Abstract
Streptococcus pyogenes secretes several proteins that influence host-pathogen interactions. A tissue-culture model was used to study the influence of the secreted cysteine protease streptococcal erythrogenic toxin B (SPE B) on the interaction between S. pyogenes strain NZ131 (serotype M49) and mammalian cells. Inactivation of the speB gene enhanced fibronectin-dependent uptake of the pathogen by Chinese hamster ovary (CHO-K1) cells compared to that in the isogenic wild-type strain. Preincubation of the NZ131 speB mutant with purified SPE B protease significantly inhibited fibronectin-dependent uptake by both CHO-K1 and CHO-pgs745 cells. The effect was attributed to an abrogation of fibronectin binding to the surface of the bacteria that did not involve either the M49 protein or the streptococcal fibronectin-binding protein SfbI. In contrast, pretreatment of the NZ131 speB mutant with SPE B did not influence sulfated polysaccharide-mediated uptake by CHO-pgs745 cells. The results indicate that the SPE B protease specifically alters bacterial cell surface proteins and thereby influences pathogen uptake.
The severity of human infection with the group A streptococcus (GAS) Streptococcus pyogenes ranges from asymptomatic colonization and uncomplicated pharyngitis to severe invasive diseases, including streptococcal toxic shock syndrome and necrotizing fasciitis. In addition, two postinfection sequelae, rheumatic fever and acute glomerulonephritis, may develop. The broad spectrum of clinical manifestations reflects the complexity of the interaction between S. pyogenes and humans.
S. pyogenes secretes many proteins that contribute to virulence. One of these proteins, streptococcal erythrogenic toxin B, also known as streptococcal pyrogenic toxin B (SPE B), is a secreted cysteine protease that contributes to virulence in murine models of infection (23, 27). The speB gene encodes a polypeptide consisting of 398 amino acids (16), the crystal structure of which was recently solved at 1.6 Å (19). Following secretion, the 40-kDa SPE B zymogen is activated by reduction and proteolysis to form a 28-kDa sulfhydryl protease (13, 14, 26). In contrast to several other virulence-associated genes, all strains of S. pyogenes have the speB gene (46). In strain NZ131 (serotype M49), SPE B production is associated with nutrient depletion during the stationary phase of growth (6). However, the proteolytic activity of SPE B does not provide amino acids or peptides that are essential for growth in amino-acid-depleted medium (35). Several biochemical properties of the SPE B protease are consistent with its contribution to virulence, including the ability to activate human interleukin 1β (20) and a 66-kDa human matrix metalloprotease (4). In addition, SPE B degrades human vitronectin, cleaves fibronectin (Fn) (21), induces apoptosis (24), and releases active kinins from kininogen (17).
Although typically considered to be an extracellular mucosal pathogen, LaPenta et al. (25) showed that S. pyogenes is internalized by cultured mammalian cells. Polypeptides that participate in GAS uptake include the streptococcal Fn-binding protein SfbI (29) and its structural variant F1 (18, 29, 31) and the M1 protein (9). The M6 protein also contributes to uptake; however, its role is not well defined (18). Bacterial surface proteins often mediate internalization by direct binding to mammalian cell surface receptors or eukaryotic glycoproteins, such as Fn. Fn can interact with mammalian cell integrins and, perhaps in conjunction with additional molecules, stimulate actin rearrangement and uptake of the bacteria into an intracellular vacuole. Recently, an alternative mechanism of pathogen uptake was described that is based on the ability of several bacteria, including S. pyogenes, to bind sulfated polysaccharides such as dextran sulfate (DS) or heparan sulfate (10). In this system, sulfated polysaccharides form a molecular bridge between the bacterium and different types of eukaryotic glycoproteins, including Fn and vitronectin (11), to enable bacterial uptake without the synthesis of specific bacterial Fn-binding proteins.
Inactivation of the speB gene in serotypes M1 and M49 decreased GAS uptake by A549 human respiratory cells (43). In contrast, inactivation of speB in M2 and M3 serotypes increased streptococcal uptake by A549 cells (3). The results indicate that the secreted protease influences pathogen uptake. Nonetheless, the molecular basis for SPE B alteration of streptococcal internalization remains unknown. The purpose of our study was to investigate the ability of SPE B to influence specific pathways of GAS uptake by cultured mammalian cells.
MATERIALS AND METHODS
Cell lines and bacterial strains.
The bacterial strains, cell lines, and plasmids used in this study are described in Table 1. CHO-K1 and CHO-pgs745 were cultured in RPMI 1640 medium supplemented with 5% fetal calf serum (FCS; Life Technologies, Gaithersburg, Md.) at 37°C in an atmosphere of 5% CO2. Unless stated otherwise, S. pyogenes was grown at 37°C in 15-ml polypropylene tubes (Corning, New York, N.Y.) containing 10 ml of Todd-Hewitt broth medium (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% yeast extract (THY) or with Trypticase soy agar containing 5% sheep blood (Becton Dickinson, Cockeysville, Md.) at 37°C in 10% CO2. Exponential- and stationary-phase cultures corresponded to A600s of ≈0.2 and ≈0.6, respectively. Escherichia coli cells were grown at 37°C in Luria-Bertani (LB) broth or on LB agar plates containing, when appropriate, kanamycin (80 μg/ml).
TABLE 1.
Bacterial strains, cell lines, and plasmids used in this study
| Bacterial strain, cell line, or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. pyogenes | ||
| NZ131 | M49 | D. R. Martin, Porirua, New Zealand |
| NZ131 speB | speB::ermAM, erythromycin resistant | 5 |
| NZ131 speB emm49 | speB::ermAM; emm49::pRML500; erythromycin and kanamycin resistant | This study |
| 86-858 | M12 | WHO Center for Reference and Research on Streptococci at the University of Minnesota, Minneapolis |
| E. coli DH10B | recA1 | Bethesda Research Laboratories, Inc., Gaithersburg, Md. |
| Cell lines | ||
| CHO-K1 | ATCC CCL 61 | American Type Culture Collection, Manassas, Va. |
| CHO-pgs745 | pgsA-745; neither heparan sulfate nor chondroitan sulfate produced | Jeffrey Esko, University of California—San Diego, La Jolla |
| Plasmids | ||
| pCR2.1 | Cloning vector | Invitrogen |
| pSF141 | Suicide vector; Kanr | 41 |
| pRML500 | 900-bp internal fragment of emm49 cloned into pSF141 | This study |
Isolation of SPE B.
S. pyogenes NZ131 was grown without agitation for 48 h in 50 ml of dialyzed THY medium (5) at 37°C in 5% CO2. The bacteria were collected by centrifugation for 15 min at 6,430 × g at 4°C, and the supernatant fluid was concentrated 120-fold with a Centricon-10 microconcentrator (Amicon, Inc., Beverly, Mass.). The concentrated supernatant fluid was reduced with 0.1% (vol/vol) 2-mercaptoethanol (Sigma) and stored at 4°C. The preparation was greater than 95% pure as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Western blot analysis with antiserum to SPE B (provided by Dieter Gerlach, Jena, Germany) confirmed that the protein isolated was SPE B. A similar procedure was used to prepare SPE B consisting predominantly of the zymogen form of the protein, except that prior to reduction with 2-mercaptoethanol, the preparation was dialyzed overnight against 10 mM sodium acetate buffer. The preparation remained stable for several months, as assessed by SDS-PAGE and the level of protease activity.
Internalization assay.
CHO-K1 and CHO-pgs745 cells were grown on 12-mm-diameter glass coverslips in 24-well plates in RPMI 1640 containing 5% FCS until nearly confluent. Prior to the addition of bacteria, the cells were rinsed in serum-free Dulbecco's modified Eagle's medium (DMEM; Life Technologies), and 1 ml of DMEM, containing FCS or human Fn (Becton Dickinson) when appropriate, was added to each well. Approximately 107 streptococci in 10 μl of DMEM were added to each well at an infection ratio of 100:1, and the plates were incubated at 37°C with 5% CO2. Following 2 h of incubation, the cells were washed four times with 1 ml of Dulbecco's phosphate-buffered saline (DPBS; 140 mM NaCl, 2.5 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 1 mM MgCl2, 1 mM CaCl2 [pH 7.4]) and fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS; 140 mM NaCl, 2.5 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 1 mM MgCl2, 1 mM CaCl2 [pH 7.4]). Extracellular bacteria were distinguished from intracellular bacteria by immunogold-silver staining with polyclonal antiserum specific for GAS (dilution, 1/500; Biodesign, Kennebunk, Maine), as previously described (10, 45). The number of intracellular streptococci in each of 50 randomly selected cells was determined for each well by light microscopy. The data are reported as the mean ± standard error of the mean from at least three independent experiments. Sulfated-polysaccharide-mediated internalization assays were done as previously described (10). Briefly, approximately 108 bacteria were preincubated in 100 μl of HEPES-buffered saline (HBS; 10 mM HEPES, 145 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose [pH 7.2]) containing 5 μg of DS per ml (average molecular weight, 500,000; Sigma Chemical Co., St. Louis, Mo.). After 10 min on ice, the bacteria were collected by centrifugation for 5 min at 16,000 × g at 20°C, washed twice with HBS, and resuspended in 100 μl of HBS containing 5 μg of Fn per ml (Becton Dickinson), as previously described (10). Following a 10-min incubation on ice, the bacteria were pelleted and resuspended in 100 μl of DMEM, and 10 μl (107 bacteria) was added to each well. The infection assay was carried out as described above.
Protease activity assay and inhibition.
The proteolytic activity of SPE B was determined with azocasein (Sigma) as a substrate. Briefly, 1 to 5 μl of SPE B was added to 100 μl of 2.7 mg of azocasein/ml in 50 mM sodium acetate buffer (pH 5.5). The mixture was incubated at 37°C for 1 h prior to the addition of 25 μl of 15% ice-cold trichloroacetic acid. After 15 min on ice, the mixture was centrifuged for 15 min at 16,000 × g at 4°C, and the A445 of the supernatant fluid was measured with a Beckman DU-65 spectrophotometer (Beckman Instruments, Somerset, N.J.). One unit of activity was defined as the amount of enzyme required to result in a 0.1 change in A445 following a 60-min incubation with azocasein at 37°C. The peptide inhibitor of the SPE B protease, Z-Leu-Val-Gly-diazomethylketone (LVG), was purchased from BACHEM California, Inc. (Torrance, Calif.).
Preincubation of NZ131 speB with SPE B.
NZ131 speB (108 CFU) was suspended in 100 μl of spent medium prepared from the speB mutant and supplemented with purified SPE B for 1 h at 37°C. Following preincubation, the bacteria were collected by centrifugation and washed three times with DMEM. The bacteria were resuspended in 100 μl of DMEM, and 10 μl (107 bacteria) was added to wells containing either CHO-K1 or CHO-pgs745 cells to assess internalization, as described above. Preincubation with 0.3 and 1.0 U of protease activity corresponded to an SPE B concentration that was 25% of that present in the supernatant fluid of NZ131 after 18 h of growth in dialyzed THY broth medium. The two levels of protease activity resulted from variation in the conversion of the zymogen to the protease. The SPE B zymogen preparation contained 0.1 U of proteolytic activity and corresponded to 120% of the concentration of SPE B present in the supernatant fluid of an 18-h culture of NZ131.
Inactivation of emm49.
The chromosomal emm49 gene of NZ131 was insertionally inactivated by electrotransformation of NZ131 speB with the suicide plasmid pRML500. To construct pRML500, an approximately 900-bp internal region of the emm49 gene was amplified by PCR. Oligonucleotide primers (9) emmF (5′-GGCGGGAATCCACTATTCGCTTAGA-3′) and emmR (5′-GGCGGGAATTCAGTTCTTCAGCTTGT-3′) were purchased from Genemed Biotechnologies, Inc. (San Francisco, Calif.). Thirty cycles of amplification were carried out with a Perkin-Elmer 9600 DNA thermocycler with strand denaturation (15 s at 94°C), annealing (30 s at 45°C), and elongation (1 min at 72°C). The total volume of the PCR mixture was 50 μl and consisted of 1 μl of chromosomal DNA isolated from NZ131 (5), 0.4 μM each primer, 20 μM deoxynucleoside triphosphates (dNTPs), 4 mM Mg2+, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer, Branchburg, N.J.). Following amplification, the PCR mixture was separated by agarose gel electrophoresis, and the approximately 900-bp amplicon was purified with a Qiaex purification kit (Qiagen, Chatsworth, Calif.), as described by the manufacturer. The purified amplicon was cloned into pCR2.1 (Invitrogen, San Diego, Calif.) as described by the manufacturer. The recombinant plasmid was digested with EcoRI, and an approximately 900-bp fragment, containing an internal region of the emm49 gene, was purified with a Qiaex purification kit (Qiagen) following agarose gel electrophoresis. The fragment was then ligated with T4 DNA ligase (New England Biolabs, Inc., Beverly, Mass.) to the suicide vector pSF141 (41), which had previously been cleaved with EcoRI and purified by agarose gel electrophoresis. The pSF141 vector confers resistance to kanamycin and does not replicate in S. pyogenes (41). Following confirmation of the composition of the recombinant plasmid, designated pRML500, the plasmid was introduced into NZ131 speB by electroporation, essentially as previously described (38). Transformants were selected on THY agar plates containing 200 μg of kanamycin per ml. Insertion of pRML500 into the emm49 locus was confirmed by Southern hybridization analysis.
Southern blot analysis.
Southern blot analysis was used to confirm the insertion of pRML500 into the emm49 locus. Streptococcal chromosomal DNA was isolated from NZ131 speB and one kanamycin-resistant (Kanr) transformant, as previously described (5). DNA was digested with various restriction endonucleases, including SspI (New England Biolabs). Following digestion, the fragments were separated by agarose gel electrophoresis and transferred to Nytran (Schleicher & Schuell, Keene, N.H.), as previously described (36). The emm49 probe consisted of the purified EcoRI fragment isolated from pRML500. The emm49 probe and EcoRI-digested pSF141 were labeled with biotin with the NEBlot Phototope kit, as described by the manufacturer (New England Biolabs). Hybridization was done under stringent conditions (65°C, 0.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS). Two fragments of SspI-digested DNA isolated from the Kanr transformant hybridized to the emm49 probe, as predicted by the additional SspI site present in pSF141 (data not shown). In contrast, only a single fragment of SspI-digested DNA from NZ131 speB hybridized to the emm49 probe (data not shown). In addition, the pSF141 probe hybridized to two fragments, similar in size to the fragments that hybridized to the emm49 probe, of SspI-digested DNA from the Kanr transformant. The pSF141 probe did not hybridize to DNA isolated from NZ131 speB. The results confirmed that pRML500 had inserted into the emm49 locus.
SDS-PAGE and immunoblot analysis.
Inactivation of emm49 was verified by Western blotting with M49 specific antiserum (provided by J. B. Dale, Department of Medicine, University of Tennessee, Memphis). Briefly, whole-cell lysates from approximately 108 streptococci were analyzed by SDS-PAGE with a 12.5% acrylamide resolving gel. Following electrophoresis, proteins were transferred to Immobilon-NC nitrocellulose membranes (Millipore Corp., Bedford, Mass.) with a Bio-Rad transfer apparatus and Towbin's buffer (42). The nitrocellulose membrane was blocked with 5% skim milk in PBS containing 0.02% Tween 20 (PBST; 10 mM Na2HPO4, 10 mM NaH2PO4, 150 mM NaCl, 0.1% (vol/vol) polysorbate [Tween 20]) for 45 min. The membrane was washed with PBST, and rabbit antiserum against an M49 synthetic peptide diluted 1:500 in PBST was added to the blot and incubated for 1 h at room temperature. Protein A-horseradish peroxidase conjugate (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) was added at a 1:10,000 dilution in PBST, and the blot was incubated for 1 h. The antibody-antigen complexes were visualized with the ECL (enhanced chemiluminescence) Western blotting detection system, as described by the manufacturer (Amersham). A similar immunoblot procedure was used to detect SfbI or its structural variant, F1, with rabbit antiserum to SfbI (provided by S. Talay and G. Molinari, GBF-National Research Center for Biotechnology, Braunschweig, Germany). The antiserum recognizes both the SfbI protein and F1.
Fn binding to NZ131.
NZ131 speB (107 CFU) was incubated for 1 h at 37°C in either sterile spent medium prepared from the speB mutant or spent medium containing purified SPE B protease (0.3 U of proteolytic activity). Following preincubation, the bacteria were collected by centrifugation, washed three times with HBS, and resuspended in 100 μl of HBS containing purified human Fn (0.5 μg; Becton Dickinson). Following a 10-min incubation on ice, the bacteria were washed three times with HBS. To assess Fn binding, the bacteria were suspended in SDS-PAGE loading buffer and analyzed by SDS-PAGE. Fn associated with the bacteria was detected by Western blotting with the Fn-specific antibody HB91 (44) and horseradish peroxidase-conjugated protein A (Sigma) as described above.
RESULTS
Inactivation of the speB gene enhances uptake of NZ131 by CHO-K1 cells.
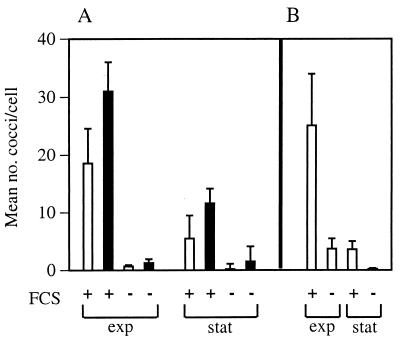
The role of bacterial extracellular products in the uptake of S. pyogenes by cultured mammalian cells was assessed by infecting CHO-K1 monolayers with isogenic NZ131 wild-type and speB mutant strains. After 2 h of infection, the host cells were extensively washed and then fixed, and the number of intracellular bacteria was determined microscopically. Both logarithmic- and stationary-phase bacteria were efficiently internalized in an FCS-dependent fashion (Fig. 1A). The speB mutant was internalized more efficiently than the wild-type strain (Fig. 1A). For comparison, the uptake of strain 86-858 (M12) was also assessed. No significant difference was observed between the magnitude of internalization of NZ131 (M49) and that of strain 86-858 (M12) (Fig. 1B). Similar results were obtained with CHO-pgs745 cells, which lack cell-surface proteoglycans (data not shown), indicating that internalization did not require this class of surface molecules. Together, the results suggested that the expression of the extracellular cysteine protease SPE B inhibited internalization of S. pyogenes by CHO-K1 cells.
FIG. 1.
Inactivation of the speB gene in NZ131 enhanced internalization by CHO-K1 cells. S. pyogenes NZ131 was grown to the mid-exponential (exp) and stationary (stat) phases in THY broth. Aliquots were centrifuged, and the bacteria were resuspended in DMEM prior to the addition of 107 CFU to preconfluent CHO-K1 cell monolayers with (+) or without (−) 5% FCS. The infection proceeded for 2 h prior to the assessment of internalization by differential immunogold-silver staining. (A) Internalization of M49 strain NZ131 (open bars) and NZ131 speB (solid bars). (B) Internalization of M12 strain 86-858. The data represent the mean number of streptococci per cell ± standard error from at least three separate experiments.
Evidence that SPE B abrogates internalization of NZ131 by CHO-pgs745 cells.
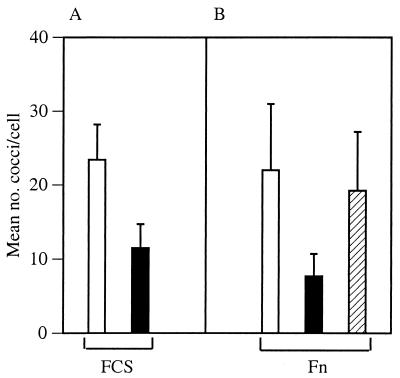
The target of the putative internalization regulatory activity of SPE B was investigated by comparing the uptake of NZ131 speB preincubated with either sterile spent medium prepared from NZ131 speB or with the same medium containing purified SPE B (0.3 U of caseinolytic activity) for 1 h at 37°C. The preincubation was done in sterile spent medium to mimic conditions associated with speB expression during the stationary phase of growth. Following preincubation, the bacterial cells were collected by centrifugation, resuspended in DMEM, and added to CHO-pgs745 monolayers. Preincubation of NZ131 speB with spent medium containing the SPE B protease (0.3 U) inhibited internalization in the presence of 5% FCS by approximately 50% compared to treatment with spent medium alone (Fig. 2A). The results suggested that SPE B altered a bacterial cell surface property that was essential for internalization.
FIG. 2.
SPE B protease decreased Fn-mediated internalization. NZ131 speB was suspended in sterile spent medium prepared from the speB mutant. The zymogen preparation (hatched bars) or activated SPE B protease (solid bars) was added or not (open bars), and the bacteria were incubated for 1 h at 37°C. Following preincubation, the bacteria were collected by centrifugation and resuspended in DMEM. Approximately 107 CFU were added to each well that contained CHO-pgs745 cells, and internalization was assessed. (A) Internalization of NZ131 speB in the presence of 5% FCS. Preincubation with the SPE B zymogen preparation (hatched bar in panel B) was not done. (B) Internalization of NZ131 speB in the presence of 2 μg of human Fn per ml. The data represent the mean number of streptococci per cell ± standard error from at least three separate experiments.
To determine if serum factors influenced SPE B inhibition of uptake, pathogen uptake was assessed under serum-free conditions. Previous reports showed that the glycoproteins Fn and vitronectin could substitute for FCS to mediate efficient internalization of Neisseria gonorrhoeae by CHO-K1 cells (10, 44). As a consequence, internalization of NZ131 speB by CHO-K1 cells in the presence of either purified vitronectin or Fn was assessed in serum-free infection assays. The results showed that NZ131 speB was efficiently internalized when as little as 0.25 μg of purified human Fn per ml was present; purified vitronectin alone did not mediate uptake (data not shown).
To determine if there was a correlation between the amount of SPE B protease activity used to pretreat the bacteria and inhibition of uptake, NZ131 speB was preincubated with either activated SPE B protease or an SPE B preparation consisting primarily of the zymogen. Bacteria preincubated with the SPE B zymogen preparation, which contained minimal protease activity (0.1 U; see Materials and Methods), were internalized nearly as efficiently as bacteria that were preincubated with spent medium alone (Fig. 2B). In contrast, treatment with proteolytically active SPE B (0.3 U) markedly reduced bacterial internalization (Fig. 2B). To confirm the results, additional internalization assays were done with bacteria that had been preincubated with the purified protease (1.0 U) in the absence or presence of the tripeptide LVG (20 μg/ml), a potent inhibitor of SPE B proteolytic activity (2). The tripeptide completely inhibited the effect of SPE B on internalization (data not shown). This effect was not observed when the inhibitor (LVG; 20 μg/ml) was present during the infection assay, instead of during preincubation, excluding the possibility that residual bacterial cell surface-associated enzymatic activity was transferred to the wells and inhibited internalization by altering eukaryotic cell components.
SPE B protease abrogated Fn binding by NZ131.
The results presented above suggested that SPE B moderated internalization by cleaving one or more streptococcal Fn-binding proteins. To test this hypothesis, the binding of purified human Fn to NZ131 speB was investigated. NZ131 speB was preincubated with spent medium alone or with spent medium containing the SPE B protease. After removal of enzyme by washing, the speB mutant was incubated with Fn for 10 min on ice. Fn bound to the bacteria was detected by Western blotting with the Fn-specific antibody HB91. Bacteria preincubated with spent medium alone bound Fn (Fig. 3, lane 1). In contrast, bacteria preincubated with purified SPE B protease (0.3 U) did not bind Fn (Fig. 3, lane 2). The results strongly suggest that SPE B-associated inhibition of bacterial internalization is caused by an abrogation of Fn binding to the bacterial cell surface.
FIG. 3.
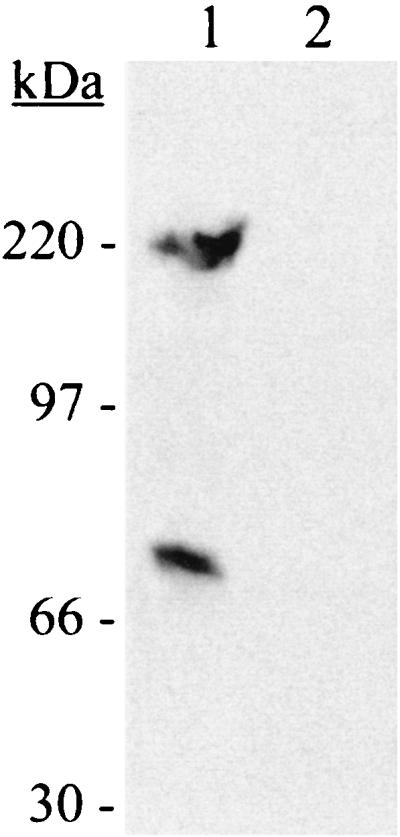
SPE B protease abrogated Fn binding to NZ131. NZ131 speB was grown to mid-exponential phase, collected by centrifugation, and incubated with spent medium alone (lane 1) or spent medium containing the SPE B protease (lane 2), as described for the internalization experiments. After being washed, the bacteria were incubated with purified human Fn. Fn associated with the bacteria was detected by immunoblotting with an Fn-specific antibody. The experiment was repeated twice, and a representative result is shown. The migration and size of the molecular mass standards are indicated. The lower band in lane 1 probably represents the 70-kDa N-terminal fragment of Fn that is frequently observed in commercial Fn preparations.
Fn-dependent cell entry of NZ131 speB is not mediated by SfbI, F1, or M49.
Previously identified Fn-binding proteins that facilitate internalization of S. pyogenes include SfbI (29); its structural variant, F1 (18, 31, 34); and M1 (9). The SPE B protease degrades M1 from the bacterial surface (1), which suggested a possible mechanism for SPE B abrogation of Fn-dependent uptake. In an attempt to identify the target of SPE B that influenced internalization, we determined whether SfbI, F1, or M49 facilitated NZ131 speB entry into CHO-K1 and CHO-pgs745 cells. Western blotting with antiserum that recognized both SfbI and F1 showed that NZ131 does not synthesize either protein (data not shown). To determine if the M49 protein was cleaved by SPE B, Western blots were performed with whole bacteria preincubated with the SPE B protease under the same conditions used for the internalization experiments. Probing of the blots with M49-specific antisera showed that SPE B (1.0 U) degraded M49 from the bacterial cell surface (data not shown).
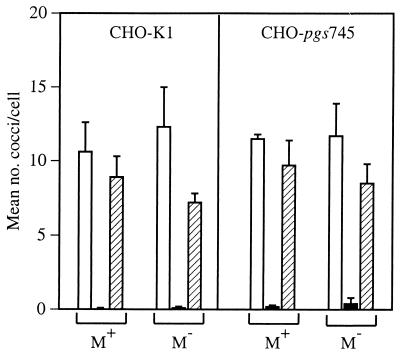
To definitively assess the contribution of the M49 protein to internalization, the emm49 gene was inactivated in NZ131 speB. Inactivation of the gene was confirmed by Southern and Western blots (see Materials and Methods). Internalization was subsequently assessed with both CHO-K1 and CHO-pgs745 cells. The degree of internalization of the NZ131 speB emm49 mutant was similar to that of the isogenic parent strain (NZ131 speB) after preincubation with either the SPE B zymogen preparation (0.1 U) or spent medium alone (Fig. 4). Furthermore, internalization of the NZ131 speB emm49 mutant was diminished by preincubation with the SPE B protease (1.0 U), similar to the results obtained with NZ131 speB (Fig. 4). The data indicated that M49 was not required for Fn-mediated internalization of NZ131 by CHO-pgs745 cells and that SPE B acted on a different target to moderate uptake.
FIG. 4.
M49 protein was not required for Fn-mediated internalization of NZ131. The internalization of NZ131 speB and NZ131 speB emm49 by CHO-K1 and CHO-pgs745 cells was determined following preincubation with the SPE B protease (solid bars), SPE B zymogen preparation (hatched bars), or spent medium alone (open bars). Data represent the mean number of streptococci per cell ± standard error from at least three separate experiments.
SPE B affects specific pathways of Fn-mediated entry of S. pyogenes.
To address the specificity of SPE B on the bacterial cell surface, we took advantage of the recent finding that sulfated polysaccharides form a molecular bridge between bacterial surface proteins and various ligands, including Fn and vitronectin, to promote internalization of pathogenic bacteria (10, 44). This event does not involve direct binding of Fn to the bacterial cell surface and thus may serve as a tool to determine the specificity of the SPE B effect on pathogen uptake. To test the effect of SPE B on polysaccharide-mediated internalization, NZ131 speB was treated with spent medium alone or with spent medium containing the purified SPE B protease (1.0 U). Following centrifugation, the bacteria were suspended and preincubated with DS prior to incubation with Fn (10). The bacteria were then added to CHO-pgs745 cells, and internalization was assessed. Preincubation with the SPE B protease had no significant influence on sulfated polysaccharide-mediated internalization compared to that in sham-treated controls (5.6 ± 0.9 and 5.3 ± 0.6 streptococci per cell, respectively). Control experiments showed that preincubation with the protease ablated Fn-mediated internalization compared to that in the sham-treated controls (0.0 ± 0.0 and 8.8 ± 1.7, respectively). The binding of DS to NZ131 speB after preincubation with the SPE B protease (1.0 U) was confirmed microscopically with DS conjugated to fluorescein (data not shown). The results showed that the internalization of NZ131 speB by a sulfated-polysaccharide-mediated mechanism was not sensitive to the proteolytic activity associated with SPE B, in contrast to uptake by direct Fn binding. Thus, SPE B altered only a subset of streptococcal surface proteins.
DISCUSSION
S. pyogenes secretes several proteins that influence the host-pathogen interaction. We used a tissue culture model to detect and characterize SPE B-mediated changes in the uptake of S. pyogenes by cultured mammalian cells. Our results with strain NZ131 indicate that SPE B inhibits Fn-dependent internalization into CHO-K1 and CHO-pgs745 cells by cleaving distinct bacterial surface receptors. In contrast, SPE B had virtually no effect on the Fn-dependent sulfated-polysaccharide-mediated mechanism of internalization. Thus, SPE B can be considered as an important determinant of the interactions between the pathogen and host cells.
Infection experiments using an speB mutant and reconstitution with purified enzyme indicated that SPE B negatively influences bacterial uptake. Further study of the mechanism behind this effect indicated that the observed effects of SPE B were caused by specific alterations in streptococcal surface properties. Preincubation of the microorganisms with purified protease decreased internalization, and this effect was completely abolished when the proteinase inhibitor peptide LVG was included in the preincubation step. Furthermore, when the inhibitor was present during the interaction of pretreated microorganisms with the mammalian cells, similar inhibition of internalization was observed, excluding the possibility that alterations in mammalian cell properties (if they exist) accounted for the result. Additional evidence that SPE B affected the bacterial cell surface was provided by the abrogation of Fn binding to the bacterial cell surface following treatment with the SPE B protease.
Several Fn-binding proteins, including M1 (8, 9), SfbI (29), and F1 (18, 31, 34), can facilitate Fn-dependent uptake of S. pyogenes by cultured mammalian cells. Neither SfbI nor F1 was detected in strain NZ131, consistent with a previous report that M49 strains do not possess the genes encoding these proteins (30). The M protein of strain NZ131 (M49) was cleaved by SPE B. However, construction of an speB emm49 double mutant indicated that M49 was not required for NZ131 uptake by CHO-K1 cells. These data point to the presence of an additional Fn-binding protein on the surface of NZ131. This is not improbable, because many of the genes are present only in specific strains. The repertoire of genes encoding Fn-binding proteins in strain NZ131 is not known, and at this time, we do not know which protein or proteins are required for Fn-dependent uptake of NZ131 speB by CHO-K1 cells.
Internalization of S. pyogenes is a complex process that involves a variety of bacterial and host cell factors. Protein F1 binds Fn, which interacts with host-cell β1 integrins to promote uptake by HeLa cells (34). Similarly, the M1 protein binds Fn, which interacts with α5β1 integrins to promote entry into A549 cells (8). M1 also facilitates laminin-dependent uptake through an alternate β1 integrin—that is, α1β1, α2β1, α3β1, α6β1, or α7β1 (8). Strain NZ131 binds sulfated polysaccharides, which bind vitronectin or Fn to promote entry by CHO-K1 cells (10; this study). Finally, oligopeptides containing the sequence RGD stimulated the uptake of an M1 strain independently of M1 (8). Our preincubation experiments showed that the SPE B protease abrogated Fn-mediated internalization of NZ131 speB by CHO-K1 cells, while it did not significantly alter Fn-dependent sulfated-polysaccharide-mediated uptake. This suggests that SPE B specifically degrades certain streptococcal receptors and that the protease redirects rather than abrogates streptococcal entry into host cells. However, SPE B cleaves Fn (21) and thus can affect polysaccharide-mediated uptake by direct cleavage of the glycoprotein. Thus, the protease may influence uptake both by degradation of Fn-binding proteins on the bacterial cell surface and by direct degradation of eukaryotic glycoproteins.
Similar to our results with CHO-K1 cells, speB mutants in serotypes M2 and M3 were internalized more efficiently by both human umbilical vein endothelial cells and A549 cells (3). In contrast, inactivation of speB in NZ131 was previously reported to inhibit internalization by A549 cells (43). CHO-K1 cells were used in the current study to distinguish between two pathways of pathogen uptake. CHO-K1 cells required the addition of Fn for Fn-mediated uptake of S. pyogenes (10; this paper), and CHO-pgs745 required exogenous sulfated polysaccharides to mediate uptake through a polysaccharide bridge. The pathways of uptake measured in previous studies were not characterized; thus, we can only speculate that the disparity between our results and those of Tsai et al. (43) results from different effects of the SPE B protease on alternative pathways of internalization.
The contribution of internalization to the virulence of S. pyogenes is not known. Internalized GAS were detected in pharyngeal epithelial cells in 93% of patients with tonsillitis (33). In addition, M1 isolates from invasive disease are internalized more efficiently in vitro, suggesting a correlation between the efficiency of in vitro internalization and severe outcomes of infection (7, 25). S. pyogenes does not replicate intracellularly in the presence of antibiotics that prohibit extracellular growth (15, 37, 43). However, viable intracellular GAS have been recovered 7 days after uptake by epithelial cells (32), although additional reports indicate that GAS persist intracellularly for 24 h after uptake (15, 37). Interestingly, removal of the barrier to extracellular growth results in viable externalized GAS (32). Internalization of GAS by epithelial cells may transiently sequester the pathogen from host immune defenses and promote the persistence of infection. The ability to maintain infection is likely to increase the chance of an infected individual developing severe disease.
Eukaryotic cells regulate a variety of biological activities by specific proteolysis of cell surface proteins (22). Proteolytic modification of bacterial surface proteins is an emerging theme among both gram-negative and gram-positive bacterial pathogens. For example, the plasminogen activator or coagulase of Yersinia pestis degrades bacterial outer membrane proteins and is associated with both dissemination and virulence (39, 40). Staphylococcus aureus is internalized by epithelial cells in an Fn-dependent manner (12) and secretes V8 protease. The V8 protease decreases Fn binding to S. aureus (28) and is thus likely to decrease Fn-dependent internalization and adherence. Similarly, SPE B-mediated remodeling of the bacterial cell surface may enable S. pyogenes to respond rapidly to changing environmental conditions. SPE B is secreted in response to specific environmental signals. The protein is detected in culture supernatant fluid of strain NZ131 during the stationary phase of growth, and production in rich medium correlates with the depletion of nutrients (6). Although the regulation of speB expression is associated with nutrient starvation, the protease does not provide peptides or amino acids for catabolism (35). We speculate that SPE B remodeling of the bacterial cell surface decreases adherence and internalization to promote dissemination, in concert with direct degradation of extracellular matrix proteins.
ACKNOWLEDGMENTS
We thank J. Dale, D. Gerlach, G. Molinari, and S. Talay for providing the antiserum used in this work and N. P. Hoe, J. M. Musser, and L. A. Smoot for critical review of the manuscript.
REFERENCES
- 1.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 2.Björck L, Åkesson P, Bohus M, Trojnar J, Abrahamson M, Olafsson I, Grubb A. Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature. 1989;337:385–386. doi: 10.1038/337385a0. [DOI] [PubMed] [Google Scholar]
- 3.Burns E H, Jr, Lukomski S, Rurangirwa J, Podbielski A, Musser J M. Genetic inactivation of the extracellular cysteine protease enhances in vitro internalization of group A streptococci by human epithelial and endothelial cells. Microb Pathog. 1998;24:333–339. doi: 10.1006/mpat.1998.0204. [DOI] [PubMed] [Google Scholar]
- 4.Burns E H, Jr, Marciel A M, Musser J M. Activation of a 66-kilodalton human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine protease. Infect Immun. 1996;64:4744–4750. doi: 10.1128/iai.64.11.4744-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee M S, Gerlach D, Yu C-E, Ferretti J J. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect Immun. 1993;61:3719–3723. doi: 10.1128/iai.61.9.3719-3723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee M S, Phillips E R, Ferretti J J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary P P, LaPenta D, Vessela R, Lam H, Cue D. A globally disseminated M1 subclone of group A streptococci differs from other subclones by 70 kilobases of prophage DNA and capacity for high-frequency intracellular invasion. Infect Immun. 1998;66:5592–5597. doi: 10.1128/iai.66.11.5592-5597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cue D, Dombek P E, Lam H, Cleary P P. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect Immun. 1998;66:4593–4601. doi: 10.1128/iai.66.10.4593-4601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dombek P E, Cue D, Sedgewick J, Lam H, Ruschkowski S, Finlay B B, Cleary P P. High-frequency intracellular invasion of epithelial cells by serotype M1 group A streptococci: M1 protein-mediated invasion and cytoskeletal rearrangements. Mol Microbiol. 1999;31:859–870. doi: 10.1046/j.1365-2958.1999.01223.x. [DOI] [PubMed] [Google Scholar]
- 10.Duensing T D, Wing J S, van Putten J P M. Sulfated polysaccharide-directed recruitment of mammalian host proteins: a novel strategy in microbial pathogenesis. Infect Immun. 1999;67:4463–4468. doi: 10.1128/iai.67.9.4463-4468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duensing T D, van Putten J P M. Vitronectin binds to gonococcal adhesin OpaA through a glycosaminoglycan molecular bridge. Biochem J. 1998;334:133–139. doi: 10.1042/bj3340133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziewanowska K, Patti J M, Deobald C F, Bayles K W, Trumble W R, Bohach G A. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infect Immun. 1999;67:4673–4678. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott S D. A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J Exp Med. 1945;81:191–196. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott S D, Dole V P. An inactive precursor of streptococcal proteinase. J Exp Med. 1947;85:305–320. doi: 10.1084/jem.85.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greco R, Martino L D, Donnarumma G, Conte M P, Seganti L, Valenti P. Invasion of cultured human cells by Streptococcus pyogenes. Res Microbiol. 1995;146:551–560. doi: 10.1016/0923-2508(96)80561-4. [DOI] [PubMed] [Google Scholar]
- 16.Hauser A R, Schlievert P M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J Bacteriol. 1990;172:4536–4542. doi: 10.1128/jb.172.8.4536-4542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herwald H, Collin M, Müller-Esterl W, Björck L. Streptococcal cysteine proteinase releases kinins: a virulence mechanism. J Exp Med. 1996;184:665–673. doi: 10.1084/jem.184.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jadoun J, Ozeri V, Burstein E, Skutelsky E, Hanski E, Sela S. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J Infect Dis. 1998;178:147–158. doi: 10.1086/515589. [DOI] [PubMed] [Google Scholar]
- 19.Kagawa T F, Cooney J C, Baker H M, McSweeney S, Liu M, Gubba S, Musser J M, Baker E N. Crystal structure of the zymogen form of the group A Streptococcus virulence factor SpeB: an integrin-binding cysteine protease. Proc Natl Acad Sci USA. 2000;97:2235–2240. doi: 10.1073/pnas.040549997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapur V, Majesky M W, Li L-L, Black R A, Musser J M. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapur V, Topouzis S, Majesky M W, Li L-L, Hamrick M R, Hamill R J, Patti J M, Musser J M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 22.Kiessling L L, Gordon E J. Transforming the cell surface through proteolysis. Chem Biol. 1998;5:R49–R62. doi: 10.1016/s1074-5521(98)90056-4. [DOI] [PubMed] [Google Scholar]
- 23.Kuo C-F, Wu J-J, Lin K-Y, Tsai P-J, Lee S-C, Jin Y-T, Lei H-Y, Lin Y-S. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect Immun. 1998;66:3931–3935. doi: 10.1128/iai.66.8.3931-3935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C-F, Wu J-J, Tsai P-J, Kao F-J, Lei H-Y, Lin M-T, Lin Y-S. Streptococcal pyrogenic exotoxin B induces apoptosis and reduces phagocytic activity in U937 cells. Infect Immun. 1999;67:126–130. doi: 10.1128/iai.67.1.126-130.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaPenta D, Rubens C, Chi E, Cleary P P. Group A streptococci efficiently invade human respiratory epithelial cells. Proc Natl Acad Sci USA. 1994;91:12115–12119. doi: 10.1073/pnas.91.25.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu T-Y, Elliott S D. Activation of streptococcal proteinase and its zymogen by bacterial cell walls. Nature. 1965;206:33–34. doi: 10.1038/206033a0. [DOI] [PubMed] [Google Scholar]
- 27.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Investig. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGavin M J, Zahradka C, Rice K, Scott J E. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621–2628. doi: 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molinari G, Talay S R, Valentin-Weigand P, Rohde M, Chhatwal G S. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun. 1997;65:1357–1363. doi: 10.1128/iai.65.4.1357-1363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Natanson S, Sela S, Moses A E, Musser J M, Caparon M G, Hanski E. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J Infect Dis. 1995;171:871–878. doi: 10.1093/infdis/171.4.871. [DOI] [PubMed] [Google Scholar]
- 31.Okada N, Tatsuno I, Hanski E, Caparon M, Sasakawa C. Streptococcus pyogenes protein F promotes invasion of HeLa cells. Microbiology. 1998;144:3079–3086. doi: 10.1099/00221287-144-11-3079. [DOI] [PubMed] [Google Scholar]
- 32.Österlund A, Engstrand L. Intracellular penetration and survival of Streptococcus pyogenes in respiratory epithelial cells in vitro. Acta Oto-Laryngol. 1995;115:685–688. doi: 10.3109/00016489509139387. [DOI] [PubMed] [Google Scholar]
- 33.Österlund A, Popa R, Nikkilä T, Scheynius A, Engstrand L. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope. 1997;107:640–647. doi: 10.1097/00005537-199705000-00016. [DOI] [PubMed] [Google Scholar]
- 34.Ozeri V, Rosenshine I, Mosher D F, Fässler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–637. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 35.Podbielski A, Woischnik M, Kreikemeyer B, Bettenbrock K, Buttaro B A. Cysteine protease SpeB expression in group A streptococci is influenced by the nutritional environment but SpeB does not contribute to obtaining essential nutrients. Med Microbiol Immunol. 1999;188:99–109. doi: 10.1007/s004300050111. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 37.Schrager H M, Rheinwald J G, Wessels M R. Hyaluronic acid capsule and the role of streptococcal entry into keratinocytes in invasive skin infection. J Clin Investig. 1996;98:1954–1958. doi: 10.1172/JCI118998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon D, Ferretti J J. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol Lett. 1991;66:219–224. doi: 10.1016/0378-1097(91)90336-9. [DOI] [PubMed] [Google Scholar]
- 39.Sodeinde O A, Sample A K, Brubaker R R, Goguen J D. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect Immun. 1988;56:2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodeinde O A, Subrahmanyam Y V B K, Stark K, Quan T, Bao Y, Goguen J D. A surface protease and the invasive character of plaque. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 41.Tao L, LeBlanc D J, Ferretti J J. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene. 1992;120:105–110. doi: 10.1016/0378-1119(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 42.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai P-J, Kuo C-F, Lin K-Y, Lin Y-S, Lei H-Y, Chen F-F, Wang J-R, Wu J-J. Effect of group A streptococcal cysteine protease on invasion of epithelial cells. Infect Immun. 1998;66:1460–1466. doi: 10.1128/iai.66.4.1460-1466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Putten J P M, Duensing T D, Cole R L. Entry of OpaA+ gonococci into HEp-2 cells requires concerted action of glycosaminoglycans, fibronectin and integrin receptors. Mol Microbiol. 1998;29:369–379. doi: 10.1046/j.1365-2958.1998.00951.x. [DOI] [PubMed] [Google Scholar]
- 45.van Putten J P M, Weel J F L, Grassmé H U C. Measurements of invasion by antibody labeling and electron microscopy. Methods Enzymol. 1994;236:420–437. doi: 10.1016/0076-6879(94)36031-6. [DOI] [PubMed] [Google Scholar]
- 46.Yu C-E, Ferretti J J. Frequency of the erythrogenic toxin B and C genes (speB and speC) among clinical isolates of group A streptococci. Infect Immun. 1991;59:211–215. doi: 10.1128/iai.59.1.211-215.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]