Abstract
Purpose:
To investigate the antitumor activity of a mitochondrial-localized HSP90 inhibitor, Gamitrinib, in multiple glioma models, and to elucidate the antitumor mechanisms of Gamitrinib in gliomas.
Experimental Design:
A broad panel of primary and temozolomide (TMZ)-resistant human glioma cell lines were screened by cell viability assays, flow cytometry, and crystal violet assays to investigate the therapeutic efficacy of Gamitrinib. Seahorse assays were used to measure the mitochondrial respiration of glioma cells. Integrated analyses of RNA sequencing (RNAseq) and reverse phase protein array (RPPA) data were performed to reveal the potential antitumor mechanisms of Gamitrinib. Neurospheres, patient-derived organoids (PDO), cell line–derived xenografts (CDX), and patient-derived xenografts (PDX) models were generated to further evaluate the therapeutic efficacy of Gamitrinib.
Results:
Gamitrinib inhibited cell proliferation and induced cell apoptosis and death in 17 primary glioma cell lines, 6 TMZ-resistant glioma cell lines, 4 neurospheres, and 3 PDOs. Importantly, Gamitrinib significantly delayed the tumor growth and improved survival of mice in both CDX and PDX models in which tumors were either subcutaneously or intracranially implanted. Integrated computational analyses of RNAseq and RPPA data revealed that Gamitrinib exhibited its antitumor activity via (i) suppressing mitochondrial biogenesis, OXPHOS, and cell-cycle progression and (ii) activating the energy-sensing AMP-activated kinase, DNA damage, and stress response.
Conclusions:
These preclinical findings established the therapeutic role of Gamitrinib in gliomas and revealed the inhibition of mitochondrial biogenesis and tumor bioenergetics as the primary antitumor mechanisms in gliomas.
Introduction
Gliomas are the most commonly occurring intrinsic brain tumors, with an estimated 100,000 newly diagnosed cases each year worldwide (1). Moreover, there are approximately 6 cases per 100,000 individuals diagnosed each year in the United States (2). In nearly 50% of patients with glioma, the disease presents as a WHO grade IV, glioblastoma (GBM), which is the most aggressive subtype of gliomas (2). Although survival has been improved due to addition of temozolomide (TMZ) to the standard-of-care treatment for newly diagnosed gliomas, the 5-year survival rate is still limited (3, 4). Other therapies, including a VEGF antibody bevacizumab and the recombinant oncolytic poliovirus PVSRIPO, have been administered in patients with recurrent gliomas, but efficacy remains quite limited (5, 6). Notably, there have been no new drugs approved by the FDA for patients with primary and recurrent gliomas since bevacizumab was approved in 2009. Therefore, it is urgent to identify novel therapies for gliomas.
Metabolic reprogramming is a hallmark of gliomas, which represents a shift of intracellular metabolism to meet energetic demands of rapidly proliferating tumor cells (7). Like many other cancers, metabolic reprogramming in glioma cells is mainly characterized as aerobic glycolysis (known as the Warburg Effect; ref. 8). With that being said, mitochondrial processes, including oxidative phosphorylation (OXPHOS), continue to be the main source of tumor-derived ATP, which exerts a critical impact toward tumor energy metabolism, tumor cell proliferation, and treatment resistance (9, 10).
Heat Shock Protein 90 (HSP90) proteins are highly conserved molecular chaperones that function predominantly to promote the folding of neo-synthesized proteins and prevent their aggregation (11). Accordingly, a variety of diverse client proteins depends on HSP90 proteins for their activity and stability, which involve cellular functions that are crucial for cell signaling, cycle, and differentiation (11-13). Previous studies have investigated the functions of HSP90 proteins and their related homologue, TNF receptor-associate protein-1 (TRAP-1) localized to the mitochondria of tumor (though not most normal) cells (9, 10, 14-16). Their roles are critical for regulating the metabolic switch between OXPHOS and aerobic glycolysis in response to energetic changes that occur in both tumors and tumor microenvironment (11, 17, 18). Previous studies have shown that HSP90 proteins located in mitochondria are essential for regulating bioenergetics in tumor cells but not normal cells (10, 16), making them therapeutic candidates in cancers. Gamitrinib (GA mitochondrial matrix inhibitor) is a mitochondrial-localized HSP90 inhibitor, which was a derivative from the HSP90 inhibitor 17-(allylamino)-17-demethoxy-geldanamycin (17-AAG) and contained another benzoquinone ansamycin backbone (16). This design allows Gamitrinib to specifically target the mitochondrial HSP90 but not cytosolic HSP90, which makes it possible to selectively affect tumor cells, while simultaneously avoiding normal tissues (10, 16). Gamitrinib has shown the antitumor activity in preclinical models for multiple cancers, including prostate cancer, melanoma, lung cancer, colon cancer, breast cancer, and GBM (9, 10, 14-16, 19). Kang and colleagues (14) first described the efficacy of Gamitrinib in a GBM cell line U87MG. Subsequent studies reported the therapeutic efficacy of Gamitrinib in other GBM cell lines, including T98G, LN229, U251MG, and A172 (9, 10, 20, 21). Unfortunately, studies that systematically investigate the effect of Gamitrinib are lacking and the antitumor mechanisms in gliomas are still mostly unknown.
Toward this goal, we investigated the therapeutic efficacy of Gamitrinib as a single agent in a broad panel of 17 primary human glioma cell lines, 6 TMZ-resistant glioma cell lines, 4 neurospheres, 3 patient-derived organoids (PDO), 1 cell line–derived xenograft (CDX), and 1 patient-derived xenograft (PDX) models. We also explored the therapeutic efficacy of Gamitrinib combined with standard-of-care therapies in glioma cell lines, neurospheres and PDOs. In addition, we performed RNA sequencing (RNAseq) and reverse phase protein array (RPPA) in glioma cells treated with or without Gamitrinib. This demonstrated that Gamitrinib exhibited its antitumor activity via inhibiting mitochondrial biogenesis and tumor bioenergetics, blocking cell-cycle progression and inducing DNA damage. In this study, we established the therapeutic role of Gamitrinib in gliomas and provided strong scientific rationale for propelling this mitochondrial-directed HSP90 inhibitor into clinical trials.
Materials and Methods
Human subjects
Patients’ tissues and blood were collected following ethical guidelines after written informed consent was obtained from each patient. The study protocols have been approved by the Institutional Review Boards (IRB) of the University of Texas Southwestern Medical Center (IRB#: STU022011-070) and West China Hospital (IRB#: 2022363). The clinical information and molecular profiles of included patients are shown in Supplementary Table S2.
Cell lines
The glioma cell line, U251MG, was kindly provided by Dr. Yiping He (Department of Pathology, Duke University, Durham, NC). The other glioma cell lines (A172, M059K, H4, Hs683, M059J, LN18, LN229, U87MG, U118MG, U138MG, DBTRG-05MG, and T98G) used in this study were obtained from the ATCC. Dr. Darell Bigner (Duke University, Durham, NC) kindly provided 13.0302 neurospheres and patient-derived glioma cell lines (D2363PXA, D645PXA, D2159MG, and D2224MG). TS-603 neurospheres were kindly provided by Dr. Hai Yan (Duke University, Durham, NC) who received the permission from MSKCC. The normal human astrocytes (NHA) were ordered from Duke Cell Culture Facility. U87MG, T98G, TS-603, and 13.0302 neurospheres grew in stem cell media (Stemcell Technologies, 5751) supplemented with proliferation supplement (Stemcell Technologies, 5751), 20 ng/mL H-EGF (Stemcell Technologies, 78006), 10 ng/mL H-bFGF (Stemcell Technologies, 78003), and 2 μg/mL Heparin (Stemcell Technologies, 7980). The rest of glioma cell lines were cultured in DMEM (Thermo Fisher Scientific, 11995073) supplemented with 10% FBS (Hyclone, SH30071.03). Astrocytes were cultured in Astrocyte Growth Medium (Sigma, 821–500). Cells were incubated in a humidified atmosphere with 5% CO2 at 37° C. For all cell lines used in this study, cell line authentication was performed by short tandem repeat fingerprinting and Mycoplasma testing was performed by use of the MycoAlert PLUS Mycoplasma Detection Kit (Lonza, LT07–703). The number of passages between thawing and use in the previously described experiments was less than 5.
Chemicals and reagents
Gamitrinib (HY-102007A) was purchased from MedChemExpress. Temozolomize (TMZ; 14163), Sodium pyruvate (S8636), Ethidium bromide (E1385), and Phenformin (P7045) were provided by MilliporeSigma. IACS-010759 (407917) was purchased from MedKoo Biosciences. Uridine (20300) and 17-AGG (11039) were brought from Cayman Chemical. CellTrace Violet (C34557) was purchased from Thermo Fisher Scientific. Propidium iodide (PI) solution (25535-16-4) was purchased from Alfa Aesar, and PSVue 643 (P-1006) was provided by Molecular Targeting Technologies. MitoTracker Green FM (9074) was ordered from Cell Signaling Technology.
Cell viability assay
Cells were seeded into 96-well plates with 3,000–5,000 cells/well for 24 hours before treatment with Gamitrinib at titrated doses for 72 hours. Three replicates were assigned for each dose condition. Ten μL Cell Count Kit-8 (APEBIO, K1018) reagent was added into each well and incubated with cells for 3 hours at 37°C. Then absorbance was measured at 450 nm by Tecan plate reader. Dose–response curves were generated and IC50 values were calculated with GraphPad Prism. Two independent experiments have been performed.
Apoptosis and cell death assay
Adherent cells were harvested with 0.25% Trypsin-EDTA (Gibco, 25200072), and washed once with 1x PBS (Gibco, 10010–023). Cells were then pelleted and stained with PSVue 643 (MTTI, P-1006) at 5 μmol/L and PI (Alfa Aesar, 25535–16–4) at 10 μg/mL diluted in 1x PBS for 5 minutes in the dark. Cells were then immediately analyzed by FACS using a BD flow cytometer and at least 5,000 cells per sample were acquired. Two to three independent experiments have been performed.
CellTrace violete assay
Adherent M059K, LN229, D2363PXA, and D2224MG cells were harvested with 0.25% Trypsin-EDTA (Gibco, 25200072), and washed once with 1x PBS (Gibco, 10010–023). Cells were labeled with CellTrace Violet (Invitrogen, C34557) according to the manufacturer’s protocol and were allowed to adhere to tissue culture plates overnight following 5 μmol/L Gamitrinib treatment. Adherent cells were collected and detected the fluorescent intensity of CellTrace Violet by FACS at days 0, 1, 3, and 5. FACS was performed by using a BD flow cytometer and at least 5,000 cells per sample were acquired. Two or three independent experiments were performed.
Immunofluorescence assay
Adherent M059K and LN229 cells were harvested with 0.25% Trypsin-EDTA (Gibco, 25200072), washed once with 1x PBS (Gibco, 10010–023), and seeded into a 24-well plate. Cells were treated with a titrated dose of Gamitrinib after adherence. Seventy-two hours after treatment, cells were fixed with 4% paraformaldehyde (Thermo Fisher Scientific, 28908) for 15 minutes at room temperature. Cells were rinsed three times with 1x PBS for 5 minutes each, permeabilized with PBS containing 0.3% Triton X-100 (Thermo Fisher Scientific, 9002–93–1) for 10 minutes, and blocked with 5% normal goat serum (Cell Signaling Technology, 5425) for 1 hour. Then, cells were incubated with anti-Ki67 fluorescent antibody (Cell Signaling Technology, 11882) overnight at 4°C in the dark. Cells were rinsed three times with 1x PBS for 5 minutes each, stained with DAPI (Cell Signaling Technology, 8961) for 5 minutes, and then washed four times with 1x PBS. Images were obtained by a fluorescence microscope. Two independent experiments have been performed.
Western blotting and antibodies
Protein was extracted from cells cultured in 6-well plates using RIPA lysis buffer (Thermo Fisher Scientific, 89900) with protease inhibitors (Thermo Fisher Scientific, 78440). A total of 30-μg protein lysates was separated by 4%–12% NuPage Bis-Tris gels (Thermo Fisher Scientific, NP0336) and then transferred to polyvinylidene fluoride membranes (Bio-Rad, 1620264). The membranes were blocked with 5% BSA (Sigma-Aldrich, A7906–100G), probed with primary antibodies overnight at 4°C, and then incubated with the relevant secondary antibody for 1 hour at room temperature. Bands were visualized using ECL (Thermo Fisher Scientific, 34080) with Bio-Rad imaging system. The antibodies of anti-PARP (9532), anti-Rb (9309), anti-pRbSer807/811 (8516), anti-γH2AX (9718), anti-TFAM (8076), anti-AMPKα (5831), anti-pAMPKαThr172 (2535), anti-ERRα (13826), anti-VDAC (4661), anti-PHB1 (2426), anti-HSP60 (12165), anti-NRF2 (12721), anti-SDHA (11998), anti–β-actin (12620), anti–rabbit-IgG-HRP (7074), and anti–mouse-IgG-HRP (7076) were purchased from Cell Signaling Technology. The antibodies of anti-PGC1α (66369), anti-UQCRC1 (21705), anti-ATP5A1 (14676), anti-COXIV (11242), anti-NDUFB8 (14794), and anti-TUFM (26730) were purchased from Proteintech. Two or three independent experiments were performed.
RT-qPCR
Total RNA was extracted from cultured cells by the miRNeasy Mini Kit (Qiagen, 217004) according to the manufacturer’s instructions. One mg of total RNA was reverse transcribed into complementary DNA(cDNA) using the cDNA synthesis kit (Thermo Fisher Scientific, K1651). Amplification of cDNA product was performed using specific primers with the Fast SYBR Green Master Mix (Thermo Fisher Scientific, 4385610) on a real-time PCR detection system (Bio-Rad). Samples were analyzed in triplicate, and β-tubulin levels were used for normalization. The following are the primers sequences for RT-qPCR: SDHB: forward primer, 5′-AAGCATCCAATACCATGGGG-3′, and reverse primer, 5′-TCTATCGATGGGACCCAGAC-3′; COX7B: forward primer, 5′-CTTGGTCAAAAGCGCACTAAATC-3′, and reverse primer, 5′-CTATTCCGACTTGTGTTGCTACA-3′; ATP5B: forward primer, 5′-CAAGTCATCAGCAGGCACAT-3′, and reverse primer, 5′-TGGCCACTGACATGGGTACT-3′; NDUFA8: forward primer, 5′-CTCCTTGTTGGGCTTATCACA-3′, and reverse primer, 5′-GCCCACTCTAGAGGAGCTGA-3′; UQCRCB: forward primer, 5′-ACTGGGGTTAATGCGAGATG-3′, and reverse primer, 5′-GTCCAGTGCCCTCTTAATGC-3′; and β-tubulin: forward primer, 5′-TGGACTCTGTTCGCTCAGGT-3′, and reverse primer, 5′-TGCCTCCTTCCGTACCACAT-3′.
Mitochondrial DNA copy-number detection
Genomic DNA (gDNA) was extracted from cultured cells using DNA Mini Kit (Qiagen, 51304) following the manufacturer’s instructions. Twenty ng of gDNA was used to conduct qPCR. Mitochondrial DNAs (mtDNA) were amplified using specific primers with the Fast SYBR Green Master Mix (Thermo Fisher Scientific, 4385610). The following are the primers sequence for qPCR: mtDNA: forward primer, 5′-CACCCAAGAACAGGGTTTGT-3′, and reverse primer, 5′-TGGCCATGGGTATGTTGTTA-3′; and β2-microglobulin: forward primer, 5′-TGCTGTCTCCATGTTTGATGTATCT-3′, and reverse primer, 5′-TCTCTGCTCCCCACCTCTAAGT-3′. Two independent experiments have been performed.
Crystal violet staining
Cells were plated into 12-well tissue culture plates at a density of 2,500 cells/well as biological duplicates in drug-free medium. Once cells were adherent, cells were treated with drug, including TMZ, IACS-010759, Phenformin, and Gamitrinib. The medium supplemented with drugs was refreshed every 3 or 4 days for 8–14 days. Cells in plates were then stained with methanol containing 0.05% crystal violet for 30 minutes at room temperature. After extensive washing with distilled water, cells were air-dried and subjected to image acquisition using Bio-Rad Imager. Two or three independent experiments were performed.
Neurosphere formation and treatment
T98G, U87MG, TS-603, and 13.0302 cells were cultured in neurobasal medium (Stemcell Technologies, 5751) containing 10% proliferation supplement (Stemcell Technologies, 5751), 20 ng/mL H-EGF (Stemcell Technologies, 78006), 10 ng/mL H-bFGF (Stemcell Technologies, 78003), and 2 μg/mL Heparin (Stemcell Technologies, 7980). Once neurospheres were formed, they were dissociated by Accutase (VWR, 490007–741) into single cells. Single cells were seeded into 12-well plates (5,000 cells/well) and treated with control medium or Gamitrinib at 1, 2.5, and 5 μmol/L for 12 days. Phase contrast images of neurospheres were acquired by ECHO Revolve Microscope at days 3, 6, 9, and 12. At day 12, neurospheres were stained using Live Dead Cell Viability Assay Kit (Sigma-Aldrich, CBA415) and imaged by ECHO Revolve Microscope. Two independent experiments were performed.
Seahorse assay
Mitochondrial respiration was measured with Agilent Seahorse XF96 FluxPak according to the manufacturer’s instructions. Adherent cells were treated with different chemicals for 48 hours, and then 1.5–2×104 cells were plated in each well of a Seahorse XF96 cell culture plate (80-mL volume) with the same treatment overnight. The culture media were then exchanged with Agilent Seahorse XF Media and equilibrated in a 37°C incubator without CO2 for 1 hour before the assay. Cellular oxygen consumption was monitored at basal condition and after injection of oligomycin, FCCP, and Rotenone plus Antimycin A. Two independent experiments were performed.
Whole exome sequencing (WES) and RNAseq
Genomic DNA was extracted from patients’ tissues and blood by the DNeasy Blood & Tissue Kit (Qiagen, 69504). DNA sequencing was performed as previously described (22). Total RNA was purified from M059K, LN229, U87MG, and A172 cells by the miRNeasy Mini Kit (Qiagen, 217004) according to the manufacturer’s instructions. The complemental DNA (cDNA) libraries were constructed and sequenced by Genewiz. Sequencing was carried out using a 150 paired-end configuration on the Illumina sequencing platform (HiSeq 2500).
RPPA
The RPPA assay was performed by the MD Anderson Cancer Center core facility using 80-μg protein per sample. The RPPA data can be accessed in Supplementary Table S1. To determine a pathway score, the protein Z-scores were calculated and all positively associated predictors were summed minus the predictors that are negatively associated with the pathway. The result was then divided by the sum of the weight of each predictor (23, 24).
PDOs and staining
PDOs were treated with either DMSO, 2.5, or 5 μmol/L gamitrinib for 6 days. On day 6, organoids were either fixed in 10% formalin for histopathological analysis or harvested for protein or RNA. For protein harvest, organoids were washed in ice-cold PBS and lysed in RIPA lysis buffer containing protease inhibitor (Sigma-Aldrich, 11836153001) and phosphatase inhibitor (Sigma-Aldrich, 04906845001). Lysed organoids were homogenized using a TissueLyser (Qiagen) for one minute. Homogenized organoids were centrifuged at 12,000 × g for 15 minutes at 4°C and supernatant was stored at −80° C for subsequent protein analysis. For RNA harvest, organoids were washed in ice-cold PBS and homogenized in 1 mL per 100 mg TRizol (Thermo Fisher Scientific, 10296028) using a TissueLyser (Qiagen) for one minute. RNA was isolated from homogenized organoids using an RNeasy Mini Kit (Qiagen, 74104) and stored at −80°C for subsequent analysis. PDOs’ FFPE slides were stained with hematoxylin–eosin (H&E) first and then immunohistochemistry (IHC) for anti-Ki67 according to the manufacturer’s instructions. All stained tissues were blindly evaluated by a pathologist at UT Southwestern Medical Center.
CDX and PDX models
The study protocol of animals in this study was approved by Animal Ethics Committee of West China Hospital (No. 20211298A). All animal procedures followed the Institutional Animal Care and Use Committee’s guidelines.
In the CDX models, female athymic mice (6–8 weeks old) were subcutaneously injected with 7 × 106 U87MG cells. When tumors grew to an average of 100 mm3, mice were randomly assigned to be injected intraperitoneally with DMSO-PBS (vehicle), Gamitrinib at 10 mg/kg, TMZ at 30 mg/kg, and Gamitrinib 10 mg/kg plus TMZ at 30 mg/kg every other day. Tumor lengths and widths were measured every other day with hand-held vernier calipers. Tumor volumes were calculated with the following formula: volume = (length × width × width)/2. Mice were sacrificed after 10-days treatment of Gamitrinib, and tumors were harvested from mice and analyzed by histology.
In the PDX models, female NCG mice (6–8 weeks old) were subcutaneously implanted with GBM PDX tissue. After tumor establishment with an average volume of 100 to 200 mm3, mice were randomly assigned to be injected intraperitoneally with vehicle or Gamitrinib at 10 mg/kg every other day (5 mice in each group). Subcutaneous tumors were measured every other day with hand-held vernier calipers. After 10-days treatment of Gamitrinib, mice were sacrificed, and tumors were harvested for histology analysis.
For orthotopic GBM xenograft models, 6- to 8-week-old female athymic mice were stereotactically injected with 1 × 105 U87MG or PDX cells into the right cerebral striatum. One week (U87MG) or 3 weeks (GBM PDX#823) after orthotopic implantation, mice were randomly assigned to be injected intraperitoneally with vehicle or Gamitrinib at 10 mg/kg every other day. Animal survival was calculated with GraphPad Software (version 8.0). The endpoint of experiment was a moribund state or death of the animal in accordance with the Institutional Animal Welfare Regulations. At the end of the experiment, brains were harvested and analyzed by histology.
Statistical analysis
The two-sided Welch t test was conducted to compare control and treatment samples. Heatmap was used to visualize data. Hierarchical clustering used “Canberra” as distance measurement method and “Complete” as the agglomeration method. Survival analysis was conducted with the Kaplan–Meier method. The log-rank test was used to determine statistical significance associated with survival analysis. The quality of Fastq sequence data were checked by Fastqc (v0.11.9; ref. 25). Then the reads were aligned to human genome GRCH38 by STAR (v2.7.3a; ref. 26). The quality of BAM files were checked by Picard CollectRnaSeqMetrics (v2.20.8; ref. 27). Then BAM files were sorted by name using Samtools (v1.10; ref. 28). Counting was performed by HTSeq (v0.11.2; ref. 29). DESeq2 (v1.28.1; ref. 30) in R (v4.0.2) was used for differential expression analysis. Then the pre-ranked GSEA (gene set enrichment analysis; gene score enrichment analysis) was performed by using fgsea (v1.14.0; ref. 31) in R.
Data availability statement
RNAseq data were deposited into Gene Expression Omnibus (accession number GSE165819 for M059K and LN229 cells; accession number GSE180872 for U87MG and A172 cells). WES data will be available upon request. RPPA data are available in Supplementary Table S1.
Results
Gamitrinib inhibits cell viability and induces cell death in glioma cell lines
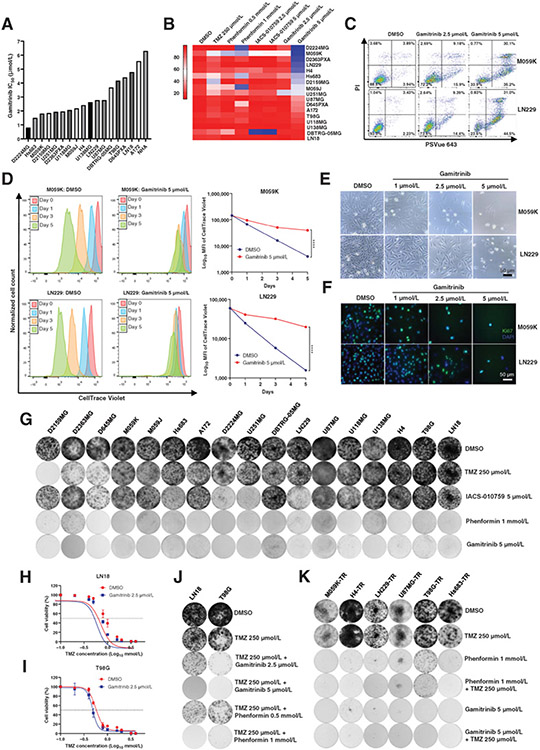
To investigate the therapeutic efficacy of Gamitrinib in gliomas, we first performed cell viability assay in a panel of 17 human glioma cell lines and calculated IC50 value of Gamitrinib when administered for 72 hours (Fig. 1A). The NHAs were included as a control. The median IC50 value to Gamitrinib for glioma cell lines was 2.46 μmol/L as opposed to the IC50 value being more than 6 μmol/L for NHA that was the most insensitive cell line (Fig. 1A). To further evaluate the impact of Gamitrinib on cell viability, we performed a FACS-based screening of 17 glioma cell lines to determine cell apoptosis and cell death (Fig. 1B). TMZ is a current standard-of-care therapy for patients with glioma (32). IACS-010759 and Phenformin are OXPHOS inhibitors that have shown antitumor activity in gliomas (33-35). Therefore, these inhibitors were chosen as controls. Cell lines were treated with TMZ at 250 μmol/L, IACS-010759 at 2.5 and 5 μmol/L, Phenformin at 0.5 and 1 mmol/L, and Gamitrinib at 2.5 and 5 μmol/L for 96 hours, then analyzed with flow cytometry (Fig. 1B). Among 17 glioma cell lines, D2224MG, M059K, D2363PXA, LN229, and H4 cell lines were more sensitive to Gamitrinib with cell viability lower than 30%. T98G, U118MG, U138MG, DBTRG-05MG, and LN18 were relatively insensitive to Gamitrinib with cell viability greater than 80% (Fig. 1B). Overall, these results indicated that Gamitrinib is effective in a subset of glioma cell lines by significantly inhibiting their cell viabilities.
Figure 1.
Therapeutic efficacy of Gamitrinib in glioma cells. A, The IC50 value of Gamitrinib for 17 human glioma cell lines and normal human astrocyte (NHA). B, Heatmap of cell viability for 17 glioma cell lines following treatment with the DMSO, TMZ at 250 μmol/L, Phenformin at 0.5 and 1 mmol/L, IACS-010759 at 2.5 and 5 μmol/L, and Gamitrinib at 2.5 and 5 μmol/L for 4 days. C, FACS data of M059K and LN229 cells. Cells were treated with indicated drugs and doses for 4 days and stained with PI and PSVue 643, and then analyzed by FACS. D, M059K and LN229 cells were labeled with CellTrace Violet and then treated with Gamitrinib at 5 μmol/L. The fluorescence intensity of CellTrace Violet was measured by FACS at days 0, 1, 3, and 5. Data were representative of 3 biological replicates. E, Phase contrast images of M059K and LN229 cells treated with Gamitrinib at 0, 1, 2.5, and 5 μmol/L for 4 days. F, Immunofluorescence staining images of M059K and LN229 cells treated with or without Gamitrinib at 5 μmol/L for 4 days. Blue indicated the DAPI staining whereas green indicated the Ki67 staining. G, Crystal violet staining images for 17 glioma cell lines. Cells were treated with DMSO, TMZ at 250 μmol/L, Phenformin at 1 mmol/L, IACS-010759 at 5 μmol/L, and Gamitrinib at 5 μmol/L for 8–14 days. Then cells were fixed and stained with crystal violet. H and I, The IC50 value of TMZ for LN18 and T98G cell lines treated with DMSO or Gamitrinib at 2.5 μmol/L. J and K, Crystal violet staining images for LN18 and T98G cell lines (J) or TMZ-resistant (TR) cell lines (K) treated with indicated drugs for 8–14 days; ****, P < 0.0001.
Subsequently, we focused on two representative cell lines, M059K and LN229. We treated M059K and LN229 with Gamitrinib at titrated doses, and observed efficacy of Gamitrinib in inducing cell apoptosis and death in a dose-dependent manner (Fig. 1C; Supplementary Fig. S1A and S1B). To further determine whether Gamitrinib inhibits cell division of glioma cells, we labeled M059K and LN229 cells with a cell membrane dye, CellTrace Violet, then treated them with DMSO or Gamitrinib at 5 μmol/L, and monitored the fluorescence intensity over 120 hours (Fig. 1D). In line with the inhibition of cell viability, Gamitrinib significantly inhibited cell division of these two cell lines (Fig. 1D). To further investigate the effect of Gamitrinib on cell proliferation, we treated these two glioma cell lines with Gamitrinib at titrated doses for 72 hours. Gamitrinib resulted in a significant reduction of cell numbers in a dose-dependent manner in both M059K and LN229 cell lines (Fig. 1E). Immunofluorescence staining with Ki67, a proliferative marker, demonstrated decreased expression in both M059K and LN229 cell lines treated with Gamitrinib in a dose-dependent manner (Fig. 1F). Collectively, these results suggested that Gamitrinib effectively induces cell apoptosis and death, and inhibits cell division and proliferation in glioma cells.
To further gain insights into the long-term efficacy of Gamitrinib, we treated the same panel of 17 glioma cell lines with TMZ at 250 μmol/L, IACS-010759 at 5 μmol/L, Phenformin at 1 mmol/L, and Gamitrinib at 5 μmol/L for 8 to 14 days followed by crystal violet staining (Fig. 1G). In line with experiments conducted for a short term, most of glioma cells were intrinsically resistant to TMZ and IACS-010759. Interestingly, long-term treatment of glioma cells with Phenformin or Gamitrinib impaired cell proliferation in all glioma cell lines (Fig. 1G).
Furthermore, we chose two representative cell lines, LN18 and T98G, which were intrinsically resistant to TMZ to explore the efficacy of the combination therapy of TMZ plus Gamitrinib (Fig. 1H and I). We treated these two cell lines with DMSO or Gamitrinib at 2.5 μmol/L plus TMZ at titrated concentrations for 72 hours and then assessed cell viability (Fig. 1H and I). We observed that Gamitrinib enhanced cell sensitivity to TMZ in both LN18 (IC50 893.7 μmol/L vs. 727.9 μmol/L) and T98G (IC50 604.3 μmol/L vs. 505.3 μmol/L) cell lines (Fig. 1H and I). Long-term combination therapy of TMZ plus Gamitrinib also inhibited cell proliferation in both LN18 and T98G cell lines (Fig. 1J). We also generated six TMZ-resistant (denoted as TMZ-R) glioma cell lines in culture and assessed the therapeutic efficacy of monotherapy (Gamitrinib or Phenformin) or combination therapy (TMZ plus Gamitrinib or Phenformin; Fig. 1K). We noticed that cell viabilities of TMZ-R cells were impaired by either monotherapy or combination therapy (Fig. 1K). Taken together, we demonstrated that Gamitrinib is effective in TMZ-R glioma cells, and combination of TMZ plus Gamitrinib might be a potential therapeutic strategy for patients with recurrent glioma.
Gamitrinib exhibits antitumor activity in neurospheres
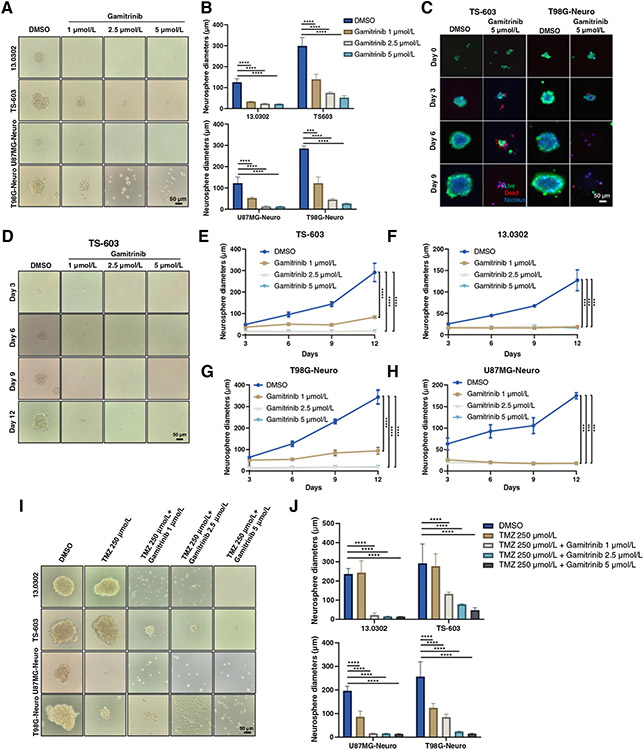
In addition to the adherent cells grown in two-dimensional culture system, we also investigated the therapeutic efficacy of Gamitrinib in neurospheres grown in a three-dimensional culture system. We generated four glioma neurospheres, including 13.0302, TS-603, U87MG, and T98G and treated them with Gamitrinib at titrated doses for 6 days. Interestingly, Gamitrinib, even administered at very low doses, significantly reduced the diameters and inhibited the growth of these four neurospheres (Fig. 2A and B). To further evaluate living and deceased cells within neurospheres, we stained them with Calcein-AM and PI dyes, respectively. We observed that Gamitrinib not only inhibited growth but also induced cell death in neurospheres (Fig. 2C; Supplementary Fig. S1C and S1D). Furthermore, we investigated the impact of Gamitrinib on the formation of neurospheres. Neurospheres were digested into single cells, which were then treated with Gamitrinib at titrated doses. As expected, even a relatively low dose of Gamitrinib at 1 μmol/L significantly prevented the formation of neurospheres (Fig. 2D-H; Supplementary Fig. S1E-S1G). In addition, we tested the efficacy of TMZ plus Gamitrinib at titrated doses in these neurospheres. Interestingly, T98G and U87MG neurospheres were highly sensitive to TMZ administered at 250 μmol/L for 6 days; however, 13.0302 and TS-603 neurospheres were highly resistant to TMZ (Fig. 2I and J). Surprisingly, Gamitrinib significantly enhanced the therapeutic efficacy of TMZ in these neurospheres (Fig. 2I and J).
Figure 2.
Antitumor activity of Gamitrinib in neurospheres. A and B, Phase contrast images of neurospheres that were treated with titrated doses of Gamitrinib for 6 days (A) and diameters of 4 to 5 representative neurospheres were measured (B). C, Immunofluorescence staining images of TS-603 and T98G neurospheres. Cells were treated with Gamitrinib at 5 μmol/L for 9 days and live/dead staining was performed at days 0, 3, 6, and 9. Green indicated live cells; red indicated dead cells; blue indicated nucleus. D, Phase contrast images of TS-603 neurospheres treated with or without Gamitrinib at indicated doses. E–H, Neurosphere growth curves for TS-603 (E), 13.0302 (F), U87MG (G), and T98G (H) treated with or without Gamitrinib at 5 μmol/L. I, Phase contrast images of neurospheres that were treated with indicated drugs for 6 days. J, Diameters of 4 to 5 representative neurospheres; ***, P < 0.001; ****, P < 0.0001.
Taken together, Gamitrinib as a monotherapy demonstrated therapeutic efficacy in both primary and TMZ-R glioma cell lines as well as glioma neurospheres. In continuation, Gamitrinib also enhanced the therapeutic efficacy of TMZ in both glioma cell lines and neurospheres.
Gamitrinib activity requires mtDNA
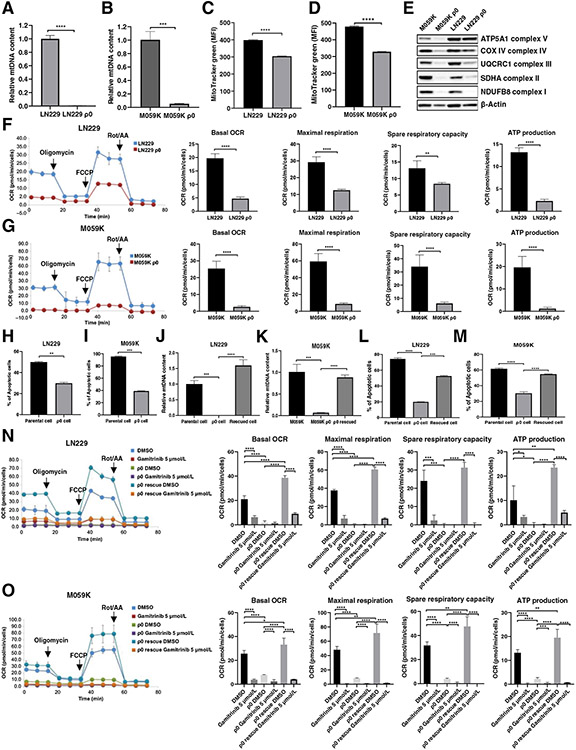
The depletion of mtDNA is usually associated with defective OXPHOS and increased susceptibility to stress. To further ensure the specificity of Gamitrinib, we derived M059K and LN229 ρ0 cell lines in which mtDNA was depleted by chronic exposure to ethidium bromide, pyruvate, and uridine for roughly 12 weeks in culture. We first confirmed the phenotype of depleted mtDNA by determining the relative mtDNA copy number in both parental and ρ0 cells. We observed a significant depletion of mtDNA copy numbers in these two ρ0 cell lines (Fig. 3A and B). Accompanying the depletion of mtDNA, the fluoresce intensity of MitoTracker Red was decreased which measured mitochondrial mass (Fig. 3C and D). Western blotting showed that the expression of OXPHOS complex subunits was decreased in these two ρ0 cells (Fig. 3E). To further elucidate the suppression of mitochondrial metabolism, we performed a mitochondrial stress test for both LN229 and M059K parental and ρ0 cells. As expected, in both cases, M059K and LN229 ρ0 cells demonstrated elevated extracellular acidification rate and a declination of basal oxygen consumption rate (OCR), accompanied by decreases in maximal respiration, spare respiratory capacity, ATP production, and proton leak (Fig. 3F and G; Supplementary Fig. S2A and S2B). These results indicated that ρ0 cells, in which mtDNA was depleted, became less dependent on mitochondrial respiration and reprogrammed the metabolic network toward glycolysis. To test the hypothesis that ρ0 cells are less sensitive to Gamitrinib, we treated both parental and ρ0 cells with Gamitrinib. We observed a significant decrease in the induction of apoptosis and cell death in both M059K and LN229 ρ0 cells treated with Gamitrinib as compared with their parental cells (Fig. 3H and I). Furthermore, we attempted to rescue the phenotype of mtDNA depletion exhibited by two ρ0 cell lines via with drawing ethidium bromide from the culture media for 8 weeks. Upon the restoration of mtDNA of ρ0 cells (Fig. 3J and K), we observed a significant increase in apoptosis and cell death in ρ0 rescued cells treated with Gamitrinib (Fig. 3L and M). We also noticed increases in OCR, maximal respiration, spare respiratory capacity, ATP production, and proton leak in ρ0 rescued cells (Fig. 3N and O; Supplementary Fig. S2C and S2D). Taken together, these findings indicated that the antitumor activity of Gamitrinib in gliomas is directly selective to mitochondrial activity.
Figure 3.
The activity and specificity of Gamitrinib requires mitochondrial DNA. A and B, Relative mtDNA copy number in paired LN229 and LN229 ρ0 cells (A), and M059K and M059K ρ0 cells (B). Data were representative of 3 biological replicates. C and D Fluorescence intensity of Mitotracker Green in LN229 and LN229 ρ0 cells (C) and M059K and M059K ρ0 cells (D). Data were representative of 3 biological replicates. E, Immunoblot analysis of OXPHOS proteins in paired parental and ρ0 cells. F and G, Mitochondrial respiration, indicated as the oxygen comsumption rate (OCR) of LN229 (F) and M059K (G) parental and ρ0 cells, was measured by a Seahorse XF96 Analyzer. Maximal respirations were determined by the sequential addition of oligomycin, FCCP, and rotenone and antimycin A, respectively. Data were representative of 4 to 5 biological replicates. H and I, The percentage of apoptosis and cell death in parental and ρ0 cells treated with or without Gamitrinib at 5 μmol/L for 3 days. Data were representative of 3 biological replicates. J and K, Relative mtDNA copy number of parental, ρ0 and ρ0 rescued cells. Data were representative of 3 biological replicates. L and M, The percentage of apoptosis and dead cells in parental, ρ0 and ρ0 rescued cells treated with or without Gamitrinib at 5 μmol/L for 3 days. Data were representative of 3 biological replicates. N and O, Mitochondrial respirations of parental, LN229 (N) and M059K (O) ρ0 and ρ0 rescued cells treated with or without 5 μmol/L Gamitrinib, were measured by a Seahorse XF96 Analyzer. Data were representative of 4 to 5 biological replicates; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Gamitrinib inhibits mitochondrial biogenesis and tumor bioenergetics in glioma cells
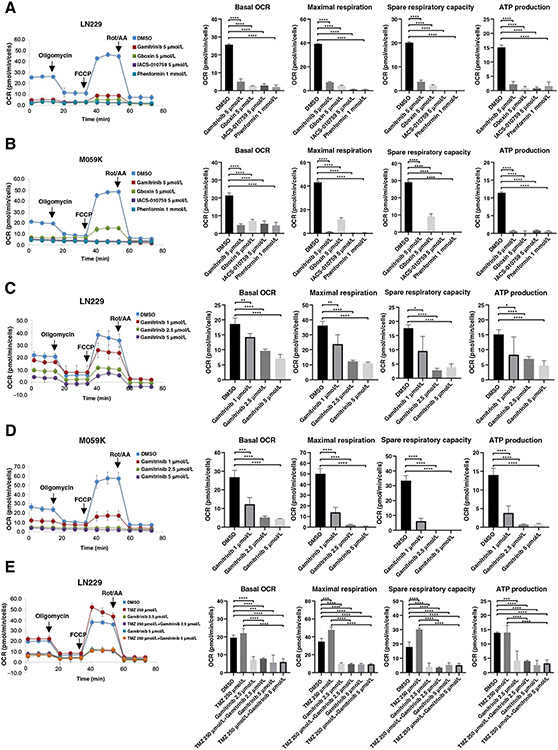
To further determine whether inhibition of mitochondrial respiration contributes to the antitumor efficacy in gliomas, we first evaluated Gamitrinib in comparison with other known OXPHOS inhibitors, including IACS-010759, Phenformin, and Gboxin (36). We performed the mitochondrial stress assay and observed all of these OXPHOS inhibitors significantly reduced the basal OCR, maximal respiration, ATP production, spare respiratory capacity, and proton leak as expected (Fig. 4A and B; Supplementary Fig. S2E and S2F). We also treated glioma cells with the cytosolic HSP90 inhibitor 17-AGG (37) from which Gamitrinib was derived and assessed the mitochondrial respiration. As expected, 17-AGG did not inhibit the mitochondrial respiration, thus indicating that Gamitrinib’s antitumor efficacy is exhibited by targeting both mitochondrial HSP90 and their client proteins (Supplementary Fig. S2G). To further investigate the effect of Gamitrinib on mitochondrial respiration, we treated M059K and LN229 cells with Gamitrinib at titrated doses and performed mitochondrial stress assay. The results indicated that Gamitrinib inhibited the mitochondrial respiration of both M059K and LN229 cell lines in a dose-dependent manner (Fig. 4C and D; Supplementary Fig. S2H and S2I). Furthermore, we explored the effect of the combination therapy of Gamitrinib plus TMZ on the mitochondrial respiration by performing mitochondrial stress test in LN229 cells. We found that TMZ alone had no effect on mitochondrial respiration. In contrast, either Gamitrinib as a monotherapy or the combination therapy of Gamitrinib plus TMZ led to a decrease in the basal OCR, maximal respiration, and ATP production in LN229 cells (Fig. 4E; Supplementary Fig. S2J). Taken together, these results suggested that Gamitrinib inhibits mitochondrial respiration and bioenergetics in glioma cells.
Figure 4.
Inhibition of mitochondrial respiration by Gamitrinib in gliomas. A and B, Mitochondrial respirations of LN229 (A) and M059K (B) cells treated with DMSO, IACS-010759, Phenformin, Gboxin, and Gamitrinib, respectively, were measured by a Seahorse XF96 Analyzer. Data were representative of 4 to 5 biological replicates. C and D, Mitochondrial respirations of LN229 (C) and M059K (D) cells treated with DMSO, Gamitrinib 1 μmol/L, Gamitrinib 2.5 μmol/L, and Gamitrinib 5 μmol/L, respectively, were measured by a Seahorse XF96 Analyzer. Data were representative of 4 to 5 biological replicates. E, Mitochondrial respirations of LN229 cells treated with DMSO, Gamitrinib, TMZ or Gamitrinib plus TMZ, respectively, were measured by a Seahorse XF96 Analyzer. Data were representative of 4 to 5 biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Integrated computational analyses of transcriptomic and targeted proteomic data reveal the antitumor mechanisms of Gamitrinib in gliomas
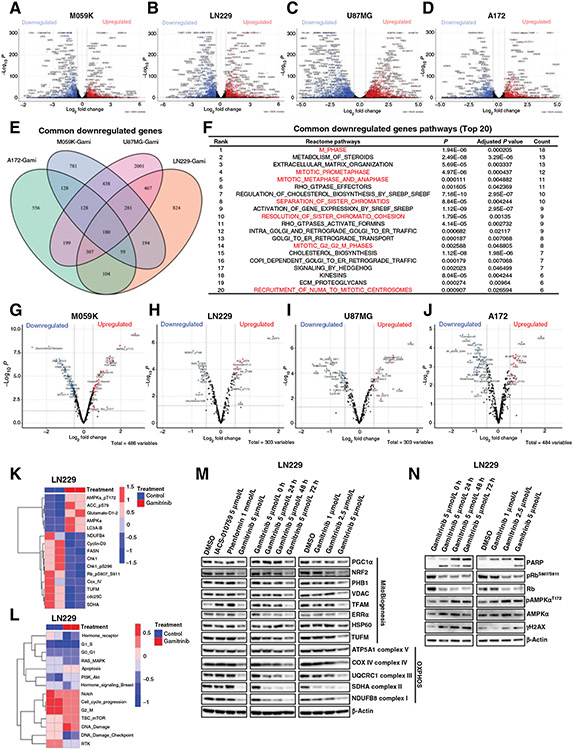
To further gain insights into antitumor mechanisms of Gamitrinib in gliomas in an unbiased way, we first performed RNAseq for M059K, LN229, U87MG, and A172 cells treated with DMSO and Gamitrinib, respectively. The analysis of RNAseq data identified 4,036 differentially expressed genes (DEG) in M059K cells treated with Gamitrinib, including 1,847 upregulated and 2,189 downregulated genes (adjusted P < 0.05; log2 fold change (FC) > ∣0.5∣; Fig. 5A). A total of 4,572 DEGs, including 2,156 upregulated and 2,416 downregulated genes were identified in LN229 cells (adjusted P < 0.05; log2 FC > ∣0.5∣; Fig. 5B). In U87MG cells, we observed 7,610 DEGs with 3,609 upregulated and 4,001 downregulated genes (adjusted P < 0.05; log2 FC > ∣0.5∣; Fig. 5C). A total of 3,376 DEGs were identified in A172 cells, including 1,715 upregulated and 1,661 downregulated genes (Fig. 5D). Furthermore, GSEA identified top-ranked pathways that were enriched in upregulated and downregulated genes, respectively (Supplementary Fig. S3A-S3H).
Figure 5.
Integrated computational analyses of transcriptomic and proteomic data in glioma cells treated with Gamitrinib. A and D, Volcano plots of differentially expressed genes in M059K (A), LN229 (B), U87MG (C), and A172 (D) cells treated with or without Gamitrinibat 5 μmol/L for 72 hours. x and y axes represent log2 fold change (FC) and −Log10 P, respectively. Gray dots indicated non-significant genes; blue dots indicated significantly downregulated genes; and red dots indicated significantly upregulated genes. E, Venn plot indicated the common downregulated genes across 4 cells treated with Gamitrinib. F, Gene set enrichment analysis indicated the top 20 ranked common pathways that were downregulated by Gamitrinib. G–J, Volcano plots of differentially expressed proteins in M059K (G), LN229 (H), U87MG (I), and A172 (J) cells treated with or without Gamitrinib at 5 μmol/L for 72 hours. K and L, Heatmaps of differentially expressed proteins indicated mitochondrial biogenesis, OXPHOS, apoptosis, and cell-cycle signaling pathway (K) and RPPA pathway scores (L) in LN229 cells treated with or without Gamitrinib for 72 hours. M and N, Immunoblot analysis of mitochondrial biogenesis and OXPHOS proteins (M) and cell cycle, apoptosis, and DNA damage proteins (N) in LN229 cells treated with or without 5 μmol/L Gamitrinib.
To investigate common antitumor mechanisms of Gamitrinib in gliomas on the transcriptional level, we first performed an integrated analysis of DEGs across all 4 glioma cell lines. Two hundred and two common upregulated genes and 180 common downregulated genes were identified across all 4 glioma cell lines (Fig. 5E; Supplementary Fig. S3I). Next, we performed GSEA based on these common upregulated and downregulated genes to identify which signaling pathways were enriched (Fig. 5F; Supplementary Fig. S3J). Interestingly, among top 20 common upregulated signaling pathways, there were 4 pathways related to cell death, including (i) transcriptional regulation by TP53, (ii) programmed cell death, (iii) death receptor signaling, and (iv) apoptosis, and 3 stress response pathways, including (i) cellular response to starvation, (ii) AUTOPHAGY, and (iii) ATF4 activates genes in response to endoplasmic reticulum stress, which were significantly enriched in common upregulated genes. In contrast, 7 signaling pathways related to cell cycle, including (i) M phase, (ii) mitotic prometaphase, (iii) mitotic metaphase and anaphase, (iv) separation of sister chromatids, (v) resolution of sister chromatid cohesion, (vi) mitotic G2 G2–M phases, and (vii) recruitment of numa to mitotic centrosomes, were significantly enriched in common downregulated genes (Fig. 5F). These results further suggested that Gamitrinib exhibits its antitumor activity via inducing cell death and engaging stress response, and inhibiting cell-cycle progression in glioma cells.
In addition, we performed RPPA for M059K, LN229, U87MG, and A172 cells to further elucidate the antitumor mechanisms of Gamitrinib on the protein level (Supplementary Table S1). The analyses of RPPA data identified 257 differential expressed proteins between Gamitrinib and control groups in M059K (a panel of 486 proteins), with 121 upregulated proteins and 136 downregulated proteins (Fig. 5G; Supplementary Fig. S4A). A total of 142 differential expressed proteins were identified in LN229 (a panel of 303 proteins), with 57 upregulated proteins and 85 downregulated proteins (Fig. 5H; Supplementary Fig. S4B). One hundred and fifty-nine differential expressed proteins were observed in U87MG (a panel of 303 proteins), with 74 upregulated proteins and 85 downregulated proteins (Fig. 5I; Supplementary Fig. S4C). A total of 189 differential expressed proteins were observed in A172 (a panel of 484 proteins) with 85 upregulated proteins and 104 downregulated proteins (Fig. 5J; Supplementary Fig. S4D).
To identify common antitumor mechanisms of Gamitrinib in gliomas at protein level, we performed a comprehensive analysis of differentially expressed proteins across all 4 cell lines. We identified 9 common downregulated proteins and 2 common upregulated proteins across all 4 cell lines (Supplementary Fig. S4E and S4F). There were 33 common downregulated proteins and 19 common upregulated proteins across more than 3 cell lines (Supplementary Fig. S4E-S4H). Among common downregulated proteins, expression levels of cdc25C, Cyclin-D3, and Chk1 were significantly inhibited in all 4 cell lines treated with Gamitrinib (Fig. 5K; Supplementary Fig. S4G and S4I-S4K), and the expression level of pRbS807/S811 was significantly decreased in LN229, U87MG, and A172 cells treated with Gamitrinib (Fig. 5K; Supplementary Fig. S4G and S4J and S4K). Furthermore, we computationally calculated scores for 14 tumor-intrinsic signaling pathways by analyzing RPPA data. We observed that DNA damage and apoptosis signaling pathways were triggered by Gamitrinib, and cell-cycle signaling pathways were substantially inhibited in these 4 cells treated with Gamitrinib (Fig. 5L; Supplementary Fig. S4L-S4N). Taken together, these findings further strengthened the notion that cell-cycle progression was inhibited in glioma cells treated with Gamitrinib.
In addition, the expression level of TUFM, a regulator of mitochondrial biogenesis, was significantly decreased in M059K, LN229, and A172 cells, indicating that Gamitrinib exhibits its antitumor activity via inhibition of mitochondrial biogenesis (Fig. 5K; Supplementary Fig. S4G and S4I and S4K). Furthermore, the energy stress sensor pAMPKaThr172 was significantly increased in M059K, LN229, and A172 cells after Gamitrinib treatment (Fig. 5K; Supplementary Fig. S4H and S4I and S4K).
Finally, we validated the findings based on the analysis of RPPA data of LN229 cells treated with Gamitrinib using western blotting. Two OXPHOS inhibitors were also included as controls such as IACS-010759 and Phenformin. Western blotting experiments verified that Gamitrinib inhibited the expression level of mitochondrial biogenesis and OXPHOS proteins to a greater extent as compared with other OXPHOS inhibitors (Fig. 5M). Moreover, we confirmed that Gamitrinib decreased the expression level of pRbS807/S811, and increased the expression levels of cleaved PARP, γH2AX, and pAMPKαT172, suggesting that Gamitrinib triggered apoptosis, DNA damage, and energy stress (Fig. 5N).
In summary, our findings strongly suggest that Gamitrinib induces cell death via inhibiting mitochondrial biogenesis and blocking cell cycle in gliomas.
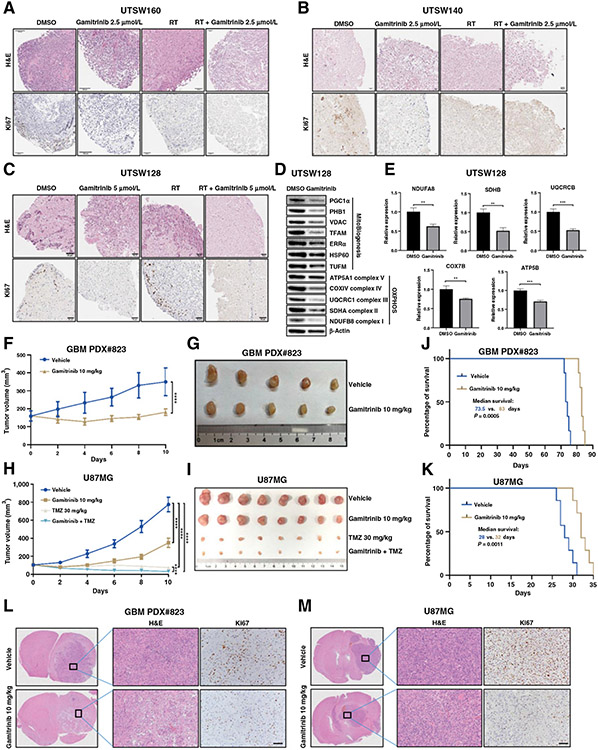
Gamitrinib inhibits the cell proliferation in GBM PDOs models
PDOs models maintain many key features of primary tumors and can be used as preclinical models to investigate therapeutic efficacy (22). As a proof-of-principle study, we further investigated the therapeutic efficacy of Gamitrinib in patient-derived GBM organoids. We generated 3 GBM PDOs (UTSW128, UTSW140, and UTSW160) and treated them with DMSO, Gamitrinib at 2.5 or 5 μmol/L, radiotherapy, and Gamitrinib plus radiotherapy, respectively. The IHC results showed that Gamitrinib at both 2.5 and 5 μmol/L doses significantly reduced the expression of Ki67 (Fig. 6A-C; Supplementary Fig. S5A-S5C). Although radiotherapy also inhibited the proliferation of PDOs, the addition of Gamitrinib to radiotherapy further reduced the proliferation of PDOs (Fig. 6A-C; Supplementary Fig. S5A-S5C). To further validate the antitumor mechanisms of Gamitrinib in PDOs models, we, respectively, performed RT-qPCR and western blotting with the RNA and protein samples derived from UTSW128 PDOs as a representative. As expected, Gamitrinib inhibited the expression of OXPHOS complex proteins and mitochondrial biogenesis at the protein level (Fig. 6D). Furthermore, the mRNA expressions of OXPHOS complex subunits were significantly decreased with Gamitrinib treatment (Fig. 6E). Taken together, these results suggested that Gamitrinib is effective in PDOs models and exhibits its antitumor activity by inhibiting OXPHOS process and mitochondrial biogenesis.
Figure 6.
Antitumor activity of gamitrinib in GBM PDOs, CDX, and PDX models. A–C, The hematoxylin and eosin (H&E) and Ki67 IHC staining of the GBM organoids UTSW 160 (A), UTSW140 (B), and UTSW128 (C). PDOs were treated with the DMSO control, Gamitrinib at 2.5 or 5 μmol/L, radiotherapy at 10 Gy, and Gamitrinib at 2.5 or 5 μmol/L plus radiotherapy at 10 Gy. D, Immunoblot analysis of mitochondrial biogenesis and OXPHOS proteins in GBM PDOs (UTSW128) treated with DMSO control or Gamitrinib at 5 μmol/L. E, Relative mRNA expression of OXPHOS genes in GBM PDOs (UTSW128) treated with DMSO or Gamitrinib. F and G, The tumor growth curve (F) and tumor images (G) of GBM PDX#823 tumor. Tumor-bearing mice were treated with control or Gamitrinib at 10 mg/kg every other day. Mice were sacrificed 10 days after Gamitrinib treatment. H and I, The tumor growth curve (H) and tumor images (I) of U87MG tumor. Tumor-bearing mice were treated with control, Gamitrinib at 10 mg/kg, TMZ 30 mg/kg, and Gamitrinib at 10 mg/kg plus TMZ 30 mg/kg every other day. Mice were sacrificed 10 days after Gamitrinib treatment. J and K, Kaplan–Meier survival curves of orthotopic PDX#823 (J) and U87MG (K) models. Six mice for each group in orthotopic PDX#823 models, and 7 mice for each group in orthotopic U87MG models; Mantel–Cox log-rank test, two-tailed test. L and M, The H&E and Ki67 IHC staining of the orthotopic PDX#823 (L) and U87MG (M) models; scale bars, 50 μm. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Gamitrinib delays tumor growth and improves survival of mice in GBM CDX and PDX models
To further determine the efficacy of Gamitrinib in vivo, we first generated a GBM PDX model (#823), which is an ideal preclinical model to evaluate the therapeutic efficacy of drugs in gliomas (38). Tumor-bearing mice were treated with the vehicle or Gamitrinib at the dose of 10 mg/kg. The results indicated that Gamitrinib significantly delayed the tumor growth of GBM PDX#823 (Fig. 6F and G). In addition, we xenografted U87MG cells subcutaneously and treated tumor-bearing mice with vehicle, Gamitrinib 10 mg/kg, TMZ 30 mg/kg, and Gamitrinib 10 mg/kg plus TMZ 30 mg/kg. As expected, both Gamitrinib and TMZ as monotherapy significantly delayed the tumor growth, and Gamitrinib plus TMZ combination therapy further inhibited the tumor growth (Fig. 6H and I). Mice were tolerant to the treatment of Gamitrinib (Supplementary Fig. S5D and S5E). Furthermore, the Ki67 IHC staining of tumor tissues indicated that Gamitrinib inhibited cell proliferation in both U87MG CDX and GBM PDX#823 models (Supplementary Fig. S5F and S5G).
To further investigate the therapeutic efficacy of Gamitrinib in orthotopic GBM models, intracranial implantation of U87MG cells and PDX cells were performed, respectively, and vehicle or Gamitrinib at the dose of 10 mg/kg were administrated subsequently. Indeed, Gamitrinib significantly improved survival of mice in both orthotopic U87MG and GBM PDX#823 models (Fig. 6J and K), which was accompanied by the reduction of the expression of Ki67 in tumor tissues (Fig. 6L and M; Supplementary Fig. S5H and S5I).
Taken together, our CDX and PDX models in which tumors were either subcutaneously or intracranially implanted demonstrated the therapeutic efficacy of Gamitrinib in gliomas as a monotherapy.
Discussion
In this study, we systematically investigated the therapeutic efficacy of Gamitrinib in a broad panel of 17 primary human glioma cell lines, 6 TMZ-resistant glioma cell lines, 4 neurospheres, 3 GBM PDOs models, and 1 GBM CDX and 1 GBM PDX models. Gamitrinib, as a single agent, inhibited cell proliferation and induced apoptosis and cell death. When combined with TMZ or radiotherapy, Gamitrinib further enhanced the efficacy of standard-of-care therapies. Mechanistically, comprehensive analyses of RNAseq and RPPA data indicated that Gamitrinib exhibited its antitumor activity via inhibition of mitochondrial biogenesis and tumor bioenergetics, blocking cell-cycle progression and inducing DNA damage.
Gamitrinib was first described in 2009 and has shown the antitumor activity in preclinical models for multiple cancers, including prostate cancer, melanoma, lung cancer, colon cancer, breast cancer, and GBM (9, 10, 16-18, 21). Among these studies, Chae and colleagues (9) investigated the antitumor activity in breast, prostate, lung, and brain tumors, and demonstrated that mitochondrial, but not cytosolic, HSP90s is a global integrator of tumor bioenergetics, autophagy, and interorganelle stress-response signaling (10). In another study of Chae and colleagues (9), they determined the role of mitochondrial HSP90 protein folding on cellular homeostasis and demonstrated that mitochondrial HSP90 is required for disease maintenance. Their two studies provide a rationale that mitochondrial HSP90 would be a tractable target for cancer therapy. In the subsequent studies, Ishida and colleagues (22) and Zhang and colleagues (23) have attempted the combination of Gamitrinib and other small-molecular inhibitors (JQ1, OTX015, and crizotinib) in GBM, and have achieved promising results. In current study, we systematically evaluated the therapeutic efficacy of Gamitrinib in gliomas, and we also investigated the combination of Gamitrinib and standard-of-care therapies in gliomas. More importantly, we performed integrated analyses of RNAseq and RPPA data to elucidate the antitumor mechanisms of Gamitrinib. To the best of our knowledge, this is the first study that comprehensively assesses the therapeutic efficacy and the antitumor mechanisms of Gamitrinib in gliomas.
Mitochondria harbor their own genome consisting of an approximately 16.6 kb double-stranded circular DNA (mtDNA) encoding 2 rRNAs, 22 tRNAs, and 13 polypeptides subunits of the respiratory chain complexes (39). Previous studies have used cells in which mtDNA was depleted (Rho0 or ρ0 cells) to examine the roles of mtDNA and particularly the roles of mtDNA in regulating apoptosis (40, 41). In our study, to demonstrate the specificity of Gamitrinib, we generated two GBM ρ0 cells after long-term exposure to ethidium bromide (EtBr) administered at very low concentrations. As expected, the expression levels of respiratory chain complexes were decreased and the mitochondrial respiration was diminished in these ρ0 cells as compared with parental cells. Upon the depletion of mtDNA, glioma cells escaped from the apoptosis induced by Gamitrinib, which can be further rescued by restoring mtDNA in ρ0 cells. These findings might partially explain the specificity of Gamitrinib to mitochondria. To further prove this possible specificity, we treated glioma cells with Gamitrinib in comparison with the cytosolic HSP90 inhibitor 17-AGG and assessed the mitochondrial respiration. As expected, Gamitrinib, but not 17-AGG, inhibited the mitochondrial respiration, thus indicating that Gamitrinib exhibits its antitumor efficacy by targeting both mitochondrial HSP90 proteins and their client proteins. Collectively, these findings underscore the specificity of Gamitrinib in terms of its ability to target mitochondrial HSP90 subcellularly, which might provide tangible therapeutic implications for gliomas.
For the first time, we performed integrated computational analyses of RNAseq and RPPA data to elucidate the antitumor mechanisms of Gamitrinib in the current study. Integrated analysis of DEGs across the 4 cells identified a total of 220 common upregulated genes and 180 common downregulated genes. Among top 20 common upregulated signaling pathways, 4 pathways related to cell death and 3 stress-response pathways were significantly enriched. Interestingly, Shi and colleagues (36) and Chae and colleagues (10) also reported the activation of stress-response pathways in cancers treated with OXPHOS inhibitors. In contrast, 7 signaling pathways related to cell cycle, were significantly enriched in common downregulated genes. Among significantly enriched pathways, several cell death–related pathways and cell-cycle–related pathways were identified. More importantly, comprehensive analysis of RPPA data across the 4 cells identified 9 common downregulated proteins, including cdc25C, Cyclin-D3, and Chk1, which are generally associated with cell-cycle progression. These comprehensive analyses of RNAseq and RPPA data highly suggested that Gamitrinib exhibited its antitumor activity via inhibition of cell-cycle progression in glioma cells. In addition, we also observed suppression of mitochondrial biogenesis and OXPHOS, and activation of the energy-sensing AMPK and DNA damage in glioma cells treated with Gamitrinib. Previous studies have also demonstrated that targeting TRAP1-directed protein folding in mitochondria with Gamitrinib inhibits both mitochondrial biogenesis and OXPHOS in several cancer types (9, 15). However, how Gamitrinib inhibits mitochondrial biogenesis and OXPHOS in cancer cells is not fully known and warrants further investigation in future studies.
PDOs are novel and three-dimensional ex vivo models that can accurately represent the diverse genetic, molecular, morphological, architectural, and functional pathophysiological hallmarks of cancer (42-46). PDOs models have provided the opportunity to facilitate the principles of 3Rs in research and have been extensively used as a preclinical model to predict the clinical outcome and as a drug-screening platform to identify personalized medicine (42). In this study, we further investigated the therapeutic efficacy of Gamitrinib by using 3 GBM PDOs. We demonstrated that Gamitrinib is effective in this PDO model and exhibits its antitumor activity via inhibiting OXPHOS process and mitochondrial biogenesis. In addition to GBM PDO models, Gamitrinib significantly delayed the tumor growth in subcutaneous GBM models and improved survival of mice in orthotopic GBM models, exhibiting its antitumor activity in vivo. Notably, a previous study also demonstrated that Gamitrinib as a monotherapy significantly improved survival of mice in an orthotopic U87MG model (47). Taken together, these findings might provide a potential rationale for propelling Gamitrinib into clinical trials to provide patients with primary and recurrent gliomas a novel therapeutic strategy.
Several limitations exist in this study, which warrants further investigation. First, because the main purpose of this study is to investigate the antitumor activity of Gamitrinib as a monotherapy in experimental models, we just combined Gamitrinib with standard-of-care therapies (such as TMZ or radiotherapy) when possible. Further studies are warranted to systematically assess the therapeutic efficacy of Gamitrinib combined with standard-of-care therapies in gliomas. Second, to determine the antitumor activity of Gamitrinib through a broad screen of gliomas, we investigated the therapeutic efficacy of Gamitrinib in both established glioma models and patient-derived models. The long-term cultured cells might not recapitulate the actual disease very well; therefore, the results from these established models need to be interpreted with caution. Third, there is a possibility that the procedure of orthotopic implantation may artificially disrupt the blood–brain barrier (BBB). Therefore, we do not have direct evidence to support that Gamitrinib could penetrate the BBB, even though Gamitrinib inhibited the proliferation of tumor cells as well as improved survival of mice in two orthotopic GBM models. Actually, the ability and degree of Gamitrinib to cross the BBB has not been formally documented (48). Further studies are warranted to pinpoint the BBB penetrance based on the compound profiles of Gamitrinib, including size, polarity, transporter, efflux, etc., using LC/MS assay. Fortunately, a first-in-human phase I clinical trial that evaluates the feasibility and safety of Gamitrinib in patients with advanced cancers is currently ongoing (ClinicalTrials.gov Identifier: NCT04827810), which we expect will soon provide us with more information to document pharmacokinetics of Gamitrinib in patients in the clinical setting (49).
In conclusion, our study systematically evaluated the therapeutic efficacy of Gamitrinib and established its therapeutic role in gliomas. Our study not only demonstrated Gamitrinib’s ability to effectively inhibit mitochondrial biogenesis and respiration, but it also suppresses cell-cycle progression and activates DNA damage in gliomas. Our findings are based on a large panel of glioma cell lines and patient-derived models and provide a proof-of-principle study for demonstrating the efficacy of Gamitrinib in the preclinical study setting. Further studies are warranted to bring this mitochondria-directed HSP90 inhibitor into clinical trials to benefit patients with glioma.
Supplementary Material
Translational Relevance.
There have been no new drugs approved by the FDA for patients with primary and recurrent gliomas since bevacizumab was approved in 2009. There is an urgent need to identify novel therapies for patients with primary and recurrent gliomas. In this study, we established the therapeutic role of a mitochondrial-directed HSP90 inhibitor, Gamitrinib, in a broad panel of TMZ-sensitive and -resistant glioma cells, as well as neurospheres, patient-derived organoids, and patient-derived xenografts. Mechanistically, Gamitrinib exhibits its antitumor activity via suppressing mitochondrial biogenesis and OXPHOS, and activating the energy-sensing AMP-activated kinase, as well as suppressing cell-cycle progression. These preclinical findings highlight the potential therapeutic role of Gamitrinib in gliomas, and provide a scientific rationale for propelling Gamitrinib into clinical trials to treat patients with primary and recurrent gliomas.
Acknowledgments
Y. Chen was funded by National Natural Science Foundation of China (31771549). S. Wei and S. Yu were funded by China Scholarship Council. The research was funded by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation to M. Herlyn, K.T. Flaherty, and G.B. Mills. The authors also thank Dr. D.C. Altieri at The Wistar Institute for the discussion of this work.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclaimer
The Editor-in-Chief of Clinical Cancer Research is an author on this article. In keeping with AACR editorial policy, a senior member of the Clinical Cancer Research editorial team managed the consideration process for this submission and independently rendered the final decision concerning acceptability.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol 2020;22: iv1–iv96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho VK, Reijneveld JC, Enting RH, Bienfait HP, Robe P, Baumert BG, et al. Changing incidence and improved survival of gliomas. Eur J Cancer 2014;50:2309–18. [DOI] [PubMed] [Google Scholar]
- 4.Ostrom QT, Cote DJ, Ascha M, Kruchko C, Barnholtz-Sloan JS. Adult glioma incidence and survival by race or ethnicity in the United States from 2000 to 2014. JAMA Oncol 2018;4:1254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van den Bent MJ, Klein M, Smits M, Reijneveld JC, French PJ, Clement P, et al. Bevacizumab and temozolomide in patients with first recurrence of WHO grade II and III glioma, without 1p/19q co-deletion (TAVAREC): a randomised controlled phase 2 EORTC trial. Lancet Oncol 2018;19:1170–9. [DOI] [PubMed] [Google Scholar]
- 6.Desjardins A, Gromeier M, Herndon JE II, Beaubier N, Bolognesi DP, Friedman AH, et al. Recurrent glioblastoma treated with recombinant poliovirus. N Engl J Med 2018;379:150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bi J, Chowdhry S, Wu S, Zhang W, Masui K, Mischel PS. Altered cellular metabolism in gliomas—an emerging landscape of actionable co-dependency targets. Nat Rev Cancer 2020;20:57–70. [DOI] [PubMed] [Google Scholar]
- 8.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11:325–37. [DOI] [PubMed] [Google Scholar]
- 9.Chae YC, Angelin A, Lisanti S, Kossenkov AV, Speicher KD, Wang H, et al. Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat Commun 2013;4:2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chae YC, Caino MC, Lisanti S, Ghosh JC, Dohi T, Danial NN, et al. Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell 2012;22:331–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condelli V, Crispo F, Pietrafesa M, Lettini G, Matassa DS, Esposito F, et al. HSP90 molecular chaperones, metabolic rewiring, and epigenetics: impact on tumor progression and perspective for anticancer therapy. Cells 2019;8:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall JA, Forsberg LK, Blagg BS. Alternative approaches to Hsp90 modulation for the treatment of cancer. Future Med Chem 2014;6:1587–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen MT, Csermely P, Soti C. Hsp90 chaperones PPARgamma and regulates differentiation and survival of 3T3-L1 adipocytes. Cell Death Differ 2013;20:1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang BH, Plescia J, Song HY, Meli M, Colombo G, Beebe K, et al. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J Clin Invest 2009;119:454–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Frederick DT, Wu L, Wei Z, Krepler C, Srinivasan S, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J Clin Invest 2016;126:1834–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegelin MD, Dohi T, Raskett CM, Orlowski GM, Powers CM, Gilbert CA, et al. Exploiting the mitochondrial unfolded protein response for cancer therapy in mice and human cells. J Clin Invest 2011;121:1349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida S, Tsutsumi S, Muhlebach G, Sourbier C, Lee MJ, Lee S, et al. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc Nat Acad Sci USA 2013;110:E1604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Q, Tu J, Dou C, Zhang J, Yang L, Liu X, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer 2017;16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang BH, Tavecchio M, Goel HL, Hsieh CC, Garlick DS, Raskett CM, et al. Targeted inhibition of mitochondrial Hsp90 suppresses localised and metastatic prostate cancer growth in a genetic mouse model of disease. Br J Cancer 2011;104:629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishida CT, Shu C, Halatsch ME, Westhoff MA, Altieri DC, Karpel-Massler G, et al. Mitochondrial matrix chaperone and c-myc inhibition causes enhanced lethality in glioblastoma. Oncotarget 2017;8:37140–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Nguyen TTT, Shang E, Mela A, Humala N, Mahajan A, et al. MET inhibition elicits PGC1alpha-dependent metabolic reprogramming in glioblastoma. Cancer Res 2020;80:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdullah KG, Bird CE, Buehler JD, Gattie LC, Savani MR, Sternisha AC, et al. Establishment of patient-derived organoid models of lower grade glioma. Neuro Oncol 2021. Nov 26. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizuka SS, Mills GB. New era of integrated cancer biomarker discovery using reverse-phase protein arrays. Drug Metab Pharmacokinet 2016;31:35–45. [DOI] [PubMed] [Google Scholar]
- 24.Labrie M, Kim TB, Ju Z, Lee S, Zhao W, Fang Y, et al. Adaptive responses in a PARP inhibitor window of opportunity trial illustrate limited functional interlesional heterogeneity and potential combination therapy options. Oncotarget 2019;10:3533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrews S FastQC: a quality control tool for high throughput sequence data. 2010. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- 26.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toolkit P. Broad institute, GitHub repository. 2019. Available from: http://broadinstitute.github.io/picard/. [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Nat Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stupp R, Hegi ME, Gilbert MR, Chakravarti A. Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol 2007;25:4127–36. [DOI] [PubMed] [Google Scholar]
- 33.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med 2018;24:1036–46. [DOI] [PubMed] [Google Scholar]
- 34.Jiang W, Finniss S, Cazacu S, Xiang C, Brodie Z, Mikkelsen T, et al. Repurposing phenformin for the targeting of glioma stem cells and the treatment of glioblastoma. Oncotarget 2016;7:56456–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Meng Y, Zhang S, Wu H, Yang D, Nie C, et al. Phenformin and metformin inhibit growth and migration of LN229 glioma cells in vitro and in vivo. Onco Targets Ther 2018;11:6039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi Y, Lim SK, Liang Q, Iyer SV, Wang HY, Wang Z, et al. Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature 2019;567:341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauvageot CM, Weatherbee JL, Kesari S, Winters SE, Barnes J, Dellagatta J, et al. Efficacy of the HSP90 inhibitor 17-AAG in human glioma cell lines and tumorigenic glioma stem cells. Neuro Oncol 2009;11:109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaubel RA, Tian S, Remonde D, Schroeder MA, Mladek AC, Kitange GJ, et al. Genomic and phenotypic characterization of a broad panel of patient-derived xenografts reflects the diversity of glioblastoma. Clin Cancer Res 2020;26: 1094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet 2005;6:815–25. [DOI] [PubMed] [Google Scholar]
- 40.Yan H, Li Y, Peng X, Huang D, Gui L, Huang B. Resistance of mitochondrial DNA-depleted cells against oxidized low-density lipoprotein-induced macrophage pyroptosis. Mol Med Rep 2016;13:4393–9. [DOI] [PubMed] [Google Scholar]
- 41.Moro L, Arbini AA, Yao JL, di Sant’Agnese PA, Marra E, Greco M. Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2. Cell Death Differ 2009;16:571–83. [DOI] [PubMed] [Google Scholar]
- 42.Porter RJ, Murray GI, McLean MH. Current concepts in tumour-derived organoids. Br J Cancer 2020;123:1209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 2020;180:188–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogawa J, Pao GM, Shokhirev MN, Verma IM. Glioblastoma model using human cerebral organoids. Cell Rep 2018;23:1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Hora CC, Schweiger MW, Wurdinger T, Tannous BA. Patient-derived glioma models: from patients to dish to animals. Cells 2019;8:1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linkous A, Balamatsias D, Snuderl M, Edwards L, Miyaguchi K, Milner T, et al. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep 2019;26:3203–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh JC, Siegelin MD, Vaira V, Faversani A, Tavecchio M, Chae YC, et al. Adaptive mitochondrial reprogramming and resistance to PI3K therapy. J Natl Cancer Inst 2015;107:dju502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen TTT, Zhang Y, Shang E, Shu C, Quinzii CM, Westhoff MA, et al. Inhibition of HDAC1/2 along with TRAP1 causes synthetic lethality in glioblastoma model systems. Cells 2020;9:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayat U, Elliott GT, Olszanski AJ, Altieri DC. Feasibility and safety of targeting mitochondria for cancer therapy—preclinical characterization of gamitrinib, a first-in-class, mitochondrial-targeted small-molecule Hsp90 inhibitor. Cancer Biol Ther 2022;23:117–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data were deposited into Gene Expression Omnibus (accession number GSE165819 for M059K and LN229 cells; accession number GSE180872 for U87MG and A172 cells). WES data will be available upon request. RPPA data are available in Supplementary Table S1.