environmental factors of disease. Mol Interv. 2010;10:271–281. doi: 10.1124/mi.10.5.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan P.L., Bowes Rickman C., Katsanis N. AMD and the alternative complement pathway: genetics and functional implications. Hum Genomics. 2016;10:23. doi: 10.1186/s40246-016-0079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan L.X., Germer C.J., La Cunza N., Lakkaraju A. Complement activation, lipid metabolism, and mitochondrial injury: converging pathways in age-related macular degeneration. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geerlings M.J., de Jong E.K., den Hollander A.I. The complement system in age-related macular degeneration: a review of rare genetic variants and implications for personalized treatment. Mol Immunol. 2017;84:65–76. doi: 10.1016/j.molimm.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khandhadia S., Cipriani V., Yates J.R., Lotery A.J. Age-related macular degeneration and the complement system. Immunobiology. 2012;217:127–146. doi: 10.1016/j.imbio.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 10.Zhang M., Jiang N., Chu Y., et al. Dysregulated metabolic pathways in age-related macular degeneration. Sci Rep. 2020;10:2464. doi: 10.1038/s41598-020-59244-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson D.H., Mullins R.F., Hageman G.S., Johnson L.V. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 12.Demirs J.T., Yang J., Crowley M.A., et al. Differential and altered spatial distribution of complement expression in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2021;62:26. doi: 10.1167/iovs.62.7.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voigt A.P., Mulfaul K., Mullin N.K., et al. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc Natl Acad Sci U S A. 2019;116:24100–24107. doi: 10.1073/pnas.1914143116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colijn J.M., den Hollander A.I., Demirkan A., et al. Increased high-density lipoprotein levels associated with age-related macular degeneration: evidence from the eye-risk and European Eye Epidemiology Consortia. Ophthalmology. 2019;126:393–406. doi: 10.1016/j.ophtha.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 15.van Leeuwen E.M., Emri E., Merle B.M.J., et al. A new perspective on lipid research in age-related macular degeneration. Prog Retin Eye Res. 2018;67:56–86. doi: 10.1016/j.preteyeres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Fan Q., Maranville J.C., Fritsche L., et al. HDL-cholesterol levels and risk of age-related macular degeneration: a multiethnic genetic study using Mendelian randomization. Int J Epidemiol. 2017;46:1891–1902. doi: 10.1093/ije/dyx189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Wang M., Zhang X., et al. The association between the lipids levels in blood and risk of age-related macular degeneration. Nutrients. 2016;8:663. doi: 10.3390/nu8100663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaisar T., Pennathur S., Green P.S., et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon S.M., Deng J., Lu L.J., Davidson W.S. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J Proteome Res. 2010;9:5239–5249. doi: 10.1021/pr100520x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.HB G., Rao V.S., Kakkar V.V. Friend turns foe: transformation of anti-inflammatory HDL to proinflammatory HDL during acute-phase response. Cholesterol. 2011;2011 doi: 10.1155/2011/274629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavanya R., Jeganathan V.S.E., Zheng Y., et al. Methodology of the Singapore Indian Chinese cohort (SICC) eye study: quantifying ethnic variations in the epidemiology of eye diseases in Asians. Ophthalmic Epidemiol. 2009;16:325–336. doi: 10.3109/09286580903144738. [DOI] [PubMed] [Google Scholar]
- 22.Foong A.W.P., Saw S.M., Loo J.L., et al. Rationale and methodology for a population-based study of eye diseases in Malay people: the Singapore Malay eye study (SiMES) Ophthalmic Epidemiol. 2007;14:25–35. doi: 10.1080/09286580600878844. [DOI] [PubMed] [Google Scholar]
- 23.Majithia S., Tham Y.C., Chee M.L., et al. Cohort profile: the Singapore Epidemiology of Eye Diseases study (SEED) Int J Epidemiol. 2021;50:41–52. doi: 10.1093/ije/dyaa238. Published correction appears in Int J Epidemiol. 2021;50:1401. [DOI] [PubMed] [Google Scholar]
- 24.Klein R., Davis M.D., Magli Y.L., et al. The Wisconsin age-related maculopathy grading system. Ophthalmology. 1991;98:1128–1134. doi: 10.1016/s0161-6420(91)32186-9. [DOI] [PubMed] [Google Scholar]
- 25.Soininen P., Kangas A.J., Würtz P., et al. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8:192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- 26.Ellul S., Wake M., Clifford S.A., et al. Metabolomics: population epidemiology and concordance in Australian children aged 11-12 years and their parents. BMJ Open. 2019;9(suppl 3):106–117. doi: 10.1136/bmjopen-2017-020900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nusinovici S., Zhang L., Chai X., et al. Machine learning to determine relative contribution of modifiable and non-modifiable risk factors of major eye diseases. Br J Ophthalmol. 2022;106:267–274. doi: 10.1136/bjophthalmol-2020-317454. [DOI] [PubMed] [Google Scholar]
- 28.Stephens M. False discovery rates: a new deal. Biostatistics. 2017;18:275–294. doi: 10.1093/biostatistics/kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glickman M.E., Rao S.R., Schultz M.R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol. 2014;67:850–857. doi: 10.1016/j.jclinepi.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Eren E., Yilmaz N., Aydin O. High density lipoprotein and it’s dysfunction. Open Biochem J. 2012;6:78–93. doi: 10.2174/1874091X01206010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betzler B.K., Rim T.H., Sabanayagam C., et al. High-density lipoprotein cholesterol in age-related ocular diseases. Biomolecules. 2020;10:645. doi: 10.3390/biom10040645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navab M., Reddy S.T., Van Lenten B.J., et al. The role of dysfunctional HDL in atherosclerosis. J Lipid Res. 2009;50(suppl):S145–S149. doi: 10.1194/jlr.R800036-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McPherson P.A., Young I.S., McKibben B., McEneny J. High density lipoprotein subfractions: isolation, composition, and their duplicitous role in oxidation. J Lipid Res. 2007;48:86–95. doi: 10.1194/jlr.M600094-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Smith J.D. Dysfunctional HDL as a diagnostic and therapeutic target. Arterioscler Thromb Vasc Biol. 2010;30:151–155. doi: 10.1161/ATVBAHA.108.179226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kontush A., Chapman M.J. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol Rev. 2006;58:342–374. doi: 10.1124/pr.58.3.1. [DOI] [PubMed] [Google Scholar]
- 36.Corsetti J.P., Sparks C.E., James R.W., et al. Low serum paraoxonase-1 activity associates with incident cardiovascular disease risk in subjects with concurrently high levels of high-density lipoprotein cholesterol and C-reactive protein. J Clin Med. 2019;8:1357. doi: 10.3390/jcm8091357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viktorinova A., Svitekova K., Stecova A., Krizko M. Relationship between selected oxidative stress markers and lipid risk factors for cardiovascular disease in middle-aged adults and its possible clinical relevance. Clin Biochem. 2016;49:868–872. doi: 10.1016/j.clinbiochem.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Gordon S.M., Xi H., et al. HDL subclass proteomic analysis and functional implication of protein dynamic change during HDL maturation. Redox Biol. 2019;24 doi: 10.1016/j.redox.2019.101222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamilton K.K., Zhao J., Sims P.J. Interaction between apolipoproteins A-I and A-II and the membrane attack complex of complement. Affinity of the apoproteins for polymeric C9. J Biol Chem. 1993;268:3632–3638. [PubMed] [Google Scholar]
- 40.Delcourt C., Michel F., Colvez A., et al. Associations of cardiovascular disease and its risk factors with age-related macular degeneration: the POLA study. Ophthalmic Epidemiol. 2001;8:237–249. doi: 10.1076/opep.8.4.237.1613. [DOI] [PubMed] [Google Scholar]
- 41.Rosenfeld S.I., Packman C.H., Leddy J.P. Inhibition of the lytic action of cell-bound terminal complement components by human high density lipoproteins and apoproteins. J Clin Invest. 1983;71:795–808. doi: 10.1172/JCI110833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scantlebury T., Maslowska M., Cianflone K. Chylomicron-specific enhancement of acylation stimulating protein and precursor protein C3 production in differentiated human adipocytes. J Biol Chem. 1998;273:20903–20909. doi: 10.1074/jbc.273.33.20903. [DOI] [PubMed] [Google Scholar]
- 43.Jenne D.E., Lowin B., Peitsch M.C., et al. Clusterin (complement lysis inhibitor) forms a high density lipoprotein complex with apolipoprotein A-I in human plasma. J Biol Chem. 1991;266:11030–11036. [PubMed] [Google Scholar]
- 44.Tschopp J., Chonn A., Hertig S., French L.E. Clusterin, the human apolipoprotein and complement inhibitor, binds to complement C7, C8 beta, and the b domain of C9. J Immunol. 1993;151:2159–2165. [PubMed] [Google Scholar]
- 45.Nissilä E., Hakala P., Leskinen K., et al. Complement factor H and apolipoprotein E participate in regulation of inflammation in THP-1 macrophages. Front Immunol. 2018;9:2701. doi: 10.3389/fimmu.2018.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haapasalo K., van Kessel K., Nissilä E., et al. Complement factor H binds to human serum apolipoprotein E and mediates complement regulation on high density lipoprotein particles. J Biol Chem. 2015;290:28977–28987. doi: 10.1074/jbc.M115.669226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roizenblatt M., Naranjit N., Maia M., Gehlbach P.L. The question of a role for statins in age-related macular degeneration. Int J Mol Sci. 2018;19:3688. doi: 10.3390/ijms19113688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peponis V., Chalkiadakis S.E., Bonovas S., Sitaras N.M. The controversy over the association between statins use and progression of age-related macular degeneration: a mini review. Clin Ophthalmol. 2010;4:865–869. doi: 10.2147/opth.s12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwig C.A., Vail D., Rajeshuni N.A., et al. Statins and the progression of age-related macular degeneration in the United States. PLoS One. 2021;16 doi: 10.1371/journal.pone.0252878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuo J.Y., Wiens M., Etminan M., Maberley D.A. Use of lipid-lowering agents for the prevention of age-related macular degeneration: a meta-analysis of observational studies. Ophthalmic Epidemiol. 2007;14:367–374. doi: 10.1080/09286580701421684. [DOI] [PubMed] [Google Scholar]
- 51.Yu Z., Kastenmüller G., He Y., et al. Differences between human plasma and serum metabolite profiles. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.