Abstract
Background
The budding yeast Komagataella phaffii (Pichia pastoris) is widely employed to secrete proteins of academic and industrial interest. For secretory proteins, signal peptides are the sorting signal to direct proteins from cytosol to extracellular matrix, and their secretion efficiency directly impacts the yields of the targeted proteins in fermentation broth. Although the α-mating factor (MF) secretion signal from S. cerevisiae, the most common and widely used signal sequence for protein secretion, works in most cases, limitation exists as some proteins cannot be secreted efficiently. As the optimal choice of secretion signals is often protein specific, more secretion signals need to be developed to augment protein expression levels in K. phaffii.
Results
In this study, the secretion efficiency of 40 α-MF secretion signals from various yeast species and 32 endogenous signal peptides from K. phaffii were investigated using enhanced green fluorescent protein (EGFP) as the model protein. All of the evaluated α-MF secretion signals successfully directed EGFP secretion except for the secretion signals of the yeast D. hansenii CBS767 and H. opuntiae. The secretion efficiency of α-MF secretion signal from Wickerhamomyces ciferrii was higher than that from S. cerevisiae. 24 out of 32 endogenous signal peptides successfully mediated EGFP secretion. The signal peptides of chr3_1145 and FragB_0048 had similar efficiency to S. cerevisiae α-MF secretion signal for EGFP secretion and expression.
Conclusions
The screened α-MF secretion signals and endogenous signal peptides in this study confer an abundance of signal peptide selection for efficient secretion and expression of heterologous proteins in K. phaffii.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13068-022-02243-6.
Keywords: Komagataella phaffii, α-Mating factor, Peptide signal, Protein secretion
Introduction
Komagataella phaffii (also referred to as Pichia pastoris) is a methylotrophic yeast, which can utilize methanol as sole carbon and energy source. After its failure as single-cell protein (SCP) production, K. phaffii was subsequently developed into a heterologous protein expression host for the production of recombinant proteins [1]. Over the past 30 years, K. phaffii has become one of the most popular expression hosts attributed to its various advantages: its ability to reach high cell densities on defined media, the presence of strong and tightly methanol-regulated Alcohol Oxidase I (AOX1) promoter, high protein expression levels and low incidence of hyperglycosylation. More than 5000 heterologous proteins have been reported to be successfully expressed in the K. phaffii system [2]. The recombinant proteins expressed in K. phaffii involved in industrial enzymes, vaccine, antibody fragments, cytokines and membrane proteins [3–8].
The heterologous proteins expressed in K. phaffii are generally secreted into the culture medium. One of the important reasons is that K. phaffii has a secretory pathway consisted of the endoplasmic reticulum (ER) and Golgi apparatus to ensure proper protein folding, processing and modification including disulfide bond formation, glycosylation and oligomerization. Compared to the secretory pathway of S. cerevisiae, the secretory pathway of K. phaffii are more similar to that of higher eukaryotes in having stacked Golgi cisternae [9–11]. Secretory proteins are released to the extracellular medium as the soluble forms, which are more similar to the native proteins in structure and have higher physiological activity. Another reason is that K. phaffii secretes few endogenous proteins out of the cell, which facilitates the purification of recombinant proteins [12].
For secretory proteins, signal peptides are the sorting signal to direct proteins from cytosol to extracellular matrix [13, 14]. To produce the recombinant proteins in expression systems, the secretion efficiency of signal peptides directly impacts the yields of the targeted proteins in fermentation broth [15]. The α-mating factor (MF) secretion signal from S. cerevisiae is the most common and widely used signal sequence for recombinant protein secretion in K. phaffii. The α-MF secretion signal of S. cerevisiae consists of 85 amino acids and contains two regions: a pre-peptide (signal peptide) consisting of N-terminal 19 amino acids and a pro-peptide consisting of 66 amino acids from position 20 to 85 [16]. Pre-peptide mediates targeting the secretory proteins into the endoplasmic reticulum, and pro-peptide is believed to be involved in mediating secretory proteins into endoplasmic reticulum-derived COPII transport vesicles and enhances secretion efficiency of recombinant proteins [17, 18]. Although α-MF secretion signal of S. cerevisiae has been successfully used for the secretion of a large number of heterologous proteins in K. phaffii, some proteins were unsuccessfully expressed when using the α-MF secretion signal [19]. In recent years, endogenous signal peptides of K. phaffii were developed to mediate secretion of heterologous proteins. Several endogenous signal peptides were reported to yield much more efficient secretion than α-MF secretion signal of S. cerevisiae [20, 21].
In addition to S. cerevisiae’s, 39 α-MF genes from other yeast species can be found in the NCBI database. It is unknown whether their α-MF secretion signal can also efficiently mediate protein secretion in K. phaffii so far. After sequencing of the K. phaffii genome in 2009, Schutter et al. analyzed signal sequences of K. phaffii according to the homologs of functionally annotated secreted proteins in S. cerevisiae and revealed a multitude of endogenous signal peptides [22], which can allow screening high efficiency secretion signals for augmenting protein expression levels in K. phaffii. In this study, we systematically evaluated secretion efficiency of 40 α-MF secretion signals from various yeast species and 32 endogenous signal peptides from K. phaffii with a D-score≥ 0.95 using EGFP as the model protein.
Results
Protein secretion with the α-MF secretion signals from S. cerevisiae, K. phaffii and K. lactis
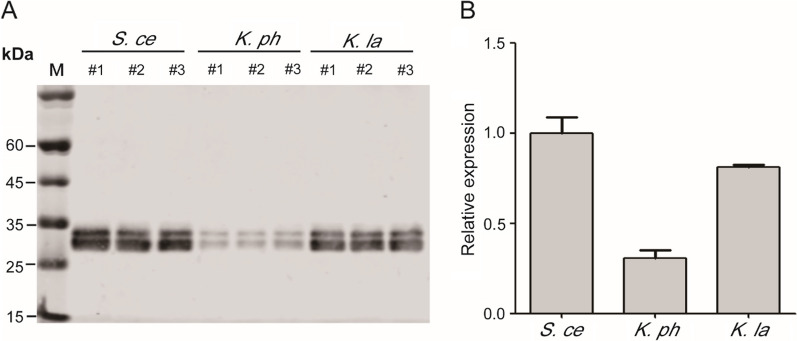
The secretion of most proteins produced in K. phaffii is mediated by the α-MF secretion signal from S. cerevisiae. In the yeast Kluyveromyces lactis expression system (New England BioLabs Inc.), K. lactis α-MF secretion signal, not S. cerevisiae α-MF secretion signal, is employed to secrete recombinant proteins. It is possible that the α-MF secretion signal from K. lactis works better than that from S. cerevisiae in K. lactis cells. The genome sequence from K. phaffii reveals a α-MF gene in K. phaffii GS115 strain. The α-MF secretion signal from S. cerevisiae works in most cases in K. phaffii, although there have been no studies to compare it to α-MF secretion signals from other yeast species. Using EGFP as a reporter, the secretion efficiency of the three α-MF secretion signals in K. phaffii were compared. The results showed that the secretion efficiency of S. cerevisiae α-MF secretion signal is the highest followed by K. lactis’s and K. phaffii’s (Fig. 1), indicating that the secretion efficiencies of α-MF secretion signals from different yeast species on protein expression were different in K. phaffii system.
Fig. 1.
Secretion and expression of EGFP mediated by the α-MF secretion signals from S. cerevisiae, K. phaffii and K. lactis. A: The expression analysis of recombinant EGFP by Western blot using anti-EGFP primary antibody. B: The quantitative analysis to A. #1, #2 and #3 represent three different recombinant strains. M: protein marker; S. ce: S. cerevisiae; K. ph: K. phaffii; K. la: K. lactis
Evaluation of α-MF secretion signals on the effect of protein secretion
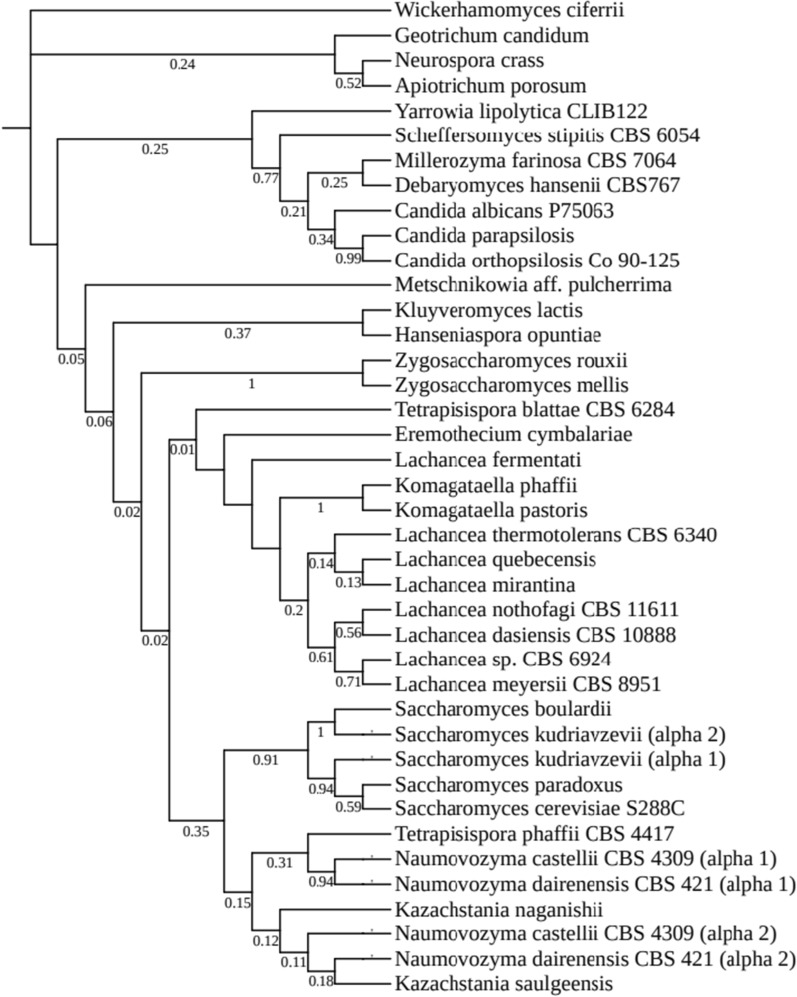
Searching NCBI database, 40 α-MF genes from different yeast species including S. cerevisiae, K. lactis and K. phaffii were found (Additional file 1: Table S1). Whether their secretion signals also work well like S. cerevisiae’s in K. phaffii has not been evaluated. The α-MF precursors were used to construct a phylogenetic tree (Fig. 2). The constructed phylogenetic tree showed several distinct clusters of α-MF precursors in yeasts. A highly close relation between K. pastoris and K. phaffii was revealed from the phylogenetic tree.
Fig. 2.
Phylogenetic tree of the α-MF precursors. The tree was constructed using the amino acid sequences of α-MF precursors. The numbers at the forks indicate the bootstrap confidence values
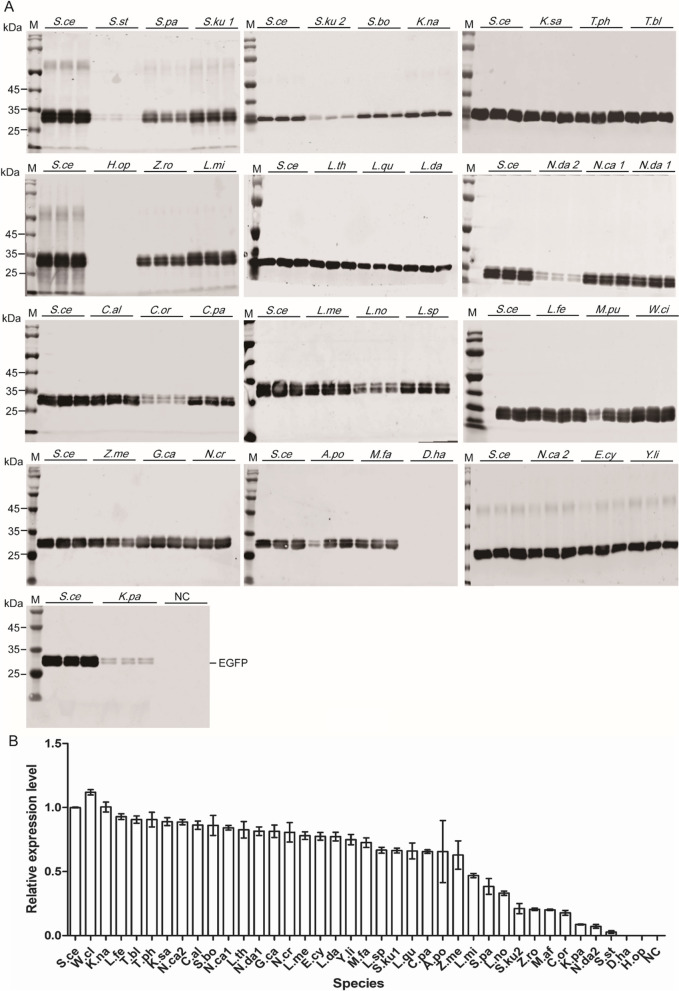
In order to eliminate codon bias on the effect of translation efficiency when these α-MF secretion signals were used to mediate EGFP secretion, coding sequences of α-MF secretion signals were optimized according to the method established in previous study (Additional file 1: Table S2) [23]. The secretion efficiencies of α-MF secretion signals were evaluated using S. cerevisiae α-MF secretion signal as a control. Almost all of the evaluated α-MF secretion signals successfully mediated EGFP secretion. Only the α-MF secretion signals of D. hansenii CBS767 and H. opuntiae failed to mediate EGFP secretion. Except for W. ciferrii α-MF secretion signal, the secretion efficiency of other α-MF secretion signals to EGFP was lower than that of S. cerevisiae (Fig. 3). The 3-D structures of α-MF secretion signals were predicted using Alphafold 2.0 AI system [24]. Most of α-MF secretion signals showed a conservative structure with a 2-stranded anti-parallel β-sheet followed by an α-helix on C-terminus (Additional file 2: Fig. S2).
Fig. 3.
The secretion and expression of EGFP mediated by α-MF secretion signals from different yeast species. A The expression analysis of recombinant EGFP by Western blot. B The quantitative analysis to A. M: protein marker; S.ce: S. cerevisiae; W.cl: W. ciferrii; K.na: K. naganishii; L.fe: L. fermentati; T.bl: T. blattae CBS 6284; T.ph: T. phaffii CBS 4417; K.sa: K. saulgeensis; N.ca2: N. castellii CBS 4309 (alpha 2); C.al: C. albicans P75063; S.bo: S. boulardii; N.ca1: N. castellii CBS 4309 (alpha 1); L.th: L. thermotolerans CBS 6340; N.da1: N. dairenensis CBS 421 (alpha 1); G.ca: G. candidum; N.cr: N. crass; L.me: L. meyersii CBS 8951; E.cy: E. cymbalariae; L.da: L. dasiensis CBS 10888; Y.li: Y. lipolytica CLIB122; M.fa: M. farinosa CBS 7064; L.sp: Lachancea sp. CBS 6924; S.ku1: S. kudriavzevii (alpha 1); L.qu: L. quebecensis; C.pa: C. parapsilosis; A.po: A. porosum; Z.me: Z. mellis; L.mi: L. mirantina; S.pa: S. paradoxus; L.no: L. nothofagi CBS 11611; S.ku2: S. kudriavzevii (alpha 1); Z.ro: Z. rouxii; M.pu: M. aff. pulcherrima; C.or: C. orthopsilosis Co 90-125; K.pa: K. pastoris; N.da2: N. dairenensis CBS 421 (alpha 2); S.st: S. stipitis CBS 6054; D.ha: D. hansenii CBS767; H.op: H. opuntiae; NC: negative control. Three independent strains were used to evaluate EGFP expression
Secretion and expression of EGFP mediated by endogenous signal peptides
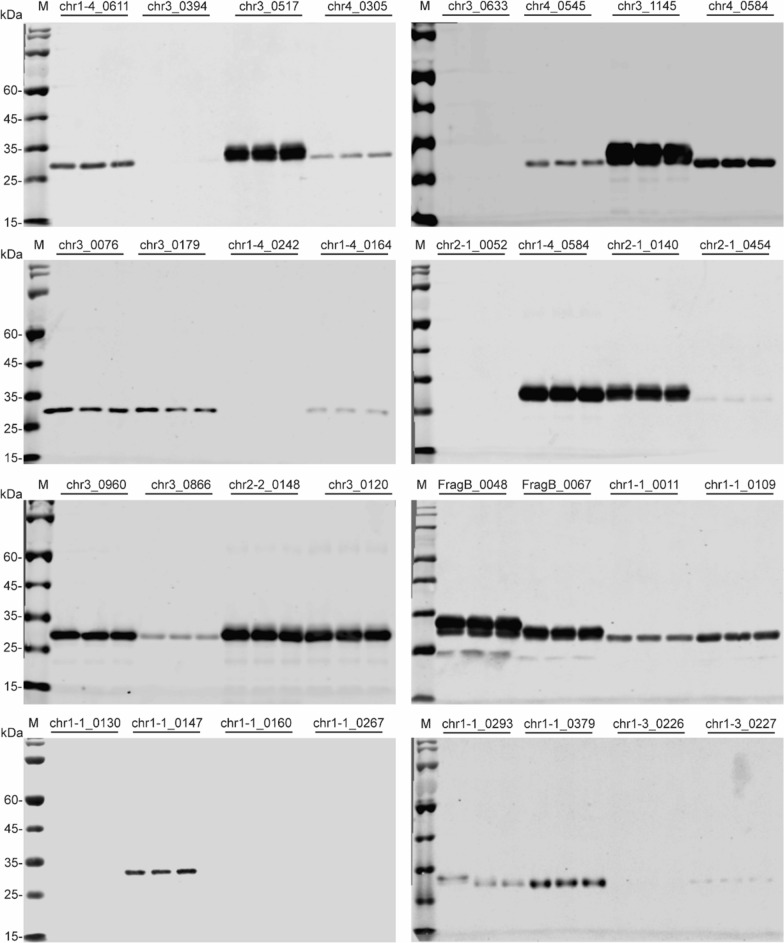
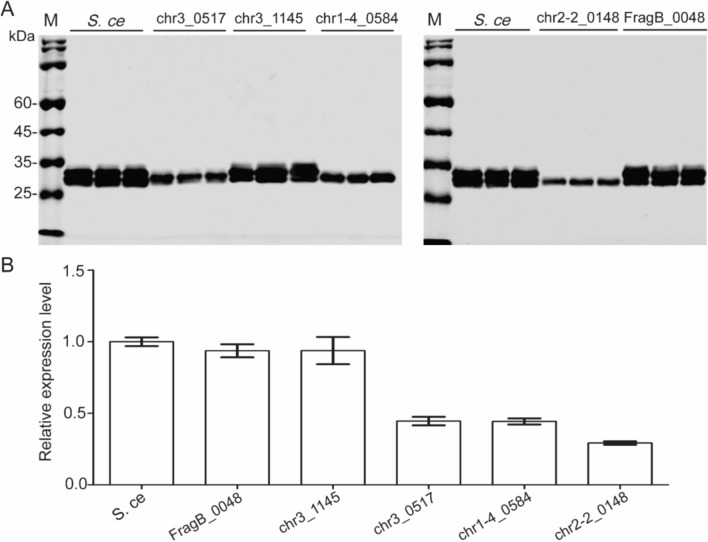
After sequencing K. phaffii genome in 2009, the genome sequence revealed a total of 54 endogenous signal peptides, which derived from homologs of functionally annotated secretory proteins of S. cerevisiae [22]. These predicted endogenous signal peptides will allow screening for functional signal peptides in K. phaffii. The D-score of 54 endogenous signal peptides were analyzed using SignalP 5.0. In this study, 32 endogenous signal peptides with D-score values greater than or equal to 0.95 were selected to evaluate EGFP secretion (Additional file 1: Table S3). 24 out of 32 endogenous signal peptides successfully mediated EGFP secretion, and the signal peptides of chr3_0517, chr3_1145, chr1-4_0584, chr2-1_0140, chr3_0960, chr2-2_0148, chr3_0120, FragB_0048 and FragB_0067 directed strong EGFP secretion and expression (Fig. 4).
Fig. 4.
The secretion and expression of EGFP mediated by endogenous signal peptides. The expression of recombinant EGFP was detected by Western blot. M: protein marker. Three independent strains were used to evaluate EGFP expression
Comparison of endogenous signal peptides with S. cerevisiae α-MF secretion signal on EGFP secretion
The S. cerevisiae α-MF secretion signal is the most used signal sequence and has high efficiency for protein secretion. In this study, several endogenous signal peptides with high secretion efficiency were successfully screened (Fig. 4). Whether did these endogenous signal peptides perform better on protein secretion than S. cerevisiae α-MF secretion signal? Five endogenous signal peptides with the highest secretion efficiency were selected to compare with S. cerevisiae α-MF secretion signal for expressing EGFP. The results showed that signal peptides of chr3_1145 and FragB_0048 had similar efficiency to S. cerevisiae α-MF secretion signal for EGFP secretion and expression (Fig. 5).
Fig. 5.
Comparison of endogenous signal peptides with S. cerevisiae α-MF secretion signal on EGFP expression. A The expression analysis of recombinant EGFP by Western blot. B The quantitative analysis to A. M: protein marker; S. ce: S. cerevisiae. Three independent strains were used to evaluate EGFP expression
Discussion
In this study, 40 α-MF secretion signals from different yeast species were tested for secretion expression of EGFP in K. phaffii. 38 out of 40 α-MF secretion signals successfully directed the secretion of EGFP, suggesting that their secretory pathways appear to be conservative in the yeast family. Yeasts are outstanding hosts to produce recombinant proteins for industrial or medical applications [25]. The yeasts including S. cerevisiae, K. phaffii, H. polymorpha, Y. lipolytica, A. adeninivorans, K. lactis, and S. pombe are commonly employed as expression hosts for production of recombinant proteins [25]. Few secretion signals have been developed for use in these yeasts. The frequently used signal sequence is the S. cerevisiae α-MF secretion signal. The α-MF secretion signals developed in this study will greatly enrich the selection of signal sequences for these yeast expression systems. At the same time, the secretion efficiency of α-MF secretion signal from W. ciferrii was higher than that from S. cerevisiae, suggesting it can be used to substitute α-MF secretion signal from S. cerevisiae for promoting secretion of heterologous proteins in K. phaffii.
The α-MF secretion signal contains two regions: a pre-peptide followed by a pro-peptide. The pre-peptide helps the nascent protein translocate to the ER. The pro-peptide is believed to play a significant role in secretion efficiency [26, 27]. The mutation or deletion on pro-peptide of S. cerevisiae α-MF changed secretion efficiency of reporter proteins [26, 28]. The deletion of K. pastoris pro-peptide significantly increased secretion of reporter proteins in our study (data not shown). The pro-peptide of S. cerevisiae α-MF with 66 amino acids forms a certain secondary structure. Lin-cereghino et al. predicted the secondary structure of S. cerevisiae α-MF pro-peptide based on a Jpred secondary structure program and knob–socket modeling of tertiary structure. The pro-peptide is consisted of a large loop region framed by two interacting helices [28]. Based on the analysis of the circular dichroism, Chahal et al. released a new structure model of S. cerevisiae α-MF pro-peptide with five beta strands and one alpha helix [26]. In this work, we used AphaFold2 model to predict the 3-D structure of α-MF pro-peptides [24]. The structure of S. cerevisiae α-MF pro-peptide is consisted of a 2-stranded anti-parallel β-sheet followed by an α-helix on C-terminus (Additional file 2: Fig. S2). Amino acids 50–56 and 60–67 constitute two β-sheet while amino acids 68–78 are present in an α-helix. Studies showed that deletion of amino acids 57–70 located within the secondary structure of S. cerevisiae α-MF pro-peptide increased secretion of recombinant protein [27, 28]. Most of α-MF pro-peptides from various yeasts have the same secondary structure like S. cerevisiae α-MF pro-peptide, indicating the structure possibly plays a functional role in expression regulation of α-MF pheromone in yeasts.
Although S. cerevisiae α-MF secretion signal works in most cases, the native signal peptides from heterologous proteins or endogenous signal peptides from K. phaffii are another viable option [20, 21, 29, 30]. Several studies showed that endogenous signal peptides were found to exhibit high secretory activity to reporter proteins [20, 21, 31]. After sequencing the genome of K. phaffii, a multitude of endogenous signal sequences were revealed [22]. Few of them have been experimentally tested to mediate secretion of target proteins. In this study, 32 endogenous signal peptides were evaluated for secretory activity, and 24 out of 32 endogenous signal peptides successfully directed EGFP secretion and expression. As the optimal choice of signal peptides is often protein specific, testing different signal peptides should influence overall yield. These endogenous signal peptides provide an abundance of choices for efficient secretion and expression of heterologous proteins in K. phaffii system. The α-MF secretion signal mediates posttranslational translocation across the ER membrane, so recombinant proteins that can fold in the cytosol may be inefficiently translocated and thus poorly secreted [32]. Barrero et al. used the peptide signal of OST1 gene and α-MF pro-peptide from S. cerevisiae to engineer a hybrid secretion signal, which yielded efficient secretion for proteins that can fold in the cytosol and for oligomeric proteins [18]. The α-MF secretion signals and endogenous signal peptides screened out in this study can also be used to construct the hybrid secretion signal library for secretion of heterologous proteins which can fold or oligomerize in the yeast cytosol.
Conclusions
In this study, the secretion efficiency of 40 α-MF secretion signals from various yeast species and 32 endogenous signal peptides from K. phaffii were evaluated. Thirty-eight α-MF secretion signals and 24 endogenous signal peptides successfully mediated the secretion and expression of the reporter protein. The screened α-MF secretion signals and endogenous signal peptides can allow screening for the optimal signal-ORF combination, which may result in augmented protein expression levels in K. phaffii.
Materials and methods
Plasmids, strains and culture media
The plasmid pPIC9K was used as backbones to construct expression vectors containing different signal sequences for secretory expression. E. coli Top10′ was used as a host for genetic manipulation and K. phaffii GS115 strain was employed as a host for expression of heterologous proteins. Luria–Bertani (LB) liquid medium (0.5% yeast extract, 1% Peptone, and 0.5% NaCl) with 100 μg/mL ampicillin was used in the growth of bacteria. GS115 strain was grown in YPD agar plates (1% yeast extract, 2% peptone, 2% dextrose and 1.5% agar). MD agar (2% dextrose, 1.34% yeast nitrogen base, 4 × 10−5% biotin and 1.5% agar) were used as selected medium to screen transformants integrated into pPIC9K plasmids. BMGY [1% yeast extract, 2% tryptone, 1.34% YNB, 1% glycerol, 100 mM potassium phosphate (pH 6.0) and 4 × 10−5% biotin] and BMMY [1% yeast extract, 2% tryptone, 1.34% YNB, 1% methanol, 100 mM potassium phosphate (pH 6.0)] media were employed to express the reporter. Top10′ was cultured at 37 ℃ and GS115 at 30 ℃ with stirring rate 220 rpm in a shaker.
The selection for α-MF secretion signals and endogenous signal peptides
For collecting the information of α-MF secretion signals, we searched NCBI protein database using “alpha mating factor” as the key word. The results of this search were analyzed using the Protein Blast tool of NCBI to filter out the identical protein sequences from different species. The collected protein sequences were further evaluated using SignalP 5.0 software to confirm that there is a signal peptide in the sequence.
According to the homology of functionally annotated secretory proteins of S. cerevisiae, De Schutter et al. analyzed the genome sequence of K. phaffii and revealed a total of 54 endogenous signal peptides in K. phaffii [22]. The D-score of 54 endogenous signal peptides were analyzed using SignalP 5.0. In this study, 32 endogenous signal peptides with D-score values greater than or equal to 0.95 were selected to evaluate the reporter secretion.
Construction of expression vectors
To evaluate the α-MF secretion signals from different yeasts and endogenous signal peptides from K. phaffii on the effect of protein secretion, The EGFP was used as the reporter gene. The EGFP was amplified from pEGFP-N1 plasmid by PCR and cloned into the pPIC9K expression vector between SnaB I and EcoR I sites for construction of pPIC9K-EGFP. The coding sequences of α-MF secretion signal and endogenous signal peptides were synthesized by gene company (Wuhan GeneCreate Biological Engineering Co., Ltd.) and cloned into the BamH I and SnaB I restriction sites of pPIC9K-EGFP, keeping the secretion signal and endogenous signal peptide coding sequence with EGFP gene in the same reading frame (Additional file 2: Fig. S1).
Electroporation of K. phaffii
Electroporation of plasmids into K. phaffii were performed as described previously [33]. Briefly, the purified plasmids were digested with restriction enzyme recommended by Pichia Expression Kit manual to obtain linear DNA. The 5–10 μg of linear plasmid DNA was used for electroporation. The transformed cells were spread on MD agar plates. The plates were incubated at 29 ℃ for 2–3 days until colonies appeared.
EGFP expression
Three colonies from MD plates were picked and cultured in BMGY at 29 ℃ at 220 rpm broth in a shaking incubator until the culture reaches an OD600 = 2–4. Then, the cells were harvested by centrifuging at 3000 ×g for 5 min at room temperature. The cell pellet was resuspended to an OD600 of 1.0 in BMMY medium with 1% methanol. The cells were cultured at 29 ℃ for 72 h and added 100% methanol to a final concentration of 1% methanol every 24 h to maintain induction. Centrifugation was performed to collect the supernatant at 12,000 ×g at 4 ℃ for 10 min. the supernatant was stored at – 80 ℃ until ready to assay.
Western blot
The expression of EGFP was evaluated by Western blot. The 10 μL of supernatant was loaded into each well of 10% SDS-PAGE gel. After finishing the electrophoresis, the proteins in the gel were transferred to Hybond-C nitrocellulose membrane (Amersham Bioscience). The transfer was done at 100 V for 2 h. Anti-EGFP antibody (Proteintech, China, Cat no. 50430-2-AP) and IRDye 800CW-conjugated goat anti-rabbit secondary antibodies (LI–COR Biosciences, Lincoln, NE, USA; cat. no. C60607-15) were employed as the primary and secondary antibody, respectively. The hybridization signals were detected and measured using LICOR Odyssey system (LI–COR, Nebraska, USA).
Phylogenetic analysis
In the phylogenetic analysis, the amino acids sequences of α-MF were aligned using MUSCLE, and the Maximum Likelihood (ML) tree was constructed by MEGA X, bootstrap was set to 1 and the other parameters were defaulted. Then, the ML tree was adjusted for presentation through the interactive tree of life (iTOL, version 6.5.2).
Supplementary Information
Additional file 1: Table S1. The information of α-MF secretion signals using in this study. Table S2. Optimized coding sequences of α-mating factor secretion signals. Table S3. The information of endogenous signal peptides.
Additional file 2: Fig. S1. The construction schematic of expression vectors with different α-MF secretion signal or endogenous signal peptide. The EGFP gene was cloned into pPIC9K between SnaB I and EcoR I sites. The pPIC9K-EGFP was digested with BamH I and SnaB I to remove the α-MF secretion signal of S. cerevisiae, and then an α-MF secretion signal from yeast specie or endogenous signal peptide was inserted to replace the α-MF signal leader of S. cerevisiae. Fig. S2. The structures of pro-peptides of α-MF secretion signals predicted by alphafold2 model. S.ce: S. cerevisiae; W.cl: W. ciferrii; K.na: K. naganishii; L.fe: L. fermentati; T.bl: T. blattae CBS 6284; T.ph: T. phaffii CBS 4417; K.sa: K. saulgeensis; N.ca2: N. castellii CBS 4309 (alpha 2); C.al: C. albicans P75063; S.bo: S. boulardii; N.ca1: N. castellii CBS 4309 (alpha 1); L.th: L. thermotolerans CBS 6340; N.da1: N. dairenensis CBS 421 (alpha 1); G.ca: G. candidum; N.cr: N. crass; L.me: L. meyersii CBS 8951; E.cy: E. cymbalariae; L.da: L. dasiensis CBS 10888; Y.li: Y. lipolytica CLIB122; M.fa: M. farinosa CBS 7064; L.sp: Lachancea sp. CBS 6924; S.ku1: S. kudriavzevii (alpha 1); L.qu: L. quebecensis; C.pa: C. parapsilosis; A.po: A. porosum; Z.me: Z. mellis; L.mi: L. mirantina; S.pa: S. paradoxus; L.no: L. nothofagi CBS 11611; S.ku2: S. kudriavzevii (alpha 1); Z.ro: Z. rouxii; M.pu: M. aff. pulcherrima; C.or: C. orthopsilosis Co 90-125; K.pa: K. pastoris; N.da2: N. dairenensis CBS 421 (alpha 2); S.st: S. stipitis CBS 6054; D.ha: D. hansenii CBS767; H.op: H. opuntiae.
Acknowledgements
Not applicable.
Author contributions
YH conceived and designed the experiments. Chenwei Z and LL performed the majority of the laboratory work. SW and Chenshan Z constructed plasmids. XC analyzed the phylogenetic tree. LL and YH wrote the manuscript. YL revised the manuscript. All authors have read and approved the final manuscript.
Funding
This project is financed by Fujian provincial health technology project (2020CXA004) and Fujian Normal University, Fujian Provincial Science Fund for Distinguished Young Scholar (2020J01311402).
Availability of data and materials
The data supporting the conclusions of this article are included with the article and its Additional files 1 and 2.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chenwei Zou and Lingfang Lu contributed equally to this work
References
- 1.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad M, Hirz M, Pichler H, Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goncalves AM, Pedro AQ, Maia C, Sousa F, Queiroz JA, Passarinha LA. Pichia pastoris: a recombinant microfactory for antibodies and human membrane proteins. J Microbiol Biotechnol. 2013;23:587–601. doi: 10.4014/jmb.1210.10063. [DOI] [PubMed] [Google Scholar]
- 4.Huang J, Xia J, Yang Z, Guan F, Cui D, Guan G, Jiang W, Li Y. Improved production of a recombinant Rhizomucor miehei lipase expressed in Pichia pastoris and its application for conversion of microalgae oil to biodiesel. Biotechnol Biofuels. 2014;7:111. doi: 10.1186/1754-6834-7-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lunsdorf H, Gurramkonda C, Adnan A, Khanna N, Rinas U. Virus-like particle production with yeast: ultrastructural and immunocytochemical insights into Pichia pastoris producing high levels of the hepatitis B surface antigen. Microb Cell Fact. 2011;10:48. doi: 10.1186/1475-2859-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navone L, Vogl T, Luangthongkam P, Blinco JA, Luna-Flores CH, Chen X, von Hellens J, Mahler S, Speight R. Disulfide bond engineering of AppA phytase for increased thermostability requires co-expression of protein disulfide isomerase in Pichia pastoris. Biotechnol Biofuels. 2021;14:80. doi: 10.1186/s13068-021-01936-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nylen A, Chen MT. Production of full-length antibody by Pichia pastoris. Methods Mol Biol. 2018;1674:37–48. doi: 10.1007/978-1-4939-7312-5_3. [DOI] [PubMed] [Google Scholar]
- 8.Xu H, Shi M, Shao C, Li H, Wu J, Yu Y, Fang F, Guo Y, Xiao W. Development of IL-15/IL-15Ralpha sushi domain-IgG4 Fc complexes in Pichia pastoris with potent activities and prolonged half-lives. Microb Cell Fact. 2021;20:115. doi: 10.1186/s12934-021-01605-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bevis BJ, Hammond AT, Reinke CA, Glick BS. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- 10.Mogelsvang S, Gomez-Ospina N, Soderholm J, Glick BS, Staehelin LA. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol Biol Cell. 2003;14:2277–2291. doi: 10.1091/mbc.e02-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O'Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karbalaei M, Rezaee SA, Farsiani H. Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J Cell Physiol. 2020;235:5867–5881. doi: 10.1002/jcp.29583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolhuis A, Matzen A, Hyyrylainen HL, Kontinen VP, Meima R, Chapuis J, Venema G, Bron S, Freudl R, van Dijl JM. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J Biol Chem. 1999;274:24585–24592. doi: 10.1074/jbc.274.35.24585. [DOI] [PubMed] [Google Scholar]
- 14.Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol. 2018;97:422–441. doi: 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Aw R, McKay PF, Shattock RJ, Polizzi KM. A systematic analysis of the expression of the anti-HIV VRC01 antibody in Pichia pastoris through signal peptide optimization. Protein Expr Purif. 2018;149:43–50. doi: 10.1016/j.pep.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julius D, Blair L, Brake A, Sprague G, Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983;32:839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- 17.Brake AJ, Merryweather JP, Coit DG, Heberlein UA, Masiarz FR, Mullenbach GT, Urdea MS, Valenzuela P, Barr PJ. Alpha-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1984;81:4642–4646. doi: 10.1073/pnas.81.15.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrero JJ, Casler JC, Valero F, Ferrer P, Glick BS. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb Cell Fact. 2018;17:161. doi: 10.1186/s12934-018-1009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Dong W, He J, Ren X, Yan W. Expression and purification of natural N-terminal recombinant bovine pancreatic trypsin inhibitor from Pichia pastoris. Biol Pharm Bull. 2008;31:1680–1685. doi: 10.1248/bpb.31.1680. [DOI] [PubMed] [Google Scholar]
- 20.Liang S, Li C, Ye Y, Lin Y. Endogenous signal peptides efficiently mediate the secretion of recombinant proteins in Pichia pastoris. Biotechnol Lett. 2013;35:97–105. doi: 10.1007/s10529-012-1055-8. [DOI] [PubMed] [Google Scholar]
- 21.Duan G, Ding L, Wei D, Zhou H, Chu J, Zhang S, Qian J. Screening endogenous signal peptides and protein folding factors to promote the secretory expression of heterologous proteins in Pichia pastoris. J Biotechnol. 2019;306:193–202. doi: 10.1016/j.jbiotec.2019.06.297. [DOI] [PubMed] [Google Scholar]
- 22.De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouze P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Lin T, Lu L, Cai F, Lin J, Jiang YE, Lin Y. Codon pair optimization (CPO): a software tool for synthetic gene design based on codon pair bias to improve the expression of recombinant proteins in Pichia pastoris. Microb Cell Fact. 2021;20:209. doi: 10.1186/s12934-021-01696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baghban R, Farajnia S, Rajabibazl M, Ghasemi Y, Mafi A, Hoseinpoor R, Rahbarnia L, Aria M. Yeast expression systems: overview and recent advances. Mol Biotechnol. 2019;61:365–384. doi: 10.1007/s12033-019-00164-8. [DOI] [PubMed] [Google Scholar]
- 26.Chahal S, Wei P, Moua P, Park SP, Kwon J, Patel A, Vu AT, Catolico JA, Tsai YF, Shaheen N, et al. Structural characterization of the alpha-mating factor prepro-peptide for secretion of recombinant proteins in Pichia pastoris. Gene. 2017;598:50–62. doi: 10.1016/j.gene.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 27.Aggarwal S, Mishra S. Differential role of segments of alpha-mating factor secretion signal in Pichia pastoris towards granulocyte colony-stimulating factor emerging from a wild type or codon optimized copy of the gene. Microb Cell Fact. 2020;19:199. doi: 10.1186/s12934-020-01460-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin-Cereghino GP, Stark CM, Kim D, Chang J, Shaheen N, Poerwanto H, Agari K, Moua P, Low LK, Tran N, et al. The effect of alpha-mating factor secretion signal mutations on recombinant protein expression in Pichia pastoris. Gene. 2013;519:311–317. doi: 10.1016/j.gene.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vadhana AK, Samuel P, Berin RM, Krishna J, Kamatchi K, Meenakshisundaram S. Improved secretion of Candida antarctica lipase B with its native signal peptide in Pichia pastoris. Enzyme Microb Technol. 2013;52:177–183. doi: 10.1016/j.enzmictec.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Latiffi AA, Salleh AB, Rahman RN, Oslan SN, Basri M. Secretory expression of thermostable alkaline protease from Bacillus stearothermophilus FI by using native signal peptide and alpha-factor secretion signal in Pichia pastoris. Genes Genet Syst. 2013;88:85–91. doi: 10.1266/ggs.88.85. [DOI] [PubMed] [Google Scholar]
- 31.Massahi A, Calik P. Endogenous signal peptides in recombinant protein production by Pichia pastoris: from in-silico analysis to fermentation. J Theor Biol. 2016;408:22–33. doi: 10.1016/j.jtbi.2016.07.039. [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald I, Glick BS. Secretion of a foreign protein from budding yeasts is enhanced by cotranslational translocation and by suppression of vacuolar targeting. Microb Cell Fact. 2014;13:125. doi: 10.1186/s12934-014-0125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Long Y, Li S, Lin T, Wu J, Zhang Y, Lin Y. Investigation on the processing and improving the cleavage efficiency of furin cleavage sites in Pichia pastoris. Microb Cell Fact. 2018;17:172. doi: 10.1186/s12934-018-1020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The information of α-MF secretion signals using in this study. Table S2. Optimized coding sequences of α-mating factor secretion signals. Table S3. The information of endogenous signal peptides.
Additional file 2: Fig. S1. The construction schematic of expression vectors with different α-MF secretion signal or endogenous signal peptide. The EGFP gene was cloned into pPIC9K between SnaB I and EcoR I sites. The pPIC9K-EGFP was digested with BamH I and SnaB I to remove the α-MF secretion signal of S. cerevisiae, and then an α-MF secretion signal from yeast specie or endogenous signal peptide was inserted to replace the α-MF signal leader of S. cerevisiae. Fig. S2. The structures of pro-peptides of α-MF secretion signals predicted by alphafold2 model. S.ce: S. cerevisiae; W.cl: W. ciferrii; K.na: K. naganishii; L.fe: L. fermentati; T.bl: T. blattae CBS 6284; T.ph: T. phaffii CBS 4417; K.sa: K. saulgeensis; N.ca2: N. castellii CBS 4309 (alpha 2); C.al: C. albicans P75063; S.bo: S. boulardii; N.ca1: N. castellii CBS 4309 (alpha 1); L.th: L. thermotolerans CBS 6340; N.da1: N. dairenensis CBS 421 (alpha 1); G.ca: G. candidum; N.cr: N. crass; L.me: L. meyersii CBS 8951; E.cy: E. cymbalariae; L.da: L. dasiensis CBS 10888; Y.li: Y. lipolytica CLIB122; M.fa: M. farinosa CBS 7064; L.sp: Lachancea sp. CBS 6924; S.ku1: S. kudriavzevii (alpha 1); L.qu: L. quebecensis; C.pa: C. parapsilosis; A.po: A. porosum; Z.me: Z. mellis; L.mi: L. mirantina; S.pa: S. paradoxus; L.no: L. nothofagi CBS 11611; S.ku2: S. kudriavzevii (alpha 1); Z.ro: Z. rouxii; M.pu: M. aff. pulcherrima; C.or: C. orthopsilosis Co 90-125; K.pa: K. pastoris; N.da2: N. dairenensis CBS 421 (alpha 2); S.st: S. stipitis CBS 6054; D.ha: D. hansenii CBS767; H.op: H. opuntiae.
Data Availability Statement
The data supporting the conclusions of this article are included with the article and its Additional files 1 and 2.