Abstract
The ten–eleven translocation (TET) family of dioxygenases consists of three members, TET1, TET2, and TET3. All three TET enzymes have Fe+2 and α-ketoglutarate (α-KG)-dependent dioxygenase activities, catalyzing the 1st step of DNA demethylation by converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), and further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). Gene knockout studies demonstrated that all three TET proteins are involved in the regulation of fetal organ generation during embryonic development and normal tissue generation postnatally. TET proteins play such roles by regulating the expression of key differentiation and fate-determining genes via (1) enzymatic activity-dependent DNA methylation of the promoters and enhancers of target genes; and (2) enzymatic activity-independent regulation of histone modification. Interacting partner proteins and post-translational regulatory mechanisms regulate the activities of TET proteins. Mutations and dysregulation of TET proteins are involved in the pathogenesis of human diseases, specifically cancers. Here, we summarize the research on the interaction partners and post-translational modifications of TET proteins. We also discuss the molecular mechanisms by which these partner proteins and modifications regulate TET functioning and target gene expression. Such information will help in the design of medications useful for targeted therapy of TET-mutant-related diseases.
Keywords: TETs, Mutations, Interaction partners, Post-translational modifications, Gene expression
Introduction
Lineage commitment and differentiation of tissue stem/progenitor cells are tightly controlled by transcriptional programing1,2 and are delicately regulated by an ordered, stepwise reconfiguration of the DNA methylome and histone modifications.3–8 Dysregulation of either transcriptional programing or the epigenetic machinery will cause diseases such as cancers by disrupting cell fate determination and differentiation. Thus, a more complete understanding of how transcriptional programing and epigenetic functioning collaboratively regulate lineage fate and differentiation of stem/progenitor cells will provide information that will improve our understanding of disease pathogenesis and can point the way toward the development of novel medications for the treatment of diseases.
Transcription factors (TFs) regulate target gene expression by binding to specific consensus motifs in their enhancers and promoters.9 The binding motifs of most TFs contain CpG dinucleotides. Such TFs have different sensitivities to methyl-CpG (mCpG) motifs for DNA binding. Many genes have CpG-rich (CpG islands or CGIs) promoters. Methylation of these promoters is associated with target gene repression due to the condensation of local chromatin.8,10,11 Removing methyl groups from these promoters is required for TF binding and gene expression. In genes with non-CGI promoters and enhancers, TF-regulated expression of such genes is determined by the methylation status of CpG within the binding motifs.12–14
The dynamic methylation of DNA is regulated by a balance of DNA methyltransferases (including DNMT1, DNMT3a, and DNMT3b) and the ten–eleven translocation (TET) family of dioxygenases (including TET1, TET2, and TET3).15,16 The methylation state of DNA sequences regulates the accessibility of key TFs to genetic regulatory elements including promoters and enhancers of target genes, which in turn determines cell fate.8,17 Disruption of the dynamic methylation programming of DNA has been observed in almost all types of hematopoietic malignancies and has emerged as a hallmark of various types of hematological cancers, including myelodysplastic syndromes (MDS), acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), diffuse large B-cell lymphomas (DLBCLs), and peripheral T-cell lymphoma (PTCL).18–42 Consistently, somatic mutations of several key regulators of DNA methylation including DNMT3A, isocitrate dehydrogenase (IDH1), IDH2, and TET2 have been detected in almost all types of hematopoietic cancers.43 Detailed studies demonstrated that somatic mutations of DNMT3A and TET2 are also frequently detected in small clones in the hematopoietic tissue of healthy people, specifically those > 50 years old. The frequency of such mutations increases during aging and has been called age-related clonal hematopoiesis (ARCH).44–46 The selective acquisition and expansion of DNMT3A- or TET2-mutant clones during aging suggest that ARCH might be a consequence of compensatory hematopoiesis against the pressure of aging. In support of such a concept, it was found that hematopoietic stem and progenitor cells (HSPCs) showing either DNMT3A or TET2 mutations display growth advantages in response to treatment with interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α), respectively.47,48 Nevertheless, people with ARCH showed a 10–12-fold increased risk for developing hematopoietic malignancies than age-matched ARCH-negative populations.49–51 Thus, as is the case with somatic DNMT3A mutations, somatic TET2 mutations are founder mutations for almost all types of hematopoietic malignancies, and occur in hematopoietic stem cells (HSCs) during aging, and are selected under the pressure of aging-associated inflammation. Additional genetic mutations are required for the full malignant transformation of TET2-mutant HSCs, which drive the abnormal proliferation, lineage commitment, differentiation, and survival of HSPCs. In addition, TET1 is frequently mutated in B-cell malignancies and TET3 is down-regulated in HSPCs during aging as well as in the malignant cells of many types of hematopoietic cancers.52 Thus, all three members of the TET family are involved in the pathogenesis of hematopoietic cancers. In this review, we summarize the research on TET–protein interaction partners and translational modifications of TETs in the regulation of TET function. We also discuss the molecular mechanism by which TET proteins regulate target gene expression.
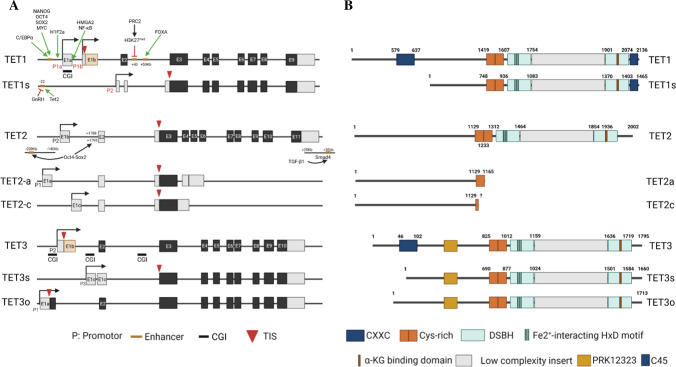
The three TET genes and their isoforms
The human TET1 gene is located on chromosome 10q21.3. It expresses two transcriptional isoforms owing to the use of alternate promoters (Fig. 1a).53 Transcription starting from promoter 1a or 1b (distal) produces a 2,136 a.a. full-length TET1 protein (2039 a.a. for the mouse), while transcription starting from promoter 2 (proximal) in front of exon 2 gives rise to a 1465 a.a. short isoform of TET1 (TET1s, 1386 a.a. for the mouse).53,54 TET1s lacks a large portion of the TET1 N-terminus, including the CXXC (CXXC5) domain. Both TET1 and TET1s have enzymatic activity. In mice, TET1 is primarily expressed in the embryo and is replaced by TET1s in adult tissues53.
Fig. 1.
TET genes and TET proteins. A TET1, TET2, and TET3 have 2, 3, and 3 transcriptional products, respectively, due to the use of alternative promoters, which are regulated by the alternative activation of enhancers. The green arrow indicates induction of expression; the red cross depicts inhibition of expression. B. The corresponding protein isoforms of TET1, TET2, and TET3. The structural domains of the proteins are indicated
The human TET2 gene is located on chromosome 4q24. In contrast to the TET1 and TET3 genes, the ancient TET2 gene was split during evolution into two genes, IDAX (also called CXXC4) and TET2. The IDAX gene is located 700 kb upstream of TET2 and is transcribed in the opposite direction; it encodes the CXXC domain-containing IDAX protein.55 The TET2 gene produces three protein isoforms, TET2-a, TET2-b, and TET2-c, because of the alternative use of three promoters and associated transcriptional initiation sites (TIS) (Fig. 1a). TET2-b utilizes the second promoter in front of exon 1b and produces a 2002 a.a. full-length TET2 protein (TET2 hereafter, 1912 a.a. for the mouse). TET2-a uses the first promoter, which is located upstream of the second promoter and produces a truncated 1165 a.a. protein terminating at a poly-A site in the fourth intron, while TET2-c utilizes the 3rd promoter in front of exon 1c and produces a much shorter truncated protein terminating within the 3rd exon. Both TET2-a and TET2-c lack enzymatic activity and might function as dominant-negative forms of TET2. TET2 is abundant in most normal human tissues, while TET2-a is primarily expressed in the human spleen, and TET2-c is weakly expressed in most tissues, with the highest levels observed in human spleen, bone marrow, fetal brain, and embryoid bodies. The dynamic switching of active promoters and enhancers regulates TET2-a, TET2, and TET2-c expression during cell state transitions between pluripotency and differentiation.56
The human TET3 gene is located on chromosome 2p13.1. The CXXC10-1 ORF is about 13 kb upstream of the annotated TSS of TET3 with the same orientation as the TET3 ORF. TET3 gene encodes three isoforms owing to the alternative use of promoters and alternative splicing (Fig. 1a). A 1795 a.a. full-length TET3 protein (TET3 hereafter; 1803 a.a. for the mouse) is transcribed starting from promoter 2 in front of exon 1b, and a 1660 a.a. TET3 short isoform (TET-3 s; 1668 a.a. for the mouse) is transcribed starting from promoter 3 in front of exon 2. A 1713 a.a. oocyte-specific isoform of TET3 (TET-3o) has been identified in the mouse, which is transcribed starting from promoter 1 in front of exon 1a approximately 5 kb upstream of the start codon with skipping of exon 1b.57 Human TET-3o has not been reported.
TET1, TET2, and TET3 share a conserved dioxygenase domain at their C-termini (Fig. 1b).58–60 The dioxygenase domain is composed of a cysteine (Cys)-rich domain and a double-stranded β-helix fold (DSβH) domain that are compactly arranged to mediate catalytic activity. The DSβH domain consists of 3 Fe2+-binding sites and one α-ketoglutarate (α-KG)-binding site. In addition, full-length TET1 and TET3 proteins contain N-terminal CXXC-type zinc-finger domains. CXXC regulates the recruitment and binding of TET1 and TET3 to DNA sequences, and provides a unique regulation of the methylation signature for genes associated with embryogenesis, gametogenesis, and neuronal development.53,61–63 TET2 protein lacks a DNA recognition domain and depends upon other DNA-binding proteins for interaction with DNA. In addition, the short forms of TET1 and TET3, including TET1s, TET-3 s, and TET-3o (an oocyte-specific isoform), all lack CXXC domains.54,63 Therefore, TET1s, TET-3o, and TET-3 s are primarily dependent on interaction with other DNA-binding proteins for DNA binding.
The mutations and expression of TET genes in the pathogenesis of cancer
The expression of TET genes in cancers
Compared to non-cancerous surrounding tissues, reduced levels of 5-hydroxymethylcytosine (5hmC) have been reported in multiple types of human cancers, such as hematopoietic malignancies, melanoma, lung cancers, pancreatic cancers, hormone-receptor-positive breast cancers, colon cancers, liver cancers, and glioblastoma multiforme, which are all associated with loss-of-function TET mutations or decreased expression levels of TET proteins.64–74 The reduction of 5hmC results in the aberrant methylation of tumor suppressor genes leading to tumor formation, progression, and invasion. Studies suggested that low 5hmC is an important marker for early diagnosis and predicts poor prognosis in some cancer types.73–80 However, in some other cancer types, including gastric cancers, lung cancers, triple-negative breast cancer, human epidermal growth factor receptor-enriched breast cancers, ovarian cancers, and gliomas, levels of TET proteins and 5-hmC are increased.81–84 TET proteins in such cancers function as oncoproteins, which promote cell proliferation and tumor progression. Thus, the roles of TET proteins in cancer pathogenesis might be tissue- and cell-type-specific.85
The mutations of TET genes in cancers
Loss-of-function TET2 mutations are commonly found in blood cells from healthy individuals over 50 years old. TET2 mutations in ARCH lead to a premalignant condition in hematopoietic tissue, which predisposes to leukemia/lymphoma transformation. TET2 mutations are commonly detected in almost all types of hematopoietic malignancies including MDS, myeloproliferative neoplasms, AML, PTCL, and DLBCL.30–42,86 Loss-of-function TET1 and TET3 mutations are detected in non-Hodgkin B-cell lymphoma, including DLBCL, and follicular lymphoma.87–91 In addition, TET1 is also mutated in 12–15% of T-ALL and 1–5% of AML patients.92,93 TET3 mutations are very rarely identified in PTCL37 and chronic lymphocytic leukemia.94 However, mutations of the TET1/2/3 genes are infrequent in solid cancers and their significance in such cases is unknown.66 In prostate cancers, TET2 mutations are detected in 6% of primary tumors and 20% of metastatic lesions.95 Whether TET2 mutations contribute to the metastatic advantage of prostate cancer needs to be determined experimentally.
Transcriptional regulation of TET1 gene in cancers (Fig. 1a)
In embryonic stem (ES) cells, pluripotent genes OCT4, NANOG, MYC, and SOX2 are strongly enriched in a super-enhancer upstream of promoter 1 of the TET1 gene, stimulating the expression of TET1 but not TET1s.53 During differentiation, TET1 is down-modulated by PRC2 binding of the super-enhancer, localized + 40 kb downstream of the TET1 TIS.96 HIF-2α binds at − 158 to − 1 bp upstream of the TIS of TET1 and induces TET1 expression in response to hypoxic conditions.97 In lung epithelial cells, p53 binds at − 192 bp/ + 29 bp of the promoter and represses TET1 expression.84 FOXA1 occupies the TET1 enhancer + 50 kb downstream of TIS and induces TET1 gene expression.98 During the prepubertal period, gonadotropin-releasing hormone (GnRH) stimulates luteinizing hormone-β polypeptide expression and differentiation of gonadotropic cells by repressing TET1s expression. GnRH plays such a role by inactivating a distal enhancer located − 20 to 22 kb upstream of the TIS.99 TET2 binds to this enhancer to maintain TET1s expression.99 TET1 is down-regulated in many types of cancers, such as breast cancer, pancreatic cancer, rectal cancer, oral squamous cell carcinoma, lymphoma, multiple myeloma, bladder cancer, liver cancer, and non-small-cell lung cancer, implying a tumor repressive activity for TET1.78,85,87,100,101 A CGI has been identified in the TET1 promoter and exon 1 region. In many types of cancers, down-regulation of TET1 might be mediated by HMGA2 and PRC2 via epigenetic methylation of the CGI promoter.96,102 C/EBPα directly binds to the TET1 promoter and regulates TET1 expression.103 In lung cancer and glioblastoma multiforme, epidermal growth factor receptor and MAPK activation silence TET1 expression by down-regulating C/EBPα. In basal-like breast cancer, thyroid carcinoma, skin cutaneous melanoma, and lung adenocarcinoma, TNFα stimulates NF-κB activation, which represses TET1 expression by binding to the TET1 promoter.104 In both cellular and animal models, inhibition of EGFR signaling restores TET1 expression.103 In colon cancers, BRAFV600E downregulates TET1 and TET2 expression which results in a hypermethylation phenotype in the cancer cells.105 TET1 down-regulation is involved in disease initiation and cancer invasiveness/metastasis, and is associated with a poor prognosis. In breast cancers, down-regulation of TET1 results in HOXA9/HOXA7 repression, which leads to breast cancer growth and metastasis.102 In prostate cancers, TET1 suppresses cancer invasiveness by activating the tissue inhibitors of metalloproteinases.106 In rectal cancers, TET1 inhibits the WNT signaling pathway by up-regulating WNT inhibitors DKK3 and DKK4. Downregulation of TET1 promotes cancer development due to the activation of WNT signaling. Interestingly, a study suggests that TET1 is overexpressed in 40% of triple-negative breast cancer patients. In these types of cancers, TET1 expression is involved in cancer activation pathways including EGFR, PI3K, and PDGF, and is correlated with cell migration, cancer stemness, tumorigenicity, and poor survival.107–109 It suggests that TET1 might function as an oncoprotein and a therapeutic target in these types of cancers.85 Furthermore, TET1s is aberrantly expressed in multiple cancer types including breast, uterine, and glioblastoma. The predominant TET1s activation in cancer cells results in dynamic site-specific demethylation outside of CGIs, which is associated with worse overall survival in breast, uterine, and ovarian cancers.54
Transcriptional regulation of TET2 and TET3 genes in cancer
Compared to TET1, the transcriptional regulation of TET2 and TET3 genes has been studied in much less detail (Fig. 1a). In ES cells, OCT4 binds to the promoter at 1788/1795 bp (relative to the TSS) of the TET2 gene and promotes TET2 expression.110 In addition, OCT4-SOX2-binding elements are identified at ∼− 140 kb and − 200 kb of the TET2 TSS.111 In response to hypoxia, HIF1α was found to repress TET2 expression in melanoma cells.112 In pancreatic cells, TGF1β induces the expression of TET2 by stimulating SMAD4 binding of an enhancer proximal to the distal 3’ region of the TET2 gene.70 Decreased TET2 and 5-hmC were found in ovarian carcinoma tissues and colorectal cancer patients, which was associated with high tumor grade, pathologic stage, lymph-node metastasis, and vascular thrombosis as well as chemoresistance and poor clinical outcomes.113–115 GATA6 is a key TF for the differentiation of pancreatic progenitors. In aggressive squamous-like PDAC subtypes, TET2 is down-regulated due to the loss of SMAD4, which is correlated, with a reduction of 5hmC and GATA6. Metformin and Vitamin C restore 5hmC and GATA6 levels by enhancing TET2 stability, reverting squamous-like tumor phenotypes and WNT-dependence both in vitro and in vivo.70
CGIs have also been identified in the promoter, intron 1, and intron 2 of the TET3 gene. TET3 is epigenetically repressed in gliomas due to the methylation of these CGIs.116 Loss of TET3 expression was identified in 32% of GCs and 28% of CRCs.117 TET3 was down-regulated in ovarian cancer cells during TGF-β1-induced epithelial–mesenchymal transition (EMT) and was correlated with pathological grade. TET3 over-expression was found to suppress ovarian cancer by up-regulating miR-30d, which then blocks TGF-β1-induced EMT.118 However, a study suggested that increased TET3 levels in ovarian carcinoma are associated with poor clinical-pathological status and poor prognosis.119
MicroRNAs regulate the expression of TET genes
The expression of TET proteins is also regulated by microRNA (miR)-mediated post-transcriptional repression.120 Approximately 30 miRNAs have been identified that repress TET2 expression, including miR-7, miR-125b, miR-29b/c, miR-26, miR-101, miR142, and Let-7.121 TET1 expression is regulated by miR-29 family members including miR-26a, miR-767, miR-494, and miR-520b.122–125 In hematopoietic tissues, miR-22 promotes HSC self-renewal and leukemic transformation by repressing TET2.126 In inflamed mouse epithelial cells, inflammatory cytokines such as IL-1β and TNF-α repress the expression of TET proteins by inducing NF-κB signaling-mediated miR20a, miR26b, and miR29c expression.127 In gastric carcinogenesis, miR-26 represses TET1/2/3 expression.81 In hepatocellular carcinomas, miR29a promotes SOCS1–MMP9 signaling axis-mediated tumor metastasis by repressing TET proteins.72 In models of type 1 diabetes, miR142-3p targets TET2 and impairs Treg differentiation and stability.128 In macrophages, Let-7 promotes IL-6 by repressing Tet2 expression.129
TETs–TDG–BER system regulates DNA demethylation
TET1, TET2, and TET3 are Fe2+ and α-KG-dependent dioxygenases. TETs catalyze the 1st step of demethylation by the hydroxylation of 5-methylcytosine (5mC) to 5hmC, and further oxidize 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC).16 To fully complete the demethylation process, 5fC and 5caC, the products of TETs, can be replaced by cysteine via either replication-dependent dilution/passive DNA demethylation or thymine DNA glycosylase (TDG) and base excision repair (BER)-mediated active DNA demethylation.59,130 Both 5fC and 5caC are substrates for TDG. TDG catalyzes the excision of 5fC and 5caC to generate an apyrimidinic site (AP site). By coordinating with BER enzymes, TDG mediates the replacement of 5fC and 5caC with cysteine. Studies suggest that TDG is essential in protecting CpG-rich promoters from hypermethylation and collaborating with key TFs by actively removing methyl groups from enhancers and promoters of target genes.131 Thus, active dynamic DNA demethylation is primarily mediated by an IDHs–TETs–TDG–BER-driven cytosine modification system. In addition, it was found that activation-induced cytidine deaminase (AID)/APOBEC mediates an alternative oxidative deamination–demethylation pathway. AID/APOBEC is required for DNA demethylation during reprogramming of somatic cells and B-cell maturation.132,133 AID catalyzes cytidine deaminases primarily at 5hmC sites to generate 5-hydroxymethyluracil (5hmU). 5hmU is subsequently cleaved by TDG, a single-strand-selective monofunctional uracil-DNA glycosylase 1 (SMUG1), Nei-like DNA Glycosylase 1 (NEIL1), or methyl-CpG-binding protein 4 (MBD4), and can be replaced by cytosine, as mediated by BER enzymes.134 Thus, AID mediates TET-dependent DNA demethylation.132,135,136 Furthermore, it was reported that both growth arrest and DNA damage-inducible protein 45a (GADD45a)137,138 and GADD45b play critical roles in the demethylation of specific promoters,137–139, and BER plays essential roles in genome-wide active DNA demethylation in primordial germ cells (PGCs).140 Further study demonstrated that TDG, AID, and GADD45a form a ternary complex in regulating the methylation state of promoters and enhancers within the genome. Thus, it is most likely that GADD45a/b–TDG–AID–BER altogether mediate active DNA demethylation.131
AID is a key enzyme that mediates DNA methylation dynamics in germinal center B cells.141–143 AID initiates the somatic hypermutation process through deamination of cytidine to uridine in the recombined variable region, followed by removal of the uracil base by uridine DNA glycosylase and DNA repair by several error-prone BER and mismatch-repair enzymes.135 AID further induces the second step of antibody diversification, class-switch recombination, through deamination of bases in the switch region, causing double-strand breaks and recombination.136 AID is a key regulator of myeloid and erythroid differentiation and DNA methylation in HSPCs.133,144 The demethylation activity of AID is severely impaired in the absence of TET2, without impairment of AID mutability, suggesting that AID is dependent on TET2 for its demethylating capacity. This explains an AID-dependent hyper-mutagenesis feature and tumor development in TET2-deficient animals.
DNA 5-hmC is an epigenetic mark of gene activation
It should be clarified that 5hmC, 5fC, and 5caC are not only intermediates of passive and active DNA demethylation but also serve as stable epigenetic marks145,146 and have distinct epigenetic regulatory functions because they are distributed genome-wide and can be recognized by specific reader proteins.147,148 For example, several selective 5-hmC readers have been identified, such as MeCP2, the MBD3/NURD complex, E3 ubiquitin-protein ligases (UHRF1 and UHRF2), DNA glycosylases (MPG and NEIL3), SALL1/SALL4, Thy28, PRMT1 (CHTOP)-methylome complex, Recql helicase, RBM14, PRP8, RPL26, MSH6, PNKP, and WDR76.148–152 Only three of them, NP95/UHRF1, MeCP2, and MBD3, have been confirmed in more than one study.149 These proteins bind to 5hmC-DNA and regulate gene expression by recruiting co-activators or co-repressors. 5hmC is present in high amounts at active enhancers and the gene bodies of highly transcribed genes.153 5-hmC is associated with the activating histone marks H3K4me1, H3K4me3, and H3K27ac153–157. This explains, in many cases, that gene expression is closely related to the 5hmC/5mC ratio of enhancers and/or promoters.158–160 In addition, some of these 5-hmC reader proteins bind to 5hmC-DNA and recruit TETs, which further recruit TDG-BER complexes for completing the remaining steps of DNA demethylation.
The selective DNA binding of TET proteins
Although all 3 TET family members and their isoforms have similar catalytic activity as demonstrated by certain levels of functional redundancy,161,162 the distinct phenotypes of Tet1, Tet2, and Tet3 knockout mice, as well as the distinct 5hmC/5mC patterns of Tet1, Tet2, and Tet3-deficient cells, suggest significant non-redundant functions for the Tet proteins.163–168 Such distinct roles of the three Tets are partially explained by their distinct expression profiles within developmental tissues. For example, Tet1 and Tet2 mRNAs levels are abundant in ES cells and PGCs,169,170 while Tet3 is the only Tet gene expressed at substantial levels in oocytes and zygotes.171,172 Tet1 is expressed in fetal heart, lung, and brain, and adult skeletal muscle, thymus, and ovary, but not in adult heart, lung, or brain. Tet2 is primarily expressed in hematopoietic tissues.100,173 Tet3 is highly expressed in neural progenitor cells where it preferentially binds to TSSs and regulates cellular identity and genes associated with the lysosomes, autophagy, and base excision repair pathways.57,174 The non-redundant functions of the three TETs and their isoforms are also determined by their selective binding to genomic DNA regions. TET1 has a high affinity for a high density of CpG promoters, while TET2 is more commonly located at low CpG density promoters.168,174 In mouse ES cells, Tet1 primarily regulates 5hmC levels at gene promoters and TSSs, whereas Tet2 mainly regulates 5hmC levels in gene bodies and exon boundaries of highly expressed genes and exons, respectively.165,175 In induced pluripotent stem cells (iPSCs), TET1 and TET2 appear to target different genomic regions and promote opposing functions in reprogramming-mediated erasure of imprints and naïve pluripotent state transitions. TET1 promotes a primed state of pluripotency, while TET2 regulates a naïve state of pluripotency.165,176,177
DNA binding by TET1 and TET3 is primarily mediated by their CXXC domains, while the DNA binding of TET2 and the short isoforms of TET1 and TET3 is mediated by interactions with partner proteins. Both TET1 and TET3 regulate DNA methylation specifically at CpG sites within and around CGIs, appearing to show more flexible substrate specificity.178,179 The TET1-CXXC domain binds CpG-rich DNA irrespective of methylation status, while the TET3-CXXC domain binds methylated CpG sites with relatively low affinity compared to a non-methylated CpG dinucleotide, with the highest affinity toward 5caC sequences.61,180,181 In addition, TET1-CXXC also binds to TFs FOXA1 and HIF2α, selectively mediating active epigenetic modifications at FOXA1 and HIF2α-dependent enhancers, respectively.98
TET1 binds CGI chromatin globally via its CXXC to protect CpG sites within and around CGIs from gaining aberrant methylation,178 while TET1s preferentially binds to CpG sites at non-CGIs and some targets CGI chromatin.54 Due to the selective binding of DNA regulatory regions, the roles of TET1 and TET3 are not always the same as their short isoforms and in many cases are the opposite. For example, compared to neurons, TET1 is highly expressed in glial cells, while TET1s is down-regulated. TET1 and TET1s expression has opposing effects on synaptic transmission and hippocampal-dependent memory.182 In mice, Tet1 is restricted to early embryos, ES cells, and PGCs, whereas Tet1s is preferentially expressed in somatic cells. The expression of Tet1 and Tet1s switches during development and regulates epigenetic memory erasure.53 TET1s is overexpressed in multiple cancer types including breast, uterine, and glioblastoma, which is associated with worse overall survival.54
The interaction partner proteins of TETs
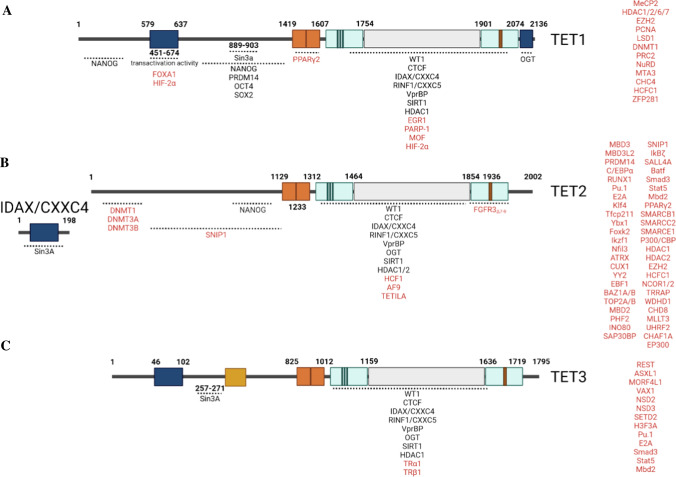
Many partner proteins of TET1, TET2, and TET3 have been identified; however, the interaction regions have been defined only for some of them (Fig. 2). Based on available information, most of the partner proteins bind to the C-terminal fragment including the DSβH enzymatic domain of TETs; only a few of them bind to the N-terminal fragment. However, the details of the interaction sites are only well known for Sin3A on TET1 and TET3, and O-linked GlcNAc transferase (OGT) on TET1. Sin3A interacts with the Sin3-interaction domain (SID) on TET1 (a.a. 889–903) and TET3 (a.a. 257–271). Although SID is absent from TET2 and its dimeric partner, CXXC4 might mediate the Sin3A–TET2 interaction.183,184 All three TETs interact with OGT through its C-terminal fragment.185–187 Detailed analysis demonstrated that the last 45 a.a. of the C-terminus (C45) of TET1 mediates OGT binding.188 However, the detailed binding sites of OGT on TET2 and TET3 have not been determined.
Fig. 2.
Interaction partner proteins of TET proteins. The interacting partner proteins of TET1, TET2, and TET3 are listed in a, b, and c, respectively. The partner proteins for which the interaction regions have been identified are listed under each TET at the corresponding regions. The partner proteins for which the interaction regions have not yet been defined are listed on the right side of each TET. The partners that are shared by all three TETs are listed in black font, while the partners that are specific for one or 2 TETs are listed in red font
Partner proteins for all 3 TETs
Among all partner proteins, some of them can interact with all three TETs. For example, CTCF can interact with all three TETs and recruit them to the CTCF-binding sites outside of CGIs, regulating DNA methylation and gene expression.189–192 CXXC4 and CXXC5 interact with the catalytic domain of TET2 as well as short isoforms of TET1 and TET3, recruiting TETs to DNA.193 As is true for the CXXC domain of TET1 and TET3, the CXXC domain in CXXC4 and CXXC5 proteins preferentially bind to unmethylated CGIs in gene promoter regions to maintain hypomethylation of CGIs.193,194 CXXC5 forms a complex with NANOG, OCT4, TET1, and TET2 and positively regulates the transcription of pluripotency genes and TET enzymes.195 Interestingly, CXXC4 negatively regulates TET2 activity by promoting caspase-mediated degradation of TET2 protein.193 WT1 physically interacts with TET2 and selectively regulates TET2-dependent expression of target genes such as RUNX1.196,197 WT1 also interacts with TET1 and TET3 for target gene expression.198 In addition, some histone modifiers such as SIRT1, histone deacetylases (HDACs) 1/2, and OGT as well as a variety of factors of the BER–DNA glycosylase pathway, including PARP1, MBD4, NEIL1, NEIL2, NEIL3, TDG, SMUG1, PARP1, LIG3, and XRCC1, also interact with all three TETs.199–201 All these shared interaction partners might partially explain the overlapping and compensatory functions of the three TET molecules.
Partner proteins that have been identified for certain TET proteins
Many of the partner proteins selectively bind to one or two of the TETs and their isoforms. Several partner proteins for TET1 have been identified; these include MeCP2, EZH2, LSD1, hMOF, and PCNA.151,156,202–205 TET3 interacts with TFs including REST, ASXL1, MORF4L1, VAX1, and thyroid hormone nuclear receptor (TR), as well as the H3K36 methyltransferases NSD2, NSD3, and SETD2, as determined by immunoprecipitation and LC–MS/MS.192 Significantly more TET2-interacting partners have been identified, including TFs (C/EBPα, PU.1, Klf4, Tfcp2l1, MBD3, MBD3L2, YBX1, FOXK2, IKZF1, NFIL3, ATRX, CUX1, YY2, WT1, EBF1, SNIP1, PML, and IκBζ158,197,206–211), histone modifiers (SMARCB1, SMARCC2, SMARCE1, P300/CBP, HDAC1, HDAC2, SIN3A EZH2, HCFC1, NCOR1/2, BAZ1A/B, TOP2A/B, MBD2, PHF2, INO80, SAP30BP, TRRAP, WDHD1, CHD8, MLLT3, UHRF2, and CHAF1A148,158,207,212–216), and signaling regulators (AMPK, JAK2, 14–3-3Z/D, and 14–3-3E proteins202). These selective interacting partners of TETs determine the TETs’ functional specificity. For example, Mbd3/NURD recruits TET1 to genomic sites to regulate the expression of 5-hmC-marked genes in ES cells.151 Lin28A binds to active promoters and recruits TET1 to regulate gene expression.217 EGR1 recruits TET1s to target genes and selectively regulates the expression of EGR1 target genes by DNA demethylation.218 In iPSCs, ZFP281 drives TET1 to the promoter of target genes including TET2 to promote primed pluripotency. SNIP1 selectively interacts with TET2 (but not TET1 nor 3), bridging TET2 to TFs, including C-MYC, CDC5L, and BCLAF1.208 SNIP1 recruits TET2 to C-MYC target genes and regulates C-MYC target gene expression. TET2–SNIP1–cMYC ternary complex regulates target gene expression, playing a crucial role in DNA damage response and cellular apoptosis.208 REST recruits neuronal TET3 to mediate 5hmC formation and transcriptional activation.192 TET-3s also interacts with NSD2, NSD3, and SETD2 to regulate gene expression by mediating H3K36 trimethylation. In addition, TET3 interacts with TR to stabilize it and enhance its function independent of TET3 catalytic activity.219 TET3 also interacts with histone variant H3F3A, regulating chromatin modification.192
Functional subgroups of the partner proteins
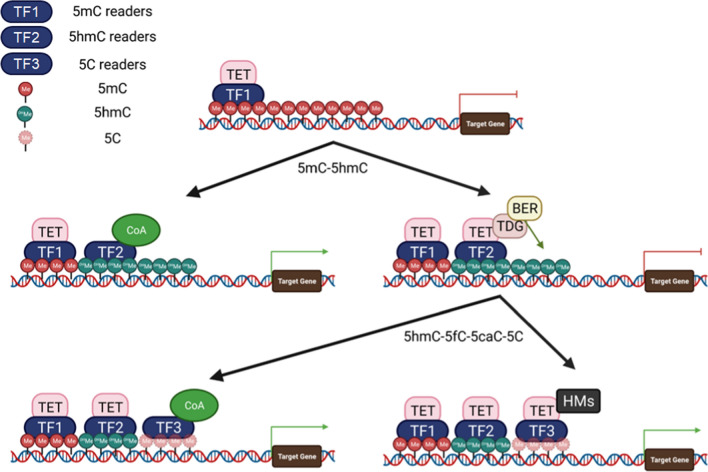
Based on their functions, the partner proteins of TETs can be divided into four groups: TFs, histone modifiers, signaling molecules, and factors of the BER–DNA glycosylase pathway. Most TFs such as NANOG, RUNX1, PU.1, and PPARγ bind to regulatory regions of their target genes and recruit TETs to regulate target gene expression.203,215,216,220 The binding motifs of ~ 66% TFs contain CpG dinucleotides. The binding of these TFs may be affected by CpG methylation.7 Based on the binding affinity of methylated CpG (mCpG) motifs, TFs can be divided into four types: MethylPlus TFs (TF1, preferred to bind to mCpG), 5hmC-DNA readers (TF2, preferred to bind to 5hmCpG), methylminus TFs (TF3, preferred to bind to CpG), and methylation-insensitive TFs (TF4, little affected by methylation)7,8 (Fig. 3). The TF1 proteins (such as CEBPB, MBD1, MBD3, MeCP2, MBD3L2, GATA3, GATA5. WT1, PRDM14, Nanog, ZFP57/KAP1, OCT4, SOX2, HOXB13, KLF4, FOXA1, EBF1, and EGR2) preferentially bind to 5mCpG motifs and function as pioneer factors to recruit TETs for converting 5mC into 5hmC.9,197,221–229 The TF2 molecules (such as MeCP2, MBD3/NURD complex, UHRF1, UHRF2, MPG, NEIL3, and SALL1/SALL4) preferentially bind to 5hmC-DNA sequences to either recruit TET–BER–DNA glycosylase complexes for fully demethylating DNA or recruit co-activators for activation of gene expression,201 while TF3 (such as AP-1, C-MYC /MAX, N-MYC, ETS-2, C-MYB, NF-κB, PAX5, RUNX1/2/3, NRF1, CTCF, CEBPα, CREB, and PU.1) bind to CpG motifs to regulate gene expression by recruiting TET-histone modification complexes.230,231 On the unmethylated DNA sequences, TETs might also play a role in maintaining the unmethylated state. Interestingly, the IDAX protein binds to unmethylated CpGs and inhibits TET2 binding to the demethylated regions through activation of caspase-mediated degradation, which might help to stop the demethylation process.193 Thus, it is most likely that the TFs form a hierarchy, which sequentially binds to DNA sequences and cooperates with TET proteins and histone modifiers to regulate target gene expression. Consequently, cell-type-specific TFs mediate a cell-type-specific binding pattern of TET proteins (Fig. 3). For example, in mouse ES cells, Tet1 uses its CXXC domain to bind to enhancers with 5mCpG islands and converts 5mC into 5hmC. Sall4a binds to 5hmC at enhancers and facilitates further oxidation of 5hmC at its binding site by recruiting Tet2.232 MBD3/NURD binds to 5hmC and recruits TET1 to genomic sites to regulate the expression of 5-hmC-marked genes.151 Such TET protein-associated sequential binding of TFs to DNA sequences is observed in almost all cellular processes by regulating the epigenetic landscape and inducing the expression of fate-determining genes.
Fig. 3.
Subgroups of TET-interacting TFs. The TET-interacting TFs can be divided into TF1, TF2, and TF3 based on their binding affinity for 5mC, 5hmC, and 5C, respectively. TF1 can bind 5mC DNA and recruits TET to initiate the first step of DNA demethylation by converting 5mC to 5hmC. TF2 can bind to 5hmC promoters/enhancers to turn on gene expression by recruiting co-activators (CoA) or to further complete the DNA demethylation elements by recruiting TET–TDG–BER complexes. TF3 binds 5C promoters/enhancers to promote gene expression by recruiting CoA or regulating gene expression by recruiting TET-histone modifiers (HMs)
Methylation serves as a barrier to reprogramming and differentiation.233,234 During induced reprograming of epiblast-like cells to PSCs, PRDM14 induces TET1/2-demethylation-mediated recruitment of OCT3/4 to the enhancers of pluripotent genes such as Klf2.235 During the specification of PSCs to primordial germ cells, PRDM14, Nanog, and OCT4 are capable of binding to 5mCpG sites to initiate the stepwise epigenetic modification by recruiting TET1/2 proteins and other epigenetic modifiers.223 During induced reprograming of B cells or embryonic fibroblasts to generate PSCs, Tet2 is recruited by Klf4 and Tfcp2lƒ1 respectively to drive active enhancer demethylation of chromatin and induce pluripotency-related genes.209 Thus, most TF1s are fate-instructive pioneer factors that initiate the cellular processes such as lineage commitment and differentiation by establishing epigenetic configurations,236–240 specifically when they collaborate with non-pioneer TFs.241–243
However, such mGpC-binding affinity-based sub-classification of TFs is not always accurate because binding affinity can be influenced by the surrounding sequence context. In addition, many TFs have more than one consensus-binding motif, while methylation only influences the binding of TFs to certain motifs. Thus, many TF3s can also function as pioneer factors, especially in collaboration with other TFs. For example, during the differentiation of fibroblasts to adipocytes, C/EBPα and CREB heterodimerize and bind half-CRE (CGTCA) and half-C/EBP (CGCAA) sequences of the tissue-specific methylated promoters to initiate DNA demethylation by recruiting TET2.244 This allows the binding of other TF3s (such as CEBPα/β, c-Jun, JunD, ATF2, or PU.1) for transcriptional activation.7 During differentiation of pro-B progenitors to pre-B progenitors, PU.1 and E2A bind to the 5mC enhancers of target genes and recruit TET2 and TET3 for stepwise DNA methylation. This is followed by the binding of other key B-cell-specific TFs to turn on the B-cell differentiation process.214 During induced pre-B-cell-to-macrophage trans-differentiation, CEBPα collaborates with PU.1 to induce the myeloid cell fate by regulating two types of enhancers on myeloid genes, pre-existing ones and de novo ones. The pre-existing enhancers are primed by PU.1, which maintains chromatin accessibility for the binding of CEBPα. In de novo promoters, CEBPα acts as a pioneer factor to initiate TET2-mediated demethylation followed by PU.1 recruitment.209,245 Therefore, the functional identification of pioneer factor(s) for different cellular processes is needed to elucidate transcriptional-epigenetic regulatory mechanisms for each cellular process.
Among the histone modifiers, most of them are negative transcriptional regulators such as the SIN3A complex, the NuRD complex, HDAC1, HDAC2, and EZH2, which mediate target gene repression,156,207,246 while some others are positive transcriptional regulators, including CBP, hMOF, NSD2, NSD3, and SETD2 that promote gene expression via modulating H3K27ac, H4K16ac, and H3K36Me on promoters.247 Such TET-related histone modification is independent of the catalytic activity of TETs. Furthermore, some of the partner proteins such as OGT, PARP1, and VprBP regulate the functions of TETs through post-transcriptional modifications (see the following section).
Post-translational regulation of TET proteins
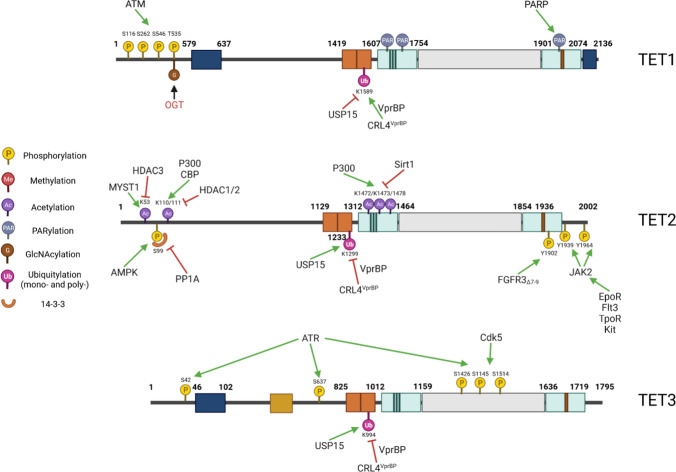
The N-terminal sequence of the TET proteins plays a critical role in regulating TET activity by interacting with their catalytic domains. Mammalian TETs undergo a plethora of post-translational modifications (PTMs). However, the functional significance of some of these modifications is not yet well understood. Some of the well-known PTMs that are commonly found on TETs are GlcNAc, phosphorylation, ubiquitylation, acetylation, and proteolysis210,212,248–251 (Fig. 4).
Fig. 4.
Post-translational modifications of TET proteins. The post-translational modifications of TET1, TET2, and TET3 are listed in a, b, and c, respectively. The green arrow depicts the addition of modifications. The Red Cross indicates removal modifications
Phosphorylation regulates the activities of TET proteins
Mass spectrometric analysis identified over 10–20 residues that can be phosphorylated in each of the TET proteins.248 However, the role of phosphorylation has only been functionally studied for a few of these residues. During DNA damage repair, ATM phosphorylates TET1 on S116, S262, and S546, regulating DNA repair.252 The energy sensor, AMPK (AMP-activated protein kinase), phosphorylates human TET2 on Ser99 (murine Tet2, Ser97), protecting TET2 protein from calpain-mediated degradation. Thus, active AMPK promotes TET2 stability and facilitates its tumor-suppressive function.250,253,254 Several members of the 14-3-3 group of adaptor proteins bind to Ser99 phosphorylated TET2 and protect it from phosphatase 2A (PP2A)-mediated dephosphorylation.255,256 The association of 14-3-3 proteins is impaired in some leukemia-related TET2-mutants (around residue Ser99), explaining the reduced protein stability of these mutant TET2 proteins.255 In diabetic mice, high glucose levels impede the tumor-suppressive activity of TET2 and accelerate tumor development by blocking AMPK-mediated phosphorylation of TET2 and reducing TET2 protein levels as demonstrated in xenograft tumor models.250 This explains why diabetic patients have an increased risk for cancer and cancer patients with diabetes have a poor prognosis as observed in epidemiological studies.257,258 The anti-diabetic drug metformin and other AMPK activators such as A769662 display antitumor activity by activating AMPK-mediated phosphorylation of TET2 Ser99 and increasing 5hmC levels. Diabetes risk reduction diets improve the survival of cancer patients.259 In addition, in erythroid progenitor cells, hematopoietic cytokines such as erythropoietin (EPO) stimulate JAK2-mediated phosphorylation of TET2 on Tyr1939 and Tyr1964 residues, which enhances TET2 binding of the TF KLF1 and increases TET2 activity for the proliferation and differentiation of erythroid progenitor cells.260 Consistently, in primary samples from patients with myeloproliferative neoplasms, JAK2V617F increases TET2 activity and 5-hmC with genome-wide loss of cytosine methylation, leading to increased expression of several oncogenic transcripts, such as MEIS1 and HOXA9.260 In hepatocellular carcinoma patients, FGFR3∆7–9, a splicing mutant of FGFR3, directly interacts with TET2 and phosphorylates TET2 on its Y1902 site, leading to the ubiquitination and proteasome-mediated degradation of TET2.261 Such phosphorylation-related down-regulation of TET2 enhances cancer cell proliferation through repression of PTEN and upregulation of AKT signaling. Interestingly, in CML cell lines, the BCR–ABL fusion protein interacts with TET2 and sequesters the latter by cytoplasmic compartmentalization in a complex tethered by FOXO3a.262 Imatinib treatment releases TET2 from the complex and imports TET2 into the nucleus together with FOXO3a to activate BIM expression by binding to the BIM promoter.262 Whether TET2 is phosphorylated by BCR-ABL kinase needs to be determined.
During neuronal differentiation, CDK5 phosphorylates TET3 on residues Ser1310 and Ser1379 (Ser1318 and Ser1387 for the mouse) within its catalytic domain, changing its dioxygenase activity.251 Phosphorylated TET3 promotes the expression of the neuron-specific TF BRN2, as well as neuronal differentiation, through enhancing the enrichment of 5hmC and H2A.Z occupancy at the promoter of the BRN2 gene. Non-phosphorylated TET3 promotes the expression of genes that are linked to metabolic processes.251 In response to DNA damage, ataxia-telangiectasia and Rad3-related kinase (ATR) phosphorylates TET3 on residues Ser42, 637, and 1426. TET3 phosphorylated in this way mediates DNA oxidation which promotes the ATR-dependent DNA damage response.263
GlcNAc regulates the activity of TET proteins
All three TETs interact with OGT.185–187 OGT regulates their stability and activities by catalyzing GlcNAc and thereby regulating the phosphorylation of the proteins at their N-termini and low-complexity insert regions.248,264,265 OGT is also involved in the regulation of the binding of TETs to some genomic sites.266 At least eight GlcNAc sites have been reported for TET1 and up to 20 such have been identified for both TET2 and TET3.185,248,264,266 Many of these GlcNAc sites, such as Ser97 and Ser374 of TET2, Ser362 and Ser557 of TET3, and Ser950 and Ser2016 of TET1, could also be phosphorylated. Thus, GlcNAc represses the phosphorylation of the corresponding sites and regulates the binding of TETs with other partners.248 In addition, the GlcNAc site Thr535 on TET1 enhances this protein's stability,266 while GlcNAc of TET3 promotes its cytoplasmic relocation,265 and GlcNAc of TET2 reduces its enzymatic activity by enhancing its nuclear export.187 Furthermore, OGT regulates the expression of TET target genes by GlcNAc and several other epigenetic modifiers and histones (see the following section).
Ubiquitination regulates the activities of the TET proteins
VprBP binds the cysteine-rich, dioxygenase domain of all three proteins, exerting a critical regulatory function on TET dioxygenases in normal tissue development and tumor suppression. VprBP induces CRL4VprBP (VprBP-DDB1-CUL4-ROC1) E3 ubiquitin ligase-mediated monoubiquitylation of TET1 on Lys1589 (Lys1537 in the mouse), of TET2 on Lys1299 (Lys1212 in the mouse), and of TET3 on Lys994 (Lys983 in the mouse). Such monoubiquitylation facilitates TET binding to chromatin and enhances 5hmC in corresponding genomic regions.212 TET2 mutations in leukemic cells on either Lys1299 or residues essential for VprBP binding result in reduced chromatin binding and activity of TET2.212 In addition, mutation of Lys983 of TET3, but of neither TET1 nor TET2, also alters the enzyme’s subcellular localization from almost exclusively nuclear to mostly cytoplasmic. Whether monoubiquitylation selectively regulates the subcellular localization of TET3 needs to be determined. Interestingly, during HIV infection, the viral protein Vpr induces CRL4VprBP–mediated poly-ubiquitination of TET2 Lys1299, inducing the degradation of TET2 to sustain IL-6 expression and enhance HIV-1 replication.212,267
Acetylation regulates the activity of TET proteins
During oxidative stress, transcriptional co-activator p300 acetylates TET2 on Lys110/111 residues to enhance the enzymatic activity of TET2 and to protect the protein against proteasomal degradation by the inhibition of TET2 ubiquitination on certain residues in the C-terminal DSBH domain.210 TET2 acetylation enhances DNMT1 binding to promote protein stability. Consequently, TET2, along with TDG, is recruited to chromatin by DNMT1 to prevent abnormal DNA methylation. TET2 Lys110/111 deacetylation is mediated by HDAC1/2.207 TET1 and TET3 are also acetylated by p300; however, the detailed sites for such modifications have not been determined.210 In MDS, SIRT1 interacts with the TET2 C-terminal domain (a.a. CD1129–2002) and deacetylates it on Lys1472, 1473, and 1478 in CD34+ HSPCs, regulating the stability and function of TET2 protein. SIRT1-deficient MDS HSPCs exhibit enhanced cell growth and self-renewal due to the reduction of TET2 levels.268 The SIRT1 activator SRT1720 inhibits colony formation in MDS HSPCs and in vivo engraftment in NSGS mice by enhancing the tumor repressive activity of TET2.
Other post-translational modifications of TET proteins
The stability of TET proteins is regulated by calpains.269 TET1 and TET2 are degraded by calpain 1 in mouse ES cells, whereas TET3 is degraded by calpain 2 during ES cell differentiation.269 TET1 interacts with PARP1/ARTD1 and is targeted by both noncovalent and covalent PARylation in TET1’s catalytic domain. The noncovalent binding of ADP-ribose polymers decreases TET1’s hydroxylase activity, while covalent PARylation stabilizes the TET1 enzyme and enhances its activity.270–272 In addition, PARP1 also promotes TET1 gene expression by regulating DNA and histone modifications on the TET1 promoter.271
Metabolic regulation of TET protein activity
Fe2+ and α-KG, together with O−2 and vitamin C, function as TET co-factors and are required for their dioxygenase activity.273–276 Both Fe2+ and α-KG bind to the catalytic domain of TETs facilitating the insertion of 5mC into their catalytic pocket and providing accommodation to the oxidized derivatives of 5mC including 5hmC, 5fC, and 5caC.179,277,278 Thus, the dioxygenase activity of TET proteins is dependent on the availability of α-KG, Fe2+, and O2. α-KG is a product of IDHs, a family of metabolic enzymes. The IDHs, IDH1, IDH2, and IDH3, catalyze the oxidative decarboxylation of isocitrate to α-KG, which is an essential step in the tricarboxylic acid cycle. IDH1/2 mutations are commonly detected in gliomas and hematopoietic malignancies which lead to the production of the oncometabolite 2-hydroxyglutarate (2HG). Mutations in other genes encoding for the metabolic enzymes succinate dehydrogenase (SDH) and fumarate hydratase (FH) are prevalent in gliomas, cholangiocarcinomas, renal cell carcinomas, and acute myeloid leukemias, among others. SDH and FH mutations lead to the production of the oncometabolites succinate and fumarate, respectively.279–281 An overabundance of these oncometabolites influences the catalytic activities of TET1/2/3 by competitive inhibition of their α-KG-binding site. In activated macrophages, itaconic acid, a metabolic product of the IRG1 enzyme, also inhibits the catalytic activity of TET2 via inhibition of TET2/α-KG binding.282 Thus, TET-mediated DNA demethylation is tightly regulated by glucose metabolism. Reactive oxygen species (ROS) and metal chelators can impede TETs’ oxidizing activities by reducing the Fe2+ availability.283 Vitamin C convents inactive Fe3+ to active Fe2+ which promotes TET1/2/3 enzymatic activity.284 Thus, in addition to its antioxidant properties, Vitamin C regulates gene expression and genomic stability by increasing TET-mediated 5hmC formation and promoting DNA demethylation.274,285–287
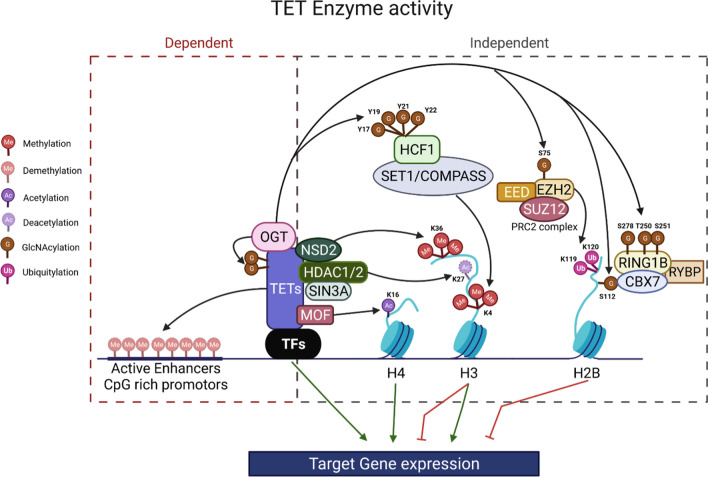
TETs regulate gene expression by both enzyme-dependent and -independent mechanisms
TETs play critical roles in organ generation during embryonic development and tissue regeneration during postnatal life.161,175 TETs play such roles by regulating the timed expression of the key genes that determine cell identity and control cell differentiation. TETs and lineage-specific TFs cooperate to influence chromatin accessibility and regulate gene expression by (1) promoting site-specific DNA demethylation (mainly enhancers and CGI-rich promoter elements) and enzymatic-dependent activity;16,58,156,172,175,277 and (2) regulating histone modifications via an enzymatic-independent activity by forming chromatin regulatory complexes with OGT, HDACs, and/or histone acetyltransferases (HATs)187,207,288,289 Fig. 5.
Fig. 5.
TETs regulate gene expression by both enzyme-dependent and -independent mechanisms. After binding to DNA regulatory regions, TETs regulate target gene expression by (1) enzymatic demethylation of 5mC; and (2) recruiting histone modifiers, including OGT, Sin3A/HDACs, or HATs.
TETs regulate gene expression by enzymatic activity-dependent site-specific DNA demethylation
TET proteins collaborate with lineage-specifying TFs in cells, promoting the expression of cell-type-specific genes by demethylation of enhancers of target genes. ChIP-seq assays demonstrated that the top enriched binding motifs of TET proteins in DNA are enriched with binding sites for lineage-specifying TFs of the respective cell types, maintaining 5hmC and demethylation state in the active enhancers of target genes in an enzyme-dependent fashion.12,30,175,222,290,291 For example, in ES cells, TET1/2 together with master self-renewal TFs, including SOX2, KLF4, ESRRG, POU5F1, and NANOG, bind enhancers of the target genes that are essential for the maintenance of self-renewal.14 In myeloid cells, TET2, together with key myeloid differentiation TFs such as ERG, RUNX1, CEBPA, and GATA1 bind enhancers of genes that are necessary for myeloid lineage commitment and differentiation.14 Loss of TET2 causes down-regulation of cell-type-specific genes due to the widespread reduction of 5hmC and increased methylation of their enhancers, altering cell fate.14,292
In many types of cancers, TET proteins function as tumor suppressors by activating the expression of tumor repressive genes. For example, in gastric cancer, TET1 inhibits the AKT and FAK signaling pathways by demethylation of the PTEN gene promoter.68 In colon cancer, TETs suppress the proliferation of cancer cells by demethylating DKK gene promoters, inhibiting the Wnt signaling cascade.293 In pancreatic cancer, TET1 restricts the cell cycle of cancer cells by up-regulating negative cell cycle regulators such as p16.69 In these types of cancers, restoration of the enzymatic activity of TETs is a potent treatment strategy. However, in some other types of cancers, TET proteins function as putative oncoproteins and promote stemness in the cancer stem-like subpopulation that drives aggressiveness and chemoresistance. For example, in ovarian cancers, TET1 induces the expression of cancer stem cell genes, which reprograms epithelial cancer cells into a cancer stem-like state.82 In gliomas, TET1 and TET3 promote stemness and self-renewal of tumor cells by regulating the expression of core stem cell genes.294,295 In breast cancers, TET1 and TET3 cooperatively induce cancer stem-like cells by activating the TNFα–p38–MAPK signaling axis.296 In such types of cancers, high levels of TET proteins promote a subpopulation of the slow-growing chemoresistant stem-like cells that are associated with disease relapse and poor prognosis.297 In addition, TET proteins also regulate EMT and cancer metastasis in a context-dependent manner.97,118,296,298 Thus, targeting TET enzymes for cancer therapy must also be strategized for context dependence.
TETs regulate gene expression via enzymatic activity-independent histone modifications
Through their enzymatic-independent activities, TET proteins primarily repress gene expression. For example, TET proteins recruit OGT to histones at the promoters of target genes and regulate target gene expression by mediating OGT-dependent GlcNAc of TFs, epigenetic regulators, and histones.89,90,299 GlcNAc of H2B on Ser112 is required for subsequent Lys120 monoubiquitination and PRC1-mediated gene silencing.300 GlcNAc of RING1B on Ser278 and Thr250/Ser251 residues promotes the binding of RING1B to CBX7 and RYBP to form PRC1, resulting in H2BK118 ubiquitination and silencing of a specific subset of genes.301 In the PRC2 complex, EZH2 is modified by GlcNAc on Ser75, which results in its being stabilized, thus negatively regulating tumor suppressor genes.302 In addition, TET proteins recruit Sin3A/HDAC1/2 to target genes, repressing target gene expression by deacetylation of histone H3K27.205,207 However, TET proteins also promote some target gene expression via enzymatic-independent activity. For example, via OGT-mediated GlcNAc of HCF1 (Tyr17, 19, 21, and 22), the key component of H3K4 methyltransferase SET1/COMPASS complexes, TET proteins promote chromatin binding and H3K4me3, inducing target gene expression.185–187,264,266,303,304 TET1 also upregulates the expression of proliferation and DNA damage repair genes by recruiting the HAT protein MOF to promoters, acetylating H4K16.247 TET3 promotes transcriptional activation in neurons by recruiting NSD2, NSD3, and SETD2, thus mediating H4K36 trimethylation.
Prospective
The activity of TET proteins shapes the local chromatin environment at enhancers and promoters to facilitate the binding of TFs and affect gene expression patterns.14 Mutations or dysregulations of TET proteins lead to abnormal DNA methylation patterns and epigenetic chromatin modifications, driving disease development. Although TET1 and TET3 have their DNA-binding CXXC domains which mediate region-specific DNA binding and demethylation, TET2 and the short forms of TET1 and TET3 rely on interactions with TFs for DNA binding. Thus, the regions of DNA binding of these TETs must conform to cell-type specificity, which is controlled by cell-type-specific TFs.
Based on their binding affinity to methylated DNA, TFs form a hierarchy in DNA binding, chromatin modification, and gene regulation. First, the methylation-insensitive pioneer TFs bind to methylated DNA to initiate DNA demethylation by recruiting TET proteins to convert 5mC to 5hmC.305 Next, the secondary level of TFs (5hmC readers) binds to 5hmC-DNA to activate gene expression by recruiting co-activators or to further complete DNA demethylation by recruiting a TET–TDG–BER complex. Finally, the third-level TFs (methylation-sensitive) occupy the unmethylated DNA to control gene expression by recruiting co-activators or co-repressors. Such sequential and cooperative binding of TFs and TETs leads to a relatively large open region of chromatin, which forms a super-enhancer in the target genes to determine the fate of cells.231 Many TFs can bind TET proteins. However, the hierarchy of these TFs has not yet been well characterized. Thus, the manner in which these TFs cooperate with TET proteins in the regulation of target gene expression needs to be better elucidated in the future.
Both positive and negative regulatory effects of TET proteins on target gene expression have been reported. TET proteins activate the expression of target genes primarily by enzymatic activity-mediated DNA demethylation and/or by OGT-regulated SET1/COMPAS-mediated H3K4 trimethylation, while they repress the expression of target genes by recruiting SIN3A/HDAC1/2 or OGT-regulated PRC1. It is still unknown how such positive and negative regulatory mechanisms are coordinated in regulating target gene expression. Furthermore, the activity of TET proteins is regulated by many types of post-translational modifications. Detailed study of the molecular mechanisms by which the activities of TET proteins are regulated and how TET proteins selectively regulate target gene expression will provide useful information for designing medications for a new generation of TET-related disease treatments.
Abbreviations
- TET
Ten–eleven translocation
- α-KG
α-Ketoglutarate
- 5mC
5-Methylcytosine
- 5hmC
5-Hydroxymethylcytosine
- 5fC
5-Formylcytosine
- 5caC
5-Carboxylcytosine
- TFs
Transcription factors
- ARCH
Age-related clonal hematopoiesis
- MDS
Myelodysplastic syndromes
- AML
Acute myeloid leukemia
- ALL
Acute lymphoblastic leukemia
- DLBCLs
Diffuse large B-cell lymphomas
- PTCL
Peripheral T-cell lymphoma
- IFN-γ
Interferon-γ
- TNF-α
Tumor necrosis factor-α
- TIS
Transcriptional initiation sites
- DSBH
Double-stranded beta-helix domain
- TDG
Thymine DNA glycosylase
- BER
Base excision repair
- ES
Embryonic stem cells
- IDHs
Isocitrate dehydrogenases
- AP site
Apyrimidinic site
- AID
Activation-induced cytidine deaminase
- 5hmU
5-Hydroxymethyluracil
- GC
Germinal center
- CSR
Class-switch recombination
- DSBs
Double-strand breaks
- PGCs
Primordial germ cells
- CGIs
CpG islands
- OGT
O-linked GlcNAc transferase
- SID
Sin3A interacts with the Sin3-interaction domain
- CoA
Coactivators
- PTM
Post-translational modifications
- AMPK
AMP-activated protein kinase
- C/EBPα
CCAAT/enhancer-binding protein alpha
- KLF4
Kruppel-like factor-4
- TFCP2l1
Transcription factor CP2 like-1
- YBX1
Y box-binding protein-1
- FOXK2
Forkhead box protein K-2
- KZF1
DNA-binding protein Ikaros 1
- NFIL3
Nuclear factor interleukin-3-regulated protein
- ATRX
Alpha-thalassemia/mental retardation syndrome X-linked transcriptional regulator
- CUX1
Homeobox protein cut-like-1
- YY2
Yin and yang-2 transcription factor
- IκBζ
Inhibitory-kappa-B-zeta
Author contributions
Kanak Joshi and Shanhui Liu drafted the first version of this review. All of the authors contributed to the writing of this manuscript. Peter Breslin did the final editing. All authors read and approved the final manuscript.
Funding
This work was supported by NIH grants R01 HL133560 and R01 CA223194 through Loyola University Chicago, as well as Loyola program development funds to Jiwang Zhang.
Availability of data and materials
This is not applicable for this review.
Code availability
This is not applicable for this review.
Declarations
Conflicts of interest
The authors declare that they have no competing financial or professional interests.
Ethics approval
This is not applicable for this review.
Consent to participate
This is not applicable for this review.
Consent for publication
This is not applicable for this review.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kanak Joshi and Shanhui Liu have contributed equally to this work.
References
- 1.McKinney-Freeman S, et al. The transcriptional landscape of hematopoietic stem cell ontogeny. Cell Stem Cell. 2012;11:701–714. doi: 10.1016/j.stem.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulis M, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barwick BG, Scharer CD, Bally APR, Boss JM. Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat Immunol. 2016;17:1216–1225. doi: 10.1038/ni.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 6.Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Y, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017 doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 9.Hu S, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 11.Greenberg MVC, Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki J, et al. TET2 mutations affect non-CpG island DNA methylation at enhancers and transcription factor-binding sites in chronic myelomonocytic leukemia. Cancer Res. 2015;75:2833–2843. doi: 10.1158/0008-5472.CAN-14-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lea AJ, et al. Genome-wide quantification of the effects of DNA methylation on human gene regulation. Elife. 2018 doi: 10.7554/eLife.37513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasmussen KD, et al. TET2 binding to enhancers facilitates transcription factor recruitment in hematopoietic cells. Genome Res. 2019;29:564–575. doi: 10.1101/gr.239277.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Zhang Y. Role of mammalian DNA methyltransferases in development. Annu Rev Biochem. 2020;89:135–158. doi: 10.1146/annurev-biochem-103019-102815. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 17.Broome R, et al. TET2 is a component of the estrogen receptor complex and controls 5mC to 5hmC conversion at estrogen receptor cis-regulatory regions. Cell Rep. 2021;34:108776. doi: 10.1016/j.celrep.2021.108776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz-Rodriguez N, Combita AL, Zabaleta J. Epigenetics in hematological malignancies. Methods Mol Biol. 2018;1856:87–101. doi: 10.1007/978-1-4939-8751-1_5. [DOI] [PubMed] [Google Scholar]
- 19.Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30:2021–2041. doi: 10.1101/gad.284109.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, Dominguez PM, Melnick AM. The many layers of epigenetic dysfunction in B-cell lymphomas. Curr Opin Hematol. 2016;23:377–384. doi: 10.1097/MOH.0000000000000249. [DOI] [PubMed] [Google Scholar]
- 21.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 23.Smith AE, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 24.Kosmider O, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swierczek SI, et al. Extent of hematopoietic involvement by TET2 mutations in JAK2V(6)(1)(7)F polycythemia vera. Haematologica. 2011;96:775–778. doi: 10.3324/haematol.2010.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tefferi A, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tefferi A, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jankowska AM, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langemeijer SM, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 30.Asmar F, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98:1912–1920. doi: 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez PM, et al. TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation, and promotes B-cell lymphomagenesis. Cancer Discov. 2018;8:1632–1653. doi: 10.1158/2159-8290.CD-18-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 35.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 36.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz R, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378:1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soucie E, et al. In aggressive forms of mastocytosis, TET2 loss cooperates with c-KITD816V to transform mast cells. Blood. 2012;120:4846–4849. doi: 10.1182/blood-2011-12-397588. [DOI] [PubMed] [Google Scholar]
- 40.Tefferi A, et al. Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23:900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coltro G, et al. Clinical, molecular, and prognostic correlates of number, type, and functional localization of TET2 mutations in chronic myelomonocytic leukemia (CMML)-a study of 1084 patients. Leukemia. 2020;34:1407–1421. doi: 10.1038/s41375-019-0690-7. [DOI] [PubMed] [Google Scholar]
- 42.Yao WQ, et al. Angioimmunoblastic T-cell lymphoma contains multiple clonal T-cell populations derived from a common TET2 mutant progenitor cell. J Pathol. 2020;250:346–357. doi: 10.1002/path.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 44.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang CRC, et al. Inflammatory cytokines promote clonal hematopoiesis with specific mutations in ulcerative colitis patients. Exp Hematol. 2019;80:36–41. doi: 10.1016/j.exphem.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hormaechea-Agulla D, et al. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNgamma signaling. Cell Stem Cell. 2021 doi: 10.1016/j.stem.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buscarlet M, et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–762. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- 50.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019 doi: 10.1126/science.aan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, et al. Isoform switch of TET1 regulates DNA demethylation and mouse development. Mol Cell. 2016;64:1062–1073. doi: 10.1016/j.molcel.2016.10.030. [DOI] [PubMed] [Google Scholar]
- 54.Good CR, et al. A novel isoform of TET1 that lacks a CXXC domain is overexpressed in cancer. Nucleic Acids Res. 2017;45:8269–8281. doi: 10.1093/nar/gkx435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- 56.Sohni A, et al. Dynamic switching of active promoter and enhancer domains regulates Tet1 and Tet2 expression during cell state transitions between pluripotency and differentiation. Mol Cell Biol. 2015;35:1026–1042. doi: 10.1128/MCB.01172-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N, et al. Intrinsic and extrinsic connections of Tet3 dioxygenase with CXXC zinc finger modules. PLoS One. 2013;8:e62755. doi: 10.1371/journal.pone.0062755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hahn MA, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin SG, et al. Tet3 reads 5-carboxylcytosine through Its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep. 2016;14:493–505. doi: 10.1016/j.celrep.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shekhawat J, et al. Ten-eleven translocase: key regulator of the methylation landscape in cancer. J Cancer Res Clin Oncol. 2021;147:1869–1879. doi: 10.1007/s00432-021-03641-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kunimoto H, Nakajima H. TET2: a cornerstone in normal and malignant hematopoiesis. Cancer Sci. 2021;112:31–40. doi: 10.1111/cas.14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bray JK, Dawlaty MM, Verma A, Maitra A. Roles and regulations of TET enzymes in solid tumors. Trends Cancer. 2021;7:635–646. doi: 10.1016/j.trecan.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 67.Dziaman T, et al. Characteristic profiles of DNA epigenetic modifications in colon cancer and its predisposing conditions-benign adenomas and inflammatory bowel disease. Clin Epigenetics. 2018;10:72. doi: 10.1186/s13148-018-0505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pei YF, et al. TET1 inhibits gastric cancer growth and metastasis by PTEN demethylation and re-expression. Oncotarget. 2016;7:31322–31335. doi: 10.18632/oncotarget.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu J, et al. TET1-mediated DNA hydroxymethylation activates inhibitors of the Wnt/beta-catenin signaling pathway to suppress EMT in pancreatic tumor cells. J Exp Clin Cancer Res. 2019;38:348. doi: 10.1186/s13046-019-1334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eyres M, et al. TET2 drives 5hmc marking of GATA6 and epigenetically defines pancreatic ductal adenocarcinoma transcriptional subtypes. Gastroenterology. 2021;161:653–668. doi: 10.1053/j.gastro.2021.04.044. [DOI] [PubMed] [Google Scholar]
- 71.Spans L, et al. Genomic and epigenomic analysis of high-risk prostate cancer reveals changes in hydroxymethylation and TET1. Oncotarget. 2016;7:24326–24338. doi: 10.18632/oncotarget.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Q, et al. MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET-SOCS1-MMP9 signaling axis. Cell Death Dis. 2017;8:e2906. doi: 10.1038/cddis.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lian CG, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–1146. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J, et al. Global DNA 5-hydroxymethylcytosine and 5-formylcytosine contents are decreased in the early stage of hepatocellular carcinoma. Hepatology. 2019;69:196–208. doi: 10.1002/hep.30146. [DOI] [PubMed] [Google Scholar]
- 75.Zahid OK, et al. Solid-state nanopore analysis of human genomic DNA shows unaltered global 5-hydroxymethylcytosine content associated with early-stage breast cancer. Nanomed Nanotechnol Biol Med. 2021;35:102407. doi: 10.1016/j.nano.2021.102407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haffner MC, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kraus TF, et al. Loss of 5-hydroxymethylcytosine and intratumoral heterogeneity as an epigenomic hallmark of glioblastoma. Tumour Biol. 2015;36:8439–8446. doi: 10.1007/s13277-015-3606-9. [DOI] [PubMed] [Google Scholar]
- 78.Thomson JP, et al. Loss of Tet1-associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 2016;76:3097–3108. doi: 10.1158/0008-5472.CAN-15-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang H, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai KW, et al. Reduction of global 5-hydroxymethylcytosine is a poor prognostic factor in breast cancer patients, especially for an ER/PR-negative subtype. Breast Cancer Res Treat. 2015;153:219–234. doi: 10.1007/s10549-015-3525-x. [DOI] [PubMed] [Google Scholar]
- 81.Deng M, et al. TET-mediated sequestration of miR-26 drives EZH2 expression and gastric carcinogenesis. Cancer Res. 2017;77:6069–6082. doi: 10.1158/0008-5472.CAN-16-2964. [DOI] [PubMed] [Google Scholar]
- 82.Chen LY, et al. TET1 reprograms the epithelial ovarian cancer epigenome and reveals casein kinase 2α as a therapeutic target. J Pathol. 2019;248:363–376. doi: 10.1002/path.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Filipczak PT, et al. p53-suppressed oncogene TET1 prevents cellular aging in lung cancer. Cancer Res. 2019;79:1758–1768. doi: 10.1158/0008-5472.CAN-18-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muller T, et al. Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol. 2012;181:675–683. doi: 10.1016/j.ajpath.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 85.Good CR, et al. TET1-mediated hypomethylation activates oncogenic signaling in triple-negative breast cancer. Cancer Res. 2018;78:4126–4137. doi: 10.1158/0008-5472.CAN-17-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lio CJ, Yuita H, Rao A. Dysregulation of the TET family of epigenetic regulators in lymphoid and myeloid malignancies. Blood. 2019;134:1487–1497. doi: 10.1182/blood.2019791475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cimmino L, et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat Immunol. 2015;16:653–662. doi: 10.1038/ni.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morin RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pasqualucci L, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Okosun J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46:176–181. doi: 10.1038/ng.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H, et al. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cancer Genome Atlas Research N et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. New Eng J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Quesada V, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 95.Nickerson ML, et al. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene. 2017;36:2172–2183. doi: 10.1038/onc.2016.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neri F, et al. TET1 is controlled by pluripotency-associated factors in ESCs and downmodulated by PRC2 in differentiated cells and tissues. Nucleic Acids Res. 2015;43:6814–6826. doi: 10.1093/nar/gkv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tsai YP, et al. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014;15:513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang YA, et al. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res. 2016;44:8153–8164. doi: 10.1093/nar/gkw498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yosefzon Y, et al. An epigenetic switch repressing Tet1 in gonadotropes activates the reproductive axis. Proc Natl Acad Sci USA. 2017;114:10131–10136. doi: 10.1073/pnas.1704393114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li L, et al. Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Sci Rep. 2016;6:26591. doi: 10.1038/srep26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Agirre X, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015;25:478–487. doi: 10.1101/gr.180240.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun M, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Forloni M, et al. Oncogenic EGFR represses the TET1 DNA demethylase to induce silencing of tumor suppressors in cancer cells. Cell Rep. 2016;16:457–471. doi: 10.1016/j.celrep.2016.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Collignon E, et al. Immunity drives TET1 regulation in cancer through NF-kappaB. Sci Adv. 2018;4:eaap7309. doi: 10.1126/sciadv.aap7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Noreen F, et al. DNA methylation instability by BRAF-mediated TET silencing and lifestyle-exposure divides colon cancer pathways. Clin Epigenetics. 2019;11:196. doi: 10.1186/s13148-019-0791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hsu CH, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 107.Teslow EA, et al. Obesity-induced MBD2_v2 expression promotes tumor-initiating triple-negative breast cancer stem cells. Mol Oncol. 2019;13:894–908. doi: 10.1002/1878-0261.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang H, et al. MiR-29b/TET1/ZEB2 signaling axis regulates metastatic properties and epithelial-mesenchymal transition in breast cancer cells. Oncotarget. 2017;8:102119–102133. doi: 10.18632/oncotarget.22183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen N, et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu Y, et al. Oct4 and the small molecule inhibitor, SC1, regulates Tet2 expression in mouse embryonic stem cells. Mol Biol Rep. 2013;40:2897–2906. doi: 10.1007/s11033-012-2305-5. [DOI] [PubMed] [Google Scholar]
- 111.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fischer AP, Miles SL. Silencing HIF-1alpha induces TET2 expression and augments ascorbic acid induced 5-hydroxymethylation of DNA in human metastatic melanoma cells. Biochem Biophys Res Commun. 2017;490:176–181. doi: 10.1016/j.bbrc.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 113.Zhang LY, Li PL, Wang TZ, Zhang XC. Prognostic values of 5-hmC, 5-mC and TET2 in epithelial ovarian cancer. Arch Gynecol Obstet. 2015;292:891–897. doi: 10.1007/s00404-015-3704-3. [DOI] [PubMed] [Google Scholar]
- 114.Tucker DW, et al. Epigenetic reprogramming strategies to reverse global loss of 5-hydroxymethylcytosine, a prognostic factor for poor survival in high-grade serous ovarian cancer. Clin Cancer Res. 2018;24:1389–1401. doi: 10.1158/1078-0432.CCR-17-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rawluszko-Wieczorek AA, et al. Clinical significance of DNA methylation mRNA levels of TET family members in colorectal cancer. J Cancer Res Clin Oncol. 2015;141:1379–1392. doi: 10.1007/s00432-014-1901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carella A, et al. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. Int J Cancer. 2020;146:373–387. doi: 10.1002/ijc.32520. [DOI] [PubMed] [Google Scholar]
- 117.Mo HY, An CH, Choi EJ, Yoo NJ, Lee SH. Somatic mutation and loss of expression of a candidate tumor suppressor gene TET3 in gastric and colorectal cancers. Pathol Res Pract. 2020;216:152759. doi: 10.1016/j.prp.2019.152759. [DOI] [PubMed] [Google Scholar]
- 118.Ye Z, et al. TET3 inhibits TGF-beta1-induced epithelial-mesenchymal transition by demethylating miR-30d precursor gene in ovarian cancer cells. J Exp Clin Cancer Res. 2016;35:72. doi: 10.1186/s13046-016-0350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao T, Pan W, Sun X, Shen H. Increased expression of TET3 predicts unfavorable prognosis in patients with ovarian cancer-a bioinformatics integrative analysis. J Ovarian Res. 2019;12:101. doi: 10.1186/s13048-019-0575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Song SJ, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng J, et al. An extensive network of TET2-targeting microRNAs regulates malignant hematopoiesis. Cell Rep. 2013;5:471–481. doi: 10.1016/j.celrep.2013.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fu X, et al. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci USA. 2013;110:17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Loriot A, et al. A novel cancer-germline transcript carrying pro-metastatic miR-105 and TET-targeting miR-767 induced by DNA hypomethylation in tumors. Epigenetics. 2014;9:1163–1171. doi: 10.4161/epi.29628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chuang KH, et al. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology. 2015;62:466–480. doi: 10.1002/hep.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang W, et al. MiR-520b suppresses proliferation of hepatoma cells through targeting ten-eleven translocation 1 (TET1) mRNA. Biochem Biophys Res Commun. 2015;460:793–798. doi: 10.1016/j.bbrc.2015.03.108. [DOI] [PubMed] [Google Scholar]
- 126.Song SJ, et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Takeshima H, et al. TET repression and increased DNMT activity synergistically induce aberrant DNA methylation. J Clin Invest. 2020;130:5370–5379. doi: 10.1172/JCI124070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Scherm MG, et al. miRNA142-3p targets Tet2 and impairs Treg differentiation and stability in models of type 1 diabetes. Nat Commun. 2019;10:5697. doi: 10.1038/s41467-019-13587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jiang S, Yan W, Wang SE, Baltimore D. Dual mechanisms of posttranscriptional regulation of Tet2 by Let-7 microRNA in macrophages. Proc Natl Acad Sci USA. 2019;116:12416–12421. doi: 10.1073/pnas.1811040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nawy T. Dynamics of DNA demethylation. Nat Methods. 2013;10:466. doi: 10.1038/nmeth.2506. [DOI] [PubMed] [Google Scholar]
- 135.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 136.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat Rev Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 138.Schmitz KM, et al. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 139.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hajkova P, et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dominguez PM, et al. DNA methylation dynamics of germinal center B cells are mediated by AID. Cell Rep. 2015;12:2086–2098. doi: 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lio CJ, et al. TET enzymes augment activation-induced deaminase (AID) expression via 5-hydroxymethylcytosine modifications at the Aicda superenhancer. Sci Immunol. 2019 doi: 10.1126/sciimmunol.aau7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Delker RK, Fugmann SD, Papavasiliou FN. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat Immunol. 2009;10:1147–1153. doi: 10.1038/ni.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kunimoto H, et al. Aid is a key regulator of myeloid/erythroid differentiation and DNA methylation in hematopoietic stem/progenitor cells. Blood. 2017;129:1779–1790. doi: 10.1182/blood-2016-06-721977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bachman M, et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem. 2014;6:1049–1055. doi: 10.1038/nchem.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bachman M, et al. 5-Formylcytosine can be a stable DNA modification in mammals. Nat Chem Biol. 2015;11:555–557. doi: 10.1038/nchembio.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu H, Wu X, Shen L, Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 149.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Takai H, et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014;9:48–60. doi: 10.1016/j.celrep.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 151.Yildirim O, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen R, et al. The 5-hydroxymethylcytosine (5hmC) reader UHRF2 is required for normal levels of 5hmC in mouse adult brain and spatial learning and memory. J Biol Chem. 2017;292:4533–4543. doi: 10.1074/jbc.M116.754580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsagaratou A, et al. Dissecting the dynamic changes of 5-hydroxymethylcytosine in T-cell development and differentiation. Proc Natl Acad Sci U S A. 2014;111:E3306–3315. doi: 10.1073/pnas.1412327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Madzo J, et al. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Rep. 2014;6:231–244. doi: 10.1016/j.celrep.2013.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guilhamon P, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mellen M, Ayata P, Heintz N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci USA. 2017;114:E7812–E7821. doi: 10.1073/pnas.1708044114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.He B, et al. Tissue-specific 5-hydroxymethylcytosine landscape of the human genome. Nat Commun. 2021;12:4249. doi: 10.1038/s41467-021-24425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dawlaty MM, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li C, et al. Overlapping requirements for Tet2 and Tet3 in normal development and hematopoietic stem cell emergence. Cell Rep. 2015;12:1133–1143. doi: 10.1016/j.celrep.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dawlaty MM, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hu L, et al. Structural insight into substrate preference for TET-mediated oxidation. Nature. 2015;527:118–122. doi: 10.1038/nature15713. [DOI] [PubMed] [Google Scholar]
- 165.Huang Y, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Putiri EL, et al. Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 2014;15:R81. doi: 10.1186/gb-2014-15-6-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 172.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hon GC, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell. 2014;56:286–297. doi: 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fidalgo M, et al. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell. 2016;19:355–369. doi: 10.1016/j.stem.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Piccolo FM, et al. Different roles for Tet1 and Tet2 proteins in reprogramming-mediated erasure of imprints induced by EGC fusion. Mol Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jin C, et al. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014;42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Frauer C, et al. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang H, et al. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 182.Greer CB, et al. Tet1 isoforms differentially regulate gene expression, synaptic transmission, and memory in the mammalian brain. J Neurosci. 2021;41:578–593. doi: 10.1523/JNEUROSCI.1821-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chandru A, Bate N, Vuister GW, Cowley SM. Sin3A recruits Tet1 to the PAH1 domain via a highly conserved Sin3-Interaction Domain. Sci Rep. 2018;8:14689. doi: 10.1038/s41598-018-32942-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhu F, et al. Sin3a-Tet1 interaction activates gene transcription and is required for embryonic stem cell pluripotency. Nucleic Acids Res. 2018;46:6026–6040. doi: 10.1093/nar/gky347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Vella P, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 186.Deplus R, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hrit J, et al. OGT binds a conserved C-terminal domain of TET1 to regulate TET1 activity and function in development. Elife. 2018 doi: 10.7554/eLife.34870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Teif VB, et al. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res. 2014;24:1285–1295. doi: 10.1101/gr.164418.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dubois-Chevalier J, et al. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res. 2014;42:10943–10959. doi: 10.1093/nar/gku780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wiehle L, et al. DNA (de)methylation in embryonic stem cells controls CTCF-dependent chromatin boundaries. Genome Res. 2019;29:750–761. doi: 10.1101/gr.239707.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Perera A, et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep. 2015;11:283–294. doi: 10.1016/j.celrep.2015.03.020. [DOI] [PubMed] [Google Scholar]
- 193.Ko M, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ayaz G, et al. CXXC5 as an unmethylated CpG dinucleotide binding protein contributes to estrogen-mediated cellular proliferation. Sci Rep. 2020;10:5971. doi: 10.1038/s41598-020-62912-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ravichandran M, et al. Rinf regulates pluripotency network genes and TET enzymes in embryonic stem cells. Cell Rep. 2019;28:1993–2003. doi: 10.1016/j.celrep.2019.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Abbas S, Erpelinck-Verschueren CA, Goudswaard CS, Lowenberg B, Valk PJ. Mutant Wilms' tumor 1 (WT1) mRNA with premature termination codons in acute myeloid leukemia (AML) is sensitive to nonsense-mediated RNA decay (NMD) Leukemia. 2010;24:660–663. doi: 10.1038/leu.2009.265. [DOI] [PubMed] [Google Scholar]
- 197.Wang Y, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57:662–673. doi: 10.1016/j.molcel.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rampal R, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep. 2014;9:1841–1855. doi: 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pan F, Weeks O, Yang FC, Xu M. The TET2 interactors and their links to hematological malignancies. IUBMB Life. 2015;67:438–445. doi: 10.1002/iub.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lazarenkov A, Sardina JL. Dissecting TET2 regulatory networks in blood differentiation and cancer. Cancers (Basel) 2022 doi: 10.3390/cancers14030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Muller U, Bauer C, Siegl M, Rottach A, Leonhardt H. TET-mediated oxidation of methylcytosine causes TDG or NEIL glycosylase dependent gene reactivation. Nucleic Acids Res. 2014;42:8592–8604. doi: 10.1093/nar/gku552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cartron PF, et al. Identification of TET1 partners that control Its DNA-demethylating function. Genes Cancer. 2013;4:235–241. doi: 10.1177/1947601913489020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Costa Y, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zheng L, et al. Modification of Tet1 and histone methylation dynamics in dairy goat male germline stem cells. Cell Prolif. 2016;49:163–172. doi: 10.1111/cpr.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zheng L, et al. The modification of Tet1 in male germline stem cells and interact with PCNA, HDAC1 to promote their self-renewal and proliferation. Sci Rep. 2016;6:37414. doi: 10.1038/srep37414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang X, Yang C, Peng X, Chen X, Feng Y. Acute WT1-positive promyelocytic leukemia with hypogranular variant morphology, bcr-3 isoform of PML-RARalpha and Flt3-ITD mutation: a rare case report. Sao Paulo medical journal = Revista paulista de medicina. 2017;135:179–184. doi: 10.1590/1516-3180.2016.020104102016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang Q, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chen LL, et al. SNIP1 recruits TET2 to regulate c-MYC target genes and cellular DNA damage response. Cell Rep. 2018;25:1485–1500. doi: 10.1016/j.celrep.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sardina JL, et al. Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell. 2018;23:905–906. doi: 10.1016/j.stem.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zhang YW, et al. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol Cell. 2017;65:323–335. doi: 10.1016/j.molcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Song C, et al. PML recruits TET2 to regulate DNA modification and cell proliferation in response to chemotherapeutic agent. Cancer Res. 2018;78:2475–2489. doi: 10.1158/0008-5472.CAN-17-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nakagawa T, et al. CRL4(VprBP) E3 ligase promotes monoubiquitylation and chromatin binding of TET dioxygenases. Mol Cell. 2015;57:247–260. doi: 10.1016/j.molcel.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Qiao Y, et al. AF9 promotes hESC neural differentiation through recruiting TET2 to neurodevelopmental gene loci for methylcytosine hydroxylation. Cell Discovery. 2015;1:15017. doi: 10.1038/celldisc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lio CW, et al. Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. Elife. 2016 doi: 10.7554/eLife.18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.de la Rica L, et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Suzuki T, et al. RUNX1 regulates site specificity of DNA demethylation by recruitment of DNA demethylation machineries in hematopoietic cells. Blood Adv. 2017;1:1699–1711. doi: 10.1182/bloodadvances.2017005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zeng Y, et al. Lin28A binds active promoters and recruits Tet1 to regulate gene expression. Mol Cell. 2016;61:153–160. doi: 10.1016/j.molcel.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sun Z, et al. EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity. Nat Commun. 2019;10:3892. doi: 10.1038/s41467-019-11905-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Guan W, et al. Methylcytosine dioxygenase TET3 interacts with thyroid hormone nuclear receptors and stabilizes their association to chromatin. Proc Natl Acad Sci USA. 2017;114:8229–8234. doi: 10.1073/pnas.1702192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fujiki K, et al. PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat Commun. 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- 221.Baubec T, Ivanek R, Lienert F, Schubeler D. Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell. 2013;153:480–492. doi: 10.1016/j.cell.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 222.Peng L, et al. MBD3L2 promotes Tet2 enzymatic activity for mediating 5-methylcytosine oxidation. J Cell Sci. 2016;129:1059–1071. doi: 10.1242/jcs.179044. [DOI] [PubMed] [Google Scholar]
- 223.Okashita N, et al. PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells. Development. 2014;141:269–280. doi: 10.1242/dev.099622. [DOI] [PubMed] [Google Scholar]
- 224.Soufi A, et al. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Boller S, et al. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B Cell programming. Immunity. 2016;44:527–541. doi: 10.1016/j.immuni.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 226.Liu Y, et al. Structural basis for Klf4 recognition of methylated DNA. Nucleic Acids Res. 2014;42:4859–4867. doi: 10.1093/nar/gku134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hashimoto H, et al. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev. 2014;28:2304–2313. doi: 10.1101/gad.250746.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vanzan L, et al. High throughput screening identifies SOX2 as a super pioneer factor that inhibits DNA methylation maintenance at its binding sites. Nat Commun. 2021;12:3337. doi: 10.1038/s41467-021-23630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wang D, et al. MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res. 2017;45:2396–2407. doi: 10.1093/nar/gkw1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Domcke S, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 232.Xiong J, et al. Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol Cell. 2016;64:913–925. doi: 10.1016/j.molcel.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 233.Looney TJ, et al. Systematic mapping of occluded genes by cell fusion reveals prevalence and stability of cis-mediated silencing in somatic cells. Genome Res. 2014;24:267–280. doi: 10.1101/gr.143891.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Polo JM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Okashita N, et al. PRDM14 drives OCT3/4 recruitment via active demethylation in the transition from primed to naive pluripotency. Stem Cell Reports. 2016;7:1072–1086. doi: 10.1016/j.stemcr.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Iwafuchi-Doi M, Zaret KS. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–1837. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mayran A, Drouin J. Pioneer transcription factors shape the epigenetic landscape. J Biol Chem. 2018;293:13795–13804. doi: 10.1074/jbc.R117.001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Larson ED, Marsh AJ, Harrison MM. Pioneering the developmental frontier. Mol Cell. 2021;81:1640–1650. doi: 10.1016/j.molcel.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Balsalobre A, Drouin J. Pioneer factors as master regulators of the epigenome and cell fate. Nat Rev Mol Cell Biol. 2022 doi: 10.1038/s41580-022-00464-z. [DOI] [PubMed] [Google Scholar]
- 240.Iwafuchi-Doi M. The mechanistic basis for chromatin regulation by pioneer transcription factors. Wiley Interdisciplin Rev. 2019;11:e1427. doi: 10.1002/wsbm.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Chronis C, et al. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mayran A, et al. Pioneer and nonpioneer factor cooperation drives lineage specific chromatin opening. Nat Commun. 2019;10:3807. doi: 10.1038/s41467-019-11791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sonmezer C, et al. Molecular co-occupancy identifies transcription factor binding cooperativity in vivo. Mol Cell. 2021;81:255–267. doi: 10.1016/j.molcel.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Rishi V, et al. CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc Natl Acad Sci USA. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.van Oevelen C, et al. C/EBPα activates pre-existing and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Reports. 2015;5:232–247. doi: 10.1016/j.stemcr.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Xue S, et al. TET3 inhibits type I IFN production independent of DNA demethylation. Cell Rep. 2016;16:1096–1105. doi: 10.1016/j.celrep.2016.06.068. [DOI] [PubMed] [Google Scholar]
- 247.Zhong J, et al. TET1 modulates H4K16 acetylation by controlling auto-acetylation of hMOF to affect gene regulation and DNA repair function. Nucleic Acids Res. 2017;45:672–684. doi: 10.1093/nar/gkw919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bauer C, et al. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-Linked N-acetylglucosamine transferase (OGT) J Biol Chem. 2015;290:4801–4812. doi: 10.1074/jbc.m114.605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Li X, et al. SIRT1 deacetylates TET2 and promotes its ubiquitination degradation to achieve neuroprotection against parkinson's disease. Front Neurol. 2021;12:652882. doi: 10.3389/fneur.2021.652882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wu D, et al. Glucose-regulated phosphorylation of TET2 by AMPK reveals a pathway linking diabetes to cancer. Nature. 2018;559:637–641. doi: 10.1038/s41586-018-0350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Rao VK, et al. Phosphorylation of Tet3 by cdk5 is critical for robust activation of BRN2 during neuronal differentiation. Nucleic Acids Res. 2020;48:1225–1238. doi: 10.1093/nar/gkz1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Coulter JB, et al. TET1 deficiency attenuates the DNA damage response and promotes resistance to DNA damaging agents. Epigenetics. 2017;12:854–864. doi: 10.1080/15592294.2017.1359452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Zhang T, et al. Phosphorylation of TET2 by AMPK is indispensable in myogenic differentiation. Epigenetics Chromatin. 2019;12:32. doi: 10.1186/s13072-019-0281-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Fiedler EC, Shaw RJ. AMPK regulates the epigenome through phosphorylation of TET2. Cell Metab. 2018;28:534–536. doi: 10.1016/j.cmet.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 255.Chen H, et al. TET2 stabilization by 14-3-3 binding to the phosphorylated Serine 99 is deregulated by mutations in cancer. Cell Res. 2019;29:248–250. doi: 10.1038/s41422-018-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kundu A, et al. 14–3-3 proteins protect AMPK-phosphorylated ten-eleven translocation-2 (TET2) from PP2A-mediated dephosphorylation. J Biol Chem. 2020;295:1754–1766. doi: 10.1074/jbc.RA119.011089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Giovannucci E, et al. Diabetes and cancer: a consensus report. Cancer J Clin. 2010;60:207–221. doi: 10.3322/caac.20078. [DOI] [PubMed] [Google Scholar]
- 258.Brower V. Illuminating the diabetes-cancer link. J Natl Cancer Inst. 2012;104:1048–1050. doi: 10.1093/jnci/djs322. [DOI] [PubMed] [Google Scholar]
- 259.Wang T, et al. Diabetes risk reduction diet and survival after breast cancer diagnosis. Cancer Res. 2021;81:4155–4162. doi: 10.1158/0008-5472.CAN-21-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Jeong JJ, et al. Cytokine-regulated phosphorylation and activation of TET2 by JAK2 in hematopoiesis. Cancer Discov. 2019;9:778–795. doi: 10.1158/2159-8290.CD-18-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Jin Z, et al. FGFR3 big up tri, open7-9 promotes tumor progression via the phosphorylation and destabilization of ten-eleven translocation-2 in human hepatocellular carcinoma. Cell Death Dis. 2020;11:903. doi: 10.1038/s41419-020-03089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mancini M, et al. Cytoplasmatic compartmentalization by Bcr-Abl promotes TET2 loss-of-function in chronic myeloid leukemia. J Cell Biochem. 2012;113:2765–2774. doi: 10.1002/jcb.24154. [DOI] [PubMed] [Google Scholar]
- 263.Jiang D, Wei S, Chen F, Zhang Y, Li J. TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response. EMBO Rep. 2017;18:781–796. doi: 10.15252/embr.201643179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ito R, et al. TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells. 2014;19:52–65. doi: 10.1111/gtc.12107. [DOI] [PubMed] [Google Scholar]
- 265.Zhang Q, et al. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT) J Biol Chem. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Shi FT, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lv L, et al. Vpr targets TET2 for degradation by CRL4(VprBP) E3 ligase to sustain IL-6 expression and enhance HIV-1 replication. Mol Cell. 2018;70:961–970. doi: 10.1016/j.molcel.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sun J, et al. SIRT1 activation disrupts maintenance of myelodysplastic syndrome stem and progenitor cells by restoring TET2 function. Cell Stem Cell. 2018;23:355–369. doi: 10.1016/j.stem.2018.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wang Y, Zhang Y. Regulation of TET protein stability by calpains. Cell Rep. 2014;6:278–284. doi: 10.1016/j.celrep.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Bamezai S, et al. TET1 promotes growth of T-cell acute lymphoblastic leukemia and can be antagonized via PARP inhibition. Leukemia. 2021;35:389–403. doi: 10.1038/s41375-020-0864-3. [DOI] [PubMed] [Google Scholar]
- 271.Ciccarone F, et al. Poly(ADP-ribosyl)ation is involved in the epigenetic control of TET1 gene transcription. Oncotarget. 2014;5:10356–10367. doi: 10.18632/oncotarget.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ciccarone F, Valentini E, Zampieri M, Caiafa P. 5mC-hydroxylase activity is influenced by the PARylation of TET1 enzyme. Oncotarget. 2015;6:24333–24347. doi: 10.18632/oncotarget.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Thienpont B, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537:63–68. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Cimmino L, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170:1079–1095. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chen J, et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet. 2013;45:1504–1509. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- 276.Yin R, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 277.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hu L, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 279.Sciacovelli M, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Bledea R, et al. Functional and topographic effects on DNA methylation in IDH1/2 mutant cancers. Sci Rep. 2019;9:16830. doi: 10.1038/s41598-019-53262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Morin A, et al. TET-mediated hypermethylation primes SDH-deficient cells for HIF2α-driven mesenchymal transition. Cell Rep. 2020;30:4551–4566. doi: 10.1016/j.celrep.2020.03.022. [DOI] [PubMed] [Google Scholar]
- 282.Chen LL, et al. Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses. Nat Cell Biol. 2022;24:353–363. doi: 10.1038/s41556-022-00853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Jakubek M, et al. Hydrazones as novel epigenetic modulators: correlation between TET 1 protein inhibition activity and their iron(II) binding ability. Bioorg Chem. 2019;88:102809. doi: 10.1016/j.bioorg.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 284.Vissers MCM, Das AB. Potential mechanisms of action for vitamin C in cancer: reviewing the evidence. Front Physiol. 2018;9:809. doi: 10.3389/fphys.2018.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Brabson JP, Leesang T, Mohammad S, Cimmino L. Epigenetic regulation of genomic stability by vitamin C. Front Genet. 2021;12:675780. doi: 10.3389/fgene.2021.675780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Lee Chong T, Ahearn EL, Cimmino L. Reprogramming the epigenome with vitamin C. Front Cell Dev Biol. 2019;7:128. doi: 10.3389/fcell.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Agathocleous M, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017;549:476–481. doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Chrysanthou S, et al. The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Res. 2022;50:3169–3189. doi: 10.1093/nar/gkac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ito K, et al. Non-catalytic roles of Tet2 are essential to regulate hematopoietic stem and progenitor cell homeostasis. Cell Rep. 2019;28:2480–2490. doi: 10.1016/j.celrep.2019.07.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Rasmussen KD, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Neri F, et al. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene. 2015;34:4168–4176. doi: 10.1038/onc.2014.356. [DOI] [PubMed] [Google Scholar]
- 294.Prasad P, Mittal SA, Chongtham J, Mohanty S, Srivastava T. Hypoxia-mediated epigenetic regulation of stemness in brain tumor cells. Stem Cells. 2017;35:1468–1478. doi: 10.1002/stem.2621. [DOI] [PubMed] [Google Scholar]
- 295.Herrmann A, et al. Integrin alpha6 signaling induces STAT3-TET3-mediated hydroxymethylation of genes critical for maintenance of glioma stem cells. Oncogene. 2020;39:2156–2169. doi: 10.1038/s41388-019-1134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wu MZ, et al. Hypoxia drives breast tumor malignancy through a TET-TNFα-p38-MAPK signaling axis. Cancer Res. 2015;75:3912–3924. doi: 10.1158/0008-5472.CAN-14-3208. [DOI] [PubMed] [Google Scholar]
- 297.Puig I, et al. TET2 controls chemoresistant slow-cycling cancer cell survival and tumor recurrence. J Clin Invest. 2018;128:3887–3905. doi: 10.1172/JCI96393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Su PH, et al. TET1 promotes 5hmC-dependent stemness, and inhibits a 5hmC-independent epithelial-mesenchymal transition, in cervical precancerous lesions. Cancer Lett. 2019;450:53–62. doi: 10.1016/j.canlet.2019.01.033. [DOI] [PubMed] [Google Scholar]
- 299.Hart GW, Housley MP, Slawson C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 300.Fujiki R, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Maury JJ, et al. RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 2015;15:182–189. doi: 10.1016/j.scr.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 302.Chu CS, et al. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci USA. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Capotosti F, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 304.Li HJ, et al. Roles of ten-eleven translocation family proteins and their O-linked β-N-acetylglucosaminylated forms in cancer development. Oncol Lett. 2021;21:1. doi: 10.3892/ol.2020.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev. 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is not applicable for this review.
This is not applicable for this review.