Abstract
Significant progress in previous decades has led to several methodologies developed to facilitate the design of optimal antimicrobial dosing. In this review, we highlight common pharmacokinetic/pharmacodynamic (PKPD) modeling techniques and their roles in guiding rational dosing regimen design. In the early drug development phases, dose fractionation studies identify the PKPD index most closely associated with bacterial killing. Once discerned, this index is linked to clinical efficacy end points, and classification and regression tree analysis can be used to define the PKPD target goal. Monte Carlo simulations integrate PKPD and microbiological data to identify dosing strategies with a high probability of achieving the established PKPD target. Results then determine dosing regimens to investigate and/or validate the findings of randomized controlled trials. Further improvements in PKPD modeling could lead to an era of precision dosing and personalized therapeutics.
Keywords: dose fractionation studies, classification and regression tree analysis, dosing regimen design, Monte Carlo simulations, optimal drug dosing
This review highlights common PKPD modeling techniques that provide insights to facilitate the design of antimicrobial dosing regimens. Results from these studies enable rational dosing to optimize clinical outcomes and can lead to an era of precision dosing.
The rise of broad-spectrum antimicrobial resistance is a critical situation that clinicians are facing today. With limited options available, there is a growing need to judiciously utilize and optimize our current arsenal of antimicrobials to improve patient outcomes. In previous decades, several advances were made to characterize the concentration–effect relationships of antimicrobials to facilitate optimal drug dosing. In general, pharmacokinetic and pharmacodynamic (PKPD) analyses allow the exploration of the time course of an observed drug effect, which provide a means to relate the effects to drug concentrations in the body [1, 2]. For most nonantimicrobials, their effects can either be measures of clinical response (eg, analgesia) or biomarkers of drug effects (eg, international normalized ratio) [3, 4]. As a result, the dose–response relationship can be elucidated by directly monitoring the physiological and/or biochemical variables of the host [5]. However, these principles do not apply to antimicrobials as their main target is the pathogen (as opposed to receptor targets in the host).
Dosing of antimicrobials differs greatly from other drugs, as the evaluation of its efficacy relies on 2 factors: (1) drug exposure achieved in the host and (2) pathogen susceptibility to the drug [6]. Due to this distinction, clinical response measures of the host are only indirect aspects of antimicrobial effectiveness. To circumvent the challenge of integrating the above factors, surrogate indices have been proposed to predict clinical success [7]. These measures relate the extent of drug exposure in the host and the susceptibility of the pathogen to the drug (commonly represented by the minimum inhibitory concentration [MIC]). Consequently, these indices represent a complex interplay between factors of the host and pathogen, which help to inform dose selection to achieve favorable outcomes [8, 9]. In an ideal setting, antimicrobial dosing should be individualized based on patient (eg, body weight, renal/hepatic function, plasma albumin) and microbial (eg, MIC, resistance mechanism genotypes) characteristics (ie, precision dosing).
Over the years, many approaches have been developed to facilitate the design of optimal dosing regimens. The objectives of this review are to describe the most common PKPD techniques from an end user's perspective and to facilitate clinician interpretation of studies using these modeling tools. Similar techniques can also be applied to minimizing adverse effects associated with antimicrobial therapy (ie, toxicodynamics). The statistical basis and theory behind these approaches are beyond the scope of this review. For brevity’s sake, we will focus on the efficacy of antibacterials in this review for the purpose of illustration.
DOSE FRACTIONATION STUDIES AND HOW THEY INFORM PKPD PARAMETER SELECTION
Background
In the early stages of nonantimicrobial drug development, dose-finding studies are essential to inform dose selection for testing in clinical trials [10, 11]. In phase II studies, the effectiveness and safety of an experimental drug are determined, typically through a dose escalation design. Doses in proportion to the maximum tolerated dose, in addition to host clinical responses, are studied to establish the dose–effect relationship of the drug. Study results are used to formulate the most optimal dosing regimen to be investigated in phase III clinical trials [12]. However, this study design is not directly applicable to antimicrobials as efficacy is also affected by pathogen susceptibility to the antimicrobial. More robust surrogate indices (ie, PKPD parameters) have thus been proposed as better predictors of clinical and microbiological success. To identify the PKPD index most closely correlated to bacterial killing (ie, efficacy), dose fractionation studies have commonly been utilized.
Experimental Design
Dose fractionation experiments involve studying various dosing regimens with different dosing intervals and fractions of the total daily dose in such a way that the total daily drug exposure is kept constant (Table 1) [8, 13]. This design can be adopted in different preclinical infection models (eg, chemostat, hollow-fiber, neutropenic murine thigh infection/pneumonia models). In the simplest design with a single pathogen, different PKPD indices (eg, Cmax/MIC, T > MIC, area under the curve [AUC]/minimum inhibitory concentration [MIC]) could be compared relying on a specific MIC as the common denominator, as shown in Table 1.
Table 1.
Exposure Profiles of a Hypothetical Antimicrobial Exhibiting Linear Kinetics (Volume of Distribution of 40 L, Clearance of 28 L/h, Elimination Half-life of 1 Hour, and Negligible Protein Binding); the Target Pathogen Is Assumed to Have an MIC of 1 mg/L
| Dosing Regimen | fCmax, mg/L | fCmin, mg/L | fAUC, mg•h/L | T > MIC, % |
|---|---|---|---|---|
| 2000 mg every 24 h | 50 | Negligible | 72 | 23 |
| 1000 mg every 12 h | 25 | 0.01 | 72 | 38 |
| 500 mg every 6 h | 12.5 | 0.2 | 72 | 60 |
Different antimicrobial pharmacodynamic characterizations may be inferred based on efficacy observed for various dosing regimens. For example, if the regimen given the least frequently (every 24 h) is the most effective, Cmax/MIC is thought to be the PKPD index most closely linked to an antimicrobial’s efficacy. In contrast, T > MIC is most associated with the regimen given most frequently (every 6 h). If there is no difference in efficacy, the AUC/MIC is the index linked to bacterial killing.
Abbreviations: AUC/MIC, area under the curve/minimum inhibitory concentration; CFU, colony-forming units; PKPD, pharmacokinetic/pharmacodynamic.
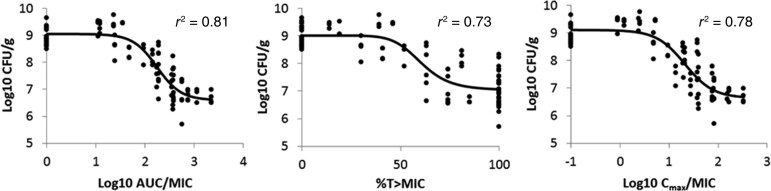
Alternatively, a scatter plot can be utilized to discriminate antimicrobial effect (typically represented by the bacterial count following 24 hours of treatment) from the PKPD indices (Figure 1). Typically, a regression curve (representing an inhibitory sigmoidal Emax model) that best fits all the data points is generated. The selection of the PKPD index most closely related to bacterial killing is derived from the regression model with the best coefficient of determination (denoted as r2). Of note, the free, unbounded fraction of a drug is often used in the calculation of PK parameters and is denoted by the prefix f, as this represents the active component of the drug [14, 15]. Results from these studies identify the PKPD index that best predicts antimicrobial efficacy and have historically been derived from 1 of the 3 PKPD indices described above. The methods used to elucidate the PKPD of an antimicrobial can be made even more robust when tested against a range of MICs from a single or multiple pathogens.
Figure 1.
Relationship between PKPD indices of minocycline and Acinetobacter baumannii burden in mouse lung tissue at 24 hours [19]. Figure reproduced with permission from the American Society for Microbiology and Zhou et al. [19]. Each data point represented observations of 1 animal. Solid lines represent the best-fit regression curve. In this context, a reduction in log CFU/g (bacterial burden) is associated with bacterial killing. AUC/MIC best depicts A. baumannii killing of minocycline; its correlation with bacterial killing has the highest r2 value. Abbreviations: AUC/MIC, area under the curve/minimum inhibitory concentration; CFU, colony-forming units; PKPD, pharmacokinetic/pharmacodynamic.
Many researchers use this regression approach to discern the PKPD index that is most closely linked to bacterial killing [16–19]. For instance, in a study by Zhou and colleagues, an in vivo neutropenic murine pneumonia model was utilized to characterize the PKPD of minocycline against Acinetobacter baumannii [19]. Briefly, the relationship between minocycline exposure and antimicrobial efficacy was described following administration of varying doses of minocycline, and the PK of the drug was elucidated through analysis of serum and epithelial lining fluid concentrations. Through regression modeling, the AUC/MIC ratio was deemed the PKPD index most closely correlated to the antimicrobial activity of minocycline against A. baumannii, due to AUC/MIC having the highest coefficient of determination among the 3 PKPD indices studied.
Expert Commentary
Traditional PKPD indices may provide a simplified characterization of antimicrobial efficacy and are used for screening purposes in the early drug development stages. However, they may not fully capture bacterial killing comprehensively [20, 21]. There is an implicit assumption that the characterization of antimicrobial efficacy can be classified into 1 of only 3 distinct PKPD indices (mentioned previously), but there may be other indices that could also accurately describe the bacterial killing of antimicrobials. For instance, the minimum concentration-to-MIC ratio (Cmin/MIC) may potentially be a better PKPD parameter compared with T > MIC, due to a ceiling effect of 100% with the latter [22, 23]. As a result, Cmin/MIC may offer greater flexibility for antimicrobial exposures to be quantified, especially when the efficacy breakpoint is beyond T > MIC of 100%.
In all of the PKPD indices defined above, these measures rely on the MIC to describe antimicrobial PD. Although widely used due to its practicality, key limitations exist with utilizing MIC in PKPD analyses. The MIC represents the drug concentration that prevents visible growth of bacteria over a fixed time frame [24]. The result can be interpreted as a binary outcome that states that an antimicrobial given at a concentration above the MIC leads to inhibition of bacterial growth, while concentrations below the MIC will fail to prevent bacterial growth. Under these conditions, the MIC does not quantitatively define the rate and extent of bacterial killing, and thus may not reflect the antimicrobial effects comprehensively [25]. Another limitation is that MIC measurements may fail to detect any potential resistant subpopulation(s) present in minute amounts (ie, below the limit of detection within the time frame in which MIC results are evaluated) [26]. Finally, MIC measurements could be subjected to inconsistencies due to laboratory variations [15, 25]. An identical bacterial strain may exhibit different MIC values in 2 different institutions utilizing 2 different types of lab equipment, which may have significant clinical implications if the true MIC lies close to the threshold between susceptible and nonsusceptible interpretations.
Due to the inherent drawbacks of MICs, innovations in non-MIC-based indices might provide further insights into antimicrobial PKPD. Researchers have utilized more objective parameters in PKPD analysis, such as minimum bactericidal concentration (MBC), mutant prevention concentration (MPC), and kill rate–based PKPD integration models [27, 28]. For instance, MPC-based PKPD indices have been used in the study of fluoroquinolones because of their propensity to select drug-resistant mutants that occur gradually through genetic mutations [27]. Although nonconventional PD parameters have been developed to describe antimicrobial effects beyond what the MIC can provide, no single PD parameter may fully characterize bacterial killing. Therefore, a combination of nontraditional and traditional PKPD indices can be used to determine the entire spectrum of antimicrobial mechanisms of action [24, 29, 30]. Further advances in the study of antimicrobial PKPD are needed to distinguish what truly defines their efficacy. Dose–response relationships may be more realistically described as a continuous (nonlinear) function as opposed to discrete categories. Different approaches to ascertain these nonlinear functions are beyond the scope of this review [31, 32].
CLASSIFICATION AND REGRESSION TREE ANALYSIS AND PARTITIONING TECHNIQUES TO IDENTIFY GOAL TARGETS
Background
Following the identification of the optimal PKPD index associated with bacterial killing, the next step is to link the index to clinically meaningful efficacy outcomes. Many study end points assessing antimicrobial therapy encode outcomes as a set of discrete states, such as whether a patient survived (mortality) or whether a pathogen continued to be recovered on repeat cultures (persistence). Lower drug exposures in a subject are typically associated with less favorable outcomes, while higher drug exposures are associated with a higher likelihood of favorable outcomes. When a trend is suspected, one may attempt to correlate different magnitudes of the PKPD index through regression analysis. This technique estimates the quantitative impact of an independent variable (eg, magnitude of drug exposure) on the dependent variable (eg, hospital mortality) [33]. Due to the dichotomy of most outcome variables, logistic regression is traditionally used.
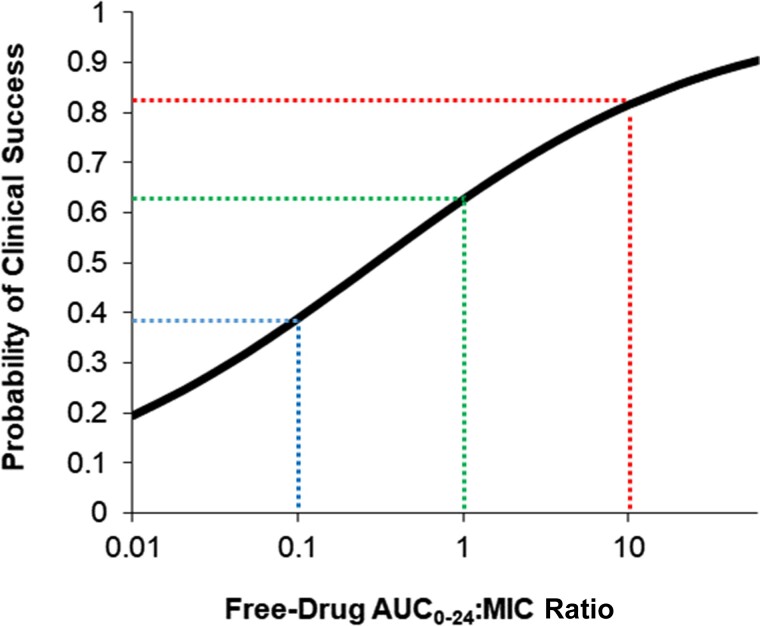
In clinical studies, logistic regression analysis is a powerful tool that yields a probability of achieving the research end points [34]. Furthermore, the analysis output is a prediction equation with coefficients that can be related to the odds ratio. However, the interpretation of these results may be subjective as the goal threshold that describes the most optimal probability of achieving a clinical outcome could be different among practitioners. As opposed to providing probabilities of an outcome based on the exposure variable (eg, AUC/MIC = 10 is associated with 82% clinical success) (Figure 2), clinicians often prefer a target exposure that has the most reliable likelihood of clinical success. For these reasons, recursive partitioning techniques have been used to provide an objective method to determine a single drug exposure value most significantly associated with an outcome of interest.
Figure 2.
Association of clinical success and tigecycline fAUC0–24:MIC ratio in patients with hospital-acquired pneumonia. Figure reproduced with permission from data reported from Bhavnani et al. [81]. The probability of clinical success associated with an fAUC/MIC of 0.1, 1, and 10 was 39% (blue), 63% (green), and 82% (red), respectively. This approach allows dosing selection to accommodate various clinical circumstances with different benefit-to-risk ratios. Abbreviations: AUC, area under the curve; MIC, minimum inhibitory concentration.
Analytical Design
Recursive partitioning techniques have been used in epidemiological studies to explore distinct patient subpopulations and identify risk factors associated with a study outcome [35, 36]. The name of this technique refers to the fact that the data set can be split into sections (partitioning) repeatedly (recursive) until the remaining data points cannot be split further or the program reaches a user-specified stopping criterion [33, 37]. Classification and regression tree (CART) analysis is a type of recursive partitioning technique that produces a clinical decision tree that is easier to interpret compared with logistic regression, making this more practical in clinical settings [38]. Researchers have utilized this methodology to relate the study outcome to antimicrobial PKPD parameters for the identification of breakpoint thresholds associated with clinical success.
Briefly, CART analysis begins with ranking the magnitude of the independent variable in ascending (or descending) order, which can be branched out into 2 subgroups. The group containing the entire sample is designated the parent node, while the subgroups are termed child nodes [33]. In many antimicrobial studies, the splitting criterion is based on the magnitude of drug exposure with the greatest statistical significance with respect to the outcome variable. This would then be designated the breakpoint threshold after further refinement. For example, the relationship between the clinical outcome (eg, mortality) observed for each individual and their drug exposure achieved (eg, serum concentration or PKPD index) can be assessed through a 2-by-2 table. Comparisons are made between patients who did and did not achieve the outcome of interest above a single drug exposure value to outcomes of patients who were below that drug exposure value. Through repeated analysis of each threshold, the magnitude of the PKPD index found to be most significantly associated with the study outcome is deemed the breakpoint (Table 2).
Table 2.
Reanalysis of Data by Aitken et al. [36] for Illustrating CART Analysis
| Cmin/MIC Ratio | Clinical Outcome | P Value | |
|---|---|---|---|
| Success, No. | Failure, No. | ||
| Threshold 1 | |||
| ≤0.7 | 0 | 1 | .364 |
| >0.7 | 21 | 11 | |
| Threshold 2 | |||
| ≤1.5 | 2 | 1 | 1 |
| >1.5 | 19 | 11 | |
| Threshold 3 | |||
| ≤2.1 | 2 | 6 | .015a |
| >2.1 | 19 | 6 | |
| Threshold 4 | |||
| ≤4 | 9 | 8 | .282 |
| >4 | 12 | 4 | |
Briefly, plasma concentrations of cefepime before the next scheduled dose (Cmin) in 33 patients were normalized to MIC of the pathogen recovered from the same subject. The PKPD exposures were linked to the clinical outcomes observed. The CART program generates a series of 2-by-2 tables correlating every observed Cmin/MIC value and clinical outcome. For illustrative purposes, this table comprises of a subset of 4 tables from the CART analysis. The Fisher exact test was utilized for statistical comparison. In this representative series of 2-by-2 tables, the most statistically significant difference in outcome was found using Cmin/MIC = 2.1 as the threshold.
Abbreviations: CART, classification and regression tree; MIC, minimum inhibitory concentration; PKPD, pharmacokinetic/pharmacodynamic.
In several instances, CART analysis has been used in the literature to help clinicians identify goal targets for antimicrobials to maximize patient outcomes [23, 39–42]. For example, Kullar and colleagues determined that patients receiving vancomycin for methicillin-resistant Staphylococcus aureus (MRSA) bacteremia with an AUC/MIC <421 have significantly higher rates of clinical failure [43]. This study, along with other evidence, provided the basis for the recommendation to target AUC/MIC ≥400 when utilizing vancomycin for MRSA infections [44–46].
Expert Commentary
CART analysis has several advantages over logistic regression. It is an objective method for predicting an outcome through classification, which leads to a clinical decision tree that is easily interpreted by clinicians [33, 37, 38]. Moreover, the breakpoint threshold is determined based on the statistical analysis of all PKPD index values observed in a given sample population [47]. CART analysis is a nonparametric technique, so no assumptions need to be made regarding the distribution of the values of the predictor variables [33].
Despite these advantages, there are several limitations of CART analysis. The breakpoints defined from CART analysis are focused on reproducibility, as opposed to being the absolute breakpoint clinicians should target to predict clinical success. If there is an absolute breakpoint, this creates perfectly separable classes that easily predict clinical success and failure. However, there may be instances where patients may still do well clinically below the identified efficacy threshold. In this scenario where a data set is nonseparable, evaluating the steepness of the logistic curve in regression analysis may be helpful in order to assess the benefits vs risks of targeting above the threshold (Figure 2). Second, the data set can only be split in between observed magnitudes of the PKPD index. Thus, the resolution of the threshold is inherently dependent on the distribution of the observed drug exposures. Lastly, CART analysis usually evaluates 1 single end point, but not all patients have end points that align with clinical success. For example, a patient who survived but had microbiological persistence in subsequent cultures might be classified differently depending on the end point used in a CART analysis. This instance illustrates a fundamental disadvantage of CART analysis as it cannot account for multiple clinical end points. Recent clinical studies have gravitated toward the desirability of outcome ranking (DOOR) analytic approach, which assesses the overall clinical outcome of a patient by evaluating the benefits/risks of antimicrobial therapy through ordinal categorization of outcomes [48–50].
MONTE CARLO SIMULATIONS TO EVALUATE DOSING STRATEGIES
Background
With a PKPD target established, the next step is to identify the dosing strategies most likely to attain that goal. During drug development, PKPD models have been utilized to facilitate the design of clinical trials and propose dosing regimens to maximize efficacy outcomes [51]. Quantitative models to describe biological (pharmacological) processes can be deterministic or stochastic [52]. Models widely used in systems pharmacology are generally deterministic, where the output is determined from a given set of conditions and attempts to predict the average population effect of the system [51]. On the other hand, stochastic processes assume that the inputs in a system are subject to randomness and that the same set of conditions may lead to variable outcomes weighted by their probability. In the study of antimicrobials, Monte Carlo simulations (a form of stochastic forecasting) utilize random sampling to generate a virtual population to predict the probability of an outcome based on prior input data [6, 53, 54]. The premise of Monte Carlo simulations is that if an experiment is repeated multiple times, we can reliably infer that the results of the simulation will be close to the expected outcome [55]. Integrating PK variability and microbiological data, such an analysis can then be utilized to evaluate the likelihood that a particular dosing regimen will achieve an established PKPD target [56, 57]. Furthermore, results can then identify dosing strategies that could be used to guide the design or validate findings of randomized controlled trials [54].
Monte Carlo simulations have been used extensively in antimicrobial development phases for several reasons. It is costly and time-consuming to test different dosing regimens comprehensively in human subjects, and thus it may be more practical to simulate a virtual clinical trial of thousands of subjects. For example, given a simple study design, to assess 3 total daily doses (eg, 2000, 1000, and 500 mg), given in 3 dosing frequencies (eg, every 24, 12, and 6 hours), and infused over 4 infusion times/modalities (eg, over 1, 2, 4, and 24 hours of continuous infusion), 36 different regimens would need to be tested in appropriately sized human trials. Additionally, most dosing adjustments of nonantimicrobials for end-organ dysfunctions are based on normalizing the overall drug exposure (AUC), which may not always apply to antimicrobials (since bacterial killing could depend on a PKPD index other than AUC/MIC). More appropriate antimicrobial dosing adjustments can be obtained through Monte Carlo simulations using the most relevant PKPD index [58, 59]. Similar simulations can also help to inform optimal dosing regimens in special populations, including patients with augmented renal clearance, critically ill patients, and obese patients [32, 60–63]. The literature has shown that these patients have PK variations that would alter the efficacy of antimicrobials at doses known to be effective in the general population. Lastly, studies have integrated Monte Carlo simulations into other analyses such as pharmacoeconomic evaluations used for formulary decision-making [64–66]. In this instance, simulations are able to identify the dosing regimens that are the most cost-effective by balancing drug acquisition cost and the likelihood of PKPD target attainment.
Analytical Design
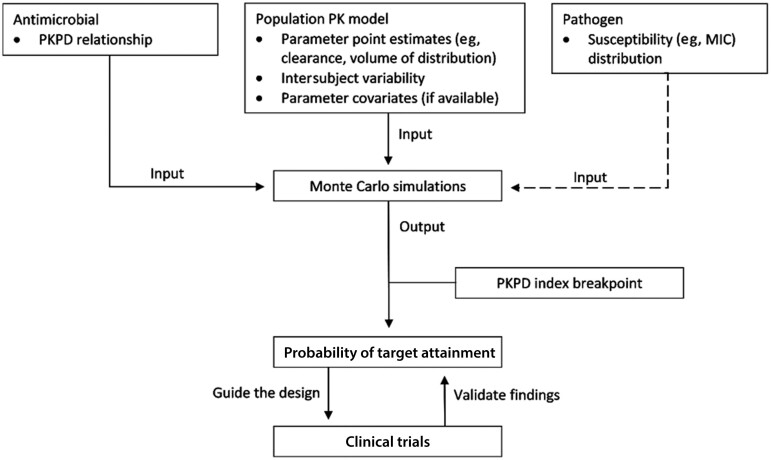
Generally, the prerequisites to perform a Monte Carlo simulation include a robust (population) PK model with well-defined PK parameters (eg, clearance, volume of distribution) in the target patient population, ideally with a covariate(s) model that informs any PK variations based on patient demographics and characteristics (eg, clearance expressed as a function of glomerular filtration rate) and a PKPD model that defines the relationship between drug exposure and outcome [6, 67]. Once these inputs are introduced, the program creates a simulated population from the PK model where each subject will be randomly assigned PK variables based on the parameter distribution of the model. A concentration–time profile for each subject is constructed and evaluated against specific MICs of a pathogen. The ultimate result would be the likelihood of achieving the predefined PKPD target, more commonly referred to as the probability of target attainment (PTA), for the entire sample population (Figure 3). Some experts believe that a PTA of 90% or higher is an appropriate threshold to support the use of a particular dosing regimen at a given MIC [53]. A weighted distribution of drug susceptibility could also be incorporated based on recent local/regional surveillance data to guide the use of empiric therapy [67]. When actual PK (eg, drug exposure, penetration) and drug susceptibility patterns (eg, MIC, resistance genotypes) are known, these factors may pave the way toward precision dosing of antimicrobial therapy [68, 69].
Figure 3.
Simple schematic depicting the process of antimicrobial dose design involving Monte Carlo simulations. The necessary a priori input models for Monte Carlo simulations include the antimicrobial PKPD relationship, a robust population PK model with well-defined parameter point estimates, intersubject variability of the parameters, and any available parameter covariates (which informs parameter variation based on subject characteristics) and microbiological data, if needed. As represented by the dashed line, the susceptibility distribution for the pathogens of interest (based on local surveillance data) may be incorporated to evaluate dosing regimens against a range of MICs. The output of simulations is then tested against the PKPD index breakpoint to generate the probability of attaining the PKPD index target specified by the user. Results allow clinicians to deduce dosing strategies that help guide the design or validate the findings of clinical trials. Abbreviations: MIC, minimum inhibitory concentration; PKPD, pharmacokinetic/pharmacodynamic.
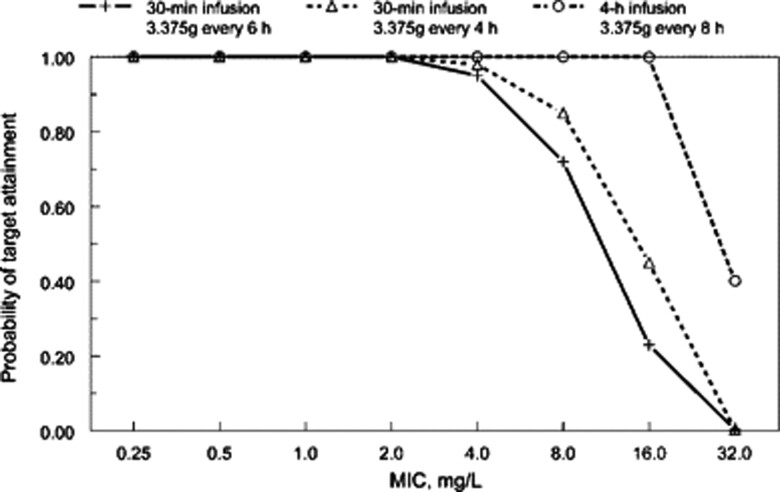
Multiple studies have utilized Monte Carlo simulations to facilitate the design of optimal dosing strategies for various antimicrobials [70–73]. For instance, extended infusions of piperacillin/tazobactam have shown higher PTAs to a variety of Enterobacterales and Pseudomonas aeruginosa, especially at an MIC range of 8–16 mg/L, despite having a lower total daily dose compared with standard regimens, as shown in Figure 4 [72, 73]. When the pathogen MIC is >16 mg/L, the PTAs for piperacillin/tazobactam decrease dramatically, regardless of the dosing strategy used. Two studies presented clinical evidence that validated the findings of the Monte Carlo simulations. Tam et al. found a higher 30-day mortality rate in patients with P. aeruginosa bacteremia receiving piperacillin/tazobactam when the MIC was >16 mg/L (Figure 5) [74]. Similarly, when compared with intermittent infusion regimens, continuous-infusion piperacillin/tazobactam was associated with increased clinical cure of ventilator-associated pneumonia when the pathogen MIC ranged from 8 to 16 mg/L (Figure 6) [75]. Thus, optimized PK/PD-based dosing provides the most clinical impact when used against pathogens expressing low- to intermediate-level resistance.
Figure 4.
Probability of target attainment analysis for piperacillin/tazobactam therapy. Figure reproduced with permission from the Oxford University Press and Lodise et al. [72]. Both intermittent and extended infusions of piperacillin/tazobactam have high probabilities of target attainment when the MIC is low (≤4 mg/L). Within the MIC range of 8–16 mg/L, the probability of target attainment for prolonged-infusion piperacillin/tazobactam is considerably higher compared with standard-infusion administrations. Beyond a certain MIC (16 mg/L in this scenario), clinically relevant dosing strategies are unlikely to achieve optimal exposures needed for bacterial killing. Abbreviation: MIC, minimum inhibitory concentration.
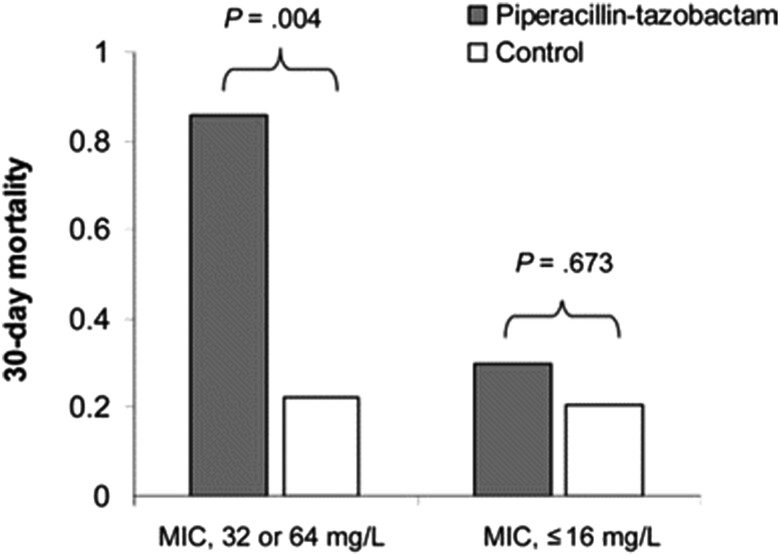
Figure 5.
Thirty-day mortality rates for patients with bacteremia due to P. aeruginosa. Figure reproduced with permission from the Oxford University Press and Tam et al. [74]. In bacteremic patients due to P. aeruginosa with reduced susceptibility to piperacillin/tazobactam (MIC = 32 or 64 mg/L), patients who received piperacillin/tazobactam experienced higher mortality compared with those who received alternative agents. The difference observed was less dramatic if the MIC was ≤16 mg/L. Abbreviation: MIC, minimum inhibitory concentration.
Figure 6.
Clinical cure of patients with ventilator-associated pneumonia due to gram-negative bacteria. Figure reproduced with permission from Lorente et al. [75]. Continuous infusion of piperacillin/tazobactam was associated with higher clinical cure rates, compared with standard-infusion administration. The benefit was most prominent when the pathogen MIC was between 8 and 16 mg/L while controlling for patient comorbidities such as age, chronic obstructive pulmonary disease, and APACHE II scores. Abbreviation: MIC, minimum inhibitory concentration.
These studies have (at least in part) led the Clinical and Laboratory Standards Institute (CLSI) to revise the susceptibility breakpoint in 2012; the CLSI more recently recommended extended-infusion piperacillin/tazobactam dosing in patients with Enterobacterales infections with MICs of 16 mg/L [76].
Expert Commentary
Despite the widespread use of Monte Carlo simulations in antimicrobial development, limitations exist for this methodology. Clinical outcome data from high-quality randomized controlled trials are still required to support the findings of Monte Carlo simulations [54]. For example, Fish and colleagues found that PKPD modeling did not accurately predict clinical or microbiological success in patients with P. aeruginosa pneumonia [77]. Therefore, factors other than PKPD targets may have to be considered when applying simulation results in clinical practice. Debates regarding the modeling tools and exposure targets that best translate preclinical data to clinical practice are ongoing. Several population modeling tools aiming to characterize specific patient populations of different physiologies (eg, neutropenia, nosocomial pneumonia) have been developed; however, assumptions on PK parameter distribution may still not apply to a specific patient [78].
A fundamental limitation of simulations is that results are only as valid as the model assumptions used for the input data [67, 79]. As mentioned previously, Monte Carlo simulations require a robust PK model used to generate PK profiles for a simulated population. For instance, antimicrobial serum concentrations used to formulate a population PK model may not appropriately predict PKPD target attainment in meningitis; using cerebrospinal fluid data as inputs would be more appropriate in this scenario. Also, there is currently no consensus on what sample size is required for a predictive model. Power analysis is typically used to determine the sample size needed in clinical studies to compare an outcome in 2 groups with appropriate statistical power. However, this approach is not directly applicable as the PK model aims to describe the behavior of the drug in the target population (eg, intersubject distribution of PK parameters, etc.). Thus, simulations based on population PK models involving a small sample size may not capture the realistic PK variability of the target population [67].
To improve the validity of results obtained from Monte Carlo simulations, new PK (or outcome) data involving additional subjects may be incorporated sequentially into the original data set [80]. This longitudinal process of merging more observed data to further refine output functions is referred to as stochastic feedback. In general, this is an iterative process that involves continually adding new data as they become available to the previous PK model and validating the simulation findings in order to produce a more informative Monte Carlo analysis. Such processes would potentially overcome the limited robustness of the initial simulation, and more optimal dosing strategies could be identified. Further advances in mathematical modeling are needed to integrate PKPD principles into the clinical setting, with an emphasis on validating simulations with experimental findings.
CONCLUSIONS
Several methodologies used to facilitate the design of optimal antimicrobial dosing in drug development were reviewed. Future advances in the application of more comprehensive modeling tools for dosing regimen design can lead to an era of precision dosing and personalized therapeutics.
Acknowledgments
We thank Sujata M. Bhavnani, PharmD (Institute for Clinical Pharmacodynamics, Inc., Schenectady, NY, USA), for data to reproduce Figure 2.
Financial support. V.H.T. is supported by the National Institutes of Health (R01AI140287-05).
Author contributions. Both authors drafted, revised, and approved the manuscript for submission.
Patient consent. This study does not include factors necessitating patient consent.
Contributor Information
Hubert C Chua, Department of Pharmacy, CHI Baylor St. Luke’s Medical Center, Houston, Texas, USA; Department of Pharmacy Practice and Translational Research, University of Houston College of Pharmacy, Houston, Texas, USA.
Vincent H Tam, Department of Pharmacy Practice and Translational Research, University of Houston College of Pharmacy, Houston, Texas, USA.
References
- 1. Wright DFB, Winter HR, Duffull SB. Understanding the time course of pharmacological effect: a PKPD approach. Br J Clin Pharmacol 2011; 71:815–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Negus SS, Banks ML. Pharmacokinetic-pharmacodynamic (PKPD) analysis with drug discrimination. Curr Top Behav Neurosci 2018; 39:245–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dumas EO, Pollack GM. Opioid tolerance development: a pharmacokinetic/pharmacodynamic perspective. AAPS J 2008; 10:537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hamberg AK, Dahl ML, Barban M, et al. A PK-PD model for predicting the impact of age, CYP2C9, and VKORC1 genotype on individualization of warfarin therapy. Clin Pharmacol Ther 2007; 81:529–38. [DOI] [PubMed] [Google Scholar]
- 5. Daryaee F, Tonge PJ. Pharmacokinetic-pharmacodynamic models that incorporate drug-target binding kinetics. Curr Opin Chem Biol 2019; 50:120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodríguez-Gascón A, Solinís MÁ, Isla A. The role of PK/PD analysis in the development and evaluation of antimicrobials. Pharmaceutics 2021; 13:833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Minichmayr IK, Aranzana-Climent V, Friberg LE. Pharmacokinetic/pharmacodynamic models for time courses of antibiotic effects. Int J Antimicrob Agents 2022; 60:106616. [DOI] [PubMed] [Google Scholar]
- 8. Nielsen EI, Cars O, Friberg LE. Pharmacokinetic/pharmacodynamic (PK/PD) indices of antibiotics predicted by a semimechanistic PKPD model: a step toward model-based dose optimization. Antimicrob Agents Chemother 2011; 55:4619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Onufrak NJ, Forrest A, Gonzalez D. Pharmacokinetic and pharmacodynamic principles of anti-infective dosing. Clin Ther 2016; 38:1930–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scmidt R. Dose-finding studies in clinical drug development. Eur J Clin Pharmacol 1988; 34:15–9. [DOI] [PubMed] [Google Scholar]
- 11. Ursino M, Zohar S, Lentz F, et al. Dose-finding methods for phase I clinical trials using pharmacokinetics in small populations. Biom J 2017; 59:804–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhavnani SM, Hammel JP. Clinical pharmacokinetic-pharmacodynamic analyses: a critical element for developing antibacterial agents. Curr Opin Pharmacol 2017; 36:124–9. [DOI] [PubMed] [Google Scholar]
- 13. Tam VH, Nikolaou M. A novel approach to pharmacodynamic assessment of antimicrobial agents: new insights to dosing regimen design. PLoS Comput Biol 2011; 7:e1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chua HC, Tse A, Smith NM, Mergenhagen KA, Cha R, Tsuji BT. Combatting the rising tide of antimicrobial resistance: pharmacokinetic/pharmacodynamic dosing strategies for maximal precision. Int J Antimicrob Agents 2021; 57:106269. [DOI] [PubMed] [Google Scholar]
- 15. Wicha SG, Märtson A-G, Nielsen EI, et al. From therapeutic drug monitoring to model-informed precision dosing for antibiotics. Clin Pharmacol Ther 2021; 109:928–41. [DOI] [PubMed] [Google Scholar]
- 16. Bergen PJ, Bulitta JB, Forrest A, Tsuji BT, Li J, Nation RL. Pharmacokinetic/pharmacodynamic investigation of colistin against Pseudomonas aeruginosa using an in vitro model. Antimicrob Agents Chemother 2010; 54:3783–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakamura R, Ito-Horiyama T, Takemura M, et al. In vivo pharmacodynamic study of cefiderocol, a novel parenteral siderophore cephalosporin, in murine thigh and lung infection models. Antimicrob Agents Chemother 2019; 63:e02031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zykov IN, Samuelsen O, Jakobsen L, et al. Pharmacokinetics and pharmacodynamics of fosfomycin and its activity against extended-spectrum-β-lactamase-, plasmid-mediated AmpC-, and carbapenemase-producing Escherichia coli in a murine urinary tract infection model. Antimicrob Agents Chemother 2018; 62:e02560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou J, Ledesma KR, Chang KT, Abodakpi H, Gao S, Tam VH. Pharmacokinetics and pharmacodynamics of minocycline against Acinetobacter baumannii in a neutropenic murine pneumonia model. Antimicrob Agents Chemother 2017; 61:e02371-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rao GG, Landersdorfer CB. Antibiotic pharmacokinetic/pharmacodynamic modelling: MIC, pharmacodynamic indices and beyond. Int J Antimicrob Agents 2021; 58:106368. [DOI] [PubMed] [Google Scholar]
- 21. Nielsen EI, Friberg LE. Pharmacokinetic-pharmacodynamic modeling of antibacterial drugs. Pharmacol Rev 2013; 65:1053–90. [DOI] [PubMed] [Google Scholar]
- 22. Tam VH, Schilling AN, Neshat S, Poole K, Melnick DA, Coyle EA. Optimization of meropenem minimum concentration/MIC ratio to suppress in vitro resistance of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49:4920–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tam VH, Chang K-T, Zhou J, et al. Determining β-lactam exposure threshold to suppress resistance development in gram-negative bacteria. J Antimicrob Chemother 2017; 72:1421–8. [DOI] [PubMed] [Google Scholar]
- 24. Wen X, Gehring R, Stallbaumer A, Riviere JE, Volkova VV. Limitations of MIC as sole metric of pharmacodynamic response across the range of antimicrobial susceptibilities within a single bacterial species. Sci Rep 2016; 6:37907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landersdorfer CB, Nation RL. Limitations of antibiotic MIC-based PK-PD metrics: looking back to move forward. Front Pharmacol 2021; 12:770518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol 2019; 17:479–96. [DOI] [PubMed] [Google Scholar]
- 27. Zhang L, Xie H, Wang Y, Wang H, Hu J, Zhang G. Pharmacodynamic parameters of pharmacokinetic/pharmacodynamic (PK/PD) integration models. Front Vet Sci 2022; 9:860472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blondeau JM. New concepts in antimicrobial susceptibility testing: the mutant prevention concentration and mutant selection window approach. Vet Dermatol 2009; 20:383–96. [DOI] [PubMed] [Google Scholar]
- 29. Chauzy A, Gaelzer Silva Torres B, Buyck J. et al. Semimechanistic pharmacodynamic modeling of aztreonam-avibactam combination to understand its antimicrobial activity against multidrug-resistant gram-negative bacteria. CPT Pharmacometrics Syst Pharmacol 2019; 8:815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Smith NM, Lenhard JR, Boissonneault KR, et al. Using machine learning to optimize antibiotic combinations: dosing strategies for meropenem and polymyxin B against carbapenem-resistant Acinetobacter baumannii. Clin Microbiol Infect 2020; 26:1207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drusano GL, Shields RK, Mtchedlidze N, et al. Pharmacodynamics of ceftazidime plus avibactam against KPC-2-bearing isolates of Klebsiella pneumoniae in a hollow fiber infection model. Antimicrob Agents Chemother 2019; 63:e00462-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maseda E, Grau S, Luque S, et al. Population pharmacokinetics/pharmacodynamics of micafungin against Candida species in obese, critically ill, and morbidly obese critically ill patients. Crit Care 2018; 22:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemon SC, Roy J, Clark MA, Friedmann PD, Rakowski W. Classification and regression tree analysis in public health: methodological review and comparison with logistic regression. Ann Behav Med 2003; 26:172–81. [DOI] [PubMed] [Google Scholar]
- 34. Lever J, Krzywinski M, Altman N. Logistic regression. Nat Methods 2016; 13:541–2. [Google Scholar]
- 35. Nelson LM, Bloch DA, Longstreth WT, Shi H. Recursive partitioning for the identification of disease risk subgroups: a case-control study of subarachnoid hemorrhage. J Clin Epidemiol 1998; 51:199–209. [DOI] [PubMed] [Google Scholar]
- 36. Pirkle CM, Wu YY, Zunzunegui MV, Gomez JF. Model-based recursive partitioning to identify risk clusters for metabolic syndrome and its components: findings from the International Mobility in Aging Study. BMJ Open 2018; 8:e018680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lewis RJ. An Introduction to Classification and Regression Tree (CART) Analysis. Citeseer; 2000. [Google Scholar]
- 38. Krzywinski M, Altman N. Classification and regression trees. Nat Methods 2017; 14:757–8. [Google Scholar]
- 39. Aitken SL, Altshuler J, Guervil DJ, et al. Cefepime free minimum concentration to minimum inhibitory concentration (fCmin/MIC) ratio predicts clinical failure in patients with gram-negative bacterial pneumonia. Int J Antimicrob Agents 2015; 45:541–4. [DOI] [PubMed] [Google Scholar]
- 40. Ingram PR, Lye DC, Tambyah PA, Goh WP, Tam VH, Fisher DA. Risk factors for nephrotoxicity associated with continuous vancomycin infusion in outpatient parenteral antibiotic therapy. J Antimicrob Chemother 2008; 62:168–71. [DOI] [PubMed] [Google Scholar]
- 41. Li C, Du X, Kuti Joseph L, Nicolau DP. Clinical pharmacodynamics of meropenem in patients with lower respiratory tract infections. Antimicrob Agents Chemother 2007; 51:1725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kashuba AD, Nafziger AN, Drusano GL, Bertino JS Jr. Optimizing aminoglycoside therapy for nosocomial pneumonia caused by gram-negative bacteria. Antimicrob Agents Chemother 1999; 43:623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kullar R, Davis SL, Levine DP, Rybak MJ. Impact of vancomycin exposure on outcomes in patients with methicillin-resistant Staphylococcus aureus bacteremia: support for consensus guidelines suggested targets. Clin Infect Dis 2011; 52:975–81. [DOI] [PubMed] [Google Scholar]
- 44. Rybak MJ, Le J, Lodise TP, et al. Therapeutic monitoring of vancomycin for serious methicillin-resistant Staphylococcus aureus infections: a revised consensus guideline and review by the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, the Pediatric Infectious Diseases Society, and the Society of Infectious Diseases Pharmacists. Am J Health Syst Pharm 2020; 77:835–64. [DOI] [PubMed] [Google Scholar]
- 45. Zasowski EJ, Murray KP, Trinh TD, et al. Identification of vancomycin exposure-toxicity thresholds in hospitalized patients receiving intravenous vancomycin. Antimicrob Agents Chemother 2018; 62:e01684-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Casapao AM, Lodise TP, Davis SL, et al. Association between vancomycin day 1 exposure profile and outcomes among patients with methicillin-resistant Staphylococcus aureus infective endocarditis. Antimicrob Agents Chemother 2015; 59:2978–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muller R, Mockel M. Logistic regression and CART in the analysis of multimarker studies. Clin Chim Acta 2008; 394:1–6. [DOI] [PubMed] [Google Scholar]
- 48. Evans SR, Rubin D, Follmann D, et al. Desirability of outcome ranking (DOOR) and response adjusted for duration of antibiotic risk (RADAR). Clin Infect Dis 2015; 61:800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Howard-Anderson J, Dai W, Yahav D, et al. A desirability of outcome ranking analysis of a randomized clinical trial comparing seven versus fourteen days of antibiotics for uncomplicated gram-negative bloodstream infection. Open Forum Infect Dis 2022; 9:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Williams DJ, Creech CB, Walter EB, et al. Short- vs standard-course outpatient antibiotic therapy for community-acquired pneumonia in children: the SCOUT-CAP randomized clinical trial. JAMA Pediatr 2022; 176:253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Irurzun-Arana I, Rackauckas C, McDonald TO, Troconiz IF. Beyond deterministic models in drug discovery and development. Trends Pharmacol Sci 2020; 41:882–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor HM, Karlin S. An Introduction to Stochastic Modeling. 3rd ed. Academic Press; 1998. [Google Scholar]
- 53. Trang M, Dudley MN, Bhavnani SM. Use of Monte Carlo simulation and considerations for PK-PD targets to support antibacterial dose selection. Curr Opin Pharmacol 2017; 36:107–13. [DOI] [PubMed] [Google Scholar]
- 54. Bradley JS, Garonzik SM, Forrest A, Bhavnani SM. Pharmacokinetics, pharmacodynamics, and Monte Carlo simulation: selecting the best antimicrobial dose to treat an infection. Pediatr Infect Dis J 2010; 29:1043–6. [DOI] [PubMed] [Google Scholar]
- 55. Kroese DP, Brereton T, Taimre T, Botev ZI. Why the Monte Carlo method is so important today. WIREs Comput Stat 2014; 6:386–92. [Google Scholar]
- 56. Bonate PL. A brief introduction to Monte Carlo simulation. Clin Pharmacokinet 2001; 40:15–22. [DOI] [PubMed] [Google Scholar]
- 57. Tennant SJ, Burgess DR, Rybak JM, Martin CA, Burgess DS. Utilizing Monte Carlo simulations to optimize institutional empiric antipseudomonal therapy. Antibiotics (Basel) 2015; 4:643–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shotwell MS, Nesbitt R, Madonia PN, et al. Pharmacokinetics and pharmacodynamics of extended infusion versus short infusion piperacillin-tazobactam in critically ill patients undergoing CRRT. Clin J Am Soc Nephrol 2016; 11:1377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xiao AJ, Caro L, Popejoy MW, Huntington JA, Kullar R. PK/PD target attainment with ceftolozane/tazobactam using monte carlo simulation in patients with various degrees of renal function, including augmented renal clearance and end-stage renal disease. Infect Dis Ther 2017; 6:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sjovall F, Alobaid AS, Wallis SC, Perner A, Lipman J, Roberts JA. Maximally effective dosing regimens of meropenem in patients with septic shock. J Antimicrob Chemother 2018; 73:191–8. [DOI] [PubMed] [Google Scholar]
- 61. Fratoni AJ, Mah JW, Nicolau DP, Kuti JL. Imipenem/cilastatin/relebactam pharmacokinetics in critically ill patients with augmented renal clearance. J Antimicrob Chemother 2022; 77:2992–9. [DOI] [PubMed] [Google Scholar]
- 62. He CY, Ye PP, Liu B, Song L, van den Anker J, Zhao W. Population pharmacokinetics and dosing optimization of vancomycin in infants, children, and adolescents with augmented renal clearance. Antimicrob Agents Chemother 2021; 65:e0089721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Crass RL, Dunn R, Hong J, Krop LC, Pai MP. Dosing vancomycin in the super obese: less is more. J Antimicrob Chemother. 2018; 73:3081–6. [DOI] [PubMed] [Google Scholar]
- 64. Tam VH, Adams S, LaRocco MT, Gerard LN, Gentry LO, Garey KW. An integrated pharmacoeconomic approach to antimicrobial formulary decision-making. Am J Health Syst Pharm 2006; 63:735–9. [DOI] [PubMed] [Google Scholar]
- 65. Turco NJ, Kane-Gill SL, Hernandez I, Oleksiuk LM, D'Amico F, Pickering AJ. A cost-minimization analysis of dalbavancin compared to conventional therapy for the outpatient treatment of acute bacterial skin and skin-structure infections. Expert Opin Pharmacother 2018; 19:319–25. [DOI] [PubMed] [Google Scholar]
- 66. McComb MN, Collins CD. Comparative cost-effectiveness of alternative empiric antimicrobial treatment options for suspected enterococcal bacteremia. Pharmacotherapy 2014; 34:537–44. [DOI] [PubMed] [Google Scholar]
- 67. Roberts JA, Kirkpatrick CM, Lipman J. Monte Carlo simulations: maximizing antibiotic pharmacokinetic data to optimize clinical practice for critically ill patients. J Antimicrob Chemother 2011; 66:227–31. [DOI] [PubMed] [Google Scholar]
- 68. Perez F, Bonomo Robert A. Precision medicine and mysteries in clinical microbiology: rationalizing epidemiology, genotype, and phenotype to guide therapeutics. Antimicrob Agents Chemother 2020; 64:e02264-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wenzler E, Adeel A, Wu T, et al. Inadequate cerebrospinal fluid concentrations of available salvage agents further impedes the optimal treatment of multidrug-resistant Enterococcus faecium meningitis and bacteremia. Infect Dis Rep 2021; 13:843–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tam VH, Louie A, Lomaestro BM, Drusano GL. Integration of population pharmacokinetics, a pharmacodynamic target, and microbiologic surveillance data to generate a rational empiric dosing strategy for cefepime against Pseudomonas aeruginosa. Pharmacotherapy 2003; 23:291–5. [DOI] [PubMed] [Google Scholar]
- 71. Zasowski E, Bland CM, Tam VH, Lodise TP. Identification of optimal renal dosage adjustments for high-dose extended-infusion cefepime dosing regimens in hospitalized patients. J Antimicrob Chemother 2015; 70:877–81. [DOI] [PubMed] [Google Scholar]
- 72. Lodise TP Jr, Lomaestro B, Drusano GL. Piperacillin-tazobactam for Pseudomonas aeruginosa infection: clinical implications of an extended-infusion dosing strategy. Clin Infect Dis 2007; 44:357–63. [DOI] [PubMed] [Google Scholar]
- 73. Shea KM, Cheatham SC, Smith DW, Wack MF, Sowinski KM, Kays MB. Comparative pharmacodynamics of intermittent and prolonged infusions of piperacillin/tazobactam using Monte Carlo simulations and steady-state pharmacokinetic data from hospitalized patients. Ann Pharmacother 2009; 43:1747–54. [DOI] [PubMed] [Google Scholar]
- 74. Tam VH, Gamez EA, Weston JS, et al. Outcomes of bacteremia due to Pseudomonas aeruginosa with reduced susceptibility to piperacillin-tazobactam: implications on the appropriateness of the resistance breakpoint. Clin Infect Dis 2008; 46:862–7. [DOI] [PubMed] [Google Scholar]
- 75. Lorente L, Jiménez A, Martín MM, Iribarren JL, Jiménez JJ, Mora ML. Clinical cure of ventilator-associated pneumonia treated with piperacillin/tazobactam administered by continuous or intermittent infusion. Int J Antimicrob Agents 2009; 33:464–8. [DOI] [PubMed] [Google Scholar]
- 76. CLSI . Piperacillin-Tazobactam Breakpoints for Enterobacterales. 1st ed. CLSI Rationale Document MR14. Clinical and Laboratory Standards Institute; 2022. [DOI] [PubMed] [Google Scholar]
- 77. Fish DN, Kiser TH. Correlation of pharmacokinetic/pharmacodynamic-derived predictions of antibiotic efficacy with clinical outcomes in severely ill patients with Pseudomonas aeruginosa pneumonia. Pharmacotherapy 2013; 33:1022–34. [DOI] [PubMed] [Google Scholar]
- 78. Drusano GL. Antimicrobial pharmacodynamics: critical interactions of ‘bug and drug.’ Nat Rev Microbiol 2004; 2:289–300. [DOI] [PubMed] [Google Scholar]
- 79. Harrison RL. Introduction to Monte Carlo simulation. AIP Conf Proc 2010; 1204:17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Balice G, Passino C, Bongiorni MG, et al. Daptomycin population pharmacokinetics in patients affected by severe gram-positive infections: an update. Antibiotics (Basel) 2022; 11:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bhavnani SM, Rubino CM, Hammel JP, et al. Pharmacological and patient-specific response determinants in patients with hospital-acquired pneumonia treated with tigecycline. Antimicrob Agents Chemother 2012; 56:1065–72. [DOI] [PMC free article] [PubMed] [Google Scholar]