Abstract
Helicases couple the hydrolysis of nucleoside triphosphates (NTPs) to the unwinding of double-stranded nucleic acids and are essential in DNA metabolism. Thus far, no inhibitors are known for helicases except heliquinomycin isolated from Streptomyces sp. As the three-dimensional structure of the hexameric replicative DNA helicase RepA encoded by the broad host-range plasmid RSF1010 is known, this protein served as a model helicase to search for inhibitory compounds. The commercially available flavone derivatives luteolin, morin, myricetin and dimyricetin (an oxidation product of myricetin) inhibited the ATPase and double-stranded DNA unwinding activities of RepA. Dimyricetin was the most effective inhibitor for both activities. Single-stranded DNA-dependent RepA ATPase activity is inhibited non-competitively by all four compounds. This finding contrasts the inhibition of phosphoinositide 3-kinase by flavones that fit into the ATP binding pocket of this enzyme. Myricetin also inhibited the growth of a Gram-positive and a Gram-negative bacterial species. As we found other hexameric and non-hexameric prokaryotic helicases to be differentially sensitive to myricetin, flavones may provide substructures for the design of molecules helpful for unraveling the mechanism of helicase action and of novel pharmacologically useful molecules.
INTRODUCTION
DNA helicases are motor proteins essential in key biological processes, which require single-stranded DNA (ssDNA) such as DNA replication, transcription, translation, repair and recombination. The unwinding of double-stranded DNA (dsDNA) by helicases is strictly processive either in 5′ → 3′ or in 3′ → 5′ direction for the first three above processes (1,2) and fuelled by hydrolysis of nucleoside 5′-triphosphates (NTPs). A large number of helicase proteins (Escherichia coli alone contains at least 12 different helicases), which are involved in many aspects of metabolism in bacterial, viral and eukaryotic systems have now been characterized in vitro. In humans, malfunction of certain DNA helicases is associated with several severe diseases including Bloom’s syndrome, xeroderma pigmentosum and Werner’s syndrome (3–6), with the development of cancer (7) and with aging (8).
The mechanism of dsDNA unwinding by helicases is still under debate (9,10), and little is known about helicase inhibitors except for non-hydrolyzable NTP analogs. In addition, the non-nucleotide polyketide heliquinomycin (HQ) isolated from the culture broth of Streptomyces sp. MJ1929-SF2 was shown to inhibit DNA helicases from HeLa cells which, however, were not purified so that these studies must be considered preliminary (11).
HQ is composed of a naphthoquinone and a coumarin moiety linked by a spiroketal system (Fig. 1). Its chemical structure inspired us to look for related, commercially available compounds containing the naphthoquinone system and to test their inhibitory action on the replicative hexameric DNA helicase RepA. The latter was chosen as a model helicase for these studies as it is biochemically well characterized (12,13) and it is the only helicase the structure of which has been determined at high resolution using protein that crystallized as hexamers from full-length subunits (14). Therefore, RepA was used for inhibitor studies, which will open the door for subsequent co-crystallization studies and structure-based mutational analysis. RepA is encoded by the broad host-range plasmid RSF1010, an 8684 bp multicopy plasmid that replicates in a wide variety of Gram-negative bacteria and also in Gram-positive actinomyces (15). RepA has 5′ → 3′ polarity with optimal dsDNA unwinding and ssDNA stimulated ATPase activity at slightly acidic pH of 5.5–6.0 (12,13).
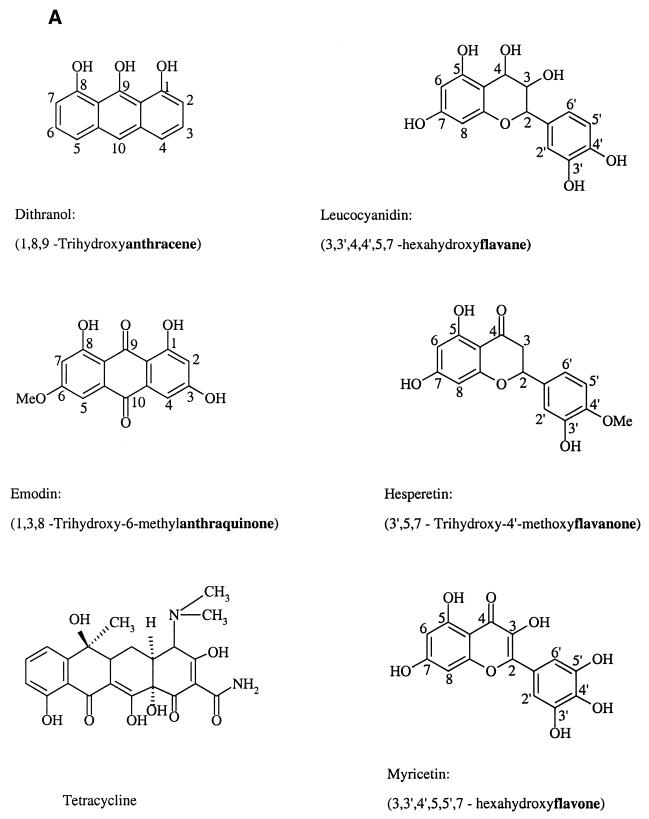
Figure 1.
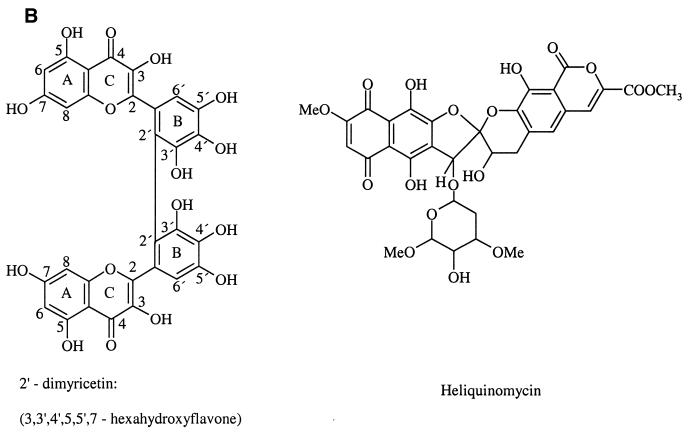
Structures of the six classes (in bold face) of compounds used in this study and of HQ. (A) Six classes of compounds analogous to substructures of HQ. (B) Dimyricetin and HQ.
Our studies showed that of the eight commercially available compounds tested (Fig. 1), the flavones luteolin, morin, myricetin and dimyricetin (an oxidation product of myricetin) inhibit the ATPase activity of RepA in the micromolar range. For both the ATPase and helicase activities of RepA, dimyricetin is the most effective inhibitor. We have shown that myricetin blocks cell growth and also inhibits several other prokaryotic helicases.
MATERIALS AND METHODS
Reagents and buffers
All chemicals used in this study were of pro analysi quality. Myricetin, leucocyanidin and tetracycline hydrochloride were purchased from Aldrich; Hesperetin and ATP were from Sigma; the other chemicals used for screening were from Lancaster. Dimyricetin was synthesized according to Lang (16). Stock solutions of inhibitors were made 10 mM (except for dimyricetin 1 mM) in dimethyl sulfoxide (DMSO) and stored at room temperature. All solutions were prepared with Milli-Q deionized water. Buffer A used for ATPase activity assays contained 40 mM Mes/NaOH pH 5.6, 10 mM MgCl2, 60 mM NaCl, 5% (vol/vol) DMSO. Buffer B used for helicase activity measurements contained 40 mM Mes/NaOH pH 5.6, 10 mM MgCl2, 1 mM dithiothreitol, 1 mM ATP, 50 µg/ml bovine serum albumin, 0.02% (wt/wt) Brij-58, 5% (vol/vol) DMSO. Buffer C used for inhibitor binding tests contained 40 mM Mes/NaOH pH 5.6, 60 mM NaCl, 5% (vol/vol) DMSO.
Purification of RepA protein
The E.coli RSF1010-encoded RepA protein was purified as described (17). The protein concentration was determined spectrophotometrically using an extinction coefficient of ɛ280 = 25 180 M–1 cm –1 (monomer).
Steady-state kinetics and determination of inhibition constants
Kinetic parameters of ATPase activity were determined in buffer A using acidic ammonium molybdate and malachite green to monitor the release of inorganic phosphate at 30°C as described (15). Before adding 100 nM (dT)20 for the stimulation of ATPase activity, 80 nM RepA (hexamer) and the inhibitors at concentrations indicated were incubated at 30°C for 10 min. Reactions were started by the addition of ATP at increasing concentrations. Each initial velocity was determined at least in duplicate, and at least four different ATP concentrations were examined. Initial reaction velocity data obtained at various substrate concentrations [S] in the presence of varying concentrations of inhibitor [I] were analyzed by plotting 1/v versus 1/[S]. The experimental data for kinetic inhibition were analyzed using equation 1 for non-competitivity (18):
where Km and Ki are the substrate binding and inhibition constants, [S] and [I] denote the concentrations of substrate and inhibitor, and v and Vmax the initial and maximum velocity of the reaction, respectively.
Oligo(dT)20 used to stimulate the ssDNA-dependent ATPase activity was synthesized at the Department of Biochemistry of FU Berlin. The concentrations were determined spectrophotometrically using an extinction coefficient of ɛ260 = 8100 M–1 cm–1 per nucleotide of oligo(dT).
DNA unwinding assay (helicase assay)
A forked helicase substrate was used similar to that described by Crute et al. (19). To viral M13mp18 DNA, a 5′-32P-labeled 53mer oligodeoxynucleotide was annealed, resulting in a double-stranded segment of 31 bp and 22 unpaired nucleotides at the 3′ end. Helicase assays were performed in 20 µl buffer B containing 45 fmol helicase substrate, 6 pmol RepA (monomer) and inhibitors as indicated.
In the absence of ATP and helicase substrate, RepA was pre-incubated with inhibitor in buffer B for 10 min on ice. Then ATP and helicase substrate in buffer B were added and the mixture was transferred to 30°C. After incubation for 10 min, the reactions were terminated by adjusting the mixture to 25 mM EDTA, 0.25% SDS and proteinase K at 0.1 mg/ml. Following incubation for an additional 5 min at 30°C, the displaced 53mer oligodeoxynucleotides were separated from helicase substrate by electrophoresis on 10% polyacrylamide gels in 89 mM Tris-base, 89 mM boric acid and 1 mM EDTA for 90 min at constant voltage of 120 V. Products were visualized by storage phosphor autoradiography and analyzed with the ImageQuant software version 5.1 (Molecular Dynamics).
Mean inhibitory concentration (IC50)
In the present study, IC50 is defined as the inhibitor concentration at which the ssDNA stimulated ATPase activity or helicase activity decreased to 50% of the control reaction without inhibitors.
Fluorescence binding assay
Fluorescence titrations were performed using a Shimadzu RF-5000 spectrofluorometer thermostated at 25°C with a temperature bath (Lauda). The intrinsic binding of inhibitors to RepA was followed by monitoring the quenching of the TNS fluorescence (λex = 400 nm; λem = 440 nm). Solutions of 2 µM RepA (monomer) and 100 µM TNS in buffer C were equilibrated for 30 min at 25°C and titrated under stirring in 1 min intervals with aliquots of stock solutions containing inhibitors. To avoid possible artifacts due to dilution and inner filter effects, the excitation wavelength was set at a spectral bandwidth of 3 nm and the emission bandwidth at 15 nm. One milliliter quartz cuvettes with 1 cm path length were used. All assays were performed in duplicate, titration points were corrected as described (15) and binding constants were determined according to:
Q = Qmax Kd / (Kd + [I]) 2
where Q is the TNS fluorescence quenching signal, and Qmax represents the maximum quenching signal. Kd is the dissociation constant of the binding site for inhibitor, [I] denotes the total inhibitor concentration.
Determination of the minimal inhibitory concentration
Overnight cultures of E.coli SCS1 (Stratagene) or Bacillus subtilis SB19 (20) were diluted 1000-fold with YT medium. Two aliquots of the dilutions, the respective flavones dissolved in DMSO, were added. In all assays the overall DMSO concentration was adjusted to 2.5%. Following incubation at 37°C for 18 h, the minimal inhibitory concentration (MIC) was determined visually.
RESULTS
Inhibitor screening
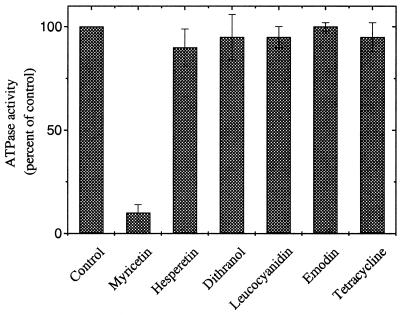
As for all replicative helicases known, the ATPase activity of RepA is stimulated by ssDNA (21). There is a strict correlation between optimal ssDNA binding at acidic pH to RepA and stimulation of ATPase activity, which drives efficient unwinding of dsDNA (21,22). The method used to measure ATPase activity allows screening for ATPase inhibitors by monitoring the ATP hydrolysis in the presence of ssDNA (15). Compounds of six representative classes of flavone derivatives (Fig. 1), which contain substructures of the DNA helicase inhibitor HQ (11), were tested for ATPase inhibition up to a final concentration of 250 µM. Myricetin was the most potent of these compounds, inhibiting RepA ATPase activity by ∼90% at 250 µM concentration. The other five classes of compounds tested did not significantly affect enzyme activity at test conditions (Fig. 2).
Figure 2.
Inhibition of the RepA ATPase activity by the six classes of compounds shown in Figure 1A. Reactions were performed in buffer A as described in the Materials and Methods using 80 nM RepA protein, 100 nM (dT)20 800 µM ATP and 250 µM compounds in buffer A. The values are normalized to the activity in the absence of inhibitor.
The ATPase activity of RepA is inhibited by flavone derivatives
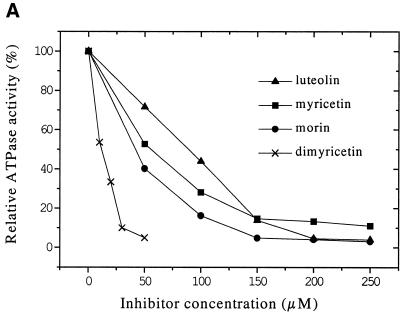
To gain insight into the influence of polyhydroxylation and the methylation of flavones, we compared the inhibitory effects of several different flavone compounds, the substitutions of which are shown in Table 1. The residual ATPase activities measured indicate that the inhibitory effect is dependent on the number and position of hydroxyl groups. Indeed, the higher hydroxylated flavones like dimyricetin, myricetin, morin and luteolin are inhibitors (Fig. 3A), whereas less hydroxylated or methylated flavone derivatives do not inhibit ATP hydrolysis. Among the compounds used, dimyricetin is the most effective inhibitor with an IC50 of 15 µM, whereas luteolin is the least effective compound with an IC50 of 90 µM (Table 2).
Table 1. Structure–activity comparisons of flavones on RepA ATPase activity.
| Flavones | Substitutions | Residual ATPase activity (percentage of control) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 3 | 5 | 7 | 8 | 2′ | 3′ | 4′ | 5′ | ||
| Reactions were performed in buffer A as described in the Materials and Methods using 80 nM RepA protein, 100 nM (dT)20 800 µM ATP and 250 µM of the flavone compounds (dimyricetin 50 µM) in buffer A. ATPase activity is expressed relative to activity without inhibitor. OCH3 indicates methoxy substitutions. | |||||||||
| Apigenin |
|
|
OH |
|
|
|
OH |
OH |
>95 |
| Galangin |
OH |
OH |
OH |
|
|
|
|
|
>95 |
| Luteolin |
|
OH |
OH |
|
|
OH |
OH |
|
5 |
| Hydroxyflavonea |
|
|
OH |
OH |
|
OH |
OH |
|
50 |
| Morin |
OH |
OH |
OH |
|
OH |
|
OH |
|
5 |
| Quercetin |
OH |
OH |
OH |
|
|
OH |
OH |
|
>90 |
| Trimethyletherb |
OH |
OH |
OH |
|
|
OCH3 |
OCH3 |
OCH3 |
>95 |
| Myricetin |
OH |
OH |
OH |
|
|
OH |
OH |
OH |
10 |
| Dimyricetin | OH | OH | OH | OH | OH | OH | 2 | ||
a3′,4′,7,8-Tetrahydroxyflavone.
bMyricetin trimethylether.
Figure 3.
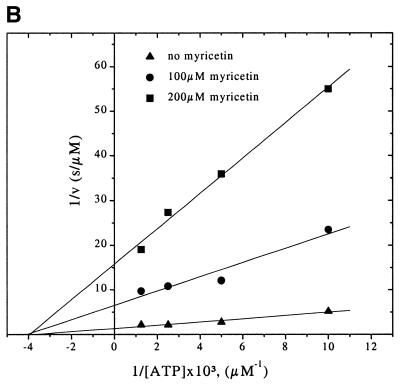
Inhibition of RepA ATPase activity by flavone derivatives. (A) Inhibitory effect of flavone compounds on RepA ATPase activity stimulated by ssDNA. Reactions were performed in buffer A as described in the Materials and Methods. ATPase activity is normalized to the activity in the absence of inhibitor. (B) Double-reciprocal Lineweaver–Burke plot describing the kinetic inhibition of ssDNA-dependent RepA ATPase activity by myricetin. The assays were carried out as described in the Materials and Methods except that myricetin was added in indicated amounts to reaction mixtures containing 100 or 200 µM of ATP.
Table 2. IC50, Ki, Kd and MIC values of different flavone derivatives.
| Compound | IC50 (µM) | Ki (µM) | Kd (µM) | MIC (mg/ml) E.coli | MIC (mg/ml) B.subtilis |
|---|---|---|---|---|---|
| IC50 and Ki values are from inhibition of the ATPase activity experiments. | |||||
| Luteolin | 90 | 38.2 ± 7.0 | 26.5 ± 1.9 | >1 | >1 |
| Morin | 45 | 18.1 ± 3.2 | 20.2 ± 4.8 | >1 | >1 |
| Myricetin | 50 | 22.5 ± 5.1 | 24.0 ± 1.7 | 0.50 | 0.25 |
| Dimyricetin | 15 | 6.7 ± 1.1 | 2.6 ± 0.9 | >1 | >1 |
The data suggest that the major structural requirements for the inhibition of RepA by flavone compounds are the presence of the C2–C3 double bond, the 4-ketone and the OH groups in positions 5, 7 and 4′ respectively. The saturation of the C2–C3 bond at the pyrone ring of flavones (i.e. flavanones) or removal/methylation of either OH group on the aromatic A and B rings (see Fig. 1) nearly abolishes inhibition of RepA, suggesting that formation of hydrogen bonds between functional groups of the flavone compounds and RepA is of importance. In the absence of an ssDNA effector, even at 250 µM myricetin the intrinsic ATPase activity of RepA was inhibited only slightly to a relative activity of ≥80% (data not shown).
Kinetic inhibition of ATPase activity by flavone derivatives
To determine whether flavones are competitive inhibitors with respect to nucleotide triphosphates, we analyzed the kinetic inhibition of RepA ATPase activity by varying the concentrations of ATP and inhibitors in the reaction mixture. ATP hydrolysis is inhibited non-competitively not only by myricetin (Fig. 3B) but also by morin, luteolin and dimyricetin (data not shown). The Ki and IC50 values of the four flavones are in accordance, dimyricetin having the lowest Ki of 6.7 µM. The Ki values are summarized in Table 2. The results indicate that the flavones do not primarily bind to the NTP binding pocket blocking the entrance of NTPs.
Inhibition of RepA helicase activity by flavone derivatives
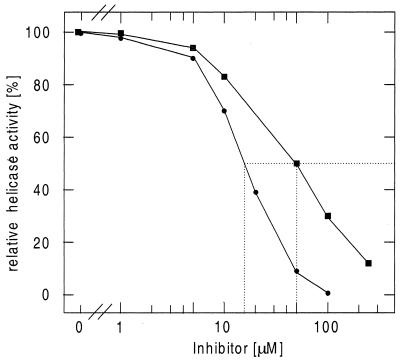
We further tested the inhibition of RepA helicase (DNA unwinding) activity by myricetin and dimyricetin (Fig. 4). Dimyricetin inhibited the helicase activity more efficiently than myricetin, with an IC50 of ∼15 and 50 µM, respectively. This is consistent with the results obtained for the inhibition of ATP hydrolysis.
Figure 4.
Inhibitory effects of myricetin and dimyricetin on RepA helicase DNA unwinding activity. The assays were carried out as described in the Materials and Methods. Filled circles, dimyricetin; filled squares, myricetin. IC50 values are indicated by the dotted lines.
Fluorescence measurements on inhibitor binding to RepA
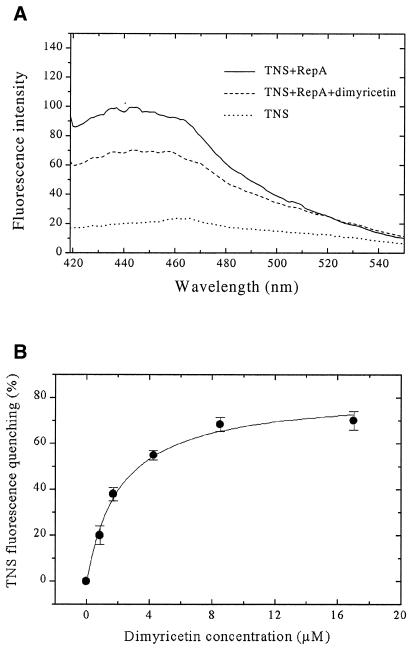
As flavones do not block the NTP binding site of RepA, the compounds may bind somewhere else to the protein. Therefore, binding of the inhibitors to RepA was followed by fluorescence spectroscopy using TNS as an extrinsic fluorescent probe. In the presence of 2 µM RepA (monomer), the fluorescence emission maximum of TNS (100 µM) is shifted from 465 to 440 nm (excited at 400 nm), and the fluorescence intensity increases ~6-fold (at 440 nm). Upon addition of inhibitors to the above mixture of RepA and TNS, the fluorescence decreases and is reduced ∼70–90% at saturating concentrations of inhibitors (Fig. 5A). The obtained titration curves were analyzed with the ligand binding equation to give dissociation constants for luteolin, morin, myricetin and dimyricetin (26.5, 20.2, 24.0 and 2.6 µM, respectively; see Fig. 5B and Table 2). The data indicate that these molecules bind to RepA. Dimyricetin interacts with RepA 10-fold stronger than the three other compounds.
Figure 5.
Results of fluorescence studies on dimyricetin binding to RepA. (A) Fluorescence emission spectra: dotted curve, 100 µM TNS alone; continuous curve, in the presence of 2 µM RepA (monomer); dashed curve, in the presence of RepA and 2 µM dimyricetin. (B) Fluorescence titration studies on binding of dimyricetin to RepA. Titrations in buffer C at 25°C with increasing amounts of dimyricetin to the mixture of RepA and TNS. The measurements were monitored by the quenching of TNS (100 µM) fluorescence at λex = 400 nm, λem = 440 nm. RepA concentration was 2 µM (monomer).
Myricetin inhibits bacterial growth
Compounds that inhibit RepA ATPase activity the most efficiently in vitro, i.e. luteolin, morin, myricetin and dimyricetin, were studied for inhibition of bacterial growth of a Gram-negative and a Gram-positive strain, E.coli SCS1 and B.subtilis SB19, respectively. Only myricetin prevented cell growth, the MIC being ∼0.50 mg/ml for SCS1 and 0.25 mg/ml for SB19. Even in the presence of 1 mg/ml of the other substances, both strains still grew. Hence, the number of the hydroxyls of the phenyl ring (ring B, Fig. 1) seems to be essential not only for inhibiting the ATPase activity, but also for bacterial growth inhibition. Dimyricetin, which inhibits RepA ATPase and helicase most efficiently, but not bacterial growth, probably does not reach the potential target(s) within the cell.
Myricetin inhibits helicases involved in various roles of nucleic acid metabolism
As myricetin inhibits bacterial growth, we were interested whether this substance also inhibits helicases other than RepA. Hexameric and non-hexameric unwinding enzymes were studied that play a role in a variety of processes including DNA repair and conjugative DNA transfer. Concerning the amino acid sequence, P1 Ban and SPP1 G40P belong to the family of DnaB-like helicases, whereas PcrA, E.coli Rep and E.coli UvrD belong to superfamily I despite being essential in various parts of nucleic acid metabolism. The eight helicases tested were all inhibited by myricetin with IC50 values ranging from 0.3 µM for G40P of B.subtilis phage SPP1 to 50 µM for RepA. Neither the oligomerization state of the enzymes nor their polarity of unwinding had a particular influence on the sensitivity to myricetin (Table 3). However, the 100-fold difference in IC50 indicates that for certain helicases myricetin is a more efficient inhibitor than for others.
Table 3. Inhibition of helicase activity of different prokaryotic helicases by myricetin.
| Helicase | IC50 (µM) | Association state | Polarity | Function | References |
|---|---|---|---|---|---|
| IC50 values are averages of at least two independent experiments. | |||||
| RepA | 50 | Hexameric | 5′ → 3′ | RSF1010 replication | (14) |
| SPP1 G40P | 0.3 | Hexameric | 5′ → 3′ | SPP1 replication | (37) |
| P1 Ban | 2.5 | Hexamerica | 5′ → 3′ | P1 and E.coli replication | (36) |
| P4 α | 5 | ≥Dimeric | 3′ → 5′ | P4 replication | (38) |
| B.subtilis PcrA | 2 | Monomeric | 3′ → 5′ | Unclear | (39) |
| E.coli Rep | 2 | Mono-/dimericb | 3′ → 5′ | Rolling-circle replication | (40,41) |
| E.coli UvrD (Helicase II) | 1.5 | Mono-/dimeric | 3′ → 5′ | DNA repair | (42) |
| R1 TraI | >10 | Monomeric | 5′ → 3′ | Conjugative DNA transfer | (43) |
aZiegelin and Lanka, unpublished data.
bCrystallized as monomer.
Using electron microscopy, we did not detect any influence of up to 70 µM myricetin on the global structures of hexameric proteins RepA, the true replicative E.coli helicase DnaB, phage P1 Ban and G40P (data not shown). Hence, the inhibitory effect of myricetin is not associated with the disruption of the hexamers.
DISCUSSION
In contrast to a variety of enzymes involved in the metabolism of nucleic acids, e.g. RNA polymerase or DNA gyrase, no easily available inhibitors for helicases are known. Such inhibitors would be helpful in understanding the mechanisms of unwinding. Furthermore, the rapid rise in the number of human-pathogenic bacteria, which have acquired resistance to all commonly used drugs, requires action to develop novel drugs. As helicases play essential roles in the metabolism of DNA and RNA and the replicative helicases of bacteria and eukaryotes differ substantially, helicase inhibitors may offer a feasible route towards this goal. Therefore, replicative prokaryotic DNA helicases constitute an attractive and yet unexplored target for development of new antibiotics as conditionally lethal DnaB mutants lead to stop of DNA synthesis and therefore unviable cells (23–25). As the three-dimensional structure of the smallest known replicative hexameric DNA helicase RepA of the broad host plasmid RSF1010 is the only one that has been determined from full-length subunits (14), it is an ideal tool to the study inhibition of helicases by natural compounds and their synthetically derived analogs using co-crystallization and X-ray analysis.
Flavones are naturally occurring polyphenolic compounds ubiquitously found in plants. They have been reported to possess widespread biological activities, are known as inhibitors for a variety of enzymes and inhibit growth of several cancerous cell lines (26–31). The cellular mechanisms underlying these anti-carcinogenic effects are still unclear but are thought to be linked to the inhibition of enzymes involved in the transduction of mitogenic signals (32).
The kinetic inhibition study of ssDNA-stimulated ATPase activity of RepA shows that the flavones inhibit RepA non-competitively with respect to ATP. As myricetin did neither disrupt RepA and other hexameric helicases tested, nor prevent hexamerization, we hypothesize that certain flavones may block binding of ssDNA to RepA rather than binding of ATP, thereby preventing RepA from translocation on ssDNA. This hypothesis is supported by our observation that myricetin caused little inhibition of the intrinsic RepA ATPase activity in the absence of ssDNA effector. The fact that flavones are known to bind weakly to ssDNA might also add to the inhibitory effect. Additionally, it is known that these compounds bind to dsDNA by intercalating due to the planar structure of the molecules (33). Such intercalation might be one reason for cytotoxicity (33). Therefore, chemical engineering might help to develop molecules with reduced ability for intercalation, but increased inhibition property. Furthermore, little is known whether intercalators stimulate or inhibit unwinding in terms of strand displacement by helicases.
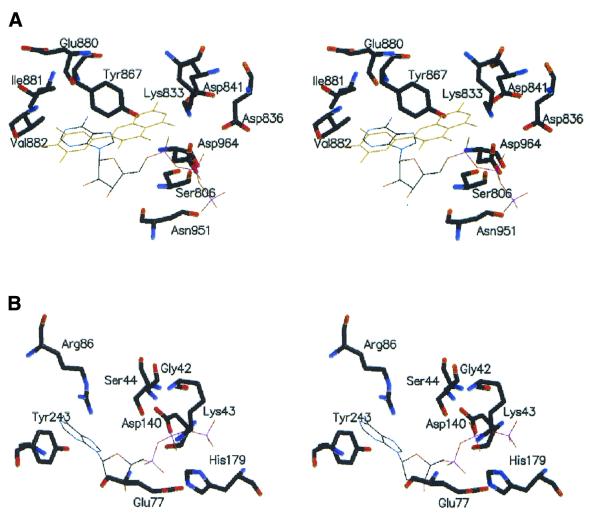
Walker et al. (33) recently published structural information on the inhibition of phosphoinositide 3-kinase (PI3K) by flavones and other inhibitors. In PI3K, myricetin and quercetin were found to be located in the ATP binding pocket. There is a salient difference, however, in the ATP active site of PI3K and RepA. In the ATP active site of PI3K, the kβ3-kβ4 loop 804MASKKKP810 (P loop) contains no glycine, and the side chain of Ser806 interacts with the β-phosphate and Lys833 at the end of β-strand kβ5 interacts with the α-phosphate of bound ATP. The loop between strands kβ7 and kβ8 forms the bottom of the ATP binding pocket and provides two hydrophobic contacts with the adenine moiety of ATP (34). In the complex formed between myricetin and PI3K, the phenyl ring of myricetin is partially overlapping and co-planar with the space occupied by the adenine moiety, and other flavones nearly occupy the ATP binding site in a comparable fashion. In contrast, the six ATP binding sites of RepA are each located at the interfaces between two adjacent monomers. They are defined by the consensus sequence for the P loop 40GAGKS44 and residues Asp140, Glu77, His179 (belonging to the same monomer) and Arg207 (from the adjacent monomer). Residues from the phosphate-binding loop (P loop), contact all three phosphates of ATP that was modeled into the structure of RepA. The adenine base is sandwiched between Arg86 of the same (in cis) and Tyr243 of the adjacent (in trans) monomer (12).
By superimposing the ATPase active sites of PI3K and RepA, we learned that the structure of the ATP binding motif of PI3K is rather different compared with that of the RepA helicase. The ATP binding site in RepA is much smaller than that in PI3K (Fig. 6) and would be unable to accommodate flavone compounds if no major structural rearrangements would occur. The flavones studied here appear to select a different way to regulate the RepA helicase function. This is further supported by the finding that wortmannin, an inhibitor which is covalently bound with high affinity at the ATP active site of PI3K (33), has nearly no inhibitory effect on RepA ATPase activity (data not shown).
Figure 6.
Comparison of the ATPase active site of PI3K and RepA. (A) The catalytic domain of PI3K with bound ATP and myricetin (36). (B) ATPase active site of RepA hexamer showing the position of ATP in the cleft between two neighboring RepA monomers. ATP was modeled into the RepA hexamer as described (14).
The target(s) of myricetin, the only one of the here investigated compounds that inhibited cell growth, may be localized within the cytoplasm or within the bacterial membrane. We suggest that myricetin inhibits at least one cytoplasmic enzyme essential for bacterial viability. One target may be a replicative helicase, as in vitro myricetin not only inhibited RepA helicase, but also a variety of helicases of different function and origin, including the phage P1 Ban protein which can functionally replace the E.coli DnaB replicative helicase (35). The specific sensitivity of each of these helicases towards myricetin suggests the design of molecules by molecular modeling with enhanced specificity for a given helicase once the site of binding is known by crystallographic studies. Of the compounds studied in vitro, dimyricetin was shown to be the most active molecule against RepA, but it did not inhibit bacterial growth, probably because dimyricetin does not cross the membrane.
Although the inhibition of RepA by luteolin, morin, myricetin and dimyricetin is only in the micromolar range, these compounds are advantageous as they are commercially available. The discovery of naturally occurring helicase inhibitors provides information regarding the type of small molecules that may be used to inhibit helicase action, and our results suggest that these compounds provide lead compounds for the design of novel drugs.
Work along these lines is in progress. As the flavone compounds are achiral, their binding affinity to RepA can certainly be enhanced by introducing chiral centers that augment their interaction with RepA. These studies will be guided by crystal structure analyses of complexes between RepA and the flavone compounds with the aim to synthesize better inhibitors with higher specificity. As the inhibition of helicase activity by these flavones is not restricted to RepA, further studies will provide insight in the mechanism of inhibition and help to clarify in general the DNA unwinding reaction by helicases.
Acknowledgments
ACKNOWLEDGEMENTS
We thank T. M. Lohman and D. Wigley for providing Rep, UvrD and PcrA DNA heicases. Work in our laboratories was supported by the European Commission grant QLK2-CT-2000-00634 to J.C.A., E.L. and W.S., by DFG-Sonderforschungsbereich 344, DFG-grant Sa196/38-1. and by Fonds der Chemischen Industrie to W.S.
REFERENCES
- 1.Matson S.W. and Kaiser-Rogers,K.A. (1990) DNA helicases. Annu. Rev. Biochem., 59, 289–329. [DOI] [PubMed] [Google Scholar]
- 2.Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- 3.Hanawalt P.C. (1994) Transcription-coupled repair and human disease. Science, 266, 1957–1958. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A. (1994) Mechanisms of DNA excision repair. Science, 266, 1954–1957. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg E.C. (1992) Xeroderma pigmentosum, Cockayne’s syndrome, helicases, and DNA repair: what’s the relationship? Cell, 71, 887–889. [DOI] [PubMed] [Google Scholar]
- 6.Yu C.-E., Oshima,J., Fu,Y.-H., Wijsman,E.M., Hisama,F., Alisch,R., Mattews,S., Nakura,J., Miki,T., Ouais,S., Martin,G.M., Mulligan,J. and Schellenberg,G.D. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 7.Egelman E.H. (1996) Homomorphous hexameric helicases: tales from the ring cycle. Structure, 4, 759–762. [DOI] [PubMed] [Google Scholar]
- 8.Bowles J.T. (1998) The evolution of aging: a new approach to an old problem of biology. Med. Hypotheses, 51, 179–221. [DOI] [PubMed] [Google Scholar]
- 9.Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- 10.Marians K.J. (2000) Crawling and wiggling on DNA: structural insights to the mechanism of DNA unwinding by helicases. Struct. Fold Des., 8, 227–235. [DOI] [PubMed] [Google Scholar]
- 11.Chino M., Nishikawa,K., Umekita,M., Hayashi,C., Yakako,Y., Tsuchida,T. and Sawa,T. (1996) Heliquinomycin, a new inhibitor of DNA helicase, produced by Streptomyces sp. MJ929-SF2. J. Antibiot., 49, 752–757. [DOI] [PubMed] [Google Scholar]
- 12.Scherzinger E., Ziegelin,G., Barcena,M., Carazo,J.M., Lurz,R. and Lanka,E. (1997) The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J. Biol. Chem., 272, 30228–30236. [DOI] [PubMed] [Google Scholar]
- 13.Xu H., Frank,J., Niedenzu,T. and Saenger,W. (2000) DNA helicase RepA: cooperative ATPase activity and binding of nucleotides. Biochemistry, 39, 12225–12233. [DOI] [PubMed] [Google Scholar]
- 14.Niedenzu T., Röleke,D., Bains,G., Scherzinger,E. and Saenger,W. (2001) Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol., 306, 479–487. [DOI] [PubMed] [Google Scholar]
- 15.Scherzinger E., Haring,V., Lurz,R. and Otto,S. (1991) Plasmid RSF1010 DNA replication in vitro promoted by purified RSF1010 RepA, RepB and RepC proteins. Nucleic Acids Res., 19, 1203–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang B. (1971) Inaugural-Dissertation zur Erlangung der Doktorwürde im Fachbereich Pharmazie der Freien Universität Berlin.
- 17.Röleke D., Hoier,H., Bartsch,C., Umbach,P., Scherzinger,E., Lurz,R. and Saenger,W. (1997) Crystallization and preliminary X-ray crystallographic and electron microscopic study of a bcterial DNA helicase (RSF1010 RepA). Acta Crystallogr. Sec. D, 53, 213–216. [DOI] [PubMed] [Google Scholar]
- 18.Stryer L. (1975) Biochemistry. W.H. Freeman and Company, San Francisco, CA. [Google Scholar]
- 19.Crute J.J., Mocarski,E.S. and Lehman,I.R. (1988) A DNA helicase induced by herpes simplex virus type 1. Nucleic Acids Res., 16, 6585–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nester E.W. and Lederberg,J. (1961) Linkage of genetic units of Bacillus subtilis in DNA transformation. Proc. Natl Acad. Sci. USA, 47, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H., Frank,J., Holzwarth,J.F., Saenger,W. and Behlke,J. (2000) Interaction of different oligomeric states of hexameric DNA-helicase RepA with single-stranded DNA studied by analytical ultracentrifugation. FEBS Lett., 482, 180–184. [DOI] [PubMed] [Google Scholar]
- 22.Xu H., Frank,J., Trier,U., Hammer,S., Schröder,W., Behlke,J., Schäfer-Korting,M., Holzwarth,J.F. and Saenger,W. (2001) Interaction of fluorescence labeled single-stranded DNA with hexameric DNA-helicase RepA: a photon and fluorescence correlation spectroscopy study. Biochemistry, 40, 7211–7218. [DOI] [PubMed] [Google Scholar]
- 23.Bonhoeffer F. (1966) DNA transfer and DNA synthesis during bacterial conjugation. Z. Vererbungls., 98, 141–149. [DOI] [PubMed] [Google Scholar]
- 24.Carl P.L. (1970) Escherichia coli mutants with temperature-sensitive synthesis of DNA. Mol. Gen. Genet., 109, 107–122. [DOI] [PubMed] [Google Scholar]
- 25.Wechsler J.A. and Gross,J.D. (1971) Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol. Gen. Genet., 113, 273–284. [DOI] [PubMed] [Google Scholar]
- 26.Bormann H. and Melzig,M.F. (2000) A DNA helicase induced by herpes simplex virus type 1. Pharmazie, 55, 129–132.10723772 [Google Scholar]
- 27.Robak J., Shridi,F., Wolbis,M. and Krolikowska,M. (1988) Screening of the influence of flavonoids on lipoxygenase and cyclooxygenase activity, as well as on nonenzymic lipid oxidation. Pol. J. Pharmacol. Pharm., 40, 451–458. [PubMed] [Google Scholar]
- 28.Hagiwara M., Inoue,S., Tanaka,T., Nunoki,K., Ito,M. and Hidaka,H. (1988) Differential effects of flavonoids as inhibitors of tyrosine protein kinases and serine/threonine protein kinases. Biochem. Pharmacol., 37, 2987–2992. [DOI] [PubMed] [Google Scholar]
- 29.Shinozuka K., Kikuchi,Y., Nishino,C., Mori,A. and Tawata,S. (1988) Inhibitory effect of flavonoids on DNA-dependent DNA and RNA polymerases. Experientia, 44, 882–885. [DOI] [PubMed] [Google Scholar]
- 30.Ono K., Nakane,H., Fukushima,M., Chermann,J.C. and Barre-Sinoussi,F. (1990) Differential inhibitory effects of various flavonoids on the activities of reverse transcriptase and cellular DNA and RNA polymerases. Eur. J. Biochem., 190, 469–476. [DOI] [PubMed] [Google Scholar]
- 31.Gamet-Payrastre L., Manenti,S., Gratacap,M.-P., Tulliez,J., Chap,H. and Payrastre,B. (1999) Flavonoids and the inhibition of PKC and PI 3-kinase. Gen. Pharmacol., 53, 1649–11657. [DOI] [PubMed] [Google Scholar]
- 32.Middleton E.,Jr and Kandaswami,C. (1993) The impact of plant flavonoids and mammalian biology: implications for immunity, inflammation and cancer. In Harborne,J.H. and Liss,A.R. (eds), The Flavonoids: Advances in Research Since 1986. Chapman and Hall, London, UK, pp. 619–652.
- 33.Austin C.A., Patel,S., Ono,K., Nakane,H. and Fisher,L.M. (1992) Site-specific DNA cleavage by mammalian DNA topoisomerase II induced by novel flavone and catechin derivatives. Biochem. J., 282, 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker E.H., Perisic,O., Ried,C., Stephens,L. and Willams,R.L. (1999) Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature, 402, 313–320. [DOI] [PubMed] [Google Scholar]
- 35.Lanka E., Mikolajczyk,M., Schlicht,M. and Schuster,H. (1978) Association of the prophage P1 Ban protein with the DnaB protein. J. Biol. Chem., 253, 4746–4753. [PubMed] [Google Scholar]
- 36.Walker E.H., Pacold,M.E., Perisic,O., Stephens,L. and Hawkins,P.T. (2000) Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin and staurosporine. Mol. Cell, 6, 909–919. [DOI] [PubMed] [Google Scholar]
- 37.Bárcena M., Martin,C.S., Weise,F., Ayora,S., Alonso,J.C. and Carazo,J.M. (1998) Polymorphic quarternary organization of the Bacillus subtilis bacteriophage SPP1 replicative helicase (G40 P). J. Mol. Biol., 283, 809–819. [DOI] [PubMed] [Google Scholar]
- 38.Ziegelin G., Scherzinger,E., Lurz,R. and Lanka,E. (1993) Phage P4 α protein is multifunctional with origin recognition, helicase and primase activities. EMBO J., 12, 3703–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanya H.S., Bird,L.E., Brannigan,J.A. and Wigley,D.B. (1996) Crystal structure of a DExx box DNA helicase. Nature, 384, 379–383. [DOI] [PubMed] [Google Scholar]
- 40.Chao K. and Lohman,T.M. (1991) DNA-induced dimerization of the Escherichia coli Rep helicase. J. Mol. Biol., 221, 1165–1182. [DOI] [PubMed] [Google Scholar]
- 41.Korolev S., Hsieh,J., Gauss,G.H., Lohman,T.M. and Waksman,G. (1997) Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell, 90, 635–647. [DOI] [PubMed] [Google Scholar]
- 42.Runyon G.T., Wong,I. and Lohman,T.M. (1993) Overexpression, purification, DNA-binding, and dimerization of the Escherichia coli uvrD gene product (helicase II). Biochemistry, 32, 602–612. [DOI] [PubMed] [Google Scholar]
- 43.Lahue E.L. and Matson,S.W. (1988) Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction. J. Biol. Chem., 263, 3208–3215. [PubMed] [Google Scholar]