Abstract
The nucleic acid polymerase-catalyzed nucleotidyl transfer reaction associated with polymerase active site closure is a key step in the nucleotide addition cycle (NAC). Two proton transfer events can occur in such a nucleotidyl transfer: deprotonation of the priming nucleotide 3′-hydroxyl nucleophile and protonation of the pyrophosphate (PPi) leaving group. In viral RNA-dependent RNA polymerases (RdRPs), whether and how active site residues participate in this two-proton transfer reaction remained to be clarified. Here we report a 2.5 Å resolution crystal structure of enterovirus 71 (EV71) RdRP in a catalytically closed pre-chemistry conformation, with a proposed proton donor candidate K360 in close contact with the NTP γ-phosphate. Enzymology data reveal that K360 mutations not only reduce RdRP catalytic efficiency but also alter pH dependency profiles in both elongation and pre-elongation synthesis modes. Interestingly, mutations at R174, an RdRP-invariant residue in motif F, had similar effects with additional impact on the Michaelis constant of NTP (KM,NTP). However, direct participation in protonation was not evident for K360 or R174. Our data suggest that both K360 and R174 participate in nucleotidyl transfer, while their possible roles in acid–base or positional catalysis are discussed in comparison with other classes of nucleic acid polymerases.
INTRODUCTION
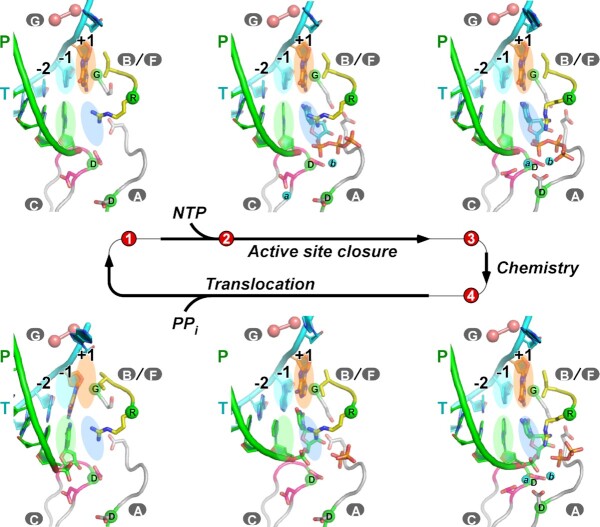
RNA-dependent RNA polymerases (RdRPs) encoded by RNA viruses represent a unique class of nucleic acid polymerases that play key roles in viral genome replication and transcription and are ideal targets for developing antivirals. Similar to other classes of polymerases, the nucleic acid biosynthesis by RdRP comprises thousands of ‘nucleotide addition cycles’ (NACs) to incorporate nucleoside monophosphate (NMP) building blocks into the nucleic acid product. A classic four-reference-state model of NAC includes a state one (S1) ‘open’ complex with a vacant active site, a nucleoside triphosphate (NTP) substrate-bound state 2 (S2) with the active site remaining open, a catalytically competent ‘closed’ state 3 (S3) with necessary conformational changes occurring after NTP binding and a post-chemistry ‘closed’ state 4 (S4) awaiting polymerase translocation to the next register that finishes the NAC (1–3) (Figure 1). Numerous three-dimensional structures of polymerase catalytic complexes have been captured at or in between these reference states. Translocation intermediates, bridging S4 and S1 of the next NAC, have been structurally captured in RdRPs and DNA-dependent RNA polymerases (DdRPs), depicting an asymmetric movement of the template–product RNA duplex, with the template strand being relatively constrained by the polymerase (4–7) (Figure 1). These NAC-related structures, if organized in a sequential manner, provide a framework to understand the dynamic process of the entire cycle.
Figure 1.
Viral RdRP NAC depicted by a series of EV71 RdRP EC crystal structures. Six RdRP EC models are shown in a sequential manner from top left to bottom left. Coloring scheme: template in cyan (+1 nucleotide in orange), product in green, palm in gray (YGDD sequence in magenta) and ring finger subdomain in yellow. Uppercase letters with gray background indicate corresponding RdRP motifs. The α-carbons of the four invariant residues are shown as green spheres. Magnesium ions (symbolized as ‘a’ and ‘b’ for metal A and metal B) are shown as cyan spheres. The hurdle residues interacting with the template +1/+2 junction (T114 and S115 in EV71 RdRP) in motif G are shown as pink spheres. Four colored ovals are used as references for in-register locations for nucleosides at positions –1 and +1. Numbers with a red background indicate four reference NAC states, while EC models placed in between these reference states represent intermediate states. PDB entries from top left to bottom left (clockwise): 5F8G (S1), 5F8I (between S2 and S3), S3 = 7W9S (S3; this work), 5F8M (S4); 5F8N (between S4 and S1, early stage during translocation); S6M = 6LSH (between S4 and S1, late stage during translocation).
The phosphoryl transfer is one of the rate-limiting steps in NAC employing a two-metal-ion mechanism (8,9). One divalent metal ion, metal A, increases the nucleophilicity of the priming 3′-OH by lowering its pKa, while the other, metal B, facilitates pyrophosphate (PPi) release (10). Both metal ions contribute to re-orientating the triphosphate moiety of rNTP/dNTP and maintaining a closed polymerase active site for catalysis. Deprotonation of the 3′-hydroxyl nucleophile and protonation of the PPi leaving group could occur prior to phosphoryl transfer in all nucleic acid polymerases (11,12). Proton donor candidate residues, K560 of bacteriophage RB69 DdDP, K631 of bacteriophage T7 DdRP, K220 of human immunodeficiency virus 1 (HIV-1) reverse transcriptase (RT) and K359 of poliovirus (PV) RdRP, were proposed in a comparative study (13). Structural evidence is also available in DdDPs and DdRPs with the candidate lysine amine group captured in close contact with the NTP triphosphate moiety or PPi (1,14–15). However, questions remain about whether PV RdRP K359 and its equivalents in other RdRPs have such a role in common. Besides the lack of structural evidence of its direct interaction with NTP, K359 is only conserved in some RNA viruses including those from Picornavirales. In contrast and among very few invariant RdRP residues, there is an arginine in RdRP motif F (corresponding to PV RdRP R174) that may also be a proton donor candidate based on RdRP structures captured with a ‘closed’ active site (S3 or S4) (3,16).
In this study, a CTP-derived S3 RdRP elongation complex (EC) of enterovirus 71 (EV71), a close relative of PV, was captured by X-ray crystallography. Besides a previously identified R174-NTP α,β-phosphate linkage interaction, K360, the PV K359 equivalent, was found interacting with the γ-phosphate of CTP bound in the RdRP active site, thus supporting its direct involvement in nucleotidyl transfer. Furthermore, Michaelis–Menten kinetics and pH dependency comparison of R174 and K360 mutants and wild-type (WT) EV71 RdRP suggests the contribution of both residues in such a catalytic process, providing a refined understanding of viral RdRP nucleotidyl transfer reaction.
MATERIALS AND METHODS
Plasmid construction and protein preparation
A pET26b-Ub vector-based plasmid containing the WT EV71 3Dpol (RdRP) (strain SK-EV006-LPS1, or strain SK) gene was used as the original cloning template to construct the mutant plasmids following previously described methods (4,17–18). Cell growth, isopropyl-β-d-thiogalactopyranoside (IPTG) induction, cell harvesting, cell lysis, protein purification and protein storage were performed as described previously (4,19). Briefly, protein was expressed in Escherichia coli strain BL21(DE3)pCG1, induced by 0.5 mM IPTG and purified by sequentially passing through nickel-affinity, anion exchange and gel filtration columns.
RNA preparation and 3Dpol EC assembly
The 35-mer (T35) and 33-mer (T33) template RNAs used in crystallography and enzymology characterizations, respectively, were prepared by T7 RNA polymerase-glmS ribozyme-based approaches described previously (20,21), and the 10-mer RNA primer (P10) was chemically synthesized (Integrated DNA Technologies). RNA construct assembly and EC assembly, purification by anion exchange chromatography and storage were as previously described (5).
EC crystallization and EC crystal soaking trials
EC crystallization was performed as described previously (4). Briefly, crystals were grown by sitting-drop vapor diffusion at 16°C using 10 mg/ml EC samples. Crystals grew in a precipitant solution containing 0.17 M ammonium sulfate, 0.085 M MES (pH 6.5), 25.5% (w/v) polyethylene glycol (PEG) 5000 monomethyl ether and 15% (v/v) glycerol. Crystal soaking trials were done using the precipitant solution supplemented with 5 mM CTP, 5 mM UTP, 10 mM MgCl2 and 5 mM tetrasodium pyrophosphate (PPi). Crystals were directly cooled and stored in liquid nitrogen prior to data collection.
Crystallographic data processing and structure determination
X-ray diffraction data was collected at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1 (wavelength: 0.9792 Å) at 100 K. A full 180° of data were collected in 0.2° oscillation steps. Reflections were integrated, merged and scaled using HKL2000 (Table 1) (22). The initial structure solution was obtained using the molecular replacement program PHASER (23) with coordinates derived from a native EV71 EC structure (PDB entries 5F8G, chains A–C) as the search model (4). Manual model building and structure refinement were done using Coot and PHENIX, respectively (24,25). The 3500 K composite simulated-annealing (SA) omit 2Fo–Fc electron density maps were generated using PHENIX. Unless otherwise indicated, protein structure superimposition was done using the maximum likelihood-based structure superpositioning program THESEUS (26).
Table 1.
X-ray diffraction data collection and structure refinement statistics
| PDB | 7W9S |
|---|---|
| Data collection a | |
| Space group | P 21 21 21 |
| Cell dimensions | |
| a, b, c (Å) | 64.0, 77.7, 150.8 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å)b | 50.00–2.53 (2.62–2.53) |
| No. of unique reflections | 25127 |
| R merge | 0.063 (0.77) |
| R meas | 0.072 (0.89) |
| CC1/2 | 0.989 (0.645) |
| I /σI | 18.1 (2.2) |
| Completeness (%) | 95.2 (90.6) |
| Redundancy | 4.5 (3.6) |
| Structure refinement | |
| Resolution (Å) | 2.53 |
| No. of unique reflections | 24670 |
| R work/Rfreec (%) | 20.0/24.1 |
| No. atoms | |
| Protein/RNA | 3526/471 |
| Ligand/Ion/Water | 29/19/15 |
| B-factors (Å2) | |
| Protein/RNA | 68.2/81.3 |
| Ligand/Ion/Water | 62.8/69.7/64.4 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.992 |
| Ramachandran stat.d | 91.4/8.3/0.0/0.2 |
aOne crystal was used for data collection.
bValues in parentheses are for highest resolution shell.
c5% of data are taken for the Rfree set.
dValues are in percentages and are for most favored, additionally allowed, generously allowed and disallowed regions in Ramachandran plots, respectively.
The stopped-flow fluorescence-based single nucleotide elongation assay
This assay was performed according to protocols described previously (5). Briefly, the 33-mer RNA template T33-Fint with an internal fluorescein label (Integrated DNA Technologies) was annealed with a 10-mer RNA primer (P10) at a 1.1:1 molar ratio to produce a T33-Fint/P10 construct. The EC was assembled using T33-Fint/P10 and EV71 RdRP upon incorporation of a G–A–G–A tetranucleotide with GTP and ATP as the only NTP substrates, producing a 14-mer (P14)-containing EC (EC14). The fluorescence signal change was measured at 22.5°C after rapid mixing of EC14 and CTP samples, producing a final solution with 25 nM RNA, 400 nM RdRP, 0.5 μM ATP/GTP and different concentrations of CTP in SF buffer containing 50 mM HEPES (pH 7.0), 30 mM NaCl, 50 mM KCl, 5 mM MgCl2 and 5 mM Tris(2-carboxyethyl)phosphine (TCEP). A rapid fluorescence signal decrease was observed when CTP was mixed with EC14 using a stopped-flow instrument (Applied PhotoPhysics Chirascan SF3). The fluorescence excitation monochromator bandwidth was set at 4 nm. For pH dependency trials, the final RdRP concentration was 150 nM, CTP was provided at 300 μM and buffering agent [NaOAc pH 4.5, 5.0, 5.5; Na3citrate pH 4.5, 5.0, 5.5; MES pH 5.5, 6.0, 6.5; HEPES pH 6.5, 7.0, 7.5, 8.0; Tris pH 7.0, 7.5, 8.0, 8.5, 9.0; glycine pH 8.5, 9.0, 9.5, 10.0, 10.5; and N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) pH 10.0, 10.5, 11.0] was provided at 5 mM.
Data analysis and fitting were done as described previously (19). Briefly, the data of signal decrease observed for CMP incorporation were fitted into a single exponential decay equation Y = amplitude × exp(–rate × t) + offset, where t is the reaction time. The single-turnover incorporation rates under various CTP concentrations were then fitted to the Michaelis–Menten-type equation, rate = kcat × [CTP]/([CTP] + KM,CTP), to determine the kcat and apparent KM,CTP values for calculating kcat/KM,CTP to assess catalytic efficiency. For pH dependency analyses, Tris-derived data were fit to either a one-ionizable-group model: rate = rateind/(1 + 10pKa,1 – pH) or a two-ionizable-group model: rate = rateind/(1 + 10pKa,1 – pH + 10 pH – pKa,2), with pKa,2 set as fixed values (10.5 or 12.0, only for the WT) (27).
Gel-based primer-dependent single nucleotide incorporation assay
To identify the optimal pH value for WT and mutant RdRPs, a typical 20 μl reaction mixture containing 5 mM buffering agent (the same as the pH dependency stopped-flow trials, pH range: 4.5–11.0), 50 mM KCl, 20 mM NaCl, 5 mM MgCl2, 4 mM TCEP, 4 μM T33/P10 RNA, 6 μM 3Dpol was pre-incubated at 22.5°C for 1 h, and then GTP was provided at 300 μM to allow the P10 to P11 conversion at 22.5°C. The T33 RNA has a similar sequence to T33-Fint except for several downstream nucleotides (5′-GGGAGAUGAAAGUCUCCAGGUCUCUCGUCGAAA-3′, different nucleotides are underlined). An equal volume of stop solution containing 95% (v/v) formamide, 20 mM EDTA (pH 8.0) and 0.02% (w/v) xylene cyano was added to quench the reaction. The RNA species were analyzed by denaturing (7 M urea) 20% (w/v) polyacrylamide gel electrophoresis (PAGE) followed by Stains-All staining (Sigma-Aldrich), and band intensity quantification was performed using ImageJ (https://imagej.nih.gov/ij) as previously described .
RESULTS
A crystal structure of an EV71 RdRP state 3 complex reveals triphosphate contact by the proton donor candidate K360
Using a previously established EV71 RdRP EC crystal form that permits multiple in-crystal NACs (4,5), we attempted to obtain S3-like structures using NTP substrate-containing solutions in crystal soaking trials. Besides cognate NTP (CTP in this case) and magnesium ion (Mg2+), a non-cognate NTP (UTP) and/or PPi were also included in short duration soaking to possibly capture the polymerase just before the chemistry step. An S3 crystal structure was solved at 2.5 Å resolution with CTP, UTP, Mg2+ and PPi all present in the soaking solution and a 5 min soaking time (Table 1; Supplementary Figure S1A). In combination with previously reported native and NTP-derived EC structures using the same crystal form, a relatively complete structural framework of NAC has been sketched (28) (Figure 1; Supplementary Video S1). In this structure, the RdRP active site adopts the closed conformation with the side chain guanidinium group of R174 motif F interacting with the α,β-phosphate linkage in an identical manner to available RdRP S3 and S4 structures (3–4,16,29). Very interestingly, the side chain amine group of K360 interacts with the γ-phosphate, with a 3.3 Å distance between the amine nitrogen atom and one of the phosphate oxygen atoms (Figure 2A–C). The close contact between residue K360 and the triphosphate of substrate NTP observed in this catalytically competent conformation supports the proton donor candidacy of K360 equivalent residues in RdRPs, even though similar close contacts have not been observed in other RdRPs (Figure 2D, E). Located at the outer rim of the NTP entry channel, K360 is adjacent to RdRP motif D that undergoes a backbone shift in coordination with motif A upon active site closure. Hence, the intrinsic backbone dynamics and the flexible nature of the lysine side chain may make the K360–γ-phosphate contact difficult to observe.
Figure 2.
A crystal structure of EV71 RdRP captured in a catalytically competent state. (A) Stereo-pair images of the EV71 RdRP EC S3 structure with 3500 K composite SA omit electron density maps (contoured at 1.2 σ) overlaid. The coloring scheme is the same as in Figure 1, with electron densities of metal ions in black. The letters in the black circles indicate the names of the catalytic motifs. The black dashed lines indicates key interactions (hydrogen bond, metal ion coordination and electrostatic interaction). (B) A schematic diagram of interactions involving the substrate CTP in the active site of the S3 structure. Two metal ions (denoted as MeA and MeB) are in cyan and the CTP is in blue. (C–E) EV71 RdRP K360-equivalent residues have not been captured in direct contact with NTP in S3 (or S3-like) structures in other viral RdRPs. 2Fo–Fc electron density maps (contoured at 1.2 σ) are overlaid with RdRP structures (labeled with PDB entries) from EV71 (C, this work), norovirus (NV) (D) and hepatitis C virus (HCV) (E). Three structures were superimposed using the main chain atoms of their motif C residues.
Among the structures with the side chain fully resolved through our soaking trials, the distance between CTP γ-phosphate and K360 side chain amine nitrogen varies between 3.3 and 4.0 Å for (Supplementary Figure S1A–C). The presence of PPi in the soaking solution probably plays a critical role in capturing this pre-chemistry state. Previously reported enterovirus RdRP structures either derived from the same crystal form but obtained after a comparable soaking duration (Supplementary Figure S1D, 5 min 10 s versus 5 min in this work), obtained after an overnight soaking (Supplementary Figure S1E) or captured after three NAC events all represent post-chemistry states (Supplementary Figure S1F) (3,4). Moreover, the reorientation of γ-phosphate allows its interactions not only with the K360 side chain but also with catalytic metal B, and therefore is probably a late event in active site closure. Accordingly, a similar triphosphate conformation is only observed in the NV S3 structure or our EV71 S3 structures obtained using a similar soaking strategy, but not in any other pre- and post-chemistry RdRP structures (3,16) (Supplementary Figure S1).
Mutations at both K360 and R174 sites impair EV71 RdRP catalysis
While R174 is invariant in RdRPs and RTs, glutamic acid (E) and aspartic acid (D) also exist at K360-equivalent positions in RdRPs (Figure 3A). We therefore hypothesize that R174-equivalent residues may also serve as proton donors and may be universal to RdRPs. In order to further dissect the function of K360 and R174 in EV71 RdRP catalysis, we made five mutants, K360R, K360A, R174K, R174A and K360A-R174A, and compared them with WT RdRP (EV71 strain SK) in a previously established stopped-flow fluorescence assay (5). The RNA construct used in this assay contains a fluorescently labeled template. An RdRP EC was assembled upon primer-dependent incorporation of a G–A–G–A tetranucleotide, and was then subjected to stopped-flow trials to mix with CTP for a single-nucleotide elongation triggering an immediate fluorescent signal decrease (Figure 3B). The catalytic rate constant kcat and Michaelis constant KM,CTP were determined for each RdRP construct. All four single-point mutants greatly impaired kcat, retaining only 0.4–4.3% of the activity of the WT enzyme (Figure 3C). Due to very low fluorescent signal change, we were not able to obtained usable data for the K360A-R174A mutant. While the K360 mutants exhibited 0.3- to 1.4-fold the WT KM,CTP value (K360R, 10.0 μM; K360A, 51.3 μM versus WT, 36.6 μM), the R174 mutants had much higher KM,CTP values that are ∼5.7- to 26.7-fold the WT level (R174K, 217 μM; R174A, 979 μM) (Figure 3C). These data suggest that both K360 and R174 are likely to contribute to the nucleotidyl transfer, but only R174 plays an important role in NTP binding. If using kcat/KM,CTP to assess catalytic efficiency, the K360A and R174A mutants had ∼260- and 2700-fold efficiency reduction in the WT level, respectively. These data are consistent with the structural observation that R174-equivalent residues interact with NTP triphosphate moiety in both ‘open’ and ‘closed’ active site conformations, while K360-equivalent residues are rarely and only captured in close proximity to NTP triphosphate moiety in the ‘closed’ conformation (Figure 2A).
Figure 3.
Enzymology characterization of EV71 RdRP and its mutants by a stopped-flow fluorescence-based approach. (A) A structure-based sequence alignment of RdRP motifs F (C-terminal part) and D (with C-terminal extension). Two RT sequences are listed for comparison. +ssRNA/–ssRNA/dsRNA: positive-strand/negative-strand/double-stranded RNA viruses. PICO (Picornaviridae): CV, coxsackievirus B3; HRV, human rhinovirus A1B; FMDV, foot-and-mouth disease virus. FLAV (Flaviviridae): CSFV, classical swine fever virus; JEV, Japanese encephalitis virus. PERM (Permutotetraviridae): TaV, Thosea asigna virus. LEVI (Leviviridae): QBETA, bacteriophage Qβ. CORO (Coronaviridae): SARS2, severe acute respiratory syndrome coronavirus 2. PBIR (Picobirnaviridae): HPBV, human picobirnavirus. CYST (Cystoviridae): PHI6, bacteriophage φ6. BIRN (Birnaviridae): IBDV, infectious bursal disease virus. REO (Reoviridae): MORV3, mammalian orthoreovirus 3. RHAB (Rhabdoviridae): VSV, vesicular stomatitis virus. PNEU (Pneumoviridae): HMPV, human metapneumovirus. PERI (Peribunyaviridae): LACV, La Crosse bunyavirus. PHEN (Phenuiviridae): SFTSV, severe fever with thrombocytopenia syndrome virus. ORTH (Orthomyxoviridae): IAV, influenza virus A. RETR (Retroviridae): HIV-1. TERT, telomerase reverse transcriptase. (B) The sequences of the T33-Fint/P10 (Fint denotes a fluorescein-labeled nucleotide at an internal position) construct used in the single-nucleotide elongation assay and the data from the WT and K360R constructs showing faster fluorescence signal change with higher CTP concentration. Each trace reflects the average signal of 5–8 replicates. A 14-mer-containing EC was assembled using GTP and ATP and then used in stopped-flow trials by mixing with CTP and monitoring the conversion of the 14-mer to the 15-mer. (C) Michaelis–Menten curve fittings of WT RdRP and its mutants. For the right panel, fitted WT curves were shown for comparison with data points omitted. kcat, KM and kcat/KM values with fitting errors or propagated fitting errors are listed for comparison.
Mutations at both K360 and R174 sites modulate pH dependency for EV71 RdRP elongation
To further explore whether mutations at K360 and R174 sites affect the pH dependency of EV71 RdRP EC, we measured the rate of single-nucleotide incorporation under a single CTP concentration of 300 μM and various pH values for WT RdRP and all single-point mutants (Figure 4; Supplementary Table S1). Seven buffering agents were used to cover a wide pH range of 4.5–11.0 in a single buffering agent setting, and the final agent concentration of 5 mM was chosen to allow enough fluorescence signal change for the majority of pH conditions. The RNA construct and EC assembly procedures are the same as those for determining kcat and KM,CTP parameters. The assembled EC was then used in stopped-flow trials for rate determination. Rates were readily determined for all RdRP constructs at all pH values, except for measurements at very low or high pH values due to reasons including protein aggregation (low pH) and low fluorescence signal change (both low and high pH) (Figure 4; Supplementary Table S1). For all measurable rates, an increasing trend upon pH increase was observed for every RdRP construct within each buffering agent (Figure 4, compare all data points in each agent set). Different agents at identical pH values yielded different rates, reflecting their difference in both chemical nature and enzyme regulation (Figure 4, compare data points at the same pH but from different agents). We should also note that rates could be quite different with buffering agent provided at different concentrations. For example, the kcat value is 23.5/s for the WT at 50 mM HEPES (7.0) but a rate of ∼40/s was determined at 5 mM HEPES (7.0) at 300 μM CTP concentration. We also tested the pH dependency in a pH range of 7–11 using multiple buffering agents for the WT and K360A mutant. For the Tris–CAPS and Tris–glycine–CAPS combinations tested, we still did not observe a rate decrease upon pH increase in this pH range (Supplementary Figure S2).
Figure 4.
The pH dependency of WT RdRP and its mutants in the elongation mode. (A–E) Using the T33-Fint/P10 construct, and following the same EC assembly and stopped-flow operation as in Figure 3, the single-nucleotide incorporation rates for the WT (A) and its mutants (B–E) were determined in a pH range of 4.5–11.0. Data from valid measurements are shown by filled squares, while open and filled circles shown at rate ‘0’ (indicated by the horizontal gray line) represent measurements not suitable for rate determination (see also Supplementary Table S1). Tris-derived WT data points (blue squares) were fitted to either a one-ionizable-group model or a two-ionizable-group model but with a constant pKa of the second ionization group (pKa,2: 10.5 or 12.0 for the WT), while only the one-ionizable-group model was used for all mutants. The WT curve generated using the one-ionizable-group model (red) in (A) is provided as a reference in (B–E).
Although we were not able to observe a rate decrease upon pH increase for any single agent or agent combinations tested, the pH-dependent rate profile is indeed different between the WT and all its mutants, in particular for Tris-derived data points. While the rate starts to level off beyond pH 8.0 for the WT, it keeps increasing for all four single-point mutants (Figure 4, dark blue points). Similar difference was observed in the PV study when comparing WT RdRP with K359L and K359R mutants (27) (PV K359 is equivalent to EV71 K360). If only considering acid–base catalysis, two ionizable groups yield a bell-shaped pH profile, from which the two pKa values can be derived (pKa,1 < pKa,2). However, our data only cover the ascending part of the profile and we therefore use the Tris-derived WT data and compare fitting to a single-pKa model or fitting to the two-pKa model with a fixed value of pKa,2 to estimate the pKa,1 and the pH-independent rate (rateind). For WT enzyme, the pKa,2 of 10.5 (corresponding to the pKa of lysine) yielded a pKa,1 of 7.0 and an rateind of 71/s, while higher pKa,2 (12.0 tested) or fitting with pKa,1 only did not affect these values much (Figure 4A).
We therefore used the single-pKa fitting for the analysis of the mutants, and consistently obtained pKa,1 values of 8.0–8.2 for all single-point mutants (Figure 4B–E). These data suggest that K360 and R174 contribute similarly to the pH dependency profile, but the profile alteration by the mutations is not necessarily related to PPi protonation. On the other hand, the rateind values are consistent with mutation type (K360R/R174K, 10–11/s; K360A/R174A, 1.5–1.6/s) but not mutation site, suggesting that both basic side chains also contribute to pH-independent (e.g. positional) catalysis.
Mutations at only the K360 site alter the optimal pH range of EV71 RdRP in the pre-elongation stage
RdRP of enteroviruses (e.g. PV and EV71) typically enters the elongation phase after incorporation of one or more nucleotides in nucleic acid primer-driven assays (30–32). Therefore it is worthwhile assessing pH dependency of RdRP catalysis also at the pre-elongation stage. A 33 nt template (T33) and a 10 nt primer (P10) were used to assemble the T33/P10 RNA construct. With GTP provided as the only NTP substrate, EV71 RdRP is able to convert P10 to an 11 nt product (P11) in pre-elongation mode (Figure 5). A pre-incubation time of 1 h was applied prior to the reaction to minimize the effect of possible slow association of RdRP and RNA, and the buffering agent concentration, pH range and single-agent setting were consistent with those in the EC pH dependency trials. For each construct, a single reaction time was set in the range of 8 s to 15 min for >80% production of P11 at its optimal pH. All RdRP constructs exhibited an optimal pH range when assessed by denaturing PAGE (Figure 5). WT RdRP had an optimal pH range of 6.0–7.0 as assessed by MES- and HEPES-derived data (Figure 5A). Both K360R and K360A mutants had the optimal range moderately but clearly shifted toward the high pH direction (Figure 5B, C: 6.5–7.0 and 6.5–7.5), while such a shift was not observed for both R174 mutants (Figure 5D, E). We were also able to test the K360A-R174A mutant in this gel-based assay and it had a similar behavior to the K360 single-point mutants. The obvious differences in optimal pH range and R174 contribution between elongation and pre-elongation modes indicate that fundamental differences in RdRP catalysis may exist between these two modes. Although K360-related mutants do shift the optimal pH towards high pH values in the pre-elongation mode assay, the optimal pH is still near neutral. Hence these data do not prove participation of K360 in PPi protonation that occurs presumably at much higher pH.
Figure 5.
The pH dependency of WT RdRP and its mutants in the pre-elongation mode. (A) the T33/P10 RNA construct was used in a single-nucleotide incorporation assay (see the Materials and Methods). When GTP is provided as the only substrate, the 10 nt primer (P10) is extended to a 11 nt product (P11). (A–F) Denaturing PAGE analysis of post-reaction RNA species for the WT and its mutants. The percentages of 11-mer intensity are used to estimate the optimal pH range (boxed pH values) of each RdRP construct. Data derived from different buffering agents are separated by vertical lines. M: P10 is provided as the migration marker. For each RdRP construct, the optimal pH range is assigned for lanes with the highest 11-mer intensity values and among which the intensity difference between neighboring lanes is not greater than 5%.
DISCUSSION
The residue boundaries of motif D are not defined consistently in different viral RdRPs, probably due to low sequence conservation at the level of all RNA viruses (28,33). If using structural homology and equivalence as a guide, motif D of EV71 RdRP can be localized to residues 352–356 that form antiparallel β-type interactions with residues 233–235 of motif A. K360 is within the C-terminal extension of motif D that is usually a loop with variations in length, structure and sequence (Figure 3A). Upon active site closure, motif D and this loop move together with motif A towards the active site (3,29). However, in previously reported RdRP structures, K360 or its equivalents have not been observed in close contact with NTP substrate even for structures with a closed active site. Previous studies suggest that K359R mutation in PV RdRP and K360R mutation in coxsackievirus B3 (CV B3) reduce the kcat (defined as kpol or maximum elongation rate in these studies) by ≥20-fold, indicating that this lysine may indeed participate in forming the transition state of the nucleotidyl transfer reaction (27,34). By capturing an S3 structure with the K360 side chain interacting with NTP γ-phosphate, we obtained structural evidence to support the direct participation of this residue in phosphoryl transfer. In one of the PV studies, K359R and K359H both change the pH dependency of catalysis, by shifting the pKa,1 to a higher value (27), consistent with our observation for both K360 and R174 single-point mutants in the elongation mode tests. Collectively, a general acid from RdRP could participate in the phosphoryl transfer, but this role can be fulfilled by not just one residue. Considering the fact that R174 is invariant but K360 is not, only the former's role as a general acid may be generally applied to RdRPs. Although our data support that both R174 and K360 are involved in nucleotidyl transfer, we were not able to prove a contribution in PPi protonation for either of them. Further attempts probing basic active residues in viral RdRPs are worthwhile to prove their possible role in PPi protonation.
The direct involvement of K360 in RdRP catalysis is also compatible with the behaviors of RdRPs or RNA viruses bearing mutations in motif D and its C-terminal extension. PV K359R, T362I and CV B3 F364W mutant viruses exhibited an attenuated phenotype (34–36), while mutations or modulating interactions in this region resulted in alteration in RdRP catalysis and fidelity in PV and CV B3 (34–39). Note that multiple processes, including NTP recruitment, peptide backbone shift of motifs A/D during active site closure and NTP triphosphate moiety alignment, may affect RdRP catalysis and fidelity. K360 and its neighboring residues in motifs A/D, together with mostly side-chain-level conformational changes in motifs B/C/F, contribute to the reshaping of the active site during the induced-fit process of active site closure. It has been pointed out that the general acid–base catalysis may not be the only mechanism to achieve phosphoryl transfer in nucleic acid polymerases (40). As shown in the case of multisubunit DdRPs, the active site trigger loop/helix (TL/TH) acts as a positional rather than as an acid–base catalyst (41,42). In single-subunit nucleic acid polymerases, the NTP triphosphate moiety may form three sets of interactions upon active site closure (Figure 6). The first set is the most conservative and involves the two invariant aspartic acid residues of motifs A and C and the two catalytic divalent metal ions. The second set of interactions are on the opposite side of the divalent metal ions and involve basic residues (usually arginines and lysines) of motif F in RdRPs or motif B in DdRPs (1,3,14,43). EV71 RdRP R174 participates in this set of interactions and is structurally equivalent to T7 RNA polymerase K631 that is highly conserved in A-family polymerases. The third set of interactions, probably the least conservative in residue type, are likely to be from the distal end of the triphosphate offered by the EV71 RdRP K360-like residues located near the entrance of the NTP/dNTP entry channel. Typically, there is a positively or negatively charged residue at the structurally identical or similar position (Figure 6). Although not yet structurally captured as in EV71 RdRP, such a residue may form direct or indirect contact with the triphosphate when forming the transition state of phosphoryl transfer and therefore may cooperate with residues involved in the other sets of interactions, for either acid–base or positional catalysis. While we were not able to determine optimal pH ranges in our elongation-mode single-buffering-agent experiments, these ranges are detectable at near neutral pH for pre-elongation trials under the same buffering settings. We propose that pre-elongation RdRP complexes may have a ‘loose’ active site with positional catalysis and acid–base catalysis having different weights if compared with the ECs. Such pre-elongation complexes with a less optimal active site could be more susceptible to solution environment changes and respond differently upon active site residue mutation.
Figure 6.
A comparison of S3 or S3-like complexes of four classes of single-subunit nucleic acid polymerases. Labeling, symbols and coloring scheme are the same as in Figure 1 except that the fingers domain is in pink. The side chain of key residues surrounding the substrate NTP (or NTP analog) are shown as sticks. Magnesium ions are shown as cyan spheres. PDB entries from left to right: 7W9S, 1RTD, 1S76 and 3KTQ. All structures were superimposed using the main-chain atoms of their motif C residues.
By solving a catalytically closed RdRP EC structure, we observed a general acid candidate lysine residue in close contact with the NTP triphosphate moiety. Together with enzymology data characterizing this lysine and an invariant motif F arginine, we collected evidence to support the contribution of both residues in RdRP phosphoryl transfer. As this residue is not universally conserved in RdRPs, the participation of its structural equivalents and other residues in motifs D and F as acid–base or positional catalysts remains to be clarified, while a structural comparison among all four classes of single-subunit nucleic acid polymerases depicts similarity of the interaction framework surrounding the NTP/dNTP triphosphate moiety in their catalytically competent state.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession number 7W9S.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Olve Peersen for pointing out the possibility that PV RdRP R174 may act as a proton donor in nucleotidyl transfer. We thank Dr Zhiyong Lou for providing the cloning materials for EV71 polymerase gene (strain SK), Qiaojie Liu and Yao Zhong for laboratory assistance, synchrotron SSRF (beamline BL17U1, Shanghai, China) for access to the beamline, the Institutional Center for Shared Technologies and Facilities, Wuhan Institute of Virology, for access to instruments, and the Core Facility and Technical Support, Wuhan Institute of Physics and Mathematics, Chinese Academy of Sciences for access to the stopped-flow instrument.
Author contributions: P.G. conceived the research; R.L. and M.W. performed the experiments; all authors analyzed the data, contributed to manuscript writing and approved the manuscript content.
Contributor Information
Rui Li, Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, No. 44 Xiao Hong Shan, Wuhan, Hubei, 430071, China; University of Chinese Academy of Sciences, Beijing, 100049, China.
Meihua Wang, Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, No. 44 Xiao Hong Shan, Wuhan, Hubei, 430071, China; University of Chinese Academy of Sciences, Beijing, 100049, China.
Peng Gong, Key Laboratory of Special Pathogens and Biosafety, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences, No. 44 Xiao Hong Shan, Wuhan, Hubei, 430071, China; University of Chinese Academy of Sciences, Beijing, 100049, China; Drug Discovery Center for Infectious Diseases, Nankai University, Tianjin, 300350, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFA0507200]; National Natural Science Foundation of China [32070185 and 31802147]; Key Biosafety Science and Technology Program of Hubei Jiangxia Laboratory [JXBS001]; and Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory, Chinese Academy of Sciences [2021ACCP-MS10]. Funding for open access charge: National Key Research and Development Program of China [2018YFA0507200] and National Natural Science Foundation of China [32070185 and 31802147].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yin Y.W., Steitz T.A.. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004; 116:393–404. [DOI] [PubMed] [Google Scholar]
- 2. Kornberg R.D. The molecular basis of eukaryotic transcription. Proc. Natl Acad. Sci. USA. 2007; 104:12955–12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gong P., Peersen O.B.. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA. 2010; 107:22505–22510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shu B., Gong P.. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl Acad. Sci. USA. 2016; 113:E4005–E4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M., Li R., Shu B., Jing X., Ye H.Q., Gong P.. Stringent control of the RNA-dependent RNA polymerase translocation revealed by multiple intermediate structures. Nat. Commun. 2020; 11:2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang J.Y., Mishanina T.V., Bellecourt M.J., Mooney R.A., Darst S.A., Landick R.. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell. 2018; 69:802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weixlbaumer A., Leon K., Landick R., Darst S.A.. Structural basis of transcriptional pausing in bacteria. Cell. 2013; 152:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steitz T.A., Steitz J.A.. A general 2-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA. 1993; 90:6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arnold J.J., Cameron C.E.. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry. 2004; 43:5126–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steitz T.A. A mechanism for all polymerases. Nature. 1998; 391:231–232. [DOI] [PubMed] [Google Scholar]
- 11. Castro C., Smidansky E., Maksimchuk K.R., Arnold J.J., Korneeva V.S., Gotte M., Konigsberg W., Cameron C.E.. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl Acad. Sci. USA. 2007; 104:4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balbo P.B., Wang E.C., Tsai M.D.. Kinetic mechanism of active site assembly and chemical catalysis of DNA polymerase beta. Biochemistry. 2011; 50:9865–9875. [DOI] [PubMed] [Google Scholar]
- 13. Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A., Götte M., Konigsberg W., Cameron C.E.. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009; 16:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y., Korolev S., Waksman G.. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998; 17:7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franklin M.C., Wang J., Steitz T.A.. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001; 105:657–667. [DOI] [PubMed] [Google Scholar]
- 16. Zamyatkin D.F., Parra F., Alonso J.M., Harki D.A., Peterson B.R., Grochulski P., Ng K.K.. Structural insights into mechanisms of catalysis and inhibition in norwalk virus polymerase. J. Biol. Chem. 2008; 283:7705–7712. [DOI] [PubMed] [Google Scholar]
- 17. Papworth C.B., Bauer J.C., Braman J., Wright D.A.. Site-directed mutagenesis in one day with >80% efficiency. Strategies. 1996; 9:3–4. [Google Scholar]
- 18. Gohara D.W., Ha C.S., Kumar S., Ghosh B., Arnold J.J., Wisniewski T.J., Cameron C.E.. Production of ‘authentic’ poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expression Purif. 1999; 17:128–138. [DOI] [PubMed] [Google Scholar]
- 19. Shi W., Ye H.Q., Deng C.L., Li R., Zhang B., Gong P.. A nucleobase-binding pocket in a viral RNA-dependent RNA polymerase contributes to elongation complex stability. Nucleic Acids Res. 2019; 48:1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Batey R.T., Kieft J.S.. Improved native affinity purification of RNA. RNA. 2007; 13:1384–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu J.Q., Lu G.L., Zhang B., Gong P.. Perturbation in the conserved methyltransferase–polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J. Virol. 2015; 89:249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W.. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. 1997; NY: Academic Press. [DOI] [PubMed] [Google Scholar]
- 23. Mccoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 25. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Theobald D.L., Wuttke D.S.. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006; 22:2171–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castro C., Smidansky E.D., Arnold J.J., Maksimchuk K.R., Moustafa I., Uchida A., Gotte M., Konigsberg W., Cameron C.E.. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009; 16:212–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gong P. Structural basis of viral RNA-dependent RNA polymerase nucleotide addition cycle in picornaviruses. Enzymes. 2021; 49:215–233. [DOI] [PubMed] [Google Scholar]
- 29. Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D. 3rd, Wetmore D.R., McGrath M.E., Ray A.S.et al.. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015; 347:771–775. [DOI] [PubMed] [Google Scholar]
- 30. Hobdey S.E., Kempf B.J., Steil B.P., Barton D.J., Peersen O.B.. Poliovirus polymerase residue 5 plays a critical role in elongation complex stability. J. Virol. 2010; 84:8072–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi W., Ye H.Q., Deng C.L., Li R., Zhang B., Gong P.. A nucleobase-binding pocket in a viral RNA-dependent RNA polymerase contributes to elongation complex stability. Nucleic Acids Res. 2020; 48:1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnold J.J., Cameron C.E.. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer–template substrate (sym/sub). J. Biol. Chem. 2000; 275:5329–5336. [DOI] [PubMed] [Google Scholar]
- 33. Poch O., Sauvaget I., Delarue M., Tordo N.. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989; 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Campagnola G., McDonald S., Beaucourt S., Vignuzzi M., Peersen O.B.. Structure–function relationships underlying the replication fidelity of viral RNA-dependent RNA polymerases. J. Virol. 2015; 89:275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weeks S.A., Lee C.A., Zhao Y., Smidansky E.D., August A., Arnold J.J., Cameron C.E.. A polymerase mechanism-based strategy for viral attenuation and vaccine development. J. Biol. Chem. 2012; 287:31618–31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu X., Yang X., Lee C.A., Moustafa I.M., Smidansky E.D., Lum D., Arnold J.J., Cameron C.E., Boehr D.D.. Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity. J. Biol. Chem. 2013; 288:32753–32765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi J., Perryman J.M., Yang X., Liu X., Musser D.M., Boehr A.K., Moustafa I.M., Arnold J.J., Cameron C.E., Boehr D.D.. Rational control of poliovirus RNA-dependent RNA polymerase fidelity by modulating Motif-D loop conformational dynamics. Biochemistry. 2019; 58:3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang X., Smidansky E.D., Maksimchuk K.R., Lum D., Welch J.L., Arnold J.J., Cameron C.E., Boehr D.D.. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure. 2012; 20:1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald S., Block A., Beaucourt S., Moratorio G., Vignuzzi M., Peersen O.B.. Design of a genetically stable high fidelity coxsackievirus B3 polymerase that attenuates virus growth in vivo. J. Biol. Chem. 2016; 291:13999–14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Belogurov G.A., Artsimovitch I.. The mechanisms of substrate selection, catalysis, and translocation by the elongating RNA polymerase. J. Mol. Biol. 2019; 431:3975–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mishanina T.V., Palo M.Z., Nayak D., Mooney R.A., Landick R.. Trigger loop of RNA polymerase is a positional, not acid–base, catalyst for both transcription and proofreading. Proc. Natl Acad. Sci. USA. 2017; 114:E5103–E5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang J., Palangat M., Landick R.. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat. Struct. Mol. Biol. 2010; 17:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang H., Chopra R., Verdine G.L., Harrison S.C.. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998; 282:1669–1675. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data Bank under accession number 7W9S.