Abstract
The Escherichia coli chromatin protein FIS modulates the topology of DNA in a growth phase-dependent manner. In this study we have investigated the global effect of FIS binding on DNA architecture in vitro. We show that in supercoiled DNA molecules FIS binds at multiple sites in a non-random fashion and increases DNA branching. This global DNA reshaping effect is independent of the helical phasing of FIS binding sites. We propose, in addition to the previously inferred stabilisation of tightly bent DNA microloops in the upstream regions of certain promoters, that FIS may perform the distinct architectural function of organising branched plectonemes in the E.coli nucleoid.
INTRODUCTION
In contrast to eukaryotic chromatin, only a part of the bacterial DNA is constrained by proteins, while the rest is present, most probably, in a plectonemically supercoiled form (1,2). Measurements of the torsional tension and long-range mobility of DNA in living Escherichia coli cells suggest that the nucleoid contains 40 or more topologically isolated domains (1,3–5) that may correspond physically to branched projections of the nucleoid visualised by immunostaining techniques (6,7). These protrusions are thought to constitute transcriptionally active DNA loops organised in domains whose boundaries may change according to the physiological state of the cell (5,7–11).
In E.coli the DNA architectural proteins involved in the structural–functional organisation of the nucleoid are synthesised in a growth phase-dependent manner (12–18). The order of abundance of several major chromatin proteins in the exponential phase of growth is FIS > HU > H-NS > IHF, whereas on entry into the stationary phase this order changes to Dps > IHF > HU > H-NS > FIS (19). This orderly change in the chromosomal protein composition on transition from exponential growth to stationary phase is accompanied by silencing of genome functions. Thus, the nucleoid proteins not only define the architecture of the bacterial chromosome, but also correlate with the pattern of gene expression during the growth cycle. The understanding of the mode of DNA organisation by these proteins is pivotal for the elucidation of structural transitions between the transcriptionally active and repressed chromatin architectures.
FIS is thought to constitute the major protein component of bacterial chromatin during the exponential growth phase (20). FIS is also implicated in coupling DNA topology with physiological changes during the outgrowth of cells from the stationary phase (21–23). This latter effect is in part due to the modulation of DNA gyrase activity, and in part due to changes in DNA architecture induced by the direct binding of FIS. Much evidence indicates that the binding of this architectural protein to helically phased high-affinity sites in regulatory DNA elements, such as upstream activating sequences (UAS) of certain promoters and enhancers of site-specific recombination systems, stabilises a specifically bent DNA geometry (24–28). In contrast, the global effect of FIS on chromatin architecture is poorly understood. FIS binds DNA in vitro with affinities varying by several orders of magnitude depending on both the ‘bendability’ of DNA and the context of the poorly conserved 15 bp ‘core’ binding site (29,30). Depending on the sequence bound, the measured extent of DNA bending by FIS varies from 50 to 90° (30). As the maximal concentration of FIS in the cell is sufficient to interact with 104 to 105 potential binding sites scattered in the E.coli chromosome (14,31) and binding of FIS modulates the superhelical density of DNA in vivo (21), we suggested that the protein may affect the global DNA architecture by stabilising writhe (32).
In this study we have investigated the direct effects of FIS binding on DNA architecture in vitro and also compared the FIS nucleoprotein complexes with those formed by H-NS, another chromatin protein often implicated in gene silencing (12,33). We show that in vitro FIS can change the overall shape of supercoiled DNA molecules. This global effect on supercoiled DNA involves multiple binding of FIS to non-random sites in DNA and results in its branching. We show that in these complexes FIS constrains a low negative superhelical density. These FIS–DNA structures provide an initial insight into the possible role of FIS in determining the architecture of chromatin in exponentially growing bacterial cells.
MATERIALS AND METHODS
Plasmids and proteins
The plasmid ptyrTlac contains the tyrT promoter cloned upstream of a lacZ gene (22,34). The plasmid pGE101 containing two SfiI cleavage sites separated by ∼900 bp was provided by G. Weinzierl [Max-Planck Institute (MPI) for terrestrial Microbiology, Marburg; unpublished]. Relaxed pUC18 DNA was obtained from commercial sources (Sigma). The FIS protein was purified as described previously (35). Purified H-NS was kindly provided by C. Gualerzi (University of Camerino).
Analysis of DNA supercoiling by FIS
Single-strand nicks were introduced in supercoiled pBR322 (5 µg) using DNase I (1 µg/ml) in the presence of 300 µg/ml ethidium bromide in 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2, 0.2 mg/ml BSA for 15 min at 30°C. The reaction was stopped by adding phenol/chlorofom/isoamyl alcohol (25:24:1), the DNA precipitated with ethanol, dissolved in distilled water and loaded on a 1% TBE gel. The open circular DNA was excised and eluted with the Qiaquick gel-extraction kit (Qiagen). Nicked pBR322 (100 ng) was incubated in a 20 µl volume with increasing amounts of FIS as indicated in the T4-DNA-Ligase buffer [66 mM Tris–HCl pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM ATP] for 15 min at 30°C. Then 2 U of T4-DNA-Ligase (Boehringer Mannheim) were added and incubation continued for 1 h. The reaction was stopped by adding EDTA, to a final concentration of 50 mM, and heat inactivation for 10 min at 65°C. After adding SDS to 0.1% and digestion with proteinase K, the reactions were loaded on 1% agarose gel containing 3 µg/ml chloroquine. High-resolution gel electrophoresis was carried out as described earlier (21). After electrophoresis the gels were stained with ethidium bromide and photographed under UV-light.
Electron microscopy
For electron microscopy (EM) the complexes between the proteins and ptyrTlac were formed in 10 µl of a buffer containing 10 mM TEA/HCl pH 8.0, 20 mM NaCl, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 30 ng of DNA and proteins as indicated. After incubation for 10 min at room temperature the complexes were fixed for 15 min in 0.2% glutaraldehyde. After fixation the samples were diluted with 20 µl of binding buffer and adsorbed to mica as described (36). Micrographs were taken using a Philips EM400T or CM100 electron microscope on 35 mm film (Agfa, RA711P) or 6.5 × 9 cm sheets (Agfa Scientia 23D56). The positions of the bound proteins were determined on 16× enlarged negatives using a LM4 digitizer (Brühl, Nuremberg, Germany).
Atomic force microscopy
For atomic force microscopy (AFM) (37,38) imaging, the plasmid DNA (pUC18 or ptyrTlac) was incubated in 100 µl buffer solution (100 mM NaCl, 20 mM HEPES pH 7.7, 5 mM NiCl) (39) on a freshly cleaved mica disk for ∼1 min in the presence or absence of FIS protein. After rinsing the samples with buffer to remove weakly attached DNA, the sample was mounted in the atomic force microscope (Nanoscope IIIa, Digital Instruments). The fluid cell of the atomic force microscope was sealed with a silicon O-ring to prevent evaporation of the buffer solution. The atomic force microscope was equipped with a piezo scanner with a maximum range of 160 µm, which allowed an estimation of the overall sample quality, before zooming into smaller regions to image the adsorbed DNA. Prior to the experiments, the scanner was calibrated against a grid of known dimensions. Silicon nitride cantilevers with integrated oxide sharpened tips (DNP-S-TT) from Digital Instruments (nominal spring constant 120 mN/m) were used for imaging. The experiments were conducted at ambient temperature (25°C) and were taken in tapping-mode (40) to minimise lateral forces between tip and sample. Scan rates varied between 1 and 5 Hz (lines/s).
Probing with restriction endonucleases
The probing with FokI was carried out at 37°C in the same buffer as described above for EM experiments. The cleavage by SfiI was carried out at 37°C in the same buffer but containing 200 mM NaCl. The proteins were incubated for 10 min with ptyrTlac DNA before the addition of the restriction endonucleases. The reactions were stopped by the addition of 50 mM EDTA and 0.2% SDS (final concentration), digested with proteinase K for 30 min at 50°C and loaded on agarose gels. After staining with ethidium bromide, the DNA fragments were visualised under UV light.
RESULTS
Analysis of DNA supercoiling induced by FIS
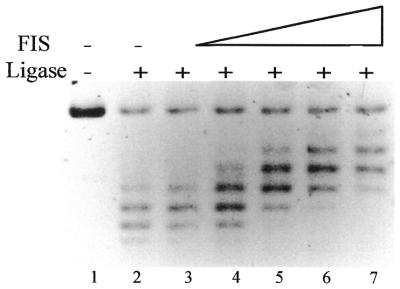
We have shown previously that on binding to supercoiled DNA, FIS alters the equilibrium distribution of plasmid topoisomers in the presence of DNA-relaxing topoisomerases (21). To test whether FIS could constrain supercoils in relaxed DNA, we incubated nicked pBR322 DNA with increasing concentrations of FIS in the presence of DNA ligase. Gel electrophoresis in high-resolution agarose gels containing chloroquine demonstrated that binding of FIS to nicked pBR322 DNA increases the number of negative supercoils trapped on ligation of the DNA (Fig. 1). We estimate that the protein changes the linking number of pBR322 DNA by 2–3. Directionally, this effect is similar to that of other DNA architectural proteins, such as HU, H-NS and HMG-D (41–44). HMG-D was much more efficient in constraining supercoils than FIS (R.Schneider and G.Muskelishvili, unpublished results) and the estimated FIS-dependent change in linking number is also much less than that reported for HU and H-NS (41,42,44). This suggests that the mode of constraint by FIS may differ from that by the other proteins in being less efficient in compacting the DNA.
Figure 1.
FIS introduces negative supercoils in DNA. Lane 1, open circular pBR322 DNA substrate; lane 2, only ligase was added. The FIS to DNA ratio in lanes 3–7 was ∼1 dimer per 800, 325, 160, 80 and 40 bp respectively. Under the conditions of electrophoresis used the more negatively supercoiled topoisomers migrate more slowly.
Visualisation of FIS–DNA complexes on supercoiled DNA
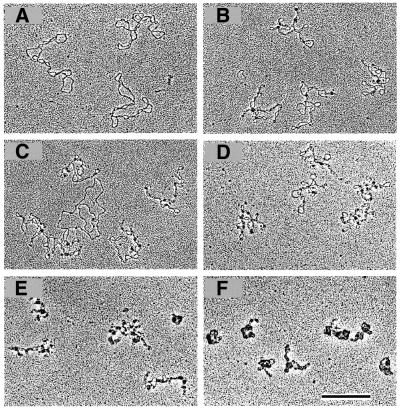
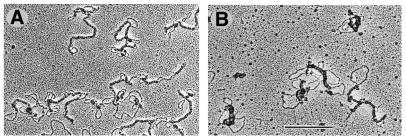
The constraint of negative supercoils by FIS indicates that the protein could alter the configuration of plasmid DNA. To investigate this possibility, complexes between FIS and supercoiled ptyrTlac DNA were formed at different protein to DNA ratios and visualised by EM. As well as a small number of isolated high-affinity FIS binding sites this plamid DNA contains three helically phased FIS binding sites in the UAS of the tyrT promoter. Already at the lowest FIS to DNA ratios used (∼1 dimer per 325 bp DNA) there was a significant alteration in the overall shape of the plasmids, such that they exhibited a more interwound and branched appearance in comparison to free plasmid DNA (Fig. 2A and B). We found that in the presence of FIS the number of branched molecules (the branch was defined as a separate DNA lobe containing at least one intrinsic crossover) increased from 22 to 94% (n = 37) and the FIS aggregates were localised preferentially at the crossovers and apical loops (in 81% of visually inspected FIS–DNA complexes, n = 250). These structures strongly suggest that FIS may facilitate branching of supercoiled DNA. At the DNA to protein ratios corresponding to ∼1 FIS dimer per 160 bp DNA, the branching and ‘tightening‘ of DNA became even more apparent (Fig. 2D). With increasing FIS to DNA ratios (≥1 dimer per 80–40 bp DNA) the DNA became more extensively occupied by FIS eventually leading to formation of compact structures (Fig. 2E and F). Similar compact structures were observed at higher FIS concentrations (∼1 dimer per 10 bp DNA).
Figure 2.
Binding of FIS leads to branching of supercoiled ptyrTlac DNA. EM images are shown. (A) Free supercoiled ptyrTlac DNA. FIS stabilises branches at a FIS to DNA ratio of ∼1 dimer per 325 bp DNA (B and C). Note that FIS, in agreement with a previous report (21), does not bind efficiently the relaxed (presumably nicked) DNA molecule in (C). The FIS to DNA ratio in (D), (E) and (F) was ∼1 dimer per 160, 80 and 40 bp, respectively. Note the ‘tightening’ of plasmid molecules with increasing FIS concentrations and the formation of compact structures at higher FIS to DNA ratios (scale bar represents 1 kb).
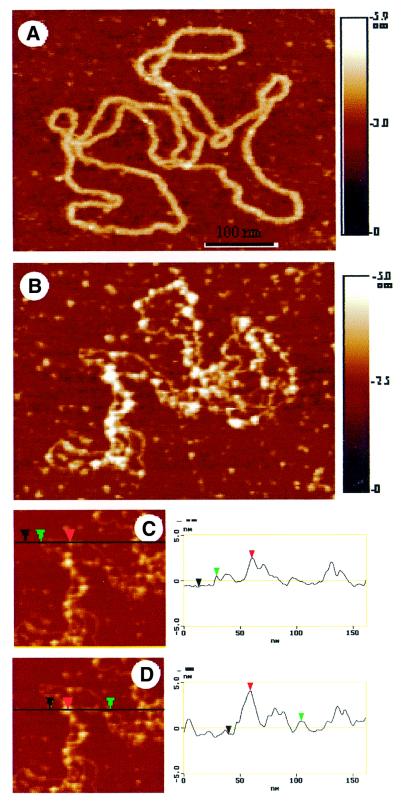
We investigated this extensive binding pattern in more detail using AFM. Visualisation of FIS nucleoprotein complexes at high FIS to DNA ratios (≥1 dimer per 100 bp) demonstrated alternating regions of ptyrTlac DNA either extensively occupied by FIS or almost devoid of bound protein (compare Fig. 3A and B). The profile plots (Fig. 3C and D) performed with the complex shown in Figure 3B showed that the profile height in the region of protein binding was 4.7 nm, exceeding that of the two crossed DNA duplexes (2 × 1.3 nm). The clustering of closely spaced FIS aggregates in Figure 3B suggests that FIS may bind DNA in a non-random manner and could also explain the formation of compact nucleoprotein structures observed by EM.
Figure 3.
FIS forms clusters on binding supercoiled ptyrTlac DNA. AFM images are shown. (A) Free supercoiled ptyrTlac. (B) Complex formed at the FIS to ptyrTlac DNA ratio of ∼1 dimer per 80–100 bp DNA. (C and D) The profile plots are shown. The height of the DNA in these images is 1.3 ± 0.1 nm. The height of the protein is deduced from the height of a DNA–protein complex formed on a portion of the plasmid without crossover. The height of the protein is then 2.0 ± 0.1 nm. Considering this height of the protein and the height of the DNA, it is possible to analyse a crossover. This is shown in the profile plot (D). The height of the crossover (4.7 ± 0.2 nm) corresponds to the doubling of the DNA height and also the height of the protein. An apparent difference in the width of the DNA between (A) and (B) is due to a difference in the tip radii. Because AFM images are always a convolution between the tip shape and the real topography, the measured size of small features (in the range of the tip size) will vary from tip to tip.
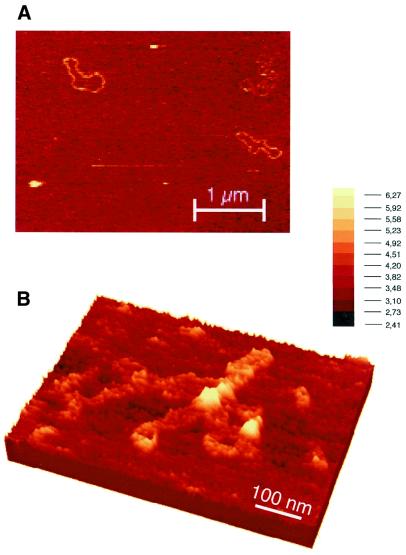
We also used AFM to investigate the FIS nucleoprotein complexes formed with relaxed pUC18 plasmid DNA. As with ptyrTlac, with pUC18 DNA FIS was associated with the crossover nodes at the bases of loops and branch points (Fig. 4B) demonstrating that these structures are not sample preparation artefacts. Similar structures with relaxed pUC18 were observed by EM (data not shown). As pUC18 DNA lacks phased FIS binding sites comparable with those in the tyrT UAS, we infer that the formation of branched DNA structures does not require this arrangement of sites.
Figure 4.
FIS is associated with crossovers and branch points in pUC18 DNA. AFM images are shown (A). An overview of free pUC18 DNA molecules. The picture shown represents baseline corrected raw data, with no filtering. (B) A complex formed at the FIS to pUC18 DNA ratio of ∼1 dimer per 325 bp DNA. The image shown represents a three-dimensional rendering of baseline corrected raw data.
Visualisation of complexes of H-NS with plasmid DNA
To test whether the FIS nucleoprotein complexes visualised by EM specifically differ from those involving other chromatin proteins of E.coli, we prepared complexes between the chromatin protein H-NS and ptyrTlac under the same conditions. We found that in contrast to FIS, H-NS formed extended ‘filaments’ associated with DNA loops of different sizes often located at the extremities (Fig. 5). The overall appearance of the majority of complexes suggested a nucleation of binding and a ‘zipper’-like propagation of H-NS along the DNA. Contour length measurements of free and packaged plasmid molecules showed that if the length of the filament in the latter was multiplied by two, the lengths of the samples were very comparable (2.20 ± 0.038 versus 2.22 ± 0.05 µm without and with H-NS, respectively, n = 25 each). Thus, the packaging of the entire circular DNA molecule in the H-NS filament would result in a 2-fold compaction of DNA along its contour length. We infer that the H-NS filaments contained two DNA duplexes packaged alongside each other. This mode of packaging supercoiled DNA clearly differs from that mediated by FIS.
Figure 5.
Electron micrographs of complexes of H-NS with ptyrTlac DNA. Elongated complexes (filaments) are formed presumably containing two DNA duplexes. Ratio of H-NS to DNA was ∼1 molecule per 10 bp (A) and ∼1 molecule per 3 bp (B). The scale bar represents 1 kb.
Probing of the binding pattern of FIS and H-NS to supercoiled DNA with restriction enzymes
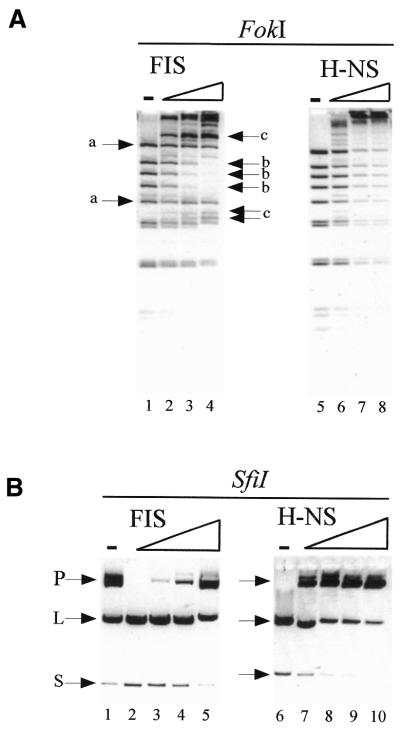
The distinct architectural features of the FIS and H-NS nucleoprotein complexes imply different modes of protein–DNA interactions. To investigate the distinct patterns of DNA occupation in solution we probed the nucleoprotein complexes assembled on ptyrTlac with frequently cutting restriction endonucleases. The rationale of this experiment is that depending on whether multiple binding of protein to DNA occurs in a random or non-random fashion, the cleavage of multiple restriction sites scattered throughout the DNA molecule will be affected differentially. For optimal resolution, high concentrations of the enzyme FokI and short incubation times (5 min) were used. In the presence of increasing FIS concentrations the abundance of some restriction fragments remained essentially unchanged (Fig. 6A, left panel, arrow ‘a’), whereas the amount of others was diminished (arrow ‘b’) and also some new fragments indicative of partial digestion appeared (arrow ‘c’). The pattern obtained implies selectivity in the inhibition of cleavage at different restriction sites and indicates that FIS occupied the binding sites in ptyrTlac sequentially and in a non-random fashion. Similar results were obtained with other frequently cutting restriction endonucleases (data not shown). In contrast, increasing concentrations of H-NS generally inhibited FokI cleavage activity without noticeable selectivity, as expected if extended parts of DNA molecules were randomly packaged by H-NS (Fig. 6A, right panel). This latter observation corroborates the notion that H-NS binding is essentially sequence independent (44,45).
Figure 6.
Probing of protein–DNA complexes with restricrtion endonucleases. (A) Probing with FokI. The FIS to DNA ratio in lanes 2–4 corresponded to 1 FIS dimer per 325, 100 and 40 bp, respectively. The H-NS to DNA ratio in lanes 6–8 corresponded to 1 molecule per 100, 30 and 10 bp, respectively. Electrophoresis was in 3% agarose gels. (A) Probing with SfiI. Low amounts of SfiI were used in the left panel (see lane 1) to reveal the stimulation of cleavage by FIS. The FIS to DNA ratio in lanes 2–5 corresponded to 1 FIS dimer per 325, 160, 80 and 40 bp, respectively. A 2-fold higher concentration of SfiI was used in the right panel (compare lanes 1 and 6) to reveal the inhibition of cleavage by H-NS. The H-NS to DNA ratio in lanes 7–10 corresponded to 1 molecule per 300, 100, 30 and 10 bp, respectively. The substrate plasmid DNA (P), the large (L) and 900 bp small (S) cleavage products are indicated. Electrophoresis was in 1% agarose gels.
In a second approach, we used the cleavage reaction by SfiI endonuclease. The cleavage at two recognition sites by SfiI requires the binding of a tetrameric enzyme in cis to two distant copies of recognition sequence and looping of the intervening DNA (46). Intramolecular interactions are generally enhanced by supercoiling (47,48) and branching of DNA. As the FIS–DNA structures observed demonstrate an interwound and branched DNA conformation, we tested the effect of FIS on the efficiency of SfiI cleavage at two sites separated by 0.9 kb in the plasmid pGE101. Under the conditions used, the SfiI cleavage was inefficient but was significantly enhanced at the FIS to DNA ratios, which lead to the formation of branched structures observed by EM (Fig. 6B, left panel, compare lanes 1 and 2). At very high FIS concentrations (lane 5) this enhancing effect was lost, probably due to an extensive occupation of the DNA by protein and resultant impediment of simultaneous binding of SfiI at two sites. With H-NS no enhancement of SfiI cleavage was observed (data not shown). In contrast, the addition of H-NS gradually reduced the cleavage by SfiI in a concentration-dependent manner (Fig. 6B, right panel).
The results of probing with restriction enzymes are thus consistent with the disparate structural features of the FIS and H-NS nucleoprotein complexes observed by EM.
DISCUSSION
In this study we present direct physical evidence that the non-random occupation of multiple FIS binding sites in supercoiled plasmids results in DNA branching. As FIS is the major component of the bacterial chromatin during the exponential growth phase (19,20), we propose that these structures may reflect some architectural features of the transcriptionally active bacterial chromatin.
A structural role for FIS in E.coli chromatin
Our EM and AFM data indicate that FIS stabilises a branched DNA conformation. The visualisation by AFM excludes the possibilities that the structures observed by EM could be artefacts of fixation or drying. In both cases the DNA was adsorbed to mica and therefore the possibility remains that adsorption might deform the structure. However, the changes in the pattern of FokI and SfiI cleavage in the presence of FIS support the notion that FIS alters the organisation of the plasmid DNA in solution.
FIS constrains negative supercoils in vitro and changes the linking number of relaxed pBR322 DNA by 2–3. This low value suggests that FIS does not constrain DNA supercoils by toroidal wrapping. The level of supercoiling induced by FIS in vitro is 3–5-fold less than that induced by HU (41). According to our data the main difference between free and FIS-bound supercoiled DNA is in the organisation of branches. Therefore, we would not necessarily expect a substantial compaction of DNA on binding FIS, whereas binding of HU leads to a strong compaction by wrapping DNA in nucleosome-like particles (41,42,49). We note however, that the fis-dependent changes in DNA topology in vivo are most probably a consequence of both direct binding effects and repression of gyrase production by FIS (22).
The strain consequent on DNA bending is thought to induce loops and branch points in supercoiled DNA (50). Thus, the stabilisation of branched DNA conformation observed by both EM and AFM can be explained by the bending of DNA by FIS, although the involvement of protein interactions between FIS molecules, especially at high FIS concentrations, cannot be excluded. The results of probing FIS–DNA complexes with SfiI strongly suggest that FIS increases the frequency of the encounters between remote DNA segments (Fig. 7). This latter feature is characteristic of supercoiled molecules containing intrinsic bends that affect the local concentration of one binding site in the proximity of the other (50). It is thus conceivable that the stabilisation of writhe and organisation of branches consequent on DNA bending by FIS modulates the global reshaping dynamics of supercoiled DNA (47,51). This would also explain the relatively low level of superhelical density constrained by FIS in vitro.
Figure 7.
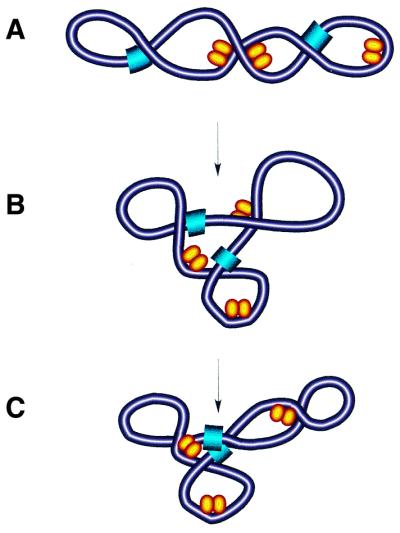
Model of DNA reshaping by FIS. (A) Binding and bending of isolated DNA regions in plectonemically supercoiled DNA by single FIS dimers (yellow ellipsoids) facilitates the branching of DNA with resultant multiplication of apical loops. (B) Note that the FIS-dependent branching of DNA locates the two remote sites (blue) in close spatial proximity (C).
We have also shown that at high protein concentrations the non-random binding of FIS to supercoiled plasmids leads to formation of clusters over extended regions of DNA. Formation of such structures in vivo would require high local FIS concentrations or clusters of high-affinity FIS binding sites within limited chromatin regions serving as ‘sinks’ for FIS. Such a possibility is suggested by recent findings using immunofluorescence microscopy and showing selective binding of the bulk of FIS protein to only a few regions of the chromosome (52).
Packaging of DNA in the H-NS nucleoprotein complexes
We observed using EM that another abundant DNA architectural protein H-NS forms structures that are distinctly different from FIS nucleoprotein complexes. The results of probing with restriction enzymes also indicate differences in the organisation of FIS and H-NS nucleoprotein complexes in solution. These disparate architectures could account for the ‘conformational competition’ implicated in the growth phase-dependent regulation of stable RNA synthesis by FIS and H-NS (53). Our EM data confirm previous observations (54) that H-NS forms a filament containing two DNA duplexes by acting as a ‘zipper’. This mode of binding compacts the DNA resulting in a 2-fold reduction in the DNA contour length. Similar propagation of H-NS polymerisation, but on a single DNA duplex, has been observed by Tupper et al. (44). It is possible that in supercoiled molecules the polymerisation of H-NS on single DNA duplexes proceeds concomitantly with their intertwining leading to the formation of an interwound filament containing two DNA duplexes joined by protein bridges. Whatever the exact mechanism, the observed polymerisation of H-NS along the DNA is consistent with the ‘DNA occlusion’ model for transcriptional silencing by H-NS (33).
Relevance of in vitro FIS–DNA complexes to in vitro structures
Any interpretation of the biological relevance of the observed FIS–DNA complexes is subject to the caveat that these complexes are formed under different conditions from those prevailing in vivo. In particular the DNA concentration is considerably lower and the system is much simplified by, for example, the omission of competing DNA-binding proteins. The FIS concentrations used are largely within the range of those occurring in vivo.
Although the relevance of the observed FIS–DNA structures for the organisation of bacterial chromatin remains to be clarified, there are clear parallels to the structure of transcriptionally active DNA. The branching of DNA multiplies the apical loops, while RNA polymerase is found to be preferentially associated with apical loops of supercoiled DNA (55). Furthermore, as a curved region in supercoiled DNA preferentially locates at the apex of a loop (50,56), the previously inferred stabilisation of a tightly bent UAS DNA microloop by FIS (27,32) could locate the FIS-dependent promoter in the vicinity of an apex and thus direct its interaction with RNA polymerase.
FIS and H-NS are among the most highly abundant of the DNA architectural proteins in E.coli. However, the concentration of FIS in the cell varies strongly being maximal (∼50 µM) during early to mid-exponential growth phase, when the transcriptional activity of the cell is highest (14,15). The variation in the concentration of H-NS with growth phase is much less (19). The branched structure stabilised on binding of FIS to supercoiled DNA remains relatively ‘open’ in comparison to DNA compacted by H-NS. In vivo, while H-NS is evenly distributed within the nucleoid, FIS even at high intracellular concentrations is localised to specific loci (52), possibly corresponding to a strong clustering of FIS binding sites on either side of the replication terminus (57). We therefore propose that the degree of compaction of the E.coli nucleoid depends on the ratio of FIS to other nucleoid-associated proteins, including H-NS. In this model the branching of DNA would maintain chromatin in an ‘open’ state accessible to transcriptional machinery during early and mid-exponential phase and the chromatin would become more compacted and less transcriptionally active on the transition to the stationary phase.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Prof. H. E. Gaub, Lehrstuhl fuer Angewandte Physik, and Digital Instruments, Veeco Metrology Group, for providing the MultiMode™ Scanning Probe Microscope used in this work. We thank Andrea Schultz for excellent technical assistance and Claudio Gualerzi for the purified H-NS protein. This work was supported by Deutsche Forschungsgemeinschaft through SFB 190 and SFB 397.
REFERENCES
- 1.Sinden R.R., Carlson,J.O. and Pettijohn,D.E. (1980) Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell, 21, 773–783. [DOI] [PubMed] [Google Scholar]
- 2.Boles T.C., White,J.H. and Cozzarelli,N.R. (1990) Structure of plectonemically supercoiled DNA. J. Mol. Biol., 213, 931–951. [DOI] [PubMed] [Google Scholar]
- 3.Worcel A. and Burgi,E. (1972) On the structure of the folded chromosome of Escherichia coli.J. Mol. Biol., 71, 127–147. [DOI] [PubMed] [Google Scholar]
- 4.Sinden R.R. and Pettijohn,D.E. (1981) Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl Acad. Sci. USA, 78, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staczek P. and Higgins,N.P. (1998) Gyrase and topo IV modulate chromosome domain size in vivo. Mol. Microbiol., 29, 1435–1448. [DOI] [PubMed] [Google Scholar]
- 6.Bohrmann B., Villiger,W., Johansen,R. and Kellenberger,E. (1991) Coralline shape of the bacterial nucleoid after cryofixation. J. Bacteriol., 173, 3149–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinow C. and Kellenberger,E. (1994) The bacterial nucleoid revisited. Microbiol. Rev., 58, 211–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dürrenberger M., Bjornsti,M.-A., Uetz,T., Hobot,J.A. and Kellenberger,E. (1988) Intacellular location of the histonelike protein HU in Escherichia coli.J. Bacteriol., 170, 4757–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pettijohn D. (1990) Bacterial chromosome structure. Nucleic Acids Mol. Biol., 4, 152–162. [Google Scholar]
- 10.Reich Z., Wachtel,E.J. and Minsky,A. (1994) Liquid-crystalline mesophases of plasmid DNA in bacteria. Science, 264, 1460–1463. [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch B.J. and Bendich,A.J. (1997) The bacterial nucleoid visualized by fluorescence microscopy of cells lysed within agarose: comparison of Escherichia coli and spirochetes of genus Borrelia.J. Bacteriol., 179, 2228–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spassky A., Rimsky,S., Garreau,H. and Buc,H. (1984) H1a, an E. coli DNA-binding protein which accumulates in stationary phase. Nucleic Acids Res., 12, 5321–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmid M.B. (1990) More than just ‘Histone-like’ proteins. Cell, 63, 451–453. [DOI] [PubMed] [Google Scholar]
- 14.Ball C.A., Osuna,R., Ferguson,K.C. and Johnson,R.C. (1992) Dramatic changes in FIS levels upon nutrient upshift in Escherichia coli.J. Bacteriol., 174, 8043–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ninnemann O., Koch,C. and Kahmann,R. (1992) The E. coli fis promoter is subject to stringent control and autoregulation. EMBO J., 11, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dersch P., Schmidt,K. and Bremer,E. (1993) Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol., 8, 875–889. [DOI] [PubMed] [Google Scholar]
- 17.Ditto M.D., Roberts,D. and Weisberg,R.A. (1994) Growth phase variation of integration host factor level in Escherichia coli. J. Bacteriol., 176, 3738–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Claret L. and Rouvière-Yaniv,J. (1996) Regulation of HUa and HUb by CRP and FIS in Escherichia coli.J. Mol. Biol., 263, 126–139. [DOI] [PubMed] [Google Scholar]
- 19.Talukder A.A., Iwata,A., Nishimura,A., Ueda,S. and Ishihama,A. (1999) Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol., 181, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishihama A. (1999) Modulation of the nucleoid, the transcriptional apparatus and the translation machinery for stationary phase survival. Genes Cells, 4, 135–143. [DOI] [PubMed] [Google Scholar]
- 21.Schneider R., Travers,A.A. and Muskhelishvili,G. (1997) FIS modulates growth-phase dependent topological transitions of DNA in E. coli.Mol. Microbiol., 26, 519–530. [DOI] [PubMed] [Google Scholar]
- 22.Schneider R., Kutateladze,T., Travers,A.A. and Muskhelishvili,G. (1999) A DNA architectural protein couples cellular physiology and DNA supercoiling in Escherichia coli.Mol. Microbiol., 34, 953–964. [DOI] [PubMed] [Google Scholar]
- 23.Schneider R., Travers,A. and Muskhelishvili,G. (2000) The expression of the Escherichia coli fis gene is strongly dependent on the superhelical density of DNA. Mol. Microbiol., 38, 167–176. [DOI] [PubMed] [Google Scholar]
- 24.Hübner P. and Arber,W. (1989) Mutational analysis of a prokaryotic recombinational enhancer with two functions. EMBO J., 8, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hübner P., Haffter,P., Iida,S. and Arber,W. (1989) Bent DNA is needed for recombinational enhancer activity in the site-specific recombination system Cin of the bacteriophage P1. The role of FIS protein. J. Mol. Biol., 205, 493–500. [DOI] [PubMed] [Google Scholar]
- 26.Lazarus L.R. and Travers,A.A. (1993) The E. coli FIS protein is not required for the activation of tyrT transcription on simple nutritional upshift. EMBO J., 12, 2483–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muskhelishvili G., Travers,A.A., Heumann,H. and Kahmann,R. (1995) FIS and RNA polymerase holoenzyme form a specific nucleoprotein complex at a stable RNA promoter. EMBO J., 14, 1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perkins-Balding C., Dias,D.P. and Glasgow,A.C. (1997) Location, degree and direction of DNA bending associated with the Hin recombinational enhancer sequence and Fis-enhancer complex. J. Bacteriol., 179, 4747–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailly C., Waring,M. and Travers,A.A. (1995) Effects of base substitutions on the binding of a DNA-bending protein. J. Mol. Biol., 253, 1–7. [DOI] [PubMed] [Google Scholar]
- 30.Pan C.Q., Finkel,S.E., Cramton,S.E., Feng,J., Sigman,D.S. and Johnson,R.C. (1996) Variable structures of FIS-DNA complexes determined by flanking DNA-protein contacts. J. Mol. Biol., 264, 675–695. [DOI] [PubMed] [Google Scholar]
- 31.Theis K. (1996) Untersuchungen zur struktur und DNA-bindung des proteins FIS aus Escherichia coli. Doctoral Thesis, Freie Universitaet Berlin, Berlin, Germany.
- 32.Muskhelishvili G. and Travers,A.A. (1997) The stabilization of DNA microloops by FIS – a mechanism for torsional transmission in transcription activation and DNA inversion. Nucleic Acids Mol. Biol., 11, 179–190. [Google Scholar]
- 33.Ussery D.W., Hinton,J.C.D., Jordi,B.J.A.M., Granum,P.E., Seirafi,A., Stephen,R.J., Tupper,A.E., Berridge,G., Sidebotham,J.M. and Higgins,C.F. (1994) The chromatin associated protein H-NS. Biochimie, 76, 968–980. [DOI] [PubMed] [Google Scholar]
- 34.Lazarus L.R. (1992) The role of FIS in tyrT transcriptional regulation. PhD thesis, University of Cambridge, Cambridge, UK.
- 35.Koch C. and Kahmann,R. (1986) Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J. Biol. Chem., 261, 15673–15678. [PubMed] [Google Scholar]
- 36.Spiess E. and Lurz,R. (1988) Electron microscopic analysis of nucleic acids and nucleic acid-protein complexes. Methods Microbiol., 20, 293–323. [Google Scholar]
- 37.Binnig G., Quate,C.F. and Gerber,C. (1986) Atomic force microscope. Phys. Rev. Lett., 56, 930–933. [DOI] [PubMed] [Google Scholar]
- 38.Hansma H.G., Vesenka,J., Kelderman,G., Morrett,H., Sinsheimer,R.L., Elings,V., Bustamante,C. and Hansma,P.K. (1992) Reproducible imaging and dissection of plasmid DNA under liquid with the AFM. Science, 256, 1180–1184. [DOI] [PubMed] [Google Scholar]
- 39.Hansma H.G. and Laney,D.E. (1996) DNA binding to mica correlates with cationic radius: assay by atomic force microscope. Biophys. J., 68, 1672–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansma P.K., Cleveland,J.P., Radmacher,M., Walters,D.A., Hillner,P.E., Bezanilla,M., Fritz,M., Vie,D., Hansma,H.G., Prater,C.B., Massie,J., Fukunaga,L., Gurley,J. and Elings,V. (1994) Tapping mode atomic force microscopy in liquids. Appl. Phys. Lett., 64, 1738–1740. [Google Scholar]
- 41.Rouvière-Yaniv J., Yaniv,M. and Germond,J.-E. (1979) E. coli DNA binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell, 17, 265–274. [DOI] [PubMed] [Google Scholar]
- 42.Broyles S.S. and Pettijohn,D.E. (1986) Interaction of the Escherichia coli HU protein with DNA. Evidence for the formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol., 187, 47–60. [DOI] [PubMed] [Google Scholar]
- 43.Payet D. and Travers,A. (1997) The acidic tail of the high mobility group protein HMG-D modulates the structural selectivity of DNA binding. J. Mol. Biol., 266, 66–75. [DOI] [PubMed] [Google Scholar]
- 44.Tupper A.E., Owen-Hughes,T., Ussery,D.W., Santos,D.S., Ferguson,D.J., Sidebotham,J.M., Hinton,J.C. and Higgins,C.F. (1994) The chromatin-associated protein H-NS alters DNA topology in vitro. EMBO J., 13, 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falconi M., Higgins,N.P., Spurio,R., Pon,C. and Gualerzi,C. (1993) Expression of the gene encoding the major bacterial nucleoid protein H-NS is subject to transcriptional autorepression. Mol. Microbiol., 10, 273–282. [DOI] [PubMed] [Google Scholar]
- 46.Nobbs T.J. and Halford,S.E. (1995) DNA cleavage at two recognition sites by the SfiI restriction endonuclease: salt dependence of cis and trans interactions between distant DNA sites. J. Mol. Biol., 252, 399–411. [DOI] [PubMed] [Google Scholar]
- 47.Vologodskii A.V., Levene,S.D., Klenin,K.V., Frank-Kamenetskii,M.D. and Cozzarelli,N.R. (1992) Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol., 227, 1224–1243. [DOI] [PubMed] [Google Scholar]
- 48.Langowski J. (1997) Modelling large DNA molecules: long-range interactions and regulation of transcription. Nucleic Acids Mol. Biol., 11, 219–237. [Google Scholar]
- 49.Tanaka H., Yasuzawa,K., Kohmo,K., Goshima,N., Kano,Y., Saiki,T. and Imamoto,F. (1995) Role of HU proteins in forming and constraining supercoils of chromosomal DNA in Escherichia coli.Mol. Gen. Genet., 248, 518–526. [DOI] [PubMed] [Google Scholar]
- 50.Rippe K., von Hippel,P. and Langowski,J. (1995) Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci., 20, 500–506. [DOI] [PubMed] [Google Scholar]
- 51.Jian H., Schlick,T. and Vologodskii,A. (1998) Internal motion of supercoiled DNA: Brownian dynamics simulations of site juxtaposition. J. Mol. Biol., 284, 287–296. [DOI] [PubMed] [Google Scholar]
- 52.Talukder A.A., Hiraga,S. and Ishihama,A. (2000) Two types of localisation of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells, 5, 613–626. [DOI] [PubMed] [Google Scholar]
- 53.Afflerbach H., Schröder,O. and Wagner,R. (1998) Conformational changes in the upstream DNA mediated by H-NS and FIS regulate E. coli rrnB P1 promoter activity. J. Mol. Biol., 286, 339–353. [DOI] [PubMed] [Google Scholar]
- 54.Dame R.T., Wyman,C. and Goosen,N. (2000) H-NS mediated compaction of DNA visulised by atomic force microscopy. Nucleic Acids Res., 28, 3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. ten Heggeler-Bordier B., Wahli,W., Adrian,M., Stasiak,A. and Dubochet,J. (1992) The apical localisation of transcribing RNA polymerase on supercoiled DNA prevents their rotation around the template. EMBO J., 11, 667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laundon C.H. and Griffith,J.D. (1988) Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell, 52, 545–549. [DOI] [PubMed] [Google Scholar]
- 57.Ussery D., Larsen,T.S., Wilkes,K.T., Friis,C., Worning,P., Krogh,A. and Brunak,S. (2001) Genome organisation and chromatin structure in Escherichia coli.Biochimie, 83, 200–212. [DOI] [PubMed] [Google Scholar]