NLE1 limits de novo protein biosynthesis and the tumorigenic potential of advanced colorectal cancer cells, suggesting NLE1 could be targeted to improve the treatment of metastatic colorectal cancer.
Abstract
Growth and metastasis of colorectal cancer is closely connected to the biosynthetic capacity of tumor cells, and colorectal cancer stem cells that reside at the top of the intratumoral hierarchy are especially dependent on this feature. By performing disease modeling on patient-derived tumor organoids, we found that elevated expression of the ribosome biogenesis factor NLE1 occurs upon SMAD4 loss in TGFβ1-exposed colorectal cancer organoids. TGFβ signaling-mediated downregulation of NLE1 was prevented by ectopic expression of c-MYC, which occupied an E-box–containing region within the NLE1 promoter. Elevated levels of NLE1 were found in colorectal cancer cohorts compared with normal tissues and in colorectal cancer subtypes characterized by Wnt/MYC and intestinal stem cell gene expression. In colorectal cancer cells and organoids, NLE1 was limiting for de novo protein biosynthesis. Upon NLE1 ablation, colorectal cancer cell lines activated p38/MAPK signaling, accumulated p62- and LC3-positive structures indicative of impaired autophagy, and displayed more reactive oxygen species. Phenotypically, knockout of NLE1 inhibit.ed proliferation, migration and invasion, clonogenicity, and anchorage-independent growth. NLE1 loss also increased the fraction of apoptotic tumor cells, and deletion of TP53 further sensitized NLE1-deficient colorectal cancer cells to apoptosis. In an endoscopy-guided orthotopic mouse transplantation model, ablation of NLE1 impaired tumor growth in the colon and reduced primary tumor-derived liver metastasis. In patients with colorectal cancer, NLE1 mRNA levels predicted overall and relapse-free survival. Taken together, these data reveal a critical role of NLE1 in colorectal cancer growth and progression and suggest that NLE1 represents a potential therapeutic target in colorectal cancer patients.
Significance:
NLE1 limits de novo protein biosynthesis and the tumorigenic potential of advanced colorectal cancer cells, suggesting NLE1 could be targeted to improve the treatment of metastatic colorectal cancer.
Introduction
Progression of microsatellite stable and chromosomally instable colorectal cancer, which represents the most frequently occurring subtype of this disease prone to develop distant metastasis, is accompanied by pro-oncogenic mutations in driver genes affecting Wnt/MYC, EGFR/KRAS/MAPK, TGFβ/SMAD, and TP53 signaling. Regarding the molecular make-up of colorectal cancer, different stratifications have been suggested such as the consensus molecular subtypes (CMS) and the colorectal cancer-intrinsic subtypes (CRIS; refs. 1, 2). Furthermore, histologic comparison between normal and cancerous colorectal tissues suggested that colorectal cancer, similar to its benign counterpart, displays a hierarchical architecture composed of nondifferentiated, stem-like cells (a.k.a. cancer stem cells; CSC) and a predominant mass of less potent differentiated tumor cells (3). With the identification of the intestinal stem cell markers LGR5 and EPHB2, the purification and functional characterization of colorectal CSCs, which also display these surface receptors, became feasible (reviewed in ref. 4). Independent studies from different laboratories demonstrated an enhanced tumorigenic potential and a critical role of LGR5+ colorectal cancer cells in self-renewal and metastasis (5–7). Intriguingly, cellular fitness and aggressiveness of colorectal cancer were tissue context-dependent in these studies and differed between subcutaneous and orthotopic (colonic) mouse transplantation models (reviewed in ref. 8). In addition, late mutagenic alterations in the tumor suppressor gene TP53 and loss of the TGFβ signaling pathway component SMAD4 were shown to increase the percentage of tumor- and metastasis-initiating CSCs in vivo (6). Because many human tumors contain either no or only few LGR5+ cells, or LGR5− cells convertible to tumor-initiating cells upon genetic ablation of the LGR5+ colorectal cancer cell subpopulation, it has been speculated that rather other molecular traits might account for functional hierarchy and tumor cell potency in colorectal cancer (3, 6, 7).
In a recent study, Smit and colleagues reported an elevated protein-biosynthetic capacity associated with increased ribogenesis in advanced colorectal cancer cells carrying mutations in APC, KRAS, SMAD4, and TP53 when compared with TP53/SMAD4 wild-type tumors (9). Shortly after, Morral and colleagues demonstrated that only a limited subset of colorectal cancer cells within a tumor displays highest ribosomal DNA transcription and protein biosynthesis rates that functionally specifies colorectal CSCs independent of their LGR5 status (10). Interestingly, the Wnt and TGFβ/SMAD signaling-regulated proto-oncogene c-MYC, which represents a global regulator of cancer-relevant processes such as transcription, growth, and proliferation (reviewed in ref. 11), plays a critical role in protein biosynthesis by driving the expression of genes involved in ribosome biogenesis (12, 13). Hence, components of the ribosomal machinery whose expression becomes deregulated upon mutational alteration of key driver pathways during colorectal cancer progression might represent attractive targets for new effective anti–colorectal cancer therapies.
Here, we identified the preribosomal particle component NLE1 as a TGFβ/SMAD4/c-MYC–regulated factor whose expression is enriched in colorectal cancer. By exploring the role of NLE1 in protein biosynthesis, growth and survival, and in colorectal cancer progression in vivo, we demonstrate that NLE1 represents a limiting factor for colorectal cancer cell fitness and disease progression.
Materials and Methods
Patient-derived fresh tissues and tissue selection for formalin-fixed paraffin-embedded samples
Biological samples of fresh normal and cancerous tissue specimen were received from individuals undergoing curative colectomy or partial hepatectomy at the Hospital Großhadern, LMU Munich (Munich, Germany). Samples were taken by a pathologist from residual resected tissue, which was not needed for diagnostic purposes, and irreversibly anonymized. This procedure has been classified as uncritical and was approved for our studies by the ethical committee (institutional review board) of the Ludwig Maximilian University of Munich (project-No. 591–16-UE and 17–771-UE). Furthermore, the Institutional Review Board of the Ludwig Maximilian University of Munich determined that informed consent was unnecessary for these samples regarding the conducted experiments and experimental data shown in this study.
Mouse strain and handling of animals
NSG (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ) immunodeficient mice were purchased from Charles River Laboratories and approximately 12 weeks old when used for xenotransplantation. Animals were housed in the animal facility at the Pathology Institute of the Ludwig-Maximilians-University (Munich, Germany) in individually ventilated cages (IVC). Mice identifications, health controls (scoring) and cage changings were performed under a laminar flow hood. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Government of Upper Bavaria, Germany (approval number: ROB-55.2–2532.Vet_02–20–136).
Colorectal cancer cell line and patient-derived tumor organoid culture
Cell lines SW620 (CCL-227) and HT29 (HTB-38; ATCC, LGC Standards) were maintained in DMEM supplemented with 10% FBS (both Gibco, Thermo Fisher Scientific) and 1× penicillin/streptomycin (Gibco, #15140122, Thermo Fisher Scientific). HCT116 cells (CCL-247; ATCC, LGC Standards) were maintained in McCoy 5A medium (Sigma, Merck) supplemented with 10% FBS and 1× penicillin/streptomycin (both Gibco, Thermo Fisher Scientific). HCEC-1CT immortalized human colonic epithelial cells (Evercyte GmbH) were maintained in DMEM supplemented with 2% FBS, 1× N2 Supplement (contains insulin, apo-transferin, and sodium-selenite; Thermo Fisher Scientific), 20 ng/mL epidermal growth factor (EGF; AF-100–15, PeproTech, part of Thermo Fisher Scientific), 1 μg/mL hydrocortisone (Sigma, Merck), and 1× penicillin/streptomycin (Gibco, Thermo Fisher Scientific). Cultures were kept at 37°C, 5% CO2 and subcultured as needed. Patient-derived tumor organoids (PDTO) were isolated from freshly dissected colorectal cancer specimen and established as we had described previously (14). For maintenance and expansion via serial passaging, PDTOs were disaggregated using 0.025% Trypsin (Gibco, Thermo Fisher Scientific) and subsequently passed through a 0.8-mm needle (21G Sterican, B. Braun) to obtain small cell aggregates or single cells. Next, PDTO cells were embedded in growth factor–reduced Matrigel (356231, BD Corning) and overlaid with tumor organoid culture (TOC) medium [advanced DMEM/F12 (ADF), supplemented with 10 mmol/L HEPES, Glutamax, 1× B27 (all Thermo Fisher Scientific], 1 mmol/L N-acetylcysteine (Sigma, Merck), 50 ng/mL recombinant human EGF (AF-100–15, PeproTech), 0.015 μmol/L prostaglandin E2 (PGE2, Sigma, Merck), 25 ng/mL human Noggin (120–10C, PeproTech), 7.5 μmol/L SB202190 and 0.5 μmol/L LY2157299 (both Selleckchem), and 50 μg/mL Normocin (Invitrogen, Thermo Fisher Scientific). 10 μmol/L Y27632 (Selleckchem) was added to TOC medium for 48 hours after plating to avoid anoikis. Medium was replaced every 2 to 3 days. Prior to treatment of PDTO cultures with 20 ng/mL recombinant TGFβ1 (100–21, PeproTech), the TGFβ inhibitor LY2157299 was omitted from the culture medium. For conditional expression of ectopic cDNAs (c-MYC and NLE1 encoding), stably transduced cell and organoid lines were treated with 500 ng/mL doxycycline (Sigma, Merck) directly after plating cells. All cell and organoid lines tested negative for Mycoplasma contamination (latest April, 2022) by using the LookOut Mycoplasma PCR Detection Kit (Sigma, Merck). Before performing experiments, cell lines and organoids were allowed to recover for at least two passages.
Plasmids and cloning
For stable, doxycycline-inducible ectopic expression of NLE1 and c-MYC genes in PDTOs and colorectal cancer cell lines, a lentiviral pTz gateway vector was generated as previously described (14). c-MYC and NLE1 open reading frame (ORF) was PCR amplified from human cDNA with attB overhang primers and subsequently reamplified using attB universal primers (Gateway Technology protocol, Invitrogen, Thermo Fisher Scientific) and Phusion High Fidelity Polymerase (NEB). The attB-flanked ORFs were cloned into pDONR221 via Gateway BP Clonase II (Thermo Fisher Scientific) and sequences were confirmed by Sanger Sequencing (M13 fw/rv primer, Eurofins). Finally, sequence-verified cDNAs were transferred from pDONR221 to pTz gateway plasmid via Gateway LR Clonase II (Thermo Fisher Scientific).
RNA isolation, cDNA preparation, and quantitative real-time PCR
Total RNA was isolated with the High Pure RNA Isolation Kit (Roche) and transcribed into complementary DNA (cDNA) using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific). Both kits were used according to manufacturer's instructions. qRT-PCR was performed using the PrimaQuant CYBR qPCR Master Mix (Steinbrenner Laborsysteme GmbH) on a LightCycler 480 (Roche). Relative expression values were normalized to PPIA, ACTB, and/or GAPDH expression and calculated using the ΔΔCt method. Oligonucleotide pairs used for CYBR green–based qRT-PCR reactions are listed in the Supplementary Materials.
In addition, qRT-PCR was performed using the PrimaQuant Probe qPCR Master Mix (Steinbrenner Laborsysteme GmbH) and Taqman Gene Expression Assays (Applied Biosystems, Thermo Fisher Scientific, see Supplementary Materials for probe IDs) following manufacturer's instructions and using the LightCycler 480 (Roche). Relative expression values were normalized to PPIA and ACTB housekeeper gene expression and calculated using the ΔΔCt method (data shown in Fig. 1F; Supplementary Figs. S1C and S5A).
Figure 1.
Effect of SMAD4 ablation in colorectal cancer organoids on tumor growth and gene expression. A, Mutation detection assay on genomic DNA obtained from PDTO cells edited via CRISPR/Cas9 to achieve SMAD4 KO (S4) or on a SMAD4 wild-type parental control (P). M, marker. B, Immunoblot analysis of SMAD4 protein levels in parental SMAD4 wild-type (WT) and CRISPR/Cas9-targeted (S4-KO) PDTO lines 2 and 4. β-Actin served as a loading control. C, Enhanced focal images of 50 μL Matrigel droplets containing SMAD4 wild-type (S4-WT) or SMAD4 knockout (S4-KO) organoid lines maintained in tumor organoid culture (TOC) medium with (+TGFβ) or without (Ctrl) 20 nmol/L recombinant TGFβ1 for 7 (PDTO2) or 10 (PDTO4) days. D, ATP content measurement–based cell viability assessment (CellTiter-Glo 3D) of SMAD4 wild-type and SMAD4 knockout (S4-KO) PDTO lines maintained in TOC medium (Ctrl) or TOC medium supplemented with 20 nmol/L recombinant TGFβ1 for 7 (PDTO2) or 10 (PDTO4) days. Statistical significance between all samples was assessed by one-way ANOVA plus Tukey multiple comparisons test. ****, P ≤ 0.0001. Shown is the mean ± SD (n = 4). E, GSEA on gene signatures derived from RNA-sequencing data generated from SMAD4 knockout versus SMAD4 wild-type colorectal cancer organoids (PDTO4S4 vs. PDTO4) maintained in TGFβ1-containing TOC medium. Shown are enrichments of the gene sets c-MYC targets V2, TGFβ signaling (MSigDB Collections, Broad Institute) and EPBH2high human colonic stem cells (HsCoSC; ref. 27). NES, normalized enrichment score; NOM P value, nominal P value; FDR-q, false discovery rate q value. F, qRT-PCR analysis of LGR5, SMOC2, and PMEPA1 gene expression in SMAD4 wild-type and SMAD4 knockout (S4-KO) PDTO lines maintained in TOC medium (Ctrl) or TOC medium supplemented with 20 nmol/L recombinant TGFβ1 (TGFβ) for 72 hours. G, qRT-PCR analysis of NLE1 and c-MYC gene expression in SMAD4 wild-type and SMAD4 knockout PDTO lines maintained in TOC medium (Ctrl) or TOC medium supplemented with 20 nmol/L recombinant TGFβ1 (TGFβ) for 72 hours. Statistical significance between all samples in F and G was assessed by ordinary two-way ANOVA plus Tukey multiple comparison test (***, P <0.001; ****, P ≤ 0.0001) for the PDTOS4 versus PDTO comparison in the presence of TGFβ1. Shown is the mean ± SD (n = 3).
Mutation detection assay
For genotyping of SMAD4 knockout PDTOs, genomic DNA was isolated using the GenElute Mammalian Genomic Kit (Sigma-Aldrich, Merck) and PCR amplification according to the Q5 HF Master Mix protocol (NEB) was performed. Primers for PCR amplification are shown in the Supplementary Materials. Analysis of amplicons was conducted using the Alt-R Genome Editing Detection Kit (IDT) according to the manufacturer's protocol.
Quantitative chromatin immunoprecipitation analysis
Chromatin immunoprecipitation (ChIP) analysis of SW620 cells was performed according to the manufacturer's protocol (SimpleChIP Plus Enzymatic Chromatin IP Kit, #9005, Cell Signaling Technologies). Ten micrograms of fragmented DNA were incubated with 1.6 μg c-MYC antibody (Cell Signaling Technology, #9402) or 1.6 μg control rabbit IgG (Cell Signaling Technology, #2729) overnight. After purification of the chromatin fragments, samples were analyzed via qRT-PCR using primer pairs listed in the Supplementary Materials.
ATP content–based cell viability assessment
Four-thousand cells of SMAD4 wild-type or knockout PDTOs were plated in 20 μL Matrigel droplets and overlaid with TOC medium containing either 0.5 μmol/L LY2157299 (Selleckchem) or 20 ng/mL recombinant human TGFβ1 (#100–21, PeproTech) was added. Ten microliters of Y27632 (SelleckChem) was added to TOC medium for 48 hours after plating to avoid anoikis. Every 2–3 days, the TOC medium was replaced. At the indicated time points after seeding, ATP content as a surrogate for cell viability of PDTOs was analyzed using the CellTiter-Glo 3D assay (Promega). In brief, TOC medium was removed, Matrigel droplets were resuspended in 35 μL Advanced DMEM/F-12 medium (Gibco, Thermo Fisher Scientific) plus 85 μL CellTiter-Glo 3D (Promega) and incubated on a shaker for 5 minutes. Next, cell lysates were resuspended using a pipette and incubated for another 15 minutes on an orbital shaker. Finally, a volume of 100 μL was transferred to a white 96-well plate (Nunc, #236105, Thermo Fisher Scientific) and measured on a Berthold Orio II Microplate Luminometer (Berthold Technologies). Cell viability of cell lines was measured using CellTiter-Glo (Promega) according to the manufacturer's instructions.
CRISPR/Cas9-mediated knockout of SMAD4, NLE1, and TP53 in colorectal cancer cell lines and PDTOs
Knockout of SMAD4 in PDTOs was performed by electroporation of the pSpCas9(BB)-2A-GFP vector (Addgene, #48138) containing a guide RNA for targeting SMAD4 in exon 8 (originally published by Drost and colleagues; ref. 15). First, PDTOs were disaggregated with 0.025% Trypsin-EDTA (Gibco, Thermo Fisher Scientific) for 5 minutes at 37°C and passed through a 0.8-mm needle by a syringe to obtain a single-cell suspension. A total of 2 × 105 cells were resuspended in 100 μL OptiMEM (Gibco, Thermo Fisher Scientific) supplemented with 10 μmol/L Y-27632 and 2 μg of the vector. For electroporation, the suspension was transferred to an electroporation cuvette (EC-002S, 2-mm gap width) and electroporated using the NEPA21 electroporator (all Nepa Gene). The following conditions were used: two poring pulses of 160 V, 5 ms, with a pulse interval of 50 ms, followed by 5 transfer pulses of 20 V, for 50 ms, with an interval of 50 ms. Afterwards, 300 μL Advanced DMEM/F-12 (Gibco, Thermo Fisher Scientific) was added to the cells and incubated for 15 minutes at room temperature. Finally, cells were embedded in Matrigel (Corning) and cultured in TOC medium plus 10 μmol/L Y27632. After 48 hours, selection with 20 ng/mL TGFβ1 (PeproTech) was started. To perform CRISPR/Cas9-mediated gene knockout of NLE1 in colorectal cancer cell lines and PDTOs, two different guide RNAs targeting NLE1 in Exon 2 were designed using a CRISPR design tool (Synthego). Cloning of guide RNA–encoding eCas9 lentiviral vectors, containing an ORF for Cas9 with enhanced specificity (16), was performed according to Ran and colleagues (17) by annealing of 20 nt guide oligonucleotides (listed in Supplementary Materials) and subsequent ligation into the lentiviral eCas9 expressing vector pLentiCRISPR-E (gift from Phillip Abbosh, Addgene plasmid #78852). TP53 knockout derivatives of HCT116 cells were obtained by a CRISPR/Cas9 approach employing the pSpCas9(BB)-2A-GFP vector (Addgene #48138). The guide RNA sequence targeting the TP53 locus in exon 3 was derived from Drost and colleagues (15). After transient transfection of cells with pSpCas9(BB)-2A-GFP vectors using FuGENE HD transfection reagent (Promega), TP53-deficient HCT116 pools were selected for 3 weeks in culture medium containing 20 μmol/L nutlin-3 (TOCRIS), a known antagonist of the TP53 inhibitor MDM2. Oligonucleotides used for annealing and subsequent cloning of guide RNA–encoding plasmids are listed in the Supplementary Materials. Sequences were verified by Sanger sequencing (Eurofins).
Stable transduction of cell lines and organoids with lentiviral particles
HEK293T cells were transfected with lentiviral vectors encoding the guide RNA of interest and second-generation packaging vectors psPAX2 and pMD2.G (kindly provided by Prof. Andreas Trumpp, DFKZ Heidelberg). Virus containing supernatant was collected 24 hours and 48 hours posttransfection and concentrated 20-fold using Lenti-X concentrator solution (Clontech, Takara Bio) according to the manufacturer's protocol. For stable transduction, colorectal cancer cells were supplemented with 8 μg/mL polybrene (Sigma-Aldrich, Merck) and directly exposed to a 20-fold dilution of the concentrated lentivirus particles. In case of PDTOs, a single-cell suspension was obtained by incubation in 0.025% Trypsin-EDTA (Gibco, Thermo Fisher Scientific) for 5 minutes at 37°C and disaggregation by passing through a syringe-attached 0.8-mm needle. Afterwards, single PDTO cells were seeded in Matrigel-coated 24-well plates and incubated for 2 hours prior to lentiviral particle exposure. For stable transduction, PDTO cultures were supplemented with 8 μg/mL polybrene (Sigma-Aldrich) and a 20-fold dilution of the concentrated virus particles. After 16 hours, transduced PDTO cells were reembedded in Matrigel (Corning). For selection, colorectal cancer cell lines and PDTOs were cultured in medium containing puromycin (1–2 μg/mL, Sigma, Merck) 24–48 hours postinfection. The selection process lasted at least 4–5 days for cell lines and 8–12 days for PDTOs and was accompanied by an appropriate killing control.
Colony formation assay
Five-hundred cells were seeded in 6-well plates and cultured for 14 days. The cell culture medium was replaced twice a week. For staining, cells were washed with PBS and fixed with 4% PFA (Santa Cruz Biotechnology) for 20 minutes at room temperature. Next, cells were washed twice with PBS and 1 mL 5 mg/mL crystal violet solution (Sigma, Merck) in 20% methanol per well was added. After 20-minute incubation at room temperature, cells were washed with PBS again until background staining was gone. Then plates were air dried for one week and pictures were taken using a Nikon D5100 reflex camera. For quantification, crystal violet staining was destained with 1 mL 10% acetic acid in ddH2O per well for 20 minutes with washing on an orbital shaker. Next, destained solution was diluted 1:10 in ddH2O and measured on a Orion II Microplate luminometer (Berthold Technologies) at a wavelength of 595 nm.
Soft agar assay
Twelve-well plates were coated with 0.8% agar (low melting SeaPlaque Agarose, Lonza) in DMEM supplemented with 10% FBS (Gibco, Thermo Fisher Scientific) and 1× penicillin/streptomycin (Gibco, Thermo Fisher Scientific). On top, 1,500 cells per well in 0.4% agar (low melting SeaPlaque Agarose, Lonza) in DMEM with 10% FBS and 1× penicillin/streptomycin were seeded and air dried for 45 minutes at room temperature. After incubation at 37°C and 5% CO2 for 24 hours, 500 μL medium was added. Medium was changed twice a week and after 14–16 days, colonies were stained with 500 μL 0.005% Crystal Violet (Sigma, Merck) for 1 hour. Then destaining by washing three times with PBS was performed and pictures of colonies were taken on a AZ100 Zoom Microscope (Nikon).
Transwell migration and invasion assays
One day before seeding, cells were washed with PBS and serum-starved for 24 hours. For migration assay, 150,000 cells were seeded in transwell chambers (8.0-μm pore size membrane; Greiner Bio-One) in serum-free medium. For invasion assay, transwell membranes (8.0-μm pore size; Greiner Bio-One) were first coated with 300 μg/mL Matrigel in coating buffer (0.01 mol/L Tris, pH 8.0, 0.7% NaCl) for 2 hours at 37°C, in line with the Matrigel manufacturer's (Corning Life Sciences) guidelines. Then, 150,000 cells were seeded into coated transwells in serum-free medium. As a chemoattractant, medium supplemented with 10% FBS was placed in the bottom chamber. Forty-eight hours later, cells on the top side of the transwell membrane were removed and transwells were washed with PBS. Next, cells were fixed with ice-cold methanol for 10 minutes at −20°C. After the membrane was air dried, cells were stained with 100 ng/mL DAPI (Sigma, Merck) in PBS and analyzed with a LSM 700 confocal microscope (Zeiss). Three random fields per membrane were captured, and migrated or invaded cells were counted with ImageJ.
xCELLigence proliferation assay
HCEC-1CT, SW620, and HT29 cells (2×103 cells in 200 μL medium/well) were seeded in E16-Plates (Acea Biosciences, Inc) according to the xCelligence Real Time Cell Analyzer (RTCA) DP manufacturer's instructions (Roche/Acea Biosciences). The growth was monitored with one sweep/hour for 145 hours in total. Analysis was performed using the RTCA 1.2.1.1002 software (Roche). The RTCA detects changes in the electrical impedance of gold electrodes on the well surface and thereby generates the so-called cell index. An increase in the cell index over time is a surrogate for active cell proliferation.
Flow cytometry–assisted cell-cycle analysis
Cell-cycle analysis was performed using the Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit in combination with the FxCycle Far Red Stain (both Invitrogen/Thermo Fisher Scientific). —Twenty-four to 72 hours after counting and plating colorectal cancer cell lines in 6-well tissue culture dishes, EdU was added to subconfluent cells at a final concentration of 10 μmol/L for 2 hours. After EdU labeling, cells were detached using 0.025% Trypsin (Gibco, Thermo Fisher Scientific) for 5 minutes at 37°C, washed with DMEM/10% FBS and PBS, and further processed according to the manufacturer's instructions. After the Click-iT reaction, cells were resuspended in 400 μL 1× Click-iT saponin-based permeabilization and wash reagent. For DNA content staining, FxCycle Far Red Stain and RNase A were added to final concentrations of 200 nmol/L and 100 μg/mL, respectively. Finally, cells were analyzed on a BD Accuri C6 Flow Cytometer (BD Biosciences) or a MACSQuant X Flow Cytometer (Miltenyi Biotec).
Annexin V/propidium iodide staining
For quantification of cells actively undergoing apoptosis, the FITC Annexin V Apoptosis Detection Kit I (BD Biosciences) was used. Prior staining, cell culture medium was collected in 15-mL tubes and 1 mL Accutase (Sigma-Aldrich, Merck) per well (6-well plate) was added. After incubation for 2 minutes at room temperature, 2 mL medium per well was added and the cells were carefully resuspended and transferred to the same 15-mL tubes. Then cells were centrifuged and supernatant was discarded. Finally, staining was performed according to manufacturer's protocol. For analysis, a BD Accuri C6 Flow Cytometer (BD Biosciences) or a MACSQuant X Flow Cytometer (Miltenyi Biotec) was used.
Flow cytometry–assisted protein synthesis assay
For detection of nascent protein synthesis in cells, colorectal cancer cell lines were labeled with 20 μmol/L O-propargyl-puromycin (OPP; Thermo Fisher Scientific) for 30 minutes, PDTOs were labeled for 60 minutes. As a positive control for inhibition of protein de novo synthesis, cells were treated with 50 μg/mL cycloheximide (Sigma/Merck) for 20 minutes prior staining with OPP. After OPP labeling, PDTOs were incubated with 0.025% Trypsin-EDTA (Gibco, Thermo Fisher Scientific) for 5 minutes at 37°C and passed through a 0.8-mm needle by a syringe to obtain a single-cell suspension. HCEC-1CT, HT29, and SW620 cells were harvested with Accutase (Sigma-Aldrich, Merck). After washing with 1% BSA (Sigma-Aldrich, Merck) in PBS (Sigma-Aldrich, Merck), single-cell suspensions were fixed and stained according to the Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay Kit Protocol (Thermo Fisher Scientific). After the Click-iT reaction, cells were washed with 1x Click-iT saponin-based permeabilization and wash reagent (Thermo Fisher Scientific), resuspended in PBS (Sigma-Aldrich, Merck) and analyzed on a BD Accuri C6 Flow Cytometer (BD Biosciences) or a MACSQuant X Flow Cytometer (Miltenyi Biotec).
Flow cytometry–assisted ROS detection assay
To detect reactive oxygen species (ROS) levels in living cells, colorectal cancer cell lines were stained using the CellROX Green Flow Cytometry Assay Kit (Thermo Fisher Scientific). In brief, colorectal cancer cell lines were labeled with 1 μmol/L CellROX reagent (Thermo Fisher Scientific) for 45 minutes according to the manufacturer's protocol. As a positive control for ROS generation, cells were treated with 400 μmol/L Tert-butyl hydroperoxide (TBHP) for 1 hour prior to labeling with the CellROX reagent. During the last 15 minutes of CellROX labeling, 5 nmol/L SYTOX Red Dead Cell stain solution was added. After staining, HT29 and SW620 cells were harvested with Accutase (Sigma-Aldrich, Merck) and washed in DMEM supplemented with 10% FBS and 1× penicillin/streptomycin (all Gibco, Thermo Fisher Scientific). Finally, cells were resuspended in DMEM and analyzed using a MACSQuant X Flow Cytometer (Miltenyi Biotec).
Endoscopy-guided orthotopic organoid transplantation into NSG mice
Orthotopic transplantation of colorectal cancer organoids was essentially performed as described previously (18). In brief, tumor organoids were recovered from Matrigel by 5 minutes incubation with 1× TrypLE-Select (Thermo Fisher Scientific) at 37°C, mechanically dissociated into 5- to 10-cell clusters with a syringe (0.8-mm needle) and resuspended in DPBS (Invitrogen, Thermo Fisher Scientific), 10% Matrigel (BD Corning) and 10 μmol/L Y-27632 (SelleckChem). For every injection (1–2 per mouse), 150 dissociated organoids in a volume of 100 μL were prepared, quantified by microscopy, and the ATP content as a surrogate for cell viability was determined for additional normalization.
Next, the colon of isoflurane-anesthetized mice was gently rinsed with Hanks' Balanced Salt Solution (Gibco, Thermo Fisher Scientific) by using a syringe and a straight gavage needle. A rigid endoscope from Karl Storz (1.9 mm in diameter) with a linear Hopkins lens optics (ColoView System) was used to perform mouse colonoscopy (Karl Storz SE & Co.KG). For injections of the organoid suspensions into the submucosa of the colon, a custom-made flexible fine needle (33-gauge, custom length of 16 inches, custom point style of 4 at 45°; Hamilton) was used. Injections that were optimally applied into the submucosa led to formation of a bubble that closed the intestinal lumen for at least 15 seconds. Only those transplanted animals that had fulfilled this quality criterion were considered for further analysis.
Next-generation sequencing (RNA sequencing) analysis and bioinformatic data processing
For RNA sequencing, total RNA was isolated from SMAD4 wild-type and syngeneic SMAD4-deficient tumor organoid lines (PDTO2 and PDTO4) using the High Pure RNA Isolation Kit (Roche Life Science) according to the manufacturer's protocol. Quality of isolated RNA was confirmed with the Agilent BioAnalyzer 2100 (Agilent). Sequencing libraries were prepared using the TrueSeq Stranded mRNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer's instructions. 50 bp single-read sequencing was performed on a HiSeq 2000 v4 (Illumina) according to the manufacturer's protocol. For bioinformatic data analysis, base calling was done with bcl2fastq 2.19.0.316. For all samples, low quality bases were removed with Fastq_quality_filter from the FASTX Toolkit 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/index.html) with 90% of the read needing a quality phred score > 20. Homertools 4.7 (19) were used for PolyA-tail trimming, and reads with a length < 17 were removed. Genomic mapping was performed with TOPHAT2 (20) for filtered reads with human genome assembly 38 and PicardTools 1.78 CollectRNASeqMetrics (https://broadinstitute.github.io/picard/). Count data were generated by htseq-count (21) using the gencode.v31.annotation.gtf (https://www.gencodegenes.org/) file for annotation. For the comparison with DESeq2 (22), the input tables containing the replicates for the groups to compare were created by a custom perl script. In the count matrix, rows with an average count number < 10 were removed. Then, DESeq2 (version 1.4.1) was run with default parameters. The results tables were annotated with gene information (gene symbol, gene type) derived from the gencode.v31.annotation.gtf file.
Cohort expression data, molecular subtypes, and geneset enrichment analysis
Gene expression data [upper quartile (UQ) normalized fragments per kilobase of exon per million mapped fragments (FKPM)] of colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) were obtained from GDC-The Cancer Genome Atlas (TCGA) datasets (23). Expression data of cohorts used for CMS and CRIS analysis were downloaded from NCBI Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo). Expression and clinical data of cohorts used for relapse-free survival analyses were obtained from NCBI GEO (www.ncbi.nlm.nih.gov/geo). Another data set covering 566 colorectal cancer cases was derived from Marisa and colleagues (GSE35982; ref. 24) and used for overall survival analysis. For CRIS classification, 515 transplanted human tumor samples (patient-derived xenografts) from 244 patients were included from the Isella and colleagues (GSE76402) data set (2). Information to assign the samples with the CMS subtypes are described in Guinney and colleagues (1) and the CRIS categories are described in Isella and colleagues (2).
NLE1 gene signatures were derived from GDC-TCGA COAD and READ datasets (n = 638 colorectal cancer tissue samples and 591 patients with available survival data; ref. 23). Gene set enrichment analysis (GSEA) was performed as described in Subramanian and colleagues (25) using the GSEApreRanked tool of the GSEA 3.0 Desktop Application (Broad Institute, http://www.gsea-msigdb.org/gsea/index.jsp) and the MSigDB hallmark gene set (26).
Immunoblot analysis and antibodies
For PDTOs, the Matrigel was dissolved by using Cell Recovery Solution (Corning) for 30 minutes on ice and organoids were washed twice in PBS. For colorectal cancer cell lines, lysates were prepared from subconfluent cultures by scraping in lysis buffer. Whole protein cell lysates were prepared using RIPA buffer supplemented with protease inhibitors, NaVO3 and PMSF (Santa Cruz Biotechnology), and phosphatase inhibitor cocktails 2 and 3 (Sigma/Merck). Lysates were incubated on ice for 30 minutes, then sonicated 3×5 seconds with 75% amplitude (HTU Soni130, G. Heinemann, Essen, Germany), spun for 20 minutes at 12,000 × g, and protein concentration was determined using the Micro BCA Protein Assay Kit (Thermo Fisher Scientific). Lysates were separated on 10%–12% SDS-acrylamide gels and proteins transferred to Immobilon PVDF membranes (Millipore, Merck). The membranes were blocked with 5% milk TBS/0.2% Tween20 for 1 hour at room temperature. For immunodetection, membranes were incubated with the following primary antibodies: anti-NLE1 (1:500, G-5, sc-377142, Santa Cruz Biotechnology), anti-p38α MAPK (L53F8; 1:1,000, mAB #9228), anti-phospho-p38 MAPK (Thr180/Tyr182; D3F; 1:1,000, mAb #4511), anti-SMAD4 (clone D3R4N, 1:1,000, mAb#46535), anti-phospho-SAPK/JNK (Thr183/Tyr185; 81E11; 1:500, mAb #4668), anti-LC3A/B (D3U4C; 1:1,000, mAb #12741), anti-PARP (1:500, pAb #9542), anti-cleaved PARP (Asp214; D64E10; 1:500, mAB #5625; all Cell Signaling Technologies), anti-α-tubulin (1:2,000, T9026. Sigma/Merck), anti-β-actin (1:2,000, A2066, Sigma/Merck), and anti-c-MYC (1:1,000, 10828–1-AP, ProteinTech). Enhanced chemiluminescence signals from horseradish peroxidase (HRP)-coupled secondary antibodies (1:10,000, Jackson ImmunoResearch) were generated using Immobilon Western HRP Substrate (Merck/Millipore) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and detected using the LI-COR Odyssey Fc imaging system (LI-COR).
IHC
For IHC staining of MKI67, 2 μm whole-tissue sections of formalin-fixed and paraffin-embedded (FFPE) tumor samples were stained using a Ventana Benchmark (Ventana Medical Systems) according to the manufacturer's instructions. Cell Conditioning Solution (CC1) was used as a pretreatment and antibody binding was visualized using the Ventana UltraView DAB IHC Detection Kit (all Ventana Medical Systems). The antibody directed against MKI67 [Clone MIB-1, Agilent (Dako)] was used at a dilution of 1:100. For detection of cleaved caspase-3 on 2-μm–thick FFPE sections, antigen-retrieval was achieved using Target Retrieval Solution Citrate pH 6 (Agilent Technologies, catalog no. S2369), and slides were incubated with the primary antibody (cleaved caspase-3, #9661, Cell Signaling Technology) at a 1:100 dilution for 1 hour at room temperature. As detection system, the ImmPRESS anti-rabbit IgG Polymer Kit (MP-7401, Vector Laboratories) was used. Samples were developed via exposure to 3,3'-diaminobenzidine (DAB+, K3468, Agilent Technologies) and counterstained with hematoxylin Gill formula (H-3401, Vector Laboratories).
Immunofluorescence detection of LC3B
SW620 and HT29 cells were grown on glass coverslides (12-mm diameter) and treated with 50 μmol/L chloroquine (CQ) 24 hours prior to analysis. Cells were washed with PBS and fixed with 4% paraformaldehyde (Santa Cruz Biotechnology) for 10 minutes at room temperature. Permeabilization of cells was achieved with 0.2% Triton X-100 (VWR) for 20 minutes at room temperature. Next, cells were washed two times with PBS and then blocked for 30 minutes in 100% FBS at room temperature. Cells on coverslides were then transferred onto parafilm and incubated with primary antibody (anti-LC3B, pAb #2775, Cell Signaling Technology, 1:200 dilution) in 50% FBS and 0.05% Tween 20 (PanReac Applichem) overnight at 4°C. After washing with 0.05% Tween 20 (PanReac Applichem) in PBS, cells were incubated with secondary antibody anti-Rabbit-Cy3 (#711–165–152, Jackson Immuno-Research Europe LTD.), diluted 1:500 in 50% FBS and 50% of 0.05% Tween 20 in PBS, for 30 minutes at room temperature in the dark. Next, cells were washed three times with 0.05% Tween 20 in PBS and stained with 100 ng/mL DAPI (Sigma, Merck) in PBS for 5 minutes at room temperature. ProLong Gold antifade DAPI mounting medium (#8961S, Cell Signaling Technology) was used for mounting. Images were captured with a LSM700 confocal microscope (Zeiss) using the 63× oil immersion objective.
Statistical analysis and survival analysis
GraphPad Prism software (v7.01) was used for statistical analyses. For calculation of significant differences between two groups of biological replicates from in vitro experiments, a Student t test (unpaired, two-tailed, Holm–Sidak method, with α level = 0.05) was applied. In case of biological replicates from colorectal cancer organoid xenotransplantation experiments, an unpaired t test with Welch correction was applied to account for different unequal SDs between groups. For the comparison of three or more groups, a multiple comparison one-way ANOVA test was applied. For unpaired data, one-way ANOVA in combination with a Tukey multiple comparisons test was performed. A Dunnett test was performed if more than one sample was compared with a nontreated control. For comparison of data with two different parameters, a two-way ANOVA with either a Tukey (when all samples were compared with each other) or Sidak (when only certain treatments were compared to each other) multiple comparison test was performed. For calculation of correlation coefficients, Pearson correlation analysis was applied. Statistical significance is indicated by asterisks in the figures and further detailed in the figure legends.
The optimal cut-off point for the continuous variable “NLE1 gene expression” (TCGA-colorectal cancer and Marisa and colleagues colorectal cancer cohorts; ref. 24) was determined by using the maximally selected rank statistics (maxstat) provided in the R package “survminer” (R package version 0.4.9; https://CRAN.Rproject.org/package=survminer). KM estimate curves and the log-rank test P values were calculated using “survival” package in R (R package version 3.2–13; https://CRAN.R-project.org/package=survival).
Imaging
Processed FFPE tissue slides from IHC staining for MKI67 and cleaved caspase-3 were scanned using a Vectra Polaris Automated Quantitative Pathology Imaging System (Akoya Biosciences). PDTO extended focal images (EFI) were generated using an AZ100 multizoom microscope (Nikon) and NIS Elements Imaging Software (Version 5.00.00, Nikon). Macroscopic images of dissected mouse organs and tumor burden was documented with a Zeiss Stemi 2000-C Zoom Stereomicroscope with CL6000 LED illumination and a Zeiss AxioCam ERc 5s (Carl Zeiss). Immunofluorescence images were taken on a Zeiss LSM700 confocal microscope using the 63× oil immersion objective (Carl Zeiss).
Data availability
Gene expression data of COAD (colorectal adenocarcinoma) and READ (rectal adenocarcinoma) used for comparison of NLE1 gene expression between cancerous and normal tissues were obtained from GDC-TCGA datasets (23) and NCBI GEO (www.ncbi.nlm.nih.gov/geo).
Gene expression changes in CRISPR/Cas9-engineered SMAD4-negative PDTO derivatives (PDTOS4 lines) compared with parental PDTOs are detailed in Supplementary Tables S2–S4. Counting files and annotation of all technical and biological replicates are provided in Supplementary Table S2.
The next-generation sequencing raw data that support the findings of this study are available on request from the corresponding author (P. Jung). These data are not publicly available due to them containing information that could compromise research participant privacy or consent. Explicit consent to deposit raw-sequencing data was not obtained from patients. Because all samples had been irreversibly anonymized prior to study start, the patients cannot be asked to provide their consent for deposit of their comprehensive genetic or transcriptomic data.
Results
SMAD4 mutation in colorectal cancer organoids prevents TGFβ-mediated downregulation of vulnerability genes
To study gene transcriptional alterations upon SMAD4 loss in colorectal cancer, we employed a CRISPR/Cas9 approach targeting SMAD4: electroporation of two different patient-derived tumor organoid (PDTO) lines (PDTO2 and PDTO4), both wild-type for genetic core components of the TGFβ signaling pathway according to whole exome sequencing analysis (see Supplementary Table S1 for PDTO characteristics), were electroporated with a guide RNA-expressing CRISPR/Cas9 (pSpCas9–2A-GFP) plasmid targeting the SMAD4 genomic locus (Fig. 1A). After selection with recombinant TGFβ1-containing culture medium, which inhibited the growth of parental organoids, SMAD4-knockout PDTO-derivatives (referred to as PDTO2S4 and PDTO4S4) were obtained, which lacked both SMAD4 protein and sensitivity toward TGFβ1-mediated growth inhibition (Fig. 1B–D). Next-generation sequencing (RNA-seq) on PDTOS4 versus parental PDTO lines after treatment with TGFβ1 for 72 hours revealed the spectrum of genes deregulated by TGFβ signaling in a SMAD4-dependent manner (Supplementary Tables S2–S4). In agreement with our observation on organoid growth capacity (Fig. 1C and D), expression of proliferation gene sets (c-MYC and E2F targets, MSigDB database (25, 26)) and a human colonic stem cell (27) gene set were enriched in TGFβ1-exposed PDTOS4 lines, while expression of a TGFβ signaling (MSigDB database) gene set was reduced when compared with parental PDTOs exposed to TGFβ1 (Fig. 1E). By quantitative real-time PCR (qRT-PCR), we confirmed that loss of SMAD4 in PDTOs largely diminished the induction of the well-known TGFβ target gene PMEPA1 (28) and impaired downregulation of intestinal stem cells markers LGR5 and SMOC2 by TGFβ1 (Fig. 1F; Supplementary Fig. S1A). Next, we asked which of the genes enriched in SMAD4-deficient PDTOs under the influence of TGFβ1 might confer a critical gain of tumor cell fitness and could therefore represent a genetic vulnerability for colorectal cancer cells. To answer this question, we made use of the Cancer Dependency Map project (DepMap), which aims to identify tumor background-specific essential genes via genome-wide CRISPR/Cas9 and shRNA screens across hundreds of established cancer cell lines (29, 30). Among the 284 genes that showed a log2-fold enrichment of > 0.6 in both PDTOS4 lines (Supplementary Table S3), only 11 genes were reported essential for colorectal cancer cells by DepMap (cut-off < −0.8 in two independent CRISPR/Cas9 screens): MYC, IFITM3, NLE1, NAT10, FBL, POLR3K, PSMG4, RRP1, LSM6, GINS2, and PRMT1 (Supplementary Table S3). Interestingly, four of these genes (NLE1, NAT10, FBL, and RRP1) encode critical factors of ribosome biogenesis and functionality and might contribute to the elevated biosynthetic capacity found in late stage colorectal cancer cells and colorectal cancer stem cells (9). Because NLE1 showed the most consistent enrichment within this functional group and scored as a dependency gene in both CRISPR/Cas9- and siRNA-based screens (DepMap project, Supplementary Fig. S1B), we focused our work on the ribosome assembly factor NLE1 and its role in colorectal cancer.
c-MYC binds to the NLE1 promoter and rescues downregulation of NLE1 by TGFβ/SMAD signaling
To study the effect of TGFβ/SMAD signaling on NLE1, we first analyzed NLE1 gene expression levels in PDTO and PDTOS4 organoid lines in the absence and presence of TGFβ pathway activation. Treatment of tumor organoids with recombinant TGFβ1 reduced NLE1 levels in PDTO but not in PDTOS4 lines (Fig. 1G; Supplementary Fig. S1C). As expected, also c-MYC gene expression was downmodulated by TGFβ1 exclusively in parental PDTO lines (Fig. 1G; Supplementary Fig. S1C). Analysis of publicly available transcriptomics data from experiments in which c-MYC had been either ectopically overexpressed or suppressed via RNA interference in cancer cell lines indicated modulation of NLE1 expression by c-MYC (Fig. 2A). To examine the potential of c-MYC to rescue TGFβ signaling–mediated NLE1 downregulation, we stably transduced TGFβ-responsive colorectal cancer organoids with lentiviruses encoding doxycycline-inducible c-MYC (PDTO4 pTz-MYC) or, as a control, an empty virus (PDTO4 pTz-Empty). After treatment with TGFβ1 for 72 hours, colorectal cancer organoids were treated with doxycycline in the presence of TGFβ1 for 48 hours or, as a control, were exposed to TGFβ1 alone. Indeed, conditional expression of ectopic c-MYC in the presence of recombinant TGFβ1 rescued NLE1 mRNA and protein expression (Fig. 2B and C). However, c-MYC overexpression was not able to drive NLE1 expression beyond the levels observed in nontreated control PDTOs (Fig. 2B and C), suggesting that endogenous c-MYC–driven NLE1 expression had already reached a plateau in this cellular background. As expected, expression levels of the stem cell marker LGR5, which does not represent a c-MYC target gene, were hardly affected by c-MYC induction (Fig. 2B). Analysis of MYC ChIP-seq data sets from various cancer cell lines (available via the ENCODE consortium; ref. 31) suggested binding of c-MYC to the promoter region of NLE1 (Fig. 2D). By performing chIP with subsequent qRT-PCR analysis (qChIP), we could confirm binding of endogenous c-MYC to the E-box (CACGTG)-containing promoter of the NLE1 gene in colorectal cancer cells, while an amplicon matching a region within NLE1 but located ∼5 kb downstream of the c-MYC binding site was not enriched (Fig. 2E). As a positive control, we also confirmed localization of endogenous c-MYC to promoters of well-described target genes DKC1 (32) and NPM1 (33), which are also involved in ribosome functionality (Fig. 2E; refs. 34, 35). In agreement with these findings, NLE1 expression correlated with expression of c-MYC targets and inversely correlated with TGFβ signaling gene set expression in a cohort of patients with colorectal cancer (GDC-TCGA_COAD+READ; Fig. 2F). Taken together, these data demonstrate that TGFβ pathway activation leads to downregulation of NLE1 expression in a SMAD4-dependent manner, and the TGFβ/SMAD signaling-repressed oncogene c-MYC directly binds to the NLE1 promoter and is able to rescue NLE1 expression in this context. Hence, a TGFβ/SMAD4/c-MYC signaling axis regulates NLE1 levels in colorectal cancer cells.
Figure 2.
Regulation of NLE1 expression by c-MYC and TGFβ signaling in colorectal cancer. A, Meta-analysis of NLE1 mRNA expression in public datasets (NCBI GEO) representing studies with c-MYC overexpression (OE; red bars) or c-MYC knockdown by RNA interference (siRNA; blue bars) in indicated cell lines. GEO accession numbers for each of the depicted experiments are indicated. B, qRT-PCR analysis of NLE1 gene expression in PDTO4 cells stably transduced with lentiviral particles encoding for a doxycycline-inducible c-MYC allele (pTz-MYC) or with a control lentivirus (pTz-Empty). Cells were left untreated or treated with either 20 nmol/L recombinant TGFβ1 (TGFβ) for 5 days alone or 500 ng/mL doxycycline was added simultaneously with 20 nmol/L recombinant TGFβ1 (βDOX) 48 hours prior to analysis. Statistical significance between all samples was assessed by ordinary two-way ANOVA plus Tukey multiple comparison test. ****, P ≤ 0.0001. Shown is the mean ± SD (n = 3). n.s., nonsignificant. C, Immunoblot analysis of c-MYC and NLE1 protein levels in PDTO4 cell lines described and treated as in B. Note that for pTz-MYC virus–transduced PDTOs, whole protein lysates of two independent experiments were analyzed. β-Actin served as a loading control. D, Binding of c-MYC to the NLE1 promoter region in the indicated cell lines as represented by c-MYC ChIP-seq signals. Note that the genomic NLE1 sequence covered by ChIP seq peaks contains one consensus c-MYC binding sequence (E-box, CACGTG). Numbers on the y-axis indicate ChIP-Seq reads. Source: The Encyclopedia of DNA Elements (ENCODE) Consortium. Visualization of the ChIP-seq peaks located in the NLE1 promoter region was done with the Integrative Genomics Viewer (IGV, Broad Institute). E, ChIP combined with qRT-PCR (qChIP) analysis of genomic DNA from SW620 cells. The amount of DNA immunoprecipitated with anti-MYC antibody or a rabbit IgG-control in each sample is shown as percentage of chromatin input. Note enrichment of the NLE1 gene promoter (NLE1p) amplicon in contrast to an amplicon located approximately 5 kb downstream within NLE1 (NLE1_ctrl). Amplified regions of the DKC1 and NPM1 promoters have been shown previously to contain c-MYC binding sites (33) and served as positive controls. Statistical significance between c-MYC IgG and rabbit control IgG groups was assessed by multiple t tests corrected for multiple comparison using the Holm–Sidak method. **, P < 0.01; ****, P < 0.0001. Mean ± SD (n = 3). F, GSEA on NLE1 colorectal cancer gene signatures derived from GDC-TCGA-COAD plus GDC-TCGA-READ RNA-seq data sets (n = 638 colorectal cancer samples; see Materials and Methods for details). Shown are enrichments of c-MYC targets V1 and TGFβ signaling gene sets (MSigDB collections, Broad Institute). NES, normalized enrichment score; NOM P value, nominal P value; FDR-q, false discovery rate q value.
NLE1 ablation diminishes de novo protein biosynthesis and inhibits colorectal cancer growth, migration/invasion, and survival
Because NLE1 is critical for ribosome maturation (36–38), we set out to study the effect of NLE1 loss on de novo protein biosynthesis in colorectal cancer cells. To achieve this, we first generated targeted NLE1 knockout derivatives via stable transduction of colorectal cancer cell lines and PDTOs (PDTOS4 lines) with lentiviral particles encoding a Streptococcus pyogenes Cas9 derivative designed for improved specificity (16) and two different guide RNAs targeting exon 2 of NLE1 (Fig. 3A and B). Next, we employed pulsed incorporation of the puromycin analog OPP into nascent polypeptide chains followed by detection of fluorophore-coupled OPP at the single cell level via flow cytometry. As a positive control, tumor cells were treated with the translation elongation inhibitor cycloheximide prior to OPP exposure, which almost completely abolished OPP incorporation (Fig. 3C–F). Indeed, targeted NLE1 knockout in HT29 and SW620 cells and in colorectal cancer organoids substantially reduced OPP incorporation, which indicated a lower de novo protein biosynthesis rate upon NLE1 loss in colorectal cancer cells (Fig. 3C–F). Presumably as a consequence of the diminished biosynthetic capacity, tumor cell proliferation, colony formation, and anchorage-independent growth in soft agar were markedly reduced upon NLE1 ablation (Fig. 4A–D; Supplementary Fig. S2A–S2D). In addition, NLE1 knockout colorectal cancer cells arrested either in the G1 (HT29) or G2 (SW620) cell-cycle phase (Supplementary Fig. S2E) and showed an elevated frequency of cells undergoing apoptosis, as indicated by increased Annexin V/propidium iodide staining and PARP cleavage (Fig. 4E and F; Supplementary Fig. S2F and S2G). Furthermore, transwell migration and invasion assays showed that loss of NLE1 reduced the HT29 and SW620 migration and invasion capacity in vitro (Fig. 4G and H; Supplementary Fig. S2H and S2I). Targeting NLE1 in SMAD4-mutant tumor organoid lines (PDTOS4) led to an overall decreased organoid formation capacity and a shift toward formation of smaller diameter PDTOs (Fig. 4I–K; Supplementary Fig. S3A–S3F). Interestingly, the effect of NLE1 ablation on tumor organoid growth rate and reseeding capacity, although substantial, was overall less pronounced when compared with classic colorectal cancer cell lines. Taken together, these data suggest that NLE1 supports protein biosynthesis in colorectal cancer cells, and NLE1 represents a limiting factor for colorectal cancer clonogenicity, growth, migration/invasion, and survival.
Figure 3.
NLE1 is important for de novo protein biosynthesis in colorectal cancer cell lines and PDTOs. A, Immunoblot analysis of NLE1 protein levels in SW620 (left) and HT29 (right) cells transduced with either pLentiCRISPR-E (Empty) control lentiviral particles or lentiviral particles encoding for two different guide RNAs targeting NLE1 (NLE1g1, NLE1g2). β-Actin served as a loading control. B, Immunoblot analysis of NLE1 protein levels in PDTO4 SMAD4-knockout cells (PDTO4S4) transduced with either pLentiCRISPR-E (Empty) control lentiviral particles or lentiviral particles encoding for two different guide RNAs targeting NLE1 (NLE1g1, NLE1g2). β-Actin served as a loading control. C, Representative flow cytometry plot showing OPP incorporation in SW620 empty cells (blue), SW620 empty cells treated with cycloheximide (CHX; gray), and SW620 NLE1 knockout cells (NLE1g1, orange; NLE1g2, red). D, Mean fluorescence intensity (MFI) of OPP incorporation in SW620 and HT29 control cells (Empty; blue), control cells treated with cycloheximide (gray) and NLE1 knockout cells (NLE1g1, orange; NLE1g2, red). MFI was normalized to control cells and served as a control (relative MFI = 1.0). n = 2 replicates per cell line. E, Representative flow cytometry plot showing OPP incorporation in PDTO4S4 control organoids (Empty; blue), PDTO4S4 control organoids treated with cycloheximide (gray), and PDTO4S4 NLE1 knockout organoids (NLE1g2; red). F, MFI of OPP incorporation in PDTO4S4 empty cells (blue), empty-treated cells with cycloheximide (gray) and NLE1 knockout organoids (NLE1g2; red). MFI was normalized to empty organoids and served as a control (relative MFI = 1.0).
Figure 4.
Effects of NLE1 ablation on growth and clonogenicity in colorectal cancer cell lines and PDTOs. A, Representative graph from xCELLigence system comparing the growth curve of SW620 empty (black), NLE1g1 (red), and NLE1g2 (blue) cells. Statistical significance between all samples at each analyzed time point was assessed by two-way ANOVA plus Tukey multiple comparisons test and is indicated for the latest time point. ****, P ≤ 0.0001. Shown is the mean ± SD (n = 8). B, Top, representative images of colony formation assays performed with SW620 empty, NLE1g1, and NLE1g2 cells. Bottom, colony formation of SW620 empty (black), NLE1g1 (red), and NLE1g2 (blue) cells was quantified and normalized to empty cells. Statistical significance between samples was assessed by an unpaired t test. ***, P ≤ 0.001; ****, P ≤ 0.0001. Shown is the mean ± SD (n = 3). C, Representative images of soft agar assays of SW620 empty, NLE1g1, and NLEg2 cells at day 14. Images were taken on a Nikon AZ100 zoom microscope. Scale bars, 1 mm. D, Grouped dot plot showing the number of colonies per image (from C) of soft agar assays performed with SW620 empty (black), NLE1g1 (red), and NLE1g2 (blue) cells. Statistical significance between samples was assessed by an unpaired t test. ****, P ≤ 0.0001. Shown is the mean ± SD (9 images taken from n = 3 replicates). E, Flow cytometry analysis of Annexin V and propidium iodide (PI) staining, which indicates apoptotic cells in SW620 empty (black), NLE1g1 (red), and NLE1g2 (blue) cells. Bar chart shows the percentages of Annexin V and propidium iodide double-positive cells. Shown is the mean ± SD (n = 2). F, Immunoblot analysis of PARP and cleaved PARP protein levels in SW620 empty, NLE1g1, and NLE1g2 cells. β-Actin served as a loading control. G, Grouped dot plot showing the number of migrating SW620 empty (black), NLE1g1 (red), and NLE1g2 (blue) cells in a transwell migration assay (uncoated membrane). Shown is the mean ± SD (18 images taken from n = 6 replicates). H, Grouped dot plot showing the number of invasive SW620 empty (black), NLE1g1 (red), and NLE1g2 (blue) cells in a transwell invasion assay (Matrigel-coated membrane). Shown is the mean ± SD (18 images taken from n = 6 replicates). Statistical significance between samples in G and H was assessed by one-way ANOVA plus Dunnett multiple comparisons test. ****, P ≤ 0.0001. I, Microscopy images of PDTO4S4 empty, NLE1g1, and NLE1g2 organoids on day 7 after plating of 4,000 cells per Matrigel droplet. Images were taken on a Nikon AZ100 zoom microscope. Scale bars, 200 μm. J, Stacked bar chart showing the diameter distribution (in percentages) of PDTO4S4 empty and NLE1g2 organoids in G. n = 300 organoids per genotype (n = 100 in three different Matrigel droplets) were measured and divided into groups of smaller than 50 μm, between 50 and 100 μm, and bigger than 100 μm organoid diameter. K, Number of organoids per image (from I). PDTO4S4 empty (black) and NLE1g2 (blue) organoid lines. Statistical significance between samples was assessed by an unpaired t test. **, P ≤ 0.01; ***, P ≤ 0.001. Shown is the mean ± SD (n = 3). See Supplementary Fig. S3 for additional data.
Notably, conditional overexpression of NLE1 in tumor organoids and in normal human colonic cells (HCEC-1CT) was not able to augment proliferation (Supplementary Fig. S4A and S4B). Besides this, HCEC-1CT cells overexpressing NLE1 did not display elevated de novo protein biosynthesis (Supplementary Fig. S4C and S4D). In line with these observations, ectopic expression of NLE1 was not able to rescue the growth-inhibitory effect of recombinant TGFβ1 on PDTOs (Supplementary Fig. S4E and S4F). These results show that additional factors beside NLE1 limit cellular proliferation and protein biosynthesis in human colonic epithelial cells, and elevated levels of NLE1 alone are insufficient to overcome TGFβ1-mediated growth inhibition of colorectal cancer organoids.
Loss of NLE1 leads to p38/MAPK activation, impaired autophagy, and elevated ROS generation in colorectal cancer cells
Agents that compromise ribosome function and protein biosynthesis, such as the translational inhibitors anisomycin and cycloheximide, have been reported to elicit a ribotoxic stress response, which can lead to activation of MAPK p38 and JNK signaling (39). While JNK phosphorylation was not detectable downstream of NLE1 loss in HT29 and SW620 colorectal cancer cells, ablation of NLE1 led to phosphorylation of p38/MAPK (Thr180/Tyr182) in this cellular context (Fig. 5A). Importantly, p38/MAPK signaling balances cancer cell autophagy and apoptosis in response to different types of cellular stress (reviewed in ref. 40), and inhibition of de novo protein synthesis, in contrast to nutrient starvation, impairs autophagic degradation (41). Because colorectal cancer cells underwent apoptosis in response to NLE1 knockout–mediated inhibition of protein biosynthesis (Fig. 4E and F; Supplementary Fig. S2F and S2G), we wondered if autophagy was also affected in this scenario. Indeed, tumor cells deficient for NLE1 displayed elevated levels of the autophagy receptor and substrate p62/SQSTM-1 (Fig. 5B), which is indicative of compromised autophagy (42). Furthermore, loss of NLE1 led to accumulation of the autophagosomal marker LC3B and an overall increase in the number and area of LC3B-positive subcellular structures, presumably due to impaired formation of degradative autophagolysosomes (Fig. 5B and C; refs. 41, 42). In concordance, treatment of colorectal cancer cells with the autophagy inhibitor chloroquine (CQ), which blocks autophagolysosome formation, induced similar but exaggerated effects (Fig. 5B and C). Notably, autophagy-impaired cancer cells contain higher ROS levels than their autophagy-proficient counterparts (43). Indeed, elevated levels of ROS occurred in HT29 and SW620 colorectal cancer cells upon NLE1 deletion (Fig. 5D and E). To which extent the concerted alteration of the processes described above contributes to the reduced fitness and elevated apoptosis observed in NLE1-deficient colorectal cancer cells needs further investigation.
Figure 5.
Loss of NLE1 affects p38/MAPK signaling, autophagy, and levels of reactive oxygen species in colorectal cancer cells. A, Immunoblot analysis of p38/MAPK (p38), phospho-p38 (p-p38) and phospho-SAPK/JNK in HT29 (left) and SW620 (right) cells transduced with either pLentiCRISPR-E (Empty) control lentiviral particles or lentiviral particles encoding for two different guide RNAs targeting NLE1 (NLE1g1, NLE1g2). α-Tubulin served as a loading control. B, Immunoblot analysis of the autophagy receptor protein p62 and autophagosome proteins LC3A/B in NLE1 wild-type (Empty) and NLE1 knockout (NLE1g1 and NLE1g2) HT29 (top) and SW620 (bottom) cells. β-Actin served as a loading control. C, Confocal microscopy images visualizing the indirect immunofluorescence staining of LC3B (red signal) as a surrogate for the occurrence of autophagosomes in NLE1 wild-type (Empty) and NLE1 knockout (NLE1g1 and NLE1g2) HT29 and SW620 cells. As a positive control for autophagosome accumulation, cells were treated with 50 μmol/L of the autophagy inhibitor chloroquine (CQ) 24 hours prior to analysis. D, Flow cytometry analysis of ROS generation in NLE1 wild-type (Empty) and NLE1 knockout (NLE1g1 and NLE1g2) HT29 and SW620 cells. As a positive control, cells were treated with the ROS-inducing agent tert-butyl hydroperoxide (+THBP) 1 hour prior to analysis. Histograms indicate cell count (normalized to mode) at different fluorescence levels originating from ROS-oxidized CellROX Green Reagent. E, Mean fluorescence intensity (MFI) of ROS-mediated CellROX oxidization in HT29 and SW620 control cells (Empty) and NLE1 knockout cells (NLE1g1 and NLE1g2). MFI was normalized to controls (relative MFI = 1.0). n = 2 replicates per cell line.
NLE1 knockout inhibits colorectal cancer growth and metastasis in an orthotopic mouse transplantation model
We next set out to clarify whether and to which extent NLE1 represents a limiting factor for colorectal cancer growth and distant metastasis in vivo. To study colorectal cancer development and progression in its natural tissue context, we employed endoscopy-guided orthotopic transplantation of genetically modified human tumor organoids into the colonic wall of immunodeficient mice (Fig. 6A; Supplementary video file). For these experiments, NLE1 knockout (KO) derivatives were generated from a highly aggressive human colorectal cancer organoid line, which efficiently forms liver metastasis in this experimental setting (kindly provided by Moritz Jesinghaus, Technical University of Munich, TUM, Munich, Germany). Prior to transplantation, NLE1 wild-type and NLE1 knockout tumor organoids were released from Matrigel, quantified by microscopy, and the ATP content as a surrogate for cell viability was determined for additional normalization. Approximately 150 viable organoids were delivered per injection site, and 1–2 injections into the submucosal space were performed per animal. Only if endoscope-guided PDTO delivery was achieved optimally (see Materials and Methods section for details), animals were chosen for further analysis. 3.5 weeks after injection, random sample colonoscopy showed massive colorectal cancer formation in two animals transplanted with NLE1 wild-type PDTOs, while NLE1 knockout PDTOs had formed a smaller visible tumor mass protruding into the colonic lumen (Fig. 6B). Because of nontolerable weight loss in three out of four mice initially transplanted with NLE1 wild-type colorectal cancer organoids, all animals were sacrificed simultaneously 5 weeks after transplantation and the primary tumor burden and occurrence of macroscopically visible distant metastasis was analyzed. Indeed, mice transplanted with PDTOs expressing wild-type NLE1 had formed on average larger primary tumors in the colon when compared to animals injected with NLE1 knockout PDTOs (Fig. 6C–E). In accordance, the percentage of MKI67-positive cells was lower in NLE1-deficient tumors (Fig. 6F and G), which indicated reduced proliferation. Furthermore, NLE1 knockout tumor glands showed an overall elevated abundance of cleaved caspase-3 (Fig. 6H), which is in agreement to what we had observed in colorectal cancer cell lines ex vivo upon NLE1 ablation. Of note, all NLE1 wild-type colonic tumor-bearing mice had developed 4–5 macroscopically visible liver metastases, while 4 of 6 mice with a NLE1 knockout primary tumor burden showed either no or only one liver metastasis, and only 2 of 6 mice had formed either 3 or 4 metastases, respectively (Fig. 6I and J). These data show that NLE1 loss in colorectal cancer organoids inhibits primary tumor growth and reduces the liver metastatic burden of xenotransplanted animals.
Figure 6.
NLE1 ablation reduces colorectal cancer primary tumor formation and liver metastasis in vivo.A, Schematic representation of endoscopy-guided PDTO orthotopic transplantation, colonoscopy follow-up, and organ analysis of sacrificed immunodeficient (NSG) mice. At time point t = 0, the process of PDTO needle injection, as seen via the endoscope camera, is depicted while the injection bubble is about to form. Control endoscopy was performed at day 25 to control for primary tumor occurrence and size and to estimate the end point of the experiment when animals need to be sacrificed due to excessive tumor burden. Mice were sacrificed 35 days after orthotopic PDTO transplantation, colonic tumors were documented, and the liver was scrutinized for occurrence of macroscopically visible metastatic foci. B, Colonoscopy of immunodeficient mice was performed 3.5 weeks after orthotopic transplantation of NLE1 wild-type (WT) or NLE1 knockout (KO) colorectal cancer organoids into the colonic wall. Note the more pronounced protrusion of NLE1-WT tumors into the colonic lumen when compared to tumors grown from NLE1-KO PDTOs. C, Scatter plot showing the areas of primary tumors grown in the colon of xenotransplanted mice. Four mice (n = 6 primary tumors) had been transplanted with NLE1 wild-type (WT) colorectal cancer organoids and 6 mice (n = 12 primary tumors) had been transplanted with NLE1 knockout (NLE1-targeting guide RNAs 1 (red dots) or 2 (blue dots) colorectal cancer organoids. All mice were sacrificed for analysis 5 weeks after xenotransplantation. Statistical significance between the NLE1-WT and NLE1-KO groups was assessed by an unpaired t test with Welch correction to account for the observed unequal SDs within the two experimental groups. *, P < 0.05. Shown is the mean ± SD.D, Macroscopic images of primary tumors formed in the colon of four mice orthotopically transplanted with 150 tumor organoids wild-type for NLE1 per injection site. All mice were sacrificed for analysis 5 weeks after xenotransplantation. Scale bars, 0.5 cm. E, Macroscopic images of primary tumors formed in the colon of six mice orthotopically transplanted with 150 tumor organoids knockout for NLE1 per injection site. All mice were sacrificed for analysis 5 weeks after xenotransplantation. Scale bars, 0.5 cm. F, Representative microscopy images of IHC staining of the proliferation marker MKI67 on FFPE tissue sections prepared from NLE1 wild-type and NLE1 knockout PDTO-derived primary tumors. Scale bars, 100 μm. G, Quantification of MKI67-positive tumor cells (in percentages) in FFPE tissue sections from NLE1 wild-type (n = 4) and NLE1 knockout (n = 6) primary tumors. An unpaired t test was performed to assess significance. ***, P < 0.001. Shown is the mean ± SD. H, Representative microscopy images of IHC staining of the apoptosis marker cleaved caspase-3 on FFPE tissue sections prepared from NLE1 wild-type and NLE1 knockout primary tumors. Scale bars, 100 μm. I, Exemplary macroscopic images of resected livers and colorectal cancer–derived liver metastases formed in mice orthotopically transplanted with NLE1 wild-type or NLE1 knockout tumor organoids. Arrows, macroscopically visible liver metastasis. J, Scatter plots showing the quantification of macroscopic metastatic foci detected in the liver of mice five weeks after endoscope-guided, orthotopic transplantation of colorectal cancer organoids either wild-type or knockout [NLE1-targeting guide RNAs 1 (red squares) or 2 (black squares)] for NLE1. Statistical significance between the NLE1-WT and NLE1-KO groups was assessed by an unpaired t test with Welch correction to account for the observed unequal SDs within the two experimental groups. **, P < 0.01. Shown is the mean ± SD. CRC, colorectal cancer.
NLE1 is enriched in Wnt/MYC molecular subtypes of colorectal cancer and predicts survival in patients with colorectal cancer
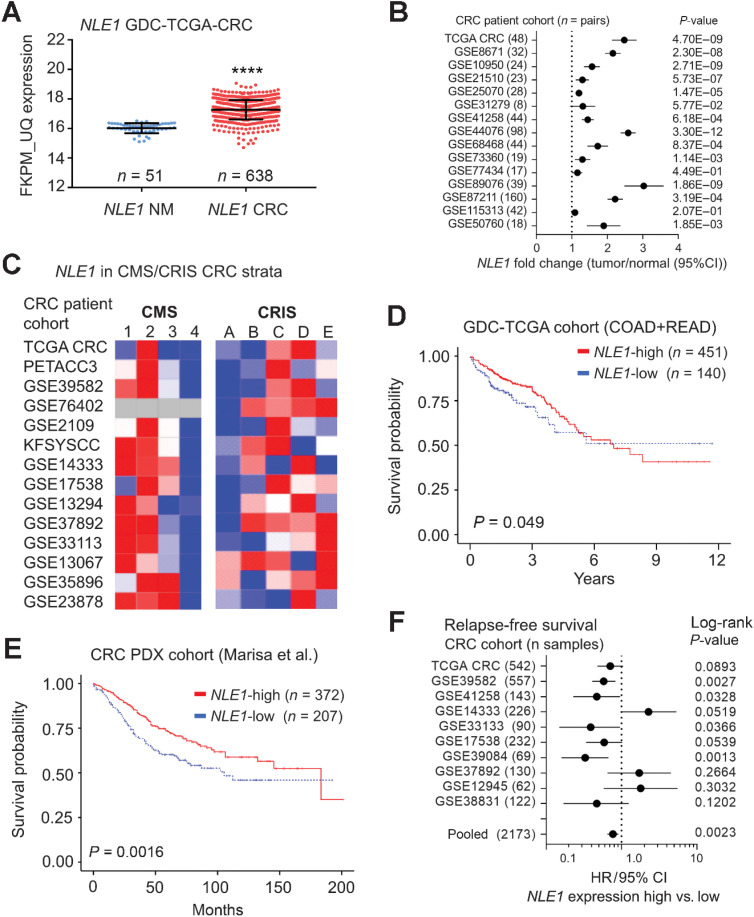
Examination of NLE1 gene expression data obtained from the TCGA (via the GDC Data Portal; refs. 23, 44) revealed elevated NLE1 mRNA levels in colorectal cancer when compared with normal tissues (Fig. 7A). Publicly available gene expression data from patient-matched pairs of colorectal cancer and normal tissues (n = 644 in total) from 15 different colorectal cancer cohorts also showed NLE1 upregulation in colorectal tumors (Fig. 7B). Considering the enrichment of c-MYC and colonic stem cell gene expression in NLE1-high expressing tumors (Fig. 2F), we wondered if NLE1 expression was associated with the cancer molecular subtype (CMS; ref. 1) and colorectal cancer intrinsic subtype (CRIS; ref. 2) specified by these molecular traits. Indeed, NLE1 levels were highest in CMS2 and in CRIS C/D characterized by high Wnt/MYC activity and intestinal stem cell gene expression (Fig. 7C). In agreement with the downregulation of NLE1 gene expression by TGFβ/SMAD signaling described above, NLE1 mRNA levels were relatively low in colorectal cancer subtypes CMS4 and CRIS-B, which display strong TGFβ signaling activity (Fig. 7C; refs. 1, 2). Notably, NLE1-high subtypes (CMS2 and CRIS-C/D) are associated with good patient prognosis, while NLE1-low subtypes (CMS4 and CRIS-B) predict poor survival of patients with colorectal cancer (1, 2). In line with these data, we found that NLE1 levels correlate with good overall survival in two independent cohorts of patients with colorectal cancer (Fig. 7D and E; refs. 24, 44). In addition, relatively high expression of NLE1 was associated with increased relapse-free survival in 8 of 11 analyzed colorectal cancer cohorts overall representing 2,173 individuals (Fig. 7F). Therefore, we suggest that NLE1 serves as a predictive biomarker for overall and progression-free survival in patients with colorectal cancer.
Figure 7.
NLE1 mRNA levels are increased in colorectal cancer, correlate with Wnt/MYC-expressing colorectal cancer molecular subtypes, and predict patient survival. A,NLE1 gene expression analysis on human GDC TCGA-COAD plus READ datasets (FKPM-UQ data): comparison of normal human mucosa samples and tumor tissues. ****, P < 0.0001; n = 638 tumor samples and n = 51 normal mucosa samples. B, Forest plot showing fold changes of NLE1 mRNA expression between colorectal cancer tumors and matched adjacent normal colonic mucosa for indicated cohorts. Data were derived from publicly available data sets. The GEO database accession number for each analyzed data set is indicated. The number of patient-matched tumor/normal pairs is indicated in brackets. Dots represent fold changes and horizontal lines show 95% confidence intervals (CI). P values were calculated using a paired t test. C, Heatmaps showing the relative abundance of NLE1 mRNA in cancer molecular subtypes (CMS; ref. 1) and colorectal cancer intrinsic subtypes (CRIS; ref. 2) of various publicly colorectal cancer cohort data sets. The GEO database accession number for each analyzed data set is indicated. Blue, relatively low expression; red, high expression levels of NLE1. D, Kaplan–Meier estimate curves of overall survival from 591 patients with colorectal cancer (GDC-TCGA COAD plus READ cohorts) in relation to NLE1 gene expression (FKPM-UQ normalized data). Patients were classified according to either low (≤ 16.001, n = 140, blue line) or high (> 16.001, n = 451, red line) NLE1 FKPM-UQ expression scores. E, Kaplan–Meier estimate curves of overall survival from 577 patients with colorectal cancer (24) in relation to NLE1 gene expression scores. Patients were classified according to either low (n = 207; blue line) or high (n = 372; red line) NLE1 expression scores. In D and E, Censored values indicate the last known follow-up time for those subjects still alive after initial diagnosis and are depicted as tick marks. F, Forest plot showing the hazard ratios (HR) for relapse-free survival of patients with colorectal cancer expressing either high or low levels of NLE1 mRNA expression in public datasets (TCGA colorectal cancer and NCBI GEO). GEO accession numbers for each of the depicted cohorts are indicated. Dots represent HRs and horizontal lines show 95% confidence intervals (CI). Log-rank P values are indicated for each analyzed data set. CRC, colorectal cancer.
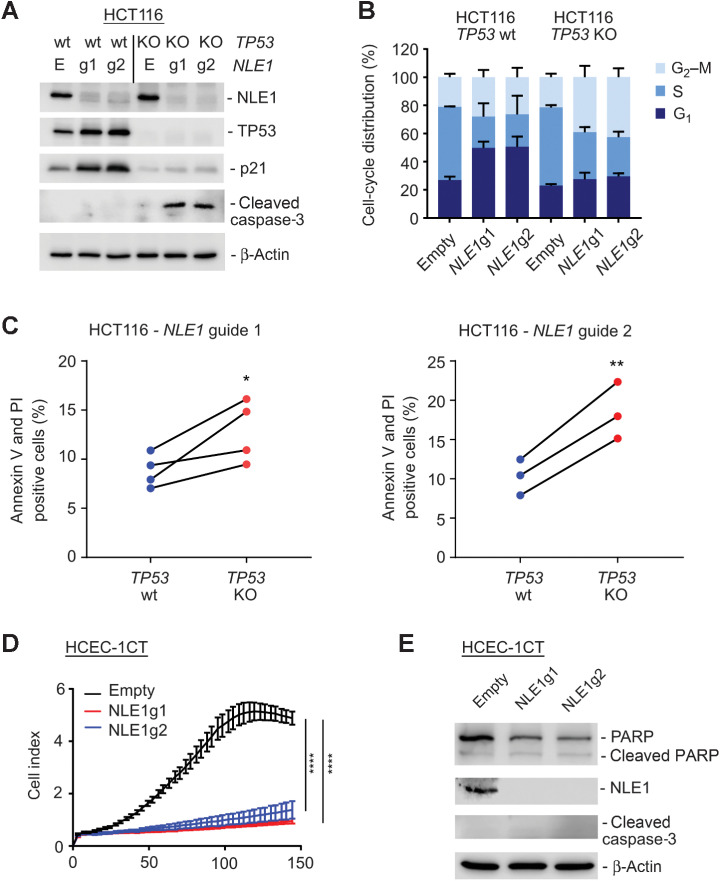
Wild-type p53 protects microsatellite instable colorectal cancer cells from NLE1 loss–mediated apoptosis
Nucleolar stress has been reported to activate the tumor suppressor protein TP53 (p53; ref. 45), which is either lost or mutated in the majority of chromosomally instable colorectal cancer cases but more frequently retained wild-type in microsatellite instable (MSI) colorectal cancer (46). Because our data suggested that NLE1 represents a potential target for colorectal cancer therapy rather than a prognosticator of unfavorable disease progression, we asked whether loss of NLE1 function might exert a differential effect on cancer cells dependent on the p53 status. To achieve this, we first performed CRISPR/Cas9 targeting of the wild-type p53 genomic locus in MSI HCT116 colorectal cancer cells, which represent a widely used and well-known cell line model for studying p53/p21 axis-dependent cell cycle effects and the determinants of apoptosis in colorectal cancer (47, 48). Next, NLE1 knockout derivatives were generated via CRISPR/Cas9 from pools of HCT116 parental and HCT116 p53 knockout (KO) cells. Indeed, loss of NLE1 in HCT116 parental colorectal cancer cells led to p53 stabilization, induction of the p53 target gene product CDKN1A (p21), and a G1 cell-cycle arrest (Fig. 8A and B). In contrast, HCT116 p53 KO cells failed to increase p21 levels, likely due to the complete lack of p53, and showed accumulation of cells in G2–M upon NLE1 loss (Fig. 8A and B). Of note, NLE1 loss in the absence of p53 induced cleavage of caspase-3, which was not detectable in a p53 wild-type situation (Fig. 8A). In concordance, a higher fraction of HCT116 p53 KO cells underwent apoptosis upon knockout of NLE1 compared to parental HCT116 (Fig. 8C). This shows that ablation of NLE1 function in MSI colorectal cancer cells leads to activation of p53/p21 signaling and G1 arrest, and complete loss of p53 sensitizes NLE1-deficient HCT116 cells to apoptosis. Notably, loss of NLE1 led to wild-type p53-independent induction of p21 mRNA and protein in CIN HT29 and, although to a much lower extent, SW620 colorectal cancer cells, both expressing mutant variants of p53 (HT29: R273H; SW620: R273H;P309S; Supplementary Fig. S5A and S5B). This suggests that also other, wild-type p53-independent mechanisms may to some extent confer a tumor cell context–dependent protection of colorectal cancer cells upon loss of NLE1. Besides this, NLE1 mRNA levels in 6 analyzed colorectal cancer cohorts were comparable in p53 wild-type and p53-mutant tumors (Supplementary Fig. S5C), suggesting that genomic alterations in p53 do not give rise to elevated expression of NLE1 in colorectal cancer.
Figure 8.
Effect of NLE1 loss in p53-proficient HCT116 MSI colorectal cancer cells and HCEC-1CT benign human colonic epithelial cells. A, Immunoblot analysis of NLE1, TP53, CDKN1A (p21) and cleaved caspase-3 protein levels in HCT116 TP53 wild-type (wt) and TP53 knockout (KO) cells stably transduced with either pLentiCRISPR-E (Empty) control lentivirus particles or lentiviral particles encoding for two different guide RNAs targeting NLE1 (NLE1g1, NLE1g2). β-Actin served as a loading control. B, Stacked bar chart showing the percentage of cell-cycle distribution of HCT116 TP53 wild-type and TP53 knockout cells stably transduced with either pLentiCRISPR-E (Empty) control lentiviral particles or lentiviral particles encoding for two different guide RNAs targeting NLE1 (NLE1g1, NLE1g2). Shown is the mean ± SD (n = 3). C, Quantification of apoptotic cell fractions, as assessed by Annexin V/propidium iodide (PI) staining and flow cytometry analysis of HCT116 TP53 wild-type and TP53 knockout cells transduced with lentiviral particles encoding Cas9 and guide RNAs targeting NLE1 (left, NLE1g1; right, NLE1g2). Statistical significance was assessed by a ratio paired t test. *, P ≤ 0.05; **, P ≤ 0.01. Each two dots (blue-red) connected by solid lines represent independent experiments. D, Representative graph from xCELLigence system comparing the growth curve of HCEC-1CT human colonic epithelial cells stably transduced with either pLentiCRISPR-E (Empty; black) control lentiviral particles or lentiviral particles encoding for two different guide RNAs targeting NLE1 (NLE1g1, red; NLE1g2, blue). Statistical significance between all samples at each analyzed time point was assessed by two-way ANOVA plus Tukey multiple comparisons test and is indicated for the latest time point. ****, P ≤ 0.0001. Shown is the mean ± SD (n = 7). E, Immunoblot analysis of NLE1, (cleaved) PARP, and cleaved caspase-3 protein levels in HCEC-1CT Empty (black), NLE1g1 (red), and NLE1g2 (blue) cells. β-Actin served as a loading control.
NLE1 loss in immortalized human colonic epithelial cells induces cell-cycle arrest rather than apoptosis
De novo protein biosynthesis, similar to DNA synthesis and RNA synthesis, represents an essential trait of cellular functionality. Hence, targeting NLE1 in patients with colorectal cancer might also affect normal proliferating cells critical for tissue self-renewal and homeostasis, thereby provoking toxicity. To address the dependency of nonmalignant cells on NLE1, we generated NLE1-deficient derivatives of immortalized HCEC-1CT human colonic epithelial cells via CRISPR/Cas9 editing. As expected, NLE1-deficient HCEC-1CT cells almost halted proliferation, underwent cell-cycle arrest, and displayed compromised de novo protein biosynthesis (Fig. 8D; Supplementary Fig. S5D–S5F). However, HCEC-1CT NLE1 knockout cells did not show increased cleavage of the apoptotic markers caspase-3 and PARP (Fig. 8E). In addition, Annexin V/PI staining did not indicate an elevated proportion of apoptotic cells upon NLE1 loss (Supplementary Fig. S5G). These data suggest that loss of NLE1 provokes cell-cycle arrest rather than programmed cell death in nonmalignant human colonic epithelial cells. Therefore, a therapeutic inhibition of NLE1 in patients with colorectal cancer may have manageable side-effects.
Discussion
Deregulated ribosome biogenesis accompanied by an augmented protein biosynthetic capacity represents a hallmark of colorectal cancer stemness, growth and metastasis (10, 49). The alteration of key drivers in colorectal cancer, such as Wnt/MYC signaling, TGFβ/SMAD signaling, and TP53 function, is closely linked to the malignant exaggeration of biosynthetic processes, and their contribution to cancer progression and metastasis represents an area of intensive research (reviewed in refs. 49, 50). The c-MYC proto-oncogene, whose expression levels are regulated by Wnt- and TGFβ signaling (51, 52), acts on ribosome biogenesis by inducing ribosomal RNA transcription and expression of factors involved in ribosomal subunit composition and nuclear-cytoplasmic export (reviewed in ref. 13). Directly targeting c-MYC function in the context of compromised Wnt- and TGFβ signaling in cancer has been extremely challenging, and there are currently no therapeutic approaches approved for the clinic. Hence, identification and characterization of c-MYC–regulated factors involved in cancer-relevant processes can open up new avenues for cancer therapy.
By performing CRISPR/Cas9-mediated knockout of SMAD4 in TGFβ-responsive colorectal cancer PDTO lines and transcriptomic analysis of isogenic SMAD4 wild-type and knockout pairs, we modelled the enrichment of human colonic stem cell markers and c-MYC target genes upon SMAD4 loss in the context of an extrinsic TGFβ1 stimulus. Interestingly, only 11 out of 284 upregulated genes in SMAD4-mutant and LGR5 positive colorectal cancer organoids represented cancer cell-autonomous dependency factors according to the DepMAP screening project (29, 30). This suggests that the majority of genes enriched in a colorectal cancer stem cell phenotype encode for either redundant functions or factors mediating the manifold interactions of cancer cells with their adjacent microenvironment. By focusing on cancer dependency genes involved in ribosome maturation and functionality, we found that expression of the ribosomal assembly component NLE1 is regulated by the TGFβ/SMAD4/c-MYC axis in colorectal cancer. NLE1 (Notchless, Rsa4 in yeast) was first described in yeast to participate in ribosome assembly and maturation, and together with the Rix1 sub-complex it is bound to the so-called Rix1 pre-60S intermediate (38). The AAA ATPase Midasin (MIDAS; Rea1 in yeast) physically contacts these two factors to mediate their removal (36, 38). This in turn triggers nuclear maturation and nuclear export competence to the pre-60S subunit (38). Importantly, site-specific mutations affecting the MIDAS-NLE1 interaction lead to a 60S subunit export defect, which ultimately reduces the 60S/40S ratio of mature ribosomal particles and leads to occurrence of “half-mer” polysomes in the cytoplasm (38). Mass spectrometry analysis on HeLa cell-derived nucleolar extracts identified NLE1 as a component of the nucleolar compartment, which indicated a similar function of NLE1 in human cells (53). Indeed, release of NLE1, together with a PeBoW (Pes1, Bop1, and WDR12) complex, by the large motor protein Midasin is a well-conserved step in eukaryotic ribosome maturation (37). We could show that the targeted knockout of NLE1 in human colorectal cancer cells reduced global de novo protein biosynthesis, which might be attributable to lower numbers of functional ribosomes and/or an accumulation of dysfunctional polysomes. Interestingly, while exponentially proliferating colorectal cancer cell lines in two-dimensional petri dishes were massively affected by NLE1 ablation with regard to proliferation, clonogenicity, anchorage-independent growth, and cell death, the NLE1 knockout showed a milder effect in three dimensionally grown, Matrigel-embedded colorectal cancer organoids. We speculate that the overall slower and more heterogeneous growth kinetics in tumor cell clusters and/or the cancer stemness-promoting, growth factor-rich PDTO (TOC) culture medium might have rendered colorectal cancer cells relatively less vulnerable to NLE1 loss. Furthermore, the relatively early passages (∼10–20) of PDTOs used in this study might exhibit a larger genetic diversity and ribosomal heterogeneity (reviewed in ref. 49) when compared to longer-term cultured classic colorectal cancer cell lines. Hence, loss of NLE1 could select for certain genetic and/or ribosomal traits present in PDTO cell subpopulations that confer partial tolerance to NLE1 ablation. Nevertheless, NLE1 knockout colorectal cancer-PDTOs showed a decreased self-renewal and growth capacity in vitro and formed smaller tumors after orthotopic transplantation into the colonic wall of immune-deficient mice. More important, primary tumors grown from NLE1 knockout PDTOs developed fewer distant metastases in the liver when compared with their NLE1 wild-type counterparts. Metastatic colonization of colorectal cancer relies on a niche-dependent plasticity of tumor cells with respect to their LGR5 status (54) and different (intermediate) epithelial-to-mesenchymal (EMT) states (reviewed in ref. 55). An enhanced biosynthetic capacity augments plasticity of cancer cells and might equip them with a survival advantage during their journey through the metastatic cascade (reviewed in (56)). Hence, ablation of NLE1 in tumor organoids, which reduced de novo protein biosynthesis in vitro, might have lowered the success rate of colorectal cancer cells to spawn distant metastasis. Future studies should address the translational adaptation programs of colorectal cancer cells at different stages of metastasis and their dependency on elevated NLE1 expression.
Although we observed a compromised migration and invasion capacity of NLE1-deficient colorectal cancer cells in vitro, which might have contributed to the lower liver metastatic load observed in vivo, the reduced primary tumor burden in mice transplanted with NLE1 knockout PDTOs represents a strong confounder variable. Due to this limitation of our in vivo approach, we cannot exclude that besides the reduced invasive and metastatic capability inherent to NLE1-targeted colorectal cancer cells, the overall lower amount of colorectal cancer cells available for primary tumor dissemination might at least partially explain the reduced liver metastatic burden observed in mice orthotopically transplanted with NLE1-deficient colorectal cancer organoids.
While we could demonstrate that NLE1 is critical for colorectal cancer cell fitness and disease progression, elevated NLE1 mRNA levels predict good overall and relapse-free survival in patients with colorectal cancer. This is in accordance with our data showing that NLE1 mRNA levels positively correlate with c-MYC target gene set expression in colorectal cancer cohorts. Furthermore, NLE1 itself was induced by the c-MYC oncogene, which itself has been reported to predict better survival in patients with colorectal cancer (57, 58). Notably, NLE1 expression levels were lowest in molecular strata of colorectal cancer associated with active TGFβ signaling and poor patient prognosis (CMS4 and CRIS-B; refs. 1, 2), which fits our data on NLE1 enrichment in TGFβ1 exposed tumor organoids lacking SMAD4. CMS4 classified colorectal cancer cells were shown to resist 5-fluoruracil (5-FU) and oxaliplatin-based chemotherapies (59), which also provoke ribosomal stress (60, 61). The relatively low levels of NLE1 in this colorectal cancer patient subgroup might indicate that these tumors rely less on elevated de novo protein biosynthesis for their survival under chemotherapy. Interestingly, CMS4-type metastatic colorectal cancer has been suggested to respond to irinotecan-based combination therapies rather than to an oxaliplatin regimen (62). If the magnitude of NLE1 expression can be used as a surrogate marker for the responsiveness of patients with colorectal cancer to different types of chemotherapy should be studied further.
Although ribosome biogenesis represents a key feature for cellular survival, the nucleolus with its diverse components and regulatory pathways might reveal effective and cancer cell-selective therapeutic targets. For example, well established chemotherapeutic agents, such as oxaliplatin and 5-FU, act on ribosomal RNA transcription and evoke ribosomal stress (60, 61). However, targeted therapeutic inhibition of NLE1 may be challenging. A good understanding of the pre-ribosomal complex interfaces between interacting assembly factors, such as Midasin-NLE1, might allow the structure-guided development of interfering small-molecule or peptide drugs able to inhibit ribosome maturation. Specific peptides that interfere with yeast Erb1 and Ytm1 protein interaction and thereby prevent NOP7 (mammals: PeBoW) complex formation and ribosome biogenesis have been reported (63). Notably, classic small-molecule drugs follow the occupancy-driven pharmacologic model, which implies the need for designing high-affinity compounds able to exert their on-target activity at low and tolerable doses. This represents a challenge for many disease targets and could prevent efficacious targeting of NLE1 in patients with colorectal cancer. Protein-targeting chimeras (PROTAC), which mediate specific ubiquitination and degradation of a desired target via the ubiquitin-proteasome pathway, follow an event-driven pharmacology to circumvent this problem. PROTACs have been designed to target oncogenes previously considered as “undruggable”, such as mutant KRAS (reviewed in ref. 64), and could also proof effective to tackle abnormally high levels of NLE1 in metastatic colorectal cancer. Furthermore, suppression of elevated NLE1 expression in colorectal cancer cells might be feasible with NLE1-specific short interfering RNAs or microRNAs. A nanoparticle-based delivery of these molecules effectively mediates RNA interference in cancer cells, and nano-carriers can be optimized to meet the therapeutic requirements of a specific disease (reviewed in ref. 65), such as colorectal cancer liver metastasis (66).
Nucleolar stress activates the tumor suppressor protein TP53 (45), which is lost in approximately 80% of chromosomally instable colorectal cancer cases but retained wild-type in many microsatellite instable (MSI) tumors (46). Sapio and colleagues could demonstrate that inhibition of ribosome biogenesis by silencing of either BOP1 or RPS19 confers cytotoxic drug resistance to cancer cells in a p53-dependent manner, and a therapeutic approach combining ribosomal inhibition and cytotoxic treatment selectively killed p53-negative tumor cells (67). In concordance, we found that deletion of TP53 prevented a CDKN1A/p21-mediated G1 cell-cycle arrest and increased the apoptotic fraction of NLE1-deficient MSI HCT116 colorectal cancer cells. Together with the lack of apoptosis induction upon NLE1 ablation observed in immortalized benign human colonic epithelial cells, our data suggest the possibility that there might exist a therapeutic window for engaging NLE1 function in patients with colorectal cancer. However, to which extent therapeutic targeting of NLE1 in a clinical setting shows tumor selectivity and affects benign tissue homeostasis at a tolerable level warrants further investigation.
Supplementary Material
Supplementary Figures S1-5 S1: Effect of SMAD4 loss on intestinal stem cell, c-MYC, and NLE1 gene expression in CRC organoids S2: Effects of NLE1 ablation on growth and clonogenicity of CRC cell lines S3: Effects of NLE1 ablation on growth and clonogenicity of CRC PDTOs S4: Effects of NLE1 over-expression in human colonic epithelial cells and CRC organoids S5: NLE1 loss-mediated cell cycle arrest and apoptosis in HCT116 CRC cells and HCEC-1CT normal human colonic epithelial cells
Supplementary Materials Oligonucleotides, qRT-PCR primer pairs, and Taqman probe assays used in this study.
Characteristics of tumor organoid lines used for CRISPR/Cas9-mediated SMAD4 deletion.
RNA Seq_count matrix
Upregulated genes in SMAD4 mutant tumor organoid lines and DepMAP Project dependency scores.
Differentially expressed genes in PDTOS4 versus PDTO cells in the presence of recombinant TGF-beta1
Endoscopy-guided orthotopic injection of colorectal cancer organoids into the colonic wall of immune-deficient mice.
Acknowledgments
The authors are grateful to Dr. Nassim Bouznad for technical advice on analyzing autophagy in colorectal cancer cells and to Anja Heier and Andrea Sendelhofert for their technical support and advice on immunohistochemistry (IHC). They also thank Mrs. Yuki Schneider-Kimoto for preparing the FFPE tissue sections used for IHC staining. This work was further supported by the European Union's Framework Programme for Research and Innovation Horizon 2020 under the Marie Skłodowska-Curie Grant Agreement No. 753058 (to M. Tschurtschenthaler) and EMBO Long-Term Fellowship (ALTF 1290–2016 to M. Tschurtschenthaler).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
T. Kirchner reports other support from Amgen, AstraZeneca, BMS, Boehringer Ingelheim, Merck, Novartis, Pfizer, Qiagen, Roche, and Takeda outside the submitted work. F. Klauschen reports personal fees and other support from Aignostics GmbH and personal fees from MSD, Roche, BMS, Lilly, Provitro AG, Novartis, and Merck outside the submitted work. D. Saur reports grants from DFG, European Research Council, and Wilhelm Sander-Stiftung during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
L.P. Loevenich: Conceptualization, formal analysis, investigation, visualization, methodology, writing–original draft. M. Tschurtschenthaler: Investigation, methodology, writing–review and editing. M. Rokavec: Formal analysis, investigation, methodology. M.G. Silva: Methodology. M. Jesinghaus: Methodology. T. Kirchner: Resources, supervision, writing–review and editing. F. Klauschen: Resources, supervision, writing–review and editing. D. Saur: Methodology, writing–review and editing. J. Neumann: Resources, validation, methodology, writing–review and editing. H. Hermeking: Resources, supervision, methodology, writing–review and editing. P. Jung: Conceptualization, resources, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration.
References
- 1. Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Isella C, Brundu F, Bellomo SE, Galimi F, Zanella E, Porporato R, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun 2017;8:15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011;8:511–24. [DOI] [PubMed] [Google Scholar]
- 4. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med 2017;23:1124–34. [DOI] [PubMed] [Google Scholar]
- 5. Cortina C, Turon G, Stork D, Hernando-Momblona X, Sevillano M, Aguilera M, et al. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol Med 2017;9:869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017;543:676–80. [DOI] [PubMed] [Google Scholar]
- 7. Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 2017;545:187–92. [DOI] [PubMed] [Google Scholar]
- 8. Dieter SM, Glimm H, Ball CR. Colorectal cancer-initiating cells caught in the act. EMBO Mol Med 2017;9:856–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smit WL, Spaan CN, de Boer RJ, Ramesh P, Garcia TM, Meijer BJ, et al. Driver mutations of the adenoma-carcinoma sequence govern the intestinal epithelial global translational capacity. Proc Natl Acad Sci U S A 2020;117:25560–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morral C, Stanisavljevic J, Hernando-Momblona X, Mereu E, Alvarez-Varela A, Cortina C, et al. Zonation of ribosomal DNA transcription defines a stem cell hierarchy in colorectal cancer. Cell Stem Cell 2020;26:845–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol 2022;19:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 2005;7:311–8. [DOI] [PubMed] [Google Scholar]
- 13. van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010;10:301–9. [DOI] [PubMed] [Google Scholar]
- 14. Dietinger V, de Durango CRG, Wiechmann S, Boos SL, Michl M, Neumann J, et al. Wnt-driven LARGE2 mediates laminin-adhesive O-glycosylation in human colonic epithelial cells and colorectal cancer. Cell Commun Signal 2020;18:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015;521:43–7. [DOI] [PubMed] [Google Scholar]
- 16. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016;351:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roper J, Tammela T, Akkad A, Almeqdadi M, Santos SB, Jacks T, et al. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat Protoc 2018;13:217–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N Engl J Med 2016;375:1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, Rossell D, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med 2011;17:1225–7. [DOI] [PubMed] [Google Scholar]
- 28. Brunschwig EB, Wilson K, Mack D, Dawson D, Lawrence E, Willson JK, et al. PMEPA1, a transforming growth factor-beta-induced marker of terminal colonocyte differentiation whose expression is maintained in primary and metastatic colon cancer. Cancer Res 2003;63:1568–75. [PubMed] [Google Scholar]
- 29. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell 2017;170:564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Behan FM, Iorio F, Picco G, Goncalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019;568:511–6. [DOI] [PubMed] [Google Scholar]
- 31. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alawi F, Lee MN. DKC1 is a direct and conserved transcriptional target of c-MYC. Biochem Biophys Res Commun 2007;362:893–8. [DOI] [PubMed] [Google Scholar]
- 33. Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 2003;4:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nachmani D, Bothmer AH, Grisendi S, Mele A, Bothmer D, Lee JD, et al. Germline NPM1 mutations lead to altered rRNA 2'-O-methylation and cause dyskeratosis congenita. Nat Genet 2019;51:1518–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, BX L-R, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014;159:148–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Suzuki H, Kobayashi Y, Wang AC, DiMaio F, Kawashima SA, et al. Structural insights into Mdn1, an essential AAA protein required for ribosome biogenesis. Cell 2018;175:822–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Romes EM, Sobhany M, Stanley RE. The crystal structure of the Ubiquitin-like domain of Ribosome assembly factor Ytm1 and characterization of its interaction with the AAA-ATPase Midasin. J Biol Chem 2016;291:882–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulbrich C, Diepholz M, Bassler J, Kressler D, Pertschy B, Galani K, et al. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 2009;138:911–22. [DOI] [PubMed] [Google Scholar]
- 39. Vind AC, Genzor AV, Bekker-Jensen S. Ribosomal stress-surveillance: three pathways is a magic number. Nucleic Acids Res 2020;48:10648–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sui X, Kong N, Ye L, Han W, Zhou J, Zhang Q, et al. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett 2014;344:174–9. [DOI] [PubMed] [Google Scholar]
- 41. Lawrence BP, Brown WJ. Inhibition of protein synthesis separates autophagic sequestration from the delivery of lysosomal enzymes. J Cell Sci 1993;105:473–80. [DOI] [PubMed] [Google Scholar]
- 42. Runwal G, Stamatakou E, Siddiqi FH, Puri C, Zhu Y, Rubinsztein DC. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep 2019;9:10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009;137:1062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 2010;29:4253–60. [DOI] [PubMed] [Google Scholar]
- 46. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated analysis of TP53 gene and pathway alterations in The Cancer Genome Atlas. Cell Rep 2019;28:3010. [DOI] [PubMed] [Google Scholar]
- 47. Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest 1999;104:263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Polyak K, Waldman T, He TC, Kinzler KW, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev 1996;10:1945–52. [DOI] [PubMed] [Google Scholar]
- 49. Elhamamsy AR, Metge BJ, Alsheikh HA, Shevde LA, Samant RS. Ribosome biogenesis: a central player in cancer metastasis and therapeutic resistance. Cancer Res 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 2018;18:51–63. [DOI] [PubMed] [Google Scholar]
- 51. He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of c-MYC as a target of the APC pathway. Science 1998;281:1509–12. [DOI] [PubMed] [Google Scholar]
- 52. Alexandrow MG, Kawabata M, Aakre M, Moses HL. Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of transforming growth factor beta 1. Proc Natl Acad Sci U S A 1995;92:3239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, et al. Functional proteomic analysis of human nucleolus. Mol Biol Cell 2002;13:4100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fumagalli A, Oost KC, Kester L, Morgner J, Bornes L, Bruens L, et al. Plasticity of Lgr5-negative cancer cells drives metastasis in colorectal cancer. Cell Stem Cell 2020;26:569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bornes L, Belthier G, van Rheenen J. Epithelial-to-mesenchymal transition in the light of plasticity and hybrid E/M States. J Clin Med 2021;10:2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Micalizzi DS, Ebright RY, Haber DA, Maheswaran S. Translational regulation of cancer metastasis. Cancer Res 2021;81:517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB, Choe G, et al. Favorable prognosis in colorectal cancer patients with co-expression of c-MYC and ss-catenin. BMC Cancer 2016;16:730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Toon CW, Chou A, Clarkson A, DeSilva K, Houang M, Chan JC, et al. Immunohistochemistry for myc predicts survival in colorectal cancer. PLoS One 2014;9:e87456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Linnekamp JF, Hooff SRV, Prasetyanti PR, Kandimalla R, Buikhuisen JY, Fessler E, et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ 2018;25:616–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bruno PM, Liu Y, Park GY, Murai J, Koch CE, Eisen TJ, et al. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat Med 2017;23:461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takimoto CH, Tan YY, Cadman EC, Armstrong RD. Correlation between ribosomal RNA production and RNA-directed fluoropyrimidine cytotoxicity. Biochem Pharmacol 1987;36:3243–8. [DOI] [PubMed] [Google Scholar]
- 62. Del Rio M, Mollevi C, Bibeau F, Vie N, Selves J, Emile JF, et al. Molecular subtypes of metastatic colorectal cancer are associated with patient response to irinotecan-based therapies. Eur J Cancer 2017;76:68–75. [DOI] [PubMed] [Google Scholar]
- 63. Orea-Ordonez L, Masia S, Bravo J. Peptides targeting the interaction between Erb1 and Ytm1 ribosome assembly factors. Front Mol Biosci 2021;8:718941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bekes M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov 2022;21:181–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xin Y, Huang M, Guo WW, Huang Q, Zhang LZ, Jiang G. Nano-based delivery of RNAi in cancer therapy. Mol Cancer 2017;16:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sendi H, Yazdimamaghani M, Hu M, Sultanpuram N, Wang J, Moody AS, et al. Nanoparticle delivery of miR-122 inhibits colorectal cancer liver metastasis. Cancer Res 2022;82:105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sapio RT, Nezdyur AN, Krevetski M, Anikin L, Manna VJ, Minkovsky N, et al. Inhibition of post-transcriptional steps in ribosome biogenesis confers cytoprotection against chemotherapeutic agents in a p53-dependent manner. Sci Rep 2017;7:9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-5 S1: Effect of SMAD4 loss on intestinal stem cell, c-MYC, and NLE1 gene expression in CRC organoids S2: Effects of NLE1 ablation on growth and clonogenicity of CRC cell lines S3: Effects of NLE1 ablation on growth and clonogenicity of CRC PDTOs S4: Effects of NLE1 over-expression in human colonic epithelial cells and CRC organoids S5: NLE1 loss-mediated cell cycle arrest and apoptosis in HCT116 CRC cells and HCEC-1CT normal human colonic epithelial cells
Supplementary Materials Oligonucleotides, qRT-PCR primer pairs, and Taqman probe assays used in this study.
Characteristics of tumor organoid lines used for CRISPR/Cas9-mediated SMAD4 deletion.
RNA Seq_count matrix
Upregulated genes in SMAD4 mutant tumor organoid lines and DepMAP Project dependency scores.
Differentially expressed genes in PDTOS4 versus PDTO cells in the presence of recombinant TGF-beta1
Endoscopy-guided orthotopic injection of colorectal cancer organoids into the colonic wall of immune-deficient mice.
Data Availability Statement
Gene expression data of COAD (colorectal adenocarcinoma) and READ (rectal adenocarcinoma) used for comparison of NLE1 gene expression between cancerous and normal tissues were obtained from GDC-TCGA datasets (23) and NCBI GEO (www.ncbi.nlm.nih.gov/geo).
Gene expression changes in CRISPR/Cas9-engineered SMAD4-negative PDTO derivatives (PDTOS4 lines) compared with parental PDTOs are detailed in Supplementary Tables S2–S4. Counting files and annotation of all technical and biological replicates are provided in Supplementary Table S2.
The next-generation sequencing raw data that support the findings of this study are available on request from the corresponding author (P. Jung). These data are not publicly available due to them containing information that could compromise research participant privacy or consent. Explicit consent to deposit raw-sequencing data was not obtained from patients. Because all samples had been irreversibly anonymized prior to study start, the patients cannot be asked to provide their consent for deposit of their comprehensive genetic or transcriptomic data.



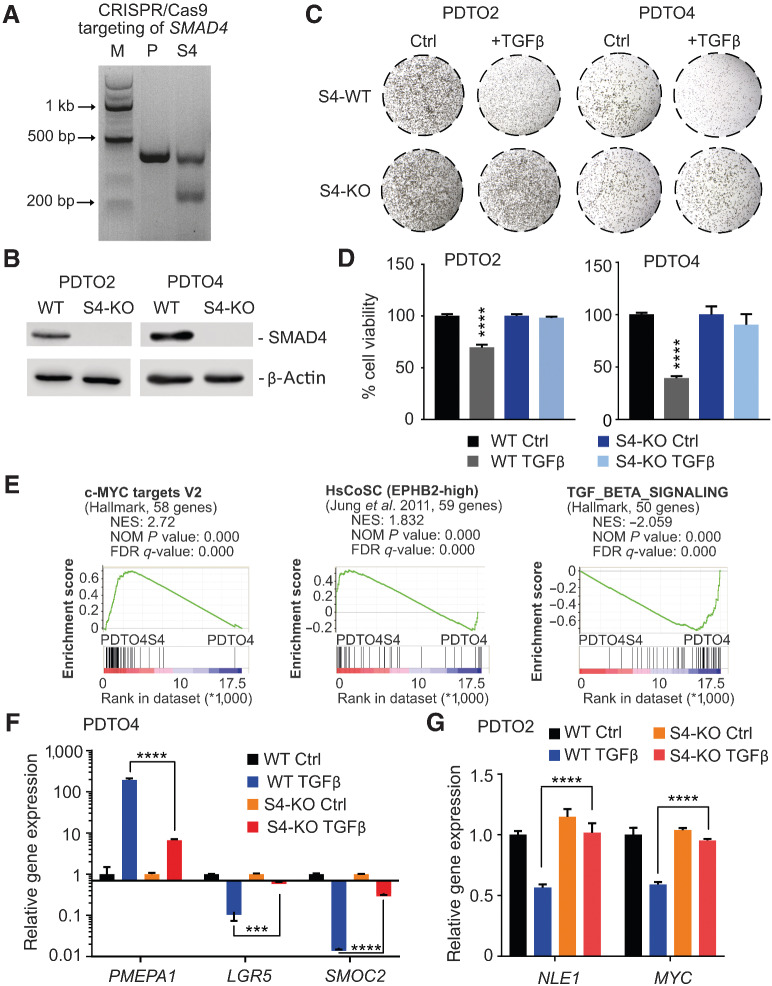
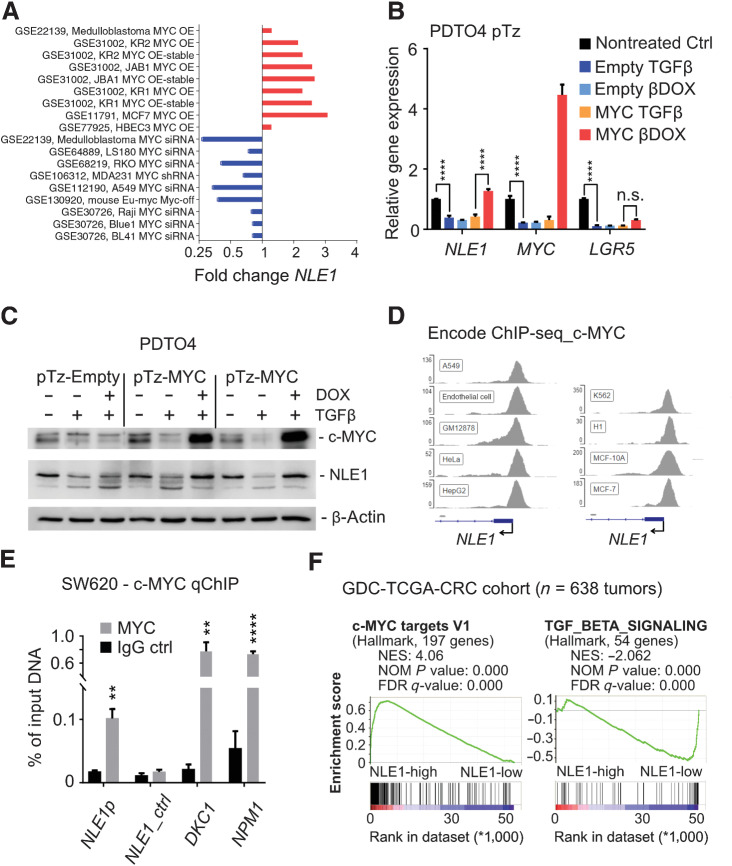
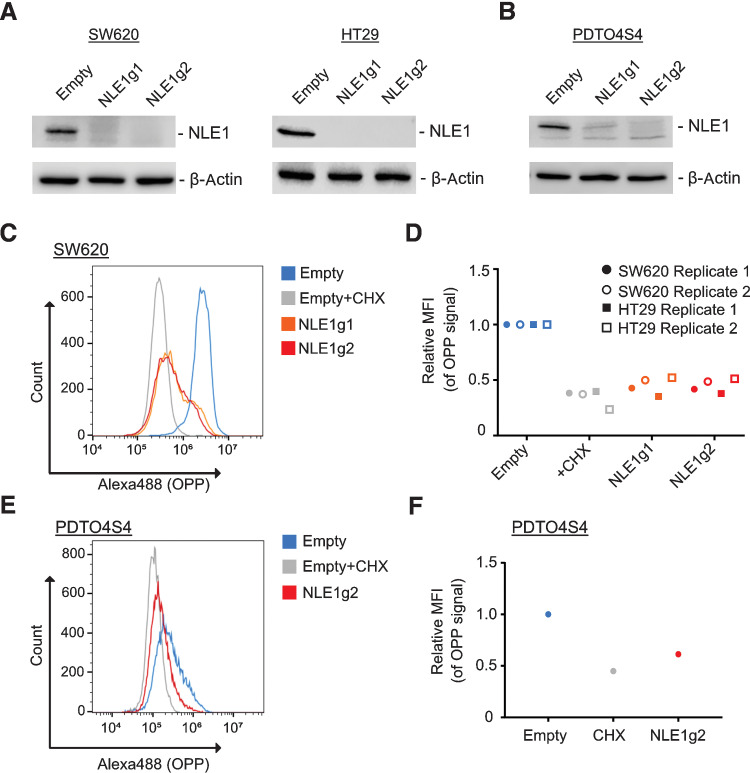
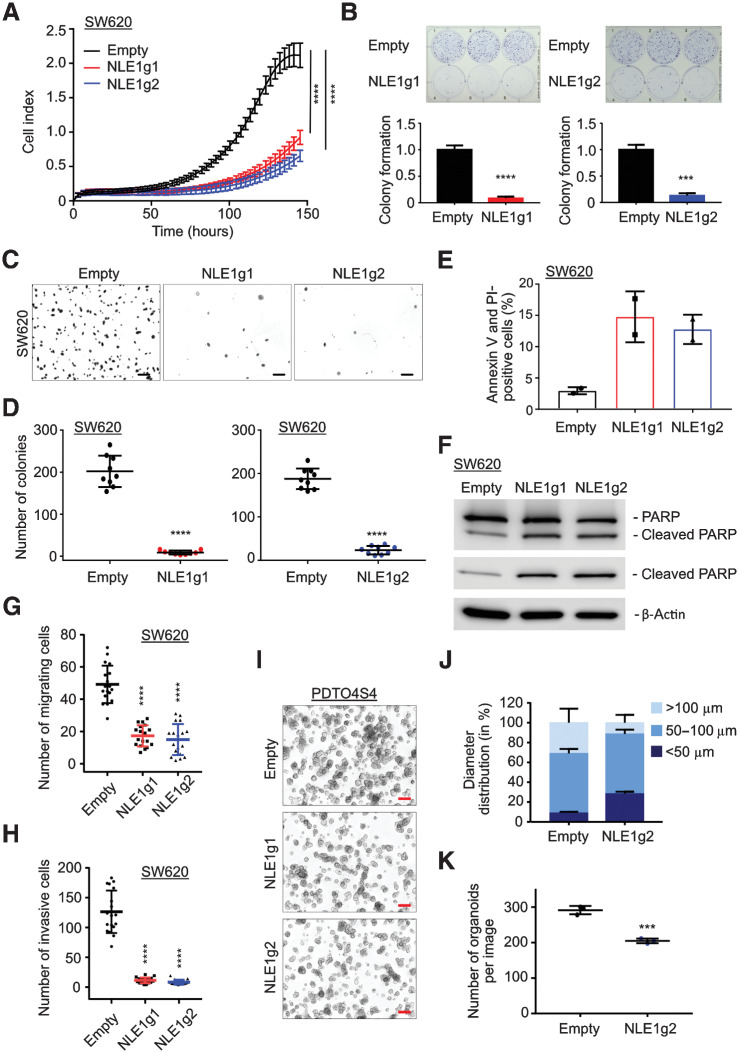
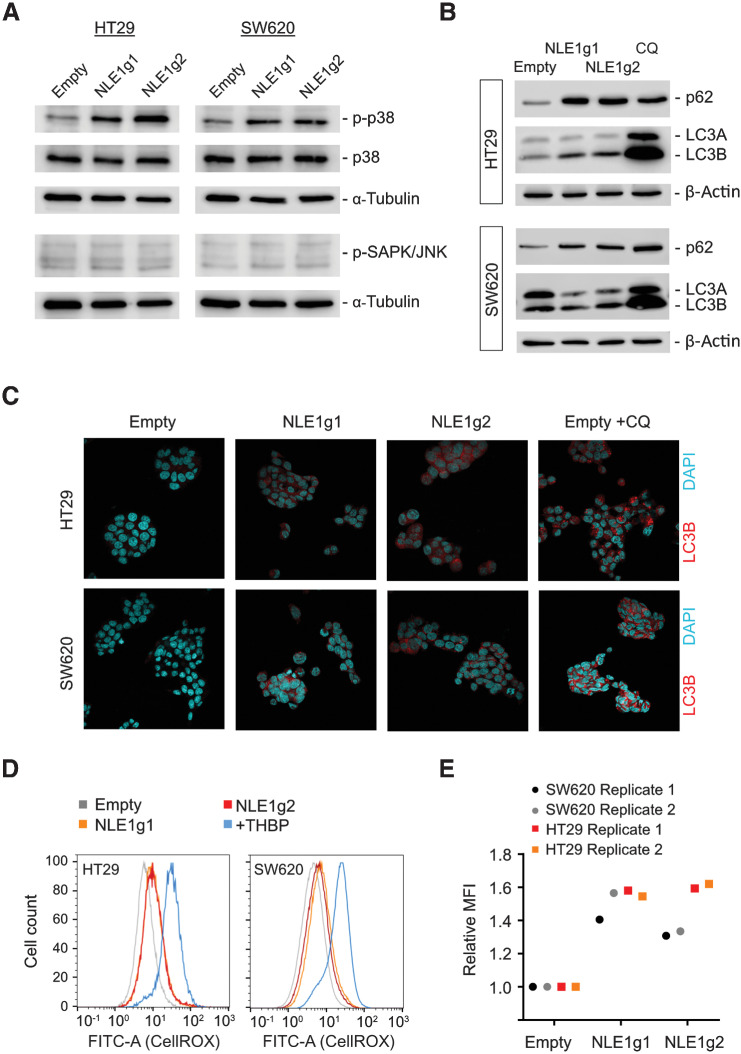
![Figure 6. NLE1 ablation reduces colorectal cancer primary tumor formation and liver metastasis in vivo. A, Schematic representation of endoscopy-guided PDTO orthotopic transplantation, colonoscopy follow-up and organ analysis of sacrificed immunodeficient (NSG) mice. At time point t = 0, the process of PDTO needle injection, as seen via the endoscope camera, is depicted while the injection bubble is about to form. Control endoscopy was performed at day 25 to control for primary tumor occurrence and size and to estimate the end point of the experiment when animals need to be sacrificed due to excessive tumor burden. Mice were sacrificed 35 days after orthotopic PDTO transplantation, colonic tumors were documented, and the liver was scrutinized for occurrence of macroscopically visible metastatic foci. B, Colonoscopy of immunodeficient mice was performed 3.5 weeks after orthotopic transplantation of NLE1 wild-type (WT) or NLE1 knockout (KO) colorectal cancer organoids into the colonic wall. Note the more pronounced protrusion of NLE1-WT tumors into the colonic lumen when compared to tumors grown from NLE1-KO PDTOs. C, Scatter plot showing the areas of primary tumors grown in the colon of xenotransplanted mice. Four mice (n = 6 primary tumors) had been transplanted with NLE1 wild-type (WT) colorectal cancer organoids and 6 mice (n = 12 primary tumors) had been transplanted with NLE1 knockout (KO, NLE1-targeting guide RNAs 1 (red dots) or 2 (blue dots) colorectal cancer organoids. All mice were sacrificed for analysis 5 weeks after xenotransplantation. Statistical significance between the NLE1-WT and NLE1-KO groups was assessed by an unpaired t test with Welch correction to account for the observed unequal SDs within the two experimental groups and is indicated by asterisks (*, P < 0.05). Shown is the mean ± SD. D, Macroscopic images of primary tumors formed in the colon of four mice orthotopically transplanted with 150 tumor organoids wild-type (WT) for NLE1 per injection site. All mice were sacrificed for analysis 5 weeks after xenotransplantation. Scale bars, 0.5 cm. E, Macroscopic images of primary tumors formed in the colon of six mice orthotopically transplanted with 150 tumor organoids knockout for NLE1 per injection site. All mice were sacrificed for analysis 5 weeks after xenotransplantation. Scale bars, 0.5 cm. F, Representative microscopy images of IHC staining of the proliferation marker MKI67 on FFPE tissue sections prepared from NLE1 wild-type (WT) and NLE1 knockout (KO) PDTO-derived primary tumors. Scale bars, 100 μm. G, Quantification of MKI67-positive tumor cells (in %) in FFPE tissue sections from NLE1 wild-type (WT, n = 4) and NLE1 knockout (KO, n = 6) primary tumors. An unpaired t test was performed to assess significance (***, P < 0.001). Shown is the mean ± SD. H, Representative microscopy images of IHC staining of the apoptosis marker cleaved caspase 3 on FFPE tissue sections prepared from NLE1 wild-type (WT) and NLE1 knockout (KO) primary tumors. Scale bars, 100 μm. I, Exemplary macroscopic images of resected livers and colorectal cancer-derived liver metastases formed in mice orthotopically transplanted with NLE1 wild-type (WT) or NLE1 knockout (KO) tumor organoids. Arrows indicate macroscopically visible liver metastasis. J, Scatter plots showing the quantification of macroscopic metastatic foci detected in the liver of mice five weeks after endoscope-guided, orthotopic transplantation of colorectal cancer organoids either wild-type (WT) or knockout [KO, NLE1-targeting guide RNAs 1 (red squares) or 2 (black squared)] for NLE1. Statistical significance between the NLE1-WT and NLE1-KO groups was assessed by an unpaired t test with Welch correction to account for the observed unequal SDs within the two experimental groups and is indicated by asterisks (**, P < 0.01). Shown is the mean ± SD.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d0ca/9755967/d0f4a025fb18/4604fig6.jpg)