Abstract
CISD-1/mitoNEET is an evolutionarily conserved outer mitochondrial membrane [2Fe-2S] protein that regulates mitochondrial function and morphology. The [2Fe-2S] clusters are redox reactive and shown to mediate oxidative stress in vitro and in vivo. However, there is limited research studying CISD-1/mitoNEET mediation of oxidative stress in response to environmental stressors. In this study, we have determined the X-ray crystal structure of Caenorhabditis elegans CISD-1/mitoNEET homologue and evaluated the mechanisms of oxidative stress resistance to the pro-oxidant paraquat in age-synchronized populations by generating C. elegans gain and loss of function CISD-1 models. The structure of the C. elegans CISD-1/mitoNEET soluble domain refined at 1.70-Å resolution uniquely shows a reversible disulfide linkage at the homo-dimeric interface and also represents the N-terminal tail domain for dimerization of the cognate kinesin motor protein KLP-17 involved in chromosome segregation dynamics and germline development of the nematode. Moreover, overexpression of CISD-1/mitoNEET in C. elegans has revealed beneficial effects on oxidative stress resistance against paraquat-induced reactive oxygen species generation, corroborated by increased activation of the p38 mitogen-activated protein kinase (MAPK) signaling cascade.
Keywords: CISD-1, Kinesin, Crystal structure, Paraquat, Environmental stress, Oxidative stress, Nematode
1. Introduction
CISD-1/mitoNEET is a homo-dimeric outer mitochondrial membrane protein harboring unique [2Fe-2S] clusters coordinated by three cysteines and one histidine, thus belonging to the CDGSH iron sulfur containing protein family [1-6]. Human isoforms of this protein family include homo-dimeric CISD-2/Miner1/NAF1 [7], localized mainly in the endoplasmic reticulum membrane and having significant structural similarity to CISD-1/mitoNEET, and monomeric CISD-3/Miner2/MiNT [8], located at the mitochondrial inner membrane and harboring two oxygen-labile [2Fe-2S] clusters per single polypeptide chain. All of these iron–sulfur clusters are redox active, but substantial differences in their oxygen sensitivities across different isoforms imply some differences in their physiological functions in regulating mitochondrial metabolism, morphology and dynamics [9-13]. Gain and loss of function studies have shown that the mammalian CISD-1/CISD-2/CISD-3 isoforms play critical roles in oxidative stress and the etiology of multiple disease states including diabetes, breast cancer, and neurodegenerative diseases such as Parkinson’s disease [14].
Oxidative stress is a phenomenon that occurs when there is an imbalance between reactive oxygen species (ROS) production and antioxidant defense mechanisms. Mitochondria are the primary producers of ROS, mainly in the form of superoxide free radicals (O2•−), which are generated as by-products of oxidative phosphorylation along the electron transport chain (ETC) [15]. During oxidative phosphorylation, the ETC transfers electrons from respiratory substrates to molecular oxygen, with concomitant formation of the proton motive force to generate ATP. During this process, respiratory complexes I and III have been proposed to be the major sites of action for O2•− formation in mammalian mitochondria, as redox reaction by-products [15]. Although O2•− is the most prominent ROS found within the cell, other forms of ROS include hydrogen peroxide (H2O2) and hydroxyl free radicals (OH•−), which can interact with free iron via Fenton chemistry reactions to induce DNA damage, lipid peroxidation, and ferroptosis [16-18].
ROS are important signaling molecules to maintain physiological homeostasis via activation of transcription of antioxidant enzymes to neutralize these ROS. Previous in vitro CISD-1/mitoNEET inhibition/knockout studies have reported elevated ROS production and oxidative stress levels are increased compared to wild-type controls; these changes simultaneously affected antioxidant defense mechanism. Furthermore, in vivo CISD-1/mitoNEET knockout mouse models displayed increased ROS production, especially in the brain [19], collectively suggesting that CISD-1/mitoNEET is crucial to mediation of oxidative stress. Knocking down/out mammalian CISD-1/mitoNEET leads to significant dysregulation of the mitochondrial bioenergetics, causing increased ROS production including O2•− and H2O2 in addition to nitric oxide. In aging, a possible role of CISD isoforms in the fission and mitophagy processes may have a beneficial effect of removing older less efficient mitochondria and allowing for newly formed mitochondria to support more efficient bioenergetics [20].
The CISD/NEET protein homologues have been found in a variety of phylogenetically distantly related organisms [21]. In free-living transparent nematode Caenorhabditis elegans, which serves as a simple multicellular model eukaryotic system, one cisd-1 homologue (annotated as W02B12.15) and two cisd-3 homologues (namely cisd-3.1 and cisd-3.2) have been detected: the cisd-1 gene putatively encodes two alternative protein isoforms CISD-1a and CISD-1b, with lengths of 103 or 134 amino acids, respectively, depending on the choice of initiation codon [22], whereas no cisd-2 homologue was detected. Interestingly, C. elegans CISD-1/mitoNEET is shown to play an important role in mitochondrial health and bioenergetics, as seen in mammalian cells, and loss of CISD-1/mitoNEET in C. elegans has led to an increase in ROS, a decrease in ATP production, and changes in the mitochondrial ultrastructure in larval stages with a higher number of hyperfused mitochondria indicative of its potential role in the mitophagy process [22-24].
While C. elegans homologues of cisd1 have been detected, the protein structure of CISD-1/mitoNEET in C. elegans and its implications on oxidative stress resistance has yet to be widely evaluated. Therefore, this study aims to determine the X-ray crystal structure of C. elegans CISD-1/mitoNEET and the potential role of this protein in oxidative stress resistance to environmental stressors. As a prototypical inducer of oxidative stress, an herbicide and pro-oxidant, paraquat (PQ, 1,1’-dimethyl-4,4’-bipyridinium dichloride) [25,26], was applied to induce ROS generation in aged C. elegans populations, and the effect of CISD-1/mitoNEET overexpression on oxidative stress development was analyzed. Previous studies have shown that agriculture workers exposed to chronic PQ use have been linked to elevated oxidative stress in the brain and ultimately developing neurodegeneration as seen with Parkinson’s and Alzheimer’s diseases [27-29]. Upon membrane potential-driven uptake of the PQ2+ dication by the energized mitochondria, PQ is largely retained within the mitochondria and interacts with the ETC, accepting an electron to generate PQ•+ [25,30]. PQ•+ then interacts with O2 to consequentially generate O2•− and the PQ2+ dication is regenerated, allowing for this process to repeat and thereby ultimately leads to oxidative stress within the cells [25]. Our present study is aimed at filling the gap of knowledge in the field by using CISD-1/mitoNEET gain and loss of function C. elegans models to evaluate ROS generation and mediation under high-dose acute PQ exposure [31-33].
2. Materials and methods
Sample preparation
Escherichia coli strain JM109 (TaKaRa, Japan) used for cloning was cultured in Luria-Bertani Lennox (LB) medium, with 50 μg/mL kanamycin when required. Chemicals mentioned in this study were of analytical grade in addition to using Milli-Q water purified via Millipore Milli-Q purification system. Optical absorption spectra of recombinant protein samples were measured by using a Beckman DU7400 spectrophotometer.
The protein sequence of C. elegans CISD-1a/mitoNEET homologue was identified from WormBase (W02B12.15), and the codon-optimized, entire W02B12.15 cDNA clone in pET3a vector was synthesized (pNU190 WGEL020101 T7p-T7tag-6HIS-mitoNEET(W02B12_15synth)-T7u, Entelechon GmbH, Germany). In efforts to unveil the structural details of this worm protein, a portion of the W02B12.15 cDNA clone coding for the water-soluble domain (residues 32-103) of C. elegans mitoNEET/CISD-1a was amplified by polymerase chain reaction (PCR) using the synthetic W02B12.15 cDNA clone and sets of the following PCR primers (designed based on the reported nucleotide sequences): 5′-GCT AGC GGA AAG AAG TTC TAC GCT AAA TTC GGT CAG CGT TCT G′ and 5′-CTC GAG CTA TTT TTT TTC AGA TTT AAC GAT CAG CG-3′. The PCR primers included twenty-one extra bases (coding for an engineered Ser-Gly linker and Lys-Lys-Phe-Tyr-Ala sequence from the Rattus norvegicus (rat) heart mitoNEET/CISD-1 sequence [34]) immediately upstream of the codon for conserved Lys32 of the target gene: the engineering of these extra amino acid sequences significantly improved heterologous overexpression of the reddish recombinant protein amenable for the subsequent structural studies. The amplified PCR product was digested with NheI and XhoI, and then inserted into the NheI/XhoI site of a pET28a vector. The pET28aCENEET-SD2 expression vector was transformed into the host strain, E. coli CodonPlus(DE3)-RIL (Stratagene), and used for the following sample preparations.
For preparation of the C. elegans mitoNEET/CISD-1a soluble domain sample, the transformants were grown overnight at 25 °C in a 3 L culture of LB medium containing 50 μg/mL kanamycin and 0.2 mM FeCl3 (Wako Pure Chemicals, Tokyo, Japan), and the recombinant holoprotein was overproduced with 1 mM isopropyl β-D-thiogalactopyranoside for 19 h at 25 °C. The cells were pelleted by centrifugation, and the recombinant C. elegans mitoNEET (residues 32-103, see below) having a hexa-histidine-tag plus a thrombin cleavage site and an engineered extra linker sequence was purified essentially as reported previously for archaeal Rieske [2Fe-2S] proteins [35], except that the entire purification was performed at 4°C using buffers adjusted at pH 8.0 and that the heat treatment of the crude cell lysate was omitted. After proteolytic removal of the hexa-histidine tag from the purified recombinant protein for 16–22 h at 4 °C using a thrombin cleavage capture kit (Novagen) according to the manufacturer’s instructions, the sample was further purified by gel-filtration chromatography (Sephadex G-75; Amersham Pharmacia Biotech), eluted at room temperature with 10 mM HEPES-NaOH buffer, pH 8.0, containing 500 mM NaCl, and then concentrated with Centriprep-10 and Microcon-YM10 apparatuses (Amicon) to ~4 mM (per protomer), rapidly frozen in liquid nitrogen, and stored at −80 °C until use. The resulting purified protein has the following deduced amino acid sequence: GSHMASSGKKFYA 32KFGQRSARCN YK-IQLDSNKI VDTVDIEDIG EKKAFCRCWK SEKWPYCDGS HGKHNKETGD NVGPLIVKSE K103K.
Crystallization, diffraction data collection, and refinement
The purified recombinant C. elegans mitoNEET/CISD-1a soluble domain, containing fifteen extra linker residues (GSHMASSGKKFYAKF) at the N-terminus, which was engineered to improve overproduction and facilitate rapid purification and is absent in the CISD-1a sequence (WormBase accession number W02B12.15), was crystallized under oxygenic conditions by the standard hanging drop vapor diffusion technique. The final conditions were droplets consisting of 1–2 μl of purified protein solution and 1-2 μl of the reservoir solution (100 mM CHES-NaOH, pH 9.5, and 30%(w/v) PEG 3000 (Emerald Biostructures Wizard Classic 1, #41), to which 5%(v/v) of 1 M Bicine, pH 8.5, was added (i.e., 95 mM CHES-NaOH, 50 mM Bicine, 28.5% (w/v) PEG 3000 in the final concentrations)) in the protein well equilibrated against the reservoir solution. Reddish single-crystals (see Fig. 2, inset) were obtained in 1-3 weeks at 4 °C (under oxygenic conditions without any reducing reagents). They were transferred to a cryoprotective solution containing 32.5%(w/v) PEG3000 (Rigaku, Ltd) and frozen in liquid nitrogen.
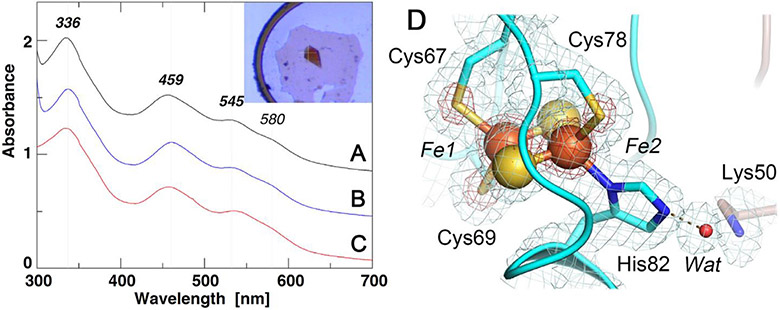
Fig. 2.
UV-visible absorption spectra of the selected NEET proteins (A-C) and an expanded view of the [2Fe-2S] cluster site of C. elegans CISD-1/mitoNEET (D). A-C) UV-visible absorption spectra of rat CISD1/mitoNEET [34] (A), C. elegans CISD-1/mitoNEET (inset, its single crystal) (B), and a thermophile homologue, TthNEET, from Thermus thermophilus HB8 [56] (C) (shown with an offset interval of +0.4 absorbance unit for clarity); D) An expanded view of the [2Fe-2S] cluster site of C. elegans CISD-1/mitoNEET together with the observed 2Fo-Fc electron density map contoured at 1.0 σ (gray) and 5.0 σ (red).
X-ray diffraction data were collected from flash-frozen, small crystals at the SPring-8 beamline BL41XU [36] using a beam size of 25(V) x 9(H) (μm2) and PILATUS3 6M detector with a detector distance of 250 mm. The data collection was performed at a wavelength of 1.0 Å with a total rotation range of 105° and rotation step of 0.5°/frame. The data was processed up to 1.70-Å resolution by using the XDS package [37]. The crystal belongs to the tetragonal space group P41212, with unit cell parameters a = b = 37.798 Å and c = 82.260 Å (Table 1). There is a single protein molecule per asymmetric unit.
Table 1.
Crystallographic data processing and refinement statistics.
| parameters | C. elegans CISD-1/mitoNEET |
|---|---|
| Data processing | |
| space group | P41212 |
| cell dimensions a = b, c (Å) | 37.798, 82.26 |
| resolution range (Å) | 34.35 - 1.70 (1.76 - 1.70) |
| total number of reflections | 49841 (4986) |
| number of unique reflections | 7056 (686) |
| completeness (%) | 99.77 (99.85) |
| redundancy | 7.1 (7.3) |
| <I/σ(I)> | 19.83 (3.50) |
| Wilson B-factor (Å2) | 25.59 |
| R merge | 0.04914 (0.472) |
| R meas | 0.05317 (0.5081) |
| R pim | 0.01976 (0.1846) |
| CC(1/2) | 0.999 (0.907) |
| Refinement | |
| number of reflections used in refinement | 7055 (685) |
| number of TLS groups used in refinement | 1 |
| R work | 0.1688 (0.2117) |
| R free | 0.1901 (0.2537) |
| number of non-hydrogen atoms | 510 |
| protein | 487 |
| ligand/ion | 4 |
| water | 19 |
| number of protein residues | 63 |
| average B-factor (Å2) | 34.03 |
| protein | 34.03 |
| ligand/ion | 22.57 |
| water | 36.58 |
| RMS (bonds) (Å) | 0.014 |
| RMS (angles) (°) | 1.47 |
| Ramachandran favored (%) | 93.44 |
| allowed (%) | 6.56 |
| outliers (%) | 0.00 |
| rotamer outliers (%) | 0.00 |
| clashscore | 0.00 |
Statistics for the highest-resolution shell are shown in parentheses.
The structure was determined by the molecular-replacement method with the program Phaser [38] using the atomic model of rat mitoNEET (manuscript in preparation) after editing by a program Sculptor [39]. The structure was corrected manually with Coot [40] and initially refined by restraint refinement (with TLS (default with one TLS group), partial occupancy settings) with the Phenix 1.1192.2_4158 program suite [41]. The resulting early model gave multiple conformations for Cys40 with an occupancy of ~0.7 for conformer A and ~0.3 for conformer B, respectively, located at the dimer interface within a cutoff distance of 3.0 Å for the possible Sγ-Sγ, bond length [42,43] in the lattice. Because the chirality of the disulfide linkage is a stereo-electronic consequence of the four free electron pairs on the two sulfide atoms that interact by repulsive forces with the neighboring β-carbon-containing groups, two energetically favorable, mirror-image and equally populated conformations for the average C1β-S1γ-C2γ-C2β torsion angle around −87° (left-handed) and +97° (right-handed) have been reported for 1505 native structures [42-44]. Close inspection of early structural models suggested Cys40Sγ(conformer A) from one monomer and Cys40Sγ(conformer B) from the other as the most likely combination to fulfill these stereochemical criteria for potential disulfide bond formation. Therefore, an early model with a (chemically reasonable) fixed occupancy of 0.5 for both conformers A and B of Cys40, was further refined by restraint refinement (with 7 cycles of simulated annealing (default Cartesian) and TLS (default with one TLS group) settings, followed by 5 cycles of TLS (default with one TLS group) settings) with the Phenix program suite [41]. The residue number for the refined mitoNEET structure quoted in this article refers to that of the C. elegans CISD-1a isoform in WormBase (W02B12.15) [22]. The refined structural model includes the CISD-1a residues 38-100 (see Fig. 4A), with Cys40 forming a possible reversible disulfide bond with the SAγ-SBγ bond length of 2.5 Å and CAβ-SAγ-SBγ-CBβ torsion angle of +96° (right-handed) at the dimer interface (see Fig. 3C), one [2Fe-2S] cluster and 19 water molecules, and does not contain the engineered extra linker residues (GSHMASSGKKFYAKF) from the pET28aCENEET-SD2 expression vector which are disordered and not resolved in the electron density. The final R factor and Rfree factor were 0.169 and 0.190, respectively (Table 1). Analyses of the stereochemical quality of the models were accomplished using MolProbity [45] in the Phenix program suite and wwPDB validation server [46] prior to deposition, and the final structure was deposited in the Protein Data Bank (PDB)[46] with the accession code 7YVZ. The structural figures presented in this article were generated with PyMOL (Schrödinger, LLC).
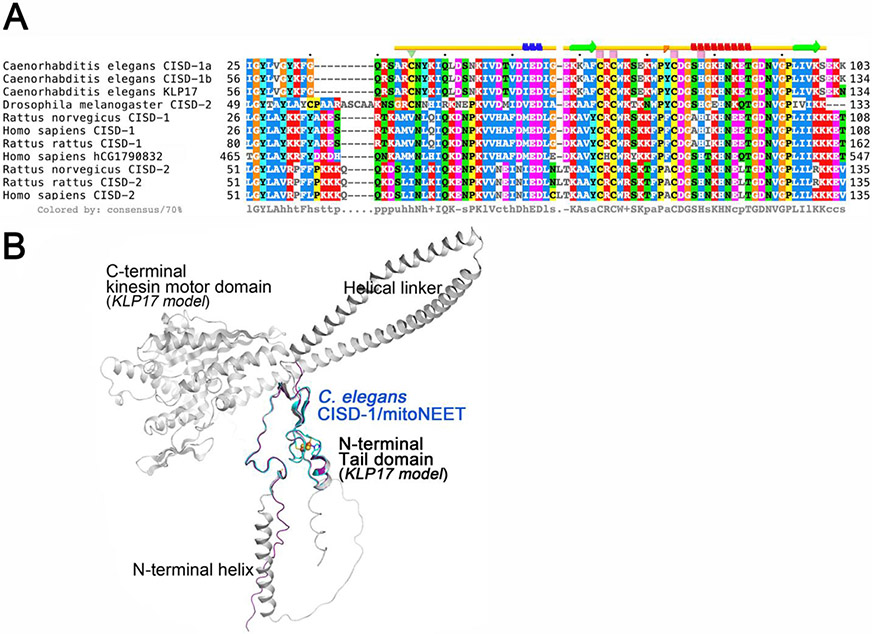
Fig. 4.
Multiple sequence alignment of selected CISD-1/CISD-2 and related proteins (A) and the superposition of the crystal structure of C. elegans CISD-1/mitoNEET protomer (cyan) on the predicted structural models by ColabFold [53] of the cognate kinesin motor protein KLP17 [61, 62] (gray) and the entire amino acid sequence region of purified C. elegans CISD-1/mitoNEET used for crystallization (purple) (B). A) Multiple amino acid sequence alignments of the selected proteins were performed by using the EMBL-EBI Clustal Omega server [80], and the secondary structural elements in the crystal structure of C. elegans CISD-1/mitoNEET (top) were analyzed by using the STRIDE server [81]; B) Note that the N-terminal tail domain of KLP17 (gray) exhibits the identical structure as found for the crystal structure (cyan) and the ColabFold [53]-predicted structural model (purple) of the C. elegans CISD-1/mitoNEET soluble domain.
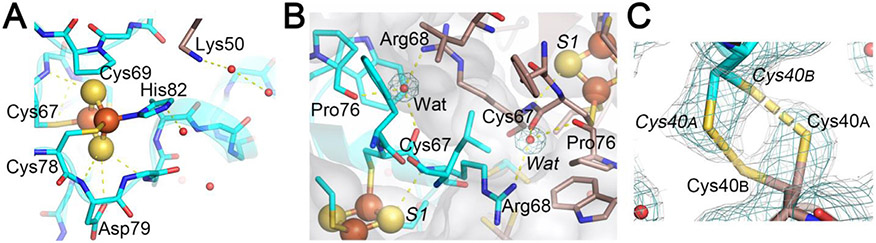
Fig. 3.
Expanded views of the N-H…S hydrogen-bonding interactions around the [2Fe-2S] cluster (A), the interprotomer hydrogen-bonding interactions between the two [2Fe-2S] clusters (B), and a reversible disulfide linkage (right-handed) contributed from Cys40 conformer A from one protomer and conformer B from the other protomer, together with the observed 2Fo-Fc electron density map contoured at 1.0 σ (cyan) and 0.6 σ (gray) (C) in the C. elegans CISD-1/mitoNEET structure. In C), the electron density map contoured at 0.6 σ (gray) in PyMOL (Schrödinger, LLC) resembles that observed contoured at 1.0 σ in Coot [40] used for the refinement (not shown).
C. elegans strains and maintenance
The following C. elegans strains were used in this study: Bristol N2 acquired from the CGC (Minneapolis, MN), and two transgenic strains sourced from Knudra (now NemaLife Inc.) a transgenic mitoNEET knockout (KO) and a transgenic mitoNEET over-expresser (TG). All strains were maintained at 20 °C on Nematode Growth Medium (NGM) agar plates seeded with 24 h culture of E. coli OP50 as described in WormBook, and the NGM-dead method was followed as previously described [47,48]. Age-synchronized populations were generated via bleaching as described in WormBook [47], transferred onto NGM agar plates at the L3/L4 larval stage containing 100 μg/mL FuDR [49] (Sigma Aldrich, Cat No. F0503) to prevent egg laying and were collected at the 5th day of adulthood for all assessments in this study.
Oxidative stress induction
Paraquat (PQ, 1,1’-dimethyl-4,4’-bipyridinium dichloride, Sigma Aldrich, Cat No. 856177) was used to induce oxidative stress. In brief, age-synchronized populations were collected and exposed to 100 mM PQ in M9 buffer for 2 h [26]. After PQ exposure, worms were washed 3 times in M9 buffer to remove excess PQ to prepare for processing as described in the methods below.
Oxidative stress assessment
Total oxidative stress was assessed via CellROX Green staining (Invitrogen, Cat No. C10444). In brief, worms were fixed for 1 h in 2% paraformaldehyde in PBS, washed 3 times in 1X phosphate buffered saline pH 7.4 (PBS), stained with 5 μM CellROX Green for 30 min, and washed an additional 3 times in PBS prior to mounting onto microscope slides [50]. Rinsed worms were mounted onto microscope slides in PBS and green fluorescence was captured using a Leica DM6B microscope. Images for CellROX Green quantification were taken at the 2.5X objective while representative images shown in this paper were taken at the 10X objective.
Mitochondrial ROS assessment
Mitochondrial ROS assessments were conducted as previously described [51] with brief modifications. Approximately 2000 worms per strain from both basal and PQ-challenged groups were collected, washed 3 times with M9 buffer, and pelleted via centrifugation at 13,000 x g for 10 min. After centrifugation, excess supernatant was removed, and the pellets were frozen in liquid nitrogen for at least 15 min or stored at −80 °C until ready to be processed. To generate worm lysates, 1 mL of ice cold M9 buffer was added to the frozen pellet and sonicated on ice at 70% amplitude for 3 min using 10 s on/off pulse intervals. After sonication, worm lysates were centrifuged at 12,000 x g for 20 min at 4 °C. Then, 50 μL of worm lysates were added to the wells of a black, opaque bottom 96-well plate, incubated with 5 μM mitoSOX Red (Invitrogen, Cat No. M36008) for 30 min in the dark at room temperature, and the fluorescence was quantified via plate reader (Synergy 4, BioTek) at Ex/Em 510/580 nm.
Hydrogen peroxide assessment
Hydrogen peroxide (H2 O2) formation was assessed via Amplex Red Hydrogen Peroxide/Peroxidase (Thermo Fisher, Cat. No A22188). Worm populations, oxidative stress induction, and lysates followed procedures as described above with slight modifications. Approximately 2000 worms per strain from both basal and PQ-challenged groups were collected, washed 3 times in 1X PBS, and pelleted via centrifugation at 13,000 x g for 10 min. After centrifugation, excess supernatant was removed, and the pellets were frozen in liquid nitrogen for at least 15 min or stored at −80 °C until ready for use. Lysates were generated via sonication by adding 1 mL of ice cold 1X PBS to the frozen pellets and sonicated on ice as described above. Amplex Red Hydrogen Peroxidase/Resofurin assessment was conducted according to manufacturer’s protocols, incubated in the dark at room temperature for 30 min, and fluorescence was quantified via plate reader (Synergy 4, BioTek) at Ex/Em 530/590 nm.
Immunoblotting and protein analysis
Oxidative stress was induced to populations of approximately 500 worms as described above. Worms from PQ-challenged groups were washed 3 times with M9 buffer to remove excess PQ and centrifuged to form a pellet with excess supernatant removed as described above. Worm pellets were flash-frozen in liquid nitrogen for at least 15 min or frozen at −80 °C until ready for processing. Protein extraction was conducted via boiling in sample buffer containing 2% SDS and 2.5% β-mercaptoethanol for 10 min in a 95 °C water bath as described previously [52]. Equal volumes of extracted protein from the samples were loaded into 4-20% SDS-PAGE gels (Bio-rad, Cat No. 4560193), transferred onto nitrocellulose membranes, and blocked for 1 h at room temperature in 5% bovine serum albumin (BSA, fatty acid free) in 0.1% Tween-20/PBS pH 7.4. mSOD2 (Cell Signaling Technology, Cat No. D3X8F), p38 MAPK (PMK1; Cell Signaling Technologies, Cat No. D13E1), phoshpo-p38 MAPK (pPMK1; Cell Signaling Technologies, Cat No. D3F9), and β-actin (Proteintech, Cat No. 66009) were used at 1:1000 dilutions and incubated overnight at 4 °C. Goat Anti-Rabbit IgG (H+L)-HRP Conjugate (Bio-rad, Cat No. 1706515) and Goat Anti-Mouse IgG (H+L)-HRP Conjugate (Bio-rad, Cat No. 1706516) secondary antibodies were used at 1:2000 dilutions and visualized via chemiluminescence using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Cat No. PK209238). Membranes were imaged via the Amersham Imager AI680. Band intensities were quantified via ImageJ. mSOD2 band intensities were normalized to β-actin while phosphorylated p38-MAPK bands intensities were normalized to p38-MAPK.
Image analysis
CellROX Green fluorescence pixel intensity was quantified via ImageJ. In brief, the integrated density of CellROX Green fluorescence was normalized to the area of individual worms corrected for background autofluorescence by subtracting the calculated difference from unstained worms.
Statistical analysis
Statistical analysis was conducted via one-way ANOVA followed by Dunnet’s post-hoc test using GraphPad Prism 9.2.
3. Results and discussion
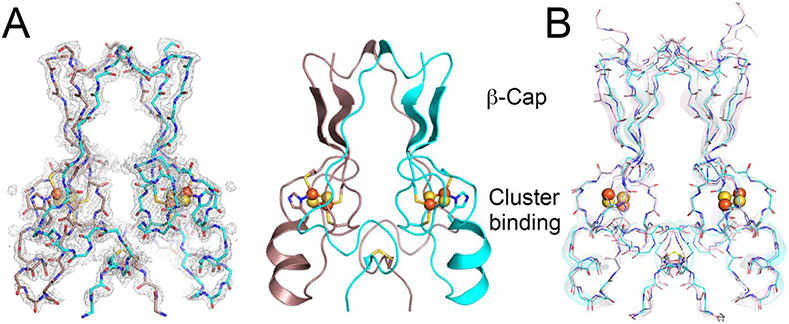
Primordial CISD-1/mitoNEET of C. elegans, encoded by the W02B12.15 gene, is a mitochondrial outer membrane protein whose X-ray crystal structure has not been reported previously [22,24]. The 1.70-Å structure of the soluble-domain of the worm CISD-1 successfully refined to an R factor of 16.9% (Rfree = 19.0%) in this study (Table 1) reveals a tightly packed homo-dimeric structure in the lattice, with each protomer related via a 180° rotation along the crystallographic dyad axis (Fig. 1). Each protomer is folded into two domains, namely, the β-cap domain and the cluster-binding domain, respectively (Fig. 1A). The β-cap domain of C. elegans CISD-1 consists of two antiparallel β-strands and a 310 helix, but does not contain the third parallel β-strand coming from the other protomer found in the closely related, “NEET” fold structures of human CISD-1/mitoNEET [3-6] and CISD-2/Miner1/NAF1 [7] (Fig. 1A,B; see also Fig. 4A, top). Superposition of these three NEET protein backbone structures showed key variations around the 310 helix and adjacent loop regions in the β-cap domain of C. elegans CISD-1, resulting in a backbone conformational shift unfavorable for forming the third parallel β-strand at the corresponding positions, while the cluster-binding domain and the [2Fe-2S] cluster core of the protomers are remarkably superimposable (Fig. 1B). Structure prediction of C. elegans CISD-1 using ColabFold that combines the fast homology search of MM-seqs2 with AlphaFold2 or RoseTTAFold [53], also indicated the presence of two anti-parallel β-strands in the β-cap domain of the predicted model, being in line with the refined structure (see Fig. 4).
Fig. 1.
Overall structure and domain topology of C. elegans CISD-1/mitoNEET homodimer (W02B12.15) refined at 1.70-Å resolution. A) The backbone tracing (left) and the cartoon representation (right) of the C. elegans CISD-1/mitoNEET soluble domain. Also shown is the observed 2Fo-Fc electron density map (left, gray) contoured at 1.5 σ. Protomers colored in dark pink and cyan, iron in red, sulfur in yellow; B) The backbone superposition of C. elegans CISD-1/mitoNEET (cyan) on human CISD1/mitoNEET (PDB code 2QH7 [3], gray) and human CISD2/Miner1 (PDB code 3FNV [7], pink).
The cluster-binding domain of C. elegans CISD-1 consists of a cluster-binding loop with the consensus CXC-//-CDGSH motif, followed by a single α-helix, and harbors the NEET-type [2Fe-2S] cluster coordinated by one histidine and three cysteine residues, exhibiting the characteristic UV–visible absorption spectra (Fig. 2, 3A). The [2Fe-2S] cluster of C. elegans CISD-1 soluble domain is significantly more stable than regular protein-bound [4Fe-4S] clusters [54,55], as evident by the well-defined electron density for the [2Fe-2S] cluster core (Fig. 2D), even after weeks of the hanging drop vapor diffusion setups for crystallization under oxygenic conditions. In the homo-dimeric structure in the lattice, the two [2Fe-2S] clusters are ~14.6 Å apart as measured by the closest, innermost Fe1-Fe1 distance (coordinated by Cys67 and Cys69) and ~17.8 Å by the outermost Fe2-Fe2 distance (coordinated by Cys78 and His82), suggesting that a rapid interprotomer electron transfer equilibrium between them would occur. Interestingly, the bridging sulfide (S1) of the cluster engages in N-H…S hydrogen-bonding interaction with Arg68Nα, whose guanidium group also interacts with the mainchain oxygen atoms of Cys67 (one of the ligand residues to the innermost Fe1 site) and Pro76 (in a cis conformation) of the other protomer via a structurally well-conserved, interior water molecule (Wat) by the interprotomer hydrogen bond network (Fig. 3B). Pro56, along with Phe66, Trp70, Trp75, Leu96, also constitute the hydrophobic core at the homodimer interface. Our previous pulsed electron paramagnetic resonance (EPR) analyses clearly showed that the outermost Fe2 site of the [2Fe-2S] cluster is reduced upon chemical reduction in rat mitoNEET and its thermophile homologue [34,56]. In this regard, although C. elegans CISD-1 was crystallized under oxygenic conditions in the absence of any reducing reagent, exposure of the resulting microcrystals to very high flux X-radiation at the SPring-8 beamline BL41XU resulted in conversion to the photo-reduced form, so the outcome probably represents a mixture of redox states: it should be kept in mind that redox state is actually very difficult to ascertain solely from crystal structures, and that complications due to X-ray photoreduction and methods to generate a specific redox state in crystals can often occur as well [57-59].
One of the most unique structural features of C. elegans CISD-1 is the presence of a reversible disulfide linkage [42-44] contributed from Cys40Sγ of each protomer, with the SAγ-SBγ bond length of 2.5 Å and CAβSAγ-SBγ-CBβ torsion angle of +96° (right-handed) at the homo-dimeric interface in the crystal lattice (Fig. 3C). The equivalent cysteine residue is also conserved in the Drosophila melanogaster CISD-2 sequence but not in mammalian CISD1/mitoNEET and CISD2/Miner1/NAF1 homologues [22-24] (Fig. 4A). Thus, this is the first example of a eukaryotic NEET protein of this kind. Previously, Conlan et al. [60] have reported substantial flexibility in the N-terminal tethering arms of the human CISD1/mitoNEET soluble domain structure, despite little variations in the β-cap domain and the cluster-binding domain structures. The presence of a reversible disulfide linkage at the interprotomer interface in C. elegans CISD-1 (Fig. 3C) implies a redox-linked control of multiple orientations on the outer mitochondrial membrane that may be functionally important for partner-protein interactions.
Unexpectedly, the NCBI Protein BLAST search against databases of selected eukaryotic species reveals that C. elegans CISD-1 sequence is highly homologous to not only the CISD-1/mitoNEET and CISD-2/Miner1/NAF1 sequences but also the N-terminal tail domain of the cognate kinesin motor protein KLP17 (W02B12.7 in WormBase, 605 amino acids) [61,62] and the C-terminal domain of human hCG1790832 gene product of poorly defined function (547 amino acids) (Fig. 4A). This was overlooked in the previous studies [22-24] presumably because of their substantially larger sizes of the predicted proteins than the CISD1/CISD2 protein orthologues (typically ~100–150 amino acids). Most strikingly, residues 38–100 covered in the refined structure of C. elegans CISD-1 are completely identical to residues 60-122 in the KLP17 tail domain. Thus, the refined structure of C. elegans CISD-1 soluble domain reported herein also represents that of the homo-dimeric N-terminal tail domain (residues 60-122) of the cognate kinesin motor protein KLP17 [61,62] (Fig. 4). Structure prediction of the entire C. elegans KLP17 protomer using ColabFold [53] suggests an N-terminal signal sequence plus an α-helix region, followed by the CISD-1 tail domain for homo-dimerization, which is further connected to the C-terminal globular kinesin motor protein domain with an ATP binding cassette motif, via two long α-helices linking these two functional domains (Fig. 4B). In the previous studies, Siddiqui and coworkers have reported that the klp-17 transcripts are confined specifically to the cell nucleus during early development, from the one-cell stage of embryogenesis until early larval stages, and that loss of klp-17 via RNAi results in embryonic lethality at the one- or two-cell stage with disorganized mitotic spindles and polyploid nucleus [61,62]. Our present results indicate that C. elegans KLP17 is a unique member of the C-terminal kinesin motor protein family that contains the N-terminal CISD-1 dimerization tail domain harboring the NEET-type [2Fe-2S](His)1 (Cys)3 cluster (Fig. 4) and likely participates in chromosome segregation dynamics in the cell nucleus and germline development. The striking structural similarity of the N-terminal tail domain of KLP17 and CISD-1/mitoNEET (Fig. 4) also implies a possible inter-organellar cross-talk between the cell nucleus and mitochondria by certain redox and/or metabolic status signals in the germline development of C. elegans. Further studies on the worm KLP17 will clarify the structure and physiological function of this new NEET-kinesin motor fusion protein.
C. elegans physiology is sensitive to changes in ROS signaling, where alterations in these signaling pathways can affect mitochondrial hormesis. A possible physiological role of C. elegans CISD-1/mitoNEET in stress response was considered by using transgenic (TG) cisd-1 overexpression and knockout (KO) strains upon exposure to an environmental stressor, PQ. Acute exposure to high concentrations of PQ is known to induce oxidative stress in C. elegans populations [26,63,64]. While many of these studies were conducted at the C. elegans developmental larval stages to eliminate age as a confounding variable, the oxidative theory of aging suggests that, as individuals age, ROS levels increase concurrent with decreases in antioxidant defense mechanisms, leading to reductions in oxidative stress resistance [65]. In this regard, it should be added that mammalian CISD-1/mitoNEET plays a protective role in aging tissues, with both liver and cardiovascular health [66] improved by CISD-1/mitoNEET protein expression via reductions in inflammation: conversely, knocking down/out mitoNEET leads to significant dysregulation of the mitochondrial energetic systems, with increased production of superoxide, hydrogen peroxide and nitric oxide implicated in the dysregulation of cellular processes [16,19,67,68]. The latter in part, seems to be related to the fission/fusion and mitophagy process that has a beneficial effect of removing older less efficient mitochondria and allowing for newly formed mitochondria to support more efficient bioenergetics, and may function as a novel drug target [6,11,14,69-71].
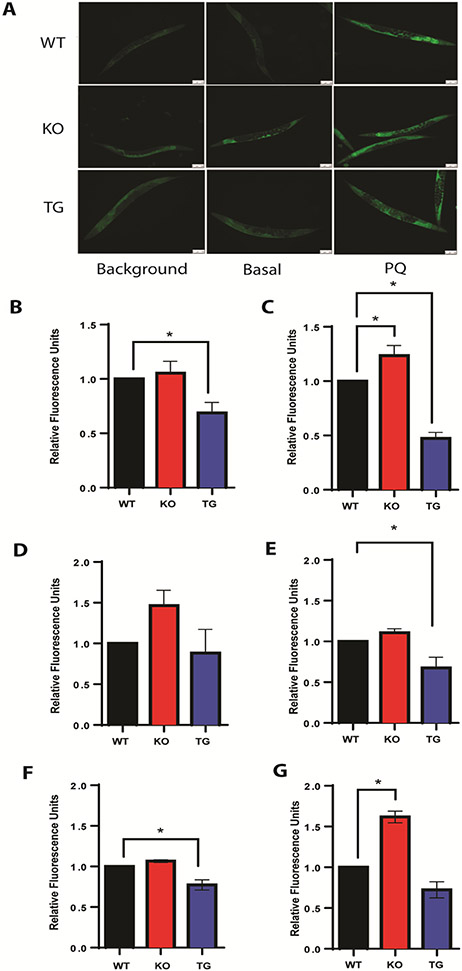
Assessment of overall oxidative stress at basal conditions and PQ-challenged groups in age-synchronized C. elegans populations indicated that PQ-challenged cisd-1 KO populations showed increased overall oxidative stress (Fig. 5A, middle column), while this effect was attenuated with the cisd-1 overexpression in our transgenic over-expresser (TG) model (Fig. 5A, right column), compared to wild-type (WT) controls (Fig. 5A, left column). Under these conditions, compared to the WT controls, cisd-1 overexpression attenuated the induction of oxidative stress by more than half, while the loss of CISD-1/mitoNEET increased overall oxidative stress by 1.25-fold (Fig. 5C), as judged by quantification of the pixel intensities of CellROX Green fluorescence visualized in Fig. 5A.
Fig. 5.
Acute paraquat (PQ) exposure induces oxidative stress in C. elegans. Day 5 Adult worms were exposed to 100 mM PQ for 2 hours. A) Fluorescence microscopy of CellROX Green staining assessing oxidative stress. Compared are the N2 (WT) in the top row, mitoNEET knockout (KO) in the middle row, and mitoNEET over-expressor (TG) in the bottom row under basal (middle column) and stressed conditions (right column). Background images (left column) are shown as controls for auto fluorescence. Images shown are taken at 10X objective and scale bars are 100 μm. B) Quantification of CellROX Green fluorescence intensity in basal populations from (A) shows protection against oxidative stress upon mitoNEET overexpression (TG); C) Quantification of CellROX Green fluorescence intensity in PQ-challenged populations from (A) shows protection against PQ-induced oxidative stress upon mitoNEET overexpression (TG). Data for panels B and C are fluorescence intensities with background subtracted and normalized to respective WT controls. N=33-46 worms. D) Assessment of mitoSOX Red fluorescence intensity in basal populations shows no statistically significant differences in mitochondrial superoxide free radical levels between KO or TG, compared to WT controls; E) Assessment of mitoSOX Red fluorescence intensity in PQ-challenged populations shows reduction of mitochondrial superoxide free radical levels upon mitoNEET overexpression (TG), compared to WT controls; F) Assessment of Amplex Red fluorescence intensity in basal populations shows no statistically significant differences in hydrogen peroxide levels between KO or TG, compared to WT controls; G) Assessment of Amplex Red fluorescence intensity in PQ-challenged populations shows a statistically significant increase in hydrogen peroxide levels upon mitoNEET knockout (KO), compared to WT controls. Each biological replicate consisted of 3-4 technical replicates. All data are normalized to WT controls prior to statistical analysis. Data represented are mean ± SEM. Data are analyzed via One-way ANOVA followed by Dunnet’s post hoc test; * p<0.05.
One of the primary mechanisms that acute, high-dose PQ exposure induces oxidative stress is by increasing mitochondrial O2•− (mO2•−) and ROS production [25]. Although MitoSOX Red has been used to evaluate mO2•− formation in C. elegans models [24,51,72], there is debate within the redox biology field regarding the accuracy of the mitoSOX fluor [73,74]. With this in mind, we used a combination of commercially available fluorescent probes to evaluate ROS production, with mitochondrial ROS detected with MitoSOX Red (Fig. 5D and 5E), and H2O2 detected with Amplex Red Hydrogen Peroxide/Peroxidase (Fig. 5G and 5E) fluorescent probes, similar to previously published studies [51,75]. From these assessments, cisd-1 expression levels influenced basal ROS levels in the worms, with greater levels of mitochondrial ROS in the KO than in the WT or TG (Fig. 5D), whereas a statistical difference was observed for PQ-challenged groups (Fig. 5E). Furthermore, H2O2 levels showed a statistically significant increase in the PQ-challenged KO group compared to WT controls (Fig. 5G), with a small decrease observed in the cisd-1 TG group compared to WT controls under basal conditions (Fig. 5F). The complexity of mO2•− generation will need to be fully elucidated in future studies using orthogonal methods including, but not limited to, EPR spectroscopy.
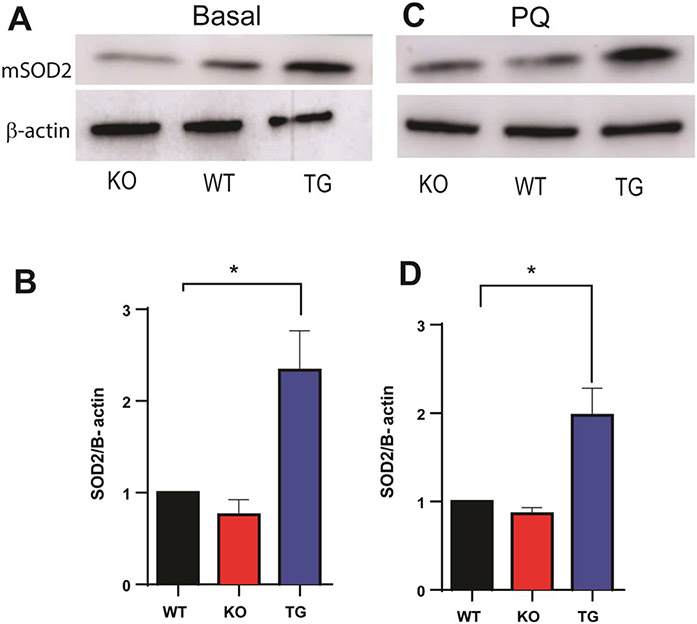
In C. elegans, PQ exposure induces elevated ROS production, which was attenuated in the TG worms. Interestingly, a statistically significant increase in mitochondrial superoxide dismutase (mSOD2) protein expression was observed in the basal and PQ-challenged TG worms, whereas only a small decrease in the KO worms was detected compared to WT controls (Fig. 6). Given the known physiological function of mSOD2 in detoxifying the deleterious ROS production that may affect mitochondrial function and potentially cause secondary cellular problems including lipid peroxidation, this in part could explain a possible mechanism for the reduced ROS level under basal and PQ-challenged conditions in the TG worms, compared to the KO worms, leading to enhanced resistance of the TG worms to the ROS-mediated induction by PQ. Previous studies showed a statistically significant increase in mSOD2 protein expression upon PQ-challenge, corroborating this finding [76,77].
Fig. 6.
Effect of mitoNEET expression on mSOD2 protein level under basal (A, B) and PQ-challenged (C, D) conditions. A and B) mSOD2 protein expression increases with mitoNEET overexpression (TG); C and D) mitoNEET overexpression (TG) leads to an increase in mSOD2 protein expression that persists with PQ. N = 3 biological replicates. Each biological replicate consists of 500 worms. Data are mean ± SEM. Data are analyzed via One-way ANOVA followed by Dunnet’s post hoc test; *p<0.05.
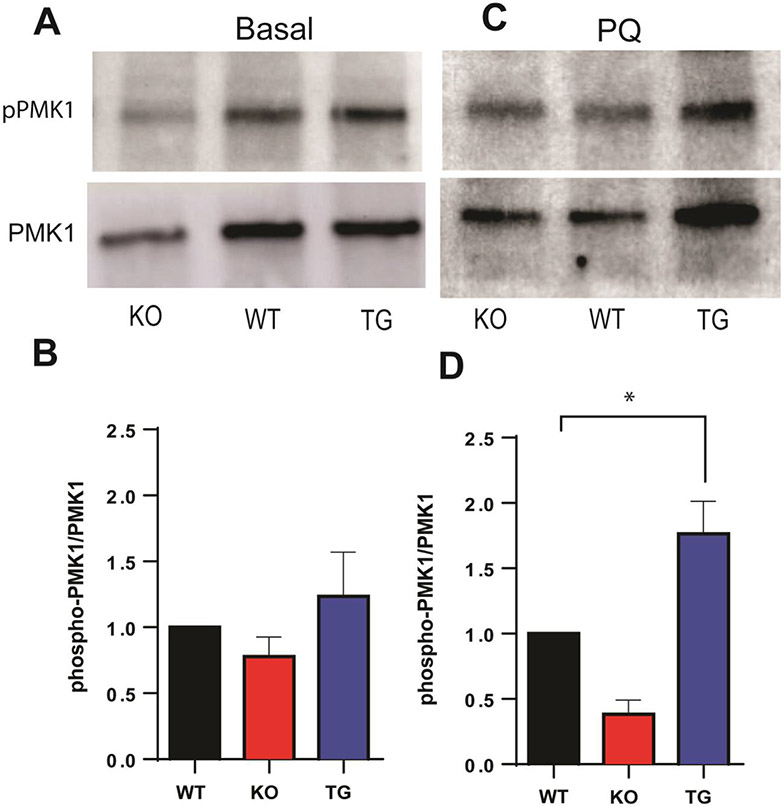
Previous studies have revealed that the evolutionarily conserved, p38 mitogen-activated protein kinase (MAPK) signaling cascade serves as a secondary protective mechanism against environmental stressors; in C. elegans, the p38-MAPK (PMK-1) signaling cascade supports immunity and defense against pathogens and oxidative stress via activation of the Nrf2/Skn-1 antioxidant defense mechanism [77-79]. Therefore, activation of PMK-1 was first analyzed by measuring the phosphorylated p38-MAPK (pPMK-1) protein expression levels under basal conditions, which showed minimal differences amongst cisd-1 KO and TG overexpression strains, compared to WT groups (Fig. 7A, B). When challenged with acute high-dose PQ, a robust increase in phosphorylation level was observed for TG overexpression strains compared to the WT groups as expected, whereas no increase was detected for the KO worms (Fig. 7C, D). This could be in part attributed to a more efficient energy metabolism via oxidative phosphorylation for ATP synthesis upon cisd-1 overexpression in C. elegans mitochondria, and is in line with the role of PQ in promoting mitochondrial ROS production and initiating the p38-MAPK phosphorylation, eventually leading to mitochondrial dysfunction in the worm model system.
Fig. 7.
Effect of mitoNEET expression on p38-MAPK (PMK1) phosphorylation under basal (A, B) and PQ-challenged (C, D) conditions. A and B) no statistically significant differences in phosphorylated p38-MAPK (pPMK1) protein expression levels between mitoNEET knockout (KO) or mitoNEET over-expressor (TG), compared to WT controls; C and D) mitoNEET overexpression (TG) leads to an increase in phosphorylated p38-MAPK (pPMK1) protein levels that persists with PQ. N=3 biological replicated where each replicate consists of 500 worms per strain. Data are mean ± SEM. Data are analyzed via One-way ANOVA followed by Dunnet’s post hoc test; *p<0.05.
4. Conclusions
The [2Fe-2S](His)1(Cys)3 cluster-containing mitochondrial outer membrane protein CISD-1/mitoNEET from C. elegans has been proposed to play versatile roles ranging from mitochondrial metabolism and bioenergetics to overall fitness and germline viability. The 1.70-Å structure of the CISD-1 soluble domain shows a tightly packed homo-dimeric structure in the crystal lattice, each protomer having the “NEET” fold structures of human homologues, yet containing a reversible disulfide linkage at the homodimer interface which is apparently conserved in the Drosophila homologues but not in mammalian orthologues. Surprisingly, the refined CISD-1 structure also appears to represent that of the N-terminal tail domain of the cognate kinesin motor protein, KLP-17, involved in chromosome segregation dynamics and germline development of the worms [61,62]. When C. elegans are exposed to the environmental stressor paraquat, a transgenic cisd-1 overexpression model of the worms showed a protective effect, like those observed in mammalian rodent studies. Under basal conditions, the worms with the CISD-1/mitoNEET KO showed an increase in ROS, while overexpression led to a decrease in ROS generation. After exposure to paraquat, TG worms showed not only increased levels of mitochondrial superoxide dismutase (mSOD2) corroborated by the reduction of deleterious ROS, but also increased phosphorylation of p38-MAPK that drives transcription of antioxidant response genes. Based on these findings, CISD-1/mitoNEET appears to play an important role in aging-related responses to cellular stressors. Future studies will help clarify better understanding of the role CISD-1 and other isoforms in C. elegans cellular metabolism and mechanistic aspects mediating ROS.
Acknowledgments
We gratefully acknowledge the support of the National Institutes of Health (NIH) for completion of this work: 5U54GM104942-03; R41 NS110070 & P20 GM109098 and T32 grant (5T32AG0523755) to J.B. Other support includes a grant from the Stark Community Parkinson’s disease Fund (Canton OH, USA). The structural study reported in this paper was supported in part by the International Collaborations in Chemistry Grant from JSPS (T.I.), JSPS Grants-in-Aid 24659202 and 26670215 (T.I.), 26·04415 (T.I. and A.T.T.), the Nagase Science and Technology Foundation Research Grant (T.I.), and the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED Grant JP21am0101070, and the synchrotron radiation experiments at the SPring-8 beamline BL41XU were approved by the Japan Synchrotron Radiation Research Institute (Proposal No. 2014A1464). A.T.T. gratefully acknowledges support as a JSPS Postdoctoral Fellow (P14415). Some of the worm strains used in this study were acquired from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Colca JR, McDonald WG, Waldon DJ, Leone JW, Lull JM, Bannow CA, Lund ET, Mathews WR, Identification of a novel mitochondrial protein ("mitoNEET") cross-linked specifically by a thiazolidinedione photoprobe, Am. J. Physiol. Endocrinol. Metab 286 (2) (2004) E252–E260. [DOI] [PubMed] [Google Scholar]
- [2].Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE, MitoNEET is an iron–containing outer mitochondrial membrane protein that regulates oxidative capacity, Proc. Natl. Acad. Sci. USA 104 (13) (2007) 5318–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Paddock ML, Wiley SE, Axelrod HL, Cohen AE, Roy M, Abresch EC, Capraro D, Murphy AN, Nechushtai R, Dixon JE, Jennings PA, MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone, Proc. Natl. Acad. Sci. USA 104 (36) (2007) 14342–14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lin J, Zhou T, Ye K, Wang J, Crystal structure of human mitoNEET reveals distinct groups of iron sulfur proteins, Proc. Natl. Acad. Sci. USA 104 (37) (2007) 14640–14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hou X, Liu R, Ross S, Smart EJ, Zhu H, Gong W, Crystallographic studies of human MitoNEET, J. Biol. Chem 282 (46) (2007) 33242–33246. [DOI] [PubMed] [Google Scholar]
- [6].Geldenhuys WJ, Long TE, Saralkar P, Iwasaki T, Nunez RAA, Nair RR, Konkle ME, Menze MA, Pinti MV, Hollander JM, Hazlehurst LA, Robart AR, Crystal structure of the mitochondrial protein mitoNEET bound to a benze-sulfonide ligand, Commun. Chem 2 (2019) 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Conlan AR, Axelrod HL, Cohen AE, Abresch EC, Zuris J, Yee D, Nechushtai R, Jennings PA, Paddock ML, Crystal structure of Miner1: the redox-active 2Fe-2S protein causative in Wolfram Syndrome 2, J. Mol. Biol 392 (1) (2009) 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lipper CH, Karmi O, Sohn YS, Darash-Yahana M, Lammert H, Song L, Liu A, Mittler R, Nechushtai R, Onuchic JN, Jennings PA, Structure of the human monomeric NEET protein MiNT and its role in regulating iron and reactive oxygen species in cancer cells, Proc. Natl. Acad. Sci. U SA 115 (2) (2018) 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li X, Wang Y, Tan G, Lyu J, Ding H, Electron transfer kinetics of the mitochondrial outer membrane protein mitoNEET, Free Rad. Biol. Med 121 (2018) 98–104. [DOI] [PubMed] [Google Scholar]
- [10].Kusminski CM, Holland WL, Sun K, Park J, Spurgin SB, Lin Y, Askew GR, Simcox JA, McClain DA, Li C, Scherer PE, MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity, Nat. Med 18 (10) (2012) 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kusminski CM, Chen S, Ye R, Sun K, Wang QA, Spurgin SB, Sanders PE, Brozinick JT, Geldenhuys WJ, Li WH, Unger RH, Scherer PE, MitoNEET-Parkin effects in pancreatic alpha- and beta-cells, cellular survival, and intrainsular cross talk, Diabetes 65 (6) (2016) 1534–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Landry AP, Cheng Z, Ding H, Reduction of mitochondrial protein mitoNEET [2Fe-2S] clusters by human glutathione reductase, Free Radic. Biol. Med 81 (2015) 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stauch KL, Villeneuve LM, Totusek S, Lamberty B, Ciborowski P, Fox HS, Quantitative proteomics of presynaptic mitochondria reveal an overexpression and biological relevance of neuronal MitoNEET in postnatal brain development, Dev. Neurobiol 79 (4) (2019) 370–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Geldenhuys WJ, Leeper TC, Carroll RT, mitoNEET as a novel drug target for mitochondrial dysfunction, Drug Discov. Today 19 (10) (2014) 1601–1606. [DOI] [PubMed] [Google Scholar]
- [15].Li X, Fang P, Mai J, Choi ET, Wang H, Yang X.-f., Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers, J. Hematol. Oncol 6 (1) (2013) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yuan H, Li X, Zhang X, Kang R, Tang D, CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation, Biochem. Biophys. Res. Commun 478 (2) (2016) 838–844. [DOI] [PubMed] [Google Scholar]
- [17].Lei P, Bai T, Sun Y, Mechanisms of ferroptosis and relations with regulated cell death: a review, Front. Physiol 10 (2019) 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weng MS, Chang JH, Hung WY, Yang YC, Chien MH, The interplay of reactive oxygen species and the epidermal growth factor receptor in tumor progression and drug resistance, J. Exp. Clin. Cancer Res 37 (1) (2018) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Geldenhuys WJ, Benkovic SA, Lin L, Yonutas HM, Crish SD, Sullivan PG, Darvesh AS, Brown CM, Richardson JR, MitoNEET (CISD1) knockout mice show signs of striatal mitochondrial dysfunction and a parkinson’s disease phenotype, ACS Chem. Neurosci 8 (12) (2017) 2759–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hung CM, Lombardo PS, Malik N, Brun SN, Hellberg K, Van Nostrand JL, Garcia D, Baumgart J, Diffenderfer K, Asara JM, Shaw RJ, AMPK/ULK1-mediated phosphorylation of Parkin ACT domain mediates an early step in mitophagy, Sci. Adv 7 (15) (2021) eabg4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lin J, Zhang L, Lai S, Ye K, Structure and molecular evolution of CDGSH iron-sulfur domains, PLoS One 6 (9) (2011) e24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].King SD, Gray CF, Song L, Mittler R, Padilla PA, The mitochondrial localized CISD-3.1/CISD-3.2 proteins are required to maintain normal germline structure and function in Caenorhabditis elegans, PLoS One 16 (2) (2021) e0245174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].King SD, Gray CF, Song L, Nechushtai R, Gumienny TL, Mittler R, Padilla PA, The cisd gene family regulates physiological germline apoptosis through ced-13 and the canonical cell death pathway in Caenorhabditis elegans, Cell Death Differ. 26 (1) (2019) 162–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hsiung KC, Liu KY, Tsai TF, Yoshina S, Mitani S, Chin-Ming Tan B, Lo SJ, Defects in CISD-1, a mitochondrial iron-sulfur protein, lower glucose level and ATP production in Caenorhabditis elegans, Biomed. J 43 (1) (2020) 32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cocheme HM, Murphy MP, Complex I is the major site of mitochondrial superoxide production by paraquat, J. Biol. Chem 283 (4) (2008) 1786–1798. [DOI] [PubMed] [Google Scholar]
- [26].Hu Q, D’Amora DR, MacNeil LT, Walhout AJM, Kubiseski TJ, The oxidative stress response in caenorhabditis elegans requires the GATA transcription factor ELT-3 and SKN-1/Nrf2, Genetics 206 (4) (2017) 1909–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Di Monte DA, Lavasani M, Manning-Bog AB, Environmental factors in Parkinson’s disease, Neurotoxicology 23 (4-5) (2002) 487–502. [DOI] [PubMed] [Google Scholar]
- [28].Richardson JR, Quan Y, Sherer TB, Greenamyre JT, Miller GW, Paraquat neurotoxicity is distinct from that of MPTP and rotenone, Toxicol. Sci 88 (1) (2005) 193–201. [DOI] [PubMed] [Google Scholar]
- [29].Kuter K, Smialowska M, Wieronska J, Zieba B, Wardas J, Pietraszek M, Nowak P, Biedka I, Roczniak W, Konieczny J, Wolfarth S, Ossowska K, Toxic influence of subchronic paraquat administration on dopaminergic neurons in rats, Brain Res. 1155 (2007) 196–207. [DOI] [PubMed] [Google Scholar]
- [30].Gray JP, Heck DE, Mishin V, Smith PJ, Hong JY, Thiruchelvam M, Cory-Slechta DA, Laskin DL, Laskin JD, Paraquat increases cyanide-insensitive respiration in murine lung epithelial cells by activating an NAD(P)H:paraquat oxidoreductase: identification of the enzyme as thioredoxin reductase, J. Biol. Chem 282 (11) (2007) 7939–7949. [DOI] [PubMed] [Google Scholar]
- [31].Kim H, Perentis RJ, Caldwell GA, Caldwell KA, Gene-by-environment interactions that disrupt mitochondrial homeostasis cause neurodegeneration in C. elegans Parkinson’s models, Cell Death Dis. 9 (5) (2018) 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gaeta AL, Caldwell KA, Caldwell GA, Found in translation: the utility of C. elegans alpha-synuclein models of Parkinson’s disease, Brain Sci. 9 (4) (2019) 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Griffin EF, Scopel SE, Stephen CA, Holzhauer AC, Vaji MA, Tuckey RA, Berkowitz LA, Caldwell KA, Caldwell GA, ApoE-associated modulation of neuroprotection from Abeta-mediated neurodegeneration in transgenic Caenorhabditis elegans, Dis. Model Mech 12 (2) (2019) dmm037218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Iwasaki T, Samoilova RI, Kounosu A, Ohmori D, Dikanov SA, Continuous-wave and pulsed EPR characterization of the [2Fe-2S](Cys)3(His)1 cluster in rat MitoNEET, J. Am. Chem. Soc 131 (38) (2009) 13659–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kounosu A, Li Z, Cosper NJ, Shokes JE, Scott RA, Imai T, Urushiyama A, Iwasaki T, Engineering a three-cysteine, one-histidine ligand environment into a new hyperthermophilic archaeal Rieske-type [2Fe-2S] ferredoxin from Sulfolobus solfataricus, J. Biol. Chem 279 (13) (2004) 12519–12528. [DOI] [PubMed] [Google Scholar]
- [36].Hasegawa K, Shimizu N, Okumura H, Mizuno N, Baba S, Hirata K, Takeuchi T, Yamazaki H, Senba Y, Ohashi H, Yamamoto M, Kumasaka T, SPring-8 BL41XU, a high-flux macromolecular crystallography beamline, J. Synchrotron Radiat 20 (2013) 910–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kabsch W, XDS, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software, J. Appl. Crystallogr 40 (2007) 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L, The importance of alignment accuracy for molecular replacement, Acta Crystallogr. D Biol. Crystallogr 60 (2004) 1229–1236. [DOI] [PubMed] [Google Scholar]
- [40].Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot, Acta Crystallogr. D Biol. Crystallogr 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix, Acta Crystallogr. D Struct. Biol 75 (2019) 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Craig DB, Dombkowski AA, Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins, BMC Bioinform. 14 (2013) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sun MA, Wang Y, Zhang Q, Xia Y, Ge W, Guo D, Prediction of reversible disulfide based on features from local structural signatures, BMC Genom. 18 (1) (2017) 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wiedemann C, Kumar A, Lang A, Ohlenschlager O, Cysteines and disulfide bonds as structure-forming units: insights from different domains of life and the potential for characterization by NMR, Front. Chem 8 (2020) 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, Jain S, Lewis SM, Arendall WB, Snoeyink J 3rd, Adams PD, Lovell SC, Richardson JS, Richardson DC, MolProbity: more and better reference data for improved all-atom structure validation, Protein Sci. 27 (1) (2018) 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Berman H, Henrick K, Nakamura H, Announcing the worldwide protein data bank, Nat. Struct. Biol 10 (12) (2003) 980. [DOI] [PubMed] [Google Scholar]
- [47].Stiernagle T, Maintenance of C. elegans, WormBook 2006, pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zheng SQ, Ding AJ, Li GP, Wu GS, Luo HR, Drug absorption efficiency in Caenorhbditis elegans delivered by different methods, PLoS One 8 (2) (2013) e56877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nass J, Abdelfatah S, Efferth T, Induction of stress resistance and extension of lifespan in Chaenorhabditis elegans serotonin-receptor knockout strains by withanolide A, Phytomedicine 84 (2021) 153482. [DOI] [PubMed] [Google Scholar]
- [50].Tao J, Wu Q-Y, Ma Y-C, Chen Y-L, Zou C-G, Antioxidant response is a protective mechanism against nutrient deprivation in C. elegans, Sci. Rep 7 (1) (2017) 43547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang W, Hekimi S, A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans, PLoS Biol. 8 (12) (2010) e1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Angeli S, Foulger A, Chamoli M, Peiris TH, Gerencser A, Shahmirzadi AA, Andersen J, Lithgow G, The mitochondrial permeability transition pore activates the mitochondrial unfolded protein response and promotes aging, eLife 10 (2021) e63453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M, Colab-Fold: making protein folding accessible to all, Nat. Methods 19 (6) (2022) 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Iwasaki T, Watanabe E, Ohmori D, Imai T, Urushiyama A, Akiyama M, Hayashi-Iwasaki Y, Cosper NJ, Scott RA, Spectroscopic investigation of selective cluster conversion of archaeal zinc-containing ferredoxin from Sulfolobus sp. strain 7, J. Biol. Chem 275 (33) (2000) 25391–25401. [DOI] [PubMed] [Google Scholar]
- [55].Beinert H, Holm RH, Münck E, Iron-sulfur clusters: nature’s modular, multipurpose structures, Science 277 (5326) (1997) 653–659. [DOI] [PubMed] [Google Scholar]
- [56].Taguchi AT, Miyajima-Nakano Y, Fukazawa R, Lin MT, Baldansuren A, Gennis RB, Hasegawa K, Kumasaka T, Dikanov SA, Iwasaki T, Unpaired electron spin density distribution across reduced [2Fe-2S] cluster ligands by 13Cβ-cysteine labeling, Inorg. Chem 57 (2) (2018) 741–746. [DOI] [PubMed] [Google Scholar]
- [57].Garman E, ‘Cool’ crystals: macromolecular cryocrystallography and radiation damage, Curr. Opin. Struct. Biol 13 (5) (2003) 545–551. [DOI] [PubMed] [Google Scholar]
- [58].Sommerhalter M, Lieberman RL, Rosenzweig AC, X-ray crystallography and biological metal centers: is seeing believing? Inorg. Chem 44 (4) (2005) 770–778. [DOI] [PubMed] [Google Scholar]
- [59].Corbett MC, Latimer MJ, Poulos TL, Sevrioukova IF, Hodgson KO, Hedman B, Photoreduction of the active site of the metalloprotein putidaredoxin by synchrotron radiation, Acta. Crystallogr. D Biol. Crystallogr 63 (2007) 951–960. [DOI] [PubMed] [Google Scholar]
- [60].Conlan AR, Paddock ML, Axelrod HL, Cohen AE, Abresch EC, Wiley S, Roy M, Nechushtai R, Jennings PA, The novel 2Fe-2S outer mitochondrial protein mitoNEET displays conformational flexibility in its N-terminal cytoplasmic tethering domain, Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun 65 (2009) 654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ali MY, Siddiqui SS, cDNA cloning and expression of a C-terminus motor kinesin–like protein KLP-17, involved in chromosomal movement in Caenorhabditis elegans, Biochem. Biophys. Res. Commun 267 (2) (2000) 643–650. [DOI] [PubMed] [Google Scholar]
- [62].Siddiqui SS, Metazoan motor models: kinesin superfamily in C. elegans, Traffic 3 (1) (2002) 20–28. [DOI] [PubMed] [Google Scholar]
- [63].Senchuk MM, Dues DJ, Van Raamsdonk JM, Measuring oxidative stress in caenorhabditis elegans: paraquat and juglone sensitivity assays, Bio Protoc. 7 (1) (2017) e2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Possik E, Pause A, Measuring oxidative stress resistance of Caenorhabditis elegans in 96-well microtiter plates, J. Vis. Exp (99) (2015) e52746, doi: 10.3791/52746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Lin MT, Flint Beal M, The oxidative damage theory of aging, Clin. Neurosci. Res 2 (5–6) (2003) 305–315. [Google Scholar]
- [66].Hua J, Gao Z, Zhong S, Wei B, Zhu J, Ying R, CISD1 protects against atherosclerosis by suppressing lipid accumulation and inflammation via mediating Drp1, Biochem. Biophys. Res. Commun 577 (2021) 80–88. [DOI] [PubMed] [Google Scholar]
- [67].Geldenhuys WJ, Piktel D, Moore JC, Rellick SL, Meadows E, Pinti MV, Hollander JM, Ammer AG, Martin KH, Gibson LF, Loss of the redox mitochondrial protein mitoNEET leads to mitochondrial dysfunction in B-cell acute lymphoblastic leukemia, Free Radic. Biol. Med 175 (2021) 226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Arnett D, Quillin A, Geldenhuys WJ, Menze MA, Konkle M, 4-hydroxynonenal and 4-oxononenal differentially bind to the redox sensor MitoNEET, Chem. Res. Toxicol 32 (6) (2019) 977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hubbard WB, Spry ML, Gooch JL, Cloud AL, Vekaria HJ, Burden S, Powell DK, Berkowitz BA, Geldenhuys WJ, Harris NG, Sullivan PG, Clinically relevant mitochondrial-targeted therapy improves chronic outcomes after traumatic brain injury, Brain 144 (12) (2021) 3788–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Saralkar P, Mdzinarishvili A, Arsiwala TA, Lee YK, Sullivan PG, Pinti MV, Hollander JM, Kelley EE, Ren X, Hu H, Simpkins J, Brown C, Hazlehurst LE, Huber JD, Geldenhuys WJ, The mitochondrial mitoNEET ligand NL-1 is protective in a murine model of transient cerebral ischemic stroke, Pharm. Res 38 (5) (2021) 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lee S, Lee S, Lee SJ, Chung SW, Inhibition of mitoNEET induces Pink1-Parkin-mediated mitophagy, BMB Rep. 55 (7) (2022) 354–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dingley S, Polyak E, Lightfoot R, Ostrovsky J, Rao M, Greco T, Ischiropoulos H, Falk MJ, Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans, Mitochondrion 10 (2) (2010) 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zielonka J, Kalyanaraman B, Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth, Free Radic. Biol. Med 48 (8) (2010) 983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kalyanaraman B, Pitfalls of reactive oxygen species (ROS) measurements by fluorescent probes and mitochondrial superoxide determination using MitoSOX, in: Berliner LJ, Parinandi NL (Eds.), Measuring Oxidants and Oxidative Stress in Biological Systems, 2020, pp. 7–9. Cham (CH). [PubMed] [Google Scholar]
- [75].Karakuzu O, Cruz MR, Liu Y, Garsin DA, Amplex red assay for measuring hydrogen peroxide production from caenorhabditis elegans, Bio Protoc. 9 (21) (2019) e3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Van Raamsdonk JM, Hekimi S, Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans, PLos Genet. 5 (2) (2009) e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K, The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response, Genes Dev. 19 (19) (2005) 2278–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM, A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity, Science 297 (5581) (2002) 623–626. [DOI] [PubMed] [Google Scholar]
- [79].Galbadage T, Shepherd TF, Cirillo SL, Gumienny TL, Cirillo JD, The Caenorhabditis elegans p38 MAPK Gene plays a key role in protection from mycobacteria, Microbiologyopen 5 (3) (2016) 436–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R, Analysis tool web services from the EMBL-EBI, Nucleic Acids.Res 41 (2013) W597–W600 Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Heinig M, Frishman D, STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins, Nucleic Acids Res. 32 (2004) W500–W502 Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]