Abstract
Inflammatory breast cancer (IBC), the most aggressive breast cancer subtype, is driven by an immunosuppressive tumor microenvironment (TME). Current treatments for IBC have limited efficacy. In a clinical trial (NCT01036087), an anti-EGFR antibody combined with neoadjuvant chemotherapy produced the highest pathological complete response rate ever reported in patients with IBC having triple-negative receptor status. We determined the molecular and immunological mechanisms behind this superior clinical outcome. Using novel humanized IBC mouse models, we discovered that EGFR-targeted therapy remodels the IBC TME by increasing cytotoxic T cells and reducing immunosuppressive regulatory T cells and M2 macrophages. These changes were due to diminishing immunosuppressive chemokine expression regulated by transcription factor EGR1. We also showed that induction of an immunoactive IBC TME by an anti-EGFR antibody improved the antitumor efficacy of an anti–PD-L1 antibody. Our findings lay the foundation for clinical trials evaluating EGFR-targeted therapy combined with immune checkpoint inhibitors in patients with cancer.
In IBC, targeting EGFR made the tumor microenvironment more immunoactive, enhancing immune checkpoint inhibitor efficacy.
INTRODUCTION
Immune checkpoint inhibitors (ICIs) have revolutionized breast cancer treatment. Two randomized trials, KEYNOTE-522 (1) and IMpassion031 (2), using the combination of an anti–programed death-1 (PD-1) antibody (pembrolizumab) or an anti–programed death ligand-1 (PD-L1) antibody (atezolizumab) with neoadjuvant chemotherapy (NAC; immunochemotherapy) have shown substantial improvement in pathological complete response (pCR) rate in patients with breast cancer. The findings of KEYNOTE-522 led to pembrolizumab approval as first-line therapy for patients with early-stage triple-negative breast cancer (TNBC) by the U.S. Food and Drug Administration. However, in KEYNOTE-522, the pCR rate was only 13.6% higher in the immunochemotherapy group than in the chemotherapy-alone group. Limited benefit of ICIs is also evident in other cancers for which ICIs have been used as frontline treatments, including melanoma and lung cancer (3–7). Furthermore, recent evidence revealed that a portion of patients with non–small cell lung cancer who initially responded to ICIs went on to develop recurrences that were resistant to ICIs (8, 9), a finding that is likely to be replicated in TNBC once the new standard of care is widely implemented in the clinic. This leads us to consider that the critical, near-term need for breast cancer therapeutics is to improve or maximize patients’ responses to ICIs.
Mechanistic studies into the interplay between the immune system and tumor cells identified the critical role of the tumor microenvironment (TME) in dictating the impact of ICIs on tumor progression (10). TMEs characterized by immunosuppressive components, including M2 macrophages, regulatory T cells (Tregs), myeloid-derived suppressor cells (MDSCs), and immunosuppressive cytokines/chemokines, are associated with ICI failure (11, 12). Preclinical and clinical studies have indicated that targeted therapy affects immune aspects of the TME and improves the clinical benefits of ICIs (13, 14). Therefore, identifying readily available pharmacological strategies that can modify the immune profile of the breast tumor and make it more susceptible to ICIs is a critical research goal in translational breast oncology.
Among the aggressive forms of breast cancer, inflammatory breast cancer (IBC) is the most lethal and least treatable (15). IBC accounts for 2 to 4% of breast cancer cases but causes 8 to 10% of breast cancer deaths. Despite significant improvement in patient survival with a multimodal therapeutic approach, the median overall survival of patients with IBC is still poor. In addition, there is a critical need to develop new, IBC-tailored breast cancer therapeutic strategies (15–18). Several IBC research groups have demonstrated that the TME is a critical driver of the IBC clinical phenotype and promotes IBC metastasis (19–21). Immune profiling in patients with stage III or de novo stage IV IBC who received NAC demonstrated an increase in tumor-infiltrating lymphocytes in the tumors of patients who had a pCR (22, 23), suggesting that the presence of cytotoxic immune cells is closely related to IBC tumor response to chemotherapy. For this reason, we set out to identify clinic-ready treatments that can immune modulate the IBC TME and prime the tumor for ICI treatment.
Our laboratory has established epidermal growth factor receptor (EGFR) as an independent prognostic marker for recurrence and survival (24, 25) and as a critical therapeutic target in IBC. This work led to a phase 2 clinical trial for patients with IBC (NCT01036087) with an anti–EGFR humanized antibody, panitumumab, combined with NAC. Panitumumab and NAC induced the highest rate (42%) of pCR yet achieved in patients with triple-negative IBC (26). This success suggested that EGFR-targeting treatments should be part of IBC-specific treatment strategies, but which mechanisms were involved in this response remains unclear.
Given the critical role of the TME in driving IBC aggressiveness and the promising clinical benefits of panitumumab, we asked whether EGFR-targeted treatments exert their clinical efficacy by modulating the TME in IBC. To answer this question, we embarked on a multifaceted mechanistic study using tumor samples from a clinical trial and a combination of in vitro and in vivo studies. In particular, we used novel humanized IBC mouse models to understand the immunological and biological impact. The work presented here demonstrates that panitumumab regulated the global expression of tumor-associated chemokines, which led to changes in immune cell infiltration and the subsequent antitumor effect. We also identified the transcription factor early growth response protein 1 (EGR1) as the putative mediator between EGFR signaling and the expression of immunosuppressive chemokines.
Furthermore, we show that patients who achieved a pCR following panitumumab and NAC treatment had an increased presence of CD8+ T cells and reduced presence of Tregs and M2 macrophages in tumor tissues after panitumumab treatment but before chemotherapy, suggesting that panitumumab alone induced the switch in the TME immune status. Critically, we demonstrated that panitumumab enhanced the efficacy of an anti–PD-L1 antibody in reducing IBC tumor growth in vivo. Our results reveal a previously undescribed role of EGFR in immune remodeling of the IBC TME and support developing a new rational therapeutic strategy based on combining EGFR-targeted therapy with ICIs. While the work presented here is IBC specific, we further discuss the likely generalizable mechanisms that may affect future therapeutic development in other “cold” cancers by suppressing the EGFR pathway.
RESULTS
Panitumumab converts the IBC TME from immunosuppressive to immunoactive
To understand the superior antitumor effect of panitumumab in patients with triple-negative IBC (NCT01036087) (26), we established IBC SUM149 and BCX010 xenografts in humanized mouse models, which allowed us to directly study the interactions between the human immune microenvironment and tumor cells in mice (27, 28). We confirmed the presence of human CD45+, CD3+, CD4+, and CD8+ cells in tumor tissue (fig. S1). Panitumumab substantially reduced SUM149 (Fig. 1A) and BCX010 (Fig. 1C) tumor growth and inhibited EGFR signaling (Fig. 1, B and D) in these humanized mouse models. To achieve an unbiased and comprehensive assessment of the impact of panitumumab on the TME in vivo, we performed a single-cell RNA sequencing (scRNA-seq) analysis using tumor cells isolated from control immunoglobulin G2 (IgG2)– or panitumumab-treated SUM149-hu-NSG-SGM3 mice (Fig. 1E and fig. S2A). Panitumumab decreased the percentage of epithelial cells but increased the percentages of CD8+ T cells, fibroblasts, and mast cells in tumor tissue (fig. S2B). In addition, differential gene expression analysis showed that panitumumab reduced the gene expression of immunosuppressive cytokines and chemokines, including TGFβ1, CCL26, CSF1, TGFβ2, TGFβ3, CCL2, CXCL3, CCL20, CXCL8 (IL-8), IL1B, CXCL2, and CXCL14, but increased the gene expression of chemokines that function as antitumor immune modulators, including CCL19, CX3CL1, and CXCL10 (Fig. 1F and fig. S3A).
Fig. 1. Panitumumab treatment remodels the TME in IBC humanized mouse models.
(A) Tumor growth curve of SUM149 xenograft in humanized mice (SUM149-hu-NSG-SGM3) treated with control IgG2 and panitumumab (five mice per group). Data are summarized as means ± SEM. *P < 0.01. (B) Immunohistochemistry (IHC) staining (left) of phosphorylated EGFR (pEGFR) and Ki67 and quantification of staining intensity (right) in tumor tissues from IgG2- and panitumumab-treated SUM149-hu-NSG-SGM3 mice. *P < 0.01. (C) Tumor growth curve of BCX010 xenograft in humanized mice (BCX010-hu-NSG-SGM3) treated with control IgG2 and panitumumab (six mice per group). Data are summarized as means ± SEM. *P < 0.05. (D) IHC staining (left) of pEGFR and Ki67 and quantification of staining intensity (right) in tumor tissues from IgG2- and panitumumab-treated BCX010-hu-NSG-SGM3 mice. *P < 0.01. (E) Uniform manifold approximation and projection (UMAP) plot of all tumor-resident cells from SUM149-hu-NSG-SGM3 tumors treated with IgG2 and panitumumab (two tumor samples per group). Only cells containing at least 100 gene features and mitochondrial gene counts of less than 20% were used (13,931 cells in IgG2-treated tumor samples and 11,593 cells in panitumumab-treated tumor samples). Clusters denoted by color are labeled with the inferred cell types. (F) Heatmap of pooled gene expression within the epithelial cells among samples treated with IgG2 and panitumumab. These genes are involved in the immune response. (G) Enrichment of pathways associated with epithelial-mesenchymal transition, TNF-α signaling via NF-κB, inflammatory response, IL-6/JAK/STAT3 signaling, and IL-2/STAT5 signaling in IgG2-treated compared to panitumumab-treated SUM149 epithelial tumor cells in humanized mice. (H) Volcano plot of significant differentially expressed genes in CD8+ T cells after panitumumab treatment in SUM149-hu-NSG-SGM3 tumors. PmAb, panitumumab; RBC, red blood cells; FC, fold change; n.s., not significant; FDR, false discover rate.
Gene set enrichment analysis revealed that compared to IgG2-treated tumors, panitumumab-treated tumors had suppressed genes related to epithelial-mesenchymal transition, tumor necrosis factor–α (TNF-α) signaling via nuclear factor κB (NF-κB), inflammatory response, interleukin-6 (IL-6)/Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling, and IL-2/STAT5 signaling (Fig. 1G, fig. S3B, and table S1) but an enriched interferon-α response pathway (fig. S3C and table S1) (29–31). In CD8+ T cells, panitumumab decreased the expression of immunosuppressive molecules, including KLRC1 (NKG2A) (32), CSF1 (33), HAVCR2 (TIM3), S100A7 (34), and S100A8 (35), but increased the expression of molecules that can boost cytotoxic T lymphocyte response or enhance T cell activation, proliferation, and differentiation, including APOE (36), ISG15 (37), and LMNA (Fig. 1H and table S2) (38). In summary, scRNA-seq results indicate that panitumumab induces changes in IBC tumor cells and the tumor immune microenvironment that may increase antitumor immune response.
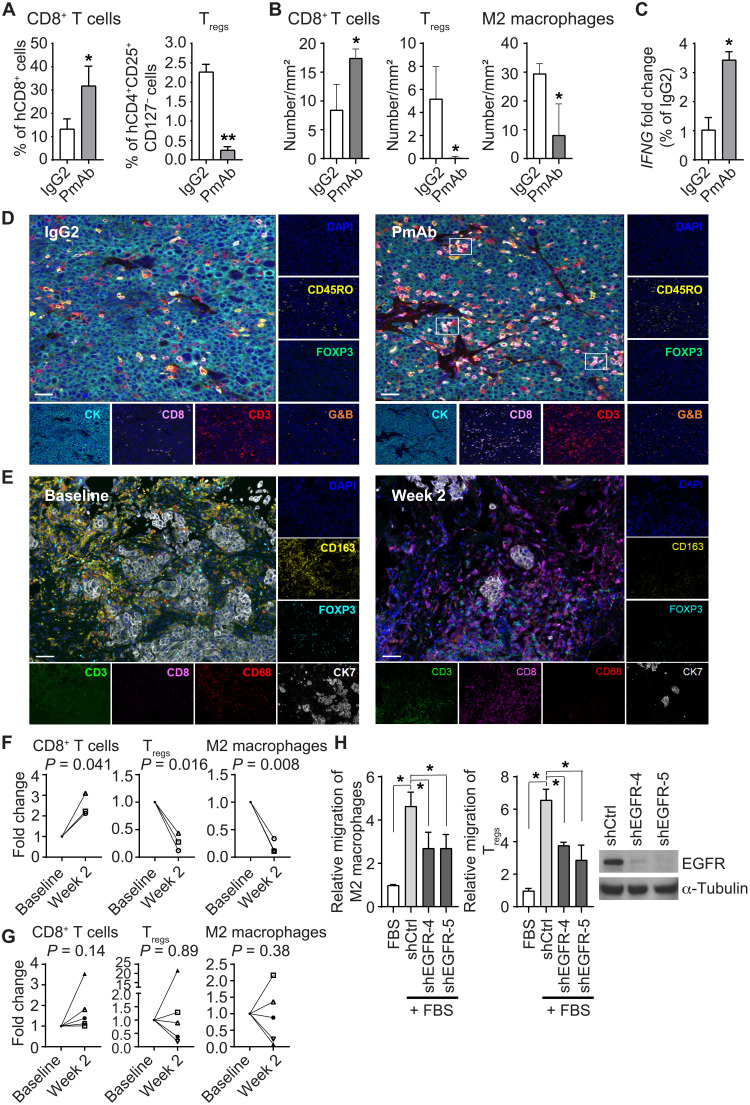
Next, we examined the functional consequences of the changes in the tumor immune microenvironment after panitumumab treatment using flow cytometry and multiplexed immunofluorescence staining. In SUM149-hu-NSG-SGM3 mice, panitumumab increased CD8+ T cells and decreased Tregs in the peripheral blood (fig. S4A) and tumor tissues (Fig. 2A). Panitumumab also increased CD8+ T cells in tumors of BCX010-hu-NSG-SGM3 mice (fig. S4B), suggesting that our finding was not tumor cell line dependent. Multiplexed immunofluorescence staining revealed more CD3+CD8+ T cells but fewer Tregs and M2 macrophages in panitumumab-treated SUM149 and BCX010 tissues than in IgG2-treated tissues (Fig. 2B and fig. S4C). In addition, increased IFNG gene expression and more CD8+CD3+granzyme B+ cells in panitumumab-treated tissues (average of 20.42 cells/mm2 versus 1.38 cells/mm2) revealed stronger cytotoxic T cell activity in panitumumab-treated than in IgG2-treated SUM149 tumors (Fig. 2, C and D). These findings indicate the increased presence of cytotoxic T cells in vivo after EGFR inhibition.
Fig. 2. Panitumumab treatment affects the TME in IBC tumors.
(A and B) Changes in CD8+ T cells, Tregs, and M2 macrophages in tissues of IgG2- and panitumumab-treated SUM149-hu-NSG-SGM3 mice analyzed by flow cytometry (A) and multiplexed immunofluorescence staining (B). (A) *P = 0.05 and **P < 0.001. (B) *P < 0.05. (C) Panitumumab-treated tissues have increased IFNG gene expression compared to IgG2-treated tissues in SUM149-hu-NSG-SGM3 mice. *P < 0.05. (D) Panitumumab-treated tissues have more CD3+CD8+G&B+ cells than IgG2-treated tissues in SUM149-hu-NSG-SGM3 mice as analyzed by multiplexed immunofluorescence staining. Scale bars, 50 μm. (E to G) Changes in CD8+ T cells, M2 macrophages, and Tregs after panitumumab treatment in matched tissues from three patients with IBC having a pCR (F) or five patients with IBC without a pCR (G) to panitumumab/NAC in primary HER2-negative IBC (NCT01036087). (E) Representative images of multiplexed immunofluorescence staining of CD3, CD8, CD68, CD163, FOXP3, and CK7 in IBC patient tissues at baseline (before panitumumab treatment, left) and week 2 (after panitumumab treatment, right). The numbers of CD8+ T cells (CD3+CD8+), M2 macrophages (CD68+CD163+), and Tregs (CD3+FOXP3+) in five randomly selected areas of each slide were calculated. Each symbol represents the same patient. Scale bars, 50 μm. (H) EGFR knockdown SUM149 clones shEGFR-4 and shEGFR-5 induce less migration of M2 macrophages and Tregs than control knockdown clone shCtrl in in vitro coculture transwell assays. *P < 0.01. G&B, granzyme B; DAPI, 4′,6-diamidino-2-phenylindole; FBS, fetal bovine serum. Experiments in (C) and (H) were independently repeated with three replicates each time.
To assess the clinical implications of panitumumab-induced tumor immune microenvironment changes, we performed multiplexed immunofluorescence staining on eight pairs of matched patient tissues collected before and after panitumumab treatment from a phase 2 trial of panitumumab combined with NAC in primary human epidermal growth factor receptor-2 (HER2)-negative IBC (NCT01036087; Fig. 2, E to G) (26). The clinicopathological characteristics of these patients are shown in table S3. In the three patients with a pCR, we observed an increase in CD8+ T cells and a decrease in Tregs and M2 macrophages after panitumumab treatment, suggesting a change toward an immunoactive TME (Fig. 2F). However, this pattern of changes was not observed in the five patients without a pCR (Fig. 2G). The samples were obtained following panitumumab treatment but before the initiation of chemotherapy, and thus, the immune changes can be ascribed only to EGFR inhibition. These results suggest that a pCR to panitumumab/NAC in patients with IBC may be associated with an immunoactive TME induced by panitumumab.
The fact that EGFR inhibition by panitumumab modulated the tumor-associated immune profile suggests that EGFR is involved in maintenance of the immunosuppressive microenvironment in IBC. To confirm this, we knocked down EGFR expression in SUM149 cells and examined the effect on the migration of immunosuppressive M2 macrophages and Tregs. As shown in Fig. 2H, SUM149 EGFR knockdown clones, shEGFR-4 and shEGFR-5, induced less migration of M2 macrophages and Tregs than control knockdown clone shCtrl, suggesting that EGFR signaling is necessary for mediation of immunosuppressive signals and subsequent recruitment of immunosuppressive cells.
Together, these results indicate that panitumumab inactivated the EGFR pathway, enhanced the infiltration of cytotoxic T cells, and reduced the infiltration of Tregs and M2 macrophages in IBC tumors. These changes in the tumor immune microenvironment suggest that EGFR mediates tumor immunosuppression and that panitumumab may improve the antitumor immune response.
Panitumumab alters chemokine expression relevant to TME immune status in IBC
To explore the mechanism by which EGFR modulates the TME in IBC, we measured cytokine expression in tumor tissues from IgG2- and panitumumab-treated SUM149-hu-NSG-SGM3 mice using a cytokine antibody array (fig. S4D). We found that panitumumab reduced the expression of CCL20, CXCL5, CCL2, and IL-8 in SUM149-hu-NSG-SGM3 mice [Fig. 3A (left); fig. S4, D and E; and table S4] and BCX010-hu-NSG-SGM3 mice (fig. S4F), which have been reported to suppress the antitumor immune response and are linked to various cancers (39–43). In contrast, panitumumab increased the expression of several chemoattractants for T cells and natural killer cells, including CCL4, CCL5, CXCL9, and CXCL10, which we validated by reverse transcription polymerase chain reaction (RT-PCR) [Fig. 3A (right), fig. S4D, and table S4]. The concept that the EGFR pathway regulates the expression of these chemokines was further supported by our findings that EGFR knockdown and panitumumab in SUM149 cells resulted in the same cytokine and chemokine expression changes (Fig. 3, B and C).
Fig. 3. Panitumumab treatment affects the TME by regulating chemokine expression in IBC cells.
(A) Expression of chemokines in SUM149-hu-NSG-SGM3 tumors treated with IgG2 and panitumumab by quantitative RT-PCR (qRT-PCR). Three tumor samples per group. *P < 0.01. (B and C) EGFR knockdown (B) or panitumumab treatment (C) affects chemokine expression in SUM149 cells by qRT-PCR. *P < 0.005 and **P < 0.05. (D) Recombinant proteins CCL4, CCL5, CXCL9, and CXCL10 individually or in combination increase the migration of CD8+ T cells. *P < 0.01 and **P < 0.05. (E) Kaplan-Meier metastasis-free survival curves in patients with IBC according to high (N = 70) and low (N = 51) metagene scores of CCL4, CCL5, CXCL9, and CXCL10 mRNA expression. P = 2.62 × 10−2. (F) Box plot of metagene score of CCL2/CCL20/CXCL5/IL-8 mRNA expression according to 252 non-IBC patients and 137 patients with IBC. P = 5.51 × 10−3. (G) Kaplan-Meier overall survival curves in patients with IBC according to high and low CCL20 mRNA expression. P = 2.36 × 10−2. (H) Recombinant protein CCL20 reduces the migration of CD8+ T cells. *P < 0.05. (I) Recombinant proteins CCL2, CCL20, CXCL5, and IL-8 individually or in combination induce the migration of M2 macrophages. *P < 0.001 and **P < 0.01. (J) Recombinant proteins CCL2 and CCL20 induce the migration of Tregs. *P < 0.01. (D) and (H) to (J) were transwell migration assays. Data are summarized as means ± SD in (A) to (D) and (H) to (J). Experiments in (A) to (D) and (H) to (J) were independently repeated three times with three replicates each time.
We further confirmed that chemokines CCL4, CCL5, CXCL9, and CXCL10 individually or in combination increased the migration of CD8+ T cells (Fig. 3D), which is consistent with previous reports (44, 45). Our analysis of these chemokines in IBC clinical samples revealed that high expression of the CCL4/CCL5/CXCL9/CXCL10 metagene correlated with better 5-year metastasis-free survival of patients with IBC (Fig. 3E) and triple-negative IBC (fig. S4G), implying that this cadre of cytokines plays a critical role in IBC tumor progression and tumor response. Panitumumab–down-regulated chemokines CCL2, CCL20, CXCL5, and IL-8 are known to suppress the antitumor immune response (39–42). We found that patients with IBC had significantly higher expression of the CCL2/CCL20/CXCL5/IL-8 metagene than patients with non-IBC (Fig. 3F). Furthermore, high CCL20 expression correlated with worse overall survival of patients with IBC (Fig. 3G). To understand the role of CCL20 in the immune profile of IBC, we investigated the impact of CCL20 on the migration of CD8+ T cells. We found that CD8+ T cell motility was down-regulated following CCL20 treatment (Fig. 3H). Conversely, CCL2, CCL20, CXCL5, and IL-8 individually or in combination increased the migration of M2 macrophages (Fig. 3I). CCL2 and CCL20 also increased the migration of Tregs (Fig. 3J), suggesting that these chemokines may participate in the recruitment of immunosuppressive cells to the tumor. In support of this interpretation, we found that patients with IBC having a pCR to panitumumab/NAC had decreased CCL2 gene expression after panitumumab treatment, whereas patients with IBC without a pCR to panitumumab/NAC had similar CCL2 gene expression before and after treatment (fig. S4H). Together, these results demonstrate that panitumumab modulates the IBC TME by increasing the secretion of chemoattractants for cytotoxic T cells and reducing the secretion of immunosuppressive chemokines. These results point to the critical roles of tumor-secreted chemokines in maintaining the immunosuppressive microenvironment in IBC.
The EGFR pathway regulates immunosuppressive chemokines CCL2, CCL20, CXCL5, and IL-8 through EGR1 in IBC
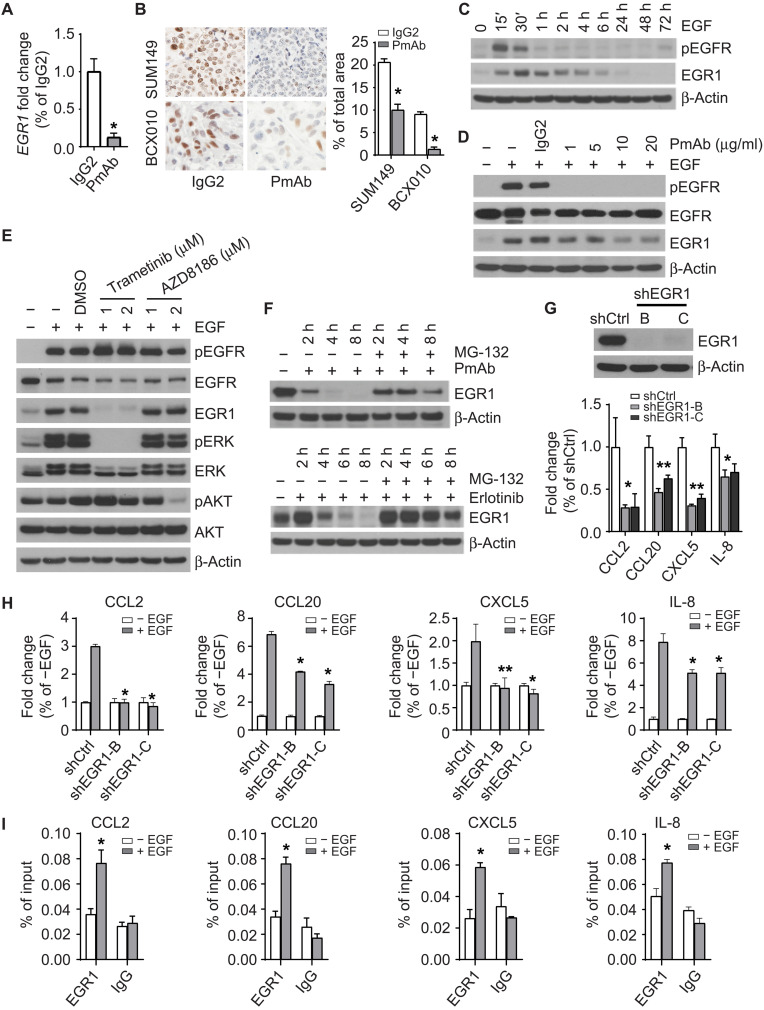
We next asked how EGFR regulates the expression of immunosuppressive chemokines CCL2, CCL20, CXCL5, and IL-8 in IBC. EGR1 is a transcription factor up-regulated in patients with IBC (46). In a nonbiased analysis of human clinical samples obtained before and after panitumumab exposure, panitumumab substantially down-regulated EGR1 gene expression in tumors of patient with IBC from the panitumumab/NAC clinical trial (26). Consistent with these results, we found that EGR1 expression was down-regulated by panitumumab in SUM149 and BCX010 cells (Fig. 4A and fig. S5A). EGR1 plays a critical role in regulating growth, differentiation, survival, and immune response (47, 48). Therefore, we hypothesized that EGR1 mediates CCL2, CCL20, CXCL5, and IL-8 expression in response to EGFR signaling in IBC.
Fig. 4. The EGFR pathway regulates the expression of CCL2, CCL20, CXCL5, and IL-8 through EGR1 in IBC.
(A) Panitumumab treatment reduces the gene expression of EGR1 in SUM149 cells. *P < 0.001. (B) Panitumumab treatment reduces the expression of EGR1 protein in SUM149-hu-NSG-SGM3 and BCX010-hu-NSG-SGM3 mice. Left: Representative images. Right: Quantitative data. *P < 0.001. (C) EGF (20 ng/ml) stimulates the expression of EGR1 in SUM149 cells. (D) Pretreatment with panitumumab mitigates the up-regulation of EGR1 by EGF stimulation in SUM149 cells. (E) Pretreatment with MEK inhibitor trametinib, but not PI3K inhibitor AZD8186, inhibits the up-regulation of EGR1 by EGF stimulation in SUM149 cells. (F) MG-132 (5 μM) treatment mitigates the decrease of EGR1 expression induced by panitumumab (20 μg/ml; top) and erlotinib (1 μM; bottom) treatments in SUM149 cells. (G) EGR1 knockdown reduces the expression of CCL2, CCL20, CXCL5, and IL-8 genes in SUM149 cells. *P < 0.05 and **P < 0.01. (H) Effect of EGF stimulation on the expression of CCL2, CCL20, CXCL5, and IL-8 genes in shCtrl and shEGR1-B and shEGR1-C clones of SUM149 cells. P value of shEGR1 clones compared to shCtrl upon EGF stimulation: *P < 0.001 and ** P = 0.01. (I) EGR1 binds to the promoter region of CCL2, CCL20, CXCL5, and IL-8 genes in SUM149 cells upon EGF stimulation. * P < 0.05. Data are summarized as means ± SD in (A) and (G) to (I). DMSO, dimethyl sulfoxide. Experiments were independently repeated three times with three replicates each time.
To test this hypothesis, we investigated whether EGFR regulates EGR1 in IBC cells. Panitumumab reduced EGR1 protein expression in tumor samples from SUM149-hu-NSG-SGM3 and BCX010-hu-NSG-SGM3 mice (Fig. 4B) and in SUM149 and BCX010 cells (fig. S5B and S5C). EGF stimulation activated EGFR signaling and up-regulated the expression of EGR1, but this effect was abrogated by pretreatment with panitumumab in SUM149 cells (Fig. 4, C and D) and BCX010 cells (fig. S5, D and E). Pretreatment with mitogen-activated protein kinase kinase (MEK) inhibitor trametinib, but not phosphatidylinositol 3-kinase (PI3K) inhibitor AZD8186, inhibited the up-regulation of EGR1 by EGF stimulation in both SUM149 (Fig. 4E) and BCX010 cells (fig. S5F), suggesting that the EGFR pathway regulates EGR1 expression through extracellular signal–regulated kinase (ERK) but not AKT. To understand how the EGFR pathway modulates EGR1 protein levels, we used the proteasome inhibitor MG-132 to examine whether EGFR promotes EGR1 protein stability. We found that MG-132 mitigated the down-regulation of EGR1 expression by panitumumab or EGFR kinase inhibitor erlotinib in SUM149 cells, suggesting that the EGFR pathway regulates the protein stability of EGR1 (Fig. 4F).
Next, we examined whether EGR1 contributes to EGFR-regulated chemokine expression. Depletion of EGR1 in SUM149 and BCX010 cells reduced the expression of CCL2, CCL20, CXCL5, and IL-8 (Fig. 4G and fig. S5G). EGR1 knockdown also reduced the up-regulation of these genes upon EGF stimulation (Fig. 4H), confirming that EGFR promotes the expression of immunosuppressive chemokines via EGR1. Chromatin immunoprecipitation assay confirmed that EGF stimulated the binding of EGR1 to the promoter region of CCL2, CCL20, CXCL5, and IL-8 in SUM149 cells (Fig. 4I), suggesting that EGR1 transcriptionally regulates the expression of these genes in response to EGFR signaling. Together, these results indicate that EGR1 is, at a minimum, one of the major transcription factors involved in creating the immunosuppressive cytokine milieu in IBC and provide a mechanistic link between EGFR signaling and the immunosuppressive TME.
Combination of panitumumab with anti–PD-L1 antibody enhances inhibition of triple-negative IBC tumor growth in humanized mouse models
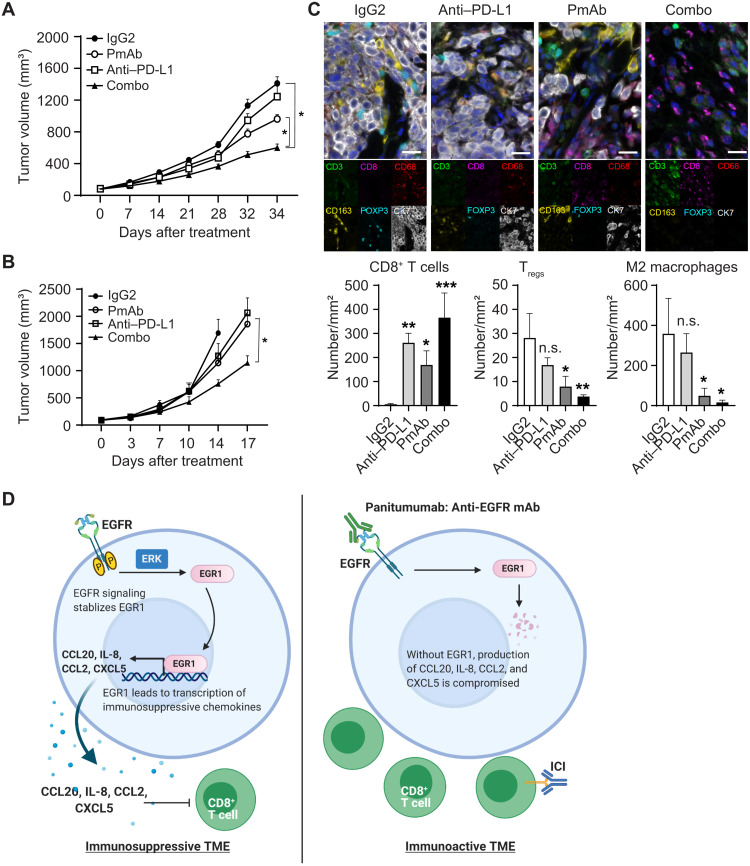
Given that panitumumab induced a shift of the IBC TME from immunosuppressive to immunoactive by regulating chemokine secretion, we reasoned that EGFR inhibition might prime IBC tumors for immune checkpoint inhibition. To test this hypothesis, we combined panitumumab with an anti–PD-L1 antibody and measured the impact of the combination on SUM149 tumor growth in immunocompetent humanized mice. We chose the anti–PD-L1 antibody because we observed more PD-L1–positive macrophages in panitumumab-treated SUM149 humanized mouse tissue than in IgG2-treated SUM149 humanized mouse tissue (fig. S5H). We used a suboptimal therapeutic dose of panitumumab to avoid masking any combinatorial enhancement. The combination treatment inhibited the growth of SUM149 tumors more than either single-drug treatment (Fig. 5A). We observed similar results in BCX010-hu-NSG-SGM3 mice (Fig. 5B). These results suggest that the modulation of the TME by panitumumab enhanced the antitumor efficacy of the ICI in IBC. Analysis of tumor tissue from SUM149-hu-NSG-SGM3 mice showed that panitumumab decreased EGR1 protein expression (fig. S5I) and reduced expression of CCL2, CCL20, CXCL5, and IL-8 (fig. S5J), confirming our in vitro and clinical findings. Multiplexed immunofluorescence staining confirmed increased CD8+ T cells and decreased Tregs and M2 macrophages in tumor tissues from panitumumab-treated SUM149-hu-NSG-SGM3 mice (Fig. 5C), mirroring the findings from the panitumumab/NAC clinical trial (Fig. 2F). Critically, the combination of panitumumab and anti–PD-L1 antibody resulted in a markedly increased presence of CD8+ T cells and a decreased presence of Tregs and M2 macrophages compared with either treatment alone (Fig. 5C). As shown in the proposed model (Fig. 5D), our results confirm that panitumumab converts the IBC TME from immunosuppressive to immunoactive and enhances the efficacy of immune checkpoint inhibition, providing the critical evidence necessary for future clinical trials.
Fig. 5. Synergistic antitumor effect of panitumumab combined with anti–PD-L1 antibody in humanized mouse models.
(A) Tumor growth curves of SUM149 xenograft in humanized mice (SUM149-hu-NSG-SGM3) treated with control IgG2, panitumumab, anti–PD-L1 antibody, and combination. Eight mice per group. *P < 0.05. (B) Tumor growth curves of BCX010 xenograft in humanized mice (BCX010-hu-NSG-SGM3) treated with control IgG2, panitumumab, anti–PD-L1 antibody, and combination. Eight mice per group. *P < 0.001. (C) Changes in CD8+ T cells, Tregs, and M2 macrophages in tissues of each group in SUM149-hu-NSG-SGM3 mice were analyzed by multiplexed immunofluorescence staining. Scale bars, 50 μm. *P < 0.05, **P < 0.01, and ***P < 0.001. (D) Proposed mechanism by which panitumumab treatment remodels the IBC TME. Left: The activation of EGFR signaling stabilizes EGR1, which regulates the expression of chemokines CCL2, CCL20, CXCL5, and IL-8. The secretion of these chemokines creates an immunosuppressive TME, which attracts Tregs and M2 macrophages but inhibits the recruitment of T cells. Right: Panitumumab inhibits EGFR signaling and induces EGR1 degradation, which reduces the secretion of CCL2, CCL20, CXCL5, and IL-8 from IBC cells, changing the TME from immunosuppressive to immunoactive. Data are summarized as means ± SEM in (A) and (B) and means ± SD in (C).
DISCUSSION
This is the first report of a clinically actionable mechanism of sensitization of IBC tumors to ICIs. We show that EGFR is one of the central signaling molecules involved in the maintenance of the immunosuppressive TME in IBC (Fig. 5D). We show that inhibiting EGFR with the humanized antibody panitumumab, which is already in clinical trials for patients with IBC (26), remodels the TME by regulating the expression of chemokines in IBC cells. Furthermore, we present evidence that these chemokines are responsible for the recruitment of immune cells into the tumor milieu, as demonstrated by increased infiltration of CD8+ T cells and decreased infiltration of M2 macrophages and Tregs in vivo following panitumumab treatment. We also provide additional evidence that the EGFR-mediated expression of immunosuppressive chemokines is regulated by EGR1, the stability of which is dependent on EGFR activation.
IBC remains one of the most challenging breast cancers to treat, as the standard-of-care therapies for identical molecular subtypes produce markedly worse outcomes in patients with IBC than in non-IBC patients (49). While the recent approval of pembrolizumab as a frontline treatment for early-stage TNBC will likely shift the therapeutic picture, it is unlikely to markedly affect treatment for patients with IBC, as 60% of them do not have triple-negative disease. Furthermore, preliminary results of our ongoing study of atezolizumab combined with chemotherapy in IBC (NCT03202316) showed that among seven patients treated with atezolizumab and anti-microtubule agent eribulin, only one patient demonstrated a tumor response. This evidence highlights the need to identify new combination strategies for patients with progressive disease and nonresponders. The critical clinical question for IBC, at this moment, based on discovery, is whether strategies are clinically available for maximizing the effect of ICIs and turning IBC into an immunoresponsive tumor. Given the historically high pCR rate in patients with triple-negative IBC following panitumumab/NAC therapy (NCT01036087), a combination of panitumumab with an ICI is the logical next step in the clinical setting. Our findings that panitumumab enhances the therapeutic efficacy of an ICI in a preclinical model warrant further clinical evaluation of this combination strategy in patients with triple-negative IBC and other cancer types.
Up-regulation of EGR1 in IBC patient samples compared to non-IBC patient samples was previously reported (46). However, the role of EGR1 in IBC pathobiology was unclear. The present study partially answers this question by demonstrating that the EGFR pathway regulates the stability of EGR1 and that EGR1 regulates the expression of CCL2, CCL20, CXCL5, and IL-8 in IBC. Because we found that these chemokines play a role in recruiting immunosuppressive M2 macrophages and Tregs, we speculate that EGR1 may play a critical role in IBC tumor progression and possibly tumor resistance by regulating the expression of immunosuppressive chemokines. Identifying additional molecular factors upstream and downstream of EGR1 and dissecting the underlying mechanism of how EGR1 modulates the tumor immune microenvironment will help us further define the role of this pathway in IBC. In addition to providing a mechanistic link between EGFR and the immunosuppressive TME, our finding that EGR1 is the putative transcription factor for the immunosuppressive cytokines offers clinically actionable information for identifying patients likely to benefit from EGFR inhibition. It is critical that EGR1 be studied as a potential biomarker for patients with IBC and for any other patients with cancer being considered for EGFR-targeted therapy.
To our knowledge, we are the first to report that EGFR mediates immunosuppression in IBC and that this effect is largely dependent on EGR1-mediated transcription of immunosuppressive chemokines. This mechanism aligns with previous research implicating tumor-derived chemokines as the critical mediators of tumor immune evasion. For example, CCL2, CCL20, CXCL5, and IL-8 are known to suppress the antitumor immune response by recruiting tumor-associated macrophages, neutrophils, and MDSCs into the TME (39–43, 50). We identified a decrease in expression of these chemokines following panitumumab treatment, which was correlated to impaired recruitment of M2 macrophages and Tregs into the tumor. High expression of immunosuppressive CCL20 is associated with worse overall survival of patients with IBC. Although not significant, we did find a trend that suggests a correlation between the change in CCL2 levels and pCR status following panitumumab/NAC treatment (fig. S4H). Equally important, the work presented here also allows us to identify a group of effector molecules for EGFR efficacy. Our finding that increased CCL4, CCL5, CXCL9, and CXCL10 levels after panitumumab treatment promote migration of CD8+ T cells suggests that targeting EGFR could remodel the TME and consequently enhance tumor control by ICIs. Using clinical samples, we found that high tumor expression of CCL4, CCL5, CXCL9, and CXCL10 is associated with improved metastasis-free survival of patients with IBC (Fig. 3E). In melanoma, the expression of tumor-derived CCL4, CCL5, CXCL9, and CXCL10 was correlated with tumor-infiltrating lymphocytes (45), and the same chemokine signature was associated with infiltration of CD8+ T cells in solid tumors (44, 51). Previous work also showed that EGFR signaling decreased CXCL10 and CCL5 expression and thus reduced CD8+ T cell infiltration in the EGFR-mutated lung adenocarcinomas (52). While further studies are needed on the exact role of individual or combinations of chemokines in IBC, we can envision our results having an immediate effect in clinical research across the spectrum of cancers as we try to identify signatures that may predict the efficacy of ICIs.
Here, we describe the first humanized mouse models of IBC. Until now, the lack of a syngeneic IBC mouse model has prevented examination of the immune system effect in IBC and prevented researchers from studying clinically relevant immunotherapy or immunotherapy combinations before proceeding to clinical trials. The humanized mouse models of IBC that we describe here replicate the clinical effects of panitumumab on the immune cell profile of tumor tissue. Widespread use of these models may allow more rapid identification of the molecular mechanisms that drive IBC and effective therapeutic combinations. That said, the fact that these models were established using allogeneic human leukocyte antigen (HLA) partially matched CD34+ human pluripotent stem cell (HPSC) donors, and IBC tumors may affect the evaluation of antigen-specific T cell responses (53). However, these are the only models available now that allow us to study the critical cross-talk between the IBC tumor and its immune microenvironment. We are establishing a humanized mouse model using CD34+ HPSCs with complete major histocompatibility complex matches with IBC tumor and an IBC transgenic mouse model. We anticipate that these next-generation IBC animal models will address the current limitations and help us further decipher the role of the immune system and the TME in the progression and treatment response of IBC.
With respect to translational implications for other solid tumors, our work aligns with previous reports (52) but provides a likely mechanism by which targeting EGFR can lead to changes in the TME. For example, previous work has shown that EGFR inhibitor erlotinib improves the efficacy of anti–PD-1 antibody in EGFR-mutated non–small cell lung cancer by decreasing CD4+ effector Treg infiltration in the TME (52). In colorectal cancers, anti-EGFR therapy in combination with chemotherapy results in increased cytotoxic T cell infiltration (54), leading some to argue that anti-EGFR treatments should be combined with ICIs in this cancer (55). That said, we are the first to offer a likely mechanism of action for these observations, including the central role of immunosuppressive chemokines and EGR1, both of which can serve as potential biomarkers for patient selection in other tumors. EGR1 specifically seems to have a highly context-dependent role in tumor progression, with both tumor-promoting (56, 57) and tumor-suppressing (58, 59) activities reported in the literature. Therefore, as this strategy is tested in the clinic, a careful analysis of this downstream pathway for both IBC and other cancers will be necessary.
In conclusion, our data identified a previously unknown biological mechanism by which panitumumab remodels the immunosuppressive TME and boosts the antitumor immune response. We reveal that the EGFR signaling–mediated immunosuppressive TME may be one of the reasons for ICI resistance. The combination of panitumumab with an ICI has a more substantial inhibitory effect on tumor growth than ICI alone, warranting further investigation in clinical trials. While the work presented here is IBC specific, inspired by the high need for new therapeutic strategies in this patient population, this mechanism will likely have broad implications across ICI-resistant or ICI-nonresponsive tumors. Last, the chemokines identified in our report warrant further investigation for their roles as predictive biomarkers and therapeutic candidates. Therefore, our work provides critical insights for developing novel combination approaches with enhanced therapeutic efficacy of ICIs.
MATERIALS AND METHODS
Experimental design
The objective of this study was to (i) understand the molecular and immunological mechanisms behind the clinical outcome of an anti-EGFR antibody, panitumumab, combined with NAC in patients with IBC having triple-negative receptor status (NCT01036087) and (ii) evaluate whether EGFR-targeted therapy can prime the tumor for ICI by modulating the IBC TME. IBC SUM149 and BCX010 cells were xenografted in humanized mice to study the impact of panitumumab on human immune cells in vivo. Mice were randomly allocated into control and drug treatment groups, and n = 5, 6, or 8 mice per group were analyzed. scRNA-seq analysis of SUM149 tumors treated with IgG2 and panitumumab in humanized mice was conducted to achieve an unbiased and comprehensive assessment of the impact of panitumumab on the TME in vivo. The changes in the TME in humanized mouse models were analyzed by flow cytometry and multiplexed immunofluorescence staining. The effect of panitumumab on the TME was examined in matched patient tissues collected before and after panitumumab treatment from a phase 2 trial of panitumumab combined with NAC in primary HER2-negative IBC (NCT01036087). The study was approved by the Institutional Review Board of The University of Texas MD Anderson Cancer Center. The association of chemokine gene expression with IBC patient outcomes was analyzed using the mRNA expression data of 137 IBC and 252 non-IBC clinical samples collected within the World IBC Consortium.
Cell lines
Human IBC cell line SUM149 was purchased from Asterand Bioscience Inc. Human IBC cell line BCX010 was provided by F. Meric-Bernstam (MD Anderson Cancer Center). These cells were validated using Short Tandem Repeat (STR) DNA fingerprinting at MD Anderson, and tests for mycoplasma contamination were negative. SUM149 and BCX010 cells were grown in Ham’s F-12 medium supplemented with 10% fetal bovine serum (FBS), insulin (5 μg/ml), hydrocortisone (1 μg/ml), and 1% antibiotic-antimycotic.
Animal studies
Humanized CD34+ NSG-SGM3 female mice aged 14 to 16 weeks were purchased from the Jackson Laboratory. All animal experiments were performed with approval from the MD Anderson Institutional Animal Care and Use Committee. The amount of human CD45+ cells in peripheral blood was measured and confirmed by the Jackson Laboratory. Tumor cells, 4 × 106 SUM149-Luc or 5 × 105 BCX010 cells, mixed with 50% Matrigel matrix, were injected into the mammary fat pad of mice. Mice were randomly grouped when tumor size reached 100 to 150 mm3 and subjected to drug treatment. For single panitumumab treatment, mice were treated with IgG2 or panitumumab at 4 mg/kg via intraperitoneal injection once a week. For panitumumab and anti–PD-L1 antibody combination treatment, mice were treated with control IgG, panitumumab (100 μg/kg), anti–PD-L1 antibody (200 μg per mouse), or the combination at the same doses via intraperitoneal injection once a week. Tumor volume and body weight were measured once a week. Tumor volume was calculated by the formula (L × W2) × 0.5, where L represents tumor length and W represents tumor width. When drug treatment was completed, mice were euthanized, and tumor samples were subjected to single-cell dissociation for fluorescence-activated cell sorting (FACS) analysis and scRNA-seq or collected and flash-frozen and embedded in paraffin blocks.
Single-cell RNA sequencing
SUM149-hu-NSG-SGM3 mice treated with IgG2 and panitumumab were euthanized, and tumor tissues were dissociated into single-cell suspension using the Tumor Dissociation Kit in combination with gentleMACS dissociators according to the manufacturer’s instructions. The 10x Genomics Chromium Single Cell 3′ Library, Gel Bead Kit v3, and Chromium Chip B Single Cell Kit were used to capture cells on the controller, aiming for cell recovery in a range of 5000 to 10,000 cells. According to the manufacturer’s protocol, captured cells were subjected to single-cell gel bead-in-emulsions, reverse transcription, cDNA amplification, and purification. Twenty-five percent of the cDNA was used to generate the library using the Chromium i7 Multiplex Kit. The barcoded libraries were cleaned using AMPure beads and assessed using Agilent D1000 ScreenTape. Ten libraries were pooled at a final concentration of 10 nM and sequenced on a NovaSeq6000 S2 sequencer at the Advanced Technology Genomics Core at MD Anderson.
Raw sequencing data were preprocessed using the Cell Ranger pipeline from 10x Genomics to align and generate quality control metrics. Only cells containing at least 100 gene features and mitochondrial gene counts of less than 20% were used. The R package Seurat was used to normalize unique molecular identifier counts and cluster the RNA expression data (60). The dimensionality was further reduced using the uniform manifold approximation and projection method (61). Cells were classified into specific types on the basis of predefined marker gene expression in the clustered data (62–66). Expression analysis was performed on individual cell types using cells expressing a minimum of 500 genes. Differential expression analysis was performed between treatment groups using DESeq2 (67) after pooling counts across the samples. Significantly differentially expressed genes were defined on the basis of a false discover rate (FDR) cutoff of 0.05 and log fold change of 1. Further downstream pathway exploration was performed by preranked gene set enrichment analysis based on log2 fold change among treatment groups across the cell types using the Hallmark pathway database (68).
FACS analysis of tumor-infiltrating immune cells
Dissociated single cells from tumor tissues were treated with RBC Lysis Buffer to lyse red blood cells. Cells were stained using the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit and stained with fluorochrome-conjugated cell surface markers, including mouse CD45 (mCD45), human CD45 (hCD45), hCD3, hCD4, hCD8, hCD25, and hCD127. The cell pellets were resuspended with 1% FBS in phosphate-buffered saline for FACS on Gallios (Beckman Coulter) and analyzed using Kaluza software (version 2.1.00001.20653). hCD45+ cells were gated from the mCD45− population. CD8+ T cells were defined as CD3+CD8+, and Tregs were defined as CD3+CD4+CD25+CD127−.
Multiplexed immunofluorescence staining and imaging
Tissues from patients with primary HER2-negative IBC were collected from a phase 2 trial of NAC with panitumumab, nab-paclitaxel, and carboplatin followed by an anthracycline-containing regimen (NCT01036087) (26). The expression of human CD3, CD8, CD68, CD163, Forkhead box P3 (FOXP3), CD45RO, granzyme B, and cytokeratin 7 (CK7) in SUM149-hu-NSG-SGM3 or BCX010-hu-NSG-SGM3 tissues or IBC patient tissues was analyzed using the Opal Polaris 7 Color immunohistochemistry kit following the manufacturer’s instructions. Multiplex stained slides were imaged using Vectra 3.0 (PerkinElmer) at ×20 magnification. inForm Tissue Analysis Software version 3.0 (Akoya) was used to analyze images, including cell segmentation, phenotyping, and cell quantitation. Staining and imaging are described in detail in the Supplementary Materials.
In vitro immune cell migration assay
To obtain peripheral blood mononuclear cells (PBMCs), healthy donors’ buffy coats were purchased from Gulf Coast Regional Blood Center. PBMCs were isolated from the buffy coats by density gradient centrifugation with Ficoll-Paque PLUS and then seeded in cell culture plates with RPMI 1640 medium supplemented with 10% FBS. The purification of CD8+ T cells, Tregs, and macrophages from PBMCs and the procedure of migration assay are described in detail in Supplementary Materials.
Cytokine antibody array
The fresh-frozen SUM149 tumors treated by IgG2 and panitumumab in SUM149-hu-NSG-SGM3 mice were dissociated and lysed with radioimmunoprecipitation assay (RIPA) buffer complemented with protease inhibitor and phosphatase inhibitor cocktails. Cytokines and chemokines in lysed tumor samples were detected using a human cytokine antibody array, which contains 80 cytokines and chemokines, following the manufacturer’s protocol. The intensities of signals were quantified by densitometry with ImageJ software (v1.52), and positive controls were used to normalize the results from different membranes. The expression of cytokines/chemokines was quantified and normalized relative to IgG2 control. Cytokines or chemokines with more than 1.5-fold change were subjected to validation with quantitative RT-PCR (qRT-PCR).
RNA isolation and qRT-PCR
Total RNA was extracted from cells or frozen tumor tissues using the RNeasy Mini Kit, and cDNA was generated using the SuperScript II Reverse Transcriptase Kit according to the manufacturer’s instructions. qRT-PCR was performed using Power SYBR Green PCR Master Mix, and transcript levels were normalized to housekeeping gene RPL3 or GAPDH. Primer sequences are described in the Supplementary Materials.
Enzyme-linked immunosorbent assay
SUM149 tumor tissues treated with IgG2 or panitumumab were mechanically dissociated and lysed with RIPA buffer complemented with protease inhibitor and phosphatase inhibitor cocktails. According to the manufacturer’s instructions, the levels of CCL2, CCL20, CXCL5, and IL-8 were quantitatively measured using R&D Systems Human ELISA kits.
EGFR or EGR1 stable or transient knockdown in IBC cells
EGFR stable knockdown clones in SUM149 cells have previously been generated (69). Mission lentiviral transduction particles targeting EGR1 were purchased from Sigma-Aldrich. EGR1 stable knockdown clones in SUM149 cells were generated according to the manufacturer’s instructions. EGR1 expression in BCX010 cells was transiently knocked down using Lipofectamine RNAiMAX reagent according to the manufacturer’s instructions. The detailed information on shRNA and small interfering RNA is described in the Supplementary Materials.
Analysis of expression of chemokines in patients with IBC
We analyzed the mRNA expression data of 137 IBC and 252 non-IBC clinical samples (N = 389) collected within the World IBC Consortium (70). IBC was defined clinically according to international consensus criteria (71). Collection criteria, sample characteristics, and gene expression profiling have been previously described (70), and more details are described in the Supplementary Materials. The expression level of chemokine genes, including CCL4, CCL5, CXCL9, CXCL10, CCL2, CCL20, CXCL5, and IL-8, was analyzed as a continuous value and as a discrete value using median expression level as a cutoff to define the high and low expression. Combined chemokine gene expression was analyzed using a metagene approach defined by the mean of standardized expression of included genes. Combined chemokine metagenes were analyzed as continuous values and discrete values using median value as the cutoff to define high and low expression levels.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8. Data were analyzed by a two-tailed Student’s t test. P < 0.05 was considered significant. The error bars represent the SD or SEM as indicated in each figure legend.
Acknowledgments
We thank the patients and their families for participating in clinical trial NCT01036087. We thank Amgen and Celgene for supporting the clinical trial. Amgen and Celgene reviewed and approved the manuscript and agreed with the decision to submit the manuscript for publication. We thank the staff of the Morgan Welch Inflammatory Breast Cancer Research Program and Clinic for conducting clinical trial NCT01036087. We thank Y. Du for constructive discussion of the project and editing of the manuscript. We thank A. Faust for constructive discussion and editing of the manuscript and for helping us generate Fig. 5D. We thank S. P. Deming and S. C. Patterson (Research Medical Library, The University of Texas MD Anderson Cancer Center) for expert editorial assistance.
Funding: This work was funded by Breast Cancer Research Foundation grant BCRF-17-161 (N.T.U.), NIH grants 1R01CA205043-01A1 and 1R01CA258523-01A1 (N.T.U.), Nylene Eckles Distinguished Professor in Breast Cancer Research (N.T.U.), the Morgan Welch Inflammatory Breast Cancer Research Program and Clinic (N.T.U.), the State of Texas Rare and Aggressive Breast Cancer Research Program (N.T.U.), the IBC Network Foundation (N.T.U.), Emerson Collective Cancer Research Fund grant 689199 (X.W.), Cathy R Smith Immunotherapy Acceleration Award (X.W.), Grand Prix Ruban Rose 2020 (F.B.), and Label Ligue Cancer du Sein 2019 and 2022 (F.B.). The multiplexed imaging scanning and analysis and flow cytometry were performed in the MD Anderson Flow Cytometry and Cellular Imaging Core Facility, which is supported, in part, by the NIH through MD Anderson’s Cancer Center Support Grant, CA016672, and by NCI Research Specialist Award 1R50 CA243707-01A1 (J.K.B.). scRNA-seq analysis was conducted at the CPRIT SINGLE CORE at MD Anderson, supported by CPRIT SINGLE CORE Facilities Grant RP180684.
Author contributions: Conceptualization: X.W., T.S., and N.T.U. Methodology: X.W., T.S., F.B., S.K., T.P., S.J.V.L., J.K.B., E.N.C., J.M.R., X.Z., and N.T.U. Investigation: X.W., T.S., G.C.M., J.W., S.S., F.B., P.F., S.K., L.T.H.P., E.N.C., F.Y., H.M., N.N., V.N.T., T.I., H.B., and Y.S. Visualization: X.W., T.S., and N.T.U. Supervision: X.W., J.W., F.B., X.Z., and N.T.U. Writing (original draft): X.W., T.S., G.C.M., F.B., and N.T.U. Writing (review and editing): X.W., T.S., G.C.M., F.B., E.N.C., J.M.R., D.T., and N.T.U.
Competing interests: N.T.U. has received research funding from Amgen and Celgene. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Upon reasonable request, noncommercially available materials can be provided pending scientific review and a completed material transfer agreement. Requests for the materials should be submitted to N.T.U. scRNA-seq data have been deposited in the NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/; accession number: GSE190559; access code: efwvkqeozjohfaj).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S5
Tables S1 to S4
References
REFERENCES AND NOTES
- 1.P. Schmid, J. Cortes, L. Pusztai, H. McArthur, S. Kummel, J. Bergh, C. Denkert, Y. H. Park, R. Hui, N. Harbeck, M. Takahashi, T. Foukakis, P. A. Fasching, F. Cardoso, M. Untch, L. Jia, V. Karantza, J. Zhao, G. Aktan, R. Dent, J. O’Shaughnessy; KEYNOTE-522 Investigators ,Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020). [DOI] [PubMed] [Google Scholar]
- 2.E. A. Mittendorf, H. Zhang, C. H. Barrios, S. Saji, K. H. Jung, R. Hegg, A. Koehler, J. Sohn, H. Iwata, M. L. Telli, C. Ferrario, K. Punie, F. Penault-Llorca, S. Patel, A. N. Duc, M. Liste-Hermoso, V. Maiya, L. Molinero, S. Y. Chui, N. Harbeck,Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): A randomised, double-blind, phase 3 trial. Lancet 396, 1090–1100 (2020). [DOI] [PubMed] [Google Scholar]
- 3.L. Gandhi, D. Rodriguez-Abreu, S. Gadgeel, E. Esteban, E. Felip, F. De Angelis, M. Domine, P. Clingan, M. J. Hochmair, S. F. Powell, S. Y. Cheng, H. G. Bischoff, N. Peled, F. Grossi, R. R. Jennens, M. Reck, R. Hui, E. B. Garon, M. Boyer, B. Rubio-Viqueira, S. Novello, T. Kurata, J. E. Gray, J. Vida, Z. Wei, J. Yang, H. Raftopoulos, M. C. Pietanza, M. C. Garassino; KEYNOTE-189 Investigators ,Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018). [DOI] [PubMed] [Google Scholar]
- 4.L. Paz-Ares, A. Luft, D. Vicente, A. Tafreshi, M. Gumus, J. Mazieres, B. Hermes, F. Cay Senler, T. Csoszi, A. Fulop, J. Rodriguez-Cid, J. Wilson, S. Sugawara, T. Kato, K. H. Lee, Y. Cheng, S. Novello, B. Halmos, X. Li, G. M. Lubiniecki, B. Piperdi, D. M. Kowalski; KEYNOTE-407 Investigators ,Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N. Engl. J. Med. 379, 2040–2051 (2018). [DOI] [PubMed] [Google Scholar]
- 5.C. Robert, G. V. Long, B. Brady, C. Dutriaux, M. Maio, L. Mortier, J. C. Hassel, P. Rutkowski, C. McNeil, E. Kalinka-Warzocha, K. J. Savage, M. M. Hernberg, C. Lebbe, J. Charles, C. Mihalcioiu, V. Chiarion-Sileni, C. Mauch, F. Cognetti, A. Arance, H. Schmidt, D. Schadendorf, H. Gogas, L. Lundgren-Eriksson, C. Horak, B. Sharkey, I. M. Waxman, V. Atkinson, P. A. Ascierto,Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372, 320–330 (2015). [DOI] [PubMed] [Google Scholar]
- 6.C. Robert, L. Thomas, I. Bondarenko, S. O’Day, J. Weber, C. Garbe, C. Lebbe, J. F. Baurain, A. Testori, J. J. Grob, N. Davidson, J. Richards, M. Maio, A. Hauschild, W. H. Miller Jr., P. Gascon, M. Lotem, K. Harmankaya, R. Ibrahim, S. Francis, T. T. Chen, R. Humphrey, A. Hoos, J. D. Wolchok,Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N. Engl. J. Med. 364, 2517–2526 (2011). [DOI] [PubMed] [Google Scholar]
- 7.J. S. Weber, S. P. D’Angelo, D. Minor, F. S. Hodi, R. Gutzmer, B. Neyns, C. Hoeller, N. I. Khushalani, W. H. Miller Jr., C. D. Lao, G. P. Linette, L. Thomas, P. Lorigan, K. F. Grossmann, J. C. Hassel, M. Maio, M. Sznol, P. A. Ascierto, P. Mohr, B. Chmielowski, A. Bryce, I. M. Svane, J. J. Grob, A. M. Krackhardt, C. Horak, A. Lambert, A. S. Yang, J. Larkin,Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 16, 375–384 (2015). [DOI] [PubMed] [Google Scholar]
- 8.V. Anagnostou, K. N. Smith, P. M. Forde, N. Niknafs, R. Bhattacharya, J. White, T. Zhang, V. Adleff, J. Phallen, N. Wali, C. Hruban, V. B. Guthrie, K. Rodgers, J. Naidoo, H. Kang, W. Sharfman, C. Georgiades, F. Verde, P. Illei, Q. K. Li, E. Gabrielson, M. V. Brock, C. A. Zahnow, S. B. Baylin, R. B. Scharpf, J. R. Brahmer, R. Karchin, D. M. Pardoll, V. E. Velculescu,Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 7, 264–276 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. N. Gettinger, L. Horn, L. Gandhi, D. R. Spigel, S. J. Antonia, N. A. Rizvi, J. D. Powderly, R. S. Heist, R. D. Carvajal, D. M. Jackman, L. V. Sequist, D. C. Smith, P. Leming, D. P. Carbone, M. C. Pinder-Schenck, S. L. Topalian, F. S. Hodi, J. A. Sosman, M. Sznol, D. F. McDermott, D. M. Pardoll, V. Sankar, C. M. Ahlers, M. Salvati, J. M. Wigginton, M. D. Hellmann, G. D. Kollia, A. K. Gupta, J. R. Brahmer,Overall survival and long-term safety of nivolumab (anti-programmed death 1 antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J. Clin. Oncol. 33, 2004–2012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. M. Fares, E. M. Van Allen, C. G. Drake, J. P. Allison, S. Hu-Lieskovan,Mechanisms of resistance to immune checkpoint blockade: Why does checkpoint inhibitor immunotherapy not work for all patients? Am. Soc. Clin. Oncol. Educ. Book 39, 147–164 (2019). [DOI] [PubMed] [Google Scholar]
- 11.S. L. Highfill, Y. Cui, A. J. Giles, J. P. Smith, H. Zhang, E. Morse, R. N. Kaplan, C. L. Mackall,Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 6, 237ra267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.B. Ruffell, L. M. Coussens,Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. Ribas, D. Lawrence, V. Atkinson, S. Agarwal, W. H. Miller Jr., M. S. Carlino, R. Fisher, G. V. Long, F. S. Hodi, J. Tsoi, C. S. Grasso, B. Mookerjee, Q. Zhao, R. Ghori, B. H. Moreno, N. Ibrahim, O. Hamid,Combined BRAF and MEK inhibition with PD-1 blockade immunotherapy in BRAF-mutant melanoma. Nat. Med. 25, 936–940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. J. Sullivan, O. Hamid, R. Gonzalez, J. R. Infante, M. R. Patel, F. S. Hodi, K. D. Lewis, H. A. Tawbi, G. Hernandez, M. J. Wongchenko, Y. Chang, L. Roberts, M. Ballinger, Y. Yan, E. Cha, P. Hwu,Atezolizumab plus cobimetinib and vemurafenib in BRAF-mutated melanoma patients. Nat. Med. 25, 929–935 (2019). [DOI] [PubMed] [Google Scholar]
- 15.M. Cristofanilli, A. U. Buzdar, G. N. Hortobagyi,Update on the management of inflammatory breast cancer. Oncologist 8, 141–148 (2003). [DOI] [PubMed] [Google Scholar]
- 16.S. Dawood, N. T. Ueno, V. Valero, W. A. Woodward, T. A. Buchholz, G. N. Hortobagyi, A. M. Gonzalez-Angulo, M. Cristofanilli,Differences in survival among women with stage III inflammatory and noninflammatory locally advanced breast cancer appear early: A large population-based study. Cancer 117, 1819–1826 (2011). [DOI] [PubMed] [Google Scholar]
- 17.K. W. Hance, W. F. Anderson, S. S. Devesa, H. A. Young, P. H. Levine,Trends in inflammatory breast carcinoma incidence and survival: The surveillance, epidemiology, and end results program at the National Cancer Institute. J. Natl. Cancer Inst. 97, 966–975 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.H. Yamauchi, W. A. Woodward, V. Valero, R. H. Alvarez, A. Lucci, T. A. Buchholz, T. Iwamoto, S. Krishnamurthy, W. Yang, J. M. Reuben, G. N. Hortobagyi, N. T. Ueno,Inflammatory breast cancer: What we know and what we need to learn. Oncologist 17, 891–899 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. G. Allen, Y. C. Chen, J. M. Madden, C. L. Fournier, M. A. Altemus, A. B. Hiziroglu, Y. H. Cheng, Z. F. Wu, L. Bao, J. A. Yates, E. Yoon, S. D. Merajver,Macrophages enhance migration in inflammatory breast cancer cells via RhoC GTPase signaling. Sci. Rep. 6, 39190 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L. Lacerda, B. G. Debeb, D. Smith, R. Larson, T. Solley, W. Xu, S. Krishnamurthy, Y. Gong, L. B. Levy, T. Buchholz, N. T. Ueno, A. Klopp, W. A. Woodward,Mesenchymal stem cells mediate the clinical phenotype of inflammatory breast cancer in a preclinical model. Breast Cancer Res. 17, 42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. R. Wolfe, N. J. Trenton, B. G. Debeb, R. Larson, B. Ruffell, K. Chu, W. Hittelman, M. Diehl, J. M. Reuben, N. T. Ueno, W. A. Woodward,Mesenchymal stem cells and macrophages interact through IL-6 to promote inflammatory breast cancer in pre-clinical models. Oncotarget 7, 82482–82492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.F. Bertucci, N. T. Ueno, P. Finetti, P. Vermeulen, A. Lucci, F. M. Robertson, M. Marsan, T. Iwamoto, S. Krishnamurthy, H. Masuda, P. Van Dam, W. A. Woodward, M. Cristofanilli, J. M. Reuben, L. Dirix, P. Viens, W. F. Symmans, D. Birnbaum, S. J. Van Laere,Gene expression profiles of inflammatory breast cancer: Correlation with response to neoadjuvant chemotherapy and metastasis-free survival. Ann. Oncol. 25, 358–365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. M. Reddy, A. Reuben, S. Barua, H. Jiang, S. Zhang, L. Wang, V. Gopalakrishnan, C. W. Hudgens, M. T. Tetzlaff, J. M. Reuben, T. Tsujikawa, L. M. Coussens, K. Wani, Y. He, L. Villareal, A. Wood, A. Rao, W. A. Woodward, N. T. Ueno, S. Krishnamurthy, J. A. Wargo, E. A. Mittendorf,Poor response to neoadjuvant chemotherapy correlates with mast cell infiltration in inflammatory breast cancer. Cancer Immunol. Res. 7, 1025–1035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N. Cabioglu, Y. Gong, R. Islam, K. R. Broglio, N. Sneige, A. Sahin, A. M. Gonzalez-Angulo, P. Morandi, C. Bucana, G. N. Hortobagyi, M. Cristofanilli,Expression of growth factor and chemokine receptors: New insights in the biology of inflammatory breast cancer. Ann. Oncol. 18, 1021–1029 (2007). [DOI] [PubMed] [Google Scholar]
- 25.B. Corkery, J. Crown, M. Clynes, N. O’Donovan,Epidermal growth factor receptor as a potential therapeutic target in triple-negative breast cancer. Ann. Oncol. 20, 862–867 (2009). [DOI] [PubMed] [Google Scholar]
- 26.N. Matsuda, X. Wang, B. Lim, S. Krishnamurthy, R. H. Alvarez, J. S. Willey, C. A. Parker, J. Song, Y. Shen, J. Hu, W. Wu, N. Li, G. V. Babiera, J. L. Murray, B. K. Arun, A. M. Brewster, J. M. Reuben, M. C. Stauder, C. M. Barnett, W. A. Woodward, H. T. C. Le-Petross, A. Lucci, S. M. DeSnyder, D. Tripathy, V. Valero, N. T. Ueno,Safety and efficacy of panitumumab plus neoadjuvant chemotherapy in patients with primary HER2-negative inflammatory breast cancer. JAMA Oncol. 4, 1207–1213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.F. Ishikawa, M. Yasukawa, B. Lyons, S. Yoshida, T. Miyamoto, G. Yoshimoto, T. Watanabe, K. Akashi, L. D. Shultz, M. Harada,Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chainnull mice. Blood 106, 1565–1573 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.L. D. Shultz, M. A. Brehm, J. V. Garcia-Martinez, D. L. Greiner,Humanized mice for immune system investigation: Progress, promise and challenges. Nat. Rev. Immunol. 12, 786–798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. P. Huber, J. D. Farrar,Regulation of effector and memory T-cell functions by type I interferon. Immunology 132, 466–474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.A. Le Bon, D. F. Tough,Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14, 432–436 (2002). [DOI] [PubMed] [Google Scholar]
- 31.B. S. Parker, J. Rautela, P. J. Hertzog,Antitumour actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 16, 131–144 (2016). [DOI] [PubMed] [Google Scholar]
- 32.P. Andre, C. Denis, C. Soulas, C. Bourbon-Caillet, J. Lopez, T. Arnoux, M. Blery, C. Bonnafous, L. Gauthier, A. Morel, B. Rossi, R. Remark, V. Breso, E. Bonnet, G. Habif, S. Guia, A. I. Lalanne, C. Hoffmann, O. Lantz, J. Fayette, A. Boyer-Chammard, R. Zerbib, P. Dodion, H. Ghadially, M. Jure-Kunkel, Y. Morel, R. Herbst, E. Narni-Mancinelli, R. B. Cohen, E. Vivier,Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 175, 1731–1743.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Y. Zhu, B. L. Knolhoff, M. A. Meyer, T. M. Nywening, B. L. West, J. Luo, A. Wang-Gillam, S. P. Goedegebuure, D. C. Linehan, D. G. DeNardo,CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 74, 5057–5069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L. Padilla, S. Dakhel, J. Adan, M. Masa, J. M. Martinez, L. Roque, T. Coll, R. Hervas, C. Calvis, L. Llinas, S. Buenestado, J. Castellsague, R. Messeguer, F. Mitjans, J. L. Hernandez,S100A7: From mechanism to cancer therapy. Oncogene 36, 6749–6761 (2017). [DOI] [PubMed] [Google Scholar]
- 35.P. Sinha, C. Okoro, D. Foell, H. H. Freeze, S. Ostrand-Rosenberg, G. Srikrishna,Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J. Immunol. 181, 4666–4675 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.M. F. Tavazoie, I. Pollack, R. Tanqueco, B. N. Ostendorf, B. S. Reis, F. C. Gonsalves, I. Kurth, C. Andreu-Agullo, M. L. Derbyshire, J. Posada, S. Takeda, K. N. Tafreshian, E. Rowinsky, M. Szarek, R. J. Waltzman, E. A. McMillan, C. Zhao, M. Mita, A. Mita, B. Chmielowski, M. A. Postow, A. Ribas, D. Mucida, S. F. Tavazoie,LXR/apoe activation restricts innate immune suppression in cancer. Cell 172, 825–840.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. D’Cunha, E. Knight Jr., A. L. Haas, R. L. Truitt, E. C. Borden,Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. U.S.A. 93, 211–215 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.V. Rocha-Perugini, J. M. Gonzalez-Granado,Nuclear envelope lamin-A as a coordinator of T cell activation. Nucleus 5, 396–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. L. Chang, J. Miska, D. A. Wainwright, M. Dey, C. V. Rivetta, D. Yu, D. Kanojia, K. C. Pituch, J. Qiao, P. Pytel, Y. Han, M. Wu, L. Zhang, C. M. Horbinski, A. U. Ahmed, M. S. Lesniak,CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory T cells and myeloid-derived suppressor cells. Cancer Res. 76, 5671–5682 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.K. W. Cook, D. P. Letley, R. J. Ingram, E. Staples, H. Skjoldmose, J. C. Atherton, K. Robinson,CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa. Gut 63, 1550–1559 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Z. G. Fridlender, J. Sun, I. Mishalian, S. Singhal, G. Cheng, V. Kapoor, W. Horng, G. Fridlender, R. Bayuh, G. S. Worthen, S. M. Albelda,Transcriptomic analysis comparing tumor-associated neutrophils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PLOS ONE 7, e31524 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. Mei, Y. Liu, N. Dai, M. Favara, T. Greene, S. Jeyaseelan, M. Poncz, J. S. Lee, G. S. Worthen,CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity 33, 106–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.K. O. Osuala, B. F. Sloane,Many roles of CCL20: Emphasis on breast cancer. Postdoc. J. 2, 7–16 (2014). [PMC free article] [PubMed] [Google Scholar]
- 44.D. Dangaj, M. Bruand, A. J. Grimm, C. Ronet, D. Barras, P. A. Duttagupta, E. Lanitis, J. Duraiswamy, J. L. Tanyi, F. Benencia, J. Conejo-Garcia, H. R. Ramay, K. T. Montone, D. J. Powell Jr., P. A. Gimotty, A. Facciabene, D. G. Jackson, J. S. Weber, S. J. Rodig, S. F. Hodi, L. E. Kandalaft, M. Irving, L. Zhang, P. Foukas, S. Rusakiewicz, M. Delorenzi, G. Coukos,Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell 35, 885–900.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.H. Harlin, Y. Meng, A. C. Peterson, Y. Zha, M. Tretiakova, C. Slingluff, M. McKee, T. F. Gajewski,Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 69, 3077–3085 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.I. Bieche, F. Lerebours, S. Tozlu, M. Espie, M. Marty, R. Lidereau,Molecular profiling of inflammatory breast cancer: Identification of a poor-prognosis gene expression signature. Clin. Cancer Res. 10, 6789–6795 (2004). [DOI] [PubMed] [Google Scholar]
- 47.M. Gururajan, A. Simmons, T. Dasu, B. T. Spear, C. Calulot, D. A. Robertson, D. L. Wiest, J. G. Monroe, S. Bondada,Early growth response genes regulate B cell development, proliferation, and immune response. J. Immunol. 181, 4590–4602 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.S. B. McMahon, J. G. Monroe,The role of early growth response gene 1 (egr-1) in regulation of the immune response. J. Leukoc. Biol. 60, 159–166 (1996). [DOI] [PubMed] [Google Scholar]
- 49.H. Masuda, T. M. Brewer, D. D. Liu, T. Iwamoto, Y. Shen, L. Hsu, J. S. Willey, A. M. Gonzalez-Angulo, M. Chavez-MacGregor, T. M. Fouad, W. A. Woodward, J. M. Reuben, V. Valero, R. H. Alvarez, G. N. Hortobagyi, N. T. Ueno,Long-term treatment efficacy in primary inflammatory breast cancer by hormonal receptor- and HER2-defined subtypes. Ann. Oncol. 25, 384–391 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. M. Lesokhin, T. M. Hohl, S. Kitano, C. Cortez, D. Hirschhorn-Cymerman, F. Avogadri, G. A. Rizzuto, J. J. Lazarus, E. G. Pamer, A. N. Houghton, T. Merghoub, J. D. Wolchok,Monocytic CCR2+ myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 72, 876–886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.J. P. Bottcher, E. Bonavita, P. Chakravarty, H. Blees, M. Cabeza-Cabrerizo, S. Sammicheli, N. C. Rogers, E. Sahai, S. Zelenay, E. S. C. Reis,NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.E. Sugiyama, Y. Togashi, Y. Takeuchi, S. Shinya, Y. Tada, K. Kataoka, K. Tane, E. Sato, G. Ishii, K. Goto, Y. Shintani, M. Okumura, M. Tsuboi, H. Nishikawa,Blockade of EGFR improves responsiveness to PD-1 blockade in EGFR-mutated non-small cell lung cancer. Sci. Immunol. 5, eaav3937 (2020). [DOI] [PubMed] [Google Scholar]
- 53.L. D. Shultz, Y. Saito, Y. Najima, S. Tanaka, T. Ochi, M. Tomizawa, T. Doi, A. Sone, N. Suzuki, H. Fujiwara, M. Yasukawa, F. Ishikawa,Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r gamma(null) humanized mice. Proc. Natl. Acad. Sci. U.S.A. 107, 13022–13027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Van den Eynde, B. Mlecnik, G. Bindea, T. Fredriksen, S. E. Church, L. Lafontaine, N. Haicheur, F. Marliot, M. Angelova, A. Vasaturo, D. Bruni, A. Jouret-Mourin, P. Baldin, N. Huyghe, K. Haustermans, A. Debucquoy, E. Van Cutsem, J. F. Gigot, C. Hubert, A. Kartheuser, C. Remue, D. Leonard, V. Valge-Archer, F. Pages, J. P. Machiels, J. Galon,The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell 34, 1012–1026.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 55.T. Liang, W. Tong, S. Ma, P. Chang,Standard therapies: Solutions for improving therapeutic effects of immune checkpoint inhibitors on colorectal cancer. Onco. Targets. Ther. 9, 1773205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.L. Li, A. H. Ameri, S. Wang, K. H. Jansson, O. M. Casey, Q. Yang, M. L. Beshiri, L. Fang, R. G. Lake, S. Agarwal, A. N. Alilin, W. Xu, J. Yin, K. Kelly,EGR1 regulates angiogenic and osteoclastogenic factors in prostate cancer and promotes metastasis. Oncogene 38, 6241–6255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Z. Ma, X. Gao, Y. Shuai, X. Wu, Y. Yan, X. Xing, J. Ji,EGR1-mediated linc01503 promotes cell cycle progression and tumorigenesis in gastric cancer. Cell Prolif. 54, e12922 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.T. Mohamad, N. Kazim, A. Adhikari, J. K. Davie,EGR1 interacts with TBX2 and functions as a tumor suppressor in rhabdomyosarcoma. Oncotarget 9, 18084–18098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L. Su, H. Cheng, A. V. Sampaio, T. O. Nielsen, T. M. Underhill,EGR1 reactivation by histone deacetylase inhibitors promotes synovial sarcoma cell death through the PTEN tumor suppressor. Oncogene 29, 4352–4361 (2010). [DOI] [PubMed] [Google Scholar]
- 60.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck III, Y. Hao, M. Stoeckius, P. Smibert, R. Satija,Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.E. Becht, L. McInnes, J. Healy, C. A. Dutertre, I. W. H. Kwok, L. G. Ng, F. Ginhoux, E. W. Newell,Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 62.M. A. Al Barashdi, A. Ali, M. F. McMullin, K. Mills,Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J. Clin. Pathol. 74, 548–552 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.C. Chen, D. Nadal, S. A. Cohen, E. Schläpfer, B. K. Mookerjee, A. Vladutiu, M. W. Stinson, P. L. Ogra, B. Albini,Direct demonstration of engraftment of human peripheral blood leukocytes in SCID mice. Int. Arch. Allergy Immunol. 97, 295–300 (1992). [DOI] [PubMed] [Google Scholar]
- 64.F. O. Martinez, S. Gordon,The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.I. Tirosh, B. Izar, S. M. Prakadan, M. H. Wadsworth II, D. Treacy, J. J. Trombetta, A. Rotem, C. Rodman, C. Lian, G. Murphy, M. Fallahi-Sichani, K. Dutton-Regester, J. R. Lin, O. Cohen, P. Shah, D. Lu, A. S. Genshaft, T. K. Hughes, C. G. Ziegler, S. W. Kazer, A. Gaillard, K. E. Kolb, A. C. Villani, C. M. Johannessen, A. Y. Andreev, E. M. Van Allen, M. Bertagnolli, P. K. Sorger, R. J. Sullivan, K. T. Flaherty, D. T. Frederick, J. Jane-Valbuena, C. H. Yoon, O. Rozenblatt-Rosen, A. K. Shalek, A. Regev, L. A. Garraway,Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S. T. Trifonova, J. Zimmer, J. D. Turner, C. P. Muller,Diurnal redistribution of human lymphocytes and their temporal associations with salivary cortisol. Chronobiol. Int. 30, 669–681 (2013). [DOI] [PubMed] [Google Scholar]
- 67.M. I. Love, W. Huber, S. Anders,Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.A. Liberzon, C. Birger, H. Thorvaldsdottir, M. Ghandi, J. P. Mesirov, P. Tamayo,The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.X. Wang, M. E. Reyes, D. Zhang, Y. Funakoshi, A. P. Trape, Y. Gong, T. Kogawa, B. L. Eckhardt, H. Masuda, D. A. Pirman Jr., P. Yang, J. M. Reuben, W. A. Woodward, C. Bartholomeusz, G. N. Hortobagyi, D. Tripathy, N. T. Ueno,EGFR signaling promotes inflammation and cancer stem-like activity in inflammatory breast cancer. Oncotarget 8, 67904–67917 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. J. Van Laere, N. T. Ueno, P. Finetti, P. Vermeulen, A. Lucci, F. M. Robertson, M. Marsan, T. Iwamoto, S. Krishnamurthy, H. Masuda, P. van Dam, W. A. Woodward, P. Viens, M. Cristofanilli, D. Birnbaum, L. Dirix, J. M. Reuben, F. Bertucci,Uncovering the molecular secrets of inflammatory breast cancer biology: An integrated analysis of three distinct affymetrix gene expression datasets. Clin. Cancer Res. 19, 4685–4696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.S. Dawood, S. D. Merajver, P. Viens, P. B. Vermeulen, S. M. Swain, T. A. Buchholz, L. Y. Dirix, P. H. Levine, A. Lucci, S. Krishnamurthy, F. M. Robertson, W. A. Woodward, W. T. Yang, N. T. Ueno, M. Cristofanilli,International expert panel on inflammatory breast cancer: Consensus statement for standardized diagnosis and treatment. Ann. Oncol. 22, 515–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R. A. Irizarry, B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, T. P. Speed,Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003). [DOI] [PubMed] [Google Scholar]
- 73.W. E. Johnson, C. Li, A. Rabinovic,Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 74.J. Taminau, D. Steenhoff, A. Coletta, S. Meganck, C. Lazar, V. de Schaetzen, R. Duque, C. Molter, H. Bersini, A. Nowé, D. Y. W. Solís,inSilicoDb: An R/Bioconductor package for accessing human Affymetrix expert-curated datasets from GEO. Bioinformatics 27, 3204–3205 (2011). [DOI] [PubMed] [Google Scholar]
- 75.B. D. Lehmann, J. A. Bauer, X. Chen, M. E. Sanders, A. B. Chakravarthy, Y. Shyr, J. A. Pietenpol,Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.I. Sicking, K. Rommens, M. J. Battista, D. Bohm, S. Gebhard, A. Lebrecht, C. Cotarelo, G. Hoffmann, J. G. Hengstler, M. Schmidt,Prognostic influence of cyclooxygenase-2 protein and mRNA expression in node-negative breast cancer patients. BMC Cancer 14, 952 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.X. Wang, H. Saso, T. Iwamoto, W. Xia, Y. Gong, L. Pusztai, W. A. Woodward, J. M. Reuben, S. L. Warner, D. J. Bearss, G. N. Hortobagyi, M. C. Hung, N. T. Ueno,TIG1 promotes the development and progression of inflammatory breast cancer through activation of Axl kinase. Cancer Res. 73, 6516–6525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Y. Ding, S. Sumitran, J. Holgersson,Direct binding of purified HLA class I antigens by soluble NKG2/CD94 C-type lectins from natural killer cells. Scand. J. Immunol. 49, 459–465 (1999). [DOI] [PubMed] [Google Scholar]
- 79.V. M. Braud, D. S. Allan, C. A. O’Callaghan, K. Soderstrom, A. D’Andrea, G. S. Ogg, S. Lazetic, N. T. Young, J. I. Bell, J. H. Phillips, L. L. Lanier, A. J. McMichael,HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 80.S. Lazetic, C. Chang, J. P. Houchins, L. L. Lanier, J. H. Phillips,Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J. Immunol. 157, 4741–4745 (1996). [PubMed] [Google Scholar]
- 81.N. Lee, M. Llano, M. Carretero, A. Ishitani, F. Navarro, M. Lopez-Botet, D. E. Geraghty,HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. U.S.A. 95, 5199–5204 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.P. Cheng, C. A. Corzo, N. Luetteke, B. Yu, S. Nagaraj, M. M. Bui, M. Ortiz, W. Nacken, C. Sorg, T. Vogl, J. Roth, D. I. Gabrilovich,Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 205, 2235–2249 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.S. D. Blackburn, H. Shin, W. N. Haining, T. Zou, C. J. Workman, A. Polley, M. R. Betts, G. J. Freeman, D. A. Vignali, E. J. Wherry,Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10, 29–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.L. Monney, C. A. Sabatos, J. L. Gaglia, A. Ryu, H. Waldner, T. Chernova, S. Manning, E. A. Greenfield, A. J. Coyle, R. A. Sobel, G. J. Freeman, V. K. Kuchroo,Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 415, 536–541 (2002). [DOI] [PubMed] [Google Scholar]
- 85.L. Dan, L. Liu, Y. Sun, J. Song, Q. Yin, G. Zhang, F. Qi, Z. Hu, Z. Yang, Z. Zhou, Y. Hu, L. Zhang, J. Ji, X. Zhao, Y. Jin, M. A. McNutt, Y. Yin,The phosphatase PAC1 acts as a T cell suppressor and attenuates host antitumor immunity. Nat. Immunol. 21, 287–297 (2020). [DOI] [PubMed] [Google Scholar]
- 86.R. Toribio-Fernandez, V. Zorita, V. Rocha-Perugini, S. Iborra, G. Martinez Del Hoyo, R. Chevre, B. Dorado, D. Sancho, F. Sanchez-Madrid, V. Andres, J. M. Gonzalez-Granado,Lamin A/C augments Th1 differentiation and response against Vaccinia virus and Leishmania major. Cell Death Dis. 9, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.D. Bogunovic, M. Byun, L. A. Durfee, A. Abhyankar, O. Sanal, D. Mansouri, S. Salem, I. Radovanovic, A. V. Grant, P. Adimi, N. Mansouri, S. Okada, V. L. Bryant, X. F. Kong, A. Kreins, M. M. Velez, B. Boisson, S. Khalilzadeh, U. Ozcelik, I. A. Darazam, J. W. Schoggins, C. M. Rice, S. Al-Muhsen, M. Behr, G. Vogt, A. Puel, J. Bustamante, P. Gros, J. M. Huibregtse, L. Abel, S. Boisson-Dupuis, J. L. Casanova,Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.V. Iglesias-Guimarais, T. Ahrends, E. de Vries, K. P. Knobeloch, A. Volkov, J. Borst,IFN-stimulated gene 15 is an alarmin that boosts the CTL response via an innate, NK cell-dependent route. J. Immunol. 204, 2110–2121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S5
Tables S1 to S4
References