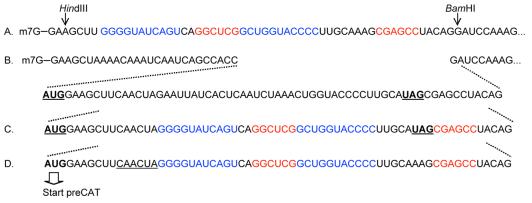Abstract
The efficiency of reinitiation in mammalian translation systems depends in part on the size and arrangement of upstream open reading frames (upORFs). The gradual decrease in reinitiation as an upORF is lengthened, confirmed here using a variety of sequences, might reflect time-dependent loss of protein factors required for reinitiation. Consistent with the idea that the duration of elongation is what matters, reinitiation was nearly abolished when a pseudoknot that causes a pause in elongation was inserted into a short upORF. Control experiments showed that this transient pause in elongation had little effect on the final protein yield when the pseudoknot was moved from the upORF into the main ORF. Thus, the deleterious effect of slowing elongation is limited to the reinitiation mode. Another aspect of reinitiation investigated here is whether post-termination ribosomes can scan backwards to initiate at AUG codons positioned upstream from the terminator codon. Earlier studies that raised this possibility may have been complicated by the occurrence of leaky scanning along with reinitiation. Re-examination of the question, using constructs that preclude leaky scanning, shows barely detectable reinitiation from an AUG codon positioned 4 nt upstream from the terminator codon and no detectable reinitiation from an AUG codon positioned farther upstream. These experiments carried out with synthetic transcripts help to define the circumstances under which reinitiation may be expected to occur in the growing number of natural mRNAs that deviate from the simple first AUG rule.
INTRODUCTION
Translation in eukaryotes is usually constrained by the position of the AUG initiator codon. Because the small ribosomal subunit engages the mRNA at the 5′ end and then migrates linearly, translation usually initiates at the AUG codon nearest the 5′ end (1). In higher eukaryotes, this ‘first-AUG rule’ holds strictly only when the 5′ proximal AUG codon resides in a favorable context (2,3). When the first AUG resides in a suboptimal context, ribosomes may initiate at the first and second AUG codons via leaky scanning (1,4).
Reinitiation is another mechanism that allows ribosomes to reach and initiate at downstream AUG codons. Studies from many laboratories have shown that, when the first AUG codon is followed shortly by an in-frame terminator codon, post-termination ribosomes apparently resume scanning and may reinitiate at a downstream site (reviewed in 1,5,6).
Genetic studies have provided some insights into the mechanism of reinitiation. Seminal experiments involving the yeast GCN4 gene revealed a central role for eIF2, the protein factor that binds Met-tRNAi (7,8). The functional concentration of eIF2 is critical because, following termination of the upstream open reading frame (upORF), the scanning ribosome must re-acquire Met-tRNAi before arriving at the intended reinitiation site downstream. Other initiation and termination factors have also been implicated in the reinitiation process (9).
Insights into the mechanism of reinitiation have also been obtained by manipulating the structure of mRNAs. The critical role of eIF2 was actually predicted, in advance of the GCN4 studies, from the fact that reinitiation became more efficient when the distance between the upORF and the next AUG codon was lengthened (10). This effect requires expanding the intercistronic sequence without introducing a significant amount of secondary structure. Besides intercistronic length, other structural features in mRNAs have been shown to modulate reinitiation in yeast (11–13). However, because some aspects of initiation differ between yeast and mammals—yeast ribosomes are less sensitive to context and more sensitive to (more readily inhibited by) secondary structure (14,15)—answers about reinitiation obtained in yeast cannot be assumed to extrapolate to higher eukaryotes. By the same token, the answers obtained herein using a mammalian translation system might not extrapolate precisely to yeast systems. Studies carried out in plants suggest that reinitiation in that system also resembles what is seen in mammals in most, but not all, respects (16).
A major constraint on reinitiation in eukaryotes is thought to be the size of the 5′ proximal ORF. This was deduced from classical studies of plant and animal viruses that produce dicistronic mRNAs in which the 3′ cistron is translationally silent (reviewed in 17). Few studies have explored systematically the effect of varying the size of the 5′ ORF, however (18,19). That is one of the questions examined here. Another question raised by earlier studies (20,21) and reinvestigated here is whether eukaryotic ribosomes can slide backwards and thus reinitiate upstream from the site of termination.
The parameters studied herein using test transcripts are not the sole determinants of whether a given natural mRNA will support reinitiation. A few interesting cases have been described in which reinitiation is precluded by inhibitory effects of the peptide encoded in the upORF (reviewed in 5). In other circumstances, translation of a short ORF may trigger degradation of the mRNA, a process critical for eliminating defective transcripts that result from mutations or incomplete splicing (22,23). Notwithstanding the importance and widespread occurrence of this degradation mechanism linked to the translation of short ORFs, the frequent occurrence of small upORFs in vertebrate mRNAs that are not rapidly degraded suggests that reinitiation plays a major role in regulating translation per se. Thus, it seems useful to explore structural features that can modulate the efficiency of reinitiation in mammalian translation systems.
MATERIALS AND METHODS
Plasmid construction
Riboprobe vector pSP64, in which the gene for chloramphenicol acetyltransferase (CAT) is preceded by a promoter for T7 RNA polymerase was described previously (24). For the experiments described herein, the structure of the mRNA leader region was varied by deleting and inserting sequences between HindIII and BamHI restriction sites that occur uniquely upstream from the CAT coding domain. The particular mRNA leader sequences devised to test aspects of reinitiation are depicted in each figure alongside the experimental results.
Transcription and translation assays
AvaI-linearized plasmid DNAs were used as templates for transcription by T7 RNA polymerase as described previously (3). All transcripts were capped with m7GpppG (Amersham Pharmacia Biotech Inc.). Transcripts were trace-labeled with [3H]UTP to facilitate purification and quantification. mRNAs were extracted with phenol and purified by application to pre-spun Sephadex G50 columns (Roche Molecular Biochemicals).
Translation was carried out using the Flexi rabbit reticulocyte lysate system from Promega Corp. supplemented with [3H]leucine (150 Ci/mmol, New England Nuclear). Reaction mixtures were supplemented with KCl and Mg(CH3COO)2 to give final concentrations of 100 and 2 mM, respectively. Each 25 µl translation reaction typically contained 0.4 µg of mRNA. Under these reaction conditions (most importantly the use of at least 2 mM Mg2+) reticulocyte translation systems show the same dependence on context and other aspects of mRNA structure as is seen in vivo (25,26). Because mRNA is degraded much faster at 30°C (the temperature recommended by the supplier) than at 25°C, all incubations in reticulocyte translation systems were carried out at the lower temperature.
Following incubation at 25°C for 40–60 min, one-tenth of each translation reaction was mixed with Laemmli sample buffer (Bio-Rad Laboratories) and analyzed by polyacrylamide gel electrophoresis (PAGE) using 15% polyacrylamide-sodium dodecyl sulfate–6 M urea gels. The gels were impregnated with autoradiographic enhancer (New England Nuclear). Autoradiograms of dried gels were obtained by exposure of X-omat AR film at –70°C for 12–24 h.
RESULTS AND DISCUSSION
Reinitiation declines as the upORF is lengthened
If the yield of CAT protein from a capped mRNA with no inhibitory features in the 5′ untranslated region (Fig. 1, lane 1) is set at 100%, the yield falls to 30–35% when a very short upORF is introduced in a way that forces CAT to be translated by reinitiation (Fig. 1, lane 2). In all the constructs described herein, the upstream AUG codon resides in a context that precludes leaky scanning. The absence of leaky scanning was confirmed by toeprinting assays (as in ref. 27), which showed that when elongation was inhibited by antibiotics, 80S initiation complexes were limited to the upstream AUG codon. The sensitivity of the toeprinting assay was adequate to rule out even low-level initiation (e.g. 10%) at the downstream site. The absence of leaky scanning is also evident from the control experiments discussed in Figures 2 and 5. Thus, these are appropriate constructs for studying reinitiation.
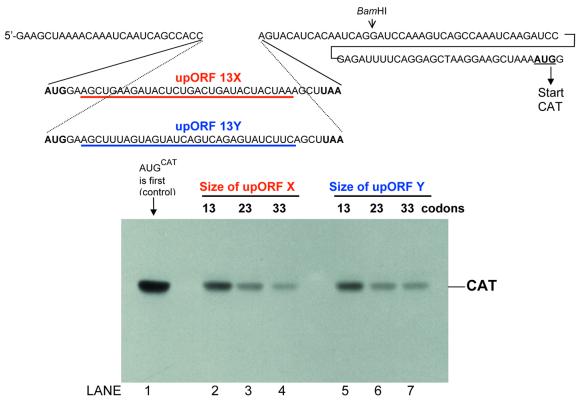
Figure 1.
The size of the upstream minicistron affects the efficiency of reinitiation. A 13-codon upORF designated 13X was inserted into the basic CAT mRNA sequence (topmost line) at the point shown. Constructs in which the minicistron was lengthened to 23 or 33 codons were obtained by reiterating a 30 nt segment (underlined in red) of upORF 13X. Lanes 2–4 in the polyacrylamide gel show the yield of CAT protein from this set of mRNAs. In similar fashion, an alternative upORF, designated 13Y, was expanded by reiterating the 30 nt segment underlined in blue, producing the mRNAs tested in lanes 5–7. For comparison, the yield of CAT from an mRNA that has no upORF is shown in lane 1.
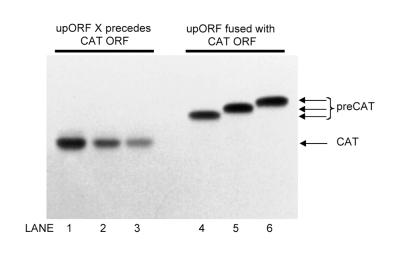
Figure 2.
The sequence used to expand the upstream minicistron does not impair elongation per se. Translation of mRNAs from series X (described in Fig. 1) in which the upORF is 13, 23 or 33 codons long is shown in lanes 1–3, respectively. In the accompanying control mRNAs, 13 (lane 4), 23 (lane 5) or 33 codon (lane 6) upORF has been fused with the CAT coding domain. This was accomplished by changing the terminator codon of the upORF from UAA to UAC and inserting one extra base to adjust the reading frame. The resulting N-terminally extended forms of CAT (labeled preCAT) are distinguishable by PAGE under the conditions described in Materials and Methods.
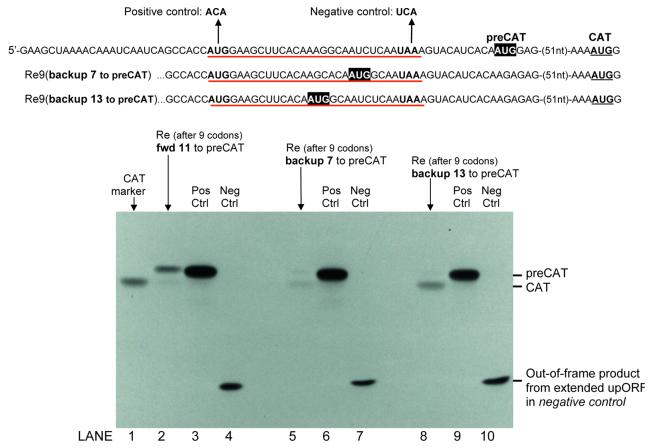
Figure 5.
Test of whether ribosomes can backup to reinitiate. In addition to a nine-codon upORF (underlined in red), each mRNA has an AUG codon (AUGpreCAT highlighted in black) upstream from and in the same reading frame as CAT. Ribosomes that engage the mRNA depicted in the first line would be expected to translate the upORF and then move forward 11 nt to reinitiate translation at AUGpreCAT. This mRNA was tested in lane 2 of the autoradiogram. With the mRNAs depicted in the second and third lines, ribosomes that translate the upORF would have to backup 7 or 13 nt to reach AUGpreCAT. Translation of these mRNAs was tested in lanes 5 and 8. The relative yields of CAT and preCAT proteins indicate the extent to which ribosomes move forward or backward after translating the upORF. For the positive controls in lanes 3, 6 and 9, the start codon of the upORF was mutated, thus making AUGpreCAT the first AUG in the mRNA. In the negative controls (lanes 4, 7 and 10), the indicated mutation of the terminator codon from UAA to UCA causes the upORF (in reading frame –2) to overlap the CAT coding domain for a distance of 133 nt. With reinitiation thus precluded in the negative controls, their failure to produce CAT or preCAT proteins establishes that there is no leaky scanning: all ribosomes recognize the upORF start codon. Thus, reinitiation is the sole mechanism for producing preCAT and/or CAT proteins in lanes 2, 5 and 8. Electrophoresis conditions were adjusted to ensure that the small ‘out-of-frame’ polypeptides (lanes 4, 7 and 10) were retained on the gel. This might be why the various forms of preCAT, differing in length by 7–9 amino acids, are not resolved from one another.
Preliminary experiments showed no change in CAT yields when the length of the upORF was increased from three to nine codons or from nine to 13 codons (data not shown). Production of CAT decreased, however, as the upORF was lengthened further. Figure 1 (lanes 2–4) shows a 3-fold decrease in the efficiency of reinitiation when the upORF was expanded from 13 to 33 codons. This was accomplished by reiterating a 30 nt segment, thereby changing the length without introducing new sequences that might inadvertently affect reinitiation. Inasmuch as nearly identical results were obtained with two different expansion sequences (Fig. 1, lanes 2–4 and lanes 5–7), it seems reasonable to conclude that the decline in reinitiation in Figure 1 results simply from lengthening the upstream minicistron.
To determine whether elongation in general might have been impaired when sequence X was reiterated—as might happen, for example, if tRNAs became limiting—I fused each version of the upORF with the CAT coding domain and compared the yields of the resulting fusion proteins. Figure 2 (lanes 4–6) shows nearly equal yields of 13X-CAT, 23X-CAT and 33X-CAT fusion proteins. Thus, the gradual reduction in CAT yields when the same sequences were used to expand the upORF (Fig. 2, lanes 1–3) reflects a problem specific to reinitiation.
The magnitude of inhibition observed here is within the range seen by other investigators who have explored the dependence of reinitiation on the length of the upORF. Hwang and Su (19), for example, saw a 2-fold decrease in reinitiation as the upORF was lengthened from seven to 18 codons and a gradual decrease as the upORF was further lengthened. Luukkonen et al. (18) saw a gradual diminishment and predicted that reinitiation should be precluded entirely when the length of the upORF reached 55 codons. The latter study of reinitiation within a viral mRNA relied on a biological assay. Hwang and Su (19) monitored protein yields directly, but their system was somewhat complicated by the occurrence of leaky scanning along with reinitiation. For the most part, however, the results of the three studies, using very different test systems, are compatible.
The reason for the inverse relationship between reinitiation and the length of the upstream cistron might be related to loss of initiation factors. One possibility suggested previously (10,28) is that certain factors required for reinitiation might remain loosely bound to the 80S ribosome and dissociate only gradually in the course of elongation. Whether this is true, and what the key factors might be, remains to be established.
Reinitiation declines when elongation is slowed by a structural constraint in the mRNA
If reinitiation declines when the upORF is lengthened because factors required for reinitiation are gradually lost in the course of elongation, one might expect reinitiation to be inhibited even by a short upORF if translation through the upORF is slowed. This was tested by inserting into the upORF a pseudoknot (Fig. 3) derived from viral mRNA. In the viral transcript, the pseudoknot was shown to cause a transient delay in elongation (29) and thus to promote frameshifting.
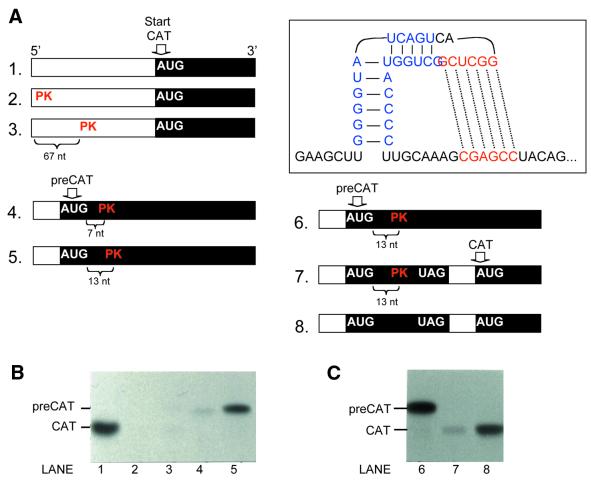
Figure 3.
Sequences of pseudoknot-containing mRNAs used in this study. Downstream from the BamH1 site (line 1) these mRNAs are identical to the transcript shown in full in Figure 1. In line A, the sequences highlighted in blue and red form a pseudoknot close to the 5′ end of the mRNA and far upstream from the CAT coding domain (construct 2 in Fig. 4A). The mRNAs depicted in lines B and C contain a 17-codon upORF defined by the underlined AUG and UAG codons. Within this upORF, a pseudoknot forms in the mRNA depicted in line C (construct 7 in Fig. 4A). The potential for base pairing within the upORF has been minimized in line B (construct 8 in Fig. 4A), which thus serves as a control for construct 7. Line D depicts another control in which the pseudoknot occurs, not within an upORF, but within the main coding domain (construct 5 in Fig. 4A). This was achieved by mutating the terminator codon of the upORF. As a result, ribosomes that initiate translation at the upstream AUG codon continue uninterrupted through the CAT coding domain, producing an N-terminally extended ‘preCAT’ protein. Because the pseudoknot in lines C and D begins 13-nt downstream from the AUG codon, assembly of an initiation complex is not impeded; the pseudoknot thus positioned is a barrier to only the elongation phase of translation. Deletion of 6 nt (underlined in line D) moves the pseudoknot closer to the start codon and thus blocks the initiation (scanning) phase of translation (construct 4 in Fig. 4A).
Just as the transient slowing of elongation has no negative effect on translation of the natural viral mRNA, there was little negative effect when the pseudoknot was inserted into the coding domain of a test transcript that encodes an isoform of CAT (Fig. 4B, lane 5). Under the conditions used for this experiment the pseudoknot does form, as evidenced by the strong inhibition seen in lanes 2–4. The control mRNA tested in lane 2, for example, has the base-paired structure very close to the cap, in which position the structure prevents 40S ribosomal subunits from binding (27). The control mRNA tested in lane 3 has enough room between the cap and the pseudoknot for a 40S subunit to bind, but the subsequent scanning step is blocked: the 40S subunit/initiation factor complex stalls on the 5′ side of the base-paired structure (27). The mRNA tested in lane 4 presumably fails for the same reason: although the pseudoknot has been moved into the coding domain, its proximity to the AUG codon would still require disruption of the structure by scanning 40S ribosomes. In contrast, the mRNA tested in lane 5 has enough room between the AUG codon and the pseudoknot so that the scanning 40S ribosomal subunit need not disrupt the structure. There is adequate room for an 80S ribosome to assemble at the AUG codon of mRNA 5; and 80S elongating ribosomes, unlike 40S initiation complexes, can penetrate stable base-paired structures (30–32). In short, the control mRNAs used in Figure 4B establish that the pseudoknot forms under our test conditions and that it does not preclude translation when positioned in the main ORF of a simple monocistronic transcript.
Figure 4.
Reinitiation is inhibited by a pseudoknot in the upORF that slows elongating ribosomes. The pseudoknot depicted in the boxed insert was positioned in the 5′ untranslated region (constructs 2 and 3), in the main coding domain (constructs 4 and 5) or in an upORF (construct 7). The polyacrylamide gels in (B) and (C) show protein yields from these structure-containing mRNAs and from unstructured control transcripts (constructs 1 and 8). The numeral preceding each mRNA in (A) matches the number of the lane in which that mRNA was tested (B and C). Construct 6 is the same as construct 5. Coding domains, which are not drawn to scale, are shown as filled boxes. The actual sequences of the mRNAs are given in Figure 3. Capped mRNAs were translated as usual at 25°C in reticulocyte lysate supplemented with [3H]leucine and 2 mM Mg2+.
In contrast with that result, which is repeated in lane 6 of Figure 4C, translation was inhibited strongly when the pseudoknot resided in an upORF (Fig. 4C, lane 7). An upORF of the same size (17 codons) that was devoid of secondary structure allowed translation of CAT (Fig. 4C, lane 8), indicating that reinitiation occurs fairly efficiently in this system. Thus, the simplest explanation for the low yield of CAT in lane 7 is that the documented slowing of elongation by the pseudoknot (29) adversely affects the ability to reinitiate. In future experiments, the pseudoknot-containing mRNA used in lane 7 might permit isolation of ribosomes that could be analyzed to detect which initiation factors are, or are no longer, associated with the stalled elongation complexes.
While the experiments in Figure 1 establish that the length of the upORF matters, Figure 4 shows that a relatively short upORF in which elongation is slowed can inhibit disproportionately. In other words, there is no strict answer regarding what size ORF is compatible with reinitiation. The experiments herein nevertheless provide some guidelines.
Post-termination ribosomes show negligible ability to migrate backwards
The mRNAs in Figure 5 were designed to test the ability of ribosomes to reinitiate at AUG codons positioned close to the termination site of the upORF. In addition to a nine-codon upORF, each of these constructs has an upstream AUG codon in-frame with the CAT coding domain. The N-terminally extended forms of CAT initiated from these upstream start codons are designated ‘preCAT.’ Controls in which the upORF was eliminated by changing its start codon from AUG to ACA show that all three forms of preCAT protein are stable and translated efficiently when AUGpreCAT is the first AUG in the mRNA (lanes 3, 6 and 9). When the upORF was present, however, access to AUGpreCAT differed markedly among the three constructs.
With the mRNA depicted in line 1, ribosomes perform as expected, translating the nine-codon upORF and then moving forward 11 nt to reinitiate at AUGpreCAT (Fig. 5, lane 2). The yield of preCAT protein is rather low but this mRNA clearly supports preCAT translation. In contrast, mRNAs that require ribosomes to scan backwards to reach AUGpreCAT produce little or no preCAT protein. In lane 8, reinitiation occurred exclusively in the forward direction at AUGCAT. In lane 5 there is a barely visible preCAT band, suggesting that some ribosomes were able to backup 7 nt to reinitiate at AUGpreCAT. The preCAT band in lane 5 is too faint to be quantified and this underscores the main point: that reinitiation occurs to a negligible extent when AUGpreCAT precedes the terminator codon (lanes 5 and 8 versus 2).
The sequence GCCACC . . . G flanking the first AUG codon in these constructs precludes leaky scanning, as confirmed by the complete absence of preCAT protein in lanes 4, 7 and 10. Given these clean negative controls as well as the complete absence of preCAT protein in lane 8, the very low yield of preCAT protein in lane 5 must be attributed to ribosomes moving backward to reinitiate, but the experiment illustrates the limitations: a 4 nt space between the terminator codon and upstream AUGpreCAT codon allows reinitiation (lane 5) but a 10 nt space does not (lane 8); and the positive result in lane 5 is very, very weak.
A notable difference between studies in which ribosomes were unable to reinitiate efficiently at sites upstream from the terminator codon of the 5′ ORF (33,34; Fig. 5) and studies in which ‘backwards scanning’ appeared to occur (20,21) is that, in the latter cases, an unfavorable context at the upstream start site may have allowed leaky scanning. Evidence suggests that access to a downstream start site via leaky scanning can be suppressed by 80S elongating ribosomes advancing from the upstream initiation site (35). As a result of this ‘elongational occlusion,’ access to a downstream start site via leaky scanning might be augmented when a terminator codon is introduced in a way that minimizes the overlap between upstream and downstream ORFs. In short, one cannot conclude that a reinitiation mechanism is operative just because access to an internal start codon depends on the termination of the upstream ORF. Reinitiation can be studied unambiguously only by using constructs that preclude leaky scanning. The negligible ability of eukaryotic ribosomes to reinitiate upstream from the site of termination, as demonstrated when leaky scanning is precluded, differs significantly from what is seen in prokaryotes (6,36).
That eukaryotic ribosomes display little backwards migration in the reinitiation mode reinforces the evidence from earlier studies in which the direction of ribosome movement in the primary scanning mode appeared to be strictly 5′ to 3′ (35). (The ‘primary scanning mode’ refers to migration from the 5′ end of the mRNA to the first AUG codon.) Initiation factors associated with the 40S ribosomal subunit might be important for achieving this forward movement. It is conceivable, for example, that the 40S subunit inherently can slide in both directions, but that backsliding is prevented by clamping of eIF3 onto the mRNA. In the primary scanning mode, the 40S subunit carries eIF3 from the outset. Following translation of an upORF, however, very limited backward movement of ribosomes might be possible (Fig. 5, lane 5) while waiting for eIF3 to regain its position on the 40S subunit, whereupon the strict 5′ to 3′ bias would be re-imposed.
Role of reinitiation in modulating translation
cDNA sequencing has uncovered a growing list of genes in which the major coding domain is preceded by small upORFs. Some of these encumbered cDNAs do not correspond to functional mRNAs; i.e. many cDNAs with upstream AUG codons have been traced to incompletely spliced transcripts or other misinterpretations (37–39). There are, however, many genuine examples of mammalian mRNAs in which the 5′ untranslated sequence includes one or two (or occasionally more) small upORFs. The frequent presence of upstream AUG codons in transcripts that encode growth factors, cytokines, transcription factors, kinases and other potent proteins has long been noted (4).
mRNAs thus structured are likely to be translated by leaky scanning and/or reinitiation. Reinitiation is inefficient when synthetic mRNAs are tested, as shown herein, and the presence of an upORF indeed reduces the translation of a wide variety of natural mRNAs (16,33,40–48). In some of these cases, an alternative form of mRNA devoid of upstream AUG codons is produced when more efficient translation is required. The next paragraph explains why this is not seen in every case.
Because reinitiation is usually inefficient, the presence of upORFs is sometimes employed to limit the translation of cytokines, transcription factors and other potent proteins that are required in only small quantities and would be harmful if overproduced. The thrombopoietin (TPO) gene provides a striking illustration. The normal gene produces a mixture of mRNAs with different leader sequences, all of which translate TPO poorly due to small upORFs (49). The most inhibitory of the upORFs is one that initiates at a moderately strong AUG codon and overlaps the TPO start codon. A remarkable study by Wiestner et al. (50) showed that some patients develop hereditary thrombocythemia because a mutation alters the pattern of splicing of TPO mRNA in a way that eliminates the inhibitory upORF, thereby elevating translation of the cytokine. The disease results from overriding a built-in constraint on translation.
Whereas the presence of upORFs and consequent imposition of a reinitiation mechanism reduces downstream translation in the foregoing examples, there are special cases in which an upORF has a facilitating effect; i.e. the upORF promotes initiation from a particular downstream site by helping ribosomes to dodge a strongly inhibitory upstream AUG codon (19,42,51,52). Inspection of these and other mRNAs (49,53–55) reveals that the strongest inhibition is caused by an upORF that overlaps or terminates very close to the start of the downstream ORF. This reinforces the aforementioned conclusion that eukaryotic ribosomes have negligible ability to scan backwards in order to reinitiate translation.
Acknowledgments
ACKNOWLEDGEMENT
Research in my laboratory is supported by grant GM33915 from the National Institutes of Health.
REFERENCES
- 1.Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- 2.Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. (1997) Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J., 16, 2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kozak M. (1991) An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol., 115, 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy J.E.G. (1998) Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev., 62, 1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abastado J.-P., Miller,P.F., Jackson,B.M. and Hinnebusch,A.G. (1991) Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol., 11, 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnebusch A.G. (1997) Translational regulation of yeast GCN4. J. Biol. Chem. 272, 21661–21664. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Barrio M.T., Naranda,T., Vazquez de Aldana,C.R., Cuesta,R., Hinnebusch,A.G., Hershey,J.W.B. and Tamame,M. (1995) GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev., 9, 1781–1796. [DOI] [PubMed] [Google Scholar]
- 10.Kozak M. (1987) Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol., 7, 3438–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant C.M. and Hinnebusch,A.G. (1994) Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol., 14, 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant C.M., Miller,P.F. and Hinnebusch,A.G. (1995) Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res., 23, 3980–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilela C., Linz,B., Rodrigues-Pousada,C. and McCarthy,J.E.G. (1998) The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res., 26, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vega Laso M.R., Zhu,D., Sagliocco,F., Brown,A.J.P., Tuite,M.F. and McCarthy,J.E.G. (1993) Inhibition of translation initiation in the yeast Saccharomyces cerevisiae as a function of the stability and position of hairpin structures in the mRNA leader. J. Biol. Chem., 268, 6453–6462. [PubMed] [Google Scholar]
- 15.Koloteva N., Müller,P.P. and McCarthy,J.E.G. (1997) The position dependence of translational regulation via RNA-RNA and RNA–protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40S ribosomes in translational initiation. J. Biol. Chem., 272, 16531–16539. [DOI] [PubMed] [Google Scholar]
- 16.Wang L. and Wessler,S.R. (1998) Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell, 10, 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M. (1978) How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell, 15, 1109–1123. [DOI] [PubMed] [Google Scholar]
- 18.Luukkonen B.G.M., Tan,W. and Schwartz,S. (1995) Efficiency of reinitiation of translation on human immunodeficiency virus type 1 mRNAs is determined by the length of the upstream open reading frame and by intercistronic distance. J. Virol., 69, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang W.-L. and Su,T.-S. (1998) Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol., 79, 2181–2189. [DOI] [PubMed] [Google Scholar]
- 20.Peabody D.S., Subramani,S. and Berg,P. (1986) Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol. Cell. Biol., 6, 2704–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas K.R. and Capecchi,M.R. (1986) Introduction of homologous DNA sequences into mammalian cells induces mutations in the cognate gene. Nature, 324, 34–38. [DOI] [PubMed] [Google Scholar]
- 22.Welch E.M. and Jacobson,A. (1999) An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J., 18, 6134–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frischmeyer P.A. and Dietz,H.C. (1999) Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet., 8, 1893–1900. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. (1994) Features in the 5′ non-coding sequences of rabbit α and β-globin mRNAs that affect translational efficiency. J. Mol. Biol., 235, 95–110. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. (1989) Context effects and (inefficient) initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol., 9, 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozak M. (1990) Evaluation of the fidelity of initiation of translation in reticulocyte lysates from commercial sources. Nucleic Acids Res., 18, 2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. (1998) Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res., 26, 4853–4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. (1992) Regulation of translation in eukaryotic systems. Ann. Rev. Cell. Biol., 8, 197–225. [DOI] [PubMed] [Google Scholar]
- 29.Somogyi P., Jenner,A.J., Brierley,I. and Inglis,S.C. (1993) Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol., 13, 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak M. (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol., 9, 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lingelbach K. and Dobberstein,B. (1988) An extended RNA/RNA duplex structure within the coding domain of mRNA does not block translational elongation. Nucleic Acids Res., 16, 3405–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liebhaber S.A., Cash,F. and Eshleman,S.S. (1992) Translation inhibition by an mRNA coding region secondary structure is determined by its proximity to the AUG initiation codon. J. Mol. Biol., 226, 609–621. [DOI] [PubMed] [Google Scholar]
- 33.Bates B., Hardin,J., Zhan,X., Drickamer,K. and Goldfarb,M. (1991) Biosynthesis of human fibroblast growth factor-5. Mol. Cell. Biol., 11, 1840–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao J. and Geballe,A.P. (1995) Translational inhibition by a human cytomegalovirus upstream open reading frame despite inefficient utilization of its AUG codon. J. Virol., 69, 1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. (1995) Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl Acad. Sci. USA, 92, 2662–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adhin M.R. and van Duin,J. (1990) Scanning model for translational reinitiation in eubacteria. J. Mol. Biol., 213, 811–818. [DOI] [PubMed] [Google Scholar]
- 37.Kozak M. (1996) Interpreting cDNA sequences: some insights from studies on translation. Mamm. Genome, 7, 563–574. [DOI] [PubMed] [Google Scholar]
- 38.Kozak M. (2000) Do the 5′ untranslated domains of human cDNAs challenge the rules for initiation of translation (or is it vice versa)? Genomics, 70, 396–406. [DOI] [PubMed] [Google Scholar]
- 39.Kubu C.J., Goldhawk,D.G., Barker,P.A. and Verdi,J.M. (2000) Identification of the translational initiation codon in human MAGED1. Genomics, 70, 150–152. [DOI] [PubMed] [Google Scholar]
- 40.Brown C.Y., Mize,G.J., Pineda,M., George,D.L. and Morris,D.R. (1999) Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene, 18, 5631–5637. [DOI] [PubMed] [Google Scholar]
- 41.Child S.J., Miller,M.K. and Geballe,A.P. (1999) Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem., 274, 24335–24341. [DOI] [PubMed] [Google Scholar]
- 42.Mittag M., Eckerskorn,C., Strupat,K. and Hastings,J.W. (1997) Differential translational initiation of lbp mRNA is caused by a 5′ upstream open reading frame. FEBS Lett., 411, 245–250. [DOI] [PubMed] [Google Scholar]
- 43.Ren H. and Stiles,G.L. (1994) Posttranscriptional mRNA processing as a mechanism for regulation of human A1 adenosine receptor expression. Proc. Natl Acad. Sci. USA, 91, 4864–4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steel L.F., Telly,D.L., Leonard,J., Rice,B.A., Monks,B. and Sawacki,J.A. (1996) Elements in the murine c-mos messenger RNA 5′-untranslated region repress translation of downstream coding sequences. Cell Growth Differ., 7, 1415–1424. [PubMed] [Google Scholar]
- 45.Wang X.-Q. and Rothnagel,J.A. (2001) Post-transcriptional regulation of the GLI1 oncogene by the expression of alternative 5′ untranslated regions. J. Biol. Chem., 276, 1311–1316. [DOI] [PubMed] [Google Scholar]
- 46.Nomura A., Iwasaki,Y., Saito,M., Aoki,Y., Yamamori,E., Ozaki,N., Tachikawa,K., Mutsuga,N., Morishita,M., Yoshida,M., Asai,M., Oiso,Y. and Saito,H. (2001) Involvement of upstream open reading frames in regulation of rat V1b vasopressin receptor expression. Am. J. Physiol. Endocrinol. Metab., 280, E780–E787. [DOI] [PubMed] [Google Scholar]
- 47.Shishido-Hara Y., Hara,Y., Larson,T., Yasui,K., Nagashima,K. and Stoner,G.L. (2000) Analysis of capsid formation of human polyomavirus JC (Tokyo-1 strain) by a eukaryotic expression system: splicing of late mRNAs, translation and nuclear transport of VP1. J. Virol., 74, 1840–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petty I.T.D., Edwards,M.C. and Jackson,A.O. (1990) Systemic movement of an RNA plant virus determined by a point substitution in a 5′ leader sequence. Proc. Natl Acad. Sci. USA, 87, 8894–8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghilardi N., Wiestner,A. and Skoda,R.C. (1998) Thrombopoietin production is inhibited by a translational mechanism. Blood, 92, 4023–4030. [PubMed] [Google Scholar]
- 50.Wiestner A., Schlemper,R.J., van der Maas,A.P.C. and Skoda,R.C. (1998) An activating splice donor mutation in the thrombopoietin gene causes hereditary thrombocythaemia. Nature Genet., 18, 49–52. [DOI] [PubMed] [Google Scholar]
- 51.Calkhoven C.F., Müller,C. and Leutz,A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev., 14, 1920–1932. [PMC free article] [PubMed] [Google Scholar]
- 52.Mudge S.J., Williams,J.H., Eyre,H.J., Sutherland,G.R., Cowan,P.J. and Power,D.A. (1998) Complex organisation of the 5′-end of the human glycine tRNA synthetase gene. Gene, 209, 45–50. [DOI] [PubMed] [Google Scholar]
- 53.Babik J.M., Adams,E., Tone,Y., Fairchild,P.J., Tone,M. and Waldmann,H. (1999) Expression of murine IL-12 is regulated by translational control of the p35 subunit. J. Immunol., 162, 4069–4078. [PubMed] [Google Scholar]
- 54.Lee Y.C., Chang,C.-W., Su,C.-W., Lin,T.-N., Sun,S.H., Lai,H.-L. and Chern,Y. (1999) The 5′ untranslated regions of the rat A2A adenosine receptor gene function as negative translational regulators. J. Neurochem., 73, 1790–1798. [DOI] [PubMed] [Google Scholar]
- 55.Byrne P.C., Sanders,P.G. and Snell,K. (1995) Translational control of mammalian serine hydroxymethyl-transferase expression. Biochem. Biophys. Res. Commun., 214, 496–502. [DOI] [PubMed] [Google Scholar]