Summary
Background
Afucosylated IgG1 responses have only been found against membrane-embedded epitopes, including anti-S in SARS-CoV-2 infections. These responses, intrinsically protective through enhanced FcγRIIIa binding, can also trigger exacerbated pro-inflammatory responses in severe COVID-19. We investigated if the BNT162b2 SARS-CoV-2 mRNA also induced afucosylated IgG responses.
Methods
Blood from vaccinees during the first vaccination wave was collected. Liquid chromatography-Mass spectrometry (LC-MS) was used to study anti-S IgG1 Fc glycoprofiles. Responsiveness of alveolar-like macrophages to produce proinflammatory cytokines in presence of sera and antigen was tested. Antigen-specific B cells were characterized and glycosyltransferase levels were investigated by Fluorescence-Activated Cell Sorting (FACS).
Findings
Initial transient afucosylated anti-S IgG1 responses were found in naive vaccinees, but not in antigen-experienced ones. All vaccinees had increased galactosylated and sialylated anti-S IgG1. Both naive and antigen-experienced vaccinees showed relatively low macrophage activation potential, as expected, due to the low antibody levels for naive individuals with afucosylated IgG1, and low afucosylation levels for antigen-experienced individuals with high levels of anti-S. Afucosylation levels correlated with FUT8 expression in antigen-specific plasma cells in naive individuals. Interestingly, low fucosylation of anti-S IgG1 upon seroconversion correlated with high anti-S IgG levels after the second dose.
Interpretation
Here, we show that BNT162b2 mRNA vaccination induces transient afucosylated anti-S IgG1 responses in naive individuals. This observation warrants further studies to elucidate the clinical context in which potent afucosylated responses would be preferred.
Funding
LSBR1721, 1908; ZonMW10430012010021, 09150161910033, 10430012010008; DFG398859914, 400912066, 390884018; PMI; DOI4-Nr. 3; H2020-MSCA-ITN 721815.
Keywords: mRNA Vaccine, Antibodies, Glycosylation, Fucosylation, COVID-19
Research in context.
Evidence before this study
Antibodies are crucial for protective immunity, which depends on both the amount of IgG and on its Fc N-glycosylation. The conserved N-glycan at position N297 has a core structure that can be modified with a fucose, a bisecting N-acetylglucosamine, galactose residues and sialic acids. Both galactose and fucose have been described to modulate the activity of complement, or natural killer (NK) and myeloid cell IgG Fc gamma receptors (FcγR), respectively. Antibodies lacking core fucose (afucosylated IgG), have an increased affinity to the FcγRIII family. Afucosylated antibodies have been found against membrane-embedded epitopes in a wide range of infectious diseases such as dengue, malaria, and COVID-19, but also against alloantigens on platelets and red blood cells. As mRNA vaccines encode for spike protein with a transmembrane region, we investigated if these induce afucosylated responses.
Added value of this study
We show that afucosylated IgG responses are induced after BNT162b2 mRNA vaccination, which likely requires the expression of the target antigen on host cells. In the case of BNT162b2 mRNA vaccination, this surprisingly mirrors our findings in natural infection with SARS-CoV-2, with a transient afucosylation phenotype in the first two weeks after seroconversion.
Implications of all the available evidence
While this work shows that afucosylated IgG can be induced upon vaccination, it is also short-lived. Further work should elucidate the molecular requirements necessary to generate afucosylated IgG response and investigate their protective capacity and/or inflammatory potential. This could aid the production of next-generation vaccines able to induce stable afucosylated IgG responses to reach possible optimal protection in settings requiring efficient FcγRIII-mediated effector functions, such as malaria.
Introduction
Immunoglobulin G (IgG) antibodies (Abs) are crucial for protective immunity in coronavirus disease 2019 (COVID-19) through both fragment antigen binding (Fab)-mediated neutralization and fragment crystallizable (Fc)-mediated effector functions. The IgG Fc-mediated effector functions mainly depend on IgG subclass and Fc N-glycosylation, of which the latter has shown to correlate with COVID-19 disease exacerbation.1, 2, 3, 4 Human IgG contains a single, conserved biantennary N-linked glycan at N297 of the Fc portion. This N-glycan has a common pentasaccharide core that can further be modified with a fucose, a bisecting N-acetylglucosamine (GlcNAc), as well as one or two galactose residues, each of which can be capped by a sialic acid. Of these glycan residues, galactose and fucose have been described to modulate the activity of complement, or natural killer (NK) and myeloid cell IgG Fc gamma receptors (FcγR), respectively5, 6, 7 (Fig. 1a).
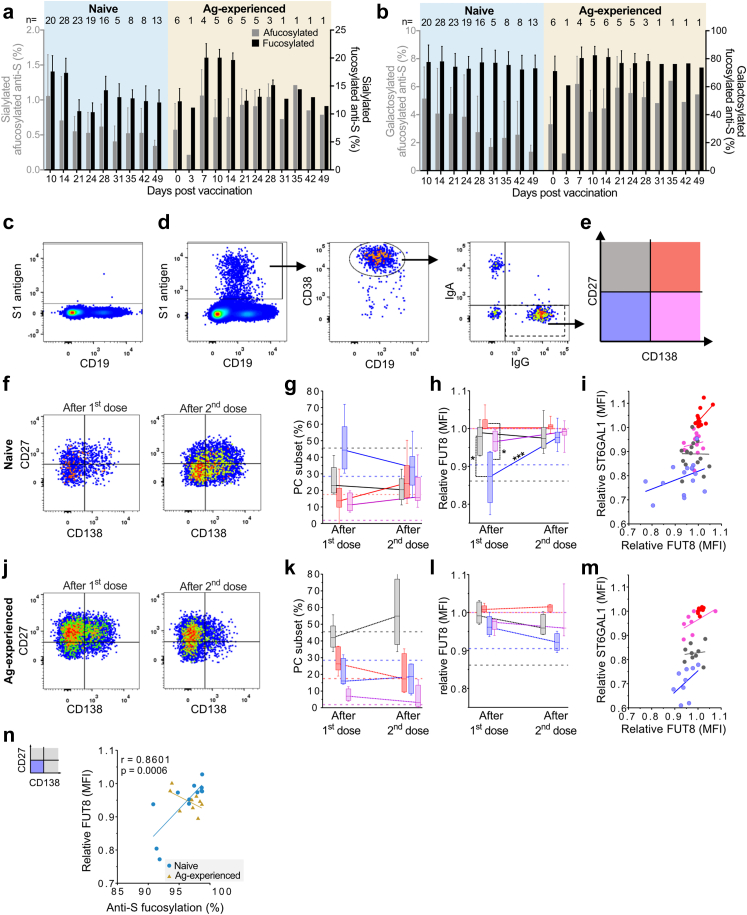
Fig. 1.
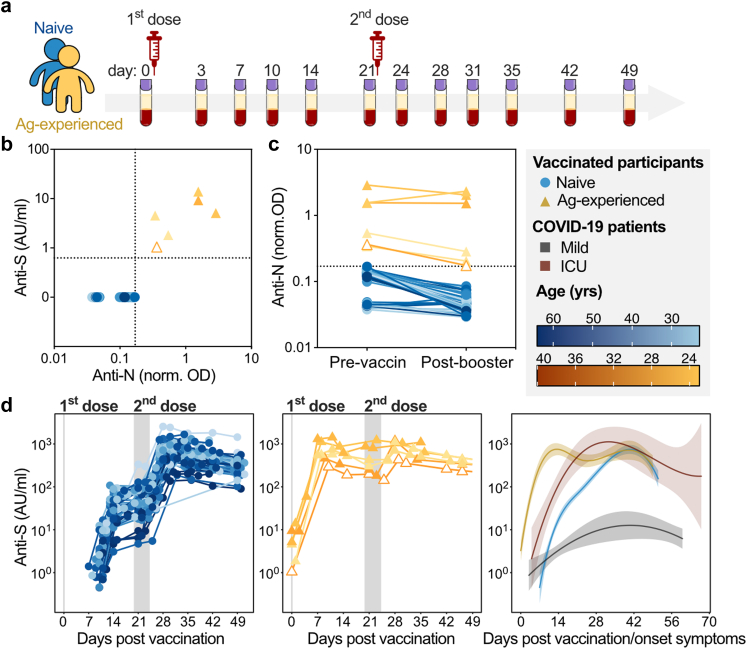
Naive and antigen-experienced individuals show divergent responses to the BNT162b2 mRNA vaccine. (a) Schematic depiction of extensive sampling of SARS-CoV-2 naive (blue) and antigen-experienced (yellow) prior to, and after the first and second dose of the BNT162b2 mRNA vaccine (b–d) SARS-CoV-2 naive (blue circles) and antigen-experienced vaccines (yellow, solid triangle: positive PCR test prior to the study, empty triangle: no positive PCR test prior to the study) IgG levels against (b) anti-spike (S) and anti-nucleocapsid (N) IgG levels prior to vaccination and (c) anti-nucleocapsid IgG levels during the sampling period. (d) Longitudinal anti-S IgG levels for naive (left, pooled cohort 1 (n = 33) and 2 (n = 9)) and antigen-experienced (middle, cohort 1 (n = 6) and 2 (n = 0)) vaccinees (detection limit ∼0.47 AU/ml) and corresponding dynamics in comparison to mild (grey) and ICU hospitalized (red) COVID-19 patients (right). No anti-S antibodies were found above detection levels before day ∼7 for naive vaccinees. Similar data for cohort 3 and 4 are plotted in Figs. S1 and S5, respectively.
Fc-galactosylation levels are highly variable (40–60%), with decreased levels being found in inflammatory diseases such as various infectious, cardiovascular, and autoimmune diseases as well as cancer.8, 9, 10, 11, 12 In contrast, increased Fc-galactosylation has been shown to characterize IgG after vaccination13,14 and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.2, 3, 4 Elevated Fc-galactosylation promotes IgG Fc–Fc interaction, leading to hexamerization, which enables docking of complement component 1q (C1q), the first component of the classical complement cascade, and ensuing complement activation.15,16
In healthy conditions, the majority of IgG found in plasma is fucosylated (∼94%),17,18 but afucosylated, antigen-specific IgG responses have been described in various pathologies, including alloimmune responses to blood cells,19, 20, 21 as well as immune responses to Plasmodium (P) falciparum antigens expressed on erythrocytes22 and to foreign proteins of enveloped viruses, including human immunodeficiency virus (HIV),23 dengue virus,24 and SARS-CoV-2.2,3 The common characteristic of such responses is that the corresponding pathogen-specific antigens are generally expressed on the host cell membrane, unlike most foreign antigens. Afucosylated IgG has an enhanced binding of up to ∼40 times to FcγRIII in comparison to its fucosylated counterpart. This results in increased cytokine production and cellular responses, such as Ab-dependent cellular phagocytosis (ADCP) and cytotoxicity (ADCC). These responses by far exceed the ∼40 times enhancement of binding affinity of afucosylated IgG to FcγRIII, presumably due to increased avidity between IgG-opsonized targets and FcγRIII-expressing effector cells.5, 6, 7,25 Intriguingly, pathogen-specific afucosylated IgG1 responses can be favourable, such as the protection seen in malaria22 and HIV,23 but can in turn cause massive inflammation via FcγRIII-mediated pathologies in patients with severe dengue fever,24 and has been shown to correlate with severe COVID-19.1,3,5,26 Full enhancement of this inflammatory response in COVID-19 also requires activation of various TLR members, contributing to triggering of a pro-inflammatory environment,27 including cytokine release such as IL-6, which is also only found systemically elevated in patients with severe COVID infections.1,2 In contrast, non-enveloped viruses, bacteria, and soluble protein-subunit vaccines, all lacking the host cell membrane context, induce virtually no afucosylated IgG responses. These include recombinant hepatitis B virus (HBV) and Plasmodium falciparum-proteins.2,22 On the contrary, when these proteins are expressed during natural infection on host cells, afucosylated IgG responses have been observed.2,22 This led us to the hypothesis that antigen presentation on the surface of host cells, possibly together with host co-factors, is required for the induction of afucosylated IgG responses.2 The new mRNA- and adenovirus-based SARS-CoV-2 vaccines induce host cell production of the SARS-CoV-2 spike (S) protein and subsequent presentation on the cell membrane, unlike traditional soluble protein-subunit vaccines.28 We hypothesize that similar to attenuated enveloped-viral vaccines,2 mRNA- and adenoviral-based vaccines might therefore also induce an afucosylated IgG response.
Here, we investigated anti-S IgG glycosylation in both naive and antigen-experienced participants after the first and second dose of the BNT162b2 mRNA vaccine against SARS-CoV-2, and show that mRNA vaccines are capable of inducing a transient afucosylated IgG response, which has clinical implications for vaccine-induced protection. Additionally, we evaluated glycosyltransferase expression in antigen-specific IgG+ plasma cell (PC) subsets to obtain insights into the generation of anti-S IgG glycosylation phenotypes. We furthermore studied the potential contribution of anti-S IgG afucosylation to inflammatory responses using an in vitro macrophage activation assay.
Methods
This study was designed to investigate the effect of the BNT162b2 BioNTech/Pfizer mRNA vaccine on anti-Spike IgG1 Fc glycosylation and PC subsets. We obtained serum, plasma and/or PBMC samples from vaccinated participants from 1) healthcare works at the Amsterdam UMC, The Netherlands (n = 39), 2) The Fatebenefratelli-Sacco Infectious Diseases Physicians Group (n = 9), 3) the University Medical Center of Schleswig-Holstein, Lübeck, Germany (n = 40), and 4) the Dutch blood bank Sanquin, the Netherlands. The discrimination between vaccinated SARS-CoV-2 naive and antigen-experienced participants was made by serology (anti-Spike and anti-Nucleocapsid IgG) and positive PCR-tests before vaccination. No other selection criteria were used and participants were selected at random.
Vaccination study cohorts and control individuals
Cohort 1. Amsterdam UMC cohort
Subjects were part of the S3 cohort study (S3 cohort; NL 73478.029.20, Netherlands Trial Register NL8645), a prospective serologic surveillance cohort study among hospital healthcare workers in the Amsterdam University Medical Center (Amsterdam UMC). Between January and March 2021, 39 cohort participants received their first dose of BioNTech/Pfizer mRNA vaccine (BNT162b2, 30 μg) (Table S2). A second dose was administered approximately 21 days after the first dose. Samples were obtained directly before and 3, 7, 10 and 14 days after the first dose, and directly before and 3, 7, 10, 14, 21 and 28 days after the second dose (Table S2).
Cohort 2. The Fatebenefratelli-Sacco Infectious Diseases Physicians Group
Nine healthcare workers at the Luigi Sacco Infectious Diseases Hospital, Milano, Italy were immunized with BioNTech/Pfizer mRNA vaccine (BNT162b2, 30 μg) and received a 2nd dose 21 days after the 1st dose. Blood samples were obtained directly before the 1st dose, and twice a week for six weeks from December 2020 to February 2021 (Table S3).
Cohort 3. Lübeck cohort
Forty subjects were recruited at the University Medical Center of Schleswig-Holstein, Lübeck, Germany from December 2020 (including samples (participants 1–22; Table S4) described in Lixenfeld et al.29): 1) 32 individuals immunized with the BioNTech/Pfizer vaccine BNT162b2 (30 μg) without or with known SARS-CoV-2 infection history (19 of these 32 individuals (analysed in Fig. S1) received the 2nd dose between day 32 and 37 after the 1st) and 2) and 8 unvaccinated individuals without SARS-CoV-2 infection history as negative control (Table S4).
Cohort 4. Convalescent plasma donors
Sanquin blood donors (n = 22) found seropositive for SARS-CoV-2 prior to vaccination were included in the study (Table S5). All participants provided written informed consent.
Ethics
All participants were included through informed consent and all studies were in accordance with the ethical principles set out in the declaration of Helsinki. The Cohort 1 study was approved by the Academic Medical Center Institutional Medical Ethics Committee of the Amsterdam UMC (reference number 2020.182). The Ethics Committee of the University of Lübeck, Germany approved the Cohort 3 study (reference number 19-019(A) and 20–123). The samples from Cohort 4 were collected only from voluntary, adult donors after written informed consent as part of routine donor selection and blood collection procedures. This study was approved by the Ethics Advisory Council of Sanquin Blood Supply Foundation, as described previously.30
Anti-SARS-Cov2 Ab levels
Cohort 1, 2 and 4
Anti-S IgG Abs levels were measured by coating MaxiSorp NUNC 96-well flat-bottom plates (Thermo Fisher Scientific, Roskilde, Denmark) overnight with 1 μg/ml recombinant, in-house produced trimerized spike protein in PBS, as described before.31 The following day, plates were washed five times with PBS supplemented with 0.02% polysorbate-20 (PBS-T) and incubated for 1 h with a dilution range of plasma from the Amsterdam UMC cohort in PBS-T supplemented with 0.3% gelatine (PTG). A serially diluted plasma pool, obtained by combining plasma from a collection of convalescent COVID-19 donors,32 was used as a calibrant. After incubation, plates were washed five times with PBS-T and incubated with 1 μg/ml anti-human IgG-horseradish-peroxidase (HRP) (clone: MH16.1, Sanquin, Amsterdam, the Netherlands). After washing, Ab binding was evaluated by adding 50% diluted tetramethylbenzidine substrate (1-step ultra TMB, #34029, Thermo Scientific). The reaction was terminated by adding equal amounts of 0.2 M H2SO4 (Merck, Darmstadt, Germany) and absorbance was measured at 450 and 540 nm. The calibrant plasma pool was assigned the value of 100 arbitrary units (AU), which corresponds to approximately 21 μg/ml.33
Anti-Receptor Binding Domain (RBD) and anti-Nucleocapsid (N) antibody levels were measured as by an RBD and N-based bridging assay, respectively, as described previously.32,33
Cohort 3
To detect anti-S1 IgG as well as anti-NCP IgG Abs, serum samples were collected on the indicated days (Table S4) and EUROIMMUN SARS-CoV-2 S1 IgG (EUROIMMUN, Luebeck, Germany; #EI 2606-9601-2 G) and EUROIMMUN SARS-CoV-2-NCP IgG (#EI 2606-9601-2 G) ELISA were performed according to the manufacturer's instructions, respectively.
To detect anti-S1 and -S2 IgG and IgG subclass (IgG1-4) Abs, 96-well ELISA plates were coated alternatively with 4 μg/ml of SARS-CoV-2-S1 (ACROBiosystems, Newark, DE 19711, USA; #S1N-C52H3) or -S2 (ACROBiosystems; #S2N-C52H5) antigen per well (HL-1 ELISA29). The plates were washed with PBS-T. Subsequently, sera (diluted 1/100 or 1/1000 in 0.05% Tween-20, 3% BSA in PBS) were added. Bound Abs were detected with HRP-coupled polyclonal goat anti-human IgG Fc (#A80-104P, RRID:AB_67064)-specific Abs purchased from Bethyl Laboratories (Montgomery, TX, USA), or monoclonal anti-human IgG1 (clone HP-6001, RRID:AB_2796627), IgG2 (clone HP-6014, RRID:AB_2796646), IgG3 (clone HP-6050, RRID:AB_2796699), or IgG4 (clone HP-6025, RRID:AB_2796691)-specific Abs purchased from Southern Biotech (Birmingham, AL, USA) in 0.05% Tween 20, 3% BSA in PBS. After incubation with the tetramethylbenzidin (TMB) substrate (BD Biosciences, San Diego, CA, USA) and terminating of the reaction with the addition of H2SO4, the optical density (OD) was measured at 450 nm. The specificities of the secondary Abs have been verified recently.29
IgG Fc glycosylation analysis by mass spectrometry
Anti-S IgG Abs were affinity-captured from plasma or sera using recombinant, in-house produced trimerized spike protein-coated plates (Thermo Fisher Scientific, Roskilde, Denmark) followed by a 100 mM formic acid elution step, as described elsewhere.2,31 Total IgG Abs were affinity-captured from plasma or sera using a Protein G AssayMAP Cartridge Rack on the Bravo (Agilent Technologies, Santa Clara, CA) or Protein G Sepharose 4 Fast Flow beads (GE Healthcare, Uppsala, Sweden) in a 96-well filter plate (Millipore Multiscreen, Amsterdam, Netherlands), respectively, as described elsewhere.2,34,35
Eluates from both anti-S and total IgG affinity-purification were dried by vacuum centrifugation and subjected to tryptic cleavage followed by LC-MS analysis as described previously.2,35
LC-MS data processing
Raw LC-MS spectra were converted to mzXML files. LaCyTools, an in-house developed software was used for the alignment and targeted extraction of raw data.36 Alignment was performed based on average retention time of at least three high abundant glycoforms. The analyte list for targeted extraction of the 2+ and 3+ charge states was based on manual annotation as well as on literature reports.2,37 Inclusion of an analyte for the final data analysis was based on quality criteria including signal-to-noise (higher than 9), isotopic pattern quality (less than 25% deviation from the theoretical isotopic pattern), and mass error (within a ±20 parts per million range) leading to a final analyte list (Table S9). Relative intensity of each glycan species in the final analyte list was calculated by normalizing to the sum of their total areas. Normalized intensities were used to calculate fucosylation, bisection, galactosylation and sialylation (Table S10).
Complement ELISAs
Pierce™ Nickel Coated Clear 96-well plates (Thermo Fisher Scientific, #15442) were incubated with 100 μL of 1 μg/ml purified RBD-protein for 1 h at RT. Hereafter, the plates were washed five times with 0.05% PBS-Tween20 and incubated with 100 μL glycoengineered COVA1-18 (2C1) hIgG1 mAbs for 1 h at RT.1,31 A two-fold dilution series was used, with a starting concentration of 20 μg/ml. Subsequently the plates were washed and 100 μL of 1:35 pooled human serum in Veronal Buffer5 with 0.1% poloxamer 407, 2 mM MgCl2 and 10 mM CaCl2 was added and incubated for 1 h at RT, as described previously.15 Consequently, the plates were washed and 100 μL 1/1000 anti-C1q-HRP38, 39, 40 was added and incubated for 1 h at RT. Lastly, the plates were washed and developed with 100 μL 0.1 mg/ml TMB solution with 0.11 M NaAc and 0.003% H2O2. The reaction was terminated with 100 μL 2 M H2SO4 and the absorbance was measured using the Biotek Synergy™ 2 Multi-Detection Microplate Reader at 450–540 nm.
The binding capacity of the in house glycoengineered COVA1-18 (2C1) hIgG1 mAbs was tested by directly coating Nunc MaxiSorp flat-bottom 96-well plates (Thermo Fisher Scientific) O/N at 4 °C with 100 μL 1 μg/ml purified SARS-CoV-2 RBD-protein. The plates were washed with PBS-T and incubated with 100 μL glycoengineered COVA1-18 (2C1) hIgG1 mAbs for 1 h at RT. A two-fold dilution series was used, with a starting concentration of 1 μg/ml. Hereafter, the plates were washed and incubated with 100 μL of 1/1000 Mouse Anti-Human IgG Fc-HRP (Southern-Biotech) for 1 h at RT. Lastly, the plates were washed and developed with TMB solution. The reaction was terminated with 2 M H2SO4 and the absorbance was measured using at 450–540 nm.
IL-6 ELISA
Supernatants of stimulated alveolar-like monocyte-derived macrophages (MDMs) were harvested after 24 h to determine cytokine production. IL-6 levels in the supernatant were measured by enzyme-linked immunosorbent assay (ELISA) using IL-6 CT205-c and CT205-d antibody pair (U-CyTech, Utrecht, the Netherlands) as described previously.1
Alveolar-like monocyte-derived macrophage differentiation
Buffy coats from healthy donors were obtained from Sanquin Blood Supply (Amsterdam, the Netherlands). Monocytes were isolated from buffy coat by density gradient centrifugation using Lymphoprep™ (Axis-Shield, Dundee, Scotland) followed by CD14+ selection via magnetic cell separation using MACS CD14 MicroBeads and separation columns (Miltenyi Biotec, Bergisch Gladbach, Germany), as previously described.41 Alveolar-like MDMs were generated by differentiating CD4+ monocytes on tissue culture plates into macrophages in the presence of 50 ng/ml of human M-CSF (Miltenyi Biotec, Bergisch Gladbach, Germany) for 6 days, followed by 24-h incubation in culture medium supplemented with 50 ng/ml IL10 (R&D System, Minneapolis, MN, USA). The resulting MDMs were then detached for stimulation using TrypLE Select (Gibco, Waltham, MA).
Cell stimulation
96-well high affinity plates were coated with 2 μg/ml soluble perfusion stabilized Spike protein as described previously.1 After overnight incubation, plates were blocked with 10% FCS in PBS for 1 h at 37 °C. Diluted heat-inactivated serum (Table S1, 1:50 dilution) was added for 1 h at 37 °C. 50,000 cells/well were stimulated in the pre-coated plates in culture medium (Iscoves's Modified Dulbecco's Culture Medium (IMDM) (Gibco) containing 5% FBS (Capricorn Scientific, Ebsdorfergrund, Germany) and 86 μg/ml gentamicin (Gibco) without or supplemented with 20 μg/ml polyinosinic:polycytidylic acid (poly(I:C)) (Sigma–Aldrich, Darmstadt, Germany).
Flow cytometric analysis of blood samples
Blood samples were collected at the indicated days in EDTA-tubes and processed or frozen within the next 3 h for flow cytometric analysis (Attune Nxt; Thermo Fisher Scientific) of different B cell populations.29 Peripheral blood mononuclear cells (PBMCs) were obtained by gradient centrifugation in Ficoll. The following fluorochrome-coupled Abs were used for surface staining: anti-CD19 (Biolegend; clone HIB19), anti-CD38 (Biolegend: HIT2), anti-IgG Fc (Biolegend; M1310G05), anti-CD27 (Biolegend: 0323) and anti-CD138 (Biolegend: MI15) as well as LIVE/DEAD Fixable Near-IR stain (Thermofisher; L34976). For additional intracellular staining, samples were fixed with Cytofix/Cytoperm according to the manufacturer's instructions (BD Biosciences) followed by permeabilization (0.05% saponin, 0.1% BSA in 0.05 × PBS) and additional staining with anti-IgG, anti-human ST6GAL1 (R&D Systems, polyclonal goat IgG Ab; #AF5924, RRID:AB_2044637), or isotype goat control IgG (R&D Systems), or anti-human FUT8 (R&D Systems, polyclonal sheep IgG; #AF5768, RRID:AB_2105499), or isotype sheep control IgG (R&D Systems), as well as SARS-CoV-2-S1 (biotin-coupled; Acro; #S1N-C82E8) and fluorochrome-coupled streptavidin (Biolegend). The anti-ST6GAL1 and anti-FUT8 Abs were labelled with Alexa Fluor 488 labelling kit (Life Technologies GmbH; #A20181). 20 million cells were recorded per sample. Flow Cytometry Standard (FCS) 3.0 files were analysed with FlowJo software version X 0.7 (BD Biosciences).
Statistical analysis
For the demographic's comparison of the different cohorts, a one-way ANOVA with Tukey's correction was carried out (Table S6). To compare the naive and antigen-experienced vaccinees per cohort, a t-test with Welch's correction and a Chi-square test was carried out for sex and age, respectively (Table S7).
To test differences in anti-S antibody fucosylation between naive and antigen-experienced vaccinees in cohort 1 (Fig. 2g), normality was tested by D'Agostino and a Wilcoxon matched pairs signed-rank test and a paired t-test was carried out for naive and antigen-experienced vaccinees, respectively. Anti-S IgG1 fucosylation between naive and antigen-experienced was compared by Kolmogorov–Smirnov test. For cohort 4 (Fig. 2g), a paired t-test was carried out following normality testing (D'Agostino test). For the sensitivity analysis adjusting for age, a stratified analysis by age subgroup (naïve ≤ 40y/o, naïve >40y/o, antigen-experienced ≤ 40y/o) was carried out with ordinary one-way ANOVA (Table S8).
Fig. 2.
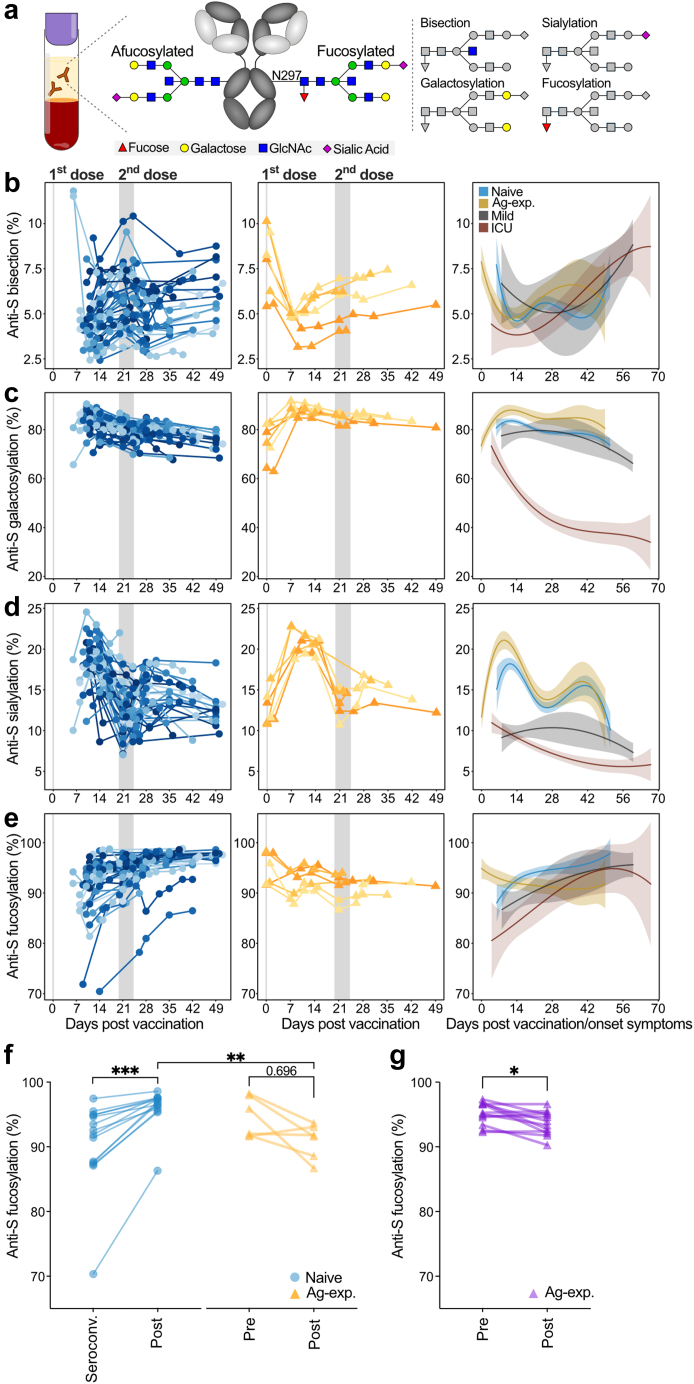
Anti-Spike IgG1 glycosylation is dynamic. (a) Schematic depiction of IgG from plasma or serum and its conserved N-glycan at position N297 with bisection, galactosylation, sialylation and fucosylation glycosylation traits displayed. Longitudinal anti-S IgG1 Fc (b) bisection, (c) galactosylation, (d) sialylation, and (e) fucosylation for naive (left, blue, cohort 1 (n = 33) and 2 (n = 9)), antigen-experienced (middle, yellow, pooled cohort 1 (n = 6) and 2 (n = 0)) in comparison to mild (grey) and ICU-admitted (red) COVID-19 patients' (right) anti-S IgG1 from our previous study.2 Similar data for cohort 3 are plotted in Fig. S2. Anti-S IgG1 fucosylation (f) of naive (blue, cohort 1, n = 14) vaccinees at seroconversion and after the second dose (Post) compared by Wilcoxon matched pairs signed rank test, and antigen-experienced (yellow, cohort 1, n = 6) before vaccination (Pre) and after the first dose (Post) compared by paired t-test. Comparison between naive and antigen-experienced vaccinees was carried out by Kolmogorov–Smirnov test. (g) Antigen-experienced (purple, cohort 4, n = 12) before (Pre) and after (Post) the first dose compared by paired t-test. Normality was tested by D'Agostino. ∗,∗∗,∗∗∗: p-value <0.05, 0.01 and 0.001, respectively.
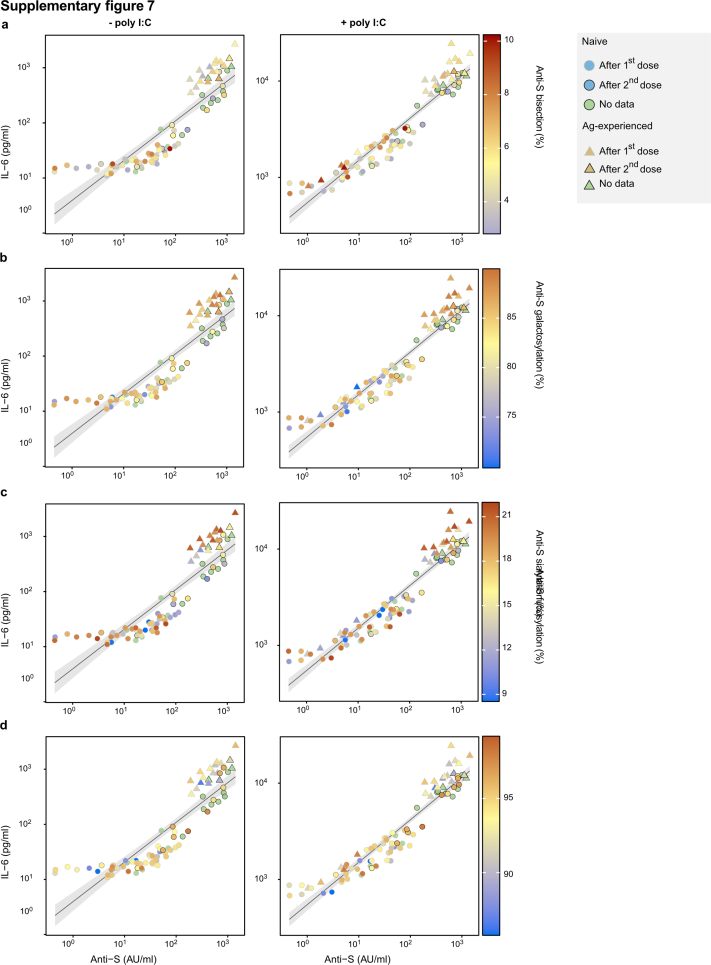
The log10 values of the anti-spike IgG levels (Fig. 3d) were used for the correlation analyses between log10 values of the measured concentrations of IL-6 (in pg/ml). The percentages of anti-S IgG1 glycosylation traits were used for the color overlay (Fig. S7). The Pearson correlation coefficients (R) and associated p-values are stated in each graph. For the comparison of the IL-6 concentration produced by alveolar-like macrophages, an unpaired t-test was performed after normality (log-normal distribution) was confirmed by D'Agostino. The correlations between anti-S IgG1 level and afucosylation (Fig. 5, Fig. S13) was determined by Pearson's correlation for datasets following normality testing (D'Agostino test). Otherwise, Spearman's correlation was performed. For adjusting clustering by cohort, mixed effect models, using cohort as random effect, were applied to obtain adjusted p-values and correlation coefficients.
Fig. 3.
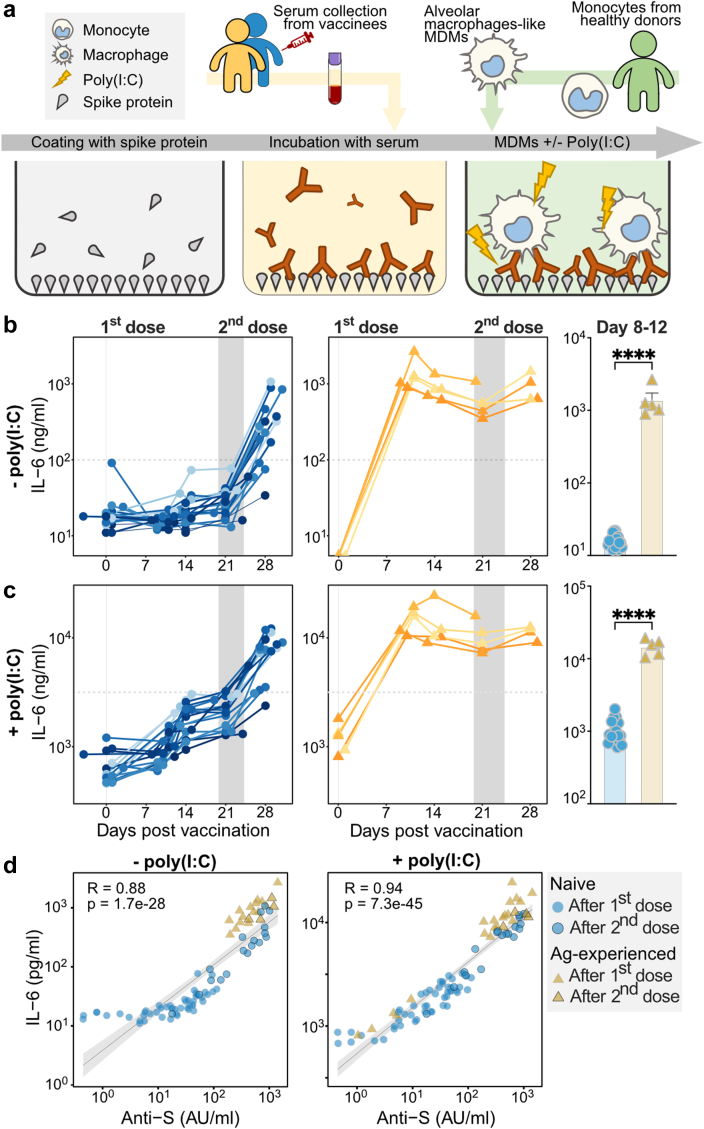
Antibody levels are primarily responsible for macrophage activation. (a) Schematic representation of the alveolar-like monocyte-derived macrophages stimulation assay with and without polyinosinic:polycytidylic acid (poly(I:C)), a double-stranded RNA analogue and TLR3 ligand. (b–c) IL-6 responses of macrophages stimulated with spike protein and naive (left, blue) and antigen-experienced (middle, yellow) vaccinee sera and a comparison of IL-6 levels for naive and antigen-experienced vaccinees around day 10 (day 8–12) by unpaired t-test with SD error bars (right) in the (b) absence or (c) presence of poly(I:C). The horizontal dashed lines indicate the IL-6 production by anti-spike monoclonal IgG1 COVA1-1831 at a concentration that represents 100 μg/mL. The COVA1-18-Fc IgG1 was 97.8% fucosylated and 19.6% galactosylated. ∗∗∗∗: p-value < 0.0001, respectively (d) Correlation between IL-6 levels and anti-S IgG levels in the absence (left) and presence (right) of poly(I:C) stimulation. All data represent a subgroup of cohort 1 (n = 23, see Table S2) and the correlation analysis was performed within only the subgroup of cohort 1.
Fig. 5.
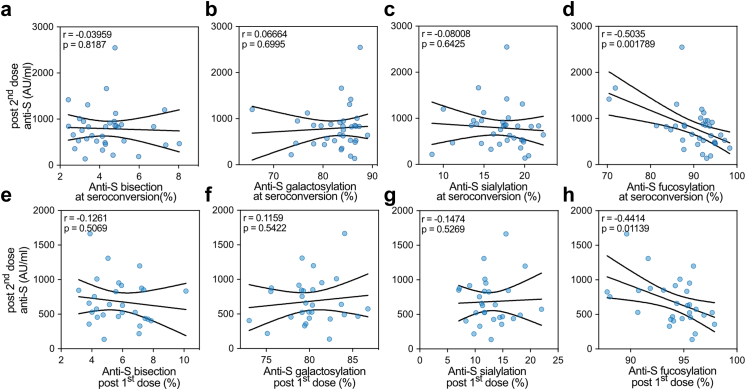
Afucosylation of anti-S IgG1 correlates with titer after the second dose. Correlation of anti-S IgG levels (highest levels after booster up to two weeks after 2nd dose) for naive vaccines from pooled cohort 1 (n = 33) and 2 (n = 9) with (a–d) anti-S IgG1 Fc glycosylation after seroconversion or (e–h) three weeks after the first dose (Post 1st dose). The correlation analysis was performed by pooling cohorts 1 and 2 and a mixed model was applied. Pearson's correlation was performed for datasets following normality (D'Agostino test). Otherwise, Spearman correlation was performed.
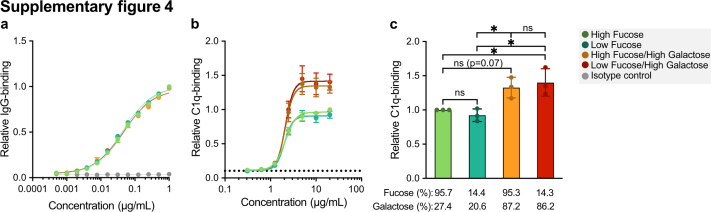
For the complement activation capacity comparison (Fig. S4) a curve fitting was performed using nonlinear regression dose–response curves with log(agonist) versus response-variable slopes. Bar graphs were visually tested for normality as n = 3, and a one-way ANOVA with Tukey's multiple-comparison test was carried out.
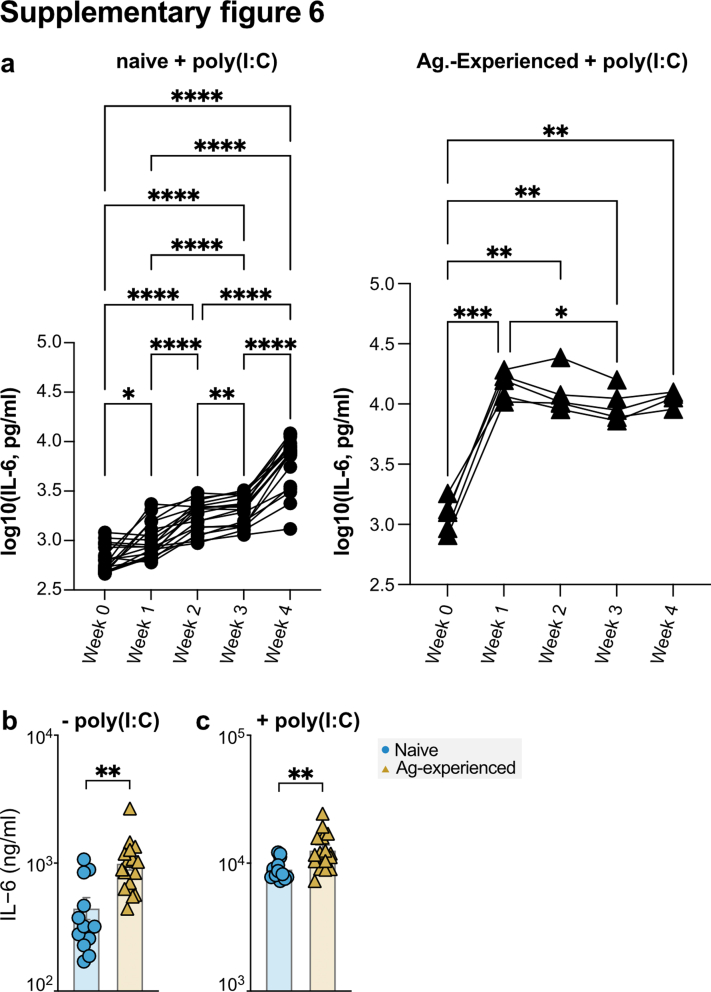
To compare IL-6 production at different time points (Fig. S6a), a repeated measures ANOVA on log-normalized levels was carried out after normality (log-normal distribution) was confirmed by D'Agostino. To compare naive and antigen-experienced vaccinees, an unpaired t-test was carried out after normality testing was carried out by D'Agostino (Fig. S6b).
In Fig. 5 and Fig. S13, cohort 1 and 2 were pooled for the analysis, as these individuals followed the same vaccine regimen, setup and sampling timeline. Otherwise, no cohorts were pooled within any analysis and each analysis was carried out within its specific cohort. These analyses were performed in either the R statistical environment (v3.6.3) or using GraphPad Prism v6.0 (GraphPad, La Jolla, CA). p-values < 0.05 were considered as significant. Asterisks indicate the degree of significance as follows: ∗, ∗∗, ∗∗∗, ∗∗∗∗: p-value < 0.05, 0.01, 0.001, 0.0001, respectively.
Role of funders
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
BNT162b2 mRNA vaccination induces transient afucosylated anti-S IgG in naive, but not antigen-experienced individuals
To analyse the immune response in naive and antigen-experienced individuals upon vaccination with the mRNA vaccine BNT162b2, blood samples were collected from healthy donors at four locations: the Amsterdam University Medical Center (UMC) in the Netherlands, the Fatebenefratelli-Sacco University Hospital in Milan in Italy, the University Medical Center of Schleswig-Holstein Lübeck in Germany, and the Dutch blood bank Sanquin in the Netherlands. As individuals in cohort 1 and 2 followed the same Pfizer vaccination regimen and sampling timeline, these cohorts were displayed together in the figures. Vaccinees participated voluntarily in this study and no prior calculation of power was carried out. Neither were individuals selected based on demographics. As a result, not all cohorts are comparable in sex and age (Table S6). However, we do believe these cohorts are representative of a healthy, adult population, slightly skew toward younger females due to the majority of these vaccinees being volunteer healthcare workers (Table 1, Fig. 1a and Tables S2–S5).
Table 1.
Cohort description.
| Cohort 1 | Cohort 2 | Cohort 3 | Cohort 4 | |
|---|---|---|---|---|
| Participants (n) | 39 | 9 | 32 | 22 |
| Of which Ag-experienced | 6 (15.38%) | 0 (0%) | 9 (28.13%) | 22 (100%) |
| Gender (female %) | 27 (69.23%) | 5 (55.55%) | 18 (56.25%) | – |
| Age | ||||
| Minimum (yrs) | 23 | 28 | 19 | 19 |
| Maximum (yrs) | 64 | 62 | 74 | 62 |
| Mean ± SD (yrs) | 41.38 ± 12.06 | 45.22 ± 13.95 | 35 ± 12.24 | 44.04 ± 13.01 |
To identify antigen-experienced individuals, anti-nucleocapsid (N) and anti-spike (S) IgG responses were investigated both prior to the first dose and during the study (Fig. 1b and c, Fig. S1). Five out of six of the antigen-experienced vaccinees had a positive PCR test before the study (Fig. 1b and c, Tables S2–S5). The detection limit for the anti-S ELISA was ∼0.47 AU/ml and vaccinated SARS-CoV-2 naive individuals showed a detectable anti-S IgG response around ten days after vaccination, which further increased upon the second dose (Fig. 1d and Fig. S1a, e, g). All vaccinated antigen-experienced individuals had anti-S IgG Abs before vaccination and levels increased fast upon the first dose of BNT162b2 (Fig. 1d and Fig. S1a and e). Antigen-experienced individuals presented a stronger humoral response after the first dose compared to naive vaccinees (Fig S1d), in line with the literature.42, 43, 44 Upon the second dose, both naive and antigen-experienced reached similar anti-S levels, which were dominated by IgG1 and IgG3 subclasses against both the S1 and S2 subunits of the S protein (Fig. S1h).3, 29 Within each cohort, SARS-CoV-2 naive and antigen-experienced vaccinees were comparable in sex, but not in age (Table S7).
To study the different responses between vaccinees and naturally infected individuals, we compared the vaccine-induced responses with the dynamics of a previously described cohort of mild and intensive care unit (ICU)-admitted COVID-19 patients. In this naturally infected cohort, the patients dynamics were based on their self-reported first day of onset, as described in detail by Larsen et al.2 (Fig. 1d, right panel).
Next, we explored anti-S and total IgG1 Fc N-glycosylation patterns over time (Fig. 2 and Figs. S2 and S3). The IgG glycoprofiling of all four cohorts was carried out using identical methods and data processing. Upon vaccination, the anti-S IgG1 levels of naive vaccinees were too low to determine IgG1 Fc glycoprofiles. Around day 10, in concurrence with rising anti-S IgG levels, IgG1 Fc glycoprofiles for naive individuals could be determined. In both naive and antigen-experienced individuals, an initial drop of anti-S IgG1 bisection levels were seen, which increased over time and dropped again upon the 2nd dose. These levels were lowered as compared to total IgG1 (Fig. 2b and Figs. S2a and S3a). An early response of highly galactosylated and sialylated anti-S IgG1 was observed in both naive and antigen-experienced individuals, both after the first and second dose (Fig. 2c and d and Fig. S2b and c, Fig. S3b and c). The anti-S IgG1 galactosylation level and time course were similar to what we previously observed in naturally infected individuals with mild symptoms. In contrast, anti-S IgG1 galactosylation dropped rapidly in ICU-admitted COVID-19 patients, as previously described (Fig. 2c).2,35 The antigen-specific galactosylation levels were remarkably increased (up to ∼20%) compared to total IgG levels. Increased IgG galactosylation is known to enhance classical complement pathway activation through enhanced hexamerization and thereby enhanced C1q-binding. To study the enhanced complement activation capacity of highly galactosylated anti-S, we produced anti-S mAb COVA1-18 (2C1) hIgG1 with and without increased galactosylation in combination with fucosylation.8,15,16,45,46 Indeed, increased C1q binding was seen for anti-S mAbs with higher galactosylation (Fig. S4), which is in line with previous reports.15 Anti-S IgG1 sialylation followed the galactosylation trend, with an increase after the first and second dose (Fig. 2d, Fig. S2c). Interestingly, anti-S IgG1 galactosylation and sialylation followed a reverse course compared to bisection.
We recently hypothesized that afucosylated IgG, hardly seen in responses to soluble protein or polysaccharide antigens, is specifically induced against foreign antigens on host cells.2 In agreement with this, up to 25% of anti-S IgG1 Fc was found to be afucosylated after vaccination with the BNT162b2 SARS-CoV-2 mRNA vaccine, in comparison to ∼6% of afucosylated total IgG1 generally found in serum or plasma (Fig. 2e and Figs. S2d and S3d). This pronounced afucosylation pattern was observed only early on in naive individuals after the first dose of BNT162b2 and decreased significantly to levels similar to total IgG1 at two to three weeks post seroconversion (Wilcoxon matched pairs signed rank test, p < 0.001) (Fig. 2e and f and Figs. S2d and S3d). This early, transient afucosylated response in naive vaccinees after the first dose was less prominent when compared to ICU-admitted COVID-19 patients and most individuals with mild symptoms (Fig. 2e). In contrast, antigen-experienced individuals had an initial anti-S IgG1 afucosylation level of ∼2–10%, and an increasing afucosylated trend was observed after vaccination (Paired t-test, p = 0.696) (Fig. 2e and f and Fig. S2e). As age differed between naive and antigen-experienced vaccinees in cohort 1 (Table S7), a sensitivity analysis was carried out (Table S8). As all antigen-experienced vaccinees in cohort 1 were under 40 years old, we stratified by age subgroups; age >40 and ≤40. When comparing the age difference between naive ≤40 and the antigen-experienced group, no difference was found (p = 0.86, unpaired t-test with Welch's correction), making these groups comparable. Furthermore, no significant difference was found for fucosylation in the naive subgroups >40 and ≤40 years old, showing that fucosylation was not influenced by age. When comparing the fucosylation levels of naive vaccinees after two immunization doses (post) with the levels of antigen-experienced vaccinees after two immunizations, being natural infection followed up with one vaccine dose (post), a significantly lowered anti-S IgG1 Fc fucosylation was shown in antigen-experienced vaccinees (Kolmogrov–Smirnov test, p > 0.0015) (Fig. 2g).
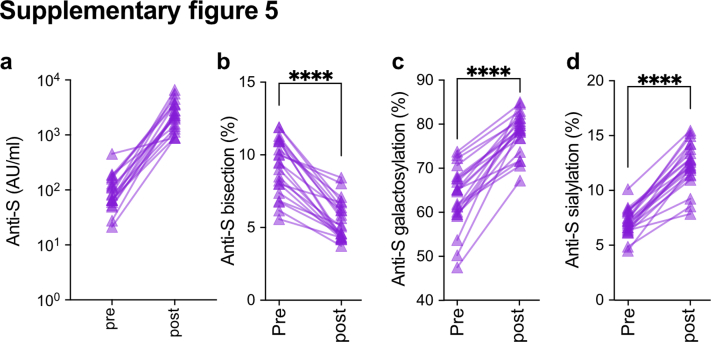
As the number of antigen-experienced vaccinees in cohort 1 was limited, we included a second antigen-experienced vaccinee group (cohort 4) to confirm the increasing afucosylation trend. Cohort 4 were blood donors of the Dutch blood bank who were previously infected with SARS-CoV-2, received one vaccine dose, and donated plasma for convalescent plasma therapy for COVID-19 patients (Fig. S5, Table S5). This antigen-experienced cohort also showed a significant drop in anti-S IgG1 Fc bisection (Wilcoxon matched-pairs signed-rank test, p < 0.0001), increased galactosylation (Wilcoxon matched-pairs signed-rank test, p < 0.0001) and sialylation (Wilcoxon matched-pairs signed rank test, p < 0.0001) after vaccination (post) (Fig. S5). As observed for the antigen-experienced vaccinees in cohort 1, a significant decrease in anti-S IgG1 Fc fucosylation was likewise seen for cohort 4 (paired t-test, p < 0.05) (Fig. 2g). Although the initial cohort (cohort 1) and this antigen-experienced cohort (cohort 4) were comparable in age, no information was available for sex in cohort 4. However, both sex and adolescent age have not been shown to influence antibody fucosylation,47 allowing such an inter-cohort comparison.
No temporal changes were observed for total IgG glycosylation (Figs. S2 and S3). Importantly, antigen-experienced individuals confirmed by PCR and/or serology all showed the same characteristic both in antibody levels and glycosylation traits, which clearly differentiated them from those identified as SARS-CoV-2 naive.
Impact of afucosylated anti-S IgG is limited because of low antibody levels
We assessed the effector function of the anti-S Abs induced by BNT162b2 mRNA vaccination by testing their capacity to induce macrophage-driven inflammatory responses. For this, we measured IL-6 production by human-derived, in vitro differentiated, alveolar-like macrophages. Stable FcγR surface expression was previously shown by Hoepel et al.1 These alveolar-like macrophages were stimulated overnight by exposure to immune complexes (ICs) generated from S protein and vaccinees’ sera in the presence and absence of virus-like co-stimuli (poly(I:C)), a double-stranded RNA analogue and TLR3 ligand.41 The anti-S monoclonal IgG1 COVA1-1831 was included as a control (Fig. 3a). Notably, anti-S ICs from antigen-experienced individuals induced significantly higher IL-6 levels compared to naive individuals for all time points after the first dose, with the most pronounced difference seen at day 10 (Fig. 3b, Fig. S6a). IL-6 induction was similar for both groups after the second dose as anti-S IgG levels became comparable (Figs. 1d and 3b and Fig. S6). Despite the clear difference between both groups, IL-6 levels were relatively low for all conditions, which is in line with various previous findings showing that IgG ICs only induce pro-inflammatory cytokine production in the presence of both high afucosylation and antibody levels in the presence of viral or bacterial co-stimulus that activates receptors such as TLRs.1,48,49 In comparison, the IL-6 levels here produced by the macrophages is an order of magnitude higher than the IL-6 produced in serum of SARS-CoV-2 infected people, with a maximum of around 100 pg/mL in the serum of severely ill COVID-19 patients.1
To further test the inflammatory capacity of anti-S IgG, we also measured IL-6 production upon TLR co-stimulation with the TLR3 ligand poly(I:C). Upon TLR co-stimulation, anti-S ICs strongly amplified IL-6 production by human macrophages in both groups (Fig. 3c). Again, the difference between the vaccinated naive and antigen-experienced individuals was most pronounced around day ten post vaccination (Fig. 3c). In both cases, the capacity of the sera to activate these macrophages seem to be explained by antibody levels (Fig. 3d). However, when anti-S IgG levels became comparable after the second dose, the sera of antigen-experienced individuals induced only slightly higher IL-6 levels both with (unpaired t-test, R = 0.88, p = 1.7e−28) and without (unpaired t-test, R = 0.94, p = 7.3e−45) poly(I:C) (Fig. 1, Fig. 3b–d and Fig. S6), which correlated with higher afucosylation levels, but not with other IgG1 glycosylation traits (Fig. S7). Our data indicate that the transient afucosylation of anti-S IgG that is produced after vaccination of naive individuals does not promote strong macrophage activation because of the concomitant low antibody levels.
Differential plasma cell responses in naive and antigen-experienced individuals
In line with the literature, a highly sialylated IgG response was observed early after vaccination, regardless of prior SARS-CoV-2 infection (Fig. 2c and Figs. S2c and S3c).14,50,51 Interestingly, these early, transient, highly sialylated anti-S were particularly fucosylated, for both naive and antigen-experienced vaccinees until day fourteen after the first dose (Fig. 1, Fig. 3a and Fig. S8). Anti-S IgG1 Fc galactosylation levels of neither naive nor antigen-experienced showed a difference between fucosylated and afucosylated anti-S IgG1 (Fig. 1, Fig. 3b and Fig. S8). This result led us to the hypothesis that early highly galactosylated and sialylated anti-S IgG1 might be produced by different PC subsets than afucosylated anti-S IgG1.
To investigate this, we analysed the anti-S1 blood-derived IgG+ CD38+ PC subset responses to assess whether they phenotypically diverge in their anti-S IgG1 glycosylation pattern.52, 53, 54 We found CD27low CD138- IgG+ CD38+ PCs to be dominant in naive individuals after both the first and second dose (Fig. 4c–g, Figs. S9a–g and S10a–f). In contrast, antigen-experienced vaccinees primarily induced CD27+ CD138- IgG+ CD38+ PCs after both doses (Fig. 4j and k, Figs. S9a–i and S10a–f), which was also the dominant subset in total IgG+ PCs of the naive, unvaccinated controls (Figs. S10a–f, S11).
Fig. 4.
Different populations of B cells express distinct levels of glycosyltransferases. (a) Sialylation and (b) galactosylation levels of all afucosylated (grey) and all fucosylated (black) anti-S IgG for naive (left, blue shading) and antigen-experienced (right, yellow shading) vaccinated participants over time of cohort 1 (n = 39) and 2 (n = 9) pooled. All afucosylated glycoforms were set to 100%, and all fucosylated glycoforms were set individually to 100% (c–n) Flow cytometry analysis of blood cells gated on single, living lymphocytes from naive and antigen-experienced vaccinees (subset of cohort 3 (n = 15), see Table S2) were analysed 7–14 days upon the first (naive: n = 6 and antigen-experienced: n = 5) or 5–8 days upon the second (naive: n = 15 and antigen-experienced: n = 4) dose (c–e) Gating strategy exemplified for a naive individual (c) pre-immunization and (d) after the first dose. S1-reactive B cells were gated and further gated for CD19int CD38+ PCs to analyse IgG+ PC subsets as defined by (e) CD27 and CD138 (f–g) Naive and (j–k) antigen-experienced vaccinees analysed according to the gating strategy (h, l) Relative fucosyltransferase 8 (FUT8; median fluorescence intensity (MFI)) expression per IgG+ PC subset compared by Mann–Whitney U test and (i, m) its correlation with relative α2,6-sialyltransferase (ST6GAL1) expression (n) Spearman correlation of relative FUT8 expression of CD27low CD138- IgG+ PCs correlated with anti-S IgG1 Fc fucosylation found in the corresponding serum and statistics of naive vaccinees (Fig. S9c). The median (MFI) of FUT8 or ST6GAL1 expression in CD138+ IgG+ S1-reactive PCs of each sample was set to 1 for inter-assay comparison. Dotted horizontal lines indicate corresponding values of total IgG+ PC subsets from untreated healthy controls (Fig. S11). ∗, ∗∗∗: p-value <0.05, 0.001, respectively. Each analysis in c–n was carried out within only cohort 3.
We found that α1,6-fucosyltransferase 8 (FUT8; the glycosyltransferase responsible for core fucosylation55) protein expression level was lowest in the CD27low CD138- IgG+ PC subset in naive individuals after the first, but not the second dose (Fig. 4h and i, l and m and Fig. S10h). The α2,6-sialyltransferase 1 (ST6GAL1; the glycosyltransferase responsible for α2,6-linked sialylation51) protein expression level was the highest in the CD27+ CD138+ and the lowest in CD27low CD138- IgG+ PC subset after both doses in naive and antigen-experienced individuals, as well as in total IgG+ PCs of unvaccinated healthy control individuals (Fig. 4i and m and Figs. S10g and S11). In naive individuals, FUT8 protein expression levels in CD27low CD138- IgG+ PCs correlated significantly (Spearman, r = 0.8061, p = 0.0006) with anti-S IgG1 fucosylation levels obtained by LC-MS (Fig. 4n).
Early afucosylated anti-S correlates with anti-S IgG titer upon the second dose
A thorough examination of individual study participants revealed that vaccinees with high initial afucosylated anti-S IgG1 often showed high anti-S IgG levels (Fig. S12). To study this possible correlation, we selected the anti-S IgG1 Fc glycosylation both at seroconversion (Fig. 5a–d and Fig. S13b–e) and after the first dose. For this, we chose the timepoint three weeks after vaccination (post 1st dose) (Fig. 5e–h and Fig. S13f–i). This was correlated with the anti-S IgG levels after the first (Fig. S13b-i) and second dose (Fig. 5). For this, we again selected the anti-S IgG level three weeks after the first dose (Post 1st dose), and the highest level reached up to two weeks post the second dose (Post 2nd dose). Cohorts 1 and 2 were pooled for these correlations and mixed effects models56 using cohort as the random effect and estimated by maximum likelihood were used to adjust for cohort clusters in these correlations.
Anti-S IgG levels after the first dose correlated with the levels after the second dose (Spearman, r = 0.7936, p < 0.0001) (Fig. S13a). Anti-S IgG1 Fc afucosylation at seroconversion (Spearman, r = −0.4387, p = 0.0087) correlated with the anti-S IgG levels after the second dose (Fig. 5d). This was also the case for afucosylation levels at the moment before the second dose (Spearman, r = −0.3936, p = 0.0346) (Fig. 5h). Anti-S IgG1 Fc afucosylation at seroconversion correlated with anti-S IgG levels before the second dose (post 1st dose) (Spearman, r = −0.4273, p = 0.0117) (Fig. S13e). However, no correlation (Pearson, r = 0.1384, p = 0.4740) was found between anti-S IgG1 fucosylation levels and anti-S IgG levels at the day of the 1st dose (post 1st dose) (Fig. S13i). No correlations were found between anti-S IgG levels and the other anti-S IgG glycosylation traits (Fig. 5a–c, e–g, and Fig. S13b–d, f–h).
Discussion
The mRNA vaccine-induced presentation of the viral S protein on the membrane of host cells mimics the S protein presentation during natural SARS-CoV-2 infections. Enveloped viruses, attenuated enveloped viral vaccines, P. falciparum-infected erythrocytes and alloantigens on blood cells express their antigens on host cells and induce persistent afucosylated IgG responses.2,21, 22, 23, 24,57 Afucosylated IgG has enhanced binding to its receptor FcγRIII, which is expressed on myeloid and NK cells. This results in increased cytokine production and cellular responses such as ADCP and ADCC. Recently, we established a method coined Fucose-sensitive ELISA for Antigen-Specific IgG (FEASI) to measure antibody fucosylation by a functional proxy. This method utilizes the enhanced affinity of FcγRIIIa for antigen-specific IgG fucosylation and translates the in vivo affinity and avidity enhancement into functional receptor binding properties.58 The readout of this functional assay correlated directly with fucosylation values obtained by MS, linking the MS-obtained antibody glyco-profiling with functional differences in FcγRIIIa binding.58 Pro-inflammatory responses of afucosylated IgG are seen in FcγRIII-mediated pathologies in patients with severe dengue fever24 and alloimmunity.20 Afucosylated antibody levels furthermore correlated with disease severity of COVID-19 patients.1, 2, 3,59 However, pathogen-specific afucosylated IgG responses seem to be protective in HIV infections23 and malaria.22 Protective functions can also be attributed to afucosylated antibodies in SARS-CoV-2 settings, as shown in NK cell-mediated SARS-CoV-2 ADCC in vaccinated and naturally infected donors.26,60,61 Here, ADCC peaked around day 11–20, coinciding with the highest levels of afucosylation seen for both SARS-CoV-2 and naturally infected donors. This emphasizes the enhanced functionality of these afucosylated antibodies, which are drivers of ADCC. Interestingly, higher ADCC was seen in severe COVID-19 patients who recovered as compared to deceased patients. Furthermore, SARS-CoV-2 infected monocytes and macrophages in COVID-19 patients have been found to go through a process of antibody-opsonized virus uptake through FcγR, which leads to inflammatory death (pyroptosis).62,63 Afucosylated antibodies could facilitate this uptake through enhanced FcγR binding and subsequently diminished viral production. Taken together, these examples illustrate the double-edged sword functionality of antigen-specific afucosylated responses, driving the balance between pro- and anti-inflammatory responses.
Here, we studied anti-S induced by the BNT162b2 mRNA vaccine. We show that the BNT162b2 mRNA vaccine induced afucosylated anti-S IgG1 responses in SARS-CoV-2 naive individuals upon seroconversion, which decreases within four weeks to the level of total IgG1. Recent work from Farkash et al. and Chakraborty et al. did not pick up this transient response due to limited sampling in time.59,64 This afucosylated response was similar but less pronounced than observed in natural SARS-CoV-2 infections.2,35 Surprisingly, this afucosylated response was transient, whereas previously long-lasting afucosylated antibodies were found in cytomegalovirus infections, malaria and alloimmunization to the red blood cell RhD antigen.2,22,57 The transient response seen upon SARS-CoV-2 mRNA vaccination suggests that a co-stimulus might be missing to induce memory B cells and long-lived IgG+ PCs producing stable anti-S IgG1 afucosylation levels. Alternatively, the lack of a local type of inflammatory signal might provide a negative feedback steering developing B cells to produce fucosylated IgG. In contrast, SARS-CoV-2 antigen-experienced vaccinees start off with low (∼2–10%) but persistent anti-S IgG1 afucosylation levels, which slightly increase upon vaccination, assuming re-activation of memory B cells generating afucosylated IgG antibodies. Next, we studied the macrophage-driven inflammatory responses of anti-S Abs induced by BNT162b2. Our study revealed that the differences in the effector functions elicited by anti-S ICs on macrophages between naive and antigen-experienced vaccinees mainly depends on the titer after the first dose, with afucosylation only being a secondary factor. Our previous work has shown that exaggerated pro-inflammatory responses were only observed with serum containing high titers of considerably afucosylated IgG1 (>10%).1,2 Such afucosylated IgG1 levels in this study were only observed early after seroconversion in naive individuals and not in combination with high titers. In line with this, anti-S ICs from vaccinee's sera induced very moderate pro-inflammatory cytokine levels in the absence of TLR co-stimulation, suggesting low inflammatory side effects in both groups after immunization with the BNT162b2 mRNA vaccine. Nevertheless, when comparing naive and antigen-experienced vaccinees after the second dose, when anti-S IgG levels were comparable, antigen-experienced individuals induced slightly higher IL-6 production in the macrophage activation assay, which is in agreement with their slightly increased levels of afucosylated IgG. Moreover, the different immune responses of naive versus antigen-experienced vaccinees were reflected in the different antigen-specific PC subsets found. Naive individuals primarily induced CD27low CD138- IgG+ CD38+ PCs, whereas antigen-experienced individuals primarily induced CD27+ CD138- IgG+ CD38+ PCs after both doses. Furthermore, the FUT8 protein expression level was reduced in naive vaccinees upon seroconversion in the CD27low CD138- IgG+ CD38+ PC subset. This FUT8 protein expression level correlated with the afucosylated IgG1 levels obtained by LC-MS observed in these individuals, thereby confirming that both the flow cytometry analysis of blood samples and LC-MS glyco-profiling confirm an early, transient afucosylated response upon vaccination in naive vaccinees.59
In accordance with previous reports on immunization14,50,51 and Fc glycosylation studies on BNT162b2 mRNA vaccination,59,64 we observed transiently, highly sialylated anti-S IgG1 one to two weeks after both the first and second dose, which has been suggested to facilitate antigen presentation in subsequent GC reactions for improving affinity maturation.14,65 Furthermore, the anti-S IgG1 for both naive and antigen-experienced vaccinees was extensively galactosylated. These high levels of IgG galactosylation showed to boost the capacity to activate the classical complement pathway, through enhanced C1q-binding. This was in line with recent findings, which showed that galactosylation promotes IgG1 hexamerization, ultimately leading to increased C1q-binding and ensuing classical complement activation.15,16,64 Interestingly, the initial drop in anti-S IgG1 bisection was inverse to the fucose levels and during the study the bisection levels were inverse to the galactosylation and sialylation levels. Previous work has shown that bisection has an inhibiting effect on core elongation and fucosylation.66,67 However, more recently, increasing IgG bisection in HEK cells did not influence IgG fucosylation levels. Furthermore, IgG bisection did not have an effect on FcγR or C1q binding, neither on its own or in combination with other glycan traits.5 We observed that early afucosylated anti-S IgG1 responses correlated significantly with anti-S IgG levels in naive individuals after the second dose.64 The mechanism behind inducing an afucosylated antigen-specific response and its subsequent consequences is unknown. One likely possibility is through a previously described FcγR-dependent mechanism, by enhanced antigen-uptake through IgG-immune complexes in professional antigen-presenting cells.68, 69, 70 Here, afucosylated IgG1 may facilitate antigen uptake, processing, and presentation through FcγRIIIa, by inducing better T-helper and therefore memory B cell responses during booster responses. At seroconversion, afucosylated anti-S IgG1 Abs in naive vaccinees might provide enhanced protection, even without high titers. Over time, when afucosylated anti-S IgG levels drop, protection in these individuals might be compensated by the increased anti-S IgG levels, which should be considered for the timing of subsequent vaccination. Furthermore, reduced levels of anti-S IgG1 afucosylation might reduce the risk of pro-inflammatory side effects, with a trade-off of dampened Fc-mediated effector functions upon pathogen contact. In antigen-experienced individuals, matters are reversed, as these individuals start off with high fucosylated anti-S IgG levels prior to vaccination, which significantly drop after vaccination. This suggests an enhanced corresponding memory B cell response, which would be in line with stronger protection in this group.71, 72, 73 Similarly, a gradual drop in fucosylation has been observed with repeated natural immunizations to antigens displayed on the membrane of P. falciparum-infected red blood cells.22 This is in contrast to alloimmunization to the red blood cell RhD antigen, where the afucosylated response in hyperimmune donors is very stable over time.20 The increased level of afucosylated anti-SARS-CoV2 IgG in vaccinated antigen-experienced individuals might have a positive impact on the therapeutic effect of convalescent plasma, especially as these donors are presently selected for clinical trials and it has been shown that increased ADCC activity of the administered antibodies positively correlates with outcome.
This was an exploratory study where we made use of samples available at the time provided voluntarily by frontline healthcare workers. Due to this, no prior sample size calculations were carried out. Together with the limited sample size, we could not stratify study participants according to sex and age. The four cohorts included in this work differ in sex and age. Furthermore, within the cohorts, SARS-CoV-2 naive and antigen-experienced vaccinees differed in age but not sex. It is known that demographics such as sex and age influence some IgG glycosylation traits.17 This is mainly the case for antibody galactosylation and both antigen-specific and total IgG1 have been shown to be confounded by sex and age.17,35,74 In contrast, total IgG fucosylation levels remain constant throughout life with the exception of an initial decrease after birth. No prior influence of gender and age on IgG fucosylation has been described.18,47 Stable afucosylated antibodies to RhD have been found to be stable for decades, independent of age.20 Furthermore, it has been recently shown that afucosylation levels of anti-S and total IgG1 are not influenced by age in natural SARS-CoV-2 infections.4 In this study, all cohorts combined spanned an age group from 19 to 74 years with a slight majority being female. Although we could not initially select our participants, we do believe they are representative of a healthy, adult population, which is overall slightly skewed towards younger females. Despite these differences, we observed similar longitudinal trends for both the anti-S titer and Fc glycosylation in all cohorts, with the exception of initial anti-S IgG1 afucosylation for naive but not antigen-experienced vaccinees. We believe this indicates that the influence of demographics on kinetics is minimal in this study. Another limitation of our study is the uneven sample size for naive and antigen-experienced vaccine recipients after the first and second dose of BNT162b2. Furthermore, it is theoretically possible that soluble, serum spike protein could lower the free anti-S IgG in our studies, however, it has been shown that spike protein is rapidly degraded in serum and becomes undetectable within ten days.75
In summary, our data demonstrate a qualitatively and quantitatively distinct IgG immune response between BNT162b2 mRNA vaccinated SARS-CoV-2 naive and antigen-experienced individuals. Transient afucosylated IgG1 responses were observed upon vaccination in naive individuals upon the first dose, which correlated with increased titer after the second vaccination. In contrast, antigen-experienced vaccinees had low levels of afucosylated anti-S, which slightly increased upon vaccination. The qualitatively distinct IgG1 glycosylation patterns might further mediate differences in protection between these two groups. Future efforts focused on studying antigen-specific, afucosylated IgG1 responses are needed to investigate their protective capacity and/or inflammatory potential in anti-viral and vaccine-induced immunity.
Contributors
Conceptualization: GV, MW, ME, JDD, APJV, ECVDS, JVC, JR; Methodology: GV, MW, WW, JDD, JN, ME, JVC, TP, HJC, JR, ASL, TLJvO, JSB; Formal analysis: JVC, TP, JR, HJC, TLJvO, TS; Investigation: JVC, TP, JR, JSB, ASL, TS, MS, WW, JN, HJC, IK, TLJvO, ELM, SL, VvK, CK, HBL; Resources: FL, RV, CG, APJV, LAVV, WH, MAS, RM, TG, MKB, JJS, Fatebenefratelli-Sacco Infectious Diseases Physicians, and the UMC COVID-19 S3/HCW study group; Data curation: JVC, TP, JR, HJC, JSB, TLJvO; Writing – Original draft: GV, MW, ME, ECVDS, JVC, TP, JR; Writing – Review and editing: GV, MW, ME, JD, ECVDS, JVC, TP, JR, HJC, CG, LAV, JSB, TS, TLJvO, MS, WH, WW, ASL, JN, SK, FL, RV, ML, ELM, IK, SL, VvK, CK, HBL, MDW, NVM, TR, RG, MAS, RM, APJV, Fatebenefratelli-Sacco Infectious Disease Physicians, and the UMC COVID-19 S3/HCW study group; Visualization: JVC, HJC, JSB, TP, TLJvO, TS; Supervision: GV, MW, ME, JVD; Project administration: GV, MW, ME, JVC; Funding acquisition: GV, MW, ME, JVD; Verified underlying data: TP, JR, HJC, MW, ME and GV. All authors had access to all data and read and approved the final version of the manuscript.
Data sharing statement
The datasets generated for this study are available on request form the corresponding authors.
UMC COVID-19 S3/HCW study group
Brent Appelman1, Diederik van de Beek2, Marije K Bomers3, Justin de Brabander1, Matthijs C Brouwer2, David TP Buis3, Nora Chekrouni2, Marit J van Gils4, Menno D de Jong4, Ayesha HA Lavell3, Niels van Mourik5, Sabine E Olie2, Edgar JG Peters3, Tom DY Reijnders1, Michiel Schinkel1, Alex R Schuurman1, Jonne J Sikkens3, Marleen A Slim5, Yvo M Smulders3, Alexander PJ Vlaar5, Lonneke A van Vught1,5, Joost Wiersinga3
1 Center for Experimental and Molecular Medicine, Amsterdam Infection & Immunity, Amsterdam University Medical Center, Amsterdam, The Netherlands
2 Neurologie, Amsterdam Neuroscience, Amsterdam University Medical Center, Amsterdam, The Netherlands
3 Interne geneeskunde, Amsterdam University Medical Center location VUmc, Amsterdam, The Netherlands
4 Medical Microbiology & Infection prevention, Amsterdam Infection & Immunity, Amsterdam University Medical Center, Amsterdam, The Netherlands
5 Intensive care medicine, Amsterdam Infection & Immunity, Amsterdam University Medical Center, Amsterdam, The Netherlands
Fatebenefratelli-Sacco infectious diseases physicians group
Spinello Antinori1,2, Cinzia Bassoli1,2, Giovanna Bestetti2, Mario Corbellino2, Alice Covizzi3, Angelica Lupo2, Laura Milazzo2, Marco Schiuma3, Alessandro Torre2
1 Department of Clinical Sciences, Fatebenefratelli, Fatebenefratelli-Sacco Hospital, Milan, Italy
2 III Division of Infectious Diseases, Fatebenefratelli-Sacco Hospital, Milan, Italy
3 Department of Clinical Sciences, Fatebenefratelli-Sacco Hospital, Milan, Italy
Declaration of interests
The authors declare that they have no conflicts of interest.
Acknowledgements
We thank the Academic Medical Centre of the University of Amsterdam, the Sanquin Blood Supply Foundation and The Fatebenefratelli-Sacco Infectious Diseases Physicians Group. We are greatly indebted to all cohort participants for their extensive participation.
Funding: Landsteiner Foundation for Blood Transfusion Research (LSBR) grants 1721 and 1908 (GV) ZonMw COVID-19 grants 1043001 201 0021 (GV). Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) 398859914 (EH 221/10-1); 400912066 (EH 221/11-1); and 390884018 (Germany's Excellence Strategies - EXC 2167, Precision Medicine in Chronic Inflammation (PMI)) (ME). Federal State Schleswig-Holstein, Germany (COVID-19 Research Initiative Schleswig-Holstein; DOI4-Nr. 3) (ME). European Union's Horizon 2020 research and innovation program H2020-MSCA-ITN grant agreement number 721815 (MW/TP). Netherlands Organisation for Health Research and Development ZonMw & the Amsterdam UMC Corona Research Fund (Amsterdam UMC COVID-19 S3/HCW study group). The Netherlands Organisation for Health Research and Development ZonMW VENI grant, Grant number 09150161910033 (LAVV). ZonMW COVID-19 grant 10430012010008 (JDD)
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104408.
Contributor Information
Manfred Wuhrer, Email: M.wuhrer@lumc.nl.
Marc Ehlers, Email: Marc.ehlers@uksh.de.
Gestur Vidarsson, Email: G.Vidarsson@sanquin.nl.
Fatebenefratelli-Sacco Infectious Diseases Physicians group:
Spinello Antinori, Cinzia Bassoli, Giovanna Bestetti, Mario Corbellino, Alice Covizzi, Angelica Lupo, Laura Milazzo, Marco Schiuma, and Alessandro Torre
UMC COVID-19 S3/HCW study group:
Brent Appelman, Diederik van de Beek, Marije K. Bomers, Justin de Brabander, Matthijs C. Brouwer, David T.P. Buis, Nora Chekrouni, Marit J. van Gils, Menno D. de Jong, Ayesha H.A. Lavell, Niels van Mourik, Sabine E. Olie, Edgar J.G. Peters, Tom D.Y. Reijnders, Michiel Schinkel, Alex R. Schuurman, Jonne J. Sikkens, Marleen A. Slim, Yvo M. Smulders, Alexander P.J. Vlaar, Lonneke A. van Vught, and Joost W. Wiersinga
Appendix A. Supplementary data
Fig. S1.
Fig. S2.
Fig. S3.
Fig. S4.
Fig. S5.
Fig. S6.
Fig. S7.
Fig. S8.
Fig. S9.
Fig. S10.
Fig. S11.
Fig. S12.
Fig. S13.
References
- 1.Hoepel W., Chen H.-J., Geyer C.E., et al. High titers and low fucosylation of early human anti-SARS-CoV-2 IgG promote inflammation by alveolar macrophages. Sci Transl Med. 2021;13(596) doi: 10.1126/scitranslmed.abf8654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen M.D., de Graaf E.L., Sonneveld M.E., et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371(6532) doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty S., Gonzalez J., Edwards K., et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol. 2019;22(1):67–73. doi: 10.1038/s41590-020-00828-7. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pongracz T., Vidarsson G., Wuhrer M. Antibody glycosylation in COVID-19. Glycoconj J. 2022:1–10. doi: 10.1007/s10719-022-10044-0. https://link.springer.com/article/10.1007/s10719-022-10044-0 [cited 2022 Feb 11] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekkers G., Treffers L., Plomp R., et al. Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc-receptor- and complement-mediated-effector activities. Front Immunol. 2017;8:877. doi: 10.3389/fimmu.2017.00877. https://pubmed.ncbi.nlm.nih.gov/28824618/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shields R.L., Lai J., Keck R., et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277(30):26733–26740. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]
- 7.Ferrara C., Grau S., Jäger C., et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA. 2011;108(31):12669–12674. doi: 10.1073/pnas.1108455108. www.pnas.org/lookup/suppl/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekkers G., Rispens T., Vidarsson G. Novel concepts of altered immunoglobulin G galactosylation in autoimmune diseases. Front Immunol. 2018;9:553. doi: 10.3389/fimmu.2018.00553. www.frontiersin.org Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudelj I., Lauc G., Pezer M. Immunoglobulin G glycosylation in aging and diseases. Cell Immunol. 2018;333:65–79. doi: 10.1016/j.cellimm.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Menni C., Gudelj I., MacDonald-Dunlop E., et al. Glycosylation profile of immunoglobulin G is cross-sectionally associated with cardiovascular disease risk score and subclinical atherosclerosis in two independent cohorts. Circ Res. 2018;122(11):1555–1564. doi: 10.1161/CIRCRESAHA.117.312174. https://pubmed.ncbi.nlm.nih.gov/29535164/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartsch Y.C., Rahmöller J., Mertes M.M.M., et al. Sialylated autoantigen-reactive IgG antibodies attenuate disease development in autoimmune mouse models of Lupus nephritis and rheumatoid arthritis. Front Immunol. 2018;9(JUN):1183. doi: 10.3389/fimmu.2018.01183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clauder A.K., Kordowski A., Bartsch Y.C., et al. IgG Fc N-glycosylation translates MHCII haplotype into autoimmune skin disease. J Invest Dermatol. 2021;141(2):285–294. doi: 10.1016/j.jid.2020.06.022. https://pubmed.ncbi.nlm.nih.gov/32653301/ Available from: [DOI] [PubMed] [Google Scholar]
- 13.Selman M.H.J., De Jong S.E., Soonawala D., et al. Changes in antigen-specific IgG1 Fc N-glycosylation upon influenza and tetanus vaccination. Mol Cell Proteomics. 2012;11(4):1–10. doi: 10.1074/mcp.M111.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T.T., Maamary J., Tan G.S., et al. Anti-HA glycoforms drive B cell affinity selection and determine influenza vaccine efficacy. Cell. 2015;162(1):160–169. doi: 10.1016/j.cell.2015.06.026. https://pubmed.ncbi.nlm.nih.gov/26140596/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Osch T.L.J., Nouta J., Derksen N.I.L., et al. Fc galactosylation promotes hexamerization of human IgG1, leading to enhanced classical complement activation. J Immunol. 2021;207(6):1545–1554. doi: 10.4049/jimmunol.2100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei B., Gao X., Cadang L., et al. Fc galactosylation follows consecutive reaction kinetics and enhances immunoglobulin G hexamerization for complement activation. MAbs. 2021;13(1) doi: 10.1080/19420862.2021.1893427. https://www.tandfonline.com/doi/abs/10.1080/19420862.2021.1893427 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baković M., Selman H., Hoffmann H., et al. High-throughput IgG Fc N-glycosylation profiling by mass spectrometry of glycopeptides. J Proteome Res. 2013;12(2):821–831. doi: 10.1021/pr300887z. https://pubmed.ncbi.nlm.nih.gov/23298168/ Available from: [DOI] [PubMed] [Google Scholar]
- 18.De Haan N., Reiding K.R., Driessen G., Van Der Burg M., Wuhrer M. Changes in healthy human IgG Fc-glycosylation after birth and during early childhood. J Proteome Res. 2016;15(6):1853–1861. doi: 10.1021/acs.jproteome.6b00038. https://pubmed.ncbi.nlm.nih.gov/27161864/ Available from: [DOI] [PubMed] [Google Scholar]
- 19.Sonneveld M.E., Koeleman C.A.M., Plomp H.R., Wuhrer M., van der Schoot C.E., Vidarsson G. Fc-Glycosylation in human IgG1 and IgG3 is similar for both total and Anti-Red-Blood cell anti-k antibodies. Front Immunol. 2018;9(JAN):31. doi: 10.3389/fimmu.2018.00129. www.frontiersin.org Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapur R., Valle L Della, Sonneveld M., et al. Low anti-RhD IgG-Fc-fucosylation in pregnancy: a new variable predicting severity in haemolytic disease of the fetus and newborn. Br J Haematol. 2014;166:936–945. doi: 10.1111/bjh.12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuhrer M., Porcelijn L., Kapur R., et al. Regulated glycosylation patterns of IgG during alloimmune responses against human platelet antigens. J Proteome Res. 2009;8(2):450–456. doi: 10.1021/pr800651j. https://pubmed.ncbi.nlm.nih.gov/18942870/ . Available from: [DOI] [PubMed] [Google Scholar]
- 22.Larsen M.D., Lopez-Perez M., Dickson E.K., et al. Afucosylated Plasmodium falciparum-specific IgG is induced by infection but not by subunit vaccination. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-26118-w. https://www.biorxiv.org/content/10.1101/2021.04.23.441082v1 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ackerman M.E., Crispin M., Yu X., et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J Clin Invest. 2013;123(5):2183–2192. doi: 10.1172/JCI65708. https://pubmed.ncbi.nlm.nih.gov/23563315/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bournazos A.S., Thi H., Vo M., et al. Antibody fucosylation predicts disease severity in secondary dengue infection. Science. 2021;372(6546):1102–1105. doi: 10.1126/science.abc7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Temming R., de Taeye S., de Graaf E., et al. Functional attributes of antibodies, effector cells, and target cells affecting NK cell–mediated antibody-dependent cellular cytotoxicity. J Immunol. 2019;203(12):3126–3135. doi: 10.4049/jimmunol.1900985. http://www.jimmunol.org/content/203/12/3126 Available from: [DOI] [PubMed] [Google Scholar]
- 26.Larsen M.D., de Graaf E.L., Sonneveld M.E., et al. Afucosylated immunoglobulin G responses are a hallmark of enveloped virus infections and show an exacerbated phenotype in COVID-19. BioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.05.18.099507v1 2020.05.18.099507. Available from: [Google Scholar]
- 27.Mabrey F.L., Morrell E.D., Wurfel M.M. TLRs in COVID-19: how they drive immunopathology and the rationale for modulation. Innate Immun. 2021;27(7–8):503. doi: 10.1177/17534259211051364. Available from:/pmc/articles/PMC8762091/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinz F.X., Stiasny K. Distinguishing features of current COVID-19 vaccines: knowns and unknowns of antigen presentation and modes of action. NPJ Vaccines. 2021;6(1):1–13. doi: 10.1038/s41541-021-00369-6. https://www.nature.com/articles/s41541-021-00369-6 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lixenfeld A.S., Künsting I., Martin E.L., et al. The BioNTech/Pfizer vaccine BNT162b2 induces class-switched SARS-CoV-2-specific plasma cells and potential memory B cells as well as IgG and IgA serum and IgG saliva antibodies upon the first immunization. medRxiv. 2021 https://www.medrxiv.org/content/10.1101/2021.03.10.21252001v1 2021.03.10.21252001Available from: [Google Scholar]
- 30.van den Hurk K., Merz E.M., Prinsze F.J., et al. Low awareness of past SARS-CoV-2 infection in healthy plasma donors. Cell Rep Med. 2021;2(3) doi: 10.1016/j.xcrm.2021.100222. https://pubmed.ncbi.nlm.nih.gov/33681828/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brouwer P.J.M., Caniels T.G., van der Straten K., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. doi: 10.1126/science.abc5902. http://science.sciencemag.org/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelzang E.H., Loeff F.C., Derksen N.I.L., et al. Development of a SARS-CoV-2 total antibody assay and the dynamics of antibody response over time in hospitalized and nonhospitalized patients with COVID-19. J Immunol. 2020;205(12):3491–3499. doi: 10.4049/jimmunol.2000767. https://www.jimmunol.org/content/205/12/3491 Available from: [DOI] [PubMed] [Google Scholar]
- 33.Steenhuis M., van Mierlo G., Derksen N.I.L., et al. Dynamics of antibodies to SARS-CoV-2 in convalescent plasma donors. Clin Transl Immunology. 2021;10(5) doi: 10.1002/cti2.1285. Available from:/pmc/articles/PMC8126762/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Falck D., Jansen B.C., de Haan N., Wuhrer M. Methods in molecular biology. Humana Press; 2017. High-throughput analysis of IgG Fc glycopeptides by LC-MS; pp. 31–47. [DOI] [PubMed] [Google Scholar]
- 35.Pongracz T., Nouta J., Wang W., et al. Immunoglobulin G1 Fc glycosylation as an early hallmark of severe COVID-19. EBioMedicine. 2021 doi: 10.1016/j.ebiom.2022.103957. https://www.medrxiv.org/content/10.1101/2021.11.18.21266442v1 2021.11.18.21266442. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jansen B.C., Falck D., De Haan N., et al. LaCyTools: a targeted liquid chromatography-mass spectrometry data processing package for relative quantitation of glycopeptides. J Proteome Res. 2016;15(7):2198–2210. doi: 10.1021/acs.jproteome.6b00171. [DOI] [PubMed] [Google Scholar]
- 37.Pucić M, Knezević A, Vidic J, et al. High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol Cell Proteomics. 2011;10(10) doi: 10.1074/mcp.M111.010090. https://pubmed.ncbi.nlm.nih.gov/21653738/ M111.010090. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGrath F.D.G., Brouwer M.C., Arlaud G.J., Daha M.R., Hack C.E., Roos A. Evidence that complement protein C1q interacts with C-reactive protein through its globular head region. J Immunol. 2006;176(5):2950–2957. doi: 10.4049/jimmunol.176.5.2950. [DOI] [PubMed] [Google Scholar]
- 39.Hack C.E., Paardekooper J., Smeenk R.J., Abbink J., Eerenberg A.J., Nuijens J.H. Disruption of the internal thioester bond in the third component of complement (C3) results in the exposure of neodeterminants also present on activation products of C3. An analysis with monoclonal antibodies. J Immunol. 1988;141(5):1602–1609. [PubMed] [Google Scholar]
- 40.Jtd Leito, Ligtenberg A.J.M., van Houdt M., van den Berg T.K., Wouters D. The bacteria binding glycoprotein salivary agglutinin (SAG/gp340) activates complement via the lectin pathway. Mol Immunol. 2011;49(1–2):185–190. doi: 10.1016/j.molimm.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Chen H.-J., Li Yim A.Y.F., Griffith G.R., et al. Meta-analysis of in vitro-differentiated macrophages identifies transcriptomic signatures that classify disease macrophages in vivo. Front Immunol. 2019 Dec 11;0:2887. doi: 10.3389/fimmu.2019.02887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatatos L., Czartoski J., Wan Y.H., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372(6549):1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tauzin A., Nayrac M., Benlarbi M., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29(7):1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. http://www.cell.com/article/S1931312821002791/fulltext Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. https://www.nature.com/articles/s41586-021-03777-9 Available from: [DOI] [PubMed] [Google Scholar]
- 45.Van Osch T.L.J., Oosterhoff J.J., Bentlage A.E.H., et al. Fc galactosylation of anti-platelet hIgG1 alloantibodies enhance complement activation on platelets. Haematologica. 2022 doi: 10.3324/haematol.2021.280493. https://pubmed.ncbi.nlm.nih.gov/35354253/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peschke B., Keller C.W., Weber P., Quast I., Lünemann J.D. Fc-galactosylation of human immunoglobulin gamma isotypes improves C1q binding and enhances complement-dependent cytotoxicity. Front Immunol. 2017;8(JUN) doi: 10.3389/fimmu.2017.00646. https://pubmed.ncbi.nlm.nih.gov/28634480/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oosterhoff J.J., Larsen M.D., van der Schoot C.E., Vidarsson G. Afucosylated IgG responses in humans—structural clues to the regulation of humoral immunity. Trends Immunol. 2022;43(10):800–814. doi: 10.1016/j.it.2022.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geyer C.E., Mes L., Newling M., Den Dunnen J., Hoepel W. Physiological and pathological inflammation induced by antibodies and pentraxins. Cells. 2021;10(5) doi: 10.3390/cells10051175. https://pubmed.ncbi.nlm.nih.gov/34065953/ Internet. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vogelpoel L.T.C., Hansen I.S., Rispens T., et al. Fc gamma receptor-TLR cross-talk elicits pro-inflammatory cytokine production by human M2 macrophages. Nat Commun. 2014;5(5444):1–8. doi: 10.1038/ncomms6444. https://www.nature.com/articles/ncomms6444 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selman M., Derks R., Bondt A., et al. Fc specific IgG glycosylation profiling by robust nano-reverse phase HPLC-MS using a sheath-flow ESI sprayer interface. J Proteomics. 2012;75(4):1318–1329. doi: 10.1016/j.jprot.2011.11.003. https://pubmed.ncbi.nlm.nih.gov/22120122/ Available from: [DOI] [PubMed] [Google Scholar]
- 51.Bartsch Y.C., Eschweiler S., Leliavski A., et al. IgG Fc sialylation is regulated during the germinal center reaction following immunization with different adjuvants. J Allergy Clin Immunol. 2020;146(3):652–666. doi: 10.1016/j.jaci.2020.04.059. https://pubmed.ncbi.nlm.nih.gov/32445838/ Available from: [DOI] [PubMed] [Google Scholar]
- 52.Garimalla S., Nguyen D., Halliley J., et al. Differential transcriptome and development of human peripheral plasma cell subsets. JCI Insight. 2019;4(9) doi: 10.1172/jci.insight.126732. https://pubmed.ncbi.nlm.nih.gov/31045577/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian Y., Wei C., Eun-Hyung Lee F., et al. Elucidation of seventeen human peripheral blood B-cell subsets and quantification of the tetanus response using a density-based method for the automated identification of cell populations in multidimensional flow cytometry data. Cytometry B Clin Cytom. 2010;78(Suppl 1) doi: 10.1002/cyto.b.20554. https://pubmed.ncbi.nlm.nih.gov/20839340/ Internet. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanz I., Wei C., Jenks S., et al. Challenges and opportunities for consistent classification of human B cell and plasma cell populations. Front Immunol. 2019;10(OCT) doi: 10.3389/fimmu.2019.02458. https://pubmed.ncbi.nlm.nih.gov/31681331/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boruah B., Kadirvelraj R., Liu L., et al. Characterizing human α-1,6-fucosyltransferase (FUT8) substrate specificity and structural similarities with related fucosyltransferases. J Biol Chem. 2020;295(50):11727–11745. doi: 10.1074/jbc.RA120.014625. https://pubmed.ncbi.nlm.nih.gov/33004438/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang B, Harhay M.O., Small D.S., Morris T.P., Li F. On the mixed-model analysis of covariance in cluster-randomized trials. arXiv. 2021 https://arxiv.org/pdf/2112.00832.pdf Available from: [Google Scholar]
- 57.Kapur R., Kustiawan I., Vestrheim A., et al. A prominent lack of IgG1-Fc fucosylation of platelet alloantibodies in pregnancy. Blood. 2014;123(4):471–480. doi: 10.1182/blood-2013-09-527978. https://pubmed.ncbi.nlm.nih.gov/24243971/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Šuštić T., Van Coillie J., Larsen M.D., et al. Immunoassay for quantification of antigen-specific IgG fucosylation. EBioMedicine. 2022;81 doi: 10.1016/j.ebiom.2022.104109. http://www.thelancet.com/article/S2352396422002900/fulltext Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chakraborty S., Med S.T., Chakraborty S., et al. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci Transl Med. 2022;7853:1–20. doi: 10.1126/scitranslmed.abm7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hagemann K., Riecken K., Jung J.M., et al. Natural killer cell-mediated ADCC in SARS-CoV-2-infected individuals and vaccine recipients. Eur J Immunol. 2022:1–11. doi: 10.1002/eji.202149470. https://onlinelibrary.wiley.com/doi/full/10.1002/eji.202149470 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu Y., Wang M., Zhang X., et al. Antibody-dependent cellular cytotoxicity response to SARS-CoV-2 in COVID-19 patients. Signal Transduct Target Ther. 2021;6(1):1–10. doi: 10.1038/s41392-021-00759-1. https://www.nature.com/articles/s41392-021-00759-1 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Junqueira C., Crespo Â, Ranjbar S., et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;6:576–584. doi: 10.1038/s41586-022-04702-4. https://www.nature.com/articles/s41586-022-04702-4 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sefik E., Qu R., Junqueira C., et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 2022;1 doi: 10.1038/s41586-022-04802-1. http://www.ncbi.nlm.nih.gov/pubmed/35483404 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Farkash I., Feferman T., Cohen-Saban N., et al. Anti-SARS-CoV-2 antibodies elicited by COVID-19 mRNA vaccine exhibit a unique glycosylation pattern. Cell Rep. 2021;37(11) doi: 10.1016/j.celrep.2021.110114. https://pubmed.ncbi.nlm.nih.gov/34883043/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lofano G., Gorman M.J., Yousif A.S., et al. Antigen-specific antibody Fc glycosylation enhances humoral immunity via the recruitment of complement. Sci Immunol. 2018;3(26):7796. doi: 10.1126/sciimmunol.aat7796. Available from: /pmc/articles/PMC6298214/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mössner E., Brünker P., Moser S., et al. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell-mediated B-cell cytotoxicity. Blood. 2010;115(22):4393–4402. doi: 10.1182/blood-2009-06-225979. https://pubmed.ncbi.nlm.nih.gov/20194898/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferrara C., Brünker P., Suter T., Moser S., Püntener U., Umaña P. Modulation of therapeutic antibody effector functions by glycosylation engineering: influence of Golgi enzyme localization domain and co-expression of heterologous beta1,4-N-acetylglucosaminyltransferase III and Golgi alpha-mannosidase II. Biotechnol Bioeng. 2006;93(5):851–861. doi: 10.1002/bit.20777. http://www.ncbi.nlm.nih.gov/pubmed/16435400 Available from: [DOI] [PubMed] [Google Scholar]
- 68.Sefik E., Qu R., Kaffe E., et al. Viral replication in human macrophages enhances an inflammatory cascade and interferon driven chronic COVID-19 in humanized mice. BioRxiv. 2021 https://www.biorxiv.org/content/10.1101/2021.09.27.461948v1 Available from: [Google Scholar]
- 69.Junqueira C., Crespo Â., Ranjbar S., et al. SARS-CoV-2 infects blood monocytes to activate NLRP3 and AIM2 inflammasomes, pyroptosis and cytokine release. Res Sq. 2021 https://www.medrxiv.org/content/10.1101/2021.03.06.21252796v1 Available from: [Google Scholar]
- 70.Röltgen K., Nielsen S.C.A., Silva O., et al. Immune imprinting, breadth of variant recognition and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022 doi: 10.1016/j.cell.2022.01.018. http://www.cell.com/article/S0092867422000769/fulltext Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamin R., Jones A.T., Hoffmann H.H., et al. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021;599(7885) doi: 10.1038/s41586-021-04017-w. https://pubmed.ncbi.nlm.nih.gov/34547765/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt F., Weisblum Y., Rutkowska M., et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600:512–516. doi: 10.1038/s41586-021-04005-0. https://www.nature.com/articles/s41586-021-04005-0 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z., Muecksch F., Schaefer-Babajew D., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021;595(7867):426–431. doi: 10.1038/s41586-021-03696-9. https://www.nature.com/articles/s41586-021-03696-9 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krištić J., Vučković F., Menni C., et al. Glycans are a novel biomarker of chronological and biological ages. J Gerontol A Biol Sci Med Sci. 2014;69(7):779–789. doi: 10.1093/gerona/glt190. https://academic.oup.com/biomedgerontology/article/69/7/779/662651 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cognetti J.S., Miller B.L. Monitoring serum spike protein with disposable photonic biosensors following SARS-CoV-2 vaccination. Sensors (Basel) 2021;21(17) doi: 10.3390/s21175857. https://pubmed.ncbi.nlm.nih.gov/34502753/ Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.