The identification of a new CD4+ T-cell metacluster that corresponds with immune status could guide effective tumor treatment by predicting response to immunotherapy using peripheral blood samples from patients.
Abstract
CD4+ T-cell immunity helps clonal proliferation, migration, and cancer cell killing activity of CD8+ T cells and is essential in antitumor immune responses. To identify CD4+ T-cell clusters responsible for antitumor immunity, we simultaneously analyzed the naïve-effector state, Th polarization, and T-cell receptor clonotype based on single-cell RNA-sequencing data. Unsupervised clustering analysis uncovered the presence of a new CD4+ T-cell metacluster in the CD62Llow CD4+ T-cell subpopulation, which contained multicellular clonotypes associated with efficacy of programmed death-ligand 1 (PD-1) blockade therapy. The CD4+ T-cell metacluster consisted of CXCR3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+ cells and was characterized by high expression of IL7 receptor and TCF7. The frequency of these cells in the peripheral blood significantly correlated with progression-free survival and overall survival of patients with lung cancer after PD-1 blockade therapy. In addition, the CD4+ metacluster in the peripheral blood correlated with CD4+ T-cell infiltration in the tumor microenvironment, whereas peripheral Th1 correlated with local CD8+ T-cell infiltration. Together, these findings suggest that CD62Llow CCR4−CCR6+ CD4+ T cells form a novel metacluster with predictive potential of the immune status and sensitivity to PD-1 blockade, which may pave the way for personalized antitumor immunotherapy strategies for patients.
Significance:
The identification of a new CD4+ T-cell metacluster that corresponds with immune status could guide effective tumor treatment by predicting response to immunotherapy using peripheral blood samples from patients.
Introduction
Immune checkpoint inhibitors (ICI), such as anti-programmed cell death-1 (PD-1)/PD-1 ligand-1 (PD-L1) antibodies, have achieved breakthrough results in long-term survival in many cancers. However, the readiness of T-cell immunity, which varies greatly from patient to patient, has led to significant differences in antitumor efficacy. It has been elucidated that antitumor T cells in tumor microenvironment (TME) to be activated by ICI are T cells continuously recruited from peripheral circulation (1–4). If T-cell clusters that are responsible for the dynamic relationship between the peripheral circulation and TME can be elucidated, it will become possible to monitor antitumor immunity by analyzing the T-cell clusters in peripheral blood, which will facilitate the selection of adequate ICI therapy for each patient.
Research in cancer immunotherapy has primarily focused on CD8+ T cells in the TME (5–7). However, it has been reported that antitumor immunity cannot be induced unless the tumor cells have the MHC class II binding neoantigens, which are recognized by CD4+ T cells (8). CD4+ T cells are likely to be the driving force of the cancer immunity cycle, allowing for a sustainable supply of CTLs to the TME (9, 10).
CD4+ T cells are multitasking to defend against a wide variety of pathogens. To perform this function most efficiently, CD4+ T cells differentiate into optimal functional types (11). The Th type 1 (Th1) promotes CD8+ T cells to kill intracellular pathogens, and the Th17 mobilizes neutrophils to eliminate fungi and extracellular bacteria. In antitumor immunity, Th1 is thought to play an important role by producing IFNγ and promoting the activity of CD8+ T cells. On the other hand, Th17 cells, which have stem cell-like properties, are believed to promote long-term antitumor immunity (12). Thus, the precise Th clusters that play roles in antitumor immunity remain to be elucidated.
We previously reported that pretreatment peripheral blood CD4+ T cells that downregulated CD62 L expression (CD62Llow) in patients with lung cancer correlated with prognosis and response to nivolumab therapy and that the tumor antigen-specific effector T cells belonged to the CD62Llow subpopulation (13–15). This study aimed to identify the CD4+ T-cell clusters responsible for antitumor immunity by simultaneously analyzing the three critical attributes, the naïve-effector state change by priming, Th polarization, and T-cell receptor (TCR) clonotype based on non–knowledge-based cell distribution according to intercellular similarity by gene and protein-level molecular expression.
Materials and Methods
Patients and treatment
Patients who had histologically or cytologically confirmed stage IV or IIIB–C non–small cell lung cancer (NSCLC) with a PD-L1 tumor proportion score of 1% or higher and were eligible for first-line pembrolizumab therapy as a clinical practice were enrolled in this observational study. PD-L1 expression was assessed using the PD-L1 IHC 22C3 pharmDx assay (DAKO). This study included 60 consecutive patients from a single institution, Saitama Medical University International Medical Center, enrolled from March 2017 to November 2018. A total of 57 of 60 patients had tumor proportion score (TPS) of 50% or greater. Peripheral blood samples for this study were collected after obtaining written informed consent. The study protocol was approved by the Internal Review Board of Saitama Medical University International Medical Center in accordance with the Declaration of Helsinki (the ethical approval number 15-221). The patients received pembrolizumab at a dose of 200 mg every 3 weeks. The second validation cohort included 36 patients with previously treated NSCLC who received every 3 weeks 200 mg pembrolizumab or every 2 weeks 240 mg nivolumab therapy as clinical practice, irrespective of PD-L1 expression.
Response assessments
Tumor imaging using CT scanning was performed every 9 weeks. The response to treatment was assessed by an independent radiological review using the RECIST, version 1.1. May 19, 2020, was set as the cut-off date for data collection.
Blood sample
Peripheral blood mononuclear cell (PBMC) samples were collected prior to the first administration of the anti-PD-1 mAb using heparinized CPT Vacutainer tubes (Becton Dickinson Vacutainer Systems) as described previously (15). The samples were frozen using Cellbanker2 (Nippon Zenyaku Kogyo Co.) in a liquid nitrogen tank. For analyses of T-cell subsets, cells were incubated for 32–48 hours in a culture medium consisting of RPMI1640 and 10% FCS. Details of mass cytometry and flow cytometry analysis are shown in Supplementary Materials and Methods (Supplementary Table S1; Supplementary Fig. S1).
Single-cell RNA sequencing
Single-cell analyses of frozen PBMCs derived from the patients before the administration of pembrolizumab therapy were performed using the Chromium Next GEM Single Cell V(D)J Reagent Kit v1.1 with Feature Barcoding technology (10x Genomics Inc.). All single-cell libraries were constructed according to the manufacturer's standard protocol. The libraries were sequenced with HiSeq 2500 (Illumina Inc.) or NovaSeq6000 (Illumina Inc.) using Illumina's standard protocol. Details of the libraries and sequence results are summarized in Supplementary Table S2A.
The sequenced reads were processed with Cell Ranger 4.0.0 using the GRCh38 reference dataset (version 2020-A for gene expression and 4.0.0 for the TCR repertoire) to derive the gene expression/feature barcode (TotalSeq-C) unique molecular identifier count matrix and TCR repertoire data (Supplementary Fig. S2). Statistical analysis and graphical drawing were performed using the Seurat 4.0 (16, 17), MAST (18), and the Monocle2 (19, 20) packages in R software. Details are shown in Supplementary Materials and Methods.
Statistical analysis
Prism 9 (GraphPad Software) was used to conduct statistical analyses. Data are expressed as the mean ± SEM unless otherwise indicated. Tests for differences between two populations were performed using Student t test. Multiple group comparisons were performed using one-way ANOVA with Tukey post hoc analysis. Survival curves were estimated using the Kaplan–Meier method. Tests for differences and HR were performed using log-rank (Mantel–Cox) test. All P values were two sided, and P < 0.05 was considered statistically significant.
Data availability
The RAW data of single-cell RNA sequencing (scRNA-seq) generated in this study are publicly available in Gene Expression Omnibus (21) at GSE168844.
Results
Clustering of peripheral CD4+ T cells
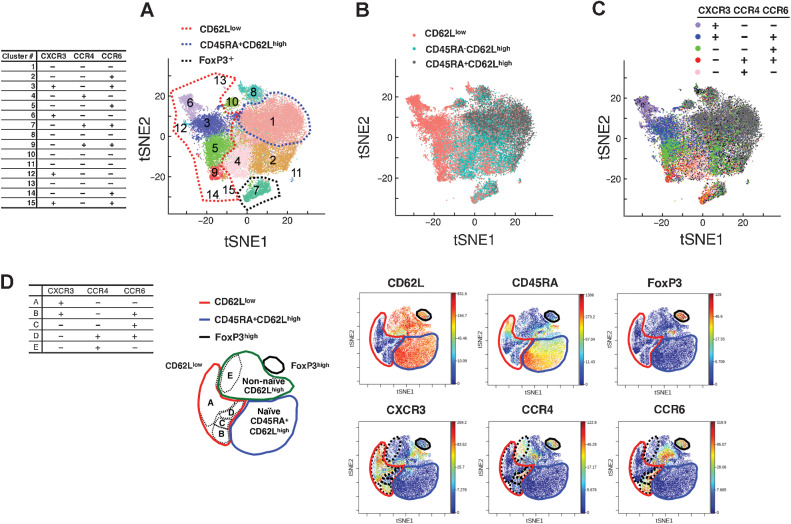
First, we collected peripheral blood samples from patients with lung cancer who were scheduled to receive pembrolizumab as first-line therapy and then performed scRNA-seq from 6 patients [2 partial responses (PR), 2 stable diseases (SD), and 2 progressive diseases (PD)] in the discovery cohort (Supplementary Tables S2A and S2B). The unsupervised clustering of cells based on RNA expression was performed and CD4+ T cells were classified into 15 clusters (Fig. 1A; Supplementary Fig. S3A). The 15 clusters were mapped by five cell-surface protein expression patterns measured as TotalSeq-C counts (CD62L, CD45RA, CXCR3, CCR4, CCR6). CD62 L and CD45RA expression patterns were used to evaluate the naïve-effector state change (Fig. 1B; Supplementary Fig. S3B and S3C). Most of the cells in clusters #3, 5, 6, 9, 12, 14, and 15 (circled in red dashed line in Fig. 1A) were composed of CD62Llow cells, which downregulated CD62 L and are considered effector T cells. In contrast, the majority of cells in cluster #1 (circled in blue dashed line in Fig. 1A) were CD62LhighCD45RA+, which met the definition of naïve T cells. The #7 cluster was considered to be regulatory T cells (Treg) owing to the presence of FoxP3 expression (circled in black dashed line in Fig. 1A; Supplementary Fig. S4A). Most of the cells belonging to the remaining clusters have a CD45RA−CD62Lhigh phenotype and will be referred to as the non-naïve CD62Lhigh fraction, which likely includes central memory T cells. Th polarization status was assessed using the expression patterns of CXCR3, CCR4, and CCR6 (22). The expression pattern of chemokine receptors at the cell level is presented in Fig. 1C (Supplementary Fig. S4B). Consequently, most cells of the #6 cluster presented the expression pattern of CXCR3+CCR4−CCR6−, which was typical for Th1, the #9 cluster cells presented the expression pattern of CXCR3−CCR4+CCR6+, which was typical for Th17, and most cells of the #4 cluster presented the expression pattern of CXCR3−CCR4+CCR6−, which was typical for Th2. The #3 cluster contained not only CXCR3+CCR4−CCR6− but also CXCR3+CCR4−CCR6+, and the #5 cluster contained CXC3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+ in addition to CXCR3−CCR4+CCD6+. CXCR3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+ cells other than the typical Th1 and Th17 will be referred to as Th1/17 and CCR6 SP, respectively, in this study.
Figure 1.
Clustering of CD4+ T cells according to scRNA-seq. A, tSNE plots derived from integrated gene expression data from scRNA-seq of 6 patients with lung cancer collected prior to pembrolizumab therapy. All 33,604 CD3+CD8−CD4+ cells were divided into 15 clusters upon unsupervised clustering. Red dashed line, the extent of clusters mostly composed of CD62Llow T cells (cluster #3, #5, #6, #9, #12, #13, #14, and #15). Blue dashed line, cluster #1—mostly composed of CD62LhighCD45RA+ T cells. The table shows the expression levels of the CXCR3, CCR4, and CCR6 proteins in each cluster as determined by the value of TotalSeq-C (C). B and C, Cell-level annotation of CD62L, CD45RA (B) and CXCR3, CCR4, and CCR6 (C) from scRNA-seq of 6 patients (same tSNE reduction as in A). Annotation was performed according to the TotalSeq-C counts of each cell in the gating method. The gating strategy is detailed in Supplementary Fig. S2. D, Representative illustrations of mass cytometry viSNE analysis for gated CD4+CD3+ cells observed after the expression of 21 molecules and the expressions of six molecules are presented. Red lines, the extent of the region representing the CD62Llow subpopulation. Blue lines, CD45RA+CD62Lhigh region. Black lines, FoxP3+ region. Green line, remaining region. Black dashed lines, the five regions mapped on the basis of CXCR3, CCR4, and CCR6 expression. The chemokine receptor expression patterns in each region are presented in the table.
Similarly, we mapped the dimensional compression diagram based on the expression of 21 molecules (CD3, CD19, CD278, CD4, CD8, CCR4, CD62L, CD27, CXCR3, CD28, CD80, CD223, CD44, CCR7, CD154, CD45RA, CD279, CCR6, FoxP3, T-bet, and CD152) by CyTOF (Standard Bio Tools K.K., Tokyo), in the discovery cohort (n = 15), and the results were in good agreement with those of unsupervised clustering by gene expression (Fig. 1D). CD4+ T cells were divided into four subpopulations according to the expression status of CD62L, CD45RA, and FoxP3: CD62Llow subpopulation, CD45RA+CD62Lhigh subpopulation, FoxP3+ Treg, and the remaining non-naïve CD62Lhigh subpopulation. The clusters of CXCR3+CCR4−CCR6− (Th1), CXCR3−CCR4+CCR6+ (Th17), CXCR3+CCR4−CCR6+ (Th1/17), and CXCR3−CCR4−CCR6+ (CCR6 SP) were observed in the CD62Llow CD4+ T-cell subpopulation, and CXCR3−CCR4+CCR6− (Th2) was observed in the non-naïve CD62Lhigh CD4+ T-cell subpopulation, which was also consistent with the annotation results of scRNA-seq.
Multicellular CD4+ T-cell clonotypes associated with PD-1 blockade therapy efficacy
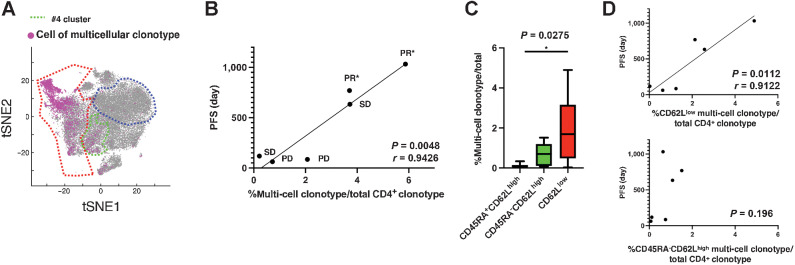
T cells that are clonally expanding in vivo are likely effector T cells that are fighting against currently existing targets (3). Therefore, we analyzed the TCR repertoire using scRNA-seq to investigate the relationship between CD4+ T cells belonging to multicellular clonotypes in the peripheral blood of patients with lung cancer and the effect of pembrolizumab therapy. First, we mapped T cells belonging to the multicellular clonotypes that presented with two or more cells with the same TCR on the t-distributed stochastic neighbor embedding (tSNE) plot (Fig. 2A). Most of these T cells were observed in the CD62Llow clusters (#3, 5, 6, and 9; Fig. 1A), albeit a small number were also observed in the Th2 type #4 cluster in the non-naïve CD62Lhigh subpopulation (circled in green dashed line in Fig. 2A).
Figure 2.
Multicellular CD4+ T-cell clonotypes associated with PD-1 blockade therapy efficacy. A, On the basis of the results of TCR repertoire analysis using scRNA-seq, cells belonging to TCR clonotypes with two or more cells detected were mapped on the same tSNE plots as in Fig. 1A–C. B, Correlation between the percentage of TCR clonotypes with ≥2 cells detected against all clonotypes and the PFS of patients with lung cancer after initial pembrolizumab treatment. PR, SD, and PD were determined by RECIST ver1.1; *, continued response. C, Percentages of TCR clonotypes that contained ≥2 cells belonging to the CD45RA+CD62Lhigh, CD45RA−CD62Lhigh, and CD62Llow T-cell subpopulations. D, Correlation analysis between the percentage of multicellular clonotypes and PFS following pembrolizumab treatment.
To scrutinize the relationship between the degree of CD4+ T-cell clonotype expansion and antitumor immunity, we examined whether the proportion of multicellular clonotypes to all detected TCR clonotypes correlated with progression-free survival (PFS) after pembrolizumab treatment. Although multicellular clonotypes only accounted for about 0.3%–6% of all CD4+ T-cell clonotypes, the multicellular clonotype percentages of all detected clonotypes were strongly positively correlated with PFS (P = 0.0048, r = 0.9426; Fig. 2B). The percentage of cells belonging to the multicellular clonotype relative to total CD4+ T cells also correlated with PFS; however, the correlation was weaker than that of the multicellular clonotype itself (Supplementary Fig. S5A). These results suggest that the CD4+ T cells responsible for the antitumor therapeutic effects of pembrolizumab belong to multicellular clonotypes, which have clonally expanded before the PD-1 blockade treatment, and that the TCR diversity of clonally expanding T cells contributes to the long-term response. Next, we investigated to which naïve-effector T-cell subpopulation this multicellular clonotype belongs. Consistent with the mapping results, almost all of the naïve CD45RA+CD62Lhigh subpopulation was composed of singletons (Supplementary Table S3). Most of the multicellular clonotypes belonged to the CD62Llow subpopulation, although a few belonged to the CD45RA−CD62Lhigh subpopulation (Fig. 2C). Interestingly, the percentage of multicellular clonotypes belonging to the CD62Llow subpopulation relative to detected CD4+ T-cell clonotypes correlated with PFS, whereas the percentage of clonotypes belonging to CD45RA−CD62Lhigh did not (Fig. 2D). The TCR diversity index of the CD62Llow subpopulation suggested that clonotypes corresponding to the antitumor effect of anti-PD-1 mAb therapy were enriched in the CD62Llow subpopulation (Supplementary Fig. S5B). However, the TCR diversity index of the CD62Llow cells did not differ between the PFS > 300 days group and the PFS < 300 days group (Supplementary Fig. S5C). The number of cells per multicellular clonotype was largest in Th1, suggesting a greater degree of clonal expansion (Supplementary Fig. S5D and S5E). On the other hand, the number of multicellular clonotypes was similar in Th1 and Th1/17. The relationship between the percentage of multicellular clonotypes per Th and PFS was examined, and it was found that only the percentage of multicellular clonotypes belonging to Th1/17·CCR6SP had a significant correlation with PFS (Supplementary Fig. S5F). The results suggest that the diversity of clonally expanded clonotypes is more important for antitumor efficacy than the degree of clonal expansion per clonotype, and that the clonotypes belonging to Th1/17·CCR6SP are the most important.
New Th clusters distinct from Th1 and Th17 found in responders
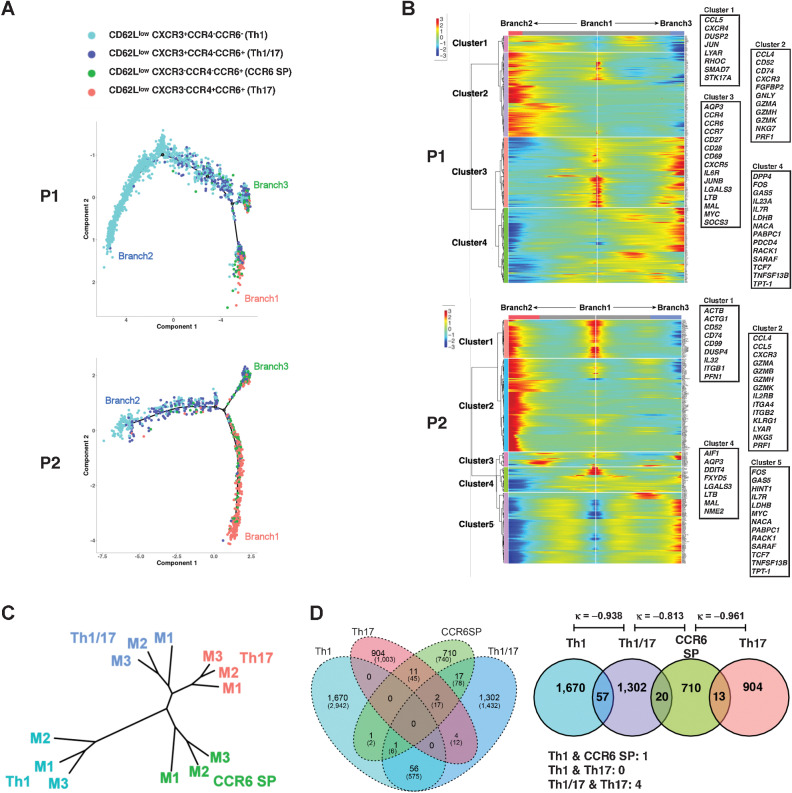
Because Fig. 1A and C show that the CD62Llow CD4+ T-cell subpopulation can be further subdivided using the property of Th polarization, we performed a pseudotime analysis to clarify the relationship between the CD62Llow Th clusters. In unsupervised clustering using gene expression, the CXCR3+CCR4−CCR6− typical Th1 cluster (#6 in Fig. 1A) and CXCR3−CCR4+CCR6+ typical Th17 cluster (#9 in Fig. 1A) were the most distantly located; therefore, we used differentially expressed genes (DEG) between Th1 and Th17 as ordering genes for pseudotime analysis (Fig. 3A; Supplementary Fig. S6A; Supplementary Table S4). In the cases that responded to pembrolizumab treatment, we observed the formation of another node (Branch3 in Fig. 3A) at a location between the typical Th17 (Branch1) and Th1 (Branch2). Branch3 contains some CXCR3−CCR4+CCR6+ cells; however, CXCR3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+ cells were the predominant components. To identify genes specific to each branch, two branched pseudotime trajectories, Branch1 to Branch2 and Branch1 to Branch3, were compared using the branched expression analysis modeling (BEAM) method (Fig. 3B). In cluster 4 of patient P1 and cluster 5 of patient P2, which were highly expressed only in Branch3, IL7 receptor (IL7R), FOS, LDHB, GAS5, RACK1, TNFSF13B, and transcriptional factor 7 (TCF7) were commonly observed. Another hierarchical clustering result based on DEGs in the four Th clusters also showed that the Euclidean distance between CXCR3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+ was the closest and furthest from Th1 and Th17, respectively (Supplementary Fig. S6B; Supplementary Table S5).
Figure 3.
Th clusters are distinct from Th1 and Th17 found in responders. A, The trajectory of the pseudotime analysis of four CD62Llow CD4+ subpopulations of PR patient (P1 and P2) calculated from DEGs between Th1-Th17 clusters of this patient. Details of DEGs are summarized in Supplementary Table S4A. B, The heatmap shows the result of BEAM. The genes representing each cluster are highlighted in the box. Details of these branch-dependent genes are summarized in Supplementary Table S4B. C, Clustering analysis based on raw β values of differentially methylated probes obtained from four isolated CD62Llow CD4+ T-cell clusters derived from three patients (M1–M3). D, TCR clonotype overlap between four subpopulations aggregated for 6 patients. Numbers represent the number of clonotypes and the numbers within parentheses represent the number of cells included in that category. κ, κ coefficient.
The differentiation status of T cells is mainly attributed to DNA methylation (23). To examine whether the differences in gene expression patterns observed with pseudotime analysis corresponded to the differentiation status, methylome analysis was performed using CXCR3+CCR4−CCR6− (Th1), CXCR3+CCR4−CCR6+ (Th1/17), CXCR3−CCR4−CCR6+ (CCR6 SP), and CXCR3−CCR4+CCR6+ (Th17) T-cell clusters sorted from CD62Llow CD4+ T cells. Clustering analysis calculated from differentially methylated probes (DMP) with significant differences by ANOVA of the four CD4+ T-cell clusters revealed that the typical Th1 and Th17 clusters were the farthest apart, while the CXCR3+CCR4−CCR6+ (Th1/17) and CXCR3−CCR4−CCR6+ (CCR6 SP) clusters were independent but relatively close to Th17 (Fig. 3C). These results revealed that in addition to classical Th1 and Th17 subtypes, there were two more CD4+ T-cell clusters, CXCR3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+, which have similar gene expression and DNA methylation patterns in the CD62Llow CD4+ T-cell subpopulation and that the patients who responded to pembrolizumab therapy tended to have more third branch consisting of CXCR3+CCR4−CCR6+ and CXCR3−CCR4−CCR6+ cells.
The gene expression patterns of Th1/17 and CCR6 SP T-cell clusters revealed significant differences from those of Th1 and Th17, which were consistent with the results of the methylation analysis. These results indicate the presence of differentiated Th cell clusters rather than reflecting a transient activation state. However, even differentiated Th cells are known to exhibit plasticity that can alter their phenotype (24). Therefore, we investigated the independence as clonotypes among the CD62Llow Th1, Th1/17, Th17, and CCR6 SP T-cell clusters. The clonotype composition of Th1, Th1/17, CCR6 SP, and Th17, respectively, of the CD62Llow CD4+ T-cell subfraction, is presented (Fig. 3D; Supplementary Fig. S6C). Overlap of clonotypes was observed between Th1 and Th1/17, Th1/17 and CCR6 SP, and CCR6 SP and Th17, albeit they were considered to be highly independent as they presented a κ coefficient close to −1. In contrast, because T-cell clusters with overlapping clonotypes may be closely related during differentiation, this clonotyping result also suggested that Th1/17 and CCR6 SP T-cell clusters were closely related clusters.
CD62Llow Th1/17 and CD62Llow CCR6 SP clusters form a functional metacluster
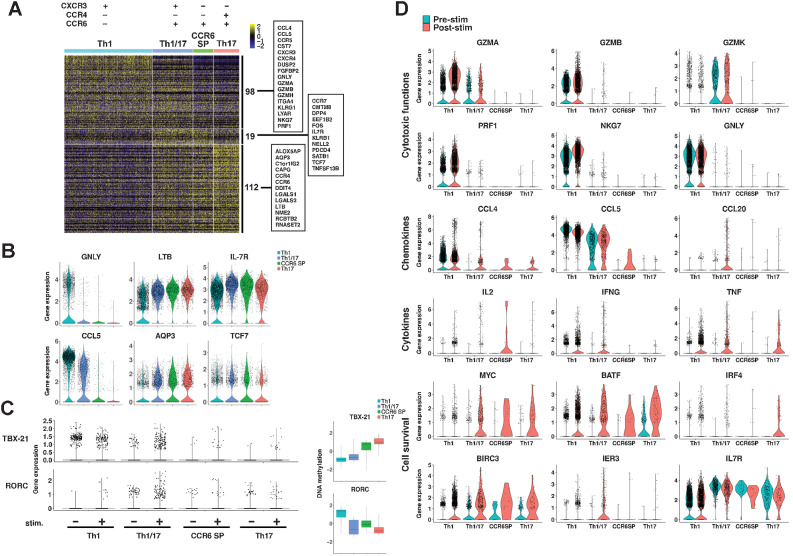
We assessed the new Th clusters in terms of molecular functions. As presented in the pseudotime analysis of gene expression and cluster analysis, the gene expression patterns were divided into three major groups: typical Th1-dominant, typical Th17-dominant, and those common to Th1/17 and CCR6 SP T-cell clusters. The heatmap of 98 Th1-dominant genes, 112 Th17-dominant genes, and 19 genes commonly expressed in Th1/17 and CCR6 SP clusters is presented in Fig. 4A (Supplementary Table S6). Genes such as GNLY, CCL5, and CCR5 revealed Th1-dominant expression in Th1 and Th1/17 cells (Fig. 4B). Genes such as LTB, and AQP3 presented Th17-dominant expression. Genes corresponding to IL7R, and TCF7 were found preferentially expressed in the Th1/17 and CCR6 SP clusters compared with those in Th1 or Th17 cells and were considered as representative of the cluster (Fig. 4B; Supplementary Fig. S7A). In addition, DPP4 (CD26), and PDCD4 were found to be preferentially expressed in Th1/17 and CCR6 SP (Supplementary Fig. S7B and S7C).
Figure 4.
Gene expression analysis of CD4+ T-cell clusters. A, Gene expression profiles of four CD62Llow CD4+ T-cell clusters. The heatmap shows DEGs among the Th1, Th1/17, CCR6 SP, and Th17 clusters using the integrated data from 6 patients; further details are presented in Supplementary Table S6. B, Gene expression of representative genes. C, Gene expression of TBX-21 and RORC was observed pre (−) and post (+) stimulation with anti-CD3/CD28 antibody-coated beads. Differences in methylation status are also shown by boxplot, which represents scaled β values of all DMPs of these genes. Details of these DMPs are summarized in Supplementary Table S7. D, DEGs of prestimulation (blue) and poststimulation (red) with anti-CD3/CD28 antibody-coated beads of each Th cluster cells that belonged to multicellular clonotypes.
Although the expression levels of transcription factor genes such as TBX-21 (T-bet) and RORC, which are important for Th differentiation, were low and no significant differences could be detected, we analyzed gene expression and methylation profiles after anti-CD3/CD28 stimulation. After CD3/CD28 stimulation, the Th1/17 cluster expressed both TBX-21 and RORC. In contrast, Th1 only expressed TBX-21 (Fig. 4C). CyTOF analysis showed that the percentage of T-bet–positive cells was highest in the Th1 cluster, followed by the Th1/17 cluster (Supplementary Fig. S7D). CCR6 SP cells expressed significantly more TBX-21 than the Th17 cluster. Consistent with these results, TBX-21 was significantly hypomethylated in the Th1 and Th1/17 clusters, followed by the CCR6 SP cluster; RORC was hypomethylated in the Th1/17, CCR6 SP, and Th17 clusters. More than half of DMPs of these genes were found upstream of the transcription start site, which is expected to be related to the regulation of gene expression. In particular, most of the DMPs of TBX-21 were found around the CpG island region. (Supplementary Table S7).
We next analyze gene expression after CD3/CD28 stimulation of CD62Llow T cells obtained from four responder patients in the first-line pembrolizumab cohort. Here, we show the representative data in Fig. 4D. The expression of genes related to cytotoxic activity was found in Th1 and Th1/17, but not in CCR6 SP and Th17. The expression of GZMB, PRF, NKG7, and GNLY was almost limited to Th1. In contrast, GZMK was more strongly expressed in Th1/17. The expression of CCL4 and CCL5, chemokines involved in activated T-cell migration, was strongest in Th1, but CCL5 expression was also observed in Th1/17. CCL20, a chemokine for CCR6, was upregulated only in Th1/17. IFNγ gene expression was found in both Th1 and Th1/17; however, Th1/17 exhibited significantly higher expression than Th1 after stimulation (average log2FC = 3.123, Padjusted = 1.87 × 10−19). Genes involved in T-cell survival, such as MYC, which is a master regulator of metabolic programming in activating T cells, and BIRC3, which encodes an inhibitor of apoptosis protein2 (IAP2), were strongly expressed in Th1/17, CCR6 SP, and Th17, while poorly expressed in Th1 (25, 26). IER3, which has an antiapoptotic function, was expressed in Th1/17 (27). IL7R was most strongly expressed in Th1/17 and CCR6 SP, and was not upregulated in Th1 even after CD3/CD28 stimulation.
Expression levels of PDCD1 (PD-1) and CTLA4 were too low to detect significant differences by ANOVA, but PD-1 expression appeared to be higher in Th1 (Supplementary Fig. S7E). Therefore, we analyzed the expression of PD-1 and CTLA4 protein levels in the four CD62Llow Th clusters by CyTOF and observed that there was a significant difference in the percentage of PD-1+ and CTLA4+ cells among the four Th clusters (Supplementary Fig. S7F). Significantly more PD-1+ cells were observed in the Th1 cluster, and more CTLA4+ cells were detected in the Th17 cluster; the Th1/17 and CCR6 SP clusters contained approximately the same number of PD-1+ and CTLA4+ cells.
Altogether, the Th1/17 and CCR6 SP clusters that had the most similar methylome profiles, and gene expression profiles shared the expression of immunologically important molecules such as IL7R, TCF7, DPP4 (CD26), and PDCD4, as well as the expression patterns of PD-1 and CTLA4, which are important for regulating T-cell function. Meanwhile, the percentage of each Th cluster in the peripheral blood showed that the Th1/17 cluster was strongly correlated with the CCR6 SP cluster, but not with the Th1 cluster (Supplementary Fig S7G). Therefore, it is suggested that the Th1/17 cluster and CCR6 SP cluster differentiate and proliferate synchronously. These results indicate that the CD62Llow Th1/17 and CCR6 SP T-cell clusters form a functional metacluster, and since they commonly had high expression levels of IL7R, we will refer to this metacluster as Th7R.
Th7R cluster correlated with antitumor efficacy of PD-1 blockade
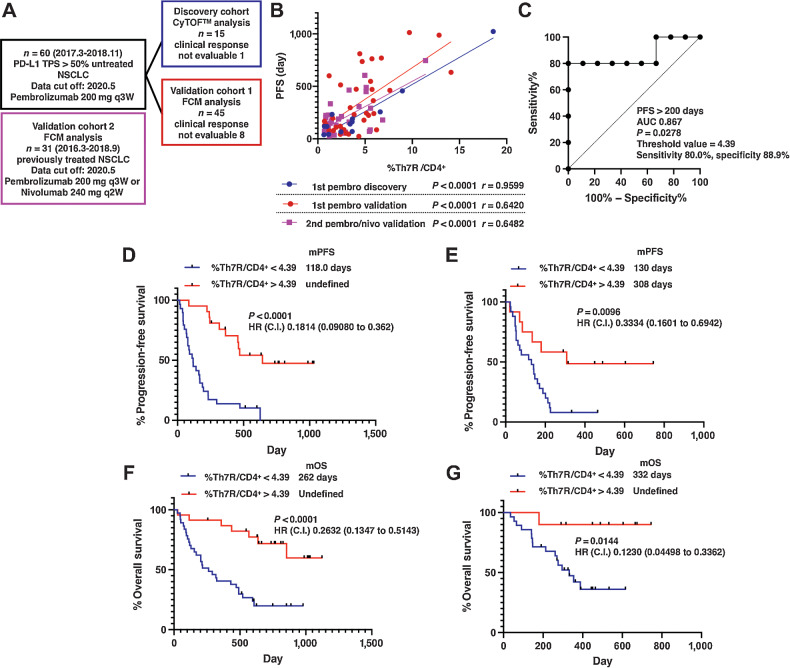
Next, we examined the predictive performance of Th7R in predicting treatment response in patients with previously untreated NSCLC with PD-L1–positive tumor treated with pembrolizumab (n = 60; Fig. 5A; Supplementary Fig. S8). A total of 57 of 60 patients had TPS of 50% or greater. The first 15 of 60 consecutively enrolled patients were analyzed as a discovery cohort (n = 15). In correlations between PFS and overall survival (OS) after pembrolizumab treatment and each CD62Llow CD4+ T-cell cluster, the FDR was set to 0.05 to avoid multiplicity and P < 0.05 was considered significant. The percentages of classical Th1 and Th17 cluster cell counts did not significantly correlate with either PFS or OS; however, the percentages of Th1/17 and CCR6 SP showed significant positive correlations with PFS and OS (Table 1A and B). CD62Llow CCR4−CCR6+ CD4+ T-cell metacluster, which represented the sum of CD62Llow Th1/17 (CXCR3+CCR4−CCR6+) and CD62Llow CCR6 SP (CXCR3−CCR4−CCR6+) T-cell clusters, presented a strong correlation with PFS (Fig. 5B; P < 0.0001, r = 0.9599). In contrast, no correlation was observed between CD62Lhigh T-cell clusters and PFS or OS.
Figure 5.
Correlation of CD4+ T-cell clusters with clinical outcomes after PD-1 blockade therapy. A, CONSORT diagram of patients with NSCLC treated with first-line pembrolizumab. B, Linear regression analysis of PFS and the percentage of pretreatment Th7R metacluster, which consisted of CD62Llow CXCR3+CCR4−CCR6+ and CD62Llow CXCR3−CCR4−CCR6+ CD4+ T-cell clusters, in the peripheral blood in the discovery cohort and the validation cohort1 treated with the first-line pembrolizumab therapy and the validation cohort 2 treated with the second-line pembrolizumab or nivolumab therapy. C, Results of ROC curve analysis of the percentage of CD62Llow CCR4CCR6+ CD4+ T-cell metacluster for the discovery cohort patients treated with first-line pembrolizumab with the indicated parameters. D and E, The Kaplan–Meier analysis of PFS after PD-1 blockade therapy in the previously untreated (validation cohort 1, n = 45; D) and treated (validation cohort 2, n = 31; E) NSCLC cohort. F and G, The Kaplan–Meier analysis of OS after PD-1 blockade therapy in the previously untreated NSCLC cohort (F) and treated NSCLC cohort (G). C.I., 95% confidence interval; HR, hazard ratio; mOS, median overall survival; mPFS, median progression-free survival.
Table 1.
Correlation between PFS (A)/OS (B) and the percentages of CD62Llow T-cell clusters based on total CD62LlowCD4+ cells or CD62Lhigh T-cell clusters based on CD45RA−CD62Lhigh CD4+ cells.
| A. | |||||
|---|---|---|---|---|---|
| CD62Llow | r | P | FDR (P = 0.05) | ||
| PFS | Th1 | NS | −0.0901 | 0.7697 | 0.0500 |
| Th17 | NS | 0.5166 | 0.0707 | 0.0375 | |
| Th1/17 | * | 0.8134 | 0.0007 | 0.0250 | |
| CCR6 SP | * | 0.8710 | 0.0001 | 0.0125 | |
| CD62Lhigh | r | P | FDR (P = 0.05) | ||
| PFS | Th1 | NS | 0.1402 | 0.6478 | 0.0500 |
| Th17 | NS | 0.1551 | 0.6129 | 0.429 | |
| Th2 | NS | −0.4195 | 0.1536 | 0.0357 | |
| Th1/17 | NS | 0.4963 | 0.0845 | 0.0286 | |
| CCR6 SP | NS | 0.5066 | 0.0773 | 0.0214 | |
| TP | NS | 0.5601 | 0.0465 | 0.0143 | |
| TN | NS | −0.6278 | 0.0216 | 0.0071 | |
| B. | |||||
| CD62Llow | r | P | FDR (P = 0.05) | ||
| OS | Th1 | NS | −0.2495 | 0.4594 | 0.0500 |
| Th17 | NS | 0.5316 | 0.0924 | 0.0375 | |
| Th1/17 | * | 0.7064 | 0.0151 | 0.0250 | |
| CCR6 SP | * | 0.7905 | 0.0038 | 0.0125 | |
| CD62Lhigh | r | P | FDR (P = 0.05) | ||
| OS | Th17 | NS | 0.0687 | 0.8410 | 0.0500 |
| Th1 | NS | 0.0838 | 0.8066 | 0.429 | |
| Th1/17 | NS | 0.3319 | 0.3187 | 0.0357 | |
| CCR6 SP | NS | 0.3326 | 0.3177 | 0.0286 | |
| Th2 | NS | −0.3757 | 0.2548 | 0.0214 | |
| TN | NS | −0.4467 | 0.1684 | 0.0143 | |
| TP | NS | 0.4503 | 0.1646 | 0.0071 | |
Note: Correlation analysis was performed using a multiple comparison test, where the FDR of Pearson correlation coefficient was estimated using the Benjamin–Hochberg procedure; cluster pairs were selected with an FDR < 5%.
Abbreviations: Th1, CXCR3+CCR4−CCR6− CD4+ T cell; Th2, CXCR3−CCR4+CCR6− CD4+ T cell; Th1/17, CXCR3+CCR4−CCR6+ CD4+ T cell; CCR6 SP, CXCR3−CCR4−CCR6+ CD4+ T cell; Th17, CXCR3−CCR4+CCR6+ CD4+ T cell; TN, CXCR3−CCR4−CCR6− CD4+ T cell; TP, CXCR3+CCR4+CCR6+ CD4+ T cell. *, P < 0.05. NS; not significant; OS, overall survival; PFS, progression-free survival.
We further examined whether Th7R metaclusters correlated with clinical outcomes after immune checkpoint inhibition therapy using two validation cohorts. One cohort consisted of 45 patients with PD-L1–positive NSCLC treated with first-line pembrolizumab therapy, and the other consisted of 31 patients with previously treated NSCLC treated with pembrolizumab or nivolumab regardless of tumor PD-L1 expression. The threshold of the Th7R metacluster was obtained from the results of ROC analysis performed in the discovery cohort to determine PFS > 200 days (Fig. 5C). In both validation cohorts 1 and 2, Kaplan–Meier analysis revealed that patients with the percentage of Th7R metacluster above the threshold had significantly better PFS (Fig. 5D and E; Supplementary Fig. S8A–S8C). The regression line presenting the correlation between Th7R metacluster and PFS obtained in the two validation cohorts did not demonstrate significant differences compared with that observed in the discovery cohort in either y intercept or slope, although they differed in terms of whether PD-1 blockade treatment was the first-line therapy and tumor PD-L1 expression (Fig. 5B; Supplementary Fig. S8D). Therefore, in addition to predicting the presence or absence of response using a certain threshold, the relationship between a greater number of Th7R metacluster and a longer PFS will be established, suggesting that the system has the performance to predict the long-term effect of PD-1 inhibitors. In validation cohort 1, there were 8 patients whose clinical response could not be assessed, and they had to be excluded from PFS analysis. Therefore, we analyzed the OS of all patients in the discovery cohort and validation cohort 1 who received the first pembrolizumab treatment (n = 60) and the validation cohort 2. Patients with a Th7R metacluster percentage above the threshold had significantly better OS in both cohorts (Fig. 5F and G).
CD4+ Th clusters associated with CD4+ or CD8+ T cells in the TME
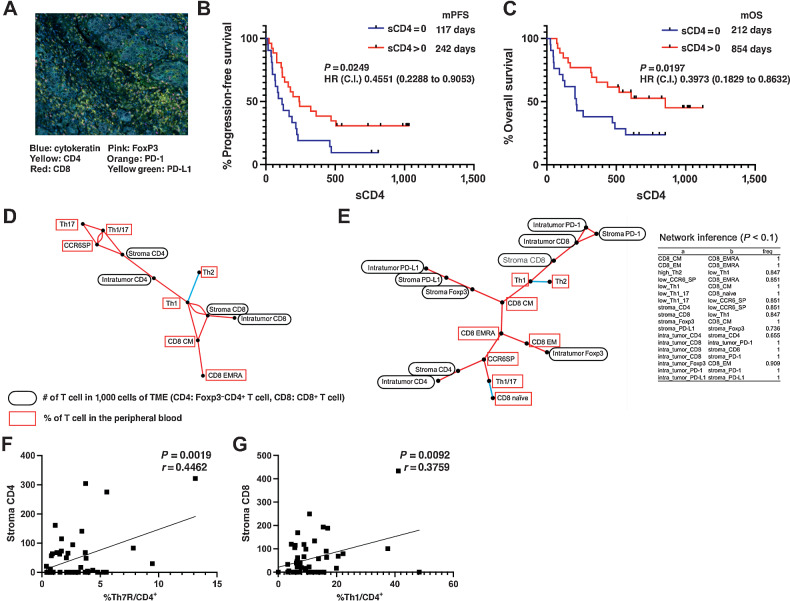
Furthermore, we examined whether T-cell infiltration into the TME correlated with CD4+ T-cell clusters in the peripheral blood. Tumor specimens obtained by core needle biopsy, transbronchial biopsy, and surgical biopsy (n = 46) were stained with cytokeratin, CD4, CD8, PD-1, PD-L1, and FoxP3. On the basis of the detection of cytokeratin plus tumor cells, 1,000 cells in the peritumor stroma and tumor were automatically counted using a multicolor analyzer. To determine the relationship between the CD4+ T-cell clusters identified in the peripheral blood that correlated with PD-1 inhibitor antitumor efficacy and the presence of other peripheral blood T-cell clusters and T cells in the TME, we performed multiple comparisons and network analyses. First, we examined the infiltration of T cells into the TME. Repeated correlation analysis of the number of immune cells in the TME revealed significant correlations between the number of CD8+ and CD4+ cells in the peritumor stroma and PFS and OS; however, in multiple comparison analyses, the most significant correlations were observed for CD4+ T cells in the peritumor stroma (Supplementary Table S8; Fig. 6A). Kaplan–Meier analysis of PFS and OS was performed in patients with at least one FoxP3−CD4+ T cell (sCD4) out of 1,000 tumor stromal cells and patients with less than one sCD4 cell (Fig. 6B and C). The results revealed that the sCD4 > 1 group presented significantly better PFS and OS than sCD4 < 1. In other words, the presence of CD4 T cells in the lung cancer stroma facilitated PD-1 inhibitor response, and the higher the number of CD4+ T cells, the longer the PFS.
Figure 6.
Correlation of T cells in the TME with clinical outcome after PD-1 blockade therapy and network analysis among T cells in the peripheral blood and TME. A, Representative multiplex IHC analysis results. Formalin-fixed and paraffin-embedded sections of biopsy or resected samples from NSCLC specimens were stained for CD4, CD8, Foxp3, PD-1, PD-L1, and cytokeratin with OPAL. Stained sections were imaged using the Vectra Automated Imaging System. The distribution of tumor cells (blue; cytokeratin) and PD-L1 (membranous; yellow-green) or infiltrating CD4+ T cells (yellow), CD8+ T cells (red), Foxp3+ T cells (pink), and PD-1+ T cells (orange) are shown. B and C, The Kaplan–Meier analysis of PFS (B) and OS (C) after pembrolizumab therapy in previously untreated NSCLC cohort for whom IHC analysis of tumor tissues was available (n = 46). Red line, group that had one or more FoxP3−CD4+ cells in the tumor stroma; blue line, group that had less than one. C.I., 95% confidence interval; HR, hazard ratio; mOS, median overall survival; mPFS, median progression-free survival. C, Results of the linear regression analysis of peritumoral stromal CD4+ cell counts and PFS of patients with lung cancer treated with pembrolizumab as first-line therapy. D, Correlation coefficients between T-cell subpopulations with FDR < 0.01 were drawn in network form. The positive and negative correlations between the subpopulations are depicted by the red and blue lines, respectively. E, The network of 18 cell clusters was inferred with a modified path consistency algorithm based on the cell fraction data in the peripheral blood and the microenvironment. The inference was performed 1,000 times, and the connection established more than 500 times was set to be significant in this analysis. The cell clusters with positive and negative correlations are connected by red and blue lines, and the fractions of established correlations by 1,000 inferences are indicated on each line. F, Correlation between peripheral blood Th7R metacluster percentage and tumor-stromal CD4+ T-cell count. G, Correlation between peripheral blood CD62Llow Th1 cell percentage and tumor-stromal CD8+ T-cell count.
To further examine whether there is an association between T-cell clusters in the peripheral blood and TME, we performed multiple comparisons of the correlation coefficients between the percentage of cell clusters with FDR < 0.01. In addition, to consider false (pseudocorrelations) correlations due to confounding effects between multiple variables, we also performed a network analysis. Consequently, partially common associations between cell clusters appeared in the two analyses (Fig. 6D and E). Interestingly, there were two groups of associations: CD62Llow Th7R cluster in peripheral blood had significant associations with peritumor stroma CD4+ T cell, and CD62Llow Th1 cluster was directly associated with CD8+ T cells in the TME (Fig. 6F and G). These results suggest that there are two association streams from immunity in the peripheral blood to that in the TME, which are governed by Th7R and Th1, respectively.
Pembrolizumab treatment resulted in PD-1 downregulation with T-bet upregulation in Th7r
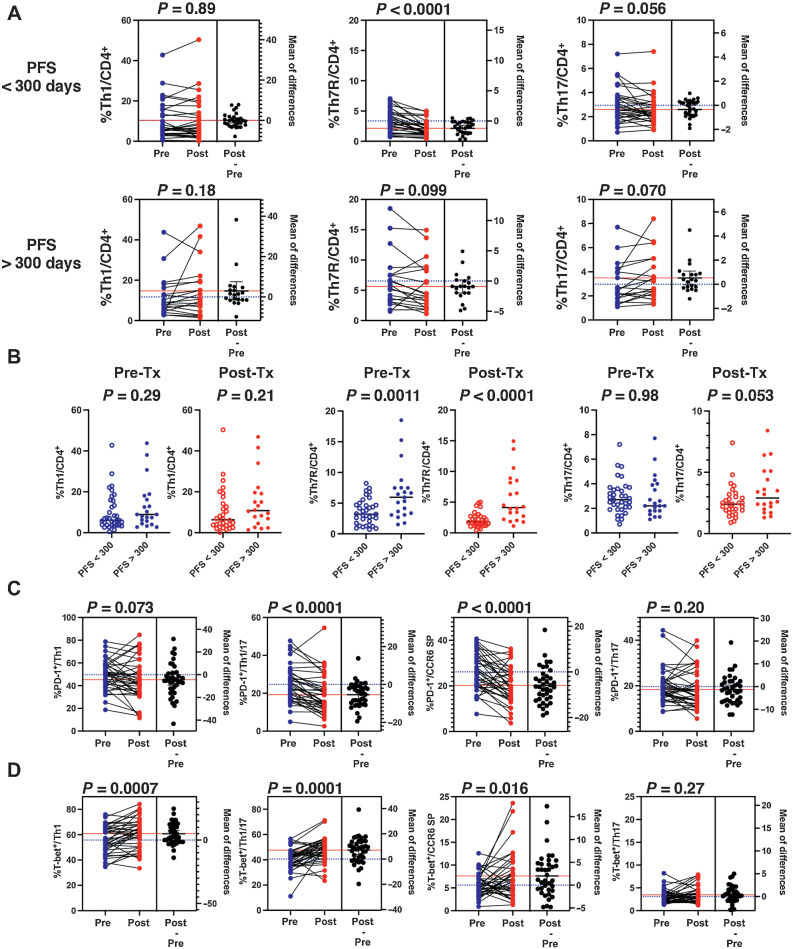
Finally, to determine how PD-1 blockade affects CD4+ T-cell clusters, we performed CyTOF analysis comparing CD4+ T-cell clusters before and after pembrolizumab treatment in 50 patients for whom blood samples were available after treatment in the first-line pembrolizumab cohort (Fig. 5A). The Kaplan–Meier curve showed that most patients with PFS longer than 300 days reached a tail plateau and achieved a long-term response. This finding is consistent with the results of previous clinical trials (Supplementary Fig. S8A). Therefore, we first examined the pretreatment and posttreatment changes that occurred in each cluster of patients with PFS < 300 days (n = 30) and PFS > 300 days (n = 20). The percentage of pretreatment Th7R in the PFS > 300 days group was significantly higher than in the healthy volunteers and the PFS < 300 days group (Supplementary Fig. S9A and S9B); no significant differences between the PFS > 300 days and PFS < 300 days groups were observed in Th1 and Th17. No differences were also observed in the percentage of CD8+ T cell and the combined percentage of IFNγ-producing Th1 and Th1/17 (Supplementary Fig. S9C). The pretreatment and posttreatment change analysis showed a decrease in the %Th7R (P < 0.0001) for patients with PFS < 300 days, whereas the %Th7R for patients with PFS >300 days did not show a significant decrease in percentages of Th7R (Fig. 7A; Supplementary Fig. S9D). As a result, the %Th7R difference between the PFS < 300 days group and the PFS > 300 days group was more noticeable after treatment than before (Fig. 7B). These changes in Th7R were thought to be caused by decrease in the number of cells belonging to multicellular clonotype, reflecting the antigen sculpting effect in the cancer immunoediting theory. In the patients with PFS > 300 days, new multicellular clonotypes were induced and the overall number of cells belonging to the multicellular clonotype was not significantly reduced (Supplementary Fig. S10). On the other hand, the change in Th-cell percentages associated with treatment was not observed for Th1 and Th17.
Figure 7.
PD-1 blockade therapy effects on CD4+ Th clusters. A, The percentages of CD62Llow Th clusters before and after pembrolizumab treatment in the lung cancer cohort treated with first-line pembrolizumab therapy divided into PFS < 300 days and PFS > 300 days groups are shown. Statistical analyses were performed using the paired Student t test. B, Comparison of the percentages of Th clusters in the PFS < 300 days and PFS > 300 days groups before and after treatment, respectively, is shown. Statistical analyses were performed using the unpaired Student t test. C and D, The changes in the percentage of PD-1–positive cells and the percentage of T-bet–positive cells in each Th cluster before and after pembrolizumab treatment are shown. Statistical analyses were performed using the paired Student t test.
Moreover, the analysis of T-bet and PD-1 expression in each cluster showed an increase in the T-bet–positive cell percentages in Th1, Th1/17, and CCR6 SP after pembrolizumab treatment (Fig. 7C). In the current study, our results showed that CD3/CD28 stimulation upregulated T-bet gene expression in Th1, Th1/17, and CCR6 SP. Because Th1, Th1/17, and CCR6 SP are clusters with high PD-1 expression, we hypothesized that this was caused by the restoration of TCR/CD28 signaling through the effect of PD-1 inhibitory therapy. Interestingly, PD-1 expression decreased in Th1/17 and CCR6 SP after treatment (Fig. 7D, P < 0.0001). This indicated the recovery from T-cell exhaustion. In contrast, no significant change was found in the in PD-1 expression of Th1 and Th17.
Discussion
Here, we found that the Th7R metacluster, consisting of Th1/17 and CCR6 SP, which forms a third subpopulation distinct from the classical Th1 and Th17 of the CD62LlowCD4+ T-cell subpopulation, is strongly associated with PD-1 blockade therapeutic efficacy. IL7R and TCF7 were identified as characteristic molecules commonly expressed. It was demonstrated that IL7R on T cell is required for the antitumor effect of anti-PD-1 and anti-CTLA4 combination therapy and that both IL7R and TCF7 contributes to survival and proliferation of antitumor effector T cells (28–30). Thus, IL7R and TCF7 expressing Th7R may play an important role as persistently functioning CD4+ T cells in antitumor immunity. Indeed, upon CD3/CD28 stimulation, Th1/17 expressed MYC, BATF, and BIRC3, which are thought to maintain T-cell survival during the antigen-stimulated state.
Apoptosis resistance, plasticity, and ability of self-renewal of cancer-associated Th17 cell is reported to mediate long-lasting antitumor immune responses (31). This study indicated that there was a trend for Th17 to decrease in PFS < 300 days (P = 0.056) and increase in PFS > 300 days (P = 0.070). Cancer and autoimmunity associated Th17 express only CCR6 and change to a Th1-like phenotype, and their reactivity is reported to be highly dependent on IFNγ production (32, 33). Thus, the CCR6 SP cluster, often referred to as Th17 because of its similarity, is likely to have the plasticity to differentiate into Th1/17, which is different from Th17 and has more Th1-like properties, and the results of this study, in which the cell numbers of Th1/17 and CCR6 SP were synchronized, support this.
A network analysis between T cells in peripheral blood and TME revealed that peripheral Th7R strongly correlated with CD4+ T cells in TME and indirectly associated with CD8+ T cells in TME. We found that the Th1 cluster expresses CCL5, whereas the Th1/17 T-cell cluster, a component of the Th7R metacluster, expresses both CCL5 and CD26. Normally, CCL5 binds to CCR1 and CCR3 to promote the migration of monocytes, eosinophils, and neutrophils, but CCL5 cleaved by CD26 (3-68) reduces binding to CCR1 and CCR3 and binds specifically to CCR5 to activate the migration and effector functions of memory T cells (34–36). The expression of CCL20, which is a chemokine for CCR6, was observed in Th1/17, but not in Th1. Thus, Th1/17 can induce more Th7R metacluster cells. However, because this is an observational study, the hypotheses described here need to be verified by genetical modification of chemokine receptor of Th7R and chemokine inhibiting intervention study in the future.
Patients who relapsed with PFS < 300 days showed a significant decrease in Th7R after pembrolizumab treatment, whereas Th7R was maintained in patients with PFS > 300 days who had a long-term response. In other words, the patients with ability to halt the decline of Th7R achieved long-term survival. Interestingly, during PD-1 blockade therapy, Th7R cells downregulated PD-1 and appeared to recover from T-cell exhaustion. These results are consistent with the findings that CD3/CD28 stimulation upregulated the expression of genes that contribute to cell survival, including MYC, BATF, IRF4, BIRC3, and IER3 in Th7R cells. Especially, BATF and IRF4 are reported to counter T-cell exhaustion (26). Clinical trials using anti-PD-1 antibodies have reported that some patients have an antitumor effect for more years after completing the PD-1 blockade therapy (37, 38). Downregulation of PD-1 in Th7R can contribute to the antitumor immunity that no longer requires PD-1 blockade therapy.
It must be cautious that T-cell frequencies obtained by scRNA-seq are based on transcriptionally active cell types. Thus, we used CyTOF analysis to verify and test numeric correlation with antitumor efficacy of the Th7R cells based on surface protein expression. The main limitation of this study is that it is an observational study. We could not determine whether the newly discovered CD4+ T-cell clusters are specifically induced by cancer antigens or whether adoptive transfer mediates the antitumor therapeutic effect. In addition, no genetically modified model experiments have been performed to directly prove the role of the characteristic molecular expression observed in scRNA-seq in the antitumor effect of CD4+ T-cell clusters, and this is a subject for future work.
As shown in this study, the expression of PD-1 and CTLA4 differs greatly among Th polarization clusters, and it is highly likely that the intensity of action of anti-PD-1/PD-L1 and anti-CTLA4 antibodies also differs among Th clusters. Accurate monitoring of circulating antitumor T-cell clusters including Th7R likely provides optimal therapeutic selection.
Supplementary Material
Legends of Supplementary Figures and Tables, and Supplementary Methods.
Supplementary Table 2, 4, 5, 6, 7.
Supplementary Table 1, 3, 8.
Supplementary Figures 1-10.
Acknowledgments
The authors thank Mrs. Koko Kodaira, Mrs. Hiroko Noguchi, and Mrs. Chieko Ono for technical assistance.
This work was supported by KAKENHI program of the Japan Society for the Promotion of Science grant 17H04184, Japan Agency for Medical Research, and Development grant 19ae0101074h0001.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
H. Kagamu reports personal fees from AstraZeneca, Bristol Myers Squibb, Chugai Pharmaceutical Co., and Ono Pharmaceutical Co. and grants from Chugai Pharmaceutical Co., Ono Pharmaceutical Co., Taiho Pharmaceutical Co., and Boeringer Ingelheim outside the submitted work; in addition, H. Kagamu has a patent for Biomarkers for predicting response to cancer therapy pending. S. Kitano reports grants and personal fees from AstraZeneca, Boehringer Ingelheim, Pfizer, MSD, Eisai, Astellas, Ono Pharmaceutical Co., Ltd., GSK, Daiichi-Sankyo, Chugai, Takeda, and Eli Lilly; personal fees from Taiho, Novartis, Sumitomo Pharma, Gilead Sciences, Bristol-Myers Squibb, Rakuten Medical, Merck Biopharma, ImmuniT Research Inc., United Immunity, and PMDA(Pharmaceuticals and Medical Devices Agency); and grants from Regeneron, Takara Bio Inc., Incyte, AMED (Japan Agency for Medical Research and Development), and JSPS(Japan Society for the Promotion of Science) outside the submitted work. O. Yamaguchi reports personal fees from Ono Pharmaceutical Co., Ltd., Bristol Myers Squibb, Chugai Pharmaceutical Co., Ltd., Takeda Pharmaceutical Company Limited, Eli Lilly and Company, and AstraZeneca outside the submitted work. K. Kaira reports grants and personal fees from AstraZeneca and personal fees from Chugai Pharmaceutical, Ono Pharmaceutical Company, Bristol-Myers Company, and Boehringer Ingelheim during the conduct of the study; and grants from AstraZeneca and Eli Lilly outside the submitted work. K. Kobayashi reports personal fees from AstraZeneca and Takeda Pharmaceutical Company outside the submitted work. Y. Kanai reports grants from Japan Agency for Medical Research and Development during the conduct of the study and grants from Japan Agency for Medical Research and Development and Japan Society for the Promotion of Science outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
H. Kagamu: Conceptualization, formal analysis, funding acquisition, visualization, writing–original draft, project administration. S. Yamasaki: Data curation, formal analysis, investigation, visualization, writing–review and editing. S. Kitano: Conceptualization, resources. O. Yamaguchi: Resources. A. Mouri: Resources. A. Shiono: Resources. F. Nishihara: Resources. Y. Miura: Resources. K. Hashimoto: Resources. H. Imai: Resources. K. Kaira: Resources. K. Kobayashi: Resources. Y. Kanai: Resources, investigation. T. Shibata: Resources, investigation. K. Horimoto: Data curation, formal analysis, visualization, writing–review and editing.
References
- 1. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 2. Osorio JC, Arbour KC, Le DT, Durham JN, Plodkowski AJ, Halpenny DF, et al. Lesion-level response dynamics to programmed cell death protein (PD-1) blockade. J Clin Oncol 2019;37:3546–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu TD, Madireddi S, de Almeida PE, Banchereau R, Chen YJ, Chitre AS, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 2020;579:274–8. [DOI] [PubMed] [Google Scholar]
- 4. Yost KE, Chang HY, Satpathy AT. Recruiting T cells in cancer immunotherapy. Science 2021;372:130–1. [DOI] [PubMed] [Google Scholar]
- 5. Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114:1537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J Clin Invest 2014;124:2246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 2018;24:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alspach E, Lussier DM, Miceli AP, Kizhvatov I, DuPage M, Luoma AM, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019;574:696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferris ST, Durai V, Wu R, Theisen DJ, Ward JP, Bern MD, et al. cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature 2020;584:624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018;18:635–47. [DOI] [PubMed] [Google Scholar]
- 11. Sallusto F, Lanzavecchia A. Heterogeneity of CD4+ memory T cells: functional modules for tailored immunity. Eur J Immunol 2009;39:2076–82. [DOI] [PubMed] [Google Scholar]
- 12. Kryczek I, Zhao E, Liu Y, Wang Y, Vatan L, Szeliga W, et al. Human TH17 cells are long-lived effector memory cells. Sci Transl Med 2011;3:104ra0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kagamu H, Shu S. Purification of L-selectin(low) cells promotes the generation of highly potent CD4 antitumor effector T lymphocytes. J Immunol 1998;160:3444–52. [PubMed] [Google Scholar]
- 14. Koyama K, Kagamu H, Miura S, Hiura T, Miyabayashi T, Itoh R, et al. Reciprocal CD4+ T-cell balance of effector CD62Llow CD4+ and CD62LhighCD25+ CD4+ regulatory T cells in small cell lung cancer reflects disease stage. Clin Cancer Res 2008;14:6770–9. [DOI] [PubMed] [Google Scholar]
- 15. Kagamu H, Kitano S, Yamaguchi O, Yoshimura K, Horimoto K, Kitazawa M, et al. CD4(+) T-cell immunity in the peripheral blood correlates with response to anti-PD-1 therapy. Cancer Immunol Res 2020;8:334–44. [DOI] [PubMed] [Google Scholar]
- 16. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 2017;14:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edgar R, Domrachev M, Lash AE. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sallusto F. Heterogeneity of human CD4(+) T cells against microbes. Annu Rev Immunol 2016;34:317–34. [DOI] [PubMed] [Google Scholar]
- 23. Chopp LB, Gopalan V, Ciucci T, Ruchinskas A, Rae Z, Lagarde M, et al. An integrated epigenomic and transcriptomic map of mouse and human alphabeta T cell development. Immunity 2020;53:1182–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 2010;327:1098–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shyer JA, Flavell RA, Bailis W. Metabolic signaling in T cells. Cell Res 2020;30:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seo H, Gonzalez-Avalos E, Zhang W, Ramchandani P, Yang C, Lio CJ, et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat Immunol 2021;22:983–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang W, Zhu J, Song X, Xu Z, Xue X, Chen X, et al. An association of Aquaporin-4 with the immunoregulation of liver pathology in mice infected with Schistosoma japonicum. Parasit Vectors 2015;8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shi LZ, Fu T, Guan B, Chen J, Blando JM, Allison JP, et al. Interdependent IL-7 and IFN-gamma signalling in T-cell controls tumour eradication by combined alpha-CTLA-4+alpha-PD-1 therapy. Nat Commun 2016;7:12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van der Leun AM, Thommen DS, Schumacher TN. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat Rev Cancer 2020;20:218–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei S, Zhao E, Kryczek I, Zou W. Th17 cells have stem cell-like features and promote long-term immunity. Oncoimmunology 2012;1:516–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu D, Notarbartolo S, Croonenborghs T, Patel B, Cialic R, Yang TH, et al. Transcriptional signature of human pro-inflammatory TH17 cells identifies reduced IL10 gene expression in multiple sclerosis. Nat Commun 2017;8:1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, et al. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 2011;12:255–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oravecz T, Pall M, Roderiquez G, Gorrell MD, Ditto M, Nguyen NY, et al. Regulation of the receptor specificity and function of the chemokine RANTES (regulated on activation, normal T cell expressed and secreted) by dipeptidyl peptidase IV (CD26)-mediated cleavage. J Exp Med 1997;186:1865–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Proost P, De Meester I, Schols D, Struyf S, Lambeir AM, Wuyts A, et al. Amino-terminal truncation of chemokines by CD26/dipeptidyl-peptidase IV. Conversion of RANTES into a potent inhibitor of monocyte chemotaxis and HIV-1-infection. J Biol Chem 1998;273:7222–7. [DOI] [PubMed] [Google Scholar]
- 36. Lim JK, Burns JM, Lu W, DeVico AL. Multiple pathways of amino terminal processing produce two truncated variants of RANTES/CCL5. J Leukoc Biol 2005;78:442–52. [DOI] [PubMed] [Google Scholar]
- 37. Gettinger S, Horn L, Jackman D, Spigel D, Antonia S, Hellmann M, et al. Five-year follow-up of nivolumab in previously treated advanced non-small-cell lung cancer: results from the CA209–003 study. J Clin Oncol 2018;36:1675–84. [DOI] [PubMed] [Google Scholar]
- 38. Reck M, Ciuleanu TE, Lee JS, Schenker M, Audigier-Valette C, Zurawski B, et al. First-line nivolumab plus ipilimumab versus chemotherapy in advanced NSCLC with 1% or greater tumor PD-L1 expression: patient-reported outcomes from CheckMate 227 Part 1. J Thorac Oncol 2021;16:665–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends of Supplementary Figures and Tables, and Supplementary Methods.
Supplementary Table 2, 4, 5, 6, 7.
Supplementary Table 1, 3, 8.
Supplementary Figures 1-10.
Data Availability Statement
The RAW data of single-cell RNA sequencing (scRNA-seq) generated in this study are publicly available in Gene Expression Omnibus (21) at GSE168844.