Using the natural variation in stamen whorl numbers in Aquilegiaspecies, we conducted QTL mapping to uncover the genetic architecture underlying variation in floral meristem termination.
Keywords: Aquilegia, floral meristem termination, QTL, stamen whorl
Abstract
Floral organs are produced by floral meristems (FMs), which harbor stem cells in their centers. Since each flower only has a finite number of organs, the stem cell activity of an FM will always terminate at a specific time point, a process termed floral meristem termination (FMT). Variation in the timing of FMT can give rise to floral morphological diversity, but how this process is fine-tuned at a developmental and evolutionary level is poorly understood. Flowers from the genus Aquilegia share identical floral organ arrangement except for stamen whorl number (SWN), making Aquilegia a well-suited system for investigation of this process: differences in SWN between species represent differences in the timing of FMT. By crossing A. canadensis and A. brevistyla, quantitative trait locus (QTL) mapping has revealed a complex genetic architecture with seven QTL. We explored potential candidate genes under each QTL and characterized novel expression patterns of select loci of interest using in situ hybridization. To our knowledge, this is the first attempt to dissect the genetic basis of how natural variation in the timing of FMT is regulated, and our results provide insight into how floral morphological diversity can be generated at the meristematic level.
Introduction
Indeterminate growth is the foundation of development in all vascular plants and is achieved by the persistent activity of stem cells in meristems (Steeves and Sussex, 1989). Apical meristems in shoots and roots are highly organized structures that maintain a delicate, yet robust, balance between the production of stem cells and organogenic cells. In the flowering plants, when a plant enters the reproductive phase, the vegetative meristem transitions to inflorescence meristem (IM) identity, which then gives rise to floral meristem (FM) identity. Although their overall cellular organization is highly similar, this shift is accompanied by a number of transitions in the properties of the meristem, including changes in the rate and patterns of primordium production, and an eventual loss of indeterminacy.
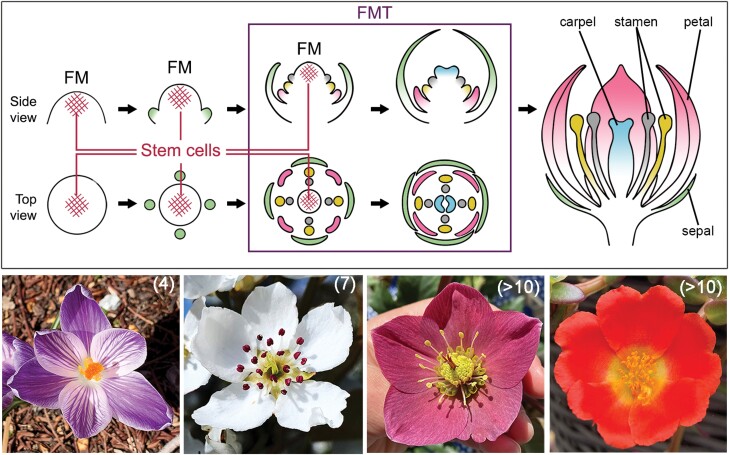
The loss of indeterminacy in the FM is a well-regulated process, termed floral meristem termination (FMT), which is crucial and universal to the development of all flowers (Fig. 1). A typical flower has four types of floral organs: sepals, petals, stamens, and carpels, which are arranged from the outermost to the innermost positions of a flower (Fig. 1). Although the central stem cells stay active during the early phases of floral organ primordia initiation, this activity will cease at a specific time point, after which all remaining meristematic cells will be incorporated into the development of the innermost organs, the carpels (Steeves and Sussex, 1989). The precise control of FMT is critical to ensuring that the flower has the correct number of organs, and variation in FMT timing is an important source of floral morphological diversity and novelty. For some species, such as Arabidopsis thaliana, FMT occurs relatively quickly, and only four whorls of floral organs are produced. In many other taxa, FM activity is maintained for a more extended period; species from the Magnoliaceae, Monimiaceae, Nelumbonaceae, Nymphaeaceae, Papaveraceae, and Ranunculaceae, for instance, can have hundreds of spirally arranged or whorled floral organs (Endress, 1990; Fig. 1). Moreover, increased numbers of floral organs can create raw materials for the evolution of new organ types, such as the sterile staminodes observed in Aquilegia (Ranunculaceae) or Mentzelia (Loasaceae) (Walker-Larsen and Harder, 2000). The diversity in floral morphology of >300 000 angiosperm species is seemingly infinite, but one of the few major evolutionary trends that is independently observed in the transition from early-diverging angiosperms to either the core eudicot or monocot lineages is the transition from variable to stable whorl numbers in a flower (Endress, 1990), which is directly determined by the timing of FMT. Therefore, understanding how FMT is regulated in different angiosperm lineages is interesting from both developmental and evolutionary perspectives.
Fig. 1.
FMT is an important and fine-tuned developmental process that occurs in all flowers. Upper panel: diagram of floral organ initiation and FMT during flower development. Organs of the same whorl share the same colors. Lower panel: example of four flowers with different whorl numbers. From left to right: Crocus vernus ‘Pickwick’, Pyrus communis, Helleborus orientalis, and Portulaca umbraticola. Numbers in parentheses indicate the number of whorls (or ranks for H. orientalis) of floral organs in each flower. Photos of C. vernus, P. communis, and P. umbraticola were taken by Ya Min, and the photo of H. orientalis was taken by Evangeline S. Ballerini.
Currently, we have relatively good knowledge of the genes that are responsible for maintaining and terminating stem cell activities in A. thaliana. In the FM, the maintenance of the stem cell population is achieved by a feedback loop between the homeodomain protein WUSCHEL (WUS) and the CLAVATA (CLV) ligand–receptor system, in which WUS promotes central stem cell activity and induces the expression of the CLV3 peptide, while activation of the CLV signaling pathway represses the expression of WUS (Schoof et al., 2000; Lenhard, 2003; Müller et al., 2006). Meanwhile, in the early developing FM, the expression of the C class organ identity gene AGAMOUS (AG) is induced by WUS, while AG specifies the identity of stamens and carpels and is also responsible for the down-regulation of WUS expression (Lenhard et al., 2001). Although the broad conservation of the WUS–CLV and WUS–AG feedback loops has been demonstrated in diverse plant taxa (Nardmann and Werr, 2006; Litt and Kramer, 2010; Whitewoods et al., 2020), the exact mechanisms by which AG controls the precise timing of WUS down-regulation have only been investigated in A. thaliana and tomato (Solanum lycopersicum). Specifically, AG activates the expression of a C2H2 zinc-finger transcription factor gene, KNUCKLES (KNU), whose protein product directly represses WUS (Payne, 2004; Sun et al., 2009; Bollier et al., 2018). Accurate timing of FMT is achieved because the activation of KNU by AG takes approximately two rounds of cell divisions, during which the stamen primordia are initiated. Once WUS down-regulation is achieved, all of the cells remaining in the center of the FM are incorporated into the carpel primordia (Sun et al., 2009). Thus, FMT is tightly connected to the expression of AG and, moreover, the production of carpels in the center of the flower. It is perhaps not surprising that when we examine variation in FMT across the angiosperms, this is primarily expressed as variation in the number of stamen whorls produced before the final production of carpels and the concomitant termination of the meristem (Endress, 2011; Ronse De Craene, 2018).
However, integral to the known mechanism of KNU activation is that it only allows for one stamen and one carpel whorl to be produced in a flower. This begs the question of whether this pathway is conserved in systems that have more than four whorls of floral organs. Unfortunately, all currently established model systems (and their close relatives) belong to lineages that exhibit no variation in their floral whorl numbers, while most of the plant taxa exhibiting such variation do not have genomic or genetic resources or functional tools. To that end, species in the genus Aquilegia, a member of the buttercup family (Ranunculaceae), are well suited for investigating this fundamental developmental process. There are ~70 Aquilegia species, which share low interspecific sequence divergence and a high degree of interfertility due to having arisen via a recent adaptive radiation (Filiault et al., 2018). Organs in Aquilegia flowers are produced in a whorl of five that are arranged in 10 orthostichies (vertical rows of organs), with alternate orthostichies positioned directly above either the sepals or the petals (Fig. 2A). Across species there is a consistent number of whorls for the sepals, petals, staminodes (with the exception of A. jonesii, which lacks staminodes; Munz, 1946), and carpels, but the number of stamen whorls varies both within and between species.
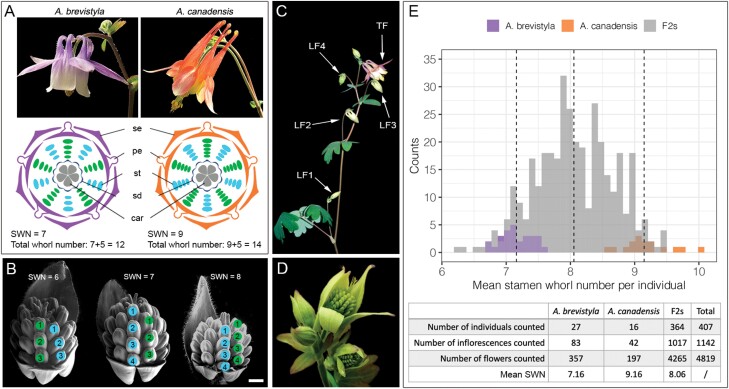
Fig. 2.
Phenotyping SWN in the parental and F2 populations. (A) Photos of flowers and floral diagrams of A. brevistyla and A. canadensis. (B) Examples of three F2 flower buds with different SWN. (C) Flowers that were sampled per inflorescence. (D) Developmental stages for which SWN was counted. (E) Histogram and summary statistics of SWN distribution in parental species and the F2s. In (A) and (B), stamen whorls positioned above the sepals are colored in blue while stamen whorls positioned above the petals are colored in green. Scale bar in (B)=100 µm. se, sepals; pe, petals: st, stamens; sd, staminodes; car, carpels.
Aquilegia brevistyla and A. canadensis are North American sister species that vary in many pollination-related traits (Fig. 2A; Bastida et al., 2010; Fior et al., 2013; Edwards et al., 2021), but also in what we term stamen whorl number (SWN). We argue here that this trait can be directly translated into variation in the timing of FMT: if the FM transitioned to carpel production and terminated earlier, there will be a smaller SWN compared with a flower in which the FM proliferates for a longer period. Using SWN as a quantitative trait, we conducted quantitative trait locus (QTL) mapping in the resultant F2 generation from crossing A. brevistyla and A. canadensis. Our results reveal a complex genetic architecture with seven QTL, which we have associated with potential candidate genes under each QTL. To our knowledge, this is the first study to dissect the genetic basis of how natural variation in the timing of FMT is regulated, and our results highlight molecular pathways that may contribute to the regulation of FMT in Aquilegia.
Materials and methods
Plant material and growth conditions
Aquilegia brevistyla and A. canadensis seeds were collected from wild populations in Alberta (Canada) and Ithaca (NY, USA), respectively. One A. canadensis (pollen recipient) was crossed with one A. brevistyla (pollen donor) to generate the F1 generation. Five F1s were self-fertilized to generate the F2 population. All F2 seeds were stratified at 4 °C in the dark for 2–4 weeks, germinated in wet soil, and transplanted in individual pots. All plants were vernalized at 4 °C for 2 months to induce flowering. The parental and F1 individuals were grown in the greenhouse of the University of California Santa Barbara, and the F2 populations were grown in the greenhouses of Harvard University. All greenhouses used the same light and temperature conditions to achieve a 16 h/8 h (day/night) photoperiod at 18 °C and 13 °C.
Seeds of Aquilegia×coerulea ‘Kiragami’ were purchased from Swallowtail Garden Seeds (Santa Rosa, CA, USA), germinated in wet soil, and grown under the same 18 °C/13 °C (day/night) condition as described above. Once the plants developed approximately six true leaves, they were transferred into vernalization conditions (16 h daylight at 6 °C and 8 h dark at 6 °C) for 3–4 weeks, and then moved back to the regular growth conditions to promote flowering.
Meristem width measurement
The entire inflorescences of at least six individuals of each parental species were collected and fixed in FAA (10% formaldehyde, 50% ethanol, 5% acetic acid), and stored at 4 °C. Samples were then dehydrated through a graded ethanol series to 100%, transferred to 100% CitriSolv, and embedded in Paraplast Plus (Sigma-Aldrich). Embedded tissues were sectioned to 8 µm thick ribbons with a rotary microtome, stained in 0.1% Toluidine Blue O solution following the protocol described in Ruzin (1999), and mounted in Permount Mounting Medium (Fisher Scientific). Sections were then imaged using the Axio Zoom Microscope at the Harvard Center for Biological Imaging. The width of each floral meristem section was measured using ImageJ. Three to six serial sections were measured for each FM, and at least three FMs were measured for each developmental stage of each species. All FMs that were measured were non-terminal flowers, and the number of non-sepal primordia captured by the section was counted.
Phenotyping
For each plant, the SWN of the terminal flower and lateral flowers 1 to 4 from the first three inflorescences were counted (Fig. 2C). If flowers of these positions in an inflorescence were damaged/undeveloped, flowers at other positions were counted to achieve a total number of five flowers per inflorescence. If an inflorescence produced less than five flowers, all flowers were counted. SWN was counted when the flowers reached ~1–2 cm in length (Fig. 2D) because, at that developmental stage, all the stamens were arranged in vertical rows, which simplified counting.
Genotyping
Detailed genotyping information can be found in Edwards et al. (2021). Briefly, the DNA of the two parents that generated the cross was extracted from flash-frozen young leaves using the NEBNext Ultra II kit (NEB) and sequenced to ~40× coverage as 150 bp reads on an Illumina MiSeq at the Biological Nanostructures Lab in the California NanoSystem Institute at UC Santa Barbara. DNA of F2s was extracted from silica-dried young leaves using Qiagen DNEasy reagents and Magattract beads (Qiagen, Inc.); libraries were prepared following the protocol of the RipTide High Throughput Rapid DNA Library Preparation kit (iGenomX, CA, USA). The F2 libraries were pooled and sequenced at the Vincent J. Coates Genomics Sequencing Laboratory (UC Berkeley) using the NovaSeq 6000 platform to generate 150 bp paired-end reads. Samples were multiplexed to generate ~1–2× coverage. All sequence data are deposited in the Sequence Read Archive under BioProject ID PRJNA720109. Scripts and genotype/phenotype data are available at: https://github.com/anjiballerini/can.x.brev/.
Sequences were aligned to the A. coerulea ‘Goldsmith’ v3.1 reference genome (https://phytozome.jgi.doe.gov) using the Burrows–Wheeler aligner (Li and Durbin, 2009), and variable sites in the parents were identified using SAMtools 0.1.19 (H. Li et al., 2009) with custom scripts used to identify the positions and genotypes at which the parents were homozygous for different alleles. These sites were used to assign reads in the F2s as having either A. canadensis or A. brevistyla ancestry. To determine the genotypes of the F2s, the genome sequences were binned into 0.5 Mb regions with moderate to high recombination frequencies and 1 Mb in regions with low or no recombination, and the frequency of reads with ancestry for each F0 parent was used to determine the genotype of the bin. These bins and genotypes were used as markers to construct a genetic map and conduct QTL mapping. This genotyping method has been implemented in Filiault et al, (2018), Ballerini et al. (2020), and Edwards et al. (2021).
Mapping
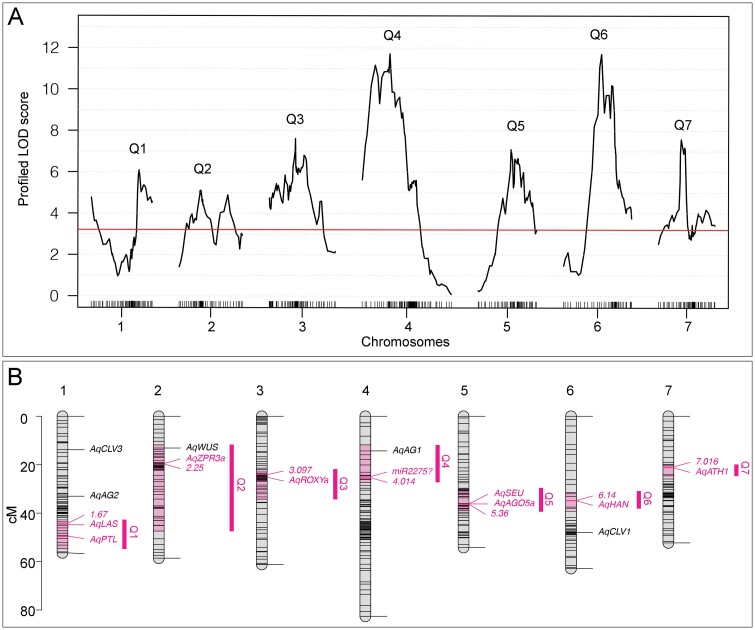
After filtering out individuals and markers with >10% of information missing due to sequencing quality, we retained a total of 366 individuals and 620 markers. A genetic map of the seven chromosomes was then constructed following the protocol of the R/qtl package v1.46-2 (Broman et al., 2003), with an error probability rate of 0.001 and ‘kosambi’ map function. Standard interval mapping with Haley–Knott regression (function ‘scanone’) was used for the initial mapping searching for potential QTL. The best multiple-QTL models are produced and selected by using function ‘stepwiseqtl’, which implements penalties on different interactions and drops one of the current main effects or interactions in each round of model comparison. Interactions among potential QTL and between QTL and covariance were detected with a two-dimensional genome scan (function ‘scantwo’). Using the estimated positions of QTL from ‘scanone’, ‘stepwiseqtl’, and ‘scantwo’ as the input, the positions of QTL were refined by using the function ‘makeqtl’ and ‘refineqtl’, which then fit with a defined multiple-QTL model (function ‘fitqtl’) with all detected interactions. F1-parent-of-origin was used as covariance in all the tested mapping models. Position and effect size of QTL were estimated using drop-one-term ANOVA in the best-fitting model. The chromosome diagram with potential candidate genes (Fig. 4) was produced by using the LinkageMapView (Ouellette et al., 2018).
Fig 4.
Genetic architecture and candidate genes. (A) LOD scores across seven chromosomes. Red line: α=0.05 genome-wide significance cut-off based on 1000 permutations. (B) Locations of QTL interval (pink regions on the chromosomes and magenta vertical bars), candidate genes, and genetic markers. All the genetic markers were named in numeric forms (e.g. 1.67 and 2.25) and only markers with the highest LOD scores under each QTL are shown.
In situ hybridization of genes of interest
Variable regions of the genes of interest were amplified by PCR (primers in Supplementary Table S1) from young inflorescence cDNA of Aquilegia×coerulea ‘Kiragami’. The PCR products were cloned into the pCR™4-TOPO vectors, sequenced to confirm identity, and reverse transcribed using T3 or T7 RNA polymerase and DIG RNA labeling mix (Sigma-Aldrich). Probe qualification and in situ hybridization steps followed Kramer, (2005). Slides were stained in calcofluor white for 5 min before imaging, and pictures were taken using the ZEISS Axio Zoom at the Harvard Center for Biological Imaging.
Statistical analysis
All statistical analyses (e.g. ANOVA andTukey’s HSD) were performed using R (version 1.1.456).
Gene trees
Homologs of AqROXYa and AqATH1 from various taxa were obtained by using BLAST on Phytozome (https://phytozome-next.jgi.doe.gov/). Multiple sequence alignments and Neighbor–Joining phylogenetic trees were constructed using Geneious Prime (v2021.1.1). We did not construct phylogenetic trees for other potential candidate genes because their homologs in A. coerulea and A. thaliana were each other’s reciprocal top BLAST hits.
Results
SWN variation in the parental species and the F2s
We counted the SWN from 357, 197, and 4265 flowers from 27, 16, and 364 A. brevistyla, A. canadensis, and F2 individuals, respectively (Fig. 2). The SWN per individual of the parental species did not overlap: the mean SWN of A. brevistyla ranges from 6.69 to 7.57; that of A. canadensis, from 8.54 to 10; and that of the F2s, which overlapped with the range of both parental species, from 6.20 to 9.50 (Fig. 2E). The mean SWN for all A. brevistyla, A. canadensis, and F2s was 7.16, 9.16, and 8.06, respectively. Subsequently, we analyzed whether the position of flowers on the inflorescence is associated with their SWN. Flower position had a significant effect on the SWN for both parental species but not for the F2s (Supplementary Fig. S1). Given lower germination rates of interspecific hybrid seeds, seeds generated by selfing five different F1 plants (all of which had the same parents) were used to obtain 364 F2 progeny that reached the flowering stage (n=53–81 per F1). One-way ANOVA revealed that the SWN of the F2s differed significantly between the F1 parents (Supplementary Fig. S2). Lastly, we also analyzed the variation of SWN among flowers of the same plants. Interestingly, a small portion of A. brevistyla (7.4%), A. canadensis (18.8%), and F2s (6%) showed no variance in the SWN across all flowers counted within an individual, and this phenomenon was dependent on neither the number of flowers counted per individual plant (Supplementary Fig. S3; Pearson’s correlation=0.052, t=0.98418, df=345, P=0.3257) nor the F1-parent-of-origin of the F2s (Supplementary Fig. S3). No significant correlation between the mean SWN per individual and the SD of SWN per individual was detected (Pearson’s correlation=0.035, t=0.70438, df=404, P=0.4816).
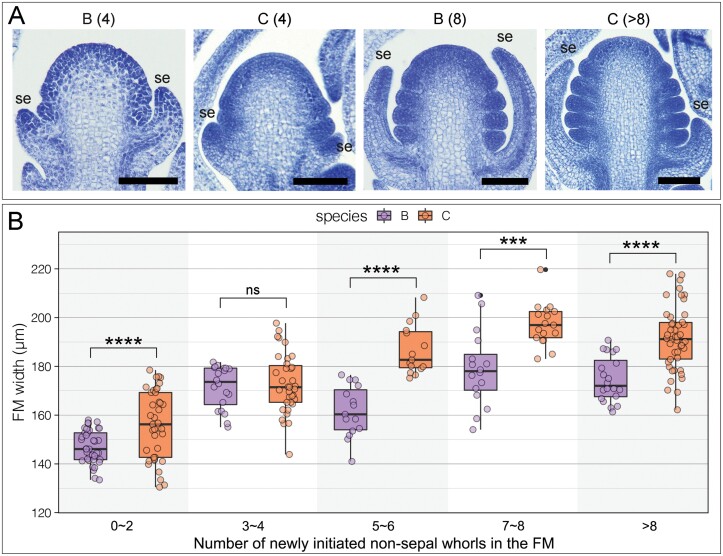
Floral meristem size
To determine whether the initial FM sizes were different between the parental species, we measured the widths of FMs of the parental species at their earliest developmental stages (Fig. 3A). In general, the FMs of A. canadensis appeared to be slightly, but significantly, wider than those of A. brevistyla throughout the early developmental stages (Fig. 3B; Supplementary Table S2). The average FM widths of A. brevistyla and A. canadensis before the initiation of carpel primordia were 174.68 µm and 191.67 µm, respectively (Supplementary Table S2). Interestingly, the temporal developmental windows for significant FM size expansion seemed to be longer in A. canadensis than in A. brevistyla (Fig. 3B; Supplementary Table S2). The significant increase in widths of A. brevistyla FMs occurred when there were 0–4 whorls of non-sepal floral organs initiating, while the significant increase in the widths of A. canadensis FMs encompassed a larger developmental period, ranging from the stages that there were 0–6 whorls of non-sepal floral organs initiating (Supplementary Table S2).
Fig. 3.
FM width measurements of the parental species during the early developmental stages. (A) Examples of FM morphologies in A. brevistyla and A. canadensis at early developmental stages. Numbers in parentheses indicate the number of newly initiated stamen whorls in each FM. Scale bars=100 µm. (B) Comparison of FM width of different developmental stages between A. brevistyla and A. canadensis. Each data point represents a measurement of an FM width from a section. Three to six continuous sections were measured for each FM, and at least three FMs were measured for each developmental stage of each species. Comparison of FM widths of each stage between the parental species was done using Tukey’s HSD. ns, not significant; ***P-value <0.001; ****P-value <0.0001; se, sepal; B, A. brevistyla; C, A. canadensis.
Genetic architecture underlying stamen whorl variation
The genetic map was constructed using a total of 620 genetic markers, which fell into seven linkage groups, matching the n=7 chromosomes in the Aquilegia genome (Supplementary Fig. S4). We recovered seven major QTL using the mean SWN per individual as a phenotype and the F1-parent-of-origin as a covariate, with one QTL on each chromosome (Fig. 4A; Table 1). The difference in LOD scores between models that included or excluded the covariate are diminutive on all chromosomes (ranging from –0.3 to 0.49; Supplementary Fig. S5A), indicating no significant interaction between the QTL and the covariate. While several LOD profiles from a single QTL model suggested that there may be two QTL on a chromosome, a two-dimensional genome scan failed to detect evidence for the presence of more than one QTL on a single chromosome (Supplementary Table S3). The presence of only one true QTL on chromosome 2 was further confirmed by controlling the two potential QTL: when the true QTL (i.e. Q2) was controlled, the presence of the second peak also disappeared (Supplementary Fig. S5B). We did detect significant interactions between two pairs of QTL: Q3 and Q7, and Q1 and Q6 (Supplementary Fig. S5C), and thus incorporated these interactions in the full QTL model (Table 1).
Table 1.
Summary statistics for QTL
| Full model result: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| y~Q1+Q2+Q3+Q4+Q5+Q6+Q7+Q1:Q6+Q3:Q7 | ||||||||||
| df | Type III SS | MS | LOD | PVE | P-value (F) | |||||
| Model | 22 | 61.87 | 2.81 | 48.04 | 44.47 | 0 | ||||
| Error | 331 | 71.27 | 0.22 | |||||||
| Total | 353 | 133.14 | ||||||||
| Drop one QTL at a time ANOVA table | ||||||||||
| Chr | Position (cM) | df | Type III SS | LOD | PVE | F-value |
P-value (F) |
Add. | Dom. | |
| Q1 | 1 | 44.233 | 6 | 5.87 | 6.1 | 4.4 | 4.6 | <0.001 | 0.05 | –0.16 |
| Q2 | 2 | 19.676 | 2 | 4.89 | 5.1 | 3.7 | 11.4 | <0.001 | –0.07 | 0.2 |
| Q3 | 3 | 24.050 | 6 | 7.43 | 7.6 | 5.6 | 5.7 | <0.001 | –0.11 | –0.01 |
| Q4 | 4 | 25.406 | 2 | 11.73 | 11.7 | 8.8 | 27.3 | <0.001 | –0.27 | 0.07 |
| Q5 | 5 | 30.155 | 2 | 6.89 | 7.1 | 5.2 | 16.0 | <0.001 | –0.25 | 0.17 |
| Q6 | 6 | 34.828 | 6 | 11.72 | 11.7 | 8.8 | 9.1 | <0.001 | –0.14 | –0.27 |
| Q7 | 7 | 20.944 | 6 | 7.38 | 7.6 | 5.6 | 5.7 | <0.001 | 0 | 0.15 |
| Q1×Q6 | 4 | 5.45 | 5.7 | 4.1 | 6.3 | <0.001 | ||||
| Q3×Q7 | 4 | 5.39 | 5.6 | 4.1 | 6.3 | <0.001 | ||||
| Estimated effects for QTL interactions | ||||||||||
| Q1 (a)×Q6 (a): 0.01 | Q1 (d)×Q6 (a): –0.23 | Q1 (a)×Q6 (d): –0.38 | Q1 (d)×Q6 (d): 0.45 | |||||||
| Q3 (a)×Q7 (a): 0.19 | Q3 (d)×Q7 (a): 0.09 | Q3 (a)×Q7 (d): –0.16 | Q3 (d)×Q7 (d): –0.24 | |||||||
Chr, chromosome; PVE, percentage variance explained; Add./(a), additive affects; Dom./(d). dominant effects.
The full QTL model had a total LOD score of 48 and explained 46.5% of the observed phenotypic variation (Table 1). Q3, Q4, and Q5 exhibit larger additive effects than dominant effects, while the remaining QTL have larger dominant effects on the phenotypic variation (Fig. 4A; Table 1; Supplementary Fig. S6). The phenotypic variation explained by each QTL and the QTL interactions was very similar, with Q4 and Q6 having the largest additive and dominant effects, respectively, and each explained 8.8% of the phenotypic variation (Table 1). This suggested that the genetic architecture of SWN is a complex trait controlled by multiple loci, each with small effect.
Potential candidate genes
In order to identify potential candidate genes underlying these QTL, we examined the genomic regions defined by markers that flanked the 95% Bayesian credible interval of each QTL, which was calculated by using the posterior distribution of 10LOD on a given chromosome. The genomic regions of Q1, Q4, Q6, and Q7 were <6 Mb in size, while those of Q2, Q3, and Q5 were >20 Mb (Supplementary Table S4). Among all the QTL, Q6 and Q2 had the smallest (1.5 Mb) and the largest (36.5 Mb) intervals, containing 226 and 3242 genes, respectively (Supplementary Table S4). We sought to identify loci for further study under each QTL based on (i) the locations of the annotated genes relative to the location of the markers with the highest LOD scores, (ii) homology to previously studied loci related to meristem function, and (iii) gene expression levels during early FM developmental stages using previously published RNA-sequencing (RNAseq) data for A. coerulea ‘Kiragami’, which sampled developmental stages covering the FMT window (Min and Kramer, 2020). Because our genotyping method used 0.5 Mb or 1 Mb binned genomic regions as the genetic markers rather than single nucleotide markers (see details in the Materials and methods), we gave the highest priority to genes located in the region of the marker with the highest LOD scores.
Under Q1, we identified a homolog of LATERAL SUPPRESSOR (LAS), AqLAS, which is interesting because mutations in the LAS orthologs in A. thaliana and tomato severely impact axillary meristem activity, suggesting that the genes may influence indeterminacy (Schumacher et al., 1999; Greb, 2003; Wang et al., 2014). One additional gene that was located 3 Mb away from AqLAS also caught our attention: AqPETAL LOSS (AqPTL). PTL is a floral organ boundary gene in A. thaliana that controls cell proliferation (Griffith et al., 1999; Brewer et al., 2004; Lampugnani et al., 2012). We considered AqPTL interesting for two reasons. First, PTL has been shown to physically interact with and be transcriptionally regulated by the C2H2 transcription factor gene JAGGED (JAG) (Sauret-Güeto et al., 2013), whose Aquilegia homolog AqJAG has been shown to be critical to maintaining the FM (Min and Kramer, 2017). Second, PTL and the gene product of HANABA TANARU (HAN) interact by sharing JAG as a direct protein partner to regulate floral morphogenesis in A. thaliana (Ding et al., 2015), and the homolog of HAN is a potential candidate gene under Q6 (Fig. 4B).
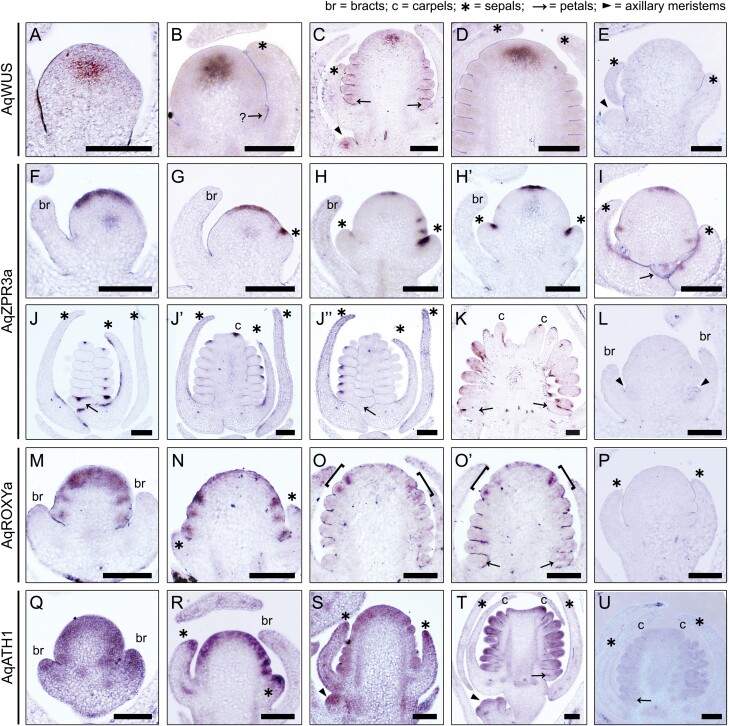
Interestingly, AqWUS is within the 95% Bayesian credible intervals of Q2, located at the edge of the interval (Fig. 4B). Since the expression of AqWUS has not been examined in situ, we analyzed its expression pattern during the early developmental stages of A. coerulea FM (Fig. 5A–E). AqWUS is expressed in a small population of cells in the center of the FM from the earliest stages (Fig. 5A–E). The expression in these central zone cells persists during the initiation of floral organs and disappears when carpel primordia start to initiate. These observations are consistent with the expression of WUS orthologs in all taxa examined to date (Nardmann and Werr, 2006; Galli and Gallavotti, 2016), suggesting functional conservation of AqWUS as well. However, the marginal position of AqWUS in Q2 makes it a less compelling candidate.
Fig 5.
In situ hybridization of AqWUS and candidate genes. (A–E) Expression patterns of AqWUS (A–C) and its sense probe (E). (A) An FM that has not produced any organs. (B) An FM that is in the process of initiating either a petal primordium or the outermost stamen primordium (and thus indicated by an arrow with a question mark). (C) An FM that has at least eight whorls of stamens initiated; AqWUS expression was also detected in an axillary meristem (arrowhead). (D) An FM that has at least 11 whorls of stamens produced. (E) Sense probe of AqWUS. (F–L) Expression patterns of AqZPR3a (F–K) and its negative control probe (L). (F) An FM that has not produced any organs subtended by a bract (br). (G) An FM that has just started to produce sepal primordia (asterisk). (H, Hʹ) Serial sections through the same FM that has only produced sepal primordia (asterisks). (I) An FM that has produced 1–2 whorls of stamens. (J, Jʹ, Jʹʹ) Serial sections through the same floral bud that has initiated all floral organs. (K) A young floral bud in which all floral organs are differentiating. (L) Sense probe of AqZPR3a. (M–P) Expression of AqROXYa (M–Oʹ) and its negative control (P). (M) An FM that has not produced any organs. (N) An FM that is in the process of initiating petal or utermost stamen primordia. (O, Oʹ) Serial sections through the same FM, with brackets indicating newly emerging primordia. (P) Sense probe of AqROXYa. (Q–U) Expression of AqATH1 (Q–T) and its negative control (U). (Q) An FM that has not produced any organs. (R) A young FM that is in the process of initiating petal and stamen primordia. (S) An FM initiating stamen primordia and an associated axillary meristem (arrowhead). (T) A floral bud with carpel primordia just initiated and an axillary meristem (arrowhead). (U) Sense probe of AqATH1. All scale bars=100 µm.
Within the highest LOD interval of Q2, which is located 8.7 Mb away from AqWUS, we identified another locus of interest, AqLITTLE ZIPPER 3a (AqZPR3a; Fig. 4B). AqZPR3a encodes a small leucine zipper-containing protein and is the homolog of the previously identified gene ZPR3 in A. thaliana (Weits et al., 2019). Homologs of the ZPR genes have been shown to regulate leaf polarity and shoot apical meristem maintenance in A. thaliana and tomato (Wenkel et al., 2007; Kim et al., 2008; Xu et al., 2019), but little is known about whether they are involved in any FM-specific functions in these plant systems. In the Aquilegia FM, AqZPR3a exhibits dynamic expression patterns (Fig. 5F–L). At the earliest stages of the FM, concentrated expression of AqZPR3a is detected across the central epidermal layer, and moderate expression is found in the central zone (Fig. 5F). This strong expression in the epidermal layer persists until the FM has initiated several whorls of floral organs, but the width of the domain seems to contract as FM development proceeds (Fig. 5F–I). In contrast, the expression in the central zone disappears rapidly after initiation of the sepal primordia (Fig. 5F, G, Hʹ, Jʹʹ). Strong expression of AqZPR3a is also detected at the adaxial boundary of all initiating floral organ primordia (Fig. 5H–Jʹʹ). However, these adaxial expression domains are restricted to the medial region of each primordium, rather than the entire abaxial surface, which can be seen in serial sections through the same FM (Fig. 5H, Hʹ, J, Jʹ, Jʹʹ). Strong but patchy expression of AqZPR3a is also detected in the adaxial epidermal layer of older lateral organs, such as the sepals (Fig. 5I, J, Jʹ), petals (Fig. 5K), stamens (Fig. 5K), and carpels (Fig. 5K). Intriguingly, expression of the ZRP genes across the epidermal layers has never been observed in any other plant systems.
Under Q3, we identified AqROXYa which codes for a thioredoxin superfamily protein and is a homolog of A. thaliana genes ROXY1 and ROXY2 (Fig. 4B; Supplementary Fig. S7). In A. thaliana, ROXY1 appears to be a negative regulator of AG, and the homolog of ROXY in maize appears to regulate shoot meristem size and phyllotaxy (Xing et al., 2005; Yang et al., 2015). In Aquilegia, expression of AqROXYa is detected across the FM at the earliest developmental stages (Fig. 5M), but this broad expression disappears once the primordia begin initiating. AqROXYa is detected in a restricted abaxial region of each emerging floral organ primordium but quickly declines once they begin to differentiate (Fig. 5M–Oʹ). For instance, in an FM with several whorls of initiated stamen primordia, the expression of AqROXYa is only detected in the abaxial side of the innermost two whorls of emerging organ primordia (Fig. 5O, Oʹ).
The confidence interval of Q4 spanned 3.8 Mb but only contained 176 annotated genes that were expressed in the RNAseq dataset (Supplementary Table S5). Tandem duplicates of the C-class gene homolog AqAG1 are located at the edge of the genomic interval, quite distant from the marker with the highest LOD score (Fig. 4B). Within the 1 Mb region (4 800 000–5 800 000) that contained the genetic marker with the highest LOD, there were only 40 expressed genes, eight of which are Aquilegia specific without any annotated A. thaliana homologs, with most of the remaining genes annotated to be involved in plant defense and basic metabolic functions (Supplementary Table S6). Since a previous study showed that miRNA2275 (miR2275) precursors and 24-PHAS loci are significantly enriched in chromosome 4 compared with other chromosomes in Aquilegia (Pokhrel et al., 2021b), we also searched for potential miRNA-encoding loci under Q4. MiR2275 is the primary miRNA that triggers phased, secondary, small interfering RNAs (phasiRNAs) of 24 nucleotides in length, and the production of 24-nt phasiRNAs requires both miR2275 copies and 24-PHAS loci as their targets (Liu et al., 2020). We found that the region with the highest LOD under Q4 overlaps with the genomic region with the highest density of miR2275 precursors and 24-PHAS loci, including three of the 11 annotated clusters of miR2275 precursors and 91 24-PHAS loci in total, comprising 31.7% of all 24-PHAS loci on chromosome 4 (~45.8 Mb) and 14.1% of such loci in the entire genome (~300 Mb) (Pokhrel et al., 2021b).
For Q5, there are two genes located within the highest LOD interval that have homologs that are known to function as FM regulators: AqSEUSS (AqSEU) and AqARGONAUTE5a (AqAGO5a). In A. thaliana, SEU is known to repress AG to regulate FM and organ patterning (Pfluger and Zambryski, 2004; Grigorova et al., 2011; Wynn et al., 2014), while in Aquilegia, AqAGO5a has been identified as a core hub gene associated with early FM development (Min and Kramer, 2020).
Under Q6, we located the homolog of HANABA TANARU (AqHAN). HAN codes for a GATA-type zinc finger transcription factor and, in A. thaliana, HAN is expressed at the organ boundaries, is known to regulate WUS expression, and directly interacts with a number of key genes in FM regulation and primordia initiation (Zhao et al., 2004; Ding et al., 2015). As mentioned above, we have detected significant interaction between Q1 and Q6 (Supplementary Fig. S5). We found it intriguing that HAN and PTL interact through JAG to control FM morphogenesis in A. thaliana (Ding et al., 2015), since their Aquilegia homologs are located within the confidence interval of Q1 and Q6, respectively.
Lastly, AqHOMEOBOX GENE 1 (AqATH1) is the only gene located within the highest LOD interval of Q7 that is annotated with a meristem-related function (Supplementary Fig. S8). In A. thaliana, ATH1 regulates the boundary between the stem and the lateral organs, but is also involved in stem cell regulation in meristems by maintaining the expression of the meristem marker gene SHOOT MERISTEMLESS via a self-activation loop (Gómez-Mena and Sablowski, 2008; Li et al., 2012; Cao et al., 2020). AqATH1 is broadly expressed across the Aquilegia FM throughout the early developmental stages (Fig. 5Q–T), in all early floral organ primordia (Fig. 5R), and at the distal tip of the young lateral organs such as the bracts (Fig. 5Q), sepals (Fig. 5R, S), and petals (Fig. 5T).
Discussion
Aquilegia is an ideal system for studying FM regulation and termination
Over recent decades, we have gained significant insight into various aspects of plant meristem development and function, but the regulation of FMT remains a poorly studied subject. This is despite the fact that FMT is an indispensable process in floral development, and variation in FMT timing is a key component of the generation of floral morphological diversity (Fig. 1). Progress in understanding the regulation and evolution of FMT is hampered due to the lack of natural variation in floral organ whorl numbers in all of our currently established model systems (and their close relatives), while taxa with such a variation generally lack the genomic and molecular resources to investigate this question further. To this end, Aquilegia can be an ideal system for studying FMT because species of Aquilegia share relatively low interspecific sequence variation combined with a high degree of interfertility thanks to its recent adaptive radiation (Hodges and Arnold, 1994; Filiault et al., 2018). At the same time, they all share a consistent floral bauplan that only varies in SWN (Munz, 1946), and possess a fully sequenced and well-annotated genome along with RNAi-based methods for functional studies (Kramer, 2009; Filiault et al., 2018). Recognizing that floral SWN is the best available quantitative trait to represent the timing of FMT, we utilized a genetic cross between two sister species differing in SWN, and sought to take the first step to explore the molecular basis of naturally occurring variation in the FMT timing. The mean SWN of A. brevistyla and A. canadensis is 7.16 and 9.16, respectively, and they do not overlap, while the mean SWN of their F2 progeny was found to encompass the entire range of the parental species (Fig. 2E).
One question we sought to explore was whether there is any difference in early FM growth dynamics between the parental species by analyzing developmental histological series of FMs (Fig. 3). Overall, we observe that the A. canadensis FMs (i) are larger in general, (ii) have a longer developmental window to increase FM width, and, yet, (iii) still make five stamens per whorl. There are numerous previous studies showing that an increase in FM diameter is often associated with an increase in floral organ number per whorl, rather than an increase in the number of whorls (e.g. Carles et al., 2004; Fan et al., 2014; Chu et al., 2019). Of course, these studies typically rely on mutagenesis or gene overexpression rather than natural variation. This suggests that natural variation in meristem size relies on a greater degree of coordination such that meristem size changes in conjunction with the size of primordia inhibition fields, allowing the number of organs per whorl to stay constant. The current data do not allow us to distinguish between whether the A. canadensis FM is growing for a longer period (e.g. perhaps plastochrons are slower, allowing more mass to be accumulated between subsequent whorls) or proliferating at a faster rate. Given what we know about the role of cell division timing in influencing FMT, answering this question is important to understanding the FMT mechanism in Aquilegia. Future studies using a recently developed live imaging technique in Aquilegia (Min et al., 2022) may allow us to compare growth rates between the initiation of successive whorls in these two species and better characterize this phenomenon.
Another curious observation regarding SWN is that we observed a small portion of individuals in both parental species as well as the F2s that exhibited no variation in SWN, regardless of how many flowers were counted on the plants. In contrast, most other individuals exhibited variation in SWN within an individual plant (Supplementary Fig. S3). This seems to suggest that there is variation in the robustness of this trait between different individuals. Unfortunately, the fact that there was no significant divergence in this pattern between the parent species meant that we could not map it in the current study, but we hope that examination of within-inflorescence SWN canalization in other Aquilegia species will allow the identification of suitable models and the dissection of its genetic basis.
Variation in the timing of Aquilegia FMT is controlled by multiple loci of small effects
We recovered seven major QTL that are responsible for variation in SWN, with one QTL located on each chromosome, and the percentage of phenotypic variance explained by each QTL ranging from 3.7% to 8.8% (Fig. 4; Table 1). These results are comparable with those of previous studies in meristem-related traits of domesticated crops, particularly maize, which also revealed multiple QTL of small effects (Vlăduţu et al., 1999; Upadyayula et al., 2006; Bommert et al., 2013; Thompson et al., 2014, 2015). Interestingly, although all the meristem-related traits measured in maize were highly heritable, the total percentage of variance explained by all the QTL was never higher than 50% (e.g. Bommert et al., 2013; Thompson et al., 2014, 2015), suggesting that there are other loci with even smaller effects that were not picked up by the QTL mapping, which is a likely scenario for our current study as well.
We have identified potential candidate genes under the QTL (Fig. 4; Supplementary Table S6), and, further, uncovered novel FM expression patterns of AqZPR3a and AqROXYa, which were the loci of interest associated with Q2 and Q3, respectively (Figs 4, 5). In A. thaliana and tomato, expression of the ZPR genes is restricted to the adaxial region of lateral organs and the central zone of the shoot meristem, and the ZPR genes function in both establishing organ polarity and restricting the stem cell domain in the meristems by acting as post-translational suppressors of the class III HD-ZIP abaxial identity genes by inhibiting their homodimerization (Wenkel et al., 2007; Kim et al., 2008; Weits et al., 2019; Xu et al., 2019). However, we have also observed strong expression of AqZPR3a in the central epidermal layer of FMs throughout their early developmental stages (Fig. 5F–I), which has not been observed in any previous studies. It will be very interesting to determine whether this expression pattern indicates a novel function or is related to known ZPR functions in modulating meristem regulation. In the case of ROXY homologs in other models, expression has been found to be restricted to incipient and newly emerged organ primordia (Xing et al., 2005; S. Li et al., 2009; Yang et al., 2015), but abaxialized expression such as what was found for AqROXYa has not been observed before. In A. thaliana, ROXY1 is known to interact with PTL to regulate floral primordium initiation while, in maize, a ROXY homolog controls meristem size primordia (Xing et al., 2005; S. Li et al., 2009; Yang et al., 2015); either of these functions could be important for controlling FMT in Aquilegia.
The Q4 locus is particularly intriguing because it explains the highest relative percentage of phenotypic variation, but it is also the QTL with the fewest obvious candidate genes to investigate (Supplementary Tables S4, S5). Chromosome 4 of Aquilegia appears to have followed a distinct evolutionary path from the rest of the genome and displays many unique features compared with the remaining six chromosomes, including having a higher proportion of genes arrayed in tandem and segmental duplicates, more genetic polymorphism and transposable elements, lower gene density, and reduced gene expression (Filiault et al., 2018; Aköz and Nordborg, 2019). Although the AqAG1 tandem duplication is included in the 95% Bayesian credible interval, it may be less likely to be the causative gene compared with other genes that were located closer to the highest LOD score marker. The lack of potential candidate genes under Q4 led us to consider other factors besides protein-coding genes, leading to the finding that the highest Q4 LOD interval overlaps with the region that harbors the most concentrated density of miR2275 precursors and 24-PHAS loci in the entire Aquilegia genome (Pokhrel et al., 2021b). As the primary miRNA that triggers 24-nt phasiRNA, a pathway that is conserved across the angiosperms, miR2275 has been shown to be expressed in the reproductive tissues of various monocot and dicot lineages, particularly in developing anthers (Zhai et al., 2015; Fei et al., 2016; Kakrana et al., 2018; Pokhrel et al., 2021a, b). However, relatively little is known about 24-nt phasiRNAs in general besides their functions in anthers. Overall, chromosome 4 remains an enigmatic component of the Aquilegia genome, so it is intriguing that the QTL is located on this structure. Certainly, it is also possible that the causal gene underlying Q4 is one of the Aquilegia-specific loci that did not have a direct A. thaliana homolog, which equally applies to the other QTL as well.
The current study is a key first step in identifying a promising list of candidate genes for regulating natural variation in FMT. The next steps in evaluating these loci will include exploration of fine-mapping, assays of gene function, conducting comparative expression analyses between A. canadensis and A. brevistyla, and examining sequence variation and patterns of allelic differentiation between populations of these species. Further areas of interest would also include exploring the potential ecological consequences of variation in SWN and FMT between these species and across the genus.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. SWN and the position of flowers on inflorescences.
Fig. S2. F1-parent-of-origin has a significant impact on the distribution of SWN in the respective F2 progeny.
Fig. S3. Distribution of the SD of SWN among the parental and F2 populations.
Fig. S4. Diagram and summary statistics of the genetic map.
Fig. S5. Confirmation of QTL underlying SWN variation.
Fig. S6. Effect plots of the markers that have the highest LOD score under each QTL.
Fig. S7. Gene phylogeny for AqROXYa.
Fig. S8. Gene phylogeny for AqATH1.
Table S1. Primers used for constructing in situ hybridization probes.
Table S2. Pairwise comparison of FM widths through early developmental stages.
Table S3. No significant evidence supporting the presence of a second QTL on any chromosome.
Table S4. Summary of number of genes under each potential QTL.
Table S5. Expressed genes under Q4.
Table S6. Information on candidate genes.
Acknowledgements
The authors would like to thank Nicole Bedford, Olivia Meyerson, and Rubén Rellán-Álvarez for discussing QTL mapping analysis; Suresh Pokhrel for providing information on miRNA in Aquilegia; Pierre Baduel and Rebecca Povilus for discussing data analysis strategies; the graduate students at the Harvard Statistics Department who volunteered their time for professional and free statistics consultation; and Karl Broman for actively answering questions about the R/qtl package on online forums.
Contributor Information
Ya Min, Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, USA.
Evangeline S Ballerini, Department of Biological Sciences, California State University, Sacramento, Sacramento, CA, USA.
Molly B Edwards, Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, USA.
Scott A Hodges, Department of Ecology & Marine Biology, University of California, Santa Barbara, CA, USA.
Elena M Kramer, Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA, USA.
Zoe Wilson, University of Nottingham, UK.
Author contributions
YM and EK: conceptualization and design; EB: collection of seeds, crossing and sequencing the parental species, and growing and pollinating the F1 individuals; ME and YM: growing the F2s; YM: conducting all phenotyping; EB, ME, and YM: preparing the libraries for sequencing of the F2 individuals and constructing the genetic map; YM: data analysis and in situ hybridization; EK and SH: supervision; YM: writing with input from the co-authors.
Conflict of interest
The authors declare no conflict of interest.
Funding
Funding has been provided by a Simmon’s Award to YM from the Harvard Center for Biological Imaging; a National Science Foundation Graduate Research Fellowship under grant no. DGE1745303 to MBE; and both a NIH Ruth L. Kirschstein National Research Service Award (F32GM103154) and a UC Santa Barbara Harvey Karp Discovery award to ESB. Sequencing was carried out by the DNA Technologies and Expression Analysis Cores at the UC Davis Genome Center, supported by NIH Shared Instrumentation Grant 1S10OD010786-01 and the Biological Nanostructures Lab at UC Santa Barbara.
Data availability
All sequence data are deposited in the Sequence Read Archive under BioProject ID PRJNA720109. Scripts and genotype/phenotype data are available upon request. All other data supporting the findings of this study can be found within the paper or within its supplementary data published online.
References
- Aköz G, Nordborg M. 2019. The Aquilegia genome reveals a hybrid origin of core eudicots. Genome Biology 20, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballerini ES, Min Y, Edwards MB, Kramer EM, Hodges SA. 2020. POPOVICH, encoding a C2H2 zinc-finger transcription factor, plays a central role in the development of a key innovation, floral nectar spurs, in Aquilegia. Proceedings of the National Academy of Sciences, USA 117, 22552–22560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastida JM, Alcántara JM, Rey PJ, Vargas P, Herrera CM. 2010. Extended phylogeny of Aquilegia: the biogeographical and ecological patterns of two simultaneous but contrasting radiations. Plant Systematics and Evolution 284, 171–185. [Google Scholar]
- Bollier N, Sicard A, Leblond J, et al. 2018. At-MINI ZINC FINGER2 and Sl-INHIBITOR OF MERISTEM ACTIVITY, a conserved missing link in the regulation of floral meristem termination in Arabidopsis and tomato. The Plant Cell 30, 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Nagasawa NS, Jackson D. 2013. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nature Genetics 45, 334–337. [DOI] [PubMed] [Google Scholar]
- Brewer PB, Howles PA, Dorian K, Griffith ME, Ishida T, Kaplan-Levy RN, Kilinc A, Smyth DR. 2004. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 131, 4035–4045 [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. [DOI] [PubMed] [Google Scholar]
- Cao X, Wang J, Xiong Y, Yang H, Yang M, Ye P, Bencivenga S, Sablowski R, Jiao Y. 2020. A self-activation loop maintains meristematic cell fate for branching. Current Biology 30, 1893–1904.e4. [DOI] [PubMed] [Google Scholar]
- Carles CC, Lertpiriyapong K, Reville K, Fletcher JC. 2004. The ULTRAPETALA1 gene functions early in Arabidopsis development to restrict shoot apical meristem activity and acts through WUSCHEL to regulate floral meristem determinacy. Genetics 167, 1893–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Jang J, Huang Z, van der Knaap E. 2019. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct 3, e00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Yan S, Jiang L, Zhao W, Ning K, Zhao J, Liu X, Zhang J, Wang Q, Zhang X. 2015. HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis). PLoS Genetics 11, e1005479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MB, Choi GPT, Derieg NJ, Min Y, Diana AC, Hodges SA, Mahadevan L, Kramer EM, Ballerini ES. 2021. Genetic architecture of floral traits in bee- and hummingbird-pollinated sister species of Aquilegia (columbine). Evolution 75, 2197–2216. [DOI] [PubMed] [Google Scholar]
- Endress PK. 1990. Patterns of floral construction in ontogeny and phylogeny. Biological Journal of the Linnean Society 39, 153–175. [Google Scholar]
- Endress PK. 2011. Evolutionary diversification of the flowers in angiosperms. American Journal of Botany 98, 370–396. [DOI] [PubMed] [Google Scholar]
- Fan C, Wu Y, Yang Q, Yang Y, Meng Q, Zhang K, Li J, Wang J, Zhou Y. 2014. A novel single-nucleotide mutation in a CLAVATA3 gene homolog controls a multilocular silique trait in Brassica rapa L. Molecular Plant 7, 1788–1792 [DOI] [PubMed] [Google Scholar]
- Fei Q, Yang L, Liang W, Zhang D, Meyers BC. 2016. Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. Journal of Experimental Botany 67, 6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiault DL, Ballerini ES, Mandáková T, et al. 2018. The Aquilegia genome provides insight into adaptive radiation and reveals an extraordinarily polymorphic chromosome with a unique history. eLife 7, e36426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fior S, Li M, Oxelman B, Viola R, Hodges SA, Ometto L, Varotto C. 2013. Spatiotemporal reconstruction of the Aquilegia rapid radiation through next-generation sequencing of rapidly evolving cpDNA regions. New Phytologist 198, 579–592. [DOI] [PubMed] [Google Scholar]
- Galli M, Gallavotti A. 2016. Expanding the regulatory network for meristem size in plants. Trends in Genetics 32, 372–383. [DOI] [PubMed] [Google Scholar]
- Gómez-Mena C, Sablowski R. 2008. ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. The Plant Cell 20, 2059–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T. 2003. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes & Development 17, 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith ME, da Silva Conceição A, Smyth DR. 1999. PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development 126, 5635–5644. [DOI] [PubMed] [Google Scholar]
- Grigorova B, Mara C, Hollender C, Sijacic P, Chen X, Liu Z. 2011. LEUNIG and SEUSS co-repressors regulate miR172 expression in Arabidopsis flowers. Development 138, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges SA, Arnold ML. 1994. Columbines: a geographically widespread species flock. Proceedings of the National Academy of Sciences, USA 91, 5129–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana A, Mathioni SM, Huang K, et al. 2018. Plant 24-nt reproductive phasiRNAs from intramolecular duplex mRNAs in diverse monocots. Genome Research 28, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-S, Kim S-G, Lee M, et al. 2008. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. The Plant Cell 20, 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer EM. 2005. Methods for studying the evolution of plant reproductive structures: comparative gene expression techniques. Methods in Enzymology 395, 617–636. [DOI] [PubMed] [Google Scholar]
- Kramer EM. 2009. Aquilegia: a new model for plant development, ecology, and evolution. Annual Review of Plant Biology 60, 261–277. [DOI] [PubMed] [Google Scholar]
- Lampugnani ER, Kilinc A, Smyth DR. 2012. PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana: PTL controls sepal boundaries. The Plant Journal 71, 724–735. [DOI] [PubMed] [Google Scholar]
- Lenhard M. 2003. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130, 3163–3173. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. 2001. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. 2009. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. The Plant Cell 21, 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pi L, Huang H, Xu, L. 2012. ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. Journal of Experimental Botany 63, 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A, Kramer EM. 2010. The ABC model and the diversification of floral organ identity. Seminars in Cell & Developmental Biology 21, 129–137. [DOI] [PubMed] [Google Scholar]
- Liu Y, Teng C, Xia R, Meyers BC. 2020. PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. The Plant Cell 32, 3059–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y, Conway SJ, Kramer EM. 2022. Quantitative live imaging of floral organ initiation and floral meristem termination in Aquilegia. Development 149, dev200256. [DOI] [PubMed] [Google Scholar]
- Min Y, Kramer EM. 2017. The Aquilegia JAGGED homolog promotes proliferation of adaxial cell types in both leaves and stems. New Phytologist 216, 536–548. [DOI] [PubMed] [Google Scholar]
- Min Y, Kramer EM. 2020. Transcriptome profiling and weighted gene co-expression network analysis of early floral development in Aquilegia coerulea. Scientific Reports 10, 19637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. 2006. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. The Plant Cell 18, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz PA. 1946. Aquilegia: the cultivated and wild columbines. Ithaca. NY: Cornell University. [Google Scholar]
- Nardmann J, Werr W. 2006. The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Molecular Biology and Evolution 23, 2492–2504. [DOI] [PubMed] [Google Scholar]
- Ouellette LA, Reid RW, Blanchard SG, Brouwer CR. 2018. LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics 34, 306–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T. 2004. KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131, 3737–3749. [DOI] [PubMed] [Google Scholar]
- Pfluger J, Zambryski P. 2004. The role of SEUSS in auxin response and floral organ patterning. Development 131, 4697–4707. [DOI] [PubMed] [Google Scholar]
- Pokhrel S, Huang K, Bélanger S, Caplan JL, Kramer EM, Meyers BC. 2021a. Pre-meiotic, 21-nucleotide reproductive phasiRNAs emerged in seed plants and diversified in flowering plants. Nature Communications 12, 4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhrel S, Huang K, Meyers, BC. 2021b. Conserved and non-conserved triggers of 24-nt reproductive phasiRNAs in eudicots. The Plant Journal 107, 1332–1345. [DOI] [PubMed] [Google Scholar]
- Ronse De Craene L. 2018. Understanding the role of floral development in the evolution of angiosperm flowers: clarifications from a historical and physico-dynamic perspective. Journal of Plant Research 131, 367–393. [DOI] [PubMed] [Google Scholar]
- Ruzin S. 1999. Plant microtechnique and microscopy. New York: Oxford University Press. [Google Scholar]
- Sauret-Güeto S, Schiessl K, Bangham A, Sablowski R, Coen E. 2013. JAGGED controls Arabidopsis petal growth and shape by interacting with a divergent polarity field. PLoS Biology 11, e1001550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. 2000. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. 1999. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proceedings of the National Academy of Sciences,USA 96, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. 1989. Patterns in plant development. Cambridge: Cambridge University Press. [Google Scholar]
- Sun B, Xu Y, Ng K-H, Ito T. 2009. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes & Development 23, 1791–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AM, Crants J, Schnable PS, Yu J, Timmermans MCP, Springer NM, Scanlon MJ, Muehlbauer GJ. 2014. Genetic control of maize shoot apical meristem architecture. G3 Genes|Genomes|Genetics 4, 1327–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AM, Yu J, Timmermans MCP, Schnable P, Crants JE, Scanlon MJ, Muehlbauer GJ. 2015. Diversity of maize shoot apical meristem architecture and its relationship to plant morphology. G3 Genes|Genomes|Genetics 5, 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadyayula N, da Silva HS, Bohn MO, Rocheford TR. 2006. Genetic and QTL analysis of maize tassel and ear inflorescence architecture. Theoretical and Applied Genetics 112, 592–606. [DOI] [PubMed] [Google Scholar]
- Vlăduţu C, McLaughlin J, Phillips RL. 1999. Fine mapping and characterization of linked quantitative trait loci involved in the transition of the maize apical meristem from vegetative to generative structures. Genetics 153, 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Larsen J, Harder LD. 2000. The evolution of staminodes in angiosperms: patterns of stamen reduction, loss, and functional re-invention. American Journal of Botany 87, 1367–1384. [PubMed] [Google Scholar]
- Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K. 2014. Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. The Plant Cell 26, 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weits DA, Kunkowska AB, Kamps NCW, et al. 2019. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 569, 714–717. [DOI] [PubMed] [Google Scholar]
- Wenkel S, Emery J, Hou B-H, Evans MMS, Barton MK. 2007. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. The Plant Cell 19, 3379–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitewoods CD, Cammarata J, Venza ZN, et al. 2020. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Current Biology 30, 2645–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn AN, Seaman AA, Jones AL, Franks RG. 2014. Novel functional roles for PERIANTHIA and SEUSS during floral organ identity specification, floral meristem termination, and gynoecial development. Frontiers in Plant Science 5, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Rosso MG, Zachgo S. 2005. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132, 1555–1565. [DOI] [PubMed] [Google Scholar]
- Xu Q, Li R, Weng L, Sun Y, Li M, Xiao H. 2019. Domain-specific expression of meristematic genes is defined by the LITTLE ZIPPER protein DTM in tomato. Communications Biology 2, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Bui HT, Pautler M, Llaca V, Johnston R, Lee B, Kolbe A, Sakai H, Jackson D. 2015. A maize glutaredoxin gene, Abphyl2, regulates shoot meristem size and phyllotaxy. The Plant Cell 27, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Zhang H, Arikit S, Huang K, Nan G-L, Walbot V, Meyers, BC. 2015. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proceedings of the National Academy of Sciences, USA 112, 3146–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Medrano L, Ohashi K, Fletcher JC, Yu H, Sakai H, Meyerowitz, EM. 2004. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. The Plant Cell 16, 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data are deposited in the Sequence Read Archive under BioProject ID PRJNA720109. Scripts and genotype/phenotype data are available upon request. All other data supporting the findings of this study can be found within the paper or within its supplementary data published online.