SOX9 facilitates therapy escape and recurrence in medulloblastoma via temporal inhibition of MYC/MYCN genes, revealing a strategy to specifically target SOX9-positive cells to prevent tumor relapse.
Abstract
Relapse is the leading cause of death in patients with medulloblastoma, the most common malignant pediatric brain tumor. A better understanding of the mechanisms underlying recurrence could lead to more effective therapies for targeting tumor relapses. Here, we observed that SOX9, a transcription factor and stem cell/glial fate marker, is limited to rare, quiescent cells in high-risk medulloblastoma with MYC amplification. In paired primary-recurrent patient samples, SOX9-positive cells accumulated in medulloblastoma relapses. SOX9 expression anti-correlated with MYC expression in murine and human medulloblastoma cells. However, SOX9-positive cells were plastic and could give rise to a MYC high state. To follow relapse at the single-cell level, an inducible dual Tet model of medulloblastoma was developed, in which MYC expression was redirected in vivo from treatment-sensitive bulk cells to dormant SOX9-positive cells using doxycycline treatment. SOX9 was essential for relapse initiation and depended on suppression of MYC activity to promote therapy resistance, epithelial–mesenchymal transition, and immune escape. p53 and DNA repair pathways were downregulated in recurrent tumors, whereas MGMT was upregulated. Recurrent tumor cells were found to be sensitive to treatment with an MGMT inhibitor and doxorubicin. These findings suggest that recurrence-specific targeting coupled with DNA repair inhibition comprises a potential therapeutic strategy in patients affected by medulloblastoma relapse.
Significance:
SOX9 facilitates therapy escape and recurrence in medulloblastoma via temporal inhibition of MYC/MYCN genes, revealing a strategy to specifically target SOX9-positive cells to prevent tumor relapse.
Graphical Abstract
Introduction
Standard treatment of medulloblastoma (MB) includes surgery, craniospinal irradiation, and chemotherapy, and is effective in up to 70% to 75% of the patients. Despite this, the harsh treatment often leads to long-term side effects or radiation-induced secondary tumors. An unfavorable outcome is almost always preceded by disease progression and relapse. Median survival for MB recurrence is less than 2 years (1). It is generally believed that recurrence progresses from a small number of tumor cells that escape surgical resection and radiotherapy and become resistant to chemotherapy (2). MB is divided into four molecular subgroups (WNT, SHH, Group 3, and Group 4) according to their gene expression signature (3). Group 3 and Group 4 MBs are biologically related groups of tumors that lack profound WNT or SHH pathway signatures. They can further be divided into distinct subtypes comprising almost two-thirds of patients where a certain Group 3 subtype has the poorest survival (4). MB relapses usually match the molecular subgroup of the primary tumor, but a considerable proportion of Group 3 and Group 4 recurrences include distant metastasis compared with SHH recurrences that emerge close to the location of the initial tumor (1, 5, 6). The data suggest that the cells giving rise to the recurrences in SHH tumors are fundamentally different from recurrences in Group 3 and Group 4 tumors.
MYC and MYCN oncogenes are frequently amplified in Group 3 and Group 4 MB and correlate with poor prognosis. In recurrent MB, MYC gene amplifications commonly occur together with TP53 mutations or loss (7). SOX9 is an HMG-box transcription factor in the SOX family of proteins and has an important role in neural stem cell (NSC) as well as glial development of the CNS (8, 9). SOX9 was recently described as a reliable marker, together with SOX2, for latent cancer cells involved in cancer metastasis (10). SOX9 has further been implicated in tumor migration and drug resistance (11, 12). We previously showed that the ubiquitin ligase FBXW7 regulates SOX9 levels posttranslationally and when FBXW7 is mutated or depleted, SOX9 promotes increased metastasis and generates more treatment-resistant MB (12, 13). An increased understanding of the process of tumor recurrence and the molecular characterization of migrating cells evading therapy is needed to develop better therapies targeting recurring tumors. Here we show how rare populations of SOX9-positive cells accumulate in a unique set of primary-recurrent patient samples. We then generated a novel mouse model in which we can follow the accumulation of these dormant cancer cells during tumor relapse at the single-cell level. We finally provide means of specific targeting of these therapy-resistant SOX9-positive cell types.
Materials and Methods
Experimental models and subject details
Patient samples
Paraffin-embedded tissue sections from paired primary and recurrent human MB samples were obtained from Dr. Ulrich Schüller and the tissue archive of the Center for Neuropathology, Hospital of University of Munich, in accordance with the Declaration of Helsinki and after patients had given their written informed consent. Approval was obtained from the institutional review boards of the hospital. Data on the DNA methylation levels of two CpGs (cg12434587 and cg12981137) in the MGMT promoter was derived from 450K methylation array (Illumina) analysis of Genomic DNA from 11 Group 3 and 19 Group 4 matched primary-recurrent MB pairs from patients treated at UK CCLG institutions and collaborating centers and generated as described previously (7). In addition, paraffin-embedded tissue sections from paired primary and recurrent human MB samples were obtained from Dr. Steven Clifford and Dr. Rebecca Hill, Newcastle University, Newcastle upon Tyne, United Kingdom. Written informed consent was obtained from all subjects and human tumor investigations were conducted in accordance with the Declaration of Helsinki and with approval from Newcastle/North Tyneside Research Ethics Committee. We also used transcriptome and DNA copy-number data for SOX9, MYC, and MYCN genes in 64 primary human MB specimens that were obtained and analyzed as described previously (14). We used transcriptome data from 763 primary human tumors. Their expression profiling with the classification of the four different subgroups has been described previously (15).
Animal models
Several different transgenic mouse lines were used in this study. The Glt1-tTA; TRE-MYCN:Luc (GTML) model and the bioluminescence imaging using luciferase has been described previously (13, 16). Mice carrying Sox9-rtTA transgene were kindly provided by Dr. Michael German (University of California San Francisco, UCSF, San Francisco, CA) and crossed with TRE-MYCN:Luc to generate the STML mice in an FVB/N background. Tg(Sox9-EGFP)EB209Gsat (pSox9-GFP) animals were obtained from Mutant Mouse Resource & Research Centers (MMRRC) facility at University of California Davis, Davis, CA. Glt1-tTA was crossed with pSox9-GFP to generate the GTML; pSOX9-GFP model and Glt1-tTA was crossed with Sox9-rtTA strains in the FVB/N background and were both kept as double heterozygotes. Athymic Nude mice were obtained from Envigo and NOD SCID gamma (NSG) mice from Jackson Laboratory. Animals were euthanized upon the development of tumor symptoms (hunched back, paralysis, reduction of fur quality, weight loss, and tilted head) or 1 year posttransplantation. Sections from tumors from Ptch+/−/Math1-SB11/T2Onc animals were obtained from Dr. Michael Taylor (University of Toronto, Toronto, Ontario, Canada) and generated as described previously (17). All experiments were performed in accordance with national guidelines and regulations and were approved by the Institutional Animal Care and Use Committee at UCSF or the Uppsala Animal Experiment Ethics Board at Uppsala University.
Orthotopic transplantation
Injections of tumor cells into mouse brains were performed as thoroughly described elsewhere (13, 18) and as follows. For allograft and in vivo transplantation studies, adult nude mice were anesthetized, and cerebellar allografts were generated by injection of 100,000 cells into the cerebellum. Mice were monitored until the phenotypic presentation of a brain tumor, at which point, they were euthanized. At sacrifice, whole brains were excised, and biopsies were taken for experimental procedures. PDX models used included BT084 (metastatic SHH MB established in Dr. Till Milde's lab), Med-211FH, Med-411FH (metastatic Gr. 3 MBs established in Dr. Jim Olson's lab), and ICb-1572MB (metastatic Gr. 3 MB established in Dr. Xiaonan Li's lab) all obtained from Dr. Robert Wechsler-Reya's lab and used as described elsewhere (18). Orthotopic PDX growth was studied in the brain and spinal cord in NSG mice that were maintained in animal facilities at Sanford Burnham Prebys Medical Discovery Institute (SBP) and the Sanford Consortium for Regenerative Medicine. All experiments were performed in accordance with national guidelines and regulations and were approved by the Institutional Animal Care and Use Committee at SBP and the University of California San Diego or the Uppsala Animal Experiment Ethics Board at Uppsala University.
Doxycycline treatments
Doxycycline (dox) was delivered ad libitum to the mice in vivo via a dox rich (625 mg/kg) diet (Harlan Laboratories/Envigo).
Cell lines
Tumor cells (GTML, GTS, and pre-GTS) were isolated as previously described from tumor-bearing mice (13) and cultured as neurospheres in serum-free conditions with neurobasal (NB) media, additional B27 supplement, 100 units penicillin, and 0.1 mg/mL streptomycin, 2 mmol/L l-glutamine, 1:500 antibiotic–antimycotic, 20 ng/mL EGF, and 20 ng/mL FGF2. Human MB002 primary cell cultures were obtained from Dr. Yoon Jae-Cho and cultured in serum-free conditions, 1:1 of neurobasal media (-vitamin A) and DMEM/F12 with sodium pyruvate, NEAA, HEPES, GlutaMAX, 100 units penicillin and 0.1 mg/mL streptomycin, B27 supplement, 20 ng/mL EGF and 20 ng/mL FGF2, 1× LIF and 1× Heparin (as previously described ref. 19). Stable doxycycline-inducible SOX9wt and SOX9CPD mutant cell lines were generated as described previously (12). Lentiviruses were first packed in HEK293T cells by the specified pTRIPZ lentivector with the packaging plasmids, psPAX2, and pMD2.G. Supernatants containing viral particles (48–96 hours posttransfection) were used to infect the target cells in the presence of freshly added hexadimethrine bromide (8 mg/mL). Seventy-two hours posttransfection, the cells were selected and maintained in an appropriate culture medium containing puromycin (0.125–0.5 μg/mL). CHLA-01-MED and CHLA-01R-MED were obtained from ATCC and cultured in l-glutamine and HEPES supplemented DMEM/F12 (Thermo Fisher Scientific, 11330057) with additional 100 units penicillin and 0.1 mg/mL streptomycin, B27 supplement, with or without growth factors; 20 ng/mL EGF and 20 ng/mL bFGF. Human cell lines were authenticated at ATCC and were used after a low number of passages in culture after being obtained. All cell lines including primary cells from mice used were used in low (maximum 15–20) passages and routinely tested for Mycoplasma (using MycoAlert Mycoplasma Detection Assays from Lonza) before use in experimental settings.
Method details
All reagents and resources used in this paper are specified in Supplementary Table S5.
Minimal residual disease
Adult nude mice were injected with 200,000 GTML2 or GTML3 cells into the cerebellum and followed for tumor development with IVIS. Three weeks posttransplantation (at tumor presentation) mice were grouped; untreated (sacrificed due to tumor burden) and dox treated. Dox was administered ad libitum for 7 days, after which, they were followed for an additional 10 days (minimal residual disease) or followed until tumor relapse. Mice were sacrificed and brains were harvested at the above-indicated time points.
Flow cytometry and cell sorting
Tumor-burdened GTML;Sox9-eGFP mice were sacrificed and tumors dissociated, cells cultured as we described above. Cells were harvested, accutased, and filtered with a 30 μm cell strainer, then sorted for GFP-positive (SOX9) cells (488_FITC-A) and GFP-negative cell population. For the cisplatin treatment experiment, cells were treated with cisplatin for 24 hours before cell sorting. The BD FACSMelody was used for sorting and quantifying in this experiment. MB002-pSOX9-GFP cells were dissociated using accutase and sorted for GFP expression (488_FITC-A). The BD FACSariaIII was used for this experiment and was analyzed using BD FACSDiVa version 8.0.
IHC
Brains were fixed in 4% PFA and embedded in a paraffin block. pre-GTS1 sphere cultures were fixed in 4% PFA for a minimum of 12 hours at 4°C and then embedded in HistoGel, followed by paraffin embedding. 6 to 7 μm sections of paraffin-embedded tissue were used for all staining. The complete protocol for the IHC was described previously (16).
Immunofluorescence
Mouse brains were isolated, dehydrated, and embedded in paraffin. Six micrometers brain sections were deparaffinized, hydrated, and antigen retrieved. Sections were blocked in 10% donkey serum with 1% BSA for 1 hour and then incubated with primary antibodies overnight at 4°C, followed by washing and secondary antibody for 1 hour protected from light. After washing nuclei were visualized with DAPI (5 mg/mL) for 5 minutes. Finally, sections were washed and mounted with Fluoromount. Images were taken using Leica DMi8 fluorescent microscope.
Lentiviral constructs
pSOX9-Luc/GFP was generated by replacing the CMV promoter of pBMN(CMV-copGFP-Luc2-Puro) with the human SOX9 promoter.
RNAscope
RNAscope Technology is a cutting-edge ISH technology based on Advanced Cell Diagnostics (ACD) unique, patented probe design strategy that enables simultaneous signal amplification and background noise suppression (www.acdbio.com). RNA scope was performed according to the manufacturer's instructions (ACD). We used specific probes detecting mouse Sox9 (ACD-401051-C2) and human MYCN (ACD-417501). The detection was performed on 6 μm sections of paraffin-embedded tissues. Sections were deparaffinized and processed according to the manufacturer's instructions. For our staining of MYCN and Sox9, we used RNAscope-Fluorescent Multiplex Detection Reagents (ACD-320821), Pretreatment Kit-FLFF (ACD-320842), RNA Scope Wash Buffer (ACD-310091), and 10× Pretreat (ACD-320043). We used recommended HybEZ oven to incubate the samples in the humidity control tray at 40°C.
Western blot analysis
Protein (20 μg) was loaded in 4% to 12% Bis-Tris gels and transferred to an iBlot nitrocellulose membrane. ECL secondary antibodies (1:5,000) were detected using Supersignal West Pico Chemiluminescent Substrate.
Doxycycline treatments
For in vitro experiments, doxycycline hyclate (Sigma) was dissolved in PBS and added to culturing medium (1 μg/mL).
Doxycycline induction or SOX9 in MB002-SOX9CPDmt
One μg/mL of doxycycline was added to MB002-SOX9CPDmt in culture media. For stable SOX9 overexpression, new DOX was added every 48 hours. For recovery experiments, cells were cultured in dox-conditioned media for 24 hours, after which, they were spun down and resuspended in fresh media for continued cell culturing in normal conditions and harvested 8, 24, 48, and 72 hours after dox removal. The cell pellet was lysed and analyzed for protein content using Western blotting.
Drug treatment and proliferation assay
To assess cell proliferation, 10,000 cells were plated in triplicates in a 96-well plate. Proliferation was measured every 24 hours for 5 days using Resazurin colorimetric assay. The MGMT inhibitor lomeguatrib was dissolved in DMSO and added to cells 24 hours post-cell plating to a final concentration of 20 μmol/L. Thiotepa was dissolved in DMSO and added similarly to a final concentration of 10 to 100 μmol/L. Doxorubicin was dissolved in H2O and added to a final concentration of 200 nmol/L. Spheres from pre-GTS1 cells were treated with cisplatin at a final concentration of 10 μmol/L. The number of SOX9-positive cells per sphere was counted in 10 spheres in treated (cisplatin) and untreated (DMSO) conditions. Statistical significance was calculated using an unpaired nonparametric Mann–Whitney test.
Long-term proliferation with doxycycline treatment
GTML and pre-GTS cells were seeded in triplicates of 10,000 cells per well in polyornithine and laminin-coated 96-well plates. Twenty-four hours post-seeding (0 hour), dox was added to cells (final concentration 1 μg/mL). Seventy-two hours after the first dose of dox, a second dose was added to fresh media. DMSO was used as a control. Fresh media was given to cells on days 7 and 10 of the experiment. Every 48 hours proliferation was measured using a resazurin-based assay.
CRISPR-CAS9 targeting Sox9
CRISPR-Cas9 gene editing of Sox9 was performed using Integrated DNA Technologies (IDT) Alt-R CRISPR-Cas9 system and protocol (Alt-R CRISPR-Cas9 System: Delivery of ribonucleoprotein complexes into Jurkat T cells using the Neon Transfection System). Three individual crRNAs targeting Sox9 were selected (GUUCACCGAUGUCCACGUCGGUUUUAGAGCUAUGCU, ACCAUGUCGGAGGACUCGGCGUUUUAGAGCUAUGCU, GAAGGGCUACGACUGGACGCGUUUUAGAGCUAUGCU). These were combined with ITDs tracrRNA ATTO 550. pre-GTS1 cells (5 × 105) were electroporated using the Neon Transfection System 10 μL Kit (setting 7; pulse voltage: 1,200, pulse width: 30, number of pulses: 1), with a combination of the three Sox9 targeting crRNAs, all at a concentration of 1.8 μmol/L, to excise a segment of the Sox9 gene, seeded as single cells in 96-well plates and visually inspected for fluorescent labeling. Single positive cells were expanded to form monoclonal cell lines. pre-GTS1 cells were electroporated resuspended in Buffer R only and then expanded from single cells to create control cell lines. Sox9 gene editing was validated using PCR (forward primer CCGGCCCCAGGAGAACACCTT, reverse primer GTGAGTGCAGCCCGCGTCCC) and protein levels were investigated using Western blot analysis (ab185966). Cell line identity was confirmed using PCR targeting the Sox9-rtTA construct specific to the GTS mouse strain.
RT-qPCR
RNA was isolated using TRizol and RNeasy Mini Kit. RNA concentration was measured with Qubit RNA BR Assay Kit. cDNA synthesis was done using SuperScript VILO cDNA Synthesis Kit. qPCR experiments were run in triplicates with 2.5 ng RNA using SYBR Green PCR Master Mix or PrimePCR (Bio-Rad Laboratories) SYBR Green Assay on StepOnePlus Real-Time PCR System or CFX96 Touch Real-Time PCR Detection System. Primer sequences for mouse GTML cells for Sox9, MYCN, and Gapdh have been used and analyzed as described previously (13). Primer sequences are found in Table 1.
Table 1.
Sequences of primers used for qPCR.
| Primer name | Primer sequence (5′ to 3′) |
|---|---|
| SOX9 (human) | TCAACGGCTCCAGCAAGAACAAG ACTTGTAATCCGGGTGGTCCTTCT |
| PrimePCR primers | Bio-Rad |
| cMYC (human) | Seq N/A cat#qHsaCID0012921 |
| GAPDH (human) | Seq N/A cat#qHsaCED0038674 |
Cytokine array
Cell lysates (200 μg) and supernatants (2 × 106 cells 48 hours in culture) were analyzed using Proteome Profiler Mouse XL Cytokine Array and instructions were followed accordingly. ECL secondary antibodies (1:5,000; GE Healthcare) were detected using SuperSignal West Femto Maximum Sensitivity Substrate. Results were analyzed with ImageJ software and negative controls were used as background. The experiment was performed twice, and the figures show representative images.
Chase assay
MB002-SOX9CPDmt and EV cells were treated with doxycycline (1 μg/mL) or DMSO for 4 hours, after which, cycloheximide (100 μg/mL) was added. Cells were harvested at the indicated time points and analyzed with Western blotting for cMYC.
Doxycycline sensitivity prediction
The 2,000 genes with the most significantly higher expression in DMSO-treated compared with doxycycline-treated GTML3 cells were considered to represent a doxycycline sensitivity score. Single-sample gene set enrichment analyses (ssGSEA) was performed with this signature on scRNA data from GTML-pSOX9-GFP cells. The ssGSEA enrichment score was then correlated to SOX9 expression in the GTML-pSOX9-GFP cell line.
Pre-GTS orthotopic allografts
Adult nude mice were injected with 200,000 pre-GTS1 or pre-GTS2 cells into the cerebellum and followed for tumor development. At tumor presentation mice were grouped; untreated (sacrificed due to tumor burden) and dox treated. Dox was administered ad libitum for 30 days, after which, they were followed until tumor relapse. Mice were sacrificed and brains were extracted at the above-indicated time points.
Whole cerebellum irradiation
Athymic Nude-Foxn1 nude mice were intracranially implanted with 1.0 × 105 GTML2 or GTML3 cells. Mice-bearing tumors were randomized into treatment groups 20 days after implantation. Image-guided whole-cerebellum irradiation was delivered with the SARRP microirradiator (XStrahl, Inc.). In brief, a 10 × 10 mm beam was delivered at 2 Gy/day, 5 days on and 2 days off, for a total of 36 Gy for both cell lines.
mRNA decay assay
MB002 CPDmt cells were treated with 1 μg/mL of dox or DMSO. After 24 hours, translation was stopped with 10 μg/mL Actinomycin D and RNA was collected after 30 minutes, 1 and 2 hours. MYC RNA levels were analyzed with qPCR using PrimePCR primers for hcMYC with HGAPDH as a control. Primer sequences are shown in Table 1.
Single-cell RNA-seq experiments
For single-cell library preparation on the 10× Genomics platform, we used the Chromium Single Cell 3′ Library & Gel Bead Kit v3 (PN-1000128), Chromium Single Cell 3′ Chip Kit v2 (PN-1000127), and Single Index Kit T Set A (PN-1000213), according to the manufacturer's instructions in the Chromium Single Cell 3′ Reagents Kits V3.1 User Guide. Just before cell capture, GTML;Sox9-eGFP, and GTS2 cell lines were harvested, accutased, and filtered with a 30 μm cell strainer, then spun at 300 g for 5 minutes followed by resuspension in PBS with 0.04%BSA on ice. A total of 17,000 cells were loaded per lane on the chip, aiming to capture 10,000 single-cell transcriptomes. All samples were processed in parallel, on the same day. The resulting cDNA libraries were quantified on an Agilent TapeStation and sequenced on an Illumina NovoSeq 6000.
Single-cell RNA-seq data analysis
The Cell Ranger v3.3.0 pipeline was used to process data generated using the 10× chromium platform. This aligns sequencing reads to the mouse reference genome mm10 v3.0.0. Following alignment, gene expression matrices were generated. Using the Seurat v3.0 package in R, we first removed cells with a low number (<200) of unique detected genes and genes (<3) with a low number of cells, then we removed the cells with unique detected genes less than 500 or great than 7,000, finally, we removed cells in which the proportion of the UMI count attributable to mitochondrial genes was greater than 20%. Umap of SOX9/KI67 and cell-cycle analysis in GTML; SOX9-GFP and GTS cells were generated using the Seurat package.
We revisited the scRNA data using the following methods to generate Fig. 4E and Supplementary Figs. S4M and S4N. For analysis of human medulloblastoma samples, previously published single-cell RNA sequencing data were acquired from the Gene Expression Omnibus (GEO), accession number GSE119926. Log-transformed TPM data were analyzed using the seurat R package (v4.1.0). Cell-cycle scoring was performed using the seurat built-in gene sets for G2–M and S phases. Variable features (n = 2,000) were identified in each sample individually and sample integration was performed. When identifying integration anchors, the two Gr3/Gr4 samples with the highest number of cells were used as reference. When integrating samples, the k.weight parameter was set equal to the cell number of the sample containing the fewest cells. After integration, data were scaled with regression for cell-cycle scores. Principal component analysis (PCA) was performed and in subsequent clustering and UMAP analyses, 30 PCAs were used.
For analysis of GTS and GTML cell line samples, raw, fastq files were aligned to a custom reference containing all mm10 transcripts as well as human MYC. Building the reference and alignment and gene expression quantification was performed using cellranger v6.1.2 (10x Genomics). For downstream analyses, filtered .h5 files were imported into Seurat (v4.1.0). Only cells with >500 UMIs and >300 genes detected were imported. For QC, the percentage of mitochondrial gene expression was calculated for each cell (%mito). Cells with %mito more than 2 SDs above the median were discarded. Cells with (log nFeature) less than 2 SDs below the median were discarded. In the next step, the DoubletFinder v2.0.3 package was used to identify putative doublets, which were filtered out. Clusters with only mitochondrial or no informative markers identified were also filtered out.
In the downstream analyses, cell-cycle scoring was performed on normalized data using the mouse orthologs of the human genes included in the seurat package. Next, SCTransform was performed while regressing for cell-cycle scores. For clustering and UMAP analyses, 30 PCAs were used and clustering was performed using a resolution level of 0.8.
For gene set enrichment analyses (GSEA) of markers identified in single-cell clusters from GTS/GTML cell lines as well as publicly available human MB data, genes with adjusted P values < 0.05 were ranked in order of increasing P values and analyzed using the online tool gProfiler (ordered query) and the following gene sets: gene ontology (GO) GO:MF, GO:CC, and GO:BP (BioMart release 2021–12–15), Kyoto Encyclopedia of Genes and Genomes (KEGG; release 2021–12–27), Reactome (BioMart release 2022–01–03), Wikipathways (release 2021–12–10), and the following MSigDB gene sets: MH (mouse-human ortholog) hallmark gene set v0.3, M2.CGP gene set v0.3, c2.CGP v7.4, and h.all.v7.4 (Hallmarks gene set).
Assay for transposase-accessible chromatin using sequencing experiments
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) with in-house Tn5 of GFP+ cells and GFP− cells from GTML;Sox9-eGFP and GTS cells was performed as described (20). Briefly, 50,000 cells were centrifuged 500 × g for 5 minutes at room temperature. The cell pellet was resuspended in 50 μL lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.01% Igepal CA-630) and centrifuged immediately at 500 × g for 10 minutes at 4°C. The cell pellet was resuspended in 50 μL transposase mixture [25 μL 2× TD buffer (20 mmol/L Tris, 10 mmol/L MgCl2, 20% dimethylformamide, 22.5 μL dH2O and 100 nmol/L in-houseTn5)] and incubated at 37°C 30 minutes. After transposition, the mixture was purified with the Qiagen Mini-Purification Kit and eluted in10 μL Qiagen EB elution buffer. Sequencing libraries were prepared following the original ATAC-seq protocol. The sequencing was performed on Illumina NovaSeq at Novogene Europe.
ATAC-seq data analysis
ATAC-seq paired-end reads were trimmed for Illumina adapter sequences and transposase sequences using an in-house script and mapped to hg19 using Bowtie2 (21) v2.1.0 with parameters—very sensitive. Over ∼11 million mapped reads were generated in each sequencing library and used for downstream data mining. Duplicate reads were removed with Picard (http://picard.sourceforge.net) v1.79. Peak calling was performed by MACS2 (22) narrow peak mode with parameters: q 0.01 –nomodel –shift 0. Overlapping peaks from all samples were merged into a consensus peak list. The number of unique and properly paired reads mapped to each peak for each sample was quantified to calculate the Pearson Correlation. The insert size of fragments was estimated from the distance between the pair-ended reads and plotted against the frequency in a histogram. To identify genes with regions of differentially opened or closed chromatin, respectively, we performed differential analyses (via the edgeR package) of ATAC-seq peaks between conditions. Subsequently, peaks were mapped to the closest gene within 20 kB and up to 2,000 genes with the closest associated peaks were selected for downstream gene-set overlap analyses.
DNA methylation profiling
DNA methylation profiling was conducted on the Illumina MethylationEPIC platform and was performed by the Uppsala SNP&SEQ Technology Platform in Uppsala (www.genotyping.se). Preprocessing of raw IDAT files was conducted in R using the MethPed (1.6.0), IlluminaHumanMethylationEPICanno.ilm10b2.hg19 (0.6.0), and IlluminaHumanMethylationEPICmanifest (0.3.0) packages. Probes were removed, if any sample had an associated P-value P > 0.01. Average β values were extracted after normalization on red-green channels using the preprocessNoob scheme. Average β after normalization with the preprocessNoob function from the minfi (version 1.24.0).
Sequencing data preparation
Total RNA was sequenced using the IonProton System in the Uppsala Genome Center, SciLifeLab, Uppsala University. For the dox-treated GTML and pre-GTS cells, raw sequencing reads were mapped to the mm9 genome assembly according to facility standards, and the according gene-specific read counts were used directly. For GTS and GTML tumors, we mapped raw sequencing reads to the mm9 mouse genome using a two-round approach of the STAR alignment algorithm, followed by a subsequent alignment of unmapped reads using BOWTIE2 in local mode. Gene-specific read counts were then obtained via the featureCounts function from the Subread package.
Differential expression analysis
Differential gene expression analyses were conducted in R using the edgeR package, including a normalization for differences between raw library sizes via the calcNormFactors function in conjunction with the TMM method. GTS and GTML tumor samples were processed in two mixed batches and displayed batch-related biases. To account for those batch effects, differential expression analyses between GTS and GTML tumors were conducted by including the batch as a factor in the edgeR design matrix. Genes were considered significantly differentially expressed if their FDR corrected P value q < 0.05.
Removal of batch effects.
Although differential expression analyses were conducted by including batches as a factor in the design matrix, various types of data visualization required the removal of batch effects among the GTML and GTS datasets before figure plotting. For this purpose, we applied the removeBatchEffect function from the R package limma.
Mapping of orthologs and translation of human gene symbols
All translation between different identifiers within the same species or between ortholog genes in mice and humans were conducted via the biomaRt package in R. Specifically, where necessary, Ensembl gene ids obtained after mapping to the Gencode mm9 annotation track were mapped to official mouse gene symbols. Mouse gene symbols (GTML and pre-GTS cells) and Ensembl gene ids (GTML and GTS tumors) were further mapped to their human orthologs and translated to HGNC symbols to facilitate cross-species analyses and GSEA.
GSEAs
All GSEAs were performed in the GenePattern (https://genepattern.broadinstitute.org/) platform and using the GSEA preranked method. Specifically, for every comparison, genes were scored as S = −sign(logFC) × log10(P-value), where the logFC and P-value were obtained from the respective edgeR differential expression analysis, and the score was used as the gene ranks. Subsequently, the ranked gene lists were analyzed using the GSEAPreranked framework on seven different gene set databases (H, hallmark gene sets; CGP, chemical and genetic perturbations; CP:KEGG, KEGG gene sets; CP:REACTOME, reactome gene sets; TFT, transcription factor targets; BP, GO biological process; C6, oncogenic signatures) and using the classical scoring scheme. Gene sets were considered significantly enriched if their associated FDR corrected P value q < 0.05.
MB subtype classification
The possible MB subtype affiliation of the mouse samples was estimated using the Metagene code for cross-platform, cross-species projection of transcription profiles (23). As an MB reference set, we merged three gene expression data sets, that is, 763 samples from (15), 123 from CBTTC (24), and 170 from (25). The merged data were batch-normalized using ComBat (26) and samples that could not be robustly classified into their annotated subgroup were discarded, resulting in a total of 997 MB samples. Using the metagene software, GTML and GTS expression profiles were then projected onto the MB-specific metagene space, subsequently classified via a support vector machine, and processed via a principal component analysis. To confirm the resulting classification, we simultaneously also projected a set of 20 MB PDX samples (18) onto the same MB data set and processed them alongside the GTML and GTS samples.
Differential expression and survival analysis in MB samples
To compare the expression of SOX9 and SOX2 between primary and metastatic MB tissues, we used expression data of 22 samples from paired primary and metastatic compartments comprising 6 patients classified as Group 4, 2 patients classified as Group 3, and 1 patient classified as SHH (27). The processed expression of both the SOX9 and SOX2 genes was downloaded from the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). For patients with multiple metastatic compartments linked to the same primary tumor, the mean value over all metastatic compartments was used to represent the corresponding metastatic expression. The significance of differences between primary and metastatic expression means was calculated using the paired Student t test. We also used the Delattre set in R2 (https://r2.amc.nl) containing 57 samples analyzed using the Human Genome U133 Plus 2.0 Array (u133p2) arrays but excluded typical nodular/desmoplastic SHH tumors, as well as apparent β-catenin, mutated WNT tumors from the analysis when comparing primary with posttreated samples. When calculating expression differences between different groups they were considered significantly different when P < 0.05, as calculated using the one-way analysis of variance (ANOVA) test included in the R2 online software. For survival analyses in Kaplan–Meier plots, we used a 10-year/120-month time point as a cut-off for survival due to the potential risk to include secondary tumors and further excluded nodular/desmoplastic MB samples from the Group 3 and Group 4 analysis from the Cavalli set to not risk including tumors from typically desmoplastic SHH MBs (15). The P value for survival curves is calculated using a log-rank test of reported patient survival data as described in R2 (https://r2.amc.nl). Only a priori fixed cut-offs (median, average, first or last quartiles) were used to calculate P values in the Kaplan–Meyer survival analysis.
Data availability
Raw and processed sequencing data have been deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) and are available via the accession number GSE162080.
Quantification and statistical analysis
Animal survival statistics
All animal experiments were conducted once. Animal survival was graphically shown as a Kaplan–Meier curve, made and assessed using GraphPad Prism 7 software. The P value for survival curves is calculated using a log-rank test.
IHC and immunofluorescence quantification
All micrographs shown are representative images of the respective mouse strain tumors. Where appropriate for statistical analysis and IHC quantification, at least three representative micrographs from at least three individual tumors were analyzed in Fiji ImageJ and QuPath bioimage analysis softwares. Staining positivity was expressed as a % of the total cell population.
Quantification of metastatic dissemination
Quantification of metastatic dissemination was assessed via histological examination of hematoxylin and eosin (H&E)–stained transverse sections of the spinal cord. Each spinal cord was divided into several smaller, transverse regions before being processed, and multiple sections were taken per spinal cord. Metastatic dissemination was considered either positive or negative—where positivity was determined as a clear aggregate of 10 or more tumor cells. The number of analyzed animals is shown in the respective figures.
Reporting of P values, sample size, and statistical significance
Significant P values are reported for appropriate figures either in the figure legend or in the respective figure panel. Sample size (n) can be found in the figure legend or the respective figure panel. Error bars represent the standard error of the mean.
Randomization and inclusion/exclusion criteria
Animals included in the allograft studies had no inclusion/exclusion criteria before being stratified into treatment or vehicle groups. After sacrifice, the presence of a physical tumor was confirmed by pathologic examination and H&E staining in the brain or spinal cord. Animals that were found dead without obvious signs of tumor (primary tumor penetrance studies) were marked as censored. No other data were excluded from the analysis and reporting in this study. No blinding was carried out at any stage of the study.
Results
SOX9 is induced in therapy-resistant brain tumor cells and accumulates during relapse
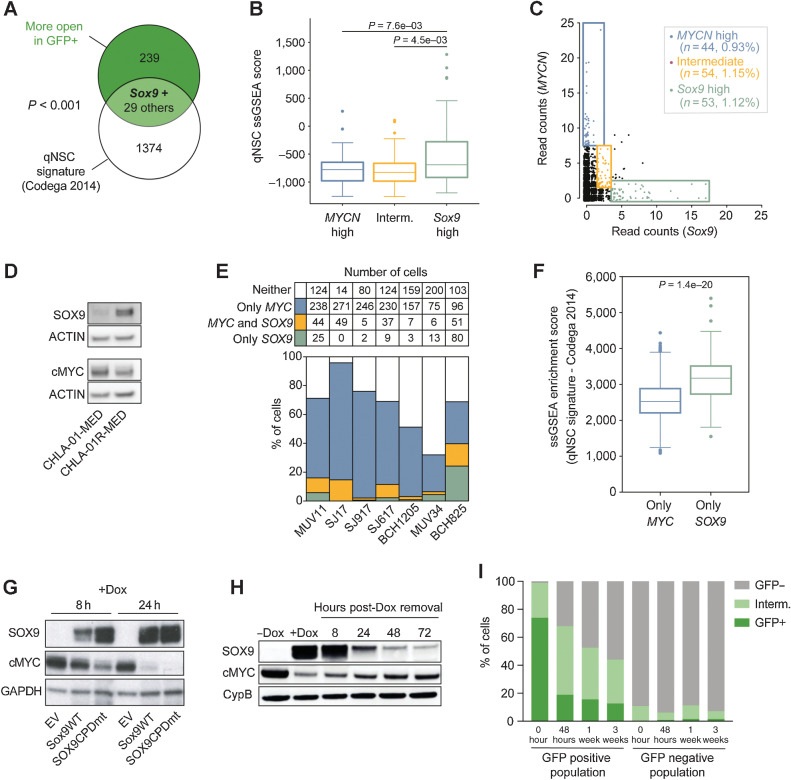
We first characterized SOX9 expression changes following MYCN depletion, using dox treatment of cultured tumor cells derived from our TetOFF-inducible Group 3 MB model (GTML), where the Glt1 promoter drives MYCN and luciferase expression (16). We previously showed that long-term (30 days) MYCN depletion from dox treatment almost always leads to recovery and complete treatment response of GTML animals that developed large brain tumors. After 48 hours of dox treatment of GTML tumor cells in vitro, MYCN levels were suppressed, and we detected a significant increase in Sox9 expression (Fig. 1A; Supplementary Fig. S1A). Although SOX9-positive cells are rare (2–3%) in GTML tumor biopsies, we observed an increased number of SOX9-positive cells after only 6 hours of dox treatment in vivo (Fig. 1B, top). Regardless of dox treatment duration, SOX9 and the proliferation marker Ki67 rarely co-stained in GTML tumors, whereas SOX9-negative tumor cells were significantly more proliferative [Fig. 1B (bottom); Supplementary Fig. S1B].
Figure 1.
SOX9 is induced in therapy-resistant brain tumor cells and accumulates during relapse. A, Expression of Sox9 in GTML tumor cells after 48 hours dox treatment (Student t test; **, P = 0.0059). B, Top, IHC showing SOX9 in GTML tumors after 6 hours, 7 days, and 30 days of dox administration in vivo, as well as an untreated GTML tumor. Bottom, IF showing SOX9 and Ki67 in GTML tumors after 6 hours, 7 days, and 30 days of dox administration in vivo, as well as an untreated GTML tumor. The analysis shows an increase of nonproliferating SOX9 expressing cells in the GTML model (see also quantification in Supplementary Fig. 1B). C, Quantification of GFP-positive cells in of GTML-pSOX9-GFP using cell sorting. The analysis shows that 24 hours cisplatin treatment increases the number of SOX9-positive cells compared with DMSO control. Student t test comparing the percentage of GFP-positive cells between DMSO and cisplatin treatment; ****, P < 0.0001. D, Quantification of SOX9 positive cells in tumors from transplanted GTML2 cells, irradiated with 2 Gy/day for 3.5 weeks (total 36 Gy). Tumors from mice that took the longest to relapse had more SOX9-positive cells. One-way ANOVA; ****, P < 0.0001. See also Supplementary Fig. 1I for representative IHC images. E, Representative images of IHC SOX9 staining of primary and recurrent tumors of all four MB subgroups. F, Quantification of SOX9-positive cells in WNT (n = 1), SHH (n = 6 as well as one re-operated relapse), Group 3 (n = 6 as well as one re-operated relapse), and Group 4 (n = 6) MB. The number of SOX9-positive cells is significantly higher in recurrent Group 3 (****, P < 0.0001) and Group 4 MB (**, P = 0.00113) compared with the primary tumor [Student t test, recurrence II (n = 1) is not included in the analysis]. G, Kaplan–Meier plot showing 10-year overall survival in Group 3 MB with high SOX9 levels (n = 28) and low SOX9 levels (n = 78; from ref. 15) at last quartile cut-off, excluding MB with extensive nodularity or tumors with desmoplastic histology. High SOX9 levels correlate to poor overall survival of Group 3 MB. H, Percentage of SOX9 levels in low (M0–M1) and high (M3) stage metastatic Group 3 MB as well as recurrent tumors of the same subgroup from ref. 12. SOX9 protein levels are higher in recurrent Group 3 MB than in primary M0–M1 stage tumor (Student t test; *, P = 0.0161). See also Supplementary Fig. S1.
We next wanted to study SOX9 expression following radiation and chemotherapy used in standard treatment. MB tumor cell lines isolated from crosses of GTML with mice expressing GFP under control of the SOX9 promoter (GTML-pSOX9-GFP) showed GFP activity in 2% to 5% of all cells (Supplementary Figs. S1C–S1F). First, we treated GTML-pSOX9-GFP cells with cisplatin and found a significant increase of GFP after 24 hours (Fig. 1C). The GFP-positive population was less sensitive to dox treatment (Supplementary Figs. S1G and S1H), in agreement with our previous results showing that SOX9 confers treatment resistance in tumor cells (12).
Next, we treated mice transplanted with GTML tumor cells in the cerebellum, with fractionated hindbrain radiotherapy in vivo over 3.5 weeks with 36 Gy. All irradiated tumors returned but relapses with longer latency had an increased number of SOX9-positive cells as compared with early recurrences or non-irradiated transplants (Fig. 1D; Supplementary Fig. S1I). Apart from being more SOX9-positive, the late recurrence had more dividing SOX9-positive cells compared with the non-irradiated GTML tumors, seen as a higher percentage of PCNA and SOX9 co-stained cells (Supplementary Fig. S1J). Further, although non-irradiated transplants did not present metastatic lesions in the spinal cord, all irradiated tumors did (Supplementary Fig. S1K).
Moving on, we asked if SOX9 is increased also in patient samples after treatment and if it is associated with recurrence. We, therefore, matched primary-recurrent patient biopsies (n = 21) from MBs of all four subgroups and counted cells positive for SOX9. SOX9-positive cells were significantly enriched in recurrences of both Group 3 and Group 4 samples (Fig. 1E and F). In addition, SOX9 was found to correlate with poor survival in Group 3 MB (Fig. 1G), but not in Group 4 and SHH subgroups (Supplementary Figs. S1L and S1M). There was no significant difference in SOX9 levels in SHH tumor pairs and too few matched WNT samples with recurrences for us to draw any conclusions (Fig. 1F).
Patients with M3 MB (i.e., macroscopic metastatic disease with spinal cord dissemination) have a worse prognosis as compared with patients with M0–M1 status (no metastases or micrometastases in the CSF; ref. 28). By studying our previous dataset of SOX9-immunostained MB biopsies (12), we found an increase of SOX9-positive cells in Group 3 M3 tumors and recurrent Group 3 MB, as compared with primary M0–M1 cases (Fig. 1H). There was an accumulation of SOX9-positive cells in recurrent Group 4 tumors and a significant increase of SOX9-positive cells in M2–M3 as compared with M0–M1 Group 4 tumors (Supplementary Fig. S1N), which is in line with M status being a predictor of poor prognosis in Group 4 MB (15). In addition, we found that SOX9 was elevated in the metastatic compartment of Group 3 and Group 4 patients from a smaller collection of paired primary/metastatic resections (Supplementary Fig. S1O).
As SOX2-positive cells are known to drive hierarchical growth and relapse in SHH tumors (29), we also studied SOX9 levels in several Ptch-deficient mouse tumors (2). All tumor cells in primary SHH tumors and their metastatic lesions were strongly positive for SOX9 (Supplementary Fig. S1P), consistent with our previous observation of high SOX9 activity in human SHH tumors (13). Immunostainings only revealed a partial overlap of SOX9 and SOX2 in GTML tumors (Supplementary Fig. S1Q) and SOX2 levels did not correlate with poor survival in Group 3 samples (Supplementary Fig. S1R).
When combining all the findings we conclude that SOX9 is correlating with poor prognosis in Group 3 MB and that SOX9-positive cells are accumulating after standard treatment, during malignant progression, and following relapse in both mouse models and patients.
SOX9 anticorrelates with MYC and marks quiescent cells in Group 3 patients with MB
To characterize differences in the SOX9-positive and SOX9-negative tumor cell population in the GTML model, we first used ATAC-seq to study GFP-sorted cells from GTML-pSOX9-GFP tumors. GFP-positive cells showed significant enrichment in SOX9 and other SOX (SOX2/3/6/10) and POU (OCT2/4/6/11) family members from HOMER motif analysis. Genes including Sox9 with regions of differentially opened chromatin that correlated with a quiescent molecular cell signature (30) in GFP-positive as compared with GFP-negative cells were shown using ssGSEAs against gene sets from quiescent and actively dividing NSCs (Fig. 2A; Supplementary Table S1). Sox9 high expressing cells in GTML tumors using scRNA-seq (Fig. 2B) also correlated with nondividing cells residing in the G0–G1 cell-cycle phase (Supplementary Figs. S2A and S2B). Further, the scRNA-seq data of GTML tumor cells showed two populations with either high expression of Sox9 or MYCN (Fig. 2C). However, there was also a smaller population with an intermediate expression of both Sox9 and MYCN (Fig. 2B and C). To confirm that the cell types identified are tumor-associated cells and to overcome the problems with poor MYCN antibodies for IHC, we used the RNAscope technique to stain-specific RNA sequences for mouse Sox9 mRNA and human MYCN mRNA (in the TML transgene) in situ. GTML tumors stained strongly for MYCN and as expected, fewer cells were positive for Sox9 (Supplementary Fig. S2C).
Figure 2.
SOX9 anticorrelates with MYC and marks quiescent cells in Group 3 patients with MB. A, Genes with higher ATAC-seq peaks in GFP-positive GTML-pSOX9-GFP cells significantly overlap with a quiescent signature (Fisher exact test; P < 0.05). B, Boxplot comparing the ssGSEA scores for a quiescent NSC signature between the groups from C, indicating that Sox9 high GTML cells appear significantly more quiescent than MYCN-high and intermediate cells (Welch t test; P < 0.05). C, Single-cell sequencing of GTML cells indicates that there exist three populations with regards to Sox9 and MYCN expression; MYCN exclusive/Sox9 exclusive/intermediate population with an expression of both MYCN and Sox9. The top ∼1% of cells with the highest MYCN and Sox9 expression, respectively, and ∼1% of cells with intermediary expression were selected for the analysis in B. D, Western blot showing protein levels of SOX9 and MYC, with actin as loading control in CHLA-01-MED and CHLA-01R-MED. SOX9 is higher in the recurrent cell line. E, The total numbers and percentages of cells with any (>0) expression of either MYC, SOX9, both, or neither in diagnostic Group 3 MBs (data from ref. 32). F, Cells from E with only SOX9 expression display a significantly higher ssGSEA score for a quiescent NSC signature as compared with cells with only MYC expression (Welch t test; P < 0.05). G, MB002 cells were transduced with lentiviral vectors encoding inducible EV, SOX9WT, or SOX9CPDmt. The transduced cell lines were treated with dox for 8 or 24 hours and analyzed for protein levels of SOX9, cMYC, and GAPDH. After the induction of SOX9, cMYC levels decreased. The decrease was more pronounced when the stabilized SOX9CPDmt was induced. H, MB002-SOX9CPDmt was treated with dox for 24 hours (for SOX9 induction), after which, they were resuspended in fresh media. SOX9, cMYC, and cyclophilin B (CypB) protein were analyzed with Western blot at 8, 24, 48, and 72 hours after dox removal. After 72 hours, protein levels decreased but neither SOX9 nor cMYC was restored to start levels. I, Quantification by FACS of GFP-positive (+), negative (−), and intermediate cells in FACS sorted GTML-pSOX9-GFP cell line, directly after sorting, 48 hours, 1 week, and 3 weeks after sorting. The number of GFP-positive cells in the positive population quickly dropped while it slowly increased in the negative population. See also Supplementary Fig. S2.
We next wanted to investigate this heterogeneity in cells from patients with MB. We started by determining the protein levels of SOX9 and MYC in the only publicly available MB primary-recurrent cell line pair, the Group 4 CHLA-01-MED and CHLA-01R-MED tumor cells (31). Again, decreased MYC expression and higher levels of SOX9 were specifically found in the recurrent cell line (Fig. 2D). In a cohort of 64 human MB samples (14), we found SOX9 mRNA levels in MYC/MYCN amplified cases to be significantly lower than the levels in MYC/MYCN non-amplified cases (Supplementary Fig. S2D, top). This was further validated in a larger cohort of 470 MB Group 3 and Group 4 samples (15), in which SOX9 expression was anti-correlated with MYC (Supplementary Fig. S2D, bottom). We finally analyzed scRNA-seq data of human MB samples (32) and found a few SOX9-positive cells in human Group 3 MB biopsies compared with the MYC-positive population that made up most cells (Fig. 2E). By comparing ssGSEA SOX9-positive cells in patient samples also presented with a significantly more quiescent signature as compared with MYC-positive populations (Fig. 2F).
We then asked if SOX9 regulates MYC expression in human brain tumors. We overexpressed inducible wild-type (WT) SOX9 and mutationally stabilized (CPDmt) SOX9 (12) in MYC-amplified MB002 cells. SOX9 induction, especially the CPDmt, led to downregulated MYC protein after 8 hours and brought it to almost undetectable levels after 24 hours (Fig. 2G). However, following dox washout, SOX9 subsequently diminished and MYC levels increased again (Fig. 2H), suggesting that SOX9 temporally regulates MYC in this setting. We further investigated the SOX9–MYC interaction in GTML-pSOX9-GFP cells after sorting out SOX9-positive and SOX9-negative cells and leaving them in normal stem cell cultures. Two days after cell sorting, the number of highly GFP-positive cells dropped to 70% whereas the intermediate (GFP low) and GFP-negative cells increased. After a few weeks most cells were negative (Fig. 2I). Interestingly, GFP-negative cells were shown to generate a few (2%) GFP-positive cells after a week in culture (Fig. 2I). These results suggest cell plasticity and that sorted SOX9-positive cells quickly become negative without any selection pressure in culture. There seemed to be a balance in favor of the SOX9-negative, MYCN high stage, in agreement with the notion that few SOX9-positive cells reside in GTML tumors or human tumors.
To see if MYC was directly regulated by SOX9, we checked its mRNA levels and methylation status in the SOX9-inducible MB002 cell lines (12). The MYC promoter (0–1.5 kb upstream of TSS) was not methylated upon SOX9 induction as seen using EPIC array analyses (Supplementary Fig. S2E). Both 8 and 24 hours of SOX9 induction (Supplementary Fig. S2F) significantly downregulated MYC mRNA levels (Supplementary Fig. S2G). By comparison, the more obvious suppression of MYC at the protein level suggests an indirect regulation or perhaps a posttranslational modification. Still, neither an Actinomycin D treatment nor a cycloheximide chase assay showed any difference in the RNA or protein stability of MYC upon SOX9 induction (Supplementary Figs. S2H and S2I). SOX9 overexpression did however lead to suppressed cMYC phosphorylation (at residues T58 and S62) and reduced proliferation (PCNA-positivity; Supplementary Fig. S2J).
In summary, we found that the rare SOX9-positive cells in GTML and human biopsies likely make up a quiescent tumor cell population distinct from the MYC/MYCN expressing majority of cells. We also identified an intermediate population expressing both transcription factors and that the tumor cells could move between these three populations.
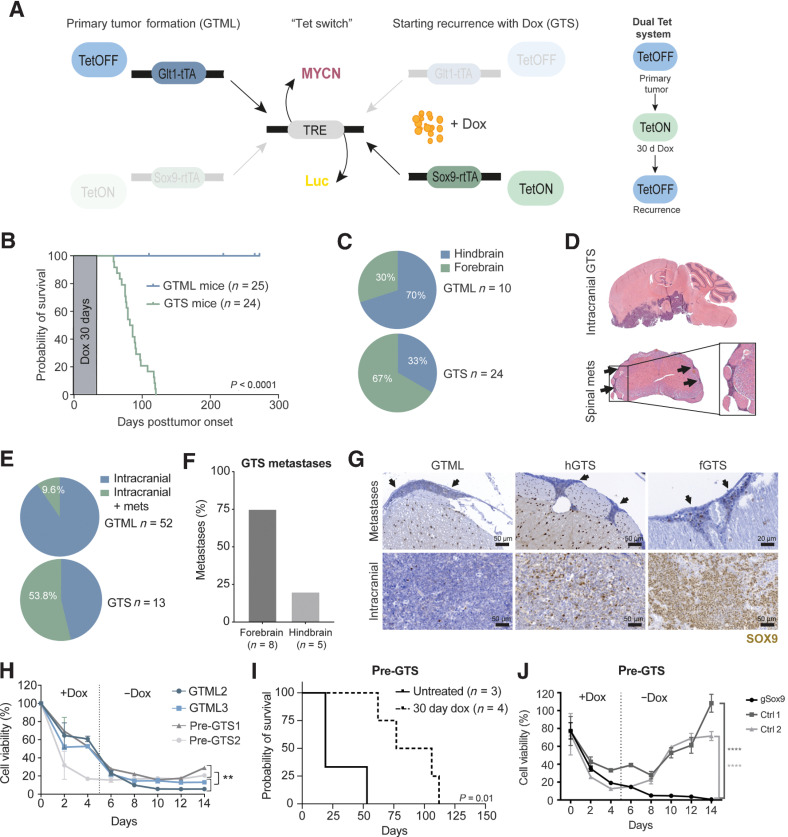
The recurrence process is SOX9-dependent and reproduced in a novel dual tet transgenic GTS model
Knowing that SOX9-positive cells are more prevalent in metastatic recurrent tumors and accumulate upon standard treatment, we next wanted to see if SOX9 cells are present in minimal residual disease (MRD) that can be detected in patients with MB (33). To generate a mouse model for MRD, we transplanted mice with two GTML cell lines (GTML2 and GTML3) with different levels of SOX9 (Supplementary Fig. S3A) and did a short dox treatment of 7 days. GTML3 tumors that were more SOX9 positive were less sensitive to dox and presented with luciferase signals as high as treatment start already 7 days post-dox treatment (Supplementary Fig. S3B). GTML2 cells that were less SOX9 positive, however, responded well to dox treatment, leaving 1% of the luciferase signal (photons/sec) relative to treatment start in GTML2, and slowly recovering (Supplementary Fig. S3B). This dynamic is similar to a residual disease seen in patients where relapses arise several months or years after initial standard treatment (33). Dox significantly extended the overall survival of GTML2 mice to 16.5 weeks, and all mice presented with relapses (Supplementary Fig. S3C). We found more SOX9 positive cells both at MRD (defined as a time point 10 days post-dox treatment) and in recurrent tumors, compared with untreated GTML2 controls (Supplementary Fig. S3D), which is similar to what we found in relapses following long-term irradiation of these cells (see Fig. 1D).
To specifically study and target SOX9-expressing cell populations in vivo, we employed a Sox9-rtTA transgene (34) driving reverse tTA (TetON) from the Sox9 promoter (Fig. 3A). First, we asked if MYCN can induce tumors from SOX9-positive cells. We crossed mice carrying Sox9-rtTA with mice carrying TML (STML) and thus created a TetON system where MYCN is activated in Sox9-expressing cells with dox. We followed 56 mice that were put on a dox diet starting from various time points: embryonal day 0, newborn (P0), and 3 weeks old animals (P21). None of these STML mice developed any tumors on continuous dox administration over 6 months (Supplementary Fig. S3E), suggesting SOX9-positive cells are likely not cells of origin for these tumors.
Figure 3.
The recurrence process is SOX9-dependent and reproduced in a novel dual Tet transgenic GTS model. A, Schematics of the GTS recurrence model. Left, primary tumor formation (GTML). In the absence of dox, the TML (TRE-MYCN-Luc) transgene is expressed in Glt1-positive cells (bidirectional expression), leading to MB primary tumor formation in the GTML model. Dox administration will halt MYCN expression (TetOFF) in the Glt1-positive cell population. Right, starting recurrence with dox (GTS). When combining the GTML TetOFF model with the TetON model, dox will simultaneously force the expression of the TML transgene in Sox9-expressing cells. When the primary tumor is presented, mice are given a 30-day dox regimen (the Tet-switch), at which time, the TetON system is active. Mice are then taken off dox to reactivate the TetOFF system and followed for tumor recurrence. B, Kaplan–Meier plot and scheme of dox treatment showing mouse survival in the GTS recurrence model. Primary tumor (GTML, TetOFF-driven) was treated with and cured after 30 days of dox administration. Tumors recurred after 45 to 120 days post-primary tumor onset/start of dox treatment (Log-rank Mantel–Cox test; P < 0.0001). C, Quantification of tumor localization in the GTML and GTS models. Seventy percent of GTS recurrences were located in the mouse forebrain in contrast to GTML tumors that most often are found in the hindbrain. D, H&E staining of a whole mouse brain with relapsed GTS tumor. The tumor recurrences were most often situated in the forebrain. Spinal cord metastasis can be found in GTS-relapsed tumors. Arrows, metastatic cells surrounding the spinal cord. E, Only 9.6% of H&E-stained sectioned spinal cords from GTML samples presented with spinal metastases (mets) as compared with 53.8% in GTS samples. F, Histogram comparing the occurrence of spinal metastases in GTS recurrences in the hindbrain or forebrain region. The majority of spinal metastases were found in forebrain relapse specimens. G, SOX9 in disseminated tumor cells in corresponding spinal cords from representative individual mice with metastatic spread (top; arrows) as compared with SOX9-positive cells in primary GTML, hGTS, and fGTS tumors (bottom). H, Cell viability (Alamar blue) of GTML versus pre-GTS cells under long-term dox treatment. Pre-GTS cells started to recover after dox treatment in vitro, but GTML cells did not (Mann–Whitney; **, P < 0.01). I, Kaplan–Meier plot showing survival probability of nude mice injected with pre-GTS1 and 2. Pre-GTS cells gave rise to primary tumors that were treated with dox for 30 days. The tumors later relapsed (log-rank Mantel–Cox test; P = 0.01). J, Cell viability (Alamar blue) of CRISPR-Cas9 edited and control pre-GTS1 cells under long-term dox treatment in vitro. Control cells started to recover after dox treatment, but Sox9-edited pre-GTS1 cells (sgSox9) did not. Unpaired t test; ****, P < 0.0001. See also Supplementary Fig. S3.
The increased expression of Sox9 in our residual disease model (see Supplementary Figs. S3A–S3D) as well as in human recurrences (see Fig 1E–H) suggest that SOX9 is involved in a relapse mechanism rather than in tumor initiation. We then crossed the GTML (TetOFF) model with the above Sox9 (TetON) model and generated a dual Tet system in a triple transgenic Glt1-tTA; TML; Sox9-rtTA (GTS) model. In the absence of dox, GTS mice are identical to GTML mice as the rtTA molecules produced under the control of the Sox9 promotor are inactive (TetOFF). However, when GTS mice are given dox, MYCN is no longer driven by the Glt1 promoter but by the Sox9 promoter (TetON). As dox is removed, MYCN is again driven from the Glt1 promoter (TetOFF; Fig. 3A). At tumor presentation, the GTML mice (n = 25) were given a dox diet for 30 days and the tumors subsequently regressed (Fig. 3B) as previously shown (16). GTS mice (n = 24) developed similarly large primary tumors and recovered after a 30-day dox diet (Supplementary Fig. S3F). However, 45 to 120 days after dox removal, all the GTS mice presented with recurrences (Fig. 3B; Supplementary Fig. S3F). Recurrent GTS tumors showed no significant differences in apoptosis or cell proliferation as compared with GTML tumors (Supplementary Figs. S3G and S3H) and resembled a large cell anaplastic or classic MB histology. Typically, the recurrent tumors stained strongly for Ki67 compared with remnant primary tumors that silently remained in the brain of mice months after sufficiently curative 30-day dox treatment, as previously described in the GTML model (Supplementary Fig. S3I; ref. 16). Recurrent tumors in GTS mice were more metastatic and mainly localized to the forebrain as compared with tumors in GTML mice that were most often found in the hindbrain (Fig. 3C–E). GTS forebrain recurrences (fGTS) most often had spinal metastases as compared with recurrences preferentially located in the hindbrain or the cerebellum (hGTS; Fig. 3F). GTML and GTS metastases presented with SOX9-positive cells at similar numbers as compared with their corresponding intracranial tumors (Fig. 3G). Overall, fGTS recurrences had significantly more SOX9-positive cells than both hGTS and GTML tumors (Fig. 3G; Supplementary Fig. S3J). Taken together, the pathology, distant location, and dissemination of recurrent tumors induced using the GTS system resemble that of metastatic or relapsed human MB (1), which was also seen following transplantation of three human Group 3 PDX MB cells that similarly disseminated and were presented with significantly less dividing (Ki67+), SOX9-positive cells in their metastatic compartment (Supplementary Fig. S3K and S3L).
Culturing GTS cells in dox, forcing overexpression of MYCN from the Sox9 promoter slows down the proliferation of GTS tumor cells in vitro (Supplementary Fig. S3M). To know if this suppression is enough to block tumor relapse in vivo, we introduced a cohort of GTS mice treated with dox for a prolonged period of 6 months, resulting in tumor regression and no relapses (Supplementary Fig. S3N). This implies that transient expression of the tumor driver MYC in quiescent SOX9-positive tumor cells is a necessity for relapse in MYC-driven tumors as similarly shown in SOX9-driven breast cancer progression (35).
We then hypothesized that the few SOX9-positive cells in the primary GTML tumor were the cause of the GTS recurrences. We started by testing if we could mimic the GTS relapses in vitro. GTML, pre-GTS1, and pre-GTS2 cells (from two primary, non-dox treated GTS tumors) were treated with dox for 5 days. After 10 days in culture, GTML cells had completely stopped proliferating whereas pre-GTS cells instead started to recover similarly to what we saw in vivo (Fig. 3H; Supplementary Fig. S3F). The latter could be continuously passaged and showed proliferation rates comparable with parental pre-GTS cell lines. Pre-GTS1 and 2 cells also generated primary tumors in mice and relapsed after 30 days of dox treatment (Fig. 3I). These data ruled out the possibility that SOX9-positive cells were nontumorigenic cells that had infiltrated the brain tumor.
Next, we used CRISPR knockout and depleted Sox9 in pre-GTS1 cells. A clonal gSox9 cell line (pre-GTS1 gSox9) was compared with two clonal control lines (pre-GTS1 Ctrl1–2; Supplementary Fig. S3O). All three lines generated brain tumors after around 3 weeks posttransplantation in vivo. However, when culturing these three lines only the pre-GTS1 gSox9 line failed to recover after dox removal (Fig. 3J) providing that SOX9 is essential for rescuing cells in the GTS recurrence model.
Overall, MB recurrence is recapitulated in the novel GTS mouse model of MB relapse in which relapse is dependent on a small population of SOX9-expressing tumor cells.
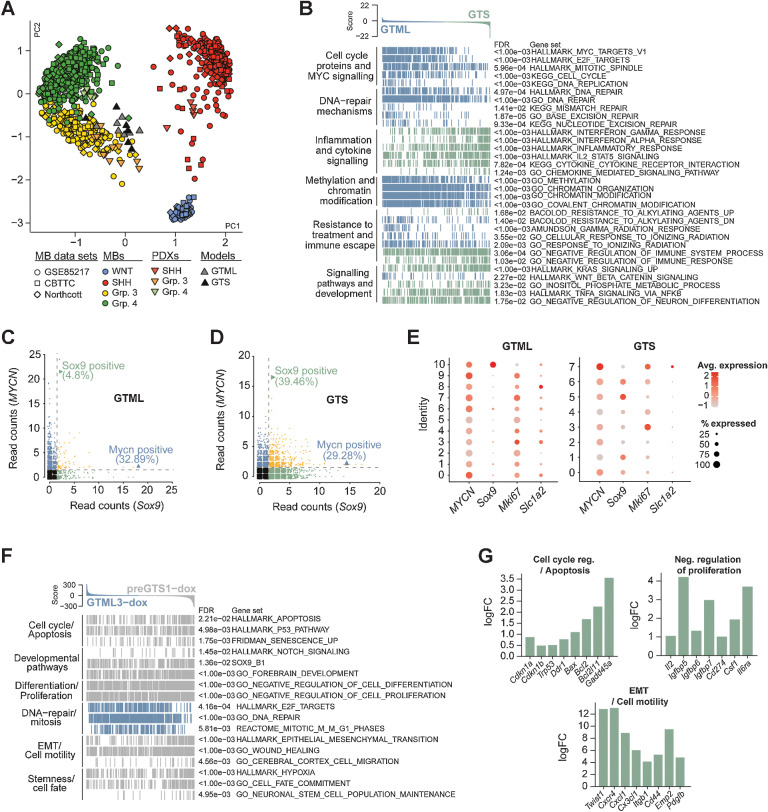
Recurrent MBs molecularly resemble primary MBs but are more inflammatory and immune evasive
To analyze the molecular differences between primary and recurrent tumors, we performed RNA-seq analysis and started with a cross-species comparison against various types of brain tumors, demonstrating a clear affiliation of both GTML and GTS tumors with MB (Supplementary Fig. S4A). Subsequently, we employed a cross-species analysis against MB patient data. The recurrent GTS tumors (n = 6) clustered near the primary GTML tumors (n = 7) and both were strongly aligned with Group 3 MB (Fig. 4A).
Figure 4.
Recurrent MBs molecularly resemble primary MBs and are more inflammatory and immune evasive. A, PCA plot showing the affiliation of primary GTML tumors and recurrent GTS tumors with expression profiles of MB subgroups (data from refs. 15, 25) Northcott and MB PDXs (18) following cross-platform projection via the Metagene (23) Tamayo method. B, GSEA results for 32 selected key gene sets from six pathway categories significantly enriched in either the GTS (green) or GTML (blue) tumors. C and D, Single-cell sequencing read counts for MYCN and Sox9 in GTML (C) and GTS (D), indicating that the percentage of MYCN-positive (reads ≥ 2) cells remained largely unchanged between the two tumor models, whereas the GTS tumor revealed an increased percentage of Sox9-expressing cells. See also Supplementary Fig. S4 and Supplementary Table S2. E, Dot plots of transgenic MYCN and murine Sox9, Mki67, and Slc1a2 gene expression in GTML (left) and GTS (right) cell lines. Cluster IDs are indicated along the y-axis. The size of the dot indicates the percentage of cells in the cluster that expressed the gene, and the color intensity indicates the average expression level in the cluster. F, Enrichment plots of pathways significantly altered in 48-hour dox-treated pre-GTS tumor cells (gray) compared with 48-hour dox-treated GTML tumor cells (blue). G, Bar plots showing the logFC for selected genes from the gene sets depicted in F as calculated by edgeR between GTML and pre-GTS cells under dox treatment. See also Supplementary Fig. S4 and Supplementary Tables S3 and S4.
Despite both tumor models affiliating with Group 3 MB, a differential gene expression analysis suggested substantial transcriptional differences between them (Supplementary Table S2). By using preranked GSEAs, we found significant enrichment of gene sets associated with MYC pathway and cell proliferation in GTML; and with gene sets associated with inflammatory responses, JAK/STAT, and NF-κB signaling in GTS (Fig. 4B). MYCN was repressed specifically in the forebrain (fGTS) tumors and there was an increase of Sox9 mRNA and protein levels in GTS tumors regardless of their location (Supplementary Figs. S4B and S4C). Typical immune escape markers like Galectin 9 (Lgals9; ref. 36) and PD-L1 (Cd274; ref. 37) were significantly upregulated in fGTS (Supplementary Fig. S4B). Further, genes implicated in extravasation and migration, like Ccl2 and Ccr2 (38), were found to be upregulated in GTS recurrences (Supplementary Fig. S4B). In line with this, we found that genes with both increased ATAC-seq signals and upregulated RNA-seq expression in GTS tumors significantly overlapped with cell locomotion (GO_LOCOMOTION; Supplementary Fig. S4D). Among the GSEA results, we also identified gene sets of cytokine pathway activation, for example, KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION that were upregulated in GTS tumors as compared with GTML tumors (Fig. 4B). A deeper analysis, by comparing fGTSs and hGTSs using GSEA, showed that the inflammatory response was a feature of the distant recurrences (Supplementary Fig. S4E). Accordingly, we used an array with more than 100 selected cytokines to identify differentially activated cytokine proteins in GTML and fGTS tumor cells. Although VEGF and IGFBP2 were dramatically reduced, IL12B and IGFBP5 were increased more than three-fold in recurrent cells with an additional accumulation of secreted IGFBP5 also in the supernatants from fGTS cell cultures (Supplementary Fig. S4F).
When performing the GSEA of hGTS and fGTS, we found increased enrichment in tumor escape from immune cells and T-cell activation. We also found significantly higher levels of Arg1 and Il10 (Supplementary Fig. S4G) in fGTS, indicating an accumulation of tumor-associated macrophages (TAM). To understand the increase, we investigated the number of macrophages and T cells in the tumors. First, Iba+ cells showed a spiky, activated morphology in all tumors with a significantly increased accumulation in GTS recurrences as compared with primary GTML tumors. Next, we found significant recruitment of Cd3+ T cells to the fGTS tumors as compared with what we found in hGTS or normal GTML tumors. Finally, to rule out that MYCN suppression from 30 dox treatment is causing this phenomenon, we also studied GTML tumors that had been treated for 30 days but we found no difference in immune cell activity to that observed in GTML tumors (Supplementary Figs. S4H and S4I).
We next used scRNA-seq to characterize the SOX9-population in both mouse models. We found that Sox9-expressing cells were more numerous in GTS cell lines (∼39%) compared with GTML cell lines (∼5%; Fig. 4C and D). As seen in a dot plot comparative analysis of scRNA-seq data, Sox9 is present in a single, distinctly defined Ki67 and Slc1a2 (Glt1) low cluster (Cluster 10) in GTML tumors. Glt1-expressing cells instead correlate with more proliferative cells (like cluster 8) that express low levels of Sox9. Given the dox switch, Slc1a2 is more downregulated in GTS clusters and only present in the cluster with the highest MYCN activity (Fig. 4E).
In contrast, several clusters with Sox9 expression exist in GTS cells (Fig. 4E). The cells with the highest level of Sox9 expression in GTS (cluster 5) showed lower expression of MYCN and Ki67 (Fig. 4E). In support of this, high Sox9 expression correlated with the G0–G1 cell-cycle phases and was mainly distinct from proliferative Ki67 expressing cells, seen as distribution in a Umap plot (Supplementary Fig. S4J). In addition, the GTS cells with the highest Sox9 expression, distinct from MYCN-expressing cells correlated better with a quiescent signature (Supplementary Figs. S4K and S4L). Still, Sox9-positive clusters in GTS have generally higher levels of Ki67, especially compared with cluster 10 in GTML cells (Fig. 4E). This was also seen when comparing the differentially upregulated genes in different clusters using GSEA and when further including scRNA data from human samples (Supplementary Fig. S4M). In Group 3 MB, SOX9 is elevated in cluster 5, which also exhibits a less proliferative phenotype (Supplementary Figs. S4M and S4N). Among other genes found commonly upregulated in both SOX9-expressing human Group 3 (cluster 5) and Sox9-expressing GTML cells (cluster 10) are Sox2, Igfbp5, and markers of mammary stem cells (Lee_Mammary_Stem_Cell_UP and Jechlinger_Epithelial_To_Mesenchymal Transition_UP; Supplementary Fig. S4M). Neither primary Group 3 MB nor GTML cells have undergone epithelial–mesenchymal transition (EMT). In contrast, Sox9-expressing GTS cells have already gone through EMT. Instead, these cells show markers mimicking E2F loss (Igelsias_E2F_Targets_UP), markers of migration (GO:BP_Locomotion), and are expressing Igfbp5. But, they also present with markers that might protect them from T cells (Supplementary Fig. S4M). One such factor is the “don´t eat me” factor Cd47, which was previously shown to be a useful target in Group 3 MB (39). They further express genes upregulated upon defective T-cell regulation (Li_Induced_T_To_Natural_Killer_UP), which was also upregulated in SOX9-positive human Group 3 MB (Supplementary Fig. S4M).
After 48-hour dox treatment, the viability of GTML tumor cells is more than halved. This should thus be a critical time point where SOX9-positive cells will start developing resistant cell clones. To detect early changes in surviving recurrent cell clones, we compared RNA-seq profiles of pre-GTS and GTML cells treated with dox for 48 hours to untreated control cells. MYCN was downregulated in both GTML and pre-GTS cells. Sox9 was upregulated in both dox-treated GTML but to a larger extent in dox-treated pre-GTS cells as expected. In contrast, Slc1a2 (Glt1) decreased in dox-treated pre-GTS compared with GTML (Fig. 4F; Supplementary Fig. S4O; Supplementary Tables S3 and S4). Gene sets differentially enriched in dox-treated pre-GTS cells included hallmarks for stemness, migration, and EMT (Fig. 4F). As SOX9-positive cells survive dox treatment, we expected to find genes involved in cell death inhibition. For example, the anti-apoptotic regulator Bcl2 was differentially upregulated in dox-treated pre-GTS cells. Again, genes involved in the negative regulation of cell proliferation (including Igfpb5) were upregulated in pre-GTS cells (Fig. 4G). As we recently reported in metastatic Group 3 MB, the Notch pathway and Twist1, drivers of dissemination of these tumors (40), were also significantly elevated in dox-treated pre-GTS cells (Fig. 4F and G). In GTML cells, cell-cycle genes/mitotic markers were instead enriched (Fig. 4F and G). All in all, our data suggest that SOX9-positive tumor cells start to reconstitute recurrent cell clones through distinct signaling pathways shortly after oncogene-targeted dox treatment.
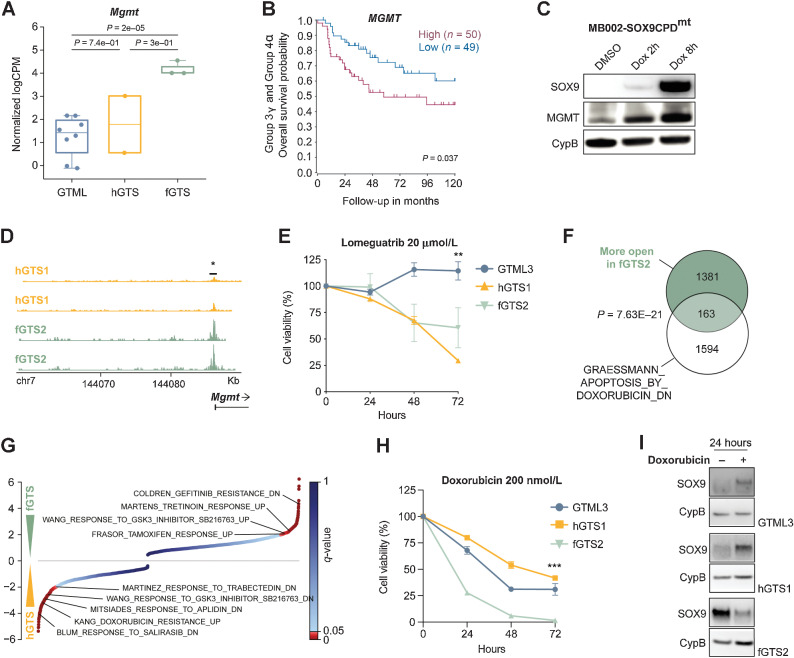
MB recurrences respond to MGMT inhibition and doxorubicin therapy
When we treated pre-GTS tumorspheres with cisplatin, we saw increased cell death but also a significant increase in the number of SOX9-positive cells surviving the treatment, similar to GTML cells (Supplementary Fig. S5A; Fig. 1C). SOX9 expression was also significantly upregulated in MB biopsies from patients given standard therapy as compared with expression in treatment-naïve patients (Supplementary Fig. S5B). We, therefore, reasoned that we could use our novel GTS model to find specific targets for treatment of relapsing tumor cells.
In the previously described GSEA between dox-treated GTML and pre-GTS, we found DNA-repair genes to be downregulated, except for Mgmt, which was upregulated (Supplementary Fig. S5C; Fig. 4F). In addition, we saw a significant downregulation of Trp53 in GTS tumors (Supplementary Fig. S5D), which has a central role in the DNA repair response and that is often lost in MB recurrence (7). Other genes involved in typical DNA repair processes like Brca2, Rad51, Parp1, Xrcc2, Fancb, and Fancc were also downregulated in our GTS model (Supplementary Figs. S5E–S5J). As mentioned, Mgmt was found to be significantly elevated in pre-GTS cells (see Supplementary Fig. S5C), but also upregulated in fGTS tumors (Fig. 5A). MGMT is a dealkylating DNA methyltransferase important in the direct reversal of DNA repair. It is implicated in treatment resistance in glioblastoma (GBM) and is significantly upregulated in recurrent GBMs (41). High MGMT levels result in significantly worse overall survival in Group 3γ and Group 4α subtypes of MB (Fig. 5B), typically driven by MYC and MYCN amplification, respectively (15). And MGMT levels are indeed highest in Group 3 MB (Supplementary Fig. S5K). When overexpressing SOX9 in human MB cells, MB002, we also found a fast upregulation of MGMT, suggesting that SOX9 is promoting its expression (Fig. 5C). We used ATAC-seq and found that the Mgmt promoter region was more open in the more SOX9-positive fGTS cells as compared with hGTS cells (Fig. 5D). We then investigated the methylation status of MGMT in a set of 30 (11 Group 3 and 19 Group 4) paired primary and relapsed MB samples. We focused on two CpG probes, previously used to study the association between MGMT promoter methylation and gene expression (41). Most patients with MB displayed hypomethylation in the primary tumors, whereas 1 patient with hypermethylated MGMT promoter underwent demethylation upon recurrence. Still, a potent MGMT inhibitor, lomeguatrib, significantly decreased the proliferation of GTS cells but did not show efficacy in GTML cells (Fig. 5E). In combination with the alkylating agent thiotepa, often used in high-risk Group 3 MB therapy, lomeguatrib decreased the proliferation of fGTS cells even further (Supplementary Fig. S5L).
Figure 5.
MB recurrences respond to MGMT inhibition and doxorubicin therapy. A, Expression levels of Mgmt in primary GTML tumors (n = 7), and recurrent hGTS (n = 2) and fGTS tumors (n = 3) from RNA-seq (Mann–Whitney test). B, High MGMT levels significantly correlated with a poor 10-year overall survival in Group 3γ and Group 4α MYC and MYCN high MB subgroups (from ref. 15 at median cut-off). C, Western blots showing induction of SOX9 in MB002-SOX9CPDmt using dox also leads to increased MGMT protein already after 2 and 8 hours. D, ATAC-seq of one GTS hindbrain cell line (hGTS1) and one GTS forebrain cell line (fGTS2) show open chromatin at the MGMT promoter in fGTS2, suggesting that MGMT expression is induced in distant recurrences (Student t test). E, Cell viability (Alamar blue) after 24, 48, and 72 hours of lomeguatrib (20 μmol/L) treatment of GTML3 versus hGTS1 and fGTS2 cell lines. The GTS cell lines are more sensitive to lomeguatrib treatment compared with GTML3. Student t test; **, P = 0.0010. F, Genes with significantly higher ATAC-Seq peaks in the fGTS2 cell line as compared with the hGTS1 line overlapped significantly (P < 0.05) with genes downregulated with doxorubicin treatment. G, Normalized enrichment scores from a GSEA comparing hGTS1 and fGTS2 expression profiles against the CGP (chemical and genetic perturbation) database of gene sets; significantly enriched drug treatment gene sets are highlighted. H, Cell viability (Alamar blue) after 24, 48, and 72 hours of doxorubicin (200 nmol/L) treatment of GTML3, hGTS1, and fGTS2. fGTS2 was significantly more sensitive to doxorubicin treatment compared with hGTS1. Student t test; ***, P < 0.0001. I, Western blots depicting SOX9 protein levels in GTML3, hGTS1, and fGTS2 after 24 hours of doxorubicin treatment (200 nmol/L). See also Supplementary Fig. S5.
Next, we wanted to find drugs that could be used to treat distant/metastatic recurrences. A GSEA against a drug database of chemical perturbations revealed several drug signatures differentially enriched between the metastatic fGTS and local hGTS tumors. It suggested that the latter might be more sensitive to doxorubicin treatment. Our prediction was strengthened by ATAC-seq data, indicating that genes with more open chromatin in fGTS cells significantly overlapped with doxorubicin-related (APOPTOSIS BY DOXORUBICIN_DN) gene sets (Fig. 5F and G). When treated with 200 nmol/L doxorubicin, fGTS cells were completely depleted after 72 hours as compared with GTML and hGTS cells where nearly half of the cells were still viable (Fig. 5H). After a shorter 24-hour exposure, doxorubicin specifically lowered SOX9 protein levels in fGTS cells whereas SOX9 levels instead increased in GTML and hGTS cells (Fig. 5I). In contrast, MYCN levels were left unaltered in all treated cell lines (Supplementary Fig. S5M).
To conclude, MGMT inhibition in combination with alkylating agents could potentially benefit patients with relapsed Group 3 MB, which manifest with hypomethylated MGMT, and doxorubicin presents as a promising future treatment for metastatic recurrent Group 3 MB.
Discussion
Recurrent tumors and metastasis are associated with poor prognosis in MB (1, 42). It was recently shown that SOX2-positive and OLIG2-positive cells driven by Ptch1 loss were capable of propagating SHH-dependent MB and inducing recurrence (29, 43). However, neither SOX2 nor OLIG2 correlates with poor prognosis in Group 3 and Group 4 MB, which are different biological entities as compared with WNT or SHH MB, as further anticipated when studying their distinct methylation profiling (15).
SOX9 is not essential for SHH-driven MB formation from granule cell precursors (44) but has recently been shown to drive radiation-induced resistance and recurrence in MB upon Ptch1 loss from contributing astrocyte-like cells (45). Non-proliferative tumor cells are defined by a SOX9-driven quiescence program dependent on the retinoic acid pathway in head and neck tumors (46) or by a WNT-driven suppression mechanism in breast tumor latency (10). By using advanced modeling and diversified profiling of mRNA and protein at the single-cell level, we identified pathways that are overrepresented in the recurrent GTS tumors as well as in Group 3 patient samples. As we expected, based on the tumor's pathology, pathways involved in cell migration, motility, and adhesion, were upregulated in the recurrent tumors. Apart from increased SOX9 expression, the elevated levels of the chemokine ligand CCL2 and its receptor CCR2 in forebrain GTS tumors that relapsed to a distant site away from the primary location are striking and correlate well with recent reports of MB dissemination patterns and metastasizing through circulating tumor cells in the blood (38). Similarly, the transformation of microglia to become tumor-associated and Arg1+ and the upregulation of typical immune escape markers like PD-L1, Lgals9, and Il10 suggests that forebrain recurrences indeed try to protect themselves against infiltrating T cells in this fully immunocompetent animal model. A similar type of macrophage-induced immune suppression has similarly been seen following irradiation of SHH tumors (47), suggesting this modulation of TAMs can be triggered following standard treatment.
The approach of combining a TetOFF system representing the bulk cells of the tumor with a TetON system representing distinct clones of dormant tumor cells can be applied for studying heterogeneity in other cancer models and especially for evaluating the properties of defined cell types within the tumor. Our data confirm that relapsing cells avoid therapy by entering a dormant quiescent state. Still, our single-cell analysis suggests that a larger cluster of intermediate SOX9-positive cells emerge that retain MYC expression in relapses. We also concluded that the SOX9-positive, resistant tumor cells are plastic and can rapidly reduce SOX9 levels and return to an MYC high state later in the relapse process when the tumor is expanding. This suggests that an upfront treatment or a treatment at the MRD state would be deemed necessary to efficiently remove these SOX9-positive therapy-escaping cells.
Downregulation of p53 and numerous DNA repair pathways observed in recurrent tumors are most likely an escape route for the tumor cell to avoid getting trapped by the repair machinery. Direct targeting of this route might thus be efficient, but it is difficult to treat something that is suppressed. In contrast, MGMT, involved in the direct reversal repair pathway, correlates with poor prognosis in Group 3 patients, was upregulated and indeed seemed to have a functional role in recurrent mouse tumors as its specific inhibition not only suppressed relapses but also successfully potentiated the effects of standard MB therapy.
Apart from MGMT targeting, our combined bioinformatics results suggested doxorubicin as a therapy for targeting SOX9-positive relapses and for downregulation of the SOX9 protein. Doxorubicin is used in the therapy of patients with hematologic and solid pediatric cancer (48), including atypical teratoid/rhabdoid tumors in the CNS but the drug cannot efficiently cross the blood–brain barrier (BBB) despite enhanced drug efficacy from pegylated liposome formulations. Still, novel techniques using aptamer-doxorubicin conjugates have shown greatly improved BBB permeability with successful treatment of brain metastases (49). When such or similar doxorubicin formulations will be approved for use in patients, they should be considered, especially for children with recurrent MYC-driven brain cancer, who currently are in great demand of better treatment options (7).
Supplementary Material
Supplementary Figure Legends
Supplementary figure 1
Supplementary figure 2
Supplementary figure 3
Supplementary figure 4
Supplementary figure 5
List of genes that are part of a quiescent cell signature and display regions of differentially opened chromatin.
Results of differential gene expression analysis between GTML tumours and GTS tumours.
Results of differential gene expression analysis between GTML3 tumor cells treated for 48 hours with DMSO or Dox.
Results of differential gene expression analysis between pre-GTS1 tumor cells treated for 48 hours with DMSO or Dox.
Resource Table
Acknowledgments
The authors thank Dr. Michael German at UCSF for generously providing them with SOX9–rtTA transgenic animals and Dr. William Weiss at UCSF for maintaining and sharing these animals with them. They acknowledge financial support from the Swedish Childhood Cancer Fund, the Swedish Cancer Society, the Swedish Research Council, the Swedish Society of Medicine, the Swedish Brain Foundation, the European Research Council - Horizon 2020 (Project No. 640275, Medulloblastoma - ERC-2014-STG), Åke Wiberg´s Foundation, Ragnar Söderberg´s Foundation, and the Worldwide Cancer Research. The authors further acknowledge technical support and radiation treatment planning at the Preclinical Cancer Treatment (PCT) Center (SciLifeLab and Uppsala University) and imaging services and support with cell sorting analyses at the BioVis facility (Uppsala University). They thank National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for assisting in massive parallel sequencing and computational infrastructure. This research was conducted using data made available by The Children's Brain Tumor Tissue Consortium (CBTTC). Work performed at NGI/Uppsala Genome Center was funded by RFI/VR and the Science for Life Laboratory, Sweden. Single-cell sequencing and methylation profiling was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. U. Schüller was further supported by the Fördergemeinschaft Kinderkrebszentrum Hamburg and XC by the Swedish Research Council (VR-2017–02074).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
S. Hutter reports a patent for Predicting cancer relapse and treatment of cancer diseases (WO 2022/050885) pending. M. Cancer reports other support from AstraZeneca AB outside the submitted work. G. Giraud reports grants from Barncancerfonden during the conduct of the study. O. Sangfelt reports grants from The Swedish Childhood Cancer Fund, The Swedish Cancer Society, The Swedish Research Council, and The Cancer Research Funds of Radiumhemmet during the conduct of the study. H. Weishaupt reports grants from The Swedish Childhood Cancer Fund and Worldwide Cancer Research (AICR) during the conduct of the study. F.J. Swartling reports grants from The Swedish Childhood Cancer Fund, The Swedish Cancer Society, The Swedish Research Council, The Swedish Society of Medicine, The European Research Council, Åke Wiberg´s Foundation, Ragnar Söderberg´s Foundation, Worldwide Cancer Research, The Science for Life Laboratory, Sweden; and other support from The Knut and Alice Wallenberg Foundation during the conduct of the study; and also has a patent for Predicting cancer relapse and treatment of cancer diseases (WO 2022/050885) pending. No disclosures were reported by the other authors.
Authors' Contributions
A. Borgenvik: Conceptualization, data curation, validation, investigation, visualization, methodology, project administration, writing–review and editing. K.O. Holmberg: Conceptualization, data curation, validation, investigation, visualization, methodology, writing–review and editing. S. Bolin: Conceptualization, data curation, validation, investigation, visualization, methodology, writing–original draft. M. Zhao: Data curation, validation, investigation, visualization, methodology, writing–review and editing. V. Savov: Data curation, investigation, visualization, methodology, writing–original draft. G. Rosén: Data curation, formal analysis, validation, investigation, visualization, methodology, project administration. S. Hutter: Data curation, validation, investigation, methodology. A. Garancher: Resources, validation, investigation, methodology. A.S. Rahmanto: Data curation, investigation, methodology. T. Bergström: Validation, investigation, methodology, project administration. T.K. Olsen: Data curation, validation, investigation, visualization, methodology. O.J. Mainwaring: Investigation, methodology. D. Sattanino: Formal analysis. A.D. Verbaan: Formal analysis. J.M. Rusert: Resources, investigation. A. Sundström: Data curation, formal analysis, validation, methodology, project administration. M.B. Bravo: Formal analysis. Y. Dang: Data curation, formal analysis, methodology. A.S. Wenz: Data curation, formal analysis, methodology. S. Richardson: Data curation, validation, investigation, methodology. G. Fotaki: Formal analysis. R.M. Hill: Resources, data curation, investigation, methodology. A.M. Dubuc: Data curation, investigation. A. Kalushkova: Formal analysis, validation, methodology. M. Remke: Data curation, investigation. M. Čančer: Investigation, methodology. H. Jernberg-Wiklund: Resources, methodology. G. Giraud: Investigation, writing–review and editing. X. Chen: Data curation, validation, methodology. M.D. Taylor: Resources, validation, methodology. O. Sangfelt: Data curation, validation, methodology. S.C. Clifford: Resources, data curation, validation. U. Schüller: Resources, validation. R.J. Wechsler-Reya: Resources, funding acquisition, investigation, methodology. H. Weishaupt: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–review and editing. F.J. Swartling: Conceptualization, resources, data curation, supervision, funding acquisition, investigation, visualization, writing-original draft, project administration, writing–review and editing.
References
- 1. Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol 2013;14:1200–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morrissy AS, Garzia L, Shih DJ, Zuyderduyn S, Huang X, Skowron P, et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016;529:351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012;123:465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hovestadt V, Ayrault O, Swartling FJ, Robinson GW, Pfister SM, Northcott PA. Medulloblastomics revisited: biological and clinical insights from thousands of patients. Nat Rev Cancer 2020;20:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar R, Smith KS, Deng M, Terhune C, Robinson GW, Orr BA, et al. Clinical outcomes and patient-matched molecular composition of relapsed medulloblastoma. J Clin Oncol 2021;39:807–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Richardson S, Hill RM, Kui C, Lindsey JC, Grabovksa Y, Keeling C, et al. Emergence and maintenance of actionable genetic drivers at medulloblastoma relapse. Neuro Oncol 2022;24:153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hill RM, Kuijper S, Lindsey JC, Petrie K, Schwalbe EC, Barker K, et al. Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease. Cancer Cell 2015;27:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 2003;17:1677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, et al. SOX9 induces and maintains neural stem cells. Nat Neurosci 2010;13:1181–9. [DOI] [PubMed] [Google Scholar]
- 10. Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell 2016;165:45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res 2012;72:1301–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suryo Rahmanto A, Savov V, Brunner A, Bolin S, Weishaupt H, Malyukova A, et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J 2016;35:2192–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 2012;21:601–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Remke M, Hielscher T, Korshunov A, Northcott PA, Bender S, Kool M, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol 2011;29:3852–61. [DOI] [PubMed] [Google Scholar]
- 15. Cavalli FMG, Remke M, Rampasek L, Peacock J, Shih DJH, Luu B, et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 2017;31:737–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, et al. Pleiotropic role for MYCN in medulloblastoma. Genes Dev 2010;24:1059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 2012;482:529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer M, Hutter S, Holmberg KO, Rosen G, Sundstrom A, Tailor J, et al. Humanized stem cell models of pediatric medulloblastoma reveal an Oct4/mTOR axis that promotes malignancy. Cell Stem Cell 2019;25:855–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res 2014;20:912–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods 2016;13:1013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamayo P, Scanfeld D, Ebert BL, Gillette MA, Roberts CW, Mesirov JP. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci U S A 2007;104:5959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Felmeister AS, Masino AJ, Rivera TJ, Resnick AC, Pennington JW. The biorepository portal toolkit: an honest brokered, modular service oriented software tool set for biospecimen-driven translational research. BMC Genomics 2016;17Suppl 4:434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 27. Wang X, Dubuc AM, Ramaswamy V, Mack S, Gendoo DM, Remke M, et al. Medulloblastoma subgroups remain stable across primary and metastatic compartments. Acta Neuropathol 2015;129:449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lafay-Cousin L, Smith A, Chi SN, Wells E, Madden J, Margol A, et al. Clinical, pathological and molecular characterization of infant medulloblastomas treated with sequential high-dose chemotherapy. Pediatr Blood Cancer 2016;63:1527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, et al. Quiescent Sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell 2014;26:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, et al. Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 2014;82:545–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu J, Margol AS, Shukla A, Ren X, Finlay JL, Krieger MD, et al. Disseminated medulloblastoma in a child with germline BRCA2 6174delT mutation and without Fanconi anemia. Front Oncol 2015;5:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019;572:74–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu APY, Smith KS, Kumar R, Paul L, Bihannic L, Lin T, et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell 2021;39:1519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krentz NAJ, van Hoof D, Li Z, Watanabe A, Tang M, Nian C, et al. Phosphorylation of NEUROG3 links endocrine differentiation to the cell cycle in pancreatic progenitors. Dev Cell 2017;41:129–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012;148:1015–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol 2005;6:1245–52. [DOI] [PubMed] [Google Scholar]
- 37. Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000;192:1027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garzia L, Kijima N, Morrissy AS, De Antonellis P, Guerreiro-Stucklin A, Holgado BL, et al. A hematogenous route for medulloblastoma leptomeningeal metastases. Cell 2018;172:1050–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gholamin S, Mitra SS, Feroze AH, Liu J, Kahn SA, Zhang M, et al. Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci Transl Med 2017;9:eaaf2968. [DOI] [PubMed] [Google Scholar]
- 40. Kahn SA, Wang X, Nitta RT, Gholamin S, Theruvath J, Hutter G, et al. Notch1 regulates the initiation of metastasis and self-renewal of Group 3 medulloblastoma. Nat Commun 2018;9:4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Klughammer J, Kiesel B, Roetzer T, Fortelny N, Nemc A, Nenning KH, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med 2018;24:1611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 2012;123:473–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang L, He X, Liu X, Zhang F, Huang LF, Potter AS, et al. Single-cell transcriptomics in medulloblastoma reveals tumor-initiating progenitors and oncogenic cascades during tumorigenesis and relapse. Cancer Cell 2019;36:302–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adolphe C, Millar A, Kojic M, Barkauskas DS, Sundstrom A, Swartling FJ, et al. SOX9 defines distinct populations of cells in SHH medulloblastoma but is not required for math1-driven tumor formation. Mol Cancer Res 2021;19:1831–9. [DOI] [PubMed] [Google Scholar]
- 45. Guo D, Wang Y, Cheng Y, Liao S, Hu J, Du F, et al. Tumor cells generate astrocyte-like cells that contribute to SHH-driven medulloblastoma relapse. J Exp Med 2021;218:e20202350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sosa MS, Parikh F, Maia AG, Estrada Y, Bosch A, Bragado P, et al. NR2F1 controls tumour cell dormancy via SOX9- and RARbeta-driven quiescence programmes. Nat Commun 2015;6:6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dang MT, Gonzalez MV, Gaonkar KS, Rathi KS, Young P, Arif S, et al. Macrophages in SHH subgroup medulloblastoma display dynamic heterogeneity that varies with treatment modality. Cell Rep 2021;34:108917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Siebel C, Wurthwein G, Lanvers-Kaminsky C, Andre N, Berthold F, Castelli I, et al. Can we optimise doxorubicin treatment regimens for children with cancer? Pharmacokinetic simulations and a Delphi consensus procedure. BMC Pharmacol Toxicol 2020;21:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Macdonald J, Denoyer D, Henri J, Jamieson A, Burvenich IJG, Pouliot N, et al. Bifunctional aptamer-doxorubicin conjugate crosses the blood-brain barrier and selectively delivers its payload to EpCAM-positive tumor cells. Nucleic Acid Ther 2020;30:117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure Legends
Supplementary figure 1
Supplementary figure 2
Supplementary figure 3
Supplementary figure 4
Supplementary figure 5
List of genes that are part of a quiescent cell signature and display regions of differentially opened chromatin.
Results of differential gene expression analysis between GTML tumours and GTS tumours.
Results of differential gene expression analysis between GTML3 tumor cells treated for 48 hours with DMSO or Dox.
Results of differential gene expression analysis between pre-GTS1 tumor cells treated for 48 hours with DMSO or Dox.
Resource Table
Data Availability Statement
Raw and processed sequencing data have been deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) and are available via the accession number GSE162080.



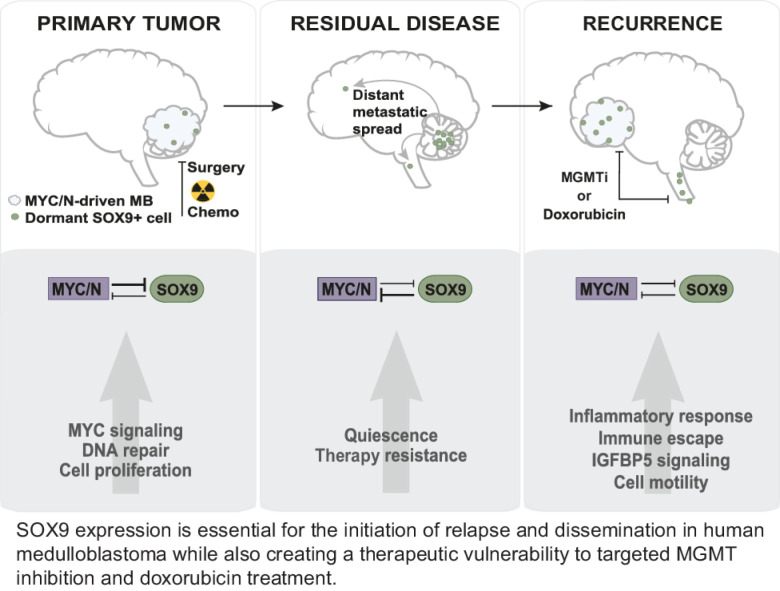
![Fig. 1. SOX9 is induced in therapy-resistant brain tumor cells and accumulates during relapse. A, Expression of Sox9 in GTML tumor cells after 48 hours dox treatment (Student t test; P = 0.0059**). B, (top) IHC showing SOX9 in GTML tumors after 6 hours, 7 days, and 30 days of dox administration in vivo, as well as an untreated GTML tumor. (Bottom) IF showing SOX9 and Ki67 in GTML tumors after 6 hours, 7 days, and 30 days of dox administration in vivo, as well as an untreated GTML tumor. The analysis shows an increase of non-proliferating SOX9 expressing cells in the GTML model (see also quantification in Supplementary Fig. 1B). C, Quantification of GFP-positive cells in of GTML-pSOX9-GFP using cell sorting. The analysis shows that 24 hours cisplatin treatment increases the number of SOX9-positive cells compared with DMSO control (Student t test comparing the percentage of GFP-positive cells between DMSO and cisplatin treatment; ****, P < 0.0001). D, Quantification of SOX9 positive cells in tumors from transplanted GTML2 cells, irradiated with 2 Gy/day, for 3.5 weeks (total 36 Gy). Tumors from mice that took the longest to relapse had more SOX9 positive cells (one-way ANOVA; ****, P < 0.0001; see also Supplementary Fig. 1I for representative IHC images). E, Representative images of IHC SOX9 staining of primary and recurrent tumors of all four MB subgroups. F, Quantification of € SOX9-positive cells in WNT (n = 1), SHH (n = 6 as well as one re-operated relapse), Group 3 (n = 6 as well as one re-operated relapse), and Group 4 (n = 6) MB. The number of SOX9-positive cells is significantly higher in recurrent Group 3 (****, P < 0.0001) and Group 4 MB (**, P = 0.00113) compared with the primary tumor [Student t test, recurrence II (n = 1) is not included in the analysis]. G, Kaplan–Meier plot showing 10-year overall survival in Group 3 MB with high SOX9 levels (n = 28) and low SOX9 levels (n = 78; from ref. 15) at last quartile cut-off, excluding MB with extensive nodularity or tumors with desmoplastic histology. High SOX9 levels correlate to poor overall survival of Group 3 MB. H, Percentage of SOX9 levels in low (M0–M1) and high (M3) stage metastatic Group 3 MB as well as recurrent tumors of the same subgroup from (12). SOX9 protein levels are higher in recurrent Group 3 MB than in primary M0–M1 stage tumor (Student t test; *, P = 0.0161). See also Supplementary Fig. S1.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/2cb6/9755969/0383799e4faf/4586fig1.jpg)