Abstract
Listeria monocytogenes is a facultative intracellular gram-positive bacterium capable of growing in the cytoplasm of infected host cells. Bacterial escape from the phagosomal vacuole of infected cells is mainly mediated by the pore-forming hemolysin listeriolysin O (LLO) encoded by hly. LLO-negative mutants of L. monocytogenes are avirulent in the mouse model. We have developed a genetic system with hly as a reporter gene allowing the identification of both constitutive and in vivo-inducible promoters of this pathogen. Genomic libraries were created by randomly inserting L. monocytogenes chromosomal fragments upstream of the promoterless hly gene cloned into gram-positive and gram-negative shuttle vectors and expressed in an LLO-negative mutant strain. With this hly-based promoter trap system, combined with access to the L. monocytogenes genome database, we identified 20 in vitro-transcribed genes, including genes encoding (i) p60, a previously known virulence gene, (ii) a putative new hemolysin, and (iii) two proteins of the general protein secretion pathway. By using the hly-based system as an in vivo expression technology tool, nine in vivo-induced loci of L. monocytogenes were identified, including genes encoding (i) the previously known in vivo-inducible phosphatidylinositol phospholipase C and (ii) a putative N-acetylglucosamine epimerase, possibly involved in teichoic acid biosynthesis. The use of hly as a reporter is a simple and powerful alternative to classical methods for transcriptional analysis to monitor promoter activity in L. monocytogenes.
Listeria monocytogenes is a gram-positive bacterium widespread in nature and responsible for sporadic severe infections in humans and other animal species (reference 3 and references therein). This pathogen is a facultative intracellular microorganism capable of invading most host cells, including epithelial cells, hepatocytes, fibroblasts, endothelial cells, and even macrophages. Each step of intracellular parasitism by L. monocytogenes is dependent upon the production of virulence factors (30, 34). Among the virulence factors, listeriolysin O (LLO) is an exotoxin encoded by the hly gene which plays a crucial role in the escape of bacteria from the phagosomal compartment. Disruption of hly in wild-type L. monocytogenes leads to a loss of hemolytic activity and a loss of virulence in the mouse model of infection (10, 13, 26). The virulence genes (hly, plcA, plcB, mpl, actA, inlA, and inlB) are clustered into two distinct loci on the chromosome and are controlled by a single pleiotropic regulatory activator, PrfA, which is required for virulence (6, 19, 23).
Transposon mutagenesis is the only successful strategy used so far to identify virulence genes in L. monocytogenes. However, it is reasonable to assume that some genes that are important in the infection process are specifically induced during host cell infection. Indeed, recent studies have shown that most PrfA-regulated virulence genes are more efficiently expressed during intracellular growth (24), and in particular, the promoters for hly and plcA are predominantly activated within the phagosomal compartment (4). This led us to investigate the use of in vivo expression technology (IVET) to identify new virulence genes of L. monocytogenes specifically induced within infected host cells. The general principle of IVET consists of using a promoterless reporter gene fused to random chromosomal DNA fragments of the pathogen of interest. Different types of reporter genes were developed, such as biosynthetic genes, genes conferring antibiotic resistance, genes encoding recombination enzymes, and the gene encoding green fluorescent protein. IVET has been successfully used with several bacterial pathogens (5, 12, 14, 20, 21, 22, 31, 37, 38).
The genetic system developed in this work allowed the identification of L. monocytogenes promoters that are either constitutive (i.e., active in bacteria grown under standard laboratory conditions and in host tissues) or specifically induced upon infection in the mouse model (in vivo). This system utilizes the plasmid-borne hly-encoded LLO both as an indicator of protein expression and as a promoter trap. This work was undertaken while the Listeria genome-sequencing project was in progress. The sequence of the genome is now complete. The availability of this source of information allowed us to identify rapidly and unambiguously all the genes corresponding to the sequences that were determined.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and DNA techniques.
Bacterial strains are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium, and L. monocytogenes strains were grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) at 37°C. The wild-type virulent strain of L. monocytogenes EGD (denoted EGDwt below) belongs to the serovar 1/2a (10). EGDΔhly serotype 1/2a (kindly provided by T. Chakraborty) is a derivative of EGDwt which contains an in-frame chromosomal deletion of 1,080 bp in the hly gene (11). EGDΔhly was transformed with the different recombinant plasmids by electroporation as previously described (25). Antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 50 μg ml−1; and spectinomycin (SPC), 60 μg ml−1.
TABLE 1.
Bacterial strains
| Strain | Characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| TOP10 | High-transformation-efficiency cells | Invitrogen |
| TOP10/pCR-hly | TOP10 transformed with pCR-hly | This work |
| TOP10/pCR-phly-hly | TOP10 transformed with pCR-phly-hly | This work |
| TOP10/pAT28-hly | TOP10 transformed with pAT28-hly | This work |
| TOP10/pTCV-hly | TOP10 transformed with pTCV-hly | This work |
| TOP10/pAT28-phly-hly | TOP10 transformed with pAT28-phly-hly | This work |
| TOP10/pTCV-phly-hly | TOP10 transformed with pTCV-phly-hly | This work |
| L. monocytogenes | ||
| EGDwt | Wild-type strain, serovar 1/2a | 10 |
| EGDΔhly | EGD derivative (serovar 1/2a) with a deletion of 1,080 bp in the hly gene | 11 |
| EGDΔhly/pAT28-hly | EGDΔhly transformed with pAT28-hly | This work |
| EGDΔhly/pAT28-phly-hly | EGDΔhly transformed with pAT28-phly-hly | This work |
| EGDΔhly/pTCV-hly | EGDΔhly transformed with pTCV-hly | This work |
| EGDΔhly/pTCV-phly-hly | EGDΔhly transformed with pTCV-phly-hly | This work |
Chromosomal DNA and plasmid isolation, restriction enzyme analyses, and amplification by PCR were performed according to standard protocols (2, 29).
Construction and analysis of L. monocytogenes genomic libraries. (i) PCR amplification of the hly gene.
The wild-type hly gene preceded by its ribosome binding site was amplified by PCR from L. monocytogenes (EGDwt) genomic DNA using the following primer pair: 5′CCGGATCCTGTAGAAGGAGAGTGAAACCCATG3′ (the ATG start codon is in boldface characters) and 5′CCCTGCAGACAATTATTCGATTGGATTATCTAC3′. The wild-type hly gene with its upstream promoter region (denoted hlyp) was amplified using the following primer pair: 5′CCGGATCCCTTAAAGTGACTTTTATGTTGAGGCA3′ and 5′CCCTGCAGACAATTATTCGATTGGATTATCTAC3′. The primers were designed to generate cohesive restriction sites (underlined) for BamHI (5′ end) and PstI (3′ end), respectively. Oligonucleotides were synthesized by Genset (Paris, France). The ampliTaq DNA polymerase of Thermus aquaticus from Perkin-Elmer (Branchburg, N.J.) was used. The amplified double-stranded DNA fragments were first cloned into the pCR cloning vector using the TA cloning kit (Invitrogen Corp., San Diego, Calif.). The clones with the inserted PCR product were selected by restriction analysis, using standard procedures. Plasmid DNA from the selected pCR recombinant was prepared using Qiagen (Chatsworth, Calif.) kits.
(ii) Subcloning into pTCV-lac or pAT28 E. coli-L. monocytogenes shuttle vectors. (a) pTCV-lac.
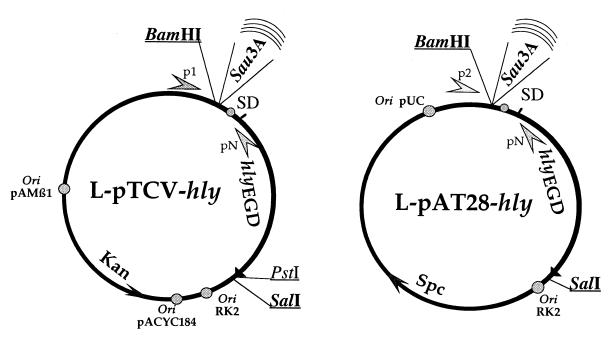
The recombinant plasmids pCR-hly and pCR-phly-hly were cut simultaneously with BamHI and PstI restriction enzymes (NEN, Beverly, Mass.). The BamHI-PstI fragments containing the hly gene were purified on agarose gels and subcloned into the low-copy-number vector pTCV-lac (27). For this, the BamHI-PstI fragment of pTCV-lac, containing the lacZ gene, was replaced by the BamHI-PstI fragment containing hly, yielding plasmids pTCV-hly and pTCV-phly-hly (Fig. 1).
FIG. 1.
Construction of libraries. The arrows flanking the BamHI cloning site, denoted p1-pN for the library in pTCV-hly and p2-pN for the library in pAT28-hly, correspond to the pairs of primers used to amplify and sequence the chromosomal DNA fragments inserted upstream of hly. The BamHI and SalI sites flanking the hly gene are underlined.
(b) pAT28.
The hly gene (with or without the promoter) was then subcloned into the high-copy-number E. coli–gram-positive-bacteria shuttle vector pAT28 (36). For this, the two pTCV recombinant plasmids were digested with BamHI and SalI (the SalI site lies directly downstream of the PstI site), and the BamHI-SalI fragment was inserted into the BamHI–SalI sites in the polylinker of pAT28, yielding plasmids pAT28-hly and pAT28-phly-hly) (Fig. 1).
(iii) Generation of the libraries.
Two banks were constructed, in pTCV-hly and in pAT28-hly. In each case, L. monocytogenes (EGDwt) chromosomal DNA was partially digested by Sau3A. DNA fragments (between 0.5 and 2 kb) were sized on TAE-agarose gels and cloned into the dephosphorylated BamHI site of either pTCV-hly or pAT28-hly. Each bank was electroporated into E. coli TOP10 cells (Invitrogen Corp.). Recombinant E. coli clones (5 × 103 to 104) were obtained. Restriction analysis of 20 different clones from each transformation confirmed that >85% of the clones had an inserted chromosomal fragment of an average size ranging between 0.8 and 2 kb. For each bank, the transformants were pooled, and the pools were grown overnight at 37°C with agitation in Luria-Bertani–SPC medium. A plasmid preparation was made from each culture. The two plasmid banks were then transferred to L. monocytogenes strain EGDΔhly by electroporation using 1 μg of plasmid per electroporation. Approximately 104 recombinant Listeria were obtained for each bank. The transformants were pooled, and the two pools were grown in BHI-SPC overnight at 37°C without agitation.
(iv) Screening of the libraries for in vivo-inducible genes.
Two strategies were tested to identify in vivo-inducible promoters. First, each bank was subjected to successive passages in mice. With the two banks, the initial frequency of hemolytic clones (i.e., constitutive promoters) was low (1 to 5%). This frequency rose after each passage in the animal, indicating an obvious link between the expression of LLO and the capacity to persist and multiply in the infected organs. For example, with the pTCV-hly bank, the initial frequency of hemolytic clones was 1%. This frequency rose after each passage in the animal (to 3, 7, and 14% after the first, second, and third passages, respectively), reaching 23% after the fourth passage.
A series of nonhemolytic clones were recovered from the brains of infected mice and were further analyzed. However, none of them corresponded to putative in vivo-inducible promoters (data not shown). We therefore used a second strategy to identify in vivo-inducible promoters and focused our efforts on the bank constructed in pAT28-hly. On horse blood agar, the hly-positive clones of this bank were significantly more hemolytic than in the previous one, due to the higher number of hly gene copies per cell, yielding a higher rate of LLO expression. Indeed, when hly preceded by its promoter, phly, was carried on the low-copy-number vector (pTCV), no detectable hemolytic activity could be recorded in bacterial culture supernatants, while with the high-copy-number vector (pAT28), the hemolytic activity recorded was fourfold higher than that in EGDwt. Two classes of clones could be visualized on blood agar: nonhemolytic or very weakly hemolytic clones and highly hemolytic clones. Only the nonhemolytic or very weakly hemolytic clones were chosen for the in vivo screening. One hundred pools of 10 different clones each (i.e., a total of 1,000 clones) were prepared and tested in vivo. For each pool, 2 × 108 bacteria (from nonagitated overnight cultures grown at 37°C) were used per inoculation (two mice were infected per pool). Each clone within the pool was thus represented approximately 2 × 107 times. Only 9 pools out of 100 tested killed the infected mice or made them visibly ill within 3 to 10 days after injection. These nine pools were further analyzed. For this, the 10 clones from each pool were individually inoculated into mice (two mice per clone at 2 × 108 bacteria per mouse). The nine clones inducing death or severe illness 3 to 4 days after injection were further studied.
The hemolytic activity in culture supernatants from these nine strains was checked on horse red blood cells (see “Hemolysis” below). None of them showed any detectable activity.
(v) Control strains.
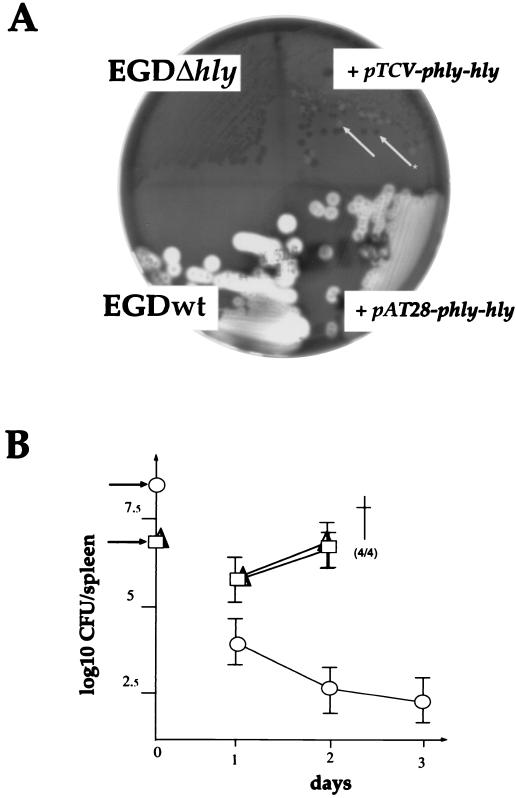
For the pTCV-hly-derived bank, we used as a positive control EGDΔhly carrying pTCV-phly-hly (hly preceded by its natural promoter). The resulting strain became weakly hemolytic on blood agar and regained in vivo virulence (Fig. 2). For the pAT28-hly-derived bank, the positive control was EGDΔhly carrying pAT28-phly-hly. The resulting strain became highly hemolytic and regained in vivo virulence (Fig. 2). The recipient strain, EGDΔhly, without a plasmid was used as a negative control.
FIG. 2.
Complementation. (A) In vitro complementation. The hemolytic phenotype of L. monocytogenes colonies was visualized on horse blood agar plates after 24 h of incubation at 37°C. The two strains denoted + pTCV-pphly-hly and + pAT28-phly-hly correspond to EGDΔhly transformed with the corresponding plasmids. The arrows show the two types of colonies (Hly+ and Hly−) observed with the strain transformed with pTCV-phly-hly in the absence of antibiotic selection (Hly− is indicated by a star). (B) In vivo complementation. The kinetics of infection was followed in the spleens of infected mice. With the negative control, EGDΔhly (○), mice were inoculated with 2 × 109 bacteria/mouse (indicated by an arrow to the left of the ordinate); with the two transformed derivatives, mice were inoculated with 2 × 107 bacteria/mouse (arrow) (□, EGDΔhly transformed with pTCV-phly-hly; ▵, EGDΔhly transformed with pAT28-phly-hly). †, death; four mice out four infected died.
Hemolysis.
Hemolytic phenotypes were visualized by spreading bacteria onto horse blood agar (BioMerieux, Marcy l'Etoile, France). Hemolytic activity was also measured by lysis of horse red blood cells on supernatants from cultures in exponential growth phase in BHI-SPC at 37°C. All the cultures were adjusted to an optical density at 600 nm of 0.6 before supernatant collection. Assays were carried out as described previously (9) at pH 6. Hemolytic activity was expressed as the reciprocal of the dilution of culture supernatant (40 μl) required to lyse 50% of horse erythrocytes.
Infection of mice.
Six- to 8-week-old pathogen-free ICR female Swiss mice (Janvier, Le Geneset St. Isle, France) were used. For the kinetics of infection, groups of five mice were inoculated intravenously in the lateral tail vein. Organs (spleen and brain) were aseptically removed and separately homogenized in 0.15 M NaCl. Bacterial numbers in the organ homogenates were determined at various intervals on BHI plates containing appropriate antibiotics. In the absence of SPC selection, PAT28-derived constructions appeared fully stable in culture (Fig. 2A). In contrast, in vivo instability was observed (not shown). Therefore, all the subsequent in vivo studies were carried out on animals pretreated with SPC (1 mg of SPC per mouse twice a day). For the pTCV-derived constructions, in vitro instability had already been observed (Fig. 2A). In vivo studies were carried out on animals that were pretreated with kanamycin (at a dose of 600 μg per mouse twice a day).
Sequence analysis of the inserted fragments and Listeria genome database.
Two different pairs of primers, flanking the BamHI cloning site and denoted p1-pN for pTCV-hly-derived plasmids and p2-pN for pAT28-hly-derived plasmids, were used to amplify and sequence the chromosomal DNA fragments inserted upstream of hly (Fig. 1). The sequences of the primers were as follows: p1, 5′GTTGAATAACACTTATTCCTATC3′; p2, 5′CAGGAAACAGCTATGACC3′; and pN, 5′TACTTTTTTTATTACGATCAAAAA3′. The PCR products were sequenced with the automated Prism 310 sequencer (Perkin-Elmer, Applied Biosystems). Then, the DNA sequence of each fragment cloned upstream of hly was launched in the complete 2,900,000-bp Listeria genome database (BLASTn search). Searches were performed via the Internet with BLAST software (1) from the National Center for Biotechnology Information home page (http://www.ncbi.nlm.gov/BLAST/). The region of the Listeria genome containing the sequenced DNA was then analyzed with the program ORFfinder (http: //www.ncbi.nlm.nih.gov/gorf/) to identify the open reading frame (ORF)—or N-terminal portions—comprised within the cloned fragment. The identified ORFs were then launched in the general databases (nonredundant BLASTp search).
RESULTS
Construction and screening of L. monocytogenes genomic libraries.
Two banks were created by fusing a promoterless copy of hly to random L. monocytogenes chromosomal fragments in gram-negative and gram-positive shuttle plasmids: pTCV-hly (low copy) and pAT28-hly (high copy). The two banks were first constructed in E. coli and then transferred by electroporation into EGDΔhly, a nonhemolytic derivative of EGDwt (Fig. 1) (see Materials and Methods). As shown in Fig. 2, the non-hemolytic control strain EGDΔhly, inoculated intravenously (2 × 109 bacteria/mouse), was avirulent and was rapidly eliminated from the spleens of infected mice. As previously reported, the LLO-negative strains are approximately 5 log units less virulent than the wild-type strains in the mouse model (8, 10, 26); the lethal infecting dose ranged between 104.5 and 105.8 for EGD and LO28, respectively (3).
Virulence was restored in EGDΔhly transformed with each plasmid vector harboring the hly gene under the control of its natural promoter (at a 100-fold-lower dose, i.e., 2 × 107 bacteria/mouse). Transformed hemolytic bacteria (Fig. 2) rapidly grew in the spleens of infected mice, ultimately resulting in the deaths of the mice within 2 to 4 days.
Constitutive promoters were screened from the two banks after spreading them onto blood agar plates and selection of hemolytic colonies. In vivo-inducible promoters were selected after passage in mice of the bank constructed in pAT28-hly. A total of 1,000 nonhemolytic or very weakly hemolytic clones were tested in the in vivo screening (see Materials and Methods for details). First, 100 pools of 10 different clones each were generated and tested in mice (for each pool, a dose of 2 × 108 bacteria was injected per mouse). Under these conditions, only 9 pools out of 100 tested killed the infected mice or made them visibly ill within 3 to 10 days after injection. These nine pools were then further analyzed: each clone was individually inoculated into mice. Nine in vivo-inducible clones inducing death or severe illness 3 to 4 days after injection were identified by this procedure.
Constitutive promoters of L. monocytogenes.
The promoters allowing secretion of LLO by bacteria grown under standard laboratory conditions were called constitutive (i.e., the hemolytic clones on blood agar plates) (Table 2). Clones expressing LLO constitutively were obtained either directly (11 of 20) or after passage of the banks in mice, spreading of the infected organs on blood agar, and selection of highly hemolytic colonies (9 of 20). The products of the 20 ORFs located downstream of the identified promoter are listed alphabetically below (designated by the name of the ORF product in the databases with the highest similarity). When no homologous protein was found, it was named Orfn, where n is the number of predicted residues. Only one sequence (denoted Iap) corresponded to a previously identified L. monocytogenes protein.
TABLE 2.
L. monocytogenes loci identified
| Clonea | Size of ORF productb | Similar proteinsc | Similarityd
|
Functione | |
|---|---|---|---|---|---|
| % Id. | % Sim. | ||||
| Constitutive promoters | |||||
| pt11 | 130 | LaaB; L. sakei | 27 | 54 | Transcriptional regulator |
| pt16 | 140 | LepS; B. subtilis | 46 | 61 | Signal peptidase |
| pt15 | 199 | Maa; E. coli | 65 | 76 | Maltose-acetyltransferase |
| pt24 | 883 | ValS; B. subtilis | 74 | 87 | Valyl-tRNA synthetase |
| pt7 | 220 | YhiD; E. coli | 46 | 61 | Mg2+ ATPase |
| pt19 | 363 | YqgU; B. subtilis | 22 | 41 | Lipoprotein |
| pt32 | 436 | YrvN; B. subtilis | 72 | 81 | Helicase |
| pt31 | 165 | YtgI; B. subtilis | 63 | 76 | Peroxidase |
| pa5 | 338 | LacD; S. aureus | 55 | 75 | Tagatose 1,6-diphosphate aldolase |
| pa308 | 80 | YtmB; B. subtilis | 60 | 76 | Unknown |
| pa4 | 243 | Orf243 | Unknown | ||
| ptS2 | 346 | Asd; B. subtilis | 69 | 82 | Aspartate semialdehyde dehydrogenase |
| ptS8 | 438 | CydA; B. subtilis | 70 | 82 | Cytochrome oxidase |
| ptS1 | 484 | Iap; L. monocytogenes | 100 | 100 | Internalin-associated protein |
| ptS3 | 244 | RluB; B. subtilis | 67 | 85 | Pseudouridine synthetase |
| ptS4 | 431 | SecY; B. licheniformis | 68 | 80 | Preprotein translocase |
| paS8 | 512 | BglI; B. subtilis | 62 | 75 | β-Glucosidase |
| paS3 | 219 | FnR; A. aeolicus | 26 | 44 | Transcriptional regulator (Crp/Fnr family) |
| paB13 | 388 | PatB; B. subtilis | 38 | 62 | Hemolysine |
| paS10 | 640 | ThrS; B. subtilis | 75 | 86 | Threonine-tRNA synthetase |
| In vivo-inducible promoters | |||||
| pa428 | 317 | PlcA; L. monocytogenes | 100 | 100 | PI-PLC |
| pa364 | 373 | YvyH; B. subtilis | 62 | 78 | N-acetyl-glucosamine epimerase |
| pa394 | 104 | Orf104 | 38 | 47 | Lipoprotein |
| pa762 | 591 | PhoR (opposite to hly) | 31 | 52 | Transcriptional regulator |
| pa303 | 301 | MviM (opposite to hly) | 37 | 48 | Virulence factor |
| pa393 | |||||
| pa669 | |||||
| pa769 | |||||
| pa896 | |||||
The clone numbers are preceded by pt for pTCV-derived or pa for pAT28-derived sequences. The pTCV-derived clones (listed alphabetically) are followed by the pAT28-derived clones. Constitutive clones were subdivided in two classes based on whether they were identified as being expressed on blood agar alone (above the line) or as selected in vivo and expressed on blood agar (below the line). A number preceded by a letter means that the clone was screened on blood agar from the spleens (S) or the brains (B) of infected animals.
Size in amino acids of the ORF product encoded by the gene located downstream of the identified promoter.
Name of the ORF product in the databases sharing the highest similarity with the identified ORF product.
Similarity with the identified ORF product. Id., identity; Sim., similarity.
Predicted (on the basis of sequence similarity) or identified function for the ORF product.
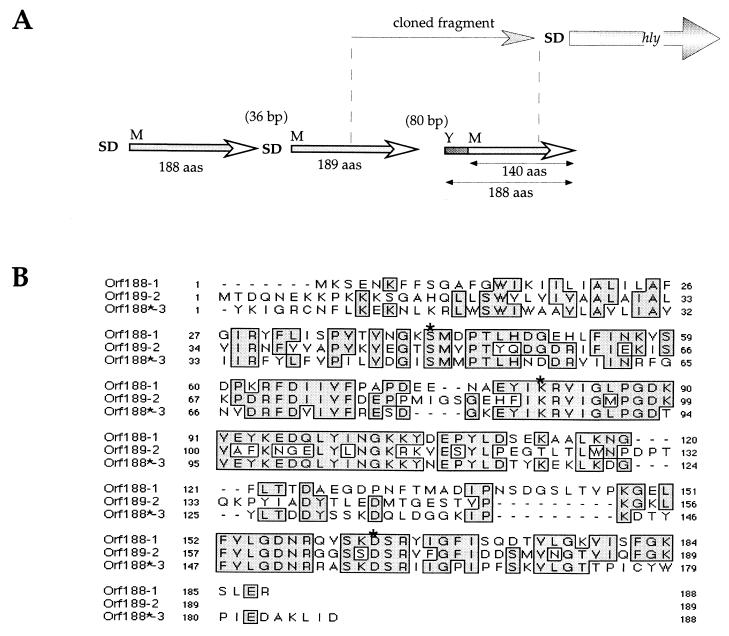
From the pTCV-hly-derived bank, the cloned fragments comprised the promoter regions of the 13 putative genes encoding the following proteins: Asd, a protein homologous to an aspartate semialdehyde dehydrogenase of Bacillus subtilis which is involved in cell wall biosynthesis and thus may be important for bacterial virulence; CydA, a protein highly similar to a putative cytochrome oxydase of B. subtilis; Iap, a major virulence-associated extracellular protein of L. monocytogenes, also reported as a murien hydrolyase (16, 39); LaaB, a protein having some similarities to a putative transcriptional regulator of Lactobacillus sakei; LepS, a protein homologous to the LepS signal peptidase (Spase) of B. subtilis (this ORF was preceded by two ORFs encoding 188 and 189 amino acids, respectively, also sharing significant similarities with one another [Fig. 3] [see Discussion]; examination of the sequence preceding the predicted AUG initiation codon of orf140 suggests that the translational start site probably lies upstream [indeed, 48 in-frame triplets precede the AUG codon, and this upstream translated portion presents similarities with the corresponding N-terminal portions of the two other ORFs]); Maa, a protein homologous to a putative maltose acetyltransferase of E. coli; RluB, a protein highly similar to a putative pseudouridylate synthase of B. subtilis; SecY, a protein homologous to the SecY preprotein translocases of Bacillus licheniformis and B. subtilis (61% identity) of identical sizes (32, 33); ValS, a protein highly similar to the valyl-tRNA synthetase of B. subtilis; YhiD, a protein homologous to a putative Mg2+ ATPase of E. coli; YqgU, a protein showing only a weak similarity to a putative lipoprotein of B. subtilis; YrvN, a protein highly similar to a putative helicase of B. subtilis; and YtgI, a protein highly similar to a putative thiol-peroxydase of B. subtilis.
FIG. 3.
The leader peptidase cluster. (A) Schematic representation of the L. monocytogenes genetic cluster. The upper part indicates the approximate size of the fragment cloned upstream of hly. The lower part represents the three consecutive ORFs. The numbers in parentheses indicate the numbers of base pairs between the stop codon of one ORF and the start codon of the following one. SD, Shine-Dalgarno consensus sequence; M, putative N-terminal methionine residue; Y, putative N-terminal tyrosine residue. For the third ORF, the N-terminal methionine residue is preceded by 48 residues (indicated by dark shading). (B) Multiple sequence alignment of the three ORF products. Multiple sequence alignment was performed using the CLUSTALw algorithm (available at http://www.infobiogen.fr/services/analyseq/cgi-bin/clustalw_in.pl). Identical residues are boxed. Orf188-1 corresponds to the product of the first ORF, Orf189-2 corresponds to the product of the second ORF, and Orf188∗-*3 corresponds to product of the third ORF (188∗ refers to the maximal predicted size, comprising the 140 amino acids starting at the first methionine plus the 48 preceding residues). The stars above residues S, K, and D indicate the conserved residues essential for leader peptidase activity.
From the pAT28-hly-derived bank, the cloned fragments comprised the promoter regions of seven putative genes encoding the following proteins: BglI, a protein highly similar to the 6-phospho-β-glucosidase of B. subtilis; FnR, a protein showing similarities to a putative transcriptional regulator from Aquifex aeolicus belonging to the Crp/Fnr family; LacD, a protein showing significant similarities to the tagatose 1,6-diphosphate aldolase of Staphylococcus aureus; PatB, a protein homologous to a putative aminotransferase of B. subtilis and to a 399-amino-acid hemolysin from Treponema denticola (32% identity; 51% similarity) (alignment of the sequences of the different amino transferases with the T. denticola hemolysin revealed 13 invariant residues [7], twelve of which are conserved in the L. monocytogenes protein [data not shown]; examination of the deduced DNA sequence of the L. monocytogenes gene and alignments with protein homologues strongly suggests that the translational initiation codon is a UUG [38] located 32 codons upstream of the first AUG [not shown]; the protein thus likely comprises 388 residues); ThrS, a protein highly similar to the threonyl-tRNA synthetase 1 of B. subtilis; YtmB, a protein similar to YtmB of B. subtilis of unknown function; and Orf243, a protein without similarity to any protein sequences in the databases.
In vivo-inducible promoters.
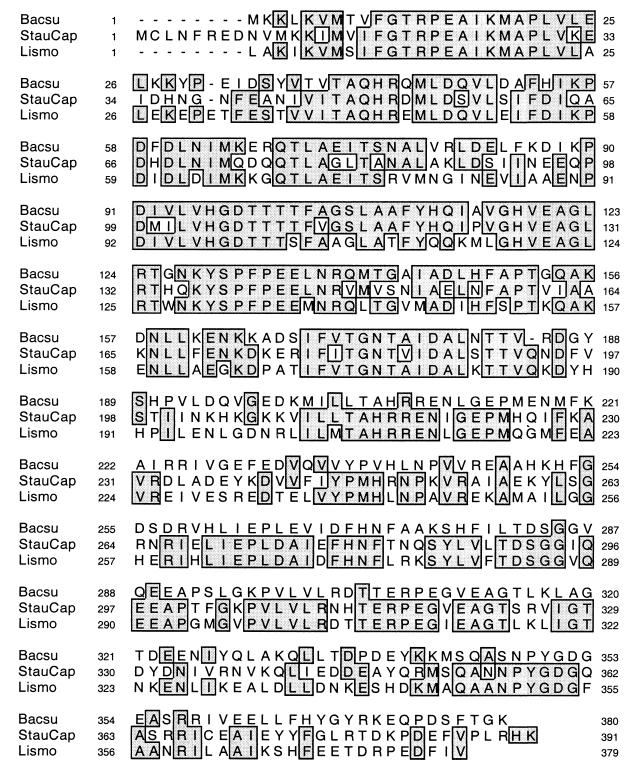
Among the in vivo-inducible sequences identified, three corresponded to promoters of genes determining identified ORFs inserted in the correct orientation with respect to the hly gene. The first clone (pa428) encompassed the promoter of plcA encoding phosphatidylinositol phospholipase C (PI-PLC), a major virulence factor of L. monocytogenes. The 1.5-kb DNA fragment cloned upstream of hly comprised the plcA promoter, the plcA gene, and the prfA promoter region, including the first 92 bp of the prfA gene. The second clone (pa364) comprised the promoter region of a protein homologous to the family of N-acetylglucosamine epimerases (Fig. 4), including YvyH of B. subtilis, Cps19ka of Streptococcus pneumoniae (58% identity), and Cap5P of S. aureus (62% identity). Examination of the deduced DNA sequence of the L. monocytogenes gene and alignments with the protein homologues strongly suggests (as for the patB sequence discussed above) that the translational initiation codon is a UUG located 6 triplets upstream of the AUG and preceded at optimal distance by a classical Shine-Dalgarno sequence (not shown). The L. monocytogenes epimerase thus likely contains 379 residues. The third clone (pa394) comprised the promoter region of a short protein of unknown function (designated Orf104) having similarities to a putative protein of the archaebacterium Aeropyrus pernix, which possesses a putative membrane lipoprotein lipid attachment site.
FIG. 4.
Multiple sequence alignment of L. monocytogenes (Lismo) sequence with those of B. subtilis (Bacsu) and S. aureus (StauCap) N-acetyl glucosamine epimerases. Alignment was performed using CLUSTALw. Identical residues are boxed. Bacsu, YvyH of B. subtilis (380 amino acids [aa]); StauCap, Cap5p protein of S. aureus (391 aa).
In two cases, the fused fragment belonged to an internal portion of a structural gene but in the direction opposite to that of hly: clone pa303 comprised the proximal part of a gene encoding a protein having similarities to the MviM virulence factor of Salmonella enterica serovar Typhimurium (18); clone pa762 comprised the proximal part of a gene encoding a protein having similarities to a PhoR-like protein of B. subtilis (30). In four constructs (clones pa393, pa669, pa769, and pa896), no ORF encoding more than 50 residues could be identified in the corresponding region of the Listeria genome. However, in those four cases, putative promoters were predicted in the fused sequences by their significant scores (0.99 or 1 of 1) with the Promoter Prediction by Neural Network pro gram (available at http://www.dot.imgen.bcm.tmc.edu:9331 /seq-search/gene-search).
Kinetics of bacterial growth in organs of mice infected by transformed Listeria harboring in vivo-inducible promoters.
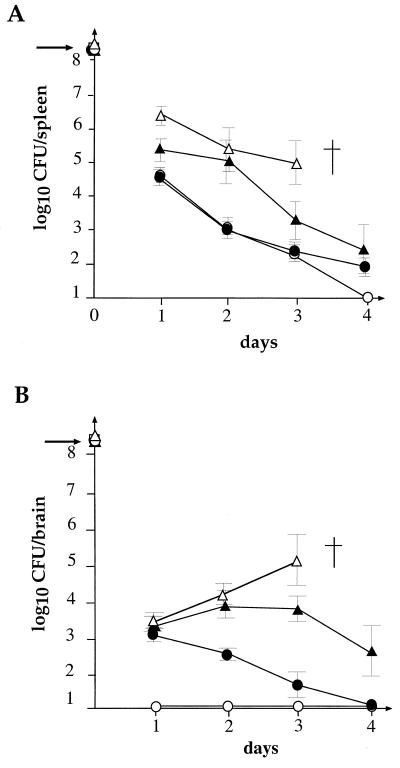
The kinetics of bacterial growth was followed in mice infected with EGDΔhly transformed with pAT28-hly harboring in vivo-inducible promoters from plcA, yvyH, or orf104 compared with EGDΔhly, used as a negative control. Mice were inoculated with 1.5 × 108 bacteria, and bacterial survival was followed in the spleen and the brain over a 4-day period (five mice per day for each strain). As shown in Fig. 5, bacteria from EGDΔhly were rapidly destroyed in the spleen and failed to infect the brain. In contrast, when hly was under the control of the yvyH promoter, the mice died 4 days after infection. When hly was under the control of the plcA (and prfA) promoter, the mice survived. However, bacterial counts were significantly higher 2 days after infection (2 to 3 log higher) in the spleen compared to the control (Fig. 5), and bacteria were found in the brain. When hly was under the control of the promoter of orf104, the mice also survived. Although bacterial counts in the spleens were almost identical to those recorded with the negative control, the mutant bacteria were able to infect the brain and persisted for 3 days before being cleared. This result clearly confirmed that these three promoters were active in vivo.
FIG. 5.
In vivo survival of the in vivo-inducible strains. The kinetics of bacterial growth was followed in organs of mice infected with EGDΔhly transformed with pAT28-hly harboring in vivo-inducible promoters from plcA or yvyH compared with EGDΔhly as a negative control. Mice were inoculated with 1.5 × 108 bacteria (indicated by an arrow to the left of the ordinate). Bacterial survival was followed in the spleen (A) and the brain (B) over a 4-day period. ○, EGDΔhly; ▴, EGDΔhly transformed with pAT28-pplcA-hly; ▵, EGDΔhly transformed with pAT28-pyvyH-hly; ●, EGDΔhly transformed with pAT28-porf104-hly; †, death. The error bars represent standard deviations.
DISCUSSION
We designed a genetic system based on the utilization of the plasmid-borne hly-encoded LLO as an indicator of protein expression and as a promoter trap in L. monocytogenes. Combined with access to the recently completed Listeria genome sequence, this hly-based system constituted (i) a simple and powerful alternative to classical methods for transcriptional analysis of constitutive promoters (hemolytic activity, which reflects hly transcription and translation, can be easily visualized on plates or quantified on erythrocytes) and (ii) a new IVET tool for the selection of in vivo-inducible loci in L. monocytogenes. For both classes of promoters, among the identified sequences were previously known virulence genes of L. monocytogenes, confirming the efficiency of the system.
Constitutive promoters.
Twenty different constitutive promoters were identified, including that of iap, encoding the internalin-associated protein p60 (in contrast to most other virulence genes of L. monocytogenes, this promoter is PrfA independent) (4, 16), and those of housekeeping genes, metabolic and biosynthetic genes, and putative transcriptional regulators. Of particular interest, a putative new L. monocytogenes hemolysin gene was also identified. The protein is similar to a hemolysin of T. denticola (7) which has significant similarities to members of the aminotransferase family. Interestingly, among the other constitutive promoters, we found two key elements of the general secretion machinery of L. monocytogenes: a cluster of leader peptidase genes and the secY gene. In B. subtilis, most genes for components of the protein secretion machinery are present in only one copy (17), despite the fact that this bacterium has a large capacity for protein secretion. The only known exception concerns the genes for type I Spases. Indeed, while in many eubacteria one Spase seems to be sufficient for the processing of secretory preproteins, in B. subtilis up to seven Spase I proteins have been identified so far. In contrast to E. coli, where it has been demonstrated that Spase activity is essential for cell growth, in B. subtilis, none of the sip genes is essential by itself. However, a specific combination of mutations in these genes is lethal (reference 35 and references therein). As shown in Table 2, orf140 encodes a protein homologous to LepS signal peptidase belonging to the Spase I family. Its promoter region directed the expression of LLO in vitro, suggesting that this protein is expressed in L. monocytogenes. Examination of the region directly upstream of orf140 revealed the presence of two genes also encoding putative Spase-like proteins. As shown in Fig. 3, the alignment of the three consecutive ORFs revealed significant amino acid conservation. Furthermore, all three ORF products possessed the conserved serine, lysine, and aspartate residues essential for Spase activity, favoring the idea that they are indeed functional. The participation of each of the three Spases in protein secretion (including secretion of virulence factors) will have to be addressed experimentally. The other gene implicated in the secretion machinery that was identified was secY, encoding the preprotein translocase SecY. The protein of L. monocytogenes is composed of 431 residues. Sequence conservation between L. monocytogenes and B. subtilis SecY proteins was uniformly distributed (data not shown), reflecting probable high functional similarities. Notably, as in B.subtilis, no other SecY homologue was found in the Listeria database.
In vivo-inducible loci.
The hly-based IVET system was specifically devised to identify promoters expressed within the host cell phagosome. Indeed, only the bacteria able to express the hly-encoded LLO in the phagosomal compartment will be able to escape from the phagosome and therefore survive and multiply in the cytoplasm of infected cells. This also represents the major limitation of the system, since promoters that would be induced in later stages of the infectious process could not be identified. Another limitation of the system is its reliance on the use of a multicopy plasmid to carry the reporter gene, which could lead, in some cases, to the titration of regulatory factors required for efficient promoter expression.
Among the nine in vivo-inducible loci identified, we found the promoter of plcA, encoding PI-PLC. The fused fragment contained both the plcA gene preceded by its promoter and the promoter of prfA. In perfect agreement with this result, it has very recently been demonstrated that the promoter for plcA was predominantly activated within the phagosomal compartment while the levels of prfA transcripts present in intracellular bacteria remained low (4). We then identified the promoter of a gene encoding a protein belonging to the family of N-acetylglucosamine epimerases (yvyH [Fig. 4]). In B. subtilis, YvyH is likely involved in the synthesis of the ManNAc-containing linkage unit between peptidoglycan and glycerol teichoic acid. In S. aureus, cap5p-encoded UDP-GlcNac2-epimerase enzymatic activity was demonstrated recently (15). This enzyme, which converts UDP-GlcNAc to N-acetylmannosamine, is involved in capsule biosynthesis. While S. aureus possesses an additional gene, mnaA, encoding UDP-GlcNac2-epimerase (15), no additional YvyH homologue could be found (by tBLASTn search) in the Listeria genome database. Another inducible promoter was located upstream of an ORF encoding 104 residues having some similarities to a putative lipoprotein. The kinetics of bacterial growth in organs demonstrated that the yvyH, plcA, and orf104 promoters allowed the in vivo-inducible expression of hly-encoded LLO (Fig. 5). Induced expression of hly under the control of the yvyH promoter was higher than under those of plcA (and prfA) and orf104. At this stage, the difference in virulence among the three constructs cannot be attributed to a difference in promoter strength and might simply be due to a less favorable positioning of the promoter sequence with respect to hly in the construct. Further characterization of the in vivo-inducible locus yvyH will be undertaken to determine its role in bacterial virulence. Five additional fusions with the in vivo-inducible phenotype were also identified. In two cases, the fragments fused to hly corresponded to the proximal portions of genes. However the polarity of transcription was in the orientation opposite to that of hly. We checked whether there was a divergently transcribed gene immediately upstream of these two promoters. In both cases, in the Listeria chromosome, the preceding gene was in the same orientation. This type of fusion has already been obtained repeatedly with other IVET systems (37), and it was speculated that such fusions might generate antisense transcripts acting to downregulate the in vivo expression of the corresponding genes. Interestingly, one of the two sequences corresponds to a PhoR homologue, a member of the two-component regulatory system PhoP-PhoR involved in the regulation of alkaline phosphatase genes in response to environmental signals. In B. subtilis, it has been shown recently (28) that the PhoP-PhoR system is involved in the control of the biosynthesis of teichoic acid, a key component of the cell walls of gram-positive bacteria. Thus, regulation of PhoR expression in vivo might be relevant for bacterial adaptation to intracellular life. In four cases, no ORF could be identified, but putative promoters were predicted within the fused sequences.
In summary, we showed that the hly-based promoter trap constituted a dual system to identify both constitutive and in vivo-inducible promoters in L. monocytogenes. This study was not exhaustive and could be extended in the future, for example, by monitoring the activity of predicted in silico promoters.
ACKNOWLEDGMENTS
We thank T. Chakraborty (Giesen, Germany) for the gift of EGDΔhly. We thank Colin Tinsley for careful reading of the manuscript.
This work was supported by CNRS, INSERM, and University Paris V.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1990. [Google Scholar]
- 3.Berche P. Bacteremia is required for invasion of the murine central nervous system by Listeria monocytogenes. Microb Pathog. 1995;18:323–336. doi: 10.1006/mpat.1995.0029. [DOI] [PubMed] [Google Scholar]
- 4.Bubert A, Sokolovic Z, Chun S K, Papatheodorou L, Simm A, Goebel W. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol Gen Genet. 1999;261:323–336. doi: 10.1007/pl00008633. [DOI] [PubMed] [Google Scholar]
- 5.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty T, Leimeister-Wachter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J Bacteriol. 1992;174:568–574. doi: 10.1128/jb.174.2.568-574.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu L, Burgum A, Kolodrubetz D, Holt S C. The 46-kilodalton-hemolysin gene from Treponema denticola encodes a novel hemolysin homologous to aminotransferases. Infect Immun. 1995;63:4448–4455. doi: 10.1128/iai.63.11.4448-4455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossart P, Vicente M F, Mengaud J, Baquero F, Perez-Diaz J C, Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989;57:3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darji A, Chakraborty T, Niebuhr K, Tsonis N, Wehland J, Weiss S. Hyperexpression of listeriolysin in the nonpathogenic species Listeria innocua and high yield purification. J Biotechnol. 1995;43:205–212. doi: 10.1016/0168-1656(95)00138-7. [DOI] [PubMed] [Google Scholar]
- 10.Gaillard J L, Berche P, Sansonetti P. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect Immun. 1986;52:50–55. doi: 10.1128/iai.52.1.50-55.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guzman C A, Rohde M, Chakraborty T, Domann E, Hudel M, Wehland J, Timmis K N. Interaction of Listeria monocytogenes with mouse dendritic cells. Infect Immun. 1995;63:3665–3673. doi: 10.1128/iai.63.9.3665-3673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathariou S, Metz P, Hof H, Goebel W. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J Bacteriol. 1987;169:1291–1297. doi: 10.1128/jb.169.3.1291-1297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kili A O, Herzberg M C, Meyer M W, Zhao X, Tao L. Streptococcal reporter gene-fusion vector for identification of in vivo expressed genes. Plasmid. 1999;42:67–72. doi: 10.1006/plas.1999.1408. [DOI] [PubMed] [Google Scholar]
- 15.Kiser K B, Bhasin N, Deng L, Lee J C. Staphylococcus aureus cap5P encodes a UDP-N-acetylglucosamine 2-epimerase with functional redundancy. J Bacteriol. 1999;181:4818–4824. doi: 10.1128/jb.181.16.4818-4824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler S, Bubert A, Vogel M, Goebel W. Expression of the iap gene coding for protein p60 of Listeria monocytogenes is controlled on the posttranscriptional level. J Bacteriol. 1991;173:4668–4674. doi: 10.1128/jb.173.15.4668-4674.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 18.Kutsukake K, Okada T, Yokoseki T, Iino T. Sequence analysis of the flgA gene and its adjacent region in Salmonella typhimurium, and identification of another flagellar gene, flgN. Gene. 1994;143:49–54. doi: 10.1016/0378-1119(94)90603-3. [DOI] [PubMed] [Google Scholar]
- 19.Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc Natl Acad Sci USA. 1990;87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 21.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 22.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland J A, Milon G, Cossart P. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol Microbiol. 1991;5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 24.Moors M A, Levitt B, Youngman P, Portnoy D A. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect Immun. 1999;67:131–139. doi: 10.1128/iai.67.1.131-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park S F, Stewart G S. High efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene. 1990;94:129–132. doi: 10.1016/0378-1119(90)90479-b. [DOI] [PubMed] [Google Scholar]
- 26.Portnoy D A, Jacks P S, Hinrichs D J. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poyart C, Trieu-Cuot P. A broad host-range mobilizable shuttle vector for the construction of transcriptional fusions to β-galactosidase in Gram-positive bacteria. FEMS Microbiol Lett. 1997;156:193–198. doi: 10.1111/j.1574-6968.1997.tb12726.x. [DOI] [PubMed] [Google Scholar]
- 28.Qi Y, Hulett F M. Role of Pho-P in transcriptional regulation of genes involved in cell wall anionic polymer biosynthesis in Bacillus subtilis. J Bacteriol. 1998;180:4007–4010. doi: 10.1128/jb.180.15.4007-4010.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Expression of cloned genes in Escherichia coli. In: Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 17.37–17.41. [Google Scholar]
- 30.Sheehan B, Kocks C, Dramsi S, Gouin E, Klarsfeld A D, Mengaud J, Cossart P. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr Top Microbiol Immunol. 1994;192:187–216. doi: 10.1007/978-3-642-78624-2_9. [DOI] [PubMed] [Google Scholar]
- 31.Staib P, Kretschmar M, Nichterlein T, Kohler G, Michel S, Hof H, Hacker J, Morschhauser J. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol Microbiol. 1999;32:533–546. doi: 10.1046/j.1365-2958.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 32.Suh J W, Boylan S A, Oh S H, Price C W. Genetic and transcriptional organization of the Bacillus subtilis spc-α region. Gene. 1996;169:17–23. doi: 10.1016/0378-1119(95)00757-1. [DOI] [PubMed] [Google Scholar]
- 33.Suh J W, Boylan S A, Thomas S M, Dolan K M, Oliver D B, Price C W. Isolation of a secY homologue from Bacillus subtilis: evidence for a common protein export pathway in eubacteria. Mol Microbiol. 1990;4:305–314. doi: 10.1111/j.1365-2958.1990.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 34.Tilney L G, Portnoy D A. Actin filaments and the growth, movement and spread of the intracellular bacterial parasite Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjalsma H, Bolhuis A, van Roosmalen M L, Wiegert T, Schumann W, Broekhuizen C P, Quax W J, Venema G, Bron S, van Dijl J M. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in Gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdivia R H, Falkow S. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol Microbiol. 1996;22:367–378. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wuenscher M D, Kohler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]