Abstract
Rationale
The current molecular classification of small-cell lung cancer (SCLC) on the basis of the expression of four lineage transcription factors still leaves its major subtype SCLC-A as a heterogeneous group, necessitating more precise characterization of lineage subclasses.
Objectives
To refine the current SCLC classification with epigenomic profiles and to identify features of the redefined SCLC subtypes.
Methods
We performed unsupervised clustering of epigenomic profiles on 25 SCLC cell lines. Functional significance of NKX2-1 (NK2 homeobox 1) was evaluated by cell growth, apoptosis, and xenograft using clustered regularly interspaced short palindromic repeats–Cas9 (CRISPR-associated protein 9)–mediated deletion. NKX2-1–specific cistromic profiles were determined using chromatin immunoprecipitation followed by sequencing, and its functional transcriptional partners were determined using coimmunoprecipitation followed by mass spectrometry. Rb1flox/flox; Trp53flox/flox and Rb1flox/flox; Trp53flox/flox; Nkx2-1flox/flox mouse models were engineered to explore the function of Nkx2-1 in SCLC tumorigenesis. Epigenomic landscapes of six human SCLC specimens and 20 tumors from two mouse models were characterized.
Measurements and Main Results
We identified two epigenomic subclusters of the major SCLC-A subtype: SCLC-Aα and SCLC-Aσ. SCLC-Aα was characterized by the presence of a super-enhancer at the NKX2-1 locus, which was observed in human SCLC specimens and a murine SCLC model. We found that NKX2-1, a dual lung and neural lineage factor, is uniquely relevant in SCLC-Aα. In addition, we found that maintenance of this neural identity in SCLC-Aα is mediated by collaborative transcriptional activity with another neuronal transcriptional factor, SOX1 (SRY-box transcription factor 1).
Conclusions
We comprehensively describe additional epigenomic heterogeneity of the major SCLC-A subtype and define the SCLC-Aα subtype by the core regulatory circuitry of NKX2-1 and SOX1 super-enhancers and their functional collaborations to maintain neuronal linage state.
Keywords: small-cell lung cancer, SCLC, NKX2-1/TTF-1, SOX1, superenhancer
At a Glance Commentary
Scientific Knowledge on the Subject
Small-cell lung cancer (SCLC) is the most aggressive form of lung cancer. Although it is clinically recognized as a single disease entity, recent progress in molecular analyses led to proposed consensus classification according to expression of four lineage transcription factors: ASCL1 (achaete-scute homolog 1) (SCLC-A), NeuroD1 (neurogenic differentiation 1), YAP1 (Yes-associated protein 1), and POU2F3 (Pou domain, class 2, transcription factor 3). Nonetheless, follow-up studies have indicated that this classification has left substantial ambiguity, especially in the major SCLC-A subtype, warranting novel approaches to develop a more precise classification of SCLC.
What This Study Adds to the Field
We applied epigenomic approaches to determine lineage heterogeneity of SCLC, subdividing SCLC-A into two clusters (SCLC-Aα and SCLC-Aσ) with distinct superenhancer landscapes. We identified NKX2-1 (NK2 homeobox 1) as one of the most differentially enriched superenhancers in SCLC-Aα and showed that it plays a critical role in SCLC development and maintenance. NKX2-1 and SOX1 (SRY-box transcription factor 1) cooccupy the SCLC-Aα genome and collaborate to control its neural lineage. This study provides functional significance of SCLC lineage heterogeneity and mechanisms through which SCLCs undergo cellular state reprogramming, laying a foundation for identification of unique vulnerabilities for developing finer personalized therapeutic approaches to benefit patients with SCLC.
Small-cell lung cancer (SCLC) is an aggressive, lethal, and rapidly progressing neuroendocrine (NE) malignancy accounting for ∼15% of lung cancer incidence and 250,000 deaths worldwide annually (1). The five-year survival rate for patients with SCLC is only 6%, compared with 19% for non–small-cell lung cancer. The lack of progress in SCLC treatment is largely attributable to inadequate molecular understanding. Advances in genomic technology have enabled new molecular characterization of SCLCs. Genome-wide methylation and transcriptomes of human SCLCs and patient-derived xenograft models have identified NE and non-NE subtypes. The NE subtype is defined by the expression of the neuronal lineage transcription factors (TFs) ASCL1 (achaete-scute homolog 1) and/or NeuroD1 (neurogenic differentiation 1) (2). Two recent studies described the non-NE SCLCs, one signified by YAP1 (Yes-associated protein 1) expression and another by POU2F3 (Pou domain, class 2, TF 3) expression (3, 4). In light of these findings, a consortium proposed a new molecular classification of SCLCs that distinguishes them into four subtypes on the basis of the expression of the four lineage-defining TFs ASCL1, NeuroD1, YAP1, and POU2F3, referred to as SCLC-A, SCLC-N, SCLC-Y, and SCLC-P, respectively (5, 6). The classification, although marking significant advances, is still a work in progress. A few new classifications have emerged on the basis of human SCLC samples or SCLC cell line RNA sequencing (RNA-seq) data: one group proposed an inflamed SCLC subtype (SCLC-I) (7), and another group identified a new subtype (NEv2) (8). In addition, not all SCLCs clearly fall into the four clusters, as they often express multiple factors at variable concentrations, especially in the SCLC-A subtype, which accounts for 70% of SCLCs (9). This wide range of heterogeneity warrants novel approaches to develop a more precise classification of SCLC on the basis of lineage subtype.
Lineage-specific TFs and the presence of super-enhancers (SEs) at their gene loci are critical determinants of cellular identity (10). In our study, we applied epigenomic approaches to investigate the differential enhancer landscapes in SCLC cell lines, human SCLC samples, and autochthonous murine SCLCs (mSCLCs) from a genetically engineered mouse model (GEMM) to reveal additional intertumoral heterogeneity, providing comprehensive evidence of distinct histone modification profiles. Furthermore, we determined a novel mechanism of subtype-specific transcriptional regulation in SCLC, integrating the core transcriptional networks and their collaborative regulation of NE target genes.
Some of the results of the study has been previously reported in the form of an abstract (11).
Methods
See the online supplement for additional details.
Cell Lines
Cell lines used in this study are listed in Table E1 in the online supplement.
Chromatin Immunoprecipitation Followed by Sequencing
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) was performed as described previously (12). Protein G magnetic beads (Dynabeads; Life Technologies) were preincubated with anti-H3K27ac (ab4729; Abcam), anti–NKX2-1 (NK2 homeobox 1) (A300-BL4000; Bethyl Laboratories), or anti–SOX1 (SRY-box transcription factor 1) (AF3369; R&D Systems) antibodies. Up to 10 ng DNA was used for the library construction using NEBNext Ultra II DNA Library Prep Kit (E7645; New England BioLabs). Sequencing was performed on NextSeq 500 (Illumina).
RNA-Seq
The procedure was performed as previously described (9, 12).
Lentiviral Transduction of Genes and Clustered Regularly Interspaced Short Palindromic Repeats–Cas9 (CRISPR-Associated Protein 9) Genome Editing
FLAG-NKX2-1 or FLAG-GFP (green fluorescent protein) open reading frame was cloned into pLEX_306 (a gift from David Root, Addgene plasmid #41391). Cells stably expressing Cas9 were generated by infection with the lentiCas9-Blast plasmid (Addgene plasmid #52962, a gift from Feng Zhang). Single-guide RNAs (sgRNAs) targeting NKX2-1 or SOX1 were selected from the Brunello library (see Table E2) (13). Additional details are provided in the online supplement.
Immunohistochemistry
Immunohistochemical analyses were performed on xenograft tumor specimens, human primary SCLC tumor specimens, and GEMM tumor specimens with anti–NKX2-1 (1:100) (sc-53136; Santa Cruz), anti-SOX1 (AF3369; R&D Systems), or anti-ASCL1 (1:100) (556604; BD Biosciences) antibody and detected with the Vectastain ABC kit (Vector Laboratories).
Coimmunoprecipitation and Liquid Chromatography–Tandem Mass Spectrometry
Chromatin fraction was prepared and coimmunoprecipitated using methods described previously (14) with modifications. Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide using the LTQ Orbitrap Velos Pro ion-trap mass spectrometer (Thermo Fisher Scientific).
Cell Proliferation Assay
Details are provided in the online supplement.
Xenograft Model
The procedure was performed as previously described (12). All xenograft studies were approved by the Institutional Animal Care and Use Committee at Icahn School of Medicine at Mount Sinai (IACUC-2018-0021).
Annexin V Staining
Details are provided in the online supplement.
In Situ Proximity Ligation Assay
In situ proximity ligation assay was performed using DuoLink in situ reagents (Sigma-Aldrich) according to the manufacturer’s instructions. Images were acquired using a Zeiss Axiocam 503 monoconfocal microscope.
Mice and Tumor Initiation
Mice harboring Rb1flox, Trp53flox, and Nkx2-1flox alleles have been previously described (15, 16). Tumors were generated by intratracheal delivery of Ad5CMV-Cre adenovirus (University of Iowa, Gene Transfer Vector Core) as previously described (17). All studies were approved by the Committee for Animal Care at the Massachusetts Institute of Technology (A-3125-01).
Data Analysis
Details are provided in the online supplement.
Statistical Analysis
Details of specific analyses are provided in the figure legends.
Data and Materials Availability
SCLC cell lines, human primary SCLC tumors, Rb1flox/flox; Trp53flox/flox (RP) and Rb1flox/flox; Trp53flox/flox; Nkx2-1flox/flox (RPN) tumors H3K27ac ChIP-seq; cell lines and RP and RPN tumors RNA-seq; NKX2-1 and SOX1 ChIP-seq are deposited with the National Center for Biotechnology Information (Gene Expression Omnibus: GSE183373).
Results
Epigenomic Profiling Subdivided SCLC-A by Differential SEs
Genome-scale chromatin organization of cells more faithfully reflects their lineage state than transcriptomics (18). To refine the current SCLC classification, we profiled genome-wide H3K27ac to define SE regions across 16 SCLC cell lines. Unsupervised hierarchical clustering on SEs on genes for transcriptional regulation with additional public data (4) revealed four subclasses (Figure 1A; see Table E3), supported by principal-component analysis (PCA) (see Figure E1A). Our clusters recapitulated aspects of the expression classification. Cluster III corresponds to SCLC-N (Figure 1A, purple). Cluster IV represents SCLC-P and SCLC-Y (Figure 1A, blue). Notably, epigenomic clustering distinguished SCLC-A into two distinct clusters, I and II (Figure 1A, red and green), referred to as SCLC-Aα and SCLC-Aσ, while ASCL1 SE is present in 92.9% (13 of 14) of SCLC-A (see Figure E1B). The H3K27ac-based SE regions are further validated by H3K4me1 sequencing data and assay for transposase-accessible chromatin with sequencing, as a proxy for overall TF engagements on a subset of different clusters of SCLCs (see Figure E1C).
Figure 1.
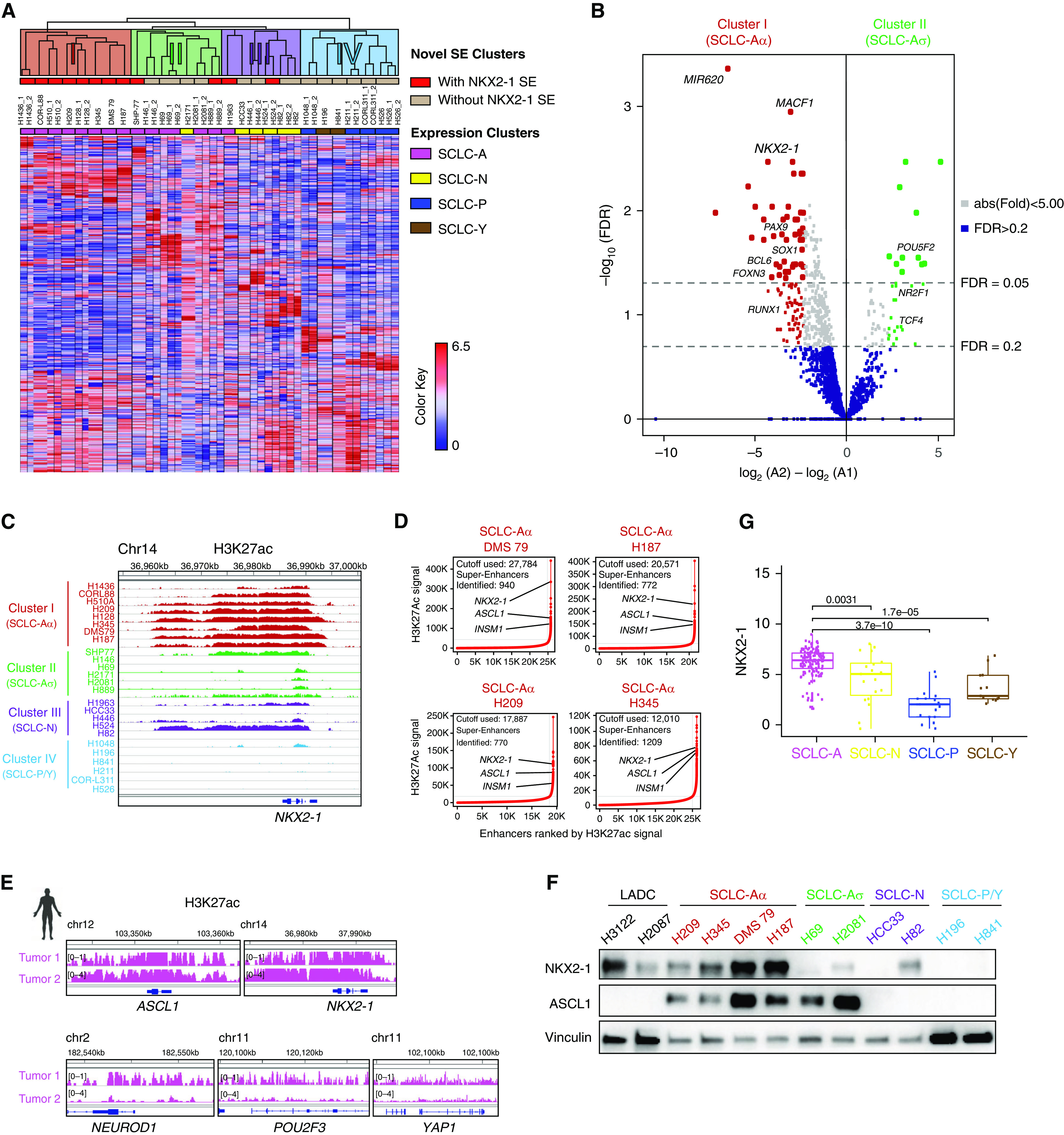
Epigenomic profiling subdivided small-cell lung cancer (SCLC) subtype SCLC-A by differential super-enhancers (SEs). (A) Unsupervised hierarchical clustering using pairwise average linkage with Pearson correlation for column and row distance measures of 25 SCLC cell lines using SE signals near transcriptional regulator genes. The color scale indicates the intensity of SE signals normalized to linear scores using the sum of squares of the values. Each row represents an SE locus (747 loci). The color shades on the dendrogram indicate four novel SE clusters, the top red columns display the presence of SE near the NKX2-1 (NK2 homeobox 1) locus, and the bottom column colors denote the consensus expression clusters. (B) Volcano plot showing 133 differentially enriched SEs (104 enriched in SCLC-Aα and 29 enriched in SCLC-Aσ) at thresholds of absolute fold change > 5 and FDR < 0.2, depicted as dotted lines. (C) Genome view tracks of H3K27ac signal at the NKX2-1 locus in four clusters of SCLC cell lines. Normalized scales are shown in Figure E1B. (D) SE plots with all enhancers rank ordered by H3K27ac signals in cell lines DMS 79, NCI-H187, NCI-H209, and NCI-H345. Dotted lines depict tangential cutoffs to define SEs. (E) Genome view tracks of H3K27ac signals at the ASCL1, NEUROD1, POU2F3, YAP1, and NKX2-1 loci in two human SCLC specimens of SCLC-A subtype. (F) Protein expression of NKX2-1, ASCL1, and vinculin as a loading control in SCLC cell lines from different clusters. NKX2-1–amplified adenocarcinoma cell lines NCI-H3122 and NCI-H2087 are shown as reference. (G) Box and jitter plots showing the distribution of NKX2-1 mRNA expression in SCLC-A, SCLC-N, SCLC-P, and SCLC-Y subtypes of 135 primary SCLC tumors (5, 29). ASCL1 = achaete-scute homolog 1; chr = chromosome; FDR = false discovery rate; INSM1 = INSM transcriptional repressor 1; LADC = lung adenocarcinoma; NEUROD1 = neurogenic differentiation 1; POU2F3 = Pou domain, class 2, transcription factor 3; YAP1 = Yes-associated protein 1.
To understand the difference underpinning SCLC-Aα and SCLC-Aσ, we identified 60 differentially enriched SEs with cutoffs set at fold change > 5 and false discovery rate < 0.05. Among 49 SEs enriched in SCLC-Aα was NKX2-1 (Figure 1B; see Table E4), where H3K27ac signal was largely absent in other subclasses (Figure 1C). To nominate possible candidates that define SCLC-Aσ, we used a more lenient cutoff at false discovery rate < 0.2. There are 29 SEs enriched in SCLC-Aσ, which included TCF4 (transcription factor 4), a TF that regulates synaptic plasticity (19) (Figure 1B; see Figure E1B). NKX2-1, also known as TTF1 (transcription termination factor 1), a lineage TF essential for the development and maintenance of lungs, brain, and thyroid, is a frequently amplified lineage-specific oncogene and a well-known marker to diagnose lung adenocarcinoma (LADC) (20–22). However, NKX2-1 is not significantly amplified in human SCLCs (23) or in the SCLC lines used (see Figure E2A). Nonetheless, SCLC-Aα cell lines harbor NKX2-1 SE, whereas SCLC-Aσ cell lines do not (Figure 1D; see Figure E2B). Local H3K27ac enrichment pattern at the NKX2-1 locus in SCLC-Aα was almost identical to LADCs with NKX2-1 amplification (see Figure E2C). In six human SCLC specimens, we found one SCLC-P tumor with POU2F3 SE and three mixed SCLC-A/SCLC-N tumors with SEs near ASCL1 and NEUROD1 loci (see Figure E2D). Notably, we observed two tumors representing SCLC-Aα with high signals at ASCL1 and NKX2-1 loci (Figure 1E).
By immunohistochemistry on another cohort of 34 human SCLCs, 65% (22 of 34) of cases were NKX2-1 positive (see Figure E2E), consistent with prior studies (24–28). In cell lines, SCLC-Aα expressed substantially higher concentrations of NKX2-1 protein than other SCLC subtypes, equivalent to NKX2-1–amplified LADCs (Figure 1F; see Figure E2F). Furthermore, RNA-seq data sets (5, 29) confirmed that NKX2-1 is highly expressed in SCLC (see Figure E2G), comparable to the concentrations in LADC (see Figure E2H), particularly in SCLC-A (Figure 1G), consistent with a previous report (24). Nonetheless, a nonbimodal distribution of NKX2-1 expression suggests that the distinction between SCLC-Aα and Aσ is not discernible from expression analysis, perhaps analogous to the findings that NKX2-1 is almost universally expressed in LADC yet amplified in only 10–15%, which we previously characterized as a distinct biological subgroup (20, 30).
NKX2-1 Survival Dependency Is Unique to SCLC-Aα
A recent study suggested that NKX2-1 promotes SCLC growth and regulates NE differentiation and antiapoptosis (31). We deleted NKX2-1 in SCLC-Aα cell lines (Figure 2A) and found that it significantly suppressed cell growth and increased annexin V–positive populations (Figure 2C; see Figure E3A). In contrast, ablating NKX2-1 in SCLC cells from other clusters (Figure 2B) had little effect. Given that acute removal of a master lineage factor in established cancer often leads to a halt in cell growth (32), these data suggest that NKX2-1 does not play a general oncogenic role in all lineage states but that its function is uniquely relevant to SCLC-Aα.
Figure 2.
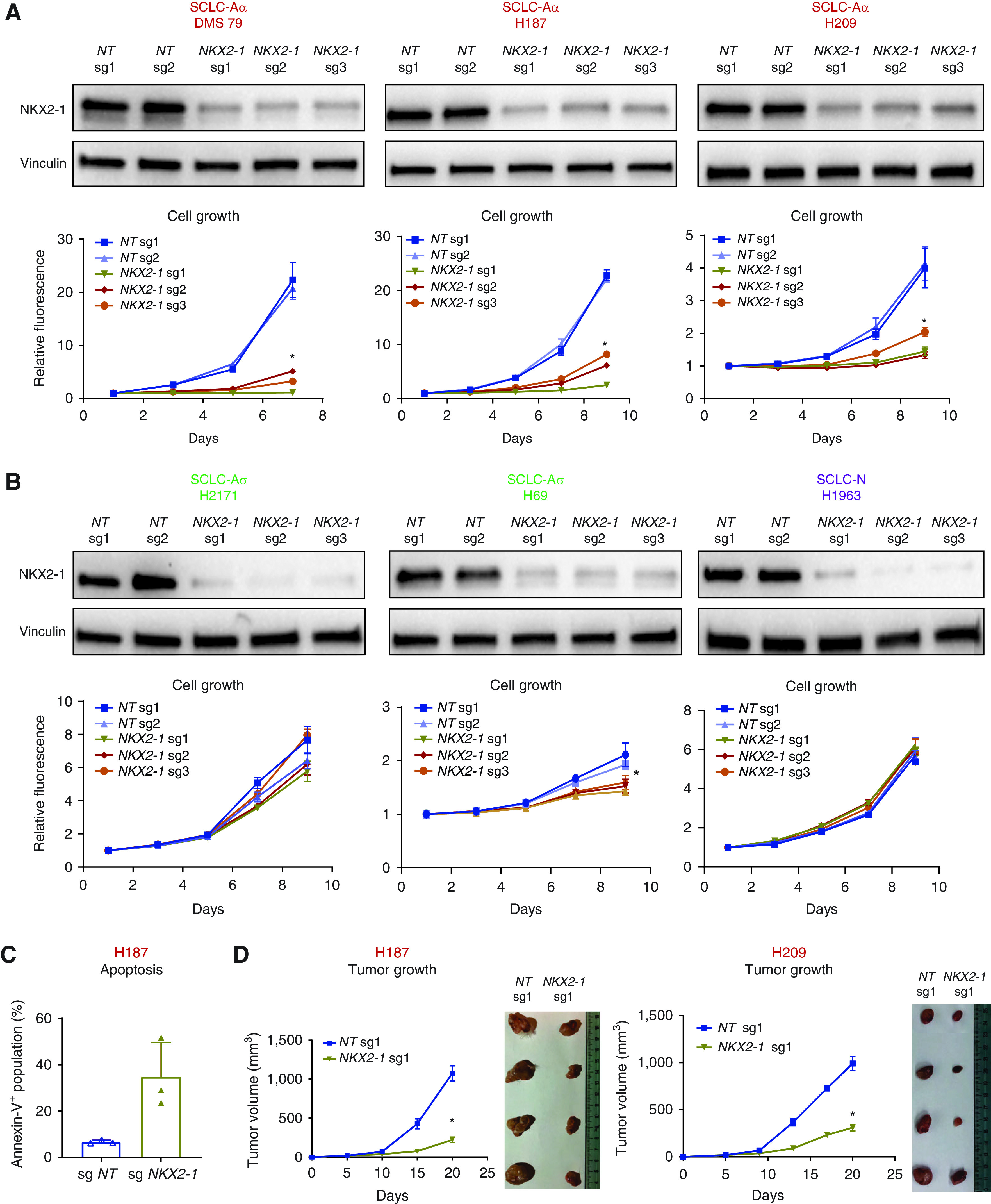
NKX2-1 (NK2 homeobox 1) survival dependency is unique to small-cell lung cancer (SCLC) subtype SCLC-Aα. (A) Top: Immunoblots showing protein expression of NKX2-1 and vinculin as a loading control in SCLC-Aα cell lines with three NKX2-1 sgRNAs or two nontarget (NT) sgRNAs. Bottom: Cell growth curve for SCLC-Aα cell lines infected with NKX2-1 sgRNAs or NT sgRNAs. Data are shown as mean ± SD (n = 3); *P < 0.001 by Kruskal-Wallis test. (B) Top: Immunoblots showing protein expression of NKX2-1 and vinculin as a loading control in SCLC-Aσ (NCI-H2171 and NCI-H69) and SCLC-N (NCI-H1963) cell lines with NKX2-1 sgRNAs or NT sgRNAs. Bottom: Cell growth curve for cell lines infected with NKX2-1 sgRNAs or NT sgRNAs. Data are shown as mean ± SD (n = 3); *P < 0.05 by Kruskal-Wallis test. (C) Bar plots showing percentage of apoptotic NCI-H187 cells infected with NT sgRNA and NKX2-1 sgRNA, measured by annexin V staining. Mean ± SD of three biological replicates is shown. Representative data for NT sgRNA 1 (sg1) and NKX2-1 sg1 can be found in Figure E3A. (D) Left: Tumor growth curve for xenograft tumors of NCI-H187 cells infected with NKX2-1 sg1 or NT sg1 in vivo. Mean ± SD of four biological replicates is shown. Xenograft tumors were resected at Day 20 after inoculation. Right: Images of resected xenograft tumors of NCI-H209 cells infected with NT sg1 or NKX2-1 sg1 in vivo at Day 20. Mean ± SD of four biological replicates is shown. *P < 0.05, Mann-Whitney test. sgRNA = single-guide RNA.
We further found a significant decrease in tumor volume for NKX2-1–deleted SCLC-Aα cells in xenograft (Figure 2D; see Figure E3B), confirming its essentiality in vivo. No obvious histological differences were observed in the cells that formed small tumors that escaped from NKX2-1 deletion (see Figure E3C). We have xenotransplanted SCLC-Aσ and SCLC-N cells with NKX2-1 deletion and observed no discernible difference in tumor growth, consistent with in vitro cell growth data (see Figure E3D). In a short period after NKX2-1 deletion in vitro, expression of NE markers was not significantly altered (see Figure E3E). Overall, these data suggest that NKX2-1 is functionally critical for SCLC-Aα in vitro and in vivo.
Genomic Occupancy of NKX2-1 in SCLC-Aα Cells Distinct from That in LADC Cells
In LADC, NKX2-1 regulates gene expression and shares its cistromes with normal lung (20); however, SCLCs typically do not express canonical NKX2-1 targets. Given NKX2-1’s roles in subsets of cortical, striatal, and pallidal neurons (33), we speculated a potential role for neuronal differentiation in SCLC-Aα. Therefore, we profiled NKX2-1 cistromes using ChIP-seq in five SCLC-Aα cell lines and compared them with that from three LADC cell lines. Pairwise correlation analysis of all NKX2-1–bound sites and PCA revealed histology-specific binding profiles (Figure 3A; see Figure E4A). Regions 4 (low-intensity signals) and 1 (high-intensity signals) are common between SCLC-Aα and LADC (Figure 3B), which included the RUNX1 locus (Figure 3C). Region 3 was unique to LADC, which included LMO3 (LIM domain only 3), an essential mediator of NKX2-1 functions in LADCs (20). Region 2 was unique to SCLC-Aα that included DCC, a receptor for netrin required for axon guidance (34) (Figure 3C). These data suggest NKX2-1 regulates distinct transcriptional programs across these two lineage states.
Figure 3.
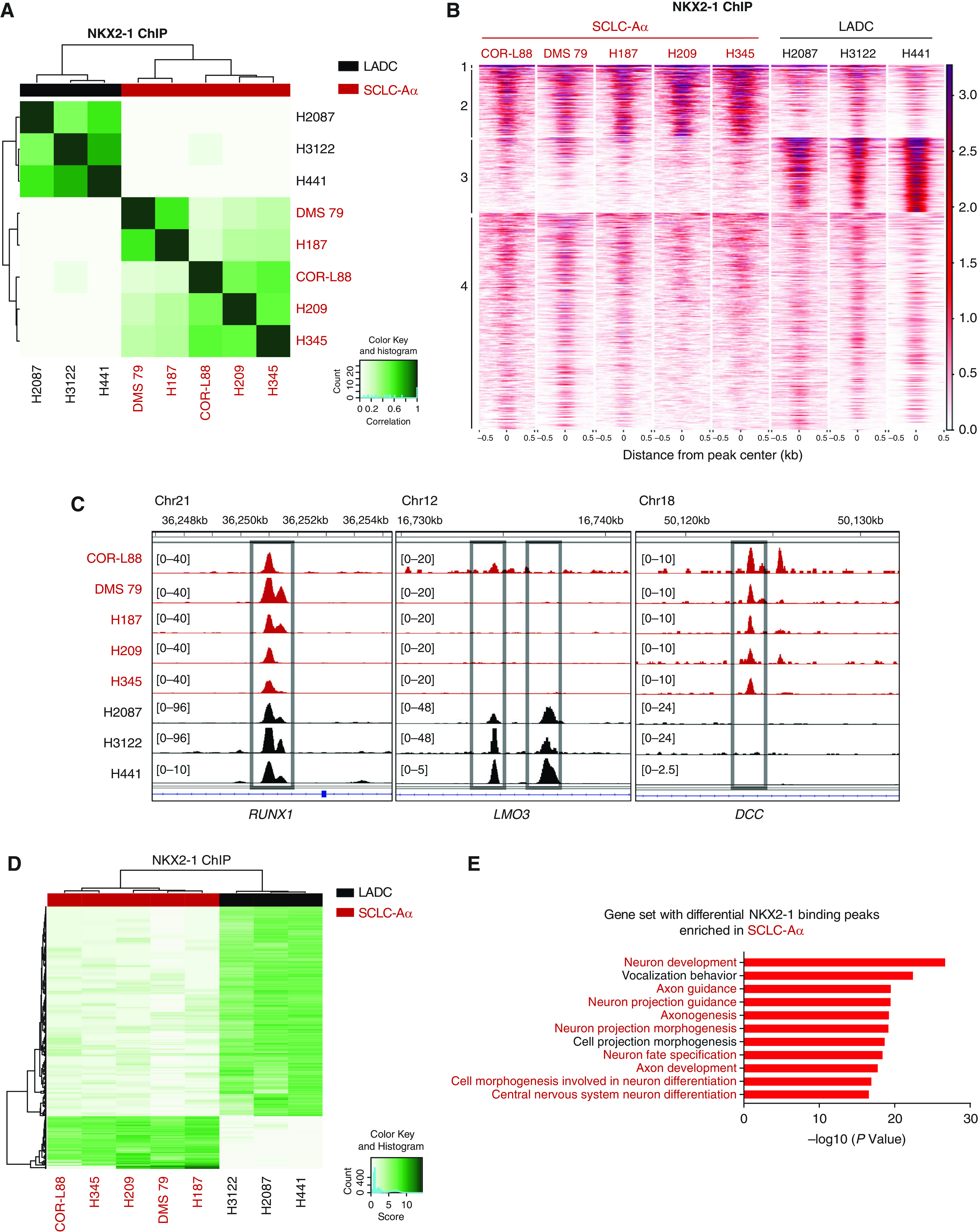
Genomic occupancy of NKX2-1 (NK2 homeobox 1) in small-cell lung cancer (SCLC) subtype SCLC-Aα cells distinct from that in lung adenocarcinoma (LADC) cells. (A) Correlation matrix depicting pairwise comparisons of identified NKX2-1 binding peaks in the five SCLC-Aα cell lines and in three LADC cell lines. Color scale represents degree of Pearson’s correlation coefficient. (B) Heatmap showing signals from NKX2-1 chromatin immunoprecipitation followed by sequencing (ChIP-seq) data in the five SCLC-Aα cell lines and three LADC cell lines. ChIP signal intensity is shown by red shading. The y-axis contains all peaks that are bound by NKX2-1 in any of SCLC-Aα and LADC (n = 37,198). Peaks are divided into four regions by k-means (k = 4) clustering. (C) NKX2-1 ChIP-seq signals in SCLC-Aα and LADC cell lines at representative loci (RUNX1, cluster 1; LMO3, cluster 3; and DCC, cluster 2). Red, SCLC-Aα cell lines; black, LADC cell lines. (D) Heatmap showing 2,671 differentially NKX2-1–bound regions (fold change ⩾ 25 and false discovery rate ⩽ 1 × 10−5), 2,010 enriched in LADC and 661 enriched in SCLC-Aα, between five SCLC-Aα cell lines and three LADC cell lines. (E) Enriched ontology by Genomic Regions Enrichment of Annotations Tool analyses for regions differentially bound by NKX2-1 in five SCLC-Aα compared with LADCs. Chr = chromosome; DCC = DCC netrin 1 receptor; LMO3 = LIM domain only 3; RUNX1 = RUNX family transcription factor 1.
Ontologies on 661 SCLC-Aα–unique regions (Figure 3D; see Figure E4B) were enriched for neuronal development, differentiation, and structure (Figure 3E). In contrast, genes regulating neuronal biology were absent in LADC-unique regions (see Figure E4C). Motif analysis revealed that SCLC-Aα–specific sites are enriched with ASCL2, Olig2 (oligodendrocyte transcription factor 2), and SOX motifs, suggesting more association with neuronal TFs. In contrast, AP-1, Ets, Runx, and Tead-binding motifs are enriched in LADC-specific regions, consistent with our previous report (20) (see Figure E4D and Table E5). These data indicate a contribution of NKX2-1 in regulating pathways for maintenance of the neural differentiation state of SCLC-Aα, distinct from its role in normal lung epithelium and LADCs.
NKX2-1 Interactome in Chromatin of SCLC-Aα Cells Includes SOX1
NKX2-1 interacts with FOXA1 (forkhead box A1) to coregulate the expression of a set of genes for lineage control in LADC (20, 21). Therefore, to identify unique partners that interact with NKX2-1 to exert distinct transcriptional program in SCLC-Aα, we performed coimmunoprecipitation followed by liquid chromatography–tandem mass spectrometry in the chromatin fraction of two SCLC-Aα cell lines ectopically expressing FLAG-NKX2-1 (Figure 4A; see Figure E5A). We found 27 overlapping NKX2-1–interacting TFs in both cell lines (Figure 4B).
Figure 4.
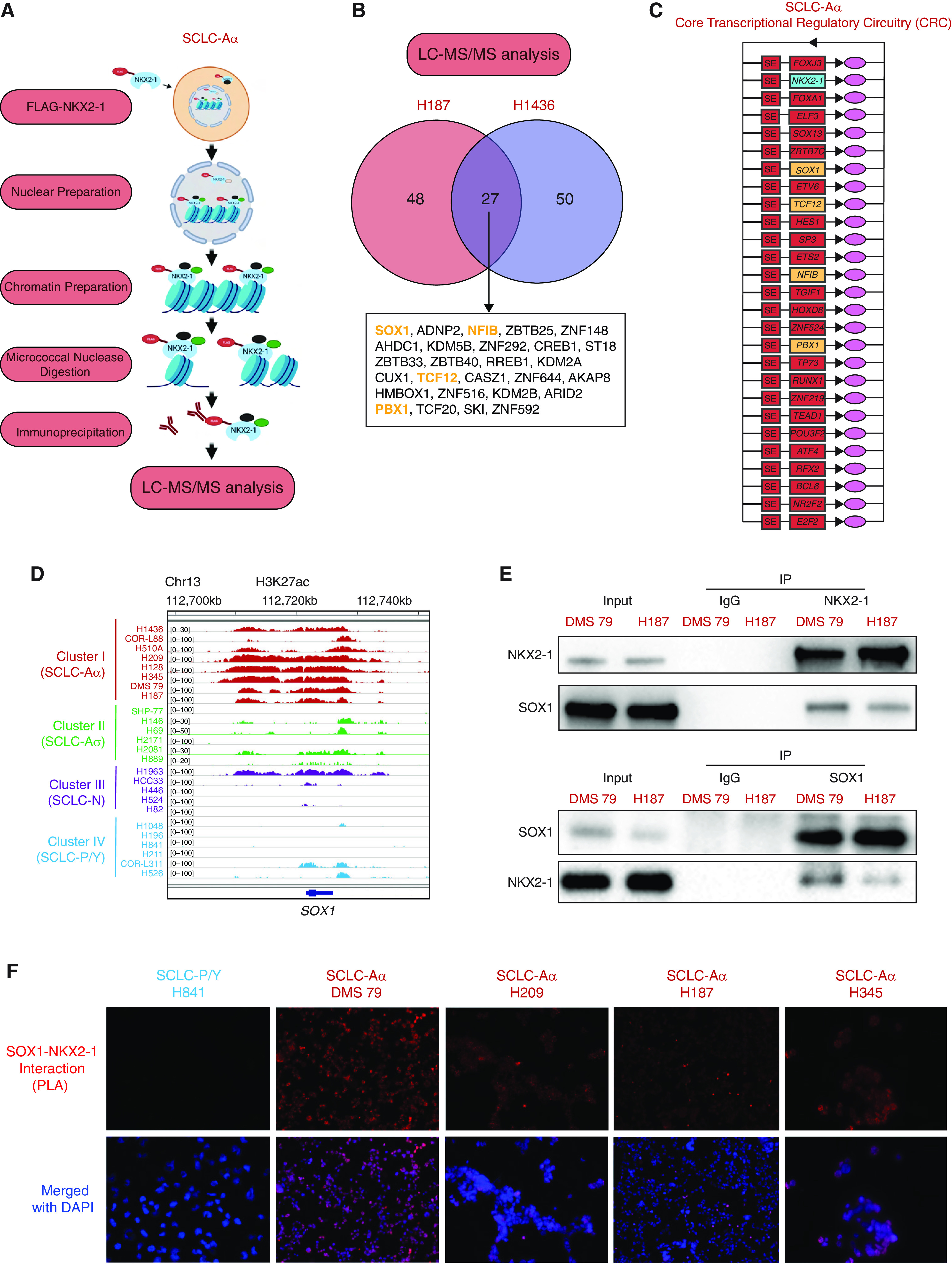
NKX2-1 interactome in chromatin of small-cell lung cancer (SCLC) subtype SCLC-Aα cells includes SOX1 (SRY-box transcription factor 1). (A) Experimental strategy to identify binding partners of NKX2-1 (NK2 homeobox 1) in SCLC-Aα. Chromatin prepared from the nuclear fraction of SCLC-Aα cell lines NCI-H187 and NCI-H1436 stably expressing FLAG-NKX2-1 was solubilized using micrococcal nuclease digestion. Coimmunoprecipitated proteins with anti-FLAG antibody from solubilized chromatin were identified using liquid chromatography–tandem mass spectrometry (LC-MS/MS). (B) Venn diagram showing the number of transcription factors identified in each of the two SCLC-Aα cell lines and the 27 overlaps found in both cell lines. The proteins indicated in yellow were also found in core transcriptional regulatory circuitry (CRC) analysis in Figure 4C. (C) CRCs commonly identified in eight SCLC-Aα cell lines. The genes in yellow were also found in LC-MS/MS analysis in Figure 4B. (D) Genome view tracks of H3K27ac signal at the SOX1 locus in four clusters of SCLC cell lines. (E) Top: Immunoblots with anti-SOX1 antibody on endogenous coimmunoprecipitates by anti–NKX2-1 antibody. Bottom: Immunoblots with anti–NKX2-1 antibody on endogenous coimmunoprecipitates by anti-SOX1 antibody as the reciprocal experiment in SCLC-Aα (DMS 79 and NCI-H187) cells. (F) In situ proximity ligation assay signals (red fluorescence) targeting endogenous SOX1 and NKX2-1 in NCI-H841, DMS 79, NCI-H209, NCI-H187, and NCI-H345 cells. Nuclei are counterstained with DAPI (blue). See Figure E5G for three SCLC-Aσ cell lines and two additional controls. ADNP2 = ADNP homeobox 2; AHDC1 = AT-hook DNA binding motif containing 1; AKAP8 = A-kinase anchoring protein 8; ARID2 = AT-rich interaction domain 2; ATF4 = activating transcription factor 4; BCL6 = BCL6 transcription repressor; CASZ1 = castor zinc finger 1; CREB1 = CAMP responsive element binding protein 1; CUX1 = cut like homeobox 1; E2F2 = E2F transcription factor 2; ELF3 = E74 like ETS transcription factor 3; ETS2 = ETS proto-oncogene 2, transcription factor; ETV6 = ETS variant transcription factor 6; FOX = forkhead box; HES1 = Hes family BHLH transcription factor 1; HMBOX1 = homeobox containing 1; HOXD8 = homeobox D8; IP = immunoprecipitation; KDM = lysine demethylase; NFIB = nuclear factor I B; NR2F2 = nuclear receptor subfamily 2 group F member 2; PBX1 = PBX homeobox 1; PLA = proximity ligation assay; POU2F3 = Pou domain, class 2, transcription factor 3; RFX2 = regulatory factor X2; RREB1 = Ras responsive element binding protein 1; RUNX1 = RUNX family transcription factor 1; SKI = SKI proto-oncogene; SP3 = Sp3 transcription factor; ST18 = ST18 C2H2C-type zinc finger transcription factor; TCF = transcription factor; TEAD11 = TEA domain transcription factor 1; TGIF1 = TGFB induced factor homeobox 1; TP73 = tumor protein P73; ZBTB = zinc finger and BTB domain containing; ZNF = zinc finger protein.
We further identified core regulatory circuitries (CRCs) (35) for SCLC-Aα and other SCLC clusters from our H3K27ac data. Common CRCs included NEUROD1 for SCLC-N, POU2F3 and ASCL2 for SCLC-P/SCLC-Y, and TCF4 in SCLC-Aσ, consistent with our differential SE analysis (Figure 1B; see Figure E5B). Common CRCs in SCLC-Aα included NKX2-1 (Figure 4C) together with SOX1, NFIB (nuclear factor I B), TCF12, and PBX1 (PBX homeobox 1), also found in the SCLC-Aα NKX2-1 interactome. Of those, H3K27ac signal at SOX1 was highly enriched in SCLC-Aα (Figures 1B and 4D), whereas NFIB, TCF12, and PBX1 SEs were present across SCLC subtypes (see Figure E5C). SOX1 is highly expressed in SCLC-A (see Figure E5D) and positively correlated with NKX2-1 mRNA expression in 135 primary SCLC tumors and 50 SCLC cell lines (5, 29) (see Figure E5E). SOX1 protein was also highly expressed specifically in SCLC-Aα cell lines (see Figure E5F).
The interaction between NKX2-1 and SOX1 was confirmed by coimmunoprecipitation, followed by Western blot in two SCLC-Aα cell lines (Figure 4E) and in addition supported by proximity ligation assay in four SCLC-Aα cell lines, three SCLC-Aσ cell lines, one SCLC-P/SCLC-Y cell line as a biological negative control, and one SCLC-Aα cell line without primary antibodies as technical negative control (Figure 4F; see Figure E5G). These data suggest robust protein–protein interaction between NKX2-1 and SOX1 uniquely in SCLC-Aα.
NKX2-1 and SOX1 Cooccupy the SCLC-Aα Genome to Collaborate in Neuronal Gene Regulation
Next, we profiled genome-wide binding profiles of NKX2-1 and SOX1 in four SCLC-Aα cell lines and found that the majority of SOX1 peaks were bound by NKX2-1 (48.6–67.8%) (Figure 5A). The 2,652 regions cobound by NKX2-1 and SOX1 in NCI-H187, 1,109 regions in DMS 79, 1,381 regions in NCI-H209, and 3,725 regions in NCI-H345 were consistently enriched for functions in neuron differentiation and development, consistent with the aggregate analysis (see Figures E6A and E6B). We defined high-confidence NKX2-1 (5,663) and SOX1 (1,101 binding sites observed in three or more cell lines) and found an overlap of 604 peaks (54.9% of SOX1 peaks) (Figure 5B), which included the NKX2-1 locus as well as neuronal genes NRXN3 (neurexin 3) (36, 37) and YWHAZ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta) (38) (Figure 5C) and ontologically enriched for functions in neuron differentiation and development (Figure 5D). These data suggest that the interaction between NKX2-1 and SOX1 elicits specific cellular identity of SCLC-Aα.
Figure 5.
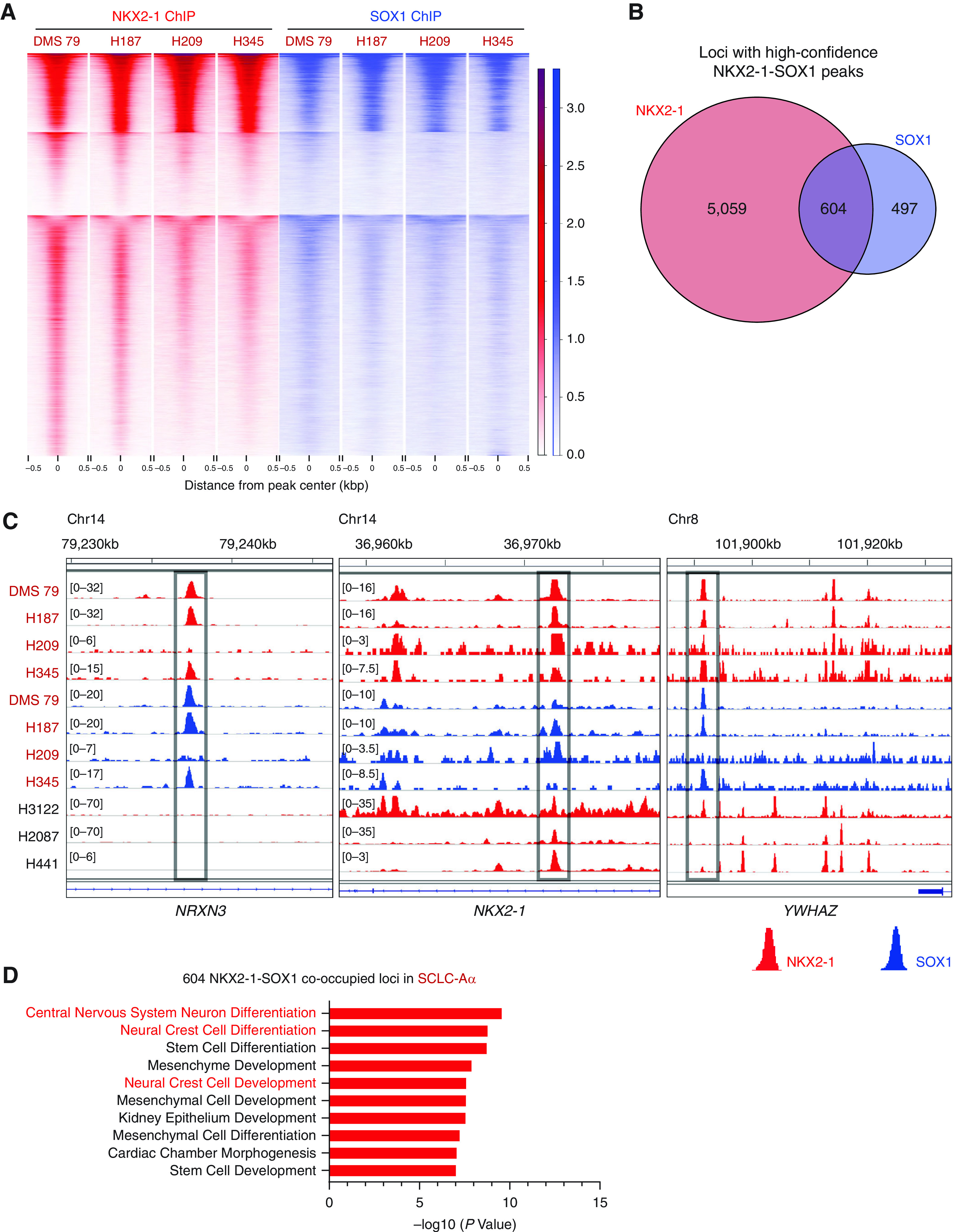
NKX2-1 (NK2 homeobox 1) and SOX1 (SRY-box transcription factor 1) cooccupy the small-cell lung cancer (SCLC) subtype SCLC-Aα genome to collaborate in gene regulation. (A) Heatmap showing k-means clustering (k = 4) of signals at the common 4,051 loci from NKX2-1 (red) and SOX1 (blue) chromatin immunoprecipitation followed by sequencing (ChIP-seq) data in the four SCLC-Aα cell lines. ChIP signal intensity is shown by shading for all NKX2-1–occupied and SOX1-occupied loci. (B) Venn diagram showing the overlap of NKX2-1 common binding sites and SOX1 common binding sites present in at least three SCLC-Aα cell lines. (C) Genome view tracks of NKX2-1 (red) and SOX1 (blue) ChIP-seq signals in four SCLC-Aα cell lines (DMS 79, NCI-H187, NCI-H209, and NCI-H345) and three LADC cell lines at the representative NKX2-1–SOX1 cooccupied loci (NRXN3 and YWHAZ, SCLC-Aα unique; NKX2-1, SCLC-Aα/LADC common). (D) Enriched ontology by Genomic Regions Enrichment of Annotations Tool analyses for regions in 604 overlapping high-confidence NKX2-1–SOX1 binding sites. Chr = chromosome; LADC = lung adenocarcinoma; NRXN3 = neurexin 3; YWHAX = tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta.
Collaboration of NKX2-1 and SOX1 Is Required to Control the Neuronal State in SCLC-Aα
We evaluated the transcriptional effect of depletion of SOX1 or NKX2-1 in NCI-H187 using RNA-seq. After the abrogation of NKX2-1 or SOX1, 1,121 and 2,499 genes were significantly downregulated, respectively, with 873 genes commonly downregulated, suggesting a substantial overlap in their transcriptional outputs (Figure 6A). We then queried the differentially codownregulated genes with the 2,652 genes with transcriptional start sites within 50 kb of SOX1–NKX2-1 cooccupied peaks in NCI-H187, which were significantly enriched for NKX2-1 or SOX1 downregulated genes by single-sample gene set enrichment analysis (Figure 6B). Leading-edge analysis of these genes revealed 289 overlaps, which we found are enriched for neuron differentiation and development (Figure 6C). Together, these data suggest that NKX2-1 and SOX1 are specifically colocalized on the SCLC-Aα genome and collaboratively contribute to controlling neuronal differentiation state. We also found that SOX1 deletion, although not affecting NKX2-1 expression, led to reduced cell growth (see Figure E7A) with increased apoptosis (see Figure E7B).
Figure 6.
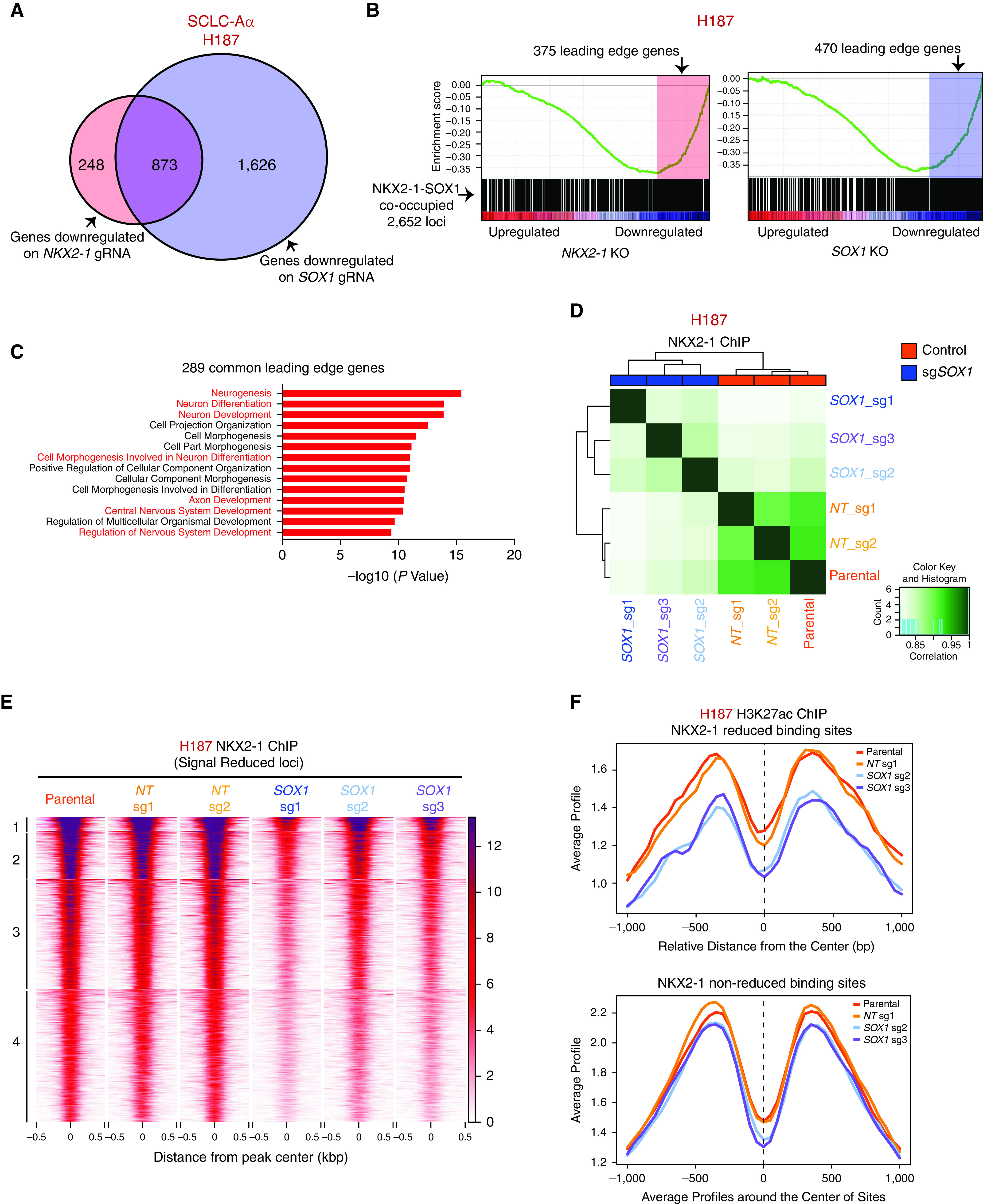
Collaboration of NKX2-1 (NK2 homeobox 1) and SOX1 (SRY-box transcription factor 1) is required to control the neuronal state in small-cell lung cancer (SCLC) subtype SCLC-Aα. (A) Venn diagram showing the overlap of downregulated genes after depletion of NKX2-1 (fold change > 1.4 and adjusted P < 0.05) and SOX1 (fold change > 2 and adjusted P < 0.05) in NCI-H187 cells. (B) Gene set enrichment analysis of genes associated with 2,652 NKX2-1–SOX1 cooccupied loci for expression data upon repression of NKX2-1 (left) or SOX1 (right) in NCI-H187 cells, with their leading-edge genes shaded. Enrichment was significant for both (NES = −2.29, P < 0.001 for NKX2-1 and NES = −2.54, P < 0.001 for SOX1). (C) Enriched gene ontologies (Molecular Signatures Database biological process) for 289 common genes in leading-edge genes for both NKX2-1 and SOX1 deletions. (D) Correlation matrix depicting pairwise comparisons of identified NKX2-1 binding regions in three SOX1 sgRNAs and three control cells (two nontarget [NT] sgRNAs, one parental cell) of NCI-H187 cell line. Color scale represents degree of Pearson’s correlation coefficient. (E) Heatmap showing k-means clustering (k = 4) of 2,067 NKX2-1 ChIP-seq loci in NCI-H187 sgSOX1 compared with control cells with reduced signal intensity. ChIP-seq signal intensity is shown by color shading. (F) Average H3K27ac ChIP-seq signals on NKX2-1 reduced binding sites and NKX2-1 nonreduced binding sites defined in Figure E7D for NCI-H187 parental, NT sgRNA 1 (sg1), SOX1 sg2, and SOX1 sg3 cells. gRNA = guide RNA; KO = knockout; NES = normalized enrichment score; sgRNA = single-guide RNA; sgSOX1 = single-guide SOX1.
We previously found that expression of Nkx2-1 was critical for global Foxa1/2 binding pattern in the LADC genome (21). To test if SOX1 is required for NKX2-1 binding to its target genes in SCLC-Aα, we performed NKX2-1 ChIP-seq in NCI-H187 cells upon SOX1 depletion. Pairwise correlation analysis of NKX2-1 peaks across cells deleted for SOX1 or control revealed a strong correlation among each group (Figure 6D), supported by PCA (see Figure E7C). Differential analysis on NKX2-1–bound sites found 2,067 peaks decreased upon SOX1 deletion (see Figure E7D). k-Means clustering of the differential sites revealed relatively uniform reduction of NKX2-1 binding after SOX1 deletion (Figure 6E), and we also found decreased H3K27ac modification compared with other sites (Figure 6F), suggesting that these NKX2-1–lost sites are no longer in active state, similar to a recent finding on Sox2 in an esophageal squamous cell cancer model for Klf5 (Kruppel like factor 1) (39). The data suggest that loss of SOX1 releases NKX2-1 from cooccupied loci in SCLC-Aα.
Nkx2-1 Is Essential for Development of SCLC-Aα Tumors in an Autochthonous Mouse Model
RP is a commonly used SCLC model that recapitulates the biology of ASCL1-positive/classic human SCLCs (15) (Figure 7A). We first profiled H3K27ac to identify SEs in mSCLC tumors from the RP model, in which Nkx2-1 is diffusely expressed. We found that all 10 tumors from two different mice harbored high H3K27ac signal around the Ascl1 locus, but none harbored SEs at the Neurod1, Yap1, or Pou2f3 locus, suggesting the SCLC-A subtype (Figure 7B; see Figure E8A), consistent with the requirement of Ascl1 for tumor formation in this model (40, 41). We found that 3 of 10 tumors simultaneously had high H3K27ac signals at the Nkx2-1 and Sox1 loci, reminiscent of SCLC-Aα, while the other 7 tumors had neither. This suggests that RP can adopt an epigenetic state resembling either SCLC-Aα or SCLC-Aσ. Consistent but ambiguous expression amounts by immunohistochemistry suggested that epigenetic profiles distinguish these lineage classes more clearly (Figure 7C).
Figure 7.
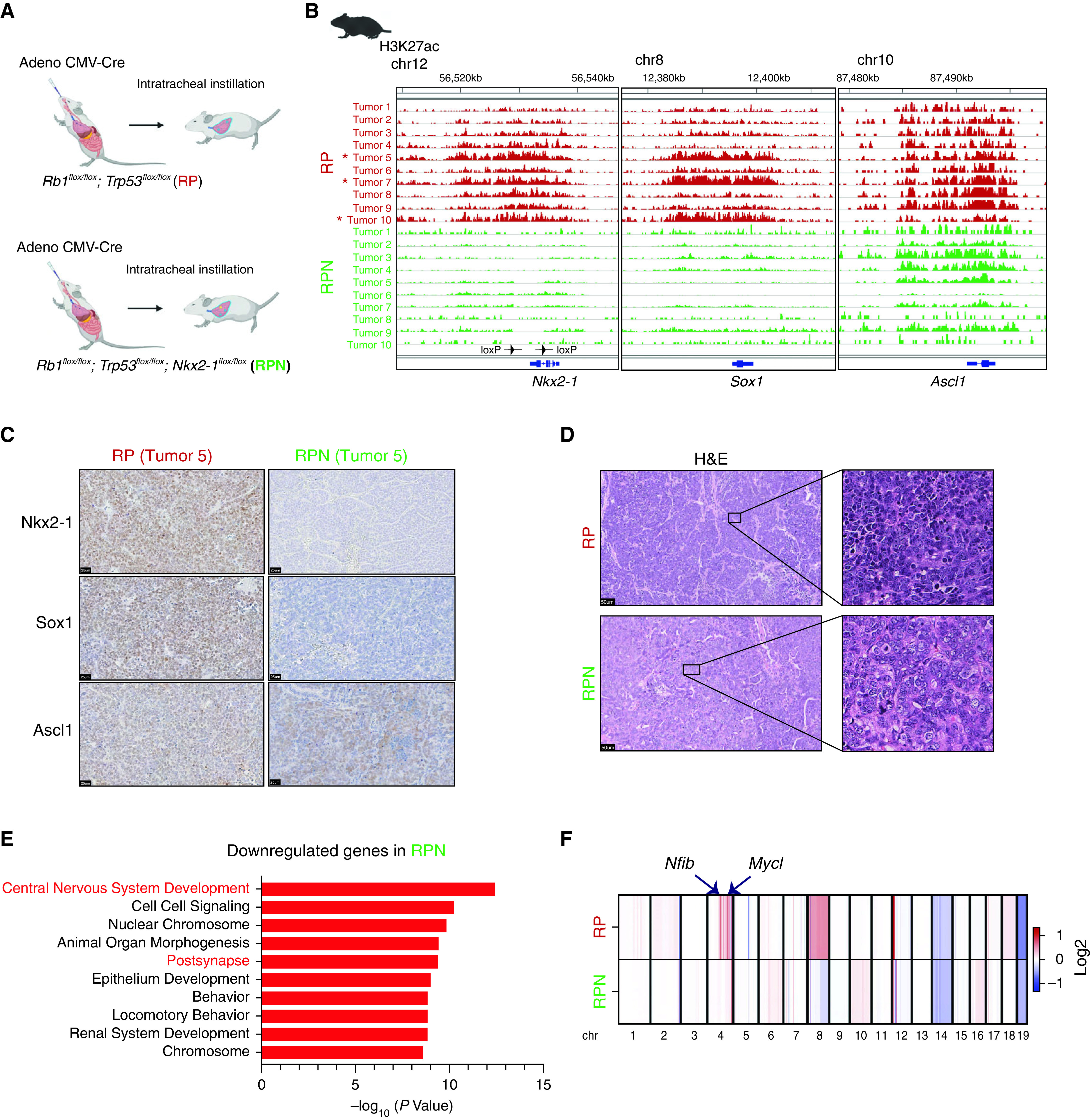
Nkx2-1 (NK2 homeobox 1) is essential for development of small-cell lung cancer (SCLC) subtype SCLC-Aα tumors in an autochthonous mouse model. (A) Genetically engineered mouse models of SCLCs. Rb1flox/flox; Trp53flox/flox (RP) mice or Rb1flox/flox; Trp53flox/flox, Nkx2-1flox/flox (RPN) mice were transduced with Adeno-CMV-Cre to initiate SCLC. (B) Genome view tracks at the Nkx2-1, Sox1 (SRY-box transcription factor 1), and Ascl1 (achaete-scute homolog 1) loci for H3K27ac chromatin immunoprecipitation followed by sequencing signals in 10 SCLC tumors from RP mice and 10 SCLC tumors from RPN mice. Asterisk indicates three tumors that have high signal at both Nkx2-1 and Sox1 loci. The arrowheads on Nkx2-1 locus indicate the loxP sites in RPN mice. (C) Representative immune-staining images for Nkx2-1, Sox1, and Ascl1 on SCLC tumors from RP and RPN mice. Scale bars, 25 μm. (D) Representative H&E images of SCLC tumors from RP and RPN mice. Scale bars, 50 μm. (E) Enriched gene ontologies (Molecular Signatures Database biological process) for the differentially downregulated genes in RPN tumors compared with RP tumors. (F) Average copy number profiles in RP and RPN models. Arrows indicate the Nfib (nuclear factor I B) and Mycl (Mycl proto-oncogene, BHLH transcription factor) loci that are specifically amplified in RP tumors (individual copy number profiles at those loci are available in Figure E8D). chr = chromosome; CMV = cytomegalovirus; H&E = hematoxylin and eosin.
To explore whether NKX2-1 contributes to mSCLC differentiation, we generated RPN mice. We confirmed that Nkx2-1 expression was absent in their tumors, whereas 80% of them were Ascl1 positive. These tumors were negative for NeuroD1 staining, suggesting that they continue to represent SCLC-A (see Figure E8B). Sox1 staining was only weakly positive in 6 of 10 cases, whereas 4 other cases, including the two Ascl1-negative tumors, were negative (Figure 7C). We observed no significant difference in overall survival between the two genotypes (see Figure E8C). Morphologically, all but one mSCLC tumor from the RP model had classical SCLC morphology (scant cytoplasm, nuclei with fine chromatin, and inconspicuous nucleoli), whereas a subset of tumors in RPN mice (15% in RPN vs. 4% in RP) exhibited a “nonclassical” histology defined by noticeably vesicular nuclei with nucleoli by two expert pathologists (E.L.S. and M.B.B.) (Figure 7D), whereas these features are not as prominent to define them as “variant” SCLCs. Of note, the nonclassical RPN tumors stain negatively for the expression of Myc, a marker frequently expressed in variant SCLC (42) (see Figure E8B). These suggest that loss of Nkx2-1 leads to stochastic propensity to form nonclassical morphology, whereas the majority of the tumors retain classical histology representing SCLC-A.
Next, we profiled H3K27ac in 10 mSCLC tumors from four RPN mice (11–13 mo postinitiation, including 5 classical and 5 nonclassical). None of the tumors from the RPN mice had high H3K27ac signal at the Neurod1, Yap1, or Pou2f3 loci, whereas most of RPN tumors (7 of 10) harbored high H3K27ac signals around the Ascl1 locus (Figure 7B; see Figure E8A). Strikingly, we found that signals at the Nkx2-1 and the Sox1 loci were absent in all RPN tumors, suggesting the SCLC-Aσ subtype (Figure 7B). These data suggest that although the RP model can develop both SCLC-Aα and SCLC-Aσ, Nkx2-1 deletion at tumor initiation precludes SCLC-Aα development.
Differential transcriptomic analysis from the tumors in RPN and RP mice also revealed significantly downregulated genes in RPN tumors enriched for nervous system development (Figure 7E), suggesting that SCLC-Aσ is hierarchically closer to the SCLC-N and SCLC-Y/SCLC-P subtypes, also supported by the data that variant morphology is more frequent in RPN. To address whether the absence of Nkx2-1 resulted in distinct tumor evolution in RPN, we profiled copy numbers of those mSCLCs and found that Nfib and Mycl (Mycl proto-oncogene, BHLH transcription factor) loci are frequently amplified in RP model (6 of 10), consistent with previous findings; however, these amplifications were absent in all tumors in RPN model, suggesting that lack of Nkx2-1 predisposed those RPN tumors to undergo a distinct path, leading to a different lineage class of tumors (Figure 7F; see Figure E8D).
Discussion
There have been studies describing new SCLC subtypes after the consensus classification, so it is important to understand how they relate to the present study. Many of those studies used RNA-seq data sets from human samples; for example, one study identified a new subtype, SCLC-I, which has low expression of ASCL1, NEUROD1, and POU2F3 (7). Although it is currently not feasible to determine how these subtypes would be represented in the epigenomic space, as the cohort in our study is different from those of the previous studies, this is an important area for future investigation. Another study identified four subtypes, NE (SCLC-A [1]), NEv2 (SCLC-A [2]), NEv1 (SCLC-N), and non-NE (SCLC-Y), from 50 SCLC cell lines (8). It appears from the membership of each subtype within SCLC-A, our epigenomic SCLC-A subdivisions are independent from those described in that study, which further implies that epigenomic features may reflect a distinct biological state of cells that may not be discerned from transcriptomic profiles.
In the current classification, SCLC-A constitutes 70% of SCLCs, while emerging evidence suggests its heterogeneity. Our study revealed distinctive epigenetic landscapes within SCLC-A that we designated SCLC-Aα and SCLC-Aσ, characterized by the presence of NKX2-1 and TCF4 SEs. NKX2-1 is required for lung development and later specification of the peripheral lung but also of bronchial NE cells (43). NKX2-1 is a well-known marker to diagnose LADCs (44), but it is also frequently expressed in SCLC. As NKX2-1 plays key roles in brain development (45–48), it has been speculated that NKX2-1 is involved in NE pathways, on the basis of its expression enriched in SCLC-A (49) and its physical interaction with ASCL1 (31). In our study, although most SCLC-A expresses NKX2-1 to variable degrees, only 48% had NKX2-1 SE. Importantly, NKX2-1 deletion in SCLC-Aσ and SCLC-N cells, in which NKX2-1 protein is still expressed, had few cell growth effects, suggesting its relevance only in SCLC-Aα.
Furthermore, our data suggest that this function is mediated through interaction with SOX1. Combinations of master TFs that assemble at tissue-specific cis-regulatory sites control distinct cellular functions essential for maintaining lineage states in many contexts. Therefore, although the role of SOX1 had never been described in the lung, it is plausible that their interaction is critical to retain a unique NE state, and it further suggests biological plasticity between glandular cell lineage and pulmonary NE lineage.
The roles of NKX2-1 in carcinogenesis are complex and remain poorly understood. Nkx2-1 deletion in neoplastic lungs causes loss of pulmonary identity and reversion to a foregut lineage. Our data suggest that Nkx2-1 contributes to maintaining a specific neural lineage state that has classical SCLC morphology. The observation that Nkx2-1 deletion is tolerated at tumor initiation in the GEMM, but not in established human SCLC-Aα cell lines, provokes a few potential interpretations. First, the timing of the deletion might be critical in determining the outcome, as evidenced in other studies of lineage factors (50, 51). Second, as cytomegalovirus-Cre targets multiple cell types, Nkx2-1 deletion may favor origination from certain cell types, thus influencing tumor evolution, as reflected by the lack of amplification at Nfib and Mycl. Third, early/in situ RPN lesions that would have become SCLC-Aα might simply evolve to an alternative identity because of lack of Nkx2-1.
Conclusions
We subdivided the major SCLC-A subtype into SCLC-Aα and SCLC-Aσ by revealing a distinct epigenetic landscape in SCLC cell lines, human SCLC tumors, and a GEMM. Collaborative NKX2-1/SOX1 regulation defines the unique lineage state for SCLC-Aα. This study will provide the foundation for future identification of vulnerabilities and enable us to develop a higher degree of personalized therapeutic approaches to benefit patients with SCLC.
Acknowledgments
Acknowledgment
The authors thank Hong Cai, Yi Ge, Ying Liu, Peipei Guo, Yifei Sun, Xiaokun Liu, Bin Zhou, Korey Kam, Mingzhe Li, Megan Vetter, Tyler Jacks, and Karen Yee for helpful discussions and technical assistance. The authors thank the NextSeq Sequencing Facility of the Department of Oncological Sciences at the Icahn School of Medicine at Mount Sinai, the Taplin Mass Spectrometry Facility at Harvard Medical School, and the Biorepository and Pathology Core Facility and Center for Comparative Medicine and Surgery at the Icahn School of Medicine at Mount Sinai.
Footnotes
Supported by American Thoracic Society Foundation Unrestricted Grant ATS-2017-24 (H.W.), American Lung Association of the Northeast Lung Cancer Discovery Award LCD-504985 (H.W.), U.S. Department of Defense grant W81XWH-19-1-0613 (H.W.), and NIH grant R01CA240342 (H.W.); Shaanxi Provincial Natural Science Foundation grant 2017JM8046 (R.K.); the Japanese Respiratory Society (T.S.), the 6th Lilly Oncology Fellowship Program (T.S.), the Uehara Memorial Foundation (T.S.), Japan Society for the Promotion of Science Kakenhi grant JP20K17192 (T.S.), the Takeda Science Foundation and MSD Life Science Foundation (T.S.); National Natural Science Foundation of China grant 82100718 (J.F.); National Key R&D Program of China grant 2018YFE0114200 (X.H.); Shanghai Pujiang Program grant 20PJ1402400 (D.Y.); NIH grant R01CA163772 (C.A.P. and H.W.); NIH grants R01CA212415 and R01CA240317 (E.L.S.) and American Lung Association Lung Cancer Discovery Award LCD-821670 (E.L.S.); and in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Author Contributions: Conceptualization and methodology, R.K. and H.W.; data collection, R.K., A.S.P., T.S., S.Y., L.B., F.J., A.S., Y.T., M.F., S. Liu, J.F., X.H., J.J., Y.M., K.G., D.Y., E.L.S., S. Li., and H.W.; resources, E.L.S., M.B.B., C.A.P., S. Li, and H.W.; data analysis, R.K., S.Y., A.S.P., S. Li, M.B.B., E.L.S., and H.W.; writing – original draft, R.K., A.S.P., and H.W.; writing – review and editing, R.K., A.S.P., E.L.S., and H.W.; supervision, C.A.P., J.Z., S. Li, and H.W.; project administration and funding acquisition, H.W.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202110-2358OC on July 18, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin . 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2. Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. DNA methylation in small-cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene . 2015;34:5869–5878. doi: 10.1038/onc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McColl K, Wildey G, Sakre N, Lipka MB, Behtaj M, Kresak A, et al. Reciprocal expression of INSM1 and YAP1 defines subgroups in small-cell lung cancer. Oncotarget . 2017;8:73745–73756. doi: 10.18632/oncotarget.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang YH, Klingbeil O, He XY, Wu XS, Arun G, Lu B, et al. POU2F3 is a master regulator of a tuft cell-like variant of small-cell lung cancer. Genes Dev . 2018;32:915–928. doi: 10.1101/gad.314815.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small-cell lung cancer. Nature . 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small-cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer . 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell . 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wooten DJ, Groves SM, Tyson DR, Liu Q, Lim JS, Albert R, et al. Systems-level network modeling of small-cell lung cancer subtypes identifies master regulators and destabilizers. PLOS Comput Biol . 2019;15:e1007343. doi: 10.1371/journal.pcbi.1007343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patel AS, Yoo S, Kong R, Sato T, Sinha A, Karam S, et al. Prototypical oncogene family Myc defines unappreciated distinct lineage states of small-cell lung cancer. Sci Adv . 2021;7:eabc2578. doi: 10.1126/sciadv.abc2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev . 2014;28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kong R, Patel AS, Sato T, Yoo S, Sinha A, Tian Y, et al. Transcriptional circuitry of NKX2-1 and SOX1 defines an unrecognized lineage subtype of small-cell lung cancer [abstract] Cancer Res . 2022;82:5709. doi: 10.1164/rccm.202110-2358OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sato T, Yoo S, Kong R, Sinha A, Chandramani-Shivalingappa P, Patel A, et al. Epigenomic profiling discovers trans-lineage SOX2 partnerships driving tumor heterogeneity in lung squamous cell carcinoma. Cancer Res . 2019;79:6084–6100. doi: 10.1158/0008-5472.CAN-19-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol . 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Invest . 2014;124:1636–1645. doi: 10.1172/JCI71545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small-cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell . 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 16. Kusakabe T, Kawaguchi A, Hoshi N, Kawaguchi R, Hoshi S, Kimura S. Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol Endocrinol . 2006;20:1796–1809. doi: 10.1210/me.2005-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc . 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng H, Xie W. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol . 2019;20:535–550. doi: 10.1038/s41580-019-0132-4. [DOI] [PubMed] [Google Scholar]
- 19. Kennedy AJ, Rahn EJ, Paulukaitis BS, Savell KE, Kordasiewicz HB, Wang J, et al. Tcf4 regulates synaptic plasticity, DNA methylation, and memory function. Cell Rep . 2016;16:2666–2685. doi: 10.1016/j.celrep.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Watanabe H, Francis JM, Woo MS, Etemad B, Lin W, Fries DF, et al. Integrated cistromic and expression analysis of amplified NKX2-1 in lung adenocarcinoma identifies LMO3 as a functional transcriptional target. Genes Dev . 2013;27:197–210. doi: 10.1101/gad.203208.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, et al. Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol Cell . 2013;50:185–199. doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Cancer Genome Atlas Research Network Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet . 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet . 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horie M, Miyashita N, Mattsson JSM, Mikami Y, Sandelin M, Brunnström H, et al. An integrative transcriptome analysis reveals a functional role for thyroid transcription factor-1 in small-cell lung cancer. J Pathol . 2018;246:154–165. doi: 10.1002/path.5109. [DOI] [PubMed] [Google Scholar]
- 25. Misch D, Blum T, Boch C, Weiss T, Crolow C, Griff S, et al. Value of thyroid transcription factor (TTF)-1 for diagnosis and prognosis of patients with locally advanced or metastatic small-cell lung cancer. Diagn Pathol . 2015;10:21. doi: 10.1186/s13000-015-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miyauchi E, Motoi N, Ono H, Ninomiya H, Ohyanagi F, Nishio M, et al. Distinct characteristics of small-cell lung cancer correlate with central or peripheral origin: subtyping based on location and expression of transcription factor TTF-1. Medicine (Baltimore) . 2015;94:e2324. doi: 10.1097/MD.0000000000002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu M, Wang B, Gil J, Sabo E, Miller L, Gan L, et al. p63 and TTF-1 immunostaining. A useful marker panel for distinguishing small-cell carcinoma of lung from poorly differentiated squamous cell carcinoma of lung. Am J Clin Pathol . 2003;119:696–702. doi: 10.1309/P5AB-R5KQ-89RN-JTFH. [DOI] [PubMed] [Google Scholar]
- 28. Ordóñez NG. Value of thyroid transcription factor-1 immunostaining in distinguishing small-cell lung carcinomas from other small-cell carcinomas. Am J Surg Pathol . 2000;24:1217–1223. doi: 10.1097/00000478-200009000-00004. [DOI] [PubMed] [Google Scholar]
- 29. Jiang L, Huang J, Higgs BW, Hu Z, Xiao Z, Yao X, et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small-cell lung cancer. PLoS Genet . 2016;12:e1005895. doi: 10.1371/journal.pgen.1005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature . 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hokari S, Tamura Y, Kaneda A, Katsura A, Morikawa M, Murai F, et al. Comparative analysis of TTF-1-binding DNA regions in small-cell lung cancer and non-small-cell lung cancer. Mol Oncol . 2020;14:277–293. doi: 10.1002/1878-0261.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol . 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Magno L, Barry C, Schmidt-Hieber C, Theodotou P, Häusser M, Kessaris N. NKX2-1 is required in the embryonic septum for cholinergic system development, learning, and memory. Cell Rep . 2017;20:1572–1584. doi: 10.1016/j.celrep.2017.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y, Bhowmick T, Liu Y, Gao X, Mertens HDT, Svergun DI, et al. Structural basis for draxin-modulated axon guidance and fasciculation by netrin-1 through DCC. Neuron . 2018;97:1261–1267.e4. doi: 10.1016/j.neuron.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saint-André V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, et al. Models of human core transcriptional regulatory circuitries. Genome Res . 2016;26:385–396. doi: 10.1101/gr.197590.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reissner C, Runkel F, Missler M. Neurexins. Genome Biol . 2013;14:213. doi: 10.1186/gb-2013-14-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature . 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Toma C, Torrico B, Hervás A, Valdés-Mas R, Tristán-Noguero A, Padillo V, et al. Exome sequencing in multiplex autism families suggests a major role for heterozygous truncating mutations. Mol Psychiatry . 2014;19:784–790. doi: 10.1038/mp.2013.106. [DOI] [PubMed] [Google Scholar]
- 39. Wu Z, Zhou J, Zhang X, Zhang Z, Xie Y, Liu JB, et al. Reprogramming of the esophageal squamous carcinoma epigenome by SOX2 promotes ADAR1 dependence. Nat Genet . 2021;53:881–894. doi: 10.1038/s41588-021-00859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep . 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olsen RR, Ireland AS, Kastner DW, Groves SM, Spainhower KB, Pozo K, et al. ASCL1 represses a SOX9+ neural crest stem-like state in small-cell lung cancer. Genes Dev . 2021;35:847–869. doi: 10.1101/gad.348295.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature . 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 43. La Rosa S, Chiaravalli AM, Placidi C, Papanikolaou N, Cerati M, Capella C. TTF1 expression in normal lung neuroendocrine cells and related tumors: immunohistochemical study comparing two different monoclonal antibodies. Virchows Arch . 2010;457:497–507. doi: 10.1007/s00428-010-0954-0. [DOI] [PubMed] [Google Scholar]
- 44. Yamaguchi T, Hosono Y, Yanagisawa K, Takahashi T. NKX2-1/TTF-1: an enigmatic oncogene that functions as a double-edged sword for cancer cell survival and progression. Cancer Cell . 2013;23:718–723. doi: 10.1016/j.ccr.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 45. Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, et al. Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol . 2000;424:409–438. doi: 10.1002/1096-9861(20000828)424:3<409::aid-cne3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 46. Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci . 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development . 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 48. Minocha S, Valloton D, Ypsilanti AR, Fiumelli H, Allen EA, Yanagawa Y, et al. Nkx2.1-derived astrocytes and neurons together with Slit2 are indispensable for anterior commissure formation. Nat Commun . 2015;6:6887. doi: 10.1038/ncomms7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cardnell RJ, Li L, Sen T, Bara R, Tong P, Fujimoto J, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small-cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget . 2017;8:73419–73432. doi: 10.18632/oncotarget.20621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Camolotto SA, Pattabiraman S, Mosbruger TL, Jones A, Belova VK, Orstad G, et al. FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer. eLife . 2018;7:e38579. doi: 10.7554/eLife.38579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zewdu R, Mehrabad EM, Ingram K, Fang P, Gillis KL, Camolotto SA, et al. An NKX2-1/ERK/WNT feedback loop modulates gastric identity and response to targeted therapy in lung adenocarcinoma. eLife . 2021;10:e66788. doi: 10.7554/eLife.66788. [DOI] [PMC free article] [PubMed] [Google Scholar]


