Abstract
Objectives. To compare 4 COVID-19 surveillance metrics in a major metropolitan area.
Methods. We analyzed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater influent and primary solids in Raleigh, North Carolina, from April 10 through December 13, 2020. We compared wastewater results with lab-confirmed COVID-19 cases and syndromic COVID-like illness (CLI) cases to answer 3 questions: (1) Did they correlate? (2) What was the temporal alignment of the different surveillance systems? (3) Did periods of significant change (i.e., trends) align?
Results. In the Raleigh sewershed, wastewater influent, wastewater primary solids, lab-confirmed cases, and CLI were strongly or moderately correlated. Trends in lab-confirmed cases and wastewater influent were observed earlier, followed by CLI and, lastly, wastewater primary solids. All 4 metrics showed sustained increases in COVID-19 in June, July, and November 2020 and sustained decreases in August and September 2020.
Conclusions. In a major metropolitan area in 2020, the timing of and trends in municipal wastewater, lab-confirmed case, and syndromic case surveillance of COVID-19 were in general agreement.
Public Health Implications. Our results provide evidence for investment in SARS-CoV-2 wastewater and CLI surveillance to complement information provided through lab-confirmed cases. (Am J Public Health. 2023;113(1):79–88. https://doi.org/10.2105/AJPH.2022.307108)
COVID-19 public health surveillance relies on multiple data sources to estimate disease burden. The number of positive clinical tests over time has served as a primary metric for tracking COVID-19 infections in North Carolina because clinical testing of individuals accurately identifies cases and is legally required for surveillance of reportable diseases, including COVID-19.1 Clinical testing is, however, costly and inefficient as a means of population-level surveillance of COVID-19.2 In addition, this metric can be limited by sensitivity,3 clinical test availability,4 and changes in testing behavior such as the rise in use of nonreportable, at-home rapid test kits.5
Surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in wastewater influent or settled wastewater solids has gained traction in public health practice.6 In addition to capturing data on symptomatic individuals who are likely to be tested, wastewater surveillance captures information on infections among asymptomatic carriers who shed the virus in feces but are less likely to be tested (Figure 1). In retrospective studies, SARS-CoV-2 RNA concentrations in wastewater have been shown to correlate positively with reported clinical COVID-19 cases.7,8 Public health officials have used wastewater surveillance trends to target public health mitigation efforts.9 Most wastewater surveillance is conducted using centralized wastewater treatment systems; wastewater surveillance is not as efficient in communities with a high proportion of people dependent on individual septic systems.
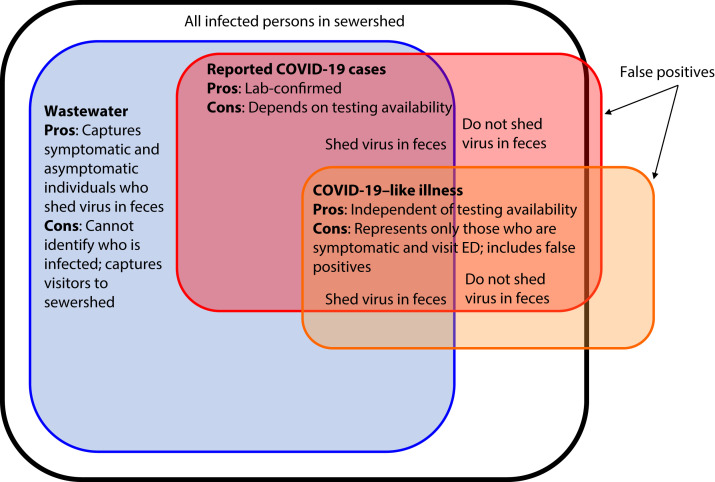
FIGURE 1—
Depiction of Populations Captured by COVID-19 Surveillance Systems
Note. ED = emergency department; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2. The black box contains all SARS-CoV-2 infections in the sewershed. Infected individuals who shed the virus in feces contribute to SARS-CoV-2 RNA levels in wastewater (blue). Data on individuals seeking diagnostic testing are captured through reportable communicable disease surveillance (red). Data on individuals exhibiting COVID-like illness (CLI) at an ED are captured through ED syndromic surveillance (orange). False positives are shown outside the black box (orange for CLI and red for diagnostic testing). All reported cases are estimates of true cases. Individuals who do not seek testing, visit an ED, or shed the virus in feces are not captured by these surveillance systems (i.e., the white space inside the black box).
Another form of surveillance used for COVID-19 response is syndromic surveillance for COVID-like illness (CLI) based on prediagnostic emergency department (ED) data not confirmed through laboratory testing. CLI captures data on individuals with serious illness and those seeking care at EDs, representing a smaller segment of the infected population. Syndromic surveillance is mandated in North Carolina10 and is routinely used for other respiratory conditions, including influenza.
Given the different segments of the population captured via wastewater, lab-confirmed case, and CLI surveillance (Figure 1), it is important to evaluate how these surveillance systems compare in a given population. Wastewater may provide more sensitive surveillance of changing infection rates in areas where there is incomplete ascertainment of cases through clinical testing.11 Increases in wastewater concentrations have sometimes preceded increases in clinical cases.12 CLI surveillance based on ED data is unlikely to be more timely than lab-confirmed case surveillance, but it may be nearly as timely. With electronic health information systems, data on CLI ascertained at EDs can be available in near real time,13 whereas laboratory testing can entail delays from sample collection to results reporting.
We compared COVID-19 surveillance data sets from a major metropolitan area and included 2 wastewater metrics. We analyzed SARS-CoV-2 RNA concentrations in wastewater influent and primary solids from a municipal wastewater treatment plant in Raleigh, NC. Subsequently, we compared wastewater levels with lab-confirmed COVID-19 and CLI counts for the sewershed to answer 3 questions: (1) Did they correlate? (2) What was the temporal alignment of the different surveillance systems? (3) Did trends (i.e., periods of significant increases or decreases) align across surveillance systems? This research can inform how public health officials look across surveillance systems to estimate COVID-19 burdens.
METHODS
Raw wastewater influent and primary clarifier solids (i.e., primary solids) were sampled from the Neuse River Resource Recovery Facility in Raleigh between April 10 and December 13, 2020. We collected 24-hour composite influent wastewater samples (100 or 500 mL) and grab samples of solids (40 mL) 2 or 3 times weekly, with some periods of daily sampling (102 dates in total; Figure A, available as a supplement to the online version of this article at https://ajph.org). This facility serves approximately 580 000 people and had average treated flows of 48 million gallons per day in 2020. Solids collected from primary clarifiers were predominantly influent solids, but waste-activated solids were also present because the facility co-settles waste-activated solids in its primary clarifiers. Although co-settling waste-activated solids in primary clarifiers is not a common wastewater treatment practice, it is a recognized practice for improved sludge thickening.14,15 The residence time of solids in the clarifiers was, on average, 2.8 days (range = 1.8–4.3 days), which is longer than typical primary clarifier residence times (on the order of hours).
Concentrations of SARS-CoV-2 N1 and N2 genes in wastewater samples were determined via reverse-transcription-droplet digital polymerase chain reaction (see Supporting Information, available as a supplement to the online version of this article at https://ajph.org). Wastewater sample processing protocols, depicted in Figure B (influent; available as a supplement to the online version of this article at https://ajph.org) and Figure C (primary solids; available as a supplement to the online version of this article at https://ajph.org), incorporated several of the current best practices.16 Normalized N1 results (Supporting Information) were used in subsequent analyses with lab-confirmed cases and CLI.
Lab-Confirmed COVID-19 Case Data
Individual-level lab-confirmed COVID-19 cases with residential addresses from the North Carolina Electronic Disease Surveillance System were provided by the North Carolina Department of Health and Human Services. Positive case counts included polymerase chain reaction–positive tests, antigen-positive tests, and a few polymerase chain reaction–negative tests determined to be positive cases based on physician case notes. Cleaned residential addresses were geocoded in ArcGIS Pro version 2.7.0 (ESRI, Redlands, CA) via the 2018 ESRI Business Analyst USA_LocalComposite locator (Supporting Information). As a means of producing daily case counts, we summed cases in the sewershed using specimen collection dates or test result report dates.
COVID-Like Illness Data
Data on individual-level CLI cases geocoded at the residential zip code level were acquired from the North Carolina Disease Event Tracking and Epidemiologic Collection Tool, a public health syndromic surveillance system capturing all civilian ED visits in North Carolina (as reporting is mandatory).13 CLI ascertained at urgent care centers was not included because NC does not share these data with external researchers.
CLI was defined according to International Statistical Classification of Diseases and Related Health Problems, 10th Revision (ICD-10; Geneva, Switzerland: World Health Organization; 1992) diagnostic codes (B97.2 or B34.2, J12.81 or J12.82, or U07.1 or U07.217) or 1 of the following conditions: a chief complaint related to coronavirus, triage notes indicating a loss of sense of taste or smell, or triage notes indicating shortness of breath with fever. CLI cases that also had diagnostic codes for influenza (J09–J11.89) were excluded unless they had 1 of the ICD-10 inclusion codes. The date for each CLI record was the ED visit date. We estimated daily CLI counts in the sewershed by summing counts in each of the 27 zip codes located entirely or partially in the sewershed, weighted by population density according to 2010 census block data.
Correlation Analysis
We used Spearman’s rank correlation to determine the relationship between wastewater SARS-CoV-2 N1 concentrations (in influent or primary solids) and lab-confirmed COVID-19 cases or CLI. To investigate temporal alignment, we compared correlation coefficients and identified the maximum coefficient as 1 data set was offset forward or backward in time relative to another data set.18–22 To reduce variation in the measurements for this analysis, we used the rolling 3-sampling-event averages of normalized SARS-CoV-2 quantities in wastewater influent and primary solids and the rolling 7-day averages for lab-confirmed cases and CLI. We used 2000 resamples with replacement to calculate bootstrap 95% confidence intervals for the correlation coefficient at each lead or lag and for all pairwise differences between correlations.23 Correlation pairs were considered significantly different if the Bonferroni-adjusted 95% confidence interval for their difference excluded 0.24
Distributed Lag Model
The distributed lag measurement error time series model is an accepted epidemiological model for time series data.25 We adapted a Bayesian distributed lag model developed previously8 as a secondary approach to investigate temporal alignment between SARS-CoV-2 RNA levels in wastewater influent or primary solids and changes in clinical case rates. The 3-day rolling average of clinical cases was predicted via wastewater measurements from 3 sampling events before the report date until 3 sampling events after. A random effect was included in the model to account for overdispersion.
Trends
Trends were classified as increasing, decreasing, or plateau through a linear regression with observations from each surveillance system as the dependent variable and date as the independent variable; trend classification was based on slope (positive, negative, or 0) and statistical significance (P < .05).6 We classified short-term and sustained trends using regressions of 3 data points (approximately 1 week in duration) and 7 data points (approximately 2 weeks), respectively.26
RESULTS
SARS-CoV-2 RNA was frequently detected in wastewater influent and solids during the 247-day study period (April 10 to Dec 13, 2020); influent samples had detectable levels of the SARS-CoV-2 N1 gene on 96 of 102 wastewater sampling dates (94%); solids samples had detectable N1 on all 102 days (Figure B). The SARS-CoV-2 N2 gene was detectable in influent on 94 of 102 days (92%) and solids on 100 of 102 days (98%). Because N1 and N2 gene concentrations were highly correlated in influent (Spearman ρ = 0.83; P < .001) and solids (ρ = 0.93, P < .001) and N1 had a slightly higher detection rate, we focused our subsequent analyses on N1.
Surveillance Data Set Correlations
SARS-CoV-2 RNA daily loads in influent and SARS-CoV-2 RNA concentrations in primary solids (Figure 2) were moderately correlated over the study period (ρ = 0.65; P < .001; C). Wastewater influent was strongly correlated with lab-confirmed cases (ρ = 0.74; P < .001; Figure 2; Figure D, available as a supplement to the online version of this article at https://ajph.org), as were wastewater primary solids (ρ = 0.71; P < .001; Figure E and Table A, available as supplements to the online version of this article at https://ajph.org). Furthermore, wastewater influent was moderately correlated with CLI (ρ = 0.61; P < .001; Figure 2; Figure F, available as a supplement to the online version of this article at https://ajph.org), whereas solids were strongly correlated with CLI (ρ = 0.71; P < .001; Figure G, available as a supplement to the online version of this article at https://ajph.org).
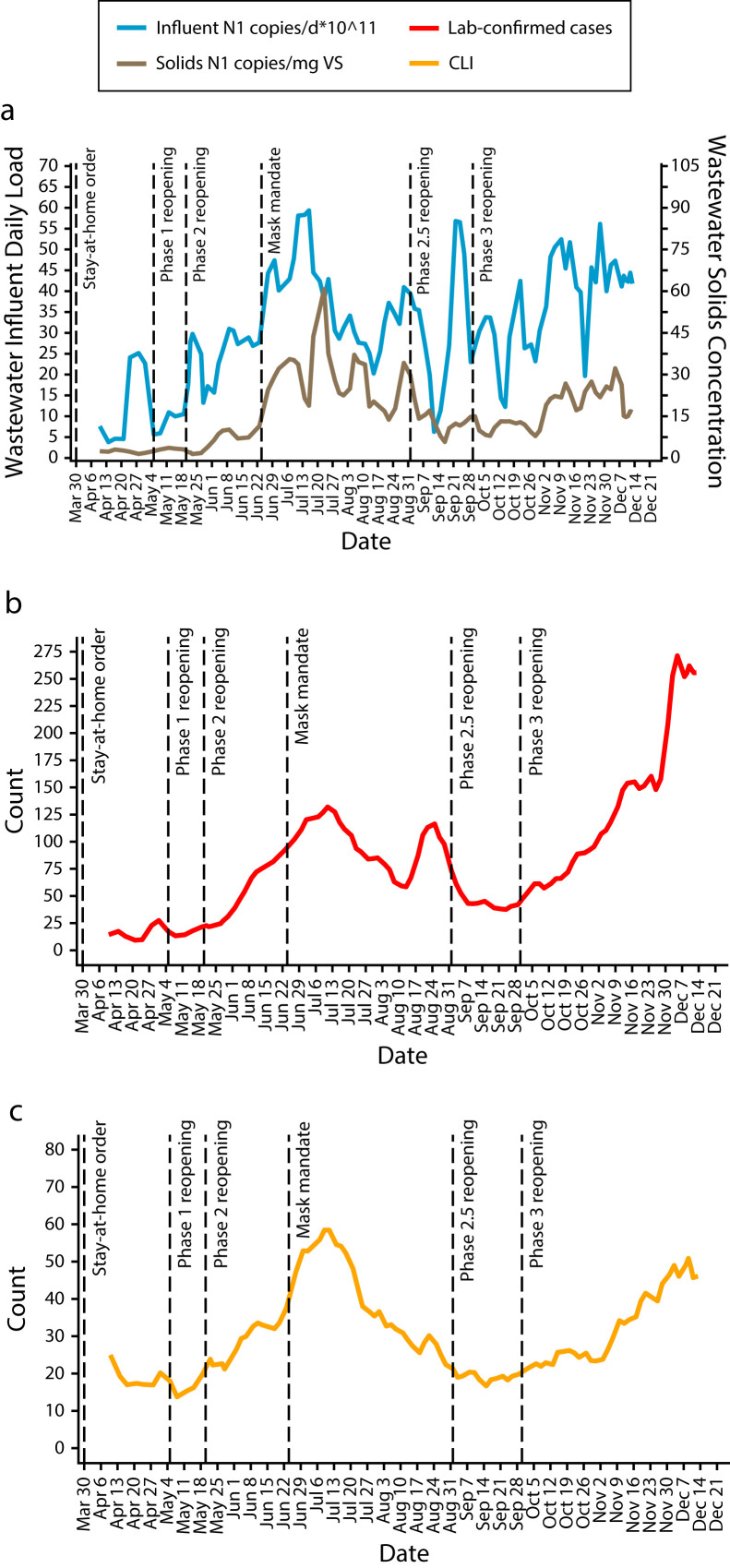
FIGURE 2—
COVID-19 Surveillance Time Series for (a) Wastewater Influent and Primary Solids, (b) Lab-Confirmed Cases, and (c) CLI Cases: Raleigh, NC, Sewershed, April 10–December 13, 2020
Note. CLI = COVID-like illness. The wastewater influent and primary solids in panel a are 3-sampling-event averages (mean ±SD duration = 5.7 ±1.2 days). Lab-confirmed (panel b) and CLI (panel c) cases are 7-day averages of daily counts. Dotted lines indicate dates of North Carolina executive orders. The specimen collection date was used for cases and the date of emergency department visit for CLI.
The strongest correlation observed was between lab-confirmed cases and CLI (ρ = 0.84; P < .001; Figure H, available as a supplement to the online version of this article at https://ajph.org); during the study period, there were 20 858 lab-confirmed COVID-19 cases and 7441 cases of CLI in the sewershed. Lab-confirmed cases and CLI were highly correlated in earlier and later portions of the study period (Table B, available as a supplement to the online version of this article at https://ajph.org). The earlier portion (April 10 through August 13, 2020) captured the first rise and fall of infections and was characterized by lower testing penetration4 and fewer ED visits (Figure I, available as a supplement to the online version of this article at https://ajph.org). The correlations between cases of CLI and wastewater (influent and primary solids) were substantially higher earlier in the study period (Table B).
Temporal Comparisons
The strongest correlation between SARS-CoV-2 N1 daily load in wastewater influent and N1 concentrations in wastewater primary solids was found for solids samples collected 2 sampling events after influent (given our sampling frequency, 2 sampling events represented 5.9 ±1.2 days; ρ = 0.65; Figure J, available as a supplement to the online version of this article at https://ajph.org).
Correlations between lab-confirmed cases and wastewater influent daily load increased slightly as cases were offset from 0 to 3 days ahead of the influent sample collection date, with the strongest correlation observed for case specimens collected 3 days before an influent sample (ρ = 0.75; Figure 3). The median duration between specimen collection date and results report date was 1 day (5th–95th percentiles: 0–4 days). When report date for case results was used instead of specimen collection date, the strongest correlation between cases and wastewater influent was observed for cases reported on the same day that influent was sampled (i.e., day 0; ρ = 0.75). For wastewater primary solids, correlations between solids concentrations and lab-confirmed cases increased gradually as cases were offset 0 days to 7 days ahead of solids, with the strongest correlation found for case specimens collected 7 days before a solids sample (ρ = 0.80). This correlation was significantly higher than correlations for case specimens collected 1 to 7 days after a solids sample.
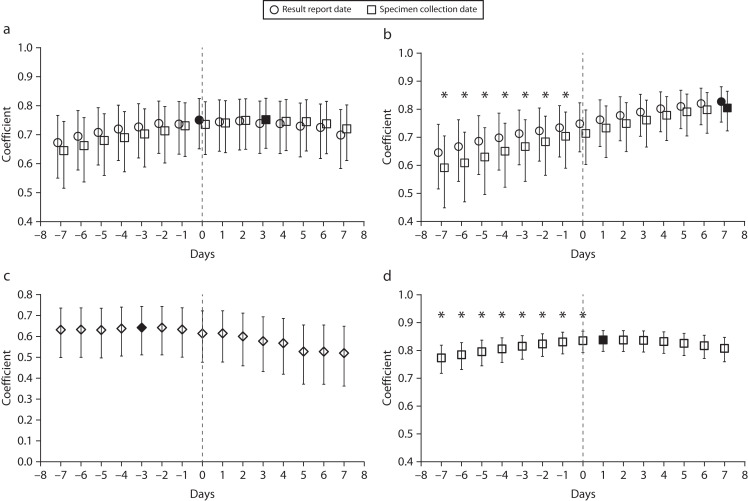
FIGURE 3—
Spearman’s Rank Correlation Coefficients for Associations Between COVID-19 Surveillance Data Sets Offset Forward or Backward in Time for (a) Lab-Confirmed Cases Offset Relative to Wastewater Influent, (b) Lab-Confirmed Cases Offset Relative to Wastewater Solids, (c) CLI Offset Relative to Wastewater Influent, and (d) Lab-Confirmed Cases Offset Relative to CLI: Raleigh, NC, Sewershed, April 10–December 13, 2020
Note. CLI = COVID-like illness. Filled-in markers indicate maximum coefficients. Asterisks indicate coefficients significantly different from the maximum after Bonferroni adjustment (the specimen collection date was used for case significance testing) according to bootstrap analyses of the distribution of coefficients (P < .05).
The strongest correlation between CLI and wastewater influent was found for CLI reported 3 days after an influent sample was collected (ρ = 0.64; Figure 3). The strongest correlation between lab-confirmed cases and CLI was found for clinical case specimens collected 1 day before the ED visit date (ρ = 0.84). This correlation was significantly higher than correlations for case specimens collected on the same day or up to 7 days after the ED visit date. Correlations were generally similar but slightly weaker for the surrounding days. Distributed lag modeling results were consistent with the correlation analysis with date offsets: wastewater influent and primary solids lagged clinical cases based on case specimen collection date (Table C, available as a supplement to the online version of this article at https://ajph.org).
Numbers of Significant Trends
Public health officials monitor for significant changes in levels of COVID-19 surveillance metrics to inform public health action.27 Short-term or weekly trend monitoring is valuable because a short-term trend can be an early indicator of a sustained trend and because, particularly at the start of the pandemic, public health officials acted as quickly as possible. Across the different surveillance data sets, we might expect the numbers of trends to be similar but the temporal alignment to be shifted. However, lab-confirmed cases exhibited substantially more short-term increases (n = 17) than CLI (n = 10), wastewater primary solids (n = 7), and wastewater influent (n = 4) over the study period. Lab-confirmed cases had a number of periods of sustained increases (n = 51) similar to that of CLI (n = 45; within 20% of each other), but wastewater primary solids (n = 21) and wastewater influent had substantially fewer (n = 20).
In terms of periods of decreasing levels of COVID-19 metrics, the numbers of short-term decreases were greatest for lab-confirmed cases (n = 8) and wastewater solids (n = 7), followed by CLI (n = 5) and wastewater influent (n = 1). Furthermore, the numbers of sustained decreases in CLI (n = 23) and cases (n = 21) were similar, whereas there were fewer decreases among wastewater solids (n = 15) and wastewater influent (n = 9).
Trend Agreement Across Surveillance Data Sets
There were 9 periods for which all data sets agreed with respect to classification of sustained trends: the periods ending June 11, July 2, July 7, July 9, July 11, and November 14 exhibited increasing trends, and the periods ending August 1, September 12, and September 15 exhibited decreasing trends (Figure 4). Not surprisingly, there were no short-term trends that agreed across the 4 surveillance data sets given the temporal shifting of the different surveillance metrics. The wastewater influent and primary solids data sets were in similar agreement with respect to sustained increases when each were compared with lab-confirmed cases.
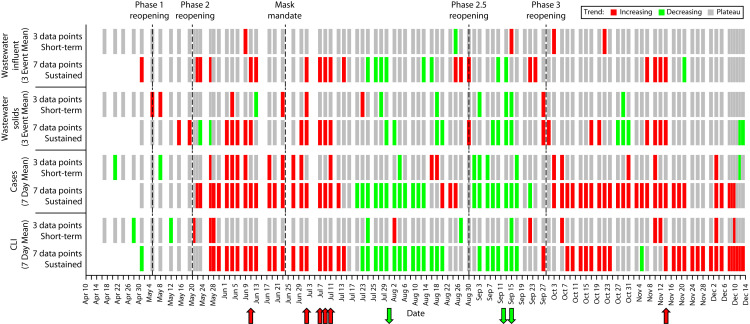
FIGURE 4—
Linear Regression Rolling Trend Classifications Illustrating Short-Term (Approximately 1 Week) and Sustained (Approximately 2 Weeks) Trends: Raleigh, NC, Sewershed, April 10–December 13, 2020
Note. CLI = COVID-like illness. Data were smoothed via rolling 7-day averages for cases and CLI and via 3-sampling-event averages for wastewater influent and solids. Statistically significant (P < .05) trends are shown in red (increasing) or green (decreasing). Color is placed on the last day of the 3- or 7-point period used in the regression. For lab-confirmed cases, specimen collection date was used. Arrows indicate periods of sustained trends for which there was agreement across the 4 surveillance data sets: the periods ending June 11, July 2, July 7, July 9, July 11, and November 14 (increases; red arrows) and the periods ending August 1, September 12, and September 15 (decreases; green arrows).
Specifically, 14 of 51 (27%) increases in cases were also increases in influent data; 16 of the 51 (31%) were increases in solids data. Five of 17 (29%) decreasing trends in case data were decreasing trends according to influent data, whereas 3 (18%) were decreases according to solids data. There was better agreement between lab-confirmed case and CLI data in sustained increases and decreases. Thirty-six of 51 (71%) sustained increases in case data were also sustained increases in CLI data, and 18 of 21 (86%) decreases in case data were also decreases in CLI data.
DISCUSSION
On the day wastewater sample collection began (April 10, 2020), there had been 206 cumulative cases and 41 new cases reported in the sewershed, although the true number of infections is unknown (Figure 1). The Raleigh sewershed had detectable levels of SARS-CoV-2 RNA in primary solids in early April, 1 month after the first lab-confirmed COVID-19 case was reported in the sewershed (March 9, 2020). Detection frequency across solids samples was high, as others have reported,28 despite the fact that primary solids in the Raleigh system also contained waste-activated solids. Monitoring wastewater primary solids was marginally more sensitive than monitoring influent.
SARS-CoV-2 RNA levels in wastewater influent were highly correlated with lab-confirmed cases, as has been reported for other wastewater–case comparisons.7,20 Despite the longer solids residence time in primary clarifiers, RNA concentrations in primary solids were also correlated highly with lab-confirmed cases, as others have reported.7,28,29
CLI being correlated with both measures of wastewater surveillance is notable given that CLI–wastewater agreement has not been widely investigated. Case or CLI correlations with wastewater becoming substantially lower later in the study period (Table B) may have been related to increasing noise in the wastewater signal. As the pandemic progressed, increases in wastewater RNA concentrations from new COVID-19 infections would have occurred in the presence of RNA contributed by individuals who were no longer test positive but continued to shed RNA in feces30 and residual RNA in the wastewater system.31,32 In addition, more individuals may have traveled in and out of the sewershed after reopening of public facilities, contributing to greater measurement error in COVID-19 burden based on wastewater.
The strongest correlations observed were between lab-confirmed cases and CLI, even early in the pandemic when there was limited test access and fewer ED visits. Although fewer ED visits would have limited the sensitivity of CLI surveillance for ascertaining infections, CLI may still have strongly correlated with lab-confirmed cases because of a larger overlap in the populations captured by diagnostic testing and CLI surveillance systems. Early in the pandemic, more testing may have been done on individuals who had severe COVID-19 and went to the ED. Noteworthy differences in case and CLI time series occurred later in the study period. A prominent peak in lab-confirmed cases in late August 2020 was not as pronounced in CLI data.
Furthermore, the extent to which cases in December 2020 exceeded previous case peaks in July and August 2020 was not represented in the other surveillance data sets and may reflect increased test access or increased testing around the winter holidays.4 Widespread COVID-19 vaccinations or a change in the predominant SARS-CoV-2 variant may affect correlations between different surveillance systems if, for example, the asymptomatic rate increases33 or the fecal shedding profile is altered.18 With COVID-19 vaccines now being widely available, wastewater surveillance can be used to identify locations where viral fecal shedding into wastewater is not declining, indicating specific locations of infection.34
Multiple surveillance metrics are used in real time by public health officials to provide a fuller COVID-19 public health picture. As such, a temporal comparison is important for the interpretation of agreement or disagreement across surveillance data sets. The reported lead time for wastewater has ranged from 0 to 2 days8,29 to as high as 235 and 336,37 weeks. In the Raleigh sewershed, trends in wastewater influent were observed earlier than lab-confirmed case trends when case results report date was used but not when specimen collection date was used, underlining that the potential for wastewater surveillance to provide an earlier warning than clinical testing depends on when test results are reported.38 The current increased availability of testing and faster turnaround times for case reporting and wastewater surveillance38 relative to the study period in 2020 may further impact temporal alignment.
Wastewater solids lagged wastewater influent likely because of long solids storage times in Neuse River Resource Recovery Facility primary clarifiers. Wastewater treatment plant design and operation is aimed at wastewater conveyance and treatment and, as such, may not provide ideal conditions for COVID-19 public health surveillance.39 Therefore, in interpreting wastewater surveillance results, the operation of the facility (with increased communication between plant operators and public health agencies) must be considered.40 Although the maximum correlation coefficient indicated that rises in CLI were a day behind rises in lab-confirmed cases, syndromic surveillance can be more timely than clinical case surveillance depending on how syndromic data are captured.41
The greater numbers of significant trends in lab-confirmed case and CLI metrics than with wastewater metrics indicated a need for public health action at times when wastewater surveillance data did not exhibit a significant change. A limiting factor for numbers of significant trends in wastewater data sets was the 95% statistical confidence requirement for trend classification, which the Centers for Disease Control and Prevention originally recommended but no longer strictly recommends for wastewater surveillance trend reporting.42 SARS-CoV-2 RNA levels in wastewater primary solids may have had less variability than influent levels as evidenced by the minimal increase in correlation between solids and rolling 7-day average of cases when crude solids data were smoothed (Table A). Therefore, solids surveillance was able to meet the statistical confidence requirement more often than influent surveillance.
PUBLIC HEALTH IMPLICATIONS
We captured COVID-19 dynamics in a major metropolitan area during the first and second waves of infections in 2020. To our knowledge, our study is the first to report agreement between CLI and wastewater surveillance and to demonstrate relationships between key COVID-19 metrics in NC.43 This study from early in the COVID-19 pandemic, when reportable testing data were better correlated with true disease incidence, supports the use of wastewater and CLI surveillance to complement lab-confirmed case surveillance, especially at times when clinical test penetration is low.
ACKNOWLEDGMENTS
This work was supported by National Science Foundation Rapid Response Research grant CBET- 2029025, the North Carolina Policy Collaboratory, and North Carolina State Center for Human Health and the Environment grant P30ES025128. David A. Holcomb was funded in part by the National Institute of Environmental Health Sciences (grant T32ES007018).
We thank Nathan Howell, Jeremy Lowe, Daniel Cockson, Emma Bolden, Victoria Ponthier, Zach Bennett, Laura Gomez, Ramya Balasubramanian, Marc Serre, Kelly Hoffman, Jeseth Delgado Vela, Adam Smith, Lauren Stadler, Krista Wigginton, Eric Johnson, and Aaron Fleischauer for their contributions to this work.
CONFLICTS OF INTEREST
There are no known potential or actual conflicts of interest.
HUMAN PARTICIPANT PROTECTION
The use of North Carolina Department of Health and Human Services health tracking data was approved by the University of North Carolina institutional review board. Because this was a secondary analysis of de-identified data collected for administrative purposes, it was not necessary or possible to obtain informed consent.
See also Keck and Berry, p. 6.
REFERENCES
- 1.North Carolina Department of Health and Human Services. NCDHHS communicable disease surveillance and reporting. 2021. https://epi.dph.ncdhhs.gov/cd/report.html
- 2.Hart OE, Halden RU. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci Total Environ. 2020;730:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogels CB, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. Previous U.S. viral testing data. Available. 2022. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/previous-testing-in-us.html
- 5.Rader B, Gertz A, Iuliano AD, et al. Use of at-home COVID-19 tests—United States, August 23, 2021–March 12, 2022. MMWR Morb Mortal Wkly Rep. 2022;71(13):489. doi: 10.15585/mmwr.mm7113e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirby AE, Walters MS, Jennings WC, et al. Using wastewater surveillance data to support the COVID-19 response—United States, 2020–2021. MMWR Morb Mortal Wkly Rep. 2021;70(36):1242–1244. doi: 10.15585/mmwr.mm7036a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham KE, Loeb SK, Wolfe MK, et al. 2022. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7737534
- 8.Peccia J, Zulli A, Brackney DE, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2022. https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/public-health-interpretation.html
- 10.North Carolina Department of Health and Human Services. 2022. https://epi.dph.ncdhhs.gov/cd/meaningful_use/syndromic.html
- 11.Cavany S, Bivins A, Wu Z, et al. Inferring SARS-CoV-2 RNA shedding into wastewater relative to the time of infection. Epidemiol Infect. 2022;150:e21. doi: 10.1017/S0950268821002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randazzo W, Truchado P, Cuevas-Ferrando E, Simón P, Allende A, Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181:115942. doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waller AE, Scholer M, Ising AI, Travers DA.2022. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7120837
- 14.Toronto Water. 2022. https://www.toronto.ca/wp-content/uploads/2018/05/8e4a-2017-TAB-Annual-Report-Final.pdf
- 15.XCG Consultants Ltd. 2022. https://www.ontario.ca/document/water-and-energy-conservation-guidance-manual-sewage-works-0
- 16.Centers for Disease Control and Prevention. 2022. https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/testing-methods.html
- 17.North Carolina Department of Health and Human Services. NCDHHS emergency department syndrome definitions. 2021. https://ncdetect.org/case-definitions
- 18.Bivins A, Bibby K. Wastewater surveillance during mass COVID-19 vaccination on a college campus. Environ Sci Technol Lett. 2021;8(9):792–798. doi: 10.1021/acs.estlett.1c00519. [DOI] [PubMed] [Google Scholar]
- 19.D’Aoust PM, Graber TE, Mercier E, et al. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci Total Environ. 2021;770:145319. doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng S, Roguet A, McClary-Gutierrez JS, et al. 2021. https://pubs.acs.org/doi/10.1021/acsestwater.1c00160
- 21.Larsen DA, Collins MB, Du Q, et al. Coupling freedom from disease principles and early warning from wastewater surveillance to improve health security. PNAS Nexus. 2022;1(1):pgac001. doi: 10.1093/pnasnexus/pgac001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maharaj AS, Parker J, Hopkins JP, et al. Comparison of longitudinal trends in self-reported symptoms and COVID-19 case activity in Ontario, Canada. PLoS One. 2022;17(1):e0262447. doi: 10.1371/journal.pone.0262447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilcox RR. Comparing Pearson correlations: dealing with heteroscedasticity and nonnormality. Commun Stat Simul Comput. 2009;38(10):2220–2234. doi: 10.1080/03610910903289151. [DOI] [Google Scholar]
- 24.Wilcox RR. Introduction to Robust Estimation and Hypothesis Testing. New York, NY: Academic Press; 2011. [Google Scholar]
- 25.Gasparrini A, Armstrong B, Kenward MG. Distributed lag non‐linear models. Stat Med. 2010;29(21):2224–2234. doi: 10.1002/sim.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Government of North Carolina. 2022. https://governor.nc.gov/news/press-releases/2020/11/17/north-carolina-introduces-covid-19-county-alert-system
- 27.Centers for Disease Control and Prevention. National Wastewater Surveillance System. Available. 2021. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance.html
- 28.Wolfe MK, Topol A, Knudson A, et al. High-frequency, high-throughput quantification of SARS-CoV-2 RNA in wastewater settled solids at eight publicly owned treatment works in northern California shows strong association with COVID-19 incidence. mSystems. 2021;6(5):e0082921. doi: 10.1128/mSystems.00829-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Aoust PM, Mercier E, Montpetit D, et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188:116560. doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Chen L, Deng Q, et al. The presence of SARS‐CoV‐2 RNA in the feces of COVID‐19 patients. J Med Virol. 2020;92(7):833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 31.Yang S, Dong Q, Li S, et al. Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J Hazard Mater. 2022;429:128358. doi: 10.1016/j.jhazmat.2022.128358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales Medina WR, D’Elia S, Fahrenfeld NL. Accumulation of SARS-CoV-2 RNA in sewer biofilms. ACS EST Water. 2022 doi: 10.1021/acsestwater.1c00345. [DOI] [Google Scholar]
- 33.Bernal JL, Andrews N, Gower C, et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on Covid-19 related symptoms, hospital admissions, and mortality in older adults in England: test negative case-control study. BMJ. 2021;373:n1088. doi: 10.1136/bmj.n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith T, Cassell G, Bhatnagar A.2021. https://jamanetwork.com/journals/jama-health-forum/fullarticle/2775225
- 35.Kumar M, Joshi M, Patel AK, Joshi CG. Unravelling the early warning capability of wastewater surveillance for COVID-19: a temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ Res. 2021;196:110946. doi: 10.1016/j.envres.2021.110946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed W, Tscharke B, Bertsch PM, et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci Total Environ. 2021;761:144216. doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karthikeyan S, Nguyen A, McDonald D, et al. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a university campus. mSystems. 2021;6(4):e0079321. doi: 10.1128/mSystems.00793-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bibby K, Bivins A, Wu Z, North D. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;202(117438) doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medema G, Been F, Heijnen L, Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr Opin Environ Sci Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClary-Gutierrez JS, Mattioli MC, Marcenac P, et al. SARS-CoV-2 wastewater surveillance for public health action. Emerg Infect Dis. 2021;27(9):1–8. doi: 10.3201/eid2709.210753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maharaj AS, Parker J, Hopkins JP, et al. 2021. https://www.medrxiv.org/content/10.1101/2021.01.15.21249879v1.full.pdf
- 42.Centers for Disease Control and Prevention. 2021. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/wastewater-surveillance/data-reporting-analytics.html
- 43.North Carolina Department of Health and Human Services; North Carolina. 2022. https://covid19.ncdhhs.gov/dashboard