Abstract
Molnupiravir (EIDD-2801) is a prodrug of a ribonucleoside analogue that is currently being used under a US FDA emergency use authorization for the treatment of mild to moderate COVID-19. We evaluated molnupiravir for efficacy as an oral treatment in the rhesus macaque model of SARS-CoV-2 infection. Twenty non-human primates (NHPs) were challenged with SARS-CoV-2 and treated with 75 mg/kg (n = 8) or 250 mg/kg (n = 8) of molnupiravir twice daily by oral gavage for 7 days. The NHPs were observed for 14 days post-challenge and monitored for clinical signs of disease. After challenge, all groups showed a trend toward increased respiration rates. Treatment with molnupiravir significantly reduced viral RNA levels in bronchoalveolar lavage (BAL) samples at Days 7 and 10. Considering the mild to moderate nature of SARS-CoV-2 infection in the rhesus macaque model, this study highlights the importance of monitoring the viral load in the lung as an indicator of pharmaceutical efficacy for COVID-19 treatments. Additionally, this study provides evidence of the efficacy of molnupiravir which supplements the current ongoing clinical trials of this drug.
Keywords: SARS-CoV-2, COVID-19, Molnupiravor, EIDD-2801, Non-human primate, Rhesus macaque
1. Introduction
The emergence of the B.1.1.529 (omicron) variant of Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2) has led to a resurgence in infection rates and created complications for disease management using existing strategies (Tian et al., 2022). The SARS-CoV-2 omicron variant has a heavily mutated receptor binding domain (RBD) that contributes to its escape from neutralizing antibodies including vaccine elicited humoral responses and monoclonal antibody therapeutics (Cao et al., 2022; Tian et al., 2022). Monoclonal antibody treatments approved for patient use against Coronavirus Infectious Disease 2019 (COVID-19) have diminished efficacy against the omicron variant (Tada et al., 2022). Furthermore, immune protection against reinfection with the omicron variant is greatly diminished compared to reinfection rates with the B.1.1.7 (alpha), B.1.351 (beta), and B.1.617.2 (delta) variants (Altarawneh et al., 2022). As a result, there has been increased interest in antiviral drugs that target the SARS-CoV-2 replication and transcription complex (RTC) which should be unaffected by the RBD mutations of the omicron variant.
SAR-CoV-2 contains 16 nonstructural proteins (NSPs) which associate to form the RNA-dependent RNA polymerase (RdRP) (Malone et al., 2022). Minimally, NSP7 and NSP8 complex with the canonical RNA polymerase NSP12 to bind viral RNA and form the holo-RdRP and serve as the minimal RTC (Malone et al., 2022; Wu et al., 2022). Some ribonucleotide analogue inhibitors of the RTC, including molnupiravir, bind in the active site and induce mutagenesis leading to an “error catastrophe” where the virus is unable to maintain its genetic element due to catastrophically high error rates during replication while others, including remdesivir-triphosphate induce delated termination rather than mutagenesis (Byléhn et al., 2021; Drake and Holland, 1999; Malone and Campbell, 2021; Wu et al., 2021, 2022). Several ribonucleotide analogue inhibitors of the RTC have been evaluated for efficacy against SARS-CoV-2 leading to the clinical development of remdesivir (sold under the brand name Veklury) (Grein et al., 2020; Rochwerg et al., 2020) and its derivative VV116 (Xie et al., 2021), nirmatrelvir–ritonavir (PF-07321332 or Paxlovid) (Hammond et al., 2022), favipiravir (Hashemian et al., 2021), dexamethasone (Caruso et al., 2020; Raju and Biatris, 2021), fluvoxamine (although the direct mechanism of action is disputed and fluvoxamine may not directly inhibit the RTC) (Calusic et al., 2022; Hashimoto et al., 2022; Wen et al., 2022), and molnupiravir (EIDD-2801 or Lagevrio) (Fischer et al., 2021; Khoo et al., 2021; Painter et al., 2021a; Painter et al., 2021b; Wen et al., 2022).
Currently, there are two United States (US) Food and Drug Administration (FDA) approved therapeutics for the treatment of COVID-19, remdesivir and the immune modulator baricitinib (Grein et al., 2020; Lin et al., 2022; Moshkovits and Shepshelovich, 2022; Rochwerg et al., 2020). The FDA emergency use authorization (EUA), which is an additional route for using experimental treatments in the context of an emerging disease with limited treatment options outside of the normal randomized clinical trial mechanism, has allowed COVID-19 patients access to treatments with monoclonal antibodies, convalescent plasma, immune modulators, nirmatrelvir–ritonavir, and molnupiravir (Bhimraj et al., 2022; Moshkovits and Shepshelovich, 2022). EUAs have also been granted and subsequently revoked for hydroxychloroquine and monoclonal antibody treatments leading some experts to question the benefits of the current EUA system (Bhimraj et al., 2022). This study was performed to provide preclinical data typically used as part of the conventional approval or licensure process.
Surges in COVID-19 have strained the capacity of US healthcare systems (Dichter et al., 2022; Griffin et al., 2020). Except for nirmatrelvir–ritonavir and molnupiravir, all antiviral and antibody treatments for COVID-19 have required skilled nursing care for administration adding to the stress on healthcare system capacity (Dal-Ré et al., 2022). Generally, early administration of pharmaceutical intervention is associated with better outcomes for antiviral therapy. Both nirmatrelvir–ritonavir and molnupiravir should be administered within five days of COVID-19 symptom onset (Dal-Ré et al., 2022). Telehealth intervention with nirmatrelvir–ritonavir or molnupiravir can be beneficial in that it allows quicker patient access to treatment, can reduce burden on the healthcare system, and ameliorates concerns of SARS-CoV-2 transmission in the healthcare setting (Lee, 2022). Additionally, molnupiravir has an advantage that it can be manufactured at a large scale much more easily than antibody treatments. Further, it does not require cold-chain storage in its capsule form which makes distribution easier (Singh et al., 2021).
In K18-hACE2 mice, the combination of molnupiravir with nirmatrelvir synergistically improved survival following SARS-CoV-2 infection (Jeong et al., 2022). The efficacy of molnupiravir against SARS-CoV-2 has been examined in numerous animal model studies (Ashour et al., 2022; Eloy et al., 2021; Lee et al., 2021). It has documented efficacy in ferret models, and an observed sex and variant biased efficacy in a hamster model (Abdelnabi et al., 2021; Cox et al., 2021; Lieber et al., 2022).
While there have been small animal studies demonstrating the effectiveness of molnupiravir against SARS-CoV-2, including recent Omicron variants, in rodents (Rosenke et al., 2022), non-human primate (NHP) models of infection are considered of upmost importance in the development of novel drugs. A rhesus macaque model of SARS-CoV-2 infection which recapitulates many aspects of COVID-19 disease in humans has become the preferred model for SARS-CoV-2 drug development (Chandrashekar et al., 2020; Munster et al., 2020; Salguero et al., 2021). Specifically, rhesus macaques experience a febrile respiratory disease which lasts 8–16 days and is characterized by pulmonary infiltrates and the detection of virus in nasal swabs and bronchoalveolar lavages (BALs) (Munster et al., 2020). Additionally, the rhesus macaque model demonstrates greater severity in aged NHPs compared to younger NHPs, similar to what has been observed in humans (Yu et al., 2020).
The ribonucleoside analog N4-hydroxycytidine (EIDD-1931) is a broadly acting antiviral that inhibits the replication of many RNA viruses (Toots et al., 2020). An isobutyric ester prodrug of EIDD-1931, molnupiravir, has increased oral bioavailability, including plasma concentrations, in an NHP model (Toots et al., 2019). Molnupiravir has undergone numerous clinical trials, many of them under the trade name molnupiravir (Caraco et al., 2022; Fischer et al., 2021; Jayk Bernal et al., 2022; Khoo et al., 2021; Mahase, 2021; Wendy P. Painter et al., 2021; Singh et al., 2021). Currently, molnupiravir is available commercially in the US as an oral prescription medication authorized under the FDA EUA mechanism (Moshkovits and Shepshelovich, 2022). The goal of this study is to test the efficacy of molnupiravir against SARS-CoV-2 infection in the rhesus macaque model. Additionally, this study provides insight into the interpretation of large-scale NHP model preclinical data for antiviral therapeutic efficacy SARS-CoV-2 considering the relatively mild disease observed in this model. Interestingly, the rapid clinical testing and the EAU use of molnupiravir due to the circumstances of COVID-19 allows for a simultaneous discussion of preclinical and clinical efficacy data for molnupiravir.
2. Materials and methods
2.1. NHP model
Specific pathogen free adult Chinese-origin rhesus macaques (Macaca mulatta, n = 20; 10 male, 10 female, 43–66 months old and individually identified by unique tattoo), were obtained from Evigo (Alice, TX, USA). All animals were evaluated by a veterinarian and determined to be healthy before being placed on study. For continuous core body temperature measurements, a DST micro-T implantable temperature logger (Star–Oddi, Gardabaer, Iceland) was surgically implanted into the peritoneal cavity of each animal prior to study initiation; data recording was set to 15 min intervals as previously described (Harris et al., 2021). Macaques were individually housed at Animal Biosafety Level 3 (ABSL3) for the duration of the study. Drinking water (reverse osmosis-purified) was provided ad libitum through an automatic watering system. Certified Primate Diet 5048 (LabDiet, St. Louis, MO, USA) and food enrichment consisting of fresh fruit and vegetables was provided to the macaques daily. Environmental enrichment including various manipulatives (Kong toys, mirrors, and puzzles) was also provided. All hands-on manipulations, including challenge, oral gavage, and biosampling, were performed on sedated animals administered ketamine (5–20 mg/kg) via intramuscular injection.
2.2. Randomization, cohorts, and treatments
Macaques were split evenly into two cohorts and then randomized by gender and body weight into three sex-balanced groups using SAS software (SAS Institute, Inc., Cary, North Carolina, United States). Unless otherwise noted, data from both cohorts were combined and analyzed together. For both cohorts, the first group of two macaques was mock-treated, the second group of four macaques received an molnupiravir dose of 75 mg/kg at each treatment, and the third group of four macaques received an molnupiravir dose of 250 mg/kg at each treatment (Table 1 ). Dosing was selected based on the reported plasma availability of molnupiravir in rhesus macaques and the inhibitory concentrations reported for SARS-CoV-2 (Holman et al., 2021; Li et al., 2022 & Toots et al., 2019). Molnupiravir powder was formulated in 1.0% methylcellulose (400 cP, SLCD9520, Sigma Aldrich, St. Louis, MO, USA) in water (GenClone, Genesee Scientific, San Diego, CA, USA) at 15 and 50 mg/mL for the second and third group, respectively, stored at 4 °C, and used within three days of reconstitution. Subjects were treated twice daily with an 8-h interval from the day of challenge to 6-days post challenge (14 total doses).
Table 1.
Subject descriptors.
| Cohort | Group | ID | Sex | Challenge Dose | Treatment Dose |
|---|---|---|---|---|---|
| 1 | 1 | RA3691 | F | 5.15 × 106 TCID50 | mock |
| RA3886 | M | ||||
| 2 | T152322 | F | 75 mg/kg EIDD-2801 | ||
| RA3621 | F | ||||
| RA3885 | M | ||||
| RA3900 | M | ||||
| 3 | RA3466 | F | 250 mg/kg EIDD-2801 | ||
| RA3626 | F | ||||
| RA3887 | M | ||||
| T151543 | M | ||||
| 2 | 1 | RA3489 | F | 6.08 × 106 TCID50 | mock |
| RA3903 | M | ||||
| 2 | RA3453 | F | 75 mg/kg EIDD-2801 | ||
| RA3675 | F | ||||
| RA3883 | M | ||||
| T152211 | M | ||||
| 3 | RA3446 | F | 250 mg/kg EIDD-2801 | ||
| RA3662 | F | ||||
| RA3882 | M | ||||
| T151807 | M |
2.3. SARS-CoV-2 challenge characterization
SARS-CoV-2 strain USA_WA-1/2020 (kindly provided by Dr. Chien-Te (Kent) Tseng at UTMB from original material provided by the Centers for Disease Control and Prevention in January 2020) was passaged once in Vero C1008 (E6) cells (BEI Resources, NR-596, Lot 3956593). Viral stocks were thawed and diluted to 1 × 106 TCID50/mL immediately prior to administration as previously described (Harris et al., 2021). The challenge strain and dose were informed by previous study of SARS-CoV-2 in rhesus macaques (Chandrashekar et al., 2020; Munster et al., 2020; Salguero et al., 2021). The challenge inoculum was administered to each subject split between 0.5 mL per nare administered using a MAD Nasal™ Intranasal Mucosal Atomization Device (Teleflex, Morrisville, North Carolina, USA). An additional 4.0 mL was administered intratracheally for a total target challenge of 5 × 106 TCID50 per subject. Following challenge, the inoculum titer was quantified as 5.15 × 106 and 6.08 × 106 TCID50 for Cohorts 1 and 2, respectively.
2.4. Clinical scoring, radiography, and euthanasia criteria
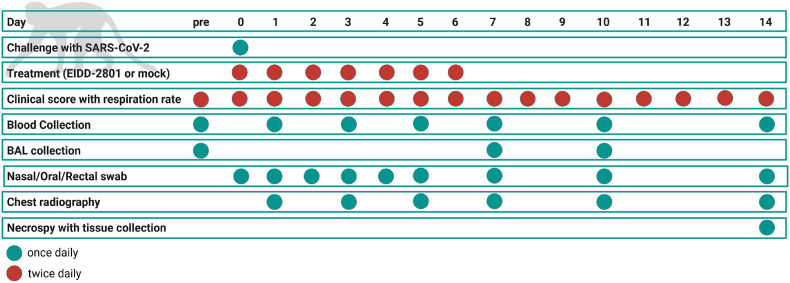
Clinical observations were conducted twice daily (morning and afternoon) during the study and included measurement of cage-side respiration rates and clinical scoring based on general appearance (e.g., hunched posture), activity, food/enrichment consumption, and outward changes in breathing patterns. Prospectively defined criteria that required immediate euthanasia included severe dyspnea and/or agonal breathing and prostate posture/reluctance to move when stimulated. No animals met endpoint criteria during the study. Ventrolateral chest radiography was performed on days 0, 1, 3, 5, 7, 10, and 14 (Fig. 1 ) using a portable GE AMX-4+ computed radiography system (General Electric Company, Boston, Massachusetts, USA) per the manufacturer's instructions. DICOM data files were independently evaluated by a veterinary radiologist via a four-pattern approach (analyses of consolidation, interstitial areas, nodules or masses, and atelectasis).
Fig. 1.
Schematic of the experimental protocol. Experimental procedures were completed on the days indicated. “pre” indicates pre-challenge procedures completed up to 1 week before challenge to collect baseline data. Green circles indicate that the procedure was done once that day. Red circles indicate that the procedure was done twice daily. Temperature was monitored at 15-min intervals during the study.
2.5. Bronchoalveolar lavage collection and processing
BAL fluid collection was performed at baseline (5–7 days prior to challenge) and on days 7, and 10. Sedated animals were treated with lidocaine or catecaine administered onto the pharynx. A laryngoscope was used to place a catheter into the trachea which was used to infuse up to 20 mL of sterile PBS into the lungs, and then aspirated to recover the sample. This process was repeated up to three times per animal. The collected BAL sample was centrifuged, and the cell pellet resuspended in 0.05 mL of PBS and processed for viral load via RT-PCR. The BAL supernatant was frozen for later titration by TCID 50 assay.
2.6. Hematology and clinical chemistry
Femoral vein peripheral blood was collected via Vacutainer® into standard collection tubes containing a clot activator (serum separator tubes) or EDTA. Clinical chemistry analyses were conducted on harvested serum using the Abaxis VetScan VS2® Chemistry Analyzer using Comprehensive Diagnostic Profile rotors (Abaxis, Inc., Union City, CA, USA). Hematology was performed on EDTA blood using the Abaxis VETSCAN® HM5 Hematology Analyzer (Abaxis, Inc., Union City, CA, USA). Serum was frozen for viral load analysis. Nasal, oral, and rectal swabs were collected on days 0–5, 7, 10, and 14 using sterile cotton-tipped medical swabs which were placed into 0.5 mL sterile phosphate-buffered saline (PBS) for viral load analysis.
2.7. Anatomical and Histopathology
At the end of the study, (day 14 post-challenge) subjects were humanely euthanized via intravenous administration of a pentobarbital-based euthanasia solution under deep anesthesia followed by bilateral thoracotomy. Necropsy was conducted and tissues (spleen, heart, liver, lung, stomach duodenum, jejunum, ileum and colon) were collected in 10% neutral buffered formalin for fixation. Fixed tissues were shipped to Experimental Pathology Laboratories, Inc. (EPL; Sterling, VA), trimmed, paraffin embedded, processed to slides, stained with H&E, and evaluated by a board-certified Veterinary Pathologist. One section each of left and right cranial and left and right caudal lung were trimmed and processed separately for evaluation; histologic grading was determined for lung as a whole and not as separate sections.
2.8. Determination of viral load
Viral load analysis was assessed by TCID50 and qRT-PCR assays as previously described (Harris et al., 2021). Briefly, TCID50 was determined with serial dilution of samples on Vero C1008 (E6) cells (BEI Resources, NR-596, Lot 3956593) which were observed for cytopathic effect following a 48-72-h incubation period. TCID50 was calculated as previously described (Ramakrishnan, 2016). RNA was isolated from supernatants of swabs and BALs in TRIzol LS reagent using Zymo Direct-zol™ RNA Mini Prep kits (Zymo Research, Irvine, CA, USA) per manufacturer instructions. RNA samples were analyzed via qRT-PCR targeting the SARS-CoV-2 E gene using a Quantifast Probe PCR kit (Qiagen, Germantown, MD, USA) on the Bio-Rad CFX96™ Real-Time PCR Detection System in conjunction with the following primer/probe set: forward primer (250 nM, 5′-ACA GGT ACG TTA ATA GTT AAT AGC GT-3′), reverse primer (250 nM, 5′-ATA TTG CAG CAG TAC GCA CAC A-3′), and probe (375 nM, 5′-6FAM-ACA CTA GCC/ZEN/ATC CTT ACT GCG CTT CG-IABkFQ-3′) (Integrated DNA Technologies, Coralville, IA, USA). Thermocycling conditions were as follows: step 1, 1 cycle, 50 °C for 10 min; step 2, 1 cycle, 95 °C for 10 min; steps 3–5, 45 cycles, 95 °C for 10 s, 60 °C for 30 s, single read. Negative controls included reaction mixtures without RNA. For quantification purposes, viral RNA extracted from the virus seed stock with a known TCID50/mL titer was used. All qRT-PCR results are expressed as genome equivalents per microliter (GEq/μL) based on comparison of standards to TCID50/mL equivalents.
2.9. Statistical analysis
All statistics were prospectively planned and conducted by a biostatistician with expertise in NHP studies. For each parameter, descriptive statistics (mean and standard deviation), including geometric means and geometric standard deviation (GSDs) when variable was log transformed, were calculated for each group and time point. Mixed models with random intercepts were used to assess if means were significantly different among groups. If there was a significant difference among groups, pairwise comparisons were performed, and both the unadjusted p-values and Tukey adjusted p-values were calculated. All statistics were performed as reported in the text using SAS software version 9.4 (SAS Institute Inc., Cary, NC, USA). Fig. 2, Fig. 3, Fig. 4, Fig. 5 were generated using GraphPad Prism version 9 (GraphPad Software, San Diego, CA, USA) and represent combined data from both cohorts. Fig. 1 was created with BioRender.com.
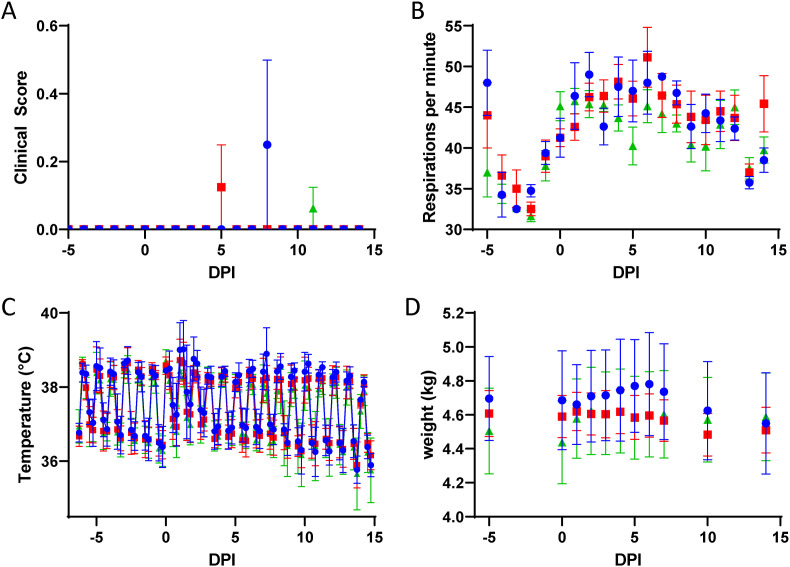
Fig. 2.
Clinical Scores, Respiration, Body Temperature, and Weight. Mock-treated (blue circles), 75 mg/kg/treatment molnupiravir treated (red squares), and 250 mg/kg/treatment molnupiravir treated (green diamonds) subjects were monitored for (A) clinical scores which were assigned based on twice daily observation of respiration, food consumption, excretion of feces and urine, activity, and appearance; (B) respiration rates; (C) body temperature as recorded every 15 min with implantable temperature loggers and transformed into 6-h rolling averages for analysis; and (D) body weight. Error bars represent standard error of the means. DPI – Days post-infection.
Fig. 3.
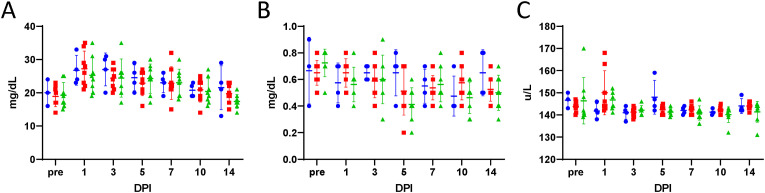
Clinical Chemistry. Clinical chemistry values were measured with a Vetscan VS2 analyzer using Comprehensive Diagnostic Profile rotors. (A) Blood urea nitrogen, (B) creatinine, and (C) sodium levels are reported for mock-treated (blue circles), 75 mg/kg/treatment molnupiravir treated (red squares), and 250 mg/kg/treatment molnupiravir treated (green diamonds) subjects. A horizontal hash-mark indicates the mean with standard deviation error bars. DPI – Days post-infection.
Fig. 4.
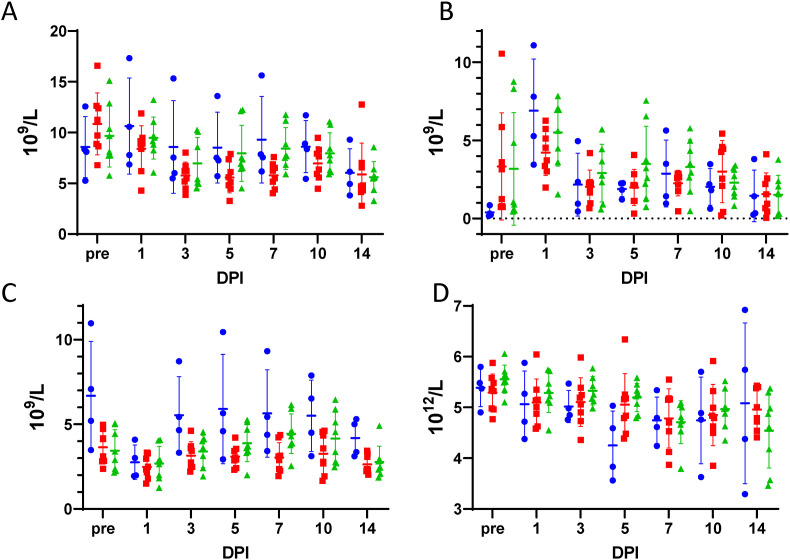
Hematology. Blood collected in sodium EDTA vacutainers was analyzed on a HM5 veterinary hematology analyzer. (A) White blood cells, (B) neutrophils, (C) lymphocytes and (D) red blood cells are reported from subjects that were mock-treated (blue circles), received 75 mg/kg/treatment molnupiravir (red squares), or received 250 mg/kg/treatment molnupiravir (green diamonds). A horizontal hash-mark indicates the mean with standard deviation error bars. DPI – Days post-infection.
Fig. 5.
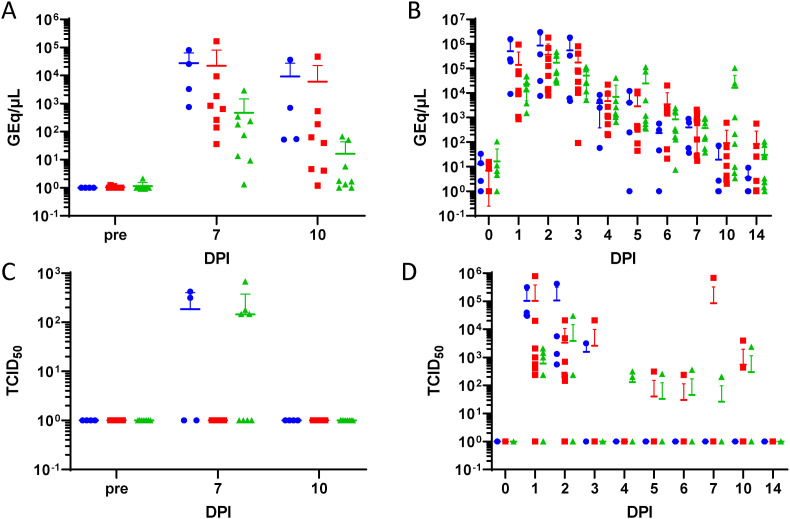
Viral Load. Quantitative reverse transcriptase PCR (qRT-PCR) targeting the SARS-CoV-2 E gene (A and B) or tissue culture 50% infectious dose (TCID50) on Vero C1008 (E6) cells (C and D) were determined for bronchoalveolar lavage (BAL) (A and C) or nasal swab (B and D) supernatants from mock-treated (blue circles), 75 mg/kg/treatment molnupiravir treated (red squares), and 250 mg/kg/treatment molnupiravir treated (green diamonds) subjects. A horizontal hash-mark indicates the mean with standard deviation error bars. DPI – Days post-infection.
3. Results
3.1. Clinical observation results
Clinical signs were not observed at any timepoint in the majority of subjects. RA3489 (Group 1 - mock-treated) on day 8 post-challenge and RA3453 (Group 2–75 mg/kg/dose molnupiravir) on day 5 post-challenge scored 1 for a mildly hunched posture. T151807 (Group 3–250 mg/kg/dose molnupiravir) also scored 1 for mildly hunched posture on Day 11 (Fig. 2A).
Respiration rates tended to increase post-challenge compared to baseline. During baseline measurement, Group 1 (mock-treated) demonstrated a greater average respiration rate (45 breaths/minute) versus Group 3 (250 mg/kg/dose molnupiravir; 36 breaths/minute); however, the difference was not significant as determined by Tukey adjusted post-hoc analysis (p = 0.0657) (Fig. 2B). Treatment groups were also compared within their respective challenge cohort. In Cohort 1, Group 1 (mock-treated) demonstrated higher respiration rates than Group 3 (250 mg/kg/dose molnupiravir) (p < 0.05, Tukey) at baseline and days 5 and 6 post-challenge. On day 6 post-challenge, Group 2 (75 mg/kg/dose molnupiravir) had higher respiration rates than Group 3 (250 mg/kg/dose molnupiravir) (p < 0.0001, Tukey). In Cohort 2 respiration rates were significantly different on days 4 and 14 post-challenge with Group 2 (75 mg/kg/dose molnupiravir) being significantly higher than either Group 1 (mock-treated) or Group 3 (250 mg/kg/dose molnupiravir) on both days (p < 0.05, Tukey).
Core body temperatures were recorded every 15 min throughout the study. Temperature data were condensed to 6-h rolling averages for statistical analysis (Fig. 2C). Group 1 (mock-treated) demonstrated a higher average temperature on Day 1 post-challenge versus Group 3 (250 mg/kg/dose molnupiravir) for morning (0600–1145) and afternoon (1200–1745) timepoints (p < 0.01, Tukey). In addition, Group 1 (mock-treated) demonstrated a higher average temperature on Day 7 post-challenge than all other groups (75 or 250 mg/kg/dose molnupiravir) for the morning and afternoon timepoints (p < 0.05, Tukey).
Individual animal body weights remained constant during the course of the study and no significant differences among the groups were measured at any time (Fig. 2D).
3.2. Clinical chemistry results
Clinical chemistry and hematological analysis were undertaken to attempt to observe biomarkers of SARS-CoV-2 infection in the NHP model. Limited differences in clinical chemistry parameters were observed between the groups. While there was not a significant difference in blood urea nitrogen (BUN) among groups over time, there was a trend for increased levels compared to baseline in all groups on days 1, 3 and 5 post-challenge (Fig. 3A). There were significant differences in creatinine levels (CRE) among groups at baseline and Day 5 post-challenge (Fig. 3B). At baseline, Group 1 (mock-treated) had a significantly lower average CRE than group 3 (250 mg/kg/dose molnupiravir). At day 5 post-challenge, Group 1 (mock-treated) demonstrated a significantly higher average CRE than Group 3 (250 mg/kg/dose molnupiravir). There were significant differences in sodium (Na+) among groups at Days 1 and 5 post-challenge. Group 1 (mock-treated) had a significantly lower level than Group 2 (75 mg/kg/dose molnupiravir) at Day 1; Group 1 had a significantly higher level than Group 3 (250 mg/kg/dose molnupiravir) at day 5 post-challenge. No significant differences in albumin, alkaline phosphatase, alanine phosphatase, amylase, calcium, globulin, glucose, potassium, phosphorous, total bilirubin, or total protein were detected (Supplemental Fig. 1).
3.3. Hematology results
Mean total white blood cell count results demonstrated a decrease from day 1 to day 3 post challenge in all groups (Fig. 4A). There were significant differences in neutrophil counts among groups at baseline versus day 1 post-challenge (Fig. 4B). Group 1 (mock-treated) demonstrated a significantly lower level of neutrophils than Groups 2 and 3 (75 and 250 mg/kg/dose molnupiravir) at baseline. Group 1 (mock-treated) had significantly higher neutrophil levels than Group 2 (75 mg/kg/dose molnupiravir) at Day 1. There were significant differences in lymphocyte counts among groups at baseline and on Days 3, 5, 7, 10 and 14 post-challenge (Fig. 4C). Group 1 (mock-treated) had consistently higher lymphocytes than Groups 2 and 3 (75 or 250 mg/kg/dose molnupiravir). There were significant differences in red blood cell (RBC) counts among groups on Day 5 post-challenge (Fig. 4D). Group 1 (mock-treated) had a significantly lower RBC counts than Groups 2 and 3 (75 or 250 mg/kg/dose molnupiravir). No significant differences in monocytes, basophils, eosinophils, or platelets were detected (Supplemental Fig. 2).
3.4. Viral load results
There were significant differences in the viral RNA levels in BAL samples among groups at Days 7 and 10 post-challenge (Fig. 5A). On both days, Group 1 (mock-treated) and Group 2 (75 mg/kg/dose molnupiravir) had significantly higher viral genome equivalents per microliter (GEq/μL) than Group 3 (250 mg/kg/dose molnupiravir) (p < 0.05, Tukey). There were significant differences in the level of viral RNA in nasal swabs among groups at Day 10 post-challenge. Specifically, Group 3 (250 mg/kg/dose molnupiravir; 1.626 × 104 GEq/μL) had significantly higher values than Group 1 (mock-treated; 1.862 × 101 GEq/μL and 2 (75 mg/mg/dose molnupiravir; 9.168 × 102 GEq/μL) (p < 0.05, Tukey) (Fig. 5B).
Supernatant from collected BAL were assayed for viable virus via TCID50 assay (Fig. 5C). There were no significant differences among groups over time, although it should be noted that minimal viable virus was detected across all collected samples. The only positive samples were: RA3886 (Group 1, mock-treated, 4.22 × 102 TCID50/mL), T151543 (Group 3, 250 mg/kg/dose molnupiravir, 6.81 × 102 TCID50/mL), RA3489 (Group 1, mock-treated, 3.16 × 102 TCID50/mL), RA3662 (Group 3, 250 mg/kg/dose molnupiravir, 1.47 × 102 TCID50/mL), and RA3882 (Group 3, 250 mg/kg/dose molnupiravir, 1.78 × 102 TCID50/mL), and T151807 (Group 3, 250 mg/kg/dose molnupiravir, 1.47 × 102 TCID50/mL). Nasal, oral and rectal swab samples were also assayed for viable virus. RA3691 (Group 1, mock-treated, day 1) and RA3886 (Group 1, mock-treated, day 1), RA3900 (Group 2, 75 mg/kg/dose molnupiravir, day 1) and RA3887 (Group 3, 250 mg/kg/dose molnupiravir, day 1 and 2) all had 3.16 × 102 TCID50/mL in the oral swabs. These were the only positive oral swab samples. There were significant differences in viral loads in nasal swab samples between groups on days 1, 2, 3 and 4 post-challenge (Fig. 5D). On days 1 and 2, Group 1 (mock-treated) demonstrated significantly higher viral loads versus Group 2 (75 mg/kg/dose molnupiravir). Similarly, Group 2 demonstrated significantly higher viral loads than Group 3 (250 mg/kg/dose molnupiravir). On day 3, Group 1 demonstrated significantly higher virus levels than Group 3. On day 4, Group 3 had significantly higher virus levels than Group 2.
3.5. Radiography results
Radiographs were read by a veterinary radiologist and the conclusions for each subject are listed in Supplemental Table 1. Because of the variability in the radiographic findings, no clear group differences could be identified; however, diffuse interstitial infiltrates characteristic of COVID-19 were observed in most Group 1 (mock-treated) subjects while findings in Groups 2 and 3 (75 and 250 mg/kg/dose molnupiravir) ranged from benign to diffuse interstitial infiltrates similar to those observed in Group 1.
3.6. Pathology results
In general, when assessing across cohorts, lung tissue collected from Groups 2 and 3 male subjects demonstrated a mild decrease in alveolar inflammation severity whereas perivascular infiltrates (mixed or mononuclear) appeared equal in severity across groups when compared to Group 1. Lung tissue collected from female subjects across the three groups presented with alveolar inflammation and perivascular infiltrates of equivalent severity. Fibrosis and/or bronchoalveolar hyperplasia was observed in Groups 1 and 3 males and/or females at minimal incidence and severity. These observations can be considered background spontaneous lesions. Therefore, due to their minimal incidence and severity, it is unclear if these findings are background lesions or related to SARS-CoV-2 infection (Chamanza 2011) (Supplemental Fig. 3). Proteinaceous alveolar fluid was observed in the lung of the majority of NHPs; however, fluid was not considered related to SARS-CoV-2 infection or treatment.
4. Discussion
The variability of disease observed in the rhesus macaque model of COVID-19, along with the mild-to-moderate disease caused, led to some issues with data analysis for this study (Munster et al., 2020; Yu et al., 2020). Treatment with the molnupiravir protected the NHPs from developing a fever after challenge in Cohort 1. However, there was no difference in Cohort 2 because the control animals did not develop a febrile response to the challenge. Radiographic analysis showed diffuse interstitial infiltrates in control animals while the findings in treated ranged from benign to diffuse interstitial infiltrates as seen in the control animals. While some evidence is presented that treatment may have impacted the clinical course of disease, overall, no systemic pattern emerged between treated and untreated NHPs. No clinical chemistry results indicated any potential issue with drug toxicity, although this model was not designed to evaluate the pharmacokinetics or adverse drug reactions. A limitation of this study is that it does not include pharmacokinetics. Neither clinical chemistry nor hematology provided parameters which appeared to be useful biomarkers for the prediction of SARS-CoV-2 related pathology. No signs of mutational pressure due to treatment were observed in terms of worsening disease compared to mock treatment or in terms of persistent active viral infection at the end of the study.
Histopathology suggested that treatment with molnupiravir may have had a mild decrease in severity of alveolar inflammation compared to non-treated control animals. However, inflammation in association with foreign material most likely introduced during gavage procedures was observed in multiple groups. This observation complicated the interpretation as inflammation caused by viral exposure cannot be separated from that caused by inhaled foreign material. In future studies, an additional control group of lavaged non-SARS-CoV2 challenged animals would assist in delineating between spontaneous background lesions and those occurring due to test article or viral exposures. Increased animal numbers in control and challenged groups may also assist in providing a greater magnitude of difference between control and treated groups. Alternatively, designating separate groups for BAL and pathology would ameliorate this issue. It is difficult to justify extensive control groups ethically and financially in NHP studies. A limitation of this studies is that given the mild course of SARS-CoV-2 disease in the NHP model, it is possible that some findings are related to treatments and sedation, which was common across all group rather than SARS-CoV-2 infection. The increased respiration rates, particularly in the mid-dose treatment group are likely not biologically relevant, especially considering the variability in this parameter observed prior to treatment and the close approximate value of the day 14 respiration rate and day −5 respiration rates in this group. The reduction in alveolar inflammation may provide some histopathological evidence in support of additional study of the early use of molnupiravir in mild to moderate COVID-19 clinical care (Grillo et al., 2021; McDonald, 2021).
Differences observed in the viral load measured by qRT-PCR between treated and untreated subjects suggests there may be subtle differences in in vivo viral replication kinetics due to molnupiravir treatment. In the case of BAL, viral load by qRT-PCR (performed on the cellular fraction recovered from BAL) may be a better indicator of SARS-CoV-2 replication than the viral load by TCID50 (performed on the acellular, fluid fraction recovered from BAL) due to dilution of the virus in the media used to perform the BAL procedure. Meta-analysis of clinical studies of molnupiravir suggest that it reduces the absolute risk of hospitalization or death from 14.1% to 7.3% in high-risk patients provided that treatment is started early in the course of infection (Singh et al., 2021). This relatively small clinical improvement is in line with the relatively small clinical improvements reported in this study. The differences in viral load from respiratory tract sampling found in this study may be a useful prognostic tool in the clinical evaluation of molnupiravir in clinical trials. It remains unclear what effect reduced viral load in the lung has on the transmission of SARS-CoV-2.
The justification for conducting this study was to provide pre-clinical regulated data in support of regulatory approval for molnupiravir in humans. This study demonstrated the efficacy of oral treatment with molnupiravir in the rhesus macaque model of SARS-CoV-2 infection. A total of twenty NHPs were successfully challenged with SARS-CoV-2 and treated with twice daily with molnupiravir. The efficacy of molnupiravir was demonstrated in the significant reduction of viral RNA levels in BAL samples from treated subjects compared to controls.
Considering the variability of clinical presentations and radiological findings in the controls between cohorts, viral load in the lung may be the most important parameter to measure for pharmaceutical efficacy. Future studies should focus on BAL sampling at more time points. Also, comparing treated and non-treated subjects to nonchallenged controls at different times post-infection may be more informative in determining differences in lung pathology amongst the groups. Ultimately, this study, although rigorously conducted, showed relatively minor differences between the treatment groups. However, molnupiravir use in humans has documented efficacy for certain indications of SAR-CoV-2 infection (Caraco et al., 2022). From this, we can conclude that smaller differences in the rhesus macaque model of SARS-CoV-2 may play an important role in the evaluation of new COVID-19 therapeutics.
Animal ethics statement
The animal research protocols used in this study were performed in strict accordance with the recommendations in the Guide for Care and Use of Laboratory Animals, Eighth Edition (National Academy Press, Washington, DC, USA, 2011). The University of Texas Medical Branch (UTMB) facility (where these studies were conducted) is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. The protocols were approved by the UTMB Institutional Animal Care and Use Committee and complied with the Animal Welfare Act, the U.S. Public Health Service Policy, and other federal statutes and regulations related to animals and experiments involving animals.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the NIH/NIAID Preclinical Models of Infectious Diseases (PMoID), Contract HHSN272201700040I/HHSN27200009.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105492.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abdelnabi R., Foo C.S., De Jonghe S., Maes P., Weynand B., Neyts J. Molnupiravir inhibits replication of the emerging SARS-CoV-2 variants of concern in a hamster infection model. J. Infect. Dis. 2021;224(5):749–753. doi: 10.1093/infdis/jiab361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarawneh H.N., Chemaitelly H., Hasan M.R., Ayoub H.H., Qassim S., AlMukdad S., Coyle P., Yassine H.M., Al-Khatib H.A., Benslimane F.M. Protection against the omicron variant from previous SARS-CoV-2 infection. N. Engl. J. Med. 2022;386:1288–1290. doi: 10.1056/NEJMc2200133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour N.A., Abo Elmaaty A., Sarhan A.A., Elkaeed E.B., Moussa A.M., Erfan I.A., Al-Karmalawy A.A. A systematic review of the global intervention for SARS-CoV-2 combating: from drugs repurposing to molnupiravir approval. Drug Des. Dev. Ther. 2022:685–715. doi: 10.2147/DDDT.S354841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhimraj A., Morgan R.L., Shumaker A.H., Lavergne V., Baden L., Cheng V.C.-C., Edwards K.M., Gandhi R.T., Gallagher J.C., Muller W.J. Therapeutic emergency use authorizations (EUAs) during pandemics: double-edged swords. Clin. Infect. Dis. 2022;74:1686–1690. doi: 10.1093/cid/ciab880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byléhn F., Menéndez C.A., Perez-Lemus G.R., Alvarado W., De Pablo J.J. Modeling the binding mechanism of remdesivir, favilavir, and ribavirin to SARS-CoV-2 RNA-dependent RNA polymerase. ACS Cent. Sci. 2021;7:164–174. doi: 10.1021/acscentsci.0c01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calusic M., Marcec R., Luksa L., Jurkovic I., Kovac N., Mihaljevic S., Likic R. Safety and efficacy of fluvoxamine in COVID‐19 ICU patients: an open label, prospective cohort trial with matched controls. Br. J. Clin. Pharmacol. 2022;88:2065–2073. doi: 10.1111/bcp.15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraco Y., Crofoot G.E., Moncada P.A., Galustyan A.N., Musungaie D.B., Payne B., Kovalchuk E., Gonzalez A., Brown M.L., Williams-Diaz A. Phase 2/3 trial of molnupiravir for treatment of Covid-19 in nonhospitalized adults. NEJM Evid. 2022;1 doi: 10.1056/EVIDoa2100043. [DOI] [PubMed] [Google Scholar]
- Caruso F., Rossi M., Pedersen J.Z., Incerpi S. Computational studies reveal mechanism by which quinone derivatives can inhibit SARS-CoV-2. Study of embelin and two therapeutic compounds of interest, methyl prednisolone and dexamethasone. J. Infect. Public Health. 2020;13:1868–1877. doi: 10.1016/j.jiph.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamanza R. Vol. 1. 2011. Non-human primates: cynomolgus (Macaca fascicularis) and rhesus (Macaca mulatta) macaques and the common marmoset (Callithrix jacchus) (Background Lesions in Laboratory Animals E-Book: A Color Atlas). 2011. [Google Scholar]
- Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nature microbiology. 2021;6(1):11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal-Ré R., Becker S.L., Bottieau E., Holm S. Availability of oral antivirals against SARS-CoV-2 infection and the requirement for an ethical prescribing approach. Lancet Infect. Dis. 2022 doi: 10.1016/S1473-3099(22)00119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichter J.R., Devereaux A.V., Sprung C.L., Mukherjee V., Persoff J., Baum K.D., Ornoff D., Uppal A., Hossain T., Henry K.N. Mass critical care surge response during COVID-19: implementation of contingency strategies–a preliminary report of findings from the Task Force for Mass Critical Care. Chest. 2022;161:429–447. doi: 10.1016/j.chest.2021.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J.W., Holland J.J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy P., Le Grand R., Malvy D., Guedj J. Combined treatment of molnupiravir and favipiravir against SARS-CoV-2 infection: one+ zero equals two? EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W.A., Eron J.J., Jr., Holman W., Cohen M.S., Fang L., Szewczyk L.J., Sheahan T.P., Baric R., Mollan K.R., Wolfe C.R. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2021;14 doi: 10.1126/scitranslmed.abl7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin K.M., Karas M.G., Ivascu N.S., Lief L. Hospital preparedness for COVID-19: a practical guide from a critical care perspective. Am. J. Respir. Crit. Care Med. 2020;201:1337–1344. doi: 10.1164/rccm.202004-1037CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo F., Barisione E., Ball L., Mastracci L., Fiocca R. Lung fibrosis: an undervalued finding in COVID-19 pathological series. Lancet Infect. Dis. 2021;21 doi: 10.1016/S1473-3099(20)30582-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., Baniecki M., Hendrick V.M., Damle B., Simón-Campos A. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N. Engl. J. Med. 2022;386:1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P.E., Brasel T., Massey C., Herst C.V., Burkholz S., Lloyd P., Blankenberg T., Bey T.M., Carback R., Hodge T. A synthetic peptide CTL vaccine targeting nucleocapsid confers protection from SARS-CoV-2 challenge in rhesus macaques. Vaccines. 2021;9:520. doi: 10.3390/vaccines9050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemian S.M., Farhadi T., Velayati A.A. A review on favipiravir: the properties, function, and usefulness to treat COVID-19. Expert Rev. Anti Infect. Ther. 2021;19:1029–1037. doi: 10.1080/14787210.2021.1866545. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Suzuki T., Hashimoto K. Mechanisms of action of fluvoxamine for COVID-19: a historical review. Mol. Psychiatr. 2022;27:1898–1907. doi: 10.1038/s41380-021-01432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman W., Holman W., Mcintosh S., Painter W., Painter G., Bush J., Cohen O. Accelerated first-in-human clinical trial of EIDD-2801/MK-4482 (molnupiravir), a ribonucleoside analog with potent antiviral activity against SARS-CoV-2. Trials. 2021;22:561. doi: 10.1186/s13063-021-05538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martín-Quirós A., Caraco Y., Williams-Diaz A., Brown M.L. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J.H., Chokkakula S., Min S.C., Kim B.K., Choi W., Oh S., Yun Y.S., Kang D.H., Lee O., Kim E., Choi J., Lee J., Choi Y.K., Baek Y.H., Song M. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. Antivir. Res. 2022;208 doi: 10.1016/j.antiviral.2022.105430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo S.H., Fitzgerald R., Fletcher T., Ewings S., Jaki T., Lyon R., Downs N., Walker L., Tansley-Hancock O., Greenhalf W. Optimal dose and safety of molnupiravir in patients with early SARS-CoV-2: a Phase I, open-label, dose-escalating, randomized controlled study. J. Antimicrob. Chemother. 2021;76:3286–3295. doi: 10.1093/jac/dkab318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Advancing your practice: molnupiravir-an oral treatment to add to the COVID-19 arsenal. AJP Aust. J. Pharm. 2022;103:74–79. [Google Scholar]
- Lee C.C., Hsieh C.C., Ko W.C. Molnupiravir—a novel oral anti-SARS-CoV-2 agent. Antibiotics. 2021;10(11):1294. doi: 10.3390/antibiotics10111294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Wang Y., Lavrijsen M., Lamers M.M., de Vires A.C., Rottier R.J., Bruno M.J., Peppelenbosch M.P., Haagmans B.L., Pan Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32:322–324. doi: 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber C.M., Cox R.M., Sourimant J., Wolf J.D., Juergens K., Phung Q., Saindane M.T., Smith M.K., Sticher Z.M., Kalykhalov A.A., Natchus M.G. SARS-CoV-2 VOC type and biological sex affect molnupiravir efficacy in severe COVID-19 dwarf hamster model. Nat. Commun. 2022;13(1):1–13. doi: 10.1038/s41467-022-32045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Niu J., Xu Y., Qin L., Ding J., Zhou L. Clinical efficacy and adverse events of baricitinib treatment for coronavirus disease‐2019 (COVID‐19): a systematic review and meta‐analysis. J. Med. Virol. 2022;94:1523–1534. doi: 10.1002/jmv.27482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. British Medical Journal Publishing Group; 2021. Covid-19: Molnupiravir Reduces Risk of Hospital Admission or Death by 50% in Patients at Risk, MSD Reports. [DOI] [PubMed] [Google Scholar]
- Malone B., Campbell E.A. Molnupiravir: coding for catastrophe. Nat. Struct. Mol. Biol. 2021;28:706–708. doi: 10.1038/s41594-021-00657-8. [DOI] [PubMed] [Google Scholar]
- Malone B., Urakova N., Snijder E.J., Campbell E.A. Structures and functions of coronavirus replication–transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L.T. Healing after COVID-19: are survivors at risk for pulmonary fibrosis? Am. J. Physiol. Lung Cell Mol. Physiol. 2021;320:L257–L265. doi: 10.1152/ajplung.00238.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovits I., Shepshelovich D. Emergency use authorizations of COVID-19–related medical products. JAMA Intern. Med. 2022;182:228–229. doi: 10.1001/jamainternmed.2021.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter Wendy P., Sheahan T., Baric R., Holman W., Donovan J., Fang L., Alabanza P., Eron J.J., Goecker E., Coombs R. Reduction in infectious SARS-CoV-2 in treatment study of COVID-19 with molnupiravir. Top. Antivir. Med. 2021:304–305. [Google Scholar]
- Painter Wendy P., Holman W., Bush J.A., Almazedi F., Malik H., Eraut N.C., Morin M.J., Szewczyk L.J., Painter G.R. Human safety, tolerability, and pharmacokinetics of molnupiravir, a novel broad-spectrum oral antiviral agent with activity against SARS-CoV-2. Antimicrob. Agents Chemother. 2021;65 doi: 10.1128/AAC.02428-20. e02428-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R., Biatris P.S. Therapeutic role of corticosteroids in COVID-19: a systematic review of registered clinical trials. Future J. Pharm. Sci. 2021;7:1–18. doi: 10.1186/s43094-021-00217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5:85. doi: 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochwerg B., Agarwal A., Zeng L., Leo Y.-S., Appiah J.A., Agoritsas T., Bartoszko J., Brignardello-Petersen R., Ergan B., Ge L. Remdesivir for severe covid-19: a clinical practice guideline. Br. Med. J. 2020;370 doi: 10.1136/bmj.m2924. [DOI] [PubMed] [Google Scholar]
- Rosenke K., Okumura A., Lewis M.C., Feldmann F., Meade-White K., Bohler W.F., Griffin A., Rosenke R., Shaia C., Jarvis M.A., Feldmann H. Molnupiravir inhibits SARS-CoV-2 variants including Omicron in the hamster model. JCI insight. 2022;7(13) doi: 10.1172/jci.insight.160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salguero F.J., White A.D., Slack G.S., Fotheringham S.A., Bewley K.R., Gooch K.E., Longet S., Humphries H.E., Watson R.J., Hunter L. Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun. 2021;12:1–14. doi: 10.1038/s41467-021-21389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.K., Singh A., Singh R., Misra A. Molnupiravir in COVID-19: a systematic review of literature. Diabetes Metab. Syndr. Clin. Res. Rev. 2021;15 doi: 10.1016/j.dsx.2021.102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Chivukula V., Herati R.S., Hubbard S.R., Mulligan M.J., Landau N.R. Increased resistance of SARS-CoV-2 Omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. EBioMedicine. 2022;78 doi: 10.1016/j.ebiom.2022.103944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Sun Y., Xu H., Ye Q. The emergence and epidemic characteristics of the highly mutated SARS‐CoV‐2 Omicron variant. J. Med. Virol. 2022;94:2376–2383. doi: 10.1002/jmv.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots M., Yoon J.-J., Cox R.M., Hart M., Sticher Z.M., Makhsous N., Plesker R., Barrena A.H., Reddy P.G., Mitchell D.G. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots M., Yoon J.-J., Hart M., Natchus M.G., Painter G.R., Plemper R.K. Quantitative efficacy paradigms of the influenza clinical drug candidate EIDD-2801 in the ferret model. Transl. Res. 2020;218:16–28. doi: 10.1016/j.trsl.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Chen C., Tang J., Wang C., Zhou M., Cheng Y., Zhou X., Wu Q., Zhang X., Feng Z. Efficacy and safety of three new oral antiviral treatment (molnupiravir, fluvoxamine and Paxlovid) for COVID-19: a meta-analysis. Ann. Med. 2022;54:516–523. doi: 10.1080/07853890.2022.2034936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Wang H., Liu Q., Li R., Gao Y., Fang X., Zhong Y., Wang M., Wang Q., Rao Z. Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Yin W., Jiang Y., Xu H.E. Structure genomics of SARS-CoV-2 and its Omicron variant: drug design templates for COVID-19. Acta Pharmacol. Sin. 2022;1–13 doi: 10.1038/s41401-021-00851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Yin W., Zhang Y., Shang W., Wang Z., Luan X., Tian G., Aisa H.A., Xu Y., Xiao G. Design and development of an oral remdesivir derivative VV116 against SARS-CoV-2. Cell Res. 2021;31:1212–1214. doi: 10.1038/s41422-021-00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z. Age‐related rhesus macaque models of COVID‐19. Anim. Models Exp. Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.