Abstract
Individual bacteria of numerous species can communicate and coordinate their actions via the production, release, and detection of extracellular signaling molecules. In this study, we used the Vibrio harveyi luminescence bioassay to determine whether Helicobacter pylori produces such a factor. Cell-free conditioned media from H. pylori strains 60190 and 26695 each induced >100-fold-greater luminescence in V. harveyi than did sterile culture medium. The H. pylori signaling molecule had a molecular mass of <10 kDa, and its activity was unaffected by heating to 80°C for 5 min or protease treatment. The genome sequence of H. pylori 26695 does not contain any gene predicted to encode an acyl homoserine lactone synthase but does contain an orthologue of luxS, which is required for production of autoinducer-2 (AI-2) in V. harveyi. To evaluate the role of luxS in H. pylori, we constructed luxS null mutants derived from H. pylori 60190 and 26695. Conditioned media from the wild-type H. pylori strains induced >100-fold-greater luminescence in the V. harveyi bioassay than did conditioned medium from either mutant strain. Production of the signaling molecule was restored in an H. pylori luxS null mutant strain by complementation with a single intact copy of luxS placed in a heterologous site on the chromosome. In addition, Escherichia coli DH5α produced autoinducer activity following the introduction of an intact copy of luxS from H. pylori. Production of the signaling molecule by H. pylori was growth phase dependent, with maximal production occurring in the mid-exponential phase of growth. Transcription of H. pylori vacA also was growth phase dependent, but this phenomenon was not dependent on luxS activity. These data indicate that H. pylori produces an extracellular signaling molecule related to AI-2 from V. harveyi. We speculate that this signaling molecule may play a role in regulating H. pylori gene expression.
In the past decade, there has been considerable progress in our understanding of intercellular communication among bacteria. In one form of intercellular communication, termed quorum sensing, bacteria release extracellular signaling molecules (autoinducers), which accumulate as the population grows. When the extracellular signaling molecule reaches a critical threshold concentration, a signal transduction cascade is triggered within each cell of the population, the final result being an alteration in gene expression (for reviews, see references 15, 19, and 21). This altered pattern of gene expression presumably increases the capacity of bacteria to survive environmental changes that accompany increased cell density. In various bacterial species, autoinducer-mediated regulation of gene expression controls diverse processes, including sporulation (27), genetic competence (2, 28), antibiotic synthesis (40), alternative sigma factor synthesis (26), the production of virulence determinants (11, 22, 48), and even the production of other quorum-sensing molecules (35).
One of the best studied of the autoinducer-controlled gene expression systems is the density-dependent bioluminescence of marine vibrios (Vibrio fischeri and V. harveyi). V. fischeri exists free in seawater, as well as in a symbiotic relationship within the light organ of squid (38). V. fischeri produces and releases an N-acyl homoserine lactone (AHL) signaling molecule. Transcription of the luciferase operon (luxCDABEGH) of V. fischeri is repressed in the absence of a threshold level of AHL. When cell density, and hence AHL level, reaches a threshold concentration, the LuxR regulator is inactivated, and bioluminescence occurs (6). Many gram-negative bacterial species produce related AHL molecules, but the AHL-mediated quorum-sensing systems are typically quite species specific. The production of AHL molecules in gram-negative bacteria is catalyzed by AHL synthases, which are usually encoded by luxI orthologues.
V. harveyi, a free-living marine bacterium, produces two different quorum-sensing molecules, designated autoinducer-1 (AI-1) and AI-2, which are each capable of regulating luciferase activity. V. harveyi AI-1 has been identified as hydroxybutanoyl-l-homoserine lactone (13). The chemical structure of the second signaling molecule (AI-2) is not known, but the luxS gene is required for its production (44). The presence of AI-2 is detected by a sensory histidine kinase (encoded by luxQ) located within the cytoplasmic membrane of V. harveyi (6). Many strains of Escherichia coli and Salmonella enterica serovar Typhimurium contain luxS orthologues and produce autoinducer molecules that are functionally similar to V. harveyi AI-2 (42–44). Surette et al. (44) have noted that luxS orthologues are present in many additional bacterial species and have suggested that many of these species may produce extracellular signaling molecules.
Helicobacter pylori is a curved, gram-negative bacterium found associated with the gastric epithelium of humans and other primates. Colonization of the human stomach with H. pylori consistently results in the development of gastric mucosal inflammation and is a risk factor for the development of peptic ulcer disease and gastric adenocarcinoma (10, 14, 25, 33). In the present report, we describe the production of an extracellular signaling molecule by H. pylori and report that H. pylori luxS is essential for this activity.
MATERIALS AND METHODS
Bacteria and culture conditions.
Wild-type H. pylori strains and all mutant strains used in this study are listed in Table 1. H. pylori strains were routinely cultured at 37°C in ambient air containing 5% CO2. V. harveyi BB152 (luxL::Tn5) is capable of producing AI-2 but not AI-1, and strain BB170 (luxN::Tn5) is capable of sensing AI-2 but not AI-1 (7, 42). V. harveyi strains were grown at 25°C on Luria-marine agar plates (5) or in autoinducer bioassay (AB) medium (20). V. harveyi strains were kind gifts of B. Bassler (Princeton University).
TABLE 1.
H. pylori strains used in this study
| Strain (parent strain) | Genotype | Urease activity | Source or reference |
|---|---|---|---|
| 60190 | Wild type | + | ATCC 49503 |
| 26695 | Wild type | + | 46 |
| L60-1 (60190) | luxS::CAT | + | This study |
| L60-2 (60190) | luxS::CAT ureA::luxS-aphA3 | − | This study |
| L26-1 (26695) | luxS::CAT | + | This study |
| L26-2 (26695) | luxS::CAT ureA::luxS-aphA3 | − | This study |
| 60190 VX-1 | vacA::xylE-aphA3 | + | 17 |
| 60190 VX-1/LC-1 | vacA::xylE-aphA3 luxS::CAT | + | This study |
Generation of cell-free CM and V. harveyi luminescence bioassay.
H. pylori strains were cultured in brucella broth supplemented with either 5% fetal bovine serum (FBS) or 0.5% charcoal (12). Cell density was monitored by readings of optical density at 600 nm (OD600). Cell-free conditioned media (CM) were prepared by centrifuging H. pylori cultures at 8,000 × g followed by filtration of the supernatant through 0.2-μm-pore-size filters. CM preparations were routinely stored frozen at −70°C. CM preparations from V. harveyi strains were prepared in the same manner, except that the cells were cultured in AB medium at 25°C.
The V. harveyi luminescence bioassay was performed essentially as described by Surette and Bassler (42). Briefly, an overnight culture of V. harveyi BB170 was diluted 1:5,000 into fresh AB medium. Experimental CM preparations were added to the diluted V. harveyi culture at a 10% (vol/vol) final concentration. Aliquots of 1 ml were removed at various time points, and total luminescence was quantified using a luminometer (ALL 2010; Analytical Luminescence Laboratory). Viable cell counts (CFU per milliliter) of V. harveyi were determined at each time point by serial dilution, and relative light units were calculated as total luminescence per 106 V. harveyi cells.
Molecular biology techniques.
Extraction of H. pylori genomic DNA, cloning, plasmid preparation, restriction enzyme digestion, repair of 5′ and 3′ overhangs, and PCR protocols were performed essentially as previously described elsewhere (4, 17, 18).
Generation of H. pylori luxS null mutants.
luxS from H. pylori 26695 genomic DNA was amplified using primers 5′ GCGGACATTGTGGCACATAGCGGC and 5′ CTATTGCCTTGCAACAAATCCCCGC, which were derived from the sequence of H. pylori 26695 (46). The 1,490-bp amplicon was cloned into pGEM-T (Promega). The chloramphenicol acetyltransferase (CAT) gene from Campylobacter coli (47) was ligated into the unique XcmI site within the cloned H. pylori 26695 luxS sequence. Disruption of the chromosomal luxS gene in H. pylori 26695 or 60190 was accomplished by natural transformation, allelic exchange, and screening for chloramphenicol-resistant H. pylori clones as previously described (17). The resulting luxS mutant strains derived from H. pylori 26695 and 60190 were designated L26-1 and L60-1, respectively. PCR analysis of genomic DNA indicated the orientation of the CAT cassettes and demonstrated that a double-crossover event had occurred in each mutant (data not shown). To assay for the presence of intracellular autoinducer, luxS H. pylori cells were disrupted by sonication (Virsonic 60; Virtis Company, Gardiner, N.Y.), using three 30-s pulses on ice with 1-min cooling intervals between pulses. The membrane fraction was removed by centrifugation at 10,000 × g for 30 min, and the resulting cytoplasmic fraction was assayed for autoinducer activity.
luxS complementation studies.
For complementation studies, we used suicide plasmid pAD-1, which the H. pylori ureA sequence with unique restriction sites for XbaI and SmaI in close proximity to the ureA ribosome binding site and initiation codon (3). This plasmid is designed such that cloned sequences can be placed within the urease locus on the H. pylori chromosome (resulting in a urease-negative phenotype), and the cloned sequence will then be transcribed under the control of the ureA promoter and translated using the ureA ribosome binding site (3). luxS was amplified from H. pylori genomic DNA using primers 5′ TGCTTAGATGAAAACACCAAAAATGAATGTAGAGAG and 5′ TCCCCCGGGTCAAACCCCCACTTCAGACCAC, which contain restriction sites for XbaI and SmaI, respectively, at the 5′ ends. The amplicon, which contained the entire luxS open reading frame (ORF), was digested with XbaI and SmaI and cloned into pAD-1. A selectable marker, aphA3 from pUC4K (Amersham Pharmacia), was then cloned into the SmaI site, and the resulting plasmid (pAD/luxS-aphA3) was used to transform H. pylori luxS null mutant strains (L60-1 and L26-1). H. pylori colonies were screened for both chloramphenicol and kanamycin resistance. The resulting urease-negative mutants were designated L60-2 and L26-2, respectively.
RESULTS
H. pylori produces an extracellular signaling molecule.
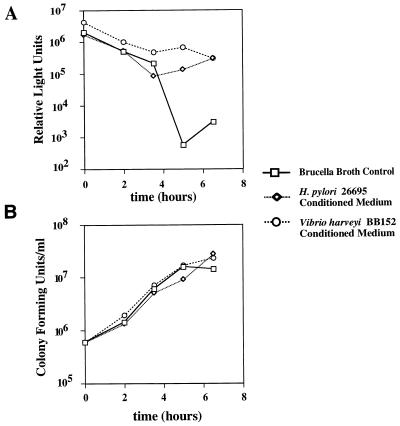
Many strains of E. coli and S. enterica serovar Typhimurium produce and release autoinducers that can induce bioluminescence in V. harveyi (7). This led us to speculate that H. pylori may produce a similar signaling molecule. To test this hypothesis, we used a V. harveyi reporter strain, BB170 (kind gift from B. Bassler), which does not have a functional sensor for AI-1 (AHL) but has an intact sensor for AI-2. V. harveyi BB152 is capable of producing AI-2, but not AI-1, and was thus used as a source of homologous AI-2 in the bioassay. Figure 1A shows the result of a typical experiment. When inoculated into AB medium containing 10% sterile brucella broth, V. harveyi BB170 produces a strong luminescent signal for about 4 h, and the levels of luminescence decrease markedly by the 5-h time point. In contrast, when inoculated into AB medium containing 10% CM from V. harveyi BB152 (a supplemental source of AI-2), V. harveyi BB170 maintains a high level of luminescence at the 5-h time point. These results confirm data previously reported by Surette and Bassler (42) and illustrate that the 5-h time point is appropriate for monitoring the effects of exogenous AI-2 in this assay.
FIG. 1.
Production of an extracellular signaling molecule by H. pylori. V. harveyi BB170 was inoculated into AB medium containing 10% CM from V. harveyi BB152, 10% CM from H. pylori 26695, or 10% sterile brucella broth. Cultures were incubated at 25°C with constant agitation, and aliquots were removed at serial time points for measurement of luminescence (A) and viable cell count (B). Luminescence is expressed in relative light units (luminescence per 106 viable V. harveyi BB170 cells). At the 5-h time point, CM from V. harveyi BB152 and H. pylori 26695 induced >100-fold-greater luminescence than did sterile brucella broth.
To determine whether H. pylori produces an extracellular signaling molecule, we inoculated V. harveyi BB170 into AB medium containing 10% CM from H. pylori 26695. Under these conditions, the level of V. harveyi luminescence was maintained at the 5-h time point (Fig. 1A). Similar results were obtained when V. harveyi BB170 was inoculated into AB medium containing 10% CM from H. pylori 60190 (data not shown). Regardless of whether prepared from brucella broth containing 5% FBS or 0.5% charcoal, H. pylori CM stimulated V. harveyi luminescence (data not shown). Differences in the luminescence of V. harveyi cultures assayed in Fig. 1A could not be attributed to variation in growth rates or bacterial cell densities (Fig. 1B). These data suggest that H. pylori produces an extracellular signaling molecule that acts on V. harveyi.
H. pylori contains a luxS orthologue.
The recent discovery by Surette et al. (44) that the luxS gene is essential for AI-2 production in V. harveyi, S. enterica serovar Typhimurium, and E. coli, prompted us to search the genomes of two sequenced strains of H. pylori (26695 and J99) for luxS orthologues. Both genomes contain luxS orthologues, designated ORF HP 0105 and JHP 0097, respectively (1, 46). The predicted primary structures of LuxS proteins from these two H. pylori strains are 95% identical. LuxS from H. pylori is most closely related to LuxS proteins found in gram-positive species (Staphylococcus aureus [67% amino acid identity], Bacillus subtilis [45% amino acid identity], and Clostridium perfringens [44% amino acid identity]) (Fig. 2). The flanking gene arrangements are widely divergent among most of these species. However, in both H. pylori and C. perfringens, luxS is linked to two genes (cysK and metB) that are involved in amino acid biosynthesis. Neither of these genes is linked to luxS loci in the other species examined to date.
FIG. 2.
Alignment of the deduced H. pylori LuxS sequence with deduced LuxS sequences from four other bacterial species. LuxS sequences from H. pylori 26695 (GenBank accession no. AE000532), S. aureus (preliminary sequence data obtained from The Institute for Genomic Research website at http://www.tigr.org/), B. subtilis (accession no. Z9919), C. perfringens (accession no. AB028629), and V. harveyi (accession no. AAD 17292) were aligned using the ClustalW algorithm. H. pylori LuxS is most closely related to LuxS from S. aureus (67% amino acid identity; 15% similarity). Positions of amino acid identity are indicated by asterisks.
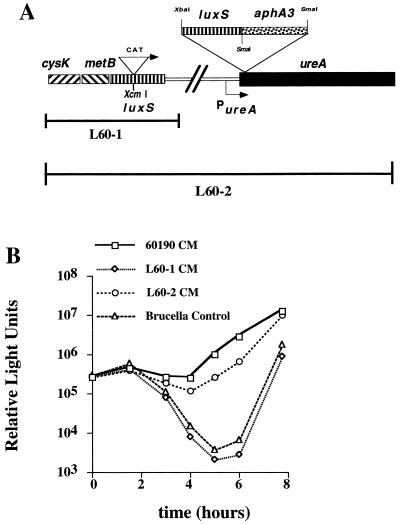
To investigate the function of the H. pylori luxS gene product, we constructed H. pylori mutants in which the luxS gene was disrupted by insertional mutagenesis. Figure 3 shows a V. harveyi bioassay comparing CM from wild-type H. pylori 60190 and CM from an isogenic luxS mutant, L60-1. CM from wild-type H. pylori 60190 induced >100-fold-higher levels of luminescence than did CM from H. pylori L60-1. Similarly, CM from wild-type H. pylori 26695 induced >100-fold-higher levels of luminescence than did CM from H. pylori 26695 containing a null mutation in luxS (data not shown).
FIG. 3.
Role of H. pylori luxS in the production of an extracellular signaling molecule. (A) H. pylori L60-1 is a luxS null mutant strain in which luxS has been disrupted by the insertion of a CAT cassette. H. pylori L60-2 contains the same luxS mutation as in L60-1, but an intact copy of luxS has been inserted within the ureA gene. (B) V. harveyi BB170 was inoculated into AB medium containing 10% CM from H. pylori 60190 (wild type), 10% CM from H. pylori L60-1 (60190 luxS::CAT), 10% CM from H. pylori L60-2 (60190 luxS::CAT ureA::luxS-aphA3), or 10% brucella broth. Comparison of V. harveyi luminescence values at the 5-h time point demonstrates that wild-type H. pylori 60190 CM induces 100-fold-greater luminescence than does either H. pylori L60-1 CM or the brucella broth control. At the 5-h time point, CM from the luxS-complemented H. pylori strain, L60-2, induces approximately 100-fold-greater luminescence than does CM from H. pylori L60-1.
In an effort to rescue the signaling phenotype in an H. pylori luxS null mutant strain under single-copy replacement conditions, we used a recently developed H. pylori suicide vector that allows the replacement of single-copy genes under the control of the strong ureA promoter (3). As shown in Fig. 3, CM from the complemented mutant strain 60190 luxS::CAT/ureA::luxS-aphA3 (L60-2) induced nearly 100-fold-greater luminescence than did CM from L60-1 or medium controls. Thus, the luxS gene is essential for the synthesis of an extracellular signaling molecule in H. pylori. The CAT cassette used for insertional mutagenesis of luxS in H. pylori L60-1 contains a sequence that is predicted to be a rho-independent terminator. Therefore, these data also suggest that genes immediately downstream from luxS are not required for production of the signaling molecule.
To test whether H. pylori luxS mutants might be defective in secretion of the signaling molecule, H. pylori wild-type and luxS mutant cells were each lysed by sonication, membrane fractions were removed by centrifugation, and the cytoplasmic fractions were tested for autoinducer activity in the V. harveyi BB170 bioassay. Autoinducer activity was detectable in the wild-type H. pylori cytoplasmic extracts but not in extracts from the luxS mutant cells (data not shown). This suggests that a functional H. pylori LuxS is required for synthesis of the signaling molecule, rather than for its secretion or release.
Characteristics of the H. pylori signaling molecule.
Experiments utilizing a 10-kDa-cutoff ultrafiltration membrane (Amicon) demonstrated that the <10-kDa fraction of H. pylori CM induced luminescence in V. harveyi BB170, whereas the >10-kDa fraction had no effect (data not shown). Incubation of H. pylori CM preparations with proteinase K (Sigma) had no effect on the autoinducer activity. The autoinducer activity in CM from H. pylori was almost completely abrogated by treatment at 100°C for 5 min. In contrast, treatment at 80°C for the same length of time had no effect on the activity of the signaling molecule (data not shown). These data indicate that the H. pylori signaling molecule is a small (<10-kDa) heat-resistant molecule that is likely not proteinaceous.
Production of the H. pylori signaling molecule is growth phase dependent.
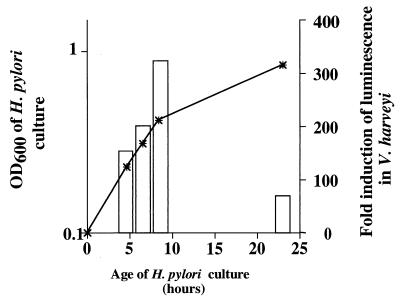
To examine the kinetics of autoinducer production in H. pylori, we prepared CM at serial time points reflecting all phases of growth from broth cultures of H. pylori 60190. The maximal autoinducer activity was detected in CM preparations from mid-logarithmic-phase cultures (Fig. 4). Levels of autoinducer activity were markedly reduced in CM prepared from stationary-phase H. pylori cultures (Fig. 4). This pattern of results was very reproducible, but the absolute values of luminescence varied considerably among individual experiments. The loss of autoinducer activity in stationary-phase cultures suggests that either the H. pylori signaling molecule is labile or H. pylori cells are capable of degrading the autoinducer.
FIG. 4.
Kinetics of H. pylori signaling molecule production. H. pylori 60190 was cultured in brucella broth containing 5% FBS, and aliquots were removed at serial time points for measurement of cell density (OD600) (asterisks) and the capacity of CM to induce luminescence in the V. harveyi BB170 bioassay system at the 5-h time point (open bars). Luminescence induction is reported as a ratio of relative light units in the presence of H. pylori 60190 CM compared to relative light units in the presence of uninoculated brucella broth. The results shown here are representative of results obtained in three independent experiments.
Growth phase-dependent regulation of vacA transcription.
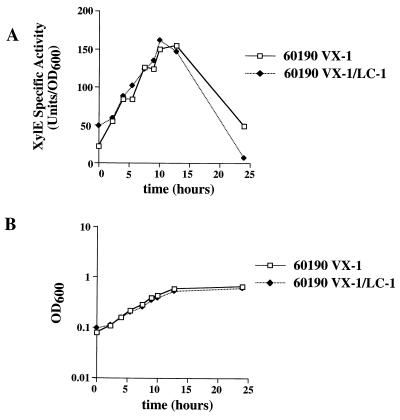
In previous studies (M. H. Forsyth and T. L. Cover, unpublished data), we have noted that the transcription of H. pylori vacA (encoding a vacuolating cytotoxin) is dependent on the bacterial growth phase. Figure 5 shows the results of a typical experiment in which vacA transcription was monitored using a reporter strain (60190 VX-1) containing a vacA::xylE transcriptional fusion (17). Levels of XylE activity are low during the early, low-cell-density portions of the growth curve. XylE activity progressively increases as cultures proceed into exponential, higher-cell-density phases of growth. Peak levels of vacA transcription are reached at the onset of stationary phase and subsequently decline over time.
FIG. 5.
Growth phase regulation of H. pylori vacA transcription. H. pylori 60190 VX-1 (containing a vacA::xylE transcriptional fusion) and H. pylori VX-1/LC-1 (containing both a vacA::xylE transcriptional fusion and a luxS::CAT mutation) were grown in brucella broth containing 5% FBS. Aliquots were removed at serial time points for measurement of XylE specific activity (A) and cell density (OD600) (B). Levels of vacA transcription, as evidenced by XylE activities, were growth phase dependent, but this phenomenon was not dependent on LuxS function.
Because this pattern of vacA transcription so closely mirrored the demonstrated kinetics of the H. pylori signaling molecule production (Fig. 4), we investigated the possibility that vacA transcription may be regulated through a luxS-dependent signaling mechanism. To test this possibility, we introduced the luxS::CAT mutation into H. pylori 60190 VX-1 (thus creating 60190 VX-1/LC-1). Both 60190 VX-1 and 60190 VX-1/LC-1 grew at approximately the same rate and to approximately the same density (Fig. 5B). XylE specific activities of these two strains did not differ significantly (Fig. 5A), which indicated that the growth phase regulation of vacA transcription is not dependent on LuxS activity.
Activity of recombinant H. pylori LuxS expressed in E. coli DH5α.
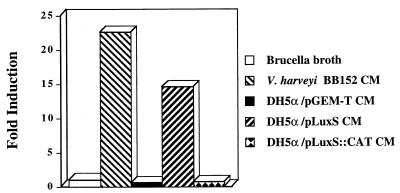
Many pathogenic E. coli strains produce an AI-2-like activity (42) that can be detected using the V. harveyi bioassay. However, the commonly used laboratory strain of E. coli, DH5α, fails to produce measurable amounts of autoinducer due to a frameshift mutation in the 3′ portion of the luxS ORF (44). To examine the function of H. pylori luxS in E. coli, we cloned the luxS orthologue of H. pylori into pGEM-T (Promega) and introduced this plasmid (pLuxS) into E. coli DH5α. Conditioned media from E. coli DH5α containing pGEM-T (without H. pylori sequence) lacked autoinducer activity in the V. harveyi bioassay, whereas CM from E. coli DH5α containing pLuxS induced much higher levels of luminescence (Fig. 6). As expected, disruption of the cloned H. pylori luxS gene, accomplished by inserting a CAT cassette into the unique XcmI site, resulted in the loss of autoinducer production. These data are further confirmation that H. pylori luxS plays an essential role in the production of an extracellular signaling molecule.
FIG. 6.
H. pylori luxS complements the luxS frameshift mutation of E. coli DH5α. E. coli DH5α, which bears a nonfunctional luxS (44), was transformed with a plasmid containing the intact H. pylori luxS gene (pLuxS), a plasmid containing a disrupted H. pylori luxS gene (pLuxS::CAT), or the cloning vector alone (pGEM-T). Each strain was grown in brucella broth to mid-logarithmic phase (OD600 of ∼0.5), and cell-free CM were prepared. The CM were tested for the capacity to induce luminescence in the V. harveyi bioassay at the 5-h time point. Sterile, pristine brucella broth was used as a control, and all induction values are relative to this sample. CM from V. harveyi BB152 was used as a positive control for the bioassay. The DH5α strain bearing a functional H. pylori luxS (pLuxS) induced high levels of luminescence in V. harveyi BB170. In contrast, DH5α strains bearing either a nonfunctional luxSHp (pLuxS::CAT) or the vector alone were incapable of inducing luminescence in this bioassay. Results shown are representative of three independent experiments.
DISCUSSION
The capacity of individual bacteria to regulate gene expression in response to changes in bacterial cell density is known as quorum sensing. Several different families of bacterial quorum-sensing molecules have been described, including AHL derivatives (AI-1 molecules), small peptides, and AI-2 molecules (2, 15, 19). In the present study, we describe the production of a quorum-sensing molecule (autoinducer) by H. pylori. Several lines of evidence suggest that the H. pylori autoinducer is closely related to the AI-2 family of signaling molecules: (i) like the AI-2 molecules of V. harveyi, E. coli, and S. enterica serovar Typhimurium, the H. pylori autoinducer is a low-molecular-mass molecule that is relatively heat resistant; (ii) the H. pylori autoinducer is detectable by a V. harveyi reporter strain that responds to AI-2, but not AI-1, molecules; and (iii) a functional luxS orthologue is essential for the production of the H. pylori signaling molecule, as well as for production of AI-2 molecules in V. harveyi, E. coli, and S. enterica serovar Typhimurium.
The AI-2 family of quorum-sensing molecules has only been recently described (44), and there are many features of this quorum-sensing pathway that are not yet completely understood. In particular, the molecular composition of AI-2 has not yet been elucidated for any bacterial species. The luxS gene clearly seems to be essential for the production of this novel autoinducer (44), but it is not known whether LuxS possesses enzymatic activity or whether it serves some other function. H. pylori LuxS does not exhibit any obvious homology to known enzymes. If LuxS is an enzyme required for AI-2 synthesis, it is not known whether luxS alone is sufficient for synthesis of AI-2 or whether other genes are required. Moreover, nothing is known about the mechanisms whereby AI-2 is released or secreted into the extracellular milieu. As noted in this study, H. pylori luxS null mutants do not contain detectable cell-associated autoinducer activity, which suggests that LuxS is not required for secretion of the autoinducer.
In both E. coli and S. enterica serovar Typhimurium (42), as well as H. pylori, production of AI-2 is influenced by the growth phase of the bacteria. The mechanism of growth phase regulation of AI-2 production is not known. However, we speculate that there may be transcriptional regulation of luxS (or possibly other genes in the AI-2 biosynthetic pathway). In addition, there may be regulation of genes in an AI-2 degradative pathway.
Many bacterial species contain luxS orthologues (Fig. 2), and production of an AI-2 molecule has now been experimentally demonstrated in V. harveyi, E. coli, S. enterica serovar Typhimurium, and H. pylori. Whether these AI-2 molecules are chemically identical or exhibit some variation is not yet known. However, the capacity of AI-2 molecules from multiple species to induce luminescence in V. harveyi suggests that there is considerable structural similarity. If LuxS is indeed a unique synthase required for AI-2 production, potentially the expression of recombinant LuxS from various bacterial species in E. coli DH5α will facilitate the characterization and comparison of these various AI-2 autoinducers.
Although it seems likely that many bacterial species produce AI-2 molecules, the function of these molecules remains poorly understood. At present, these autoinducers are known to regulate the expression of luciferase in V. harveyi (6) and the expression of a type III secretion system in E. coli O157:H7 (41). However, the full complement of genes that might be regulated by AI-2 has not yet been explored. We consider it likely that AI-2 molecules may regulate gene expression in each of the bacterial species that produce them. If this hypothesis is correct, then mechanisms must be present for sensing and responding to these autoinducers. In V. harveyi, AI-2 is sensed by a histidine kinase (LuxQ) located within the cytoplasmic membrane (6). It seems likely that similar sensing systems may be operable in E. coli, S. enterica serovar Typhimurium, and H. pylori, but these systems have not yet been identified. The H. pylori genome sequence predicts the existence of several histidine kinases, but it is not known which, if any, of these has a functional activity corresponding to V. harveyi LuxQ.
The transcription of vacA (encoding H. pylori vacuolating cytotoxin) is growth phase dependent (Forsyth and Cover, unpublished data; this study), and the growth phase induction of vacA transcription seems to resemble the production kinetics of H. pylori AI-2. To test whether the growth phase regulation of vacA is dependent on H. pylori AI-2, we compared the transcription of vacA in wild-type and isogenic luxS mutant H. pylori strains. Inactivation of luxS did not alter vacA transcription, and therefore the mechanism of vacA growth phase regulation remains unknown. Growth phase regulation has been documented for virulence determinants in multiple bacterial species (24, 31, 32, 36) and is likely to be important in pathogenesis. We speculate that yet another quorum-sensing system may operate in H. pylori and that this system is the mediator the observed induction of vacA transcription.
Quorum-sensing systems mediated by AHL signaling molecules are typically quite species specific. In contrast, the AI-2 quorum-sensing systems of E. coli, S. enterica serovar Typhimurium (42–44), and H. pylori do not appear to be species specific. One possible role for such a nonspecific bacterial sensing system could be detection of total viable bacterial biomass as an indication of competition for resources. For example, alteration of gene expression in response to increasing bacterial density might be advantageous if it resulted in decreased utilization of nutrients or if it induced utilization of alternate nutrients.
Alternatively, this H. pylori signaling molecule may not regulate H. pylori gene expression, but may act on various other bacterial species. For example, some S. aureus strains produce signal molecules which interfere with quorum sensing in other S. aureus strains (23), and S. aureus may also produce a signaling molecule identical to that produced by Enterococcus faecalis (16, 34). Yet another alternative target for H. pylori signal molecules may be the cells of the gastric system itself. For example, Pseudomonas aeruginosa produces a signal molecule, OdHL, N-(3-oxododecanoyl)-l-homoserine lactone, that possesses immunomodulatory activity (45) and alters the expression of P2Y2 and P2Y4 receptors in tracheal gland cells (39). If gastric epithelial cells were responsive to an H. pylori signaling molecule, this might represent a bacterial strategy for altering the gastric environment to allow persistent H. pylori colonization (8, 9).
H. pylori is perhaps unique in terms of its place within the gastrointestinal ecosystem. While nearly the entire length of the gastrointestinal tract has an abundant and diverse microbial flora, H. pylori is often the sole species colonizing the gastric mucosa. Given the vast number of bacteria from countless species which pass through the stomach, many of which can colonize other regions of the alimentary canal, it is remarkable that H. pylori is nearly the only species to successfully colonize this niche. This may be due to unique properties that allow H. pylori to enter the gastric mucus layer and escape the very acidic pH of the gastric environment (29, 30). In addition, H. pylori produces bacteriocins that may act to prevent colonization by other bacterial species (37). We speculate that the capacity of H. pylori to produce and detect AI-2 may be an important component of the process by which bacterial overgrowth in the gastric mucosa is prevented.
ACKNOWLEDGMENTS
We thank Bonnie L. Bassler for her kind gifts of V. harveyi strains. We also thank Dawn Israel and Martin Blaser for their gift of pAD-1 as well as Mark McClain for productive discussions.
This work was supported by grant AI 39657 from the National Institutes of Health and by the Medical Research Service of the Department of Veterans Affairs.
REFERENCES
- 1.Alm R A, Ling L L, Moir D T, et al. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Alloing G, Granadel C, Morrison D A, Claverys J P. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol. 1996;21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 3.Ando T, Israel D A, Kusugami K, Blaser M J. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J Bacteriol. 1999;181:5572–5580. doi: 10.1128/jb.181.18.5572-5580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1994. [Google Scholar]
- 5.Bassler B L, Wright M, Showalter R E, Silverman M R. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 6.Bassler B L, Wright M, Silverman M R. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x. [DOI] [PubMed] [Google Scholar]
- 7.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser M J. Ecology of Helicobacter pylori in the human stomach. J Clin Investig. 1997;100:759–762. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaser M J, Kirschner D. Dynamics of Helicobacter pylori colonization in relation to the host response. Proc Natl Acad Sci USA. 1999;96:8359–8364. doi: 10.1073/pnas.96.15.8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blaser M J, Perez-Perez G I, Kleanthous H, Cover T L, Peek R M, Chyou P H, Stemmermann G N, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 11.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck G M, Smith J S. Medium supplementation for growth of Campylobacter pyloridis. J Clin Microbiol. 1987;25:597–599. doi: 10.1128/jcm.25.4.597-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J G, Meighen E A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 14.Cover T L, Blaser M J. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996;41:85–117. [PubMed] [Google Scholar]
- 15.Dunny G M, Winans S C. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 16.Firth N, Fink P D, Johnson L, Skurray R A. A lipoprotein signal peptide encoded by the staphylococcal conjugative plasmid pSK41 exhibits an activity resembling that of Enterococcus faecalis pheromone cAD1. J Bacteriol. 1994;176:5871–5873. doi: 10.1128/jb.176.18.5871-5873.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsyth M H, Atherton J C, Blaser M J, Cover T L. Heterogeneity in levels of vacuolating cytotoxin gene (vacA) transcription among Helicobacter pylori strains. Infect Immun. 1998;66:3088–3094. doi: 10.1128/iai.66.7.3088-3094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsyth M H, Cover T L. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. J Bacteriol. 1999;181:2261–2266. doi: 10.1128/jb.181.7.2261-2266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing gene regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg E P, Hastings J W, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 21.Hastings J W, Greenberg E P. Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J Bacteriol. 1999;181:2667–2668. [Google Scholar]
- 22.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;192:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji G, Beavis R C, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 24.Karita M, Tummuru M K R, Wirth H P, Blaser M J. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect Immun. 1996;64:4501–4507. doi: 10.1128/iai.64.11.4501-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labigne A, deReuse H. Determinants of Helicobacter pylori pathogenicity. Infect Agents Dis. 1996;5:191–202. [PubMed] [Google Scholar]
- 26.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 27.Li S, Lee B U, Shimkets L J. CsgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 28.Magnuson R, Solomon J, Grossman A D. Biochemical and genetic characterization of a competence pheromone from Bacillus subtilis. Cell. 1994;77:207–221. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 29.McGowan C C, Cover T L, Blaser M J. Helicobacter pylori and gastric acid: biological and therapeutic implications. Gastroenterology. 1996;110:926–938. doi: 10.1053/gast.1996.v110.pm8608904. [DOI] [PubMed] [Google Scholar]
- 30.McGowan C C, Necheva A, Thompson S A, Cover T L, Blaser M J. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol Microbiol. 1998;30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 31.Mellies J, Rudel T, Meyer T F. Transcriptional regulation of pilC2 in Neisseria gonorrhoeae: response to oxygen availability and evidence for growth-phase regulation in E. coli. Mol Gen Genet. 1997;255:285–293. doi: 10.1007/s004380050499. [DOI] [PubMed] [Google Scholar]
- 32.Mikulskis A V, Delor I, Thi V H, Cornelis G R. Regulation of the Yersinia enterocolitica Yst gene. Influence of growth phase, temperature, osmolarity, pH and bacterial host factors. Mol Microbiol. 1994;14:905–915. doi: 10.1111/j.1365-2958.1994.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 33.Mobley H L. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am J Med. 1996;100:25–115. doi: 10.1016/s0002-9343(96)80223-3. [DOI] [PubMed] [Google Scholar]
- 34.Muscholl-Silberhorn A, Samberger E, Wirth R. Why does Staphylococcus aureus secrete an Enterococcus faecalis-specific pheromone? FEMS Microbiol Lett. 1997;157:261–266. doi: 10.1111/j.1574-6968.1997.tb12782.x. [DOI] [PubMed] [Google Scholar]
- 35.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcriptional regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 37.Putsep K, Branden C I, Boman H G, Normark S. Antibacterial peptide from Helicobacter pylori. Nature. 1999;398:671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- 38.Ruby E G, McFall-Ngai M J. Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri. Trends Microbiol. 1999;7:414–420. doi: 10.1016/s0966-842x(99)01588-7. [DOI] [PubMed] [Google Scholar]
- 39.Saleh A, Figarella C, Kammouni W, Marchand-Pinatel S, Lazdunski A, Tubul A, Brun P, Merten M D. Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone inhibits expression of P2Y receptors in cystic fibrosis tracheal gland cells. Infect Immun. 1999;67:5076–5082. doi: 10.1128/iai.67.10.5076-5082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schripsema J, de Rudder K E, van Vliet T B, Lankhorst P P, de Vroom E, Kijne J W, van Brussel A A. Bacteriocin small of Rhizobium leguminosarum belongs to the class of N-acyl-l-homoserine lactone molecules, known as autoinducers and as quorum sensing cotranscription factors. J Bacteriol. 1996;178:366–371. doi: 10.1128/jb.178.2.366-371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperandio V, Mellies J L, Nguyen W, Shin S, Kaper J B. Quorum sensing controls expression of type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surette M G, Bassler B L. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Natl Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surette M G, Bassler B L. Regulation of autoinducer production in Salmonella typhimurium. Mol Microbiol. 1999;31:585–595. doi: 10.1046/j.1365-2958.1999.01199.x. [DOI] [PubMed] [Google Scholar]
- 44.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telford G, Wheeler D, Williams P, Tomkins P T, Appleby P, Sewell H, Stewart G S, Bycroft B W, Pritchard D I. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect Immun. 1998;66:36–42. doi: 10.1128/iai.66.1.36-42.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomb J F, White O, Kerlavage A R, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 48.Winson M K, Camara C, Latifi A, et al. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]