Abstract
Initiation of human immunodeficiency virus type 1 (HIV-1) reverse transcription occurs by extension of the cellular tRNA3Lys which anneals to the primer-binding site (PBS) on the 5′ non-translated region of the viral RNA genome. The A-rich sequence (A-loop) upstream of the PBS interacts with the anticodon loop of tRNA3Lys and has been proposed to be essential for conferring specificity to tRNA3Lys for priming the initiation of HIV-1 reverse transcription. We observed that polyamide nucleic acid targeted to the A-loop sequence (PNAal) exhibits high binding specificity for its target sequence. The PNAal pre-bound to the A-loop sequence prevents tRNA3Lys priming on the viral RNA consequently blocking in vitro initiation of reverse transcription. Further, PNAal can efficiently disrupt the preformed [tRNA3Lys–viral RNA] complex thereby rendering it non-functional for reverse transcription. The endogenous reverse transcription in disrupted HIV-1 virions containing packaged tRNA3Lys and its replicating enzyme RT was significantly inhibited by PNAal, thus providing direct evidence of the involvement of the A-loop region of viral RNA genome in tRNA3Lys priming process. These findings suggest the potential of the A-loop region as a critical target for blocking HIV-1 replication.
INTRODUCTION
Currently utilized therapies for AIDS involve inhibitors which block viral maturation and reverse transcription of viral RNA into double-stranded DNA (1–3). However, the rapid emergence of drug-resistant strains has considerably overshadowed the benefits of the clinically available anti-human immunodeficiency virus type 1 (HIV-1) drugs. Selection of the dominant, pre-existing drug-resistant variants and the abundance of latently infected cells (which possess integrated proviral DNA) are the potential barriers encountered to effective drug therapy (2–4). A potentially important approach to overcome this problem is to target those regions of the viral genome that are essential for viral replication but averse to mutational changes.
The unique 5′ (R-U5-PBS) non-translated region (1–333 nt) of the HIV-1 genome containing several critical domains essential for viral replication may be an ideal target for drug intervention. These critical domains comprise of: (i) primer-binding site (PBS; nt 183–201), essential for tRNA3Lys primed initiation of reverse transcription (5–9); (ii) the A-loop region, located upstream of the PBS (nt 168–173) is essential for the selection and interaction of tRNA3Lys primer (10–12); (iii) the long terminal repeat (LTR) sequences at the 5′ and 3′ ends, essential for viral transcription and integration (13); and (iv) the trans-activation response element (TAR), essential for viral gene expression via transcriptional activation (14–16) and probably having some additional role in the initiation of reverse transcription (17,18). These regulatory sequences in the 5′ non-translated region are averse to mutational changes and, therefore, can be potential targets for arresting viral replication.
We have earlier shown that a polyamide nucleic acid (PNA) targeted to the PBS region of the viral genome, blocks the initiation of reverse transcription (19). We have further demonstrated that PNA targeted to the TAR sequence of the viral RNA genome is able to prevent Tat–TAR interaction by efficient sequestration of the TAR and block Tat-mediated trans-activation of HIV-1 LTR transcription (20). PNAs are DNA homologs containing a peptide backbone of 2-aminoethylglycine units to which purine and pyrimidine bases are linked. PNAs are resistant to degradation by nucleases and proteases (21) and are able to bind the target region in the duplex DNA by invasion and displacement of one of the DNA strands (19,22,23). The high stability of PNA–nucleic acid complexes is reduced dramatically by single base pair mismatches, suggesting that PNAs recognize their targets in a sequence-specific manner (19,21,23). PNA, with a terminal DNA nucleotide, is also recognized as a bona fide primer by HIV-1 reverse transcriptase (RT), resulting in abortive reverse transcription products (19,24,25).
In this communication, we present evidence to demonstrate that a 15mer PNA, targeted to the A-loop sequence (named as PNAAL) upstream of the PBS region, can specifically sequester the target sequence and effectively inhibit the initiation of reverse transcription. We chose the A-loop region as a target as it has been shown to be critical for stabilization of the tRNA3Lys primer on the viral genome (10–12,26–29). A scrambled PNA sequence was used in all experiments as a negative control to determine the specificity of PNAAL. Our results indicate that PNAAL either prevents the tRNA3Lys primer forming a complex with the PBS region of the viral genome or destabilizes the tRNA3Lys–PBS complex and inhibits both initiation of reverse transcription and elongation of initiated primer in vitro. The PNAAL was equally effective in blocking the process of reverse transcription in isolated HIV-1 virions. The significance of these findings may lead to advances in HIV-1 therapeutic research.
MATERIALS AND METHODS
DNA modifying enzymes were purchased from Promega or Roche Biochemicals. Tritiated dNTPs, [γ-32P]ATP and [α-32P]dNTPs were the products of Dupont-New England Nuclear Inc. The DNA oligomers were synthesized at the Molecular Resource Facility at UMDNJ. PNA oligomers were synthesized at the Applied Biosystems Inc. All other reagents were of the highest available purity grade and purchased from Fisher, Millipore Corp., and Bio-Rad.
Plasmid and clones
The expression vector pKK-RT66 was constructed in this laboratory and used to purify the wild type HIV-1 RT (19). An HIV-RNA expression clone pHIV-PBS was a generous gift from Dr M. A. Wainberg (30). This clone contains a 947 bp fragment (+473 to +1420) of pHλHXB2 HIV-1 proviral clone and was used to transcribe the 495 base-long HIV-1 RNA template.
Isolation of p66/51 HIV-1 RT
The recombinant clone pKK-RT66 encoding the wild type p66 HIV-1 RT was expressed in JM109 and purified as described before (19). The heterodimeric HIV-1 RT (p66/p51) was generated by the proteolytic cleavage of the p66/p66 homodimer as described below. The proteolytic buffer in a total volume of 2 ml contained 0.1 M potassium phosphate, pH 7.5, 1.0 M NaCl, 1 mM dithiothreitol (DTT), 1 mM EDTA, 25 µM of p66/p66 (6.1 mg) and 25 µM of HIV-1 protease (1.15 mg). The mixture was incubated at 4°C and the extent of proteolytic cleavage was monitored by SDS–PAGE followed by Coomassie blue staining. Following complete cleavage of the second p66 subunit (∼16 h incubation), the reaction mixture was diluted 20-fold with 20 mM potassium phosphate buffer, pH 8.0, and applied to a 5 ml phosphocellulose column pre-equilibrated with a buffer containing 20 mM potassium phosphate, pH 8.0, 50 mM NaCl and 1 mM DTT. After washing the column with 10 column vol of the same buffer, the heterodimeric RT was eluted with 20 ml of this buffer containing 250 mM NaCl and concentrated by precipitating with ammonium sulfate at 70% saturation. The precipitate was dissolved in a small volume of buffer consisting of 50 mM Tris–HCl pH 7.5, 100 mM NaCl and 1 mM DTT, followed by dialysis against the same buffer containing 50% glycerol. The enzyme preparation was stored at –70°C. Protein concentrations were determined by using the Bio-Rad colorimetric kit as well as by spectrophotometric measurements using É280 = 2.62 × 105 M–1 cm–1 for p66/51 heterodimers (31).
Isolation of HIV-1 virions
The lymphocyte cells, CEM CD4+ (12D7), were grown in complete RPMI-1640 medium. The cells in the log phase were harvested, and washed with equal volumes of phosphate buffered saline (PBS) without Ca2+ or Mg2+. Cells (107 cells/ml) were re-suspended in RPMI-1640 medium (without fetal calf serum, l-glutamine or penicillin/streptomycin). A portion of the cell suspension (250 µl, 2.5 × 106 cells) mixed with proviral plasmid DNA (3 µg) was electroporated at 230 V and then plated in 10 ml of complete RPMI-1640 media (32). Cells were subsequently incubated at 37°C in 5% CO2 containing humidified air for 3–4 days. The cells were centrifuged at 1200 r.p.m. for 7 min and the supernatant was used to monitor the levels of p24 antigen using the ELISA p24 antigen kit (ABBOTT Laboratories). The culture supernatant was further centrifuged at 30 000 g for 90 min to pellet the HIV-1 virions. The HIV-1 virion pellet was disrupted in a buffer containing 50 mM Tris–HCl pH 7.5, 100 mM NaCl and 0.1% NP-40 and used as a source of endogenous RT as well as tRNA3Lys-primed HIV-1 viral RNA genome.
Construction of the HIV-1 RNA expression clone pU5-PBS for gel retardation analysis
We constructed an HIV-1 RNA expression clone, pU5-PBS, in order to generate a shorter HIV-1 RNA transcript for analyzing its binding affinity to the A-loop PNA. This clone contains a 183 bp fragment corresponding to nt 473–656 of pHλHXB2 HIV-1 proviral clone comprising of PBS, U5 and part of the R region. For constructing this clone, the 183 bp fragment was amplified from the plasmid pHIV-PBS (30) using Pfu DNA polymerase and upstream and downstream primers with XbaI and XhoI restriction sites, respectively. The sequence of the upstream primer was 5′-GAT ACT CTA GAA GAT CTG AGC CT-3′ and that of the downstream primer was 5′-GAT ACC TCG AGC AGG TCC CTG TTC GG-3′. The resulting PCR product was digested with XbaI and XhoI restriction enzymes and cloned in pET-28a vector.
Preparation of R-U5-PBS HIV-1 RNA template
Two plasmids, pHIV-PBS and pU5-PBS, were used to transcribe the 495 and 200 base-long U5-PBS RNA templates, respectively. The 495 base-long RNA transcript contains 11 nt derived from the plasmid vector flanking the 5′ terminus and the remaining 484 nt correspond to part of the non-coding and gag-coding regions, the primer binding sequence, U5 and part of the R region (57 nt). The 200 base-long RNA transcript contains 183 nt corresponding to the HIV-1 region comprising of the primer-binding sequence, U5 and part of the R region. The additional 17 nt flanking the 5′ terminus are derived from the plasmid vector. The plasmid pHIV-PBS was linearized by AccI and pU5-PBS by XhoI restriction enzymes and transcribed using T7 RNA polymerase and other reaction components from Roche Biochemicals. After in vitro transcription reaction, the DNA template was removed by DNase I digestion and the RNA transcripts were purified by phenol–chloroform extraction and alcohol precipitation. The RNA products were dissolved in DEPC-treated water containing 10 mM DTT and further purified by G-50 spin column and stored at –70°C.
Preparation of 32P-labeled U5-PBS RNA template
The plasmid pU5-PBS linearized with XhoI was transcribed using T7 RNA polymerase (Roche Molecular Biochemicals) to prepare 32P-labeled 200 base-long U5-PBS RNA. The RNA transcript was labeled internally by including [α-32P]UTP (3000 Ci/mmol; Amersham Life Sciences) and purified by phenol–chloroform extraction and alcohol precipitation. The RNA product was dissolved in DEPC-treated water containing 10 mM DTT, further purified by G-50 spin column and stored at –70°C.
Gel retardation assay
The internally 32P-labeled 200 base-long U5-PBS RNA (5000 Cerenkov c.p.m.) was incubated at varying molar ratios of PNAAL or scrambled PNA for 2 h at 37°C in a binding buffer containing 30 mM Tris–HCl pH 8.0, 60 mM KCl, 5.5 mM MgCl2, 10 mM DTT, 10% glycerol, 0.01% NP-40 and 500 ng of r (I-C), in a final volume of 15 µl (20). Two microliters of RNA gel loading dye (0.27% bromophenol blue and 20% glycerol) was added to the samples and subjected to native gel retardation analysis on an 8% polyacrylamide gel in Tris–borate–EDTA (TBE) buffer containing 5% glycerol. The gels were routinely pre-run at 100 V for 30 min at 4°C in TBE buffer, pH 8.2. The RNA–PNA complexes were resolved from the free RNA at a constant voltage of 150 V at 4°C and subjected to phosphorimager analysis.
Labeling of tRNA3Lys
The high-pressure liquid chromatography-purified human placental tRNA3Lys obtained from BIOS&T was 3′ end labeled with [32P]pCp by T4 RNA ligase according to the standard protocol (33). The labeled product was extracted three times with phenol–chloroform, precipitated with alcohol, lyophilized and suspended in TE buffer. This was further purified on a NAP-10 column to remove the unincorporated radiolabeled nucleotides.
Destabilization of tRNA3Lys–U5-PBS RNA complex by PNAAL
The 495 base-long U5-PBS RNA transcript was incubated with the labeled tRNA3Lys (2 × 104 c.p.m.) at 37°C for 1 h in a buffer containing 50 mM Tris–HCl pH 7.8, 1 mM DTT and 60 mM KCl. This was followed by further incubation for another 1 h in the presence and absence of PNAAL or scrambled PNA. Two microliters of RNA gel loading dye were added to 10 µl of the reaction mixture. The tRNA3Lys–U5-PBS RNA complex formed was resolved by electrophoresis at 4°C on a 6% non-denaturing polyacrylamide gel at 150 V for 3 h in 89 mM Tris–borate buffer, pH 8.2. The gel was dried and analyzed by phosphorimager.
Reverse transcription of U5-PBS RNA template primed with 17mer DNA or tRNA3Lys
An aliquot of the 495 base-long U5-PBS RNA template was annealed with either the labeled 17mer DNA primer complementary to the PBS or with the natural tRNA3Lys. The molar ratio of RNA template to 17mer DNA primer or to tRNA3Lys was 2:1. Reverse transcription reactions were carried out by incubating 2.5 nM of U5-PBS RNA–tRNA3Lys template primer with 50 nM of HIV-1 RT in a reaction mixture containing 25 mM Tris–HCl pH 7.8, 10 mM DTT, 100 µg/ml BSA, 5 mM MgCl2 and 50 µM of dATP, dTTP, dGTP and 2 µM of [α-32P]dCTP (0.25 µCi/pmol). In experiments with U5-PBS RNA template primed with 5′-32P-labeled 17mer DNA primer, the unlabeled dNTPs were supplemented in the reaction at a final concentration of 50 µM (each). The reaction was initiated by the addition of enzyme and terminated by the addition of equal volume of Sanger’s gel loading solution (34). The products were resolved on an 8% polyacrylamide–urea gel.
The U5-PBS RNA pre-bound with tRNA3Lys or 17mer DNA primer was incubated with PNAAL complementary to the A-loop region upstream of the PBS, at varying molar ratios in a buffer containing 50 mM Tris–HCl pH 7.8, 10 mM DTT, 60 mM KCl and 5 mM MgCl2 at 37°C for 2 h. In another set of experiments, both PNAAL and tRNA were allowed to compete for binding to U5-PBS RNA. For this, U5-PBS RNA was incubated with PNAAL at varying molar ratios along with tRNA3Lys at 37°C for 2 h. The extent of DNA synthesis catalyzed by HIV-1 RT in the presence or absence of the PNAAL or scrambled PNA was assessed as described above. A similar set of experiments was performed with scrambled PNA.
Effect of PNAAL on tRNA3Lys-derived endogenous reverse transcription in disrupted HIV-1 virions
The tRNA3Lys-derived endogenous reverse transcription in disrupted HIV-1 virions was carried out as described below. An aliquot (7 µl) of disrupted virions was incubated in a reaction mixture containing 50 mM Tris–HCl pH 7.8, 10 mM DTT, 60 mM KCl, 5 mM MgCl2, 0.01% NP-40, 100 µg/ml BSA, 20 µM each of dATP, dGTP, dTTP and 1 µM of [α-32P]dCTP (0.5 µCi/pmol) in a total volume of 15 µl. The reaction was carried out at 37°C for 30 min and quenched with 20 mM EDTA. The products were alcohol precipitated and analyzed on an 8% denaturing polyacrylamide–urea gel followed by phosphorimaging.
The effect of PNAAL or scrambled PNA on the endogenous reverse transcription in the disrupted virions was assessed by pre-incubating the disrupted virions at varying concentrations of PNA at 4°C for 16 h. The control sample was incubated under identical conditions in the absence of PNA. The extent of endogenous reverse transcription in the disrupted virions was monitored as described above
RESULTS AND DISCUSSION
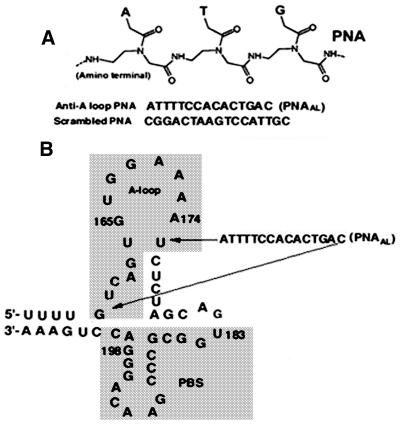
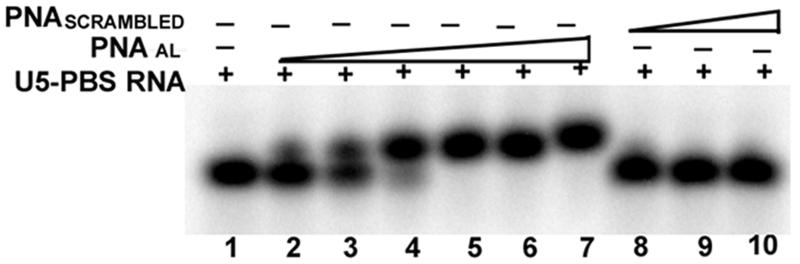
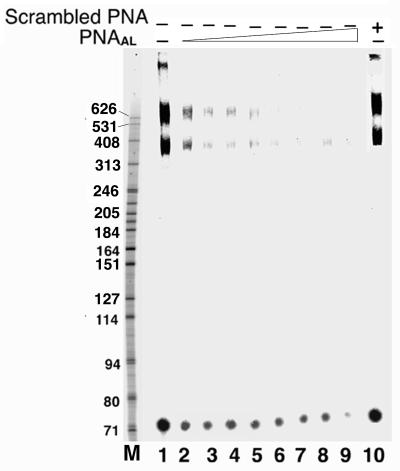
The initiation of HIV-1 reverse transcription occurs by extension of the cellular tRNA3Lys primer annealed near the 5′ non-translated region of the viral RNA genome at a site called the PBS. The 18-nt PBS sequence, located downstream of the U5 region of the 5′ LTR, spans from nt 183–201 of the viral RNA genome and is complementary to the 3′ terminal nucleotides of the primer tRNA3Lys (7). The A-loop region upstream of the PBS has been implicated in the selection and stabilization of tRNA3Lys on the viral genome (10–12). In this study, we have employed a polyamide nucleotide analog (PNAAL) to target the A-loop region in order to examine its influence on tRNA3Lys priming and on the process of reverse transcription. The sequences of PNAAL and non-specific scrambled PNA used in the experiments are shown in Figure 1A. Both PNAAL and scrambled PNA contain purine and pyrimidine bases linked with polyamide backbone in lieu of sugar–phosphate backbone. The target sequence corresponding to the stem–loop upstream of the PBS region where PNAAL binds is shown in Figure 1B. To ascertain the ability of PNAAL to interact with the A-loop region, gel retardation assays were performed with 32P-labeled 200 base-long U5-PBS RNA and PNAAL (Fig. 2). Incubation of PNAAL with U5-PBS RNA, resulted in the formation of a specific RNA–PNAAL complex that could be detected by native PAGE. A shift in U5-PBS RNA mobility was noted with the appearance of a band that ran slower on the gel (Fig. 2, lanes 2–7). This slower moving complex was not present when scrambled PNA was used in place of PNAAL (Fig. 2, lanes 8–10).
Figure 1.
Sequence and general structure of scrambled and PNAAL targeted to the A-loop region of U5-PBS HIV-1 RNA. (A) General structure of PNA. (B) Secondary structure of the HIV-1 RNA genome corresponding to the primer binding site and A-loop region.
Figure 2.
Specificity of PNAAL to its target sequence on the HIV-1 RNA. The binding specificity of PNAAL and scrambled PNA to the A-loop sequence was assessed by gel mobility shift of the 32P-labeled 200 base-long U5-PBS HIV-1 RNA as described under Materials and Methods. Lanes 1–7 represent molar ratios of U5-PBS RNA to PNAAL of 1:0, 1:0.2, 1:0.4, 1:1, 1:2.5, 1:5 and 1:10, respectively. Lanes 8–10 represent experiment with scrambled PNA at molar ratio of U5-PBS RNA to scrambled PNA of 1:2.5, 1:5 and 1:10, respectively.
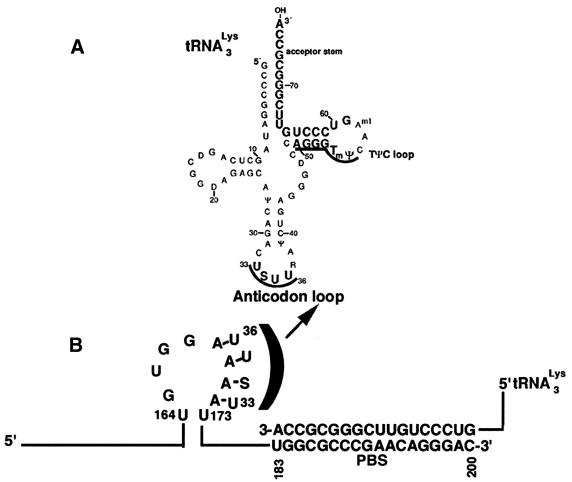
During assembly of the HIV-1 virion, several cellular tRNAs including tRNA3Lys are packaged into the virion particle but HIV-1 utilizes only tRNA3Lys for the initiation of reverse transcription (9,35,36). The sequences upstream of the PBS are complementary to the anticodon loop and the T Ψ C loop and arm of tRNA3Lys (Fig. 3), that have been proposed to be essential for conferring specificity for utilizing tRNA3Lys for the initiation of reverse transcription (12). Among these, the A-loop region upstream of the PBS seems to interact with the U-rich anticodon (USUU) of the tRNA3Lys primer (12,29,37). Mutational studies in this region have shown that the A-loop is essential for maintaining the selective use of the tRNA3Lys primer (10,12,38). We therefore reasoned that sequestering of this important region may have a strong destabilizing effect on the formation of the [enzyme–tRNA3Lys–viral RNA] complex and, in turn, may block the initiation of HIV-1 reverse transcription.
Figure 3.
Schematic representation showing the interaction of tRNA3Lys with the A-loop region of HIV-1 RNA. (A) The secondary structure of tRNA3Lys. The anticodon loop spans from nt 33 to 36. (B) Priming of the PBS region of U5-PBS RNA with the 3′ terminal 18 nt of tRNA3Lys. For clarity, the interaction of the anticodon loop of tRNA3Lys with the A-loop sequence of U5-PBS RNA is depicted by half bracket.
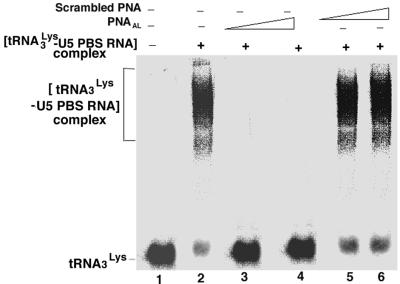
We therefore examined the formation of the [tRNA3Lys–viral RNA] complex in the presence or absence of PNAAL or scrambled PNA. Results obtained from gel retardation studies are shown in Figure 4. As seen in Figure 4, the preformed [tRNA3Lys–viral RNA] complex is disrupted by the interaction of PNAAL with its target sequence on the viral genome (Fig. 4, lanes 3 and 4). The non-specific scrambled PNA had no influence on this interaction (Fig. 4, lanes 5 and 6). It is possible that interaction between the tRNA3Lys and PBS in the virion may be stabilized to a much higher ordered structure by interacting with the A-loop region of the viral RNA genome (39), and perturbation in this loop structure due to its interaction with the complementary PNAAL may have a strong destabilizing effect on the formation of the [tRNA3Lys–viral RNA] complex. This interaction may, in turn, prevent initiation of HIV-1 reverse transcription and the transition of initiated primer to elongation.
Figure 4.
Destabilization of the interaction between tRNA3Lys and U5-PBS RNA by PNAAL. The labeled tRNA3Lys was incubated with the 495 base-long U5-PBS HIV-1 RNA in the presence or absence of PNAAL or scrambled PNA as described in Materials and Methods. The [U5-PBS RNA–tRNA3Lys] complex formed in the absence or presence of PNAAL was resolved by non-denaturing gel retardation analysis. Lane 1, 32P-labeled tRNA3Lys alone; lane 2–4, pre-formed [tRNA3Lys–U5-PBS RNA] complex incubated in the presence of PNAAL at molar ratios of U5-PBS RNA to PNAAL of 0.0, 1:5 and 1:10, respectively. Lanes 5 and 6 represent incubation of [tRNA3Lys–U5-PBS RNA] complex in the presence of scrambled PNA at molar ratios of U5-PBS RNA to scrambled PNA of 1:5 and 1:10, respectively. The positions of free tRNA3Lys as well as tRNA3Lys complexed with U5-PBS RNA are shown on the left.
In order to ascertain this possibility, reverse transcription of U5-PBS HIV-1 RNA primed with either 5′-32P-labeled 17mer DNA primer or with tRNA3Lys by HIV-1 RT was examined in the absence or presence of PNAAL. Incubation of PNAAL with U5-PBS HIV-1 RNA pre-bound with the 17mer DNA primer had no effect on the initiation of reverse transcription but caused complete blockage of elongation near the A-loop region (Fig. 5). At molar ratios of 1:2, or higher, of template to PNAAL, extension of 32P-labeled DNA primer was aborted after processive addition of 9 nt upstream of the PBS region, the starting sequence for interaction of PNAAL (Fig. 5, lanes 4–6). During reverse transcription, HIV-1 RT has the ability to displace the RNA or DNA oligomers bound upstream on the template due to its strand displacement activity (19). However, the accumulation of products at the ninth template nucleotide position shows that it is unable to displace PNAAL due to strong binding of PNAAL to its target sequence. Similar inhibition of strand displacement activity of herpes simplex DNA helicase has been noted with the PNA–DNA complex (40).
Figure 5.
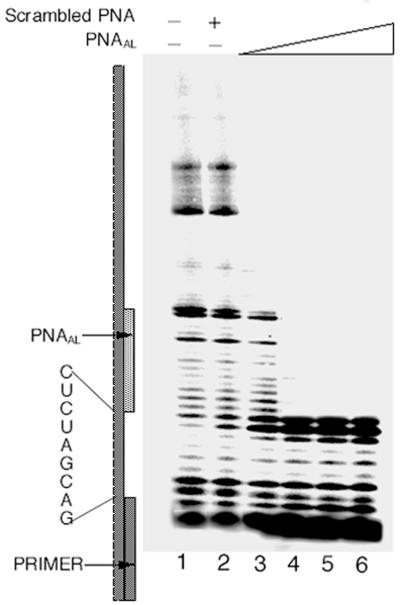
Effect of PNAAL on reverse transcription of U5-PBS HIV-1 RNA primed with the labeled 17mer DNA primer. The U5-PBS RNA template primed with the 5′-32P-labeled 17mer PBS DNA was incubated in the presence or absence of scrambled PNA or increasing concentrations of PNAAL at 37°C. After 2 h incubation, dNTP and HIV-1 RT were supplemented to initiate reverse transcription as described under Materials and Methods. The reaction products were analyzed on a denaturing 8% polyacrylamide–urea gel and subjected to phosphorimager analysis. Lane 1 represents the primer extension in the absence of PNAAL or scrambled PNA. Lane 2 shows the extension of the 17mer primer in the presence of scrambled PNA at a 1:4 molar ratio of template primer to scrambled PNA. Lanes 3–6 depict the primer extension in the presence of PNAAL at 1:1, 1:2, 1:3 and 1:4, molar ratios of template primer to PNAAL, respectively.
As tRNA3Lys is the natural primer for HIV-1 reverse transcription, we examined the effect of PNAAL on tRNA3Lys primed initiation of reverse transcription catalyzed by HIV-1 RT. As expected, reaction incubation of PNAAL with the U5-PBS HIV-1 RNA pre-bound to tRNA3Lys resulted in a significant inhibition of reverse transcription (Fig. 6, left panel, lanes 2–4). At a 1:4 ratio of template to PNAAL, complete abolition of initiation of reverse transcription was observed (Fig. 6, left panel, lane 4), thus indicating the ability of PNAAL molecules to destabilize the bound tRNA3Lys primer by invading the stem–loop target sequence on the viral RNA. Under similar conditions, scrambled PNA had no influence on the initiation and elongation process (Fig. 6, left panel, lanes 5–7). The inhibitory effect of PNAAL was more prominent when it was incubated with U5-PBS RNA along with tRNA3Lys (Fig. 6, right panel, lanes 2–4), suggesting that PNAAL can effectively inhibit reverse transcription by interfering with the priming of tRNA3Lys on the viral RNA or by displacing the primed tRNA3Lys from the template. The observed inhibition was concentration dependent with respect to PNAAL. At a 1:4 molar ratio of template to PNAAL, complete blockage of initiation of reverse transcription was noted (Fig. 6, right panel, lane 4).
Figure 6.
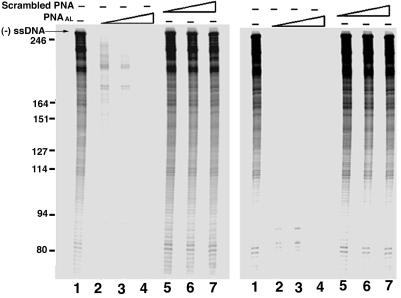
Effect of PNAAL on tRNA3Lys-derived reverse transcription of U5-PBS RNA. The U5-PBS HIV-1 RNA template pre-bound to tRNA3Lys (A) was incubated in the presence or absence of varying concentrations of PNAAL or scrambled PNA at 25°C for 6 h. In another set (B), the U5-PBS HIV-1 RNA template was pre-incubated in the presence or absence of PNAAL or scrambled PNA followed by priming with tRNA3Lys. The tRNA3Lys-derived reverse transcription was initiated by the addition of dNTP and HIV-1 RT as described in Materials and Methods. The reaction products were resolved on an 8% denaturing polyacrylamide–urea gel followed by phosphorimager analysis. Lane 1, control without PNA; lanes 2–4, show the initiation and extension of reverse transcription products in the presence of PNAAL at 1:1, 1:2 and 1:3 molar ratios of TP to PNAAL, respectively. Lanes 5–7 show the initiation and extension of reverse transcription products in the presence of scrambled PNA at 1:1, 1:2 and 1:3 molar ratios of template primer to scrambled PNA. The predicted size of tRNA3Lys-derived (–) ssDNA is 253 nt. The position of the tRNA3Lys-derived full-length (–) strand strong stop DNA (– ssDNA) is shown on the left.
As the inhibitory function of PNAAL on the initiation of tRNA3Lys primed reverse transcription and its subsequent extension was observed in vitro in the absence of other viral components/proteins, it was of interest to see whether such inhibition could also be seen with the isolated HIV-1 virion particles. The disrupted HIV-1 virions containing endogenous tRNA3Lys-primed viral RNA genome and reverse transcriptase were incubated at various concentrations of PNAAL on ice for indicated time points, and then supplemented with four dNTPs plus [α-32P]dCTP to initiate the reaction. It was observed that at all the concentration ranges of PNAAL (50 nM to 2.5 µM) the initiation of reverse transcription was significantly reduced and the elongation of initiated product was efficiently blocked (Fig. 7, lanes 2–9). As expected, scrambled PNA had no influence on the endogenous reverse transcription. These results suggest that the interaction of PNAAL with the A-loop region on the viral genome may block the base pairing interaction between the anticodon of tRNA3Lys and A-loop, thereby destabilizing the tRNA3Lys priming on the viral genome. The observation of two major products longer than 400 nt in reactions with disrupted virions suggests that these products may have been generated as a result of strand transfer during reverse transcription. Surprisingly, the labeled endogenous tRNA ran below the expected 77 nt position. It is possible that some population of the endogenous tRNA3Lys may have undergone cleavage of 5–6 nt either from the 5′ or the 3′ terminus during incubation with PNAAL prior to the initiation of reverse transcription, thus resulting in the shorter species.
Figure 7.
Effect of PNAAL on endogenous tRNA3Lys-derived reverse transcription in disrupted HIV-1 virions. The disrupted HIV-1 virions containing endogenous tRNA3Lys primed viral RNA genome, reverse transcriptase and other viral proteins were incubated at varying concentrations of PNAAL and then supplemented with four dNTPs and [α-32P]dCTP to initiate the reverse transcription process as described in Materials and Methods. The reverse transcription products were extracted with phenol–chloroform, precipitated with alcohol and analyzed on an 8% denaturing gel followed by visualization on a phosphorimager. Lane 1 is the control lane showing the endogenous reverse transcription products in the absence of PNA. Lanes 2–9 represent the reactions carried out at PNAAL concentrations of 50, 100, 250, 500 and 750 nM and 1, 2 and 2.5 µM, respectively. Lane 10 shows the reaction in the presence of 2.5 µM scrambled PNA. Molecular markers (M) are shown in the leftmost lane. The two major bands migrating above the 400 nt position represent products of strand transfer reaction.
It has been shown that dethiolation of mcm 5S2U at position 34 (26) or its modification from SUU to the suppresser anticodon CUA in the anticodon of tRNA3Lys destabilizes the interaction between tRNA3Lys and the viral RNA genome (41). The mutant tRNA3Lys with CUU anticodon was efficiently packaged in vivo but was unable to initiate reverse transcription. The inability of in vivo packaged mutant tRNA3Lys (CUU) to prime reverse transcription was suggested to be a consequence of its inability to interact with this A-rich loop of the viral genome. Wakefield et al. (9) have shown that the A-loop region is involved in primer tRNA3Lys placement. Infection of cells with HIV-1 containing a PBS complementary to tRNAHis rather than tRNA3Lys was found to quickly revert back to the original PBS complementary to tRNA3Lys. However, when the A-loop was changed such that it was complementary to tRNAHis anticodon, the tRNAHis became stabilized in the viral population (12). It is obvious that the integrity of the A-loop structure is essential for initiation of reverse transcription and could be a prime target for halting viral replication. Our results show that polyamide nucleic acid, designed to complement the A-loop region, is able to block initiation of tRNA3Lys primed reverse transcription but not with oligonucleotide primed reaction. However, the extension of oligonucleotide primed synthesis is completely terminated at the A-loop region suggesting that HIV-1 RT is unable to displace the PNA bound to the A-loop region.
The stability of PNAs in biological fluids together with its unique sequence specificity suggests that these synthetic polynucleobase molecules may have great potential as antisense agents in vivo. An antisense PNA targeted to the coding region of Ha-ras mRNA was found to interfere with translation initiation complex and arrest polypeptide chain elongation (42). Recently, this approach has been used to block telomerase activity by PNA targeted to telomerase RNA leading to progressive telomere shortening and causing immortal human breast epithelial cells to undergo apoptosis and cell death (43,44). Telomeres regained their initial length following removal of anti-telomerase PNA from the cell culture (45). The antisense and antigene activity of PNAs targeted to AUG start codon and 5′ UTR of human B-cell lymphoma (Bcl)-2 gene, were able to block (Bcl)-2 protein synthesis in a cell-free system (46). Repeated i.c.v. administration of PNA targeted to a region of the rat delta opoid receptor, significantly inhibited the antinociceptive response and locomotor response to delta opoid receptor agonists (47). Despite the extremely favorable and encouraging antisense activity of PNAs, their biodelivery into the cells remains a major obstacle. A PNA–DNA hybrid mimicking the NF-kappaB binding site of HIV-1 promoter has been reported (48). Recently, several studies have reported methods for effective biodelivery of PNAs and their antisense effects in cells. Certain peptide–PNA conjugates have been shown to be efficiently taken up by the cells and exhibit antisense activity on the targeted gene (49–56). A similar approach to improve cellular uptake of PNAs is currently in progress in our laboratory. A cocktail of such modified PNA targeted to various critical regions of the HIV-1 genome may be a potential inhibitor of HIV-1 replication.
Acknowledgments
ACKNOWLEDGEMENT
This research was supported by a grant from the NIAID (AI42520).
REFERENCES
- 1.Larder B.A. (1993) Inhibitors of HIV reverse transcriptase as antiviral agents and drug resistance. In Skalka,A.M. and Goff,S.P. (eds), Reverse Transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, p. 205.
- 2.Ho D.D., Neumann,A.U., Perelson,A.S., Chen,W., Leonard,J.M. and Markowitz,M. (1995) Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature, 373, 123–126. [DOI] [PubMed] [Google Scholar]
- 3.Wei X., Ghos,S.K., Taylor,M.E., Johnson,V.A., Emini,E.A., Deutsch,P., Lifson,J.D., Bonhoeffer,S., Nowak,M.A., Hahn,B.H., Sag,M.S. and Shaw,G.M. (1995) Viral dynamics in human immunodeficiency virus type 1 infection. Nature, 373, 117–122. [DOI] [PubMed] [Google Scholar]
- 4.Embretson J., Zupancic,M., Ribas,J.L., Burke,E., Racz,P., Tenner-Racz,K. and Haase,A.T. (1993) Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature, 362, 359–362. [DOI] [PubMed] [Google Scholar]
- 5.Muesing M.A., Smith,D.H., Cabradilla,C.D., Benton,C.V., Lasky,L.A. and Capon,D.J. (1985) Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature, 313, 450–458. [DOI] [PubMed] [Google Scholar]
- 6.Muesing M.A., Smith,D.H. and Capon,D.J. (1987) Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell, 48, 691–701. [DOI] [PubMed] [Google Scholar]
- 7.Ratner L., Haseltine,W., Patarca,R., Livak,K.J., Starcich,B., Josephs,S.F., Doran,E.R., Rafalski,J.A., Whitehorn,E.A., Baumeister,K., Ivanoff,L., Petteway,S.R.,Jr, Pearson,M.L., Lautenberger,J.A., Papas,T.S., Ghrayeb,J., Chang,N.T., Gallo,R.C. and Wong-Stall,F. (1985) Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature, 313, 277–284. [DOI] [PubMed] [Google Scholar]
- 8.Sherman P.A., Dickson,M.L. and Fyfe,J.A. (1992) Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J. Virol., 66, 3593–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakefield J.K., Wolf,A.G. and Morrow,C.D. (1995) Human immunodeficiency virus type 1 can use different tRNAs as primers for reverse transcription but selectively maintains a primer binding site complementary to tRNA(3Lys). J. Virol., 69, 6021–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang S.M., Zhang,Z. and Morrow,C.D. (1997) Identification of a sequence within U5 required for human immunodeficiency virus type 1 to stably maintain a primer binding site complementary to tRNAMet. J. Virol., 71, 207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Kang,S.M. and Morrow,C.D. (1997) Stability of HIV type 1 proviral genomes that contain two distinct primer-binding sites. AIDS Res. Hum. Retroviruses, 13, 253–262. [DOI] [PubMed] [Google Scholar]
- 12.Wakefield J.K., Kang,S. and Morrow,C.D. (1996) Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNA(His). J. Virol., 70, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vink C., Van Gent,D.C., Elgersma,Y. and Plasterk,R.H.A. (1991) Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol., 65, 4636–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen B.R. and Greene,W.C. (1990) Functions of the auxiliary gene products of the human immunodeficiency virus type 1. Virology, 178, 1–5. [DOI] [PubMed] [Google Scholar]
- 15.Fujisawa J., Seiki,M., Kiyokawa,T. and Yoshida,M., (1985) Functional activation of the long terminal repeat of human T-cell leukemia virus type I by a trans-acting factor. Proc. Natl Acad. Sci. USA, 82, 2277–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong-Staal F. and Gallo,R.C. (1985) Human T-lymphotropic retroviruses. Nature, 317, 395–403. [DOI] [PubMed] [Google Scholar]
- 17.Harrich D., Ulich,C. and Gaynor,R.B. (1996) A critical role for the TAR element in promoting efficient human immunodeficiency virus type 1 reverse transcription. J. Virol., 70, 4017–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrich D., Ulich,C., Garcia-Martinez,L.F. and Gaynor,R.B. (1997) Tat is required for efficient HIV-1 reverse transcription. EMBO J., 16, 1224–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee R., Kaushik,N., Modak,M.J., Vinayak,R. and Pandey,V.N. (1998) Polyamide nucleic acid targeted to the primer binding site of the HIV-1 RNA genome blocks in vitro HIV-1 reverse transcription. Biochemistry, 37, 900–910. [DOI] [PubMed] [Google Scholar]
- 20.Mayhood T., Kaushik,N., Pandey,P.K., Kashanchi,F., Deng,L. and Pandey,V.N. (2000) Inhibition of Tat-mediated transactivation of HIV-1 LTR transcription by polyamide nucleic acid targeted to TAR hairpin element. Biochemistry, 39, 11532–11539. [DOI] [PubMed] [Google Scholar]
- 21.Hirschman S.Z. and Chen,C.W. (1996) Peptide nucleic acids stimulate gamma interferon and inhibit the replication of the human immunodeficiency virus. J. Investig. Med., 44, 347–351. [PubMed] [Google Scholar]
- 22.Mollegaard N.E., Buchardt,O., Egholm,M. and Nielsen,P.E. (1994) Peptide nucleic acid.DNA strand displacement loops as artificial transcription promoters. Proc. Natl Acad. Sci. USA, 91, 3892–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peffer N.J., Hanvey,J.C., Bisi,J.E., Thomson,S.A., Hassman,C.F., Noble,S.A. and Babiss,L.E. (1993) Strand-invasion of duplex DNA by peptide nucleic acid oligomers. Proc. Natl Acad. Sci. USA, 90, 10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutz M.J., Benner,S.A., Hein,S., Breipohl,B. and Uhlmann,E. (1997) Recognition of uncharged polyamide-linked nucleic acid analogs by DNA polymerases and reverse transcriptases. J. Am. Chem. Soc., 119, 3177–3178. [Google Scholar]
- 25.Misra H.S., Pandey,P.K., Modak,M.J., Vinayak,R. and Pandey,V.N. (1998) Polyamide nucleic acid-DNA chimera lacking the phosphate backbone are novel primers for polymerase reaction catalyzed by DNA polymerases. Biochemistry, 37, 1917–1925. [DOI] [PubMed] [Google Scholar]
- 26.Isel C., Marquet,R., Keith,G., Ehresmann,C. and Ehresmann,B. (1993) Modified nucleotides of transfer-RNA (3 Lys) modulate primer/template loop–loop interaction in the initiation complex of HIV-1 reverse transcription. J. Biol. Chem., 268, 25269–25272. [PubMed] [Google Scholar]
- 27.Isel C., Ehresmann,C., Keith,G., Ehresmann,B. and Marquet,R. (1995) Initiation of reverse transcription of HIV-1: secondary structure of theHIV1RNA/tRNA (3Lys) (template/primer) complex. J. Mol. Biol., 247, 236–250. [DOI] [PubMed] [Google Scholar]
- 28.Isel C., Westhof,E., Massire,C., Le Grice,S.F.J., Ehresmann,B., Ehresmann,C. and Marquet,R. (1999) Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J., 18, 1038–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang C., Li,X., Rong,L., Inouye,P., Quan,Y., Kleiman,L. and Wainberg,M.A. (1997) The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J. Virol., 71, 5750–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arts E.J., Li,X., Gu,Z., Kleiman,L., Parniak,M. and Wainberg,M.A. (1994) Comparison of Deoxyoligonucleotide and t-RNAlys-3 as primers in an endogenous human immunodeficiency virus-1 in vitro reverse transcription/template-switching reaction. J. Biol. Chem., 269, 14672–14680. [PubMed] [Google Scholar]
- 31.Kati W.M., Johnson,K.A., Jerva,L.F. and Anderson,K.S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem., 267, 25988–25997. [PubMed] [Google Scholar]
- 32.Kashanchi F., Shibata,R., Ross,E.K., Brady,J.N. and Martin,M.A. (1994) Second-site long terminal repeat (LTR) revertants of replication-defective human immunodeficiency virus: effects of revertant TATA box motifs on virus infectivity, LTR-directed expression, in vitro RNA synthesis and binding of basal transcription factors TFIID and TFIIA. J. Virol., 68, 3298–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.S., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley—Intersciences, John Wiley & Sons, Inc., New York, NY, Vol. 1.
- 34.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M., Mak,J., Ladha,A., Cohen,E., Klein,M., Rovinski,B. and Kleiman,L. (1993) Identification of tRNAs incorporated into wild type and mutant human immunodeficiency virus type 1. J. Virol., 67, 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mak J., Jiang,M., Wainberg,M.A., Hammarskjold,M.L., Rekosh,D. and Kleiman,L. (1994) Role of Pr160gag-pol in mediating the selective incorporation of tRNA (Lys) into human immunodeficiency virus type 1 particles. J. Virol., 68, 2065–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isel C., Keith,G., Ehresmann,B., Ehresmann,C. and Marquet,R. (1998) Mutational analysis of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucleic Acids Res., 26, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Kang,S.M., Li,Y. and Morrow,C.D. (1998) Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA, 4, 394–406. [PMC free article] [PubMed] [Google Scholar]
- 39.Aiyar A., Ge,Z. and Leis,J. (1994) A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J. Virol., 68, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bastide L., Boehmer,P.E., Villani,G. and Lebleu,B. (1999) Inhibition of a DNA-helicase by peptide nucleic acids. Nucleic Acids Res., 27, 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y., Shalom,A., Li,Z., Wang,J., Mak,J., Wainberg,M.A. and Kleiman,L. (1996) Effects of modifying the tRNA3Lys anticodon on the initiation of human immunodeficiency virus type 1 reverse transcription. J. Virol., 70, 4700–4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dias N., Dheur,S., Nielsen,P.E., Gryaznov,S., Van Aerschot,A., Herdewijn,P., Helene,C. and Saison-Behmoaras,T.E. (1999) Antisense PNA targeted to the coding region of Ha-ras mRNA arrest polypeptide chain elongation. J. Mol. Biol., 294, 403–416. [DOI] [PubMed] [Google Scholar]
- 43.Hamilton S.E., Simmons,C.G., Kathiriya,I.S. and Corey,D.R. (1999) Cellular delivery of peptide nucleic acids and inhibition of human telomerase. Chem. Biol., 6, 343–351. [DOI] [PubMed] [Google Scholar]
- 44.Shammas M.A., Simmons,C.G., Corey,D.R. and Reis,R.J. (1999) Telomerase inhibition by peptide nucleic acids reverses ‘immortality’ of transformed human cells. Oncogene, 18, 6191–6200. [DOI] [PubMed] [Google Scholar]
- 45.Herbert B., Pitts,A.E., Baker,S.I., Hamilton,S.E., Wright,W.E., Shay,J.W. and Corey,D.R. (1999) Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl Acad. Sci. USA, 96, 14276–14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mologni L., Nielsen,P.E. and Gambacorti-Passerini,G. (1999) In vitro transcriptional and translational block of the bcl-2 gene operated by peptide nucleic acid. Biochem. Biophys. Res. Commun., 264, 537–543. [DOI] [PubMed] [Google Scholar]
- 47.Fraser G.L., Holmgren,J., Clarke,P.B. and Wahlestedt,C. (2000) Antisense inhibition of delta-opioid receptors gene function in vivo by peptide nucleic acids. Mol. Pharmacol., 57, 725–731. [DOI] [PubMed] [Google Scholar]
- 48.Mischiati C., Borgatti,M., Bianchi,N., Rutiglano,C., Tomassetti,M., Feriotto,G. and Gambari,R. (1999) Interaction of the human NF-kappaB p52 transcription factor with DNA-PNA hybrids mimicking the NF-kappaB binding sites of the human immunodeficiency virus type 1 promoter. J. Biol. Chem., 274, 33114–33122. [DOI] [PubMed] [Google Scholar]
- 49.Aldrian-Herrada G., Desarmenien,M.G., Orcel,H., Boissin-Agasse,L., Mery,J., Brugidou,J. and Rabie,A. (1998) A peptide nucleic acid (PNA) is more rapidly internalized in cultured neurons when coupled to a retro-inverso delivery peptide. The antisense activity depresses the target mRNA and protein in magnocellular oxytocin neurons. Nucleic Acids Res., 26, 4910–4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branden L.J., Mohamed,A.J. and Smith,C.I. (1999) A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nature Biotechnol., 17, 784–787. [DOI] [PubMed] [Google Scholar]
- 51.Chinnery P.F., Taylor,R.W., Diekert,K., Lill,R., Turnbull,D.M. and Lightowlers,R.N. (2000) Peptide nucleic acid delivery to human mitochondria. Gene Ther., 6, 1919–1928. [DOI] [PubMed] [Google Scholar]
- 52.Harrison J.G., Frier,C., Laurant,R., Dennis,R., Raney,K.D. and Balasubramanian,S. (1999) Inhibition of human telomerase by PNA-cationic peptide conjugates. Bioorganic Medicinal Chem. Lett., 9, 1273–1278. [DOI] [PubMed] [Google Scholar]
- 53.Ljungstrom T., Knudsen,H. and Nielsen,P.E. (1999) Cellular uptake of adamantyl conjugated peptide nucleic acids. Bioconjugate Chem., 10, 965–972. [DOI] [PubMed] [Google Scholar]
- 54.Kamagai I., Takahashi,T., Hamasaki,K., Ueno,A. and Mihara,H. (2000). Construction of HIV Rev peptides containing peptide nucleic acid that bind HIV RRE IIB RNA. Bioorganic Medicinal Chem. Lett., 10, 377–379. [DOI] [PubMed] [Google Scholar]
- 55.Pooga M., Soomets,U., Hallbrink,M., Valkna,A., Saar,K., Rezaei,K., Kahl,U., Hao,J.X., Xu,X.J., Wiesenfeld-Hallin,Z., Hokfelt,T., Bartfai,T. and Langel,U. (1998) Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo.Nature Biotech., 16, 857–861. [DOI] [PubMed] [Google Scholar]
- 56.Scarfi S., Giovine,M., Gasparini,A., Damonte,G., Millo,E., Pozzolini,M. and Benatti,U. (1999) Modified peptide nucleic acids are internalized in mouse macrophages RAW 264.7 and inhibit inducible nitric oxide synthase. FEBS Lett., 451, 264–268. [DOI] [PubMed] [Google Scholar]