ABSTRACT
Myelodysplastic syndromes (MDS) and their progression to secondary acute myeloid leukemia (sAML) are associated with an altered protein expression including extracellular matrix (ECM) components thereby promoting an inflammatory environment. Since the role of the proteoglycan biglycan (BGN) as an inflammatory mediator has not yet been investigated in both diseases and might play a role in disease progression, its expression and/or function was determined in cell lines and bone marrow biopsies (BMBs) of MDS and sAML patients and subpopulations of MDS stem cells by Western blot and immunohistochemistry. The bone marrow (BM) microenvironment was analyzed by multispectral imaging, patients’ survival by Cox regression. ROC curves were assessed for diagnostic value of BGN. All cell lines showed a strong BGN surface expression in contrast to only marginal expression levels in mononuclear cells and CD34+ cells from healthy donors. In the MDS-L cell line, CD34−CD33+ and CD34+CD33+ blast subpopulations exhibited a differential BGN surface detection. Increased BGN mediated inflammasome activity of CD34−CD33+TLR4+ cells was observed, which was inhibited by direct targeting of BGN or NLRP3. BGN was heterogeneously expressed in BMBs of MDS and sAML, but was not detected in control biopsies. BGN expression in BMBs positively correlated with MUM1+ and CD8+, but negatively with CD33+TLR4+ cell infiltration and was accompanied by a decreased progression-free survival of MDS patients. BGN-mediated inflammasome activation appears to be a crucial mechanism in MDS pathogenesis implicating its use as suitable biomarker and potential therapeutic target.
Abbreviations: Ab, antibody; alloSCT, allogenic stem cell transplant; AML, acute myeloid leukemia; BGN, biglycan; BM, bone marrow; BMB, bone marrow biopsy; casp1, caspase 1; CTLA-4, cytotoxic T lymphocyte-associated protein 4; DAMP, danger-associated molecular pattern; ECM, extracellular matrix; FCS, fetal calf serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HD, healthy donor; HSPC, hematopoietic stem and progenitor cell; HSC, hematopoietic stem cell; IFN, interferon; IHC, immunohistochemistry; IL, interleukin; MDS, myelodysplastic syndrome; MPN, myeloproliferative neoplasm; MSI, multispectral imaging; NGS, next-generation sequencing; NLRP3, NLR family pyrin domain containing 3; OS, overall survival; PBMC, peripheral blood mononuclear cell; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1, PFS, progression-free survival; PRR, pattern recognition receptor; SC, stem cell; SLRP, small leucine-rich proteoglycan; TGF, transforming growth factor; TIRAP, toll/interleukin 1 receptor domain-containing adapter protein; TLR, toll-like receptor; Treg, regulatory T cell.
KEYWORDS: AML, MDS, extracellular matrix, biglycan, inflammasome
Introduction
Myelodysplastic syndromes (MDS) comprise heterogeneous myeloid stem/progenitor cell neoplasms characterized by bone marrow (BM) failure, inflammatory deregulation1 and increased risk of blast accumulation and progression to secondary acute myeloid leukemia (sAML).2 MDS exert a high clinical diversity ranging from an indolent disease to the development of sAML in approximately 30% of MDS patients, which underlines the demand for appropriate risk stratification at diagnosis and during the disease course.
In myeloid neoplasms, the dynamic interactions between leukemic cells and the BM micromilieu including extracellular matrix (ECM) and stromal cells are frequently overlooked although this interplay plays a critical role in the development of MDS, its progression to sAML as well as in its drug response and resistance development. In cancer, including leukemia, the ECM consisting of a complex interconnected network of matrix and matrix-associated proteins becomes highly deregulated and can exert both pro- and anti-tumorigenic functions leading to the control of the innate immune response by affecting the inflammasome activity.3–5
Biglycan (BGN), a member of the small leucine rich proteoglycan (SLRP) family, is a key component of the ECM and is physiologically expressed in different tissues.6 It is involved in cell growth, adhesion, bone mineralization, migration, differentiation and cell signaling.7 Furthermore, BGN can interact with the toll-like receptor (TLR)2 and TLR4 on immune cells, which generates a proinflammatory response by inducing the NLRP3 inflammasome, the activation of caspase-1 (casp1) and the release of various cytokines and chemokines.8–11 An altered BGN expression has been reported in different diseases including multiple types of solid cancers.5,10,12 In some cancers, BGN overexpression is associated with an aggressive growth, metastasis formation and poor patients’ prognosis, while in others it exerts tumor suppressive effects accompanied by a prolonged survival and better prognosis.13,14 These data suggest tumor type-dependent role of BGN with diagnostic, prognostic and therapeutic relevance.11,15–17
So far, little information is available regarding ECM alterations in hematopoietic malignancies, despite the ECM provides a mechanical scaffold and serves as source of growth factors and cytokines,3 which be associated with a pro-inflammatory signaling and an altered tumor microenvironment (TME).18–20 Although a differential expression of proteoglycan family members including BGN have been demonstrated in hematopoietic malignancies compared to healthy controls,21–23 the role of BGN expression in relation to molecular features, innate immune activation and pro-inflammatory signaling in MDS and/or sAML has not yet been analyzed. The present study determined the expression and clinical significance of BGN in MDS and sAML and its link to the aberrant immune cell composition of the TME. These results may provide a novel perspective for the development of BGN as a therapeutic target for MDS and/or sAML thereby improving the patients’ outcomes.
Materials and methods
Cell lines and cell culture
The AML cell lines HL-60, THP-1, KG-1 and Mono-mac6 (MM6) were purchased from American Type Culture Collection. The MDS-L cell line was kindly provided by Dr. K. Tokyama (Kawaski Medical School, Kurashiki, Japan). The MPN cell lines HEL, SET-2 and the sAML cell line MOLM-13 were kindly provided by Prof. M. Haemmerle. With the exception of MDS-L cultured in RPMI supplemented with 20% FCS, 10 mM glutamine, antibiotics and 10 ng/mL human recombinant interleukin (IL)-3 (PeproTech, Hamburg, Germany), all other cell lines were cultured in RPMI supplemented with 10% fetal calf serum (FCS), 10 mM glutamine and antibiotics.
Isolation of peripheral blood mononuclear cells, magnetic separation and culture
Peripheral blood mononuclear cells (PBMCs) from five healthy donors (HD) were isolated from total blood through gradient centrifugation with lymphocyte separation media (anprotec, Bruckberg, Germany). Cells were washed with cold sorting buffer and either used for flow cytometry or for magnetic separation of CD34+ cells. The CD34+ cells were isolated using CD34 magnetic microbeads (Miltenyi, Bergisch-Gladbach, Germany). Circulating hematopoietic stem cells (HSC) and bone marrow hematopoietic cells were cultured in X–Vivo15 medium (Lonza Bioscience, Basel, Switzerland) supplemented with 10 ng/mL IL-3, 40 ng/mL thyroid peroxidase, 100 ng/mL stem cell factor and 100 ng/mL fms-like tyrosine kinase 3 (Flt3; Peprotech).
Flow cytometry and cell sorting
For characterization of different subpopulations, MDS-L cells were stained in PBS with the Zombie AquaTM Fixable Viability Kit (Biolegend, San Diego, USA) for 20 minutes at room temperature and then incubated with a 9-color antibody (Ab) panel for 30 minutes at 2°C. Flow-cytometric evaluation was performed on a BD LSR Fortessa (Becton Dickinson, Heidelberg, Germany), cell sorting on a BD Aria Fusion (BD). BGN surface expression was assessed by staining with a polyclonal rabbit anti-BGN Ab (Novus Biologicals, Littleton, USA) on ice for 30 minutes followed by incubation with an anti-rabbit Cy5 AffiniPure Fab2 (Jackson Immunoresearch, Cambridgeshire, USA) as secondary Ab for 20 minutes on ice.
Immunofluorescence
MDS-L cells were stained with a conjugated PE-Dazzle anti-CD34 Ab and the polyclonal anti-BGN Ab using an anti-rabbit IgG Fab2 Alexa Fluor® 488 (Cell Signaling Technology, Danvers, USA) as secondary Ab. Stained cells were analyzed on a fluorescence microscope (Evos Floid imaging system, Thermo Fisher Scientific, Waltham, USA). Composite images were created using the overlay feature of the Evos Floid imaging system.
Patient samples and ethics approval
BM biopsies (BMB) from healthy donors (HD), MDS and sAML patients were collected as a part of the routine diagnostic approach in the period from 2015 to 2021 at the Institute of Pathology of the Martin-Luther University Halle-Wittenberg, Halle, Germany. The study was approved by the Ethical Committee of the Medical Faculty in Halle, Germany (2017–81). Clinical data from these patients were available. BM aspirates of three sAML patients analyzed were available as part of the AML study approved by the Ethical Committee of the Medical Faculty in Halle, Germany (2014–75).
Immunohistochemistry and multispectral imaging
3 µm thick formalin-fixed and paraffin embedded tissues were subjected to immunohistochemistry (IHC) with the Bond Polymer refine detection Kit (Leica, DS9800) according to the manufacturer’s instructions on a fully automated immunohistochemistry stainer (Leica Bond) using the antibodies in Supplementary Table 3.
Multispectral imaging (MSI) was performed as recently described24 employing two established MSI Ab panels presented in Supplementary Table 2.
Live patient cells were resuspended and immobilized in Epredia™ Richard-Allan Scientific™ HistoGel™ (Thermo Fisher Scientific) according to the manufacturers’ instructions. The immobilized cells were then mounted onto a histoblock and then subjected to the MSI protocol.
Mutation analysis of bone marrow samples of MDS patients
Targeted mutation analyses were conducted using Next-Generation Sequencing (NGS; Ion GeneStudio S5 prime, Thermo Fisher Scientific) as recently described.25
Western blot analysis and subcellular fractionation
For the quantification of BGN as well as of inflammasome-related molecules.10 to 25 μg total protein were subjected to Western blot analysis as described26 using Abs listed in Supplementary Table 3.
BGN localization was determined upon subcellular fractionating into cytoplasmic, membrane and nuclear proteins using a fractionation kit (Cell Signaling) according to the manufacturer’s protocol. The nuclear cell fraction served as a loading control, while 1 μg of recombinant human BGN in RPMI 1640 (Sigma Aldrich, St. Louis, USA) served as a positive control.
Caspase 1 (Casp 1) inflammasome assay
4 x 104 sorted cell subpopulations (SC1, SC2, TLR4) were seeded into a 96-well plate and incubated for 4 h in MDS-L culture medium alone or supplemented with recombinant BGN (1 μg/mL) with or without further addition of TGF-β (1 μg/mL, Biolegend), IFN-γ (2000 U/mL), IFN-α (2000 U/mL), IL-1Ra (100 ng/mL, Sigma Aldrich) or the TIRAP inhibitor (1 μg/mL). Moreover, TLR4+ cells were co-cultured for 4 h with MDS-L-SC2 cells in the absence or presence of rabbit anti-BGN Ab (220 ng/mL, Proteintech) for 1 h at 37°C. MDS-L cells were treated with the NF-kB inhibitor SN50 (100 ng/mL, Merck, Darmstadt, Germany) or with the glyburide intermediate 16673–34-0 (1.5 mM). Casp1-specific activation was detected using the Caspase-Glo® 1 Inflammasome Assay (Promega) according to the manufacturer’s instructions. Luminescence was measured in a Costar white 96-well plate using the Tecan Infinite 200 Pro plate reader device (Tecan, Maennedorf, Switzerland).
Cytokine secretion
The cytokine concentrations in the cell supernatants were determined by the LEGENDplex™ Human-Inflammation-Panel-1 kit (Biolegend) using a BD Fortessa flow cytometer according to manufacturer’s instructions.
Statistical analysis
Paired and unpaired t-tests (two-tailed) as well as log Rank survival analyses were conducted with the GraphPad Prism 9.0.0 software. Hierarchical clustering, ROC curves and the Cox regression survival analysis the IBM SPSS 24 software was employed. Differences were considered significant for values p < .05.
Results
BGN expression in MDS/AML/MPN cell lines
The basal mRNA and protein expression of BGN was investigated in five AML/MDS/MPN cell lines using qPCR and Western blot analysis demonstrating a heterogeneous BGN transcript (data not shown) and protein expression pattern with the highest expression levels in the cell lines HL-60, KG-1 and HEL (Figure 1a), while MDS-L and SET-2 showed a slightly lower BGN expression. Furthermore, BGN was detected on the cell surface of all cell lines, but not on PBMCs from HDs (Figure 1b). Subcellular fractionation demonstrated a complete lack of BGN in the cytoplasmic and nuclear fraction, whereas BGN was found on the surface membrane, in the organelle fraction and whole-cell lysate of MDS-L cells (Figure 1c). Despite low total BGN protein levels, >90% of THP-1 cells expressed BGN on their surface, possibly due to sBGN bound to its receptor TLR4. Indeed, a strong correlation between BGN and TLR4 expression could be found in all cell lines (Figure 1d). Analysis of different subpopulations revealed that BGN expression did neither correlate to CD34 nor to CD33 expression in AML cell lines (Supplementary Figure 1), while CD33+CD34− (named CD33SP) and CD33+CD34+ (named CD34+) MDS-L subpopulations exhibit high and low BGN surface levels, respectively (Figure 2a).
Figure 1.
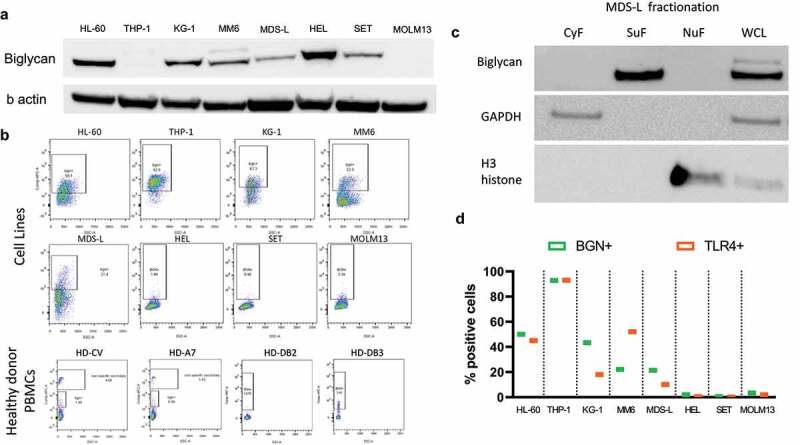
Expression and localization of BGN in AML/MDS cell lines. (a) Western blot analysis was performed as described in Materials and Methods using an anti-BGN-specific Ab. Staining with an anti-GAPDH Ab served as loading control. A differential BGN expression was found in the 4 AML (HL-60, THP-1, KG-1, MM6), 2 MPN (HEL, SET) one MDS (MDS-L) and one sAML (MOLM13) cell lines. (b) Dot plots revealing the surface expression of BGN in the 8 cell lines as well as PBMCs of four healthy donors. (c) Spatial distribution of BGN in the MDS-L cell line after cell fractionation. Three fractions are investigated (cytoplasmic fraction CyF, surface fraction SuF, nuclear fraction and cytoskeleton NuF) along with a whole cell lysate as a control. Successful separation of fractions was determined using an anti-GAPDH Ab and an anti-H3 histone Ab for the CyF and NuF fractions, respectively. (d) Comparison of the surface expression of BGN (green) and TLR4 (Orange) on the 8 cell lines. The cells were stained in separate samples for both BGN and TLR4 as described in Materials and Methods and their expression was compared afterward.
Figure 2.
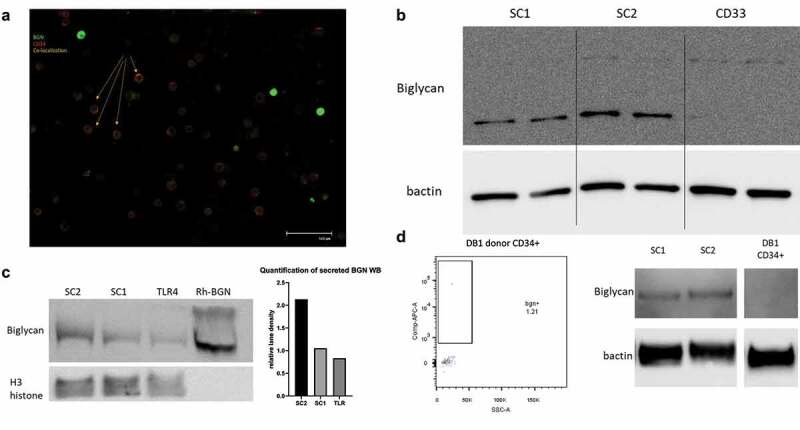
Distinct distribution of BGN in MDS-L subpopulations. (a) Fluorescence microscope picture demonstrating the differential distribution of BGN on the MDS-L cells. The MDS-L cells were stained according to protocol and a possible co-localization of their signal was investigated. Cells with strong green fluorescence (BGN+) do not express CD34 (red), while cells expressing lower amounts of BGN on their surface appear to be CD34+ (Orange color due to co-localization). (b) BGN expression in sorted subpopulations of the MDS-L cell line made in duplicate. Differential expression can be observed between the Stem Cell 1 fraction (SC1), Stem Cell 2 fraction (SC2) and the total CD33SP subpopulation. Both the non-glycosylated (bottom) and glycosylated (top) form of BGN can be detected in the sorted subpopulations, in different amounts (n = 2). (c) Identification of BGN in the supernatants of the MDS-L subpopulations of interest (SC1, SC2, TLR4) after overnight culture. The total supernatants were diluted with an SDS buffer and directly loaded on an acrylamide gel after heating. Human recombinant BGN in cell medium is used as a positive control, while 5 μL of the NuF of total MDS-L cells was used as a spike-in loading control as demonstrated by the H3 histone band below (n = 1). The relative lane density is presented for the 3 different subpopulations after calculation of the ratio of BGN to the H3 histone signal (loading spike-in control). (d) Representative surface and total BGN expression in circulating CD34+ cells of a healthy blood donor as determined by flow cytometry and Western blotting respectively. For total BGN expression, SC1 and SC2 (left) were used as positive controls for BGN, while β-actin was used as a loading control.
Distinct BGN expression in MDS stem cell subpopulations
Since different MDS-related stem cell (SC) subpopulations have been documented,27,28 BGN expression was analyzed in the CD33SP and CD34+ SC subpopulations of the MDS-L cell line (Supplementary Figure 2). For some experiments, CD33SP cells were further selected for TLR4 expression, while CD34+ SC were subdivided into two subpopulations, SC1 and SC2, based on the expression of CD123 and IL1RAcP, respectively. The SC1 and even more the SC2 cells were the main BGN producers, while the CD33SP cells only expressed marginal BGN (Figure 2b) and its TLR4+ subfraction secreted only low sBGN amounts (Figure 2c). Compared to the SC1 and TLR4+ subpopulation, SC2 cells secreted a 2-fold higher BGN concentration determined by densitometric analysis (Figure 2c). In contrast, the sorted circulating CD34+ cells from an HD totally lacked BGN expression (Figure 2d). In order to facilitate the possible connection of MDS-SC with BGN, bone marrow aspirates from three patients with progressed MDS were analyzed (P39, P54, P56, Supplementary Table 1a). At a first glance, the patients were found to have differential distinct BM infiltration, with P39 and P54 the SC1 and SC2 subpopulations, respectively, while for patient P56 almost all CD34+ cells were negative for CD123 and IL1RacP (Figure 3a). These differences were also reflected by the frequency of TLR4+ cells in the aspirate, with the lowest levels in the patient P54. BGN staining also revealed the same correlation between BGN detection and TLR4 levels for 2/3 patients, with the exception of patient P54. Furthermore, the total BGN levels were investigated in the CD34+ and CD34− fractions of the patients’ aspirate. Despite the CD34− fractions from all 3 patients showed no differences in BGN expression, CD34+ sub-population analysis revealed increased BGN levels in the SC2 rich cells of P54 (Figure 3b), while P56 also showed intermediate BGN expression levels. These data were validated by MSI analysis of the total BM aspirates showing higher levels of CD34+BGN+ cells in the aspirates of P54 and P56, while the levels of CD33+BGN+ and CD34−CD33−BGN+ were consistent for all three patients (Figure 3c). Interestingly, more CD33+NLRP3+ cells were found in the BM aspirate from P56, but only decreased CD33+TLR4+ cell levels were found in P54 compared to the other patients.
Figure 3.
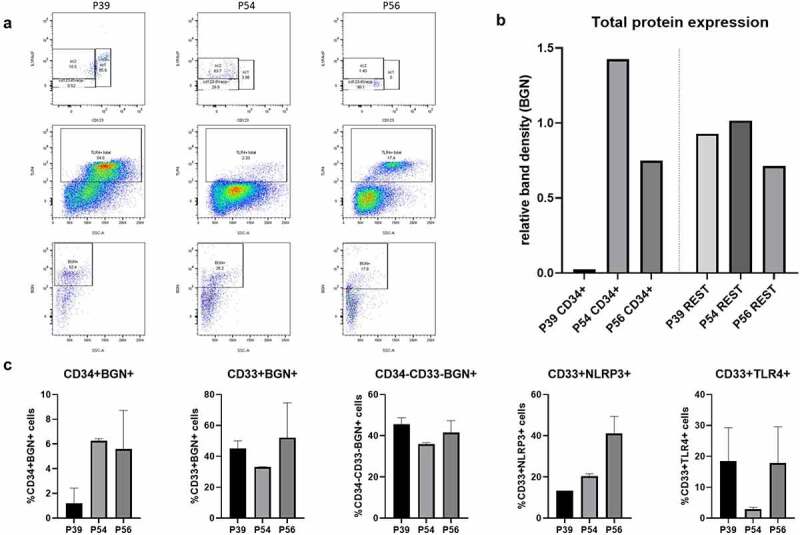
Identification of BGN expressing MDS subpopulations in BM aspirates of sAML patients. (a) Flow cytometry analysis using 9 color ab panel as well as single staining with an anti-BGN ab for detection of the different MDS subpopulations and BGN surface expression. The distribution of CD34+ MDS-SC subpopulations of the sAML patients were identified using the CD123 and IL1RAcP markers (top) and total TLR4+ cells were calculated (middle), while surface BGN expression on the total cell population was additionally analyzed (bottom). (b) Densitometry analysis after Western blot of the total protein lysates of the three sAML patients (20 μg protein) from the CD34+ fraction (left) and the CD34− fraction (right) of the BM aspirates was loaded after magnetic separation of the cells and investigated for BGN expression (n = 1) as described in Materials and Methods. β-actin was used as a loading control. The relative band densities (BGN/ β-actin) are presented in the graph. (c) Quantification of inflammasome related molecules on fixed cell blocks from the BM aspirates of the three sAML patients. Five subpopulations of interest were investigated (CD34+BGN+, CD33+BGN+, CD34−CD33−BGN+, CD33+NLRP3+, CD33+TLR4+). The observed percentages were shown along with their SD, which derived from the quantification of these populations on 3 different cuts of the histoblock for each patient (n = 3, technical replicates/patient).
BGN-mediated inflammasome activation in MDS-L subpopulations
Since soluble BGN could act as a DAMP leading to inflammation by binding to TLR4 and the P2x7 purinergic receptor,8,9,29 the effect of BGN on the inflammasome activation in the MDS-L SC1, SC2 and TLR4+ subpopulations either left untreated or treated with recombinant human BGN was determined. Only in TLR4+ cells, BGN treatment resulted in a significant increase of casp1 activity (Figure 4a), which was accompanied by a high condensation of organelles coupled with a swelling of the cytoplasmic membrane indicating pyroptotic cell death (Figure 4b). Due to the lack of TLR4 expression, both untreated and BGN-treated SC1 and SC2 subpopulations secreted no or marginal levels of inflammation-related cytokines, whereas BGN-treated TLR4+ cells showed a significant increase in IL-1 and IL-8 and a statistically non-significant increase in IL-18 (p = .0871) (Figure 4c; Supplementary Figure 3). In contrast, IFN-α2, IL-33 and the proinflammatory MCP-1 and IL-23 were constitutively expressed in TLR4+ cells, which was not significantly affected by BGN treatment.
Figure 4.
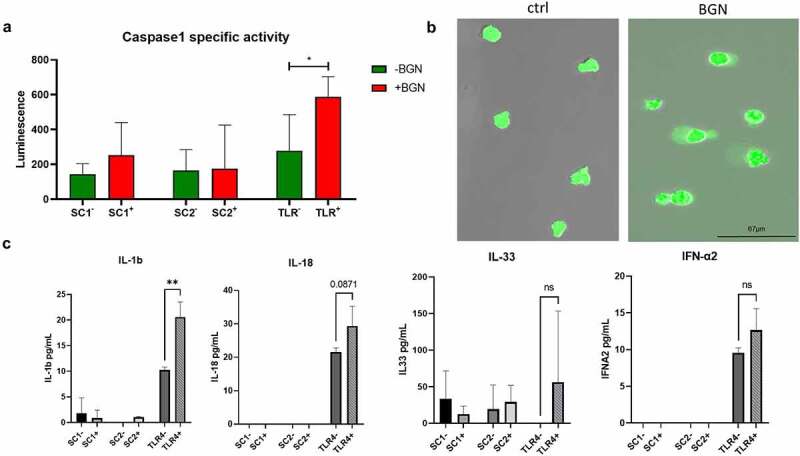
Altered casp1 activity and cytokine secretion pattern in MDS-L subpopulations. (a) Bar chart demonstrating casp1 activity after 4 h stimulation of the three subpopulations of interest (SC1, SC2, TLR4) with BGN. Casp1 activity was measured after harvesting in a luminometer. Unstimulated cells for each of the MDS-L subpopulations are depicted with green color and a minus sign (-), while cells stimulated with BGN with red and a plus (+) sign. The mean luminescence is represented with the colored bands, while the standard deviation was plotted using the black error bars adjustment to its respective mean value (whiskers). T-tests were conducted to determine statistical significance using the average of two measurements (technical replicates) in three (n = 3) independent experiments. P values <0,5 were considered significant. (b) Fluorescence microscope image of the TLR4 subpopulation of the MDS-L cell line after stimulation with BGN. The cells demonstrate cytoplasmic swelling, previously associated with inflammasome activation. TLR4 cells were additionally stained with CFSE to increase the visibility of the effect. (c) Cytokine secretion in the supernatant of the 3 subpopulations of interest before and after 4 h stimulation with BGN. The inflammatory cytokine profile of the 3 subpopulations was assessed using a multiplexed cytokine assay with flow cytometry. The inflammasome related cytokines IL-1β, IL-18 and IL-33 are investigated, along with IFN-α. The concentration of the cytokines is shown in pg/mL (y axis). The average concentration is represented with gray scale bands, while the standard deviation reported by the Whiskers. T- tests were conducted to determine statistical significance using the average of two measurements (technical replicates) in three (n = 3) independent experiments.
Inflammasome inhibition through direct targeting of BGN and NLRP3
In order to evaluate the pathways involved in the BGN-mediated inflammasome activation, TLR4+ cells were treated with IFN-γ, TGF-β1 or the toll/interleukin 1 receptor domain-containing adapter protein (TIRAP) inhibitor. The suggested interactions within this pathway are shown in Supplementary Figure 4. IFN-γ treatment significantly inhibited inflammasome activation and downregulated the purinergic receptor (Figure 5a, b), while TGF-β1 treatment caused a decrease in NLRP3 and P2x7 expression (Figure 5b). TGF-β1 treatment slightly increased the lin+ (CD34−CD33+lineage+) and decreased the TLR4+ and CD34+ subpopulations leading to a higher ratio of SC1 to SC2 cells (Figure 5c). In contrast, IFN-γ did not affect any of the subpopulations with the exception of an increase in TLR4+ cells (Figure 5c).
Figure 5.
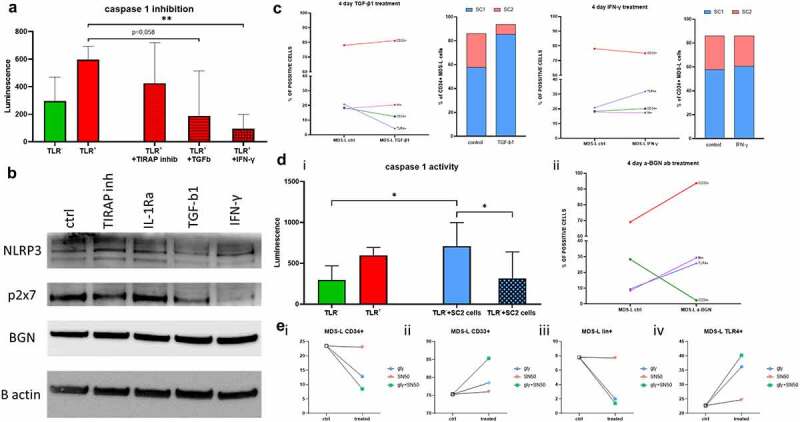
Altered casp1 activity in MDS-L subpopulations by inflammasome inhibitors. (a) Casp1 activity measured via luminescence on the TLR4 subpopulation of MDS-L cells after stimulation with BGN (left) as well as additional stimulation with TIRAP inhibitors, TGF-β1 or IFN-γ. The mean luminescence and SD are presented in the graph. T- tests were conducted to determine statistical significance using the average of two measurements (technical replicates) in three (n = 3) independent experiments. (b) Western blot analysis investigating expression of BGN and the inflammasome related proteins P2X7 and NLRP3 in total MDS-L cells. The cells were either left untreated (ctrl) or treated with potential inhibitors of the pathway for 4 days (TIRAP inhibitors, IL-1Ra, TGF-β1 or IFN-γ). β-actin was used as a loading control for the comparisons between different treatments (n = 1). (c) Flow cytometric analysis of the MDS-L subpopulations after 4 day treatment with the different inhibitors of the inflammasome, TGF-β1 and IFN-γ. The different subpopulations are depicted with different colored lines (red: total CD33; green: total CD34; purple: lin; blue: TLR4), while the bar charts show the ratios of the two stem cell like populations before and after treatment (blue: SC1; red: SC2). The numbers on the Y axis represent percentage of cells gated on the total alive cells. (d) Comparison of casp1 activity (i) in the TLR4 subpopulation in untreated (green), recombinant BGN-treated cells (red) as well as co-cultured with the high BGN-producing stem cell subpopulation SC2, either alone (blue) or together with anti-BGN antibodies (blue-crossed). (ii) Flow cytometry analysis of total MDS-L cells treated for 4 days with the anti-BGN antibodies. The different subpopulations are depicted with different colors (red: total CD33; green: total CD34; purple: lin; blue: TLR4-blue). T tests were utilized for the identification of statistical significance. Both mean luminescence and SD are shown in the graph. Technical duplicates were used to eliminate possible errors, within the 3 biological replicates performed (n = 3). (e) Flow cytometry analysis of total MDS-L cells after treatment with the glyburide intermediate (blue circle), the NF-kB inhibitor SN50 (red triangle) or their combination (green square). The (i) CD34+, (ii) CD33+, (iii) lin+ and (iv) TLR4+ subpopulations were analyzed, before and after treatment to reveal a possible synergistic effect of blockade of both parts of the pathway (n = 1).
Since the casp1 activation assay showed a BGN-mediated inflammasome activation, the biological relevance of MDS-derived BGN was investigated. The supernatant of the BGN-secreting SC2 subpopulation served as a source of BGN for stimulation of TLR4+ cells leading to a significantly increased casp1 activity in TLR4+ cells, which could be blocked by treatment with polyclonal anti-BGN Ab prior to co-culture with TLR4+ cells suggesting a direct involvement of BGN in the inflammasome activation. Treatment for 4 days with an anti-BGN Ab or the NLRP3 inhibitor 16673–34-0 decreased the number of CD34+ cells with a consequent increase in CD33SP cells followed by a lower, but still substantial increase in TLR4+ and lin+ subpopulations (Figure 5d, e). In addition, no/marginal changes were observed after treatment with the NF-kB inhibitor SN50 alone, whereas a combined treatment with both inhibitors had additive or synergistic effects.
Differential expression of BGN in bone marrow biopsies (BMBs) of patients with MDS/sAML
IHC analyses of BMBs from 49 HD, 59 MDS patients and 48 sAML patients with known clinicopathological characteristics (Supplementary Table 1b; Figure 6a) demonstrated a differential BGN expression pattern with H-scores ranging between 0 and 120 (Figure 5b). BMBs from HD had a very low BGN expression (median H-score: 6.5) compared to the MDS (median H-score = 20 95% CI 10–30, maximum H-score: 80) and sAML (median H-score = 20 95% CI 15–30, maximum H-score 120) BMBs expressed significantly higher BGN levels (Figure 5bi). As a result, the upper threshold of the 95% CI of the median (BGN h score = 30) was used to stratify the patient samples in BGNlow/BGNhigh. In addition, BGN expression was not limited in MDS to blast cells, since no correlation between the frequencies of BGN+ cells to the blast cell count in the BMBs existed (Figure 6bii).
Figure 6.
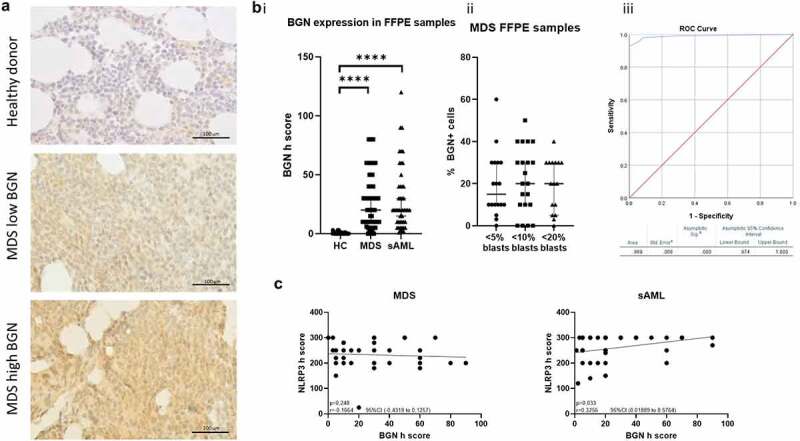
Distinct BGN expression in MDS and AML BMB. (a) Representative pictures of BGN IHC staining in FFPE BMB from a healthy donor, a low expressing MDS patient and a high expressing MDS patient. The slides were stained as mentioned in the materials and methods. A scale has been used as a reference. (b) Differential BGN expression between (i) healthy donors and MDS/sAML patients indicated by a comparison of the BGN H-scores and (ii) MDS patients with different blast counts indicated by a comparison of the percentage of BGN positive cells. (iii) Depiction of the generated ROC curve for the determination of the diagnostic value of BGN H-score. The area under curve (AOC) is presented along with the significance levels (t test), median of the patient samples and the 95% CI. (c) Correlation plots comparing the BGN (x axis) and NLRP3 (y axis) h score determined. The correlations are separately investigated in MDS (left) and sAML (right) patient samples. The linear regression is additionally seen on the graph together with the p value, r and 95%CI derived from the correlation analysis.
ROC curves demonstrated a high sensitivity and specificity for BGN in the discrimination between HD and MDS/sAML samples with a recorded AUC = 0,989 (CI 95%, Figure 6biii). A heterogeneous NLRP3 expression was found in BMBs of MDS and sAML patients (Supplementary Figure 5a), which correlated with BGN expression in BMBs from sAML, but not from MDS patients (Figure 6c, Supplementary Figure 5b).
Effects of BGN in the BM microenvironment and on the progression-free survival of MDS patients
To get insights into the impact of BGN expression on the composition of the BM microenvironment, MSI was performed using an Ab panel focusing on the CD34 and CD33 subpopulations and on selected components of the inflammasome pathway (Figure 7a). The mean fluorescence intensity (MFI) of BGN in CD34+ cells positively correlated with the MFI of NLRP3 in the CD33SP cells was found in the BMBs of the MDS patient cohort (Figure 7b), while BGNlow samples showed a significantly increased ratio of CD33SPTLR4+/CD33SPTLR4− cells (Figure 7c). Moreover, BGNhigh patients had a significantly increased number of endothelial cells producing BGN (CD34+CD31+BGN+ cells) within the CD31+ cells. In contrast, the majority of the CD31+ endothelial cells in BGNlow patients lack BGN expression (Figure 7d). MSI results demonstrated a higher infiltration of CD3+CD8− (indicated as CD4+) cells in the BMB of MDS patients with lower BGN H-scores, whereas the CD8+/CD4+ ratio increased with higher BGN expression levels in the BMBs (Figure 8aandb). High BGN expression was accompanied by an increased density of MUM1+ plasma cells, while a cluster with intermediate/high BGN H-scores showed the highest frequency of Tregs (CD3+FOXP3+). BGNhigh BMBs exhibit a higher number of Tregs in the proximity of CD8+ and CD4+ cells and a lower number of CD4+ cells within a 10 μm radius of CD8+ cells. In contrast, BGNlow patients have more CD8+ cells in the proximity of blasts, but a low number of surrounding Tregs (Figure 8b). Despite this favorable microenvironment, conventional IHC revealed that patients with CD34+ BGNhigh cells often have an increased expression of CD152 (CTLA-4), while no correlation was found with CD274 and PDCD1 (Supplementary Figure 6)
Figure 7.
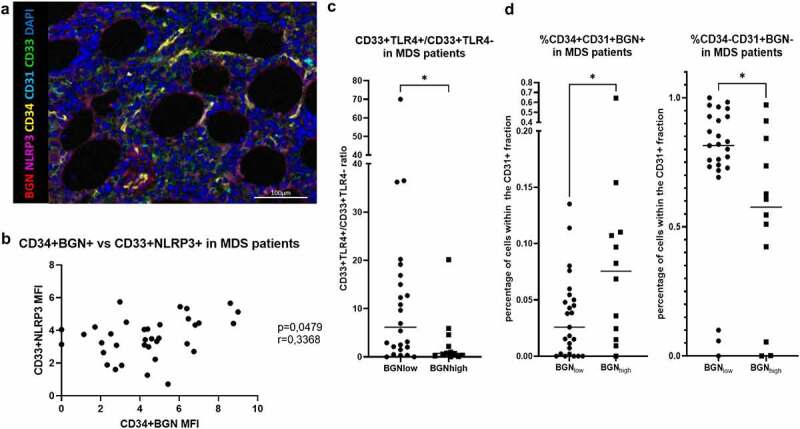
Multispectral imaging of inflammasome components on MDS BMBs. (a) Representative picture of a BMB stained with antibodies against CD34 (yellow), CD33 (green), CD31 (cyan), NLRP3 (violet), BGN (red) and DAPI (blue). (b) Correlation between the MFI of BGN within theCD34+ subpopulation of cells and NLRP3 within the CD33+ cells when analyzing the BMB of the MDS patient cohort. A significance along with the r value between the MFI of these molecules in the subpopulations of interest was found. (c) The ratio of CD33+TLR+/CD33+TLR4− cells determined by IHC are shown for BGNlow and BGNhigh patients. The median is shown for the depicted ratios within the biopsies as well as the significance (t-test). (d) The percentage of endothelial progenitor cells (characterized as CD34+CD31+ cells) expressing BGN (CD34+CD31+ BGN+, left) and the normal endothelial cells not producing BGN (CD34−CD31+ BGN−, right) were found within the BGNlow and BGNhigh patients. Median values of the reported percentages are represented by the black line, which was significant as determined by the T-test can be also observed.
Figure 8.
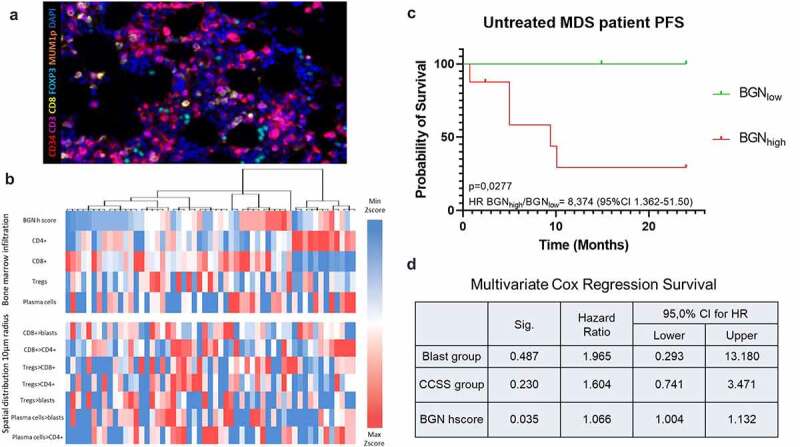
Correlation of BGN with the immune cell signature and its clinical relevance. (a) Representative picture of BMB stained with Ab against CD34 (red), CD3 (violet), CD8 (yellow), FOXP3 (cyan), MUMp1 (Orange) and DAPI (blue). To standardize the spatial distribution analysis, three MSI fields at the x20 zoom (1872 × 1404 pixel, 0.5 µm/pixel) were manually selected on each slide based on representativeness and tissue size. Since the BMBs showed a high variability in quality, only areas with preserved architecture were chosen. (b) Hierarchical clustering and heat map of the BM of MDS/sAML patients, based on their infiltrate. Blue fields show numbers lower than the 10% percentile, white fields are values close to the median, while red fields are values higher than the 90% percentile. The upper part of the table represents the frequencies of cells in the BM, while the lower part the mean number of cell type A within a 10 μm radius of the cell type B (indicated as A > B). (c-d) Log-rank progression free survival analysis (c) as well as multivariate Cox Regression survival analysis (d) of 14 MDS patients that did not receive treatment, regarding their BGN expression in the BM alone or together with the two commonly used risk factor markers (blast numbers and cytogenetic score). In the univariate analysis, BGNlow (n = 6) patients are depicted with green, while BGNhigh (n = 8) patients with red. Statistical significance as well as the hazard ratio between patients is depicted for both uni- and multivariate analysis. For the Cox regression model, the BGN H scores were used as a continuous variable, instead of stratifying the patients in high and low expressors, in order to investigate the possible effect of an increased BGN H score on the PFS of patients.
Correlating the clinicopathological characteristics and progression-free survival (PFS) with BGN expression, no significant differences in the PFS between patients with BGNlow versus BGNhigh BMBs regarding the established progression score to sAML was observed (Supplementary Figure 7a). Further analysis of the clinicopathological characteristics of BGNvery low and BGNvery high BMBs showed a tendency of BGNvery low for a lower IPSS-R scoring, which was independent of blast counts and cytogenetic score, since the BGNvery high group had favorable clinicopathological characteristics for both criteria (Supplementary Figure 7b). The common mutations found in MDS patients were not correlated with the BGN expression (Supplementary Table 4). Survival analysis of patients treated with the demethylating agent Vidaza had a comparable PFS regardless of BGN expression in the BM (Supplementary Figure 8, n = 18). In contrast, higher BGN expression levels in the BMBs of untreated MDS patients (n = 14) correlated with a significant decrease in PFS with a > 8 times higher risk of progression to sAML (Figure 8c). Combining the multivariate survival analysis using the BGN H-score with the risk scoring systems revealed that the BGN H-score represents an independent negative prognostic factor in the small cohort of patients analyzed (Figure 8d).
Discussion
Cancer-associated inflammation has been attributed to an attempt of the host immune system to eliminate tumor cells and provide tissue protection, but it can also facilitate tumorigenesis. Deciphering the function of proteoglycans in mediating cancer-associated inflammatory processes may provide novel insights into the biology of tumors including hematologic malignancies.30
In this study, a novel role of BGN in MDS and a potential mechanism, by which MDS stem cells utilize this SLRP to sustain a proinflammatory BM microenvironment, was identified. In the past, both pro-tumorigenic and tumor suppressive properties of BGN have been identified11 and related to patients’ outcomes. In the MDS/sAML context, different processes promoting inflammation have been documented, such as the downregulation of miRNAs in the del(5q) syndrome or TLR4 activation.19,31 In our model, BGN secreted by distinct MDS-L subpopulations with stem cell properties binds to mature CD33SPTLR4+ cells leading to inflammasome activation. This not only results in their inflammatory death, but also in the disruption of hematopoiesis as demonstrated by inflammasome inhibition leading to an increase of the lin+ TLR4+subpopulation. These findings were confirmed by MSI on BMB samples, where BGNhigh biopsies had less CD33+TLR4+ cells, further supporting the possible inhibitory role of BGN toward mature hematopoiesis. The importance of this BGN-mediated feedback loop in the BM of MDS patients is underlined by amplification of this effect in other TLR4+ subpopulations of the BM.32 Since chronic inflammation has been shown to contribute to various myeloid malignancies,33 the expression of BGN in AML,MDS, MPN cell lines and BMBs of MDS patients, but not in BMBs of patients without hematologic malignancies indicates a strong link between this SLRP and hematological malignancies.
In vitro experiments revealed heterogeneous effects of inhibitors of the BGN-mediated inflammasome activation on the cell differentiation. TGF-β1 and IFN-γ treatment showed promising results, although their use as treatment for MDS patients requires caution, since TGF-β can directly upregulate BGN,34 whereas IFN-γ signaling in MDS and AML was linked to an increase in PDCD1, which could be detrimental.31 Currently, a phase I clinical trial investigates the potential benefits of IFN-γ treatment in MDS patients (ClinicalTrials.gov: NCT04628338). Our results suggest that targeting of BGN by a combination of SN50 and 16673–34-0 inhibited casp1 activation causing CD34+ cell death and an increased frequency of the TLR4+ subpopulation, whereas treatment with an anti-BGN antibody increased the lin+ fraction. The importance of a dual targeting of the NLRP3 pathway was underlined by the synergistic effect upon inhibitor treatment, indicating a therapeutic potential for the combination of NF-kB inhibitors with glyburide, leading to results similar to the anti-BGN antibody treatment.
The lack of BGN expression in healthy CD34+ cells and absence of TLR4-bound BGN in total PBMCs and circulating HSCs demonstrates that BGN overexpression already occurs at the early onset of MDS and is maintained during disease progression to sAML. However, it is noteworthy that a hallmark of MDS is the activation of the NLRP3 inflammasome, which drives clonal expansion and pyroptotic cell death. Independent of the genotype, MDS hematopoietic stem and progenitor cells (HSPCs) overexpress inflammasome proteins and manifest activated NLRP3 complexes that direct activation of casp-1, generation of IL-1β, IL-18 and pyroptotic cell death.19 Interestingly, a correlation of NLRP3 with BGN expression in the sAML samples, but not in MDS was demonstrated indicating that disease progression coincides with the pyroptotic effects of BGN.
The discrepancy between the percentage of BGN-expressing cells in the BM of MDS/sAML patients and the number of blasts, as well as the analysis of the frozen BM aspirates, indicate a contribution of additional cell populations involved in this upregulation. As shown above, these could be among others, CD34+CD31+ endothelial progenitor cells, reported to produce BGN within other cancer types.35 Most importantly, despite the small patient cohort investigated, the observed effects of BGN on the PFS of untreated MDS patients suggest its use as a biomarker and potential therapeutic target for this disease. The BGN-driven alterations of the BM microenvironment include a high frequency of MUM1+ cells in BGNhigh MDS BMBs,36 but also high levels of CD8+ T and CD3+FOXP3+ cells. One could speculate that TLR4 stimulation by BGN upregulates MCP1, which was significantly elevated in the TLR4+ subset of MDS-L cells, resulting in a strong attraction of T cells into the BM.37 Furthermore, the CD3+FOXP3+ subpopulation does not necessarily represent a Treg subset, since high levels of IL-1β produced in the BGN-expressing BM could indicate Th17 differentiation, further promoting a proinflammatory microenvironment.38 Since BGN expression is epigenetically controlled and can be altered by 5-azacytidine treatment, treatment of BGNlow patients with demethylating agents might exert detrimental effects.35 Basal BGN levels in BMBs of MDS patients have clinical relevance due to the lack of progression of MDS BGNlow patients to sAML and suggesting its use as an independent prognostic factor in the patients’ cohort analyzed.
To the best of our knowledge, this study suggested for the first time BGN as a novel prognostic biomarker and therapeutic target of sAML/MDS by its direct inhibition through BGN-specific antibodies and/or inflammasome inhibitors.
Supplementary Material
Acknowledgments
We would like to thank Maria Heise and Nicole Ott for their excellent secretarial help.
Funding Statement
This work was supported by the Wilhelm-Roux-Program of the Medical Faculty of the MLU (CW) and Sander grant (2019.076.1, BS)..
Data availability statement
The data that support the findings of this study are available from the corresponding author, BS, upon reasonable request (barbara.seliger@uk-halle.de).
Disclosure statement
The authors declare that they have no conflicts of interest.
Author contribution
The corresponding author did ensure that the descriptions are accurate and all authors agreed on the final version of the manuscript. Christoforos Vaxevanis was involved in the design and execution of the experiments and writing the original draft. Marcus Bauer collected clinical and pathological data, performed the MSI analyses, Karthik Subbarayan performed the Western blot and qPCR analysis, Michael Friedrich and Chiara Massa were discussing the experimental outline and worked on the manuscript, Katharina Biehl performed sample preparation and staining for MSI analysis, Claudia Wickenhauser provided study materials and reviewed the manuscript. Barbara Seliger was mentoring the team, coordinated the manuscript, wrote together with Christoforos Vaxevanis the original draft and is responsible for the final version of this manuscript.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/2162402X.2022.2152998
References
- 1.Ganan-Gomez I, Wei Y, Starczynowski DT, Colla S, Yang H, Cabrero-Calvo M, Bohannan ZS, Verma A, Steidl U, Garcia-Manero G, et al. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 2015;29(7):1458–12. doi: 10.1038/leu.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber S, Parmon A, Kurrle N, Schnutgen F, Serve H.. The clinical significance of iron overload and iron metabolism in myelodysplastic syndrome and acute myeloid leukemia. Front Immunol. 2021;11:627662. doi: 10.3389/fimmu.2020.627662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zanetti C, Krause DS. “Caught in the net”: the extracellular matrix of the bone marrow in normal hematopoiesis and leukemia. Exp Hematol. 2020;89:13–25. doi: 10.1016/j.exphem.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Frevert CW, Felgenhauer J, Wygrecka M, Nastase MV, Schaefer L. Danger-associated molecular patterns derived from the extracellular matrix provide temporal control of innate immunity. J Histochem Cytochem. 2018;66(4):213–227. doi: 10.1369/0022155417740880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roedig H, Damiescu R, Zeng-Brouwers J, Kutija I, Trebicka J, Wygrecka M, Schaefer L. Danger matrix molecules orchestrate CD14/CD44 signaling in cancer development. Semin Cancer Biol. 2020;62:31–47. doi: 10.1016/j.semcancer.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 6.Appunni S, Rubens M, Ramamoorthy V, Anand V, Khandelwal M, Sharma A. Biglycan: an emerging small leucine-rich proteoglycan (SLRP) marker and its clinicopathological significance. Mol Cell Biochem. 2021;476(11):3935–3950. doi: 10.1007/s11010-021-04216-z. [DOI] [PubMed] [Google Scholar]
- 7.Miguez PA. Evidence of biglycan structure-function in bone homeostasis and aging. Connect Tissue Res. 2020;61(1):19–33. doi: 10.1080/03008207.2019.1669577. [DOI] [PubMed] [Google Scholar]
- 8.Babelova A, Moreth K, Tsalastra-Greul W, Zeng-Brouwers J, Eickelberg O, Young MF, Bruckner P, Pfeilschifter J, Schaefer RM, Gröne H-J, et al. Biglycan, a danger signal that activates the NLRP3 inflammasome via toll-like and P2X receptors. J Biol Chem. 2009;284(36):24035–24048. doi: 10.1074/jbc.M109.014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frey H, Schroeder N, Manon-Jensen T, Iozzo RV, Schaefer L. Biological interplay between proteoglycans and their innate immune receptors in inflammation. FEBS J. 2013;280(10):2165–2179. doi: 10.1111/febs.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roedig H, Nastase MV, Wygrecka M, Schaefer L. Breaking down chronic inflammatory diseases: the role of biglycan in promoting a switch between inflammation and autophagy. FEBS J. 2019;286(15):2965–2979. doi: 10.1111/febs.14791. [DOI] [PubMed] [Google Scholar]
- 11.Diehl V, Huber LS, Trebicka J, Wygrecka M, Iozzo RV, Schaefer L. The role of decorin and biglycan signaling in tumorigenesis. Front Oncol. 2021;11:801801. doi: 10.3389/fonc.2021.801801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng-Brouwers J, Pandey S, Trebicka J, Wygrecka M, Schaefer L. Communications via the small leucine-rich proteoglycans: molecular specificity in inflammation and autoimmune diseases. J Histochem Cytochem. 2020;68(12):887–906. doi: 10.1369/0022155420930303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niedworok C, Rock K, Kretschmer I, Freudenberger T, Nagy N, Szarvas T, Vom Dorp F, Reis H, Rübben H, Fischer JW, et al. Inhibitory role of the small leucine-rich proteoglycan biglycan in bladder cancer. PLoS One. 2013;8(11):e80084. doi: 10.1371/journal.pone.0080084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber CK, Sommer G, Michl P, Fensterer H, Weimer M, Gansauge F, Leder G, Adler G, Gress TM. Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines. Gastroenterology. 2001;121(3):657–667. doi: 10.1053/gast.2001.27222. [DOI] [PubMed] [Google Scholar]
- 15.Zhao SF, Yin XJ, Zhao WJ, Liu LC, Wang ZP. Biglycan as a potential diagnostic and prognostic biomarker in multiple human cancers. Oncol Lett. 2020;19:1673–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subbarayan K, Leisz S, Wickenhauser C, Bethmann D, Massa C, Steven A, Seliger B. Biglycan-mediated upregulation of MHC class I expression in HER-2/neu-transformed cells. Oncoimmunology. 2018;7(4):e1373233. doi: 10.1080/2162402X.2017.1373233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner W, Saffrich R, Wirkner U, Eckstein V, Blake J, Ansorge A, Schwager C, Wein F, Miesala K, Ansorge W, et al. Hematopoietic progenitor cells and cellular microenvironment: behavioral and molecular changes upon interaction. Stem Cells. 2005;23(8):1180–1191. doi: 10.1634/stemcells.2004-0361. [DOI] [PubMed] [Google Scholar]
- 18.Sallman DA, List A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 2019;133(10):1039–1048. doi: 10.1182/blood-2018-10-844654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basiorka AA, McGraw KL, Eksioglu EA, Chen X, Johnson J, Zhang L, Zhang Q, Irvine BA, Cluzeau T, Sallman DA, et al. The NLRP3 inflammasome functions as a driver of the myelodysplastic syndrome phenotype. Blood. 2016;128(25):2960–2975. doi: 10.1182/blood-2016-07-730556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee T, Calvi LM, Becker MW, Liesveld JL. Flaming and fanning: the spectrum of inflammatory influences in myelodysplastic syndromes. Blood Rev. 2019;36:57–69. doi: 10.1016/j.blre.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campo S, Campo GM, Avenoso A, D’Ascola A, Musolino C, Calabro L, Bellomo G, Quartarone E, Calatroni A. Lymphocytes from patients with early stage of B-cell chronic lymphocytic leukaemia and long survival synthesize decorin. Biochimie. 2006;88(12):1933–1939. doi: 10.1016/j.biochi.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Rydstrom K, Joost P, Ehinger M, Eden P, Jerkeman M, Cavallin-Stahl E, Linderoth J. Gene expression profiling indicates that immunohistochemical expression of CD40 is a marker of an inflammatory reaction in the tumor stroma of diffuse large B-cell lymphoma. Leuk Lymphoma. 2012;53(9):1764–1768. doi: 10.3109/10428194.2012.666541. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Liang S, Zhan Q, Yang L, Chi J, Wang L. HSPG2 overexpression independently predicts poor survival in patients with acute myeloid leukemia. Cell Death Dis. 2020;11(6):492. doi: 10.1038/s41419-020-2694-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauer M, Vaxevanis C, Bethmann D, Massa C, Pazaitis N, Wickenhauser C, Seliger B, et al. Multiplex immunohistochemistry as a novel tool for the topographic assessment of the bone marrow stem cell niche. Methods Enzymol. 2020;635:67–79. [DOI] [PubMed] [Google Scholar]
- 25.Bauer M, Vaxevanis C, Al-Ali HK, Jaekel N, Naumann CLH, Schaffrath J, Rau A, Seliger B, Wickenhauser C. Altered spatial composition of the immune cell repertoire in association to CD34+ blasts in myelodysplastic syndromes and secondary acute myeloid leukemia. Cancers. 2021;13(2):186. doi: 10.3390/cancers13020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedrich M, Vaxevanis CK, Biehl K, Mueller A, Seliger B. Targeting the coding sequence: opposing roles in regulating classical and non-classical MHC class I molecules by miR-16 and miR-744. J Immunother Cancer. 2020;8(1):e000396. doi: 10.1136/jitc-2019-000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Kao Y-R, Sun D, Todorova TI, Reynolds D, Narayanagari S-R, Montagna C, Will B, Verma A, Steidl U, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med. 2019;25(1):103–110. doi: 10.1038/s41591-018-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shastri A, Will B, Steidl U, Verma A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129(12):1586–1594. doi: 10.1182/blood-2016-10-696062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer L, Babelova A, Kiss E, Hausser H-J, Baliova M, Krzyzankova M, Marsche G, Young MF, Mihalik D, Götte M, et al. The matrix component biglycan is proinflammatory and signals through toll-like receptors 4 and 2 in macrophages. J Clin Invest. 2005;115(8):2223–2233. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Shi L, Zhao Y, Fei C, Guo J, Jia Y, Wu D, Wu L, Chang C. Cellular senescence induced by S100A9 in mesenchymal stromal cells through NLRP3 inflammasome activation. Aging. 2019;11(21):9626–9642. doi: 10.18632/aging.102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X, Wang H, Jin T, Xu Y, Mei L, Yang J, Beltrami AP. TLR4 activation promotes bone marrow MSC proliferation and osteogenic differentiation via Wnt3a and Wnt5a signaling. PLoS One. 2016;11(3):e0149876. doi: 10.1371/journal.pone.0149876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Recher C. Clinical implications of inflammation in acute myeloid leukemia. Front Oncol. 2021;11:623952. doi: 10.3389/fonc.2021.623952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heegaard AM, Xie Z, Young MF, Nielsen KL. Transforming growth factor beta stimulation of biglycan gene expression is potentially mediated by sp1 binding factors. J Cell Biochem. 2004;93(3):463–475. doi: 10.1002/jcb.20189. [DOI] [PubMed] [Google Scholar]
- 35.Maishi N, Ohba Y, Akiyama K, Ohga N, Hamada J, Nagao-Kitamoto H, Alam MT, Yamamoto K, Kawamoto T, Inoue N, et al. Tumour endothelial cells in high metastatic tumours promote metastasis via epigenetic dysregulation of biglycan. Sci Rep. 2016;6(1):28039. doi: 10.1038/srep28039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreth K, Brodbeck R, Babelova A, Gretz N, Spieker T, Zeng-Brouwers J, Pfeilschifter J, Young MF, Schaefer RM, Schaefer L, et al. The proteoglycan biglycan regulates expression of the B cell chemoattractant CXCL13 and aggravates murine lupus nephritis. J Clin Invest. 2010;120(12):4251–4272. doi: 10.1172/JCI42213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tzanakakis G, Neagu M, Tsatsakis A, Nikitovic D. Proteoglycans and immunobiology of cancer—therapeutic implications. Front Immunol. 2019;10:875. doi: 10.3389/fimmu.2019.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mailer RK, Joly AL, Liu S, Elias S, Tegner J, Andersson J. IL-1beta promotes Th17 differentiation by inducing alternative splicing of FOXP3. Sci Rep. 2015;5:14674. doi: 10.1038/srep14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, BS, upon reasonable request (barbara.seliger@uk-halle.de).


