Abstract
A compensatory mutation (M230I) in the primer grip of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) restores the replication capacity of virus having a Y115W mutation in their RT coding region. The Y115W substitution impairs DNA polymerase activity and produces an enzyme with a lower fidelity of DNA synthesis. Gel-based fidelity assays with the double mutant Y115W/M230I revealed that the M230I substitution increased the accuracy of mutant Y115W. Y115W/M230I showed wild-type misinsertion fidelity in assays performed with DNA/DNA templates. However, when present alone, M230I conferred reduced fidelity as determined in misinsertion and mispair extension fidelity assays, as well as in primer extension assays carried out with three dNTPs. The mutant M230I showed a 3.3–16-fold increase in misinsertion efficiency for G, C and T opposite T, compared with the wild-type enzyme. Its fidelity was not influenced by nucleotide substitutions in the template/primer around the incorporation site. However, its accuracy was apparently affected by the structure of the 5′-overhang of the template strand. Unlike wild-type HIV-1 RT, nucleotide selectivity of mutant M230I at dT:dG, dT:dC and dT:dT mispairs was almost exclusively dependent on the Km values for correct and incorrect dNTPs, a characteristic that has not been described for other low fidelity mutants of HIV-1 RT.
INTRODUCTION
During the retrovirus life cycle, the human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) replicates the viral genomic RNA to synthesize a double-stranded DNA which integrates into the host genome. RT is a multifunctional enzyme, possessing RNA- and DNA-dependent DNA polymerase, RNase H, strand transfer and strand displacement activities (1). HIV-1 RT is a heterodimer composed of two subunits of 66 and 51 kDa, with subdomains termed fingers, thumb, palm and connection in both subunits and an RNase H domain in the larger subunit only. The overall folding of both subunits is similar, but the spatial arrangements of their subdomains are remarkably different. Highly conserved amino acid regions in the fingers and palm subdomains of the 66 kDa subunit, together with two α-helices of the thumb subdomain, act as a clamp to position the template/primer relative to the polymerase active site. The polymerase active site resides within the palm subdomain of the 66 kDa subunit, which contains the catalytic aspartic acid residues 110, 185 and 186. Other residues in their vicinity, such as Lys65, Arg72, Asp113, Ala114, Tyr115 and Gln151, are involved in the interaction with the incoming dNTP (2).
Crystallographic data have implicated residues 227–235 (referred to as the primer grip region) in orienting the primer terminus for nucleophilic attack on an incoming dNTP (3). In agreement with this proposal, it has been shown that substitution of Ala for Met230 produces a significant decrease in the dNTP binding affinity of the enzyme (4). When introduced into a proviral HIV-1 clone, the M230A mutation produces a non-infectious virus that shows severe defects in proviral DNA synthesis (5). Alanine-scanning mutagenesis of other residues forming the primer grip also produces RTs with significant alterations in their DNA polymerase and RNase H activities. In the case of mutations W229A, G231A and Y232A, the affinity of the recombinant RT for a template/primer was substantially reduced, compromising viral infectivity (4,6,7). Several primer grip mutant RTs, such as P226A, F227A, G231A, Y232A, E233A and H235A, displayed an altered RNase H phenotype, which could result from repositioning of the template/primer in the nucleic acid-binding cleft (7,8). Also, mutations at residues 226, 227, 229, 230, 231, 232 and 235 reduced or eliminated RNA-primed minus strand synthesis from the primer-binding site (9).
The process of reverse transcription is error prone and contributes to the high degree of genetic variability of the virus. One of the consequences of the high mutation rate has been the emergence of variants escaping the host’s immune response, as well as viruses that are resistant to anti-retroviral drugs such as RT or protease inhibitors. Nucleotide selectivity depends primarily on base pair geometry (reviewed in 10,11). The interactions between the viral RT and its substrates (i.e. dNTPs and template/primer duplexes) are important determinants of the fidelity of DNA synthesis. Mutagenesis studies have shown that several amino acid substitutions at the dNTP-binding site may either increase or decrease the fidelity of DNA synthesis. Examples of mutations that may increase the accuracy of HIV-1 RT are K65R, D76I, D76R, D76V, R78A, M184V and M184I (12–16). Non-conservative substitutions of Tyr115, as well as substitutions of Ala for Arg72 or Met184, reduce the accuracy of HIV-1 RT (17–21). Residues outside the dNTP-binding site can also influence nucleotide incorporation and fidelity of DNA synthesis. For example, replacing Gly262 with Ala or Trp266 with Glu, Leu, Ala, Ile, Val, Arg or Tyr led to RT mutants with a low frameshift fidelity (22–24). Gly262 and Trp266 are located in α-helix H and are involved in interactions with the primer strand at the minor groove of the template/primer (22,25).
Mutations in the primer grip have been reported to produce a low to moderate increase in fidelity of DNA synthesis. Thus, RTs having Ala instead of Phe227, Trp229, Met230, Gly231 or Tyr232 were 40–76% less efficient than the wild-type enzyme in extending a mismatch in primer extension assays performed in the presence of all four dNTPs (26). The higher accuracy of mutants F227A and W229A compared with the wild-type RT was further confirmed in misinsertion fidelity assays measuring the incorporation of A, C, G or T opposite T (26).
In transfection experiments carried out with proviral DNA having the RT mutation Y115W, we found a compensatory substitution (M230I) in the primer grip which restored the replication capacity of the virus (27). Y115W impaired RT activity by decreasing the dNTP binding affinity of the polymerase, but acquisition of the M230I mutation compensated for the dNTP binding defect. All retroviral RTs have either Met or Leu at the equivalent position of Met230 of HIV-1 RT (28). Met230 is highly conserved among HIV-1 isolates. Leu230 has been identified in virus grown in cell culture and passaged in the presence of the non-nucleoside inhibitor delavirdine (29). In addition, mutation M230I has recently been found in the RT coding region of two viral isolates obtained from patients treated with the RT inhibitor HBY 097 (30).
In this study, we report on the effects of the M230I mutation on fidelity of DNA synthesis, either in the presence or absence of the nucleotide-binding site mutation Y115W. The reported data indicate that M230I restores wild-type fidelity of DNA-dependent DNA synthesis when the Y115W mutation is present. Interestingly, HIV-1 RT bearing the single mutation M230I showed reduced fidelity in different sequence contexts. While nucleotide substitutions at the primer positions interacting with the side chain of Met230 did not have an influence on nucleotide discrimination, the structure of the 5′-extension of the template strand appears as an important determinant of fidelity of DNA synthesis.
MATERIALS AND METHODS
Enzymes
WT RT and mutants Y115W, M230I and Y115W/M230I were expressed and purified as described previously (17,27). All RTs were purified as p66/p51 heterodimers. In this study, mutations were introduced in both subunits of the RT. The 51 kDa polypeptide was obtained with an extension of 14 amino acid residues at its N-terminal end, which includes six consecutive histidine residues to facilitate its purification by metal chelate affinity chromatography.
Fidelity assays
Misinsertion and mispair extension fidelity assays were performed essentially as described previously (31,32), using a standing-start protocol. Templates/primers used in these assays are shown in Figure 1. Gel-purified synthetic oligonucleotide primers were 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase and then annealed to their corresponding templates in 150 mM NaCl and 150 mM magnesium acetate, as described (18). Assays were performed in 10 µl of 50 mM HEPES, 15 mM NaCl, 15 mM magnesium acetate, 130 mM potassium acetate, 1 mM dithiothreitol and 5% (w/v) polyethylene glycol 6000, pH 7.0. The template/primer concentrations were 20–30 nM. The active enzyme concentration was ∼1.2–8.0 nM. Reactions were initiated by incubating the enzyme with the corresponding annealed template/primer in the absence of nucleoside triphosphates (10 min at 37°C), followed by addition of appropriate nucleotides at various concentrations. The rate of product formation was measured for 11 to 14 different concentrations of each correct or incorrect dNTP. Typically, for misinsertion fidelity assays concentrations were in the range 3 nM–20 µM for correct dNTPs and 50 µM–10 mM for incorrect dNTPs. Elongation reactions for the incorporation of correct dNTPs were carried out at 37°C for 10–30 s, depending on the template/primer used. Under these conditions, reactions were linear with respect to time and proportional to the enzyme concentration. After incubation, reactions were terminated by adding EDTA (5 mM final concentration), followed by heat denaturation. Products were resolved in 20% polyacrylamide–urea gels and quantitated with a BAS 1500 scanner. Elongation measurements were fitted to the Michaelis–Menten equation and the kcat and apparent Km values were determined as previously described (17,33).
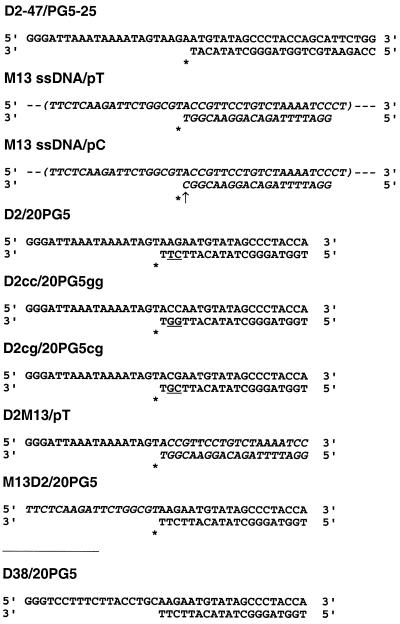
Figure 1.
Nucleotide sequences of the template/primer complexes used in the assays. Templates D2, D38 and D2-47 derive from an HIV-1BH10 gag sequence that starts at nucleotide 915 (5′-end), according to the sequence numbering of GenBank accession no. M15654. M13-derived sequences are shown in italic. Incorporation sites are indicated with an asterisk and the mismatch in template/primer M13 ssDNA/pC is marked with an arrow. Underlined nucleotides at positions –2 and –3 of primers highlight the differences between templates/primers D2/20PG5, D2cc/20PG5gg and D2cg/20PG5cg. M13 ssDNA, M13 single-stranded DNA.
Extension of primers in the presence of three dNTPs
The template/primer D38/20PG5 (Fig. 1) was used. 20PG5 was 5′-end-labeled and annealed to D38 as described above. Five microliters of a solution containing 20–50 nM enzyme and 60 nM template/primer in 100 mM HEPES, 30 mM NaCl, 30 mM magnesium acetate, 130 mM potassium acetate, 1 mM dithiothreitol and 5% (w/v) polyethylene glycol 6000, pH 7.0, were incubated at 37°C for 10 min. Primer extension was initiated by adding 5 µl of a mixture containing three or four dNTPs at a concentration of 200 µM each, in 130 mM potassium acetate, 1 mM dithiothreitol and 5% (w/v) polyethylene glycol 6000. Reactions were incubated at 37°C for 0–30 min and then stopped by adding 4 µl of 10 mM EDTA in loading buffer containing 90% formamide. DNA synthesis products were separated on a 20% denaturing polyacrylamide gel, visualized by autoradiography and quantitated by phosphorimaging.
RESULTS
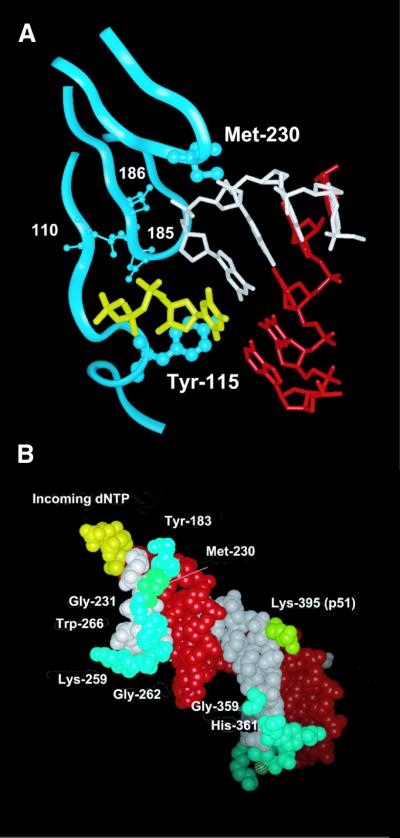
Crystal structures of HIV-1 RT complexed with double-stranded DNA (2,3,34) revealed that the side chain of Met230 interacts with the ribose of the nucleotide at position –2 of the primer (Fig. 2). Met230 together with other residues (i.e. Tyr183, Gly231, Trp266 and Gly262, among others) contact the primer in the minor groove of the template/primer complex. The distances between atoms Sδ and Cɛ of Met230 and carbon and oxygen atoms of the interacting ribose ring are in the range 3.6–4.2 Å. Modeling studies of the M230I mutation predict that atoms Cβ, Cγ1 and Cδ of Ile could occupy equivalent positions to those observed for atoms Cβ, Cγ and Sδ of Met230. In the mutant RT, the absence of an ɛ-methyl group would reduce the contact surface between this residue and the primer strand, while the γ2-methyl group of Ile230 would protrude towards the ribose ring at position –2 of the primer. As a consequence of the mutation, the free 3′-hydroxyl group of the primer could adopt a different orientation in the active site of the polymerase.
Figure 2.
Interaction between Met230 and the primer strand in an HIV-1 RT/DNA/DNA template/primer/dNTP complex. (A) View of the dNTP-binding site showing the location of Tyr115 and Met230 (represented by a ball and stick model). The template, primer and incoming dNTP are shown in red, white and yellow, respectively. Ribbons correspond to the p66 backbone at residues 108–120, 180–190 and 225–235. Catalytic aspartic acid residues at positions 110, 185 and 186 are also shown. The template strand (red), the primer strand (white) and the incoming dNTP (yellow) are shown in (B), together with residues of HIV-1 RT that interact with the primer strand, using a space filling representation. Indicated amino acids (Tyr183, Met230, Gly231, Lys259, Gly262, Trp266, Gly359 and His361 of p66 and Lys395 of p51) are located at <4 Å from the primer. Figures were made with the program Insight II v.98.0, using the Brookhaven Protein Databank coordinates 1RTD (2).
Steady-state kinetic parameters for the incorporation of nucleotides at the 3′-end of the primer showed that WT RT, the single mutants Y115W and M230I and the double mutant Y115W/M230I displayed roughly similar kcat values, although the apparent Km values for the incorporation of dNTP were higher for the single mutant Y115W than for the WT enzyme (27). The M230I mutation mitigates the polymerase activity defect shown by the Y115W mutant, probably through restoration of the proper alignment of the α-phosphate of the dNTP and the 3′-hydroxyl group of the primer terminus.
Misinsertion and mispair extension fidelity of the double mutant Y115W/M230I
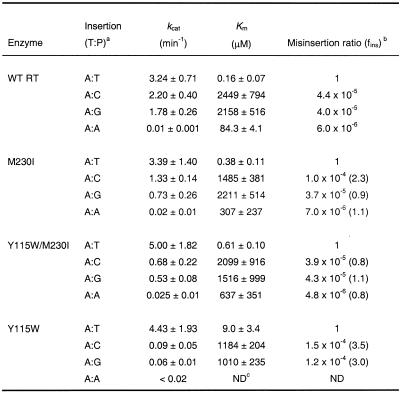
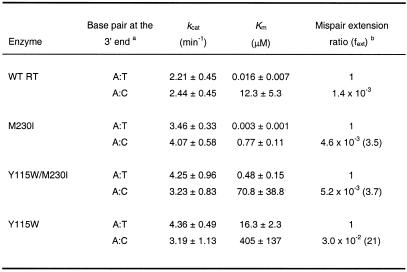
In agreement with our previous results, misinsertion fidelity assays using template/primer D2-47/PG5-25 (Table 1) revealed that the apparent Km for incorporation of the correct nucleotide (dTTP) was 15–56 times higher for mutant Y115W than for WT RT and mutants M230I and Y115W/M230I. Misinsertion ratios were very similar for WT RT and the double mutant Y115W/M230I and ranged from 4.8 × 10–6 to 4.4 × 10–5. However, Y115W misinserted C or G opposite A with a 3.0–3.5-fold higher efficiency than the WT RT. The differences between mutant M230I and WT RT were not significant.
Table 1. Misinsertion fidelity of WT and mutant RTs, as obtained using the template/primer D2-47/PG5-25.
After formation of the RT/DNA/DNA complex, D2-47/PG5-25 elongation reactions were incubated at 37°C for 20 s for dTTP and 45 s–60 min for dCTP, dGTP and dATP. The concentration of template/primer in these assays was 30 nM. Data shown are the mean values ± SD, obtained from a non-linear least squares fit of the kinetics data to the Michaelis–Menten equation. Each of the experiments was performed independently at least three times.
aThe first base indicates the template nucleotide at position +1 and the second base indicates the nucleotide incorporated in the assay.
bfins = [kcat(incorrect)/Km(incorrect)]/[kcat(correct)/Km(correct)], where incorrect nucleotides were dCTP, dGTP or dATP, while the correct nucleotide was dTTP. Numbers in parentheses represent the relative increase in the misinsertion ratio shown by the mutant RT relative to the wild-type enzyme.
cND, not determined.
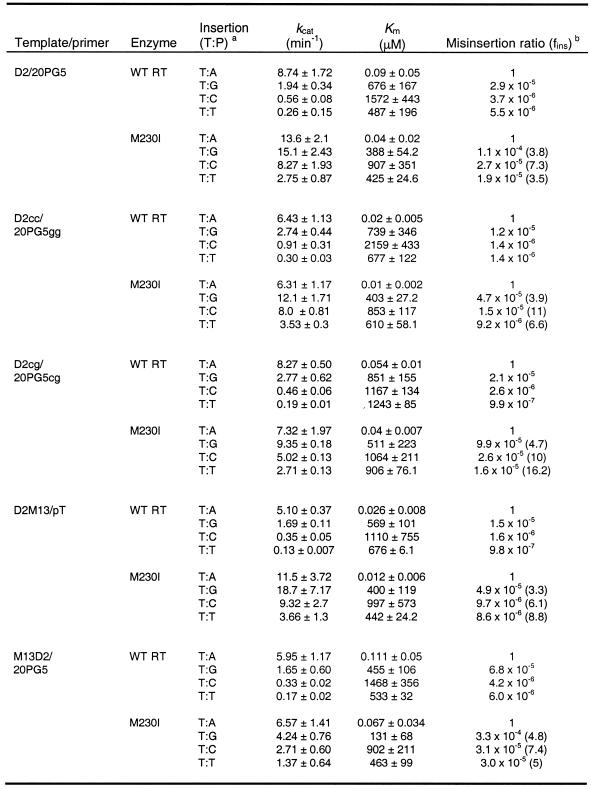
In misinsertion assays performed using the M13 ssDNA/pT complex, differences between Y115W/M230I and the WT RT were again very small (Table 2), with misinsertion ratios ranging from 2.1 × 10–5 to 4.9 × 10–5. These assays also revealed a surprisingly lower misinsertion fidelity for mutant M230I. This enzyme showed misinsertion ratios ranging from 9.3 × 10–5 to 7.6 × 10–4 and were 4.6–16 times higher than those obtained with the WT RT. Interestingly, these assays showed that mutant M230I was prone to misincorporate G opposite T, due to the low Km value for dGTP incorporation.
Table 2. Misinsertion fidelity of WT and mutant RTs, as obtained using the template/primer ssDNA M13/pT.
After formation of the RT/DNA/DNA complex, ssDNA M13/pT elongation reactions were incubated at 37°C for 30 s for dATP and 45 s–60 min for dCTP, dGTP and dTTP. The concentration of template/primer in these assays was 20 nM. Data shown are the mean values ± SD, obtained from a non-linear least squares fit of the kinetics data to the Michaelis–Menten equation. Each of the experiments was performed independently at least three times.
aThe first base indicates the template nucleotide at position +1 and the second base indicates the nucleotide incorporated in the assay.
bfins = [kcat(incorrect)/Km(incorrect)]/[kcat(correct)/Km(correct)], where incorrect nucleotides were dCTP, dGTP or dTTP, while the correct nucleotide was dATP. Numbers in parentheses represent the relative increase in the misinsertion ratio shown by the mutant RT relative to the wild-type enzyme.
The kinetics of mispair extension were studied for correctly matched base pairs (A:T) and for mismatch A:C, in the sequence context of M13 ssDNA/pT. In all cases, the incorporation of a correct T opposite A at the 3′-end of the primer was measured (Table 3). WT RT showed an A:C mispair extension efficiency of 1.4 × 10–3, which is ∼20 times lower than for mutant Y115W. In contrast, M230I and Y115W/M230I showed mispair extension efficiencies that were only 3 times higher than for the WT enzyme, thereby revealing the compensatory effect of M230I for the loss of accuracy caused by Y115W.
Table 3. Mispair extension fidelity of WT and mutant RTs, as obtained using templates/primers ssDNA M13/pT and ssDNA M13/pC.
After formation of the RT/DNA/DNA complex, mispair extension reactions were incubated at 37°C for 30 s. The concentration of template/primer in these assays was 20 nM. In all cases, we measured incorporation of A opposite T at position +1. Data shown are the mean values ± SD, obtained from a non-linear least squares fit of the kinetics data to the Michaelis–Menten equation. Each of the experiments was performed independently at least three times.
aThe first base corresponds to the template and the second base to the primer.
bfext = [kcat(mismatched)/Km(mismatched)]/[kcat(matched)/Km(matched)]. Numbers in parentheses represent the relative increase in the mispair extension ratio shown by the mutant RT relative to the wild-type enzyme. Our analysis assumes that RTs bind with roughly equal affinity to the matched and mismatched template/primer ends (10,19).
Effect of the template/primer sequence context on the misinsertion fidelity of mutant M230I
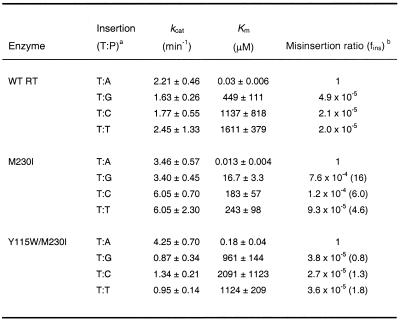
M230I showed decreased DNA synthesis misinsertion fidelity when assayed with M13 ssDNA/pT (Table 2), but not with template/primer D2-47/PG5-25 (Table 1). In addition, misincorporation of G opposite T was rather efficient with mutant M230I, due to its low Km value for dGTP (Table 2). Templates/primers D2/20PG5 and M13 ssDNA/pT show identical sequence at the incorporation site (Fig. 1). In both cases, the correct nucleotide substrate is dATP and in both DNA/DNA complexes its complementary template base (T) is flanked by G and A, at the 5′ and 3′ positions, respectively. Misinsertion fidelity assays performed using the duplex D2/20PG5 revealed that misinsertion ratios for G, C and T opposite T were 3.5–7.3 times higher for mutant M230I than for the WT enzyme (Table 4). Interestingly, M230I showed a Km value for incorporation of dGTP at the 3′-end of primer 20PG5 of 388 µM, which was more than 20 times higher than the value obtained with M13 ssDNA/pT, suggesting that the effect observed for M230I and substrates dGTP and M13 ssDNA/pT did not depend on the nucleotide to be incorporated or on any of the flanking residues in the template/primer.
Table 4. Misinsertion fidelity of WT RT and mutant M230I, as obtained using templates/primers D2/20PG5, D2cc/20PG5gg, D2cg/20PG5cg, D2M13/pT and M13D2/20PG5.
After formation of the RT/DNA/DNA complex, elongation reactions were incubated at 37°C for 10 s for dATP, and 10 s–30 min for dCTP, dGTP and dTTP. The concentration of template/primer in these assays was 20 nM. Data shown are the mean values ± SD, obtained from a non-linear least squares fit of the kinetics data to the Michaelis–Menten equation. Each of the experiments was performed independently at least three times.
aThe first base indicates the template nucleotide at position +1 and the second base indicates the nucleotide incorporated in the assay.
bfins= [kcat(incorrect)/Km(incorrect)]/[kcat(correct)/Km(correct)], where incorrect nucleotides were dCTP, dGTP or dTTP, while the correct nucleotide was dATP. Numbers in parentheses represent the relative increase in the misinsertion ratio shown by the mutant M230I relative to the wild-type enzyme.
The side chain of Met230 interacts with the ribose of the nucleotide at position –2 of the primer (Fig. 2). While positions –2 and –3 of the primer pT are occupied by two purine nucleotides (GG), there are two pyrimidines (TC) at the equivalent positions of 20PG5. In order to study the influence of these two nucleotides on the fidelity of DNA synthesis of M230I, we used templates/primers D2cg/20PG5cg and D2cc/20PG5gg. These complexes contained one or two pyrimidine to purine substitutions at positions –2 and –3 of the primer, when compared with D2/20PG5 (Fig. 1). As shown in Table 4, the results obtained with D2cg/20PG5cg and D2cc/20PG5gg were similar to those obtained with D2/20PG5. Again, misinsertion ratios of mutant M230I were consistently higher in comparison to those obtained with WT RT, but were similar to the values obtained with D2/20PG5.
The influence of the 5′-extension of the template and the double-stranded portion of the template/primer was studied using complexes D2M13/pT and M13D2/PG5 (Fig. 1). D2M13/pT contains the single-stranded DNA 5′-extension of the D2/20PG5 complex and the double-stranded DNA region of M13 ssDNA/pT. On the other hand, M13D2/20PG5 has the annealed region of D2/20PG5, with an 18 nt overhang bearing the sequence found in M13 single-stranded DNA. Using both templates/primers, WT RT showed misinsertion ratios ranging from 9.8 × 10–7 to 1.5 × 10–5 (Table 4). Misinsertion efficiencies obtained with mutant M230I were 3.3–8.8 times higher than those determined with WT RT and were similar to those found with D2/20PG5. In all cases, misinsertion ratios for M230I were higher than for the WT enzyme. The differences between the catalytic constants determined for correct and incorrect nucleotides were relatively small in the case of M230I RT. The nucleotide selectivity of this mutant was determined by the differences between the Km values for correct and incorrect dNTPs.
Interestingly, the apparent Km values for the incorporation of G opposite T obtained with all templates/primers derived from complex D2/20PG5 ranged from 131 to 851 µM, well above the value obtained using the M13 ssDNA/pT complex (16.7 µM). While D2/20PG5 and its related duplexes contain a 38 nt template DNA, M13 ssDNA is a 5.7 kb circular single-stranded DNA. Therefore, the total nucleotide concentration in assays performed with M13 ssDNA/pT is much higher. In order to test whether this factor could affect the steady-state kinetic constants for incorporation of nucleotides, we performed assays with template/primer M13D2/20PG5 in the presence of a 20 nM concentration of M13 ssDNA, which was added exogenously after the M13D2/20PG5 annealing reaction. The determination of kinetic constants for incorporation of G opposite T by M230I did not show significant variations in comparison with the data reported in Table 4. For example, under these conditions the apparent Km values for incorporation of A opposite T and G opposite T were 0.106 ± 0.04 and 541 ± 120 µM for WT RT and 0.075 ± 0.024 and 104 ± 43 µM for mutant M230I, respectively.
The differences in the kinetic parameters for incorporation of dGTP found with M230I and template/primer M13 ssDNA/pT, resulting in significant variations in misinsertion fidelity, could be attributed to the complexity of the 5′-extension of the template. Single-stranded DNA molecules have a strong tendency to fold back on themselves to form irregular double-helical hairpin loops whenever the sequence permits significant numbers of nucleotides to base pair. These structures could affect the geometry of the polymerase active site and, hence, the fidelity of DNA synthesis. DNA secondary structure predictions were carried out with nucleotides 5301–5420 of the template M13 ssDNA sequence (GenBank accession no. L08818), using the program Mfold v.3.0 (35). These predictions showed that 60 nt at the single-stranded DNA 5′-extension adjacent to the incorporation site can generate stable secondary structures (data not shown), with an estimated ΔG value at 37°C of –5.1 kcal/mol. In contrast, the potential to generate DNA secondary structures was very low for the 38 nt long templates used in misinsertion fidelity assays (ΔG values ranged from –0.1 to 0.8 kcal/mol at 37°C).
Lower fidelity of mutant M230I as revealed using primer extension assays in the presence of three dNTPs
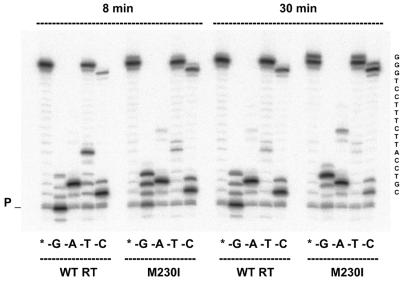
The reduced fidelity of mutant M230I was further confirmed in primer extension assays with only three of the four dNTPs complementary to template nucleotides. Under these conditions, elongation of the primer past a template nucleotide complementary to the excluded dNTP requires the insertion of an incorrect nucleotide, followed by extension of the generated mismatched primer. The results obtained using template/primer D38/20PG5 are given in Figure 3. In all reactions with WT RT and mutant M230I, a substantial accumulation of DNA products is observed at the site corresponding to the missing oligonucleotide (+2 in lanes –A; +5 in lanes –T; +1 in lanes –C), while primer elongation was very low in reactions without dGTP. After an 8 min incubation a higher elongation efficiency was observed in reactions performed with M230I in the absence of dGTP, dTTP or dCTP (lanes –G, –T and –C, respectively). Elongation products represented 76.4% of the total amount in reactions carried out in the absence of dGTP with M230I, versus only 15% as determined with the WT RT. The amount of full-length products was also higher for mutant M230I than for the WT RT in reactions performed in the absence of dTTP or dCTP (74.8 versus 40.0% in reactions without dTTP and 23.0 versus 5.2% in reactions without dCTP). In the absence of dATP, differences between the two enzymes were significant only for the longer incubation time, where the band at position +9 was barely detectable with WT RT and represented 5.6% of the total products in reactions catalyzed by the mutant M230I. Taken together, our data indicate that M230I confers reduced fidelity, particularly when kcat values have a major impact on nucleotide discrimination.
Figure 3.
Extension of primer 20PG5 by WT RT and mutant M230I in assays containing a DNA template (D38) and lacking a complementary dNTP. Reactions were incubated for 8 and 30 min, as indicated. Lanes marked with an asterisk indicate that all four nucleotides are included in the dNTP mix. Lanes marked –G, –A, –T and –C represent the missing fourth nucleotide from the dNTP mix in the respective set of experiments. The sequence of the single-stranded template extension is shown on the right. P indicates the position of the unextended primer.
DISCUSSION
A principal determinant of polymerase fidelity is geometric selection of the nucleotide for insertion into DNA (10,36). The discrimination of a correct base pair is achieved through steric complementarity between a Watson–Crick base pair and the polymerase active site. Many reports dealing with the identification of residues that control the fidelity of HIV-1 RT describe the effects of mutations on misinsertion and mispair extension fidelity (12–14,17–19,37,38), but the role of DNA sequence and structure is not yet fully understood. Comprehensive studies on the fidelity of the avian myeloblastosis virus and HIV-1 RTs revealed large differences in misinsertion ratios depending on the template/primer sequence context and suggest that error frequencies depend on the type of mispair formed, its base context and the kinetic properties of the polymerase making the error (32,33). The nascent template/primer duplex in the polymerase active site interferes with correct nucleotide incorporation by affecting the polymerase active site. The analysis of crystal structures of hotspots for polymerase slippage, including (CA)n and (A)n tracts in different intermolecular contexts, as well as DNA/DNA duplexes associated with DNA polymerases (i.e. HIV-1 RT, Bacillus stearothermophilus polymerase I large fragment, Thermus aquaticus DNA polymerase I large fragment, bacteriophage T7 DNA polymerase and human DNA polymerase β) indicate that the A-conformation of the nascent DNA duplex contributes to increased polymerase fidelity (39).
The substitution of Ile for Met230 has a dual effect on misinsertion and mispair extension fidelity of DNA synthesis. If the Y115W mutation were present, then M230I would reduce the loss of fidelity caused by the substitution in the nucleotide-binding site. In the sequence background of the WT RT (subtype B, clone BH10), the presence of Ile at position 230 produced a moderate but significant decrease in fidelity in elongation reactions carried out with DNA/DNA duplexes. This effect was more significant for misinsertions dT:dG, dT:dC and dT:dT and was clearly dependent on the structure of the template/primer. For certain mispairs (dT:dG), misinsertion efficiencies for mutant M230I were up to 16 times higher than for the WT RT. The misinsertion ratio of dT:dG pairs was roughly similar for WT RT in the sequence contexts of M13 ssDNA/pT and D2/20PG5 (4.9 × 10–5 versus 2.9 × 10–5, respectively). However, differences were larger in the case of mutant M230I (7.6 × 10–4 with M13 ssDNA/pT versus 1.1 × 10–4 with D2/20PG5). These differences result from the low Km for dGTP shown by mutant M230I in misinsertion fidelity assays carried out with M13 ssDNA/pT. Interestingly, both templates/primers show the same sequence at the incorporation site and have identical flanking nucleotides at this position. In addition, our results show that the misinsertion fidelity of mutant M230I is not affected by transitions or transversions in the primer sequence involving nucleotides that interact with the side chain at position 230.
The observed differences cannot be explained by assuming a different structure of the double-stranded DNA portion of the template/primer in complexes M13 ssDNA/pT and D2/20PG5. The steady-state kinetic parameters reported for dT:dG misinsertion on the M13 ssDNA/pT duplex using mutant M230I were different from those obtained with 38 nt templates, even though several sequence contexts were analyzed. The unique characteristics of incorporation of G opposite T in M13 ssDNA/pT complexes by mutant M230I could result from a perturbation of the polymerase active site architecture caused by potential secondary structures at the 5′-extension of the duplex.
In addition to the sequence context, the formation of secondary structures in the template strand could also provide an explanation for some inconsistencies found between fidelity estimates obtained from gel-based fidelity assays and M13 phage-based mutation assays (for example with mutant RTs L74V, E89G and M184V) (16,37,40). In the M13 gapped duplex assay, the generation of new hotspots and variable error specificities may be conditioned by the structure of the relatively long single-stranded lacZα DNA template, which is copied during the gap filling reaction. Examples of mutant HIV-1 RTs displaying similar overall error rates, but altered error specificities compared with the WT enzyme, are L74V, E89G, Q151M, M184V and the multidrug-resistant mutant A62V/V75I/F77L/F116Y/Q151M (16,40,41).
Amino acid replacements in HIV-1 RT that confer reduced fidelity of DNA synthesis include non-conservative substitutions of Tyr115 (i.e. Y115W, Y115V, Y115A and Y115G, among others) (17–19), R72A (20), M184A (21) and substitutions at positions 262 and 266 (i.e. G262A, W266A, etc.) (22–24). Misinsertion fidelity assays showed that Tyr115 mutants and R72A RT display higher apparent Km values for correct nucleotides compared with the WT RT. As a result, these enzymes show poorer discrimination against incorrect dNTPs. In the case of mutant R72A, the effect is more pronounced for misincorporations opposite T in the sequence context 5′-CTGG, due to a 1200-fold increase in the Km for the correct nucleotide (20). The lower fidelity reported for M184A results from its higher mispair extension rate as determined in steady-state fidelity assays using a 47/19mer containing A:A or A:G mispaired termini (21). Primer extension assays in the presence of three dNTPs were consistent with the lower accuracy of this mutant. However, kinetic parameter determinations were not obtained and the mechanism leading to the loss of accuracy shown by M184A awaits further clarification. Substitutions at positions 262 and 266 result in catalytically efficient RTs, but displaying a template/primer binding defect that results in a lower frameshift fidelity (22,23). The mutant M230I is a catalytically efficient DNA polymerase, with kcat and Km values for correct nucleotides similar to those reported for the WT enzyme. However, unlike the case of the WT enzyme, whose kcat values for an incorrect dNTP are very low in comparison to those obtained for the correct dNTP, the mutant M230I shows kcat values for the incorrect nucleotides which are often similar or higher than for the correct dNTP, as found with several templates/primers (see Tables 2 and 4).
These effects have not been reported for the other error-prone RTs, such as Tyr115 mutants or R72A (17–20). Unfortunately, a mechanistic interpretation of the data is problematical due to the difficulties in relating the steady-state kinetic parameters kcat and Km to steps in the mechanism of nucleotide addition (42,43). A pre-steady-state kinetic analysis would be necessary for an accurate assessment of the contribution of kpol (maximum rate of polymerization) and Kd (dNTP binding affinity) to the nucleotide selectivity displayed by the mutant RTs. To our knowledge, M184V remains the only mutant RT whose fidelity has been estimated from kpol/Kd ratios (44). Nevertheless, in these studies as well as in previous analysis with the WT RT (42,45,46), there was broad agreement between the reported selectivity values and those previously obtained using steady-state kinetic parameters.
Crystallographic studies have shown that the template/primer duplex in HIV-1 RT/DNA complexes undergoes a 45° bend that accompanies the change from A-like to B-form DNA (3) and makes ‘non-specific’ sugar and base contacts 2–6 bp upstream of the 3′-terminus of the primer. Residues that interact with the primer along the minor groove of the DNA/DNA complex are Tyr183, Met230, Gly231, Trp266, Gly262 and Gln258 (Fig. 2). At least three of them, including Met230, appear to influence fidelity of DNA synthesis (22–24; this work), revealing that this region plays a pivotal role in controlling nucleotide selectivity. Prokaryotic type I DNA polymerases and mammalian DNA polymerase β lack the primer grip motif found in HIV-1 RT and other RNA-dependent DNA polymerases.
A comparison of the crystal structures of complexes of DNA polymerases and templates/primers with or without the incoming nucleotide show that the equivalent positions in type I DNA polymerases correspond to a loop at the base of the thumb subdomain that connects β-strand 8 and α-helix K (residues 582–587 in T.aquaticus DNA polymerase, 624–629 in B.stearothermophilus polymerase and 438–442 in bacteriophage T7 DNA polymerase) (47–49). The role of most of these residues in the fidelity of DNA synthesis has not been investigated. The substitution of Ala for Arg682 in the Klenow fragment of Escherichia coli DNA polymerase I produced a mutant enzyme displaying lower mispair extension fidelity and showing a 3–14-fold higher base substitution error rate in M13-based genetic assays, compared with the WT polymerase (50). Arg682 is part of the sequence QNIPVR, which is conserved in the loop of T.aquaticus DNA polymerase I, spanning residues 582–587. In human and rat DNA polymerase β (51), residues 104–110 contact primer positions 3–4 nt upstream of its 3′-end, while Lys234, Met236, Arg254 and Asp256 are involved in interactions with primer position –2. Met236 occupies the position equivalent to Met230 of HIV-1 RT, but the importance of this residue in fidelity of DNA synthesis has not been assessed.
In summary, our results indicate that Met230 of HIV-1 RT plays a key role in controlling polymerase fidelity, probably by keeping the 3′-OH primer terminus in an appropriate orientation for the nucleophilic attack required for nucleotide incorporation into the growing DNA chain. Substitutions at the polymerase dNTP-binding pocket (i.e. Y115W) or alterations in the structure of the 5′-extension of the template strand could have an impact on the geometry of the polymerase active site. In this scenario, certain amino acid replacements, such as M230I, could be tolerated in the highly conserved primer grip of HIV-1 RT, despite reducing the ability of the polymerase to discriminate between a correct and an incorrect nucleotide. A better knowledge of the interactions involved and the mechanism leading to discrimination between correct and incorrect nucleotides is a necessary step towards designing specific inhibitors targeting the fidelity of HIV-1 RT.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Esteban Domingo for valuable suggestions. This work was supported in part by Fondo de Investigación Sanitaria grants nos 98/0054-01 and 01/0067-01 and by an institutional grant to the Centro de Biología Molecular ‘Severo Ochoa’ from the Fundación Ramón Areces. M.G.-R. was supported by a pre-doctoral fellowship from the Instituto de Salud Carlos III.
REFERENCES
- 1.Telesnitsky A. and Goff,S.P. (1997) Reverse transcription and the generation of retroviral DNA. In Coffin,J., Hughes,S.H. and Varmus,H. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 121–160. [PubMed]
- 2.Huang H., Chopra,R., Verdine,G.L. and Harrison,S.C. (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- 3.Jacobo-Molina A., Ding,J., Nanni,R.G., Clark,A.D.,Jr, Lu,X., Tantillo,C., Williams,R.L., Kamer,G., Ferris,A.L., Clark,P., Hizi,A., Hughes,S.H. and Arnold,E. (1993) Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl Acad. Sci. USA, 90, 6320–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wöhrl B.M., Krebs,R., Thrall,S.H., Le Grice,S.F.J., Scheidig,A.J. and Goody,R.S. (1997) Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66—implications for DNA synthesis and dimerization. J. Biol. Chem., 272, 17581–17587. [DOI] [PubMed] [Google Scholar]
- 5.Yu Q., Ottmann,M., Pechoux,C., Le Grice,S. and Darlix,J.-L. (1998) Mutations in the primer grip of human immunodeficiency virus type 1 reverse transcriptase impair proviral DNA synthesis and virion maturation. J. Virol., 72, 7676–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacques P.S., Wöhrl,B.M., Ottmann,M., Darlix,J.-L. and Le Grice,S.F.J. (1994) Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem., 269, 26472–26478. [PubMed] [Google Scholar]
- 7.Ghosh M., Jacques,P.S., Rodgers,D.W., Ottman,M., Darlix,J.-L. and Le Grice,S.F.J. (1996) Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry, 35, 8553–8562. [DOI] [PubMed] [Google Scholar]
- 8.Palaniappan C., Wisniewski,M., Jacques,P.S., Le Grice,S.F.J., Fay,P.J. and Bambara,R.A. (1997) Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem., 272, 11157–11164. [DOI] [PubMed] [Google Scholar]
- 9.Powell M.D., Ghosh,M., Jacques,P.S., Howard,K.J., Le Grice,S.F.J. and Levin,J.G. (1997) Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J. Biol. Chem., 272, 13262–13269. [DOI] [PubMed] [Google Scholar]
- 10.Goodman M.F., Creighton,S., Bloom,L.B. and Petruska,J. (1993) Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol., 28, 83–126. [DOI] [PubMed] [Google Scholar]
- 11.Kunkel T.A. and Bebenek,K. (2000) DNA replication fidelity. Annu. Rev. Biochem., 69, 497–529. [DOI] [PubMed] [Google Scholar]
- 12.Bakhanashvili M., Avidan,O. and Hizi,A. (1996) Mutational studies of human immunodeficiency virus type 1 reverse transcriptase: the involvement of residues 183 and 184 in the fidelity of DNA synthesis. FEBS Lett., 391, 257–262. [DOI] [PubMed] [Google Scholar]
- 13.Wainberg M.A., Drosopoulos,W.C., Salomon,H., Hsu,M., Borkow,G., Parniak,M.A., Gu,Z., Song,Q., Manne,J., Islam,S., Castriota,G. and Prasad,V.R. (1996) Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science, 271, 1282–1285. [DOI] [PubMed] [Google Scholar]
- 14.Kim B., Hathaway,T.R. and Loeb,L.A. (1998) Fidelity of mutant HIV-1 reverse transcriptases: interaction with the single-stranded template influences the accuracy of DNA synthesis. Biochemistry, 37, 5831–5839. [DOI] [PubMed] [Google Scholar]
- 15.Kim B., Ayran,J.C., Sagar,S.G., Adman,E.T., Fuller,S.M., Tran,N.H. and Horrigan,J. (1999) New human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis—accuracy, template binding and processivity. J. Biol. Chem., 274, 27666–27673. [DOI] [PubMed] [Google Scholar]
- 16.Shah F.S., Curr,K.A., Hamburgh,M.E., Parniak,M., Mitsuya,H., Arnez,J.G. and Prasad,V.R. (2000) Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem., 275, 27037–27044. [DOI] [PubMed] [Google Scholar]
- 17.Martín-Hernández A.M., Domingo,E. and Menéndez-Arias,L. (1996) Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J., 15, 4434–4442. [PMC free article] [PubMed] [Google Scholar]
- 18.Martín-Hernández A.M., Gutiérrez-Rivas,M., Domingo,E. and Menéndez-Arias,L. (1997) Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res., 25, 1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cases-González C.E., Gutiérrez-Rivas,M. and Menéndez-Arias,L. (2000) Coupling ribose selection to fidelity of DNA synthesis: the role of Tyr-115 of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem., 275, 19759–19767. [DOI] [PubMed] [Google Scholar]
- 20.Lewis D.A., Bebenek,K., Beard,W.A., Wilson,S.H. and Kunkel,T.A. (1999) Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleotide binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J. Biol. Chem., 274, 32924–32930. [DOI] [PubMed] [Google Scholar]
- 21.Pandey V.N., Kaushik,N., Rege,N., Sarafianos,S.G., Yadav,P.N.S. and Modak,M.J. (1996) Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry, 35, 2168–2179. [DOI] [PubMed] [Google Scholar]
- 22.Beard W.A., Stahl,S.J., Kim,H.-R., Bebenek,K., Kumar,A., Strub,M.-P., Becerra,S.P., Kunkel,T.A. and Wilson,S.H. (1994) Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase—alanine scanning mutagenesis of an α-helix in the thumb subdomain. J. Biol. Chem., 269, 28091–28097. [PubMed] [Google Scholar]
- 23.Bebenek K., Beard,W.A., Casas-Finet,J.R., Kim,H.-R., Darden,T.A., Wilson,S.H. and Kunkel,T.A. (1995) Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J. Biol. Chem., 270, 19516–19523. [DOI] [PubMed] [Google Scholar]
- 24.Beard W.A., Bebenek,K., Darden,T.A., Li,L., Prasad,R., Kunkel,T.A. and Wilson,S.H. (1998) Vertical-scanning mutagenesis of a critical tryptophan in the minor groove binding track of HIV-1 reverse transcriptase—molecular nature of polymerase-nucleic acid interactions. J. Biol. Chem., 273, 30435–30442. [DOI] [PubMed] [Google Scholar]
- 25.Bebenek K., Beard,W.A., Darden,T.A., Li,L., Prasad,R., Luxon,B.A., Gorenstein,D.G., Wilson,S.H. and Kunkel,T.A. (1997) A minor groove binding track in reverse transcriptase. Nature Struct. Biol., 4, 194–197. [DOI] [PubMed] [Google Scholar]
- 26.Wisniewski M., Palaniappan,C., Fu,Z., Le Grice,S.F.J., Fay,P. and Bambara,R.A. (1999) Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J. Biol. Chem., 274, 28175–28184. [DOI] [PubMed] [Google Scholar]
- 27.Olivares I., Sánchez-Merino,V., Martínez,M.A., Domingo,E., López-Galíndez,C. and Menéndez-Arias,L. (1999) Second-site reversion of a human immunodeficiency virus type 1 reverse transcriptase mutant that restores enzyme function and replication capacity. J. Virol., 73, 6293–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Y. and Eickbush,T.H. (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J., 9, 3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olmsted R.A., Slade,D.E., Kopta,L.A., Poppe,S.M., Poel,T.J., Newport,S.W., Rank,K.B., Biles,C., Morge,R.A., Dueweke,T.J., Yagi,Y., Romero,D.L., Thomas,R.C., Sharma,S.K. and Tarpley,W.G. (1996) (Alkylamino)piperidine bis(heteroaryl)piperizine analogs are potent, broad-spectrum nonnucleoside reverse transcriptase inhibitors of drug-resistant isolates of human immunodeficiency virus type 1 (HIV-1) and select for drug-resistant variants of HIV-1IIIB with reduced replication phenotypes. J. Virol., 70, 3698–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleim J.-P., Winters,M., Dunkler,A., Suarez,J.-R., Riess,G., Winkler,I., Balzarini,J., Oette,D., Merigan,T.C. and the HBY 097/2001 Study Group (1999) Antiviral activity of the human immunodeficiency virus type 1-specific nonnucleoside reverse transcriptase inhibitor HBY 097 alone and in combination with zidovudine in a phase II study. J. Infect. Dis., 179, 709–713. [DOI] [PubMed] [Google Scholar]
- 31.Boosalis M.S., Petruska,J. and Goodman,M.F. (1987) DNA polymerase insertion fidelity—gel assay for site-specific kinetics. J. Biol. Chem., 262, 14689–14696. [PubMed] [Google Scholar]
- 32.Mendelman L.V., Boosalis,M.S., Petruska,J. and Goodman,M.F. (1989) Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem., 264, 14415–14423. [PubMed] [Google Scholar]
- 33.Ricchetti M. and Buc,H. (1990) Reverse transcriptases and genomic variability: the accuracy of DNA replication is enzyme specific and sequence dependent. EMBO J., 9, 1583–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding J., Das,K., Hsiou,Y., Sarafianos,S.G., Clark,A.D.,Jr, Jacobo-Molina,A., Tantillo,C., Hughes,S.H. and Arnold,E. (1998) Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol., 284, 1095–1111. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M., Mathews,D.H. and Turner,D.H. (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In Barciszewski,J. and Clark,B.F.C. (eds), RNA Biochemistry and Biotechnology, NATO ASI Series. Kluwer Academic, Norwel, MA, Vol. 70, pp. 11–43.
- 36.Goodman M.F. (1997) Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl Acad. Sci. USA, 94, 10493–10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drosopoulos W.C. and Prasad,V.R. (1996) Increased polymerase fidelity of E89G, a nucleoside analog-resistant variant of human immunodeficiency virus type 1 reverse transcriptase. J. Virol., 70, 4834–4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutiérrez-Rivas M., Ibáñez,A., Martínez,M.A., Domingo,E. and Menéndez-Arias,L. (1999) Mutational analysis of Phe-160 within the ‘palm’ subdomain of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol., 299, 615–625. [DOI] [PubMed] [Google Scholar]
- 39.Timsit Y. (1999) DNA structure and polymerase fidelity. J. Mol. Biol., 293, 835–853. [DOI] [PubMed] [Google Scholar]
- 40.Drosopoulos W.C. and Prasad,V.R. (1998) Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J. Virol. , 72, 4224–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rezende L.F., Curr,K., Ueno,T., Mitsuya,H. and Prasad,V.R. (1998) The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J. Virol., 72, 2890–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kati W.M., Johnson,K.A., Jerva,L.F. and Anderson,K.S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem., 267, 25988–25997. [PubMed] [Google Scholar]
- 43.Johnson A.A., Ray,A.S., Hanes,J., Suo,Z., Colacino,J.M., Anderson,K.S. and Johnson,K.A. (2001) Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem., 276, 40847–40857. [DOI] [PubMed] [Google Scholar]
- 44.Feng J.Y. and Anderson,K.S. (1999) Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry, 38, 9440–9448. [DOI] [PubMed] [Google Scholar]
- 45.Zinnen S., Hsieh,J.-C. and Modrich,P. (1994) Misincorporation and mispaired primer extension by human immunodeficiency virus reverse transcriptase. J. Biol. Chem., 269, 24195–24202. [PubMed] [Google Scholar]
- 46.Kerr S.G. and Anderson,K.S. (1997) RNA dependent DNA replication fidelity of HIV-1 reverse transcriptase: evidence of discrimination between DNA and RNA substrates. Biochemistry, 36, 14056–14063. [DOI] [PubMed] [Google Scholar]
- 47.Li Y., Korolev,S. and Waksman,G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J., 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiefer J.R., Mao,C., Braman,J.C. and Beese,L.S. (1998) Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature, 391, 304–307. [DOI] [PubMed] [Google Scholar]
- 49.Doublié S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 50.Minnick D.T., Bebenek,K., Osheroff,W.P., Turner,R.M.,Jr, Astatke,M., Liu,L., Kunkel,T.A. and Joyce,C.M. (1999) Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J. Biol. Chem., 274, 3067–3075. [DOI] [PubMed] [Google Scholar]
- 51.Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]