Chu et al. identify a clinical drug, nortriptyline that can potently inhibit macropinocytosis-mediated fatty acid uptake, and further find that nortriptyline effectively suppresses tumor growth, lipogenesis, and hepatic steatosis in combination with ND-646, a selective ACC1/2 inhibitor.
Abstract
Fatty acid uptake is essential for cell physiological function, but detailed mechanisms remain unclear. Here, we generated an acetyl-CoA carboxylases (ACC1/2) double-knockout cell line, which lacked fatty acid biosynthesis and survived on serum fatty acids and was used to screen for fatty acid uptake inhibitors. We identified a Food and Drug Administration–approved tricyclic antidepressant, nortriptyline, that potently blocked fatty acid uptake both in vitro and in vivo. We also characterized underlying mechanisms whereby nortriptyline provoked lysosomes to release protons and induce cell acidification to suppress macropinocytosis, which accounted for fatty acid endocytosis. Furthermore, nortriptyline alone or in combination with ND-646, a selective ACC1/2 inhibitor, significantly repressed tumor growth, lipogenesis, and hepatic steatosis in mice. Therefore, we show that cells actively take up fatty acids through macropinocytosis, and we provide a potential strategy suppressing tumor growth, lipogenesis, and hepatic steatosis through controlling the cellular level of fatty acids.
Graphical Abstract
Introduction
In mammalian cells, palmitate is de novo synthesized from acetyl-CoA and malonyl-CoA by fatty acid synthase using NADPH as the reducing entity (Menendez and Lupu, 2007; Pietrocola et al., 2015). Then, palmitate or other fatty acid precursors are further elongated to very long-chain fatty acid (VLCFA), which predominantly takes place in the endoplasmic reticulum using malonyl-CoA as the elongation unit (Jakobsson et al., 2006; Kihara, 2016). Therefore, malonyl-CoA is critical for the biosynthesis of fatty acids, which is produced by cytosolic acetyl-CoA carboxylase 1 (ACC1), as well as mitochondrial outer membrane–bound ACC2 under some conditions (Menendez and Lupu, 2007). It is well documented that fatty acid synthesis is required for lipogenesis, tumor growth, and hepatic steatosis, and is up-regulated by multiple signaling pathways in the tissues of adipose, cancer, and liver (Liu et al., 2020; Postic and Girard, 2008; Zhu and Thompson, 2019). Therefore, elevated fatty acid synthesis is associated with cancer therapy and hepatosteatosis treatment, but compounds targeting the relevant enzymes, in particular ACC1/2 and fatty acid synthase, have shown limited efficacy in preclinical tumor models or non-alcoholic fatty liver disease clinical trials, and remain largely inapplicable for clinical setting (Chen et al., 2019; Röhrig and Schulze, 2016; Snaebjornsson et al., 2020).
Additionally, for physiological function, cells must also assimilate fatty acids and lipids from the circulation (Mallick et al., 2021; Medes et al., 1953; Röhrig and Schulze, 2016). The uptake of lipids is mainly facilitated by clathrin-mediated low density lipoprotein (LDL) receptor endocytosis (Brown and Goldstein, 1986; He et al., 2002). In contrast, fatty acid ingestion is mediated through the fatty acid translocase/cluster of differentiation 36 (CD36) or fatty acid transport proteins (FATP1-6), which may be aided by fatty acid–binding proteins (FABP1-9 and 12; Furuhashi and Hotamisligil, 2008; Kazantzis and Stahl, 2012). FATPs display similar functions to acyl-CoA synthetases. For CD36, it contains a fatty acid–binding site, and then could be internalized through endocytic pathways to transport fatty acids into cells (Hao et al., 2020); however, it is unclear if other pathways mediating fatty acid uptake exist.
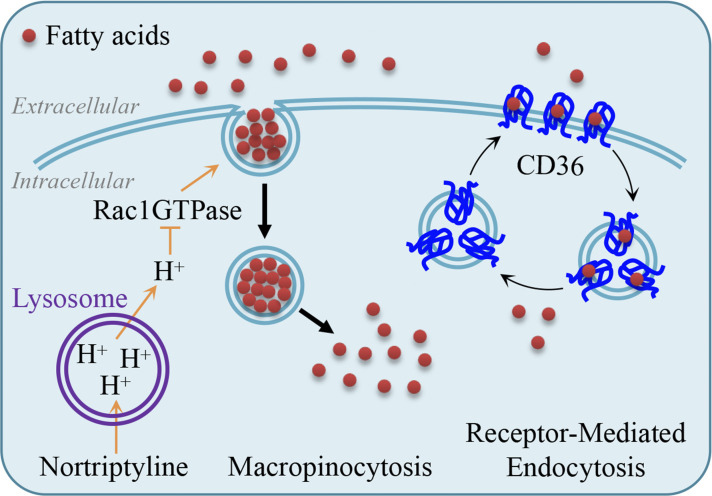
Endocytosis is a cell process where cells ingest extracellular substances, and mechanistically includes receptor-mediated endocytosis, phagocytosis, and pinocytosis. Macropinocytosis is a regulated pinocytic process and is initiated from plasma membrane ruffles, which invaginate and pinch off to form large endocytic vacuoles or macropinosomes. Macropinocytosis usually mediates the non-selective uptake of extracellular solute molecules and nutritious cargoes (Doherty and McMahon, 2009; Sivanand and Vander Heiden, 2020); thus, it is an important pathway for nutrient acquisition. It is accepted that cancer cells assimilate extracellular proteins as amino acid sources through macropinocytosis (Commisso et al., 2013; Davidson et al., 2017; Kamphorst et al., 2015; Sivanand and Vander Heiden, 2020; Zhang and Commisso, 2019). In this study, we show that cells actively take up fatty acids via macropinocytosis, and we characterize a drug that blocks this macropinocytosis-mediated fatty acid uptake to validate the suppressive effects of fatty acid synthesis inhibition on lipogenesis, tumor growth, and hepatic steatosis.
Results
Identifying nortriptyline as a fatty acid uptake inhibitor
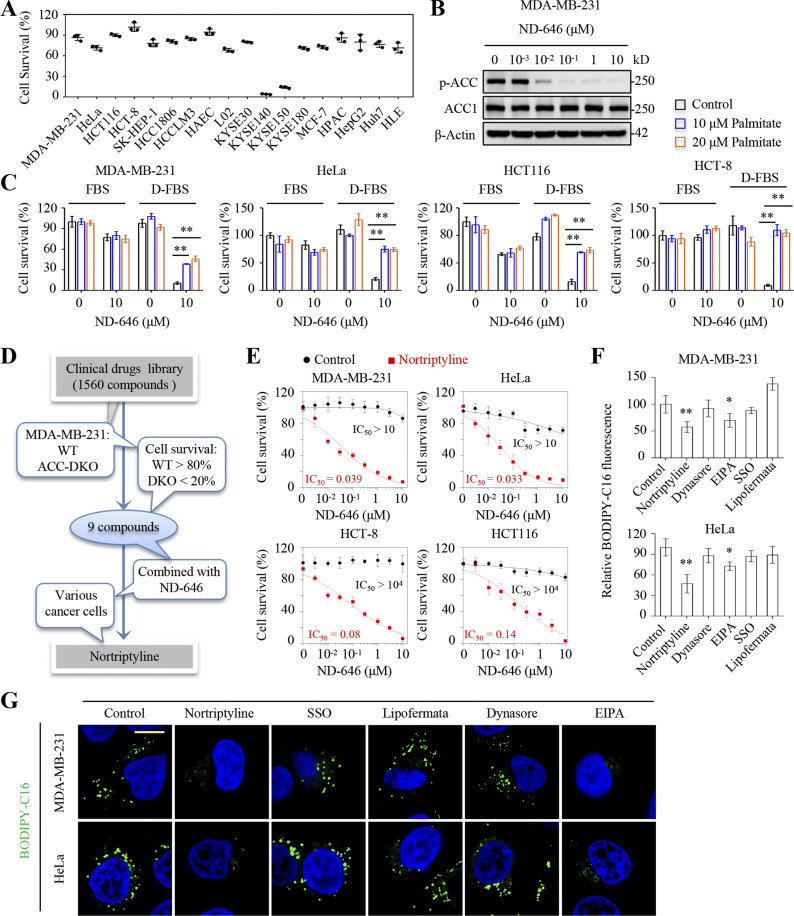
The carboxylation of acetyl-CoA to malonyl-CoA by ACC1 or ACC2 is indispensable in fatty acid biosynthesis. ND-646 is a potent selective inhibitor of human ACC1 with an IC50 of 3.5 nM and ACC2 with a half maximal inhibitory concentration (IC50) of 4.1 nM (Svensson et al., 2016). However, ND-646, even at a high concentration of 10 μM, only showed marginal inhibitory effects toward cancer cells, except for two esophageal cancer cell lines, KYSE140 and KYSE150 (Fig. 1 A). At this concentration, ND-646 disrupted ACC1 dimerization and completely blocked its phosphorylation at Ser79 (Fig. 1 B). Conditionally, 10 μM ND-646 largely killed cancer cells, including MDA-MB-231, HeLa, HCT116, and HCT-8 cell lines, in medium containing delipidized FBS, but this process was significantly reversed by the supplementation with palmitate (Fig. 1 C). These data suggest that these cancer epithelial cells had active fatty acid uptake processes. However, blocking CD36 and FATPs, the major previously reported mediators of fatty acid absorption, with, respectively, sulfosuccinimidyl oleate (SSO; Coort et al., 2002; Kuda et al., 2013) and lipofermata (Zhang et al., 2018), did not effectively potentiate ND-646’s killing ability at different concentrations (Fig. S1, A and B).
Figure 1.
Identification of nortriptyline as an inhibitor of fatty acid uptake. (A) Inhibitory effects of ND-646 on different cancer cell lines. Cells were treated with or without ND-646 (10 μM) for 4 d. Data were presented relative to control cells without ND-646 treatment. (B) Western blots showing total and phosphorylated ACC1 levels in MDA-MB-231 cells treated with different concentrations of ND-646 for 24 h. (C) Survival of cancer cells in full FBS or delipidized FBS (D-FBS)–contained medium with or without 10 or 20 μM palmitate for 5 d. (D) Schematic showing fatty acid uptake inhibitor screen. (E) Inhibitory effects of ND-646 on cancer cells in the presence or absence of 10 μM nortriptyline for 4 d. (F and G) BODIPY-C16 uptake in MDA-MB-231 cells. Cells were pretreated with vehicle, nortriptyline (10 μM), dynasore (10 μM), EIPA (10 μM), SSO (100 μM) or lipofermata (1 μM) for 8 h, and then incubated with 2 μM BODIPY-C16 for 2 h (F) or 5 μM BODIPY-C16 for 1 h (G). BODIPY-C16 fluorescence was measured by a microplate reader (F). BODIPY-C16 (green) indicates fatty acid uptake, and DAPI staining (blue) indicates nuclei (G). Images in G were representative of at least three independent experiments. Data were from triplicate experiments (A, C, E, and F), and all experimental data were verified in at least two independent experiments. Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test. Scale bar = 10 μm. Source data are available for this figure: SourceData F1.
Figure S1.
The inhibitory effects of compounds in combination with ND-646 on cancer cell growth. (A and B) Inhibitory effects of SSO (A) and lipofermata (B) on MDA-MB-231 or HeLa cells in the presence or absence of 10 μM ND-646; or inhibitory effects of ND-646 on MDA-MB-231 and HeLa cells with or without 100 μM SSO (A), 1 μM (in MDA-MB-231), or 10 μM (in HeLa) lipofermata (B). Cells were treated for 4 d. (C) Inhibitory effects of nortriptyline on MDA-MB-231, HeLa, HCT116, and HCT-8 cells with or without 10 μM ND-646. Cells were treated for 4 d. (D) Inhibitory effects of EIPA on MDA-MB-231 and HeLa cells with or without 10 μM ND-646, or inhibitory effects of ND-646 on MDA-MB-231 and HeLa cells with or without 10 μM EIPA. Cells were treated for 4 d. Data were from triplicate experiments, and all experimental data were verified in at least three independent experiments. Error bars indicate the mean ± SD.
To identify the effective fatty acid uptake inhibitors, we first knocked out both ACC1 and ACC2 in MDA-MB-231 cells using the CRISPR/Cas9 system and generated a cell line dependent on serum fatty acids for proliferation (Fig. S2 A). We then used this double knockout (DKO) model cell line to screen a library of 1560 Food and Drug Administration–approved drugs. We identified nine drugs which significantly killed DKO cells but left WT cells unaffected at 10 μM ND-646 (Fig. 1 D and Fig. S2, B–D). After verifying these compounds in combination with ND-646 in MDA-MB-231, HeLa, HCT116, and HCT-8 cells, we found that nortriptyline, fluphenazine, and perphenazine showed clear synergistic killing abilities with ND-646 across all cell lines (Fig. S2 E). However, only nortriptyline, a tricyclic antidepressant (Hershman et al., 2014), was well tolerated in all cell lines (Fig. S2 E), and thus was used for the ongoing experiments. With nortriptyline, the inhibitory effects of ND-646 on various cancer cell lines were dramatically increased (Fig. 1 E), and vice versa (Fig. S1 C).
Figure S2.
Fatty acid uptake inhibitor screen. (A) Representative Western blot analysis of CRISPR-Cas9–mediated knockout of ACC1 and ACC2 (DKO) in MDA-MB-231 clone (right). Cell growth assay for MDA-MB-231/ACC1/2-DKO clone in medium containing full FBS or delipidized FBS. (B) Inhibitory effects of drugs on MDA-MB-231 ACC1/2-DKO cells. The primary screen identified 141 drugs which inhibited cell survival at least to 70%. (C) Inhibitory effects of selected drugs in B on MDA-MB-231/WT and ACC1/2-DKO cells. The secondary screen identified nine drugs which inhibited ACC1/2-DKO cell survival at least to 80% and inhibited MDA-MB-231/WT no more than 20%. (D) The nine drugs from C. (E) Inhibitory effects of nine drugs (10 μM) on MDA-MB-231/ACC-DKO, and also MDA-MB-231, HeLa, HCT116, and HCT-8 cells with or without 10 μM ND-646. Cells were treated for 4 d. The third screen identified one drug which synergistically killed cells in combination with ND-646 across cell lines. Data were from triplicate experiments (A and E), and all experimental data were verified in at least three independent experiments. Error bars indicate the mean ± SD. Source data are available for this figure: SourceData FS2.
We next used 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY)-C16, a fluorescent palmitate analog (Huang et al., 2002), to track fatty acid uptake, and confirmed that nortriptyline significantly reduced BODIPY fluorescence in MDA-MB-231 and HeLa cells, while SSO and lipofermata did not, as detected by the microplate reader (Fig. 1 F) and confocal microscope (Fig. 1 G), when compared with control cells. Taken together, nortriptyline can potentially block fatty acid uptake.
Nortriptyline suppresses fatty acid uptake in vitro and in vivo
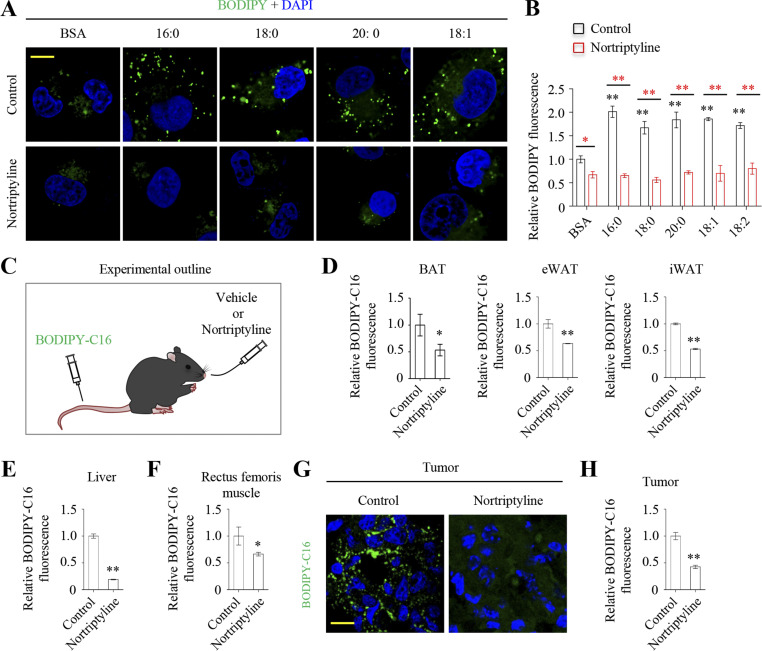
Considering that most in vivo free fatty acids are bound to proteins, mainly albumin, we treated MDA-MB-231 cells with several BSA-bound fatty acids including FA(16:0), FA(18:0), FA(20:0), FA(18:1), and FA(18:2), and measured their uptake. We stained neutral cell lipids with BODIPY and observed that with BSA-bound fatty acids supplementation significantly increased cell BODIPY fluorescence, which was almost completely repressed by nortriptyline (Fig. 2, A and B). To further investigate whether nortriptyline affected in vivo fatty acid uptake, we intravenously injected BODIPY-C16 into C57BL/6 mice that were intragastrically administered nortriptyline or vehicle (Fig. 2 C). We then removed tissues, generated single cells by digestion, and measured BODIPY-C16 fluorescence by flow cytometry. Nortriptyline significantly decreased BODIPY-C16 fluorescence in liver cells, adipocytes from inguinal white adipose tissue (iWAT), epididymal white adipose tissue (eWAT), brown adipose tissue (BAT), and rectus femoris muscle cells (Fig. 2, D–F). Furthermore, we intravenously injected BODIPY-C16 into nude mice xenografted with MDA-MD-231–derived tumors and showed that tumor tissues significantly absorbed BODIPY-C16, which was blocked by intragastric nortriptyline administration (Fig. 2, G and H). These data suggest that nortriptyline suppresses fatty acid uptake in vivo.
Figure 2.
Nortriptyline blocks fatty acid uptake in vitro and in vivo. (A and B) MDA-MB-231 cells were pretreated with serum-free medium for 8 h and nortriptyline (10 μM) for 1 h, followed by treatment with BSA-conjugated fatty acids with different chain lengths or saturation (100 μM) for 4 h with or without nortriptyline. BODIPY was used to label lipid droplets. Images were taken (A), and fluorescence were measured by flow cytometry (B). (C) Schematic showing BODIPY-C16 absorption in mice. Mice were intragastrically administered 20 mg/kg nortriptyline or vehicle every day for 3 d. Then, 100 μg BODIPY-C16 was intravenously injected. At 3 h after injection, tissues were removed and prepared. (D–F) Relative cell fluorescence from BODIPY-C16–injected tissue. Fluorescence was measured by flow cytometry. (G) Visualization of BODIPY-C16 uptake in tumor cryosection images of nude mice xenografted with MAD-MB-231–derived tumors. Mice were intravenously injected with BODIPY-C16 and intragastrically administered nortriptyline as described in C. (H) Relative cell fluorescence from BODIPY-C16–injected mice tissues. Fluorescence was measured by flow cytometry. Images (A and G) were representative of three independent experiments. Data were from triplicate experiments, and all experimental data were verified in at least two independent experiments (B). Data were from three mice (D–F and H). Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test. Black asterisks show comparison with control group. Scale bar = 10 μm.
Nortriptyline inhibits fatty acid uptake depending on cellular acidification
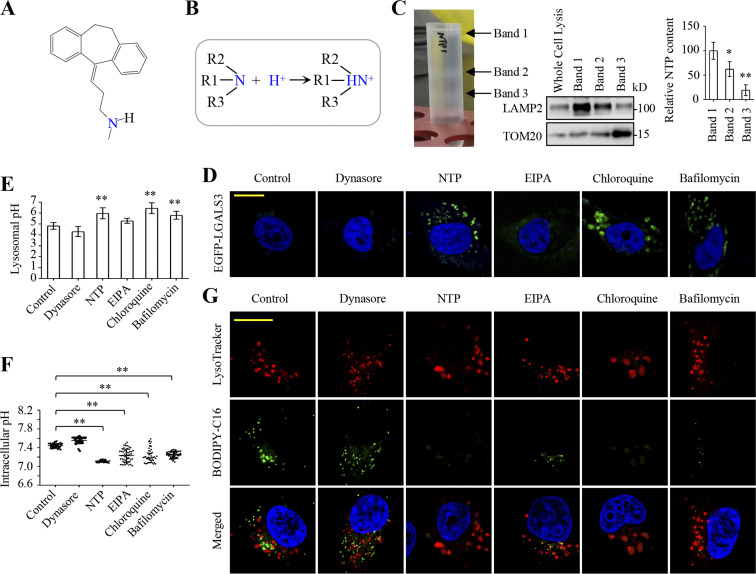
Nortriptyline (Fig. 3 A) and the other eight compounds (Fig. S2 D) were hydrophobic group–containing amines that could be protonated to ammonium salts in acidic microenvironments (Villamil Giraldo et al., 2014; Fig. 3 D). Therefore, these compounds could be trapped and accumulated in, and thus alkalize cell acid vesicles, such as lysosomes. Indeed, these drugs induced lysosomal puncta of EGFP-LGALS3 that detects sublethal lysosomal membrane permeabilization induced by alkalization (Aits et al., 2015; Fig. S3 A). To determine if nortriptyline was enriched in lysosomes, we homogenized cells treated with nortriptyline, fractionated homogenates by ultracentrifugation, and generated three fractionated bands containing lysosomes and/or mitochondria. Equal sample volumes were used to measure nortriptyline content and detect lysosomal LAMP2 and mitochondrial TOM20 proteins. We observed that nortriptyline content was proportional to lysosomes but not mitochondria (Fig. 3 C), which suggest that nortriptyline can be relatively enriched in lysosomes. We also confirmed that nortriptyline induced EGFP-LGALS3 puncta formation (Fig. 3 D), increased lysosomal pH (Fig. 3 E) and homogenously reduced cellular pH values (Fig. 3 F). These observations were consistent with lysosomotropic agents which facilitated cellular acidification through proton release from lysosomes (Cui et al., 2017). Knowing that lysosomes are associated with receptor-mediated endocytosis, we investigated relationship between lysosomes and fatty acid uptake, and showed that most BODIPY-C16 puncta were not co-localized with LysoTracker-Red staining (Fig. 3 G). We next used dynasore to block dynamin-dependent endocytosis, including LDL receptor- and CD36-mediated endocytosis (Macia et al., 2006), but dynasore neither inhibited the cellular uptake nor affected the localization of BODIPY-C16 (Fig. 1, F and G; and Fig. 3 G). These results suggest that fatty acid uptake is not mediated by lysosomes in cancer cells.
Figure 3.
Nortriptyline blocks fatty acid uptake through lysosome-mediated cellular acidification. (A) Nortriptyline structure. (B) Schematic showing amines protonation in acidic macroenvironments. (C) MDA-MB-231 cells were pretreated with nortriptyline (10 μM) for 12 h, and then cell homogenates were fractionated by ultracentrifugation. Three fractionated bands (left panel) were collected. Equal sample volumes were used to measure LAMP2 and TOM20 by Western blotting, and nortriptyline was detected by LC-MS/MS (right panel). (D) Fluorescent puncta of MDA-MB-231/EGFP-LGALS3 cells treated with or without dynasore (10 μM), nortriptyline (10 μM), EIPA (10 μM), chloroquine (10 μM), or bafilomycin A1 (200 nM) for 30 min. (E) Lysosomal pH in MDA-MB-231 cells was quantified using LysoSensor Yellow/Blue DND-160. The lysosomotropic ratiometric fluorescence was measured by a microplate reader. Cells were treated with EIPA (10 μM), nortriptyline (10 μM), chloroquine (10 μM), or bafilomycin A1 (200 nM) for 1 h. (F) The pH in MDA-MB-231 cells treated as in E for 40 min was measured using SNARF-5F fluorescence ratio. Ratiometric fluorescence was measured by microscopy. (G) The co-localization of BODIPY-C16 and lysosomes in MDA-MB-231 cells incubated with 5 μM BODIPY-C16 for 55 min prior to 50 nM LysoTracker Red treatment for another 15 min. Images (D and G) were representative of at least three independent experiments. Data were from three independent experiments (C); quintuple experiments (E); or scored from at least 50 cells/experiment (F). All experimental data (E and F) were verified in at least two independent experiments. Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test. Scale bar = 10 μm. Source data are available for this figure: SourceData F3.
Figure S3.
Cellular acidification suppresses micropinocytosis-mediated fatty acid uptake. (A) Fluorescent puncta of MDA-MB-231/EGFP-LGALS3 cells treated with or without each of selected nine drugs (10 μM) for 30 min. (B) Relative BODIPY-C16 fluorescence in MDA-MB-231 and HeLa cells treated without or with 10 μM chloroquine. Cells were pretreated with vehicle or chloroquine for 8 h and then incubated with 2 μM BODIPY-C16 for 2 h. Error bars indicate the mean ± SD of quadruple experiments. (C) Inhibitory effects of chloroquine on MDA-MB-231 and HeLa cells with or without 10 μM ND-646, or inhibitory effects of ND-646 on MDA-MB-231 and HeLa cells with or without 10 μM chloroquine. Cells were treated for 4 d. Error bars indicate the mean ± SD of triplicate experiments. (D) Cellular pH calibration in MDA-MB-231 cells using clamping buffers. MDA-MB-231 cells were loaded with SNARF-5F (20 μM) and then incubated with predetermined pH buffers. Nigericin (10 μg/ml) was used to equilibrate pH levels inside and outside cells. At least 50 cells were scored per experiment (left) and a calibration curve plotted (right). Images (A) were representative of at least three independent experiments. Data were from triplicate experiments (B and C), and all experimental data were verified in at least three independent experiments. Error bars indicate the mean ± SD. *, P < 0.05; two-tailed Student’s t test. Scale bar = 20 μm.
As a typical lysosomotropic, chloroquine was one of the nine drugs identified in our screen; it also inhibited BODIPY-C16 uptake (Fig. S3 B) and synergistically suppressed proliferation of MDA-MB-231 and HeLa cells in combination with ND-646 (Fig. S3 C). Although chloroquine induced EGFP-LGALS3 puncta formation (Fig. 3 D), it was associated with a relative increase in lysosomal swelling when compared with nortriptyline (Fig. 3, D and G). Importantly, chloroquine induced lysosomal alkalization and cellular acidification (Fig. 3, E and F). Now we tested whether cellular acidification accounted for the blockage of fatty acid uptake, and used 5-(N-ethyl-N-isopropyl)-amiloride (EIPA) to inhibit the activity of Na+/H+ exchanger (NHE) to increase the cellular acidification (Masereel et al., 2003). As expected, EIPA triggered significant cell acidification (Fig. 3 F), and together with its significant suppression of BODIPY-16 uptake (Fig. 1, F and G) and the potentiated killing ability of ND-646 to cancer cells (Fig. S1 D). EIPA did not induce the formation of EGFP-LGALS3 puncta (Fig. 3 D), indicating that nortriptyline and EIPA have distinct intracellular locations to trigger cellular acidification. Bafilomycin A1, a potent vacuolar H+-ATPase inhibitor, inhibited lysosomal acidification, and thus induced EGFP-LGALS3 puncta, enhanced lysosomal pH, decreased cellular pH, and suppressed BODIPY-C16 uptake (Fig. 3, D–G). Taken together, these data suggest that nortriptyline inhibits fatty acid uptake mainly by inducing lysosome-mediated cellular acidification.
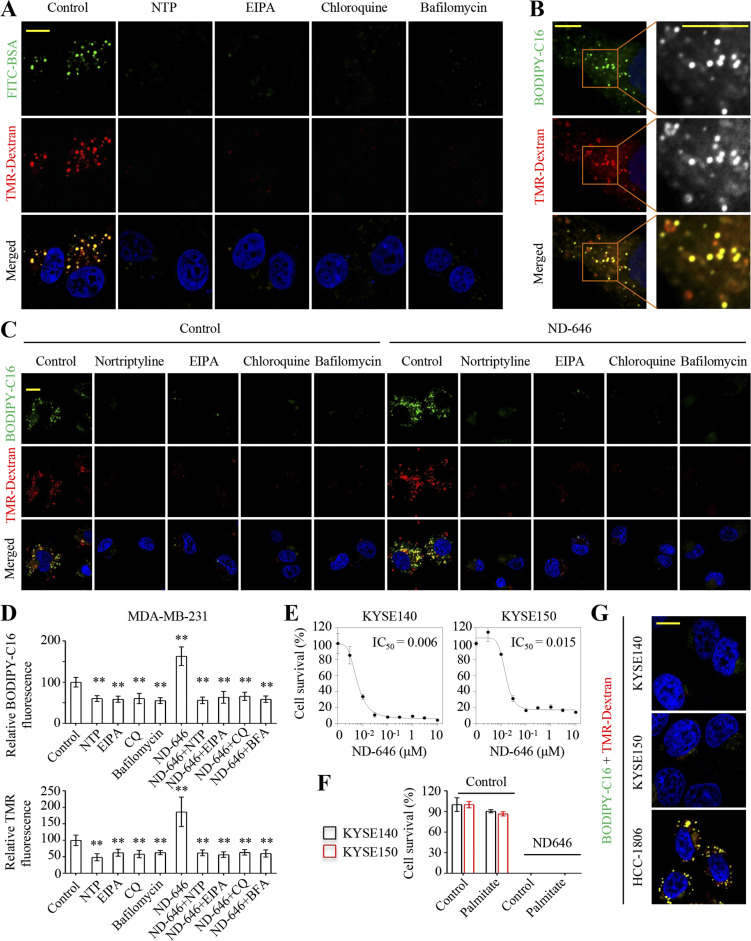
Nortriptyline inhibits macropinocytosis that accounts for fatty acid uptake
It was reported that EIPA inhibited macropinosome formation without affecting other endocytic pathways by inducing cellular acidification (Koivusalo et al., 2010; West et al., 1989). To test whether nortriptyline also suppressed macropinocytosis, we used two established markers of macropinocytosis, FITC-BSA and tetramethylrhodamine-labeled high-molecular-mass dextran (TMR-Dextran), to visualize macropinosomes upon their formation (Commisso et al., 2013). Similar to EIPA, nortriptyline potently blocked the uptake of both FITC-BSA and TMR-Dextran (Fig. 4 A). Indeed, chloroquine and bafilomycin A1 also suppressed macropinocytosis (Fig. 4 A), consistent with a previous report (Zhang et al., 2022). Therefore, macropinocytosis was most likely involved in fatty acid uptake. Also, BODIPY-C16 co-localized with TMR-Dextran in different cell lines, including MDA-MB-231, HCC-1806, HeLa, and HCT-8 cells (Fig. 4 B and Fig. S4 A). Nortriptyline, chloroquine, bafilomycin A1, and EIPA strongly reduced the fluorescence intensity of both BODIPY-C16 and TMR-Dextran (Fig. 4, C and D). In contrast, ND-646 notably promoted the uptake of BODIPY-C16 and TMR-Dextran in MDA-MB-231 (Fig. 4, C and D) and HeLa cells (Fig. S4 B), which was largely suppressed by nortriptyline, chloroquine, bafilomycin A1, or EIPA. During our cancer cell sensitivity screen cancer cells to ND-646, KYSE140 and KYSE150 cells were much vulnerable to treatments (Fig. 1 A). Now, we re-confirmed that ND-646 potently killed KYSE140 and KYSE150 cells in a concentration-dependent manner, with IC50 values of ∼6 and 15 nM, respectively (Fig. 4 E), which was not rescued by supplementation with palmitate at all (Fig. 4 F). Consistent with this observation, BODIPY-C16 and TMR-Dextran were not taken up by both cell lines (Fig. 4 G), indicating these cells lacked active macropinocytosis and hence largely lost the ability to absorb fatty acids. Taken together, these data suggest that macropinocytosis can actively mediate fatty acid uptake.
Figure 4.
Cells take up fatty acids through macropinocytosis. (A) Uptake of FITC-BSA and TMR-Dextran in MDA-MB-231 cells. Cells were serum-starved for 12 h. EIPA (10 μM), nortriptyline (10 μM), chloroquine (10 μM), or bafilomycin A1 (200 nM) were pretreated at the time of serum removal. Then, cells were incubated with 1 mg/ml TMR-Dextran and 0.5 mg/ml FITC-BSA in serum-free DMEM for 1.5 h. (B) BODIPY-C16 co-localization with TMR-Dextran in MDA-MB-231 cells. Cells were incubated with 5 μM BODIPY-C16 for 30 min prior to 1 mg/ml TMR-Dextran treatment for another 30 min. The right image represents a higher magnification of the boxed areas. (C) Uptake of BODIPY-C16 and TMR-Dextran in MDA-MB-231 cells. Cells were pretreated with vehicle, EIPA (10 μM), nortriptyline (10 μM), chloroquine (10 μM), and bafilomycin A1 (200 nM) for 8 h and then incubated with or without ND-646 (1 μM) before staining with BODIPY-C16 and TMR-Dextran. (D) Relative BODIPY-C16 and TMR-Dextran fluorescence in MDA-MB-231 cells. Cells were pretreated and incubated with or without ND-646 (10 μM) and BODIPY-C16 (2 μM) or TMR-Dextran (1 mg/ml) for 2 h. (E) Inhibitory effects of ND646 on KYSE140 and KYSE150 cells. (F) Survival of KYSE140 and KYSE150 cells treated with or without ND-646 (10 μM). The medium was supplemented with or without 10 μM palmitate. (G) Uptake of BODIPY-C16 and TMR-Dextran in KYSE140, KYSE150, and HCC-1806 cells. HCC-1806 cells were used as a positive control. Cells were treated as in B. Images (A, B, C, and G) were representative of at least three independent experiments. Data were from quintuple experiments (D–F), and all experimental data were verified in at least two independent experiments. Error bars indicate the mean ± SD. **, P < 0.01; two-tailed Student’s t test. Scale bar = 10 μm.
Figure S4.
Fatty acid uptake correlates with macropinocytic activity. (A) Co-localization of BODIPY-C16 and TMR-Dextran in HCC1806, HeLa, and HCT-8 cells. Cells were incubated with 5 μM BODIPY-C16 for 30 min prior to 1 mg/ml TMR-Dextran treatment for another 30 min. The right panel shows fluorescence intensity of BODIPY-C16 (green) and TMR-Dextran (red) across lines shown in the left panel. (B) Relative BODIPY-C16 and TMR-Dextran fluorescence in HeLa cells. HeLa cells were pretreated with vehicle, nortriptyline (10 μM), or EIPA (10 μM) for 8 h and then incubated with or without ND-646 (1 μM) and 2 μM BODIPY-C16 or 1 mg/ml TMR-Dextran for 2 h. (C) Western blotting showing CRISPR-Cas9–mediated knockout of RAC1 in MDA-MB-231 cells. (D) Relative BODIPY-C16 and TMR-Dextran fluorescence in MDA-MB-231 RAC1-KO cells. Cells were incubated with BODIPY-C16 (0.5 μM) for 1 h or TMR-Dextran (1 mg/ml) for 2 h. Fluorescence was measured by flow cytometry. (E) Inhibitory effects of ND646 on MDA-MB-231/RAC1-KO cells. (F) Western blotting showing CRISPR-Cas9–mediated knockout of CD36 in MDA-MB-231 cells. (G and H) Uptake of BODIPY-C16 (G) or TMR-Dextran (H) in MDA-MB-231/WT or CD36-KO, or KO cells re-expressing CD36. Cells were incubated with BODIPY-C16 (0.5 μM; G) for 1 h or TMR-Dextran (1 mg/ml; H) for 2 h. Fluorescence was measured by flow cytometry. Images (A) were representative of at least three independent experiments. Data were from quintuple experiments (B) or triplicate experiments (D, E, G, and H), and all experimental data were verified in at least three independent experiments. Error bars indicate the mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test. Scale bar = 10 μm. Source data are available for this figure: SourceData FS4.
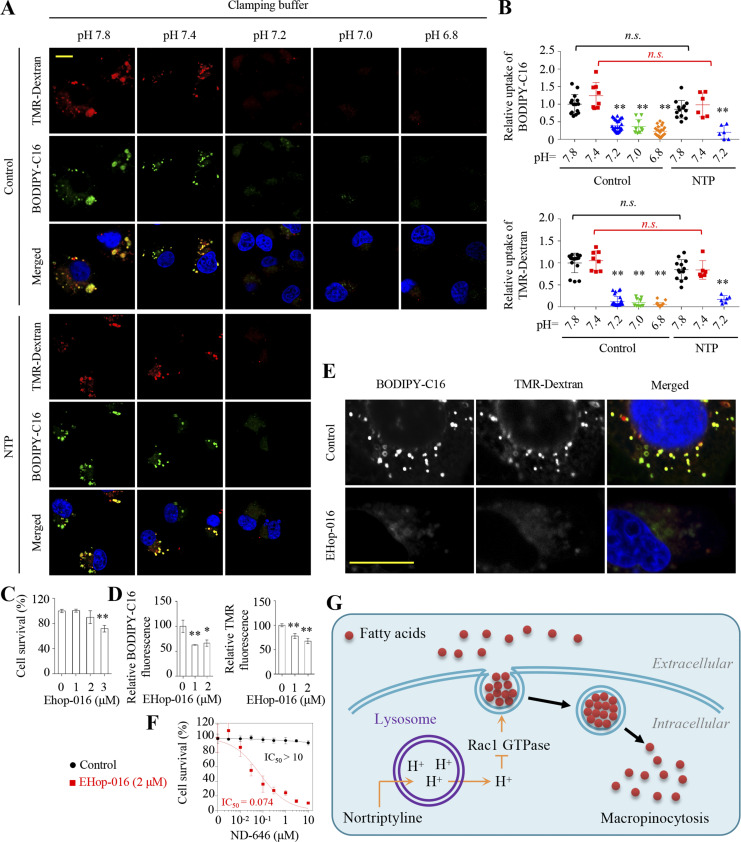
Furthermore, we used pH clamping buffers (Fig. S3 D) to confirm that an acidic intracellular pH environment sufficiently blocked the uptake of BODIPY-C16 and TMR-Dextran, while de-acidification disabled the inhibitory effects of nortriptyline toward ingesting BODIPY-C16 and TMR-Dextran (Fig. 5, A and B). It was previously reported that cellular acidification blocked the activities of GTPases (Koivusalo et al., 2010), especially Rac1, a GTPase required for the formation of macropinosomes (Yoshida et al., 2009). Therefore, we tested whether Rac1 inhibition suppressed macropinocytosis-mediated fatty acid uptake. First, we knocked out Rac1 in MDA-MB-231 cells and found it significantly prevented the uptake of both TMR-Dextran and BODIPY-C16, and dramatically enhanced the inhibitory effect of ND-646 on cancer cells (Fig. S4, C–E). Then, we used EHop-016, a Rac1 inhibitor, at concentrations that did not affect cell proliferation (Fig. 5 C), and confirmed that EHop-016 reduced the uptake of BODIPY-C16 and TMR-Dextran (Fig. 5, D and E), and sensitized cancer cells to ND-646 (Fig. 5 F). Therefore, nortriptyline inhibits fatty acid uptake most likely by inducing cellular acidification to block the formation of macropinosomes (Fig. 5 G).
Figure 5.
Cytosolic acidification blocks macropinocytosis-mediated fatty acid uptake. (A) Effect of pH on the uptake of TMR-Dextran and BODIPY-C16 in MDA-MB-231 cells with or without nortriptyline. Nigericin (10 μg/ml) was used to equilibrate pH inside and outside cells. Cells were pretreated with vehicle or nortriptyline (10 μM) for 8 h and then incubated with or without BODIPY-C16 (5 μM) for 30 min prior to TMR-Dextran (1 mg/ml) treatment for another 30 min in serum-free DMEM media with a predetermined pH. (B) BODIPY-C16 and TMR-Dextran fluorescence in A was analyzed using ImageJ. Data were normalized to pH 7.8. (C) Inhibitory effects of EHop-016 on MDAMB-231 cell survival with indicated concentrations for 4 d. (D) Relative BODIPY-C16 and TMR-Dextran fluorescence in MDA-MB-231 cells. Cells were pretreated with vehicle or EHop-016 (1 or 2 μM) for 24 h and incubated with BODIPY-C16 (2 μM) for 2 h or TMR-Dextran (1 mg/ml) for 1 h. (E) Uptake of BODIPY-C16 and TMR-Dextran in MDA-MB-231 cells. Cells were pretreated with vehicle or EHop-016 (2 μM) for 24 h and then incubated with BODIPY-C16 (5 μM) for 30 min prior to TMR-Dextran (1 mg/ml) treatment for another 30 min. (F) Inhibitory effects of ND-646 on MDA-MB-231 cells with or without EHop-016 (2 μM) for 4 d. (G) A working model showing macropinocytosis-mediated fatty acids uptake. NTP triggers lysosome-mediated cellular acidification, which blocks macropinocytosis through impaired GTPase activities. Images (A and E) were representative of at least three independent experiments. Data were from quintuple experiments (C, D, and F), and all experimental data were verified in at least three independent experiments. Error bars indicate the mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test. Scale bar = 10 μm.
It was reported that fatty acid uptake was impaired in some tissues in CD36-KO mice (Coburn et al., 2000), suggesting CD36 involvement in fatty acid uptake. Here, we knocked out CD36 in MDA-MB-231 cells (Fig. S4 F) and confirmed that the CD36-KO partially suppressed the uptake of BODIPY-C16 (Fig. S4 G). Meanwhile, we observed that CD36-knockout also partially suppressed macropinocytosis by measuring the ingestion of TMR-Dextrans (Fig. S4 H). Moreover, the uptake of BODIPY-C16 or TMR-Dextrans in KO cells was reversed by re-expression of CD36 (Fig. S4, F–H), suggesting that CD36 protein is required for robust macropinocytosis and fatty acid uptake. We further observed that nortriptyline inhibited fatty acid uptake and macropinocytosis to a similar extent in both CD36-expressing or -null MDA-MB-231 cell lines, indicating that macropinocytosis plays a critical role in taking up fatty acids in these cells.
Nortriptyline and ND-646 synergistically reduce cellular fatty acids and suppress tumor growth
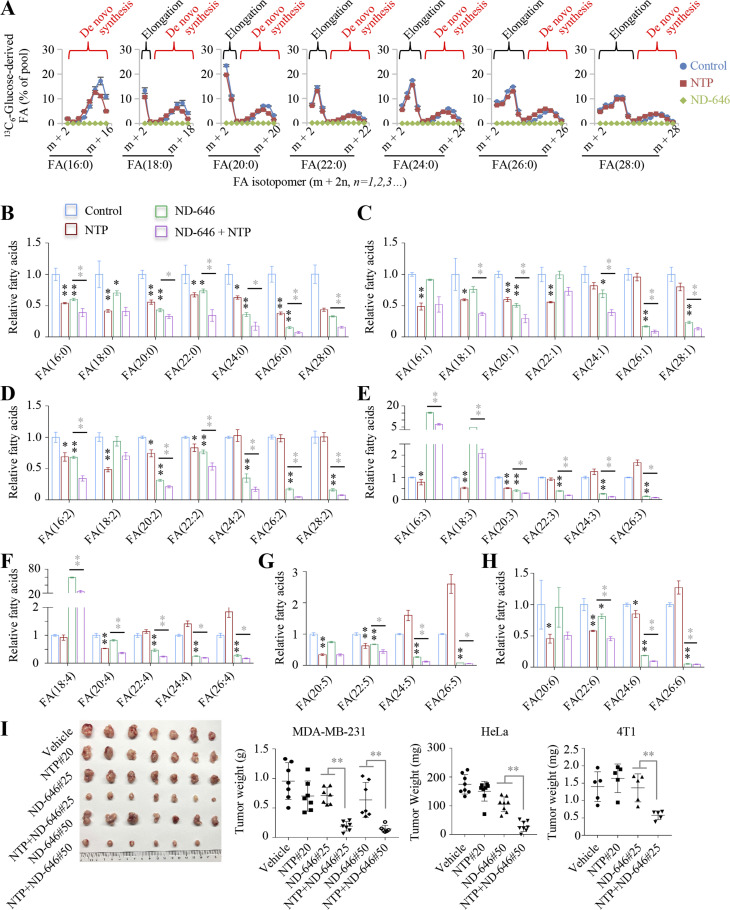
Now, we determined whether nortriptyline affected fatty acid biosynthesis using 13C6-glucose-tracing in MDA-MB-231 cells. Since acetyl-CoA or derived malonyl-CoA contained 13C from 13C6-glucose or not, labeled fractions should fit to the model of binomial distribution. VLCFAs can be produced from de novo synthesized palmitate, or elongated on the already existed palmitate or other precursors using malonyl-CoA as the two-carbon unit (Jakobsson et al., 2006); both processes can be distinguished based on the labeling model of fatty acids (Fig. 6 A). Our results showed that ND-646 (10 μM) was almost completely blocked, while nortriptyline did not affect de novo synthesis and elongation of fatty acids (Fig. 6 A and Fig. S5). Meantime, nortriptyline significantly reduced the cellular levels of all detected saturated fatty acids and most unsaturated fatty acids with ≤22-carbon (Fig. 6, B–H). Surprisingly, ND-646 did not decrease unsaturated VLCFAs with longer carbon-chains, which was most likely attributed to active elongation from precursors (Fig. 6 A and Fig. S5). This speculation was further confirmed by the results that nortriptyline, when combined with ND-646, further decreased almost all the detected fatty acids, including unsaturated VLCFAs (Fig. 6, B–H).
Figure 6.
Nortriptyline and ND-646 synergistically reduce cellular fatty acids and tumor growth. (A) Mass isotopomer analysis of fatty acids in MDA-MB-231 cells cultured with 13C6-glucose in the presence of 10 μM nortriptyline or ND-646. (B–H) Relative cellular levels of fatty acids in MDA-MB-231 cells plus 10 μM nortriptyline and/or ND-646. (I) Effects of nortriptyline and ND-646 on tumor growth after treatment for 21 d. MDA-MB-231–derived tumors in nude mice were treated with vehicle, 25 or 50 mg/kg ND-646, and/or 20 mg/kg nortriptyline (n = 7); HeLa-derived tumors (n = 8) were treated with 50 mg/kg ND-646 and/or 20 mg/kg nortriptyline; 4T1-derived tumors (n = 5) were treated with 25 mg/kg ND-646 and/or 20 mg/kg nortriptyline. Digital images show dissected tumors from MDA-MB-231 (left), and tumor weights are also shown. Data (A–H) were from three independent cultures. Error bars indicate the mean ± SD. *, P < 0.05; **, P < 0.01; two-tailed Student’s t test. Black asterisks show comparison with control group.
Figure S5.
Effects of nortriptyline and ND-646 on fatty acid biosynthesis. Mass isotopomer analysis of fatty acids in MDA-MB-231 cells cultured with 25 mM 13C6-glucose for 48 h in the presence of 10 μM nortriptyline or 10 μM ND-646. Data were from three independent cultures. Error bars indicate the mean ± SD.
We now tested the in vivo efficacy of ND-646 and/or nortriptyline in tumors. We intragastrically administered drug combinations to nude mice xenografted with MDA-MB-231, HeLa, and 4T1 cell–derived tumors, and observed that 20 mg/kg nortriptyline strongly and synergistically prevented tumor growth in combination with 25 or 50 mg/kg ND-646, but alone, they showed no or minor effects (Fig. 6 I).
Nortriptyline and ND-646 do not affect body weight in normal mice
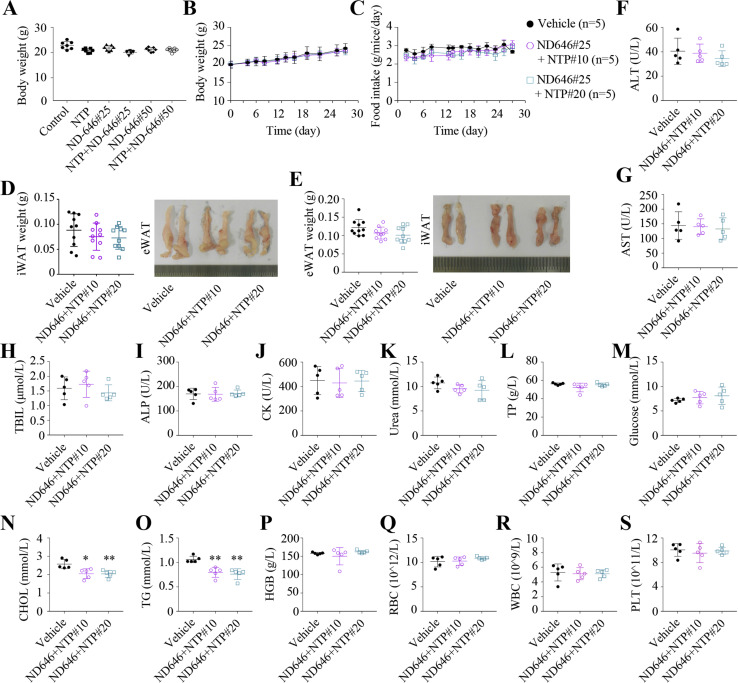
In aforementioned nude mice studies, nortriptyline and/or ND-646 did not significantly affect body weight (Fig. 7 A). Therefore, we measured the effects of the combination of ND-646 and nortriptyline on normal C57BL/6 mice. We observed that 25 mg/kg ND-646 in combination with 10 or 20 mg/kg nortriptyline did not significantly affect body weight and food intake of normal mice (Fig. 7, B and C), but slightly reduced the weight of iWAT and eWAT (Fig. 7, D and E). Furthermore, we measured several serum biochemical markers, including the liver function markers ALT (alanine transaminase), AST (aspartate transaminase), and TBIL (total bilirubin; Fig. 7, F–H), the bone marker ALP (alkaline phosphatase; Fig. 7 I), the cardiac marker CK (creatine kinase; Fig. 7 J), the renal marker urea nitrogen (Fig. 7 K), and also TP (total protein), glucose, total cholesterol, and triglycerides (TG; Fig. 7, L–O). Combined treatments did not influence the blood levels of these indexes, except total cholesterol and TG that were reduced upon treatments (Fig. 7, N and O). In addition, routine blood tests also indicated that the level of hemoglobin and the numbers of red blood cells, white blood cells and platelets were normal in mice treated with nortriptyline and ND-646 (Fig. 7, P–S). Taken together, these results suggest that nortriptyline and ND-646 combinations are well tolerated in mice.
Figure 7.
Nortriptyline combined with ND-646 is tolerated in mice. (A) The body weights of nude mice xenografted with MDA-MB-231–derived tumors from Fig. 6 I. (B and C) The body weights (B) and food intake (C) of normal C57BL/6 mice with indicated treatments. (D and E) Representative images and fat pad weight in normal C57BL/6 mice with indicated treatments. (F–S) Serum ALT (F), AST (G), TBIL (H), ALP (I), CK (J), urea (K), TP (L), glucose (M), CHOL (cholesterol; N), and TG (O) of normal C57BL/6 mice at the end of indicated treatments. (P–S) The HGB (hemoglobin; P), RBC (Q), WBC (R), and PLT (platelets; S) of normal C57BL/6 mice at the end of indicated treatment. Data were the mean ± SEM (B and C); or mean ± SD (A and F–S). *, P < 0.05; **, P < 0.01; two-tailed Student’s t test (A, n = 7; B–S, n = 5).
Nortriptyline and ND-646 synergistically suppress lipogenesis and hepatic steatosis in obese mice
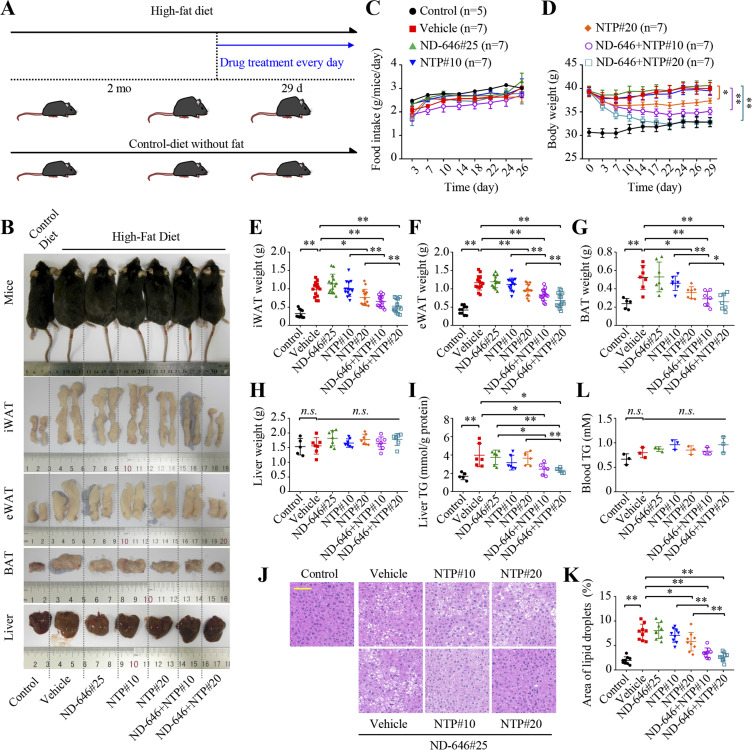
To further investigate the effects of nortriptyline and ND-646 on lipogenesis and hepatic steatosis, we fed C57BL/6 mice a high-fat diet (HFD) for 2 mo and generated obese mice with an approximate 30% increase in body weight. Mice fed the same diet without fat were used as controls (Fig. 8 A). We then intragastrically administered these obese mice with 10 or 20 mg/kg nortriptyline and/or 25 mg/kg ND-646 once every day for 29 d (Fig. 8 B). We measured concomitant food uptake and body weight and showed that nortriptyline and/or ND-646 did not affect food intake of obese mice (Fig. 8 C). Also, 25 mg/kg ND-646 did not influence body weight of obese mice at all (Fig. 8 D) and 10 mg/kg nortriptyline exerted no weight-reducing effects, but it did when combined with 25 mg/kg ND-646 (Fig. 8 D). In contrast, 20 mg/kg nortriptyline alone significantly reduced body weight of obese mice, and restored body weight to control levels in combination with 25 mg/kg ND-646 (Fig. 8 D). Thus, both synthesis and uptake of fatty acids contribute to weight gain of mice even during a HFD.
Figure 8.
Nortriptyline and ND-646 synergistically suppress lipogenesis and hepatic steatosis. (A) The protocol assessing the effects of nortriptyline and/or ND-646 on mice fed a HFD diet. Untreated mice fed the same diet without fat were used as controls. (B) Representative images showing body shape, fat pad, and liver morphology in control mice or HFD-fed mice treated with or without ND-646 and/or nortriptyline. (C and D) Food intake (C) and body weight (D) of control or HFD-fed mice (n = 5 for control or 7 for others). (E–H) Weight of iWAT (E, n = 10 for controls or 14 for others), eWAT (F, n = 10 for controls or 14 for others), BAT (G, n = 5 for controls or 7 for others) and liver (H, n = 5 for controls or 7 for others) of control or HFD-fed mice treated with or without ND-646 and/or nortriptyline. (I) Liver TG (triglyceride) content in control or HFD-fed mice treated with or without ND-646 and/or nortriptyline (n = 5 for controls or 6 for others). (J and K) Representative H&E staining of dissected liver tissue (J). Scale bar = 100 μm. Quantified liver lipid droplet areas (K, n = 9). (L) Blood TG of control or HFD-fed mice treated with or without ND-646 and/or nortriptyline (n = 3). *, P < 0.05; **, P < 0.01. Data were mean ± SEM, two-way ANOVA (C and D); or mean ± SD, two-tailed Student’s t test (E–I, K, and L).
At post-mortem, a larger volume and increased weight of iWAT, eWAT, and BAT in obese mice fed a HFD when compared with control mice (Fig. 8, B and E–G). Also, 25 mg/kg ND-646 and 10 mg/kg nortriptyline synergistically reduced the volume and weight of iWAT, eWAT, and BAT, although alone, they exerted no effect (Fig. 8, B and E–G). Nortriptyline (20 mg/kg) alone significantly suppressed fat mass gain and almost completely restored the volume and weight of iWAT, eWAT, and BAT to normal levels when combined with 25 mg/kg ND-646 (Fig. 8, B and E–G). In the meantime, although we did not observe weight change in liver tissues in both control and obese mice (Fig. 8 H), the content of liver TG became much higher in obese mice when compared with controls, which was not significantly reduced by nortriptyline or ND-646 alone but almost backed to normal under their simultaneous treatments (Fig. 8 I). Accordingly, H&E staining showed that lipid droplet size in liver tissue was much higher in obese mice but drastically decreased upon combined treatment with both ND-646 and nortriptyline (Fig. 8, J and K). Furthermore, blood TG were only slightly increased in obese mice when compared with control mice (Fig. 8 L). ND-646 and/or nortriptyline treatment only marginally enhanced these levels in obese mice, but a significant difference was not observed (Fig. 8 L). Thus, the simultaneous blockage of fatty acid synthesis and uptake can effectively suppress lipogenesis and hepatic steatosis.
Discussion
While overwhelming evidence now shows that fatty acids enter cells mainly through a regulated pathway, rather than membrane diffusion, detailed mechanisms remain largely uncharacterized (Hao et al., 2020; Ibrahim et al., 2020; Su and Abumrad, 2009). Currently, the transmembrane proteins FATPs and CD36 are reported to mediate the uptake of fatty acids (Dourlen et al., 2015; Su and Abumrad, 2009). However, they are not bona fide membrane pore transporters (Glatz et al., 2010; Luiken et al., 2016), and thus do not directly absorb extracellular fatty acids. Herein we show that cells can take up fatty acids actively through macropinocytosis, and identify an approved clinical drug, nortriptyline, which induces cellular acidification to inhibit macropinocytosis-mediated fatty acid uptake. Thus, macropinocytosis can transport fatty acids on a large scale and may be a critical pathway for fatty acid uptake of cells.
CD36 KO mice show defective uptake and utilization of fatty acids in muscle and adipose tissues but not in liver (Coburn et al., 2000), indicating that CD36 is somehow required for fatty acid uptake, at least in some tissues. We show that CD36 knockout simultaneously and partially suppresses macropinocytosis, in addition to fatty acid uptake in MDA-MB-231 cells, thus raising the possibility that CD36 could facilitate macropinocytosis-dependent fatty acid uptake. Thus, the mechanism underpinning macropinocytosis regulation by CD36 deserves further exploration. Importantly, nortriptyline suppresses fatty acid uptake in cells regardless of CD36, and it can also significantly block in vivo fatty acid uptake in mouse liver, muscle, and adipose tissue. These results imply that liver cells and adipocytes take up fatty acids mainly through macropinocytosis, which is possibly also regulated by CD36, in accordance with previous findings (Coburn et al., 2000). Similarly, our data do not rule out the possibility that CD36 is a fatty acid transporter and delivers some particular fatty acids as the signal molecules (Silverstein and Febbraio, 2009).
Fatty acid biosynthesis is frequently upregulated in tumor tissue and during hepatic steatosis; therefore, the related inhibitors have been extensively characterized in terms of their therapeutic potential against these diseases (Dorn et al., 2010; Menendez and Lupu, 2007). However, no definitive clinical trials have been reported. Here, we reveal that fatty acid synthesis blockage significantly promotes macropinocytosis-mediated fatty acid uptake, which could largely antagonize the suppressive effects of cellular fatty acids afforded by fatty acid synthesis inhibitors. This possibly accounts for the failure of fatty acid synthesis inhibitors in vitro and in vivo. Therefore, now we can simultaneously block both ACC1/2 and macropinocytosis using their relevant inhibitors, such as ND-646 and nortriptyline, to control cellular fatty acids. Combining these inhibitors exerted more suppressive effects on adipose tissue mass in obese mice when compared with normal mice. This is most likely due to obese adipose tissue having higher metabolic rates. In addition, blocking autophagy has been reported to suppress β-oxidation of fatty acids and thus enhance triglyceride storage in lipid droplets in cultured hepatocytes and mouse liver (Singh et al., 2009). We show that nortriptyline is a potential lysosomotropic agent, and msy repress autophagic cargo turnover. As such, nortriptyline may slow the breakdown of lipid droplets and attenuate its efficacy in antagonizing hepatic steatosis. However, simultaneously blocking fatty acid uptake and synthesis may eventually reduce intracellular lipid droplets, even if their breakdown slows down. This possibly explains why we can observe suppressive effects for combined nortriptyline and ND-646 on mouse hepatic steatosis, but these effects are not as significant as in adipose tissue over the same treatment time (Fig. 8). Therefore, our findings promote the development of fatty acid synthesis inhibitors for clinical use and highlights the pivotal role of lipid metabolism in disease.
On the other hand, nortriptyline is typically used as an antidepressant and is originally believed to enhance the synaptic concentration of norepinephrine and/or serotonin in the central nervous system through blocking re-uptake by presynaptic receptors (Merwar et al., 2022). Nortriptyline is also reported to inhibit the activity of histamine, 5-hydroxytryptamine, and acetylcholine, which facilitate antidepressant actions (Merwar et al., 2022). Therefore, it will be interesting to examine if nortriptyline-mediated macropinocytosis inhibition also contributes to its antidepressant efficacy, which may promote better antidepressant drugs or improve treatments for depression.
Materials and methods
Cell culture
MDA-MB-231, HeLa, HCT116, HCT-8, HCC1806, SKHEP-1, HAEC, HPAC, HepG2, Huh7, and 293T were obtained from the American Tissue Culture Collection. HCCLM3, L02, and KYSE150 were obtained from China Center for Type Culture Collection. HCT-8, HCC1806, and KYSE150 cells were cultured in 1640 (Corning), and other cells were cultured in DMEM (Corning). The above mediums were supplemented with 10% FBS (#900-108; Gemini) and 100 U/ml penicillin and streptomycin (#15140122; Gibco). All cells were cultured in a humidified atmosphere of 5% CO2 at 37°C.
For delipidized medium, cells were first seeded in the medium containing regular 10% FBS and were switched into the medium containing 10% delipidized FBS (#P30-3402; PAN) upon treatment on the following day.
Generation of knockout cell line
Both ACC1 and ACC2 were deleted form MDA-MB-231 cells using the CRISPR-Cas9 system. pCDH-Cas9-2A-GFP-BSD was used to express Cas9. Single-guide RNAs were cloned into pLentiGuide-Vector linearized with BsmBI. ACC1/2-DKO cell clones were generated with the following guide sequences: ACC1, 5′-GAAGACCTTAAAGCCAATGC-3′; and ACC2, 5′-GGGTTGGGACCTGTAGCGTC-3′. CD36-KO cells was generated with the following guide sequence: 5′-GATGTTTCAGAACCTATTGA-3′. MDA-MB-231 cells were co-transfected with Cas9-EGFP, pLentiGuide-sg sgACC1 and sgACC2, or sgC36 plasmids using lipofectamine 3000 (#L3000-015; Invitrogen). Single cells were sorted into 96-well plate based on green fluorescence and maintained until colonies formed. CD36 restoration was performed by infecting MDA-MB-231/CD36-KO cells with lentivirus expressing full-length human CD36 cDNA. For Rac1-KO pooled cells, single-guide RNAs were cloned into lentiCRISPRv2 (one vector system) linearized with BsmBI. The guide sequences used for Rac1 are 5′-ATTTAAGATACTTACACAGT-3′ and 5′-CTGTTTGCGGATAGGATAGG-3′. Then, the lentivirus was produced in 293T cells and MDA-MB-231 cells were infected with indicated lentivirus. Cells were validated for knockout of related genes by Western blotting.
Lentivirus production
The expression plasmid pCDH-CMV-GFP-LGALS3 or lentiCRISPRv2 plasmids, pCMV-Dr8.91, and pCMV-VSV-G were co-transfected into 293T cells using polyethylenimie (#23966-1; Polyscience), and then media containing the virus was collected at 48 h after transfection. Cells were infected with medium containing viruses in the presence of hexadimethrine bromide (10 μg/ml, #H9268; Sigma-Aldrich) for 48 h, and then cells were selected with puromycin.
Western blotting
After desired treatments as specified, cells were washed twice with PBS and lysed in buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycer-ophosphate, 1 mM sodium vanadate, 1 mg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride) on ice. Equal amounts of protein (30 μg) were loaded onto 8% SDS-PAGE gels. Western detection was performed using a chemiluminescence Western blot scanner (ChampGel 7000, SAGECREATION). For protein detection, the following antibodies were used: ACC1 (#21923-1-AP; Proteintech), ACC2 (#8578; Cell Signaling), phosphorylation ACC (#3661; Cell Signaling), CD36 (#18836-1-AP; Proteintech), LAMP2 (#66301-1-Ig; Proteintech), TOM20 (ab56783; Abcam), GAPDH (#60004-1-Ig; Proteintech), Lamin B1 12987-1-AP(proteintech), RAC1 (#24072-1-AP; Proteintech), β-Actin (#AC026; Abclonal), β-tubulin (#AC008; Abclonal), goat anti-rabbit IgG/HRP (#115-035-003; Jackson) and goat anti-mouse IgG/HRP (#111-035-003; Jackson).
Drug testing
The Food and Drug Administration–Approved Drug Library Mini (#HY-L022M; MCE) was used to screen fatty acid uptake inhibitors. MDA-MB-231/ACC-DKO or MDA-MB-231/WT cells were plated in 96-well plates at 2000 cells per well and were treated on the following day either with vehicle or 10 μM of candidate drugs for 4 d. Then, cell viability was measured with the use of the CellTiter-Glo Luminescent Cell Viability Assay Kit (#G7571; Promega) and the plate was read by microplate luminometer to monitor the luminescence signal.
Cell proliferation and survival assay
As for the cell proliferation, cells were plated in triplicate in 12-well plates at 104 cells per well. After the desired treatments as indicated in the figures, cells were washed twice with PBS to remove dead cells, and then were trypsinized. The cell number was determined using Muse Cell Analyzer (Luminex Cooperation). For each well, the fold-change in cell number relative to day 0 was determined.
As for the cell survival assay, 2,000 cells per well were seeded in 96-well plates and were treated on the following day either with vehicle or drugs with the indicated concentrations in the figures for 4 d. Then, cell viability was determined using the CellTiter-Glo Luminescent Cell Viability Assay Kit (#G7571; Promega).
Preparation of palmitate/BSA complex
The palmitate was prepared according to a previously described protocol (Cousin et al., 2001). Briefly, palmitate (#P0500; Sigma-Aldrich), oleate (#O1008; Sigma-Aldrich), linoleate (#L1012; Sigma-Aldrich), and linolenate (#L2376; Sigma-Aldrich) was solubilized in 0.1 M NaOH (heating at 70°C) at 100 mM, and combined with 10% (wt/vol) fatty acid–free BSA (#A8806; BSA, Sigma-Aldrich; heating at 55°C) to make a fatty acid concentration of 5 mM. Stearate (#S4751; Sigma-Aldrich) and arachidate (#A3631; Sigma-Aldrich) were solubilized in trichloromethane at 200 mM separately and combined with 10% (wt/vol) fatty acid–free BSA (heating at 55°C) to make a concentration of 2 mM. Stored fatty acid/10% BSA stock solutions were heated for 15 min at 55°C, and then cooled to room temperature before use.
Subcellular fractionation
To perform lysosome enrichment, subcellular fractionation was carried out using the Lysosome Enrichment Kit (#89839; Thermo Fisher Scientific). Briefly, cells were treated with nortriptyline for 12 h, then the cell pellet was resuspended in Lysosome Enrichment Reagent A and homogenized with a dounce homogenizer on ice (∼80 strokes). The homogenate was mixed with equal volume Lysosome Enrichment Reagent B, then subjected to centrifugation at 500 ×g for 10 min at 4°C, and the supernatant was diluted in OptiPrep gradient media to a final concentration of 15% OptiPrep. The sample was then carefully overlayered on the top of a discontinuous density gradient (17, 20, 23, 27, and 30%). The gradient was subjected to ultracentrifugation at 145,000 ×g in an MLS-50 bucket rotor (Beckman) for 2 h at 4°C. After the spin, three bands were formed, and 1 ml of each fraction band were collected from top to bottom. The fraction was mixed with two volumes of PBS and then divided into two parts. These mixtures were centrifuged at 18,000 ×g for 30 min at 4°C. The pellet from one part was used for protein detection by Western blots. The pellet from another part was resuspended with 1 ml extraction fluid (the ratio of H2O:acetonitrile:methanol is 1:1:3). Macromolecules and debris were removed by centrifugation at 14,000 ×g for 20 min at 4°C, and the metabolites-containing supernatants were dried under nitrogen gas. Dried samples were stored at −80°C and then resuspended in 50 μl 80% methanol and prepared for LC/MS analyses.
Lysosomal galectin puncta assay
MDA-MB-231 cells stably expressing GFP-LGALS3 by GFP-LGALS3 lentivirus infection were grown on 12-well plates (in Fig. S3 A) or on glass coverslips (in Fig. 3 D). After 24 h culture, cells were treated with the indicated chemicals for 30 min. Images of LGALS3 distribution were visualized with an Olympus inverted microscope equipped with a charge-couple camera (in Fig. S3, A). Cells were fixed with 4% (wt/vol) paraformaldehyde (PFA; #P1110; Solarbio) for 15 min. Then, cells were mounted with the anti-fade medium (#H-1000; Vector Laboratories) and visualized with a confocal laser-scanning microscope (LSM880, Carl Zeiss) with a 63× oil-immersion objective with 1.4 numerical aperture (Plan-Apochromat 63×/1.4 Oil DIC M27, Carl Zeiss; in Fig. 3 D). The filter set for GFP imaging was excitation/emission: 480/530 nm.
LysoTracker staining and lysosomal pH measurement
For LysoTracker imaging, the cells were stained with LysoTracker Red DND-99 (50 nM; #L7528; Thermo Fisher Scientific) for 15 min, then the fluorescence was captured using a confocal laser-scanning microscope (LSM880, Carl Zeiss) with a 63× oil-immersion objective with 1.4 numerical aperture (Plan-Apochromat 63×/1.4 Oil DIC M27, Carl Zeiss). A 594-nm laser was used to visualize the fluorescence.
For lysosomal pH value measurement, cells grown on glass bottom cell culture dish (#801002; NEST) were labeled with 2 μM LysoSensor Yellow/Blue DND-160 (#L7545; Thermo Fisher Scientific) for 20 min at cell culture condition, and excessive dye was washed out using PBS. For pH calibration, the labeled cells were treated with 10 μM monensin (#HY-N0150; MCE) and 10 μM nigericin (#HY-100381; MCE) in 2-(N-morpholino)ethanesulfonic acid buffer (5 mM NaCl, 115 mM KCl, 1.3 ml MgSO4, 25 mM 2-(N-morpholino)ethanesulfonic acid), with the pH adjusted to a range from 3.5 to 7.0. For drugs treatment, the labeled cells were treated with nortriptyline (10 μM, #HY-B1417; MCE), dynasore (10 μM, #S8047; Selleck), EIPA (10 μM, #HY-101840; MCE), chloroquine (10 μM,#C2301; TCI) or bafilomycin A1 (200 nM, #HY-81280; MCE) for 1 h in complete DMEM medium without phenol red. The samples were read in a SpectraMax iD5 (Molecular Devices) with excitation at 355 nm. The ratio of emission 460/535 nm was then calculated for each sample. The pH values were determined from the linear standard curve generated through the pH calibration samples.
Cytosolic pH measurements
Dual-emission ratio fluorescence (580/640 nm) of SNARF-5F (#S23923; Thermo Fisher Scientific) were used to measure pH. Cells grown on glass bottom cell culture dish (#801002; NEST), were loaded with 20 μM of SNARF-5F acetoxymethyl-ester for 20 min at cell culture condition. Then cells were washed with PBS three times and treated with nortriptyline (10 μM), dynasore (10 μm), EIPA (10 μM), or chloroquine (10 μM) or bafilomycin A1 (200 nM) in culture medium for 40 min. Imaging of live cells was performed at 37°C with CO2 in culture medium with inhibitor or pH-clamping buffer as indicated. pH measurements were performed using a confocal laser-scanning microscope (LSM880, Carl Zeiss) with a 63× oil-immersion objective with 1.4 numerical aperture (Plan-Apochromat 63×/1.4 Oil DIC M27, Carl Zeiss) with excitation at 488 nm. The ratio of signals obtained at 550–602 nm (for 580 nm) and 621–674 nm (for 640 nm). At least 10 fields were randomly selected from different regions across the entirety of each sample.
For pH clamp experiments, cells loaded with 20 μM of SNARF-5F were incubated with a clamping buffer (120 mM KCl, 5 mM NaCl, 20 mM HEPES, 1 mM CaCl2, 1 mM MgCl2, and 10 μg/ml nigericin) of desired pH for 10 min. In the clamping buffer, the pH in cells were determined as above described.
Fatty acid uptake assay and imaging
BODIPY-C16 (#D3821; Thermo Fisher Scientific) was dissolved in methanol and stored as small aliquots of 5 mM stock solutions at −20°C. Fatty acid uptake assay was performed as previous described (Ibrahim et al., 2020). Briefly, MDA-MB-231 or HeLa cells were plated onto 96-well black-walled, clear-bottom plates (#6005182; PerkinElmer) at 2 × 104 cells per well. After overnight culture, cells were pretreated with vehicle, nortriptyline (10 μM), dynasore (10 μM), EIPA (10 μM), SSO (100 μM, #HY-112847A; MCE), lipofermata (1 μM, #HY-116788), or EHop-016 (1 or 2 μM, MCE, #HY-12810) for the indicated time. Then, cells were given 2 μM BODIPY-C16 in the serum-free media with the indicated chemicals for 2 h. The solution of BODIPY-C16 was then completely aspirated, and the cells were washed with 0.1% BSA (in PBS) twice. To quench extracellular fluorescence, 0.08% Trypan Blue was added, and intracellular fluorescence was swiftly measured using a Spectramax microplate reader (TriStar2 S LB 942, Berthold Technologies). Reading from wells containing cells without BODIPY-C16 were used as background noise and subtracted from experimental values.
As for imaging, cells were cultured on glass coverslips. After 24 h culture, cells were pretreated without or with indicated inhibitors, and then were given 5 μM BODIPY-C16 in the serum-free media with the indicated chemicals for 1 h or the indicated time. Then, cells were washed with PBS containing 0.5% (wt/vol) BSA and preceded to fixtion with 4% (wt/vol) PFA for 15 min. Cells were mounted with the anti-fade medium (#H-1000; Vector Laboratories). Images were captured using a confocal laser-scanning microscope (LSM880, Carl Zeiss) with a 63× oil-immersion objective with 1.4 numerical aperture (Plan-Apochromat 63×/1.4 Oil DIC M27, Carl Zeiss). A 488-nm laser and a 405-nm were used to visualize BODIPY-C16 (excitation: 488 nm, emission: 512 nm) and DAPI (excitation: 405 nm, emission: 459 nm), respectively.
For free fatty acid uptake assay, cells were pretreated in serum-free medium for 8 h, and NTP (10 μM) or vehicle for 1 h, followed by treatment with 100 μM BSA-conjugated fatty acid for 4 h. Cells were then stained with 0.5 μM BODIPY 493/503 (D3922; Thermo Fisher Scientific) for 30 min at the end of BSA-conjugated fatty acid incubation period, and the fluorescence intensity was examined using BD LSRFortessa cell analyzer (BD Biosciences). For imaging, cells were stained with 2 μM BODIPY 493/503 for 30 min and images were captured as described above.
TMR-Dextran uptake assay and imaging
70 kD TMR-Dextran (#D1818; Thermo Fisher Scientific) was dissolved in PBS and stored as small aliquots of 100 mg/ml stock solutions at −20°C. The cells were pretreated with indicated inhibitors and incubated with the inhibitors-containing serum-free DMEM supplemented with 2 mg/ml of TMR-Dextran treatment for 2 h. Then, cells were washed and quenched the extracellular fluorescence as described in Fatty acid uptake assay and imaging. The intracellular fluorescence of TMR-Dextran was measured using a Spectramax microplate reader (bottom-read, excitation: 540 nm, emission: 590 nm, TriStar2 S LB 942, Berthold Technologies).
The macropinosome were imaged using confocal microscope (LSM880, Carl Zeiss) and analyzed using the “Analyze Particles” feature in ImageJ (National Institutes Health) as previous described (Commisso et al., 2013). For BODIPY-C16 and macropinosome co-localization, 24 h after cell seeding, cells were treated without or with indicated drugs for 8 h. Then, cells were incubated with the inhibitors-containing serum-free DMEM supplemented with 5 μM of BODIPY-C16 for 30 min, prior to both BODIPY-C16 and 1 mg/ml of TMR-Dextran treatment for 30 min. At the end of the incubation period, cells were washed with PBS containing 0.5% (wt/vol) BSA and preceded to fixtion with 4% (wt/vol) PFA for 15 min. Cells were DAPI-treated to stain nuclei and mounted with the anti-fade medium. Images of macropinosomes were captured using a confocal laser-scanning microscope (LSM880, Carl Zeiss) with a 63× oil-immersion objective with 1.4 numerical aperture (Plan-Apochromat 63×/1.4 Oil DIC M27, Carl Zeiss). A 543-nm laser (excitation: 543 nm, emission: 614 nm) was used to visualize TMR-Dextran.
Dialyzed FBS preparation
The 80 ml FBS was put into 20 cm dialysis membrane (#YA1076; 7 kD cut-off, Solarbio), and dialyzed against 2 liters of 0.15 M of NaCl at 4°C for 2 h. Then the NaCl solution was refreshed two to three times. The FBS was dialyzed until glucose <5 mg/dl, and then proceed to be filtered with 0.22 μm membrane under sterile condition. This dialyzed FBS was used for isotope labeling experiments.
Isotope tracing of fatty acids
Fatty acids analysis was performed on Q Exactive orbitrap mass spectrometer (Thermo Fisher Scientific). Experiments were performed in medium containing 10% FBS-Dialyzed. DMEM without glucose, glutamine, pyruvate, and phenol red was prepared from power (Caisson) by adding 3.7 g NaHCO3 per liter and adjusting the pH to 7.4, then supplemented with 10 mM [U-13C]-glucose, 2 mM glutamine, and 1 mM pyruvate. For saponified fatty acid analysis, 5 × 105 cells were seeded in 60-mm dishes overnight, and treated with indicated chemicals for 24 h and were changed into 13C-containing medium with indicated chemicals for 48 h. Samples were prepared as previously described (Kamphorst et al., 2013). The medium was aspirated completely and cells were washed with PBS 3 times quickly and gently. Cells were extracted in 1 ml 50% (vol/vol) methanol solution containing 0.1 M of HCl (prechilled to −20°C). The resulting liquid and cell debris were scraped into a glass vial. Chloroform (0.5 ml) was added, the mixture was vortexed for 1 min and then centrifuged for 15 min at 3,000 rpm, and the chloroform layer was transferred to a glass vial. Equal amounts of extracts from different treatments were dried under nitrogen gas (room temperature), reconstituted into 1 ml 90% methanol solution containing 0.3 mM of KOH, incubated at 80°C for 1 h to saponify fatty acids, acidified with 0.1 ml of formic acid, extracted twice with 1 ml of hexane, and dried under nitrogen gas. Dried samples were stored at −80°C and then resuspended in 150 μl dichloromethane: methanol and prepared for LC/MS analyses. Cortecs C18 column (2.1 × 100 mm; Waters) was applied for analysis. Mobile phase A was prepared by dissolving 0.77 g of ammonium acetate in 400 ml of HPLC-grade water, followed by adding 600 ml of HPLC-grade acetonitrile. Mobile phase B was prepared by mixing 100 ml of acetonitrile with 900 ml isopropanol. An 18-min gradient with flow rate of 250 μl/min was used. The linear gradient was as follows: 0 min, 30% B; 2.5 min, 30% B; 8 min 50% B; 10 min, 98% B; 15 min, 98% B; 15.1 min, 30% B; 18 min, 30% B. Data with mass ranges of m/z 150–600 were acquired at negative ion mode. The full scan was collected with resolution of 70,000. The source parameters are as follows: spray voltage: 3 kv; capillary temperature: 320°C; heater temperature: 300°C; sheath gas flow rate: 35 Arb; auxiliary gas flow rate: 10 Arb. Isotope-labeled fatty acids were assigned based on in-house database containing 13C-labeled medium to very long chain fatty acids according to accurate ion masses using Tracefinder 3.2 (Thermo Fisher Scientific). Mass tolerance of 10 ppm was applied for precursor mass matching. Chromatographic peak area was used for relative quantitation.
Animal studies
All procedures using animals were approved by the Capital Medical University Institutional Animal Care and Use Committee. The mice were housed in a pathogen-free animal barrier facility.
Subcutaneous tumor studies
For xenografts, 5 × 106 of MDA-MB-231 or 4 × 106 of HeLa or 106 of 4T1 cells were injected subcutaneously into the hind flanks of female athymic nude mice. Tumor prevention studies began at 14 d (MDA-MB-231) or 10 d (HeLa) or 7 d (4T1) after tumor cell injection. Tumor growth and body weight were measured every 3 d using digital calipers and electronic scale. The tumor volumes were calculated using the formula 1/2 × L × W2. The average tumor volume at the initiation of treatment was around 50 mm3. Six treatment groups were designed for MDA-MB-231–derived tumors: vehicle, 25 mg/kg of ND-646 (# PBLJ7829; PharmaBlock Science), 50 mg/kg of ND-646, 20 mg/kg of nortriptyline, 50 mg/kg of ND-646 plus 20 mg/kg of nortriptyline, and 25 mg/kg of ND-646 plus 20 mg/kg of nortriptyline. Four treatment groups were designed for HeLa and 4T1 cell–derived tumors: vehicle, 25 or 50 mg/kg of ND-646, 20 mg/kg of nortriptyline groups, 25 or 50 mg/kg of ND-646 plus 20 mg/kg of nortriptyline. Mice were randomized into their treatment groups based on similar tumor volumes. For all studies, drugs were administered by oral gavage and treated once a day for 21 d. Vehicle solution (0.9% NaCl, 1% Tween 80, and 0.5% methylcellulose), ND-646 and nortriptyline were dissolved in vehicle solution. For combination treatment, ND-646 was administrated at morning and nortriptyline at afternoon, and the time between does was ∼8 h. All the mice were killed at the end, and tumors were harvested and weighed.
Absorption of BODIPY-C16 in vivo
For BODIPY-C16 in vivo in C57 male mice, mice were dosed orally once a day with either a vehicle solution or nortriptyline at 20 mg/kg for 7 d. Mice were fasted overnight (16 h), after last does of vehicle or nortriptyline were given for 2 h and then intravenously injected with 100 μg of BODIPY-C16 diluted in PBS to a volume of 150 μl. At 3 h after injection, livers were perfused with PBS through the left ventricle. The tissues were removed, rinsed with PBS, cut into small pieces and digested in RPMI containing 0.05% collagenase/dispase (1026938001; Roche) for 1 h at 37°C. The resulting liver, muscle, and heart suspension were passed through a 70-μm Nitex filters, centrifuged at 800 ×g (10 min at 4°C), and the cell pellet were resuspended in PBS, respectively. For adipose tissue analyses, iWAT, eWAT, and interscapular brown adipose tissue were resected, digested as above tissues. The resulting adipose tissues suspension were passed through a 250-μm Nitex filters, centrifuged at 400 ×g for 5 min to separate the stromal vascular fraction from the adipocytes as described (Daquinag et al., 2021; Daquinag et al., 2011; Traktuev et al., 2008). The fluorescence intensity was examined using BD LSRFortessa cell analyzer (BD Biosciences). For BODIPY-C16in nude mice bearing tumor, when tumors derived from MDA-MB-231 reached an average volume of 400 mm3, mice were dosed orally once a day with either a vehicle solution or nortriptyline at 20 mg/kg for 3 d. Mice were fasted and intravenously injected with BODIPY-C16 as described as above. At 3 h after injection, tumors were removed, rapidly frozen in liquid nitrogen and embedded in tissue-freezing medium (#4583; SAKURA). The embedded samples were cut, mounted on glass slides and mounted with the anti-fade medium. The BODIPY-C16 uptake in vivo was determined from at least five sections per tumor or tissue with five fields analyzed per section. The images were taken as described in Fatty acid uptake assay and imaging. Part of the tumors were digested and centrifuged, and fluorescence intensity of tumor cells were examined by flow cytometry as described above. Three mice were used per treatment and analyzed.
Hepatic steatosis studies
For hepatic steatosis mice model, the 8-wk-old C57BL/6N male mice (specific pathogen–free grade) were purchased from Weitonglihua Company. Standard control diet (Cat# H10010; Huafukang Company) and sterile water were given ad libitum to control mice. Mice were given HFD (#H10060; Huafukang Company) for a total period of 12 wk to generate obese mice with about 15% increase in body weight. Six treatment groups were designed: control (n = 5, control diet group), vehicle (n = 7), 25 mg/kg of ND-646 (n = 7), 10 mg/kg of nortriptyline (n = 7), 20 mg/kg of nortriptyline (n = 7), 25 mg/kg of ND-646 plus 10 mg/kg of nortriptyline (n = 7) and 25 mg/kg of ND-646 plus 20 mg/kg of nortriptyline (n = 7). Mice were randomized into their treatment groups based on similar body weight. The drugs were dissolved and given as above tumor experiment. The body weight and food intake were monitored every 3 d. At the end of the study, the blood was drawn and euthanized 1 h after the final dose. Serum TG, ALT, and AST were measured using Roche Modular P800 Automatic Analyzer (Roche). Liver TG were assayed using a triglyceride assay kit (#E1025; Applygen Technologies). The livers were fixed in 4% PFA overnight and then embedded in paraffin wax. The embedded samples were cut and mounted on glass slides. Then the samples on glass slides were then stained with H&E. Images were captured using a scanning microscope (Leica GT450) with a 20× objective. The liver lipid droplets area was analyzed using ImageJ (National Institutes of Health).
Toxicological assessment
The 5-wk-old C57BL/6N male mice (specific pathogen–free grade) were purchased from Weitonglihua Company. Three treatment groups were designed: vehicle (n = 5), 25 mg/kg ND646 plus 10 mg/kg nortriptyline (n = 5), and 25 mg/kg ND646 plus 20 mg/kg nortriptyline (n = 5). Mice were randomized into their treatment groups based on similar body weight. The drugs were dissolved and given as above. The body weight and food intake were monitored every 2 d. At the end of the study, the blood was drawn and mice were euthanized 1 h after the final dose. Blood was collected in EDTA tubes and blood routine examination were analyzed by Tek-II automatic animal blood analyzer (TEC Technology). The other blood samples were centrifuged at 3,000 ×g for 10 min under 4°C and serum TBIL, ALP, CK, urea, TP, glucose, cholesterol, and TG were measured using 7600 Automatic Analyzer (Hitachi).
Statistics
Data are given as means ± SD or SEM. Statistical significance was calculated as appropriate using two-tailed Student’s t test or two-way ANOVA as indicated in the figure legends. Asterisks in the figures indicated statistical significances (*, P < 0.05; **, P < 0.01).
Online supplemental material
Fig. S1 shows the effects of SSO, lipofermata, nortriptyline, EIPA, and/or ND-646 on MDA-MB-231, HeLa, HCT116, and HCT-8 cells. Fig. S2 indicates screening of inhibitors for fatty acid uptake using MDA-MB-231/ACC1/2-DKO cells, identifies nine candidates, and then tests their effects on cancer cells. Fig. S3 shows the effects of identified candidates on lysosomal membrane permeabilization and the uptake of fatty acids, related to cellular acidification. Fig. S4 demonstrates the correlation of fatty acid uptake and macropinocytic activity. Fig. S5 shows the effects of nortriptyline and ND-646 on the biosynthesis of fatty acids.
Supplementary Material
contains original blots for Fig. 1.
contains original blots for Fig. 3.
contains original blots for Fig. S2.
contains original blots for Fig. S4.
Acknowledgments
We thank Dr. Xiaohui Liu (Metabolomics Facility at Tsinghua University Branch of China National Center for Protein Sciences, China) for technical help.
This work is supported by grants 82030093 and 81972567 from the Natural Science Foundation of China, grant KM202110025030 from the Scientific Research Common Program of Beijing Municipal Commission of Education, and grant Jingyiyan-2021-10 from the Beijing Municipal Institute of Public Medical Research Development and Reform Pilot Project.
Authors contributions: B. Li conceived and designed the study, and supervised the project; Q. Chu and P. Liu performed the experiments on gene knockout cells; Q. Chu and J. An performed the experiments about the effects of drugs and inhibitors on cancer cells; Q. Chu., P. Liu and J. An performed the animal experiments; Y. Song, X. Zhai,and R. Yang helped to do some constructs and cell experiments; J. Niu and C. Yang provided some advice; B. Li. Q. Chu and J. An analyzed the data; B. Li and Q. Chu wrote the paper.
References
- Aits, S., Kricker J., Liu B., Ellegaard A.M., Hämälistö S., Tvingsholm S., Corcelle-Termeau E., Høgh S., Farkas T., Holm Jonassen A., et al. 2015. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 11:1408–1424. 10.1080/15548627.2015.1063871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, M.S., and Goldstein J.L.. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232:34–47. 10.1126/science.3513311 [DOI] [PubMed] [Google Scholar]
- Chen, L., Duan Y., Wei H., Ning H., Bi C., Zhao Y., Qin Y., and Li Y.. 2019. Acetyl-CoA carboxylase (ACC) as a therapeutic target for metabolic syndrome and recent developments in ACC1/2 inhibitors. Expert Opin. Investig. Drugs. 28:917–930. 10.1080/13543784.2019.1657825 [DOI] [PubMed] [Google Scholar]
- Coburn, C.T., Knapp F.F. Jr, Febbraio M., Beets A.L., Silverstein R.L., and Abumrad N.A.. 2000. Defective uptake and utilization of long chain fatty acids in muscle and adipose tissues of CD36 knockout mice. J. Biol. Chem. 275:32523–32529. 10.1074/jbc.M003826200 [DOI] [PubMed] [Google Scholar]
- Commisso, C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., et al. 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 497:633–637. 10.1038/nature12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coort, S.L., Willems J., Coumans W.A., van der Vusse G.J., Bonen A., Glatz J.F., and Luiken J.J.. 2002. Sulfo-N-succinimidyl esters of long chain fatty acids specifically inhibit fatty acid translocase (FAT/CD36)-mediated cellular fatty acid uptake. Mol. Cell. Biochem. 239:213–219. 10.1023/A:1020539932353 [DOI] [PubMed] [Google Scholar]
- Cousin, S.P., Hügl S.R., Wrede C.E., Kajio H., Myers M.G. Jr, and Rhodes C.J.. 2001. Free fatty acid-induced inhibition of glucose and insulin-like growth factor I-induced deoxyribonucleic acid synthesis in the pancreatic beta-cell line INS-1. Endocrinology. 142:229–240. 10.1210/endo.142.1.7863 [DOI] [PubMed] [Google Scholar]
- Cui, Y., Wang Y., Liu M., Qiu L., Xing P., Wang X., Ying G., and Li B.. 2017. Determination of glucose deficiency-induced cell death by mitochondrial ATP generation-driven proton homeostasis. J. Mol. Cell Biol. 9:395–408. 10.1093/jmcb/mjx011 [DOI] [PubMed] [Google Scholar]
- Daquinag, A.C., Gao Z., Fussell C., Immaraj L., Pasqualini R., Arap W., Akimzhanov A.M., Febbraio M., and Kolonin M.G.. 2021. Fatty acid mobilization from adipose tissue is mediated by CD36 posttranslational modifications and intracellular trafficking. JCI Insight. 6:e147057. 10.1172/jci.insight.147057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daquinag, A.C., Zhang Y., Amaya-Manzanares F., Simmons P.J., and Kolonin M.G.. 2011. An isoform of decorin is a resistin receptor on the surface of adipose progenitor cells. Cell Stem Cell. 9:74–86. 10.1016/j.stem.2011.05.017 [DOI] [PubMed] [Google Scholar]
- Davidson, S.M., Jonas O., Keibler M.A., Hou H.W., Luengo A., Mayers J.R., Wyckoff J., Del Rosario A.M., Whitman M., Chin C.R., et al. 2017. Direct evidence for cancer-cell-autonomous extracellular protein catabolism in pancreatic tumors. Nat. Med. 23:235–241. 10.1038/nm.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, G.J., and McMahon H.T.. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857–902. 10.1146/annurev.biochem.78.081307.110540 [DOI] [PubMed] [Google Scholar]
- Dorn, C., Riener M.O., Kirovski G., Saugspier M., Steib K., Weiss T.S., Gäbele E., Kristiansen G., Hartmann A., and Hellerbrand C.. 2010. Expression of fatty acid synthase in nonalcoholic fatty liver disease. Int. J. Clin. Exp. Pathol. 3:505–514. [PMC free article] [PubMed] [Google Scholar]
- Dourlen, P., Sujkowski A., Wessells R., and Mollereau B.. 2015. Fatty acid transport proteins in disease: New insights from invertebrate models. Prog. Lipid Res. 60:30–40. 10.1016/j.plipres.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Furuhashi, M., and Hotamisligil G.S.. 2008. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 7:489–503. 10.1038/nrd2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz, J.F., Luiken J.J., and Bonen A.. 2010. Membrane fatty acid transporters as regulators of lipid metabolism: Implications for metabolic disease. Physiol. Rev. 90:367–417. 10.1152/physrev.00003.2009 [DOI] [PubMed] [Google Scholar]
- Hao, J.W., Wang J., Guo H., Zhao Y.Y., Sun H.H., Li Y.F., Lai X.Y., Zhao N., Wang X., Xie C., et al. 2020. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 11:4765. 10.1038/s41467-020-18565-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, G., Gupta S., Yi M., Michaely P., Hobbs H.H., and Cohen J.C.. 2002. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277:44044–44049. 10.1074/jbc.M208539200 [DOI] [PubMed] [Google Scholar]
- Hershman, D.L., Lacchetti C., Dworkin R.H., Lavoie Smith E.M., Bleeker J., Cavaletti G., Chauhan C., Gavin P., Lavino A., Lustberg M.B., et al. 2014. Prevention and management of chemotherapy-induced peripheral neuropathy in survivors of adult cancers: American society of clinical oncology clinical practice guideline. J. Clin. Oncol. 32:1941–1967. 10.1200/JCO.2013.54.0914 [DOI] [PubMed] [Google Scholar]
- Huang, H., Starodub O., McIntosh A., Kier A.B., and Schroeder F.. 2002. Liver fatty acid-binding protein targets fatty acids to the nucleus. Real time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 277:29139–29151. 10.1074/jbc.M202923200 [DOI] [PubMed] [Google Scholar]
- Ibrahim, A., Yucel N., Kim B., and Arany Z.. 2020. Local mitochondrial ATP production regulates endothelial fatty acid uptake and transport. Cell Metabol. 32:309–319.e7. 10.1016/j.cmet.2020.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson, A., Westerberg R., and Jacobsson A.. 2006. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 45:237–249. 10.1016/j.plipres.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Kamphorst, J.J., Cross J.R., Fan J., de Stanchina E., Mathew R., White E.P., Thompson C.B., and Rabinowitz J.D.. 2013. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. USA. 110:8882–8887. 10.1073/pnas.1307237110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst, J.J., Nofal M., Commisso C., Hackett S.R., Lu W., Grabocka E., Vander Heiden M.G., Miller G., Drebin J.A., Bar-Sagi D., et al. 2015. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75:544–553. 10.1158/0008-5472.CAN-14-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantzis, M., and Stahl A.. 2012. Fatty acid transport proteins, implications in physiology and disease. Biochim. Biophys. Acta. 1821:852–857. 10.1016/j.bbalip.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, A. 2016. Synthesis and degradation pathways, functions, and pathology of ceramides and epidermal acylceramides. Prog. Lipid Res. 63:50–69. 10.1016/j.plipres.2016.04.001 [DOI] [PubMed] [Google Scholar]
- Koivusalo, M., Welch C., Hayashi H., Scott C.C., Kim M., Alexander T., Touret N., Hahn K.M., and Grinstein S.. 2010. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188:547–563. 10.1083/jcb.200908086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuda, O., Pietka T.A., Demianova Z., Kudova E., Cvacka J., Kopecky J., and Abumrad N.A.. 2013. Sulfo-N-succinimidyl oleate (SSO) inhibits fatty acid uptake and signaling for intracellular calcium via binding CD36 lysine 164: SSO also inhibits oxidized low density lipoprotein uptake by macrophages. J. Biol. Chem. 288:15547–15555. 10.1074/jbc.M113.473298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M., Wang Y., Yang C., Ruan Y., Bai C., Chu Q., Cui Y., Chen C., Ying G., and Li B.. 2020. Inhibiting both proline biosynthesis and lipogenesis synergistically suppresses tumor growth. J. Exp. Med. 217:217. 10.1084/jem.20191226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luiken, J.J., Chanda D., Nabben M., Neumann D., and Glatz J.F.. 2016. Post-translational modifications of CD36 (SR-B2): Implications for regulation of myocellular fatty acid uptake. Biochim. Biophys. Acta. 1862:2253–2258. 10.1016/j.bbadis.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Macia, E., Ehrlich M., Massol R., Boucrot E., Brunner C., and Kirchhausen T.. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 10:839–850. 10.1016/j.devcel.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Mallick, R., Basak S., and Duttaroy A.K.. 2021. Fatty acids and evolving roles of their proteins in neurological, cardiovascular disorders and cancers. Prog. Lipid Res. 83:101116. 10.1016/j.plipres.2021.101116 [DOI] [PubMed] [Google Scholar]
- Masereel, B., Pochet L., and Laeckmann D.. 2003. An overview of inhibitors of Na(+)/H(+) exchanger. Eur. J. Med. Chem. 38:547–554. 10.1016/S0223-5234(03)00100-4 [DOI] [PubMed] [Google Scholar]
- Medes, G., Thomas A., and Weinhouse S.. 1953. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 13:27–29. [PubMed] [Google Scholar]
- Menendez, J.A., and Lupu R.. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7:763–777. 10.1038/nrc2222 [DOI] [PubMed] [Google Scholar]
- Merwar, G., Gibbons J.R., Hosseini S.A., and Saadabadi A.. 2022. Nortriptyline. StatPearls, Treasure Island, FL: [PubMed] [Google Scholar]
- Pietrocola, F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., and Kroemer G.. 2015. Acetyl coenzyme A: A central metabolite and second messenger. Cell Metabol. 21:805–821. 10.1016/j.cmet.2015.05.014 [DOI] [PubMed] [Google Scholar]
- Postic, C., and Girard J.. 2008. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: Lessons from genetically engineered mice. J. Clin. Invest. 118:829–838. 10.1172/JCI34275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig, F., and Schulze A.. 2016. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 16:732–749. 10.1038/nrc.2016.89 [DOI] [PubMed] [Google Scholar]
- Silverstein, R.L., and Febbraio M.. 2009. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2:re3. 10.1126/scisignal.272re3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand, S., and Vander Heiden M.G.. 2020. Transcriptional activation of macropinocytosis by the Hippo pathway following nutrient limitation. Genes Dev. 34:1253–1255. 10.1101/gad.343632.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaebjornsson, M.T., Janaki-Raman S., and Schulze A.. 2020. Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metabol. 31:62–76. 10.1016/j.cmet.2019.11.010 [DOI] [PubMed] [Google Scholar]
- Su, X., and Abumrad N.A.. 2009. Cellular fatty acid uptake: A pathway under construction. Trends Endocrinol. Metabol. 20:72–77. 10.1016/j.tem.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson, R.U., Parker S.J., Eichner L.J., Kolar M.J., Wallace M., Brun S.N., Lombardo P.S., Van Nostrand J.L., Hutchins A., Vera L., et al. 2016. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nat. Med. 22:1108–1119. 10.1038/nm.4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktuev, D.O., Merfeld-Clauss S., Li J., Kolonin M., Arap W., Pasqualini R., Johnstone B.H., and March K.L.. 2008. A population of multipotent CD34-positive adipose stromal cells share pericyte and mesenchymal surface markers, reside in a periendothelial location, and stabilize endothelial networks. Circ. Res. 102:77–85. 10.1161/CIRCRESAHA.107.159475 [DOI] [PubMed] [Google Scholar]
- Villamil Giraldo, A.M., Appelqvist H., Ederth T., and Öllinger K.. 2014. Lysosomotropic agents: Impact on lysosomal membrane permeabilization and cell death. Biochem. Soc. Trans. 42:1460–1464. 10.1042/BST20140145 [DOI] [PubMed] [Google Scholar]
- West, M.A., Bretscher M.S., and Watts C.. 1989. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J. Cell Biol. 109:2731–2739. 10.1083/jcb.109.6.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Hoppe A.D., Araki N., and Swanson J.A.. 2009. Sequential signaling in plasma-membrane domains during macropinosome formation in macrophages. J. Cell Sci. 122:3250–3261. 10.1242/jcs.053207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Di Martino J.S., Bowman R.L., Campbell N.R., Baksh S.C., Simon-Vermot T., Kim I.S., Haldeman P., Mondal C., Yong-Gonzales V., et al. 2018. Adipocyte-derived lipids mediate melanoma progression via FATP proteins. Cancer Discov. 8:1006–1025. 10.1158/2159-8290.CD-17-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M.S., Cui J.D., Lee D., Yuen V.W., Chiu D.K., Goh C.C., Cheu J.W., Tse A.P., Bao M.H., Wong B.P.Y., et al. 2022. Hypoxia-induced macropinocytosis represents a metabolic route for liver cancer. Nat. Commun. 13:954. 10.1038/s41467-022-28618-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Commisso C.. 2019. Macropinocytosis in cancer: A complex signaling network. Trends Cancer. 5:332–334. 10.1016/j.trecan.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., and Thompson C.B.. 2019. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 20:436–450. 10.1038/s41580-019-0123-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
contains original blots for Fig. 1.
contains original blots for Fig. 3.
contains original blots for Fig. S2.
contains original blots for Fig. S4.