Abstract
Immunoblotting sera from 26 patients with septicemia due to an epidemic strain of methicillin-resistant Staphylococcus aureus (EMRSA-15), 6 of whom died, revealed an immunodominant EMRSA-15 antigen at 61 kDa. There was a statistically significant correlate (P < 0.001) between survival and immunoglobulin G to the 61-kDa band. The antigen was identified by sequencing positive clones obtained by screening a genomic expression library of EMRSA-15 with pooled sera from patients taken after the septicemic episode. Eluted antibody reacted with the 61-kDa antigen on immunoblots. The amino terminus was obtained by searching the S. aureus NCTC 8325 and MRSA strain COL databases, and the whole protein was expressed in Escherichia coli TOP 10F′. The derived amino acid sequence showed homology with ABC transporters, with paired Walker A and Walker B motifs and 73% homology to YkpA from Bacillus subtilis. Epitope mapping of the derived amino acid sequence with sera from patients who had recovered from EMRSA-15 septicemia delineated seven epitopes. Three of these epitopes, represented by peptides 1 (KIKVYVGNYDFWYQS), 2 (TVIVVSHDRHFLYNNV), and 3 (TETFLRGFLGRMLFS), were synthesized and used to isolate human recombinant antibodies from a phage antibody display library. Recombinant antibodies against peptides 1 and 2 gave logarithmic reductions in organ colony counts, compared with control groups, in a mouse model of the infection. This study suggests the potential role of an ABC transporter as a target for immunotherapy.
The global spread of bacteria resistant to multiple antibiotics (43) has increased the urgency to develop new antibacterial agents. Before antibiotics became available 50 years ago, antibodies, in the form of immune serum therapy, were widely used to treat a range of bacterial infections (9). Many of the problems which led to their abandonment in favor of antibiotics can now be overcome through antibody engineering, making antibody-based therapeutics a feasible objective. Phage antibody display libraries can be produced from the mRNA of human peripheral blood antibody-secreting cells and the cDNA immunoglobulin genes encoding heavy- and light-chain variable domains linked together to produce a library of human recombinant antibody fragments or to produce single-chain Fv fragments (scFv) (27, 46). Since the displayed antibody fragment retains its antigen binding capability, it is possible to enrich for recombinant phage expressing high-affinity scFv by panning against specific antigens or their epitopes. The key factor is then to determine which bacterial antigens are associated with an antibody response and whether such an antibody is protective. This study describes the first steps toward application of this approach to methicillin-resistant Staphylococcus aureus (MRSA).
The spread of MRSA is of particular concern because of their virulence and resistance to multiple antibiotics (4). S. aureus has been described as the most frequently isolated bacterial pathogen in hospitals (3) and is the cause of osteomyelitis, endocarditis, septic arthritis, pneumonia, abscesses, and the toxic shock syndrome (26). By 1992, over 40% of S. aureus strains in large hospitals in the United States were methicillin resistant (43). The reported incidence of S. aureus bacteremia in England and Wales increased from 6,010 in 1994 to 10,237 in 1998, with the proportion due to MRSA rising fourfold (13). For 30 years, vancomycin and teicoplanin were the mainstay of treatment of serious MRSA infections; thus, reports of treatment failures in the United States and Japan, associated with intermediate resistance to these antibiotics (21, 45), raised the specter of untreatable staphylococcal infections (39).
Certain strains of MRSA have a propensity to spread, and these became called epidemic MRSA (EMRSA) in the United Kingdom (3a). One of these, EMRSA-15 (35), is currently the most prevalent strain in this country, affecting 167 hospitals. In one teaching hospital, the Central Manchester Healthcare Trust, the number of MRSA isolates rose from 10 in 1994 to 369 in 1998, and most were EMRSA-15; there were 42 septicemias and 11 deaths. Analysis of the antibody response by immunoblotting has been complicated by antigenic variation in S. aureus: so far the technique has been more valuable in demonstrating antigenic variation between strains (7). The existence of a large outbreak due to EMRSA-15 provided an opportunity to study and compare the antibody responses in different groups of patients infected by the same strain. A range of antigenic bands were delineated; one at 61 kDa was the most commonly associated with immunoglobulin G (IgG) and IgM antibodies in patients recovering from EMRSA-15 septicemia. When cloned and sequenced, this was identified by its specific motifs as an ABC transporter (16, 23, 33). Epitopes on the antigen were mapped by the Geysen technique (19), as previously described for Candida albicans Hsp90 and Streptococcus oralis PAc (8, 28); the derived amino acid sequence was synthesized as a series of overlapping oligopeptides on pins, and reactivity with patient sera was assayed by a modified enzyme-linked immunosorbent assay (ELISA). Synthetic peptides representing these B-cell linear epitopes were used to select scFv from a phage antibody display library. A preliminary assessment of therapeutic potential was carried out in a mouse model of EMRSA-15 infection.
MATERIALS AND METHODS
Antigen preparation for immunoblotting.
The antigen preparation was obtained from a clinical isolate of EMRSA-15 grown in nutrient broth no. 2 (Oxoid, Basingstoke, United Kingdom) at 37°C and fragmented as previously described (7, 10). EMRSA-15 was defined by Gram stain, positive coagulase, biochemical profile including negative urease, sensitivity to phage 75, and gel pattern on pulsed-field gel electrophoresis following SmaI digestion (35).
Sera examined by immunoblotting.
Group 1 comprised hospital inpatients with nasal carriage of EMRSA-15 but otherwise no evidence of infection (n = 8). Group 2 consisted of patients with EMRSA-15-infected wounds requiring systemic treatment with vancomycin. Blood cultures remained negative (n = 16). Group 3 contained patients with septicemia due to a methicillin-sensitive strain of S. aureus (MSSA) who were successfully treated by antibiotics (n = 8). Group 4 comprised patients with septicemia due to EMRSA-15 successfully treated by vancomycin with additional rifampin where appropriate (n = 20). Sera were available from all patients 72 h after starting therapy, and in 13 cases multiple serum samples (up to four) were available before and after the first positive blood culture. Sera for group 5 were from patients who died from EMRSA-15 septicemia with a positive blood culture within 72 h of death (n = 6). Sera were examined at a dilution of 1:10 against immunoblots of EMRSA-15 as described previously (5, 8). Blots for which the antibody response was >50 mm by reflectance densitometry (Chromoscan 3; Joyce Loebl) were regarded as positive. When multiple sequential sera were tested, a constant result was recorded if the variation in height of the trace remained within 5 mm. A rising antibody response was recorded if there was an increase of at least 30 mm in trace height. A new antibody titer was recorded if a band with a height of >50 mm, absent in the earliest serum, appeared in later sera.
Preparation and screening of a genomic library of EMRSA-15.
A genomic library was constructed in the expression vector lambda ZAP Express (Stratagene Ltd., Cambridge, United Kingdom) essentially as described by Young and Davies (48). Chromosomal DNA, from a clinical isolate of EMRSA-15, was partially digested by SauIIIa, and fragments in the size range of 2 to 9 kbp were inserted into the vector. The library was screened with pooled sera (1:100) from patients with antibodies to EMRSA-15 detected by alkaline phosphatase-conjugated goat anti-human IgG (1:5,000; Sigma, Poole, United Kingdom).
Characterization of positive clones and DNA sequencing.
Antibodies in patient sera (1:10) were affinity purified against positive recombinant plaques, and the bound antibody, eluted with glycine buffer (pH 2.8), was screened against an immunoblot of EMRSA-15 (8). DNA sequencing was performed by the chain termination method (Sequenase version 2.0 kit; United States Biochemical, Cambridge, United Kingdom). The first set of annealing reactions was done with universal primers T3 and T7; subsequent primers were derived from the sequences obtained from both the coding and noncoding strands. The DNA sequence was analyzed by a BLAST search performed using the National Center for Biotechnology Information's (BCM)BLAST Web server. The sequence produced was used to search the S. aureus NCTC 8325 genome sequence project database (www.genome.ou.edu/cgi-bin/Staph_server.p) to obtain the amino end. The sequence thus assembled was used to search a second S. aureus database, derived from the MRSA strain COL isolated in 1975 (available at the Institute for Genome Research [TIGR] web site [www.tigr.org]). The contig 4348 thus identified was put into the BCM Search Launcher (www.imgen.bcm.tmc.edu:9331/seq-search/nucleic-acid_search), and the genes were identified by the WU-BLASTX+BEAUTY program. Proteins homologous to the EMRSA-15 ABC transporter were identified on the BCM Searcher Launcher (www.imig/bcm.tmc.edu:9331/seq-search/protein-search html) by the BLASTP+BEAUTY program.
Expression of the complete ABC transporter protein.
The complete protein was expressed in Escherichia coli TOP 10F′ (Invitrogen Corp., Oxon, United Kingdom) by amplifying the gene from purified EMRSA-15 DNA by PCR using forward (5′ATGTTACAAGTAACTGAT) and reverse (5′TTTTAACGCCATTTC) primers. The gene was sequenced and cloned into the pBAD vector by means of a pBAD-TA-TOPO cloning kit (Invitrogen). The recombinant E. coli was grown at 37°C, and protein induction was initiated by 0.02% arabinose. The cells were harvested and fragmented, and the presence of the tagged recombinant protein was confirmed by probing with a monoclonal antibody to the V5 tag (1:5,000). The protein was purified by an Ni-nitrilotriacetic acid (NTA) agarose column (Qiagen, Crawley, United Kingdom) to bind the His tag on the amino end of the recombinant protein. It was eluted off the column with increasing concentrations of imidazole, giving a final protein concentration of 1 mg/ml. Identity was confirmed by immunoblotting against the V5 monoclonal antibody and direct amino acid sequencing (performed in the Biochemistry Department, University of Cambridge, Cambridge, United Kingdom).
Polyclonal antiserum was prepared by injecting a New Zealand White rabbit (Charles River, Maidstone, United Kingdom) by intravenous bolus with the recombinant protein (0.5 mg) in complete Freund's adjuvant, followed fortnightly by the protein in incomplete Freund's adjuvant until seroconversion. Seroconversion was monitored by immunoblotting pre- and postbleed sera (1:100) against EMRSA-15.
Epitope mapping the ABC transporter protein.
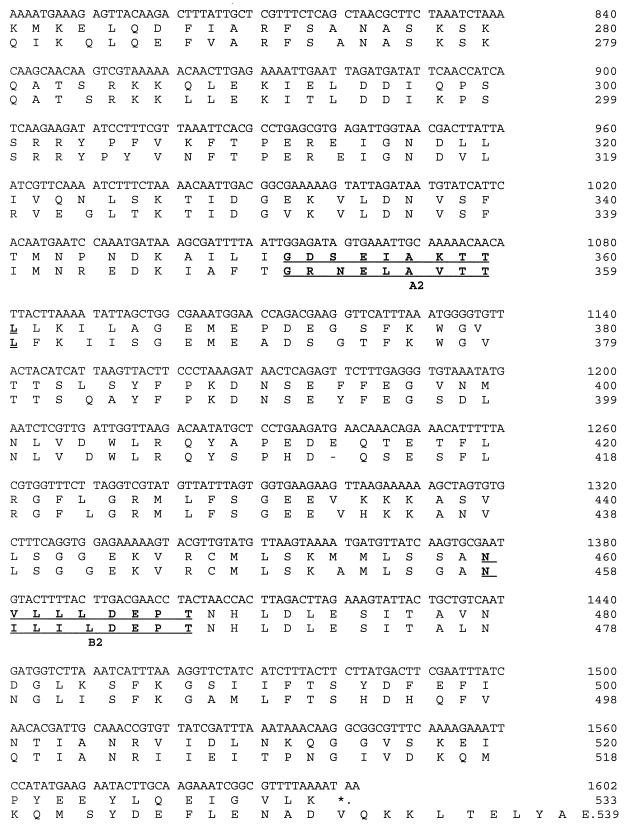
A series of overlapping nonapeptides covering the amino acid sequence derived from the antibody-positive clone (starting at residues DPT [Fig. 1]) were synthesized on polythylene pins with reagents from an epitope scanning kit (Cambridge Research Biochemicals, Cambridge, United Kingdom) as described previously by Geysen et al. (19); the first nonapeptide consisted of residues 1 to 9, the second consisted of residues 2 to 10, etc. The reactivity of each peptide with patient sera (1:200) was determined for IgG by ELISA. Data were expressed as A405 after 30 min of incubation. Sera were examined from patients with septicemia due to EMRSA-15, both survivors (n = 4) and nonsurvivors within 72 h of death (n = 4), patients with EMRSA-15 culture-positive wound swabs requiring vancomycin therapy who remained blood culture negative (n = 5), and hospitalized patient controls with no evidence of MRSA infection (n = 2) (Table 4).
FIG. 1.
Staphylococcal ABC transporter DNA and amino acid sequences. The YkpA protein amino acid sequence shown underneath for comparison. Walker A (A1 and A2) and B (B1, and B2) motifs in bold.
TABLE 4.
Epitope map values for wells where the mean OD was at least 2 standard deviations above that of the control
| Well no. | Epitope sequencea | Mean OD (SD)
|
|||
|---|---|---|---|---|---|
| Hospital inpatient controls (n = 2) | Wound-infected patients (n = 5) | Septicemic patients who died (n = 4) | Septicemic patients who survived (n = 4) | ||
| 64 | DRHFLN | 0.476 (0.393) | 0.815 (0.281) | 0.547 (0.249) | 0.823 (0.547) |
| 65 | DRHFLN | 0.480 (0.316) | 0.972 (0.329) | 0.568 (0.244) | 1.131 (0.351) |
| 66 | DRHFLN | 0.521 (0.359) | 1.051 (0.276) | 0.610 (0.243) | 1.350 (0.625) |
| 67 | DRHFLN | 0.416 (0.304) | 0.855 (0.199) | 0.511 (0.213) | 1.164 (0.545) |
| 86 | GNYD | 0.484 (0.358) | 0.932 (0.253) | 0.531 (0.206) | 1.313 (0.614) |
| 87 | GNYD | 0.490 (0.359) | 0.997 (0.292) | 0.560 (0.236) | 1.232 (0.483) |
| 88 | GNYD | 0.649 (0.427) | 0.923 (0.251) | 0.581 (0.167) | 1.308 (0.410) |
| 89 | GNYD | 0.663 (0.231) | 1.027 (0.260) | 0.780 (0.110) | 1.235 (0.479) |
| 90 | GNYD | 0.833 (0.402) | 1.057 (0.279) | 0.679 (0.109) | 1.522 (0.551) |
| 91 | GNYD | 0.843 (0.421) | 1.108 (0.272) | 0.869 (0.278) | 1.533 (0.545) |
| 153 | RRYPF | 0.670 (0.368) | 1.16 (0.179) | 0.782 (0.251) | 1.526 (0.551) |
| 154 | RRYPF | 0.578 (0.219) | 1.1189 (0.204) | 0.863 (0.287) | 1.748 (0.460) |
| 155 | RRYPF | 0.653 (0.227) | 1.216 (0.186) | 0.779 (0.254) | 1.917 (0.509) |
| 156 | RRYPF | 0.635 (0.243) | 0.98 (0.127) | 0.805 (0.230) | 1.593 (0.461) |
| 157 | RRYPF | 0.667 (0.374) | 1.176 (0.241) | 0.836 (0.292) | 1.761 (0.649) |
| 158 | RRYPF | 0.683 (0.274) | 1.147 (0.222) | 0.765 (0.191) | 1.774 (0.563) |
| 211 | KTTLLK | 0.439 (0.176) | 0.752 (0.62) | 0.495 (0.125) | 1.145 (0.502) |
| 212 | KTTLLK | 0.581 (0.207) | 0.802 (0.087) | 0.669 (0.167) | 1.360 (0.384) |
| 213 | KTTLLK | 0.582 (0.197) | 0.923 (0.127) | 0.663 (0.157) | 1.351 (0.374) |
| 214 | KTTLLK | 0.587 (0.219) | 0.949 (0.126) | 0.680 (0.186) | 1.506 (0.570) |
| 233 | GVTTSLS | 0.447 (0.257) | 0.937 (0.148) | 0.496 (0.193) | 1.091 (0.512) |
| 234 | GVTTSLS | 0.589 (0.441) | 0.970 (0.145) | 0.543 (0.186) | 1.129 (0.44) |
| 235 | GVTTSLS | 0.551 (0.341) | 1.015 (0.126) | 0.585 (0.213) | 1.448 (0.626) |
| 255 | VDWLR | 0.492 (0.357) | 0.970 (0.156) | 0.513 (0.181) | 1.280 (0.509) |
| 256 | VDWLR | 0.520 (0.407) | 1.011 (0.18) | 0.548 (0.194) | 1.219 (0.463) |
| 257 | VDWLR | 0.596 (0.488) | 1.054 (0.225) | 0.576 (0.173) | 1.296 (0.433) |
| 258 | VDWLR | 0.414 (0.326) | 1.010 (0.243) | 0.505 (0.173) | 1.046 (0.476) |
| 259 | VDWLR | 0.571 (0.538) | 0.746 (0.238) | 0.598 (0.201) | 1.308 (0.497) |
| 272 | RGFL | 0.613 (0.430) | 1.105 (0.20) | 0.640 (0.203) | 1.502 (0.582) |
| 273 | RGFL | 0.603 (0.420) | 1.059 (0.181) | 0.649 (0.234) | 1.464 (0.576) |
| 274 | RGFL | 0.752 (0.439) | 1.200 (0.306) | 0.775 (0.233) | 1.695 (0.640) |
| 275 | RGFL | 0.698 (0.444) | 1.289 (0.238) | 0.801 (0.278) | 1.699 (0.586) |
| 276 | RGFL | 0.750 (0.301) | 1.286 (0.245) | 0.876 (0.229) | 1.860 (0.696) |
| 277 | RGFL | 0.739 (0.297) | 1.272 (0.25) | 0.823 (0.261) | 1.739 (0.690) |
The overlapping amino acid sequences were derived by a comparison of first and last peptide sequences and were used to define the epitopes.
Preparation of phage antibody display library and scFv.
The phage antibody display library and scFv were produced essentially as previously described (8, 28). Briefly, mRNA was prepared from 20 ml of patient peripheral blood by separation of lymphocytes over Ficoll followed by guanidinium thiocyanate extraction and purification on an oligo(dT)-cellulose column (Quick Prep mRNA; Pharmacia, St. Albans, United Kingdom). First-strand cDNA synthesis was performed with a constant-region primer for all four subclasses of human IgG heavy chains (HulgG1 to -4) using avian myeloblastosis virus reverse transcriptase (HT Biotechnology, Cambridge, United Kingdom). The heavy-chain variable-domain genes were amplified by primary PCRs with family-based forward (HuJH1 to -6) and backward (HuVH1 1a to 6a) primers. An SfiI restriction site was introduced upstream to the VH3a back-generated product prior to assembly with a diverse pool of light-chain variable-domain genes (8, 28). The latter also introduced a linker fragment (Gly4 Ser)3 and a downstream NotI site. By use of the SfiI and NotI restriction enzyme sites, the product was unidirectionally cloned into a phagemid vector. The ligated vector was introduced into E. coli TG1 by electroporation, and phages were rescued with the helper phage M13K07 (Pharmacia). To enrich for antigen-specific scFv, the phage library was panned against peptides representing three of the epitopes delineated by epitope mapping (peptide 1, KIKVYVGNYDFWYQS; peptide 2, TVIVVSHDRHFLNNV; and peptide 3, TETFLRGFLGRMLFS) and the purified ABC transporter protein. Panning was performed in immunotubes coated with peptide (10 ng/ml) or the purified transporter (1 mg/ml). Bound phages were eluted with log-phase E. coli TGI. After rescue with M13K07, the phages were repanned against peptide a further three times. BstN1 (New England Biolabs, Hitchen, United Kingdom) DNA fingerprinting was used to confirm enrichment of specific scFv after successive rounds of panning.
Assessment in an animal model.
EMRSA-15 was grown overnight in brain heart infusion at 37°C and washed in saline, and the concentration was determined by hemocytometer and by plating of dilutions on blood agar. Next, 2 × 107 CFU were injected as a 0.1-ml bolus into the lateral tail vein of 22- to 24-g female CD1 mice (Charles River). Two hours after inoculation, randomized groups of animals were given intravenously 100 μl of the hyperimmune rabbit antiserum against the ABC transporter or control unimmunized rabbit antiserum (experiment 1) or (for experiments 2 to 5) 200 μl of phage ABC 1 or ABC 2 (against peptide 1), ABC 3, ABC 4, or ABC 5 (against peptide 2), ABC 6 or ABC 7 (against peptide 3), or ABC 8 or ABC 9 (against the whole ABC protein) or a negative control phage (Table 5). Bacterial cell counts were made from kidney, liver, and spleen and expressed as the mean log10 CFU per gram plus standard deviation.
TABLE 5.
Results of in vivo assessment of the human recombinant antibodiesa
| Expt no. | Antibody | Antibody specificity | No. of mice | Colony counts (mean log10 CFU/g ± SD)
|
||
|---|---|---|---|---|---|---|
| Kidney | Liver | Spleen | ||||
| 1 | Unimmunized rabbit | 15 | 7.13 ± 1.3 | 4.84 ± 2.32 | 4.11 ± 1.45 | |
| Hyperimmune rabbit | ABC protein | 15 | 6.61 ± 0.98b | 3.84 ± 1.28c | 3.35 ± 0.36b | |
| 2 | Negative phage | 5 | 7.86 ± 1.73 | 5.45 ± 1.73 | 5.09 ± 1.23 | |
| ABC 6 | Peptide 3 | 5 | 7.57 ± 1.49 | 5.74 ± 1.05 | 4.96 ± 1.03 | |
| ABC 7 | Peptide 3 | 5 | 7.72 ± 1.47 | 6.04 ± 1.14 | 5.74 ± 0.94 | |
| ABC 8 | ABC protein | 5 | 7.79 ± 1.79 | 4.35 ± 1.52c | 4.81 ± 2.04 | |
| ABC 9 | ABC protein | 6 | 7.32 ± 1.32b | 5.3 ± 0.59 | 4.19 ± 0.26b | |
| 3 | Negative phage | 12 | 8.23 ± 2.47 | 6.03 ± 1.35 | 6.05 ± 1.39 | |
| ABC 1 | Peptide 1 | 9 | 7.62 ± 1.99b | 4.71 ± 0.69c | 4.38 ± 1.48c | |
| ABC 3 | Peptide 2 | 9 | 8.85 ± 3.27 | 4.88 ± 0.97c | 4.15 ± 0.96c | |
| ABC 6 | Peptide 3 | 9 | 7.56 ± 1.41b | 5.6 ± 1.65b | 5.6 ± 1.67b | |
| 4 | Negative phage | 12 | 8.36 ± 1.46 | 4.84 ± 0.93 | 4.75 ± 1.06 | |
| ABC 1 | Peptide 1 | 12 | 7.35 ± 1.8c | 4.21 ± 1.38b | 3.65 ± 0.84c | |
| ABC 2 | Peptide 1 | 12 | 6.9 ± 1.01c | 3.8 ± 0.96c | 3.19 ± 0.50c | |
| ABC 4 | Peptide 2 | 12 | 6.82 ± 1.00c | 3.47 ± 1.09c | 3.75 ± 0.92c | |
| ABC 5 | Peptide 2 | 12 | 7.23 ± 1.25c | 4.25 ± 1.25b | 4.25 ± 1.34b | |
| 5 | Negative phage | 10 | 8.45 ± 2.58 | 4.98 ± 1.14 | 4.32 ± 1.41 | |
| ABC 4 | Peptide 2 | 10 | 6.86 ± 0.94c | 3.88 ± 0.91c | 3.82 ± 0.80b | |
The phage dose was 109±0.5 PFU. Mice were culled at 24 h in experiment 1 but at 48 h for all following experiments.
Semilogarithmic reduction compared to negative control.
Logarithmic reduction compared to negative control.
RESULTS
Immunoblotting.
Immunoblotting revealed bands ranging in apparent molecular mass from 18 to 260 kDa. In patients who recovered from an EMRSA-15 septicemia (group 4), bands at 30, 36, 37, 42, 45, 55, 57, and 61 kDa were recognized by ≥50%, and in IgM responses were the most pronounced (Table 1). All 20 of these patients had IgG and 17 (85%) had IgM antibody to the 61-kDa band. In the 16 patients with EMRSA-15 wound infections (group 2), the 61-kDa band was again immunodominant, 15 (94%) having IgG and 11 (69%) having IgM to this band. In contrast, only one of the six patients who died of EMRSA-15 septicemia had IgG to the 61-kDa band (17%), though three had some IgM. Relatively few EMRSA-15 bands were recognized by sera from patients with MSSA septicemia (group 3) but the 61-kDa band was still detected by four out of eight patients. A minority of nasal carriers had IgG to a range of staphylococcal antigens. The presence of IgG antibody to the 61-kDa band and survival from EMRSA-15 septicemia (group 4) was statistically significant (P = 0.0001) compared to fatal cases; the presence of the antibody in group 4 was also significantly greater than in nasal carriers (P = 0.0008) (Fisher's exact test, two tailed; P < 0.001).
TABLE 1.
Immunoblot results of patient sera against EMRSA-15
| Antigen apparent mol mass (kDa) | No. of patients with indicated antibody
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 (n = 8)
|
Group 2 (n = 16)
|
Group 3 (n = 8)
|
Group 4 (n = 20)
|
Group 5 (n = 6)
|
||||||
| IgM | IgG | IgM | IgG | IgM | IgG | IgM | IgG | IgM | IgG | |
| 260 | 4 | 5 | 1 | 7 | 2 | |||||
| 240 | 1 | 1 | 6 | 1 | 6 | 2 | ||||
| 220 | 3 | 1 | 2 | 3 | 4 | 1 | ||||
| 140 | 2 | 2 | 2 | 1 | 2 | 9 | 1 | 2 | ||
| 120 | 3 | 1 | 4 | 6 | 1 | |||||
| 97 | 2 | 2 | 2 | 6 | 2 | |||||
| 69 | 1 | 2 | 5 | 6 | 5 | 8 | 4 | |||
| 61 | 3 | 11 | 15 | 4 | 4 | 17 | 20 | 3 | 1 | |
| 59 | 6 | 11 | 1 | 5 | 4 | 2 | 2 | |||
| 57 | 2 | 5 | 3 | 2 | 9 | 16 | 3 | 2 | ||
| 55 | 4 | 2 | 5 | 3 | 1 | 1 | 6 | 10 | 3 | 2 |
| 51 | 3 | 2 | 2 | 3 | 3 | 8 | 9 | 2 | 2 | |
| 48 | 1 | 2 | 2 | 1 | 3 | 7 | ||||
| 45 | 7 | 1 | 4 | 10 | 3 | |||||
| 42 | 2 | 1 | 4 | 4 | 1 | 7 | 14 | 1 | 1 | |
| 37 | 2 | 1 | 5 | 10 | 1 | |||||
| 36 | 1 | 3 | 10 | 1 | ||||||
| 30 | 1 | 1 | 2 | 4 | 10 | 1 | 2 | |||
| 28 | 1 | 3 | ||||||||
| 26 | 1 | 1 | 2 | 5 | ||||||
| 23 | 1 | 3 | ||||||||
| 18 | 1 | 1 | ||||||||
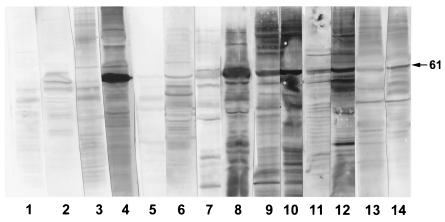
Multiple sera were available from 13 patients in group 4. These showed rising antibodies to 11 bands (Table 2), of which the most prominent was the 61-kDa band; 92% of patients produced an IgM and/or IgG response against this band. Figure 2 illustrates this response.
TABLE 2.
Immunoblot results of multiple sera against EMRSA-15 from survivors of EMRSA-15 septicemia
| Antigen apparent mol mass (kDa) | No. of patients with indicated rising or new antibody (n = 13)
|
||
|---|---|---|---|
| IgM | IgG | IgM and/or IgG | |
| 61 | 7 | 9 | 12 |
| 59 | 1 | 1 | 1 |
| 57 | 2 | 3 | 3 |
| 55 | 3 | 3 | |
| 51 | 2 | 2 | 3 |
| 48 | 1 | 1 | |
| 45 | 1 | 1 | |
| 42 | 2 | 5 | 6 |
| 37 | 1 | 1 | 2 |
| 36 | 1 | 1 | 2 |
| 30 | 1 | ||
FIG. 2.
Immunoblots of EMRSA-15 with paired sera from five patients with EMRSA-15 infection: case 1, pre- and postinfection sera showing IgM (lanes 1 and 2, respectively) and IgG (lanes 3 and 4); case 2, pre- and postinfection sera showing IgM (lanes 5 and 6, respectively) and IgG (lanes 7 and 8); cases 3, 4, and 5, preinfection (lanes 9, 11, and 13, respectively) and postinfection (lanes 10, 12, and 14, respectively) IgG. The 61-kDa band is marked.
Characterization of positive recombinant clones.
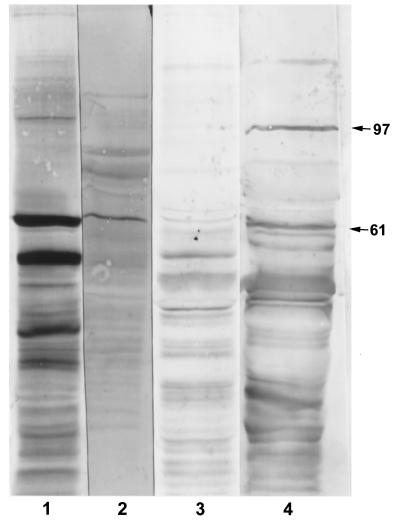
Screening the EMRSA-15 library with sera from patients infected with EMRSA-15 gave two positive clones for which affinity selection showed that the antibody bound cross-reacted with the 61-kDa band on immunoblot (Fig. 3, lane 2). These clones demonstrated a partial sequence in frame with the β-galactosidase gene; in each case, the total insert size was 4.5 kb. The derived amino acid sequences from both clones produced a protein with ATP-binding domains and a sequence homologous to the ABC transporter proteins (16, 23); this was the C-terminal fragment of the protein, starting at DPT (Fig. 1) and subsequently called the EMRSA-15 ABC protein. A search in the S. aureus NCTC 8325 genome sequence project database and produced matches with contig 1184 (also referred to as 1177 and 1158), which had sequences partially overlapping the identified sequence. This in turn allowed the synthesis of PCR primers for cloning and resequencing of the full gene. This gene was 100% homologous to a sequence derived from the MRSA strain COL (TIGR database) on contig 4348.
FIG. 3.
Immunoblots of EMRSA-15 showing one of the sera used to screen the phage expression library (lane 1), the subfraction of this serum which bound to and was then eluted from the positive clone (lane 2), the rabbit serum before (lane 3) and after (lane 4) immunization with the recombinant ABC protein. Bands at 97 and 61 kDa are marked.
The EMRSA-15 ABC protein derived amino acid sequence demonstrated 73% homology with the YkpA ABC transporter of Bacillus subtilis (SPTREMBL accession no. Q047971) (22, 33) (Fig. 1; see Table 3), 56% with an ABC transporter from Borrelia burgdorferi (EMBL accession no. AE001174) (17), 53% with an ABC transporter from Treponema pallidum (EMBL accession no.: AE001254) (18), 48% with an ABC transporter from Helicobacter pylori (EMBL accession no. AE000596 (41), and 45% with a hypothetical ABC transporter Ybit from E. coli K-12 (EMBL accession no. AE000184) (6). Lower homologies were seen with staphylococcal proteins such as Vga and VgaB, the ATP-binding protein involved in virginiamycin resistance (32.4% identify over 389-amino-acid overlap) (1, 2), MsrA protein of Staphylococcus epidermidis (27.1% identify over a 435-amino-acid overlap) (36, 37) and the MsrSA protein of S. aureus (27.1% identify over a 435-amino-acid overlap) (29).
TABLE 3.
Order of genes compared to the EMRSA-15 ABC transporter derived from the MRSA contig 4348a
| Position in contig 4348 (bp) | Homology | Size (bp) | Apparent mol mass (Da) | % Identity |
|---|---|---|---|---|
| 5224–6522 | FemA, S. aureus, EMBL accession no. X17688 | 1,302 | 50,650 | 100 |
| 6544–7800 | FemB, S. aureus, EMBL accession no. X17688 | 1,260 | 49,676 | 100 |
| 9901–10599 | OPP-2F, oligopeptide transporter putative ATPase domain, S. aureus, SPTREMBL accession no. Q92GN3 | 702 | 26,257 | 100 |
| 10648–11368 | OPP-2D, oligopeptide transporter putative ATPase domain, S. aureus, SPTREMBL accession no. Q92GN4 | 777 | 29,680 | 100 |
| 11355–12182 | OPP-2C, oligopeptide transporter putative ATPase domain, S. aureus, SPTREMBL accession no. Q92GN5 | 831 | 31,255 | 60 |
| 12178–13161 | OPP-2B, oligopeptide transporter putative ATPase domain, S. aureus, SPTREMBL accession no. Q92GN6 | 987 | 36,814 | 100 |
| 14025–15818 | YjbG, oligopeptidase F homologue, B. subtilis, EMBL accession no. Z99110 | 1,830 | 70,143 | 28 |
| 15985–16599 | PhoU, Streptococcus pneumoniae, EMBL accession no. AF118229 | 651 | 24,192 | 31 |
| 16618–17358 | PstB, ABC protein, S. pneumoniae, EMBL accession no. AF118229 | 753 | 28,108 | 64 |
| 17522–18346 | PstA, transmembrane protein, S. pneumoniae, EMBL accession no. AF118229 | 816 | 29,130 | 34 |
| 18477–19259 | PstC, transmembrane protein, S. pneumoniae EMBL accession no. AF118229 | 816 | 28,761 | 33 |
| 20080–20436 | PstS, phosphate-binding protein, S. pneumoniae, EMBL accession no. AF118229 | 876 | 30,757 | 27 |
| 19645–20415 | SPHX protein, Synechococcus sp., EMBL accession no. D26161 | 1,014 | 36,374 | 39 |
| 21555–22319 | YitL, B. subtilis, SPTREMBL accession no. 006747 | 897 | 34,053 | 37 |
| 22553–24151 | EMRSA-15 ABC transporter, YkpA homologue, B. subtilis, SPTREMBL accession no. Q04797 | 1,623 | 61,055 | 73 |
| 25029–26216 | Aspartokinase II, Bacillus sp., SPTREMBL accession no. Q59229 | 2,223 | 44,342 | 41 |
| 26292–27257 | Aspartate semialdehyde dehydrogenase, B. subtilis, Swiss-Prot accession no. Q04797 | 1,041 | 37,847 | 48 |
| 27283–28119 | Dihydrodopicolinate synthetase, Methanococcus jannaschii, EMBL accession no. U67680 | 870 | 31,579 | 42 |
| 28919–29617 | YkuQ, B. subtilis, EMBL accession no. Z99111 | 711 | 24,987 | 60 |
| 29799–30914 | Putative hippurate hydrolase, B. subtilis, EMBL accession no. Z99118 | 1,251 | 45,239 | 36 |
| 31000–31818 | Alanine racemase, S. pneumoniae, EMBL accession no. AF171873 | 1,104 | 39,857 | 29 |
| 32003–33247 | Diaminopimelate decarboxylase, B. subtilis, EMBL accession no. L09228 | 3,762 | 48,788 | 49 |
Source: (TIGR database [www.tigr.org]).
Expression of the full protein.
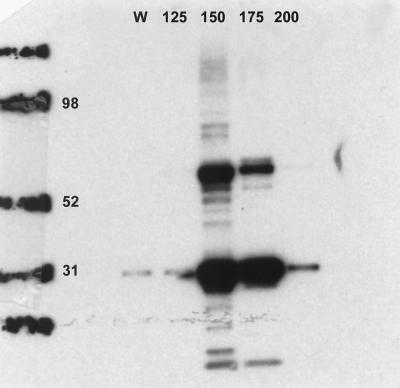
When the protein was reexpressed using the pBAD vector, it had an apparent molecular mass of 61 kDa and cross-reacted on immunoblotting with the monoclonal antibody against the V5 epitope. It also produced a second band at 32 kDa. Direct amino acid sequencing of the 32-kDa band yielded the sequence KKQLEKIEL (amino acid 305 onward [Fig. 1]). The protein was eluted from the Ni-NTA column by increasing levels of imidazole from 125 to 250 mM and, following elution, demonstrated two bands of apparent molecular weight 61 and 32 kDa when the immunoblot was probed with V5 monoclonal antibody (Fig. 4). Immunization of a rabbit with this protein produced a dominant antibody response to bands at 97 and 61 kDa (Fig. 3, lanes 3 and 4).
FIG. 4.
Elution of the ABC transporter from the Ni-NTA column by increasing levels of imidazole (125 to 200 mM fractions marked). Numbers on the left represents molecular mass markers (in kilodaltons). W, wash fraction, obtained after the flowthrough and before elution with imidazole, using 10 mM imidazole as instructed by the manufacturer.
Epitope mapping.
Epitope mapping defined seven areas where sera from patients with an EMRSA-15 septicemia who survived produced three or more consecutive wells with a mean optical density (OD) at least 2 standard deviations above that of sera from uninfected hospital inpatient controls or septicemic patients who died (Table 4). These epitopes were also positive with sera from patients with EMRSA-15 wound infections. Compared to the derived sequences from YkpA, each epitope was highly conserved, whereas only KTTLLK was significantly conserved with the MsrA, MsrSA, Vga, and VgaB sequences. Peptides 1, 2, and 3, representing epitopes GNYD, DRHFLN, and RGFL, respectively, were synthesized.
Human recombinant antibodies.
These three peptides were used to pan the phage antibody display library. BstN1 fingerprinting of the PCR-amplified scFv inserts showed that before panning, the library was highly heterogeneous. After four rounds of panning against peptide 1, two BstNI fingerprints predominated; representatives of each of these types, ABC 1 and ABC 2, were selected for animal work. Focusing was less pronounced after panning with peptides 2 and 3. Five phages (ABC 3, 4, 5, 6, and 7) were selected for animal work because in each case there were multiple representatives present after panning. Panning against the ABC transporter gave two clone types, represented by ABC 8 and ABC 9.
Preliminary assessment in mice.
Activity was assessed in terms of a logarithmic reduction in organ colony counts (kidney, liver, and spleen) compared to the negative control group (Table 5). The model was set up using the hyperimmune rabbit serum. This produced a logarithmic reduction in liver counts and a semilogarithmic reduction in kidney and spleen counts compared to the control serum. A preliminary assessment of the scFv was then carried out, the amount of scFv which could be given being limited by the toxicity of the phage itself so that dosage was restricted to ≤5 × 109 phage. The scFvs against peptides 1 and 2 showed greater activity than those panned against peptide 3 or the whole recombinant ABC protein. ABC 1 (a scFv against peptide 1) and ABC 4 (against peptide 2) each produced a logarithmic reduction in colony counts in two out of three organs in separate experiments.
DISCUSSION
This study examined the antibody response in 20 patients who survived blood culture-confirmed septicemia due to a common epidemic strain of MRSA (EMRSA-15) and 6 patients who died. Previous studies have shown a complex picture with humoral responses demonstrated against exotoxins, exoenzymes, cell wall components such as peptidoglycan and teichoic acid, and capsules (5, 42).
This study had the advantage that antigenic variation between infecting isolates was eliminated since a single epidemic strain was responsible for all infections. IgM responses were most marked in survivors of blood culture-positive septicemia, suggesting a specific antibody response to EMRSA-15 superimposed on a background of naturally occurring antibodies. Analysis of these antibody responses showed that a 61-kDa band was the most highly immunogenic in survivors of both EMRSA-15 and MSSA septicemia and in patients with EMRSA-15 wound infections. There was a statistically significant association between survival and the presence of IgG to this band (P = 0.0001). The 42-kDa band may be protein A previously shown to be immunodominant in staphylococcal infection (20).
The 61-kDa band was identified as a member of the ABC transporter group of proteins which are found in prokaryotes and eukaryotes. They are involved in the import or export of substrates across biological membranes (16, 23). There were examples of the four short motifs that are conserved with the ABC transporters of E. coli. Site A1 (Fig. 1) was a Walker A site (GXXGXGKST), while A2 was a variant (GDSEIAKTTL). Sites B1 and B2 were Walker B sites (hydrophobic, hydrophobic, hydrophobic, hydrophobic residue DEPT), while the LSGG signature was absent before B1 but present before B2 (Fig. 1, amino acid 479). The fourth motif is a conserved histidine located approximately 30 amino acids downstream of the aspartic acid of the Walker B motif preceded by four hydrophobic residues and followed by a charged residue. This occurred after the B1 but not the B2 site (23).
The highest homology score (73%) was with the YkpA ABC transporter from B. subtilis, which has been defined as a subfamily 3 extruder (22, 33). This family also includes the products of exp2, ydiF, yfmM, and yfmR. These genes encode ATPases belonging to a single transcriptional unit, with the genes encoding the putative integral membrane proteins not detected in their vicinity (33). The Walker A motifs in the C-terminal nucleoside binding domains of both YkpA and EMRSA-15 ABC were modified such that the second and third invariant glycine residues replaced by glutamic acid and alanine residues. EMRSA-15 ABC was also typical of subfamily 3 dimeric ATPases in having different and extreme distances between both Walker signatures. For YkpA, this has led to the suggestion that the N- and C-terminal domains have developed different functions (33). This may also be true for the EMRSA-15 ABC transporter.
EMRSA-15 ABC had lower homologies with Vga, VgaB (1, 2), MsrA (36, 37), and MsrSA (29). MsrA encodes a 488-amino-acid ABC transporter protein with a theoretical molecular mass of 55.9 kDa on S. epidermidis plasmid pUL5054 and has been associated with macrolide resistance by an active efflux pump (36, 37). MsrSA encodes a 488-amino-acid pump isolated from S. aureus MS 8968 on plasmid pMS97, which was 98% identical to MsrA and expressed inducible resistance to macrolides and streptogramin type B antibiotics. When MsrA was subcloned into S. aureus RN4220 (37), using plasmid pUL5054 on which the gene was located, there was constitutive resistance to erythromycin and inducible resistance to streptogramin B. This was despite there being only inducible resistance to both antibiotics in the original strain of S. epidermidis (36). In Staphylococcus xylosus, the C-terminal ATP-binding domain alone was capable of conferring resistance to erythromycin (31).
vga is a gene on S. aureus plasmid pIP680 conferring resistance to the virginiamycin A-like antibiotics (streptogramin A, pristinamycin II, and virginiamycin M) encoding a protein of 522 amino acids with a calculated molecular mass of 60 kDa (1). vgaB is a second staphylococcal gene conferring resistance to streptogramin, virginiamycin M, mikamycin A, synergistin A, and dalfopristin (1). It was isolated from S. aureus BM3385 on plasmid pIP1633 and encoded a protein of 552 amino acids with a calculated molecular mass of 61 kDa. Both of these proteins were similar to EMRSA-15 ABC in having Walker A and B motifs with no evidence of membrane-spanning domains. The EMRSA-15 strain was resistant to erythromycin and clindamycin. Other ABC transporters genes described for S. aureus have included abcA, which was in direct proximity to pbpD, which coded for penicillin-binding protein 4. A mutation in abcA was more resistant to cefoxitin and methicillin than the parent strain (15). Further ABC transporters have been shown to be encoded in an iron-regulated operon in S. epidermidis (12) and to be involved with molybdate transport in S. aureus (32).
In recent years, in vivo expression technology and signature tagged mutagenesis have been developed for identifying staphylococcal virulence features (14, 25, 30). In a murine renal abscess model, in vivo expression technology identified 45 staphylococcal genes induced during infection, including genes encoding K+/Cu2+-transporting ATPases (25); signature tagged mutagenesis identified 50 mutants with attenuated virulence in a murine model of infection following intraperitoneal inoculation (30). These included aspartate semialdehyde dehydrogenase (Asd), diaminopimelate decarboxylase (B. subtilis LysA), and transporters involved in oligopeptide transport. Searching the databases for matches to the nearby genes revealed that the EMRSA-15 ABC transporter was in close geographic proximity (Table 3). In B. subtilis, the dap operon includes asd, dapG (aspartokinase II), dapA (dihydrodipicolinate synthase), and open reading frames (ORFs) orfX and orfY (dipicolinate synthase) (10). These enzymes are key to the conversion of l-aspartate to diaminopimelic acid, which is an important component of the cell wall peptidoglycan.
Subsequently Coulter et al. (14) confirmed the importance of peptide and amino acid transporters. Eight Tn917 insertions, with similar attenuation profiles, were mapped to multiple genes of the E. coli nickel permease (Opp-1 operon) and oligopeptide permease (Opp-2 operon) (40). Four strongly attenuated in vivo mutants were mapped to the region downstream of Opp-2, near ORFs encoding FemA and FemB, which mediate pentaglycine peptidoglycan cross-linking of the S. aureus cell wall (38). The close geographic proximity of FemA, FemB, Opp-2 (represented by Opp-2B, Opp-2C, Opp-2D, and Opp-2F [Table 3]), the phosphate transport homologues of S. pneumoniae, YkpA, and homologues of the products of the dap operon of B. subtilis (Table 3) suggest that the corresponding genes are all involved either in generating the building blocks for cell wall synthesis or in the biosynthesis itself. Thus, this study demonstrated that a YkpA homologue in MRSA was a target for humoral immunity which may be important as blockade may prevent the uptake of a molecule vital to cell wall biosynthesis.
Epitope mapping demonstrated seven epitopes (Table 4). Epitope DRHFLN, included the conserved histidine located approximately 30 amino acids downstream of the aspartic acid of the Walker B motif. For E. coli ABC transporters, this has been felt to be essential for function (23). The mammalian homologue of the ABC transporter protein is the human multidrug resistance P-glycoprotein (24). The ABC transporter LmrA, from Lactococcus lactis, mediates antibiotic resistance in this bacterium and when expressed in human lung fibroblast cells conferred resistance to cytotoxic drugs (44). The functional heterologous expression of LmrA in eukaryotic cells strongly implies that its ability to confer drug resistance is independent of any auxiliary proteins. The substrate and modulator specificities of LmrA and P-glycoprotein were similar. The structure of P-glycoprotein has been subjected to molecular dissection such that the cytoplasmic, transmembrane, and extracellular parts of the molecule have been delineated (24). The epitopes defined as DRHFLN, GNYD, RRYPF, GVTTLSS, and VDWLR have no obvious homologue. KTTLLK might be represented by amino acids GTTLVL (amino acids 317 to 322, P-glycoprotein) and RGFL might be represented by amino acids RGWK (amino acids 210 to 214, P-glycoprotein), which were postulated as being on the outside of the mammalian cell (24).
Clearly any activity mediated by a scFv must be independent of the Fc component of an antibody, since it is absent from these recombinant antibody fragments. Ramisse et al. (34) showed that passive local immunotherapy with human plasma-derived immunoglobulins (IVIG) was therapeutic in a model of staphylococcal pneumonia. IVIG saturated with protein A or its F(ab′)2 fragments was as efficient as intact IVIG, suggesting that protection did not require opsonization through IgG Fc-phagocyte Fc-receptor interactions.
Targeting ABC transporters conserved between different species of bacteria could be a way in which the host's immune system maximizes therapeutic affect against a range of infections using the minimum number of antibodies. The hyperimmune rabbit serum raised against the recombinant ABC transporter cross-reacted strongly with one other staphylococcal antigen at 97 kDa which may also be an ABC transporter (Fig. 3, lane 4). The immunodominance of similar proteins has been reported in patients with endocarditis due to Enterococcus faecalis (11, 48). Perhaps ABC transporters constitute generic antigens, conserved between multiple genera, and the neutralization of these essential transporter proteins may be able to inhibit a wide variety of microbial import and export functions. The potential therapeutic activity of such antibodies is under investigation, starting with the reexpression of scFvs in E. coli in a phage-free form in order to facilitate further assessment with greater antibody concentrations.
REFERENCES
- 1.Allignet J, Loncle V, el Sohl N. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene. 1992;117:45–51. doi: 10.1016/0378-1119(92)90488-b. [DOI] [PubMed] [Google Scholar]
- 2.Allignet J, el Sohl N. Characterization of a new staphylococcal gene, vgaB, encoding a putative ABC transporter conferring resistance to streptogramin A and related compounds. Gene. 1997;202:133–138. doi: 10.1016/s0378-1119(97)00464-2. [DOI] [PubMed] [Google Scholar]
- 3.Archer G L. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 3a.Anonymous. Revised guidelines for the control of methicillin-resistant Staphylococcus aureus infection in hospitals. British Society for Antimicrobial Chemotherapy, Hospital Infection Society and the Infection Control Nurses Association. J Hosp Infect. 1998;39:253–290. doi: 10.1016/s0195-6701(98)90293-6. [DOI] [PubMed] [Google Scholar]
- 4.Ayliffe G A. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24(Suppl. 1):S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 5.Bell J A, Pennington T H, Petrie D T. Western blot analysis of staphylococcal antibodies present in human sera during health and disease. J Med Microbiol. 1987;23:95–99. doi: 10.1099/00222615-23-2-95. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Burnie J P, Matthews R C, Lee W, Murdoch D. A comparison of immunoblot and DNA restriction patterns in characterising methicillin-resistant isolates of Staphylococcus aureus. J Med Microbiol. 1989;29:255–261. doi: 10.1099/00222615-29-4-255. [DOI] [PubMed] [Google Scholar]
- 8.Burnie J P, Brooks W, Donohoe M, Hodgetts S, Al-Ghamdi A, Matthews R C. Defining antibody targets in Streptococcus oralis infection. Infect Immun. 1996;64:1600–1608. doi: 10.1128/iai.64.5.1600-1608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall A, Scharff M D. Return to the past: the case for antibody-based therapies in infectious diseases. Clin Infect Dis. 1995;21:150–161. doi: 10.1093/clinids/21.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N Y, Jiang S-Q, Klein D A, Paulus H. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem. 1993;268:9448–9465. [PubMed] [Google Scholar]
- 11.Clark I M, Burnie J P. Isolation and sequence determination of an immunodominant antigen from Enterococcus faecalis. Serodiagn Immunother Infect Dis. 1993;5:85–92. [Google Scholar]
- 12.Cockayne A, Hill P J, Powell N B L, Bishop K, Sims C, Williams P. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect Immun. 1988;66:3767–3774. doi: 10.1128/iai.66.8.3767-3774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Communicable Disease Report. Methicillin resistance in Staphylococcus aureus isolated from the blood in England and Wales: 1994 to 1998. CDR Weekly. 1999;9:65. , 68. [PubMed] [Google Scholar]
- 14.Coulter S, Schwan N W R, Ng E Y W, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 15.Domanski T L, Bayles K W. Analysis of Staphylococcus aureus genes encoding penicillin-binding protein 4 and an ABC-type transporter (peptidoglycan; cross-linking; transpeptidase; carboxypeptidase) Gene. 1995;167:111–113. doi: 10.1016/0378-1119(96)82965-9. [DOI] [PubMed] [Google Scholar]
- 16.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 18.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–88. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 19.Geysen H, Rodda S J, Mason T J, Tribbick G, Schoofs P G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 20.Greenberg D P, Bayer A S, Turner D, Ward J I. Antibody responses to protein A in patients with Staphylococcus aureus bacteremia and endocarditis. J Clin Microbiol. 1990;28:458–462. doi: 10.1128/jcm.28.3.458-462.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiramatsu K, Aritaka H, Haamaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 22.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 23.Linton K J, Higgins C F. Micro genomics: the Escherichia coli ATP-binding cassette (ABC) proteins. Mol Microbiol. 1998;28:5–13. doi: 10.1046/j.1365-2958.1998.00764.x. [DOI] [PubMed] [Google Scholar]
- 24.Loo T W, Clarke D M. Molecular dissection of the human multidrug resistance P-glycoprotein. Biochem Cell Biol. 1999;77:11–23. [PubMed] [Google Scholar]
- 25.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 26.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 27.Marks J D, Hoogenboom H R, Bonnett T P, McCafferty J, Griffiths A D, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed in phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 28.Matthews R C, Hodgetts S, Burnie J P. Preliminary assessment of a human recombinant antibody fragment to hsp90 in murine invasive candidiasis. J Infect Dis. 1995;171:1668–1671. doi: 10.1093/infdis/171.6.1668. [DOI] [PubMed] [Google Scholar]
- 29.Matsuoka M, Endou K, Kobayashi H, Inoue M, Nakajima Y. A plasmid that encodes three genes for resistance to macrolide antibiotics in Staphylococcus aureus. FEMS Microbiol Lett. 1998;167:221–227. doi: 10.1111/j.1574-6968.1998.tb13232.x. [DOI] [PubMed] [Google Scholar]
- 30.Mei M-J, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 31.Milton I D, Hewitt C L, Harwood C R. Cloning and sequencing of a plasmid-mediated erythromycin resistance determinant from Staphylococcus xylosus. FEMS Microbiol Lett. 1992;97:141–148. doi: 10.1016/0378-1097(92)90377-z. [DOI] [PubMed] [Google Scholar]
- 32.Neubauer H, Pantel I, Lindgren P, Gđtz F. Characterization of the molybdate transport system ModABC of Staphylococcus carnosus. Arch Microbiol. 1999;172:109–115. doi: 10.1007/s002030050747. [DOI] [PubMed] [Google Scholar]
- 33.Quentin Y, Fichant G, Denizot F. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J Mol Biol. 1999;287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- 34.Ramisse F, Szatanik M, Binder P, Alonos J M. Passive local immunotherapy of experimental staphylococcal pneumonia with human intravenous immunoglobulin. J Infect Dis. 1993;168:1030–1033. doi: 10.1093/infdis/168.4.1030. [DOI] [PubMed] [Google Scholar]
- 35.Richardson J F, Reith S. Characterization of a strain of methicillin resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J Hosp Infect. 1993;25:45–52. doi: 10.1016/0195-6701(93)90007-m. [DOI] [PubMed] [Google Scholar]
- 36.Ross J I, Eady E A, Cove J H, Cunliffe W J, Baumberg S, Wootton J C. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol. 1990;4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 37.Ross J I, Eady E A, Cove J H, Baumberg S. Identification of a chromosomally encoded ABC-transport system with which the staphylococcal erythromycin exporter MsrA may interact. Gene. 1995;153:93–98. doi: 10.1016/0378-1119(94)00833-e. [DOI] [PubMed] [Google Scholar]
- 38.Stranden A M, Ehlert K, Labischinski H, Berger-Bachi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabaqchali S. Vancomycin-resistant Staphylococcus aureus: apocalypse now? Lancet. 1997;350:1644–1645. doi: 10.1016/S0140-6736(05)64269-9. [DOI] [PubMed] [Google Scholar]
- 40.Tam R, Saier M H., Jr Structural, functional, and evolutionery relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 42.Traub W H, Bauer D, Leonhard B. Immunobiology of methicillin-resistant Staphylococcus aureus: immune response of rabbits and patients to systemic infection. Exp Chemother. 1996;42:118–132. doi: 10.1159/000239431. [DOI] [PubMed] [Google Scholar]
- 43.U.S. Congress, Office of Technology Assessment. Impacts of antibiotic-resistant bacteria. OTA-H-629. U.S. Washington, D.C.: Government Printing Office; 1995. [Google Scholar]
- 44.van Veen H R, Callaghan W R, Soceneantu L, Sardini A, Konings W N, Higgins C F. A bacterial antibiotic-resistance gene that complements the human multidrug-resistance P-glycoprotein gene. Nature. 1998;391:291–295. doi: 10.1038/34669. [DOI] [PubMed] [Google Scholar]
- 45.Waldvogel W. New resistance in Staphylococcus aureus. N Engl J Med. 1999;340:556–557. doi: 10.1056/NEJM199902183400709. [DOI] [PubMed] [Google Scholar]
- 46.Winter G, Mistein C. Man-made antibodies. Nature. 1991;349:293–299. doi: 10.1038/349293a0. [DOI] [PubMed] [Google Scholar]
- 47.Xu Y, Jiang L, Murray B E, Weinstock G M. Enterococcus faecalis antigens in human infections. Infect Immun. 1997;65:4207–4215. doi: 10.1128/iai.65.10.4207-4215.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young R A, Davies R W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci USA. 1983;80:1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]