Graphical abstract
Keywords: Deconvolution, Spatial transcriptome, Statistic model, Regression, Machine learning
Abstract
The spatial transcriptome has enabled researchers to resolve transcriptome expression profiles while preserving information about cell location to better understand the complex biological processes that occur in organisms. Due to technical limitations, the current high-throughput spatial transcriptome sequencing methods (known as next-generation sequencing with spatial barcoding methods or spot-based methods) cannot achieve single-cell resolution. A single measurement site, called a spot, in these technologies frequently contains multiple cells of various types. Computational tools for determining the cellular composition of a spot have emerged as a way to break through these limitations. These tools are known as deconvolution tools. Recently, a couple of deconvolution tools based on different strategies have been developed and have shown promise in different aspects. The resulting single-cell resolution expression profiles and/or single-cell composition of spots will significantly affect downstream data mining; thus, it is crucial to choose a suitable deconvolution tool. In this review, we present a list of currently available tools for spatial transcriptome deconvolution, categorize them based on the strategies they employ, and explain their advantages and limitations in detail in order to guide the selection of these tools in future studies.
1. Introduction
Cells—the basic building blocks of living organisms—are arranged in specific patterns to form complex tissues, and then form organs. Parsing cellular expression is, therefore, critical for understanding the mechanisms underlying physiological processes in living organisms. Although all normal cells in an organism share almost the same genome, their gene expression profiles and morphologies tend to be distinct. Traditional bulk analysis methods can be used to extract average population information from a group of cells, but these methods result in the loss of valuable properties of individual cells [1]. Single-cell RNA sequencing (scRNA-seq) technologies developed in recent years provide insights into cell heterogeneity; however, these methods overlook spatial characteristics, which are also important for understanding cellular fate and behavior [2]. Spatial transcriptome technologies, when combined with histopathology techniques, are powerful tools for elucidating cell heterogeneity while also retaining spatial information that can aid in the extraction of more comprehensive data about biologically significant subjects such as the spatial heterogeneity of diseases and the delineation of embryonic development.
Recently, many spatial transcriptome sequencing techniques have been developed. In general, these techniques can be divided into three main categories: microscopy-based methods, laser capture microscopy-based (LCM-based) methods, and next-generation sequencing (NGS) with spatial barcoding methods [3]. The microscopy-based methods can be further divided into fluorescent in situ hybridization-based (FISH-based) approaches (e.g., smFISH [4], osmFISH [5], MERFISH [6], and seqFISH [7]) and in situ sequencing-based (ISS-based) approaches (e.g., ISS [8], HybISS [9], FISSEQ [10], and BARseq [11]). FISH-based approaches have high gene detection efficiency, and ISS-based approaches can be applied to large tissue regions. It is possible to achieve single-cell resolution and/or subcellular transcript localization with these methods. However, both approaches require predefined probes and can only detect a limited number of genes. The LCM-based methods entail locating the region of interest and analyzing its transcriptome using gene chips or RNA-seq. The advantages of this method include its ability to obtain wide transcriptional profiles and its application to 3D tissues; however, it is challenging to scale up LCM-based methods to larger numbers of samples. In the spatial-barcoding-based methods, target RNA molecules are captured in situ and subsequently sequenced ex situ (e.g., 10X Visium, DBiT [12], Slide-seq [13], HDST [14], and Stereo-seq [15]). These methods enable high-throughput gene detection of large areas of tissue across various samples, but they cannot achieve single-cell resolution. In these methods, tissues are detected by multiple spots, so they are also known as spot-based methods. The diameter of the spot varies according to the platform, but it is typically in the range of 2–10 μm or 50–100 μm. A single spot may contain 1–100 cells, which could be of different cell types. This implies that the measured gene expression of a spot could represent a mixture of multiple cells with different gene expression patterns. As a result, it is difficult to figure out the relationship between single cells with these methods. Understanding such relationships is crucial for both the clinical treatment of disease and research on the basic mechanics of life. For example, in tumor studies, understanding the patterns of cell colocalization in the immune microenvironment is important for determining appropriate therapeutic strategies [16].
With the development of computational methods in this field, several deconvolution algorithms that can parse spot-based data into single-cell level expression profiles have been published, and these algorithms have improved the application potential of spatial barcoding-based methods [13], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]. Thus, the spot-based method is currently the most popular strategy for spatial transcriptome sequencing. To further promote the application of this method and relevant studies related to it, in this review, we have described data preprocessing and cell deconvolution tools that can be used for spatial spot-based methods in this field. This review summarizes 15 published deconvolution tools for spatial transcriptome (ST) data and categorizes them based on their primary strategy. By providing this, we hope to help new researchers in this field gain a preliminary understanding of spot-based spatial transcriptome data processing and the concept of deconvolution, in order to help them choose appropriate methods for their studies.
2. Data preprocessing for the expression profiles of spatial spots
For general scRNA-seq analysis, a central task is to generate an expression matrix that illustrates the number of transcripts observed for each gene in each cell. The spot-based spatial transcriptomics method, similar to scRNA-seq, also uses NGS readouts; hence, it shares some upstream preprocessing steps with scRNA-seq. The difference is that each barcode (or a combination of barcodes representing different dimensions) represents a spot instead of a single cell, and a spot may contain several cells. For spatial transcriptome data, deconvolution is required to determine the cell type composition of each spot based on the generated expression matrix. Therefore, the general data processing pipeline of spatial barcoding-based transcriptomics can be split into (1) upstream preprocessing to generate the expression matrix, (2) deconvolution to infer the composition of cellular subtypes for spots, and (3) visualization.
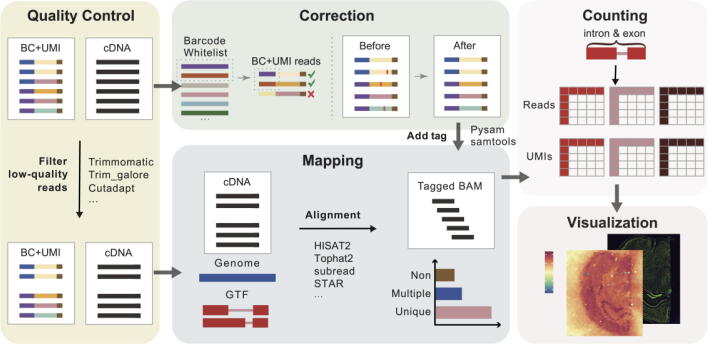
To obtain an expression matrix for spots, raw data from the sequencer are processed through quality control, correction (barcode, UMI correction, and deduplication), mapping (sequence alignment), counting, and visualization. These steps are referred to as data preprocessing steps. Several tools can be used to carry out these processes, including the vendor-specific Space Ranger created by 10X Genomics based on their prior Cell Ranger or more general analysis tools (without visualization for spatial transcriptome) such as zUMIs [31], UMI-tools [32], and scPipe [33]. These steps will be briefly introduced in this section (Fig. 1).
Fig. 1.
Framework of the typical spatial transcriptome preprocessing pipeline. First, FASTQ files are subjected to quality control treatment. Reads with low-quality barcodes or UMIs are discarded, and the remaining reads are mapped to the corresponding genome. Next, a barcode/UMI correction process is carried out to fetch reads of the same origin. Then, a spot-by-gene count matrix is generated according to cDNA and barcode/UMI reads. Finally, the expression matrix is visualized as a series of heat maps.
Quality control. The first step of preprocessing is to filter out useless reads, including reads with low-quality barcodes and/or UMIs, and to trim non-informative repetitive sequences. This step eliminates the major noise, such as spurious barcodes caused by non-specific amplification, and reduces the amount of data for subsequent quantification.
Correction. The remaining reads are carried out with barcode/UMI correction. Usually barcodes or UMIs with Hamming distance or Levenshtein distance < 2 are considered to represent the same spot/cell or the same molecule. Reads are deduplicated to avoid PCR amplification bias when two read pairs share the same UMI and barcode sequence.
Mapping. Next, reads are mapped to the genome using an aligner, such as STAR[34] (zUMIs), BWA[35] (UMI-tools), bowtie2 [36], and subread [37] (scPipe). These aligners and then generate a bam file, annotating aligned reads with the corresponding genome positions. With the provided gtf file, overlaps between the read and the intronic or exonic region can be assessed. For example, zUMIs can produce two mutually exclusive annotation files—one containing information about introns and the other containing information about exons. With featureCounts in Rsubread, the aligned reads are first assigned to exons, and the remaining reads are then assigned to introns. As a result, the expression matrix becomes more informative, and the subsequent analysis process becomes more flexible.
Counting. The quantification steps are carried out based on the bam/sam files obtained. Only high-quality mapped, non-PCR duplicates with reliable barcodes and UMIs are used to generate the gene-barcode expression matrix.
Visualization. Once the above steps are completed, an expression matrix is obtained. Unlike single-cell transcriptome results, each barcode corresponds to a spot in this matrix. The expression for each gene at each spot at different positions is visualized as a heat map based on the relationship between the barcode and spatial position. This heat map can be superimposed on the tissue image.
There are several tools available for each step, and there are also many integrated tools that can be used to directly carry out all these steps. It is more convenient to use the integrated tools instead of building up a new pipeline. However, integrated tools such as Space Ranger only work with data from 10X Genomics or data in a format that is compatible with 10X Genomics. Further, while zUMIs is more universally applicable, it cannot be used to process tissue images. Thus, there is a need for a more widely applicable and extensive integrative tool. According to existing methods, a spot may contain 1 to 100 cells. As a result, the gene profile for each spot represents mixed heterogeneous cell types, and single-cell resolution is lost. This limitation underscores the need to integrate current spatial transcriptomics platforms with scRNA-seq to maximize the resolution of spatial transcriptome. Several deconvolution algorithms have been developed to solve this problem, and 15 mainstream tools are described below.
3. Inferring the proportions of cellular subtypes for spots
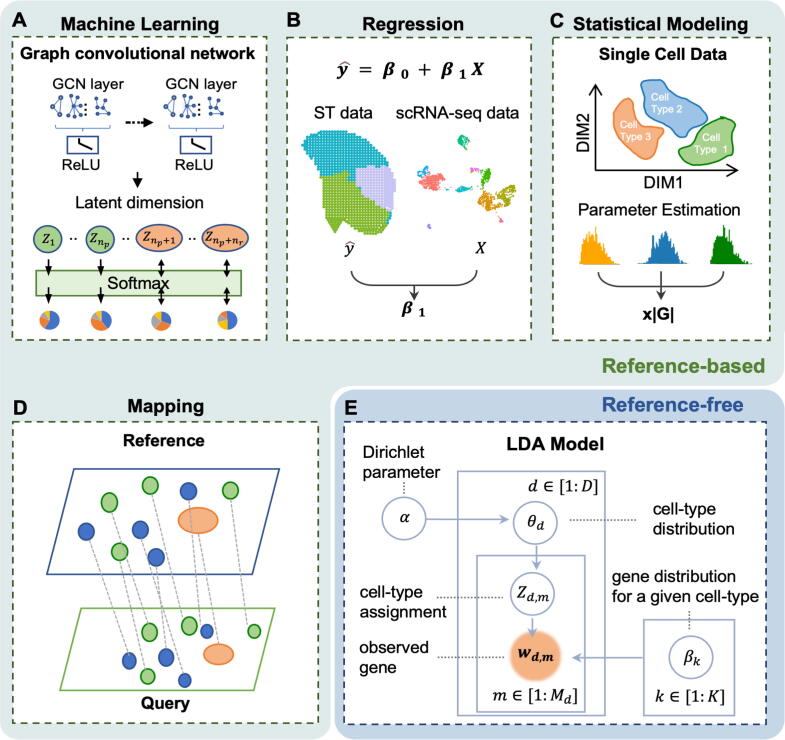
As described above, deconvolution algorithms have been developed in recent years to support the development of high-throughput single-cell resolution spatial transcriptome techniques. According to our survey, there are 15 tools that can be used for ST data deconvolution, including AdRoit [17], DSTG [23], Cell2location [19], RCTD [25], Stereoscope [26], DestVI [22], STRIDE [30], CARD [28], NMFreg [13], SpatialDecon [18], SpatialDWLS [27], SPOTlight [29], Seurat V3 [20], Tangram [21], and STdeconvolve [24]. These tools can be broadly classified as machine learning-based, statistical modeling-based, regression-based, data mapping-based, and reference-free according to the main strategies they use. Indeed, these tools usually adopt two or more strategies to construct their scheme and we categorized them according to their main innovative strategy (Fig. 2, Table 1).
Fig. 2.
A summary of ST deconvolution methods. Published tools for deconvolution can be divided into four categories according to their main strategy: regression-based, statistical-modeling-based, machine learning-based, and mapping-based. All these tools need scRNA-seq data or gene marker lists as reference. However, a recently developed tool based on the LDA model can deconvolve cell proportions without the need for reference data.
Table 1.
Summary of 15 deconvolution tools.
| Tools | Reference | Strategies | Type | Year of Publication | Refs. |
|---|---|---|---|---|---|
| STdeconvolve | Reference-free | latent Dirichlet allocation (LDA) | / | 2022/4 | [24] |
| STRIDE | scRNA-seq data | a topic-model-based method & Bayesian method | Machine learning | 2022/3 | [30] |
| DSTG | scRNA-seq data | semi-supervised graph-based convolutional network | Machine learning | 2021/9 | [23] |
| Seurat V3 | scRNA-seq data | canonical correlation analysis | Mapping | 2019/6 | [20] |
| Tangram | sn/scRNA-seq data | deep learning | Mapping | 2021/11 | [21] |
| SpatialDWLS | scRNA-seq data/marker gene list | an extension of dampened weighted least squares | Regression | 2021/5 | [27] |
| CARD | scRNA-seq data/marker gene list | conditional autoregressive model-based deconvolution | Regression | 2022/5 | [28] |
| SpatialDecon | scRNA-seq data/public scRNA-seq atlas | log-normal regression | Regression | 2022/1 | [18] |
| NMFreg | scRNA-seq data | non-negative matrix factorization regression | Regression | 2019/3 | [13] |
| SPOTlight | scRNA-seq data | seeded NMF regression & topic model & NNLS | Regression | 2021/2 | [29] |
| RCTD | scRNA-seq data | maximum-likelihood estimation (MLE) | Statistics | 2022/4 | [25] |
| Cell2location | scRNA-seq data | a Bayesian model | Statistics | 2022/5 | [19] |
| AdRoit | scRNA-seq data | a weighted regularized linear model | Statistics | 2021/10 | [17] |
| DestVI | scRNA-seq data | a conditional deep generative model | Statistics | 2022/4 | [22] |
| Stereoscope | scRNA-seq data | maximum-likelihood estimation (MLE) & maximum a posteriori (MAP) | Statistics | 2020/10 | [26] |
No potential conflict of interest was reported by the authors.
3.1. Machine learning-based algorithms
Machine learning-based approaches typically involve a learning model (Fig. 2A), such as the neural network. Models are trained based on scRNA-seq data to determine classifications or predictions for spatial data and to uncover key insights into data mining projects. DSTG [23] and STRIDE [30] are machine learning-based deconvolution strategies that implement a graph convolutional neural network and a topic model, respectively.
Graphs can be used to represent topological relationships within data, such as sample similarity. For example, DSTG [23] is a deconvolution method that utilizes a graph convolutional neural network and transforms unsupervised tasks into semi-supervised tasks. This tool first generates pseudo-ST data using scRNA-seq data by randomly selecting and mixing 2–8 cells based on known scRNA-seq data. Next, downsampling is used to ensure that the expression of pseudo spots is at the same sequencing depth level as the real-ST data. Subsequently, DTSG leverages canonical correlation analysis to reduce the dimensions of pseudo-ST and real-ST data and then uses the K-Nearest Neighbor (k-NN) method to establish the nearest neighbor relationship between all the spots. A network graph with all spots as nodes is obtained and its edges are constructed between the nodes when those spots are mutual nearest neighbors. Each spot contains the expression of 2000 highly variable genes. Based on such a graph structure, a semi-supervised graph convolutional network learns latent associations between the graph structure and gene expression patterns, allowing it to predict the cellular composition of the spots. Since there are many mixing possibilities of cells for pseudo spots, and the inferred cell composition of real spots relies on pseudo spots, the accuracy of this method may be affected by the number of generated pseudo spots, which can also strongly impact computational efficiency. This may be a tradeoff for users of this tool. As graph theory has been a hot topic in recent years and new graph structures are currently being proposed, it is possible that future graph structures will be better suited to tackling such problems.
In addition to graph models, some natural language-processing models have also gradually been attracting attention in this field. For example, STRIDE [30] is a topic-modeling-based spatial transcriptomics deconvolution method that estimates cell type proportions from ST data with the topic profile learned from scRNA-seq data. Using Bayes’ theorem, a cell-type-by-topic distribution can be inferred from known cell types in scRNA-seq data. The pre-trained topic model is then used in spatial transcriptomics to infer the topic distributions of each spot. Additionally, STRIDE provides downstream analysis functions, such as signature detection and visualization, spatial domain identification, and spatial architecture reconstruction. Such a package can make the entire spatial transcriptome data analysis process more convenient.
In short, graph convolutional neural networks and topic models are semi-supervised and unsupervised learning models, respectively, which are well integrated into the deconvolution process. Furthermore, numerous machine-learning models, such as support vector machines, and Convolutional Neural Network (CNN), have deeply penetrated various areas of biological algorithms, and we anticipate that more machine-learning-related deconvolution tools will be developed in the future.
3.2. Regression-based strategies
As the most intuitive strategy, regression-based methods are the most popular methods in the deconvolution field (Fig. 2B). Non-negative matrix factorization (NMF) is a typical algorithm. This method extracts the biological correlation coefficient of the data in the gene expression matrix, organizes the genes and samples, grasps the internal structural characteristics of the data, and groups samples into varied phenotypes [38]. This method typically generates a weighted feature matrix and a basis matrix after decomposition. At present, it is widely used in biological subtyping. In addition, the NMF-based framework is also useful for researchers to develop ST deconvolution algorithms. In the following sections, we will go through the algorithms for performing deconvolution tasks in which the NMF serves as the core framework.
NMF regression (NMFreg) was initially proposed to deal with data from Slide-seq [13]. Slide-seq is a spatial transcriptomic sequencing strategy that can achieve a 10-µm spatial resolution, with more than 60 % of the spots containing one cell and the rest containing two or more cells, which is indicative of the need for single-cell distribution optimization [13]. By borrowing the expression profile from scRNA-seq data, NMFreg manages to recompose the single cell component within each spot as a weighted mixture of cells, and this is achieved by non-negative least squares (NNLS) regression. With scRNA-seq dataset of the mouse brain [39], NMFreg successfully maps classical cell types onto spatial spots. However, the accuracy and robustness of NMFreg have not been validated for data from other ST platforms, except for Slide-seq, so this might be a major setback for its wide application.
In the recently published deconvolution method CARD [28], NMF also serves as the backbone for linking scRNA-seq data, spatial spot composition, and residual error. CARD is inspired by the conditional autoregressive model [40], and it uses pieces from neighboring spots for estimating cell type proportions. This is a significant improvement over existing spatial deconvolution approaches. Additionally, CARD has demonstrated robustness in spatial deconvolution both for spot-based technologies such as 10X Visium and non-spot-based technologies such as seqFISH and MERFISH. If known cell marker genes are provided, CARD can also use pre-determined makers instead of scRNA-seq data; in this way, its applications can be further expanded.
Another bioinformatics tool, SPOTlight [29], is developed to determine the cell composition of spatial spots through a non-smooth NMF method [41]. This method produces sparser results during the factorization, enhancing the cell-type-specific topic profile and reducing overfitting during training compared with the traditional NMF method. Further, NNLS regression can be applied to calculate the topic profile of each spot as well as the cell type composition within each spot via minimizing residuals. In the case of mouse brain ST data, SPOTlight can recover the associated tissue structure when in situ hybridization images are used as the ground truth. In the case of more concrete tumor sections, SPOTlight can also identify clinically relevant features that might help deepen the biological understanding of cancer.
The deconvolution algorithm developed for bulk RNA-seq provides foundational ideas for this field. For example, dampened weighted least squares (DWLS) is a cell-type deconvolution method designed for bulk RNA-seq based on scRNA-seq cell markers [42]. Researchers have explored DWLS as an alternative to NNLS in situations where the influence of highly expressed genes on corresponding cell types is mitigated by a second error term. This has set the stage for future work on spatial deconvolution, that is, SpatialDWLS. In SpatialDWLS, parametric analysis of gene set enrichment (PAGE) [27] is used to determine the cell types within each spatial spot using scRNA-seq data or previously determined marker genes. Inference of cell type decomposition is mainly based on the concept of DWLS, except that extra constraints are used to reduce the false-positive rate since the number of cells within a single spatial spot is limited. With ST data from the human heart, SpatialDWLS has been used to capture the spatial–temporal variations that occur during organ growth. SpatialDWLS also shows potency and efficiency when applied to various ST platforms, including seqFISH + data and 10X Visium data.
As another alternative to NMF, SpatialDecon [18] employs log-normal regression [43] in the deconvolution procedure, as a result of which it can rectify the skewness of both transcriptome and ST data as well as their unequal variance. Moreover, SpatialDecon can generate cell-type-specific profiles from public databases, so it has the ability to handle ST data when coupled scRNA-seq data are not available. Therefore, after log-normal regression is applied and the background noise in ST data is reduced, SpatialDecon can be used to achieve better performance than traditional deconvolution strategies such as NNLS, v-support vector regression, and DWLS. Further, by implementing a nuclei segmentation process, SpatialDecon can utilize tissue images from the GeoMx platform to further improve the accuracy of cell assignment. To sum up, SpatialDecon integrates the latest advances in algorithms and public data resources in order to improve the accuracy and robustness of deconvolution analysis of spatial omics data. With this method, researchers can obtain information about cell type and abundance without relying on scRNA-seq data in order to achieve a wider range of applications. Overall, these regression-based algorithms are extensions and innovative versions of traditional deconvolution algorithms.
In conclusion, the regression-based methods described above chart cell composition within ST spots by employing the concept of matrix decomposition as the major framework and are optimized via more concrete distributions or parameter settings. These methods can use either scRNA-seq or pre-defined cell markers as reference data. Further, newly developed algorithms such as CARD and SpatialDecon can further enhance the interpretability and cross-platform performance of these methods, and thus, help shed light on more complex biological tasks via the decoding of tissue textures. Among these methods, SpatialDecon is one of the first to use tissue image information for deconvolution and lays the basis for further expanding the applications of tissue imaging data.
3.3. Statistical modeling-based strategies
Statistical modeling-based strategies represent another type of deconvolution method (Fig. 2C). In these strategies, gene expression is fitted by different statistical distributions. Previous studies have shown that the gene expression of each cell type generally follows negative binomial (NB) distributions [44], [45]. Some commonly used models to estimate the parameters of the above distributions include Bayesian, maximum likelihood estimation (MLE), and maximum a posteriori (MAP). The tools in this category are briefly introduced below according to the main statistical model they employ.
Accurate and Robust Method to Infer Transcriptome Composition (AdRoit) [17] is intended for bulk RNA-seq data, but it can also process ST data. To infer the cell type composition in a bulk sample or spot, AdRoit uses scRNA-seq data as a reference. This algorithm integrates two feature selection methods to choose genes for deconvolution based on their information richness. One of the feature selection methods is based on the selection of genes that are significantly enriched in certain cell types, known as marker genes, and the other one is designed to select genes that are highly variable in expression across all cell types, known as highly variable genes. Thus, AdRoit utilizes NB distribution to fit the gene expression of each selected gene in each cell type and uses MLE to calculate mean expression and variation in expression. Then, in order to infer percent combinations in spots, AdRoit builds a weighted regularized linear model. In particular, this tool uses an adaptive learning approach to minimize sequencing platform bias and regularization to reduce collinearity among closely related cell sub-types. AdRoit has high sensitivity and accuracy as a reference-based deconvolution method. However, it is incapable of identifying unknown cell types or cell types that are rare in the reference data.
RCTD [25], or robust cell type decomposition, is another computational method that decomposes cell type mixtures using cell type profiles learned from single-cell RNA-seq while accounting for differences in sequencing technologies. RCTD first computes the average gene expression profile for each cell type using scRNA-seq data. By fitting each spatial transcriptomics spot as a linear combination of individual cell types, RCTD generates a spatial map of cell types. The gene expression of each cell type for a given spot is estimated by fitting a statistical model to observed gene counts, which are assumed to follow Poisson distributions. This model is also optimized with MLE. The RCTD is novel in that it employs a statistical model to eliminate platform effects, but it also has the limitation of treating platform effects uniformly across all cell types.
Stereoscope [26] is a method that is based on the assumption that the gene expression determined from both spatial and single-cell data follows an NB distribution pattern. While all other reference-based methods require feature selection as the first step, this pre-processing step is not required for Stereoscope [26]. Firstly from single-cell data, Stereoscope estimates parameters of the NB distribution for all genes within each cell type by using MLE. A weighted combination of the single-cell parameters can be used to form equivalent parameters for a distribution describing gene expression for a mixture of cell types. Next, the weights are estimated to best fit with spatial data by using MAP. The proportion of each cell type is then calculated based on these weights.
DestVI [22] is a multi-resolution deconvolution tool that uses a conditional deep generative model to learn a continuous cell subtype expression profile that reflects the successive change of cell states; in contrast, other deconvolution tools can only produce discrete cell type expression profiles. DestVI produces a cell-type-specific snapshot of the transcriptional state for each spot. It assumes that the expression of each gene follows a negative binomial distribution. Then, it employs two different latent variable models (LVMs, the scLVM and the stLVM), and fits the models with amortized variational inference and MAP inference, respectively. In addition to general MAP inference, DestVI uses a penalized likelihood method to infer some parameters in LVMs. Finally, the cell type composition is estimated for each spot.
Cell2location [19] is a Bayesian inference strategy that can deconvolute ST data by integrating data from scRNA-seq or single-cell nuclei RNA-seq (snRNA-seq). The first step of Cell2location is to determine the characteristic expression of the cell type from scRNA or snRNA-seq data. A negative binomial regression model is used here to fit the cell type characteristics expression and to eliminate the batch effect that causes the overdispersion of single-cell data. In the second step, these cell type characteristics are used to deconvolve the mRNA count in spatial transcriptome data in order to estimate the proportion of each cell type in each spot. Another unique feature of Cell2location is that it can use prior knowledge of the analyzed tissue to estimate absolute cell type abundance. Furthermore, variational approximate inference and GPU acceleration make it highly computationally efficient.
It is worth noting that the algorithms described above eliminate some batch effects by introducing statistical distributions that are tailored to the gene distribution. So far, the most common one is NB distribution. Because fitting statistical models makes these algorithms more robust to a wide range of noise sources, statistical fitting strategies are expected to become increasingly popular in future tool development. However, it should be noted that parameter derivation strategies play important roles in these tools and might pose some problems. For example, Bayesian inference may have low computational efficiency, and MLE is easily affected by local optimal solutions. Moreover, MAP is particularly sensitive to prior assumption errors.
3.4. Data mapping-based strategies
Apart from directly performing the deconvolution process, mapping ST data with scRNA-seq data into latent space is also a useful strategy to allocate cell types into spatial spots (Fig. 2D). Several mapping-based algorithms have been used to perform this strategy.
Tangram [21] is a comprehensive analysis framework designed for ST data that incorporates multiple platforms. In short, built on deep learning, Tangram is capable of mapping lower-resolution ST data with single-cell data while linking these maps with histological and anatomical information from the associated specimen. By aligning sn/scRNA-seq data with ST data, Tangram corrects low-quality spatial measurements and decomposes low-resolution measurements into individual cells. By focusing on the cell type deconvolution component, after deriving the gene intersection set between sn/scRNA-seq data and ST data, the framework randomly arranges the sn/scRNA-seq subset profile in the space and uses a specifically designed spatial correlation function to quantify the spatial correlation between each gene in sn/scRNA-seq and ST data. To maximize the total spatial correlation across the genes shared by the datasets, Tangram then rearranges the sn/scRNA-seq subset profile. If a tissue image is provided, as found in the 10X Visium dataset, nuclei segmentation results serve as the deterministic constraints for cell numbers in each spot. Alternatively, the output will be a matrix containing the probability of cells in each spatial spot. In addition, this deconvolution function has been extended to deal with data from multiple ST platforms, including sequencing-based (10X Visium) and non-sequencing-based (ISH, smFISH, STARmap, and MERFISH) technologies.
As a cornerstone framework, Seurat [20] has become the most widely used single-cell data analysis tool that can be applied for a wide range of processes—from raw data processing to a variety of downstream analyses. The operation commands provide friendly functions, especially for data obtained from the CellRanger or SpaceRanger process of the 10X Genomics Company. For its recent version, the research team developed it further by proposing a more comprehensive scheme to incorporate multimodal data for sc/snRNA-seq, epigenomic data, proteomic data, and ST data that might significantly help researchers interpret concrete biological phenomena. In terms of mapping single cell types into ST spots, the deconvolution pipeline can be divided into two major steps. First, canonical correlation analysis is applied to both scRNA-seq data and ST data to generate a lower dimension space with an extra L2 penalty on the canonical coefficients. Subsequently, mutual nearest neighbors are searched to capture similar anchors, and shared nearest neighbor graphs [46] are introduced to calculate the score for each anchor as the filtering parameter. This step generates a solid basis for subsequent integration analysis and can be realized by the FindTransferAnchors function in Seurat. Secondly, the probability distribution of each cell type in each spot is imputed by transfer learning based on previously generated anchors; this can be achieved via the TransferData function in Seurat. Thus, with Seurat, it is possible to reliably perform the spatial deconvolution step when integrating SMART-seq2 data with STAR-map data for mouse brain tissue.
Data mapping-based methods are generally not limited to ST data deconvolution. They can also integrate other types of omics data and, thus, correlate different levels of biomolecular information, allowing for deeper biological knowledge mining.
3.5. Reference-free strategies
A large number of deconvolution algorithms have been developed over the last few years, and we have briefly described them above. However, research in this field is ongoing and is happening at an accelerated pace. Some algorithms have used different approaches to solving this issue: for example, STdeconvolve [24] (Fig. 2E) effectively eliminates the reliance on reference data, which, like STRIDE, is a topic modeling-based method. An unusual feature of STdeconvolve is that it does not require a scRNA-seq reference. STdeconvolve performs feature selection immediately after ST data are input. And to infer the cell-type distribution in each spot, it integrated a latent Dirichlet allocation (LDA) process, which is a popular topic modeling technique. Detailly, it considers each spot to be a mixture of K cell types and represents each spot with a multinomial distribution of cell type probabilities. Each cell type is defined as a probability distribution over the genes present in the ST dataset. By estimating those latent parameters in the model using the variational expectation–maximization approach, the proportion of cell types and the expression profile of each cell type in each spot can be calculated.
No matter how similar ST data and scRNA-seq data may be, they originate from distinct tissues, so using scRNA-seq data as the reference is sometimes inaccurate. Moreover, the corresponding scRNA-seq reference is sometimes hard to get. Compared with reference-based methods, the reference-free method has the advantage that it bypasses the constraints of scRNA-seq reference. Therefore, the development of reference-free deconvolution techniques is urgently required.
4. Discussion
Spatial transcriptome technologies that can be used to study tissues in terms of both transcriptome dimensions and spatial location dimensions are a powerful aid in research on cellular regulatory networks, cancer pathogenesis, and other physiopathological phenomena. How spatial transcriptome data can be analyzed is also critical to the study of the aforementioned problem. Commercial spatial transcriptome sequencing technologies are currently incapable of reaching single-cell resolution when evaluating the entire transcriptome profile. For example, NGS with spatial barcoding is one of the most popular ST technologies, and its upstream step is similar to that of traditional scRNA-seq methods. However, the final result is a spot-by-gene expression matrix in which spots represent the mixed expression profiles of multiple cell types. As a result, inferences about the cellular composition of each spot must be made. A number of tools have been developed to infer the cell composition of spots. Most of these methods require scRNA-seq data as reference, assume that gene expression follows an NB distribution, and use different models to infer the proportion of cell types.
Although these deconvolution tools help a lot in parsing ST data, it needs to be noticed that ST data and the corresponding reference single-cell data are actually from different regions. The spatial transcriptome is generated from a tissue slide, a specific plane of a 3D tissue/organ. While single-cell RNA-seq data is generated by the dissociation of the tissue/organ with a more 3D architecture. That is to say, those reference-based deconvolution computational tools always integrate single-cell readouts coming from different parts of the 3D tissue with a single plane that has been analyzed by spatial transcriptomics. This process will introduce some bias. Besides, there are also missing and mismatched cell types, perturbations, and batch effect differences between the single-cell transcriptomes and the ST data. So there are still great demands for developing high-throughput single-cell resolution spatial transcriptomic technologies.
At the same time, the sequencing depth has a strong influence on the spatial sequencing results, which further affects the deconvolution performance. Insufficient sequencing depth will lead to low sequencing saturation and gene coverage, while too high sequencing depth will hardly provide more effective information. Thus, the optimal sequencing depth in spatial transcriptome needs further exploration. The study of STRIDE[30], which compared the performance of a series of deconvolution tools by simulation datasets with various sequencing depths from 1,000 to 20,000 reads per cell, notes that the optimal sequencing depth may be specific for each tool and some algorithms seem to be less affected by the depth (such as the CCA-based algorithm). However, only six methods are compared, and a large-scale in-depth exploration of the optimal sequencing depth is desired.
A final challenge is that the published tools for spatial transcriptome deconvolution described in this review are incapable of producing highly accurate predictions based on arbitrary data sets. One previous study assessed the performance of part of the deconvolution tools mentioned above [47], including stereoscope, Cell2location, SpatialDWLS, RCTD, STRIDE, DestVI, Tangram, and DSTG. Using simulated ST datasets, it shows that the performance of RCTD consistently ranks top. And in some datasets, SpatialDWLS, Tangram, Cell2location, stereoscope, and STRID perform better than others. In addition to performance comparison, several important steps for data preprocessing before deconvolution are also given, including 1) Removal of low-quality cells; 2) Normalization of the expression matrix; and 3) Selection of highly variable genes. Another review also benchmarked several deconvolution tools, including stereoscope, RCTD, SPOTlight, Tangram, DSTG, Cell2location, AdRoit, SpatialDWLS, DestVI, and STdeconvolve[48]. RCTD, Adroit, and Tangram perform better in this comparison. Above all, these tools perform differently with data from different platforms so it is necessary to consider the feature of each tool to choose a suitable one.
Currently, in the absence of high-throughput technologies, deconvolution of spot-based spatial transcriptome data is a crucial method for obtaining single-cell resolution spatial transcriptome profiles, which enable a variety of downstream analyses and the extraction of biological knowledge. Detailly, after obtaining the results of deconvolution, a series of downstream data analyses can be performed to detect biologically relevant features, including spatially variable genes, spatial gene patterns, and spatial regions. Many tools are developed by implementing different strategies to process these tasks, such as MULTILAYER [49], Trendsceek [50], SpatialDE [51], SPARK [52], SOMDE [53], and Giotto [54]. Among them, MULTILAYER is able to infer biologically relevant features by utilizing contiguous spots as a readout of gene co-expression patterns within the analyzed tissue, thereby making better use of the spatial information. Further, identification of cell–cell interactions, inferring gene-gene interactions, and gene expression imputation from ST data with H&E image analysis are also important in this field and have been rapidly developed in recent years. Previous publications have summarized these works in detail [3], [55]. Overall, these tools provide us with deep and meaningful insights into biological phenomena based on spatial transcriptome data.
By briefly describing the process of processing spatial transcriptome data and listing existing deconvolution methods, we hope that this review will be useful for both experimenters interested in spatial transcriptomics and researchers interested in developing new deconvolution methods. We anticipate that our review will provide interested researchers with an overview of the field and help them understand the various strategies and innovative directions in the field.
CRediT authorship contribution statement
Yingkun Zhang: Writing - Original Draft, Visualization, Writing - Review & Editing. Xinrui Lin: Writing - Original Draft, Visualization, Writing - Review & Editing. Zhixian Yao: Writing - Original Draft, Writing - Review & Editing. Di Sun: Writing - Review & Editing. Xin Lin: Visualization, Writing - Review & Editing. Xiaoyu Wang: Writing - Review & Editing. Chaoyong Yang: Writing - Review & Editing. Jia Song: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC) (22104080 to J. S., NSFC 21735004 and 21927806 to C. Y.) and the Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZLCX20212601).
References
- 1.Lin S., Liu Y., Zhang M., Xu X., Chen Y., Zhang H., et al. Microfluidic single-cell transcriptomics: moving towards multimodal and spatiotemporal omics. Lab Chip. 2021;21:3829–3849. doi: 10.1039/D1LC00607J. [DOI] [PubMed] [Google Scholar]
- 2.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moses L., Pachter L. Museum of spatial transcriptomics. Nat Methods. 2022 doi: 10.1038/s41592-022-01409-2. [DOI] [PubMed] [Google Scholar]
- 4.Haimovich G, Gerst JE. Single-molecule Fluorescence in situ Hybridization (smFISH) for RNA Detection in Adherent Animal Cells. Bio-Protoc 2018;8:e3070. 10.21769/BioProtoc.3070. [DOI] [PMC free article] [PubMed]
- 5.Codeluppi S., Borm L.E., Zeisel A., La Manno G., van Lunteren J.A., Svensson C.I., et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods. 2018;15:932–935. doi: 10.1038/s41592-018-0175-z. [DOI] [PubMed] [Google Scholar]
- 6.Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. 庄RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah S., Takei Y., Zhou W., Lubeck E., Yun J., Eng C.-H.-L., et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell. 2018;174:363–376.e16. doi: 10.1016/j.cell.2018.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wählby C., et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 9.Gyllborg D., Langseth C.M., Qian X., Choi E., Salas S.M., Hilscher M.M., et al. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res. 2020;48:e112. doi: 10.1093/nar/gkaa792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J.H., Daugharthy E.R., Scheiman J., Kalhor R., Ferrante T.C., Terry R., et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Sun Y.-C., Zhan H., Kebschull J.M., Fischer S., Matho K., et al. High-throughput mapping of long-range neuronal projection using in situ sequencing. Cell. 2019;179:772–786.e19. doi: 10.1016/j.cell.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Yang M., Deng Y., Su G., Enninful A., Guo C.C., et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell. 2020;183:1665–1681.e18. doi: 10.1016/j.cell.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriques S.G., Stickels R.R., Goeva A., Martin C.A., Murray E., Vanderburg C.R., et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vickovic S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D., et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen A., Liao S., Cheng M., Ma K., Wu L., Lai Y., et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell. 2022;185:1777–1792.e21. doi: 10.1016/j.cell.2022.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Andersson A., Larsson L., Stenbeck L., Salmén F., Ehinger A., Wu S.Z., et al. Spatial deconvolution of HER2-positive breast cancer delineates tumor-associated cell type interactions. Nat Commun. 2021;12:6012. doi: 10.1038/s41467-021-26271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T. AdRoit is an accurate and robust method to infer complex transcriptome composition 2021:14. [DOI] [PMC free article] [PubMed]
- 18.Danaher P., Kim Y., Nelson B., Griswold M., Yang Z., Piazza E., et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun. 2022;13:385. doi: 10.1038/s41467-022-28020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleshchevnikov V., Shmatko A., Dann E., Aivazidis A., King H.W., Li T., et al. Cell 2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol. 2022;40:661–671. doi: 10.1038/s41587-021-01139-4. [DOI] [PubMed] [Google Scholar]
- 20.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biancalani T. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat Methods. 2021;18:25. doi: 10.1038/s41592-021-01264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez R., Li B., Keren-Shaul H., Boyeau P., Kedmi M., Pilzer D., et al. DestVI identifies continuums of cell types in spatial transcriptomics data. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01272-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Q., Su J. DSTG: deconvoluting spatial transcriptomics data through graph-based artificial intelligence. Brief Bioinform. 2021;22:bbaa414. doi: 10.1093/bib/bbaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller B.F., Huang F., Atta L., Sahoo A., Fan J. Reference-free cell-type deconvolution of multi-cellular pixel-resolution spatially resolved transcriptomics data. Bioinformatics. 2021 doi: 10.1101/2021.06.15.448381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cable D.M., Murray E., Zou L.S., Goeva A., Macosko E.Z., Chen F., et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat Biotechnol. 2022;40:517–526. doi: 10.1038/s41587-021-00830-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson A. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography 2020:8. [DOI] [PMC free article] [PubMed]
- 27.Dong R., Yuan G.-C. SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol. 2021;22:145. doi: 10.1186/s13059-021-02362-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat Biotechnol. 2022;17 doi: 10.1038/s41587-022-01273-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elosua-Bayes M., Nieto P., Mereu E., Gut I., Heyn H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 2021;49:e50. doi: 10.1093/nar/gkab043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun D., Liu Z., Li T., Wu Q., Wang C. STRIDE: accurately decomposing and integrating spatial transcriptomics using single-cell RNA sequencing. Nucleic Acids Res. 2022;50:e42. doi: 10.1093/nar/gkac150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. zUMIs - A fast and flexible pipeline to process RNA sequencing data with UMIs. GigaScience 2018;7. 10.1093/gigascience/giy059. [DOI] [PMC free article] [PubMed]
- 32.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian L., Su S., Dong X., Amann-Zalcenstein D., Biben C., Seidi A., et al. scPipe: a flexible R/bioconductor preprocessing pipeline for single-cell RNA-sequencing data. PLOS Comput Biol. 2018;14:e1006361. doi: 10.1371/journal.pcbi.1006361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamamoto R., Takasawa K., Machino H., Kobayashi K., Takahashi S., Bolatkan A., et al. Application of non-negative matrix factorization in oncology: one approach for establishing precision medicine. Brief Bioinform. 2022;23:bbac246. doi: 10.1093/bib/bbac246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saunders A., Macosko E.Z., Wysoker A., Goldman M., Krienen F.M., de Rivera H., et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell. 2018;174:1015–1030.e16. doi: 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee D. A comparison of conditional autoregressive models used in Bayesian disease mapping. Spat Spatio-Temporal Epidemiol. 2011;2:79–89. doi: 10.1016/j.sste.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Pascual-Montano A., Carazo J.M., Kochi K., Lehmann D., Pascual-Marqui R.D. Nonsmooth nonnegative matrix factorization (nsNMF) IEEE Trans Pattern Anal Mach Intell. 2006;28:403–415. doi: 10.1109/TPAMI.2006.60. [DOI] [PubMed] [Google Scholar]
- 42.Tsoucas D., Dong R., Chen H., Zhu Q., Guo G., Yuan G.-C. Accurate estimation of cell-type composition from gene expression data. Nat Commun. 2019;10:2975. doi: 10.1038/s41467-019-10802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muggeo VMR. A note on regression with log Normal errors: linear and piecewise linear modelling in R. n.d.:6.
- 44.Svensson V. Droplet scRNA-seq is not zero-inflated. Nat Biotechnol. 2020;38:147–150. doi: 10.1038/s41587-019-0379-5. [DOI] [PubMed] [Google Scholar]
- 45.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.D., Tadmor M.D., et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B., Zhang W., Guo C., Xu H., Li L., Fang M., et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat Methods. 2022;19:662–670. doi: 10.1038/s41592-022-01480-9. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Liu W, Luo T, Yu Z, Jiang M, Wen J, et al. A comprehensive comparison on cell-type composition inference for spatial transcriptomics data. Brief Bioinform 2022:bbac245. 10.1093/bib/bbac245. [DOI] [PMC free article] [PubMed]
- 49.Moehlin J., Mollet B., Colombo B.M., Mendoza-Parra M.A. Inferring biologically relevant molecular tissue substructures by agglomerative clustering of digitized spatial transcriptomes with multilayer. Cell Syst. 2021;12(694–705):e3. doi: 10.1016/j.cels.2021.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Edsgard D., Johnsson P., Sandberg R. Identification of spatial expression trends in single-cell gene expression data. Nat Methods. 2018;15:339–342. doi: 10.1038/nmeth.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson V., Teichmann S.A., Stegle O. SpatialDE: identification of spatially variable genes. Nat Methods. 2018;15:343–346. doi: 10.1038/nmeth.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun S., Zhu J., Zhou X. Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies. Nat Methods. 2020;17:193–200. doi: 10.1038/s41592-019-0701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hao M, Hua K, Zhang X. SOMDE: A scalable method for identifying spatially variable genes with self-organizing map. Bioinformatics 2021:btab471. 10.1093/bioinformatics/btab471. [DOI] [PubMed]
- 54.Dries R., Zhu Q., Dong R., Eng C.-H.-L., Li H., Liu K., et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 2021;22:78. doi: 10.1186/s13059-021-02286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams C.G., Lee H.J., Asatsuma T., Vento-Tormo R., Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14:68. doi: 10.1186/s13073-022-01075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]