Abstract
Stress-induced neuroinflammation is considered an important mechanism in the pathogenesis of depression. As immune effector cells in the brain, microglia play an essential role in neuroinflammation under stress, but the underlying mechanism remains controversial. Here, we performed RNA-seq and ATAC-seq to study microglia-specific epigenomic changes in mice after 12 weeks of exposure to mild stress. Our study revealed that chronic stress induced pronounced anxiety and depressive-like behavioral changes. However, microglia did not manifest a state of neuroinflammatory activation; instead, they displayed morphological changes characterized by hyper-ramification. Furthermore, we revealed large-scale transcriptional repression in microglia isolated from the stressed brain, including many interferon (IFN)-regulated genes (IRGs) and some encompassing DNA repeats. GSEA showed that the down-regulated genes were enriched in the IFN-mediated neuroimmune signaling pathways. In addition, integrative analysis with a published scRNA-seq dataset revealed that these down-regulated genes were enriched in a distinct subpopulation of “Interferon microglia”. ATAC-seq analysis further showed that differential gene expression was positively correlated with the changes in chromatin accessibility, and the IFN-stimulated response element (ISRE) was enriched in the down-regulated ATAC-seq loci. Interestingly, this phenotype was not associated with the production of IFNs. Instead, the gene encoding Activating Transcription Factor 3 (ATF3) was significantly increased in the stressed microglia, which might contribute to the transcriptional repression of IRGs. Our study reported microglia-specific transcriptional repression of IRGs independent of the production of IFNs, providing some new insights into neuroimmune dysregulation under prolonged stress.
Keywords: Chronic stress, Interferon signaling, Microglia, Transcriptome, Chromatin accessibility, Activating transcription factor 3
1. Introduction
Stress is an essential and instinctive defense mechanism against environmental insults; however, it can cause adverse effects once beyond the body's limit. Chronic stress is becoming a significant risk factor in many health problems including depression, a common but severe mental disorder characterized by persistent low mood and loss of interest (Kendler et al., 1999; Tennant, 2002). Although it is widely acknowledged that chronic stress increases susceptibility to depression, the underlying mechanism remains largely elusive. In recent years, accumulating evidence has shown a positive correlation between dysregulation of the neuroimmune system and depression (Osimo et al., 2019; Howren et al., 2009). The elevation of pro-inflammatory cytokines has been detected in peripheral plasma and the central nervous system (CNS) of patients (Maes et al., 1995; Tonelli et al., 2008; Pandey et al., 2012), and stress-based animal models of depression (Pugh et al., 1999; O'Connor et al., 2003). Moreover, both preclinical and clinical studies reported promising evidence for anti-inflammation treatments for depression (Müller et al., 2006; Reynolds et al., 2004).
It is well-accepted that stress can induce neuroinflammation, but different types of stress result in the release of different groups of inflammatory cytokines (Wohleb et al., 2018; Mckim et al., 2017). Acute stress induces the release of typical pro-inflammatory cytokines (Marsland et al., 2017). Meanwhile, chronic stress exposure causes a relatively mild inflammatory response and primarily targets the IFN-signaling pathway (Rohleder, 2019). Previous studies reported the increases of serum IFN-β and interferon regulatory factor 8 (IRF8) in a mouse model of chronic stress, and systemic blockade of IFN-I signaling attenuated the depressive-like behaviors (Tripathi et al., 2021; Akagi et al., 2014). Moreover, studies on postmortem human brains reported elevated expression of IFN-regulated genes (IRGs) in the prefrontal cortex (PFC) of suicide subjects (Tripathi et al., 2021). However, studies from other groups also reported controversial results. For example, an early study reported a dose-dependent attenuation of IFN-γ production and immunocompromised state after exposure to mild electric foot-shock in rats (Sonnenfeld et al., 1992).
Microglia are the primary immune cells in the CNS and manifest various morphological features under physiological and pathological conditions. They display ramified morphology characterized by short and fine processes in the resting state and transform into an amoeboid morphology under adverse stimulations (Boche et al., 2013). Meanwhile, differential microglia morphological changes have been reported in response to acute and chronic stress (Rohleder, 2019; Hammen et al., 2009). For example, acute stress induced microglial proliferation with an increase of soma area and decrease of process length, followed by a switch to a dystrophic morphology and expression of apoptotic markers at a later stage (Kreisel et al., 2014). In contrast, hyper-ramification was observed in microglia under chronic stress, characterized by the increases in process length and number of branches (Xie et al., 2021; Hinwood et al., 2013).
Although the role of microglia in response to stress is of high interest, its underlying regulatory mechanism remains largely unknown. Sex is an essential biological factor involved in emotional behaviors (Williams et al., 2022; Labonte et al., 2017; Touchant and Labonte, 2022), and microglia functions (Han et al., 1186; Lenz and McCarthy, 2015; Bollinger, 2021). Previous studies have shown that both males and females develop anxiety and depressive-like behaviors after chronic stress, but males show more pronounced cellular phenotypes in microglia than females (Bollinger, 2021; Wohleb et al., 2018; Woodburn et al., 2021). Therefore, in the current study, we focused on the males to study stress-induced morphological and functional alterations in microglia. We first constructed a chronic mild unpredictable stress (CMUS) mouse model, evaluated the anxiety and depressive-like behaviors, and then examined the morphological changes of microglia in multiple brain regions. Meanwhile, we isolated microglia from the whole brain (olfactory bulb and cerebellum removed) and performed RNA-seq and ATAC-seq to study microglia-specific epigenomic alterations in response to prolonged stress (Fig. 1A).
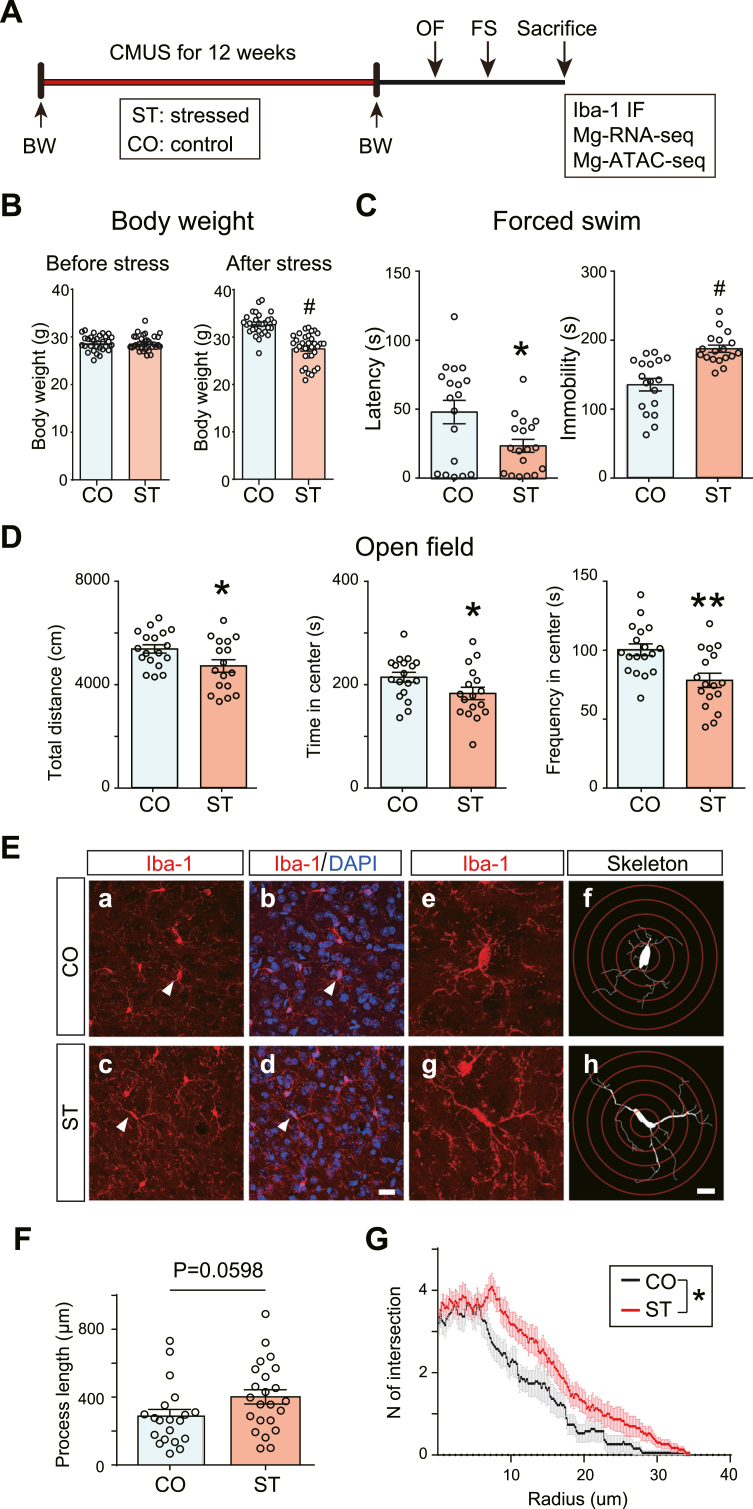
Fig. 1.
Behavioral and microglia phenotype after chronic stress. (A) Experimental design. CMUS, chronic mild unpredictable stress. ST, stressed group; CO, control group. OF, open field test. FSW, forced swim test. IHC, Immunohistochemistry. Mg-RNA-seq, microglia-specific RNA-seq. Mg-ATCA-seq, microglia-specific ATAC-seq. (B) Body weight before and after exposure to CMUS. Unpaired t-test, #P < 0.0001. N = 29–33/group. (C) Forced swim. Unpaired t-test, #P < 0.0001. N = 15–17/group. (D) Open field. Unpaired t-test, *P < 0.05, **P < 0 0.01. N = 17–18/group. (E) Representative images of anti-Iba1 immunofluorescence staining in the prefrontal cortex. Scale bar, 20 μm (a–d), 10 μm (e–h). (F) Bar plot quantifies the length of the microglia process. N = 20–23 processes/3 animals/group. Unpaired t-test. G) Sholl analysis for microglia branch complexity. N = 20–23 processes/3 animals/group. Two-way ANOVA test. *P < 0.05. All data are expressed as mean ± SEM.
2. Methods
2.1. Animals
All animal work was granted by the Animal Care and Use Committee of Shanghai Medical College, Fudan University. Up to five adult C57BL/6Slac mice (Shanghai Slake Laboratory Animal Co., Ltd, China) were housed per independent cage under constant conditions (21–24 °C, 45–55% humidity), with a 12-12-hr day/night cycle. All mice were allowed sterile water and food ad libitum. Only males were used in the current study.
2.2. Behavioral assays
2.2.1. Chronic mild unpredictable stress (CMUS)
Mice in the stress group (ST) were exposed to two or three random stressors every day for 12 weeks, according to a previous study with minor modifications (Liu et al., 2018). The detailed protocol was presented in Table S1. Age-matched mice in the control group (CO) were housed in home cages.
2.2.2. Open field test
Mice were placed in an open field box (40 cm × 40 cm × 40 cm) and allowed free exploration for 15 min. Half of the whole arena in the center was defined as the central zone. EthoVision XT 14.0 was used for track tracing and data analysis. Total distance, number of visits to the center, and time spent in the center were measured to evaluate anxiety-like behaviors.
2.2.3. Forced swim test
Mice were placed in glass beakers filled with 30 cm-deep water at 25 ± 1 °C and allowed free swimming for 6 min. EthoVision XT 14.0 was used for track tracing and data analysis. Latency to the first immobility and total immobility time in the first 5 min were measured to evaluate depressive-like behaviors.
2.3. Immunohistochemistry
Mice were perfused with 4% PFA and brains were dissected and sectioned into 30 μm coronal sections. In brief, free-floating sections were washed and incubated in the blocking solution (0.1% Triton X-100, 1% normal goat serum in 1 × PBS) at room temperature for 30 min, followed by overnight incubation with anti-Iba1 (Abcam, ab178846). The sections were then washed and incubated in the secondary antibody (goat anti-rabbit Alexa Fluor 594, Jackson, 111-585-003) at room temperature for 60 min, followed by DAPI counterstaining. Images were captured with an Olympus VS120 microscope and analyzed by ImageJ. Sholl analysis was used to evaluate cell morphology.
2.4. Microglia-specific epigenomic profiling
2.4.1. Preparation of single-cell suspension from adult brain
Single-cell suspension from the adult brain was prepared using Adult Brain Dissociation Kit (Miltenyi Biotec, 130-107-677) and Debris Removal Solution (Miltenyi Biotec, 130-109-398) according to the manufacturer's instructions. In brief, animals were anesthetized with 2% isoflurane and perfused with ice-cold 1 × PBS. Next, the whole brain was dissected (olfactory bulb and cerebellum removed), washed, finely minced, and transferred into the C-tube containing 1950 μl Enzyme mix 1 (1900 μl buffer X and 50 μl Enzyme P) and 30 μl of ice-cold Enzyme mix 2 (20 μl buffer Y and 10 μl Enzyme A). C tube was then attached to Miltenyi's gentleMACS Octo Dissociator with Heaters, and program 37 °C _ABDK_1 was launched for 30 min. After digestion, the brain homogenate was resuspended in 1 × HBSS, filtered through a 70-μm filter, and collected by centrifugation. Next, the cell pellet was resuspended with 6200 μl of iced cold 1 × HBSS, mixed with 1800 μl of ice-cold Debris Removal Solution, and 4 ml of iced cold 1 × HBSS was then gently overlaid on top of the cell suspension, followed by centrifugation at 4 °C, 3000 g, for 10 min. Three phases were formed after spin. The top two phases were discarded, and the remainder was refilled with 1 × HBSS to a final volume of 15 ml. The cells were further collected and resuspended in 1 × HBSS containing 1% fetal bovine serum. The purity and viability of the cell suspension were checked under a microscope and used for following microglia isolation.
2.4.2. Microglia isolation
Single-cell suspension was first incubated in Fc receptor blocking buffer (Miltenyi Biotec, 130-092-575) for 10 min to minimize microglia activation during handling and then incubated with anti-CD11b-MicroBeads (Miltenyi, 130-093-634) for 10 min at 4 °C in the dark. The sample then went through the LS separation column (Miltenyi Biotec, 130-042-401) for positive selection. Finally, an aliquot of purified cells was further incubated with an anti-CD11b-FITC monoclonal antibody (M1/70) (eBioscience, 11-0112-81) for 30 min at 4 °C in the dark, and the purity of CD11b + cells was analyzed using Beckman flow cytometer.
2.4.3. Total RNA extraction
Total RNA was extracted from isolated microglia with the Direct-zol RNA Microprep kit (Zymoresearch, R2062) following the manufacturer's instructions. Briefly, cells were lysed in TRI reagent, mixed with an equal volume of 100% ethanol, and then transferred to a Zymo-Spin™ IC column. After washing with RNA Wash Buffer, the samples were incubated with DNase I on the column for 15 min at room temperature. The column was then washed with Direct-zol™ RNA PreWash and RNA Wash Buffer, and RAN was eluted in DNase/RNase-free water.
2.4.4. Real-time RT-PCR
In brief, total RNA from isolated microglia was reverse-transcribed using the iScript™ cDNA Synthesis Kit (BIORAD, 1708891). Real-time qPCR (Thermo Fisher Scientific Applied Biosystems QuantStudio5) was performed using Power SYBR™ Green PCR Master Mix (Thermo, 4368702). Cell-type specific genes were selected: Aif1, Itgam, Cx3cr1, Hexb, P2ry12 (microglia); Mbp (oligodendrocytes); Grin2b, Rbfox3, and Map2 (neuron); S100β, Gfap, and Slc1a3 (astrocyte). All the primers were listed in Table S2.
2.4.5. RNA-seq library preparation
Total RNA from isolated microglia was sent to Annoroad, China, for mRNA-seq library preparation, and sequenced on Illumina XTen, 150 bp, paired-end.
2.4.6. ATAC-seq library preparation
ATAC-seq was performed using freshly isolated microglia from the same animal used for RNA-seq. In brief, isolated microglia were homogenized in the lysis buffer (0.32 M sucrose, 5 mM CaCl2, 3 mM MgAce2, 0.1 mM EDTA, 10 mM Tris-HCl, pH = 8, 0.1% NP-40), and nuclei were collected by centrifugation. ATAC-seq was performed following published protocol with some modifications (Shen et al., 2021). In brief, nuclei were incubated with 5 μl of Tn5 enzyme in 50 μl of digestion buffer (TD501-02, TruePrep DNA Library Preparation Kit V2, Vazyme, China) at 37 °C for 30 min. DNA was then purified using a MinElute gel extraction kit (Qiagen, 28604) and eluted in 24 μl of EB buffer. The PCR reaction was set up as follows: 24 μl of DNA, 5 μl of PPM, 10 μl of 5 × TAB, 5 μl of N7 primer, 5 μl of N5 primer, and 1 μl of TAE buffer, and the PCR programming was: 72 °C for 3 min, 98 °C for 30 s, 11 cycles of amplification: 98 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s. Library DNA with a size between 150 bp and 500 bp was selected. The quality of the library was evaluated by Agilent 4200 TapeStation, and the concentration was measured with Qubit4. The library was then sent for sequencing on Illumina X Ten, PE, 150 bp.
2.5. ELISA
The concentrations of IFN-α, IFN-β, and IFN-γ in mouse brain and spleen were measured using ELISA kits (Mouse, IFN-α ELISA Kit, JM-02649M1; Mouse IFN-β ELISA Kit, JM-02407M1; Mouse IFN-γ ELISA Kit, JM-02465M1, Jiangsu Jingmei Biological Technology Co., Ltd, China) following the manufacturer's instructions. Protein concentration was measured with a BCA protein quantification assay kit (JM-100009A, Jiangsu Jingmei Biological Technology Co., Ltd, China) and used for normalization.
2.6. Data analysis
2.6.1. RNA-seq
Raw data were first evaluated by FastQC for quality control. Adapter sequences, low-quality reads, and too short reads were removed from raw data using Trim-galore with parameters: -quality 20 --phred33 --stringency 1 --length 20 –paired. Next, Tophat2 v2.1.1 was used for alignment (UCSC mm10) with parameters: -max-multihits 20, --transcriptome-max-hits 60; --GTF gencode.vM20.annotation.gtf. Samtools v1.9 was used to sort and build the alignment file index. Gene counts were generated using FeatureCounts v1.6.3 with parameters: p -t exon -g gene_id; -a gencode.vM20.annotation.gtf, and FPKMs were calculated. Differential analysis was conducted using DESeq2, and significant genes were defined as FDR <0.05. Gene Set Enrichment Analyses were performed using the GSEA software (version 3.0) from the Broad Institute (Subramanian et al., 2005). ShinyGO v0.741 (“Genome” function) was used for gene clustering analysis on differential genes with the parameters of 1 Mb window size, 4 steps in a window, and FDR <0.0001. BamCoverage from Deeptools was used to generate normalized bigwig files.
2.6.2. ATAC-seq
Raw data was evaluated by FastQC and cleaned as described above. Bowtie2 v2.3.4.3 was used for alignment (UCSC mm10) using parameters: -phred33, --sensitive; multiple alignments were only reported as the best with MAPQ; --maxins 1000. After removing duplications, Samtools v1.9 was used to sort and build the alignment file index. Differential analysis was performed using diffReps with P value of 0.001 and window size of 1 Kb. Signals of “Control.avg” less than 100 in “Down” events and “Treatment.avg” less than 100 in “Up” events were considered as background and excluded. BamCoverage from Deeptools was used to generate normalized bigwig files, which were then visualized on IGV. Motif search was performed using the “findmotifs.pl” function of HOMER.
2.6.3. DNA repeats
For DNA repeats analysis, reference repeat sequences were obtained from the UCSC RepeatMasker program (http://www.repeatmasker.org. 1996–2010.), which offers a unique coordinate for individual annotated repeats. In order to assign the signal (RNA-seq or ATAC-seq reads) to individual repeats, multiple mapping reads were excluded, and unique reads were counted on individual annotated DNA repeats by FeatureCounts v1.6.3 (featureCounts -T 28 -p -t exon -g id -a ucsc.repeats.gtf -o count bam). DESeq2 was used to perform differential analysis of signals on DNA repeats between the CO and ST groups.
2.6.4. Integrative analysis of bulk RNA-seq and published scRNA-seq
Cell-type enrichment analysis was generated with differentially expressed genes (DEGs) from our bulk microglia RNA-seq and the published microglia scRNA-seq data (GSE145454) (Ndoja et al., 2020). All the DEGs were divided into “Up_genes” and “Down_genes” and analyzed as two gene sets, respectively. Firstly, UMAP information of a single cell was generated using the R package Seurat's Dimplot function, and the cell clusters were visualized by R packages ggplot2 and ggalt. The average scRNA-seq expression values of “Up_genes” and “Down_genes” gene sets were calculated, scaled by the function in R (center = T, scale = T), and then visualized on a fitting color scale of each cell in UMAP plots. Secondly, the expression value of scRNA-seq for the individual gene in “Up_genes” and “Down_genes” was visualized using Seurat's DoHeatmap function. Only those genes detected in scRNA-seq were shown. Thirdly, marker genes of each cell cluster in scRNA-seq data were identified using Seurat's FindAllMarkers with the following parameters: only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25, and enrichment scores of the “Up_genes” and “Down_genes” in each cell cluster were calculated using R package GeneOverlap (Peña et al., 2019; GeneOverlap, 2016). Finally, cell-type enrichment was calculated with the AUCell algorithm from the open-source irGSEA R package (https://github.com/chuiqin/irGSEA).
Cell-type deconvolution analysis of our bulk microglia RNA-seq was calculated by CIBERSORTx (Newman et al., 2019). Expression matrices of all genes from our bulk RNA-seq data and the published microglia scRNA-seq data (Ndoja et al., 2020) were generated, and the Cell-Fractions module from CIBERSORTx was used to perform an estimation of the proportions of distinct cell clusters in bulk RNA-seq from CO and ST groups.
2.6.5. Integrative analysis of RNA-seq and ATAC-seq
Correlation analysis between RNA-seq and ATAC-seq was conducted as previously described with modifications (Wang et al., 2021). Pearson's correlation coefficient between the fold change of differential genes or DNA repeats (RNA-seq: gene, FDR <0.05; DNA repeats, P < 0.05) and corresponding ATAC-seq signal were performed. P-value was calculated in R. Scatter plots, and a linear regression curve was produced using the R package “ggplot2".
3. Results
3.1. Behavioral and cellular phenotypes after chronic stress
We first evaluate the behavioral changes after prolonged stress in the current CMUS model. Consistent with published literature (Gao et al., 2018), we observed a significant loss of body weight in the stressed mice (Fig. 1B). In the open field test, the total distance and time spent in the center were significantly decreased in ST compared to CO (Fig. 1D), indicating hypoactivity and anxiety-like behavior. In the forced swim test, the latency to first immobility was significantly decreased, and total immobility time was significantly increased in ST (Fig. 1C), indicating elevated behavioral despair.
Accumulating evidence reported neuroinflammation after chronic stress and its contribution to mood behaviors (Walker, 2012). Microglia are the primary immune cells in the brain and vital cellular mediators during neuroinflammation. We, therefore, did anti-Iba1 staining to check the condition of microglia with a focus on PFC (Fig. 1E), a critical brain region associated with depression. It has been reported that stress can cause microglia activation, often described as swollen cell bodies with shorter and thicker processes or amoeboid-like morphology (Walker, 2012). However, in our current CMUS mouse model with prolonged exposure (12 weeks), microglia in the ST group displayed hyper-ramification morphology featured by an increased length of branches (P = 0.0598) (Fig. 1F) and an increased number of branch intersections (Fig. 1G). We also checked the dentate gyrus (DG) of the hippocampus (Figs. S1A, C, E) and amygdala (AMYG) (Figs. S1B, D, F), and observed similar hyper-ramification morphological changes of microglia in ST, although the differences in AMYG did not reach statistical significance.
3.2. Quality control for microglia isolation from adult mouse brain
Hyper-ramification of microglia in response to chronic stress was previously reported (Walker, 2012), but its underlying molecular mechanism remains largely unknown. Therefore, we performed RNA-seq and ATAC-seq on microglia isolated to study microglia-specific epigenomic alterations in response to chronic stress. To this end, we first optimized the microglia isolation protocol with stringent quality checks. We purified microglia from adult mice brains by magnetic-activated cell sorting (MACS) using anti-CD11b conjugated magnetic beads (Fig. S2A). After MACS, we performed flow cytometry analysis to evaluate the purity of isolated microglia (CD11b+) (Figs. S2B–C). As shown in Fig. S2B, around 23.69% of events were CD11b-FITC+ in the single-cell preparation before MACS, and 80.82% of the “CD11b+" cells were CD11b-FITC + after MACS. Only 1.15% of the “CD11b-" cells after MACS were CD11b-FITC+. This purity was comparable to the previously reported data (C, 2016). Moreover, real-time PCR on isolated microglia and brain homogenates showed enrichment of cell-marker genes in microglia, but not other cell types (Fig. S2C). After optimization, we applied this protocol to the CO and ST brains for subsequent molecular studies.
3.3. Transcriptional repression of IFN-regulated genes in microglia after chronic stress
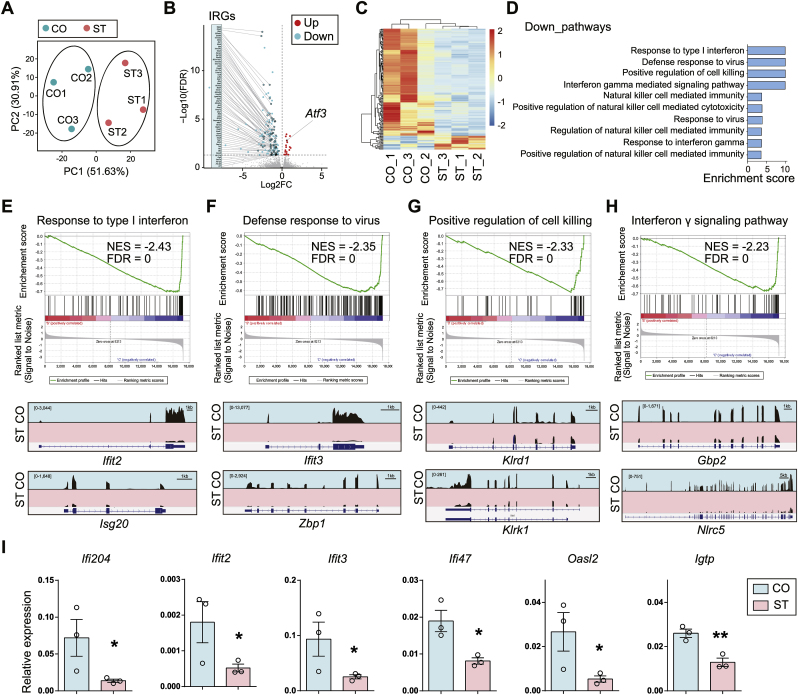
We performed RNA-seq on isolated microglia from CO and ST brains with three biological replicates. PCA analysis revealed a clear separation by treatment (PC1 51.63%, PC2 30.91%) (Fig. 2A). Differential analysis revealed a total of 158 DEGs (FDR < 0.05, ST/CO), and most of them were down-regulated (88.6%, 140/158) in ST (Fig. 2B–C, Table S4). Only 18 genes were up-regulated in ST. We noticed there were many genes associated with interferon, and indeed, 65 out of the 140 down-regulated genes belonged to the family of interferon-regulated genes (IRGs) when checked in the database of Interferome v2.0 (http://interferome.its.monash.edu.au/interferome/, “Mus musculus”, “Nerve system”, “microglial cell”) (Rusinova et al., 2013) (Fig. 2B). Moreover, GSEA revealed multiple significantly down-regulated functional pathways (ST vs. CO) related to IFN-mediated immune responses. The top four were “Response to type I interferon (Ifit2, Isg20)", “Defense response to virus (Ifit3, Zbp1)", “Positive regulation of cell kill (Klrd1, Klrk1)", and “Interferon γ signaling pathway (Gbp2, Nlrc)" (Fig. 2D). We selected two representative genes for each pathway (see genes listed above), and IGV map tracks showed robust decreases of RNA-seq signal in ST compared to CO (Fig. 2E–H). There was no significantly enriched up-regulated pathway. We further performed real-time RT-PCR on isolated microglia and validated the decrease of some down-regulated IRGs in ST compared to CO (Fig. 2I).
Fig. 2.
Microglia-specific transcriptional repression of genes in IFN-signaling pathway after chronic stress. (A) PCA plot of RNA-seq using isolated microglia from mice exposed to chronic stress (ST) and controls (CO). N = 3/group. (B) The volcano plot shows the differential gene expression in ST compared to CO. Red dot, up-regulated genes (Up); blue dot, down-regulated genes (Down); gray dot, non-significant genes. FDR <0.05. Notice the massive down-regulation of genes in ST, and many of them (65 out of 140) are IFN-regulated genes (IRGs) (Interferome v2.0). Notice the increase of Atf3 in ST. (C) Clustered heatmap shows differential gene expression (ST v.s. CO). (D) Gene Set Enrichment Analysis (GSEA) shows down-regulated functional pathways (ST vs. CO). Notice high enrichment for IFN-signaling pathways. Enrichment score = -Log10(P+10−10). (E–H) (Top) Enrichment plots and (Bottom) IGV map tracks for representative genes in the top four down-regulated pathways. Notice a decrease of RNA-seq signal for IRGs in ST compared to CO. (I) Real-time RT-PCR validation for the down-regulated IRGs. N = 3/group. Mean ± SEM. Unpaired t-test, one-tailed, *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
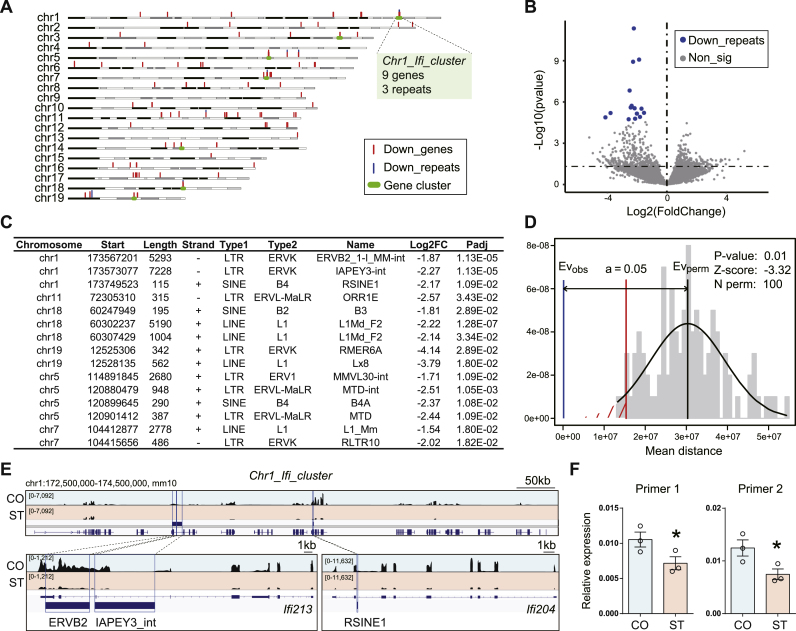
Interestingly, some down-regulated genes tend to cluster together on the genome. Indeed, we identified seven significant clusters of genes using the “Genome” function of ShinyGO v0.741 (Fig. 3A). These clustered genes were located on 7 different chromosomes, including a cluster of IFN-inducible genes (Ifi214, Ifi213, Ifi209, Ifi208, Ifi207, Ifi204, and Ifi206) on chr1:172,500,000–174,500,000, mm10 (Fig. 3A, “Chr1_Ifi_cluster”). Moreover, transcripts from DNA repeats in some of these gene clusters were also down-regulated in ST (Fig. 3A). DNA repeats comprise almost half of the mammalian genome and contribute to cell diversity in the brain, with implications for some pathological conditions (Lapp and Hunter, 2019; Jnsson et al., 2020). Alterations of transcripts from DNA repeats, for example, the long interspersed element-1 (L1), were reported in response to stress (Bachiller et al., 2017). Therefore, we performed differential analysis for the transcription of DNA repeats and revealed large-scale down-regulation in ST compared to CO (Fig. 3B). However, only 15 DNA repeats reached statistical significance (FDR <0.05), and all of them were down-regulated in ST (Fig. 3C). These down-regulated DNA repeats belong to LTR, LINE, and SINE families, including ERVK, ERVL-MaLR, L1, B2, and B4 (Fig. 3C). We performed a permutation test to assess the distance between the significantly down-regulated DNA repeats and genes and found it was significantly shorter than expected (P < 0.01, Z = −3.32) (Fig. 3D). The distance (Evobs) was almost equal to zero (Fig. 3D, also shown in Fig. 3A), indicating great overlap between down-regulated DNA repeats and genes. For example, three DNA repeats (ERVB2, IAPEY3_int, RSINE1) resided in the gene body of Ifi213 and Ifi204 in “Chr1_Ifi_cluster”. IGV map tracks showed substantial decreases in RNA-seq signal for DNA repeats and genes in ST compared to CO (Fig. 3E). We further performed real-time RT-PCR on isolated microglia and validated decreased transcription on this ERVB2 in ST using two sets of primers (Fig. 3F).
Fig. 3.
Microglia-specific transcriptional repression of DNA repeats after chronic stress. (A) Genome distribution of down-regulated genes (down_genes, red vertical bars) and DNA repeats (blue vertical bars). Gene clusters are highlighted with green horizontal bars. Notice the cluster of genes encoding IFN-induced proteins located at the end of chromosome 1 (“Chr1_Ifi_cluster”). (B) The volcano plot shows differential DNA repeats transcription after chronic stress (ST v.s. CO). Blue, down-regulated DNA repeats (“Down_repeats”); gray, non-significant. FDR <0.05. N = 3/group. The genome locations of these 15 “Down_repeats” are labeled in panel A. (C) List of significantly down-regulated DNA repeats. (D) Permutation test for the distance between significantly down-regulated DNA repeats and genes. (E) IGV map tracks show RNA-seq signal at Chr1_Ifi_cluster. Notice three DNA repeats, ERVB2, IAPEY3_int, and RSINE1, located in the gene bodies of Ifi213 and Ifi204. (F) Real-time RT-PCR validation for the ERVB2 in Chr1_Ifi_cluster using two different primers. N = 3/group. Mean ± SEM. Unpaired t-test, one-tailed, *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. Stress-induced DEGs were enriched in a distinct subpopulation of microglia
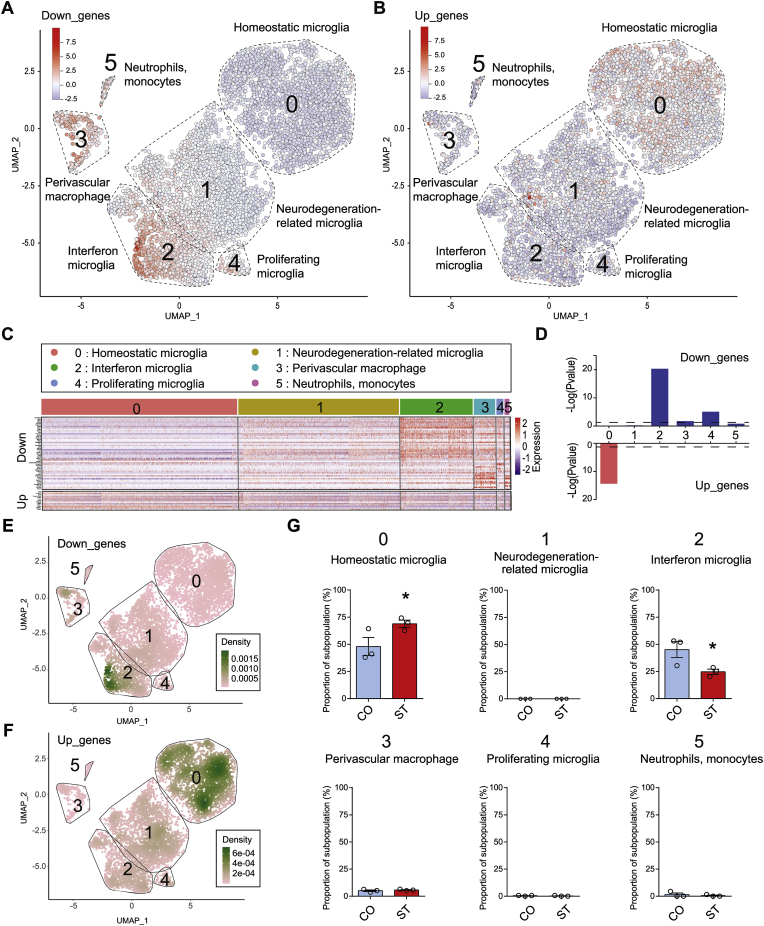
Recent studies using scRNA-seq revealed multiple sub-populations of microglia in the adult brain, especially under different pathological conditions (Hammond et al., 2018). For example, six cell clusters were identified in a published scRNA-seq dataset (GSE145454) on isolated microglia from mouse forebrain (Ndoja et al., 2020), defined as “Homoeostatic microglia” (cluster 0), “Neurodegeneration-related microglia” (cluster 1), “Interferon microglia” (cluster 2), “Perivascular macrophage” (cluster 3), “Proliferating microglia” (cluster 4), and “Neutrophils/monocytes” (cluster 5). We visualized the expression of DEGs from our bulk RNA-seq in single cells of this published scRNA-seq (GSE145454) and noticed a distinct distribution pattern in microglia subpopulations for the down- and up-regulated genes, respectively (Fig. 4A–C). Indeed, gene set enrichment analysis showed that the set of down-regulated genes was significantly enriched in “Interferon microglia” (P = 4.53e-21), “Perivascular macrophage” (P = 1.81e-2), and “Proliferating microglia” (P = 8.60e-6). In contrast, the up-regulated gene set was enriched in “Homeostatic microglia” (P = 4.29e-15) (Fig. 4D). Alternatively, cell-type enrichment analysis using the AUCell algorithm in irGSEA revealed the same results (Fig. 4E–F). We also did cell deconvolution analysis to estimate the proportions of distinct microglia subpopulations in our bulk microglia RNA-seq in CO and ST groups and found a significant increase of “Homeostatic microglia” and a decrease of “Interferon microglia” in ST (Fig. 4G). These results indicated that distinct subpopulations of microglia responded to CMUS differentially, and the transcriptional repression occurred primarily in “Interferon microglia".
Fig. 4.
Enrichment of chronic stress-induced differential expressed genes in distinct microglia subpopulations. (A, B) Scatterplots visualize the expression and distribution of (A) “Down_genes” and (B) “Up_genes” in the UMAP map of published microglia scRNA-seq data (GSE145454). The six cell clusters (cluster 0 to cluster 5) represent six subpopulations of microglia (Ndoja, A. et al., 2020, Cell). In the current study, visualized genes are differentially expressed genes (DEGs) from the microglia bulk RNA-seq. The color indicates the average expression of detectable DEGs in the published microglia scRNA-seq data. (C) Seurat's DoHeatmap shows the individual gene expression level of DEGs in each cell of the published microglia scRNA-seq. Only genes detected in scRNA-seq were shown. (D) Bar plots show the gene set enrichment score of (Top) “Down_genes” and (Bottom) “Up_genes” in each microglia subpopulation. (E, F) Density scatterplots visualize cell-type enrichment for (E) “Down_genes” and (F) “Up_genes” in AUcell (https://github.com/chuiqin/irGSEA). Notice the enrichment of down-regulated genes in cluster 2 of “Interferon microglia”, and up-regulated genes in cluster 0 of “Homeostatic microglia”. (G) Bar graphs show the estimated proportions of distinct subpopulations in the bulk microglia expression profiles in CO and ST calculated by CIBERSORTx. N = 3/group. Mean ± SEM. Unpaired t-test, one-tailed, *P < 0.05. Notice the increase of cluster 2 of “Interferon microglia” and the decrease of cluster 0 of “Homeostatic microglia” in ST. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Alterations of chromatin accessibility in microglia after chronic stress
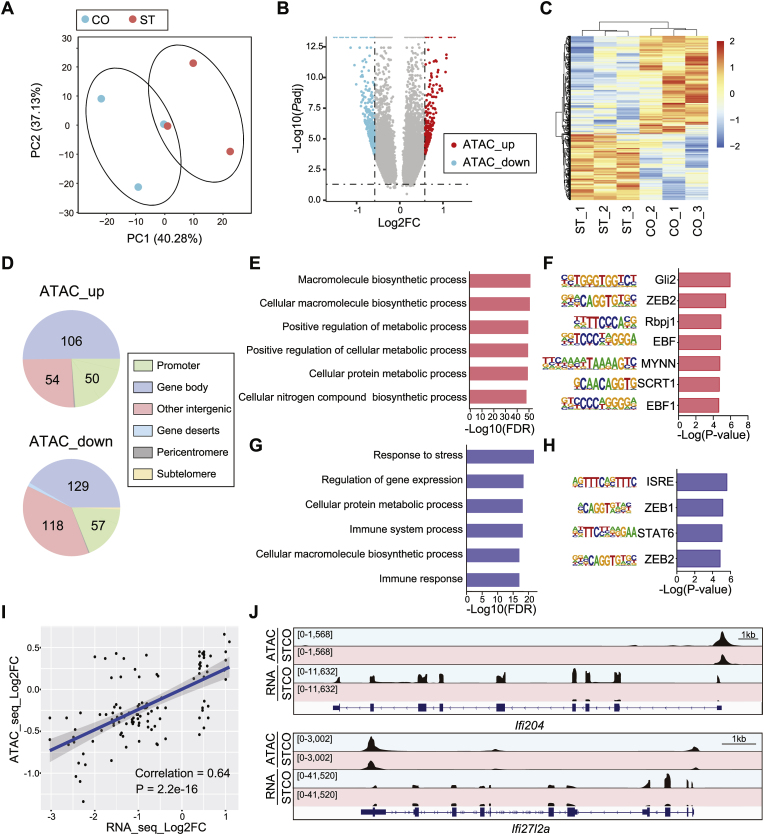
To further study the mechanism underlying chronic stress-induced differential gene expression in microglia, we performed ATAC-seq to check microglia-specific chromatin accessibility (Table S4). PCA analysis showed separation of ATAC-seq signal by treatment (CO v.s. ST) (PC1 40.28%, PC2 37.13%) (Fig. 5A). Differential analysis identified significantly altered ATAC-seq loci (FDR <0.05, ST/CO) (Fig. 5B and C). For both up- and down-regulated loci, most of which were annotated to the genic regions: “ATAC_up”, 50.24% at gene body and 23.70% at promoter; “ATAC_down”, 41.61% at gene body and 18.39% at promoter (Fig. 5D). ShinyGO analysis showed that genes associated with “ATAC_up” loci were enriched in protein biosynthetic and metabolic pathways (Fig. 5E). In contrast, genes associated with the “ATAC_down” loci were enriched in the pathways for “Response to stress” and “Immune system process” (Fig. 5G). Homer motif search further identified multiple important transcription factors (TFs) binding motifs for the “ATAC_up” and “ATAC_down” loci, respectively (Fig. 5F, H). Notably, ISRE (Interferon-Sensitive Responsive Element) motif was the top one enriched in the “ATAC_down” loci (Fig. 5H). Moreover, we performed Pearson's correlation test and revealed a significant positive correlation (correlation = 0.64, P = 2.2e-16) between ATAC-seq and RNA-seq signal of DEGs (Fig. 5I), suggesting the changes in chromatin accessibility contributed to the alterations of gene expression in microglia after chronic stress. As illustrated, ATAC-seq signals on the promoters of two IFN-inducible genes, Ifi204 and Ifi27la, were significantly decreased in ST, accompanied by decreased gene transcription (Fig. 5J).
Fig. 5.
Microglia-specific alterations of chromatin accessibility after chronic stress. (A) PCA plot of ATAC-seq using isolated microglia from mice exposed to chronic stress (ST) and controls (CO). N = 3/group. (B) The volcano plot and (C) clustered heatmap show differential ATAC-seq signals in microglia after chronic stress (ST v.s. CO). Red dot, up-regulated loci (ATAC_up); blue dot, down-regulated loci (ATAC_down); gray dot, non-significant loci. FDR <0.05. (D) Genome annotations for up-and down-regulated ATAC-seq loci. (E, G) GO analysis for genes associated with (E) up- and (G) down-regulated loci, defined as “promoter + gene body”. (F, H) Homer de novo motif prediction for (F) up- and (H) down-regulated loci. (I) Pearson's correlation test for RNA-seq and ATAC-seq signal at differential gene loci. (J) IGV map tracks show ATAC-seq and RNA-seq signals at Ifi204 and Ifi27l2a loci. Notice decreased ATAC-seq (gene promoter) and RNA-seq (coding sequence) signal in the ST compared to CO. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also looked into the alterations of chromatin accessibility on DNA repeats in ST compared with CO. Although differential analysis identified a certain number of DNA repeats with up- and down-regulated ATAC-seq signals in ST (P < 0.05) (Fig. S3A), none of them survived with the cutoff of FDR <0.05. To check whether the changes in chromatin accessibility contributed to the observed transcriptional repression of DNA repeats (Fig. 3), we performed Pearson's correlation test and revealed a significant but weakly positive correlation between ATAC-seq and RNA-seq signals (correlation = 0.217, P = 0.017) (Fig. S3B). In addition, we did a permutation test to assess the distance between DNA repeats with differential ATAC-seq and RNA-seq signals and found no significance for either the up-regulation (Fig. S3C) or down-regulation (Fig. S3D) events. As illustrated, there is a full-length L1 (L1Md_F2, chr18: 60,302,237 - 60,308,431) located on a cluster of down-regulated genes in ST (Fig. S3E, Fig. 3A). There was a significant decrease in RNA-seq signal on L1Md_F2 in ST but no change in ATAC-seq signal (Fig. S3E). Instead, decreased ATAC-seq signal was detected on the promoter of Gm4951 (Fig. S3E), which encodes an “immunity-related GTPase” and is located upstream of this L1Md_F2. Together, they suggested that alterations of chromatin accessibility did not directly contribute to the transcriptional repression of DNA repeats in microglia after chronic stress.
3.6. ATF3 activation and the repression of IRGs after chronic stress
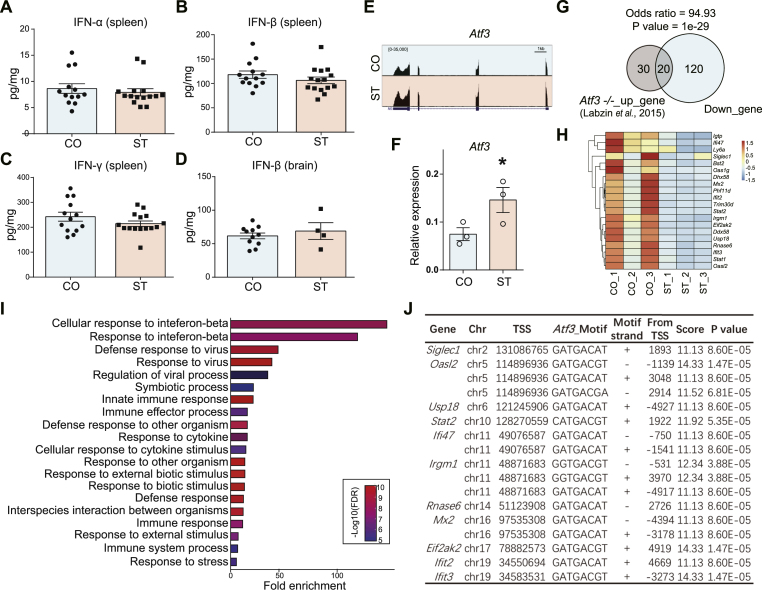
As both RNA-seq and ATAC-seq data pointed to the transcriptional repression of IRGs in ST, we speculate that the production of IFNs might be altered after chronic stress. We, therefore, performed ELISA to examine the levels of IFNs in the brain and spleen. Surprisingly, there was no difference between ST and CO (Fig. 6A–D), suggesting the suppression of IRGs in the current model of chronic stress was not mediated by IFNs.
Fig. 6.
Reverse correlation between Atf3 and IFN-regulated genes after chronic stress. (A–D) IFN levels in (A–C) spleen (N = 13–15/group) and (D) brain (N = 4–11/group) from the stressed (ST) and control (CO) mice. Unpaired t-test. Mean ± SEM. (E) IGV map track shows RNA-seq signal at Atf3 gene locus. Notice increased signal in ST. (F) Real-time RT-PCR validation for Atf3. N = 3/group. Mean ± SEM. Unpaired t-test, one-tailed, *P < 0.05. (G) Overlap between the top 50 genes up-regulated in Atf3 knockout macrophage (GSE61055) and genes down-regulated in the microglia after chronic stress. (H) Heatmap shows the expression of 20 overlapping genes in panel G. (I) GO analysis for the 20 overlapping genes. Notice the top enrichment for IFN-β pathways. (J) List of ATF3 binding motifs at the promoters (TSS ± 5 kb) from 11 of the 20 overlapping genes.
Activating Transcription Factor 3 (ATF3) plays a vital role in response to stress (Hai et al., 1999) and participates in regulating IRGs (Sood et al., 2017; Xu et al., 2021). Genetic ablation of Atf3 in macrophages resulted in the up-regulation of genes enriched in IFN-signaling pathways (Labzin et al., 2015). In the current study, Atf3 was one of the 18 genes up-regulated in the stressed microglia (Fig. 2B–C, Fig. 6E), which was validated by real-time RT-PCR (Fig. 6F). We, therefore, speculated that ATF3 might contribute to the transcriptional repression of IRGs after chronic stress. We compared the 140 microglia-specific down-regulated genes in the current study with the top 50 up-regulated genes in Atf3 knockout macrophage (Labzin et al., 2015) and found that 20 genes overlapped between these two gene sets (Odd ratio = 94.93, P = 1e-29) (Fig. 6G). Clustered heatmap showed decreased RNA-seq signals of these 20 genes in ST compared to CO (Fig. 6H), and GO analysis revealed significant enrichment in the IFN-β and anti-virus pathways (Fig. 6I). In addition, 11 of the 20 overlapping genes harbored at least one ATF3 binding motif proximal to the TSS (TSS ± 5 Kb) (Fig. 6J). Together, it suggested that ATF3 might function as a negative regulator for the transcriptional repression of IRGs in microglia after chronic stress.
4. Discussion
In the current study, we constructed a CMUS model in which male mice were exposed to multiple unpredictable environmental insults daily for 12 weeks. The stressed mice exhibited anxiety- and depressive-like behavioral changes compared to the control. We focused on microglia and observed morphological changes characterized by hyper-ramification in multiple brain regions, including PFC, DG, and AMYG. We isolated microglia from the whole brain to examine the microglia-specific alterations of transcriptome and chromatin accessibility after chronic stress. RNA-seq identified a robust down-regulation of IRGs in ST, and GSEA analysis showed that the down-regulated genes were primarily enriched in the IFN-related immunoregulatory pathways. Moreover, integrative analysis with a published microglia scRNA-seq dataset indicated that the down-regulated genes were enriched in a distinct subpopulation of “Interferon microglia”. In addition, the ISRE motif was enriched in the down-regulated ATAC-seq loci. Despite the striking transcriptional repression of IRGs, there was no change in the level of IFNs in either brain or spleen after chronic stress, suggesting an IFN-independent regulatory mechanism. We then looked into the up-regulated genes in ST and focused on a gene encoding a master transcriptional regulator, ATF3, which is of great relevance to the regulation of IRGs (Labzin et al., 2015). Indeed, there was a significant overlap between the up-regulated genes in Aft3 knockout macrophages and the down-regulated genes from our current study. These overlapping genes were highly enriched in the IFN-β pathways, most of which contained ATF3 motifs on the promoters. Together, our study reported IFN-independent transcriptional repression of IRGs in microglia after prolonged stress, providing novel molecular evidence for the adverse impacts of long-term stress on the neuroimmune system.
4.1. Morphological and functional abnormality of microglia after chronic stress
Microglia originate from primitive yolk sac macrophages and enter the brain at the early embryonic stage to become resident myeloid cells. They play an important role in the maintenance of normal CNS homeostasis. During the critical postnatal period, microglia participate in pruning dendritic spines and regulate synaptic plasticity (Hong et al., 2016). In the adult brain, microglia maintain a continuous and rapid surveillance state, monitoring the dynamics of the intracerebral microenvironment. In the presence of cellular damage or adverse external stimuli, microglia undergo significant morphological changes and exert phagocytosis to remove cellular debris or invasive exotic substances. Environmental stress can also induce microglial responses. Most evidence from previous studies has reported microglia activation in response to stress, manifested by the release of inflammatory factors, aberrant phagocytosis, and subsequent functional and structural damage of neurons (Frank et al., 2019). However, other groups also reported that microglia did not undergo a typical amoeboid transformation upon chronic stress but exhibited a hyper-ramification morphology (Boche et al., 2013; Xie et al., 2021; Hinwood et al., 2013; Smith et al., 2019; Maras et al., 2022). We observed microglia hyper-ramification in the current study and substantial repression of IRGs, suggesting an immunocompromised state after prolonged stress. We speculated that microglia's differential morphological and functional changes in response to environmental stress might be related to different types, duration, and intensity of stress, as well as the physical conditions of animals.
Accumulating evidence suggests the heterogeneity of microglia, especially during brain development and under various pathological conditions (Hammond et al., 2018). A recently published scRNA-seq study on microglia with the knockdown of COP1 revealed six distinct clusters of microglia subpopulations (Ndoja et al., 2020). COP1 is an E3 ubiquitin ligase and controls the expression of the TF CCAAT/enhancer binding protein beta (c/EBPβ), which can drive potent activation of microglia representing both pro-inflammatory and neurodegeneration signatures (Ndoja et al., 2020). Among the six microglia subpopulations, one is enriched for IRGs and thus named “Interferon microglia”. We performed an integrative analysis between our microglia bulk RNA-seq and this published microglia scRNA-seq and revealed a significant enrichment of down-regulated genes in the “Interferon microglia”, suggesting cell type specificity for the observed transcriptional repression of IRGs after chronic stress. Besides our study, other groups also reported the enrichment of differential genes under various pathological conditions in “Interferon microglia”, including viral infection, lipopolysaccharide stimulation, Alzheimer's disease, and glioma (Friedman et al., 2018), further suggesting a pathological significance of this cell population.
4.2. Dysregulation of IFN signaling pathway in microglia after chronic stress
IFN-signaling pathways are critical components of the body's immune system to fight against viral infection. They can be grouped into two significant subtypes with opposite functions: type I (IFN-α and -β) with pro-inflammatory activity and type II (IFN-γ) with anti-inflammatory activity. In recent years, multiple studies have reported that IFN-signaling pathways participate in the regulation of stress-related mood behaviors (Tripathi et al., 2021; Capuron et al., 2003). Both clinical and preclinical studies have suggested type I IFNs can promote the release of pro-inflammatory factors and lead to a negative mood state (Zheng et al., 2014; Coch et al., 2019; Tripathi et al., 2021). However, in the current study, the level of IFN-β was not changed after chronic stress. On the contrary, multiple genes responding to type I IFNs were down-regulated in the stressed microglia. This discrepancy might be attributable to the different time courses of stress, as the duration of stress in the current study was up to 12 weeks, much longer than the duration of 4–6 weeks in published works. Under prolonged stress, an organism's immune system may transform from initial inflammatory response to a suppressed state with compromised immune defense functions. In addition to the pro-inflammatory type I IFNs, dysregulation of the anti-inflammatory type II IFN is also involved in stress-related emotional dysfunction (Connor et al., 2009; Sonnenfeld et al., 1992; Curtin et al., 2009). Down-regulation of genes in the IFN-γ signaling pathway was also detected in the current stress model without a change in IFN-γ production. Our data and others supported the vital role of IFN-signaling pathways in response to stress, although different regulatory mechanisms might be involved.
Dysregulation of IFN-signaling is usually caused by the abnormal production of IFNs (Ivashkiv and Donlin, 2014). However, in the current study, despite the robust repression of IRGs, there were no alterations of IFNs in the brain or the peripheral immune organ spleen. We, therefore, looked into the literature and found that this phenotype might be related to the dysregulation of intrinsic immunity, which is a part of the constitutive mechanism of immunosurveillance. Intrinsic immunity is an IFN-independent antiviral response mediated by the constitutive expression of host restriction factors, including IRGs (Wu et al., 2018). This immune regulatory mechanism was recently reported in the stem cells and neurons (Wu et al., 2018; Cho et al., 2013). The sustained expression of IRGs is primarily mediated by the transcriptional factor complex U-ISGF3, composed of IRF9 and unphosphorylated forms of STAT1 and STAT2 (Michalska et al., 2018). In the current study, RNA-seq and ATAC-seq data revealed decreased signals on Irf9, Stat1, and Stat2, indicating abnormal intrinsic immunity in microglia independent of IFNs after chronic stress.
4.3. ATF3 in the regulation of IRGs after chronic stress
ATF3 belongs to the family of ATF/CREB transcriptional factors and is a critical stress-inducible factor (Chen et al., 1996). Clinical studies have provided evidence for ATF3 in many neuropsychological disorders (Weigelt et al., 2011; Zeng et al., 2020). In addition, multiple lines of evidence have suggested that ATF3 can act as a negative feedback regulator of IRGs during inflammatory response (Gilchrist et al., 2006). For example, knockdown of Atf3 resulted in the upregulation of type I/II IRGs in human cancer cells (Xu et al., 2021) and mouse macrophages (Labzin et al., 2015). In addition, knockdown of Atf3 in mouse neuroblastoma cells increased the expression of Stat1/2 and Irf9 (Sood et al., 2017). In line with this, Atf3 is one of the few genes significantly up-regulated in the stressed microglia in our current study. Its binding motif was present in the promoters of most down-regulated IRGs. Moreover, some IRGs distribute as gene clusters in the genome, for example, the “Chr1_Ifi_cluster”, indicating a transcriptional co-regulatory mechanism. Indeed, such a mechanism was reported at IRGs loci via chromatin interactions among enhancer-like promoters (Epromoters), defined as a collection of promoters that act as “bona fide enhancers” to induce the transcription of proximal or distal genes (Santiago-Algarra et al., 2021). Together, we speculated that ATF3 might function as a master regulator controlling the clustered IRGs in response to chronic stress.
4.4. Dysregulation of DNA repeats in microglia after chronic stress
An interesting phenomenon is that within some of the down-regulated IRGs gene clusters, the transcription of some DNA repeats was also affected accordingly, including L1. L1 is one of the most abundant DNA sequences in the human genome. The full-length L1 has DNA transposon activity, which is thought to be an important mechanism for generating genomic polymorphisms during evolution. Currently, most L1s are defective in the mammalian genome. However, previous studies have shown that active L1s are present in the brain and contribute to neural cell diversity (Suarez et al., 2018; Muotri and Gage, 2006; Muotri et al., 2005). Moreover, multiple groups reported the activation of L1s in the brain on the transcriptional level and its transposon activity in animals exposed to early life stress (Bedrosian et al., 2018; Cuarenta et al., 2020). In contrast, we observed transcriptional repression of some site-specific DNA repeats after chronic stress, most of which were located within or near the down-regulated genes. Meanwhile, the ATAC-seq signal was not altered on these DNA repeat loci, and genome-wide there was a relatively low correlation between the changes in chromatin accessibility and the transcriptional repression of DNA repeats. We, therefore, speculated that the changes of DNA repeats in microglia were not a direct effect in response to chronic stress but rather an accompanying phenomenon due to the repression of adjacent genes. It is worth mentioning that, among all the down-regulated DNA repeats, the most significant one was an L1 sequence located on chr18. Sequence analysis revealed that this was a full-length L1 containing all the functional elements (Fig. S3E). Whether this full-length LI1 has transposable activity in microglia under physiological conditions and whether its transcriptional repression after chronic stress affects microglia function are interesting questions for future study.
5. Major limitations in the current study
While our research has studied the alterations of microglia after chronic stress from multiple perspectives, it has several limitations that must be acknowledged. First, we only included male animals in the current study. There are apparent sex differences in depression regarding the clinical prevalence and symptoms (Williams et al., 2022), and RNA-seq revealed distinct patterns of transcriptomic dysregulation in the brains of female and male patients (Labonte et al., 2017; Touchant and Labonte, 2022). Meanwhile, sex difference has also been reported in the studies of microglia 28,29 30. Multiple groups have shown that males have more pronounced morphological and functional changes of microglia than females in the stress-based animal models (Bollinger, 2021; Wohleb et al., 2018; Woodburn et al., 2021). In contrast, another group reported microglia activation after 6 or 28 days of variable stress only in females (Tsyglakova et al., 2021). Our current study provided evidence for a suppressed state of microglia after prolonged stress in males. It would be necessary to further study the females using the same paradigm. Secondly, the brain region is another common confounding factor in the study of microglia after stress. Most studies focused on the PFC, considering its essential role in depression (Wohleb et al., 2018; Woodburn et al., 2021; Tsyglakova et al., 2021; Bollinger et al., 2020; Hinwood et al., 2012). However, besides PFC, stress-induced microglia changes have also been observed in other brain regions, including the nucleus accumbens, CA3 and DG of the hippocampus, AMYG, lateral septum, and more (Tsyglakova et al., 2021; Delpech et al., 2015; Ramirez et al., 2017). In the current study, we examined PFC, DG, and AMYG and observed consistent morphological changes with hyper-ramification of microglia. We, therefore, isolated microglia from the whole brain (olfactory bulb and cerebellum excluded) to collect sufficient input samples and ensure the data quality of the RNA-seq and ATAC-seq experiments. However, in this way, other than the massive repression of genes like IRGs, we might miss some subtle or region-specific gene changes that were important for stress-induced functional abnormality. Thirdly, although our current study provided some evidence showing a negative correlation between ATF3 and IRGs, we did not conduct any mechanistic studies to establish the causal effect of ATF3 on the regulation of IFNs-signaling and microglia dysfunction in response to stress.
6. Conclusion
In the current study, we performed unbiased epigenomic profiling on microglia purified from an adult mouse brain and provided valid evidence for microglia-specific transcriptional repression of IRGs after prolonged stress, which was in line with the observed hyper-ramification of microglia in PFC and other brain regions. This phenomenon was independent of the production of IFNs and might be associated with the dysregulation of intrinsic immunity.
CRediT authorship contribution statement
Yuan Zhang: Conceptualization, Investigation, Methodology, Validation, Formal analysis, Writing – original draft, Writing – review & editing, Visualization. Yuhao Dong: Investigation, Methodology, Validation, Formal analysis, Writing – review & editing, Visualization. Yueyan Zhu: Investigation, Methodology, Validation, Writing – review & editing, Visualization. Daijing Sun: Writing – review & editing, Visualization. Shunying Wang: Investigation, Validation, Writing – review & editing. Jie Weng: Investigation, Writing – review & editing. Yue Zhu: Methodology. Wenzhu Peng: Methodology, Writing – review & editing, Visualization. Bo Yu: Conceptualization, Writing – review & editing, Funding acquisition. Yan Jiang: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (No. 81971272, No.32170601) (Y.J.); the Science and Technology Innovation 2030 - Major Project (No. 2021ZD0203001) (Y.J.); the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01), ZJ Lab, and the Shanghai Center for Brain Science and Brain-Inspired Technology (Y.J.); the Pudong New Area Clinical Plateau Discipline Project (No. PWYgy2021-03) and Program for Medical Key Departments of Shanghai (No. ZK2019A10) (B.Y.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2022.100495.
Contributor Information
Bo Yu, Email: paul.yubo@gmail.com.
Yan Jiang, Email: yan.jiang@fudan.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Akagi T., et al. Interferon regulatory factor 8 expressed in microglia contributes to tactile allodynia induced by repeated cold stress in rodents. J. Pharmacol. Sci. 2014;126:172–176. doi: 10.1254/jphs.14143sc. [DOI] [PubMed] [Google Scholar]
- Bachiller S., Del-Pozo-Martín Y., Carrión Á M. L1 retrotransposition alters the hippocampal genomic landscape enabling memory formation. Brain Behav. Immun. 2017;64:65–70. doi: 10.1016/j.bbi.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Bedrosian T.A., Quayle C., Novaresi N., Gage F.H. Early life experience drives structural variation of neural genomes in mice. Science. 2018;359:1395–1399. doi: 10.1126/science.aah3378. [DOI] [PubMed] [Google Scholar]
- Boche D., Perry V.H., Nicoll J.A.R. Review: activation patterns of microglia and their identification in the human brain. Neuropathol. Appl. Neurobiol. 2013;39:3–18. doi: 10.1111/nan.12011. [DOI] [PubMed] [Google Scholar]
- Bollinger J.L. Uncovering microglial pathways driving sex-specific neurobiological effects in stress and depression. Brain Behav Immun Health. 2021;16 doi: 10.1016/j.bbih.2021.100320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger J.L., Horchar M.J., Wohleb E.S. Diazepam limits microglia-mediated neuronal remodeling in the prefrontal cortex and associated behavioral consequences following chronic unpredictable stress. Neuropsychopharmacology. 2020;45:1766–1776. doi: 10.1038/s41386-020-0720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C T. Easy to use microglia purification. 2016. https://www.biocompare.com/Product-Reviews/184718-Easy-to-use-Microglia-purification/
- Capuron L., et al. Association of exaggerated HPA axis response to the initial injection of interferon-alpha with development of depression during interferon-alpha therapy. Am. J. Psychiatr. 2003;160:1342–1345. doi: 10.1176/appi.ajp.160.7.1342. [DOI] [PubMed] [Google Scholar]
- Chen B.P., Wolfgang C.D., Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol. Cell Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., et al. Differential innate immune response programs in neuronal subtypes determine susceptibility to infection in the brain by positive-stranded RNA viruses. Nat. Med. 2013;19:458–464. doi: 10.1038/nm.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coch C., Viviani R., Breitfeld J., Münzer K., Stingl J. Interferon-beta-induced changes in neuroimaging phenotypes of appetitive motivation and reactivity to emotional salience. Neuroimage: Clinica. 2019;24 doi: 10.1016/j.nicl.2019.102020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor J.C., et al. Interferon-γ and tumor necrosis factor-α mediate the upregulation of indoleamine 2,3-dioxygenase and the induction of depressive-like behavior in mice in response to Bacillus calmette-guérin. J. Neurosci. 2009;29:4200. doi: 10.1523/JNEUROSCI.5032-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuarenta A., et al. Early life stress increases Line1 within the developing brain in a sex-dependent manner. Brain Res. 2020;1748 doi: 10.1016/j.brainres.2020.147123. [DOI] [PubMed] [Google Scholar]
- Curtin N.M., Boyle N.T., Mills K.H.G., Connor T.J. 2009. Psychological Stress Suppresses Innate IFN-γ Production via Glucocorticoid Receptor Activation: Reversal by the Anxiolytic Chlordiazepoxide. Brain, Behavior, and Immunity. [DOI] [PubMed] [Google Scholar]
- Delpech J.C., et al. Microglia in neuronal plasticity: influence of stress. Neuropharmacology. 2015;96:19–28. doi: 10.1016/j.neuropharm.2014.12.034. [DOI] [PubMed] [Google Scholar]
- Frank M.G., Fonken L.K., Watkins L.R., Maier S.F. Microglia: neuroimmune-sensors of stress. Semin. Cell Dev. Biol. 2019;94:176–185. doi: 10.1016/j.semcdb.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman B.A., et al. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of alzheimer's disease not evident in mouse models. Cell Rep. 2018;22:832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- Gao X., et al. Proceedings of the National Academy of Sciences of the United States of America; 2018. Chronic Stress Promotes Colitis by Disturbing the Gut Microbiota and Triggering Immune System Response; p. E2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeneOverlap Shen. 2016. Test and Visualize Gene Overlaps. [Google Scholar]
- Gilchrist M., et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- Hai T., Wolfgang C.D., Marsee D.K., Allen A.E., Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- Hammen C., Kim E.Y., Eberhart N.K., Brennan P.A. Chronic and acute stress and the prediction of major depression in women. Depress. Anxiety. 2009;26:718–723. doi: 10.1002/da.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Stevens B. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2018;50 doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Fan Y., Zhou K., Blomgren K., Harris R.A. Uncovering sex differences of rodent microglia. J. Neuroinflammation. 2021;18:74. doi: 10.1186/s12974-021-02124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinwood M., Morandini J., Day T.A., Walker F.R. Evidence that microglia mediate the neurobiological effects of chronic psychological stress on the medial prefrontal cortex. Cerebr. Cortex. 2012;22:1442–1454. doi: 10.1093/cercor/bhr229. [DOI] [PubMed] [Google Scholar]
- Hinwood M., et al. Chronic stress induced remodeling of the prefrontal cortex: structural re-organization of microglia and the inhibitory effect of minocycline. Cerebr. Cortex. 2013;23:1784–1797. doi: 10.1093/cercor/bhs151. [DOI] [PubMed] [Google Scholar]
- Hong S., Dissing-Olesen L., Stevens B. New insights on the role of microglia in synaptic pruning in health and disease. Curr. Opin. Neurobiol. 2016;36:128–134. doi: 10.1016/j.conb.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howren M.B., Lamkin D.M., Suls J. Associations of depression with C-reactive protein, IL-1, and IL-6: a meta-analysis. Psychosom. Med. 2009;71:171–186. doi: 10.1097/PSY.0b013e3181907c1b. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jnsson M.E., Garza R., Johansson P.A., Jakobsson J. Transposable elements: a common feature of neurodevelopmental and neurodegenerative disorders. Trends Genet. 2020 doi: 10.1016/j.tig.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Kendler K.S., Karkowski L.M., Prescott C.A. Causal relationship between stressful life events and the onset of major depression. Am. J. Psychiatr. 1999;156:837. doi: 10.1176/ajp.156.6.837. [DOI] [PubMed] [Google Scholar]
- Kreisel T., et al. Dynamic microglial alterations underlie stress-induced depressive-like behavior and suppressed neurogenesis. Mol. Psychiatr. 2014;19:699–709. doi: 10.1038/mp.2013.155. [DOI] [PubMed] [Google Scholar]
- Labonte B., et al. Sex-specific transcriptional signatures in human depression. Nat. Med. 2017;23:1102–1111. doi: 10.1038/nm.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labzin L.I., et al. ATF3 is a key regulator of macrophage IFN responses. J. Immunol. 2015;195:4446–4455. doi: 10.4049/jimmunol.1500204. [DOI] [PubMed] [Google Scholar]
- Lapp H.E., Hunter R.G. Early life exposures, neurodevelopmental disorders, and transposable elements. Neurobiol. Stress. 2019;11 doi: 10.1016/j.ynstr.2019.100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz K.M., McCarthy M.M. A starring role for microglia in brain sex differences. Neuroscientist. 2015;21:306–321. doi: 10.1177/1073858414536468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.Y., et al. Sucrose preference test for measurement of stress-induced anhedonia in mice. Nat. Protoc. 2018;13:1686–1698. doi: 10.1038/s41596-018-0011-z. [DOI] [PubMed] [Google Scholar]
- Maes M., et al. Increased plasma concentrations of interleukin-6, soluble interleukin-6, soluble interleukin-2 and transferrin receptor in major depression. J. Affect. Disord. 1995;34:301. doi: 10.1016/0165-0327(95)00028-l. [DOI] [PubMed] [Google Scholar]
- Maras P.M., et al. Differences in microglia morphological profiles reflect divergent emotional temperaments: insights from a selective breeding model. Transl. Psychiatry. 2022;12:105. doi: 10.1038/s41398-022-01821-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland A.L., Walsh C., Lockwood K., John-Henderson N.A. The effects of acute psychological stress on circulating and stimulated inflammatory markers: a systematic review and meta-analysis. Brain Behav. Immun. 2017;64:208–219. doi: 10.1016/j.bbi.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckim D.B., Weber M., Niraula A., Sawicki C.M., Godbout J.P. Microglial recruitment of IL-1?-producing monocytes to brain endothelium causes stress-induced anxiety. Mol. Psychiatr. 2017;23:1421–1431. doi: 10.1038/mp.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska A., Blaszczyk K., Wesoly J., Bluyssen H.A.R. A positive feedback amplifier circuit that regulates interferon (IFN)-Stimulated gene expression and controls type I and type II IFN responses. Front. Immunol. 2018;9:1135. doi: 10.3389/fimmu.2018.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller N., et al. The cyclooxygenase-2 inhibitor celecoxib has therapeutic effects in major depression: results of a double-blind, randomized, placebo controlled, add-on pilot study to reboxetine. Mol. Psychiatr. 2006;11:680–684. doi: 10.1038/sj.mp.4001805. [DOI] [PubMed] [Google Scholar]
- Muotri A.R., Gage F.H. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- Muotri A.R., et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Ndoja A., et al. Ubiquitin ligase COP1 suppresses neuroinflammation by degrading c/EBPβ in microglia. Cell. 2020;182:1156–1169. doi: 10.1016/j.cell.2020.07.011. e1112. [DOI] [PubMed] [Google Scholar]
- Newman A.M., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K.A., et al. Peripheral and central proinflammatory cytokine response to a severe acute stressor. Brain Res. 2003;991:123–132. doi: 10.1016/j.brainres.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Osimo E.F., Baxter L.J., Lewis G., Jones P.B., Khandaker G.M. Prevalence of low-grade inflammation in depression: a systematic review and meta-analysis of CRP levels. Psychol. Med. 2019;49:1958–1970. doi: 10.1017/s0033291719001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G.N., et al. Proinflammatory cytokines in the prefrontal cortex of teenage suicide victims. J. Psychiatr. Res. 2012;46:57–63. doi: 10.1016/j.jpsychires.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C.J., et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat. Commun. 2019;10:5098. doi: 10.1038/s41467-019-13085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh C.R., et al. Role of interleukin-1 beta in impairment of contextual fear conditioning caused by social isolation. Behav. Brain Res. 1999;106:109–118. doi: 10.1016/s0166-4328(99)00098-4. [DOI] [PubMed] [Google Scholar]
- Ramirez K., Fornaguera-Trias J., Sheridan J.F. Stress-induced microglia activation and monocyte trafficking to the brain underlie the development of anxiety and depression. Curr Top Behav Neurosci. 2017;31:155–172. doi: 10.1007/7854_2016_25. [DOI] [PubMed] [Google Scholar]
- Reynolds J.L., Ignatowski T.A., Sud R., Spengler R.N. Brain-derived tumor necrosis factor-alpha and its involvement in noradrenergic neuron functioning involved in the mechanism of action of an antidepressant. J. Pharmacol. Exp. Therapeut. 2004;310:1216–1225. doi: 10.1124/jpet.104.067835. [DOI] [PubMed] [Google Scholar]
- Rohleder N. Stress and inflammation - the need to address the gap in the transition between acute and chronic stress effects. Psychoneuroendocrinology. 2019;105:164–171. doi: 10.1016/j.psyneuen.2019.02.021. [DOI] [PubMed] [Google Scholar]
- Rusinova I., et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Algarra D., et al. Epromoters function as a hub to recruit key transcription factors required for the inflammatory response. Nat. Commun. 2021;12:6660. doi: 10.1038/s41467-021-26861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., et al. Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell. 2021;184:2843–2859. doi: 10.1016/j.cell.2021.04.020. e2820. [DOI] [PubMed] [Google Scholar]
- Smith K.L., et al. Microglial cell hyper-ramification and neuronal dendritic spine loss in the hippocampus and medial prefrontal cortex in a mouse model of PTSD. Brain Behav. Immun. 2019;80:889–899. doi: 10.1016/j.bbi.2019.05.042. [DOI] [PubMed] [Google Scholar]
- Sonnenfeld G., Cunnick J.E., Armfield A.V., Wood P.G., Rabin B.S. Stress-induced alterations in interferon production and class II histocompatibility antigen expression. Brain Behav. Immun. 1992;6:170–178. doi: 10.1016/0889-1591(92)90016-h. [DOI] [PubMed] [Google Scholar]
- Sood V., et al. ATF3 negatively regulates cellular antiviral signaling and autophagy in the absence of type I interferons. Sci. Rep. 2017;7:8789. doi: 10.1038/s41598-017-08584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez N.A., Macia A., Muotri A.R. LINE-1 retrotransposons in healthy and diseased human brain. Dev Neurobiol. 2018;78:434–455. doi: 10.1002/dneu.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant C. Life events, stress and depression: a review of recent findings. Aust. N. Z. J. Psychiatr. 2002;36:173–182. doi: 10.1046/j.1440-1614.2002.01007.x. [DOI] [PubMed] [Google Scholar]
- Tonelli L.H., et al. Elevated cytokine expression in the orbitofrontal cortex of victims of suicide. Acta Psychiatr. Scand. 2008;117:198–206. doi: 10.1111/j.1600-0447.2007.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchant M., Labonte B. Sex-specific brain transcriptional signatures in human MDD and their correlates in mouse models of depression. Front. Behav. Neurosci. 2022;16 doi: 10.3389/fnbeh.2022.845491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A., et al. Type 1 interferon mediates chronic stress-induced neuroinflammation and behavioral deficits via complement component 3-dependent pathway. Mol. Psychiatr. 2021;26:3043–3059. doi: 10.1038/s41380-021-01065-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsyglakova M., et al. Sex and region-specific effects of variable stress on microglia morphology. Brain Behav Immun Health. 2021;18 doi: 10.1016/j.bbih.2021.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker F.R. Evidence that microglia mediate the neurobiological effects of chronic psychological stress on the medial prefrontal cortex. Cerebr. Cortex. 2012;22:1442–1454. doi: 10.1093/cercor/bhr229. [DOI] [PubMed] [Google Scholar]
- Wang X., Yan J., Shen B., Wei G. Integrated chromatin accessibility and transcriptome landscapes of doxorubicin-resistant breast cancer cells. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.708066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K., et al. TREM-1 and DAP12 expression in monocytes of patients with severe psychiatric disorders. EGR3, ATF3 and PU.1 as important transcription factors. Brain Behav. Immun. 2011;25:1162–1169. doi: 10.1016/j.bbi.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Williams E.S., Mazei-Robison M., Robison A.J. Sex differences in major depressive disorder (MDD) and preclinical animal models for the study of depression. Cold Spring Harbor Perspect. Biol. 2022;14 doi: 10.1101/cshperspect.a039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohleb E.S., Terwilliger R., Duman C.H., Duman R.S. Stress-induced neuronal colony stimulating factor 1 provokes microglia-mediated neuronal remodeling and depressive-like behavior. Biol. Psychiatr. 2018;83:38–49. doi: 10.1016/j.biopsych.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodburn S.C., Bollinger J.L., Wohleb E.S. Synaptic and behavioral effects of chronic stress are linked to dynamic and sex-specific changes in microglia function and astrocyte dystrophy. Neurobiol Stress. 2021;14 doi: 10.1016/j.ynstr.2021.100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., et al. Intrinsic immunity shapes viral resistance of stem cells. Cell. 2018;172:423–438. doi: 10.1016/j.cell.2017.11.018. e425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., et al. Inhibition of phosphodiesterase-4 suppresses HMGB1/RAGE signaling pathway and NLRP3 inflammasome activation in mice exposed to chronic unpredictable mild stress. Brain Behav. Immun. 2021;92:67–77. doi: 10.1016/j.bbi.2020.11.029. [DOI] [PubMed] [Google Scholar]
- Xu L., et al. ATF3 downmodulates its new targets IFI6 and IFI27 to suppress the growth and migration of tongue squamous cell carcinoma cells. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D., He S., Ma C., Wen Y., Li H. Network-based approach to identify molecular signatures in the brains of depressed suicides. Psychiatr. Res. 2020;294 doi: 10.1016/j.psychres.2020.113513. [DOI] [PubMed] [Google Scholar]
- Zheng L.S., et al. Mechanisms for interferon-α-induced depression and neural stem cell dysfunction. Stem Cell Rep. 2014;3:73–84. doi: 10.1016/j.stemcr.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.