Abstract
The toxicity of mercury (Hg) mainly depends on its form. Whole-cell biosensors respond selectively to toxic Hg(ii), efficiently transformed by environmental microbes into methylmercury, a highly toxic form that builds up in aquatic animals. Metabolically engineered Escherichia coli (E. coli) have successfully produced rainbow colorants. By de novo reconstruction of the carotenoid synthetic pathway, the Hg(ii)-responsive production of lycopene and β-carotene enabled programmed E. coli to potentially become an optical biosensor for the qualitative and quantitative detection of ecotoxic Hg(ii). The red color of the lycopene-based biosensor cell pellet was visible upon exposure to 49 nM Hg(ii) and above. The orange β-carotene-based biosensor responded to a simple colorimetric assay as low as 12 nM Hg(ii). A linear response was observed at Hg(ii) concentrations ranging from 12 to 195 nM. Importantly, high specificity and good anti-interference capability suggested that metabolic engineering of the carotenoid biosynthesis was an alternative to developing a visual platform for the rapid analysis of the concentration and toxicity of Hg(ii) in environmentally polluted water.
Genetically controlled carotenoid biosynthesis produces a specific and sensitive biosensor for inorganic mercury.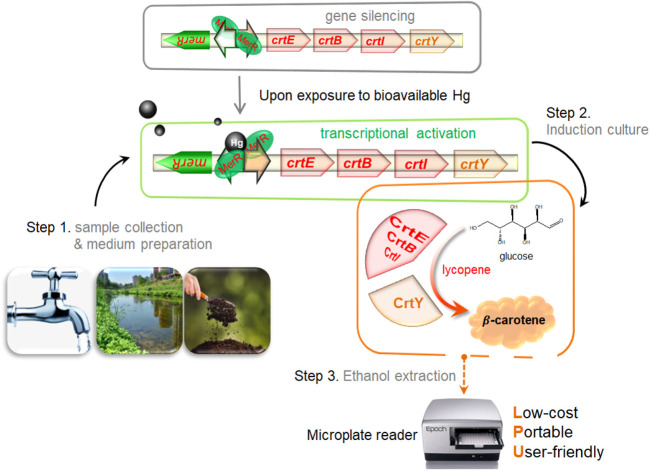
Introduction
Persistent Hg pollution poses a considerable challenge to human health globally.1 Instrumental tests are widely used for environmental heavy metal detection. However, conventional analytic methods fail to differentiate total Hg and Hg(ii).2 Elemental Hg, inorganic Hg, and organic Hg are the primary chemical forms in the environment.3 Inorganic Hg(ii) mainly originates from anthropogenic activities.3 The observed Hg species in plants is mainly inorganic Hg(ii).4 Environmental sulfate-reducing bacteria (SRB) can quickly transform bioavailable Hg(ii) into methyl Hg, which is selectively enriched in the aquatic food chain and threatens human health.3,5 Determining inorganic Hg(ii), the substrate for SRB, is vital for environmental risk assessment of polluted water.6 As a supplement to instrumental methods, various programmed whole-cell biosensors have been artificially designed to focus on detecting bioavailable heavy metals, including Hg(ii), by simulating environmental microbes for their ecological risk prediction.7–9
Whole-cell biosensors consist of a sensor and an actuator, allowing them to monitor environmental heavy metals using colorimetric, electrochemical, fluorescent, or luminescent output signals.10,11 Among traditional reporters, colorimetric rainbow colorants have recently been influential in developing mini-equipment, low-cost, and portable whole-cell biosensors for toxic metals. Cold-colored pigments, such as indigoidine,12,13 violacein,14,15 and their derivatives,16 have been successfully employed in developing optical biosensors for toxic heavy metals. However, few studies have focused on developing warm-colored pigment-based biosensors. The warm-colored system may not be sufficiently visually striking. Differential fluorescent coloration distinguished heavy metals with similar properties.17,18 Furthermore, multicolor indications showed great potential in the qualitative detection of blood zinc.19,20 More pigments, as colorful reporters, are expected to be employed in designing versatile whole-cell biosensors for use on different occasions.
Recent advances in metabolic engineering enable engineered cells to become microbial factories for the biosynthesis of natural colorants.21 The carotenoid synthetic pathway is an essential metabolic flux toward warm-colored substances, including red lycopene, orange β-carotene, and yellow zeaxanthin, which are economically valuable food additives, antioxidants, and nutraceuticals.22 Among many chassis cells, bacteria, especially E. coli, became ideal candidates because of their transparent genetic background, self-renewability, and tolerance of a harsh environment. The reconstruction of the carotenoid biosynthetic pathway in E. coli has been shown to be an efficient strategy for developing high-performance biological factories.23
In the present study, the carotenoid biosynthetic pathway was assembled de novo under the control of the Hg(ii) sensory module in E. coli. A visual platform was developed based on the production of Hg(ii)-responsive warm-colored pigment. The resultant bacterial biosensors responded to a nanomolar concentration of Hg(ii), followed by the biosynthesis of red lycopene and orange β-carotene. The sensing property, dose–response relationship, and metal selectivity were studied in detail. This novel pigment-based biosensor shows potential for predicting ecological risk by determining the bioavailable Hg(ii) in environmental samples, including surface water and soil extracts, which can be quickly introduced into the culture systems.
Materials and methods
Bacteria, vectors, and reagents
E. coli TOP10 was used as the host for gene cloning, and Rosetta (DE3) was used as the host for Hg(ii)-inducible pigment biosynthesis. The plasmids and engineered bacteria used in the study are listed in Table S1.† Luria-Bertani (LB) broth (1% tryptone, 0.5% yeast extract, 1% sodium chloride, and 50 μg per mL ampicillin) was used for recombinant bacterial culture. Metal salts were of analytical grade and dissolved in purified water at 1 mM for the test. β-Carotene (Puritan's Pride, New York, USA) was used to build the visible light scanning spectrum.
Construction of bacterial biosensors based on the carotenoid biosynthetic pathway
crtE encoding geranylgeranyl pyrophosphate synthase, crtB encoding phytoene synthase, and crtI encoding phytoene dehydrogenase are required for lycopene biosynthesis from the substrate FPP.24 The lycopene synthetic gene cluster, a tricistronic genetic unit crtEBI, was synthesized de novo (Sangon, Shanghai, China) according to the E. coli codon preference (Fig. S1†). The synthetic lycopene biosynthetic cassette was inserted into the NdeI/SacI sites of pET-21a to generate pET-crtEBI. To further assemble the β-carotene biosynthetic gene cluster, crtY encoding lycopene cyclase was genetically fused to the crtEBI cassette to generate tetracistronic crtEBIY, which was inserted into the NdeI/SacI sites of pET-21a to generate pET-crtEBIY. A Hg(ii) sensory module containing the merR gene and the mer promoter was PCR amplified from pPmer and inserted into the BglII/XbaI sites of pET-crtEBI and pET-crtEBIY to generate pPmer-crtEBI and pPmer-crtEBIY, respectively. Furthermore, an independent mer promoter was introduced into the upstream region of each pigment biosynthetic enzyme by an overlap PCR to generate pPmer-crtEBIY-4.
Comparison of Hg(ii)-driven color signals from the carotenoid biosynthetic pathway
To compare color signals derived from two carotenoid derivatives, lycopene and β-carotene, overnight cultures of R/Hg-EBI and R/Hg-EBIY were diluted in 1 : 100 fresh LB broth. After culturing at 37 °C for three hours to produce the early exponential phase, a double-dilution method was used to obtain 0–1562 nM Hg(ii)-exposed groups to ensure concentration accuracy in the nanomolar range. After another 3 h of induction, 1.0 mL cultures of R/Hg-EBI were centrifuged at 8000g for 1 min, and the biosensor cell pellets were observed. Bacterial density and a β-carotene-based signal derived from R/Hg-EBIY were determined by colorimetry.
Response patterns of a β-carotene-based signal toward Hg(ii)
Early exponential phase cultures of R/Hg-EBIY were exposed to 0, 49, and 195 nM Hg(ii). Following induction at 37 °C with shaking at 250 rpm, the cultures were sampled at one-hour intervals. Ethanol phases containing pigment were prepared and measured by colorimetry at 452 nm.
Detection selectivity assay
Early exponential phase cultures of R/Hg-EBIY were exposed to Pb(ii), Cd(ii), Zn(ii), and Hg(ii) at various concentrations. After induction at 37 °C for 3 h, bacterial density and pigment signal were determined at 600 nm and 452 nm, respectively.
Non-target metal interference assay
Early exponential phase cultures of R/Hg-EBIY were exposed to 195 nM Hg(ii) in a combination of 6 μM divalent metal ions. Bacterial density and pigment signal were determined after induction at 37 °C for 3 h.
Spectrophotometric detection
Aliquots (100 μL) of bacterial cultures were pipetted into a 96-well microplate and read using a plate reader (BioTek Epoch, USA) at 600 nm for bacterial density. The pellets were prepared from 1.9 mL of culture by centrifugation, extracted with 200 μL of ethanol, and vortexed violently for 1 min. Aliquots (150 μL) of the upper ethanol extract were transferred into the microplate, read at 452 nm for the β-carotene-derived signal, and scanned at wavelengths of 350–550 nm at intervals of 2 nm.
Results and discussion
Tuneable control of carotenoid production by metalloregulator MerR
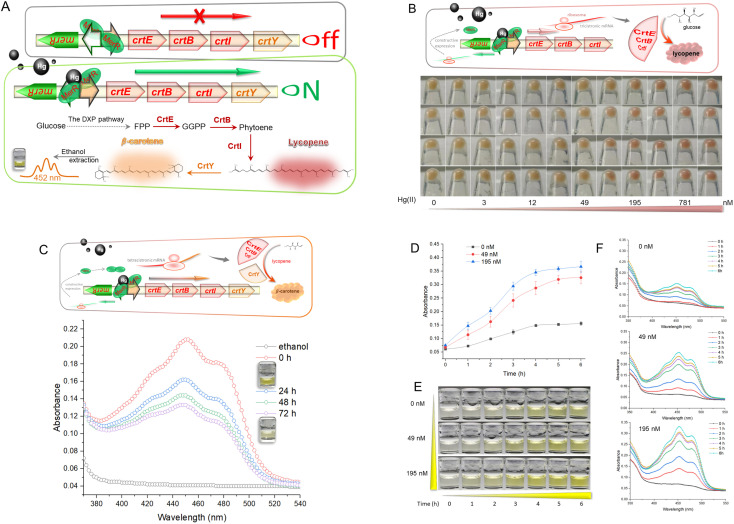
Hg-mediated toxicity has always been a great barrier against microbial survival. The mer operon is a well-evolved mechanism of Hg detoxification among microbes.25 The metalloregulator MerR efficiently utilizes three cysteine residues with a high affinity toward Hg(ii) to activate the transcription of Hg-resistant genes.26 This study cloned the mer promoter regulated by dimeric MerR into the upstream of the carotenoid biosynthetic gene clusters. As shown in Fig. 1A, dimeric MerR plays the role of a transcription repressor without direct exposure to Hg(ii), but rapidly transforms into a transcription activator under Hg(ii) exposure. Upon transcription, the products of the carotenoid biosynthetic gene cluster synthesize red lycopene and orange β-carotene. As shown in Fig. 1B, the dose–response relationship between the accumulation of lycopene and Hg(ii) exposure was observed. The red color of the cell pellet was visible upon exposure to 49 nM Hg(ii) and above. Lycopene was demonstrated to be a recognizable indicator due to its red color.
Fig. 1. (A) Mechanism of bioavailable Hg(ii) detection by metabolically engineered E. coli. DXP, 1-deoxy-d-xylulose 5-phosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl pyrophosphate. (B) Dose-dependent lycopene biosynthesis in response to toxic Hg(ii). (C) Visible absorbance curves of Hg(ii)-induced β-carotene in ethanol. (D) Dose–time–response curves of R/Hg-EBIY toward Hg(ii). The values shown are mean ± SD (n = 3). (E) Color change of ethanol phases of R/Hg-EBIY after exposure to Hg(ii) at different time intervals. (F) Representative visible absorbance curves of three concentrations of Hg(ii)-induced β-carotene in ethanol.
Because of its high antioxidant activity, the red color of soluble lycopene in organic phases fades rapidly. High-performance liquid chromatography (HPLC) is widely used to determine carotenoid extraction.27 However, colorimetric analysis facilitates the development of a mini-equipment biosensor that can be read directly by the naked eye.9 Thus, the crtY gene encoding lycopene cyclase, which transforms lycopene into more stable β-carotene, was introduced into the reporter system (Fig. 1C).
The commercial β-carotene was dissolved in ethanol and scanned at wavelengths of 350–550 nm (Fig. S2†). The Hg(ii)-induced β-carotene could also be extracted with nontoxic ethanol and analyzed colorimetrically. Although there may be some impurities in the ethanol extract of Hg(ii)-induced biosensor cell pellets, absorption spectra of commercial β-carotene (Fig. S2†) and Hg(ii)-induced pigment (Fig. 1C) were still similar and composed of three peaks. The maximum absorbance peak of β-carotene is located near 452 nm, which was chosen for pigment quantification in the following study. Accompanying the slow fading of the orange color of β-carotene dissolved in ethanol at 37 °C, the peak of the visible light scanning spectrum is also decreasing (Fig. 1C). Due to its hydrophobicity and instability, it was not easy to maintain the stability of dissolved β-carotene in previous studies.28 This will become an inherent disadvantage when β-carotene is employed as a reporter. Therefore, fast reading needs to be done following the ethanol extraction to avoid the negative influence of oxidation.
To study the time–response relationship, R/Hg-EBIY in the early exponential phase was exposed to different concentrations of Hg(ii). A dose-related increase in the β-carotene-derived signal was observed with an extension of incubation time (Fig. 1D). Upon exposure to Hg(ii), both the β-carotene-derived color signal and the background increased with the extension of induction time. It is worth noting that, including the background, the orange color was not significantly deepened after a 4 h induction (Fig. 1E). The dose–time–response pattern could also be well compared from the visible absorbance spectrum (Fig. 1F). Although the maximum production of β-carotene was observed after 4 h of induction, a 3 h induction was enough to give an apparent visual signal, and was chosen in the following study to shorten the detection time. The induction time for the accumulation of pigment signal is significantly shorter than that for fluorescent signal amplification. Overnight induction is usually done in detecting metal ions using fluorescent protein-based biosensors.18,29
Because the naked eye does not easily recognize the warm-colored pigment,30 Rosetta (DE3), a widely used bacterial host for improved heterogonous protein production, was used to enhance the expression of CrtEBIY for higher biosynthesis of β-carotene. In previous studies, although the naked eye recognized the color deepening of cell pellets, quantitative colorimetric detection was challenging in warm-colored pigment-based biosensors.19,31 An artificial mer operon with vigorous transcriptional activity, biosensor cells with active metabolic activity in the exponential phase, an optimized bacterial host, and codon-optimized pigment biosynthesis genes were believed to contribute to the satisfactory performance of the resultant biosensor in this study. The independent mer promoter was associated with enhanced expression of the downstream gene.29 However, the improved overall expression level of CrtEBIY did not increase β-carotene production (Fig. S3†). Studies have shown that balancing gene expression optimizes the carotenoid metabolic pathway.22,24 The desired strength of ribosome binding sites (RBSs) is a vital factor in optimizing the genetic circuit for improved β-carotene accumulation, which we will look at in a further study.
The metabolically engineered biosensor in response to bioavailable Hg(ii)
To systematically evaluate the response characteristics of this biosensor toward Hg(ii), R/Hg-EBIY was induced with 0, 1.5, 3, 6, 12, 24, 49, 98, 195, 391, 781, and 1562 nM Hg(ii) in the exponential phase. As shown in Fig. 2A, both toxic Hg(ii) in this concentration range and increased biosynthesis of β-carotene slightly affected bacterial growth. The limit of detection (LOD) calculated using the mean blank value plus three times the standard deviation was 12 nM (Fig. 2B). The β-carotene derived signal responsive to Hg(ii) was significantly increased between 12 and 195 nM and then remained stable (Fig. 2C). If the overexpression of the reporter gene has no significant effect on bacterial growth, the biosensing response pattern will be mainly determined by the inherent characteristics of the Hg(ii) sensory module employed. Therefore, the overall response trend is consistent with MerR-based biosensors developed previously.32,33 The colorimetric assay showed linearity over a wide range from 12 to 195 nM (Fig. 2D). Due to the low LOD, the quantitative detection range of β-carotene-based biosensors is significantly lower than that of fluorescent protein-based whole-cell biosensors.33–35 The increase in the Hg(ii)-inducible absorption spectrum is evident (Fig. 2E). It shows that absorbance with the range of 425–475 nm can be chosen to determine the biosensing signal, which also broadens the threshold for practical application. The color of the cell pellets and the ethanol extraction phase changed toward light orange with increasing Hg(ii) (Fig. 2F). This orange color gradient in the ethanol phase is visible to the naked eye.
Fig. 2. The performance of colorimetric bacterial biosensor R/Hg-EBIY exposed to increased concentrations of Hg(ii). (A) Bacterial densities of R/Hg-EBIY after exposure to increased concentrations of Hg(ii). (B) The response sensitivity of R/Hg-EBIY. The asterisk represents the LOD. (C) The dose–response curve of R/Hg-EBIY. Data shown are mean ± SD (n = 5). (D) Regression analysis of the relationship between visual signal and Hg(ii) ranging from 12 to 195 nM. The x-axis displays the Hg(ii) concentration on the log2 scale. Visible absorbance curves (E) and images (F) of various concentrations of Hg(ii)-induced β-carotene in ethanol phases.
An overall comparison of advanced whole-cell biosensors toward Hg(ii) employing various reporters is listed in Table 1. The pigment-based biosensor has the advantage of multiple biosensors. The target metal-induced pigment synthetase can catalyze the continuous accumulation of colorant reporters, amplifying the biosensing signal.36 Thus, the LOD of the β-carotene-based biosensor was significantly lower than those of previously developed fluorescent protein-based whole-cell biosensors using MerR as the Hg(ii) sensory element.34,35,37 The only disadvantage of signal amplification is that it sometimes leads to higher detection background and lower signal-to-noise ratio.15 In previous studies, only luciferase-based whole-cell biosensors could respond to Hg(ii) in the nanomolar range.38,39 Due to the signal amplification effect, the LOD of a pigment-based biosensor is comparable to a biosensor employing enzymatic reporters, including β-galactosidase and luciferase. However, its advantage over these traditional biosensors is its independence from expensive substrates and instruments.10
Comparison of Hg(ii) whole-cell biosensors using various reporters.
| Reporters | Extra substrate | Signal amplification | Signal type | Detection method |
|---|---|---|---|---|
| Fluorescent proteins | — | — | Various | Fluorometry |
| Luciferase | + | + | Single | |
| β-Galactosidase | + | + | Single | Colorimetry |
| Pigment synthetase | — | + | Tailored |
Visualization of pigment-based reporters is a true advantage for developing a potential mini-equipment biosensor. However, the current operation is too complex to meet the requirement of rapid on-site detection. Compared with the colorimetric detection of β-carotene in ethanol, direct visual observation of the color of the culture or cell pellets has the potential to become a promising semi-quantitative method. Our study presents a proof-of-concept study for a novel design of a pigment-based biosensor, which is expected to be used in the colorimetric determination of Hg(ii) in environmental water samples mixed into the culture system.
Precise response toward bioavailable Hg(ii)
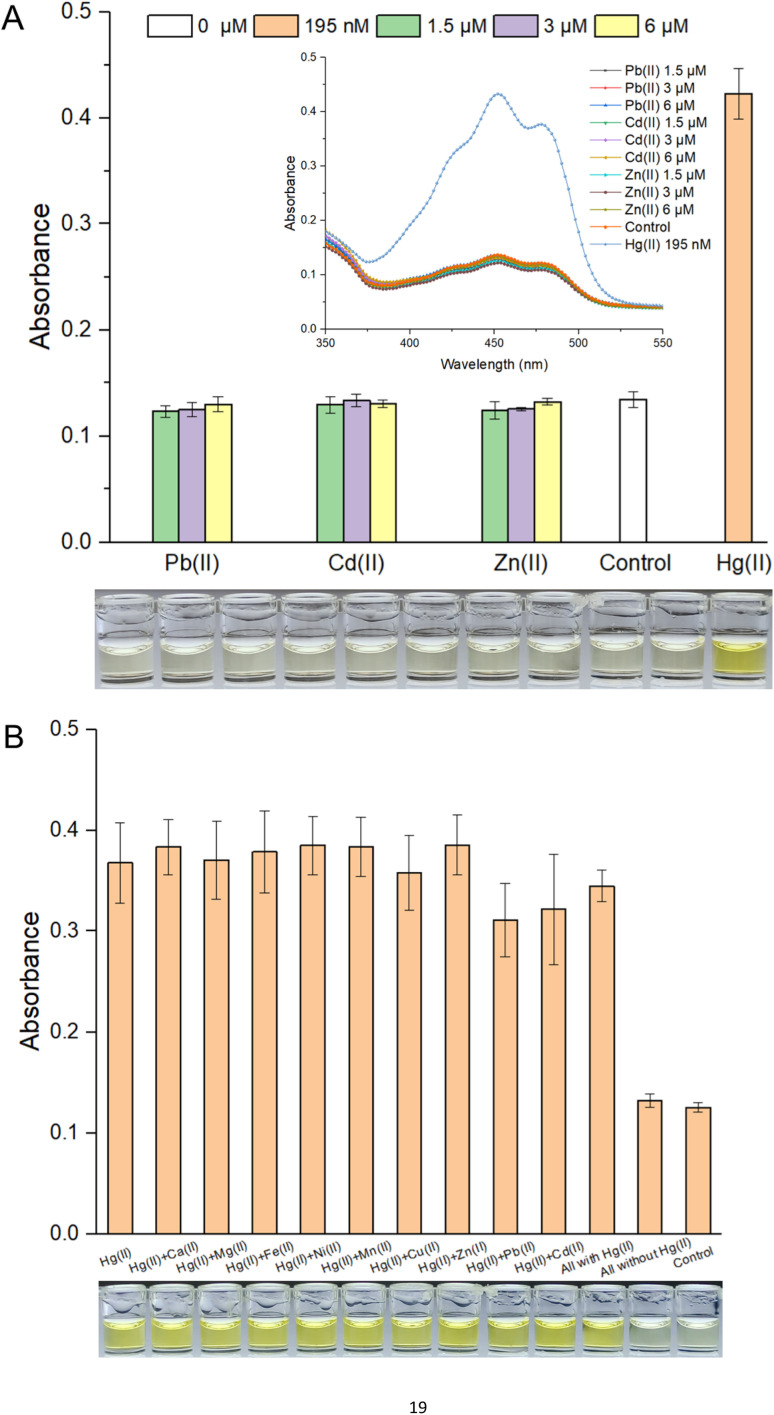
Response selectivity is another vital property of a whole-cell biosensor. The heavy metal selectivity of MerR-like metalloregulators, such as Cd(ii)-responsive CadR and Pb(ii)-responsive PbrR, is usually poor. Directed evolution40,41 and optimized genetic circuits18,42 are necessary to improve the heavy metal selectivity of the resultant biosensors. Considering the high specificity of the MerR regulator toward Hg(ii), natural MerR-based biosensors always show a high response to Hg(ii).26,33 Previous studies have shown that pigment-based actuators would not interfere with the performance of metal-responsive modules.12,20 As shown in Fig. S4A,† the bacterial densities were not significantly decreased when exposed to different concentrations of Pb(ii), Cd(ii), and Zn(ii). As expected, the whole-cell biosensor using β-carotene as the output signal showed a high specific response toward Hg(ii). The visible light absorbance and the striking orange color in ethanol are outstanding (Fig. 3A). Importantly, the responses toward Pb(ii), Cd(ii), and Zn(ii) were similar to the background in the no-exposure control group (Fig. 3A).
Fig. 3. (A) Response selectivity of colorimetric bacterial biosensor R/Hg-EBIY. Inset is the visible light scanning spectra of various ethanol extraction phases. (B) Anti-interference characteristics of R/Hg-EBIY. Data shown are mean ± SD (n = 3). A group of representative photographs of the results are shown below.
It is well known that chemically related group 12 metals (zinc, cadmium, and mercury) are not well distinguished by MerR-like transcription regulators.26 This type of biosensor commonly showed a nonspecific response toward group 12 members.17,43 The association of non-target metal ions with a MerR-like regulator was believed to block target metal ions binding to it, so it could sometimes trigger the transcription of reporter genes,40 but it could not do so in most cases.16 In this study, exposure to various mixed metals exerted no significant influence on bacterial growth (Fig. S4B†). As shown in Fig. 3B, although most of these interfering metal ions, such as Ca(ii), Mg(ii), Fe(ii), Ni(ii), Mn(ii), Cu(ii), and Zn(ii), exerted no influence on the response of a β-carotene-based biosensor toward Hg(ii), a slight decrease was still observed in the Pb(ii) and Cd(ii) coexistence group. It was exciting to see that all groups exposed to mixed metal ions containing Hg(ii) showed a striking orange color, significantly different from the two nonexposed groups (Fig. 3B).
By introducing the water samples into the culture medium inoculated with programmed bacteria, bacterial biosensors have been validated in sensing toxic, bioavailable, and bioaccessible target metal ions in environmental water samples, including potable water, surface water, and soil extracts.12,16,30,44,45 Complex interference factors, such as dissolved organic matter, trace metal ions, and pH values, slightly influenced the biosensing performance in these previous studies. However, such studies are still in the proof-of-concept stage. Further study should address the optimization of genetic circuits, simplifying the procedure, and determining the environmental sample detection range.
Conclusions
In this study, we report a novel colorimetric whole-cell biosensor toward Hg(ii) by employing the carotenoid synthetic pathway in E. coli. Upon exposure to a high Hg(ii) concentration, intracellular accumulation of red lycopene enabled bacterial cells to become novel visual biosensors independent of the instrument. Ethanol-extracted β-carotene made it quantitatively possible to detect bioavailable Hg(ii) by colorimetry. The LOD is comparable to those of luciferase-based biosensors but independent of an extra substrate and expensive devices. Furthermore, this novel biosensor is highly selective toward Hg(ii) and silent towards various metal ions. Based on this proof-of-concept study, synthetic biology and metabolic engineering are demonstrated to contribute to the emergence of novel, colorful reporter-based biosensors. A streamlined procedure will be built to make this novel biosensor applicable for practical purposes, and the platform will be extensively validated against various environmental samples in future studies.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82073517), the Natural Science Foundation of Guangdong Province (2019A1515011989, 2021A1515012472), the Science and Technology Program of Shenzhen (KCXFZ20201221173602007), the Shenzhen Key Medical Discipline Construction Fund (SZXK068), and the Shenzhen Fund for Guangdong Provincial High-level Clinical Key Specialties (SZGSP015).
Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra06764a
References
- Magos L. Clarkson T. W. Ann. Clin. Biochem. 2006;43:257–268. doi: 10.1258/000456306777695654. [DOI] [PubMed] [Google Scholar]
- Santa-Rios A. Barst B. D. Tejeda-Benitez L. Palacios-Torres Y. Baumgartner J. Basu N. Chemosphere. 2021;266:129001. doi: 10.1016/j.chemosphere.2020.129001. [DOI] [PubMed] [Google Scholar]
- Barkay T. Miller S. M. Summers A. O. FEMS Microbiol. Rev. 2003;27:355–384. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- Shahid N. M. Khalid S. Bibi I. Bundschuh J. Khan Niazi N. Dumat C. Sci. Total Environ. 2020;711:134749. doi: 10.1016/j.scitotenv.2019.134749. [DOI] [PubMed] [Google Scholar]
- Stenzler B. Hinz A. Ruuskanen M. Poulain A. J. Environ. Sci. Technol. 2017;51:9653–9662. doi: 10.1021/acs.est.7b01414. [DOI] [PubMed] [Google Scholar]
- Frances-Monerris A. Carmona-Garcia J. Acuna A. U. Davalos J. Z. Cuevas C. A. Kinnison D. E. Francisco J. S. Saiz-Lopez A. Roca-Sanjuan D. Angew. Chem. 2020;59:7605–7610. doi: 10.1002/anie.201915656. [DOI] [PubMed] [Google Scholar]
- Belkin S. Wang B. Microb. Biotechnol. 2022;15:103–106. doi: 10.1111/1751-7915.13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraskie M. Roshid M. H. O. O'Connor G. Dikici E. Zingg J. M. Deo S. Daunert S. Biosens. Bioelectron. 2021;191:113359. doi: 10.1016/j.bios.2021.113359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. Yu H. Zhang B. Liu S. Liu C. G. Li F. Song H. Biotechnol. Adv. 2022;60:108019. doi: 10.1016/j.biotechadv.2022.108019. [DOI] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Liu L. Yi J. World J. Microbiol. Biotechnol. 2021;38:9. doi: 10.1007/s11274-021-03198-w. [DOI] [PubMed] [Google Scholar]
- Van der Meer J. R. Belkin S. Nat. Rev. Microbiol. 2010;8:511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Gao C. X. Li H. Lin Y. R. Yun J. P. Chen Y. T. Yi J. Environ. Technol. Innovat. 2022:102511. doi: 10.1016/j.eti.2022.102511. doi: 10.1016/j.eti.2022.102511. [DOI] [Google Scholar]
- Hui C. Y. Guo Y. Li L. M. Liu L. Chen Y. T. Yi J. Zhang N. X. Appl. Microbiol. Biotechnol. 2021;105:6087–6102. doi: 10.1007/s00253-021-11441-5. [DOI] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Liu L. Zhang N. X. Gao C. X. Yang X. Q. Yi J. RSC Adv. 2020;10:28106–28113. doi: 10.1039/D0RA04815A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Li H. Gao C. X. Yi J. Sci. Rep. 2022;12:6898. doi: 10.1038/s41598-022-11051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Zhu D. L. Li L. M. Yi J. Zhang N. X. Biosens. Bioelectron. 2022;214:114531. doi: 10.1016/j.bios.2022.114531. [DOI] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Wu J. Liu L. Yang X. Q. Guo X. Xie Y. Yi J. Front. Microbiol. 2021;12:696195. doi: 10.3389/fmicb.2021.696195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C. Y. Guo Y. Li H. Chen Y. T. Yi J. Front. Microbiol. 2022;13:846524. doi: 10.3389/fmicb.2022.846524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNerney M. P. Michel C. L. Kishore K. Standeven J. Styczynski M. P. Nat. Commun. 2019;10:5514. doi: 10.1038/s41467-019-13454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watstein D. M. Styczynski M. P. ACS Synth. Biol. 2018;7:267–275. doi: 10.1021/acssynbio.7b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabowo C. P. S. Eun H. Yang D. Huccetogullari D. Jegadeesh R. Kim S.-J. Lee S. Y. Trends Chem. 2022;4:608–626. doi: 10.1016/j.trechm.2022.04.009. [DOI] [Google Scholar]
- Yang D. Park S. Y. Lee S. Y. Adv. Sci. 2021;8:e2100743. doi: 10.1002/advs.202100743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. Park S. Y. Park Y. S. Eun H. Lee S. Y. Trends Biotechnol. 2020;38:745–765. doi: 10.1016/j.tibtech.2019.11.007. [DOI] [PubMed] [Google Scholar]
- Ghodasara A. Voigt C. A. Nucleic Acids Res. 2017;45:8116–8127. doi: 10.1093/nar/gkx530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathema V. B. Thakuri B. C. Sillanpaa M. Arch. Microbiol. 2011;193:837–844. doi: 10.1007/s00203-011-0751-4. [DOI] [PubMed] [Google Scholar]
- Chang C. C. Lin L. Y. Zou X. W. Huang C. C. Chan N. L. Nucleic Acids Res. 2015;43:7612–7623. doi: 10.1093/nar/gkv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. C. Zhong Y. J. Liu J. Sandmann G. Chen F. Metab. Eng. 2013;17:59–67. doi: 10.1016/j.ymben.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Stutz H. Bresgen N. Eckl P. M. Free Radic. Res. 2015;49:650–680. doi: 10.3109/10715762.2015.1022539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N. X. Guo Y. Li H. Yang X. Q. Gao C. X. Hui C. Y. PLoS One. 2021;16:e0252190. doi: 10.1371/journal.pone.0252190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Huang Z. L. Zhu D. L. Hu S. Y. Li H. Hui C. Y. Front. Microbiol. 2022;13:975421. doi: 10.3389/fmicb.2022.975421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K. Inoue K. Takahashi Y. Ueda S. Isoda K. Yagi K. Maeda I. Appl. Environ. Microbiol. 2008;74:6730–6738. doi: 10.1128/AEM.00498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Hui C. Y. Liu L. Chen M. P. Huang H. Y. Sci. Rep. 2021;11:13516. doi: 10.1038/s41598-021-92878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. Shen Y. Zou Y. Sun P. Wei W. Zhao J. Zhang C. Analyst. 2018;143:630–634. doi: 10.1039/C7AN00587C. [DOI] [PubMed] [Google Scholar]
- Wei H. Cheng H. Ting M. Wen-Hui Z. Xian-Gui L. Appl. Microbiol. Biotechnol. 2010;87:981–989. doi: 10.1007/s00253-010-2548-9. [DOI] [PubMed] [Google Scholar]
- Mahbub K. R. Krishnan K. Naidu R. Megharaj M. Environ. Technol. Innovat. 2017;8:64–70. doi: 10.1016/j.eti.2017.04.003. [DOI] [Google Scholar]
- Zhao E. M. Suek N. Wilson M. Z. Dine E. Pannucci N. L. Gitai Z. Avalos J. L. Toettcher J. E. Nat. Chem. Biol. 2019;15:589–597. doi: 10.1038/s41589-019-0284-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. Wang J. Du R. Liu Y. Chi J. He X. Huang K. Luo Y. Xu W. Biosens. Bioelectron. 2020;150:111899. doi: 10.1016/j.bios.2019.111899. [DOI] [PubMed] [Google Scholar]
- Din G. Hasan F. Conway M. Denney B. Ripp S. Shah A. A. J. Appl. Microbiol. 2019;127:1125–1134. doi: 10.1111/jam.14399. [DOI] [PubMed] [Google Scholar]
- Hansen L. H. Sorensen S. J. FEMS Microbiol. Lett. 2000;193:123–127. doi: 10.1111/j.1574-6968.2000.tb09413.x. [DOI] [PubMed] [Google Scholar]
- Cai Y. Zhu K. Shen L. Ma J. Bao L. Chen D. Wei L. Wei N. Liu B. Wu Y. Chen S. Environ. Sci. Technol. 2022;56:10062–10071. doi: 10.1021/acs.est.2c00627. [DOI] [PubMed] [Google Scholar]
- Jia X. Ma Y. Bu R. Zhao T. Wu K. Amb. Express. 2020;10:67. doi: 10.1186/s13568-020-01004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. Zhao T. Liu Y. Bu R. Wu K. FEMS Microbiol. Lett. 2018:365. doi: 10.1093/femsle/fny157. [DOI] [PubMed] [Google Scholar]
- Bereza-Malcolm L. Aracic S. Franks A. E. Sensors. 2016;16:2174. doi: 10.3390/s16122174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Verma N. Singh A. K. Sensor. Actuator. B Chem. 2017;240:248–254. doi: 10.1016/j.snb.2016.08.160. [DOI] [Google Scholar]
- Chen P.-H. Lin C. Guo K.-H. Yeh Y.-C. RSC Adv. 2017;7:29302–29305. doi: 10.1039/C7RA03778C. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.