Abstract
Retinal development occurs in mice between embryonic day E11.5 and post-natal day P8 as uncommitted neuroblasts assume retinal cell fates. The genetic pathways regulating retinal development are being identified but little is understood about the global networks that link these pathways together or the complexity of the expressed gene set required to form the retina. At E14.5, the retina contains mostly uncommitted neuroblasts and newly differentiated neurons. Here we report a sequence analysis of an E14.5 retinal cDNA library. To date, we have archived 15 268 ESTs and have annotated 9035, which represent 5288 genes. The fraction of singly occurring ESTs as a function of total EST accrual suggests that the total number of expressed genes in the library could approach 27 000. The 9035 ESTs were categorized by their known or putative functions. Representation of the genes involved in eye development was significantly higher in the retinal clone set compared with the NIA mouse 15K cDNA clone set. Screening with a microarray containing 864 cDNA clones using wild-type and brn-3b (–/–) retinal cDNA probes revealed a potential regulatory linkage between the transcription factor Brn-3b and expression of GAP-43, a protein associated with axon growth. The retinal EST database will be a valuable platform for gene expression profiling and a new source for gene discovery.
INTRODUCTION
The vertebrate retina is a multilayered tissue acting at the interface of input light and visual perception. In the mouse, the formation of this complex structure takes place between embryonic day E11.5 and post-natal day P8 requiring a combination of intrinsic and extrinsic factors (1). Six neuronal cell types [retinal ganglion cell (RGC), amacrine, bipolar, horizontal, rod and cone] and a single glial cell type (Müller) are formed when actively dividing neuroblasts leave the cell cycle and commit to cell fates in a temporally ordered sequence.
Differentiation occurs along all three axes. Along the anterior–posterior axis, uncommitted cells differentiate into distinct retinal cell types and migrate to their final positions within either the ganglion, inner nuclear or outer nuclear layer. Along the dorsal–ventral axis, RGCs and other cell types acquire a dorsal–ventral pattern that can be observed by retinotectal projection maps or by molecular markers that distinguish dorsal axons from ventral axons (2,3). Along the nasal–temporal axis, differentiation proceeds as a bidirectional propagation wave from the center of the developing retina to the peripheral regions (4).
Environmental cues are particularly crucial in retinal development because naïve neuroblasts are not intrinsically programmed for any particular lineage (1,5). In the zebrafish retina, hedgehog signaling factors have been implicated as essential components of the propagation event (4), and sonic hedgehog appears to negatively regulate RGC formation behind the wave front (6). Other secreted factors have been shown to play key roles in retinal development. Fibroblast growth factors (FGFs) emanating from the surface ectoderm appear to control the decision to become neural retina or pigmented epithelium (7,8), and FGFs play roles in other aspects of retinal patterning and differentiation (9).
The Notch–Delta pathway negatively regulates the first cell-fate commitment made in the retina, which is to become a RGC (10–12). Recently, the proneural gene math5, which encodes a bHLH transcription factor and is repressed by Notch signaling, was shown to be required for RGC formation (13). Brn-3b, a Class IV POU domain protein, acts downstream of Math5 as an essential transcription factor in RGC differentiation, axon outgrowth and cell survival (14). Brn-3b may also play a role in RGC specification and may be a direct target of Math5 (15–17). Pax6 is required upstream of several bHLH factors to maintain the multipotent state of retinal progenitor cells (18), and a number of bHLH factors and other transcription factors have been implicated in the formation of RGCs, bipolar cells, amacrine cells and photoreceptor cells (1,5,19).
Progress in defining the genetic regulatory events that lead to the formation of the vertebrate retina has been substantial, particularly over the past decade. However, many of the regulatory pathways and genes found to be critical for retinal development are known to occur in the development of other vertebrate tissues and organs. What is unclear is how these pathways ultimately lead to the unique repertoire of expressed genes that define the functioning retina. As a starting point, it would be valuable to define the complete set of expressed genes for the developing retina. This would enable comparisons to be made with other neuronal tissues, and perhaps more important, would provide the basis for establishing a well-defined platform upon which gene expression profiling experiments could be based and new genes could be discovered.
At E14.5, most cells in the developing retina are either uncommitted neuroblasts or newly differentiated retinal neurons (20). It is clear that in this heterogeneous population of cells, the complete expressed gene set will provide information not only on the events occurring at E14.5 but on past and future events as well. Here we report the sequence analysis of an E14.5 retinal cDNA library and the establishment of an EST database. The database currently contains 15 268 ESTs, of which 9035 have been annotated and represent 5288 genes. From the current database, we estimated that there could be as many as 27 000 genes expressed in the E14.5 retina. A pilot microarray gene expression profiling analysis using 864 clones identified GAP-43, encoding a protein required for proper RGC axon growth and pathfinding, as a potential target of Brn-3b. The retinal EST database can therefore be used successfully to reveal novel gene regulatory linkages involved in retinal development. We expect that the embryonic retinal EST database will be an accessible, valuable resource for the biomedical research community.
MATERIALS AND METHODS
Retinal cDNA library construction
To generate an embryonic retinal cDNA library, total RNA was isolated from about 450 E14.5 manually dissected neural retinas (Swiss Webster Strain, Taconics) using TRIZOL Reagent (Life Technologies), and mRNA was subsequently purified using the mRNA Isolation Kit (Pharmacia). The cDNA library was constructed using the Uni-ZAP II XR Library Construction Kit (Stratagene). A single package of the cDNA library in lambda ZAP vector (5 × 106 p.f.u.) was mass-excised to generate the cDNA library in pBluescript SK II (+). Single bacterial colonies were grown on Luria–Bertani (LB) agar plates and arrayed into 384-well plates by a Q-bot (Gentix). The percentage of non-recombinant clones was <0.1%.
Plasmid preparation and EST sequencing
Clones from the 384-well plates were subarrayed into 96-well blocks with a Biomek 2000 robotic workstation and cultured in 2× LB at 37°C for 24 h with vigorous shaking in the presence of 100 µg/ml ampicillin. Plasmid minipreps were made with the Nucleospin Multi-96 Plasmid Purification Kit (Clonetech) on the QIAGEN BioRobot 9600 system. Sequencing reactions from the 5′ end of the cDNA clone were carried out with a T3 primer (5′-ATTAACCCTCACTAAAG-3′), using the BIG Dye Cycle Sequencing Ready reaction version 2.0 (PE Biosystem) following the manufacturer’s protocol, and the sequencing was performed on the 3700 Genetic Analyzer (PE Biosystem).
EST database analysis and management
Raw sequences were first screened by Phred (21,22) in combination with a manual inspection to assure quality. The sequences were then processed by RepeatMasker (http://repeatmasker.genome.washington.edu/) to filter repetitive and vector sequences. Sequences representing rRNA and mitochondrial DNA were discarded. BLASTN and BLASTX searches were performed locally on a Digital AlphaServer 4100 Model 5/400 against the mouse EST database, and the redundant GenPept database in the GenBank was downloaded periodically as it was updated. The EST sequences have been deposited in the GenBank database (accession nos BG799964–BG808997 and BI985056–BI991757).
Estimation of singly occurring ESTs
The fraction of singly occurring ESTs as a function of the total number of ESTs was fitted using a weighted linear regression. Numerical recipes have been described by Press et al. (23). It was assumed that the variance of the number of single occurrences was proportional to the square root of the number of single occurrences (Poisson approximation) and these values were used as weighing factors. For validation, samples were selected without replacement from all ESTs and multiple clustering runs were made showing that the Poisson approximation was acceptable.
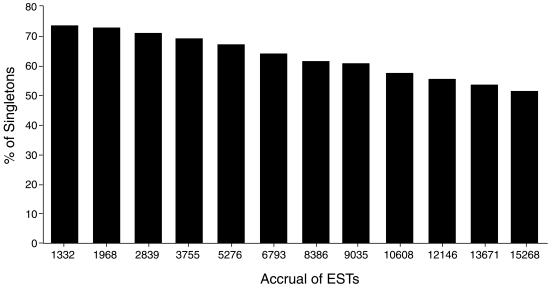
Overall, S(n) = a + bn and n’ is the value such that S(n’) = 0. Data are taken from Figure 3. For total EST = 2839, a = 0.7575, b = –1.7333e-05, n = 43 702; for total EST = 3755, a = 0.7579, b = –1.7552e-05, n = 43 180; for total EST = 5276, a = 0.7554, b = –1.6393e-05, n = 46 081; for total EST = 6793, a = 0.7574, b = –1.7192e-05, n = 44 055; for total EST = 8386, a = 0.7572, b = –1.7127e-05, n = 44 210; for EST 9035, a = 0.7565, b = –1.6885e-05, n = 44 803; for total EST = 12 146, a = 0.7543, b = –1.6337e-05, n = 46 171.
Figure 3.
Single occurring ESTs as a function of total EST accrual. The histogram shows eight cluster analyses performed as ESTs were added to the database. The percent of single occurrences decreases with increasing EST accrual.
Microarray fabrication and hybridization with retinal cDNA probes
cDNA inserts of individual clones were obtained by PCR amplification with 2 µl from an overnight bacterial culture and vector primers (T3, 5′-TTAACCCTCACTAAAGGGAAC-3′, and T7, 5′-GTAATACGACTCACTATAGGG-3′) in a total volume of 50 µl in 96-well plates. Clones from nine 96-well plates (864 clones) were used to make the pilot array. PCR was performed by preheating the samples at 94°C for 5 min, 40 cycles at 94°C for 30 s, 53°C for 30 s and 72°C for 2 min, followed by 3 min of extension at 72°C. The PCR products were electrophoresed on a 1% agarose gel to monitor the success of the reaction. The amplified cDNA inserts were cleared by centrifugation, and 20× SSC was added to each sample to a final concentration of 3×. The microarrays were fabricated by spotting the cDNA inserts onto poly-l-lysine-coated glass slides (Sigma) with the OmniGrid microarrayer (Genemachines).
Five micrograms of total RNA from wild-type or brn-3b (–/–) E14.5 retinas was used to generate fluorescence-labeled cDNA probes (Cy3 or Cy5) by reverse transcription with the 3DNA Kit (Genisphere) following the manufacturer’s procedure. The microarray slides were treated by snap heating and UV irradiation. This was followed by blocking with 70 mM succinic anhydride dissolved in 90% 1-methyl-2-pyrrolidinone and 0.1 M boric acid (pH 8.0), for 15 min at room temperature and then denaturation in 95°C water for 2 min was performed as described (24). Hybridization was performed at 65°C with the microarray under a coverslip in a humidity chamber in a total volume of 20 µl of hybridization buffer (0.25 M NaPO4, 4.5% SDS, 1 mM EDTA, 1× SSC, 25 µg/ml oligo dT blocking reagent, 25 µg/ml mouse Cot-1 DNA). Following the hybridization reaction, the slides were washed twice with 2× SSC for 10 min at room temperature followed by rinsing with ethanol and air drying. The slides were processed by scanning with a ScanArray 3000 scanner (General Scanning).
Quantitative RT–PCR
Total RNA was reverse transcribed with Superscript reverse transcriptase (Life Technologies) at 42°C. cDNA equivalent to 40 ng of total RNA was used for PCR amplification with the Hotstart Taq DNA polymerase (Qiagen) in a volume of 20 µl. The reaction mixture was preheated at 95°C for 15 min, denatured at 94°C for 30 s, annealed at 50°C for 30 s and extended at 72°C for 1 min. The optimal cycle number was determined, being 25 cycles for GAP-43, 20 for β-actin and 20 for brn-3b. The primers for GAP-43 were as follows: forward, 5′-GTGCTGCTAAAGCTACCACT-3′ and reverse, 5′-CTTCAGAGTGGAGCTGAGAA-3′. The primers for β-actin were forward, 5′-CAACGGCTCCGGCATGTGC-3′ and reverse, 5′-CTCTTGCTCTGGGCCTCG-3′. The primers for brn-3b were forward, 5′-TCTGGAAGCCTACTTCGCCA-3′ and reverse, 5′-CCGGTTCACAATCTCTCTGA-3′. The PCR products were separated on 2% agarose gels and visualized by staining with ethidium bromide.
Indirect immunofluorescence staining
Retinal tissues were collected from embryos at E13.5 resulting from matings between brn-3b (+/–) males and brn-3b (–/–) females. Retinas were cultured for 40 h in laminin-coated Petri dishes as described previously (14). Genotypes were determined as described (25). The explanted retinas were fixed with 3.2% paraformaldehyde and labeled by indirect immunofluorescence. Primary antibodies were mouse anti-GAP-43 (Zymed Laboratories, Inc.) and mouse anti-neurofilament light chain (Zymed Laboratories, Inc.). Secondary antibodies were Alexa-488-conjugated anti-mouse IgG (Molecular Probes) and Cy5-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.). Propidium iodide was used to visualize nuclei. Images were collected with an Olympus FV500 laser scanning confocal microscope. Each presented image was a projection of five optical sections with 0.5-µm intervals.
RESULTS AND DISCUSSION
Acquisition of E14.5 retinal ESTs and data management
To approach our long-range objective of a complete gene set for the developing retina, we constructed a retinal cDNA library using mRNA extracted from 450 E14.5 murine retinas. The cDNA library contained 5 million primary recombinants with an average insert size of ∼1.8 kb. Provided that the cDNA clones accurately represent the E14.5 retinal mRNA population, the complexity of the library should reflect that of the retinal transcriptome at this stage and should be sufficient for identifying all expressed genes. The ESTs were obtained by sequencing from the 5′ end of individual clones. To date, 18 816 sequencing reactions have been performed. After removal of low-quality sequences and sequences representing ribosomal, mitochondrial and repetitive sequences, 15 268 high-quality sequences with a minimum of 200 bp of continuous sequence and at least 98% accuracy were retained for further analysis. The sequences were used to search the redundant GenPept protein database and mouse EST database in GenBank with the BLASTX and BLASTN programs, respectively. The search results were archived in our mouse retinal EST database (RetinalExpress). A web interface (http://odin.mdacc.tmc.edu/RetinalExpress) was created so that keyword or sequence homology searches against the database can be performed. The ESTs have also been deposited into GenBank.
Clustering and annotation of the retinal EST database
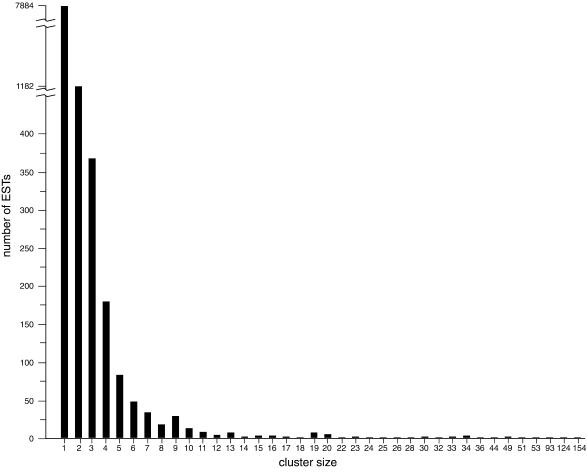
Cluster analysis was performed periodically as ESTs accumulated using the ESTate program (http://www.hgmp.mrc.ac.uk/~gslater/ESTate) to group overlapping ESTs, assuming that ESTs clustered together were from the same gene. The clustering results were used to calculate the number of genes expressed in the E14.5 retina (see below). Figure 1 shows the clustering results of the 15 268 ESTs; 7884 were single occurrences, representing 51.6% of the total ESTs analyzed to date. The rest fell into 2022 clusters with sizes ranging from two (1182 clusters) to 154 (one cluster), although ∼80% of the ESTs belonged to cluster sets of four or less (Fig. 1). Of the 9906 cluster groups (single occurrence included), 2584 (26%) have no matches in the GenBank mouse EST database. The cluster analysis indicated that the vast majority of genes expressed in the E14.5 retina fall into a low-abundance, high-complexity class and, furthermore, that the expressed gene set is not dominated by a small number of highly prevalent expressed genes. One explanation for the relatively low level of redundancy in the retinal expressed gene set is that, unlike fully differentiated cell types or tissues, the E14.5 retina contains a dynamic heterogeneous population of dividing and non-dividing cells. Retinal cells at E14.5 include actively dividing neuroblasts, biased progenitor cells that have entered into G0, and committed and differentiated neuronal cell types (21).
Figure 1.
Cluster analysis for 15 268 retinal ESTs. The histogram shows the number of ESTs in each cluster group as a function of cluster size. There are a total of 9906 individual clusters, 7884 are single occurrences and 1041 occur from two to 154 times.
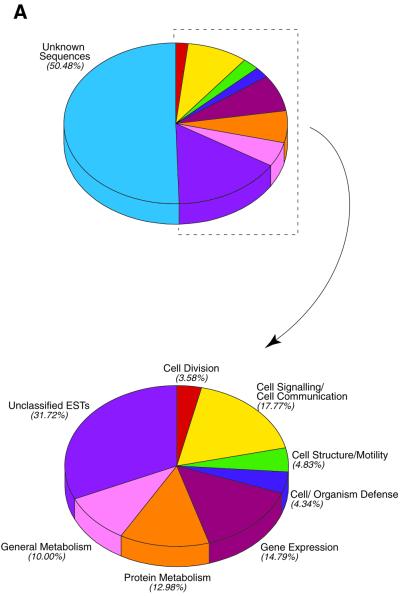
To further analyze the E14.5 transcriptome, we have annotated the first 9035 ESTs that were obtained. Clustering these 9035 ESTs resulted in 6514 individual cluster groups including single occurrences. However, the gene number represented by the 9035 ESTs is less than the number of individual cluster groups. This is because ESTs from different regions of the same gene did not fall into the same cluster. In addition, alternatively spliced forms of a single gene could also be grouped into different clusters. Using the clustering results as a guide, the ESTs in the database were annotated and classified based on their known or inferred function (Fig. 2A and see Table 2). During this process, clusters representing the same gene were merged into a single group. As a result, a total of 2147 cluster groups were found to represent 1745 unique known genes that were classified into seven known functional classes and 23 subclasses (Fig. 2A and see Table 2). Sixty-five genes were assigned to two classes, leading to a total of 1810 assignments. In addition, there were 1035 cluster groups representing known genes that could not be readily classified, and 3323 cluster groups representing unknown ESTs. As is the case for other reported EST databases, unknown ESTs made up ∼40% of the total ESTs in the E14.5 retinal database (26). Based on the gene representation in clusters that were readily categorized, a redundancy factor of 1.23 (2147/1745) was then used to calculate the number of genes represented by the unclassified and unknown cluster groups, yielding 841 and 2702 genes, respectively. Based on these considerations, the 9035 retinal ESTs represent 5288 genes.
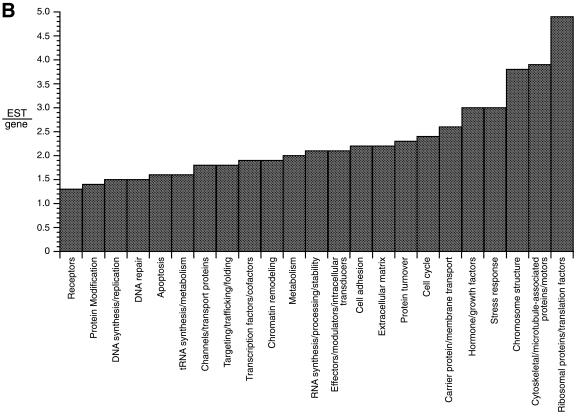
Figure 2.
Annotation of the first 9035 retinal ESTs. (A) Distribution of functional classes. The top pie chart shows the distribution with unknown ESTs included. The unknown ESTs are removed from the bottom pie chart. (B) Distribution of EST prevalence. The histogram shows 23 functional subclasses as a function of EST number per gene. The distribution ranges from 1.3 ESTs/gene (receptors) to 4.9 ribosomal proteins/translation factors.
Table 2. Retinal EST classification.
| Class | Subclass | Gene | ESTs | Average EST/gene |
|---|---|---|---|---|
| Cell division | DNA synthesis/replication | 21 | 32 | 1.5 |
| Cell cycle | 43 | 102 | 2.4 | |
| Chromosome structure | 31 | 117 | 3.8 | |
| Cell signaling/cell communication | Cell adhesion | 25 | 55 | 2.2 |
| Channels/transport proteins | 45 | 79 | 1.8 | |
| Effectors/modulators/intracellular transducers | 184 | 390 | 2.1 | |
| Hormones/growth factors | 16 | 48 | 3.0 | |
| Protein modification | 156 | 225 | 1.4 | |
| Receptors | 45 | 59 | 1.3 | |
| Cell structure/motility | Cytoskeletal/microtubule-associated proteins/motors | 116 | 449 | 3.9 |
| Extracellular matrix | 12 | 26 | 2.2 | |
| Cell/organism defence | DNA repair | 21 | 31 | 1.5 |
| Apoptosis | 32 | 52 | 1.6 | |
| Carrier protein/membrane transport | 16 | 41 | 2.6 | |
| Stress response | 46 | 136 | 3.0 | |
| Gene expression | RNA synthesis/processing/stability | 161 | 337 | 2.1 |
| Transcription factors/co-factors | 165 | 318 | 1.9 | |
| Chromatin remodeling | 66 | 123 | 1.9 | |
| Protein metabolism | Targeting/trafficking/folding | 122 | 221 | 1.8 |
| Ribosomal proteins/translation factors | 111 | 544 | 4.9 | |
| Protein turnover | 104 | 236 | 2.3 | |
| tRNA metabolism | 7 | 11 | 1.6 | |
| Metabolism | Metabolism | 265 | 541 | 2.0 |
The cluster results were also used as a guide to assess the prevalence of gene subclasses in the retinal transcriptome. As with other tissues, the most abundantly expressed genes were those for translation factors, cytoskeletal proteins and housekeeping proteins, although several regulatory genes were also highly expressed (Table 1). Among the most prevalent subclasses were genes encoding ribosomal proteins and translation factors, cytoskeletal components and chromosomal structural proteins. These were represented by average EST/gene ratios of 3.8 or greater (Fig. 2B and Table 2). The least prevalent subclasses, which contained average EST/gene ratios of 1.6 or less, encoded membrane and nuclear receptors, enzymes involved in protein modification, and proteins for RNA synthesis, DNA replication and repair, apoptosis and tRNA synthesis (Fig. 2B and Table 2). Within a subclass, the gene-prevalence distribution was usually broad. For instance, the subclass of 165 genes encoding transcription factors and co-factors had an average EST/gene ratio of 1.9, but the prevalence ranged from 1 to 13 (Fig. 2 and Tables 2 and 3).
Table 1. Abundantly expressed genes in the retina.
| Gene name | Number of ESTs |
|---|---|
| Elongation factor 1-alpha (EF-1) | 98 |
| Alpha-tubulin | 76 |
| Beta-tubulin | 53 |
| Actin A4 | 35 |
| Ubiquitin | 33 |
| Alpha-enolase | 28 |
| FGF-15 | 26 |
| Glyceraldehyde-3-phosphate dehydrogenase | 25 |
| Elongation factor 2 (EF-2) | 23 |
| Ubiquitin protein ligase (Nedd-4) | 22 |
| Histone H3.3A | 22 |
| HSP70 | 22 |
| 14-3-3 Protein epsilon | 21 |
| sFRP-2/SDF5/Sarp1 | 20 |
| Ribosomal protein L7 | 20 |
| Ribosomal protein L4 | 20 |
| Cyclin D1 | 19 |
| Prothymosin alpha | 19 |
| Tumor protein (TCTP)/lens epithelial protein | 18 |
| Ribosomal protein SA/P40 (34/67 kDa laminin receptor) | 18 |
| Peptidyl-prolyl cis–trans isomerase A | 17 |
| hnRNP-E2 | 17 |
| Acidic ribosomal protein P0 | 15 |
| Pr22/stathmin | 15 |
| Heat stable antigen (nectadrin, murine CD24 glycoprotein) | 15 |
| Calmodulin | 15 |
| Ferritin light chain | 15 |
| Ribosomal protein S3a | 15 |
| M2-type pyruvate kinase | 15 |
| Histone H2A.Z | 14 |
| KRIP-1/TIF1B | 14 |
| RAN | 14 |
| 14-3-3 Protein theta | 13 |
| Math5 | 13 |
| Histone H2A.1 | 13 |
| Ribosomal protein L3 (J1 protein) | 12 |
| SCG10 | 11 |
| Ribosomal protein S2 | 11 |
| Ribosomal protein L12 | 11 |
| Marcks-related protein (MAC-MARCKS/MRP) | 11 |
| HSP 90-beta | 11 |
| Dlxin-1/NRAGE (neurotrophin receptor-interacting MAGE homolog) | 10 |
| hnRNP-K | 10 |
| Beta catenin | 10 |
| Importin beta-1 subunit | 10 |
Table 3. Transcription factors involved in eye development.
| Transcription factor | Type | ESTs | NIA 15K |
|---|---|---|---|
| Math5 (atonal homolog 5) | bHLH | 13 | No |
| HES-6 (hairy/enhancer of split 6) | bHLH | 6 | No |
| Chx10 | Homeobox | 5 | No |
| SOX11 | HMG | 5 | Yes |
| mLHX2 | LIM-homeobox | 4 | No |
| Neurogenin-2 | bHLH | 4 | No |
| Pax6 | Paired box | 4 | No |
| RFP-1 (retina-derived POU-domain factor-1) | POU | 3 | No |
| Six3 | Homeobox | 3 | No |
| Six6 (Optx2) | Homeobox | 3 | No |
| BHF-1/NeuroD | bHLH | 2 | No |
| HES-5 (hairy/enhancer of split 5) | bHLH | 2 | No |
| HES-1 (hairy/enhancer of split 1) | bHLH | 2 | No |
| Ash1 (achaete-scute homolog 1) | bHLH | 2 | No |
| ASH2 | bHLH | 2 | Yes |
| ROR-beta/retinoid-related orphan receptor (ROR) beta | Nuclear receptor | 2 | No |
| Rx/Rax (retina and anterior neural fold homeobox gene) | Homeobox | 1 | No |
| Math3 (atonal homolog 3) | bHLH | 1 | No |
| SOX8 | HMG | 1 | Yes |
| SOX9 | HMG | 1 | Yes |
| CASP (homeobox gene Cut alternatively spliced product) | Homeobox | 1 | Yes |
| TBX3 (T-box protein) | T-box | 1 | No |
| Islet-2 | LIM-homeobox | 1 | No |
| OC-2 (ONECUT homeodomain class) | Homeobox | 1 | No |
The accrual rates for ESTs added to the database allowed us to estimate the total number of expressed genes in the retinal cDNA library. As the total number of ESTs increased, the fraction of singly occurring ESTs decreased (Fig. 3). The fraction of singly occurring ESTs as a function of the total number of ESTs was fitted using a weighted linear regression. By this analysis, the fraction of singly occurring ESTs would approach zero when the total number of ESTs was between 43 000 and 46 000. Because our analysis of the 9035 ESTs resulted in 5288 genes, assuming this ratio is maintained as additional ESTs are added to the database, an EST/gene ratio of 1.71 (9035/5288) yields an estimate of 25 000–27 000 unique genes in the E14.5 retinal cDNA library. Considerable uncertainty exists in our calculation, mostly because the relationship between low-abundance ESTs and the number of genes they represent has not been well established. The true number of expressed genes will become more apparent as more ESTs are analyzed. The number of retinal expressed genes by this calculation approaches recent estimates that have been made for the total number of genes in the human genome (27,28), even though a further analysis suggests this number is an underestimate (29). Nevertheless, our initial analysis is instructive and indicates that the developing retina contains an exceedingly complex population of expressed genes. Indeed, a substantial portion of the genes encoded in the mouse genome is likely to be expressed in the developing retina. The uniqueness of the retinal transcriptome must therefore derive from the distinct prevalence and spatial distribution of the expressed genes and from the small set of genes whose expression is restricted to the retinal cell types.
Regulatory genes expressed in the E14.5 retinal transcriptome
The retinal transcriptome at E14.5 reflects a complex regulatory network operating during retinal organogenesis and consisting of thousands of intrinsic and extrinsic factors. Although the overall representation of genes in various regulatory classes is similar to that in other EST databases (26), the developing retina represents a unique expressed gene set in terms of genes encoding regulatory proteins such as transcription factors, signaling molecules and cell cycle proteins. A complete list of the regulatory genes found in the 9035 ESTs is included (see Supplementary Material).
A closer examination of the composition of genes encoding transcription factors in the retinal database clearly demonstrates a distinct distribution when compared with non-retinal EST databases. Of the 165 known genes coding for transcription factors (see Supplementary Material), at least 24 have specific roles in eye development (Table 3). Ten of these genes were found in cluster groups larger than three, including Math5, Hes6, Chx10 and Pax6. Other transcription factor genes implicated in retinal development include Hes1, Hes5, Mash1, NeuroD, Neurogenin-2, OC-1, Six3, Sox9, Islet2 and mLHX2 (Table 3). Furthermore, there are a large number of putative transcription factors, particularly ones containing zinc-finger domains, whose functions have not been characterized (see Supplementary Material). It is reasonable to assume that many of these factors are involved in regulatory events within the developing retina.
A number of genes with regulatory functions in signaling and cell cycle pathways were also prevalent in the database (Table 4). Genes for two secreted molecules, FGF-15 and SFRP-2, were highly represented (Table 4). FGF-15 is expressed in the developing neural system, including the developing retina, where it first appears at E9.5 in the inner cell layer of the optic cup (30). FGF-15 expression is detected later in the ventricular layer where dividing neuroblasts are found (30). The prominent representation of FGF-15 in E14.5 retina may reflect a role in cell-fate commitment, dorsal–ventral retinal patterning or wave propagation along the nasal–temporal axis.
Table 4. Proteins in signaling and cell cycle pathways involved in eye development.
| Protein | ESTs | NIA 15k |
|---|---|---|
| FGF-15 | 26 | No |
| SFRP-2 (secreted frizzled-related protein 2) | 20 | No |
| Cyclin D1 | 19 | Yes |
| Calmodulin | 15 | Yes |
| SCG10 (stathmin homolog) | 11 | No |
| Beta catenin | 10 | Yes |
| GAP-43 (neuromodulin) | 6 | Yes |
| Ulip (UNC-33-like phosphoprotein) | 6 | No |
| Ulip3 (collapsin response mediator 1) | 6 | No |
| Dkk-3 (Dickkopf gene) | 4 | Yes |
| EGF repeat transmembrane protein DBI-1 (related to Notch) | 3 | Yes |
| Manic fringe | 3 | Yes |
| Robo2 (repulsive guidance receptor roundabout 2) | 3 | No |
| SMO (smoothened homolog) | 3 | No |
| MAB21L2 | 3 | No |
| CDC42 | 2 | Yes |
| Nogo-A/Reticulon 4-A (inhibitor of neurite growth) | 2 | Yes |
| MAB21L1/CAGR1 (Mab-21 homolog) | 2 | No |
| P35 (CDK5 regulatory subunit 1) | 2 | No |
| Axin 1 | 2 | Yes |
| Su(H) (suppressor of hairless) | 2 | No |
| Numb | 2 | No |
| Plexin-B1/SEP receptor | 2 | Yes |
| Plexin 2 | 1 | No |
| Sema4g (semaphorin subclass 4 member g) | 1 | Yes |
| Neuropilin-1 (semaphorin III receptor) | 1 | Yes |
| ICAT (beta catenin-interacting protein) | 1 | No |
| ELF-1/CEK7-L (EPH-related receptor tyrosine kinase ligand) | 1 | No |
| Notch4/INT-3 | 1 | No |
| Plexin 3 (transmembrane protein SEX) | 1 | Yes |
| Deltex (Notch signaling regulator) | 1 | No |
| Epherin-B1 | 1 | No |
| Chat (Cas and HEF1-associated signal transducer) | 1 | No |
| Reelin | 1 | No |
| SOS (son-of-sevenless-1) | 1 | Yes |
| mDab1 (Disabled homolog) | 1 | Yes |
| Semaphorin VIa | 1 | No |
| CDK5 (PSSALRE kinase) | 1 | Yes |
| DVL-2 (Dishevelled homolog) | 1 | No |
| Notch1/motch | 1 | No |
| Munc13–2 (UNC-13 homolog) | 1 | No |
SFRP-2 is a member of the Frizzled-related class of secreted proteins that share sequence similarity with the Wnt receptor Frizzled (31). SFRPs bind to Wnt ligands and modulate their action. Although the developmental roles of SFRPs are uncertain, two SFRPs, SFRP-2 and SFRP-5, are expressed in the retina (32). SFRP-5 is expressed in the pigmented epithelial layer and SFRP-2 is expressed in the inner nuclear layer of the mature retina (32). In the E14.5 retinal database, SFRP-2 is highly represented by 22 ESTs, but SFRP-5 has not appeared. The rarity of SFRP-5 may reflect the fact that the pigmented epithelial layer was not included when RNA was isolated from the retina during our cDNA library construction. Based on their expression patterns, it has been suggested that SFRP-2 and SFRP-5 play roles in establishing the polarity of the photoreceptor cells (32).
The gene encoding cyclin D1 was also highly represented in the E14.5 retinal database (nine ESTs, Table 4). Cyclin D1 is one of three D-type cyclins that promote progression through the G1 phase of the cell cycle by activating the cyclin-dependent kinases CDK4 and CDK6 (33). The high cyclin D1 representation is consistent with previous reports that cyclin D1 is abundantly expressed throughout the entire retina at E14.5 (34,35). Retinas of mice with targeted mutations in the cyclin D1 gene show reduced cell proliferation and enhanced apoptosis of photoreceptor cells (34–36).
Components of all the major developmental signaling pathways are present in the E14.5 retinal database. These pathways include Notch–Delta, Wnt–β-catenin, Epherin–Eph, FGF, BMP–TGF-β, neurotrophin–P75 and the several tyrosine kinase receptors. Genes involved in neurite growth and pathfinding (GAP-43, SCG10, robo2, Nogo A and genes for semaphorins and their receptors) are also prominently represented (Table 4). A complete list of annotated genes involved in signaling pathways can be found in the Supplementary Material. In addition, many of the unknown and unidentified genes in our retinal clone set may also function as unidentified components of these pathways.
Comparison to the National Institute of Aging (NIA) mouse 15K cDNA clone set
To further evaluate the specificity of the expressed gene set in E14.5 retina, we compared the occurrences of the 9035 ESTs in our data set with those in the NIA 15K mouse cDNA clone set, which was derived from multiple mouse tissues (37). ESTate clustering was performed after pooling our retinal ESTs with the sequences from the NIA 15K clones. Of 9035 ESTs, only 3257 (36%) had matches in the NIA 15K cDNA database. Although the results suggested that the two databases were largely non-overlapping, the large difference in sequence representation between the two was likely to be an overestimate because most of the ESTs in the databases represent partial sequences. Thus, different ESTs representing the same gene would not cluster together in this analysis. Nevertheless, the cluster analysis between the retinal EST and the NIA 15K databases imply large differences.
In a second analysis, we used the full-length cDNA sequences of the known genes displayed in Tables 2 and 3 to search the NIA 15K cDNA clone set database. The use of full-length cDNA sequences eliminated the uncertainties associated with ESTs. The majority of transcription factors (19/24) involved in retina development were not represented in the NIA 15K cDNA database (Table 3). Furthermore, of the 41 signaling and cell cycle genes involved in eye development, 24 were present only in the retinal database. Thus, many regulatory genes that were well represented in the E14.5 retinal cDNA library were absent in the NIA cDNA database. These results emphasize the usefulness of specialized libraries and databases thereby ensuring the presence of critical elements of the tissue or cell type in question.
Given that the set of retinal expressed regulatory genes is highly reflective of the events associated with retinal development, a major challenge now is to connect these events together into the gene regulatory network that ultimately defines the developing retina. The retinal database will serve as a resource for gene expression profiling experiments where conserved signaling pathways have been or can be perturbed by gene targeting. Patterns of gene expression can then be compared between control and perturbed retinas.
A novel regulatory link revealed by gene expression profiling with E14.5 retinal ESTs
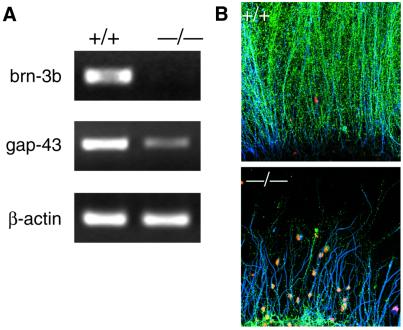
The E14.5 retinal EST database provides a platform from which gene expression profiling experiments can be performed by microarray technology (37). One of our objectives is to identify target genes of Brn-3b, a Class IV POU domain transcription factor that is required for the survival of RGCs, and their correct axon projection and pathfinding (14,25,38). We chose Brn-3b because its role appears to be specific for RGC differentiation as opposed to Math5, which plays a broader role as a proneural gene setting up a competence field from which RGCs are selected. In an initial attempt, we generated a microarray with inserts from 864 clones and hybridized it with cDNA probes derived from wild-type and brn-3b (–/–) E14.5 retina, labeled with Cy3 and Cy5, respectively. The experiment yielded several differences between the two probes, but one clone was particularly affected by the absence of Brn-3b. The clone, which encoded the growth-associated protein 43 (GAP-43), was significantly downregulated in the brn-3b (–/–) retina. The hybridization results were confirmed by quantitative RT–PCR with wild-type and brn-3b (–/–) RNAs (Fig. 4A) and by indirect immunofluorescent staining of wild-type and brn-3b (–/–) retina with an anti-GAP-43 antibody (Fig. 4B). Axons from wild-type explants showed intense GAP-43 expression whereas those from mutant explants showed little or no expression (Fig. 4B). These results implicate the gene encoding GAP-43 as a possible downstream target gene of Brn-3b.
Figure 4.
GAP-43 is downregulated in brn-3b (–/–) E14.5 retina. (A) RT–PCR with primers for brn-3b, GAP-43 and β-actin (as a normalization standard). (B) Staining of E13.5 retinal explants with anti-GAP-43 (green), propidium iodide (red) or neurofilament light chain (blue). +/+ Refers to retinas from wild-type mice and –/– to retinas from brn-3b (–/–) mice.
GAP-43 is associated with growth cones and has been suggested to be involved in axon growth (39). Axons from the optic nerve of GAP-43 knockout mice are aberrantly routed after they reach the optic chiasm (39–42). In comparison, in retinas of brn-3b (–/–) mice, most RGCs undergo apoptosis before the mice are born because of abnormal axon growth and other RGC defects (25,43). Thus, brn-3b (–/–) retinas display a more severe phenotype than those of GAP-43 knockout mice. However, like GAP-43-deficient RGCs, the residual RGCs from brn-3b (–/–) mice project axons that are abnormal (14). The surviving axons from RGCs of brn-3b (–/–) mice show defects in pathfinding at the optic chiasm, similar to the defects associated with the lack of GAP-43 (38). These results suggest that one aspect of Brn-3b’s function is to regulate the expression of genes associated with axon growth and pathfinding, and that GAP-43 represents one of the genes in this class. While GAP-43 expression is known to occur as neurons extend their axons, we have not yet established the precise timing of GAP-43 expression in RGC with respect to brn-3b expression. In addition, establishing GAP-43 as a direct downstream target of Brn-3b requires an analysis of the GAP-43 transcriptional control region. Large-scale gene expression profiling with the E14.5 retinal EST database will provide a more complete picture of the regulatory network that connects Brn-3b to the downstream, upstream and lateral events associated with the differentiation and survival of RGCs.
CONCLUSIONS
We have established an annotated database of retinal ESTs from the developing E14.5 mouse retina. To our knowledge, this is the first annotated database from developing mouse retinal tissue from which gene expression profiling and gene discovery experiments can be based. The database described here will complement existing databases that contain ESTs from the adult mouse retina (44). The database, which currently contains 15 268 ESTs, can be accessed through a web interface (http://odin.mdacc.tmc.edu/RetinalExpress) that allows for keyword and sequence homology searches. We continue to add to the database to approach our final objective of complete representation, which by our current estimation should approach 27 000 genes.
Through annotation, the retinal database provides a snapshot in time and space of the expressed gene set required for forming a functioning retina. Many retinal and eye specific regulatory genes in our database were not present in the widely used NIA 15K database, indicating the importance of creating mammalian EST databases from selective tissues. The wealth of available information within the database can now be used by interested investigators to obtain a more comprehensive picture of retinal development. The use of genetically engineered mice, in which deletion of individual regulatory genes perturb retinal development, can be linked to gene expression profiling with the complete E14.5 retinal transcriptome. In this way, global patterns of gene expression, altered by regulatory gene disruptions, can be assessed and interpreted in the context of gene regulatory networks. Changing patterns of gene expression can also be assessed in time and space, thereby providing a means to integrate genetic alterations with the dynamic events occurring in the retina as it forms during embryogenesis.
SUPPLEMENTARY MATERIAL
Supplementary material is available on NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by NEI grants (EY11930 and EY13523) and the Robert A. Welch Foundation. The University of Texas M. D. Anderson DNA Sequencing Facility is supported in part by NCI Cancer Center Support Grant CA16672.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed at: Department of Biochemistry and Molecular Biology, Box 117, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Boulevard, Houston, TX 77030, USA. Tel: +1 713 792 3646; Fax: +1 713 790 0329; Email: wklein@mdanderson.org BG799964–BG808997, BI985056–BI991757
REFERENCES
- 1.Livesey F.J. and Cepko,C.L. (2001) Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci., 2, 109–118. [DOI] [PubMed] [Google Scholar]
- 2.Koshiba-Takeuchi K., Takeuchi,J.K., Matsumoto,K., Momose,T., Uno,K., Hoepker,V., Ogura,K., Takahashi,N., Nakamura,H., Yasuda,K. et al. (2000) Tbx5 and the retinotectum projection. Science, 287, 134–137. [DOI] [PubMed] [Google Scholar]
- 3.Brown A., Yates,P.A., Burrola,P., Ortuño,D., Vaidya,A., Jessell,T.M., Pfaff,S.L., O’Leary,D.D. and Lemke,G. (2000) Topographic mapping from the retina to the midbrain is controlled by relative but not absolute levels of EphA receptor signaling. Cell, 102, 77–88. [DOI] [PubMed] [Google Scholar]
- 4.Neumann C.J. and Nusslein-Volhard,C. (2000) Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science, 289, 2137–2139. [DOI] [PubMed] [Google Scholar]
- 5.Cepko C.L. (1999) The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr. Opin. Neurobiol., 9, 37–46. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X.M. and Yang,X.J. (2001) Regulation of retinal ganglion cell production by Sonic hedgehog. Development, 128, 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pittack C., Grunwald,G.B. and Reh,T.A. (1997) Fibroblast growth factors are necessary for neural retina but not pigmented epithelium differentiation in chick embryos. Development, 124, 805–816. [DOI] [PubMed] [Google Scholar]
- 8.Hyer J., Mima,T. and Mikawa,T. (1998) FGF1 patterns the optic vesicle by directing the placement of the neural retina domain. Development, 125, 869–877. [DOI] [PubMed] [Google Scholar]
- 9.McCabe K.L. Gunther,E.C. and Reh,T.A. (1999) The development of the pattern of retinal ganglion cells in the chick retina: mechanisms that control differentiation. Development, 126, 5713–5724. [DOI] [PubMed] [Google Scholar]
- 10.Austin C.P., Feldman,D.E., Ida,J.A.,Jr and Cepko,C.L. (1995) Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development, 121, 3637–3650. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad I., Zagouras,P. and Artavanis-Tsakonas,S. (1995) Involvement of Notch-1 in mammalian retinal neurogenesis: association of Notch-1 activity with both immature and terminally differentiated cells. Mech. Dev., 53, 73–85. [DOI] [PubMed] [Google Scholar]
- 12.Dorsky R.I., Chang,W.S., Rappaport,D.H. and Harris,W.A. (1997) Regulation of neuronal diversity in the Xenopus retina by Delta signaling. Nature, 385, 67–70. [DOI] [PubMed] [Google Scholar]
- 13.Wang S.W., Kim,B.S., Ding,K., Wang,H., Sun,D., Johnson,R.L., Klein,W.H. and Gan,L. (2001) Requirement for math5 in the development of retinal ganglion cells. Genes Dev., 15, 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S.W., Gan,L., Martin,S.E. and Klein,W.H. (2000) Abnormal polarization and axon outgrowth in retinal ganglion cells lacking the POU-domain transcription factor Brn-3b. Mol. Cell. Neurosci., 16, 141–156. [DOI] [PubMed] [Google Scholar]
- 15.Liu W., Khare,S.L., Liang,X., Peters,M.A., Liu,X., Cepko,C.L. and Xiang,M. (2000) All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development, 127, 3237–3247. [DOI] [PubMed] [Google Scholar]
- 16.Liu W., Mo,Z. and Xiang,M. (2001) The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc. Natl Acad. Sci. USA, 98, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutcheson D.A. and Vetter,M.L. (2001) The bHLH factors Xath5 and XneuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev. Biol., 232, 327–338. [DOI] [PubMed] [Google Scholar]
- 18.Marquardt T., Ashery-Padan,R., Andrejewski,N., Scardigli,R., Guillermot,F. and Gruss,P. (2001) Pax6 is required for the multipotent state of retinal progenitor cells. Cell, 105, 43–55. [DOI] [PubMed] [Google Scholar]
- 19.Harris W.A. (1997) Cellular diversification in the vertebrate retina. Curr. Opin. Genet. Dev., 7, 651–658. [DOI] [PubMed] [Google Scholar]
- 20.Cepko C.L., Austin,C.P., Yano,X., Alexiades,M. and Ezzedine,D. (1996) Cell fate determination in the vertebrate retina. Proc. Natl Acad. Sci. USA, 93, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 22.Richterich P. (1998) Estimation of errors in ‘raw’ DNA sequences: a validation study. Genome Res., 8, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Press W.H., Teukolsky,S.A., Vetterling,W.T. and Flannery,B.P. (1992) Fortran, The Art of Scientific Programming, 2nd Edn. Cambridge University Press, NY.
- 24.Lashkari D.A., DeRisi,J.L., McCusker,J.H., Namath,A.F., Gentile,C., Hwang,S.Y., Brown,P.O. and Davis,R.W. (1997) Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl Acad. Sci. USA, 94, 13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gan L., Wang,S.W., Huang,Z. and Klein,W.H. (1999) POU domain factor Brn-3b is essential for retinal ganglion cell differentiation and survival but not for initial cell fate specification. Dev. Biol., 210, 469–480. [DOI] [PubMed] [Google Scholar]
- 26.Kawai J., Shinagawa,A., Shibata,K., Yoshino,M., Itoh,M., Ishii,Y., Arakawa,T., Hara,A., Fukunishi,Y., Konno,H. et al. (2001) Functional annotation of a full-length mouse cDNA collection. Nature, 409, 685–690. [DOI] [PubMed] [Google Scholar]
- 27.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 28.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon.,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 29.Hogenesch J.B., Ching,K.A., Batalov,S., Su,A.I., Walker,J.R., Zhou,Y., Kay,S.A., Schultz,P.G. and Cooke,M.P. (2001) A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell, 106, 413–415 [DOI] [PubMed] [Google Scholar]
- 30.McWhirter J.R., Goulding,M., Weiner,J.A., Chun,J. and Murre,C. (1997) A novel fibroblast growth factor gene expressed in the developing nervous system is a downstream target of the chimeric homeodomain oncoprotein E2A-Pbx1. Development, 124, 3221–3232. [DOI] [PubMed] [Google Scholar]
- 31.Rattner A., Hsieh,J.C., Smallwood,P.M., Gilbert,D.J., Copeland,N.G., Jenkins,N.A. and Nathans,J. (1997) A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc. Natl Acad. Sci. USA, 94, 2859–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang J.T., Esumi,N., Moore,K., Li,Y., Zhang,S., Chew,C., Goodman,B., Rattner,A., Moody,S., Stetten,G. et al. (1999) Cloning and characterization of a secreted Frizzled-related protein that is expressed by the retinal pigmented epithelium. Hum. Mol. Genet., 8, 575–583. [DOI] [PubMed] [Google Scholar]
- 33.Sherr C.J. (1994) G1 phase progression: cycling on cue. Cell, 79, 551–555. [DOI] [PubMed] [Google Scholar]
- 34.Sicinski P., Donaher,J.L., Parker,S.B., Li,T., Fazeli,A., Gardner,H., Haslem,S.Z., Bronson,R.T., Elledge,S.J. and Weinberg,R.A. (1995) Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell, 82, 621–630. [DOI] [PubMed] [Google Scholar]
- 35.Fantl V., Stamp,G., Andrews,A., Rosewell,I. and Dickson,C. (1995) Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev., 9, 2364–2372. [DOI] [PubMed] [Google Scholar]
- 36.Ma C., Papermaster,D. and Cepko,C.L. (1998) A unique pattern of photoreceptor degeneration in cyclin D1 mutant mice. Proc. Natl Acad. Sci. USA, 95, 9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kargul G.J., Dudekula,D.B., Qian,Y., Lim,M.K., Jaradat,S.A., Tanaka,T.S., Carter,M.G. and Ko,M.S. (2001) Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nature Genet., 28, 17–18. [DOI] [PubMed] [Google Scholar]
- 38.Erkman L., Yates,P.A., McLaughlin,T., McEvilly,R.J., Whistenhunt,T., O’Connell,S.M., Krones,A.I., Kirby,M.A., Rappaport,D.H., Bermingham,J.R. et al (2000) A POU domain transcription factor-dependent program regulates axon pathfinding in the vertebrate visual system. Neuron, 28, 779–792. [DOI] [PubMed] [Google Scholar]
- 39.Strittmatter S.M., Fankhauser,C., Huang,P.L., Mashimo,H. and Fishman,M.C. (1995) Neuronal pathfinding is abnormal in mice lacking the neuronal growth cone protein GAP-43. Cell, 80, 445–452. [DOI] [PubMed] [Google Scholar]
- 40.Kruger K., Tam,A.S., Lu,C. and Sretavan,D.W. (1998) Retinal ganglion cell axon progression from the optic chiasm to initiate optic tract development requires autonomous function of GAP-43. J. Neurosci., 18, 5692–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sretavan D.W. and Kruger,K. (1998) Randomized retinal ganglion cell axon routing at the optic chiasm of GAP-43-deficient mice: association with midline recrossing and lack of ipsilateral axon turning. J. Neurosci., 18, 10502–10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F., Lu,C., Severin,C. and Sretavan,D.W. (2000) GAP-43 mediates retinal axon interaction with lateral diencephalon cells during optic tract formation. Development, 127, 969–980. [DOI] [PubMed] [Google Scholar]
- 43.Xiang M. (1998) Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev. Biol., 197, 155–169. [DOI] [PubMed] [Google Scholar]
- 44.Livesey F.J., Furukawa,T., Steffen,M.A., Church,G.M. and Cepko,C.L. (2000) Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr. Biol., 10, 301–310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.