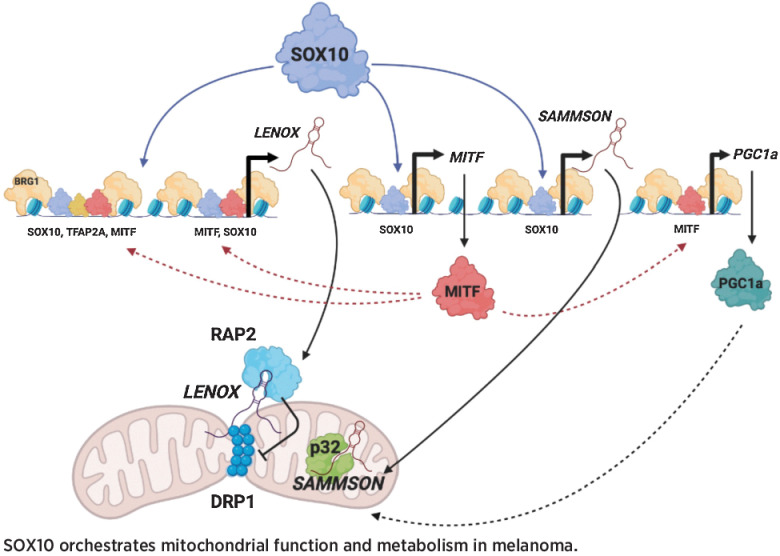
The lncRNA LENOX is a novel regulator of melanoma metabolism, which can be targeted in conjunction with MAPK inhibitors to eradicate melanoma cells.
Abstract
Tumor heterogeneity is a key feature of melanomas that hinders development of effective treatments. Aiming to overcome this, we identified LINC00518 (LENOX; lincRNA-enhancer of oxidative phosphorylation) as a melanoma-specific lncRNA expressed in all known melanoma cell states and essential for melanoma survival in vitro and in vivo. Mechanistically, LENOX promoted association of the RAP2C GTPase with mitochondrial fission regulator DRP1, increasing DRP1 S637 phosphorylation, mitochondrial fusion, and oxidative phosphorylation. LENOX expression was upregulated following treatment with MAPK inhibitors, facilitating a metabolic switch from glycolysis to oxidative phosphorylation and conferring resistance to MAPK inhibition. Consequently, combined silencing of LENOX and RAP2C synergized with MAPK inhibitors to eradicate melanoma cells. Melanomas are thus addicted to the lncRNA LENOX, which acts to optimize mitochondrial function during melanoma development and progression.
Significance:
The lncRNA LENOX is a novel regulator of melanoma metabolism, which can be targeted in conjunction with MAPK inhibitors to eradicate melanoma cells.
Graphical Abstract
Introduction
Intratumor heterogeneity is a major determinant of therapeutic resistance. Melanoma tumors are notoriously heterogeneous comprising cell populations with distinct properties and gene expression signatures (1–5). Rare vulnerabilities common to most melanoma cell states have been identified and successfully exploited to overcome therapy resistance, such as inhibition of mitochondrial translation (6).
Transcription factor SOX10 is an essential determinant of heterogeneity in melanoma. Melanocytic state cells express microphthalmia-associated transcription factor (MITF) and SOX10, whereas neural crest-like cells express SOX10, but not MITF (4, 7). In melanocytic cells, MITF and SOX10 bind together at cis-regulatory elements to promote proliferation, survival, and cell metabolism (8–11). Targeted therapies can induce the undifferentiated state expressing neither MITF nor SOX10 (4, 12–14). Upon MAP kinase inhibition, the neural crest and undifferentiated states play key roles in minimal residual disease and the emergence of drug-resistant populations (15, 16).
Long noncoding (Lnc) RNAs are regulators of virtually every process in the cell (17) in particular adaptive processes involved in tumor progression and therapy resistance (18–20). The melanoma-specific lncRNA SAMMSON (Linc01212) is a SOX10-regulated gene essential for melanoma cell proliferation and survival (21, 22). SAMMSON inhibition severely affects mitochondrial function, inducing proteotoxic stress, resulting in cell death in vitro and tumor regression in preclinical settings when combined with MAPK inhibition (21).
Here we characterize the melanoma-specific lncRNA LENOX (LINC00518) expressed in all known melanoma states and essential for proliferation and survival. LENOX interacts with the RAP2C small GTPase promoting its interaction with DRP1 and impairing mitochondrial fission through enhanced DRP1 S637 phosphorylation. The resulting increase in mitochondrial elongation and optimal oxidative phosphorylation promotes melanoma cell survival and resistance to MAP kinase inhibitors.
Materials and Methods
Analysis of RNA sequencing data
Single-cell RNA sequencing (RNA-seq) data from MEL006 patient-derived xenograft (PDX) tumors (GSE116237) were processed using Seurat and genes were divided into protein-coding and noncoding based on their biotype (Ensembl 104). Mean expression level in each melanoma population was calculated excluding cells in which the gene was not captured. SD across the melanoma populations was divided by mean expression to calculate the coefficient of variation and rank genes. The 50 protein-coding and noncoding genes with lowest coefficient of variation were identified and their expression was measured across normal tissues and melanoma samples in the genotype-tissue expression (GTEx) and The Cancer Genome Atlas (TCGA) databases, respectively. All other used RNA sequencing datasets are indicated in the text.
Cell culture and GapmeR transfections
Melanoma cell lines Sk-mel-25, Sk-mel-25R, Sk-mel-28, and 501Mel were grown in RPMI1640 medium supplemented with 10% fetal calf serum (FCS) and gentamycin; IGR-37 and IGR-39 in RPMI1640 medium supplemented with 15% FCS and gentamycin. MM011, MM117, MM047, and MM099 were grown in HAM-F10 medium supplemented with 10% FCS, 5.2 mmol/L glutamax, 25 mmol/L Hepes, and penicillin/streptomycin (7.5 μg/mL). M229, M229R, M249, and M249R were grown in DMEM medium supplemented with glucose (4.5 g/L), 5% FCS, and penicillin/streptomycin (7.5 μg/mL). A375 cells were grown in DMEM medium supplemented with glucose (4.5 g/L), 10% FCS, and gentamycin. HEK293T cells were grown in DMEM medium supplemented with glucose (1 g/L), 10% FCS, and penicillin/streptomycin (7.5 ug/mL). To assess cell growth and viability cells were stained with Trypan Blue (Invitrogen). Vemurafenib (PLX4032), trametinib (GSK1120212), and dabrafenib (GSK2118436) were purchased from Selleckchem. Sk-mel-25, Sk-mel-28, A375, and 501Mel were obtained from ATCC, all other cell lines were gifts from collaborators. All cell lines were regularly tested using the Venor GeM Mycoplasma Detection Kit, and used at less than 10 passages.
GapmeR and siRNA were transfected using Lipofectamine RNAiMAX (Invitrogen) with 20 nmol/L of GapmeR (Qiagen) or siRNA (Thermo Fisher Scientific). GapmeRs and siRNAs sequences are listed in Supplementary Table S1. For combination, GapmeR experiments cells were transfected with 15 nmol/L of LENOX GAP#2 and/or 5 nmol/L SAMMSON Gapmer. For vemurafenib/trametinib+dabrafenib-GapmeR cotreatment, cells were cultured for 3 days in presence or absence (DMSO only) of vemurafenib (1 μmol/L), transfected with 15 nmol/L of control GapmeR, LENOX GAP#2, siRAP2C or siTFAP2A, then cultured for additional 3 days before harvesting. Colony-forming ability was assessed by plating 500 cells/9.6 cm2, for 10 days and fixing in formalin and staining with 0.05% Crystal Violet solution (Sigma Aldrich).
Melanosphere formation assay
501Mel cells were plated in 10 cm petri dishes without any coating in KO DMEM medium supplemented with 25% KSR, AANE, 2.5 mmol/L Glutamax, 125 μg/mL penicillin/streptomycin, and 50 mmol/L β-mercaptoethanol. Every 3 days pictures of 10 different areas uniformly distributed across the petri were taken by light microscopy. Quantification was performed by ImageJ to calculate the mean and standard deviation for each sample.
Plasmid cloning and lentiviral transduction
GFP-RAP2B was a gift from Philip Stork (Addgene plasmid #118321; http://n2t.net/addgene:118321; RRID:Addgene 118321), whereas pLJC2-RAP2A-3xFLAG was a gift from David Sabatini (Addgene plasmid #87974; http://n2t.net/addgene:87974; RRID:Addgene_87974). LENOX isoforms and RAP2C cDNAs were synthesized by Genscript under the control of a CMV promoter. These plasmids were used for transient transfection of HEKT using PEI (Polysciences, ref. 23966). LENOX and RAP2 cDNAs were cloned into the pCW57-GFP-P2A-MCS vector (a gift from Adam Karpf; Addgene plasmid #71783; http://n2t.net/addgene:71783; RRID: Addgene_71783). A LENOX shRNA (shLENOX) or a scrambled control (shCTRL) were cloned in LT3GEPIR (a gift from Johannes Zuber; Addgene plasmid #111177; http://n2t.net/addgene:111177; RRID:Addgene_111177). Lentiviral particles were produced in HEK293T cells, purified by ultracentrifugation, and resuspended in PBS. After titration, melanoma cells were infected at MOI of 1 and selected by puromycin addition to the media (1 μg/mL).
CRISPR interference
501Mel cells were cotransfected with plasmid expressing dead Cas9 protein fused to the Kruppel-associated box (KRAB) domain-containing KAP1 (dCas9-KAP1) and the red fluorescent protein mScarlet (pX-dCas9-KRAB-Scarlet), together with a plasmid expressing GFP and three single guide RNAs targeting the transcription start site of LENOX (pcDNA3-sgRNA-GFP) or a control plasmid expressing GFP only (pCMV-GFP). Double Scarlet-GFP positive cells were sorted 24 hours after cotransfection, stained with Cell Trace Violet and cultured for additional 96 hours.
A375 xenograft model and bioluminescent imaging
Swiss nude mice were purchased from Charles River Laboratories (France) and housed under specific pathogen‐free conditions. Animal care, use, and experimental procedures were conducted in accordance with recommendations of the European Community (86/609/EEC) and Union (2010/63/UE) and the French National Committee (87/848). The ethics committee of IGBCM in compliance with institutional guidelines approved animal care and use (APAFIS#31519–2021051708529028). A375 cells expressing the Dox-inducible shLENOX or scrambled control (shCTRL) were transduced with a lentiviral vector containing the Firefly luciferase gene (kind gift of Catherine-Laure Tomasetto, IGBMC). Mice were injected on the rear flank with 5 × 106 cells resuspended in 100 μL of PBS+ Cultrex Basement Membrane Extract (ref. 3432–005–01; R&D Systems). After 4 days, shRNAs and GFP were induced by Dox administration (ref. D9891; Sigma Aldrich) in drinking water supplemented with 5% sucrose. Tumor growth was monitored by caliper measurement every 2 days and volume was calculated with the formula: 4/3 × π × length/2 × width/2 × h/2. After sacrifice, primary tumors were dissected and dissociated as single cells using the MACS Tumor Dissociation Kit (ref. 130–095–929; Milteny Biotech). Cells were stained with Zombie violet (ref. 423113; BioLegend) and with an alexa-647 anti-H2kq antibody (ref. 115106; BioLegend) to identify dead and murine cells, respectively. Bioluminescence imaging was performed at 7 and 14 days after injection. Prior to imaging, mice were injected intraperitoneally with 100 μL of Xenolight D-luciferin potassium salt (15 mg/mL, #122799; Perkin Elmer). Mice were anesthetized with 5% isoflurane and transported in a sterile cage inside the IVIS Spectrum Imager (Perkin Elmer). Bioluminescence acquisition was performed in auto-mode and expressed as radiance (photon/second).
Patient-derived xenografts
The cutaneous melanoma MEL006res is part of Trace collection (https://gbiomed.kuleuven.be/english/research/50488876/54502087/Trace). MEL006res was derived from the BRAFV600E MEL006 PDX originally sensitive to the dabrafenib–trametinib combination (DT) and rendered resistant by continuous DT treatment (23). Written informed consent was obtained from the patient and all procedures involving human samples were approved by the UZ Leuven/KU Leuven Medical Ethical Committee (S63799) and carried out in accordance with the principles of the Declaration of Helsinki and with GDPR regulations. The experiments were approved by the KU Leuven animal ethical committee under ECD P164–2019 and performed in accordance with the internal, national, and European guidelines of Animal Care and Use. Mice were maintained in IVC cages in a semi-pathogen-free facility under standard housing conditions with continuous access to food and water. The KU Leuven animal facilities comply with all appropriate standards [cages, space per animal, temperature (22°C), light, humidity, food, water], and all cages are enriched with nesting materials. Tumor pieces were implanted subcutaneously in the hip of female NMRI nude BomTac:NMRI-Foxn1nu, 10-week-old females (Taconic Biosciences). Mice were engrafted with PDX MEL006res and drug naïve tumors were grown to 100 mm3 before administration of antisense oligonucleotides (ASO; 15 mg/kg) every second day by subcutaneous injection in the back. A second cohort was treated daily with DT by oral gavage with 30 mg and 0.3 mg/kg DT, respectively, was started when the tumors became palpable. Once tumors reached 200 mm3, the mice were randomly assigned to the different cohorts and treated with daily with ASO as described above.
Analysis of oxygen consumption rate in living cells
Oxygen consumption rate (OCR) was measured in an XF96 extracellular analyzer (Seahorse Bioscience). 20,000 transfected cells per well were seeded 48 hours prior the experiment. The cells were incubated at 37°C and the medium was changed to XF base medium supplemented with 1 mmol/L pyruvate, 2 mmol/L glutamine, and 10 mmol/L glucose for 1 hour before OCR profiling with the Mitostress Test Kit sequentially exposed to 2 μmol/L oligomycin, 1 μmol/L carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and 0.5 μmol/L rotenone and antimycin A. Cells were washed with PBS, fixed with 3% PFA, permeabilized with 0.2% triton. Nuclei were counterstained with DAPI (1:500) and number of cells per well determined.
Proliferation and viability analyses by flow cytometry
To assess cell viability and proliferation, cells were stained with Cell Trace Violet (Invitrogen) on the day of transfection harvested after 72 hours and stained with Annexin V (BioLegend) and TOPRO-3 (Invitrogen) or the active caspase-3 Kit (BD Biosciences). Cells were analyzed on a LSRII Fortessa (BD Biosciences) and data were analyzed with FlowJo software (TreeStar). To assess cytochrome C content, 501Mel and A375 cells were transfected with the indicated GapmeRs or siRAP2C, permeabilized with digitonin (50 μg/mL) for 5 minutes on ice, fixed in 4% PFA for 20 minutes at room temperature, and incubated in blocking buffer (3% BSA, 0.05% saponin in PBS) for 1 hour. Cells were stained overnight at 4°C with an anti-cytochrome C mAb coupled to Alexa Fluor 647 (BioLegend, #612310). The day after, cells were stained with Zombie Violet and the anti-active caspase-3 antibody for 30 minutes at room temperature and analyzed by flow cytometry as above.
Cell-cycle progression analysis by flow cytometry
Cells were stained using the Click-it Edu Kit (Thermo Fisher Scientific) and TOPRO-3. Briefly, cells were cultured with 10 μmol/L Edu for 1.5 hours, harvested by trypsinization, washed once in 1% BSA-PBS, and fixed in 4% PFA for 15 minutes at room temperature. After a wash in 1% BSA-PBS cells were permeabilized with saponin-permeabilization buffer for 15 minutes and stained with the Click-it reaction cocktail for 30 minutes. Finally, they were washed once in 1% BSA-PBS, resuspended in 500 μL of PBS with 10 nmol/L TOPRO-3, and left for 10 minutes at room temperature.
Intracellular reactive oxygen species analysis by flow cytometry
Cells were stained in adherent conditions with CellRox Deep Red (Thermo Fisher Scientific) at final concentration of 500 nmol/L following manufacturer instructions. After harvesting, cells were stained for active caspase-3 (BD Biosciences) and analyzed on a LSRII Fortessa (BD Biosciences). To induce reactive oxygen species (ROS), cells were treated with THBP (200 μmol/L) for 30 minutes, and to inhibit ROS induction, cells were treated with NAC (1 μmol/L) for 1 hour before THBP administration. To induce apoptosis, cells were treated with staurosporine (500 nmol/L) for 16 hours.
MitoTracker analysis by flow cytometry
Cells were stained with Annexin V and after one wash in Annexin V binding buffer and PBS, diluted in PBS+5% FCS and stained with MitoTracker CMXRos Red (200 nmol/L) for 25 minutes at 37°C. After one wash in PBS, cells were stained with TOPRO-3 for 10 minutes at room temperature. For FCCP-treated samples, cells were incubated with 50 μmol/L FCCP for 30 minutes before MitoTracker staining. Samples were analyzed on a LSRII Fortessa (BD Biosciences).
RNAscope
RNAs for LENOX, MITF, and SOX10 were detected with the RNAscope assay (Advanced Cell Diagnostics, ACD) according to the manufacturer's protocols. Patient sections were deparaffinized, incubated with hydrogen peroxide at room temperature for 10 minutes, boiled with target retrieval reagent for 15 minutes, and then treated with protease plus reagent at 40°C for 30 minutes. Sections were hybridized with Hs-MITF probe (ACD, catalog no. 310951), Hs-SOX10 probe (ACD, catalog no. 484121), at 40°C for 2 hours. Probes for Hs-LENOX and were custom designed by ACD. Hybridization signals were amplified and visualized with RNAscope Multiplex Fluorescent Reagent Kit v2 (ACD, catalog no. 323100). For co-detection of RAP2 and p32 with LENOX, cells were fixed for 30 minutes with formaldehyde 3.7%, washed with PBS and incubated 10 minutes at room temperature with H2O2. After one wash in distilled water, primary antibodies diluted in co-detection diluent (1/100 for RAP2, 1/200 for p32) were added o/n at 4°C. Slides were washed in PBS+tween 0.1% (PBST), fixed in formaldehyde 3.7% for 30 minutes, and washed again in PBST. Slides were treated with protease III and washed with PBS. LENOX hybridization signals were amplified following the Multiplex Fluorescent Kit. Finally, RAP2 and p32 signals were developed by secondary antibodies incubation (diluted 1/2,000 in co-detection diluent), followed by tyramide signal amplification (TSA Plus Kit, NEL760001KT, Perkin Elmer). Images were captured with a confocal (Leica DMI6000) microscope.
Immunofluorescence of fixed cells
Cells grown on glass slides in 24-well plates, were fixed with 4% paraformaldheyde for 15 minutes. After two washes with PBS buffer they were permeabilized in PBS+triton X-100 0.1% for 5 minutes and blocked with PBS+10% FCS inactivated for 20 minutes. Primary antibodies were incubated overnight at 4°C and after three washes with PBS+Triton 0.1%, cells were stained for 1 hour at room temperature with Alexa Fluor-488 conjugated secondary antibodies (Life technologies) diluted 1/500 in PBS+10% FCS. After three washes with PBS+Triton 0.1%, cells were stained with DAPI (final concentration 1 μg/mL) and mounted on microscopy slides. Anti-TP53BP1 (NB100–304), anti-RAP2 (Santa Cruz Biotechnology, sc-515711), and anti-p32 (Bethyl, A302–863A) were diluted 1/200 in PBS+10% FCS; anti-γH2AX (Ser139, Abcam, ab11174) 1/400. Images were captured with a confocal (Leica DMI6000) microscope. RAP2-p32 colocalization and the number of gH2Ax or TP53BP1 positive nuclear foci were quantified using ImageJ. Mander's correlation coefficient of RAP2 over p32 was calculated using JACoP.
MitoTracker live imaging
Cells were cultured in 4 wells 35 × 10 mm dishes (CellView, Greiner Bio-one), stained for 1 hour with MitoTracker Red CMXRos (125 nmol/L) and Hoechst 33342 (1 μg/mL) and z-stack images acquired on a confocal (Leica DMI6000) microscope in a temperature controlled (37°C) chamber. Mitochondrial shape and network analyses were performed on ImageJ as described previously (24, 25). Briefly, projections of multiple z-stack sections into one image were performed by generating a maximum intensity composite and pre-processed using “subtract background” (radium 1um), “sigma filter plus” (radius 0.1 μm, 2.0 sigma), “enhance local contrast/CLAHE” (block size 64, slope 2.0), “gamma correction” (0.8), and tubeness (sigma 0.361). The adaptive threshold plugin (https://sites.google.com/site/qingzongtseng/adaptivethreshold#use) was used to identify mitochondria and the image post-processed using “despeckle.” The resulting binary image was used as the input for the “analyze particles” command, measuring for area, perimeter, and shape descriptors. Form factor (FF) was derived as the inverse of the circularity (26). For network connectivity analysis, the “skeletonize 2D/3D” command was applied to produce a skeleton map and the “analyze skeleton” command calculated the number of branches and branch junctions in the network.
RNA extraction and qPCR
Total mRNA isolation was performed using TRIzol and isopropanol precipitation. Isolation of cytosolic, nuclear soluble, and chromatin-associated RNA was performed as described in ref. 27. Briefly, cells were harvested and washed in PBS buffer, resuspended in 0.15% NP-40 lysis buffer, and centrifuged on a 24% sucrose cushion taking supernatant as the cytosolic fraction. Nuclei were resuspended in 1M urea, 1% NP-40 lysis buffer, and centrifuged to recover the nuclear soluble fraction in the supernatant. The chromatin pellet was finally resuspended in 1 mL of TRIzol, solubilized using a 21-gauge needle, and isolated following manufacturer instructions. Cyosolic and nuclear soluble fractions were cleared by centrifugation and RNA was isolated from 200 μL of each using 1 mL of TRIzol. Total and fractionated RNAs were treated with DNAseI following the TurboDnase Free Kit instructions (Thermo Fisher Scientific) and reversed transcribed using Superscript IV reverse transcriptase (Thermo Fisher Scientific) following manufacturer instructions. qRT-PCR was carried out with SYBR Green I (Roche) and monitored by a LightCycler 480 (Roche). Target gene expression was normalized using TBP, HBMS, GAPDH, ACTB, RPL13A as reference genes. Primers for RT-qPCR were designed using Primer3 and listed in the Supplementary Table S2.
Protein extraction and Western blotting
Whole cell extracts were prepared by freeze–thaw technique using LSDB 500 buffer [500 mmol/L KCl, 25 mmol/L Tris at pH 7.9, 10% glycerol (v/v), 0.05% NP-40 (v/v), 16 mmol/L DTT, and protease inhibitor cocktail]. Lysates were subjected to SDS-PAGE and proteins were transferred onto a nitrocellulose membrane. Membranes were incubated with primary antibodies in TBS+ 5% BSA + 0.01% Tween-20. Overnight at 4°C. The membrane was then incubated with HRP-conjugated secondary antibody (Jackson ImmunoResearch) for 1 hour at room temperature, and visualized using the ECL detection system (GE Healthcare). Antibodies used are listed in Supplementary Table S3.
Mitochondria fractionation
Mitochondria were isolated with the Mitochondria Isolation Kit (Thermo Fisher Scientific) following manufacturer instructions. Harvested cells were washed and pelleted, resuspended in buffer A, and incubated 2 minutes on ice. Buffer B was added for 5 minutes, vortexing every minute, and diluted with buffer C. Nuclei were pelleted 10 minutes at 700 × g and supernatant centrifuged for 15 minutes at 3,000 × g. Purified mitochondria were washed once in buffer C and used for RNA (TRIzol–isopropanol precipitation) or protein (TBS+CHAPS 2%) extraction.
LENOX pulldown and LC/MS-MS analysis
MM011 cells were grown in 15 cm petri dishes, harvested by trypsinization, washed, pelleted, resuspended in lysis buffer (TrisHCl 20 mmol/L pH8, NaCl 200 mmol/L, MgCl2 2.5 mmol/L, Triton 0.05%, DEPC water) supplemented with fresh DTT (1 mmol/L), protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific) and RNAsin (Thermo Fisher Scientific) and kept 20 minutes on ice. Membranes were pelleted 3,000 × g for 3 minutes at 4°C and supernatant precleared for 1 hour at 4°C with streptavidin-coated sepharose beads. The lysate was incubated 2 hours with streptavidin coated beads and 400 pmol anti-PCA3 or LENOX-specific DNA biotinylated oligonucleotides (listed in Supplementary Table S4). Beads were pelleted for 3 minutes at 3,000 × g and washed three times with lysis buffer. After final wash beads were divided for RNA and protein extraction. RNA was purified by TRIzol and isopropanol precipitation, digested with DNAse, reverse transcribed and analyzed by qPCR for LENOX and TINCR. Proteins were eluted by boiling beads in Laemli sample buffer and separated on NuPAGE Novex 4% to 12% gradient gels. Three independent experiments were performed and the entire lane was excised in seven consecutive bands and subjected to “in-gel” digestion. Proteins were reduced in 10 mmol/L DTT for 1 hour at 56ºC and alkylated with 55 mmol/L iodoacetamide for 45 minutes at room temperature. Enzymatic digestion was performed using 12.5 ng/μL trypsin overnight at 37ºC. Tryptic peptides were extracted from the gel with 3% trifluoroacetic acid (TFA) and 30% acetonitrile (ACN). The extracted peptides were concentrated onto homemade StageTips reversed phase microcolumns. Peptides were eluted in 40 μL buffer B [80% ACN, 0.1% formic acid (FA)]. ACN was evaporated using a vacuum concentrator (Speed Vac, Eppendorf) and the volume of the eluates were adjusted to 5 μL with 1% TFA for peptide separation and analysis in a reversed-phase nano-flow liquid chromatographic (nRP-LC) column using an EASY-nLC 1200 (Thermo Fisher Scientific) coupled to an Q-Exactive HF instrument (Thermo Fisher Scientific) through a nano-electrospray ion source (EASY-Spray; Thermo Fisher Scientific). The nRP-LC system was operated in one column set-up with an EasySpray PEPMAP RSLC C18 (Thermo Fisher Scientific) kept at 45°C constant. Solvent A was 0.1% FA and solvent B was 0.1% FA in 80% ACN. Samples were loaded in aqueous 0.1% (FA) solution at constant pressure (980 Bar). Peptides were separated with a gradient of 3% to 30% solvent B over 69 minutes followed by a gradient of 30% to 60% for 5 minutes and 60% to 95% over 5 minutes at a flow rate of 300 nL/min. The Q-Exactive was operated in the data-dependent acquisition (DDA) mode and MS spectra (from m/z 375–1,550) were analyzed in the Orbitrap detector with resolution R = 60,000 at m/z 200. The fifteen most intense peptide ions were isolated to a target value of 3e6 and fragmented by Higher Energy Collision Dissociation (HCD) with a normalized collision energy (NCE) setting of 28. The maximum allowed ion accumulation times was 80 milliseconds for MS-MS. The dynamic exclusion time was set to 20 seconds.
Post-acquisition MS data analysis for proteomics
Acquired raw data were analyzed with the integrated suite of algorithms MaxQuant, version 1.6.1.1, using the Andromeda search engine. FDR for both peptides and protein identifications was set to a maximum of 0.01. Carbamidomethylation of cysteine was set as a fixed modification. Uniprot Human sequence database was used for peptide identification (74470 Entries). LFQ intensity calculation was enabled requiring a minimum LFQ ratio count equal to two. The match between runs (MBR) feature was selected and a tolerance of 0.7 minutes was specified for the match time window option. The “protein groups” (.txt) output file from MaxQuant was processed by Perseus software for statistics. Briefly, no imputation for missing values was used, and the data were filtered, to have 3 valid values in at least one group. A t Student test was used to compare protein co-enriched in the LENOX- versus the PCA3- pulldown experiments and the threshold settings to select significant enriched proteins were S0 = 0 and FDR = 0.05.
Immunoprecipitation
Cells were grown in 15 cm petri dishes, harvested by scraping, resuspended in lysis buffer (20 mmol/L Tris-HCl pH 8, 200 mmol/L NaCl, 2.5 mmol/L MgCl2, 0.05% Triton, DEPC water) supplemented with DTT (1 mmol/L), protease/phosphatase inhibitor cocktail (Thermo Fisher Scientific) and RNAsin (Thermo Fisher Scientific) and kept on ice for 15 minutes, pipetting every 3 minutes. Membranes were pelleted 10 minutes at 10,000 × g at 4°C and the supernatant precleared 1 hour at 4°C with protein G magnetic beads. Lysate was quantified by Bradford protein quantification assay (Bio-Rad) and incubated overnight at 4°C with indicated antibodies. Protein G magnetic beads were added for 3 hours at 4°C to isolate RNA–protein complexes and washed five times in lysis buffer. After final wash RNA was purified by TRIzol and isopropanol precipitation and proteins eluted by boiling beads in Laemli sample buffer.
Proximity ligation assay
The PLA Duolink FarRed protocol (Sigma Aldrich, ref. DUO92008) was performed following manufacturer's instructions. Primary antibodies recognizing RAP2 (sc-515711) and DRP1 (12957–1-AP) were used at a concentration of 1:200. PLA Probe Anti-Rabbit PLUS and PLA Probe Anti-Mouse MINUS were used.
Statistical analysis
Comparison between experimental groups were mainly performed using one-way ANOVA, Dunnett, or Tukey multiple comparison test, as indicated in legends. Other statistical tests are indicated in the appropriate legends (*, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001).
Resources
All oligonucleotides and antibodies used are listed in Supplementary Tables S1 to S4.
Data availability
The datasets generated during and/or analyzed in this study are available from the corresponding authors E. Leucci or I. Davidson upon request.
Results
LENOX is expressed in all known melanoma states and is associated with melanoma progression
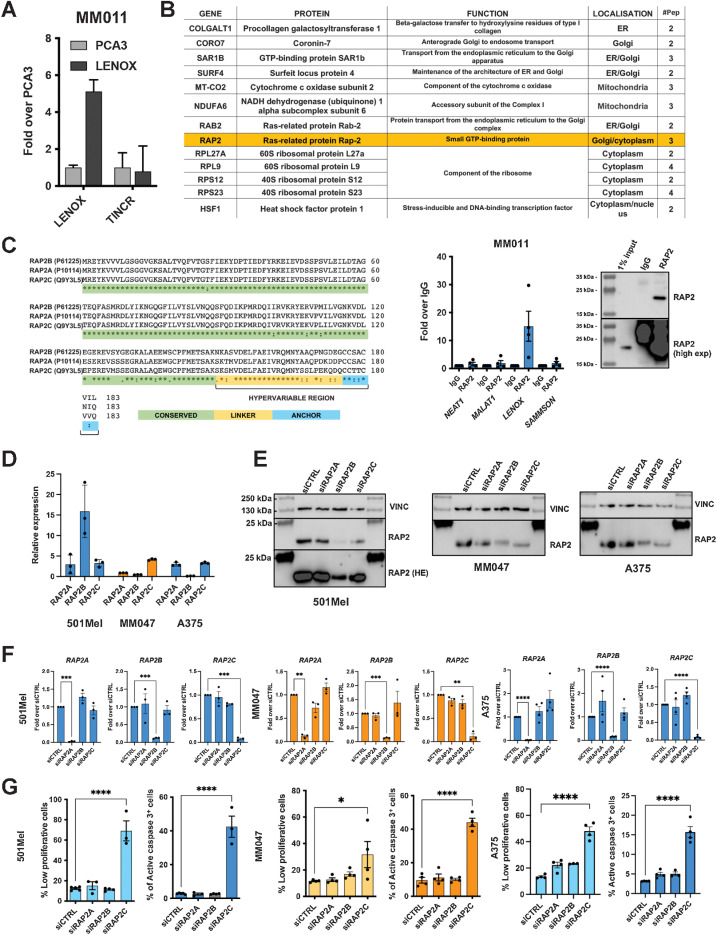
To identify melanoma-specific genes common to all cellular states, we re-analyzed singe cell (sc)RNA-seq of a melanoma PDX undergoing targeted MAP kinase inhibition (MAPKi) therapy (4). We selected the 50 mRNAs and lncRNAs with the lowest coefficient of variation across all melanoma cell states (Fig. 1A). Protein coding genes associated with housekeeping functions were expressed in the different melanoma states and across normal tissues. However, LINC00518 (hereafter LENOX) was highly expressed in human melanoma and across melanoma cell states, but poorly in normal tissues (Fig. 1A and B).
Figure 1.
LENOX is expressed in normal skin melanocytes and melanoma. A, Top 50 protein-coding and noncoding transcripts based on expression across melanoma states (GSE116237) and in GTEx and SKCM TCGA. LENOX is highlighted by dark bars. B, Expression of LENOX, SAMMSON, and TFAP2A in melanoma subpopulations. Only cells where the RNAs were captured were included in the analysis. The percentage of cells analyzed for each population is indicated. C, RNAscope detection of LENOX and MITF in normal skin and melanoma FFPE sections. D, LENOX expression in melanocytic (blue) and undifferentiated (orange) cell lines. E, LENOX expression in 501Mel cells expressing the dCas9-KAP1 protein with control or LENOX promoter-targeting sgRNAs. F–H, Proliferation, apoptosis, and colony formation following dCas9-KAP1-mediated LENOX silencing compared with negative control by one-way ANOVA (Dunnett test). I and J, Proliferation and apoptosis of LENOX or control GapmeR-transfected cells compared by one-way ANOVA (Dunnett test). K, Tumor growth of A375 cells expressing shCTRL or shLENOX (N = 5). L, Bioluminescence emission was measured on day 7 and 14 using the IVIS Spectrum Image and compared by Mann–Whitney test. M, Growth of MEL006res PDX treated with control or LENOX targeting ASO. Tumor size was measured daily for 20 days. Statistical analysis were calculated using two-way ANOVA. *, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001.
In normal tissues LENOX expression was restricted to sun-exposed and nonexposed skin and testis (Supplementary Fig. S1A), but it was strongly and specifically expressed in cutaneous and uveal melanoma (Supplementary Fig. S1B). LENOX expression in MITF-expressing melanocytes was confirmed by RNAscope on sections from normal skin (Fig. 1C) and in MITF-expressing cells of primary melanoma (Fig. 1C). LENOX was expressed in melanoma cells, but not in infiltrating immune or stromal cells (Supplementary Fig. S1C). LENOX expression was upregulated in melanoma compared with nevi (Supplementary Fig. 1D; refs. 28, 29) and increased with Clark score (Supplementary Fig. S1E) and increased copy number due to its coamplification with TFAP2A in around 6.5% to 8% of melanomas (Supplementary Fig. S1F). High LENOX expression was associated with decreased survival of patients with melanoma (Supplementary Fig. S1G). RT-qPCR on a collection of melanocytic and undifferentiated lines confirmed that LENOX was consistently expressed irrespective of cell state and driver mutation (Fig. 1D).
The LENOX promoter comprises a MLT1B ERVL-MaLR retroviral long terminal repeat conferring primate specificity analogous to SAMMSON containing the LTR1 ERV1 element (30). Juxtaposition of the MLT1B element to a SINE element (MIRb) created a SOX10 binding site and prominent SOX10 binding was observed with that of MITF, BRG1, and the H3K27ac mark (Supplementary Fig. S1H). Accordingly, LENOX was regulated by both MITF and SOX10 in 501Mel cells (Supplementary Fig. S1I; ref. 10]. However, LENOX expression across cell states suggested additional modes of regulation. Public ChIP-seq data from melanocytic and undifferentiated melanoma cells identified several potential enhancer elements (EN 1–6) in the TFAP2A/LENOX locus (Supplementary Fig. S1H; refs. 10, 31, 32). For example, EN1 was marked by H3K27ac, H3K4me1, TFAP2A, and FOSL2, whereas the LENOX promoter showed SOX10 binding and H3K4me3. EN1 and EN5 showed binding of TFAP2A that is expressed in drug-tolerant states (Fig. 1B). siTFAP2A silencing diminished LENOX expression in 501Mel and MM047 lines, but had little effect on SAMMSON (Supplementary Fig. S1J). Furthermore, LENOX and TFAP2A expression were positively correlated in TCGA and in melanoma cell lines (Supplementary Fig. S1K; refs. 2, 33). In contrast, LENOX silencing (see below) did not affect TFAP2A expression (Supplementary Fig. S1L). Combinatorial regulation by MITF, SOX10, and TFAP2A may account for LENOX expression across melanoma cell states in accordance with the observation that LENOX and TFAP2A are part of a gene signature discriminating melanomas from other tumors and positively correlating with disease aggressiveness (designated C6ORF218 in ref. 34).
LENOX is required for melanoma cell proliferation and survival
To silence LENOX, we transfected melanoma cells with a vector encoding the CRISPR/dCAS9-KAP1 fusion protein with sgRNAs specific for the LENOX promoter that reduced its expression by more than 90% compared with control (Fig. 1E), resulting in an increase in slow proliferating cells (Fig. 1F), in Annexin V–positive apoptotic cells (Fig. 1G) and decreased colony formation (Fig. 1H).
We designed two independent locked nucleic acid ASOs (LNA-GapmeRs) whose transfection reduced LENOX expression by over 80% compared with a nontargeting control (CTR; Supplementary Fig. S2A). Reduced cell numbers were detected 72 hours after LENOX silencing in melanocytic and undifferentiated lines (Supplementary Fig. S2B), but not in HEK293T cells where LENOX was not expressed. GapmeR-mediated silencing resulted in strongly reduced cell proliferation (Fig. 1I) and apoptosis (Fig. 1J; Supplementary Fig. S2C) with early and late apoptotic cells observed in flow cytometry using Annexin V labeling (Supplementary Figs. S2D and S2E). We also silenced LENOX using a doxycycline (Dox)-inducible shRNA coupled to GFP for monitoring by flow cytometry (Supplementary Fig. S3A). LENOX was strongly decreased in 501Mel and A375 cells, with reduced proliferation (Supplementary Figs. S3B and S3C).
A375 cells were injected subcutaneously in immunodeficient mice to form tumors and Dox-induced GFP confirmed in tumor cells (Supplementary Fig. S3D). Cells expressing LENOX shRNA showed strongly impaired tumor growth confirmed by bioluminescence measurements (Fig. 1K and L; Supplementary Figs. S3E and S3F). Tumors were significantly smaller and in two cases regressed. We also targeted LENOX in mice engrafted with the BRAFV600E cutaneous melanoma PDX MEL006res (23). In drug naïve conditions, the LENOX GAP#2 ASO significantly reduced LENOX expression and PDX growth compared with control GAP ASO (Fig. 1M; Supplementary Fig. S3G). All three targeting strategies therefore supported the essential role of LENOX in melanocytic, neural crest, and undifferentiated melanoma cells irrespective of driver mutation.
Databases predict several alternatively spliced LENOX isoforms sharing a common last exon with three potential polyadenylation sites mapped by 3′RACE as present in all isoforms (Supplementary Fig. S4A). Exon–exon junctions were confirmed on 501Mel RNA-seq data. Isoforms 1, 3, and 5 were most abundantly expressed in human melanomas and in melanoma cell lines (Supplementary Fig. S4B). We generated 501Mel cells with Dox-inducible expression of isoforms 1, 2 or 3, or GFP as control showing a time-dependent increase of their expression (Supplementary Figs. S4C and S4D). Dox treatment increased proliferation of LENOX overexpressing cells, their colony formation and growth as 3D melanospheres (Supplementary Figs. S4E–S4G). Sustained LENOX expression was confirmed in spheroids after 12 days of culture (Supplementary Fig. S4H). Analogous observations were made in A375 cells (Supplementary Figs. S4I and S4J). Thus, although LENOX silencing compromised melanoma cell proliferation and survival, its ectopic expression promoted growth under 2D and 3D conditions.
LENOX interacts with the RAP2 GTPases
RNAscope showed that LENOX was predominantly expressed in the cytoplasm around the nuclear periphery in melanoma cells in vivo (Fig. 1C) and in cultured melanoma cells confirmed by RT-qPCR on RNA from different cell compartments (Supplementary Figs. S5A and S5B). Reconstitution of 3D cellular images showed LENOX enrichment in the cytoplasm (Fig. 1C; Supplementary Fig. S5A).
To identify LENOX interacting proteins, we performed pulldown from cytoplasmic extracts of MM011 cells using a tiling array of LENOX-complementary biotinylated oligonucleotides, or negative control prostate cancer lincRNA PCA3 oligonucleotides followed by MS. LENOX was enriched using its cognate oligonucleotides, but not those of the PCA3 control, whereas lincRNA TINCR was not enriched under any conditions (Fig. 2A). Proteins found uniquely in the LENOX pulldown included several ribosomal proteins or endoplasmic reticulum, Golgi, or mitochondrial proteins (Fig. 2B, top). To assess their interaction with LENOX, we immunoprecipitated (IP) RAP2, SURF4, SAR1B, and NDUFA6. However, only the antibody against RAP2 efficiently precipitated its target protein from cell extracts (Fig. 2B, bottom; Supplementary Fig. S5C). RAP2 IP fractions showed strong enrichment of LENOX, but not SAMMSON, MALAT, or NEAT1. As additional controls, we performed IP of XRN2 that enriched SAMMSON and NEAT1, but not LENOX (Supplementary Fig. S5C).
Figure 2.
LENOX interacts with the RAP2 GTPases. A, Enrichment of LENOX or TINCR after oligonucleotide-mediated pulldown from MM011 cells. B, Top, proteins detected by mass spectrometry only in the LENOX pulldown from MM011 cells. Bottom left, selective enrichment of LENOX following RAP2 IP. Bottom right, IP of RAP2 with 1% input showing short and long exposures. C, RAP2A, B, and C protein sequences illustrating amino acid homology. D, RAP2A/B/C expression in indicated cells normalized over housekeeping genes. E and F, RAP2A/B/C expression in the above cells after transfection with indicated siRNAs compared with siCtrl by one-way ANOVA (Dunnett test). VINCULIN was used as a loading control in right panel. G, Proliferation and apoptosis following silencing with indicated siRNA and comparison with control by one-way ANOVA (Dunnett test). *, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001.
The RAP2 small GTPases are encoded by 3 paralogous genes RAP2A, RAP2B, and RAP2C with high amino acid identity, but distinguished by a short hypervariable C-terminal region (Fig. 2C). Consequently, the RAP2 antibody recognizes all three paralogs. Each paralog was expressed across melanoma cell states, but also in stromal and immune cells with RAP2A showing the lowest expression in melanoma patients (Supplementary Figs. S5D–S5G).
The 501Mel, A375, and MM047 cells showed variable levels of each paralog (Fig. 2D). Silencing of each with specific siRNAs (Fig. 2E and F) revealed that only silencing of RAP2C induced slow growth and apoptosis thus phenocopying LENOX silencing (Fig. 2G; Supplementary Figs. S5H–S5J). We generated cell lines with Dox-inducible expression of FLAG-tagged RAP2A, B, or C (Supplementary Fig. S5K). RAP2C protein accumulated to higher levels than RAP2A or B. FLAG-IP of each recombinant protein or pan-RAP2 IP of endogenous RAP2 from the GFP-control cells showed that LENOX, but not SAMMSON, was enriched with all RAP2 paralogues proportionate to their variable expression levels (Supplementary Fig. S5L). Thus, although LENOX interacted with all RAP2 paralogs, RAP2C was the biologically relevant form required for proliferation and survival.
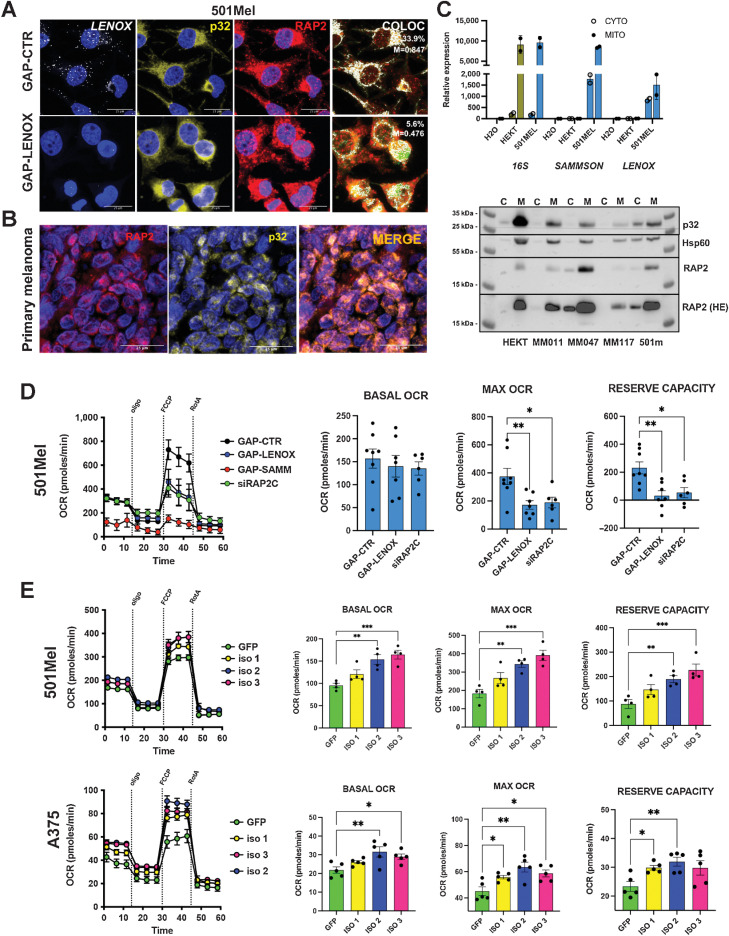
LENOX and RAP2C promote oxidative phosphorylation
Coupling RNAscope with immune-staining showing overlapping localized cytoplasmic RAP2 and LENOX (Fig. 3A). RAP2 and LENOX labeling tightly coincided with the mitochondrial p32 protein, and RAP2-p32 colocalization was further seen in primary melanoma (Fig. 3B). Purification of mitochondrial and cytoplasmic fractions showed the presence of 16S mitochondrial ribosomal RNA almost exclusively in the mitochondrial fraction, whereas LENOX and SAMMSON were present in both the cytoplasmic and mitochondrial fractions (Fig. 3C; Supplementary Fig. S6A). Mitochondrial purity was confirmed by strong enrichment of p32 and HSP60 in the mitochondrial fraction where RAP2 was also enriched even in HEK293T cells (Fig. 3C).
Figure 3.
LENOX and RAP2 modulate oxidative phosphorylation. A, Co-imaging of LENOX, p32, and RAP2 in 501Mel cells transfected with the indicated GapmeRs. B, RAP2-p32 colocalization in a human melanoma FFPE section. C, Levels of 16S rRNA, SAMMSON, and LENOX in mitochondrial and cytoplasmic fractions (top) and levels of RAP2, P32, and HSP60 proteins (bottom) D. Mitostress test of 501Mel cells 48 hours after transfection with indicated GapmeRs or siRAP2C. Experimental groups were compared by one-way ANOVA (Dunnett test). E, Mito Stress Test on cells with ectopic LENOX isoform expression. Experimental groups were compared as in D. *, P < 0.033; **, P < 0.0021; ***, P < 0.0002.
Although RAP2 and p32 colocalized in cells transfected with control GapmeR, LENOX silencing resulted in a 5-fold decrease of RAP2-p32 colocalization with RAP2 signal seen in the surrounding cytoplasm (Fig. 3A; Supplementary Fig. S6B). This was not due to decreased RAP2 levels that were unaffected by LENOX silencing (Supplementary Figs. S6C and S6D). ShLENOX knockdown strongly reduced LENOX, but not SAMMSON expression in both the cytoplasm and mitochondria fractions, reduced RAP2-p32 colocalization and reduced RAP2 levels in mitochondria (Supplementary Figs. S6E–S6G).
We investigated the effect of LENOX and RAP2C silencing on mitochondrial activity by profiling OCR as a measure of oxidative phosphorylation (OXPHOS). LENOX or RAP2C silencing did not affect basal OCR, but potently decreased maximal and reserve capacities (Fig. 3D). SAMMSON knockdown potently reduced OCR under all conditions. Ectopic LENOX expression increased basal, maximal, and reserve OCR in both 501Mel and A375 cells (Fig. 3E).
To link compromised OXPHOS to decreased cell growth and apoptosis, we asked if LENOX or RAP2C silencing and the impaired mitochondrial function and induced ROS. We performed flow cytometry on cells labeled with Annexin V, TOPRO, and MitoTracker Red CMXRos and gated on the nonapoptotic Annexin V/TOPRO-negative cells. Compared with control, LENOX or RAP2C silencing led to the appearance of higher proportions of CMXRos low cells indicative of reduced mitochondrial polarization that was strongly induced using FCCP as a positive control (Supplementary Fig. S7A).
We next stained LENOX, RAP2C, or control silenced cells with anti-caspase-3 and Cell ROX to distinguish nonapoptotic cells (Q4) from apoptotic cells (Q3) and cells with elevated ROS (Q1; Supplementary Fig. S7B). As positive control, THBP (tert-Butyl hydroperoxide) induced elevated ROS that was suppressed using the ROS scavenger N-acetyl-L-cysteine (NAC), whereas staurosporine induced apoptosis, but not ROS. LENOX or RAP2C silencing induced a large increase in apoptotic cells, but also cells with augmented ROS as well as apoptotic ROS-high cells (Q2). Increased ROS following LENOX or RAP2C silencing was associated with DNA damage observed by increased γH2AX and TP53BP1 foci, not seen following RAP2A or control silencing (Supplementary Figs. S7C and S7D). LENOX or RAP2C silencing also increased the population of cells arrested in G2–M phase (Supplementary Fig. S7E). We further gated cells negative for caspase-3 and zombie violet to select nonapoptotic cells that were analyzed for cytochrome C content. Compared with controls, LENOX or RAP2 silencing significantly increased the number of cytochrome C low cells, indicating increased cytochrome C leakage may also promote subsequent cell apoptosis (Supplementary Fig. S7F).
LENOX or RAP2C silencing therefore led to mitochondrial depolarization, generating ROS and cytochrome C leakage, inducing DNA damage, cell-cycle block and ultimately apoptosis.
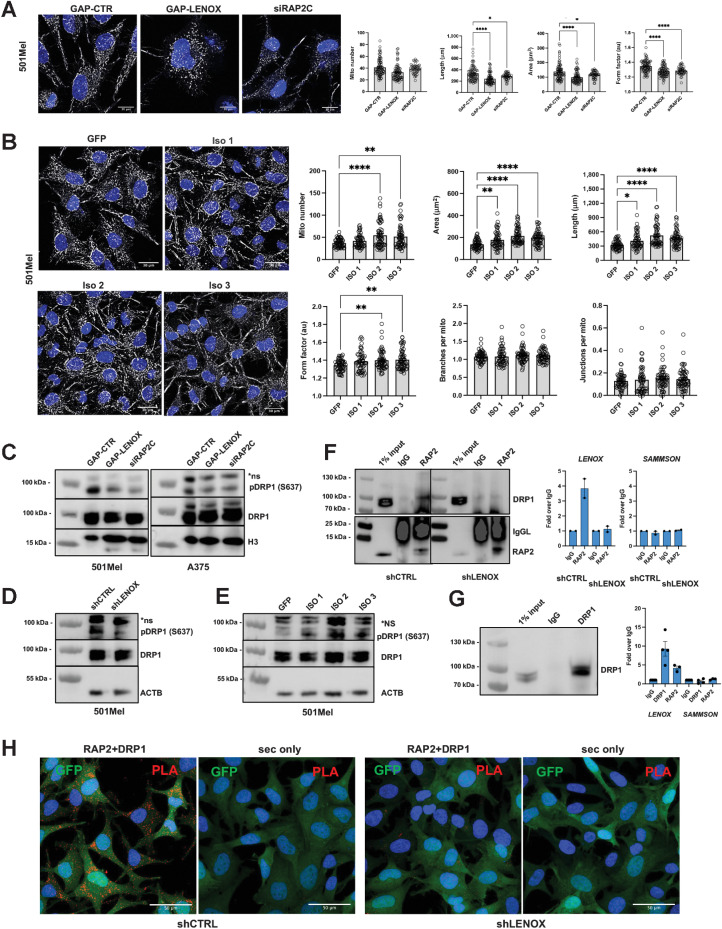
LENOX and RAP2C regulate DRP1 phosphorylation and mitochondrial fission
LENOX or RAP2C silencing did not alter the total number of mitochondria per cell, but led to smaller, shorter, and rounder mitochondria with reduced numbers of branches and junctions (Fig. 4A; Supplementary Figs. S8A and S8B). In contrast, ectopic LENOX expression increased mitochondrial size and length, but did not induce a hyperfused phenotype as the numbers of branches and junctions were unaffected (Fig. 4B).
Figure 4.
LENOX promotes RAP2 interaction with DRP1 regulating its phosphorylation on serine 637 and mitochondrial fusion. A, MitoTracker CMXROS Red staining of 501Mel cells transfected with the indicated GapmeR or siRAP2C. MitoTracker signal is shown in grayscale and Hoescht in blue. Total mitochondrial count, area, perimeter, and mean form factor were calculated and compared by one-way ANOVA (Dunnett test). B, MitoTracker CMXROS Red staining of 501Mel cells with ectopic LENOX isoform expression. Mitochondrial parameters were calculated and compared as above. C–E, Phospho-DRP1 S637 and total DRP1 levels in 501Mel and A375 cell extracts, with H3 and ACTB as loading controls. F, Left, RAP2 IP from 501Mel cells expressing shRNAs revealed with antibodies to the indicated proteins. Right, LENOX and SAMMSON levels in the indicated IP fractions. G, IP of DRP1 from 501Mel cells (left) and LENOX and SAMMSON levels in the IP (right). H, PLA-mediated detection RAP2–DRP1 interaction in 501Mel cells. Appropriate negative controls using secondary antibodies only are shown. *, P < 0.033; **, P < 0.0021; ****, P < 0.0001.
Mitochondrial homeostasis is dynamically regulated by a balance between phosphorylation of S616 of the GTPase DRP1 by ERK2 that stimulates mitochondrial fission (35, 36) and phosphorylation at S637 inhibiting DRP1 and promoting mitochondrial fusion (37–39). LENOX or RAP2C silencing reduced DRP1 S637 phosphorylation (Fig.4C and D), whereas it was increased in cells ectopically expressing LENOX (Fig. 4E) consistent with the changes in mitochondrial morphology.
We investigated interactions between DRP1 and RAP2. DRP1 was coprecipitated with RAP2 antibody, but not control IgG, in cells expressing control shRNA, but lost in shLENOX-expressing cells showing their LENOX-dependent interaction (Fig. 4F). Accordingly, LENOX, but not SAMMSON, was enriched in both the RAP2 and DRP1 IPs, but not the control IP (Fig. 4F and G). Similarly, LENOX was enriched in the RAP2 IP in control cells, but not in shLENOX cells, whereas SAMMSON was not enriched under any condition (Fig. 4F). In an independent approach, proximity ligation assay with DRP1 and RAP2 antibodies showed their cytoplasmic interaction in control cells that was strongly diminished in the shLENOX cells (Fig. 4H). LENOX therefore promotes a RAP2–DRP1 interaction that enhanced S637 phosphorylation, mitochondrial fusion, and optimized OXPHOS.
RAP2 associated with mitochondria in HEK293T cells (Fig. 3C) prompting us to investigate RAP2 function in these cells in absence of LENOX. RAP2B and RAP2C knockdown induced the strongest reductions of RAP2 protein levels, however, no effect on proliferation or apoptosis was seen upon knockdown of any RAP2 paralog (Supplementary Figs. S9A and S9B). Accordingly, LENOX-GapmeR or siRAP2C did not induce changes in mitochondrial morphology (Supplementary Fig. S9C). In contrast, ectopic LENOX expression by transient transfection, in particular of isoforms 2 and 3, induced a RAP2-DRP1 interaction not seen in GFP transfected cells (Supplementary Figs. S9D–S9F). Moreover, ectopic LENOX expression led to increased mitochondrial elongation in transfected cells and increased OCR (Supplementary Figs. S9G and S9H).
Hence, despite its mitochondrial association, RAP2 did not regulate OXPHOS and/or cell viability in HEK293T cells. In contrast, ectopic LENOX expression promoted a RAP2-DRP1 interaction, hijacking RAP2 to regulate mitochondrial homeostasis and function.
Cooperativity of LENOX and SAMMSON
Expression of LENOX and SAMMSON across melanoma cell states and their complementary roles on mitochondrial function suggested they may act cooperatively with one another. Compared with individual SAMMSON or LENOX knockdown with suboptimal concentrations of GapmeR for each lincRNA, combinatorial knockdown led to a more potent reduction in 501Mel and MM011 proliferation (Fig. 5A) and a strong increase in their apoptosis (Figs. 5B). Similarly, combinatorial knockdown in undifferentiated MM047 cells cooperatively induced slow proliferation and induced a potent increase in apoptosis (Fig. 5A and B). The synergistic effect of combinatorial silencing was evident after 10 days of culture, by which time, essentially all the MM047 cells were eliminated (Fig. 5C). LENOX and SAMMSON therefore cooperatively promoted melanoma cell proliferation and survival.
Figure 5.
LENOX and SAMMSON cooperatively promote melanoma cell survival. A and B, Proliferation and apoptosis after transfection with suboptimal doses of LENOX or SAMMSON GapmeRs as single or pairwise combinations compared between groups by one-way ANOVA. C, Crystal violet staining of cells transfected as indicated and cultured for 10 days. Percentages of area occupied in each condition were compared by one-way ANOVA. *, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001.
LENOX and RAP2C are required for increased OXPHOS upon BRAF inhibition
As LENOX and RAP2C regulated mitochondrial morphology and OXPHOS under basal growth conditions, we asked if this pathway was also important upon metabolic stress such as suppression of glycolysis by BRAF inhibitors that promote a metabolic switch to OXPHOS (40, 41).
M229 cells treated with BRAF inhibitor vemurafenib (2) showed upregulated LENOX expression after 3 days that returned to basal level at later times. Similarly, TFAP2A, SOX10, and PAX3 were all upregulated during this acute phase, whereas SAMMSON was reduced (Supplementary Fig. S10A). We confirmed upregulated LENOX, TFAP2A, SOX10, and PAX3 expression between 12 and 72 hours in vemurafenib-treated M229, Sk-Mel-25, 501Mel, and A375 cells (Supplementary Fig. S10B). Consistently, LENOX levels in mitochondria increased upon vemurafenib treatment, whereas SAMMSON displayed a progressive reduction (Supplementary Fig. S10C). LENOX, TFAP2A, and SOX10 upregulation was also seen with encorafenib, DT, and confirmed for LENOX in A375 and 501Mel cells (Supplementary Figs. S10D and S10F). Increased LENOX expression after MAPKi was also observed in MEL006 PDX treated with DT and in triple wild-type patients treated with trametinib and durvalumab (Supplementary Figs. S10G and S10H), suggesting that LENOX induction was not restricted to BRAF mutant melanoma.
Upregulation of TFAP2A protein was observed upon vemurafenib, dabrafenib, or trametinib exposure (Supplementary Fig. S10I). Consistently, TFAP2A-bound EN#1 and 5 EN#5 showed increased H3K27ac levels in vemurafenib-treated A375 cells (Supplementary Fig. S10J). The LENOX promoter showed an analogous increase and stronger BRD4 recruitment that was abrogated by concomitant THZ1 treatment. RNA-seq data confirmed that upregulation of LENOX, TFAP2A, and SOX10 was also inhibited by THZ1 (Supplementary Fig. S10K). These results highlight the role of TFAP2A and SOX10 in LENOX induction by MAPKi treatment.
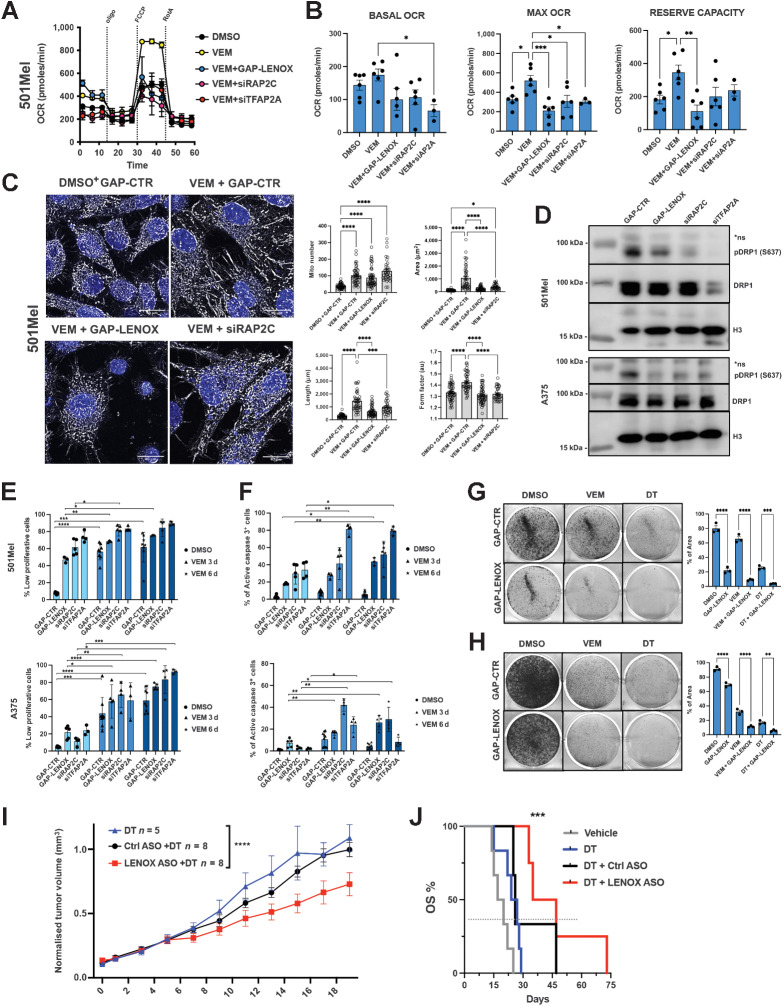
We investigated if vemurafenib-induced LENOX expression was required to stimulate OXPHOS and promote cell survival and hence an increased vulnerability to LENOX silencing. LENOX, RAP2C, or TFAP2A silencing strongly reduced the vemurafenib-mediated increase in basal, reserve, and maximal OCR compared with DMSO control cells showing the essential role of this axis in the adaptive metabolic switch (Fig. 6A and B). LENOX knockdown was comparable in DMSO- and vemurafenib-treated cells and its levels were also reduced by siTFAP2A, corroborating its role in LENOX induction (Supplementary Fig. S10L).
Figure 6.
LENOX and RAP2C promote metabolic switch upon BRAF inhibition. A and B, Mito Stress Test of 501Mel cells grown for 3 days with DMSO or vemurafenib (1 μmol/L) and transfected as indicated. Experimental groups were compared by one-way ANOVA (Tukey test). C, MitoTracker CMXROS Red staining of cells treated with DMSO or vemurafenib, transfected as indicated and analyzed by confocal microscopy as in Fig. 4A. D, Phospho-DRP1 S637 and total-DRP1 levels in cells treated as above. H3 was used as loading control. E and F, Proliferation and apoptosis of cells treated as indicated. Experimental groups were compared by one-way ANOVA. G and H, Crystal violet staining of GapmeR-transfected cells. Percentages of occupied areas were compared by one-way ANOVA. I, Growth of MEL006res PDX in mice treated with DT with or without nontargeting ASO (DT+GAP-CTR) or LENOX-targeting ASO (DT+GAP-LENOX). Tumor size was measured daily for 20 days. Statistics were calculated using two-way ANOVA (Šidák multiple comparisons test). J, Kaplan–Meier plot showing overall survival (OS) of mice described in I. DT, n = 5; DT+GAP-CTR, n = 3; DT+GAP-LENOX ASO, n = 4. Statistical analysis were calculated by log-rank (Mantel–Cox) test. *, P < 0.033; **, P < 0.0021; ***, P < 0.0002; ****, P < 0.0001.
In accordance with increased OXPHOS, vemurafenib-treated cells displayed a more complex and elongated mitochondria network compared with DMSO (Fig. 6C; Supplementary Figs. S10M and S10N). LENOX or RAP2C silencing in vemurafenib-treated cells restored the rounder and shorter morphology, reduced mitochondrial connectivity and DRP1 S637 phosphorylation (Fig. 6D). LENOX and RAP2C therefore promoted mitochondrial fusion required for adaptive increase of OXPHOS upon vemurafenib treatment.
Vemurafenib potently induced arrest of cell proliferation after 3 and 6 days, which was not enhanced by LENOX, RAP2C, or TFAP2A silencing that otherwise strongly reduced proliferation of DMSO control cells (Fig. 6E). In contrast, vemurafenib alone did not induce apoptosis during this period (Fig. 6F). Importantly, vemurafenib-treated cells displayed increased apoptosis compared DMSO controls after LENOX, RAP2C, or TFAP2A silencing that was particularly evident at 6 days. DT treated cells were also more sensitive to LENOX silencing (Fig. 6G and H). Thus, MAPKi cooperated with LENOX silencing to induce apoptosis underlining the critical role of the TFAP2A/LENOX /RAP2C axis in the metabolic adaptation to drug treatment.
We tested whether LENOX silencing could inhibit growth of the MEL006res PDX in presence of MAPKi. Mice were engrafted and once palpable, the tumors were treated with DT and the resistant lesions subsequently challenged with either control GAP ASO (DT+GAP-CTR) or LENOX GAP#2 ASO (DT+GAP-LENOX). Compared with DT alone, or DT with control GAP-ASO, tumor progression with LENOX GAP#2 ASO was strongly and significantly reduced, leading to better overall survival (Fig. 6I and J), revealing that LENOX targeting inhibited MAPKi resistant PDX tumor growth.
We assessed OXPHOS in vemurafenib-treated cells ectopically expressing LENOX. In control cells, OCR levels were stimulated by vemurafenib and further increased by ectopic LENOX expression (Supplementary Fig. S11A). As vemurafenib stimulates endogenous LENOX expression, overall LENOX levels were higher in vemurafenib-treated cells than in the DMSO controls (Supplementary Fig. S11B). Furthermore, inhibition of melanospheres by vemurafenib treatment was rescued by ectopic LENOX expression (Supplementary Fig. S11C). Loss and gain of LENOX underscore its essential role of LENOX in the adaptive response to MAPKi.
As MAPKi treatment inhibited glycolysis and increased sensitivity to LENOX silencing, we asked whether such vulnerability was observed under other conditions where glycolysis was inhibited. Growth under low glucose or in presence of galactose upregulated both TFAP2A and LENOX expression (Supplementary Figs. S11D and S11E), highlighting the role of TFAP2A as a metabolic sensor promoting compensatory gene expression changes such as increased LENOX. Increased DRP1 S637 phosphorylation and OXPHOS were also observed and reduced by LENOX silencing, particularly in cells grown in galactose (Supplementary Figs. S11F and S11H). Thus, inhibition of glycolysis was compensated by a LENOX-dependent increase in OXPHOS increasing their vulnerability to LENOX loss (Supplementary Fig. S11I).
Discussion
Metabolic reprogramming is a hallmark of cancer cells allowing them to adapt to changing and often stressful environments while balancing energy production with the availability of metabolites used to fuel anabolic process required for cell proliferation (42, 43). Emerging evidence suggests that targeting mitochondrial function and ATP generation may represent a common vulnerability across different cell states and tumor types (23). Here we describe how LENOX regulates mitochondrial dynamics and OXPHOS under basal, MAPKi, and metabolic stress conditions.
We identify the poorly characterized GTPase RAP2C as a binding partner of LENOX mediating its biological effects. RAP2 proteins have been proposed as intermediate activation molecules in the LATS1/2–YAP/TAZ signaling cascade (44). However, in melanoma cells, RAP2C has a distinct function in regulating mitochondrial fusion and metabolism. LENOX or RAP2C silencing was associated with increased fission and reduced maximal and reserve but not basal OXPHOS levels. Gain of LENOX function increased mitochondrial fusion and stimulated basal, maximal, and reserve capacities, further linking mitochondrial fusion status to altered OXPHOS capacity (45).
Mitochondrial homeostasis involves several GTPases (46) including DRP1 that forms oligomeric rings to drive fission through GTP hydrolysis, a process dynamically regulated by its posttranslational modification (46, 47). RAP2C interacted with DRP1 in a LENOX-dependent manner to enhance S637 phosphorylation and mitochondrial fusion and promote survival by limiting ROS production and cytochrome C release. DRP1 inhibition and mitochondrial fusion inhibit apoptosis induced by a variety of stress situations (38, 39, 48, 49). Moreover, fine tuning of DRP1 activity to increase mitochondrial fusion and OXPHOS has further been associated with increased oncogenic transformation (50, 51). LENOX may therefore optimize OXPHOS and survival to favor oncogenic transformation and subsequent progression in stressful cellular environments encountered during primary tumor growth and metastatic dissemination (52). We note also that increased DRP1 S637 phosphorylation by LENOX-RAP2C associated with increased melanoma cell proliferation was distinct from a P53-dependent increase in phospho-S637 observed at the onset of cellular senescence in nonmelanoma lines (53).
TFAP2A acts as a metabolic sensor that promotes OXPHOS and cell survival by upregulating LENOX in response to metabolic stress induced by MAPKi and/or glycolysis inhibition. Consequently, MAPKi or glycolysis inhibited cells showed enhanced vulnerability to LENOX silencing since reducing OXPHOS capacity under conditions where glycolysis was also impaired potently induced cell death. Moreover, ASO-mediated LENOX targeting in vivo, inhibited growth of both drug naïve melanoma PDX and MAPKi-resistant PDX suggesting LENOX inhibition as a promising therapeutic strategy for drug-resistant melanoma. Combinatorial targeting of glycolysis and mitochondrial metabolism in tumors has indeed been previously proposed as a therapeutic approach (54, 55).
Intriguingly, SAMMSON and LENOX are located adjacent to the MITF and TFAP2A loci, respectively, and are coamplified with them in 8% to 10% of melanoma. These lincRNAs may have acquired their melanoma specific functions through genomic association with these highly expressed loci. As each lincRNA regulated complementary pathways converging on mitochondria, their combinatorial silencing potently induced apoptosis, suggesting an effective therapeutic option to target undifferentiated cell states in minimal residual disease and relapse. This cooperativity highlights how critical hallmark functions of MITF, SOX10, and TFAP2A are mediated by their protein-coding targets and cooperatively acting lincRNAs. SOX10 orchestrates multiple aspects of mitochondrial function, directly regulating SAMMSON and activating MITF that drives PPARGC1A and LENOX expression promoting mitochondrial biogenesis and homeostasis (21, 40).
In conclusion, melanomas are addicted to both SAMMSON and LENOX that play distinct and complementary functions to optimize protein translation and mitochondrial function, two critical needs of cancer cells.
Supplementary Material
Legends to Supplementary Figures and Tables
LENOX is associated with melanoma clinical features and regulated by TFAP2A.
LENOX regulates melanoma cell proliferation and survival
LENOX is required for xenograft growth
Gain of LENOX function stimulates melanoma cell proliferation
RAP2 paralog expression in melanoma cells
LENOX regulates RAP2 mitochondrial localisation
LENOX and RAP2C knockdown induce mitochondrial dysfunction, release of reactive oxygen species (ROS), DNA damage and cell cycle block
LENOX regulates mitochondrial morphology
Ectopic LENOX expression increases oxidative phosphorylation in HEKT cells
MAPK inhibition increases LENOX expression downstream of TFAP2A.
LENOX overexpression promotes resistance to BRAF and glycolysis inhibition.
Acknowledgments
The authors thank J-C. Marine and G. Ghanem for the MM-series primary melanoma cells, D. Lipkser and the Dermatology Clinic of Strasbourg University Hospital for patient melanoma sections, the staff of the IGBMC common facilities. This work was supported by grants from the ITMO Cancer, the Ligue Nationale contre le Cancer, the Fondation ARC 2020 PJA3, the ANR-10-LABX-0030 and ANR-10-IDEX-0002–02. Work in the Leucci laboratory was supported by the Melanoma Research Alliance (MRA; https://doi.org/10.48050/pc.gr.80542) grants, the Amanda and Jonathan Eilian Young Investigator Award 2018, the Belgian Federation for Cancer grant FAF-F/2018/1184, and by the FWO-KOTK grant (#G097620N). Trace staff were supported by Stichting Tegen Kanker grant 2016-054. Work in the Bonaldi and Cuomo laboratory was supported by EPIC-XS, project number 823839, funded by the Horizon 2020 program of the European Union. ID is an ‘équipe labellisée’ of the Ligue Nationale contre le Cancer. G. Gambi was supported by fellowships from the Fondation pour la Recherche Médicale and the SATT-Conectus Alsace. V. Katopodi is a recipient of the FWO PhD fellowship 1S47519N.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
G. Gambi reports personal fees from FRM during the conduct of the study; personal fees from SATT Conectus outside the submitted work; and also has a patent for EP20305074.5 (Antisense oligonucleotide targeting Linc00518 for treating melanoma) pending. V. Katopodi reports grants from FWO during the conduct of the study. E. Leucci reports grants from Belgian Federation for Cancer FAF-F/2018/1184, Melanoma Research Alliance (MRA; https://doi.org/10.48050/pc.gr.80542), and FWO-KOTK grant (#G097620N) during the conduct of the study. I. Davidson reports grants from ITMO Cancer and Ligue National Contre le Cancer during the conduct of the study; in addition, I. Davidson has a patent for Antisense oligonucleotide targeting LINC00518 for treating melanoma (EP20305074.5) pending. No disclosures were reported by the other authors.
Authors' Contributions
G. Gambi: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–original draft. G. Mengus: Investigation, methodology. G. Davidson: Formal analysis, investigation. E. Demesmaeker: Investigation, methodology. A. Cuomo: Data curation, formal analysis, investigation, methodology. T. Bonaldi: Data curation, formal analysis, methodology. V. Katopodi: Investigation, methodology. G.G. Malouf: Resources, funding acquisition, methodology. E. Leucci: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. I. Davidson: Conceptualization, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 2014;5:5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J, et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 2018;33:890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ennen M, Keime C, Gambi G, Kieny A, Coassolo S, Thibault-Carpentier C, et al. MITF-high and MITF-low cells and a novel subpopulation expressing genes of both cell states contribute to intra and inter-tumoral heterogeneity of primary melanoma. Clin Cancer Res 2017;23:7097–107. [DOI] [PubMed] [Google Scholar]
- 4. Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, et al. Toward minimal residual disease-directed therapy in melanoma. Cell 2018;174:843–55. [DOI] [PubMed] [Google Scholar]
- 5. Rambow F, Marine JC, Goding CR. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev 2019;33:1295–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vendramin R, Konnova A, Adnane S, Cinque S, Katopodi V, Knezevic Z, et al. Activation of the integrated stress response in drug-tolerant melanoma cells confers vulnerability to mitoribosome-targeting antibiotics. Cancer Biology; 2020Available from:http://biorxiv.org/lookup/doi/10.1101/2020.06.26.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laurette P, Coassolo S, Davidson G, Michel I, Gambi G, Yao W, et al. Chromatin remodellers Brg1 and Bptf are required for normal gene expression and progression of oncogenic Braf-driven mouse melanoma. Cell Death Differ 2019;27:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 2011;30:2319–32. [DOI] [PubMed] [Google Scholar]
- 9. Vivas-Garcia Y, Falletta P, Liebing J, Louphrasitthiphol P, Feng Y, Chauhan J, et al. Lineage-restricted regulation of SCD and fatty acid saturation by MITF controls melanoma phenotypic plasticity. Mol Cell 2020;77:120–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H, et al. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife 2015;4:e06857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goding CR, Arnheiter H. MITF-the first 25 years. Genes Dev 2019;33:983–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Riesenberg S, Groetchen A, Siddaway R, Bald T, Reinhardt J, Smorra D, et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat Commun 2015;6:8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai X, Fisher DE, Flaherty KT. Cell-state dynamics and therapeutic resistance in melanoma from the perspective of MITF and IFNgamma pathways. Nat Rev Clin Oncol 2019;16:549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175:984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marin-Bejar O, Rogiers A, Dewaele M, Femel J, Karras P, Pozniak J, et al. Evolutionary predictability of genetic versus nongenetic resistance to anticancer drugs in melanoma. Cancer Cell 2021;39:1135–49. [DOI] [PubMed] [Google Scholar]
- 16. Boshuizen J, Vredevoogd DW, Krijgsman O, Ligtenberg MA, Blankenstein S, de Bruijn B, et al. Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat Commun 2020;11:3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Statello L, Guo C-J, Chen L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huarte M. The emerging role of lncRNAs in cancer. Nat Med 2015;21:1253–61. [DOI] [PubMed] [Google Scholar]
- 19. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aprile M, Katopodi V, Leucci E, Costa V. LncRNAs in cancer: from garbage to Junk. Cancers (Basel) 2020;12: 3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016;531:518–22. [DOI] [PubMed] [Google Scholar]
- 22. Vendramin R, Verheyden Y, Ishikawa H, Goedert L, Nicolas E, Saraf K, et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat Struct Mol Biol 2018;25:1035–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vendramin R, Katopodi V, Cinque S, Konnova A, Knezevic Z, Adnane S, et al. Activation of the integrated stress response confers vulnerability to mitoribosome-targeting antibiotics in melanoma. J Exp Med 2021;218:e20210571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nikolaisen J, Nilsson LIH, Pettersen IKN, Willems PHGM, Lorens JB, Koopman WJH, et al. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One 2014;9:e101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chaudhry A, Shi R, Luciani DS. A pipeline for multidimensional confocal analysis of mitochondrial morphology, function, and dynamics in pancreatic β-cells. Am J Physiol Endocrinol Metab 2020;318:E87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merrill RA, Flippo KH, Strack S. Measuring mitochondrial shape with ImageJ. In: Strack S, Usachev YM, editors. Techniques to investigate mitochondrial function in neurons [Internet]. New York, NY: Springer New York; 2017. [cited 2021 Nov 29]. page31–48. Available from:http://link.springer.com/10.1007/978–1-4939–6890–9_2. [Google Scholar]
- 27. Conrad T, Ørom UA. Cellular fractionation and isolation of chromatin- associated RNA. Ørom UA, editor. Enhancer RNAs: methods and protocols. New York, NY: Springer New York; 2017. [DOI] [PubMed] [Google Scholar]
- 28. Badal B, Solovyov A, Di Cecilia S, Chan JM, Chang LW, Iqbal R, et al. Transcriptional dissection of melanoma identifies a high-risk subtype underlying TP53 family genes and epigenome deregulation. JCI Insight 2017;2:e92102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kunz M, Loffler-Wirth H, Dannemann M, Willscher E, Doose G, Kelso J, et al. RNA-seq analysis identifies different transcriptomic types and developmental trajectories of primary melanomas. Oncogene 2018;37:6136–51. [DOI] [PubMed] [Google Scholar]
- 30. Babaian A, Mager DL. Endogenous retroviral promoter exaptation in human cancer. Mob DNA 2016;7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fontanals-Cirera B, Hasson D, Vardabasso C, Di Micco R, Agrawal P, Chowdhury A, et al. Harnessing BET inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene. Mol Cell 2017;68:731–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seberg HE, Van Otterloo E, Loftus SK, Liu H, Bonde G, Sompallae R, et al. TFAP2 paralogs regulate melanocyte differentiation in parallel with MITF. PLoS Genet 2017;13:e1006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wouters J, Kalender-Atak Z, Minnoye L, Spanier KI, De Waegeneer M, Bravo González-Blas C, et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat Cell Biol 2020;22:986–98. [DOI] [PubMed] [Google Scholar]
- 34. Rambow F, Job B, Petit V, Gesbert F, Delmas V, Seberg H, et al. New functional signatures for understanding melanoma biology from tumor cell lineage-specific analysis. Cell Rep 2015;13:840–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, et al. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell 2015;57:537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol Cell 2015;57:521–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 2007;282:21583–7. [DOI] [PubMed] [Google Scholar]
- 38. Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 2007;8:939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Z, Jiang H, Chen S, Du F, Wang X. The Mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012;148:228–43. [DOI] [PubMed] [Google Scholar]
- 40. Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell 2013;23:302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov 2014;4:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 43. Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between metabolism and cancer biology. Cell 2017;168:657–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meng Z, Qiu Y, Lin KC, Kumar A, Placone JK, Fang C, et al. RAP2 mediates mechanoresponses of the hippo pathway. Nature 2018; 560:655–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chan DC. Mitochondrial dynamics and its involvement in disease. Annu Rev Pathol 2020;15:235–59. [DOI] [PubMed] [Google Scholar]
- 46. Sabouny R, Shutt TE. Reciprocal regulation of mitochondrial fission and fusion. Trends Biochem Sci 2020;45:564–77. [DOI] [PubMed] [Google Scholar]
- 47. Chang C-R, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1: chang & blackstone. Ann N Y Acad Sci 2010;1201:34–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li J, Huang Q, Long X, Guo X, Sun X, Jin X, et al. Mitochondrial elongation-mediated glucose metabolism reprogramming is essential for tumour cell survival during energy stress. Oncogene 2017;36:4901–12. [DOI] [PubMed] [Google Scholar]
- 49. Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011;13:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Spurlock B, Parker D, Basu MK, Hjelmeland A, Gc S, Liu S, et al. Fine-tuned repression of Drp1-driven mitochondrial fission primes a “stem/progenitor-like state” to support neoplastic transformation. Elife 2021;10:e68394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bonnay F, Veloso A, Steinmann V, Köcher T, Abdusselamoglu MD, Bajaj S, et al. Oxidative metabolism drives immortalization of neural stem cells during tumorigenesis. Cell 2020;182:1490–507. [DOI] [PubMed] [Google Scholar]
- 52. Bergers G, Fendt S-M. The metabolism of cancer cells during metastasis. Nat Rev Cancer 2021;21:162–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim YY, Um J, Yoon J, Lee D, Lee YJ, Kim DH, et al. p53 regulates mitochondrial dynamics by inhibiting Drp1 translocation into mitochondria during cellular senescence. FASEB J 2020;34:2451–64. [DOI] [PubMed] [Google Scholar]
- 54. Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 2015;11:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol 2017;14:11–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends to Supplementary Figures and Tables
LENOX is associated with melanoma clinical features and regulated by TFAP2A.
LENOX regulates melanoma cell proliferation and survival
LENOX is required for xenograft growth
Gain of LENOX function stimulates melanoma cell proliferation
RAP2 paralog expression in melanoma cells
LENOX regulates RAP2 mitochondrial localisation
LENOX and RAP2C knockdown induce mitochondrial dysfunction, release of reactive oxygen species (ROS), DNA damage and cell cycle block
LENOX regulates mitochondrial morphology
Ectopic LENOX expression increases oxidative phosphorylation in HEKT cells
MAPK inhibition increases LENOX expression downstream of TFAP2A.
LENOX overexpression promotes resistance to BRAF and glycolysis inhibition.
Data Availability Statement
The datasets generated during and/or analyzed in this study are available from the corresponding authors E. Leucci or I. Davidson upon request.