Abstract
Artificial intelligence (AI) is an unstoppable force that is starting to permeate all aspects of our society as part of the revolution being brought into our lives (and into medicine) by the digital era, and accelerated by the current COVID-19 pandemic. As the population ages and developing countries move forward, AI-based systems may be a key asset in streamlining the screening, staging, and treatment planning of sight-threatening eye conditions, offloading the most tedious tasks from the experts, allowing for a greater population coverage, and bringing the best possible care to every patient.
This paper presents a review of the state of the art of AI in the field of ophthalmology, focusing on the strengths and weaknesses of current systems, and defining the vision that will enable us to advance scientifically in this digital era. It starts with a thorough yet accessible introduction to the algorithms underlying all modern AI applications. Then, a critical review of the main AI applications in ophthalmology is presented, including diabetic retinopathy, age-related macular degeneration, retinopathy of prematurity, glaucoma, and other AI-related topics such as image enhancement. The review finishes with a brief discussion on the opportunities and challenges that the future of this field might hold.
Keywords: Artificial intelligence, Machine learning, Deep learning, Ophthalmology, Retina, Optical coherence tomography, Age-related macular degeneration, Diabetic retinopathy, Glaucoma, Retinopathy of prematurity
1. Introduction
Artificial Intelligence (AI) has experienced unparalleled growth in recent years, excelling at cognitive tasks that computers were never thought capable of performing. In the field of ophthalmology, these techniques find a particularly good fit. Firstly, the success of AI relies on having vast amounts of data, which high-incidence conditions such as diabetic retinopathy (DR) [1] or age-related macular degeneration (AMD) readily provide. Secondly, one of the most mature AI sub-fields is image recognition, and eye fundus images or optical coherence tomography (OCT) are widely adopted, low-cost, non-intrusive imaging modalities, which show huge potential for automatic analysis and quantification.
At a global level, there are several key challenges in ophthalmology that AI can help overcome. Ongoing population aging means that the incidence of conditions such as AMD and DR (along with diabetes [2]) will only continue to rise, hence posing an ever-increasing burden on the already saturated healthcare systems of the world, with the last straw being the COVID-19 pandemic. This is especially relevant for Low- and Middle-Income Countries (LMIC) [1], where such systems are more brittle and there are not enough trained specialists [3]. Furthermore, while retinopathy of prematurity (ROP) only affects extremely premature infants in high-income countries, in LMICs it affects older children and is experiencing a rise in incidence due the better critical care for premature babies [4]. In this context, AI-based systems can be extremely useful in streamlining the screening, staging, and treatment planning of such conditions, offloading the most tedious tasks from the experts, allowing for a greater population coverage, and bringing the best possible care to every patient.
In practice, AI systems have already shown performances equal or above expert levels for DR grading [5,6,7], AMD grading [8], and general diagnosis from OCT images [9]. Not only that, in 2018 the U.S. Food and Drug Administration (FDA) approved the IDx-DR [10], an AI-based system for DR screening, and the first FDA-authorized autonomous AI diagnostic system in any field of medicine. Furthermore, the advent of genetic testing [11] and the ubiquity of Electronic Health Records (EHR) are paving the way for a fully personalized healthcare, in which an algorithm will decide the optimal treatment and dosage holistically [12] based on all the available patient information.
In the next two to five years, the field of ophthalmology (and many others) will be deeply transformed by the universal adoption of these technologies [13]. It is therefore crucial for the clinicians to have a solid understanding of the core algorithms that are fueling this revolution (as it is crucial for the data scientists to understand the underlying medical problem too). Hence, a significant effort has been made in Section 2 to introduce the key concepts and algorithms underlying most publications in the field. Section 3 will present a summary of the main lines of research, focusing on the observed trends. Finally, Section 4 will discuss the main challenges and opportunities that will likely shape the future of AI and ophthalmology.
2. The algorithms powering the AI revolution
AI is a very loosely used term, which encompasses many different fields with a shared purpose of developing systems able to manifest intelligent behaviors. Frequently, however, AI is used to refer to Machine Learning (ML), which is a sub-field of AI that studies algorithms able to learn from experience. Through this section, an attempt will be made to introduce and demystify the few ML algorithms and ideas hiding behind all the buzzwords and latest research using concrete examples from the field of ophthalmology.
2.1. Overview
Section 2.2 will begin with a very brief dive into the history and recent achievements of AI and ML in general. Then, Section 2.3 will introduce some basic ML concepts, and Sections 2.3 and 2.4 will present the Linear Regression (LR) and Logistic Regression algorithms, which are the simplest kind of ML models and the basis for NNs. As it will be explained in Section 2.5, each neuron of a NN is just like a LR followed by an activation function. Like physiological neurons, artificial neurons receive input signals and, through a simple internal calculation, generate an output signal. In the brain, many simple neurons can be connected to achieve a very complex and capable network; likewise, in artificial NNs, neurons are arranged in layers and, as more layers are added, the NN becomes deeper and the knowledge it can represent becomes more complex, hence the field now known as Deep Learning (DL).
Several modifications to this basic structure have been proposed, such as Convolutional Neural Networks (CNNs) (Section 2.6), which are specifically designed to deal with imaging data, enabling, for instance, the detection, segmentation, and classification of cells in a histopathological image as being either cancerous or not, or the diagnosis of retinal diseases from an OCT; or Recurrent Neural Networks (RNNs) (Section 2.7), which allow us to deal with sequential (e.g.: transient) data, such as that from Electronic Health Records (EHRs). For completeness, Sections 2.8 and 2.9 will present the Decision Tree (DT) algorithm, ensembling methods, and the basics of Natural Language Processing (NLP).
2.2. Brief history and recent advancements
The field of ML started around the 1950s, with the invention of the precursors of the current Neural Networks (NNs) and the Gradient Descent (GD) algorithm, which is used to train them. Over the years, several key discoveries ensued thanks to the work of researchers such as Geoffrey Hinton, Yann LeCun, and Yoshua Benigio, but it was not until the early 2010s when the true revolution began, arguably due to the confluence of three main factors: the theoretical breakthroughs that allowed to train deep NNs, the explosion of available data for training them, and the rise of Graphics Processing Units (GPU) computing which made the training procedure feasible in terms of time cost.
The pivotal point was the proposal of the AlexNet NN architecture [14], which won the 2012 ImageNet [15] image classification competition by a large margin. In 2015 Deepmind's AlphaGo [16] was able to beat the world champion in Go, a complex game requiring very-long-term planning. In 2019, OpenAI published the GPT2 [17] language model, which was trained on a huge corpus of text scraped from the internet and was able to answer general questions and generate human-indistinguishable text given a prompt. Lastly, in 2020, Deepmind's AlphaFold 2 [18] was able to achieve an unparalleled accuracy at predicting protein structure, bringing it very close to experimental techniques, and being labeled by many as the biggest discovery in computational biology in the last decades [19]. Despite their relatively recent history, today NNs are found everywhere, powering a plethora of every-day applications including text, voice and image recognition, stock market forecast, language translation, fraud prevention, autonomous driving, genetic analysis, disease diagnosis, and many more [20].
2.3. Basic concepts
Supervised Learning, the reigning paradigm inside ML, is based on the diagram presented in Fig. 1 . An input (e.g.: a fundus image) is fed to a model which transforms it into an output (e.g.: DR grade). Notice that represents the output of the model, while is the ground truth label. If the model is perfect, then .
Fig. 1.
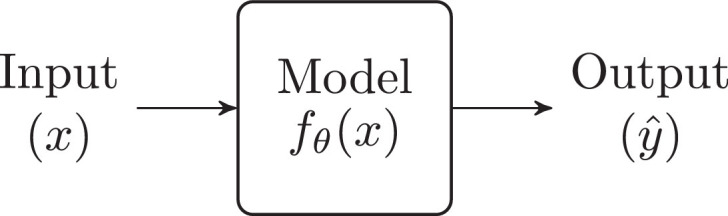
Supervised learning.
Inside the model, is a parametric function with parameters (which are just some numbers). This means that the transformation it performs on the input to build the output depends on the value of its parameters. The core concept behind Supervised Learning is the following: these parameters , rather than being defined by hand, are learned autonomously by the model, by training on many examples of (,) pairs. During the training process, is modified so that can model the transformation from to as accurately as possible. Once the training finishes, the parameters reach their optimal value, and can then be used to infer (predict) the output given any input .
2.4. Linear regression
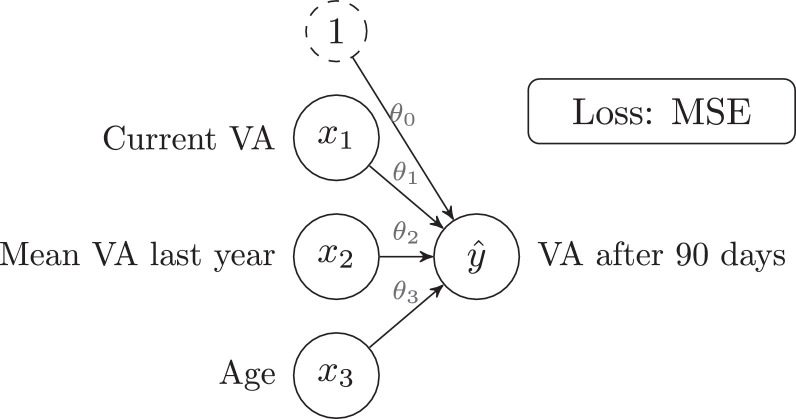
LR is one of the simplest Supervised Learning algorithms and the primary building block for NNs. As a practical example, in Rohm and coworkers [18] the authors train a LR model on the task of predicting “visual acuity (VA) after 90 days” (this is the output , measured in letters) for a patient with AMD, given some features such as “current VA”, “mean VA last year” and “zge” (these are the inputs and ):
-
•
: Current VA
-
•
: Mean VA last year
-
•
: Age
-
•
: VA after 90 days
Note that many more input features are used in the original paper, but only those three will be considered here for simplicity. It must be noted that the authors use Lasso regression, which is a regularized variant of the vanilla LR (regularization is introduced in Section 2.5).
Eq. 1 defines the LR model, where and are the parameters of the model. For instance, after training the model, if the values for the parameters were: and , then the model would have learned the following: “VA after 90 days" equals the “Current VA" minus 0.5 letters.
| (1) |
The objective of the training procedure is to find the values of that make best able to model the relationship between and . In ML, this is done by looking at many examples (653 patients are used in the paper) and learning from them. SGD [9], and its variants, is the most commonly used algorithm for training NNs. Training algorithms are usually known as optimizers, as they optimize (minimize) the error by changing the value of the parameters.
SGD starts by setting the values of the parameters to a random value. Then, it iteratively (i.e., repeatedly) takes a batch (i.e., several) of training samples computes the Mean Squared Error (MSE) between (the prediction obtained by using the model: ) and (the ground truth) and modifies the value of the parameters slightly in an attempt to reduce that error. MSE is defined as so that when equals the error is zero and, otherwise, the error grows quadratically as the distance between and increases. At the end of the training, the error should have been reduced and should be approximately equal to for all training samples.
The average performance of the model on all the samples is reported through metrics. In Rohm et al. [11], the authors report a Mean Absolute Error (MAE) of 5.5 letters and a Root Mean Square Error (RMSE) of 9 letters (when considering patients with only one previous visit). The definition and interpretation of these metrics can be found in Table 1 . Fig. 2 shows a graphical representation of the LR model defined in Eq. 1.
Table 1.
Comparison of metrics for regression, classification, and segmentation problems.
 |
TP = true positives; FP = false positives; TN = true negatives; FN = false negatives.
Fig. 2.
Linear regression model.
The simplicity of the LR model allows for a direct interpretation of the trained parameters. In this paper, and , which can be interpreted as: “VA after 90 days” will be high if “Mean VA last year” has been high, but lower than “Current VA”; also, “VA after 90 days” does not depend on “Age”.
Lastly, before training any kind of ML algorithm, a preprocessing technique known as data normalization (or standardization) is almost always employed. It consists in applying Eq. 2 to each input feature, where represents the feature of the input (e.g.: current VA), is its mean, and its standard deviation. This formula allows to equalize the relative importance of the features; otherwise, a feature with large values (such as “age”, which could have a value of 90), would have an a priori higher importance than a feature such as “current VA”, which has much smaller values.
| (2) |
2.5. Linear classification: logistic regression
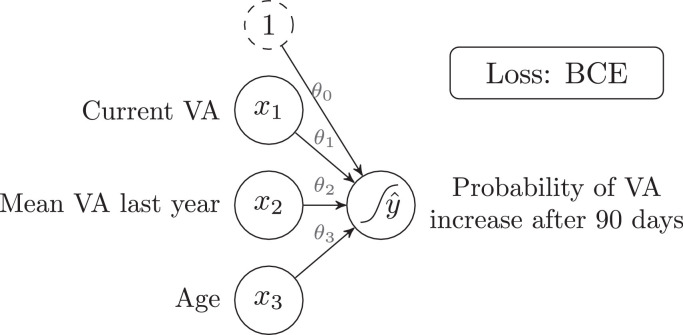
Consider again the previous problem, with the same inputs but a different output: “VA increases after 90 days". This is now a classification task with two possible outcomes: “VA increases after 90 days” or “VA does not increase after 90 days”, hence called a binary (two-class) classification task. Both these outputs can be numerically encoded as follows:
-
•
0: “VA does not increase after 90 days”
-
•
1: “VA increases after 90 days”
This encoding should be interpreted as the “Probability of VA increasing after 90 days". With this setup, LR could be used to learn to predict the output (which is now either 0 or 1). However, the output of the LR equation is unbounded ( can take any value, even above one or below zero), which is meaningless in the context of probabilities. To restrain to always take values between zero and one, a sigmoid () activation function must be applied to the output of Eq. 1, as shown in Eq. 3. This algorithm is called logistic regression, and it is essentially a LR with the addition of the sigmoid function to saturate the output between zero and one.
| (3) |
Finally, even if the MSE loss from LR could be used, a more appropriate loss for binary classification problems is Binary Cross-Entropy (BCE). Fig. 3 shows a graphical representation of the Logistic Regression model defined in Eq. 3. An S-shaped curve has been added to the output node to indicate that a sigmoid function is applied to that node. For multi-class classification problems, softmax () is used as activation function, and Multi-Class Cross-Entropy as loss.
Fig. 3.
Logistic regression model.
2.6. Non-linear regression and classification: feed forward neural networks
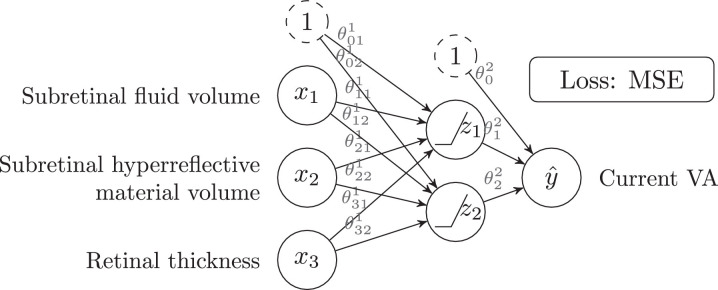
Unlike the previous algorithms, Feed Forward Neural Networks (FFNN, also known as multilayer perceptrons) can model complex non-linear relationships between inputs and outputs. By way of example, Aslam et al. [21] uses a FFNN to infer the “current VA" of a patient with AMD (output ) given some features extracted from an OCT, such as: “subretinal fluid volume”, “subretinal hyperreflective material volume” and “retinal thickness” (inputs ). Fig. 4 shows the structure of a NN similar to the one used in this paper, while Eqs. 4-6 define its behavior. A FFNN is no more than a stack of LR layers with activation functions in between. and are intermediate features that the NN has learned, and the output is just a linear regression over these intermediate features.
| (4) |
| (5) |
| (6) |
Fig. 4.
Feed forward neural network for regression with a two-neuron hidden layer.
The FFNN of Fig. 4 has one hidden layer with two neurons (or units) and uses ReLU () as activation function. The NN from the mentioned paper is similar, but instead employs a single ten-neuron hidden layer, sigmoid activation function, and considers 16 inputs (instead of just three). The number of hidden layers, the number of neurons in each layer, or the choice of the activation function (sigmoid, ReLU, etc.) are all called hyperparameters of the model. On one hand, they are similar to the parameters (also called weights) in the sense that their values influence the final performance of the model (e.g.: the larger the number of layers, the more complex the relationships that the NN will be able to learn). On the other hand, unlike the parameters, they are not trained by SGD and, instead, they must be manually chosen beforehand. Other notable examples of hyperparameters are the batch size, the learning rate (which controls the speed of the SGD algorithm), the choice of the input features, and, in general, any decision that may affect the performance of the model. NNs with many layers are known as Deep Neural Networks and define the sub-field of ML known as Deep Learning.
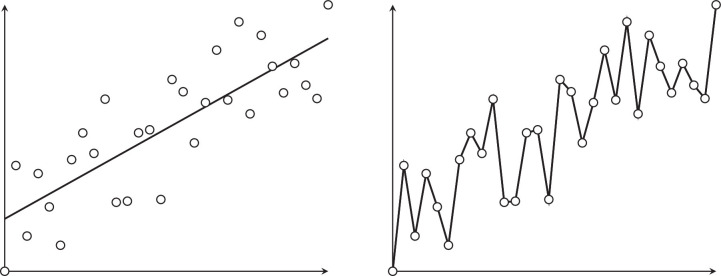
NNs have an enormous modeling power, and a sufficiently large FFNN (with many layers and many neurons per layer) could theoretically learn any dataset to perfection. However, such a model is likely to fail when used on data that it has not been trained with. This problem is known as overfitting (Fig. 5).
Fig. 5.
Good fitting (left) compared to overfitting (right).
It is of utmost importance to detect and control overfitting, otherwise, a seemingly excellent model could perform very poorly when tested on real-world data. For this reason, when developing an AI model, the available data is usually divided into three subsets:
-
•
Train set ( of the data): Used to train the parameters (weights) of the model through SGD.
-
•
Validation set ( of the data): After training on the train set, the model is evaluated on the validation set. Then, the hyperparameters of the model are manually tweaked to try to improve the performance and the model is retrained. This is an iterative process that usually ends when the performance on the validation set cannot be further improved.
-
•
Test set or external validation set ( of the data): Kept secret until both the parameters and the hyperparameters are considered final. Then, the actual performance of the model is evaluated on this set.
In Aslam and coworkers [21], out of 1210 OCTs, 847 (70%) are used for training, 182 (15%) for validation, and 182 (15%) for test. The RMSE on the train and test sets is 8.18 and 8.21 letters, respectively. The better performance on the train set may evidence a very slight degree of overfitting.
As an alternative to this three-way splitting procedure, -fold Cross-Validation (CV) consists in taking disjoint subsets of similar size, training the same model times on of them, and validating with the remaining one. The CV performance is computed as the average validation performance on the validation folds. Nevertheless, a properly independent test set should also be used in addition to this technique, unless no hyperparameter tuning is performed on the validation set (in which case there is no potential for overfitting). In any case, any papers where no such test set is employed should be interpreted with caution. CV is used in works such as Cao and coworkers [22], Arsalan and coworkers [23], Schmidt-Erfurth and coworkers [24], or von der Emde and coworkers [25].
To combat overfitting there exist a set of techniques collectively known as regularization. For instance, regularization adds the absolute value of the parameters to the loss, so that, besides minimizing the error, the training procedure also tries to minimize the magnitude of the parameters. A LR model with regularization is known as Lasso regression.
2.7. Convolutional neural networks
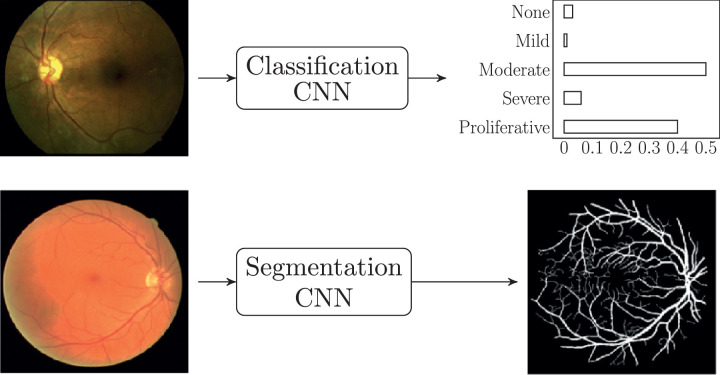
CNNs are NNs that have been modified to better deal with image data. Due to their excellent performance on image recognition tasks, and the ubiquity of imaging in the ophthalmological praxis, they are used in most of the research papers covered in this review. There are two main tasks that CNNs are designed to perform: classification and segmentation. As shown in Fig. 6, both types of CNNs take an image as input (e.g.: a fundus image); however, a classification CNN produces a categorical output (e.g.: a DR rating), while a segmentation CNN produces another image as output (e.g.: a retinal vessel mask).
Fig. 6.
Types of CNNs: classification (above) and segmentation (below). Images from Sayres et al. [69] and Arsalan et al. [23].
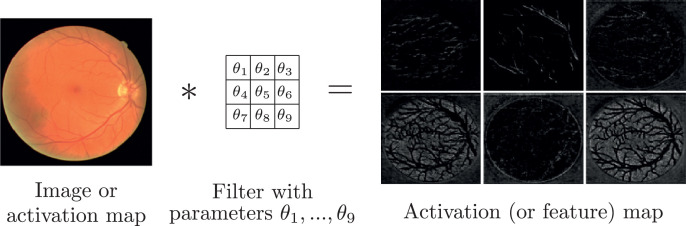
The main building block of any CNN is the convolution, which is a mathematical operation that takes an image as input and applies a filter to it to produce an activation map (also called feature map) as output (Fig. 7 ). This filter is usually a simple matrix of parameters which can be learned through SGD, thus enabling the CNN to learn by itself the filters that produce the feature maps that are optimal for solving a particular problem. Furthermore, feature maps can have several channels (just like a fundus image has three channels: red, green, and blue), each channel representing a different feature of the input image. Finally, feature maps can be further convolved with new filters, which allows to stack a series of convolutions, hence creating a whole CNN.
Fig. 7.
Convolution: an image (or an activation map) is convolved with a filter (with learnable parameters θ), to produce an activation map (also called feature map). Images from Arsalan et al. [23].
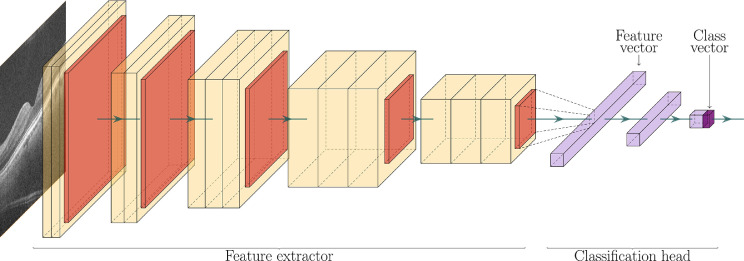
In Antony et al. [26], the authors use a CNN architecture known as VGG16 [27] to diagnose AMD from OCT A-scans (a binary classification problem). Fig. 8 summarizes this architecture. Each box represents an activation map, which is obtained as a result of applying a convolution and an activation function (ReLU in this case) to the previous feature map. The arrows in between boxes represent a downscaling operation (max-pooling in this case), which reduces the resolution of the preceding feature map. The first part of the CNN is known as the encoder since it encodes all the important features of the image into the last feature map. This last feature map is then flattened into a feature vector (a list of numbers), which goes through the classification head. This classification head is just a FFNN (Section 2.5) which takes a feature vector as input and produces a single number as output: the probability of the patient having AMD. Therefore, each layer of the classification head is just a layer from a standard NN.
Fig. 8.
VGG16 architecture. OCT image from Antony et al. [26].
To summarize, CNNs work by filtering (convolving) an input image with learnable filters and reducing its resolution gradually, until a feature vector is obtained. Then, this feature vector is fed through a FFNN to obtain the classification label. Section 2.5 used Aslam and coworkers [21] as an example where the authors employed a FFNN to infer the “current VA” of a patient with AMD given some features extracted from an OCT, such as: “subretinal fluid volume,” “subretinal hyperreflective material volume” and “retinal thickness.” However, it could be argued that this choice of features is somewhat arbitrary, and maybe other features would have yielded better results. In contrast, when using a CNN, it is the CNN that learns by itself the optimal features to choose and encodes them into the feature vector, which is then passed to a FFNN for classification. This simple example shows that the difference between classical ML and DL is more than just the use of deep NNs (NNs with many layers), but also the fact that the features are not chosen by hand, but instead learned by the algorithm.
CNN architectures can be tailor-designed by an expert by combining the basic building blocks (convolution, max-pooling, activation function, etc.). However, there exist several architectures that are known to perform very well in general, such as VGG16 [27], ResNet [28], InceptionV3 [4], and, more recently, EfficientNet [29]. When a well-known architecture is used, often a technique known as transfer learning [30] is also employed. It consists in setting the initial values of the parameters (which are otherwise random, as described in SGD), to the values of the parameters of that CNN trained on another dataset, such as ImageNet. This technique, which is also used by Antony and coworkers [26], allows the CNN to leverage the already trained filters, and just fine-tune them to the new task. For the AMD diagnosis problem, the authors report a sensitivity, specificity, and Area Under the Curve (AUC) of 0.967, 0.91, and 0.87, respectively. Table 1 provides an overview of the interpretation of these metrics.
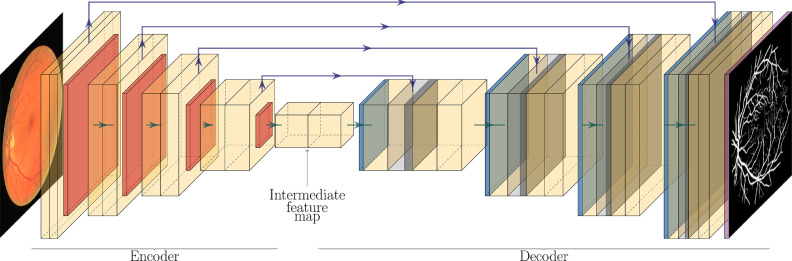
Regarding segmentation, in Arsalan and coworkers [23], the authors use a CNN to segment retinal vessels from a fundus image. Their CNN architecture, albeit custom, is heavily influenced by the very famous U-Net architecture [31], which is outlined in Fig. 9. Comparing it to the VGG16 architecture, the U-Net is comprised of an encoder block (almost identical to the encoder in VGG16), and a decoder block (which VGG16 does not have). The encoder block transforms the input image into a very feature-rich intermediate feature map. The decoder block is identical to the encoder, except that the downsampling operations have been replaced by upsampling operations. At the end of the decoder, the last activation map passes through a sigmoid activation function, and a segmentation mask is obtained. Additionally, skip connections (represented by upper arrows) transfer information from the encoder to the decoder. This primarily helps to improve the sharpness of the output, which would otherwise be poor due to all the downsampling and upsampling operations. Typical loss functions for segmentation are BCE (which the authors use) and Sørensen-Dice Similarity Coefficient (DSC) loss.
Fig. 9.
U-Net architecture. Fundus image and vessel segmentation mask from Arsalan et al. [23].
For this problem, the authors report a sensitivity, specificity, AUC, and accuracy of 0.8526, 0.9791, 0.9883, and 0.9697 respectively. Despite the very good results, reporting accuracy in a segmentation problem can be misleading, as there is often a huge imbalance between the classes. In Fig. 6, the vessel segmentation mask is mostly comprised of black pixels; hence a model that simply produces a black image as output could have very high accuracy (although the sensitivity would be zero). A good alternative metric for reporting segmentation results is the DSC (Table 1).
Although CNNs can be extremely powerful algorithms, they require vast amounts of training images to avoid overfitting, which is often a challenge with the often-scarce medical imaging data. To help alleviate this issue, data augmentation [32] allows to artificially increase the amount and variability of the training images by applying a series of random transformations (rotations, shifts, contrast and brightness modifications, etc.) to them. This method, which is employed by many authors, such as Arsalan et al. [23], can also be understood as a form of regularization.
Concerning interpretability, NNs are black boxes: unlike LR, the trained parameters are generally not interpretable, and it is difficult to understand how or why these algorithms produce a particular prediction. For classification CNNs, a technique known as Class Activation Maps (CAMs) (used in Antony et al. [26]), allows to peek at what parts of the image the CNN is looking at when it makes a particular prediction (Fig. 10). Another similar, more recent, technique is the Integrated Gradient Method, which is employed by Bellemo et al. [33].
Fig. 10.
Class activation maps for a CNN trained on AMD classification. The colored regions are important for the CNN to perform the classification task. Image from Antony et al. [26].
Three-dimensional (3D) CNNs are CNNs that can take full 3D-images (such as an OCT B-scan) as input (Fig. 11). They are identical to their two-dimensional (2D) counterparts, except that they use 3D convolutions, 3D max-pooling operations, etc. It must be noted that 3D images can also be analyzed with a 2D-CNN on a slice-by-slice basis. 3D-CNNs, however, can make better use of the contextual information provided only by the whole 3D image.
Fig. 11.
3D CNN for OCT B-scan segmentation. Images from De Fauw et al. [47].
As discussed in Section 2.4, data standardization is a typical preprocessing step in any ML pipeline, including CNNs. For images, however, Eq. 2 is not usually applied over all the images in the dataset at once, but rather over all the pixel intensities of an image, for each image independently. I. e.: after standardization, any given image will have a mean pixel intensity of 0 and a standard deviation of 1. This is a simple yet effective way of correcting intensity differences among images before feeding them to a CNN.
2.8. Recurrent neural networks
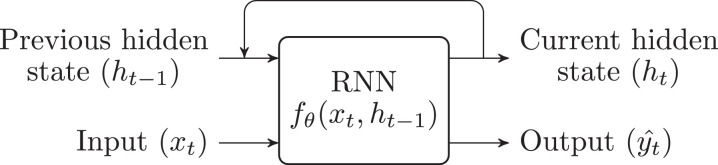
RNNs are NNs specifically designed to handle sequential (e.g.: transient) data, such as EHRs. Internally, they keep a hidden state , which can be seen as a summary of all the past information (e.g.: input values and patterns) that are relevant to the task that the network is performing. To produce a prediction, they combine the previous hidden state and the current input to generate the output (Fig. 12). The most commonly used architecture is the Long Short-Term Memory (LSTM) [18]. No uses of this kind of NNs have been found in the ophthalmology literature, yet they have been included for completeness.
Fig. 12.
Recurrent neural network.
2.9. Decision trees and ensemble methods
Decision Trees (DT) are non-linear supervised learning algorithms that encode the input-output relationship in a tree structure (Fig. 13). They can be used both for regression and classification problems and have the advantage of being easily interpretable.
Fig. 13.
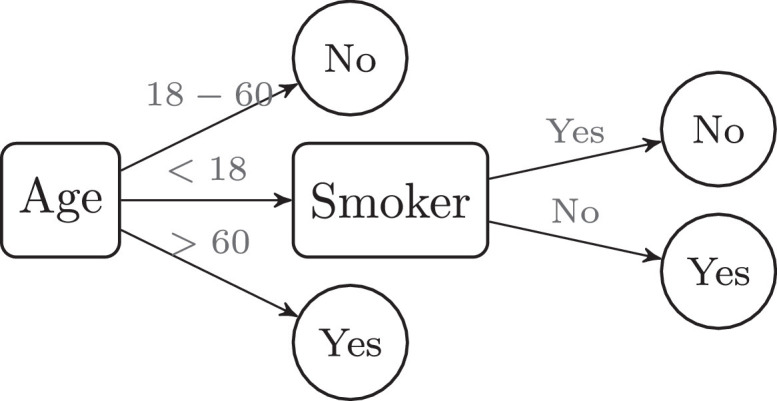
Decision tree for classification.
Very often, instead of training a single DT, an ensemble of many different DT (known as Random Forest) is trained on the same problem. Then, the majority vote of the ensemble (in classification problems), or the mean prediction (in regression problems) is taken as the final output. Ensembling can be applied to any model (even to different kinds of models). As an example, in Gargeya and Leng [34], the authors train a CNN on the task of DR detection from fundus images. Then, after training the CNN, they strip out the classification head and instead substitute it for a Gradient Boosting classifier, which is a variant of Random Forest. Thus, when given a fundus image, the CNN produces a feature vector, to which they also append patient metadata; all this information is forwarded to a Random Forest, which makes the final prediction.
2.10. Natural language processing
Unlike previous sections, Natural Language Processing (NLP) is not a specific algorithm or method, but rather a field within AI that attempts to build algorithms able to understand natural language. Even if no direct applications have been found in the context of ophthalmology, it may be used to interpret medical reports. From a technical point of view, the input to NLP algorithms is typically a series of tokens (e.g.: words or parts of words) and the output is typically either a class (for text classification, sentiment analysis, etc.) or another series of tokens (for text translation, question answering, text summarization, etc.). Although NLP models used to employ RNNs due to their ability to deal with sequences (of tokens in this case), now the Transformer [7] architecture is used almost exclusively due to its superior efficiency and scalability. Recent papers in the field, such as GPT-2 [1], have shown that impressive natural language models are achievable, but at the cost of using a prohibitively large training dataset, billions of parameters, and an immense computational budget (the latest iteration of GPT, GPT-3 [14], is estimated to have cost at least 4.6$ million only in training costs).
Human communication is extremely complex, as it implicitly assumes a shared model of the world, which machines can only begin to understand by ingesting large amounts of data. Therefore, even if NLP can still be applied successfully to many tasks without the need for such complex models, state-of-the-art NLP applications will likely be kept private and under a licensing fee for the foreseeable future, since the entry barrier to the technology has risen significantly.
3. AI in ophthalmology: past and present
Up until now, ophthalmologists have taken diagnostic, monitoring, and treatment decisions regarding various ocular diseases based on their clinical characteristics, tests run on the many devices used in this specialty, and the appreciable differences between a healthy eye versus the different stages of a diseased one. AI is a discipline on the rise, which will potentially free ophthalmologists from these monotonous tasks. At present, its main focus lies on fundus and OCT image analysis using CNNs for the classification, gradation, segmentation, and prediction of eye diseases.
This section will present an overview of the evolution, state-of-the-art, and predicted future developments of AI applied to DR (Section 3.1), AMD (Section 3.2), ROP (Section 3.3), and glaucoma (Section 3.5), as well as other AI-related applications and/or diseases, such as image enhancement or vessel segmentation (Section 3.6). To accompany the textual explanation, three tables are used: Table 2 , for models employing OCTs as inputs, Table 3 for models employing fundus images as input, and Table 3, specifically for VA prediction or estimation.
Table 2.
Summary of most relevant works where fundus images are taken as input.
| Year | Reference | Topic | Model | Dataset (patients) | Output: classes | AUC | Sens. | Spec. | Better than experts? (N) |
|---|---|---|---|---|---|---|---|---|---|
| 2020 | Varadarajan et al. [68] | DME | CNN (InceptionV3) | Thailand (4732) / EyePACS-DME (554) | Center-involved DME | 0.890 | 0.850 | 0.800 | Yes (3) |
| Intraretinal fluid | 0.810 | ||||||||
| Subretinal fluid | 0.880 | ||||||||
| 2020 | Singh and Gorantla [71] | DME | CNNs (Hierarchical Ensemble of 5) | Messidor (1200) / IDRiD (516) | DME | 0.947/ 0.979 | 0.972/ 0.945 | ||
| DME: (three-class grading) | 0.964 | 0.958 | |||||||
| 2019 | Arsalan et al. [23] | DR, Vessel seg. | CNN (Custom, Vess-Net) | DRIVE (40) / CHASE-DB1 (128) / STARE (20) | Vessel segmentation map | 0.980–0.988 | 0.802–0.853 | 0.979–0.984 | |
| 2019 | Sayres et al. [69] | DR | CNN (InceptionV4) | Patients from Krause2018 (1612) | DR: None, mild, moderate, severe, prolif. | 0.925 | 0.946 | Yes (10) | |
| 2019 | Cao et al. [22] | DR | Naive Bayes on grey coocurrence matrix | Private (1000) | DR | 0.938 | 0.949 | 0.928 | |
| 2019 | Yang et al. [67] | DR | CNN (IDx-DR) | Private (500) | DR: None, mild, moderate, severe, prolif. | 0.988 | 0.880 | ||
| 2019 | Peng et al. [70] | AMD (Bilateral images) | CNN (Inceptionv3: DeepSeeNet) | AREDS (4549) | AMD: AREDS early AMD progression risk scale (five classes) | 0.590 | 0.930 | Yes (88) | |
| Drusen: Small / none, medium, large | 0.940 | 0.718 | 0.871 | Yes | |||||
| Pigment. abnorm. | 0.930 | 0.732 | 0.957 | Yes | |||||
| Late AMD | 0.970 | 0.627/ 0.538 | 0.987/ 0.898 | No | |||||
| 2019 | Coyner et al. [50] | ROP | CNN (InceptionV3) | i-ROP (898) | ROP: Image quality assesment (2 classes) | 0.9650 | 0.939 | 0.836 | Same (6) |
| 2019 | Tan et al. [72] | ROP | CNN (ROP.AI) | Australasian ART-ROP (500) | ROP: Normal, plus-disease | 0.9770 | 0.939 | 0.807 | |
| ROP: Normal, pre-plus disease | 0.814 | 0.807 | |||||||
| 2018 | Grassmann et al. [45] | AMD | CNN (Ensemble of six) | AREDS (3654) / KORA (5555) | AMD: AREDS scale (13 classes) | 0.538/0.328 | 0.969/0.957 | Yes | |
| 2018 | Rajalakshmi et al. [73] | DR (Phone images) | CNN (EyeArt) | Private (296) | DR grade: Any, DME, prolif., referrable | 0.781–0.993 | 0.688–0.898 | ||
| 2018 | Kanagasingam et al. [74] | DR | CNN (IDx-DR) | Private (193) | DR: None, mild, moderate, severe, prolif. | 0.920 | |||
| 2018 | Krause et al. [36] | DR | CNN (InceptionV4) | EyePACS (242252) | DR: None, mild, moderate, severe, prolif. | 0.986 | 0.970 | 0.917 | Same (6) |
| 2018 | Brown et al. [52] | ROP | CNN (Vessel seg.: U-Net) + CNN (Classif.: InceptionV1) | i-ROP (898) | ROP | 0.940 | 0.930 | 0.940 | Yes (8) |
| ROP: Plus disease | 0.980 | 1,000 | 0.940 | ||||||
| 2018 | Wang et al. [51] | ROP | CNN (custom, Inception-like) | Chengdu Women Children's Central Hospital (1273) | ROP | 0.849 | 0.969 | Same (3) | |
| ROP: Minor, severe | 0,736 | 0,933 | |||||||
| 2017 | Li et al. [16] | Glaucoma | CNN (InceptionV3) | Guangdong (3970) | Glaucoma: Referable, nonreferable | 0.986 | 0.956 | 0.920 | |
| 2017 | Gargeya and Leng [34] | DR | CNN (Custom) + Decission tree (Grad. Boosting) | EyePACS (15000) / Messidor-2 (874) / E-Optha (100) | DR: Referable, Non Referable | 0.940–0.970 | 0.900–0.940 | 0.870–0.980 | |
| 2017 | Ting et al. [39] | DR | CNN (VGG-19) | SIDRP 10-15 (27979) / Guangdong (3970) + 9 others | DR: Referable, non referable | 0.889-0.983 | 0.905-1.000 | 0.733-0.916 | |
| Glaucoma | SIDRP 10-15 (27979) + 5 others | Glaucoma: Referable, non referable | 0.942 | 0.964 | 0.872 | ||||
| AMD | Idem | AMD: Referable, non referable | 0.931 | 0.932 | 0.887 | ||||
| 2017 | Burlina et al. [2] | AMD | CNN (AlexNet) | AREDS (4613) | AMD | 0.940 | 0.846 | 0.920 | Same |
| 2016 | Abràmoff et al. [75] | DR | CNN (IDx-DR vX2.1) | Messidor-2 (874) | DR: None or mild, present, vision-threatening | 0.980 | 0.968 | 0.870 | |
| 2016 | Gulshan et al. [3] | DR, DME | CNN (InceptionV3) | EyePACS (4997) / Messidor-2 (874) | DR: Referable, non referable | 0.991/ 0.990 | 0.903/ 0.870 | 0.981/ 0.985 | |
| DME: Referable, non referable | 0.974 | 0.907 | 0.938 | ||||||
| 2012 | Zheng et al. [44] | AMD | Hierarchical trees + SVM | ARIA (161) / STARE (97) | AMD | 0.994 | 1,000 | ||
| 2011 | Agurto et al. [8] | AMD, DR (3 FOV fundus) | Feature extraction + Partial Least Squares | RIST (378) / UTHSCSA (444) | DR: Normal, sight-threatening | 0.890/ 0.920 | 0.950 | 0.500 | |
| DR-related: Microaneurysms, hemorrhages, exudates, neovascularization, etc. | 0.770-0.980 | 0.830–1.00 | 0.500 | ||||||
| AMD-related: Drusen, Pigment. abnorm., GA | 0.770-0.920 | 0.88–1.000 | 0.500 |
Table 3.
Summary of most relevant works in classification and segmentation tasks, where OCT images are taken as input.
| Year | Reference | Topic | Model | Dataset (patients) | Output: classes | AUC | Sens. | Spec. | Better than experts? (N) |
|---|---|---|---|---|---|---|---|---|---|
| 2019 | Motozawa et al. [46] | AMD | CNN (Custom, VGG-like) | Private (271) | AMD | 0.995 | 1,000 | 0.918 | |
| AMD: Fluid, no fluid | 0.991 | 0.984 | 0.883 | ||||||
| 2019 | Antony et al. [26] | AMD | CNN (VGG16) | Private (384) | AMD (+ relevant B-scans) | 0.967 | 0.910 | 0.870 | |
| 2019 | Hassan et al. [41] | DME: Fluid seg., Vessel seg. (Fundus + OCT) | CNN (Custom) + Heavy processing | Rabani and Zhang (683) | DME | 0.970 | 0.920 | Yes (3) | |
| Segmentation: Hard exudates, Blood vessel, Retinal fluid | (DSCs: 0.707, | ||||||||
| 0.820, 0.902) | |||||||||
| 2019 | Kuwayama et al. [76] | AMD, DR, ERM + Others | CNN (Custom) | Private (1200) | Normal | 0.850 | 0.970 | ||
| Wet AMD | 1,000 | 0.770 | |||||||
| DR | 0.780 | 1,000 | |||||||
| ERM | 0.750 | 0.750 | |||||||
| 2018 | De Fauw et al. [47] | AMD, DR, ERM + Others | CNN (Segmentation, Custom) + CNN (Classification, Custom) | UK National Heatlh Service (NHS) (14884) | Referral: urgent, non urgent | 0.992 | Yes (8) | ||
| Normal | 0.995 | Same | |||||||
| MRE | 0.990 | Yes | |||||||
| CNV | 0.993 | Yes | |||||||
| Drusen | 0.974 | Same | |||||||
| ERM | 0.966 | Same | |||||||
| Others (GA, CSR, Full/partial thickness macular hole, VMT) | 0.980 | Same | |||||||
| 2018 | Shigueoka et al. [57] | Glaucoma (OCT + SAP) | Feature extraction + Several ML classifiers | University of Campinas, Brazil (124) | Glaucoma: Early or moderate, none | 0.931 | 0.900 | 0.800 | Same (3) |
| 2017 | Schlegl et al. [77] | AMD, DME: Fluid seg. | CNN (Segmentation, custom) | Private (1200) | Intraretinal fluid | 0.940 | |||
| Subretinal fluid | 0.933 |
3.1. Diabetic retinopathy
DR is a condition that affects the small blood vessels of the retina in the eyes of people with diabetes. It is estimated that approximately 93 million people across the world have DR and it is expected that a third of the world's diabetic population will develop it at some point [35]. Diabetic macular edema (DME) is an additional complication that can occur at any stage, and it is associated with significant visual loss. DME is characterized by the thickening of the macular region of the retina due to fluid build-up from blood leaks. There are several effective treatments available, such as laser photocoagulation, anti-vascular endothelial growth factor (VEGF) drugs, intravitreal steroid injections, and vitrectomy. Many of these can prevent vision loss, stabilize vision, and in certain cases improve it provided DME is detected and treated in the early stages. In this context, AI can boost the implementation of automated generalized screening for deferrable DR and DR staging, among several other applications.
Currently, AI-automated DR screening and staging from fundus images is one of the most promising AI tools in medicine. Over the last few years, many authors have shown that such systems can consistently match the performance of the experts, sometimes outperforming them, [20,36], while being a more cost-effective [37] and wider-reaching alternative to current screening programs. Furthermore, commercial systems for DR screening are already available, with IDx-DR being the first FDA-authorized autonomous AI diagnostic system [38] and, more recently, EyRIS SELENA+ [33,39] receiving clearance for use both in the European Union and Singapore [40].
As can be seen in Table 2, Agurto et al. [8] proposed in 2011 one of the first ML-based models to detect DR and other related pathologies (such as microaneurysms, hemorrhages, and exudates), as well as AMD-related pathologies such as drusen, pigmentation, and geographic atrophy (GA) by using RIST (378 patients) and UTHCSA (444 patients) datasets for training and validation. The AUCs ranged from 0.890 to 0.920 for detecting DR with visual impairment, achieving a sensitivity of 0.95 with a specificity set at 0.5. Also, for the determination of DR-related pathologies, they reached an AUC between 0.770 and 0.980.
However, it was not until 2016 that the DL revolution began in ophthalmology. Since, the size (the number of patients) of the datasets (see Tables 2, 3, and 4 ) has increased over time and, in current AI systems, metrics such as AUC, accuracy, sensitivity, and specificity have improved. Besides sheer dataset size, there tends to be an ever-increasing heterogeneity in the imaging data (e.g.: heterogeneities in scanners, medical centers, patient ethnicities, etc.), and a shift of focus from basic classification to grading (i.e., mere detection vs. disease staging), which are all factors that might pose a greater challenge for the algorithms, but eventually result in more robust and useful systems. Current AI models in the field are almost exclusively based on CNNs.
Table 4.
Summary of most relevant works in the task of VA prediction, where OCT images are taken as input.
| Year | Reference | Topic | Model | Dataset (patients) | Output | Results |
|---|---|---|---|---|---|---|
| 2019 | von der Emde et al. [25] | AMD, VA (Fundus Autofluor. + Infrarred Reflection) | CNN (Segmentation) + Random Forest | Private (90) | VA Fundus-controlled perimetry (mesopic, cyan, red) | MAE: 3.94, 4.9, 4.02 dB |
| 2018 | Rohm et al. [11] | AMD, VA (EHR) | Lasso LR | Private (653) | VA delta at 3 months | MAE: 0.16 logMAR |
| Private (453 of above) | VA delta at 12 months | MAE: 0.11 logMAR | ||||
| 2017 | Aslam et al. [21] | AMD, VA (Patient Age) | FFNN | Private (1210) | VA at current time | R2: 0.852 (letters) |
| 2017 | Schmidt-Erfurth et al. [24] | AMD, VA | CNN (Segmentation) + Random forest | HARBOR (614) | BCVA at 12 months | R2: 0.7 (logMAR) |
As an example of these trends, in 2018, Krause et al. [36] developed a CNN for DR and DME detection and staging: DR was classified according to the International Clinical Diabetic Retinopathy disease severity scale: no DR, mild, moderate, severe, and proliferative, while DME was classified as either referrable or not. The Inception-V4 CNN model was trained on the 1.5 million fundus images from the Eye-PACs dataset, while the 2000 images from the Eye-PACS-2 dataset were employed for validation, achieving comparable results to three retinal specialists and three US board-certified ophthalmologists. For instance, for moderate or worse DR classification, the model achieved a sensitivity and specificity of 97.1% and 92.3% (respectively), compared to a median of 75.2% and 97.9% for the ophthalmologists, and a median of 74.6% and 99.3% for the retinal specialists. For referrable DME classification, the model presented a sensitivity and specificity of 94.9% and 94.4%, compared to 91.5% and 98.7% for the median ophthalmologist. Nevertheless, the combination of the experts (by majority vote) still outperformed the model.
Most authors employ fundus images for developing DR diagnosis systems, which are generally more accessible than OCTs. Hassan et al. [41] argued that perhaps, using both fundus and OCT images the performance could be improved. Their CNN-based model achieved a sensitivity and specificity of 0.970 and 0.920, for referrable DME detection, which are similar results to those obtained in the previous model, suggesting that OCTs may provide little advantages as compared to fundus images for this task.
Besides DR diagnosis, some authors, such as Rasti and coworkers [17], focused on anti-VEGF response prediction for DME treatment. In this paper, a CNN was trained on OCT images from 127 subjects to classify them as either experimenting at least a 10% reduction in retinal thickness following the treatment, or not. The authors obtained a sensitivity and specificity of 80.1% and 85.0% on a 5-fold CV set. Even if CV is not ideal for validating a model, it shows potential for future research on the topic.
3.2. Age-related macular degeneration
AMD is the main cause of irreversible vision loss in people aged 50+ years in developed countries [42]. The estimations of prevalence indicate that it affects 9% of people aged 45-85 across the world, amounting to a total of 196 million people. It is a complex disease, associated with genetic and environmental risk factors, which is typically characterized by the appearance of drusen (yellow lipid deposits under the retina). Recently, anti-angiogenic drugs have revolutionized the treatment of wet (advanced) AMD, significantly reducing blindness and visual impairment if the condition is diagnosed early, the patient is referred, the treatment is adhered to, and guidelines and protocols are complied with. This disease is typically graded employing a 13-class system based on the AREDS [43] scale, where higher scores represent more advanced stages, and class 13 is reserved for ungradable images.
One of the first models for automated AMD classification was proposed by Zheng and coworkers [44] in 2012. Leveraging two public fundus image datasets of 258 patients, the technique was capable of classifying referrable / non-referrable AMD with an AUC of 0.994, a specificity of 1, and a sensitivity of 0.994. They also made comparisons with the four prior papers published on the topic by other research groups and confirmed the superiority of the new method. Nevertheless, 10-fold CV was employed for validation (instead of a test set, or an external validation set), which might have biased the results.
Similar to DR, since 2016 AI-based AMD diagnosis started receiving a renewed interest. In 2018, Grassmann and coworkers [45], employed an ensemble of six CNNs (using AlexNet, VGG, ResNet, etc.) to automatically grade fundus images according to the 13-class AREDS scale (which is a much more challenging problem compared to the previous binary -i.e.: referrable AMD vs. non-referrable- AMD classification). They used the AREDS dataset (with over 120,000 fundus images from over 5000 patients) for training and testing, and the Cooperative Health Research in the Region of Augsburg (KORA) dataset (5555 images) as an external validation set. External validation sets are comprised of images from a different dataset (different hospital, possibly different scanners) than those used for training, and hence can often be more representative of the real-world performance of the model (especially if several external validation sets are employed). The average (over all 13 classes) sensitivity and specificity for the AREDS test set was 0.538 and 0.969, while for the KORA external validation set, it was 0.328 and 0.957. This data, which has been taken from the supplementary material of said paper, allows us to introduce an interesting topic: the difficulty in interpreting the results. Even if the sensitivities seem low, such values are to be expected for a 13-class classification problem where the differences between adjacent classes are minimal. In fact, the authors break down the KORA results (considering only patients aged 55+) by ranges of classes: for intermediate AMD (AREDS classes 4-9) the sensitivity/specificity was 0.822/0.971, and for late-stage AMD (AREDS classes 10-12), it was 1/0.965. Researchers should carefully consider whether a 13-class system for automated AMD classification is needed, while the clinicians should take care to correctly interpret the results.
Unlike in DR, many published papers on this topic employ OCT images as input, instead of fundus images. For instance, Motozawa and coworkers [46] employ a simple CNN architecture to predict whether an OCT was normal or contained AMD-related findings (drusen, pseudo-drusen, Pigmented Epithelial Detachment (PED), geographic atrophy, etc.). The model was trained on 271 patients from a private dataset, from which the training and the validation set were extracted. Once again, this is not ideal methodologically, since there seems to be no proper test set, and hence the model might be overfitted and the results overestimated. They achieved a sensitivity/specificity of 1/0.918.
To close off the topic of AMD detection, De Fauw and coworkers [47] is hitherto one of the best articles both in terms of methodology and results. It introduced a two-stage model comprised of a CNN for segmentation (epiretinal membrane [ERM], subretinal and intraretinal fluid, etc.), followed by an ensemble of CNNs for classification. It could determine the prognosis for a large number of pathologies (such as choroidal ceovascularization [CNV], MRE, Global Serious Retinopathy [GSR], etc.) using OCTs from 14,884 patients (from two different scanners), and reaching an AUC above 0.99 for the majority of the diseases (and above 0.966 for all of them), hence equaling the performance of an expert ophthalmologist in those tasks. They also classified the OCTs in terms of referral urgency (urgent, semi-urgent, routine, and observation only), achieving an AUC of 0.9921 for the urgent class and outperforming the experts in this task. Regarding the methods, they used a proper training/validation/test set split approach, which is explained in detail both in the main paper and the supplementary material. Their two-stage model approach (segmentation + classification) not only achieves excellent results but also helps the end-user (the ophthalmologist) understand the final predictions better since these are directly based on the informative segmentations generated in the first stage.
Regarding VA prediction in the context of AMD, Table 4 summarizes the most relevant papers. For instance, in von der Emde and coworkers [25] and Aslam and coworkers [21], the authors tried to predict VA at present without measuring, while in Rohm et al. [11] and Schmidt-Erfurth et al. [24] VA is predicted three or twelve months after undergoing treatment with anti-angiogenic drugs. These studies may offer significant assistance with treatment for this pathology, which requires a high degree of adherence and compliance to maximize the resulting VA.
Finally, some authors have delved into the topic of AMD progression prediction. For instance, Bhuiyan and coworkers [48] employed the AREDS dataset to predict the incidence of late AMD from fundus images over two years, achieving an accuracy of 86.36%. Yim and coworkers [15] focused instead on predicting the progression to wet AMD in the second eye using OCT images, reaching a sensitivity/specificity of 0.80/0.55, and outperforming most of the consulted experts.
3.3. Retinopathy of prematurity
ROP is a retinovascular disease that affects both extremely premature infants in developed countries to older babies worldwide (due to the lack of appropriate screening in middle-income countries) [6,49]. Even if early treatment has shown to be effective, much is still to be known about this condition [50].
Wang et al. [51] introduced one of the first ML models based on CNNs for determining the presence and severity of ROP from fundus images. They used a dataset of 1,273 patients, attaining a sensitivity and specificity of 0.849 and 0.969. That same year, Brown and coworkers [52], attempted to identify ROP in fundus images by employing first a U-net CNN to perform capillary segmentation, which was then combined with the fundus image (as an additional channel) to perform classification (normal, pre-plus disease, plus-disease). They achieved a sensitivity of 0.930 and a specificity of 0.940 for plus-disease diagnosis, outperforming 6 out of 8 experts. Similar models have been developed as of recent (see Table 2), obtaining overall very compelling results. An AI solution for ROP screening might be particularly timely for middle-income countries, where this disease is becoming more prevalent due to better critical care for premature babies combined with the lack of experts able to detect and manage the disease.
3.4. Glaucoma
Glaucoma is a disease of the optic nerve, which becomes damaged due to the increase of pressure as aqueous humor builds up in the eye. It is a major cause of visual impairment and blindness all over the world [53], with 3.54% of people affected by it between ages 40 and 80, an estimate of 76 million people by 2020 [54]. Although it can be treated (typically with daily eye drops), it manifests itself mostly asymptomatically. Along with glaucoma, the main causes of blindness worldwide are cataracts and uncorrected refraction errors [55]. In this context, AI can play a crucial role in helping implement cost-effective generalized screening and optimizing early treatment.
In 2018, Li and coworkers [16] used fundus images from 3,970 patients to train a CNN model to detect referrable glaucomatous optic neuropathy (vertical cup-to-disc ratio of 0.7 or more), achieving an AUC of 0.986, a sensitivity of 0.956, and a specificity of 0.920. In addition to image normalization and data augmentation, a preprocessing step known as local space average color [56] was employed to improve the color consistency of the images, even if the illumination changed.
Shigueoka and coworkers [57] used OCTs and standard automated perimetry (SAP) from 124 patients as input to a ML model trained for discerning between healthy and glaucomatous individuals. From the OCTs, 17 features were extracted by measuring retinal nerve fiber layer thickness at different points of the image, while from SAP, mean deviation, pattern standard deviation, and glaucoma hemifield test features were computed. Several ML classification algorithms, such as RFs and FFNNs were applied to the problem and validated using 10-fold CV. The best performing model was a Radial Basis Function (RBF) Network, which is an algorithm similar to a FFNN, but using special RBF activation functions instead. It proved to attain a statistically similar performance when compared to glaucoma specialists and the Combined Structure-Function Index. The authors conclude the paper marking the importance of such a technique in the context of primary care as a key tool for the diagnosis, treatment, or early referral to a specialist, especially when there is no glaucoma specialist available.
3.5. Others
Besides the previously discussed conditions, there a growing field of research in several related techniques and diseases. For instance, automatic retinal blood vessel segmentation in fundus images can provide useful information for several clinical applications [58]. For instance, Arsalan et al. [23] train a residual U-Net CNN for this task using three different datasets (for a total of 200 images), achieving an AUC above 0.982 for all datasets. Even if 200 images might seem insufficient, segmentation models usually require much fewer images to train as compared to classification models, since every image contains much more information: a full segmentation mask, i.e. a label for every pixel, versus a single label for the whole image. Furthermore, as an expert, generating the segmentations for the system is much more time consuming, so datasets tend to be smaller.
Another related topic is AI image enhancement, which consists in training a CNN to improve the quality of an image, increase its resolution, remove artifacts, reduce noise, etc. Halupka and coworkers [59] employ two kinds of models to remove speckle noise from OCTs: a pure CNN, and a Generative Adversarial Network (GAN). GANs are typically comprised of two CNNs: a generator and a discriminator; both are trained in tandem: the generator tries to fool the discriminator by generating OCTs that look real, while the discriminator tries to identify whether any given OCT (generated or not) is actually real. After training, the generator should be able to generate realistic-looking OCT images. In the context of this paper, a GAN is used to enhance the features of the original OCT image. After training, the authors achieve a significant improvement in objective metrics with the pure CNN for noise reduction, while the GAN generates images that are qualitatively perceived as better by the experts. In fact, GANs can be more “creative” and bolder at adding detail, which human perception favors. In the wider context of ophthalmology, GANs could also be used to generate synthetic (artificial) images after being trained on a dataset (e.g.: to complement smaller datasets).
Keratoconus is a corneal disease resulting in irregular astigmatism and vision loss, which is typically analyzed by using a technique known as Scheimpflug imaging. Since 2010, a few authors have employed ML systems trained on features extracted from Scheimpflug maps to identify keratoconus [60,61], achieving AUCs above 0.98 in CV. Cataracts are the clouding of the lens of the eye, resulting in a decrease of vision, and representing a major cause for visual impairment in LMICs [62]. In this context, AI has contributed to the development of more accurate intraocular lens calculation formulas [63]. As a final example, Achiron et al. [64] used an RF ML algorithm to predict the outcome of refractive surgeries on a dataset of over 17,000 cases.
4. Discussion: challenges and opportunities
The global population aging is jeopardizing our sight, the sense held by humans to be most important [65]. It is estimated that DR will affect 300 million people by 2025 and 500 by 2050 [5], with up to 30% of diabetic patients presenting any degree of DR, and 10% suffering from sight-threatening DR. Similarly, AMD is already the main cause for vision loss in older patients in developed countries [42] and, as life expectancy rises, the incidence of advanced AMD will also rise. The situation is yet direr in LMICs, where the life expectancy has risen sharply over the last few decades, yet they lack the medical professionals needed to detect and manage these diseases. For instance, Hussain and coworkers [66] estimates an AMD prevalence of 0.6-1.1% in these countries by 2030.
Although eye-related AI research is broad, right now it focuses on detection and staging of diseases from either fundus or OCT images through CNNs, achieving expert-level performances in many cases. Even if the deployment of these systems is still limited, it is foreseeable that this trend will continue and that these algorithms will slowly but surely make their way into the clinical practice in the next few years. In the future it will be possible to automatically classify all ophthalmological using an image-based AI system; moreover, in a second phase, we will likely supplement these systems with additional patient information including, but not limited to, demographic data, medical history, comorbidities, or genetic indicators.
This could signify a true revolution: the automated global screening for eye diseases could mean earlier detection and referral, speeding-up treatment by adapting the treatment guidelines to the reality of each patient, encouraging adherence and compliance by patients and, ultimately, resulting in a better life for the patient. This is especially true for LMIC, where there are not enough experts to attend to their aging population (or their newborns, in the case of ROP). We are already past the time of wondering whether a certain technique will work or not. We already know it does. Instead, we should focus on developing systems that are properly validated and employ data from a wide array of sensors, medical centers, and a representative cohort of the population (in terms of age, sex, ethnicity, diseases, etc.). The lack of diversity within datasets is especially concerning from an ethical point of view, as it could exacerbate the already serious healthcare disparities between population groups. Therefore, we should actively strive to improve collaboration among centers world-wide to obtain the best datasets possible, and we should also stop to reflect on how to best bring these methods to the clinical practice. We are already on our way there, but much work is still to be done, and it depends on all the actors involved (ophthalmologists, data scientists, corporations, individuals, etc.) to do their part to make this a reality. Also, AI progress should advance in parallel with new treatment strategies to complement each other.
We are living in a digital era, accelerated with the emergence of COVID, where there is a critical need for clean (or “green”) systems, to reduce costs and inefficiencies, and to improve time management. In this situation, AI has the key for addressing some critical questions, such as: Will I have to go to a retinal specialist for diagnosis, or will I be able to do this from home using my mobile device and receive a response in real-time? Will there be a system to monitor treatment adherence and provide real-time alerts and guidance? When will technology enable all data to be integrated into a worldwide health system that will expand knowledge on a global scale? As 5G continues to expand, smartphones will provide a baseline set of devices able to run AI algorithms locally, while some of the heaviest computing tasks will be offloaded to the cloud. Eventually, AI systems might be the tools that will enable doctors to make significant improvements to patient management, achieving a better life for everyone through science and technology.
5. Conclusion
AI may be the answer to healthcare system sustainability amidst an aging world population, the quick developments of LMIC countries, and a deadly global pandemic. We are already on our way, but to succeed we require collaboration and goodwill from all actors involved. Lastly, time is one of the most valuable resources and, the more doctors and ophthalmologists are freed by AI, the more time they will have to focus on the reason for their existence: the patient.
6. Method of literature search
The literature search for this review was based on combining a set of keywords from the medical field (ophthalmology, retina, glaucoma, Retinopathy of Prematurity, Age-Related Macular Degeneration and Visual Acuity) with a set of keywords from the Machine Learning field (Artificial Intelligence, Deep Learning, and Convolutional Neural Network). All terms from each set were independently combined with all terms from the other.
The main repository for the search was PubMed, although Google Scholar was also employed for completeness. Articles deemed relevant (by inspection of the title and abstract) from January 2016 to June 2020 were reviewed, amounting to a total of around 400. The main inclusion criteria were the perceived quality of the research and the focus on Artificial Intelligence. A few select articles published before 2016 were included for historical purposes, as well as some articles dealing with closely related topics (such as keratoconus, or image enhancement). Most of the papers were in English; only the abstract was considered for those that were not.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationshipsthat could have appeared to influence the work reported in this paper.
Acknowledgments
We want to cordially thank Borja Corcóstegui and Patricia Udaondo for their help reviewing this paper. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- 1.Cheloni R., Gandolfi S.A., Signorelli C., Odone A. Global prevalence of diabetic retinopathy: protocol for a systematic review and meta-analysis. BMJ Open. 2019;9(3) doi: 10.1136/bmjopen-2018-022188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams R., Karuranga S., Malanda B., et al. 9th edition. Vol. 162. Diabetes Res Clin Pract; 2020. (Global and regional estimates and projections of diabetes-related health expenditure: results from the International Diabetes Federation Diabetes Atlas). [DOI] [PubMed] [Google Scholar]
- 3.Yang W.H., Zheng B., Wu M.N., et al. An evaluation system of fundus photograph-based intelligent diagnostic technology for diabetic retinopathy and applicability for research. Diabetes Ther. 2019;10(5):1811–1822. doi: 10.1007/s13300-019-0652-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartnett M.E. Retinopathy of prematurity: evolving treatment with anti-vascular endothelial growth factor. Am J Ophthalmol. 2020 doi: 10.1016/j.ajo.2020.05.025. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varadarajan A.V., Bavishi P., Ruamviboonsuk P., et al. Predicting optical coherence tomography-derived diabetic macular edema grades from fundus photographs using deep learning. Nat Commun. 2020;11(1):1–8. doi: 10.1038/s41467-019-13922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayres R., Taly A., Rahimy E., et al. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology. 2019;126(4):552–564. doi: 10.1016/j.ophtha.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Krause J., Gulshan V., Rahimy E., et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125(8):1264–1272. doi: 10.1016/j.ophtha.2018.01.034. [DOI] [PubMed] [Google Scholar]
- 8.Peng Y., Dharssi S., Chen Q., et al. DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126(4):565–575. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Fauw J., Ledsam J.R., Romera-Paredes B., et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–1350. doi: 10.1038/s41591-018-0107-6. Accessed: June 23, 2020. [DOI] [PubMed] [Google Scholar]
- 10.Abràmoff M.D., Lavin P.T., Birch M., Shah N., Folk J.C. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. npj Digit Med. 2018;1(1). doi:10.1038/s41746-018-0040-6 [DOI] [PMC free article] [PubMed]
- 11.Ding Y., Liu Y., Yan Q., et al. Bivariate analysis of age-related macular degeneration progression using genetic risk scores. Genetics. 2017;206(1):119–133. doi: 10.1534/genetics.116.196998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogunovic H., Waldstein S.M., Schlegl T., et al. Prediction of anti-VEGF treatment requirements in neovascular AMD using a machine learning approach. Invest Ophthalmol Vis Sci. 2017;58(7):3240–3248. doi: 10.1167/iovs.16-21053. [DOI] [PubMed] [Google Scholar]
- 13.Abramoff M.D., Keane P., Odaibo S., Ting D. Ophthalmic frontiers: AI. The ophthalmologist. Published 2019. Available at: https://theophthalmologist.com/subspecialties/ophthalmic-frontiers-ai. Accessed May 13, 2020
- 14.Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. Vol 25.; 2012. Available at: http://code.google.com/p/cuda-convnet/. Accessed January 22, 2021
- 15.Russakovsky O., Deng J., Su H., et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 16.Silver D., Huang A., Maddison C.J., et al. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529(7587):484–489. doi: 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- 17.Peters M.E., Neumann M., Iyyer M., et al. Improving language understanding by generative pre-training. OpenAI. Published online 2018:1-10. Accessed January 22, 2021. Available at: https://gluebenchmark.com/leaderboard%0Ahttps://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf
- 18.Jumper J., Evans R., Pritzel A., et al. Fourteenth Critical Assessment of Techniques for Protein Structure Prediction (Abstract Book) 2020. High Accuracy Protein Structure Prediction Using Deep Learning; pp. 22–24. Accessed November 12, 2018. Available at: https://deepmind.com/blog/article/alphafold-a-solution-to-a-50-year-old-grand-challenge-in-biology. [Google Scholar]
- 19.Callaway E. “It will change everything”: DeepMind's AI makes gigantic leap in solving protein structures. Nature. 2020;588(7837):203–204. doi: 10.1038/d41586-020-03348-4. [DOI] [PubMed] [Google Scholar]
- 20.Lecun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 21.Ebner M. Color constancy based on local space average color. Mach Vis Appl. 2009;20(5):283–301. doi: 10.1007/s00138-008-0126-2. [DOI] [Google Scholar]
- 22.EyRIS. Published 2020. Available at: https://www.eyris.io/latest_news.cfm?id=37. Accessed October 19, 2020
- 23.Flaxman S.R., Bourne R.R.A., Resnikoff S., et al. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Glob Heal. 2017;5(12):e1221–e1234. doi: 10.1016/S2214-109X(17)30393-5. [DOI] [PubMed] [Google Scholar]
- 24.Gargeya R., Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124(7):962–969. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 25.Grassmann F., Mengelkamp J., Brandl C., et al. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology. 2018;125(9):1410–1420. doi: 10.1016/j.ophtha.2018.02.037. [DOI] [PubMed] [Google Scholar]
- 26.Gulshan V., Peng L., Coram M., et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA - J Am Med Assoc. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 27.Gunasekeran D.V., Ting D.S.W., Tan G.S.W., Wong T.Y. Artificial intelligence for diabetic retinopathy screening, prediction and management. Curr Opin Ophthalmol. 2020;31(5):357–365. doi: 10.1097/ICU.0000000000000693. [DOI] [PubMed] [Google Scholar]
- 28.Halupka K.J., Antony B.J., Lee M.H., et al. Retinal optical coherence tomography image enhancement via deep learning. Biomed Opt Express. 2018;9(12):6205. doi: 10.1364/boe.9.006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassan B., Hassan T., Li B., Ahmed R., Hassan O. Deep ensemble learning based objective grading of macular edema by extracting clinically significant findings from fused retinal imaging modalities. Sensors. 2019;19(13):2970. doi: 10.3390/s19132970. Accessed June 15, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. IEEE Computer Society; 2016. Deep residual learning for image recognition; pp. 770–778. Vol 2016-Decem. [DOI] [Google Scholar]
- 31.Hussain N., Khanna R., Hussain A. Trend of retinal diseases in developing countries. Expert Rev Ophthalmol. 2008;3(1):43–50. doi: 10.1586/17469899.3.1.43. [DOI] [Google Scholar]
- 32.International Diabetes Federation, International Federation on Ageing, International Agency for the Prevention of Blindness. The Diabetic Retinopathy Barometer Report: Global Findings.; 2016. Available at: https://www.iapb.org/wp-content/uploads/DR-Global-Report-1.pdf
- 33.Islam M.M., Poly T.N., Walther B.A., Yang H.C., Li Y.-C. (Jack). Artificial intelligence in ophthalmology: a meta-analysis of deep learning models for retinal vessels segmentation. J Clin Med. 2020;9(4):1018. doi: 10.3390/jcm9041018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanagasingam Y., Xiao D., Vignarajan J., Preetham A., Tay-Kearney M.-L., Mehrotra A. Evaluation of artificial intelligence–based grading of diabetic retinopathy in primary care. JAMA Ophthalmol. 2018 doi: 10.1001/jamanetworkopen.2018.2665. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuwayama S., Ayatsuka Y., Yanagisono D., et al. Automated detection of macular diseases by optical coherence tomography and artificial intelligence machine learning of optical coherence tomography images. J Ophthalmol. 2019;2019:1–7. doi: 10.1155/2019/6319581. Accessed March 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arsalan O.w.a.i.s., Mahmood C.h.o. Park. Aiding the diagnosis of diabetic and hypertensive retinopathy using artificial intelligence-based semantic segmentation. J Clin Med. 2019;8(9):1446. doi: 10.3390/jcm8091446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z., He Y., Keel S., Meng W., Chang R.T., He M. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018;125(8):1199–1206. doi: 10.1016/j.ophtha.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 38.Bhuiyan A., Wong T.Y., Ting D.S.W., Govindaiah A., Souied E.H., Smith R.T. Artificial intelligence to stratify severity of age-related macular degeneration (AMD) and predict risk of progression to late AMD. Transl Vis Sci Technol. 2020;9(2):1–12. doi: 10.1167/TVST.9.2.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motozawa N., An G., Takagi S., et al. Optical coherence tomography-based deep-learning models for classifying normal and age-related macular degeneration and exudative and non-exudative age-related macular degeneration changes. Ophthalmol Ther. 2019;8(4):527–539. doi: 10.1007/s40123-019-00207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–1359. doi: 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 41.Perez L., Wang J. The effectiveness of data augmentation in image classification using deep learning. arXiv. Published online December 13, 2017. Available at: http://arxiv.org/abs/1712.04621. Accessed March 11, 2020
- 42.Rajalakshmi R., Subashini R., Anjana R.M., Mohan V. Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence. Eye. 2018;32(6):1138–1144. doi: 10.1038/s41433-018-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasti R., Allingham M.J., Mettu P.S., et al. Deep learning-based single-shot prediction of differential effects of anti-VEGF treatment in patients with diabetic macular edema. Biomed Opt Express. 2020;11(2):1139. doi: 10.1364/boe.379150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Retina International. Burden: AMD – Retina International's AMD Toolkit. Published 2019. Available at: http://retina-amd.org/menu/burden-of-amd-2/#incidenceandprevalence. Accessed June 21, 2020
- 45.Rohm M., Tresp V., Müller M., et al. Predicting visual acuity by using machine learning in patients treated for neovascular age-related macular degeneration. Ophthalmology. 2018;125(7):1028–1036. doi: 10.1016/j.ophtha.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 46.Ronneberger O., Fischer P., Brox T. Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Vol. 9351. Springer Verlag; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [DOI] [Google Scholar]
- 47.Bellemo V., Lim Z.W., Lim G., et al. Artificial intelligence using deep learning to screen for referable and vision-threatening diabetic retinopathy in Africa: a clinical validation study. Lancet Digit Heal. 2019;1(1):e35–e44. doi: 10.1016/S2589-7500(19)30004-4. [DOI] [PubMed] [Google Scholar]
- 48.Ruder S. An overview of gradient descent optimization algorithms. arXiv. 2016:1–14. http://arxiv.org/abs/1609.04747 Published online September 15Available at: [Google Scholar]
- 49.Achiron A., Gur Z., Aviv U., et al. Predicting refractive surgery outcome: machine learning approach with big data. J Refract Surg. 2017;33(9):592–597. doi: 10.3928/1081597X-20170616-03. [DOI] [PubMed] [Google Scholar]
- 50.Schlegl T., Waldstein S.M., Bogunovic H., et al. Fully Automated detection and quantification of macular fluid in OCT using deep learning. Ophthalmology. 2018;125(4):549–558. doi: 10.1016/j.ophtha.2017.10.031. [DOI] [PubMed] [Google Scholar]
- 51.Schmidhuber J. Long short-term memory. Neural Comput. 1997;9:1736–1780. doi: 10.1162/neco.1997.9.8.1735. https://www.mitpressjournals.org/doi/abs/10.1162/neco.1997.9.8.1735 Available at: [DOI] [PubMed] [Google Scholar]
- 52.Schmidt-Erfurth U., Bogunovic H., Sadeghipour A., et al. Machine learning to analyze the prognostic value of current imaging biomarkers in neovascular age-related macular degeneration. Ophthalmol Retin. 2018;2(1):24–30. doi: 10.1016/j.oret.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 53.Shaw J.E., Sicree R.A., Zimmet P.Z. Global estimates of the prevalence of diabetes for 2010 and 2030. Guariguata L, Nolan T, Beagley J, Linnenkamp U, Jacqmain O, eds. Diabetes Res Clin Pract. 2010;87(1):4-14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed]
- 54.Shigueoka L.S., Vasconcellos J.P.C. de, Schimiti R.B., et al. Automated algorithms combining structure and function outperform general ophthalmologists in diagnosing glaucoma. Mortazavi B, ed. PLoS One. 2018;13(12):e0207784. doi: 10.1371/journal.pone.0207784. [DOI] [PMC free article] [PubMed]
- 55.Siddiqui A.A., Ladas J.G., Nutkiewicz M.A., Chong J.K., Marquezan M.C., Hamilton D., Evaluation of New IOL formula that integrates artificial intelligence. ASCRS ASOA Annual Meeting. Published 2018. Accessed October 21, 2020. Available at: https://ascrs.confex.com/ascrs/18am/meetingapp.cgi/Paper/45603.
- 56.Simonyan K., Zisserman A. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. 2015. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- 57.Singh R.K., Gorantla R. DMENet: diabetic macular edema diagnosis using hierarchical ensemble of CNNs. Pławiak P, ed . PLoS One. 2020;15(2) doi: 10.1371/journal.pone.0220677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smadja D., Touboul D., Cohen A., et al. Detection of subclinical keratoconus using an automated decision tree classification. Am J Ophthalmol. 2013;156(2):237–246. doi: 10.1016/j.ajo.2013.03.034. e1. [DOI] [PubMed] [Google Scholar]
- 59.Souza M.B., Medeiros F.W., Souza D.B., Garcia R., Alves M.R. Evaluation of machine learning classifiers in keratoconus detection from orbscan ii examinations. Clinics. 2010;65(12):1223–1228. doi: 10.1590/S1807-59322010001200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2015. Rethinking the inception architecture for computer vision; pp. 2818–2826. 2016-Decem. [DOI] [Google Scholar]
- 61.Tan M., Le Q. V. EfficientNet: rethinking model scaling for convolutional neural networks. arXiv. Published online May 28, 2019. Available at: http://arxiv.org/abs/1905.11946.
- 62.Tan Z., Simkin S., Lai C., Dai S. Deep learning algorithm for automated diagnosis of retinopathy of prematurity plus disease. Transl Vis Sci Technol. 2019;8(6):23. doi: 10.1167/tvst.8.6.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tham Y.C., Li X., Wong T.Y., Quigley H.A., Aung T., Cheng C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 64.The International Agency for the Prevention of Blidness. Global Vision Impairment Fact. The International Agency for the Preventionof Blidness (IAPB). Published 2019. Available at: https://www.iapb.org/vision-2020/who-facts/. Accessed June 21, 2020
- 65.Ting D.S.W., Cheung C.Y.L., Lim G., et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA J Am Med Assoc. 2017;318(22):2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaswani A., Shazeer N., Parmar N., et al. Attention is all you need. In: Advances in neural information processing systems. Vol 2017-December. Neural information processing systems foundation; 2017:5999-6009. Available at: https://arxiv.org/abs/1706.03762v5. Accessed January 23, 2021
- 67.Abràmoff M.D., Lou Y., Erginay A., et al. Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning. Investig Ophthalmol Vis Sci. 2016;57(13):5200–5206. doi: 10.1167/iovs.16-19964. [DOI] [PubMed] [Google Scholar]
- 68.Agurto C., Simon Barriga E., Murray V., et al. Automatic detection of diabetic retinopathy and age-related macular degeneration in digital fundus images. Investig Ophthalmol Vis Sci. 2011;52(8):5862–5871. doi: 10.1167/iovs.10-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antony B.J., Maetschke S., Garnavi R. Automated summarisation of SDOCT volumes using deep learning: transfer learning vs de novo trained networks. Hu J, editor. Automated summarisation of SDOCT volumes using deep learning: transfer learning vs de novo trained networksPLoS One. 2019;14(5) doi: 10.1371/journal.pone.0203726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aslam T.M., Zaki H.R., Mahmood S., et al. Use of a neural net to model the impact of optical coherence tomography abnormalities on vision in age-related macular degeneration. Am J Ophthalmol. 2018;185:94–100. doi: 10.1016/j.ajo.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 71.Viberg Å. The verbs of perception: a typological study. Linguistics. 1983;21(1) doi: 10.1515/ling.1983.21.1.123. Accessed June 15, 2020. [DOI] [Google Scholar]
- 72.von der Emde L., Pfau M., Dysli C., et al. Artificial intelligence for morphology-based function prediction in neovascular age-related macular degeneration. Sci Rep. 2019;9(1):11132. doi: 10.1038/s41598-019-47565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J., Ju R., Chen Y., et al. Automated retinopathy of prematurity screening using deep neural networks. EBioMedicine. 2018;35:361-368. doi:10.1016/j.ebiom.2018.08.033 [DOI] [PMC free article] [PubMed]
- 74.WHO. Bulletin of the World Health Organiztion. 1994;1(5):1–6. Available at: https://www.ncbi.nlm.nih.gov/pmc/issues/169786/. Accessed June 21, 2020
- 75.Xie Y., Nguyen Q.D., Hamzah H., et al. Artificial intelligence for teleophthalmology-based diabetic retinopathy screening in a national programme: an economic analysis modelling study. Lancet Digit Heal. 2020;2(5):e240–e249. doi: 10.1016/S2589-7500(20)30060-1. [DOI] [PubMed] [Google Scholar]
- 76.Yim J., Chopra R., Spitz T., et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat Med. 2020;26(6):892–899. doi: 10.1038/s41591-020-0867-7. [DOI] [PubMed] [Google Scholar]
- 77.Zheng Y., Hijazi M.H.A., Coenen F. Automated “disease/no disease” grading of age-related macular degeneration by an image mining approach. Investig Ophthalmol Vis Sci. 2012;53(13):8310–8318. doi: 10.1167/iovs.12-9576. [DOI] [PubMed] [Google Scholar]
Further reading
- 78.Brown J.M., Campbell J.P., Beers A., et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmology. 2018;136:803–810. doi: 10.1001/jamaophthalmol.2018.1934. American Medical Association. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brown T.B., Mann B., Ryder N., et al. Language models are few-shot learners. arXiv. 2020;1(May):1–7. https://arxiv.org/abs/2005.14165 Available at: Accessed January 22, 2021. [Google Scholar]
- 80.Burlina P.M., Joshi N., Pekala M., Pacheco K.D., Freund D.E., Bressler N.M. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135(11):1170–1176. doi: 10.1001/jamaophthalmol.2017.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao K., Xu J., Zhao W.Q. Artificial intelligence on diabetic retinopathy diagnosis: an automatic classification method based on grey level co-occurrence matrix and naive Bayesian model. Int J Ophthalmol. 2019;12(7):1158–1162. doi: 10.18240/ijo.2019.07.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coyner A.S., Swan R., Campbell J.P., et al. Automated fundus image quality assessment in retinopathy of prematurity using deep convolutional neural networks. Ophthalmol Retin. 2019;3(5):444–450. doi: 10.1016/j.oret.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darlow B.A., Gilbert C. Retinopathy of prematurity – a world update. Semin Perinatol. 2019;43(6):315–316. doi: 10.1053/j.semperi.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 84.Davis M.D., Gangnon R.E., Lee L.Y., et al. The age-related eye disease study severity scale for age-related macular degeneration: AREDS report no. 17. Arch Ophthalmol. 2005;123(11):1484–1498. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]