Abstract
Background
Emerging studies have demonstrated the critical role of RNA m6A methylation in tumor progression, whereas lncRNA m6A modification profiles in breast cancer remain largely unknown. Our previous study has shown that METTL14 accelerates breast cancer migration and invasion in an m6A‐dependent manner, making it critical to analyze METTL14‐mediated m6A modification at a transcriptome‐wide scale in breast cancer.
Methods
Here, we performed MeRIP‐seq analysis in METTL14 overexpressed and control MDA‐MB‐231 cells. Conjoint analysis of MeRIP‐seq and RNA‐seq data was used to select lncRNAs with m6A methylation and differential expression. Finally, the screened lncRNA was verified by MeRIP‐PCR and its function was studied via transwell assay.
Results
Our results determined that high expression of METLL14 results in 3996 hypermethylation peaks from 3107 transcripts, and 4100 hypomethylation peaks from 2918 transcripts. Furthermore, conjoint analysis of MeRIP‐seq and RNA‐seq data identified 25 lncRNAs with discrepant methylation and simultaneously discrepant expression, among which the top 10 differentially expressed LncRNAs were AC026401.3, CYTOR, LINC01943, AC084125.2, FLJ20021, LINC00472, and NORAD, MALAT1, AL161431.1, and LINC01764. Moreover, over‐expressed METTL14 stimulated the m6A modification of AC084125.2, while decreasing its expression. Compared to adjacent tissues, AC084125.2 was lowly expressed in tumors and could be used as a biomarker in the diagnosis of breast cancer. Meanwhile, AC084125.2 inhibited the migration and invasion of cancer cells.
Conclusion
In conclusion, METTL14‐mediated m6A modification of lncRNAs, which might provide reference for future intervention in tumor progression.
Keywords: breast cancer, long non‐coding RNA, MeRIP‐seq, N6‐methyladenosine, RNA‐seq
Mettl4‐mediated m6A modification is involved in breast cancer development.
1. INTRODUCTION
Breast cancer is the second leading cause of cancer death in women. 1 Patients with breast cancer usually have late diagnosis, poor prognosis, and high recurrence rate. 2 At present, the diagnosis of breast cancer is usually based on mammography and histopathological evaluation. 3 However, the benefits of these detection methods were greater in older women, while younger women were more likely to have false‐positive results. 4 Therefore, it is urgent to find molecular targets to enhance the diagnostic ability and improve the prognosis of breast cancer.
N6‐methyladenosine (m6A), as an important internal RNA modification in higher eukaryotes, has become a focus of epigenetic research in recent years. M6A influences every stage of RNA metabolism, such as RNA splicing, export, translation, and stability, 5 and plays an important role in mutiple biological processes, including cancer development. 6 , 7 Currently, m6A is rarely studied in breast cancer. Previous studies have found that overexpression of m6A‐related genes (YTHDF1, YTHDF3, and KIAA1429) was associated with poor prognosis in breast cancer. 8 In addition, HBXIP promoted breast cancer progression by upregulation of methyltransferase METTL3 via depressing tumor suppressor let‐7G. 9 Similarly, we previously found that m6A modifier METTL14 may promote the invasion and metastasis of breast cancer through m6A modification. 10 The above studies indicated that m6A‐related genes are abnormally expressed in breast cancer, but how m6A modification regulates the invasion and metastasis of breast cancer is still unclear.
Long non‐coding RNAs (lncRNAs) are a group of non‐coding RNAs with a nucleotide > 200 nt. 11 Emerging researches have proved that most lncRNAs are disorderly expressed in cancer and can be used as a novel biomarker of cancer. For example, lncRNA HOXA‐AS2 was abnormally expressed in multifarious malignant tumors, such as breast cancer, gastric cancer, and gallbladder cancer. 12 LncRNA FOXCUT was overexpressed in endometrial cancer cells and promoted epithelial mesenchymal transformation of cancer cells. 13 Currently, increasing evidences demonstrating that m6A‐related genes influence tumor occurrence and development by regulating lncRNAs expression in an m6A‐dependent manner. Zuo and her colleagues certified that METTL3‐mediated upregulation of LINC00958 can be used as a therapeutic target in hepatocellular carcinoma. 14 Moreover, METTL14‐mediated m6A modification inhibited the proliferation and metastasis of colorectal cancer by decreasing the stability of XIST via YTHDF2. 15 These studies indicated that m6A modification‐mediated regulation of lncRNAs is crucial for tumor proliferation and metastasis. Dismayingly, the role and molecular mechanism of m6A modification of lncRNA in breast cancer are still less.
Our research group has previously proved that METTL14 is overexpressed in breast cancer and induces the migration and invasion of breast cancer. 10 Here, we will further explore the regulation of METTL14 on m6A modification and expression of lncRNA in breast cancer, and explore the role of METTL14‐mediated lncRNA m6A modification in the migration and invasion of breast cancer, to provide a new reference for the diagnosis and treatment of breast cancer.
2. MATERIALS AND METHODS
2.1. Cell culture and cell transfection
The normal human mammary epithelial cells (MCF‐10A) and the human breast cancer cell lines (MDA‐MB‐468, MCF‐7, SKBR3, MDA‐MB‐231, and BT474) were obtained from Procell Life Technologies Co. LTD, which were cultured in DMEM with 10% fetal bovine serum (FBS). According to previous experience, 16 the synthetic METTL14 overexpression plasmid, AC084125.2 overexpression plasmid, and corresponding negative control were transfected into cells by Lipofectamine 3000 (Invitrogen).
2.2. Quantitative real‐time PCR (qPCR)
Total RNA was isolated and extracted from cells and tissues using Trizol reagent (Invitrogen). Then, Reverse transcription of RNAs was performed with a Reverse Transcription Kit (Takara) based on the manufacturer's protocols. The ABI Q6 detection system (Applied Biosystems Inc.) was used to run qPCR. The method was employed for relative quantification of gene expression. The qPCR primers were listed in Table S1. The template of METTL14 and lncRNA was standardized by GAPDH.
2.3. Western blot analysis
RIPA lysis buffer (Thermo) was used to extract total protein products from the cells. Next, proteins were separated on 8% SDS‐PAGE and transferred to PVDF membranes. The membranes were blocked with skimmed milk for 3 h at 37°C and then incubated with primary antibody overnight at 4°C. Finally, the membranes were incubated with secondary antibody for 2 h at 37°C. The ECL System (Thermo) was used to develop the films, and then quantified with ImageJ software. The primary antibodies were as follows: anti‐METTL14 (Abcam, #ab220030, 1:1000) and anti‐GAPDH (Proteintech, #60004‐1‐Ig, 1:10000). The secondary antibodies were as: goat anti‐rabbit IgG H&L (HRP) (Beyotime, #A0208, 1:1000) and goat anti‐mouse IgG H&L (HRP) (Beyotime, #A0216, 1:1000).
2.4. MeRIP sequencing and RNA sequencing
Total RNA was extracted using TRIzol™ Reagent (Invitrogen). The concentration of total RNA was measured by Qubit RNA HS assay kit (Invitrogen). Then, the total RNA was fragmented into 100–200 nt RNA fragments using 10X RNA Fragmentation Buffer. The reaction was stopped by adding 10X EDTA. Methylated RNA immunoprecipitation was performed using EpiTM m6A immunoprecipitation kit (Epibiotek). Briefly, to acquire immunoprecipitated RNA fragments, the fragmented RNA was co‐cultured with anti‐m6A monoclonal antibody (Abcam) for 3 h at 4°C and then with protein A/G magnetic beads (Invitrogen) at 4°C for 2 h. The m6A‐enriched RNA was purified using TRIzol™ Reagent (Invitrogen). The library was prepared by KAPA Stranded RNA‐Seq Kit (Illumina). Input IP‐free and m6A IP samples were both sequenced at 150‐bp double‐ended on an Illumina NovaSeq 6000 sequencer.
2.5. MeRIP‐seq data processing
Cutadapt (v2.5) was performed for trimming adapters and filtering sequences, Hisat2 aligner (v2.1.0) was utilized to align remaining reads and the human Ensemble genome GRCh38. M6A peaks were identified using exomePeak R package (v2.13.2). The exomePeak R package was performed for identifying differential m6A peaks. Guitar R package (v1.16.0) was used to visualize m6A‐RNA‐related genomic features. Homer (v4.10.4) was utilized to selected for the de novo motif analysis of the m6A peaks with p value < 0.05.
2.6. RNA‐seq data processing
The featureCounts (v1.6.3) was utilized to calculate the reads mapped the genome. The DESeq2 R‐package was performed for analyzing differential gene expression (| log2 [fold change] | > 0.585 and p < 0.05).
2.7. Functional enrichment analysis
The miRanda (http://www.microrna.org/microrna/home.do) and RNAHybrid (http://bibiserv.techfak.uni‐bielefeld.de/rnahybrid/) were used for predicting targeted genes. Target genes were identified by the intersection of the two databases. Functional enrichment analysis of genes was performed using GO (http://www.geneontology.org) enrichment analysis and KEGG (http://www.genome.jp/kegg) database.
2.8. Data mining analysis
GEPIA database (http://gepia.cancer‐pku.cn/index.html) was performed for the differential expression of lncRNAs in breast cancer.
2.9. Clinical specimens
All patients gave written informed consent before participation in this study. A total of 35 pairs of breast cancer tissues and normal tissues adjacent to cancer were acquired from the Nanjing Drum Tower Hospital of the Affiliated Hospital of Nanjing University Medical School. This study was approved by the Human Research Ethics Committee of Nanjing University Medical School.
2.10. Transwell assay
The MDA‐MB‐231 cells were suspended in serum‐free medium and seeded into the upper chambers. The medium with 10% FBS was placed into the bottom chamber. Subsequently, the cells were incubated in a CO2 incubator for 24 h. Next, the cells migrated to the bottom of the membrane were fixed with 4% paraformaldehyde, and tinted with 1% crystal violet. Finally, a microscope is used for imaging.
2.11. Statistical analysis
Graphpad Prism 8.0. (La Jolla) was employed for statistical analysis. Mean ± standard deviation (SD) was expressed the results. Differences between the two groups were analyzed by Student's t test. p value < 0.05 was considered significant.
3. RESULTS
3.1. Overview of the m6A methylation map mediated by METTL14 in breast cancer
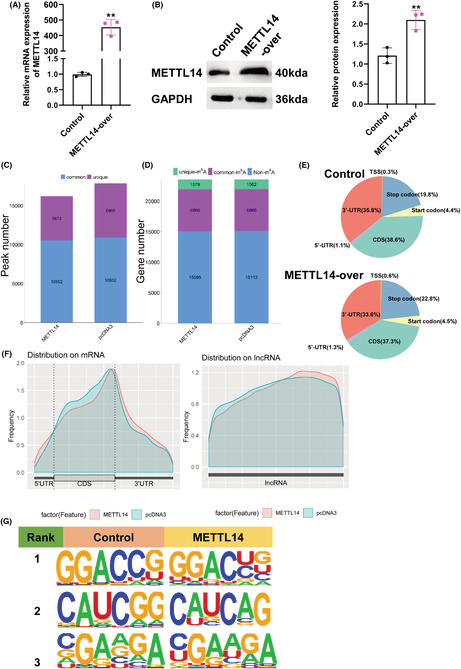
In previous research, we revealed METTL14 was overexpressed in breast cancer and promoted breast cancer migration and invasion. 10 As is well‐known, METTL14 acts as an m6A writer, which functions by influencing m6A methylation in transcripts. 15 Thus, we first transfected MDA‐MB‐231 cells with METTL14‐overexpressed plasmid and corresponding control, and qPCR and western blot were used to detect the transfection efficiency (Figure 1A,B). Then, MeRIP‐Seq analysis was performed to analyze the transcript of lncRNAs with m6A‐modified for transfected cells. In total, m6A‐seq identified 16,225 and 17,862 m6A peaks from 8445 and 8428 m6A‐modified transcripts in METTL14 overexpression and control MDA‐MB‐231 cells, respectively (Figure 1C,D). The distribution of m6A peaks in NC group exhibited that m6A peaks both in NC group and METTL14‐over group were mainly focused on the coding sequence (CDS) and 3′untranslated region (3′UTR), and the m6A residues near the stop codon were the most enriched (Figure 1E,F). Furthermore, we identified the main motifs of METTL14 group were GGACUG, CAUCAG, and RGAAGA (R is C/U), and the motifs of control group were GGACCG, CAUCGG, and KGAAGA (K is C/G/A/U) (Figure 1G).
FIGURE 1.
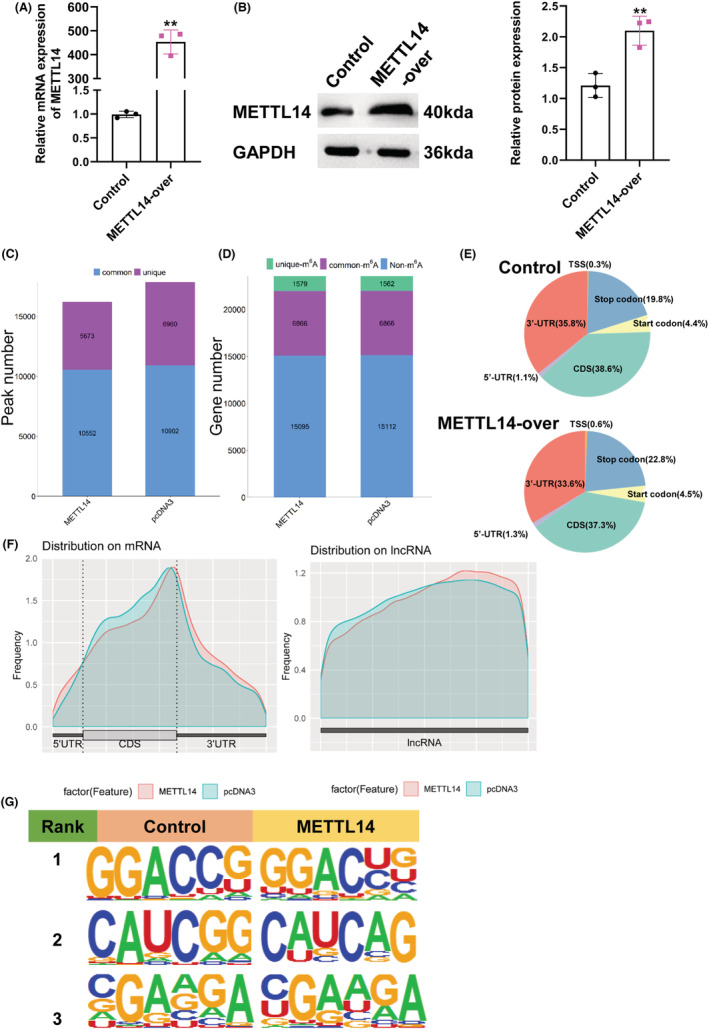
Characteristics of m6A methylation in breast cancer. Relative RNA (A) and protein (B) expression of METTL14 in MDA‐MB‐231 cells transfected with METTL14 overexpression plasmid. **p < 0.01. (C, D) PEAK and gene number analysis of MeRIP‐Seq results of MDA‐MB‐231 cells transfected with METTL14 overexpression. (E) Pie charts exhibiting the distribution of m6A peaks. (F) M6A peaks distribution of regions of m6A‐modified transcripts. (G) The m6A motif was identified by the MEME motif analysis.
3.2. Transcripts with differentially modified m6A peaks and differentially expressed transcripts
To investigate the m6A modification regulated by METTL14, transcripts with differential m6A peaks between the control and METTL14 overexpression groups were analyzed. We found that overexpression of METTL14 triggered 3996 hypermethylation peaks from 3107 transcripts, and 4100 hypomethylation peaks from 2918 transcripts (Figure 2A). GO analysis of transcripts with different modified m6A peaks indicated that they were particularly associated with DNA transcription, modification, and cell proliferation (Figure 2B). Moreover, analysis of KEGG analysis revealed that m6A modifications were enriched in a part of genes related to mTOR signaling pathway and TNF signaling pathway (Figure 2C). Next, we further detected the transcriptome profiles in control and METTL14‐over group by RNA‐seq (MeRIP‐seq input library). Differentially expressed genes were detected by DEGseq. We acquired 150 up‐regulated transcripts and 154 downregulated transcripts in METTL14 overexpression group compared to control (Figure 2D). Next, according to the association analysis of the MeRIP‐seq and RNA‐seq data, we constructed a four‐quadrant diagram of the m6A methylation transcripts and differentially expressed transcripts. As shown in Figure S1, the purple genes represented the top 10 transcripts with the largest difference multiple in m6A, and the red dots represent transcripts with m6A methylation and expression both meeting p < 0.05. Functional enrichment analysis showed they were associated with RNA translation, cell division, cancer‐related transcriptional dysregulation, and pathways (Figure 2E,F). Taken together, these information suggested that METTL14 overexpression caused abnormal m6A modification and dysregulation expression of transcripts, resulting in disorder of cell division, proliferation, and cancer‐related signaling pathways.
FIGURE 2.
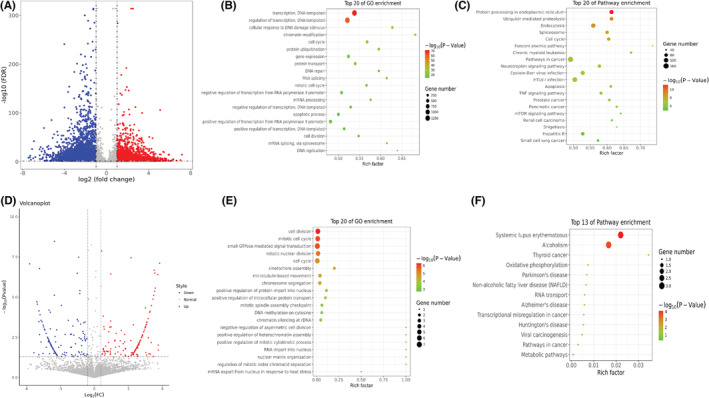
Functional enrichment analysis of transcripts. (A) Volcano plots expressed the numbers of transcripts with significantly differential m6A peaks. (B) Bubble chart represents GO terms. (C) Bubble chart represents KEGG pathway. (D) Volcano plots showed the differential expressed transcripts. (E) Bubble chart represents GO terms. (F) Bubble chart represents KEGG pathway.
3.3. lncRNAs with differential m6A peaks and differential expression induced by METTL14
Dysregulated lncRNAs were found in different cancers, which has become an important regulator of cancer metastasis and migration, such as m6A‐modified lncRNA ThAP7‐AS1 mediated by METTL3 owning oncogenic function. 17 Therefore, in order to explore lncRNA with differentially m6A peaks and differential expression by METTL14‐induced in breast cancer, we focused on lncRNAs in MeRIP‐seq and RNA‐seq data. As shown in Figure 3A, compared to control, METTL14‐over group had 40 lncRNAs with prominent upregulation of m6A peaks, and 32 lncRNAs accompanied by downregulated m6A peaks. Meanwhile, in RNA‐seq, METTL14 overexpression stimulated nine lncRNAs with significantly overexpressed and 10 lncRNAs with lower expression (Figure 3B). Next, conjoint analysis of the MeRIP‐seq and RNA‐seq data, there were 14 hyper‐methylated lncRNAs were upregulated, three hypo‐methylated lncRNAs were downregulated, and eight hypo‐methylated lncRNAs were upregulated in the two groups (Figure 3C). Considering that METTL14 mediated m6A modification mainly inhibits lncRNA expression, 18 our candidate lncRNAs that regulated by METTL14 via m6A modification need to meet: (1) m6A modification of lncRNA was enhanced after METTL14 overexpression; (2) lncRNA expression was downregulated after METTL14 overexpression; (3) lncRNA with low expression in breast cancer, since METTL14 was upregulated in breast cancer. We finally screened out seven candidate lncRNAs that met the above conditions. As shown in Figure 3D, these lncRNAs were regulated by METTL14 modification. In summary, METTL14 regulated the m6A methylation level of lncRNAs, resulting in changes in its level, suggesting that these lncRNAs may be important factors in the progression of breast cancer.
FIGURE 3.
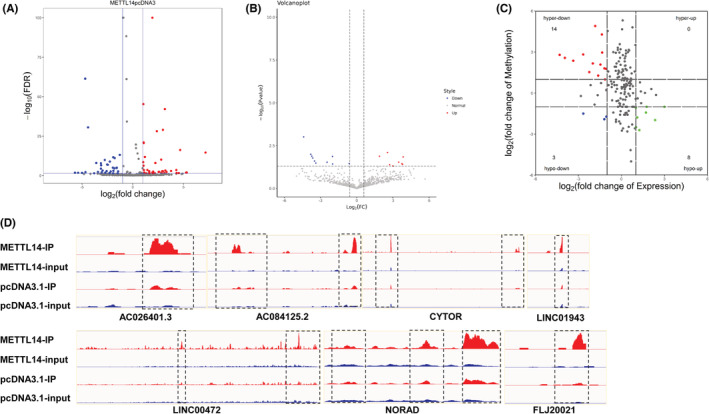
Conjoint analysis of MeRIP‐seq and RNA‐seq data. (A) Volcano plots expressed the numbers of lncRNAs with significantly differential m6A peaks. (B) Volcano plots showed the differential expressed lncRNAs. (C) The four‐quadrant diagram showed the m6A methylation lncRNAs and differentially expressed lncRNAs. The red dots expressed downregulated lncRNAs with hypermethylation, the blue dots expressed downregulated lncRNAs with hypomethylation, and the green dots expressed upregulated lncRNAs with hypomethylation. (D) Peak of lncRNAs in the METTL14‐overexpression and control group
3.4. AC084125.2 is the m6A‐modified lncRNA of METTL14
To determine which lncRNAs are involved in the metastasis and invasion of breast cancer, we further verified the selected lncRNAs. In GEPIA database, we found that three lncRNAs (AC026401.3, CYTOR, and LINC01943) were highly expressed in breast cancer and the other four (AC084125.2, FLJ20021, LINC00472, and NORAD) were lowly expressed (Figure 4A). Subsequently, we performed qPCR for validation. Compared with control, the enrichment of m6A in AC084125.2, LINC01943, and NORAD was notably risen under METTL14 overexpression (Figure 4B). In addition, compared with the control, overexpression of METTL14 substantially promoted the expression of AC026401.3, LINC00472, and LINC01943, inhibited the level of AC084125.2 (Figure 4C). Besides, inhibition of METTL14 dramatically enhanced the expression of AC084125.2 (Figure 4D). Based on the above results, we selected AC084125.2 as the subsequent target for analysis. Through the detection of clinical samples (n = 35), we found that the expression of AC084125.2 in breast cancer tissues was lower than that in para‐carcinoma tissue (Figure 4E). Furthermore, we performed receiver operating characteristic (ROC) curve analysis, and found AC084125.2 could be as a biomarker for breast cancer, and the area under curve (AUC) was 0.8853 (p < 0.0001) (Figure 4F). At the cutoff value, the sensitivity and specificity of breast cancer diagnosis were 88.57% and 85.71%, respectively. Collectively, these data indicated that AC084125.2 was the m6A‐modified target of METTL14, and might be act as a biomarker for breast cancer diagnosis.
FIGURE 4.
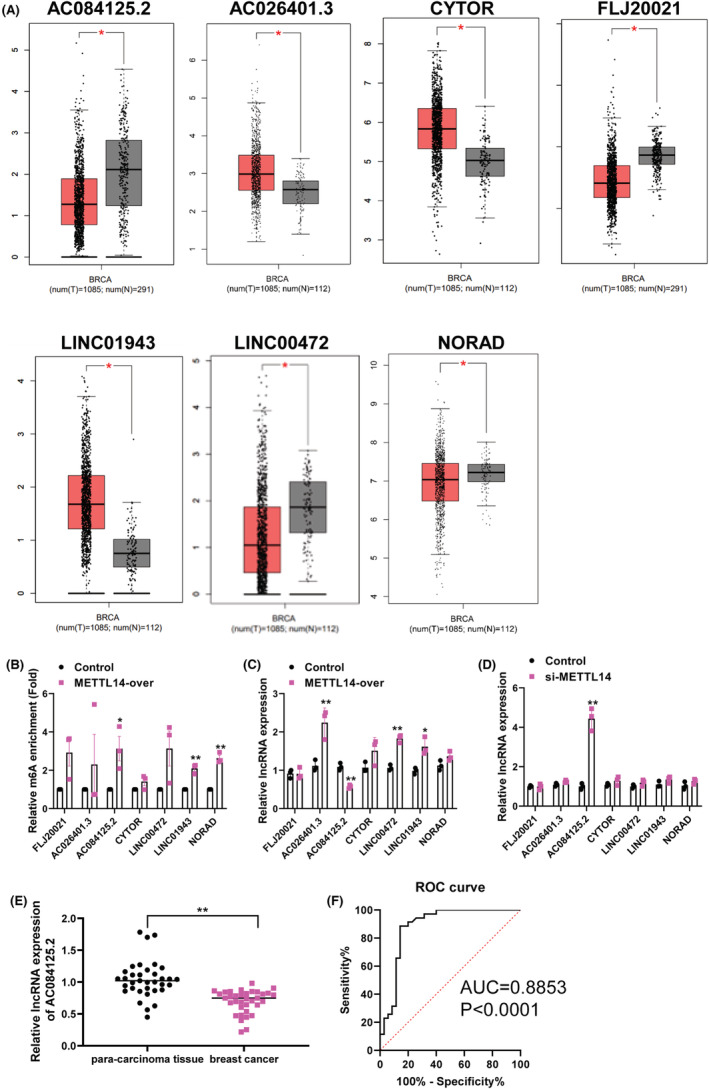
AC084125.2 is the target of METTL14 in breast cancer cells. (A) The expression of lncRNAs between tumor and normal samples in GEPIA database. (B) MeRIP‐qPCR detected the interaction between METTL14 and the candidate lncRNAs. (C) The relative expression of candidate lncRNAs was detected by qPCR after transfection with METTL14 overexpression plasmid in MDA‐MB‐231 cells. (D) The relative expression of candidate lncRNAs was detected by qPCR after transfection with si‐METTL14 in MDA‐MB‐231 cells. (E) Relative expression of AC084125.2 in breast cancer tissues and para‐carcinoma tissues (n = 35) was examined by qPCR assays. (F) The ROC curve of AC084125.2. *p < 0.05, **p < 0.01
3.5. AC084125.2 hinders breast cancer cell progression
To examine whether AC084125.2 expression inhibits breast cancer cell progression, we first detected the expression of AC084125.2 in various breast cancer cell lines. The results showed that AC084125.2 was low expressed in cells with a strong metastatic ability (MDA‐MB‐231 and MDA‐MB‐468), whereas it was high expressed in cells with a strong metastatic ability (SKBR3 and BT474) (Figure 5A). Subsequently, we used AC084125.2 overexpression plasmid to enhance AC084125.2 expression in MDA‐MB‐231 cells (Figure 5B). Cell migration results showed that overexpression of AC084125.2 significantly inhibited cell migration (Figure 5C). Similarly, high expression of AC084125.2 observably depressed cell invasion (Figure 5D). Taken together, AC084125.2 is a tumor suppressor molecular in breast cancer.
FIGURE 5.
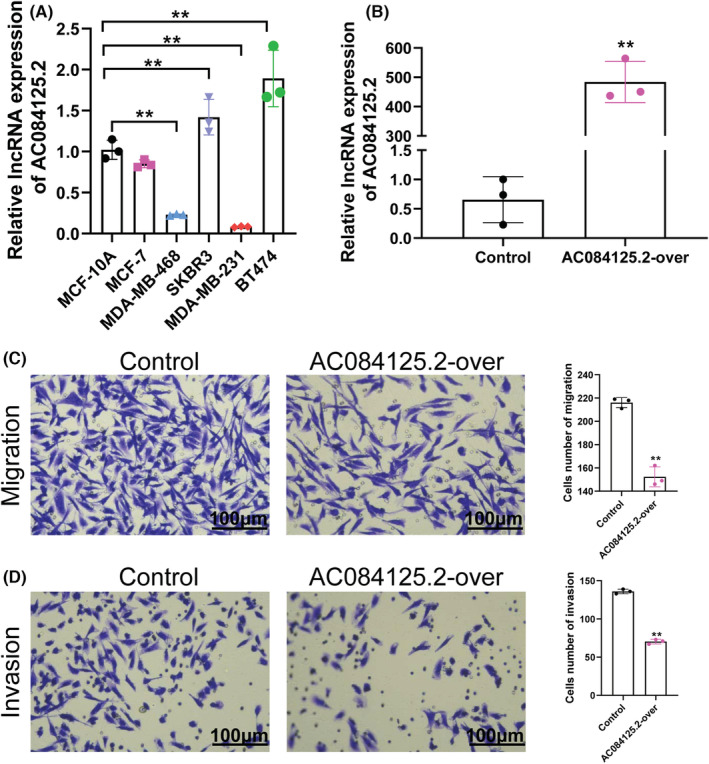
AC084125.2 suppresses breast cancer cell migration and invasion in vitro. (A) Relative expression of AC084125.2 in cell line of breast cancer. (B) Relative expression of AC084125.2 in MDA‐MB‐231 cells transfected with AC084125.2 overexpression plasmid. (C, D) Overexpression of AC084125.2 reduced the abilities of migration and invasion of MDA‐MB‐231 cells. Scale bar = 100 μm. **p < 0.01
4. DISCUSSION
M6A methylation disorder is closely related to the development of various cancers. 19 Our earlier studies have shown that METTL14 regulates the level of m6A in breast cancer, and regulates the malignant phenotype of cancer cells in an m6A‐dependent manner. 10 With the development of RNA sequencing, lncRNA has gradually become a hot spot in cancer research. However, it is not clear whether METTL14 affects the transcriptome‐wide m6A‐methylated lncRNA in breast cancer.
In this study, we found that overexpression of METTL14 resulted in changes in the methylation level of the transcriptome in breast cancer cells. In addition, we found lncRNAs with differential methylation and synchronously differential expression by combining MeRIP‐seq and RNA‐seq data. Further qPCR validation demonstrated that METTL14 increased m6A methylation level of AC084125.2, resulting in its downregulation. AC084125.2 could be used as a biomarker of breast cancer and participate in the metastasis and migration of breast cancer. Our results provided a basis for the subsequent study of the molecular mechanism of m6A‐modified lncRNAs in breast cancer, and offered a new idea for clinical treatment of breast cancer.
M6A modification is the most abundant modification in mRNA, and its level changes affect the occurrence and development of cancer. Some studies have performed MeRIP sequencing in colon cancer and found that the methylation level is abnormally expressed in cancer tissues. 20 Meantime, many studies have shown that lncRNAs m6A modification can be used as a biomarker in a variety of cancers and is related to the prognosis of cancer. 21 In view of our previous discovery that METTL14 promotes the progression of breast cancer by regulating N6‐methyladenosine, but we do not know the impact on the methylation level of transcriptome, therefore, we performed methylation sequencing on overexpressed METTL14 in breast cancer cells for the first time, and found that METTL14 overexpression affected the methylation level of lncRNAs.
M6A mainly modifies the methylation of RNA molecules, 22 thus affecting the expression and stability of RNA and affecting biological processes to varying degrees, such as cancer metastasis. 23 Lu and his colleagues discovered METTL14 relies on m6A to promote the m6A modification of SIRT1 mRNA and cause the degradation of SIRT1 mRNA. 24 In addition, other studies demonstrated that silencing METTL14 decreased the m6A level of circORC5, but increased the expression of circORC5, resulting in poor survival of gastric cancer patients. 25 METTL14 upregulation decrease the expression of PERP through m6A modification, which promotes the growth and metastasis of pancreatic cancer. 26 The above studies suggested that METTL14 reduced expression of target genes by increasing m6A modification in cancer. In view of this, we combined MeRIP‐seq and RNA‐seq data to found some lncRNAs with hypermethylation‐RNA downregulation. Then through PCR verification, it was found that METTL14 overexpression promoted the methylation level of AC084125.2 and inhibited the expression of AC084125.2. Similar to other studies, 27 we suggested METTL14‐mediated m6A methylation inhibits the expression of target RNAs, as the evidence that METTL14 decreases AC084125.2 expression in an m6A‐dependent manner.
In recent years, the importance of lncRNAs in the occurrence and metastasis of cancer has been confirmed. 28 For example, the expression of lncRNA SPINT1‐AS1 was upregulated in breast cancer, and knockdown of SPINT1‐AS1 sharply lessened the proliferation and migration ability of breast cancer cells. 29 In our results, AC084125.2 was low expressed in breast cancer patients and cell lines, and overexpression AC084125.2 tremendously declined the migration and invasion of cancer cells. Besides, other reports reported that the AUC of lncRNA SChLAP1 in breast cancer was 0.8639 and p < 0.001, and breast cancer could be diagnosed by SChLAP1. 30 However, the AUC of AC084125.2 (0.8853) was higher than that of SChLAP1, indicating that AC084125.2 is more suitable as diagnostic markers for breast cancer. Summarily, we hypothesized that high expressed METTL14 promoted the malignant phenotype of breast cancer cells by reducing AC084125.2 expression via increasing m6A modification.
5. CONCLUSION
Taken together, we performed high‐throughput sequencing to reveal transcriptome‐wide m6A‐methylated lncRNA mediated by METTL14 overexpression in breast cancer. Conjoint analysis of MeRIP‐seq and RNA‐seq data discovered the lncRNAs with m6A methylation and synchronously differential expression. Furthermore, based on the reported METTL14‐regulated RNA model, we screened AC084125.2 as the target lncRNA of METTL14. We speculated that METTL14 relies on m6A modification to reduce AC084125.2 expression, thus promoting the progression of breast cancer. However, the specific regulatory mechanism needs to be further explored in the future. In brief, our study offers a theoretical basis for clinical diagnosis and targeted therapy of breast cancer in the future.
AUTHOR CONTRIBUTIONS
Jianfeng Sang and Xianbiao Shi contributed to study concept and design. Dandan Yi and Fazhan Xu performed the experiments, analyzed the data, and wrote the article. Ru Wang, Chaoyu Jiang, Jiabo Qin, and YiHsuan Lee contributed to the experimental work. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors report there are no competing interests to declare.
PATIENT CONSENT STATEMENT
All participants' written informed consents were obtained.
Supporting information
TABLE S1 The qPCR‐Primers used in this study.
FIGURE S1 The four‐quadrant diagram showed the m6A methylation transcripts and differentially expressed transcripts. The purple name represented the top 10 transcripts with the largest difference multiple in m6A, and the red dots represent MeRIP‐seq and RNA‐seq data meeting both p < 0.05. The first quadrant is transcripts with hypermethylation‐RNA upregulation, the second quadrant is transcripts with hypermethylation‐RNA downregulation, and the third quadrant is transcripts with hypomethylation‐RNA downregulation and the fourth quadrant is transcripts with hypomethylation‐RNA upregulation.
ACKNOWLEDGMENTS
The authors appreciate all participants enrolled in the present study.
Yi D, Xu F, Wang R, et al. Deciphering the map of METTL14‐mediated lncRNA m6A modification at the transcriptome‐wide level in breast cancer. J Clin Lab Anal. 2022;36:e24754. doi: 10.1002/jcla.24754
Dandan Yi and Fazhan Xu contributed equally as first authors.
Contributor Information
Xianbiao Shi, Email: sxbnju0510@163.com.
Jianfeng Sang, Email: glyy8c@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Fahad UM. Breast cancer: current perspectives on the disease status. Adv Exp Med Biol. 2019;1152:51‐64. [DOI] [PubMed] [Google Scholar]
- 2. Katsura C, Ogunmwonyi I, Kankam HK, Saha S. Breast cancer: presentation, investigation and management. Br J Hosp Med (Lond). 2022;83(2):1‐7. [DOI] [PubMed] [Google Scholar]
- 3. Berry DA, Cronin KA, Plevritis SK, et al. Cancer Intervention and Surveillance Modeling Network (CISNET) collaborators. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784‐1792. [DOI] [PubMed] [Google Scholar]
- 4. McDonald ES, Clark AS, Tchou J, Zhang P, Freedman GM. Clinical diagnosis and management of breast cancer. J Nucl Med. 2016;57(Suppl 1):9 S‐16 S. [DOI] [PubMed] [Google Scholar]
- 5. Chen Y, Lin Y, Shu Y, He J, Gao W. Interaction between N6‐methyladenosine (m6A) modification and noncoding RNAs in cancer. Mol Cancer. 2020;19:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou Z, Lv J, Yu H, et al. Mechanism of RNA modification N6‐methyladenosine in human cancer. Mol Cancer. 2020;19(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6‐methyladenosine in cancer progression. Mol Cancer. 2020;19:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu L, Liu X, Dong Z, et al. N6‐methyladenosine‐related genomic targets are altered in breast cancer tissue and associated with poor survival. J Cancer. 2019;10:5447‐5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cai X, Wang X, Cao C, et al. HBXIP‐elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let‐7g. Cancer Lett. 2018;415:11‐19. [DOI] [PubMed] [Google Scholar]
- 10. Yi D, Wang R, Shi X, Xu L, Yilihamu Y, Sang J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6‐methyladenosine and hsa‐miR‐146a‐5p expression. Oncol Rep. 2020;43:1375‐1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9(6):e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang J, Su Z, Lu S, et al. LncRNA HOXA‐AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229‐233. [DOI] [PubMed] [Google Scholar]
- 13. Yang X, Zhao X, Cheng L, Wei R. LncRNA FOXCUT stimulates the progression of endometrial cancer. Crit Rev Eukaryot Gene Expr. 2021;31(5):59‐66. [DOI] [PubMed] [Google Scholar]
- 14. Zuo X, Chen Z, Gao W, et al. M6A‐mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang X, Zhang S, He C, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down‐regulating oncogenic long non‐coding RNA XIST. Mol Cancer. 2020;19:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li B, Liu D, Xu J, et al. Overexpression of lncRNA MAPT‐AS1 exacerbates cell proliferation and metastasis in breast cancer. Transl Cancer Res. 2022;11:835‐847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu HT, Zou YX, Zhu WJ, et al. lncRNA THAP7‐AS1, transcriptionally activated by SP1 and post‐transcriptionally stabilized by METTL3‐mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2022;29:627‐641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meng L, Lin H, Huang X, Weng J, Peng F, Wu S. METTL14 suppresses pyroptosis and diabetic cardiomyopathy by downregulating TINCR lncRNA. Cell Death Dis. 2022;13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Z, Wang Q, Zhang M, et al. Comprehensive analysis of the transcriptome‐wide m6A methylome in colorectal cancer by MeRIP sequencing. Epigenetics. 2021;16:425‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zuo L, Su H, Zhang Q, et al. Comprehensive analysis of lncRNAs N6‐methyladenosine modification in colorectal cancer. Aging (Albany NY). 2021;13:4182‐4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma S, Chen C, Ji X, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hao L, Wang JM, Liu BQ, et al. m6A‐YTHDF1‐mediated TRIM29 upregulation facilitates the stem cell‐like phenotype of cisplatin‐resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res. 2021;1868:118878. [DOI] [PubMed] [Google Scholar]
- 24. Lu Z, Liu H, Song N, et al. METTL14 aggravates podocyte injury and glomerulopathy progression through N6‐methyladenosine‐dependent downregulating of Sirt1. Cell Death Dis. 2021;12:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fan HN, Chen ZY, Chen XY, et al. METTL14‐mediated m6A modification of circORC5 suppresses gastric cancer progression by regulating miR‐30c‐2‐3p/AKT1S1 axis. Mol Cancer. 2022;21:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang M, Liu J, Zhao Y, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N6 adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng C, Zhang H, Zheng J, Jin Y, Wang D, Dai Z. METTL14 benefits the mesenchymal stem cells in patients with steroid‐associated osteonecrosis of the femoral head by regulating the m6A level of PTPN6. Aging (Albany NY). 2021;13:25903‐25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR‐153‐5p/ARHGAP18 signaling axis. Aging (Albany NY). 2018;10:3371‐3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou T, Lin K, Nie J, et al. LncRNA SPINT1‐AS1 promotes breast cancer proliferation and metastasis by sponging let‐7 a/b/i‐5p. Pathol Res Pract. 2021;217:153268. [DOI] [PubMed] [Google Scholar]
- 30. Cheng S, Zhao Q, Zheng Z. Expression and clinical significance of lncRNA‐SChLAP1 in breast cancer. J BUON. 2021;26:728‐733. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S1 The qPCR‐Primers used in this study.
FIGURE S1 The four‐quadrant diagram showed the m6A methylation transcripts and differentially expressed transcripts. The purple name represented the top 10 transcripts with the largest difference multiple in m6A, and the red dots represent MeRIP‐seq and RNA‐seq data meeting both p < 0.05. The first quadrant is transcripts with hypermethylation‐RNA upregulation, the second quadrant is transcripts with hypermethylation‐RNA downregulation, and the third quadrant is transcripts with hypomethylation‐RNA downregulation and the fourth quadrant is transcripts with hypomethylation‐RNA upregulation.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


