Abstract
Long QT syndrome (LQTS) is a cardiovascular disease characterized by QT interval prolongation that can lead to sudden cardiac death. Many mutations with heterogeneous mechanisms have been identified in KCNH2, the gene that encodes for hERG (Kv11.1), which lead to onset of LQTS type 2 (LQTS2). In this work, we developed a LQTS2-diseased tissue-on-a-chip model, using 3D coculture of isogenic stem cell–derived cardiomyocytes (CMs) and cardiac fibroblasts (CFs) within an organotypic microfluidic chip technology. Primarily, we created a hiPSC line with R531W mutation in KCNH2 using CRISPR-Cas9 gene-editing technique and characterized the resultant differentiated CMs and CFs. A deficiency in hERG trafficking was identified in KCNH2-edited hiPSC-CMs, revealing a possible mechanism of R531W mutation in LQTS2 pathophysiology. Following creation of a 3D LQTS2 tissue-on-a-chip, the tissues were extensively characterized, through analysis of calcium handling and response to β-agonist. Furthermore, attempted phenotypic rescue via pharmacological intervention of LQTS2 on a chip was investigated.
An organotypic heart on-a-chip modeling long QT syndrome type 2 was created to study effect of R531W mutation in LQTS2 pathology.
INTRODUCTION
Heart disease remains the leading cause of mortality, despite great efforts in cardiovascular research (1). Long QT syndrome (LQTS), a heart disease that affects 1:2000 of healthy births (2), is characterized by prolongation of the QT interval. An elongated QT can lead to tachyarrhythmias, typically triggered by stress, which can lead to sudden cardiac death. Many variants of LQTS exist and are classified by the gene implicated for disease onset, with type 2 (involving mutations in KCNH2) encompassing ~25 to 30% of LQTS cases (3). KCNH2 encodes for hERG (Kv11.1), the pore-forming potassium channel subunit that produces the rapid component of the delayed rectifier repolarizing current (IKr). hERG is a tetrameric channel composed of six transmembrane segments, S1 to S6, and cytoplasmic N- and C-terminal domains (Fig. 1A) (4). The voltage-sensing domain (VSD; S1 to S4) includes a voltage sensor (S4) that is enriched with positively charged amino acids and S1 to S3 containing negatively charged amino acids that aid through acting as counter charges (5), while segments S5 and S6 form the pore domain, which functions in ion selectivity and gating conductance. The S4 region of hERG, the voltage sensor, includes regularly spaced, charged amino acids that function together in response to changes in membrane potential (6). R531 is among these important amino acids within the VSD and has been suggested to have a number of functions, including gating charge movement and inactivation (7). Mutations at R531, specifically R531W, R531Q, and a deletion from R531 to L539, have been clinically identified and implicated in LQTS type 2 (LQTS2) onset (8). Many single-base mutations in hERG have been documented, which lead to LQTS2 (8), with most resulting in missense mutations because of heterologous expression (9), and are found in locations spread throughout hERG (10). The complexity of LQTS2 disease is further exacerbated by the tetrameric nature of the channel, as heterozygous mutations can result in haploinsufficiency, negatively affecting coassembly of the protein (11). Overall, it has been found that pathological mutations in KCNH2 generally reduce IKr; however, the mechanism by which the mutation affects the hERG channel varies. Four phenotypic classes of mutations that affect IKr and lead to LQTS2 exist, which include loss-of-function, gain-of-function, nonfunctional but trafficking-competent, and trafficking-deficient hERG channels (12, 13), or alternatively from a combination of these mechanisms (14, 15). Together, the varying mechanism of mutation in pathophysiology renders pharmacological treatment of LQTS2 extremely complex and highly dependent on a particular mutation. To that end, understanding the mechanism of mutation in LQTS2 is pertinent in efficient disease treatment. Much of the culminating research in LQTS2 in the past has involved studying KCNH2 mutations in heterologous expression systems. However, such systems lack the characteristics, both biologically and physiologically, of cardiomyocytes (CMs) (16–18). The advent of deriving human induced pluripotent stem cells (hiPSCs) from patient cells has resulted in a major thrust in recent in vitro research to elucidate the mechanisms of particular mutations implicated using cells from patients with LQTS2 within two-dimensional (2D) culture assays (19–28). These approaches have provided noteable advances in the understanding of the specific mechanisms of how different mutations result in LQTS2 pathology.
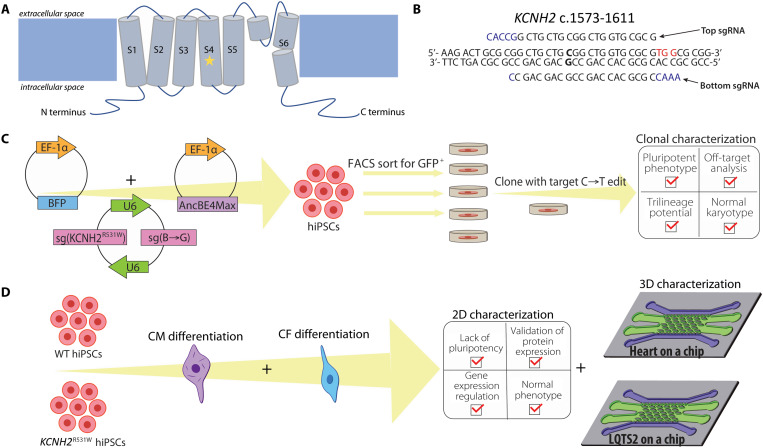
Fig. 1. hERG channel structure, design of generation of hiPSC clones, and design of cardiac lineage differentiation and subsequent characterization of hiPSC clones.
(A) Schematic of the hERG channel structure. The mutation of interest, R531W, resides in the fourth transmembrane domain, as denoted with a star. Overview of the design of (B) sgRNAs and (C) hiPSC transfection using BIG-TREE to introduce R531 mutation and select for edited hiPSCs. (D) Differentiation of resultant hiPSCs into cardiomyocytes (CMs) and cardiac fibroblasts (CFs) and subsequent characterization in 2D, and the formation and characterization of both healthy heart-on-a-chip and LQTS2-on-a-chip in 3D.
However, an inherent obstacle in the use of patient-derived cells is the innate genetic heterogeneities or unknown mutations among patients that may obscure dissection of the specific role of KCNH2 in the onset of LQTS2 (29). The development of CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 (CRISPR-associated protein 9) gene-editing strategies renders the ability to robustly induce specific gene edits in hiPSCs, allowing for well-controlled mechanistic studies of the direct effect of base-level changes on cell and tissue function. Type II CRISPR-Cas is a system discovered in prokaryotes, which provides immunity against foreign viruses through incorporation of genetic material from these pathogens (30). There is a wide family of Cas enzymes with varying functions; however, Cas9 has received much attention because of its ability, guided by the complementarity of guide RNA sequences, to cut double-stranded DNA at specific places in the genome (30). Thus, in recent years, vast amounts of research have been performed to create precise gene edits in various cell types and sources, using CRISPR-Cas9, and related modified versions (31). The Cas9 system relies on the following components: the Cas9 enzyme, noncoding CRISPR RNA (crRNA), and a transactivating crRNA (tracrRNA). The crRNA serves as the targeting RNA sequence, which will complementarily bind to the DNA sequence of the target gene. The tracrRNA serves as the RNA sequence for Cas9 loading. Cas9, in turn, binds to the protospacer adjacent motif (PAM) and cuts the DNA 3 base pairs (bp) upstream. After a double-stranded break (DSB), either nonhomologous DNA end joining or homology-directed repair (HDR) can occur. To facilitate specific gene edits, the naturally occurring nucleotide repair system, HDR, can be used if a sequence that is homologous to the specific site of edit in the DNA is supplied with delivery of Cas9.
The Cas9 enzyme has been modified to optimize its capability in gene-editing. Specifically, the Cas9 nickase (Cas9n) was created through modification of the RuvC1 domain in the Streptococcus pyogenes Cas9 (spCas9) nuclease sequence, creating an amino acid substitution from aspartate to alanine (D10A) (31). The Cas9n simply “nicks” one strand of DNA, while the nuclease creates DSBs. Cas9n requires binding to each strand of the complementary DNA (cDNA) to induce a DSB, thus greatly reducing chances of off-target mutagenesis (32). However, the success of the nickase in targeted editing hinges on efficient HDR, which results in low editing efficiencies. To overcome this limitation, base editors have been fused to Cas9n to introduce the desired base edit, negating the need for HDR (33–37). Nonetheless, efficient selection of the cells that have incorporated the Cas9 machinery and active base editing is pertinent and made particularly more difficult if the editing efficiency specific for the target site is low. Historically, methods involving introduction of antibiotic resistance and/or fluorescent reporter cassettes have been described, which are either permanently or temporarily introduced into the cell (38, 39). An innovative approach, termed BIG-TREE (40–42), has recently been developed that involves delivery of an episomal reporter with the Cas9 system that serves as a marker for cells with active editing. Briefly, the system involves delivery of a blue fluorescent protein (BFP) plasmid along with nickase Cas9D10A fused to a cytidine deaminase. Once inside the cell, the editor will change the existing genomic C’s to T’s within the editing window, as well as the C’s on the BFP plasmid [as guided by the sg(BFP→GFP) sequence], resulting in expression of a green fluorescent protein (GFP) plasmid. The BFP to GFP conversion can be used as a marker for active editing, rendering this technique particularly useful in cell sorting to create isogenic clones.
Using CRISPR-Cas9 or other gene-editing/delivery techniques, different mutations of KCNH2 have been studied in human stem cells (22, 24, 43–45). However, many of these studies relied on 2D culture, limiting the ability to elucidate cell-ECM (extracellular matrix) and tissue-level interactions, which are pertinent to better understanding the clinical pathophysiology of LQTS2. To address limitations of 2D platforms, organ-on-a-chip technologies have been developed, which enable the formation of 3D biomimetic human tissue environments within precisely engineered microfluidic platforms. To that end, we recently described an anisotropic 3D human stem cell–derived heart-on-a-chip platform, validated with three distinct cardiac cell types, namely, rat, human embryonic stem cell–derived, and hiPSC-derived (46). Notably, the cardiac tissue was composed of coculture of these CMs with interstitial, isogenic cardiac fibroblasts (CFs) within a 3D biomimetic hydrogel to better mimic the physiology of the native myocardium. In-depth characterization of our heart-on-a-chip platform revealed an enhanced maturation of the 3D tissue state, through gene expression, cellular-level and tissue-level structure, calcium handling, and drug responsiveness studies (46). Our heart-on-a-chip platform has also shown a great promise in modeling cardiac-specific diseases, such as myocardial infarction (47), due to its physiologically relevant structure and function.
Here, we describe the development of a 3D human cardiac on-chip tissue microfluidic model of LQTS2. Of identified mutations in LQTS2, to our knowledge, there does not yet exist a study that involves the use of gene-edited human stem cells to investigate the mutation within the important VSD, R531W, which has been clinically implicated in the onset of LQTS2 (8), and corresponding tissue pathophysiology. Furthermore, there lacks a complex, 3D in vitro biomimetic model of myocardial tissue that incorporates KCNH2R531W-edited stem cell–derived cardiac cells to truly mimic LQTS2 in a native-like 3D tissue environment. To address these limitations, our approach presented here involves the following: (i) creation of isogenic hiPSC lines with R531W mutation in KCNH2 induced via BIG-TREE gene-editing, (ii) differentiation and subsequent extensive characterization of CMs and CFs from the gene-edited hiPSCs, and (iii) modeling LQTS2 heart tissue in our 3D on-chip microfluidic device, recently validated for the creation of mature cardiac tissue, with KCNH2-edited iPSC-CMs and iPSC-CFs for the mechanistic studies of the role of R531W in LQTS2 pathology. To study the emanation of pathological characteristics that resemble LQTS2 from introduction of a mutation in KCNH2 into hiPSC-CMs and hiPSC-CFs, phenotypes of resultant cardiac tissues within a 3D heart-on-a-chip model were further analyzed. Specifically, to study the role of KCNH2 in emanation of these phenotypes, the contractile properties of the engineered cardiac tissue composed of KCNH2-mutated hiPSC-CMs, both in the presence and absence of “stress,” in the form of catecholamines, were observed. The resultant beating frequency and interbeat interval variability were recorded to assess chronotropic property. Similarly, calcium transients were measured to assess the different calcium handling properties of 3D human cardiac tissues with R531W in KCNH2 to elucidate the potential mechanism of the mutation in arrhythmogenicity in response to catecholamines. Overall, our study demonstrated the novel formation of a 3D LQTS2 human tissue-on-a-chip composed of cocultured, isogenic cardiac cells with R531W mutation. Furthermore, we unveiled the possibility of different pathological mechanism involved in gating of hERG via R531W mutation, therefore highlighting the capabilities of our gene-editing strategy combined with the heart-on-a-chip in the study of 3D personalized cardiac tissues.
RESULTS
Design of sgRNAs
To investigate the pathophysiology of LQTS2 within a precisely controlled environment, we incorporated CRISPR-Cas9 gene-editing techniques to introduce a point mutation in hiPSCs in KCNH2. To introduce a C→T edit at base 1591 to cause R531W in KCNH2 using BIG-TREE gene-editing technique, the single-guide RNA (sgRNA) sequence was to be designed to place the target edit 12 to 18 bp upstream of a PAM that facilitates binding of spCas9, 5′-NGG-3′, where N is any nucleotide. Therefore, we were limited to two different sgRNA sequences at this target that we could clone into the vector for base-editing, with one 13 bp and the other 18 bp upstream from a suitable PAM sequence. On the basis of previously recorded editing efficiencies in relation to PAM location (40), we moved forward with the sgRNA with PAM 13 bp downstream of the edit (Fig. 1B). Within the editing window, there are two C’s that may be affected: (i) the target C 13 bp upstream and (ii) bystander C 16 bp upstream. Editing at the target C would result in the desired change of amino acid from arginine to tryptophan (i.e., R531W), while an edit at the bystander C would be a silent mutation, as the amino acid would remain leucine (Fig. 2A).
Proof-of-principle editing R531W in KCNH2 of HEK293
Once the guide RNAs were designed, we added bases to these oligonucleotides (Fig. 1B) to create 5′ overhangs that are compatible with Bbs I digestion, as well as to allow binding of DNA polymerase III. These sgRNA oligonucleotides were duplexed and then cloned into the sg(BFP→GFP) vector that contains two U6 promoters. After verification of sgRNA incorporation, the plasmid was transformed through Escherichia coli and isolated. The plasmid (Addgene, #138270) that was used to introduce the gene edit included sequences for spCas9n [Cas9(D10A)], the cytidine base editor AncBE4Max, and inhibitors of uracil DNA glycosylases (UGI) to prevent base excision repair that would change the edited U back to a C. This Cas9 plasmid, along with the sgRNA and BFP plasmids, was transfected into human embryonic kidney (HEK) 293 cells to assess editing efficiency at the target site. Forty-eight hours after transfection, flow cytometry was performed to analyze BFP+ and BFP+/GFP+ cells (with 11.9% of cells as BFP+/GFP+). Of the sorted BFP+/GFP+ cells, 6.25 ± 2.06% incorporated the target C→T edit, and 72.5 ± 2.38 incorporated the C→T edit at the bystander C. These reported editing efficiencies were higher than those in BFP+, double-negative (DN), and unsorted (US) cells (table S1), highlighting the efficacy of using GFP expression as the reporter of transfection.
Generation and characterization of LQTS2 hiPSC line with c.C1591T in KCNH2
As the gene editing system we implemented proved capable of introducing the desired edit into HEK293 cells, we moved forward to creating an edited hiPSC line. In particular, hiPSCs were transfected through electroporation and delivery of the BFP plasmid, plasmid with the Cas9n sequence, and plasmid with the sgRNA sequences. We performed two rounds of hiPSC transfection to ensure sufficient production of hiPSC clones. With both transfected hiPSC populations, fluorescence-activated cell sorting (FACS) was performed, including bulk sorting to determine editing efficiencies, as well as single-cell sorting of BFP+/GFP+ cells into 96-well plates to create multiple different hiPSC clones. The clones were grown for 1 to 2 weeks, then passaged into 48-well plates, and routinely expanded until culture in six-well plates. The clones were cryopreserved and the genomic DNA (gDNA) was analyzed at the target site to assess frequency of editing. The editing efficiency of BFP+/GFP+ hiPSCs was 4.5 ± 2.08% for the target C→T edit and 66.25 ± 4.86% for the bystander C. The first round of transfection revealed that 1 of 12 viable clones had the target heterozygous edit (deemed clone 4), while 2 of 17 viable clones had the target heterozygous edit in the second round (deemed clones 2 and 9; fig. S1A). At the bystander C within the editing window, 22 of 29 clones had a homozygous edit, and 7 of 29 clones had a heterozygous edit, revealing the editing efficiency of Big-TREE as 100% for at least one allele at this bystander site. The difference in editing efficiency at the target site compared to the bystander site corresponds with the finding in the literature that base editing efficiency varies greatly depending on the location of edit, due to a variety of confounding factors, including surrounding sequence and target base accessibility (35). Therefore, we describe the production of three hiPSC clones, namely, clones 2, 4, and 9, with a heterozygous C→T edit at the target site, identified in literature as pathogenic (8), and a homozygous edit of C→T at the bystander site within the editing window.
Detailed analysis was performed on the generated lines to assess maintenance of hiPSC characteristics and phenotype. All three generated clonal lines were subjected to pluripotency analysis, through immunofluorescence (IF) and quantitative polymerase chain reaction (qPCR) (fig. S1, B and C), revealing up-regulation of pluripotency markers and thus hiPSC phenotype. In addition, it was observed that all three clones had a lack of expression for CM and fibroblast expression (fig. S1, D and E). Lastly, the three clones were all differentiated into CMs to ensure cardiac differentiation potential with KCNH2R531W. Immunostaining for sarcomeric α-actinin (SAA) (fig. S2A) and gene expression analysis of cardiac genes (fig. S2B) revealed successful CM differentiation of all generated hiPSC clones.
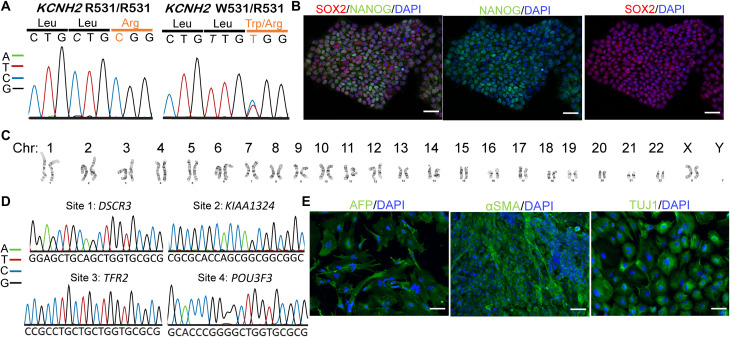
Of the edited KCNH2R531W hiPSC clones, clone 4 was used for all further experiments. We subjected this clone, here referred to as “edited hiPSCs,” to further analysis through karyotyping, trilineage differentiation potential, and potential off-target analyses. The edited hiPSCs had high expression of pluripotency markers (Fig. 2B) and a normal euploid karyotype (Fig. 2C) and spontaneously differentiated into the three layers (Fig. 2E). As CRISPR-Cas9 editing techniques have the possibility to affect genomic locations other than the target, we also sought to assess potential off-target effects in our edited hiPSCs. Using CCTop , the top four possible off-target loci were sequenced, including the following: site 1: DSCR3; site 2: KIAA1324; site 3: TFR2; and site 4: POU3F3. Primers were designed to target the particular locations on the gene where the predicted off-target editing would occur (table S2), and no off-target edits were found (Fig. 2D).
Fig. 2. Creation and validation of the hiPSC line with R531W mutation in KCNH2.
(A) Trace files of the target site in wild-type (WT) (left) and edited (right) hiPSCs, showing a heterozygous edit in c.C1591T, resulting in R531W. (B) Immunostaining of pluripotency markers (where SOX2 is in red and NANOG is in green), (C) karyotype analysis, (D) potential off-target loci sequencing, and (E) trilineage potential of the hiPSC with R531W colony, revealing maintenance of hiPSC characteristics after gene editing. Scale bars, 50 μm (B) and 100 μm (E).
Characterization of isogenic stem cell–derived cardiac cells with KCNH2R531W mutation
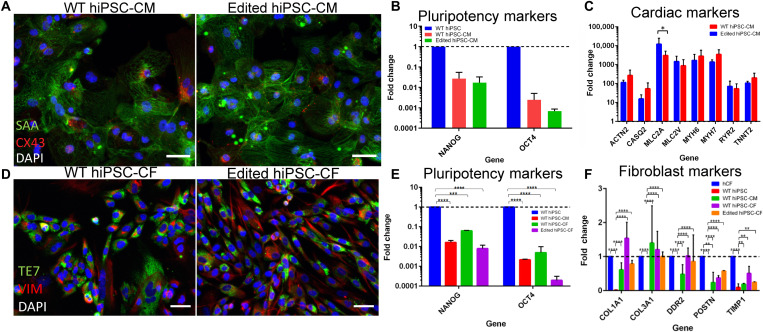
To serve as control to the edited hiPSCs, we used the hiPSCs without the introduced mutation and deemed them as wild-type (WT) hiPSCs. After extensive analysis and confirmation of editing, both sets of hiPSCs were then differentiated into CMs and CFs (Fig. 3). Both sets of CMs and CFs were subjected to characterization, via IF and qPCR, to assess the phenotype and regulation of gene expression. CMs from both WT and KCNH2R531W hiPSCs exhibit expression of striated sarcomeres and localized connexin 43 (CX43) (Fig. 3A), lack expression of pluripotency markers (Fig. 3B), and demonstrate similar up-regulation of an array of CM-specific genes (Fig. 3C), with the only significant difference observed in MLC2A. As all CM-specific genes assayed had great up-regulation, we suggest that the difference in expression observed in MLC2A may be due to variabilities inherent in CM differentiation. Regardless, the edited hiPSCs demonstrated successful CM differentiation.
Fig. 3. Characterization of isogenic cardiac cells differentiated from WT and edited hiPSCs.
(A to C) Characterization of CM differentiation of WT and edited hiPSCs, including (A) immunostaining for sarcomeric α-actinin (SAA, green) and CX43 (red), (B) lack of pluripotency marker expression, and (C) expression of CM-specific genes. (D to F) Characterization of CF differentiation of WT and edited hiPSCs, including (D) immunostaining for TE7 (green) and vimentin (red), (E) lack of pluripotency marker expression, and (F) expression of fibroblast-specific genes. Scale bars, 50 μm.
In addition, CFs were differentiated from both WT and edited hiPSCs, and their phenotypes were assessed. IF of the fibroblast markers, TE7 and vimentin (Fig. 3D), revealed the successful differentiation of CFs from both WT and edited hiPSCs, with an according lack of expression of pluripotency markers (Fig. 3E). In addition, CFs differentiated from both types of hiPSCs demonstrated up-regulation of ECM markers, especially COL1A1 and COL3A1 (Fig. 3F). We also used commercially available human cardiac fibroblasts (hCFs) (Lonza) as the positive control and found no significant differences in gene expression of ECM markers of CFs differentiated between WT and edited hiPSCs.
Electrophysiology assessment of KCNH2R531W in stem cell–derived CMs
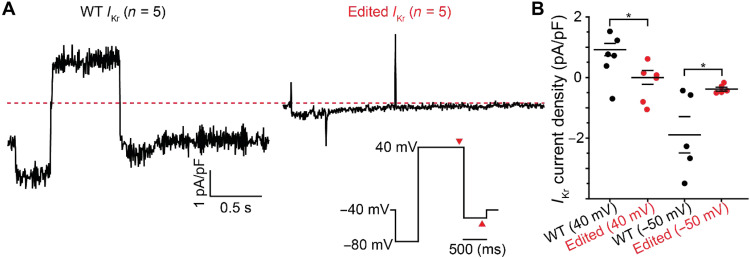
To investigate the impact of the R531W mutation on IKr currents, whole-cell patch-clamp electrophysiology experiments were conducted using ion channel blockers JNJ-303 and nifedipine to isolate IKr currents. Individual WT and edited hiPSC-CM currents were evoked by voltage-clamp measurements. After which, a particular cell was exposed to blocker E-4031 to assess the magnitude of IKr (Fig. 4A and fig. S3). Edited hiPSC-CM IKr current density was found to be significantly decreased (Fig. 4B), indicative that the impact of R531W mutation is detrimental to hERG function. While these data show a hERG-based IKr deficiency in the edited hiPSC-CMs, it requires further exploration on whether this was a result of trafficking or loss of function.
Fig. 4. Whole-cell patch-clamp experiments show decreased IKr current in edited hiPSC-derived CMs.
(A) Average whole-cell IKr current traces for WT (left) and edited (right) hiPSC-CMs show diminished IKr in edited cells. IKr is calculated as the subtraction of IK current by IK + E-4031 current. The voltage protocol is inset. (B) IKr current density measured at steady-state time points at 40 and −50 mV (as marked by red arrowheads in voltage inset). *P < 0.05; specifically at 40 mV, P = 0.017 and −50 mV, P = 0.036.
R531W and trafficking of hERG in hiPSC-derived CMs
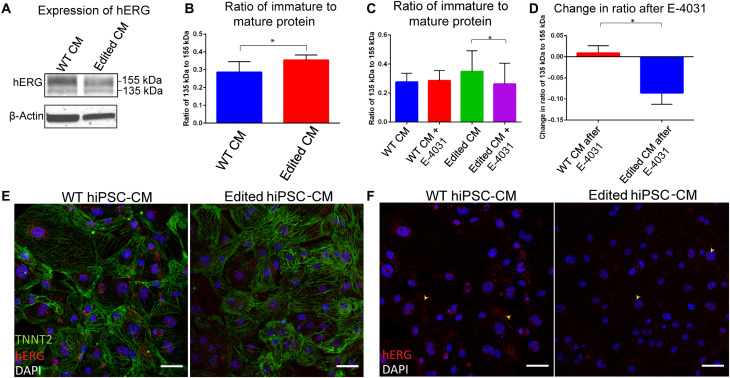
To study the effect of R531W mutation on hERG trafficking in hiPSC-derived CMs, we performed Western blotting (WB) for hERG1a on both WT and edited hiPSC-CMs. In normal hERG channel synthesis, the immature protein is synthesized and undergoes core glycosylation in the endoplasmic reticulum (ER), resulting in a weight of 135 kDa. The immature protein is then transported to the Golgi apparatus, and complex glycosylation occurs. The mature hERG channel, with a final subunit weight of 155 kDa, is then inserted into the plasma membrane (12). When performing WB for hERG channels using an anti-hERG antibody, two bands occur at weights 135 and 155 kDa, revealing levels of immature and mature protein, respectively.
WB has been used to identify protein trafficking defects, as evidenced by increased expression of 135 kDa compared to 155 kDa hERG, which occurs as a result of a mutation in KCNH2 (11, 12, 14, 15, 22, 23, 43, 48, 49). In our WB data, consistent to previous studies, we also noted a significantly increased expression ratio of 135 kDa:155 kDa of hERG in the edited CMs compared to WT CMs (Fig. 5, A and B), suggesting that the mutation affects protein trafficking. This finding was further validated by the staining of cardiac troponin T (TNNT2) and hERG of WT and edited CMs (Fig. 5, E and F), as more hERG is identified in WT CMs outside of the nucleus in the cytoplasm, while there is apparently higher expression of hERG near the nucleus of edited CMs. To further validate the hypothesis of trafficking dysregulation due to the mutation, we attempted to rescue levels of mature protein by exposing the cells to the hERG channel blocker E-4031. Exposure of hERG trafficking–deficient cells to E-4031 for 24 hours has been shown to rescue protein levels via probing WB of hERG (11), as hERG channel blocking agents have been proposed to act as chaperones to promote proper protein folding to enable trafficking and insertion into the plasma membrane (50). In control conditions, WB analysis revealed no effect on WT CMs after exposure to E-4031 (Fig. 5C). However, edited CMs exhibited a significantly decreased ratio of immature to mature protein after E-4031 exposure, compared to a baseline level of edited CMs (Fig. 5C). Furthermore, the change in ratio of immature to mature protein was quantified by subtraction of the ratio after E-4031 exposure by the baseline ratio, revealing a significantly decreased change in edited CMs (Fig. 5D). The increased expression of mature hERG that occurs after exposure to E-4031 in edited CMs further underlies the potential role of defective trafficking that arises from R531W mutation.
Fig. 5. Defective trafficking of hERG in CMs differentiated from hiPSCs with R531W mutation.
(A) Western blot for hERG1a and (B) densitometry analysis of 135 to 155 kDa bands of WT and edited hiPSC-CMs. (C) Densitometry analysis of 135 to 155 kDa bands of WT and edited hiPSC-CMs after 24 hours of exposure to E-4031, and (D) the change in ratio of 135 to 155 kDa hERG after exposure to E-4031. (E) Immunostaining of TNNT2 (green), hERG (red), and 4′,6-diamidino-2-phenylindole (DAPI) (blue), and (F) just hERG (red) and DAPI (blue) in WT and edited hiPSC-CMs. Scale bars, 50 μm. *P < 0.05.
Development of LQTS2 tissue-on-a-chip within a microfluidic platform
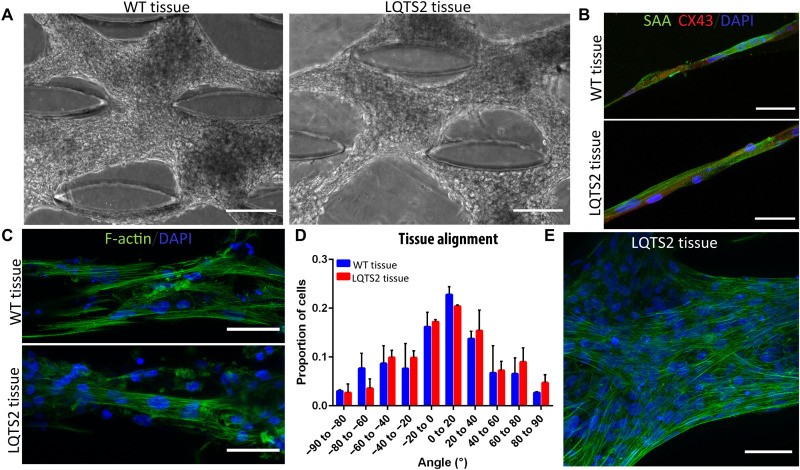
To model LQTS2-on-a-chip, we used our well-characterized and validated microfluidic heart-on-a-chip as a platform, demonstrated to create anisotropic, 3D, mature cocultured cardiac tissues (46). To our knowledge, this is the first study demonstrating the development of human iPSC-derived LQTS2 tissue within a microfluidic chip to characterize the pathological nature of R531W in KCNH2. Particularly,, hiPSC-CMs and hiPSC-CFs from WT and LQTS2 were encapsulated within a collagen-based hydrogel and injected into the microfluidic chip to form a healthy cardiac tissue- and LQTS2-on-a-chip, respectively. The tissues were cultured within the devices for a total of 14 days, forming aligned and condensed tissues around the innate microposts within the device (Fig. 6A). Analysis of alignment and cardiac markers of the tissues revealed similar tissue formation within the devices (Fig. 6, B to E). Specifically, both WT and LQTS2 tissues were present with elongated cells with striated sarcomeres and abundant CX43 staining (Fig. 6B), forming highly anisotropic tissues around the microposts within the devices (Fig. 6, C to E).
Fig. 6. Development of LQTS2 onachip.
Phase contrast images of tissue formation within microfluidic chips after 14 days of culture for isogenic (A) WT and LQTS2 tissues. Scale bars, 100 μm. (B and C) Immunostaining of WT (top) and LQTS2 (bottom) tissues formed in microfluidic chip after 14 days with (B) SAA (green), CX43 (red), and DAPI (blue) and (C) F-actin (green) and DAPI (blue). Scale bars, 50 μm. (D) Alignment analysis of nuclei within tissues at day 14. (E) Enlarged image of highly aligned (F-actin, green; DAPI, blue) LQTS2 tissue within the chip. Scale bar, 50 μm.
Assessment of calcium handling in WT and LQTS2 tissues on-a-chip
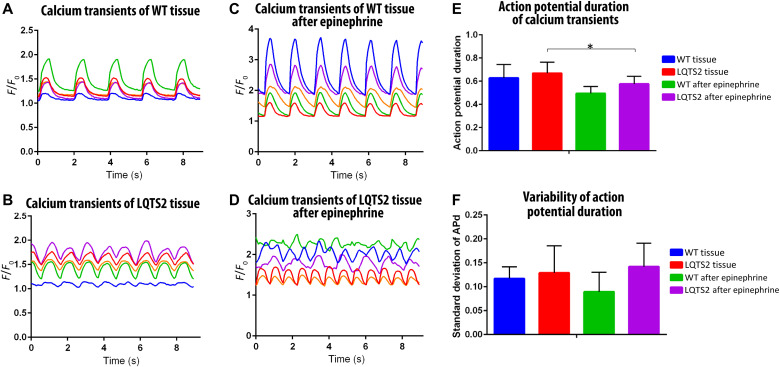
Within cardiac tissues, IKr alters action potential duration (APd) by regulating the rate of repolarization. To assess the role of hERG-based IKr deficiency caused by R531W mutation on tissue functionality and contractility, calcium transients of 3D tissues-on-a-chip at baseline and after dosing with a β-agonist, a common agent used in clinical provocation testing to aid in identification of LQTS patients (51), to probe for possible pathological response were investigated (Fig. 7). Representative traces of WT tissues before and after epinephrine and LQTS2 tissues before and after epinephrine can be seen in Fig. 7 (A to D). To begin, the APd of each calcium transient peak was determined from each of the five different regions of interest (ROIs) within each acquired video. The average APd from each ROI was then averaged with the other ROIs to form the variable of APd (Fig. 7E). The SD of the APd within each ROI was averaged with the other ROIs to form variability of APd (Fig. 7F; see the Supplementary Materials for the generated Matlab code). As analyzed from the calcium transients, the APd trends higher for LQTS2 tissues compared to WT tissues, both in comparison to baseline tissues and tissues exposed to epinephrine, albeit not to a significant extent. After treatment with epinephrine, the LQTS2 tissues exhibited a significantly decreased APd (Fig. 7E). Furthermore, the APd variability of WT tissues decreased after exposure to epinephrine. However, the variability of the APd trended higher for LQTS2 tissues compared to WT tissues, both at baseline and after epinephrine stimulation (Fig. 7F). These assessments further highlighted the potential effect of R531W mutation on aberrant calcium handling.
Fig. 7. Assessment of calcium transients of LQTS2 on-a-chip.
(A to D) Representative calcium transients of five different ROIs in one video of WT tissue before (A) and after (C) epinephrine and LQTS2 tissue before (B) and after (D) epinephrine. (E and F) Analysis of calcium transients from n = 4 biological experiments, showing (E) action potential duration (APd) and (F) variability of action potential. *P < 0.05.
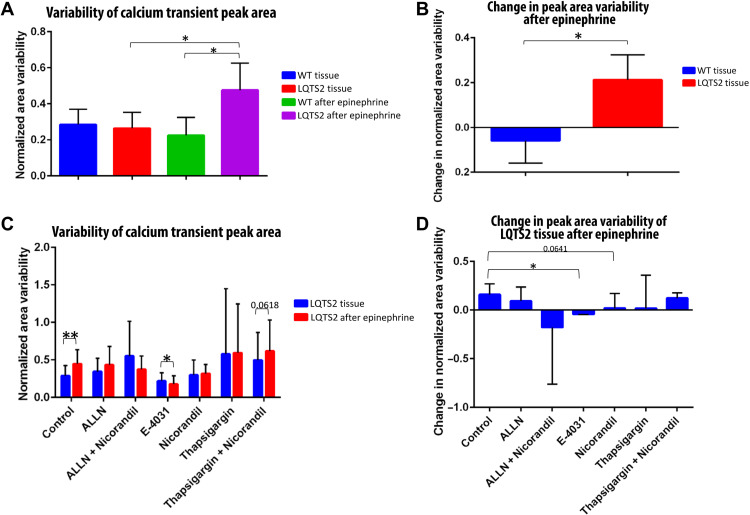
To better understand the functionality of WT and LQTS2 tissues, we analyzed the average area of the calcium transient peaks (i.e., APd multiplied by peak height). The SD of the peak area was determined for each ROI and averaged among each ROI in one video. Then, the SD was normalized to average peak height to produce the variable of “variability of calcium transient peak area” (Fig. 8). Notably, our analysis revealed a significantly increased peak area variability in LQTS2 tissues after exposure to epinephrine, compared to LQTS2 tissues at baseline. Furthermore, the variability of peak area was significantly higher in the LQTS2 tissues after epinephrine compared to WT tissues after epinephrine (Fig. 8A). We further calculated the effect of epinephrine by subtracting the peak area variability of the tissues after epinephrine by the baseline peak area variability. The change in variability of area in LQTS2 tissues is significantly higher than the change in area variability of WT tissues due to epinephrine (Fig. 8B), underlying the detrimental effect on calcium handling of R531W mutation that is exacerbated by β-agonist stimulation.
Fig. 8. Pharmaceutical rescue of LQTS2 on-a-chip.
(A and C) Variability of calcium transient peak area in (A) WT and LQTS2 tissues and in (C) LQTS2 tissues exposed to various pharmaceuticals. (B and D) Change in peak area variability after epinephrine exposure of (B) WT and LQTS2 tissues and (D) LQTS2 tissues exposed to various pharmaceuticals. *P < 0.05 and **P < 0.01.
Probing the rescue of LQTS2 tissues-on-a-chip with pharmaceuticals
To further probe the mechanism of R531W mutation in KCNH2 in mature hiPSC-derived cardiac tissues, we tested various pharmaceuticals, with different mechanisms of action, on the potential rescue of LQTS2 phenotype within the LQTS2 tissues -on-a-chip. As previously mentioned, E-4031 is a hERG channel blocker that has been identified to rescue the trafficking deficiency from analysis of WB in edited CMs. The mechanism by which E-4031 affects protein trafficking is thought to be through reduced association of ER chaperones (such as calnexin) with the immature protein, which supports more efficient processing and trafficking of hERG (50). However, the process of normal hERG trafficking is complex and involves a variety of other processes to function normally. Specifically, in other trafficking-deficient hERG channels [such as A561V (43, 52) and IVS9-28A/G (23)], ALLN, a proteasome inhibitor, has been found to aid in the rescue of hERG trafficking because of mutation in hiPSC-CMs. If rescue with ALLN is demonstrated, then there is evidence that these particular mutations result in increased activity of proteasomes that act to degrade misfolded proteins, supporting the conclusion that the mutation disrupts protein trafficking to the plasma membrane. Another pharmaceutical, thapsigargin, has been used to probe the role of KCNH2 mutations in hERG trafficking (49). Thapsigargin is a sarcoplasmic/ER Ca2+–adenosine triphosphatase (SERCA) inhibitor, which alters levels of Ca2+ in both the cytoplasm and the ER. Changes in Ca2+ levels then affect activity levels of calcium-dependent molecular chaperones in the ER, which, in turn, may affect protein folding. Therefore, KCNH2 mutations [i.e., F805C (49)] that prove responsive to thapsigargin are suggested to alter hERG trafficking through protein misfolding.
In addition to affecting KCNH2 function through trafficking deficiencies, mutations can also result in dysfunctional hERG channels that affect gating properties. Therefore, we additionally tested nicorandil, a IKATP opener, which has been previously demonstrated to normalize prolonged repolarization in LQTS2 hiPSC-CMs (25). Specifically, nicorandil both activates potassium channels to enhance K+ efflux and inhibits voltage-gated Ca2+ channels (53). Nicorandil functional rescue is considered to be consistent with loss-of-function LQTS2 CM mechanism of action, as opposed to trafficking. Given that many hERG mutations manifest as a combination of trafficking and loss-of-function deficiencies, we tested combinatorial pharmaceutical treatment to assess whether R531W mutation affects more than one mechanism that leads to reduced IKr. We tested a combination of ALLN and nicorandil, as well as thapsigargin and nicorandil, to target different processes involved in trafficking combined with hERG channel functionality.
After exposure of LQTS2 tissues to various pharmaceuticals, the calcium transients at baseline and after exposure to epinephrine were recorded. The variability of the calcium transient peak area was analyzed, demonstrating that, in control conditions, the area variability of LQTS2 tissues significantly increased after stimulation with β-agonist (Fig. 8C), as previously identified in Fig. 8A. Similarly, there is a near-significant increase in peak area variability (P = 0.0618) in LQTS2 tissues exposed to both thapsigargin and nicorandil. However, exposure to E-4031 caused a significant reduction in peak area variability in LQTS2 tissues after exposure to epinephrine, demonstrating the potential of E-4031 to rescue R531W mutation. Furthermore, the change in peak area variability due to epinephrine was calculated, and a significant difference was identified between the control LQTS2 tissues, showing increased variability, and tissues exposed to E-4031, with decreased variability after epinephrine (Fig. 8D). There was a near-significant difference (P = 0.0641) in LQTS2 tissues exposed to nicorandil compared to control, revealing that there might be an added mechanism of R531W mutation on hERG channel functionality. Overall, these pharmaceutical treatment studies demonstrate the possibility of partial rescue for patients with R531W mutation via E-4031 administration, in addition to the added potential effect of R531W on the loss of function of hERG, evidenced by near rescue of LQTS2-on-a-chip tissues with nicorandil.
DISCUSSION
LQTS2 is a highly complex, and occasionally lethal, cardiac disease (3, 54). Hence, much research has been completed on the implicated mutations in KCNH2, which encodes hERG (Kv11.1), that result in LQTS2 to shed light on the best course of treatment for the disease (10). Pathological mutations have been identified throughout the hERG protein, with differing mechanisms as to how the presence of the mutation leads to a decrease in IKr (12–14). Thorough understanding of both the identity and particular mechanism of an individual’s mutation is essential for effective pharmacological treatment in LQTS2. To add to current understandings of LQTS2, we here demonstrated the study of the missense mutation R531W in KCNH2, introduced into hiPSCs, and how it leads to LQTS2 pathophysiology.
In this work, we designed a gene-editing system, using BIG-TREE, to introduce the mutation c.C1591T both in HEK293 cells, as validation, and in hiPSCs. Extensive characterization of the generated clones of edited hiPSCs revealed maintenance of hiPSC pluripotency, phenotype, and differentiation potential, as well as a lack of off-target gene-editing effects. With the validated, edited hiPSCs, we presented successful differentiation of isogenic cardiac cells (both CMs and CFs) and further characterized the role of R531W mutation in these cardiac cells. Specifically, we identified a significant reduction in IKr in CMs with the mutation compared to WT CMs, confirming that the cause of the pathophysiology is R531W mutation dependent.
Mutations in KCNH2 can lead to loss-of-function, gain-of-function, nonfunctional, or trafficking-deficient hERG channels (12, 13). In recent works, a remarkable amount of KCNH2 mutations has been found to affect trafficking of hERG to the plasma membrane, leading to sequestering of functional and nonfunctional channels in the ER (11, 12, 15, 22, 23, 43, 48, 49). However, most of these studies have been completed in 2D and/or in heterologous systems. In this work, we present the first, to our knowledge, study of R531W mutation in KCNH2 in CMs differentiated from hiPSCs, in both 2D and 3D. We found that R531W mutation in KCNH2 in hiPSC-CMs led to significantly more immature hERG (135 kDa) in a ratio to mature hERG (155 kDa) than WT hiPSC-CMs, suggesting that the mutation causes sequestration of the immature hERG channels in the ER. In addition, we were able to rescue the trafficking deficiency identified by WB analysis by exposing the edited CMs to the hERG channel blocker, E-4031.
We expanded our study into the formation of 3D isogenic cardiac tissues within an enhanced microfluidic device to form both WT and LQTS2 tissues-on-a-chip. Successful formation of the isogenic, cocultured tissues within an aligned formation was revealed through cardiac marker and alignment analysis. To that end, this study demonstrated the formation, and extensive study, of an LQTS2 disease-on-a-chip model, composed of cocultured cardiac cells differentiated from hiPSCs with introduced R531W mutation. Specifically, the functionality of the resultant tissues was probed via calcium transient analysis, revealing the significantly increased variability in peak area in LQTS2 tissues compared to WT tissues when exposed to β-agonist. This trend in aberrant calcium handling due to epinephrine stimulation was reversed when the LQTS2 tissues were exposed to E-4031, further highlighting the effect of R531W mutation on hERG trafficking. We also identified a near-significant decrease in change in peak area variability in response to epinephrine in LQTS2 tissues exposed to nicorandil. Speculatively, we interpret this finding to suggest a possible additional effect of R531W mutation on hERG gating; however, further studies, potentially including techniques to measure calcium transient amplitude in addition to duration (55), are needed to understand such mechanism.
A previous study on R531W mutation that focused on heterologous expression of hERG in HEK293 cells found that R531W did not appear to affect the trafficking (56). However, such studies arise from using transfection with cytomegalovirus and similar promoters (or highly expressing stable cell lines), which results in much higher expression of the channel that, in turn, leads to the likely result that more channels may slip through the ER quality control to the membrane. Therefore, findings regarding LQTS2 mutations have been found to differ when expressed in biologically relevant cells at physiologic levels, such as in hiPSC-CMs, over studies previously done in heterologous systems, as such systems were hindered by the lack of accurate recapitulation of CM biology (15). For example, Paulussen et al. (15) discovered that T65P in KCNH2 not only affects channel activation, as previously found in heterologous systems, but also additionally plays a critical role in trafficking of hERG channels. Additional studies into heterologous systems have demonstrated the important role of the arginine residue at position 531 in normal gating of hERG (7, 56–60), as introducing mutations at R531 induced the greatest perturbation in channel opening than other mutations in the S4 domain. Our findings suggest that the role of R531 is crucial in hERG function, in trafficking to the plasma membrane in addition to possible effects on channel function, further underlying the disconnect between heterologous expression systems and more physiologically relevant models.
In summary, we demonstrated the development of isogenic 3D cardiac tissues with R531W mutation in KCNH2 within our established microfluidic device. The resultant anisotropic tissues have great potential in the study of LQTS2 on-a-chip.
Altogether, we described creation of an hiPSC line with R531W mutation in KCNH2 with the BIG-TREE editing technique. In addition, we thoroughly validated the hiPSC line to confirm maintenance of hiPSC characteristics, including pluripotency and trilineage differentiation potential, as well as the lack of off-target editing. Furthermore, we differentiated the edited hiPSC line into both CMs and CFs and validated the phenotypes through gene expression and IF of the differentiated cells. In addition, we investigated the effect of the base mutation on hERG channel function and localization. hERG localization was analyzed through immunofluorescent staining for hERG and WB for hERG protein expression. The function of the hERG channel was analyzed through patch-clamp techniques to measure IKr. Furthermore, the edited hiPSC-CMs and CFs were encapsulated in a 3D collagen-based hydrogel and incorporated in our innovative microfluidic platform to create both healthy and LQTS2 heart-on-a-chip tissues. The functionality of the 3D tissues was assessed, and potential rescue of the diseased phenotype was analyzed through administration of pharmaceuticals with differing mechanisms of action to further probe the biological significance of R531W in KCNH2 in LQTS2 onset. In summary, we demonstrated the development of isogenic 3D cardiac tissues with R531W mutation in KCNH2 within our established microfluidic device, with great potential in the study of LQTS2-on-a-chip.
MATERIALS AND METHODS
Plasmid construction
The sgRNAs were synthesized as oligonucleotides, and 5′ phosphates were added by incubating 1 μg of oligonucleotide in 1× T4 DNA ligase buffer (New England Biolabs) and 10 U of T4 polynucleotide kinase (New England Biolabs), incubating at 37°C overnight. The oligonucleotides were duplexed by heating the reactions to 90°C within aluminum blocks for 5 min, and then the blocks and the reaction were allowed to reach room temperature (RT) gradually. Then, the sgRNA backbone was digested, using Bbs I–HF in cutsmart buffer at 37°C for 1.5 hours, and then purified with a PCR cleanup kit (Qiagen), creating a solution (40 ng/μl) of purified, digested sgRNA vector. The vector was dephosphorylated using 1 U of rSAP in cutsmart buffer at 37°C overnight and then purified with a PCR cleanup kit (Qiagen), creating a purified, dephosphorylated plasmid DNA solution at 20 to 40 ng/μl. This prepared plasmid sgRNA vector was ligated with the oligonucleotide duplex, using T4 DNA ligase, through incubation at RT for an hour. Alongside, a no insert ligation and vector-only controls were created.
The ligated plasmid sgRNA vector was then transformed by mixing 5 μl of the mixture with 50 μl of chemically competent E. coli cells (STK300-10, GeneCopoeia) on ice. After 30 min of incubation on ice, the mixture was heat-shocked at 42°C for 30 s and then transferred back to ice for 5 min. LB medium, without antibiotic, was added, and the samples were grown in a 37°C incubator, shaking at 220 rpm, for 1 hour. The outgrowth solution was plated on LB agar/ampicillin plates overnight at 37°C. Multiple colonies were picked the following morning and grown in 4 ml of LB with ampicillin in the shaking incubator overnight. The plasmid DNA was extracted using a Miniprep kit and then sequenced, via Sanger sequencing at Genewiz, to ensure intact U6 promoters and sgRNA sequence. After sequence confirmation, the plasmid DNA was retransformed in the competent cells via incubation of 200 μl of the starter culture in 200 ml of LB with ampicillin overnight. The plasmid DNA was then extracted using a Maxiprep kit and then concentrated using alcohol precipitation with sodium acetate and an ultracentrifuge at 18,000 rpm for 33 min at 4°C. The sequence was once again verified through Sanger sequencing.
HEK transfection
HEK293 cells (LT008, GeneCopoeia) were seeded in 24-well plates at ~185,000 cells per well. The next day, the cells were transfected via addition of 500 ng total of DNA per well (300 ng of Cas9 plasmid, 100 ng of sgRNA plasmid, and 100 ng of BFP plasmid), 1 μl of P3000, 0.75 μl of Lipofectamine 3000, and 50 μl of Opti-MEM. After 24 hours, the medium was changed. Transfected HEK293 cells were imaged for BFP and GFP expression and subjected to flow cytometry 48 hours after transfection.
hiPSC transfection
hiPSCs [IMR90-4, WiCell (61)] were maintained in mTeSR and passaged with 0.5 mM EDTA at least three times after thawing before transfection. To prepare cells for transfection, the hiPSCs were seeded in Matrigel-coated 12-well plates and maintained in mTeSR1 until 70 to 80% confluent. The transfection master mix consisted of 100 μl of Opti-MEM, 4 μl of LipoStem, 300 ng of BFP plasmid, 300 ng of guide plasmid, and 900 ng of base editor plasmid per well. After medium change, the master mix was added dropwise to each well. The medium was changed the next day, and the hiPSCs were subjected to flow cytometry 48 hours after transfection.
Fluorescence-activated cell sorting
Approximately 48 hours after transfection, HEK cells or hiPSCs were bulk sorted via the BD FACSAria Ilu-Cell Sorter, with the help from the ASU Genomics Core, into DN, US, BFP+, and BFP+/GFP+ populations, directly into prepared Phire master mix. Specifically, 1000 cells were sorted into a master mix of 25 μl of Phire, 5 μl of forward and reverse primers at 10 μM, and 15 μl of NF H2O. Immediately after sorting, PCR was performed using the on-target primers listed in table S1, and products were then verified on 1% agarose gels. The products were subjected to column purification using the QIAquick PCR Purification Kit (Qiagen) before sending to Genewiz for sequencing to determine editing efficiencies.
hiPSC base editing and clonal isolation
To determine editing efficiency, the cells sorted into Phire master mix (n = 4 each for DN, BFP+, BFP+/GFP+, and US) were subjected to PCR using primers flanking the edit on KCNH2 (table S1) and then sequenced via Genewiz. The editing efficiencies were determined using MoriarityLab-EditR program. Single-cell sorting was also performed on the BD FACSAria Ilu-Cell Sorter to create hiPSC clones. Specifically, BFP+/GFP+ cells were sorted as single cells into 96-well plates into a solution of CloneR (STEMCELL Technologies), mTeSR1, and 5 μM Y-27632 (STEMCELL Technologies). The medium was changed the next day with mTeSR1, and then the medium was changed every other day until visible colonies were observed. Viable colonies, once ~50% confluent, were passaged with Accutase into one well of a 24-well plate. The cells were further expanded until culture within a six-well plate, and then the clones were subjected to genotyping analysis.
Sequence analysis at on- and off-targets
To perform genotyping, gDNA was extracted from frozen cell pellets using the DNeasy Blood and Tissue Kit (Qiagen). PCR was performed on the gDNA using Phusion High-Fidelity DNA Polymerase (New England Biolabs) for both on-target and off-target loci, with primers listed in file S1. PCR products were column-purified using the QIAquick PCR Purification Kit (Qiagen) and then sent for sequencing to Genewiz. To determine possible off-target loci, CCTop was used with input parameters set for spCas9 against human genome reference sequence hg38 (62), and the top four loci were sequenced. Similar to described above, EditR was used to quantify on- and off-target editing.
Karyotype analysis
To analyze the karyotype of the cell lines, cytogenetic analysis was performed (via Cell Line Genetics) using standard protocols for G-banding on 20 metaphase cells.
Trilineage differentiation of edited hiPSCs
Trilineage differentiation was performed on hiPSCs as embryoid bodies (EBs) to ensure maintenance of pluripotency through the editing process. To form the EBs, hiPSCs were seeded at 1.5 to 2e6 cells per well of six-well low adhesion plate and placed on orbital shaker at 100 rpm within a 37°C incubator. The hiPSCs were grown for 2 days on the orbital shaker, with the mTeSR1 medium changed each day. On day 2, the medium was changed to differentiation medium [Dulbecco’s modified Eagle’s medium (DMEM)/F12, 20% fetal bovine serum (FBS), and 1% penicillin/streptomycin], and the cells were maintained on the orbital shaker for four more days. On day 6, the EBs were plated on Matrigel-coated wells and grown for 20 more days, with medium changes using the differentiation medium every 1 to 2 days. The plated EBs were fixed on D26 and subjected to IF for trilineage markers to assess pluripotency potential.
hiPSC CM differentiation
hiPSCs were differentiated into CMs using our previously defined method (46). Briefly, hiPSCs were grown in mTeSR1 until 75 to 85% confluent, and then CM differentiation was initiated via Wnt inhibition with 7 to 10 μM CHIR99021 in RPMI + B27 − insulin. Wnt activation was initiated on day 3, with supplementation of 5 μM IWP2 in RPMI + B27 − insulin. Cardiac maturation was supported from days 7 to 13 with RPMI + B27 + insulin. On days 13 and 16, the wells were washed with 1× phosphate-buffered saline (PBS), and the CMs were purified with RPMI no glucose + B27 + insulin, supplemented with 4 mM sodium lactate. After recovery on day 19 with RPMI + B27 + insulin, the purified CMs were replated on Matrigel-coated plates to remove dead cells and debris. The CMs were used for all characterizations from day 25 onward.
hiPSC CF differentiation
hiPSCs were differentiated into CFs based on a recently developed protocol from Zhang et al. (63). hiPSCs were cultured in mTeSR until 75 to 85% confluence, and then differentiation was initiated via Wnt inhibition with 7 to 10 μM CHIR99021 in RPMI + B27 − insulin. On day 1 of differentiation, the medium was changed to RPMI + B27 − insulin. On day 2 of differentiation, the medium was changed to CF basal medium, supplemented with basic fibroblast growth factor (75 ng/μl). The medium was exchanged every other day until day 20, when the hiPSC-CFs were passaged, considered as passage 0 (P0), with 0.05% trypsin-EDTA. From P0 onward, the hiPSC-CFs were cultured in Fibroblast Growth Medium 3 (FGM3), routinely passaged with 0.05% trypsin-EDTA, and cryopreserved in 70% FGM3, 20% FBS, and 10% dimethyl sulfoxide. hiPSC-CFs were analyzed via qPCR to verify lack of expression of pluripotent markers and CM markers, as well as up-regulation of fibroblast-specific markers. hiPSC-CFs were also stained for vimentin and TE7.
3D cardiac tissue formation within a microfluidic chip
The heart-on-a-chip microfluidic devices were fabricated as previously described (46, 47, 64). Briefly, silicon wafers were coated with a thin 200 μm layer of SU-8 2075, and a transparent design of the device channel was transferred to the SU8 via ultraviolet exposure and subsequent cross-linking. The silicon wafer was used as the master template in soft lithography to create positive replicas of the microfluidic device channels, composed of polydimethylsiloxane (PDMS). These channels were bonded to coverslips via plasma bonding, and the devices were sterilized via autoclave. Isogenic cocultured cardiac tissues were formed at a 4:1 ratio of hiPSC-CMs:hiPSC-CFs and encapsulated within a collagen (2 mg/ml):20% Matrigel hydrogel, as previously described (46, 47, 64). The tissues were cultured within the microfluidic devices for a total duration of 14 days.
Pharmaceutical treatment and assessment of WT and LQTS2 tissues-on-a-chip
After formation of mature cardiac tissues within the microfluidic chip, the functionality of both WT and LQTS2 tissues was studied. Furthermore, pharmacological rescue was attempted with ALLN, thapsigargin, and nicorandil, and the resultant contractile function and calcium handling of the tissues were assessed. Specifically, ALLN (10 μM) was added on day 12 and incubated for 42 hours. Thapsigargin (1 μM) was added on day 13 and incubated for 24 hours. E-4031 (5 μM) was added on day 13 and incubated for 24 hours. Nicorandil (1 μM) was added on day 14 and incubated for 30 min. Tissue contraction was recorded before and after pharmaceutical treatment. Calcium transients were performed on day 14 for all conditions, following a previously defined protocol (46). Tissues were dosed with epinephrine (0.2 μg/ml) in RPMI + B27 + insulin for 5 min at 37°C, and then resultant contraction and calcium transients were recorded.
Microscopy
Phase contrast and IF images were acquired using Zeiss Axio Observer Z1 equipped with Apotome2 (Zeiss) and ZenPro software. Time-lapse, fluorescence imaging was recorded at 10× objective at 37°C on day 14 to analyze calcium transient characteristics of heart-on-a-chip tissues after pharmacological treatment. To extract calcium transients identified via Fluo-4 AM, we used our previously described method (46). Briefly, Fiji was used to highlight five different ROIs within each video. The fluorescent intensity was divided by the background fluorescence, deemed F/F0, and plotted for each ROI. Matlab was used to process the calcium transients and to identify the location of peaks, as well as the time interval and area of each peak. The time interval of the peaks was deemed the APd, and the SD of APd was determined for each ROI.
Immunostaining
For IF, all samples were first washed with 1× PBS, fixed with 4% paraformaldehyde for 15 to 20 min at 37°C, and then washed three times with PBS-glycine. The subsequent protocol was dependent based on the target proteins. For immunostaining involving hERG, the samples were washed two times with 1× PBS for 10 min each and then permeabilized with 0.1% Triton X-100 for 10 min at RT. The samples were incubated with 0.75 M glycine in PBS for 10 min at RT and then washed with 1× PBS. The samples were then blocked in 10% goat serum for 30 min at 37°C. Primary antibodies [mouse anti-SAA (A7811, Sigma-Aldrich), mouse anti-TNNT2 (MA512960, Thermo Fisher Scientific), mouse anti-TE7 (CBL271, Sigma-Aldrich), rabbit anti-connexin43 (ab11370, Abcam), and rabbit anti-hERG (ab196301, Abcam)] were diluted at 1:200 in 10% goat serum and added to the samples for 30 min at 37°C. Samples were washed three times with 1× PBS for 10 min each and then incubated with 1:1000 anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 594 [in 1:1000 4′,6-diamidino-2-phenylindole (DAPI)] for 30 min at 37°C. The samples were washed three times with 1× PBS before imaging.
For IF involving the trilineage differentiation markers of EBs, the samples were washed two times with PBS for 10 min each and then permeabilized for 30 min with IF buffer at 4°C. After, the samples were blocked for 1 hour with 10% goat serum at RT. Primary antibodies [mouse anti-α smooth muscle actin (SMA) (1:50; 14976082, Invitrogen), rabbit anti-α-Fetoprotein (AFP) (1:50; 15375, Santa Cruz Biotechnology), and mouse anti-TUJ1 (1:1000; 1289248, Fitzgerald)] were diluted in 10% goat serum and incubated at 4°C overnight. The next morning, samples were washed three times for 10 min each with 1× PBS. Secondary antibodies (1:500) were diluted in 1:1000 DAPI and added to the samples for 1 hour at RT. The samples were washed three to five times for 10 min each with 1× PBS before imaging.
For IF with the following antibodies: rabbit anti-Sox2 (1:100; D6D9, Cell Signaling Technology), mouse anti-Nanog (1:200; ab173368, Abcam), rabbit anti-vimentin (1:200; D21H3, Cell Signaling Technology), and mouse anti-TE7 (1:100; CBL271, Sigma-Aldrich), samples were first washed with PBS–Tween 20 for 10 min. Then, samples were permeabilized with IF buffer for 30 min at RT. Samples were blocked with 10% goat serum for 1 hour, and then primary antibodies were diluted in 10% goat serum and incubated with the samples for 1 hour at 37°C. The samples were washed three times with PBS–Tween 20 for 20 min each, and then secondary antibodies were added at 1:500 dilution in 1:1000 DAPI and incubated with samples for 45 min at 37°C. After that, samples were washed three to five times with PBS–Tween 20 for 10 min each and then imaged.
Quantitative real-time reverse transcription PCR
To perform gene expression, total RNA was extracted from cell samples with Total RNA MicroPrep Kit (Zymo). RNA quality and concentration were assessed with Epoch Microplate Spectrophotometer. cDNA was synthesized from total RNA using iScript Reverse Transcriptase Supermix (Bio-Rad), and iTaq Universal SYBR Green Supermix (Bio-Rad) was used to perform qPCR on synthesized cDNA, with 18S as the housekeeping gene. Primers were validated via melt curve analysis and PCR product size verification. For qPCR, 0.1 μl of cDNA and 8 μM dilution of forward and reverse primers were used for 10-μl reactions within 96-well plates, and the qPCR plates were analyzed with qTower.
Patch clamp
Electrophysiology recordings were performed on hiPSC-derived CMs (23 days after differentiation). Whole-cell voltage-clamp measurements were collected with an Axopatch 200B amplifier using Clampex 10.7 software (Molecular Devices). Data analysis was performed using Clampfit 11.1 (Axon Instruments). Cells were plated on 8-mm glass coverslips 2 to 6 hours before data collection. At the time of measurement, the glass coverslips were placed in an extracellular buffer solution containing 140 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5.5 mM glucose, and 5.0 mM Hepes. The extracellular solution pH was adjusted to 7.4 using NaOH, and osmolality was adjusted to 310 mOsm with sucrose. The osmolality was measured using a Vapro 5600 vapor pressure osmometer (Wescor). The extracellular solution was supplemented with JNJ-303 (1 μM) and nifedipine (5 μM) to block ICaL and IKs currents, respectively. The cells were acclimatized in the extracellular solution for 10 min before patching. Glass pipettes were pulled from borosilicate glass capillaries (World Precision Instruments) using a P-2000 laser puller (Sutter Instruments) and heat-polished with an MF-830 microforge (Narashige). Pipettes were filled with intracellular buffer solution containing 125 mM potassium gluconate, 20 mM KCl, 5 mM potassium adenosine triphosphate, 10 mM Hepes, and 10 mM EGTA. The pH of the solution was adjusted to 7.2 using KOH, and osmolality was adjusted to 300 mOsm. IK currents were recorded using the intracellular and extracellular solutions as described above. The IK + E-4031 currents were recorded after perfusion of an extracellular solution containing 1 μM E-4031 from the same cells used to measure IK. IKr currents were calculated by subtraction of the current before and after E-4031 perfusion. The current magnitudes (in picoamperes) were normalized by the cell membrane capacitance (in picofarads) to give current densities. There were five replicates for each condition, which were measured on different days.
Western blot
To assess localization of hERG channels within CMs, the cells were lysed within their six-well plates, and the supernatants were collected. Specifically, the culture plates were placed on ice, and the medium was removed. The cells were washed with ice-cold 1× PBS and then lysed in ice-cold modified radioimmunoprecipitation assay lysis buffer, containing protease inhibitor cocktail (Sigma-Aldrich), protease inhibitor cocktail 2 (Sigma-Aldrich), and dithiothreitol (Sigma-Aldrich), and collected into a chilled microcentrifuge tube. The solution was constantly agitated for 30 min at 4°C, and the insoluble material was removed by centrifugation at 12,000 rpm for 20 min at 4°C. The supernatant was transferred to a new chilled microcentrifuge tube, and the protein concentration was determined by Bradford protein assay (Bio-Rad). An n = 5 set of biologically independent WT and edited hiPSC-CMs was assessed in the control experiments analyzing hERG trafficking using hERG antibody (D1Y2J, Cell Signaling Technology). In WB with CMs after E-4031 exposure, an n = 8 set of biologically independent WT and edited hiPSC-CMs was assessed. Only samples with consistent levels of the loading control, actin, were quantified to determine densitometry ratio of 135 to 155 kDa of hERG.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6 and/or R Studio. Student’s two-tailed, unpaired t tests with the assumption that variances are equal were used for WB and electrophysiology data. Two-way analysis of variance (ANOVA), specifically Holm-Sidak’s multiple comparisons test, was used for analysis of DCt values of gene expression data, as well as in the analysis of contractile signals and tissue alignment.
Acknowledgments
Funding: We would like to thank NSF CAREER Award #1653193 (to M.N.), Arizona Biomedical Research Commission (ABRC) New Investigator Award (to M.N.), and the Flinn Foundation (to M.N.) for providing funding sources for this project. Research reported in this publication was also supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM141933 (to W.D.V.H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions: Conceptualization: J.V., R.Q.M., D.B., and M.N. Methodology: J.V., W.D.V.H., R.Q.M., D.B., and M.N. Investigation: J.V., H.F.M., and N.K. Visualization: J.V., H.F.M., N.K., W.D.V.H., D.B., R.Q.M., and M.N. Supervision: J.V., W.D.V.H., D.B., R.Q.M., and M.N. Writing—original draft: J.V., W.D.V.H., and M.N. Writing—review and editing: J.V., H.F.M., N.K., W.D.V.H., D.B., R.Q.M., and M.N.
Competing interests: J.V. and M.N. are inventors on a pending patent related to this work filed by the Arizona State University (no. US20210054321A1, filed on 15 October 2018, published on 25 February 2021) detailing creation of a 3D heart on a chip. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Tables S1 and S2
Other Supplementary Material for this : manuscript includes the following:
Movies S1 to S6
Supplemental source code
REFERENCES AND NOTES
- 1.P. A. Heidenreich, J. G. Trogdon, O. A. Khavjou, J. Butler, K. Dracup, M. D. Ezekowitz, E. A. Finkelstein, Y. Hong, S. C. Johnston, A. Khera, D. M. Lloyd-Jones, S. A. Nelson, G. Nichol, D. Orenstein, P. W. F. Wilson, Y. J. Woo,Forecasting the future of cardiovascular disease in the United States: A policy statement from the American Heart Association. Circulation 123,933–944 (2011). [DOI] [PubMed] [Google Scholar]
- 2.P. J. Schwartz, M. J. Ackerman, A. L. George Jr., A. A. M. Wilde,Impact of genetics on the clinical management of channelopathies. J. Am. Coll. Cardiol. 62,169–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Alders, H. Bikker, I. Christiaans, Long QT syndrome, GeneReviews [Internet] (2003).
- 4.J. I. Vandenberg, M. D. Perry, M. J. Perrin, S. A. Mann, Y. Ke, A. P. Hill,hERG K+ channels: Structure, function, and clinical significance. Physiol. Rev. 92,1393–1478 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Y. P. Shi, S. Thouta, T. W. Claydon,Modulation of hERG K+ channel deactivation by voltage sensor relaxation. Front. Pharmacol. 11,139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.H. J. Witchel, J. C. Hancox,Familial and acquired long qt syndrome and the cardiac rapid delayed rectifier potassium current. Clin. Exp. Pharmacol. Physiol. 27,753–766 (2000). [DOI] [PubMed] [Google Scholar]
- 7.D. R. Piper, W. A. Hinz, C. K. Tallurri, M. C. Sanguinetti, M. Tristani-Firouzi,Regional specificity of human ether-a'-go-go-related gene channel activation and inactivation gating. J. Biol. Chem. 280,7206–7217 (2005). [DOI] [PubMed] [Google Scholar]
- 8.J. D. Kapplinger, D. J. Tester, B. A. Salisbury, J. L. Carr, C. Harris-Kerr, G. D. Pollevick, A. A. Wilde, M. J. Ackerman,Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION® long QT syndrome genetic test. Heart Rhythm 6,1297–1303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. C. Hancox, A. G. Stuart, S. C. Harmer,Functional evaluation of gene mutations in Long QT Syndrome: Strength of evidence from in vitro assays for deciphering variants of uncertain significance. J. Congenit. Cardiol. 4,6 (2020). [Google Scholar]
- 10.C. L. Anderson, C. E. Kuzmicki, R. R. Childs, C. J. Hintz, B. P. Delisle, C. T. January,Large-scale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat. Commun. 5,5535 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Q. Gong, C. L. Anderson, C. T. January, Z. Zhou,Pharmacological rescue of trafficking defective HERG channels formed by coassembly of wild-type and long QT mutant N470D subunits. Am. J. Physiol. Heart Circ. Physiol. 287,H652–H658 (2004). [DOI] [PubMed] [Google Scholar]
- 12.D. Thomas, J. Kiehn, H. A. Katus, C. A. Karle,Defective protein trafficking in hERG-associated hereditary long QT syndrome (LQT2): Molecular mechanisms and restoration of intracellular protein processing. Cardiovasc. Res. 60,235–241 (2003). [DOI] [PubMed] [Google Scholar]
- 13.E. Ficker, D. Thomas, P. C. Viswanathan, A. T. Dennis, S. G. Priori, C. Napolitano, M. Memmi, B. A. Wible, E. S. Kaufman, S. Iyengar, P. J. Schwartz, Y. Rudy, A. M. Brown,Novel characteristics of a misprocessed mutant HERG channel linked to hereditary long QT syndrome. Am. J. Physiol. Heart Circ. Physiol. 279,H1748–H1756 (2000). [DOI] [PubMed] [Google Scholar]
- 14.B. P. Delisle, B. D. Anson, S. Rajamani, C. T. January,Biology of cardiac arrhythmias: Ion channel protein trafficking. Circ. Res. 94,1418–1428 (2004). [DOI] [PubMed] [Google Scholar]
- 15.A. Paulussen, A. Raes, G. Matthijs, D. J. Snyders, N. Cohen, J. Aerssens,A novel mutation (T65P) in the PAS domain of the human potassium channel HERG results in the long QT syndrome by trafficking deficiency. J. Biol. Chem. 277,48610–48616 (2002). [DOI] [PubMed] [Google Scholar]
- 16.R. Chandra, C. F. Starmer, A. O. Grant,Multiple effects of KPQ deletion mutation on gating of human cardiac Na+ channels expressed in mammalian cells. Am. J. Physiol. 274,H1643–H1654 (1998). [DOI] [PubMed] [Google Scholar]
- 17.S. Friedrichs, D. Malan, P. Sasse,Modeling long QT syndromes using induced pluripotent stem cells: Current progress and future challenges. Trends Cardiovasc. Med. 23,91–98 (2013). [DOI] [PubMed] [Google Scholar]
- 18.M. Derangeon, J. Montnach, I. Baró, F. Charpentier,Mouse models of SCN5A-related cardiac arrhythmias. Front. Physiol. 3,210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.I. Itzhaki, L. Maizels, I. Huber, L. Zwi-Dantsis, O. Caspi, A. Winterstern, O. Feldman, A. Gepstein, G. Arbel, H. Hammerman, M. Boulos, L. Gepstein,Modelling the long QT syndrome with induced pluripotent stem cells. Nature 471,225–229 (2011). [DOI] [PubMed] [Google Scholar]
- 20.E. Matsa, D. Rajamohan, E. Dick, L. Young, I. Mellor, A. Staniforth, C. Denning,Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur. Heart J. 32,952–962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. L. Lahti, V. J. Kujala, H. Chapman, A. Koivisto, M. Pekkanen-Mattila, E. Kerkelä, J. Hyttinen, K. Kontula, H. Swan, B. R. Conklin, S. Yamanaka, O. Silvennoinen, K. Aalto-Setälä,Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis. Models Mech. 5,220–230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.F. C. P. Mesquita, P. C. Arantes, T. H. Kasai-Brunswick, D. S. Araujo, F. Gubert, G. Monnerat, D. Silva Dos Santos, G. Neiman, I. C. Leitão, R. A. Q. Barbosa, J. L. Coutinho, I. M. Vaz, M. N. Dos Santos, T. Borgonovo, F. E. S. Cruz, S. Miriuka, E. H. Medei, A. C. Campos de Carvalho, A. B. Carvalho,R534C mutation in hERG causes a trafficking defect in iPSC-derived cardiomyocytes from patients with type 2 long QT syndrome. Sci. Rep. 9,19203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. Mura, A. Mehta, C. J. Ramachandra, R. Zappatore, F. Pisano, M. C. Ciuffreda, V. Barbaccia, L. Crotti, P. J. Schwartz, W. Shim, M. Gnecchi,The KCNH2-IVS9-28A/G mutation causes aberrant isoform expression and hERG trafficking defect in cardiomyocytes derived from patients affected by long QT Syndrome type 2. Int. J. Cardiol. 240,367–371 (2017). [DOI] [PubMed] [Google Scholar]
- 24.M. Bellin, S. Casini, R. P. Davis, C. D'Aniello, J. Haas, D. Ward-van Oostwaard, L. G. Tertoolen, C. B. Jung, D. A. Elliott, A. Welling, K. L. Laugwitz, A. Moretti, C. L. Mummery,Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 32,3161–3175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.G. Duncan, K. Firth, V. George, M. D. Hoang, A. Staniforth, G. Smith, C. Denning,Drug-mediated shortening of action potentials in LQTS2 human induced pluripotent stem cell-derived cardiomyocytes. Stem Cells Dev. 26,1695–1705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D. Yoshinaga, S. Baba, T. Makiyama, H. Shibata, T. Hirata, K. Akagi, K. Matsuda, H. Kohjitani, Y. Wuriyanghai, K. Umeda, Y. Yamamoto, B. R. Conklin, M. Horie, J. Takita, T. Heike,Phenotype-based high-throughput classification of long QT syndrome subtypes using human induced pluripotent stem cells. Stem Cell Rep. 13,394–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.J. Kuusela, V. J. Kujala, A. Kiviaho, M. Ojala, H. Swan, K. Kontula, K. Aalto-Setälä,Effects of cardioactive drugs on human induced pluripotent stem cell derived long QT syndrome cardiomyocytes. Springerplus 5,234 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.P. Garg, A. Oikonomopoulos, H. Chen, Y. Li, C. K. Lam, K. Sallam, M. Perez, R. L. Lux, M. C. Sanguinetti, J. C. Wu,Genome editing of induced pluripotent stem cells to decipher cardiac channelopathy variant. J. Am. Coll. Cardiol. 72,62–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Q. Li, J. Wang, Q. Wu, N. Cao, H.-T. Yang,Perspective on human pluripotent stem cell-derived cardiomyocytes in heart disease modeling and repair. Stem Cells Transl. Med. 9,1121–1128 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.P. Horvath, R. Barrangou,CRISPR/Cas, the immune system of bacteria and archaea. Science 327,167–170 (2010). [DOI] [PubMed] [Google Scholar]
- 31.L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini, F. Zhang,Multiplex genome engineering using CRISPR/Cas systems. Science 339,819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.F. A. Ran, P. D. Hsu, C.-Y. Lin, J. S. Gootenberg, S. Konermann, A. E. Trevino, D. A. Scott, A. Inoue, S. Matoba, Y. Zhang, F. Zhang,Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154,1380–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.H. A. Rees, D. R. Liu,Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19,770–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.G. Yang, X. Huang,Methods and applications of CRISPR/Cas system for genome editing in stem cells. Cell Regen. 8,33–41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.A. V. Anzalone, L. W. Koblan, D. R. Liu,Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38,824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 36.B. Yang, L. Yang, J. Chen,Development and application of base editors. CRISPR J. 2,91–104 (2019). [DOI] [PubMed] [Google Scholar]
- 37.A. Eid, S. Alshareef, M. M. Mahfouz,CRISPR base editors: Genome editing without double-stranded breaks. Biochem. J. 475,1955–1964 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.F. Ann Ran, P. D. Hsu, J. Wright, V. Agarwala, D. A. Scott, F. Zhang,Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8,2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. Ren, K. Xu, Z. Liu, J. Shen, F. Han, Z. Chen, Z. Zhang,Dual-reporter surrogate systems for efficient enrichment of genetically modified cells. Cell. Mol. Life Sci. 72,2763–2772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.N. Brookhouser, S. J. Tekel, K. Standage-Beier, T. Nguyen, G. Schwarz, X. Wang, D. A. Brafman,BIG-TREE: Base-edited isogenic hPSC line generation using a transient reporter for editing enrichment. Stem Cell Rep. 14,184–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.K. Standage-Beier, S. J. Tekel, N. Brookhouser, G. Schwarz, T. Nguyen, X. Wang, D. A. Brafman,A transient reporter for editing enrichment (TREE) in human cells. Nucleic Acids Res. 47,e120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. J. Tekel, N. Brookhouser, K. Standage-Beier, X. Wang, D. A. Brafman,Cytosine and adenosine base editing in human pluripotent stem cells using transient reporters for editing enrichment. Nat. Protoc. 16,3596–3624 (2021). [DOI] [PubMed] [Google Scholar]
- 43.A. Mehta, G. L. Sequiera, C. J. Ramachandra, Y. Sudibyo, Y. Chung, J. Sheng, K. Y. Wong, T. H. Tan, P. Wong, R. Liew, W. Shim,Re-trafficking of hERG reverses long QT syndrome 2 phenotype in human iPS-derived cardiomyocytes. Cardiovasc. Res. 102,497–506 (2014). [DOI] [PubMed] [Google Scholar]
- 44.K. O. Brandão, L. van den Brink, D. C. Miller, C. Grandela, B. J. van Meer, M. P. H. Mol, T. de Korte, L. G. J. Tertoolen, C. L. Mummery, L. Sala, A. O. Verkerk, R. P. Davis,Isogenic Sets of hiPSC-CMs harboring distinct KCNH2 mutations differ functionally and in susceptibility to drug-induced arrhythmias. Stem Cell Rep. 15,1127–1139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Y. Wang, P. Liang, F. Lan, H. Wu, L. Lisowski, M. Gu, S. Hu, M. A. Kay, F. D. Urnov, R. Shinnawi, J. D. Gold, L. Gepstein, J. C. Wu,Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J. Am. Coll. Cardiol. 64,451–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J. Veldhuizen, J. Cutts, D. A. Brafman, R. Q. Migrino, M. Nikkhah,Engineering anisotropic human stem cell-derived three-dimensional cardiac tissue on-a-chip. Biomaterials 256,120195 (2020). [DOI] [PubMed] [Google Scholar]
- 47.J. Veldhuizen, R. Chavan, B. Moghadas, J. G. Park, V. D. Kodibagkar, R. Q. Migrino, M. Nikkhah,Cardiac ischemia on-a-chip to investigate cellular and molecular response of myocardial tissue under hypoxia. Biomaterials 281,121336 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. L. Anderson, B. P. Delisle, B. D. Anson, J. A. Kilby, M. L. Will, D. J. Tester, Q. Gong, Z. Zhengfeng, M. J. Ackerman, C. T. January,Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation 113,365–373 (2006). [DOI] [PubMed] [Google Scholar]
- 49.B. P. Delisle, C. L. Anderson, R. C. Balijepalli, B. D. Anson, T. J. Kamp, C. T. January,Thapsigargin selectively rescues the trafficking defective LQT2 channels G601S and F805C. J. Biol. Chem. 278,35749–35754 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Q. Gong, M. A. Jones, Z. Zhou,Mechanisms of pharmacological rescue of trafficking-defective hERG mutant channels in human long QT syndrome. J. Biol. Chem. 281,4069–4074 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.H. Vyas, M. J. Ackerman,Epinephrine QT stress testing in congenital long QT syndrome. J. Electrocardiol. 39,S107–S113 (2006). [DOI] [PubMed] [Google Scholar]
- 52.A. Kagan, Z. Yu, G. I. Fishman, T. V. McDonald,The dominant negative LQT2 mutation A561V reduces wild-type HERG expression. J. Biol. Chem. 275,11241–11248 (2000). [DOI] [PubMed] [Google Scholar]
- 53.W. R. Kukovetz, S. Holzmann, G. Pöch,Molecular mechanism of action of nicorandil. J. Cardiovasc. Pharmacol. 20,S1–S7 (1992). [DOI] [PubMed] [Google Scholar]
- 54.E. Wallace, L. Howard, M. Liu, T. O'Brien, D. Ward, S. Shen, T. Prendiville,Long QT syndrome: Genetics and future perspective. Pediatr. Cardiol. 40,1419–1430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.H. Chen, B. Jiang, J. G. Shamul, X. He,Image entropy-based label-free functional characterization of human induced pluripotent stem cell-derived 3D cardiac spheroids. Biosens. Bioelectron. 179,113055 (2021). [DOI] [PubMed] [Google Scholar]
- 56.C. M. McBride, A. M. Smith, J. L. Smith, A. R. Reloj, E. J. Velasco, J. Powell, C. S. Elayi, D. C. Bartos, D. E. Burgess, B. P. Delisle,Mechanistic basis for type 2 long QT syndrome caused by KCNH2 mutations that disrupt conserved arginine residues in the voltage sensor. J. Membr. Biol. 246,355–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D. R. Piper, J. Rupp, F. B. Sachse, M. C. Sanguinetti, M. Tristani-Firouzi,Cooperative interactions between R531 and acidic residues in the voltage sensing module of hERG1 channels. Cell. Physiol. Biochem. 21,37–46 (2008). [DOI] [PubMed] [Google Scholar]
- 58.M. Zhang, J. Liu, G.-N. Tseng,Gating charges in the activation and inactivation processes of the HERG channel. J. Gen. Physiol. 124,703–718 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Zhang, J. Liu, M. Jiang, D.-M. Wu, K. Sonawane, H. R. Guy, G.-N. Tseng,Interactions between charged residues in the transmembrane segments of the voltage-sensing domain in the hERG channel. J. Membr. Biol. 207,169–181 (2005). [DOI] [PubMed] [Google Scholar]
- 60.R. N. Subbiah, C. E. Clarke, D. J. Smith, J. Zhao, T. J. Campbell, J. I. Vandenberg,Molecular basis of slow activation of the human ether-a-go-go related gene potassium channel. J. Physiol. 558,417–431 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.J. Yu, M. A. Vodyanik, K. Smuga-Otto, J. Antosiewicz-Bourget, J. L. Frane, S. Tian, J. Nie, G. A. Jonsdottir, V. Ruotti, R. Stewart, I. I. Slukvin, J. A. Thomson,Induced pluripotent stem cell lines derived from human somatic cells. Science 318,1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- 62.M. Stemmer, T. Thumberger, M. del Sol Keyer, J. Wittbrodt, J. L. Mateo,CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLOS ONE 10,e0124633 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.J. Zhang, R. Tao, K. F. Campbell, J. L. Carvalho, E. C. Ruiz, G. C. Kim, E. G. Schmuck, A. N. Raval, A. M. da Rocha, T. J. Herron, J. Jalife, J. A. Thomson, T. J. Kamp,Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat. Commun. 10,2238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.J. Veldhuizen, M. Nikkhah,Developing 3D organized human cardiac tissue within a microfluidic platform. J. Vis. Exp. 172,e62539 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Tables S1 and S2
Movies S1 to S6
Supplemental source code