Abstract
Mitochondrial dysfunction causes the production of reactive oxygen species (ROS) and oxidative damage, and oxidative stress and inflammation are considered key factors causing intervertebral disc degeneration (IVDD). Thus, restoring the mitochondrial dysfunction is an attractive strategy for treating IVDD. Platelet-derived extracellular vesicles (PEVs) are nanoparticles that target inflammation. Moreover, the vesicles produced by platelets (PLTs) have considerable anti-inflammatory effects. We investigate the use of PEVs as a therapeutic strategy for IVDD in this study. We extract PEVs and evaluate their properties; test their effects on H2O2-induced oxidative damage of nucleus pulposus (NP) cells; verify the role of PEVs in repairing H2O2-induced cellular mitochondrial dysfunction; and demonstrate the therapeutic effects of PEVs in a rat IVDD model. The results confirm that PEVs can restore impaired mitochondrial function, reduce oxidative stress, and restore cell metabolism by regulating the sirtuin 1 (SIRT1)-peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α)-mitochondrial transcription factor A (TFAM) pathway; in rat models, PEVs retard the progression of IVDD. Our results demonstrate that the injection of PEVs can be a promising strategy for treating patients with IVDD.
Keywords: Intervertebral disc degeneration, Nucleus pulposus, Platelets, Vesicles, Reactive oxygen species, Mitochondrial dysfunction
Graphical abstract
1. Introduction
Lower back pain (LBP) is one the most common musculoskeletal disorders in modern society [1]. Earlier epidemiological studies have confirmed that the prevalence of LBP increases with age, which can lead to significant family and social burden [1,2]. Intervertebral disc degeneration (IVDD) has been identified as a major cause of LBP. Disc degeneration is characterized by disc collapse, fracture of the fibrous annulus (AF), disc dehydration, which includes loss of proteoglycans and water in the nucleus pulposus (NP), calcification of the cartilage end plate, and changes in the extracellular matrix (ECM) [[3], [4], [5], [6]]. The current conservative treatment and surgical interventions for IVDD cannot halt or reverse the progression of disc degeneration, and they are more likely to exacerbate adjacent disc injury [[7], [8], [9], [10]]. Treatment strategies to prevent or delay the progression of degeneration at an early stage while protecting the physiological stability of the intervertebral disc (IVD) remain under investigation [11]. Although there is a wide range of pharmaceutical drugs available for treating IVDD, there are certain drawbacks, which include the absence of an abundant blood supply to the IVD [12], prolonged failure caused by extracellular expansion of the drug, failure to target delivery in a timely manner, and side effects such as the inflammation of the adjacent tissue after the local administration of the treatment [13]. In the field of basic research, several reports have been published on cellular exosome therapy for IVDD; this therapy has relatively good biocompatibility and low immunogenicity [[14], [15], [16]]. However, the active protein component in exosome therapy cannot be clarified, and it remains a great challenge to accurately, effectively, and selectively identify and isolate the exosomes [17].
Platelet-rich plasma (PRP) is an autologous cell-derived therapy [18]. At present, PRP and pure PRP without leukocytes are being used in the clinical treatment of various diseases [19,20]. Despite the large variety of applications, the efficacy of regenerative treatments using PRP remains controversial owing to the absence of large controlled clinical trials [21]. PRP comprises a complex list of components, including pro-inflammatory factors, anti-inflammatory factors, chemokines, and transforming growth factors. During the study on the specific molecular efficacy of PRP, the exact effective proteins of PRP involved in clinical treatment are unclear [22]. Moreover, the underlying mechanisms, principles, and safety of PRP therapy for clinical use remain to be elucidated [18]. An autologous product that does not specify its formulation and composition [23,24] has variations in its composition in terms of platelets (PLTs), white blood cells (WBC), and red blood cells (RBC) [25,26]. Consequently, the consensus on PRP bio-formulations is nonexistent at present. Sufficiently powered and well-documented clinical research is needed to determine the full potential and therapeutic molecular efficacy of PRP in a variety of indications [18].
Platelets (PLTs) are anucleate blood cells produced by the cytoplasmic lysis of mature megakaryocytes in the bone marrow [27]. These cells are increasingly recognized as pivotal participants in numerous pathophysiological processes, including thrombosis, inflammation, atherogenesis, and tumor growth, and metastasis [28,29]. In addition, related research has reported that PLTs can enhance the wound-healing activity of mesenchymal stem cells (MSCs) and provide them with energy for metabolism by functional mitochondria [30,31]. These studies have demonstrated that PLTs can be used as an adjuvant for improving therapeutic efficacy. However, PLTs are easily affected by various conditions (such as temperature, shake, and pollution) during storage and transportation, resulting in an inflammatory response [32]. In these states, PLTs can be positively primed to reach an activated condition [33]. Thrombin-activated PLTs release α-granules and dense granules located in the PLT cytoplasm. The α-granules contain pro-inflammatory cytokines, such as IL-1β, IL-6, and TNF-α, which can aggravate the local inflammatory response [[34], [35], [36]] and further affect the therapeutic effect of PLTs [[37], [38], [39]]. Thus, further studies are required to prevent the pro-inflammatory effects of PLTs and better utilize their therapeutic efficacy.
Nanomaterial therapy is a current trend in nanomedicine [40]. PLT-derived extracellular vesicles (PEVs), also known as PLT dust and PLT-derived particles, are a heterogeneous group of small, lipid-bound nanoparticles that play a key role in mediating pathological and physiological processes [[41], [42], [43], [44], [45]]. In recent years, PEVs have been identified as unique therapeutic modalities that demonstrate advantages in hemostasis, inflammation, and tissue repair, as well as drug delivery carriers [45]. A study has reported that PEVs secreted lower levels of inflammatory cytokines (IL-1β, IL-6, and TNF-α) compared to activated PLTs. The low level of cytokine releases from PEVs may be due to the fact that α-granules do not exist in PEVs [39,46]. Notably, PEVs can penetrate tissue barriers, such as the synovium, lymph, and bone marrow, to achieve specific therapeutic effects [37]. Moreover, the majority of PEVs are stable structures with diameters in the range of 100–200 nm; such structures retain some effective mitochondrial proteins and are more easily ingested by cells [[47], [48], [49]]. Thus, PEVs exhibit non-negligible therapeutic potential for injury, inflammation, and tissue repair [[50], [51], [52]]. However, mesenchymal stem cell (MSC)-derived EVs are currently the main research direction in nanomedicine [53,54], and limited attention has been focused on the study of PEVs in the treatment of degenerative diseases. We believe that this nanomaterial therapy may provide more potential therapeutic prospects in the future.
Oxidative stress in NP cells is a cause of irreversible IVDD [55]. Mitochondria are the target organelles of intracellular reactive oxygen species (ROS), and mitochondrial dysfunction leads to intracellular metabolic imbalance [55,56]. As the powerhouse of the cell [57], mitochondria can provide energy for a range of activities in the form of adenosine triphosphate (ATP) molecules [58,59]. The impairment of mitochondrial function is strongly associated with degenerative diseases [[60], [61], [62]]. Inflammatory degeneration is exacerbated because of an insufficient ATP supply when mitochondria are impaired [63,64]. Fen et al. demonstrated that the mitochondrial structure and function were damaged in the pathological process of IVDD [65,66]. Inflammation or mechanical injury alters the mitochondrial membrane potential, which further reduces cell proliferation and induces apoptosis; this ultimately leads to IVDD [67]. Therefore, repairing damaged mitochondria by inhibiting oxidative stress is considered a valid option for delaying IVDD.
AMP-activated protein kinase (AMPK) is a highly conserved sensor that detects elevated levels of AMP and ADP caused by mitochondrial ATP depletion [68]. It is an important molecular target for the treatment of degenerative diseases. During inflammation, AMPK activation protects mitochondrial function by promoting the expression of sirtuin 1 (SIRT1) [69,70]. In addition, peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α) is a regulatory mechanism required for AMPK in NP cells [71]. Increased PGC1α expression reverses mitochondrial biogenesis, oxidative phosphorylation, and intracellular ATP levels in degenerative cells [72]. Mitochondrial transcription factor A (TFAM) is a gene regulator that mitigate ROS production by wrapping around mitochondrial DNA complexes [73]; nuclear respiratory factors 1 and 2 (Nrf1 and Nrf2, respectively) are also essential for the mitochondrial biological function [25,54].
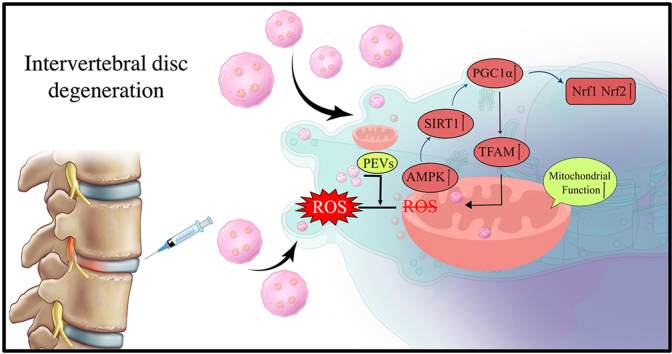
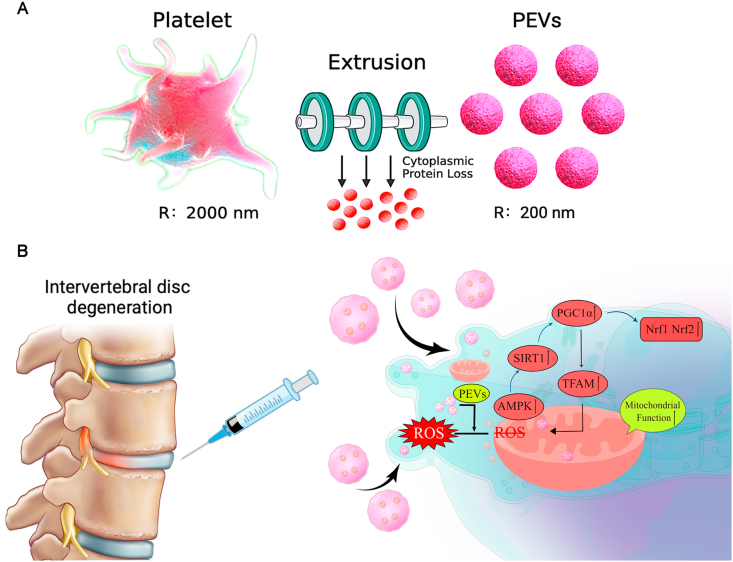
In this study, we isolated and characterized platelet-derived vesicles and investigated the role of PEVs in H2O2-induced oxidative stress and mitochondrial dysfunction in NP cells. Further, we used a rat IVDD model to verify the therapeutic effect of PEVs on IVDD (Fig. 1). We hypothesize that PEVs can restore mitochondrial function in NP cells through the SIRT1–PGC1α–TFAM pathway and prevent the progression of degenerative changes in IVDD.
Fig. 1.
(A) Illustration and technical theory of preparation of PEVs in vitro. (B) A schematic diagram of the IVDD retarding pathway by the local injection of PEVs reduce oxidative stress and activate the mitochondrial functional protein pathway.
2. Materials and methods
2.1. Materials
Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Gibco BRL (Gaithersburg, MD, USA). Aggrecan (ab3778) and Adamts5 (ab41037) were purchased from Abcam (Cambridge, USA). TSG101 (28283-1-AP), CD9 (20597-1-AP), CD63 (25682-1-AP), CD41 (24552-1-AP), Col2α (28459-1-AP), SOX9 (67439-1-Ig), MMP3 (18165-1-AP), MMP13 (17873-1-AP), SIRT1 (13161-1-AP), PGC1α (66369-1-Ig), TFAM (22586-1-AP), Nrf1 (12482-1-AP), Nrf2 (16396-1-AP), βactin (20536-1-AP), and GAPDH (10494-1-AP) were purchased from Proteintech® (Beijing, China).
2.2. Preparation and characterization of isolated PLTs and PEVs
PRP was prepared using a previously described method with minor modifications [74,75]. All blood specimens used in the present study were approved by the Ethical Committee of the Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University. Informed consent was obtained. In brief, whole blood samples were collected from healthy donors and mixed with acid citrate dextrose solution A (ACD-A) anticoagulant (1 mL ACD-A/9 mL blood). After centrifugation at 100×g for 15 min, the PLT-containing plasma was collected and centrifuged at 800×g for 20 min. The supernatant plasma was discarded, and the PLT pellet was resuspended in residual plasma to obtain PRP (SI Fig. 1). To prepare PEVs, PLTs were mixed in phosphate buffered saline (PBS) containing ethylenediamine tetraacetic acid (EDTA, 5 mM, Sigma Aldrich) and prostaglandin e1 (PGE1, 1 mM, MCE). Next, PLTs were passed through 1000 nm and 500 nm filters, and then thrice through a 200 nm filter to obtain PEVs. Finally, the supernatant was collected for ultracentrifugation, and PEVs were enriched at 100,000 rpm for 2 h. We observed the morphology and size of PLTs and PEVs using cryo-transmission electron microscopy (cryo-TEM, 200 kV, FEI Tecnai G2 F20, USA), and we quantified their particle size and zeta potential using dynamic light scattering (DLS). We tested the stability of the PLTs and PEVs by measuring the particle size changes at 0, 1, 3, 5, and 7 days using DLS. In addition, we extracted PLTs and PEVs proteins using a protein isolation kit (Invitrogen, Carlsbad, USA), and we quantified the protein concentration using a BCA kit (Beyotime, China). The protein expression levels of PLTs and PEVs were detected using SDS-PAGE and western blotting, respectively. Coomassie brilliant blue staining (Beyotime, China) was used to detect differences in protein expression between the PLTs and PEVs.
2.3. Evaluation of materials for cytotoxicity and proliferation
The toxicity and proliferation of NP cells incubated with PEVs were tested for 1, 3, 5, and 7 days using a CCK-8 detection kit (MCE, USA). The toxicity of the different concentrations of each inhibitor (chlorpromazine, filipin III, wortmannin, and cytochalasin D) in NP cells by CCK-8 kit assay. We added a 10 μL CCK-8 solution to the serum-free medium of the cells, and then incubated them at 37 °C for 2 h. The absorbance at 450 nm was measured using a Varioskan LUX microplate reader (Thermo Fisher Scientific, USA). All experiments were performed in triplicate.
2.4. Cytokine detection
Production was provided by the manufacturer; 100 μg/mL PLTs and 100 μg/mL PEVs were activated with or without thrombin, respectively. The levels of IL-1β and IL-6 were detected using ELISA assay kits (BMS224-2, BMS213-2, Thermo Fisher Scientific). All experiments were performed in triplicate.
2.5. Cell culture and handling
The NP cells were obtained from Sprague–Dawley rats purchased from Cyagen Biosciences, Inc. (Shanghai, China). First, the NP tissue was macroscopically isolated from the caudal disc of 4 weeks male SD rats. Then, the tissue was digested with collagenase II for 4 h at 37 °C. The cell mixture filtered through the cell sieve was centrifuged at 1000 rpm for 5 min to obtain primary NP cells. The NP cells were cultured in DMEM supplemented with 10% FBS in a humidified incubator with 5% CO2. The second passage of the cells was used for subsequent experiments. All cells were seeded in six-well plates and 10 cm plate. Then, the cells were divided into four groups: control group (normal control group), 200 μM H2O2 treatment for 12 h (H2O2 group), H2O2 + 100 μg/mL platelets (PLTs group), and H2O2 + 100 μg/mL PEVs (PEVs group).
2.6. ROS detection and evaluation
The ROS levels in the NP cells were detected using an ROS detection kit (Beyotime, China). The NP cells were pre-treated with 200 μM H2O2 for 6 h, and then each group was added with 100 μg/mL of PLTs or PEVs for 24 h. Then, the NP cells were incubated with 20 mM 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, Beyotime, China) for 37 °C, 20 min. Before flow cytometry analysis, the cells were washed three times with a serum-free medium. Fluorescence signal intensity was determined by the oxidative conversion of DCFH-DA. Finally, changes in intracellular ROS levels were detected by fluorescence microscopy and flow cytometry. We analyzed the results using FlowJo software. All experiments were performed with four replicates each.
2.7. Flow cytometry and TUNEL staining for cell apoptosis
The apoptosis rate of NP cells was detected using a fluorescein isothiocyanate–annexin V and propidium iodide (FITC/PI) Apoptosis Detection Kit (Yeasen, China) according to the manufacturer's instructions. NP cells were pre-treated with 100 μg/mL of PLTs and 100 μg/mL of PEVs for 24 h, respectively. Then, 400 μM H2O2 were added for 4 h to induce apoptosis in NP cells. NP cells were centrifuged and then resuspended in 300 mL of PBS supplemented with annexin V–Alexa FITC (5 μL) and PI (10 μL). Following incubation (10–15 min, 20 °C) in the dark, cell apoptosis was detected using a flow cytometer after adding 400 μL 1 × binding buffer. The total apoptosis rate is the sum of the early stage of apoptosis (Annexin V–positive/PI-negative) and late stage of apoptosis (Annexin V–positive and PI-positive). For apoptosis phenotype, NP cells were labeled with the terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) Apoptosis Detection Kit (Yeasen, China). The changes in apoptosis levels in the NP cells were detected by fluorescence microscopy, and the TUNEL-positive cells were counted. We analyzed the results using FlowJo software. All experiments were performed with four replicates each.
2.8. Senescence-associated β-gal (SA-β-gal) staining
The senescence phenotype of NP cells was analyzed using SA-β-Gal staining kit (Beyotime, China) and a microscope. Specifically, NP cells were pre-treated with 100 μM H2O2 for 48 h to induce senescence in NP cells. Subsequently, 100 μg/mL of PLTs and 100 μg/mL of PEVs were added for 24 h, respectively. NP cells were fixed with 0.2% glutaraldehyde for 15 min at room temperature. Following 3 washes with PBS, NP cells were stained with X-gal staining solution overnight. Blue-stained cells showing high SA-β-gal activity were defined as senescent cells. Representative images were captured under a microscope, and the SA-β-gal-positive cells were counted. All experiments were performed with four replicates each.
2.9. PEVs uptake and mitochondrial localization
The mitochondria of NP cells were labeled with a MitoTracker Green fluorescent probe (Beyotime, China) and cultured in DMEM for 24 h. For uptake studies, the 1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) fluorescent probe (Beyotime, China) was incubated with PEVs for 12 h and washed three times with PBS to remove unlabeled particles. DiI-loaded PEVs were incubated with NP cells for 24 h and 4,6-diamidino-2-phenylindole (DAPI) with the NP cell nuclei for 10 min. Then, the cells were fixed with 4% paraformaldehyde. Finally, the localization of DiI and MitoTrackcr × Green fluorescent probe was observed using fluorescence microscopy.
2.10. Quantitative real-time polymerase chain reaction (qPCR) analysis
Total RNA was extracted from lysed NP cells using TRIzol reagent (Invitrogen) and an RNA purification kit (CW0581, Kangwei) according to the manufacturer's instructions. The cDNA was reverse transcribed using the MMLV reverse transcription reagent (Takara). βactin was used as an internal reference. Real-time PCR was performed using a SYBR Master Mix qPCR kit (Takara). The primers used are listed in Table 1. All experiments were performed in triplicate.
Table 1.
Summary of PCR primers used.
| Gene Name | Used for | Primer sequence | Product length | Amplicon Size |
| Col2α | mRNA quantification | F: CAGGATGCCCGAAAATTAGGG | 21 | 132 |
| R: ACCACGATCACCTCTGGGT | 19 | |||
| Sox9 | mRNA quantification | F: GAGCCGGATCTGAAGAGGGA | 20 | 151 |
| R: GCTTGACGTGTGGCTTGTTC | 20 | |||
| Acan | mRNA quantification | F: CCTGCTACTTCATCGACCCC | 20 | 150 |
| R: AGATGCTGTTGACTCGAACCT | 21 | |||
| Mmp13 | mRNA quantification | F: CTTCTTCTTGTTGAGCTGGACTC | 23 | 173 |
| R: CTGTGGAGGTCACTGTAGACT | 21 | |||
| Mmp3 | mRNA quantification | F: ACATGGAGACTTTGTCCCTTTTG | 23 | 192 |
| R: TTGGCTGAGTGGTAGAGTCCC | 21 | |||
| Adamts5 | mRNA quantification | F: GGAGCGAGGCCATTTACAAC | 20 | 110 |
| R: CGTAGACAAGGTAGCCCACTTT | 22 | |||
| βactin | mRNA quantification | F: CCTCTATGCCAACACAGT | 18 | 230 |
| R: AGCCACCAATCCACACAG | 19 | |||
| P16(Cdkn2a) | mRNA quantification | F: GTGTGCATGACGTGCGGG | 18 | 127 |
| R: GCAGTTCGAATCTGCACCGTAG | 22 | |||
| P21(Cdkn1a) | mRNA quantification | F: CCAGGCCAAGATGGTGTCTT | 20 | 103 |
| R: TGAGAAAGGATCAGCCATTGC | 21 | |||
2.11. Western blot analysis
Proteins were extracted from the NP cells using a RIPA buffer containing a 1% protease inhibitor (PMSF) (Boster, China). The protein concentration in each group of cells was quantified using a BCA protein concentration kit (Beyotime, China). Proteins were separated using 10% SDS-PAGE and transferred from the gel to a polyvinylidene fluoroether membrane (Millipore, Shanghai, China). The fluoroether membrane was blocked with 5% non-fat milk for 1 h. After blocking, the membrane was washed with Tris-HCl-buffered saline (TBS) containing 0.1% Tween-20 three times for 5 min each. Subsequently, the membranes were incubated with Col2α, SOX9, Aggrecan, MMP3, MMP13, Adamts5, SIRT1, PGC1α, TFAM, Nrf1, Nrf2, and βactin at 4 °C for 12 h; the internal control was βactin. The membrane was incubated with a specific horseradish peroxidase-conjugated secondary antibody (Beyotime, China) for 1 h at 18–25 °C after washing with TBST three times for 5 min each. Excess secondary antibody was washed away with TBST three times for 5 min each. Finally, the signal intensity of the reactive bands on the membrane was visualized using the ChemiDoc Touch imaging system (Bio-Rad, USA). All above experiments were performed in triplicate.
2.12. Immunofluorescence staining
Each group of the NP cells was fixed on a control dish with 4% paraformaldehyde for 20 min, washed three times with PBS for 3 times, and the cells were permeabilized with 0.5% Triton X-100 (diluted in PBS) for 20 min. The NP cells were blocked with 5% bovine serum albumin (BSA) for 30 min Adamts5 (1:100, Proteintech, China), MMP13 (Proteintech, China), Col2α (1:100; Proteintech, China), Aggrecan (1:100, Sigma, USA), and PGC1α (1:100, Proteintech, China) were incubated with NP cells at 4 °C for 12 h. Then, the cells were incubated with a fluorescent secondary antibody (goat anti-rabbit Alexa Fluor 488 (1:500, Beyotime, China)) at room temperature for 1 h, which is followed by DAPI for 10 min. Images were obtained using a fluorescence confocal microscope (Nikon, Tokyo, Japan). All above experiments were performed with four replicates each.
2.13. Assessment of mitochondrial function
According to the manufacturer's instructions, the MitoTracker Green (Beyotime, C1048, China), MitoSOX-Red (Beyotime, C1045, China), and JC-1 fluorescent probe (Beyotime, C2006, China) were used to detect mitochondrial membrane potential changes in NP cells. The morphology of mitochondria of NP cells was observed by transmission electron microscopy (TEM; H-9500; Hitachi, Tokyo, Japan). The decrease in mitochondrial membrane potential is a hallmark event in the early stages of apoptosis [76,77]. JC-1 (5,5′,6,6′-tetrachloro1,1′,3,3′-tetramethylbenzimidazolylcarbocyanine iodide) is an ideal fluorescent probe widely used to detect mitochondrial membrane potential (△Ψm). JC-1 monomers can produce green fluorescence (△Ψm↓) while JC-1 aggregates can produce red fluorescence (△Ψm↑) [78,79]. We measured the proportion of mitochondrial depolarization by comparing the relative proportions of red and green fluorescence. Therefore, the transition of JC-1 from red fluorescence to green fluorescence can detect the decrease of cell membrane potential and serve as an early detection indicator of apoptosis. Then we used the selective SIRT1 activator (SRT-1720, Beyotime, SC0267) and the selective SIRT1 inhibitor (EX-527, MCE, HY-15452) to verify the mechanism of PEVs repairing impaired mitochondria in pathological NP cells. All experiments were performed with four replicates each.
2.14. ATP detection evaluation
The ATP levels of NP cells were assessed using the luminescent ATP detection assay kit (Abcam, Ab113849). All above experiments were performed with four replicates each.
2.15. Lactic acid detection
According to the manufacturer's instructions, the lactic acid assay kit (Nanjing Jiancheng, A019-2-1) was used to measure the lactic acid concentration in the cell culture serum to reflect the cellular metabolic state. All experiments were performed with four replicates each.
2.16. Manufacture of rat IVDD model
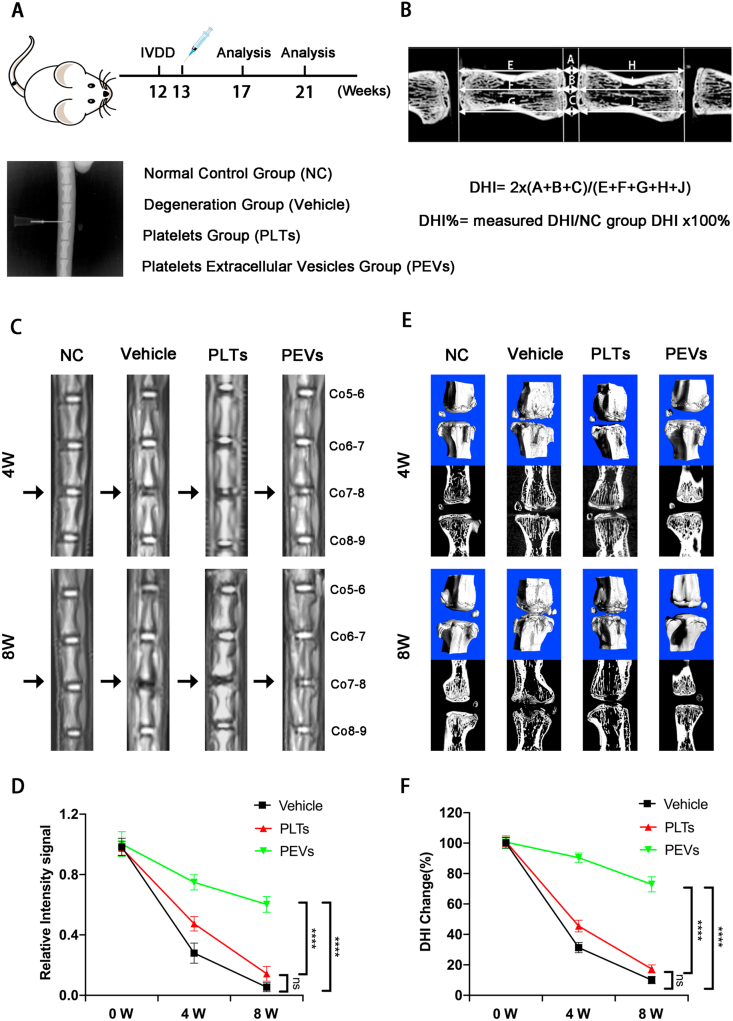
In this study, an IVDD rat model was induced via fine needle puncture. Male Sprague–Dawley rats (200–250 g, 12 weeks old) were randomly allocated into 4 groups (n = 9): NC group, Puncture + vehicle group, Puncture + PLTs group, and Puncture + PEVs group. All rats were treated according to the standard guidelines approved by the Institutional Animal Care and Use Committee of Zhejiang Center of Laboratory Animals. In detail, the rats were anesthetized via the intravenous injection of pentobarbital sodium (40 mg/kg) and then positioned prone on a platform. The IVDD rat models were established using a 26G needle (diameter = 0.45 mm) puncture to coccyx spaces Co7–8. The selected coccygeal intervertebral plane was confirmed using manual palpation and X-ray fluoroscopy. After disinfection with iodate alcohol, a 26G needle was inserted at the level of the annulus fibrosus by palpation, passing through the NP to the contralateral annulus fibrosus. After incomplete penetration, the needle was rotated by 360° twice for 30 s. The depth of needle penetration was controlled by the resistance of contralateral ring fibers. One week after the initial puncture, injured discs injected with 5 μL vehicle (PBS), 5 μL PLTs (100 μg/mL), or 5 μL PEVs (100 μg/mL) were set as the vehicle, PLTs, or PEVs group, respectively. Normal IVD Co7–8 was the NC group. Each group of segment treatments was labeled. No deaths or diseases were recorded during the experiment process. After 4 or 8 weeks, the discs were harvested and investigated. All experiments were performed under sterile conditions; the rats were transferred to a warm and ventilated environment after surgery until recovery from anesthesia. Then, the rats were given free access to water and food. The selected coccygeal intervertebral plane was confirmed by digital palpation and fluoroscopy at 4 and 8 weeks after the first surgery.
2.17. Radiological assessment analysis
The micro-computed tomography (micro-CT, Skyscan1275) and magnetic resonance imaging (MRI) were performed at 4- and 8-weeks post-operation with a GE Sigma CV/I (1.5 T) to evaluate changes in the NP signal and height of the intervertebral space. Image J software was used to measure the signal intensity of the IVD at Co7-8 on T2-weighted central images. According to the modified Thomson classification, the MRI images are classified into grades I to IV (I, normal; II, signal intensity is slightly decreased but the high signal area is significantly narrowed; III, signal intensity is moderately decreased; and IV, signal intensity is severely decreased); they were evaluated by three experienced radiologists and three orthopedic surgeons. Further, the IVD height was measured by micro-CT analysis and calculated as the disc height index (DHI) that represents the average height of the anterior, middle, and posterior discs. The change in the DHI was calculated using the value at 0 week as a reference. The CT images were measured using Data-viewer software, and DHI was calculated using the following equation: disc height index (DHI) = 2 × (A + B + C)/(D + E + F + G + H + I); DHI% = measured DHI/NC group DHI × 100% for each group).
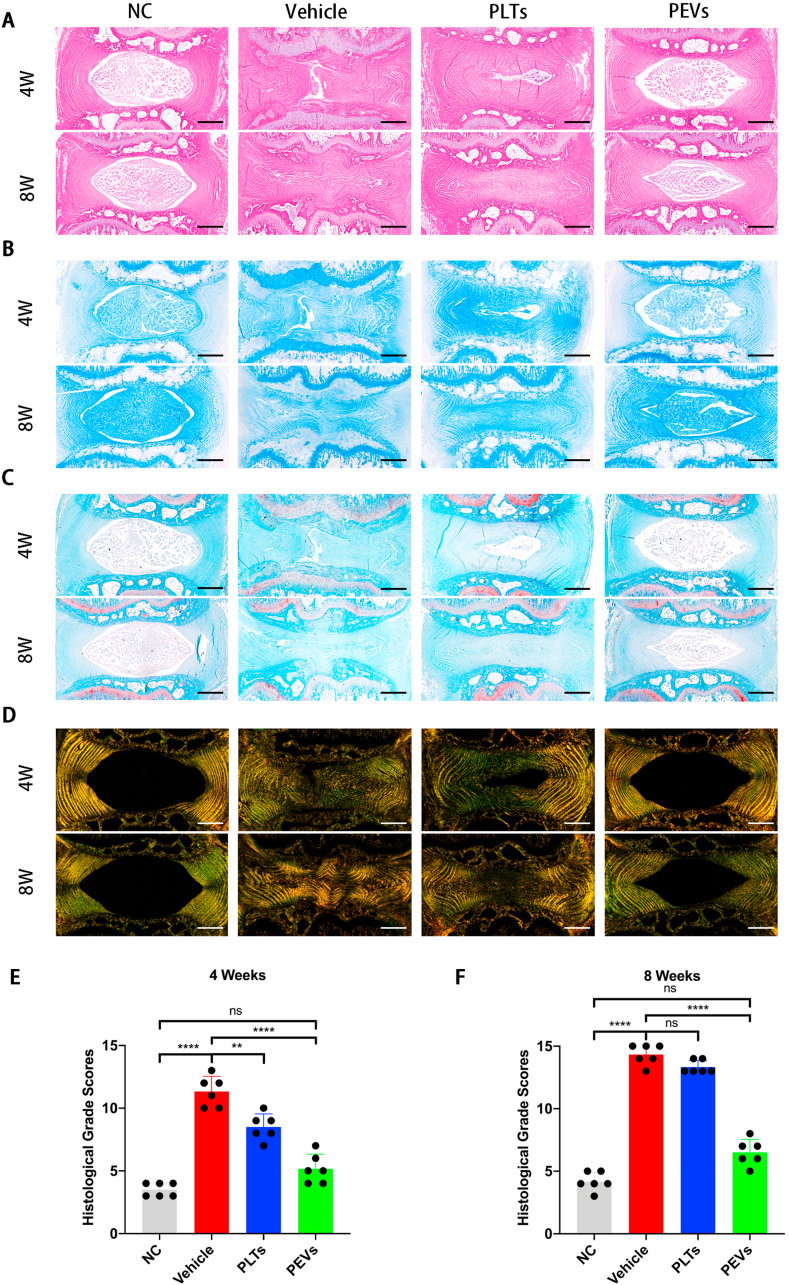
2.18. Histological analysis and immunofluorescence staining
The Digital Pathology Slide Scanner (KFBIO) were performed at rats intervertebral disc (IVD). The IVD tissues were stripped from the IVD segments of the rats. The IVDs were fixed in 4% buffered paraformaldehyde and decalcified in a decalcification solution (14% EDTA) at 18–25 °C for 4 weeks. The decalcified discs were paraffin-embedded to obtain sections; the sections were then stained with hematoxylin and eosin (H&E), Alcian blue (A&B), Safranin O-fast Green (S&O), and Sirius Red (S&R). The specimens were examined and photographed using a digital microscope (KFBIO, KF-PRO-120). According to the definition of histological grade (Table 2), the histological images of each group are classified into 0 to 15 scores. For immunofluorescence staining, discs were blocked by incubation with 5% BSA (Fdbio science); incubated with primary antibodies against Col2α, MMP13, and PGC1α; and this was followed by incubation with the corresponding secondary antibodies conjugated with Alexa Fluor 488 fluorescent dye (Proteintech) and stained with DAPI (Beyotime). Discs were blocked by incubation with 5% BSA (FDbio Science), followed by incubation with the DHE fluorescence and MitiSOX Red fluorescence probes and staining with DAPI (Beyotime). The morphology of mitochondria of IVD was observed by transmission electron microscopy (TEM; H-9500; Hitachi, Tokyo, Japan). All experiments were performed with three replicates each.
Table 2.
Definition of histological grade.
| Category | Grade |
| I. Matrix of the nucleus pulposus |
|
| |
| |
| II. Cellularity of the nucleus pulposus |
|
| |
| |
| III. Structure of anulus fibrosus |
|
| |
| |
| IV. Border between the nucleus pulposus and anulus fibrosus |
|
| |
| |
2.19. Systemic toxicity experiments in vivo
After rats were sacrificed, the main organs (heart, liver, spleen, lung, and kidney) were collected for H&E staining to evaluate systematic pathological changes.
2.20. Statistical analysis
All study results are presented as the mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism (v5.0) to perform one-way or two-way analysis of variance (ANOVA). Duplicate groups were included in all experiments. The differences between the groups were further assessed using Tukey's multiple comparison test. The difference in the results was significant when the p-value was <0.05. The significance of these numbers is indicated by asterisks: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005, and ∗∗∗∗p < 0.001. All signal intensity and fluorescence expression levels were quantified using ImageJ software.
3. Results
3.1. PEVs preparation and characterization
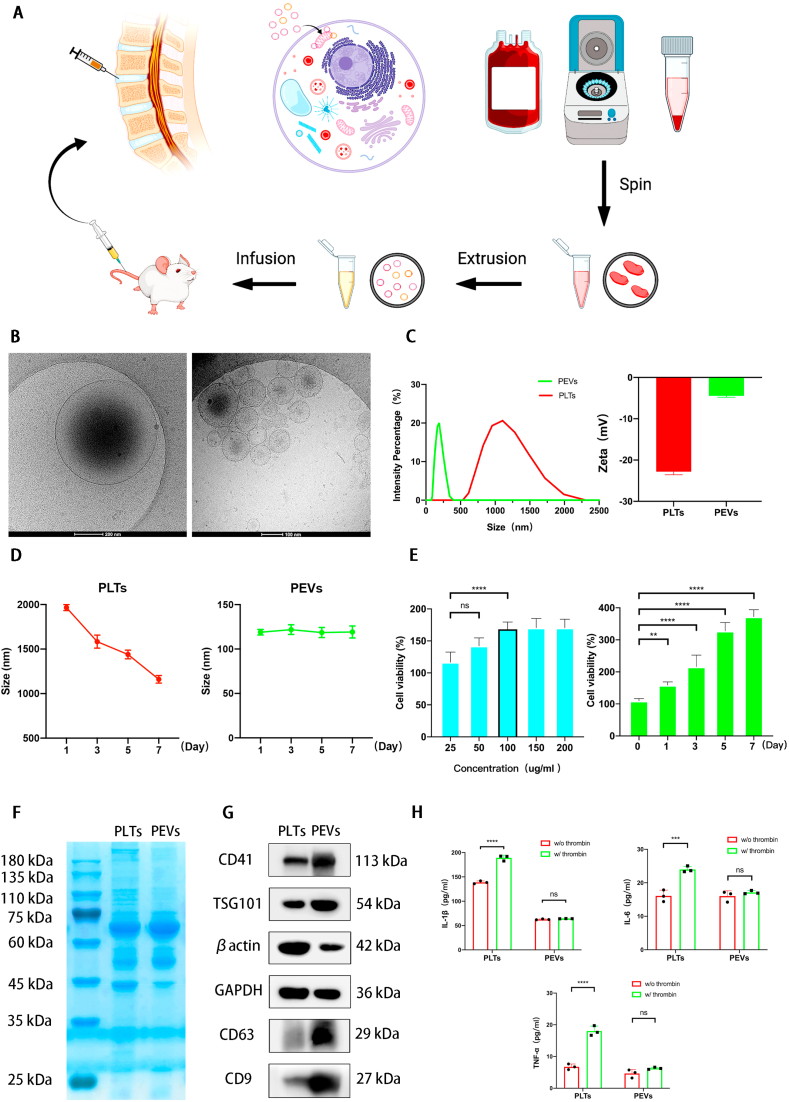
PEVs were obtained from PLTs through extrusion method (extrusion through 1000, 500, and 200 nm filters) (Fig. 1A) and were injected into the intervertebral disc of rats (Fig. 1B). A previous study reported that cells can release a large number of cell-derived extracellular vesicles (CEVs) in a short period of time through mechanical extrusion [[80], [81], [82], [83]]. The technical theory of the method is that CEVs are formed as a result of the disruption of the cell membrane using shear or frictional forces and the reorganization of lipid bilayer-forming vesicles in seconds [80,84]. Meanwhile, vesicles can randomly encapsulate surrounding cellular components or drugs during the extrusion process; thus, part of the unencapsulated protein or components would be lost [85]. Then we drew the schematic diagram of the preparation and injection of PEVs (Fig. 2A). In this study, we compared the properties of PLTs and PEVs using cryo-TEM. The PEVs have a smaller size of ∼200 nm compared with PLTs with a size of ∼1 μm (Fig. 2B). Then, we measure PEVs and PLTs using DLS, and the results show that PLTs have a particle size of 1.1 μm and a zeta potential of −22.8 mV, whereas the PEVs have a particle size of 195.6 nm and a zeta potential of −4.3 mV (Fig. 2C). Subsequently, we place PEVs and PLTs at 37 °C for 1, 3, 5, and 7 days and find that the size of PEVs remained stable during this period using DLS; the size of the PLTs gradually decrease (Fig. 2D). We then examine the proliferative effect of PEVs on NP cells using the CCK-8 kit. The results of CCK-8 depicted that the most suitable concentration of PEVs to promote NP cell proliferation is 100 μg/mL (Fig. 2E). We selected 100 μg/mL PEVs and 100 μg/mL PLTs in the subsequent cellular experiment. The cell proliferation assay elucidated that NP cells had a good proliferative capability after PEVs treatment. In addition, no apparent biological toxicity was examined in all groups at rats after administration of PEVs and PLTs after 4 and 8 weeks (SI Fig. 2), which was consistent with the outcome of CCK-8 assay.
Fig. 2.
Preparation and characterization of the platelet-derived extracellular vesicles. (A) Schematic diagram of the preparation and injection of PEVs. (B) Cryo-TEM images of PLTs and PEVs. Scale bar = 200 nm, 100 nm. (C) The size distribution analysis and surface zeta potential of PLTs and PEVs. (D) Stability of PLTs and PEVs in vitro. (E) The toxicity of different concentrations (0–200 μg/mL) and durations (0–7 days) of 100 μg/mL PEVs in NP cells by CCK-8 kit assay. (F) SDS-PAGE of PLTs and PEVs with Coomassie brilliant blue staining. (G) Western blot analysis of CD41, TSG101, βactin, GAPDH, CD63, and CD9 from the PLT and PEV lysates. Quantification of CD41 protein is in SI Fig. 3. Uncropped gel is in SI Fig. 4A. (H) The amount of IL-1β, IL-6, and TNFα cytokines from PLTs and PEVs in the absence and presence of thrombin. The significance between every two groups was calculated using a two-tailed Student's t-test (H) or one-way analysis of variance (ANOVA) with Tukey's post-hoc test (D–E). Data are presented as mean ± SD (n = 3). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
Furthermore, we determined the protein composition of PLTs and PEVs to study the reasons for this result, and we found that their compositions are similar in the Coomassie Brilliant Blue staining (Fig. 2F). The membrane marker proteins (CD9, CD41, and CD63) were significantly enriched in PEVs, and cytoplasmic proteins of PLTs (GAPDH and βactin) were lower in PEVs (Fig. 2G).
Then we compared the abilities of PLTs and PEVs to produce inflammatory cytokines following thrombin activation. We observed a significant increase in IL-1β, IL-6, and TNFα production in platelets after thrombin addition. In contrast to PLTs, PEVs exhibit no significant changes in inflammatory factor levels before and after thrombin addition (Fig. 2H). The results indicate that activated platelets can accelerate inflammation through the release of inflammatory factors (IL-1β, IL-6, and TNFα); by contrast, PEVs do not release these pro-inflammatory factors to aggravate inflammation, thus making them a more promising option for targeting inflammatory diseases. The reason why PEVs do not cause inflammation may be that their cytoplasmic proteins are reduced and the pathways for releasing inflammatory factors are disturbed. Referring to a previous study, the low level of cytokine releases from PEVs may be attributed to the fact that α-granules (particle size ranging between 200 and 300 nm) do not exist in PEVs [39,86].
3.2. PEVs alleviate oxidative stress in NP cells
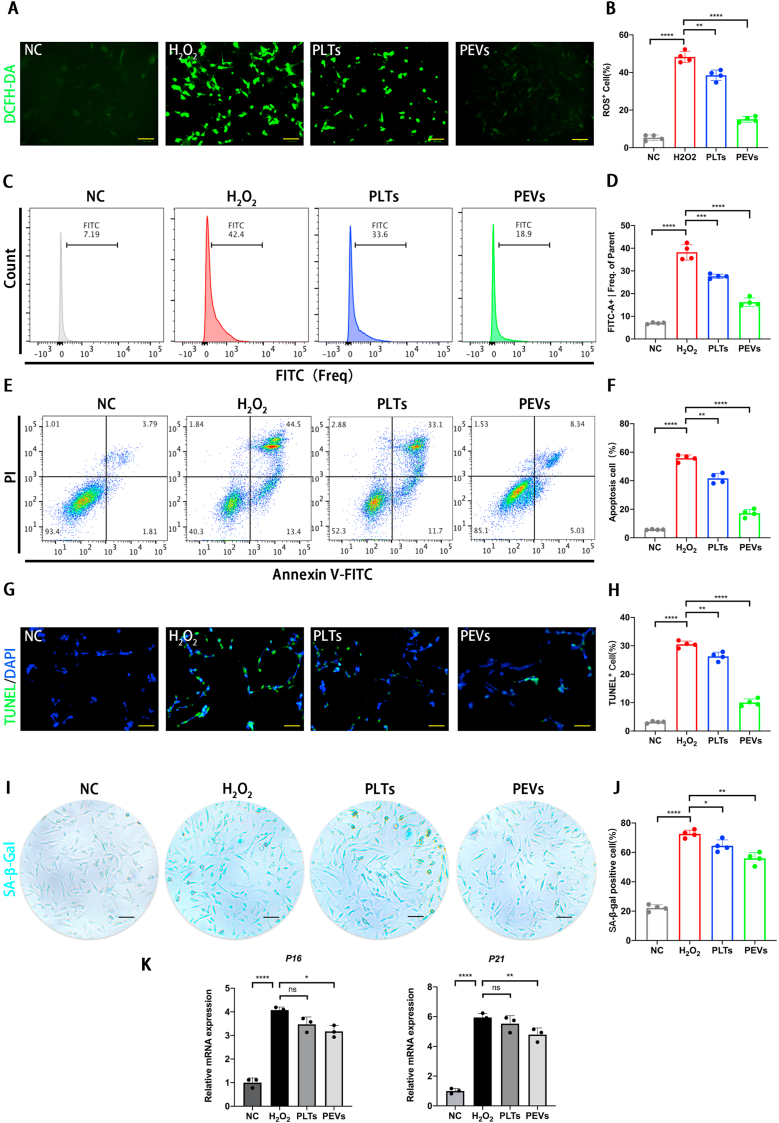
We used DCFH-DA as a probe to assess ROS levels in untreated NP cells (control group), H2O2 pretreated NP cells (H2O2 group), 100 μg/mL platelet-treated NP cells (PLTs group), and 100 μg/mL platelet vesicle-treated NP cells (PEVs group). Fig. 3A and B depicted that there is a low ROS level in the control group, whereas a high ROS level elucidated that strong green fluorescence can be clearly observed after H2O2 stimulation. The addition of PLTs resulted in a decrease in cellular fluorescence intensity. Interestingly, the effect of PEVs on reducing ROS levels was more pronounced than that of PLTs. These results indicate that PEVs have a better ability to scavenge intracellular ROS. Further, we quantified H2O2-induced ROS production by flow cytometry. The percentages of DCF-positive NP cells were 6.9%, 38.25%, 27.65%, and 16.25% in the control, H2O2, PLTs, and PEVs groups, respectively (Fig. 3C and D). Together, these results suggest that PEVs have a better antioxidant capacity than that of PLTs.
Fig. 3.
Effects of PEVs on oxidative stress, apoptosis, and senescence in degenerative NP cells. (A) Fluorescence images of NP cells with ROS staining by DCFH-DA probe. Scale bars = 50 μm. (B) ROS level of NP cells according to (A). (C) The ROS level was determined by flow cytometry with DCFH-DA probe. (D) The ROS abundance of NP cells according to (C). (E–F) Detection of NP cells apoptosis by flow cytometry. (G–H) Detection of NP cells apoptosis by fluorescence confocal microscopy. Scale bars = 50 μm. (I–J) NP cells senescence detection by SA-β-gal staining. Scale bars = 50 μm. (K) The effects of PLTs and PEVs on mRNA expression levels of P16 and P21 in NP cells subjected to 100 μM H2O2 stimulation. Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 4). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
3.3. PEVs inhibit apoptosis and senescence in NP cells
We utilized the methods of FITC-annexin V/PI apoptosis assay, TUNEL staining, SA-β-Gal staining, and P16 and P21 mRNA detection to verify the effect of PEVs on the apoptosis and senescence phenotype of NP cells. The flow cytometry results of the PI/FITC apoptosis assay yielded apoptosis rates of NP cells of 5.7%, 55.7%, 41.69%, and 17.21% in the NC, H2O2, PLTs, and PEVs groups, respectively (Fig. 3E and F) The results of TUNEL staining depicted a low apoptosis rate in the NC group, whereas a strong, clearly observable green fluorescence of H2O2 group elucidated a high apoptosis rate. The addition of PEVs and PLTs resulted in a decrease in cellular fluorescence intensity. PEVs exerted superior reducing effect on the apoptosis rate of NP cells compared with PLTs (Fig. 3G and H). Taken together, these data indicate that PEVs inhibit the apoptosis of NP cells. Regarding the result of Sa-β-Gal staining, the senescence of NP cells started to be considerably reduced by the action of PEVs, whereas the effect of PLTs was not significant (Fig. 3I and J). Additionally, the qPCR results illustrated that H2O2 stimulation increases the mRNA expression levels of cell senescence markers (P16, P21), whereas PEVs notably downregulated them (Fig. 3K). These data indicate that PEVs inhibit senescence in NP cells. Mitochondrial dysfunction is an essential part of the senescence and apoptosis phenotype, which is linked with the major building blocks of senescence and apoptosis via multiple feedback loops [[87], [88], [89], [90]]. Because PEVs contain mitochondria-related proteins such as sirtuin-1 (SIRT1), we suppose that PEVs may inhibit senescence and apoptosis in NP cells by regulating mitochondrial function.
3.4. Effect of PEVs on the metabolism proteins of NP cells
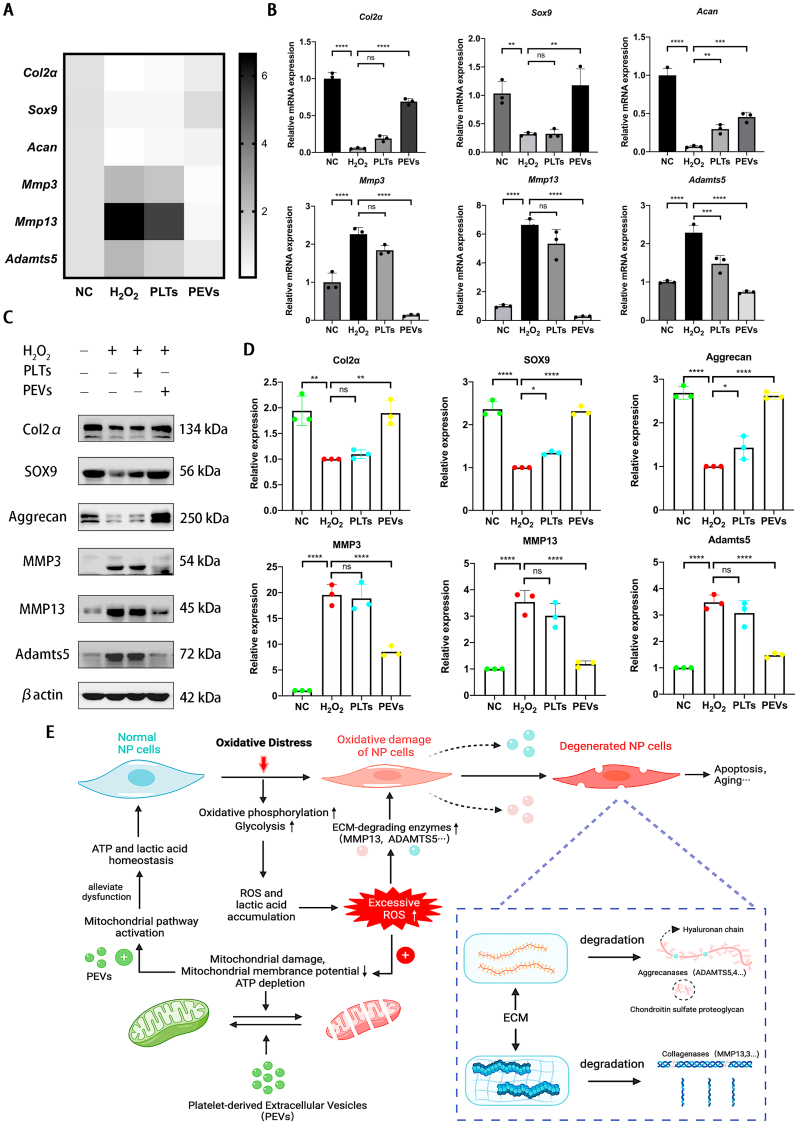
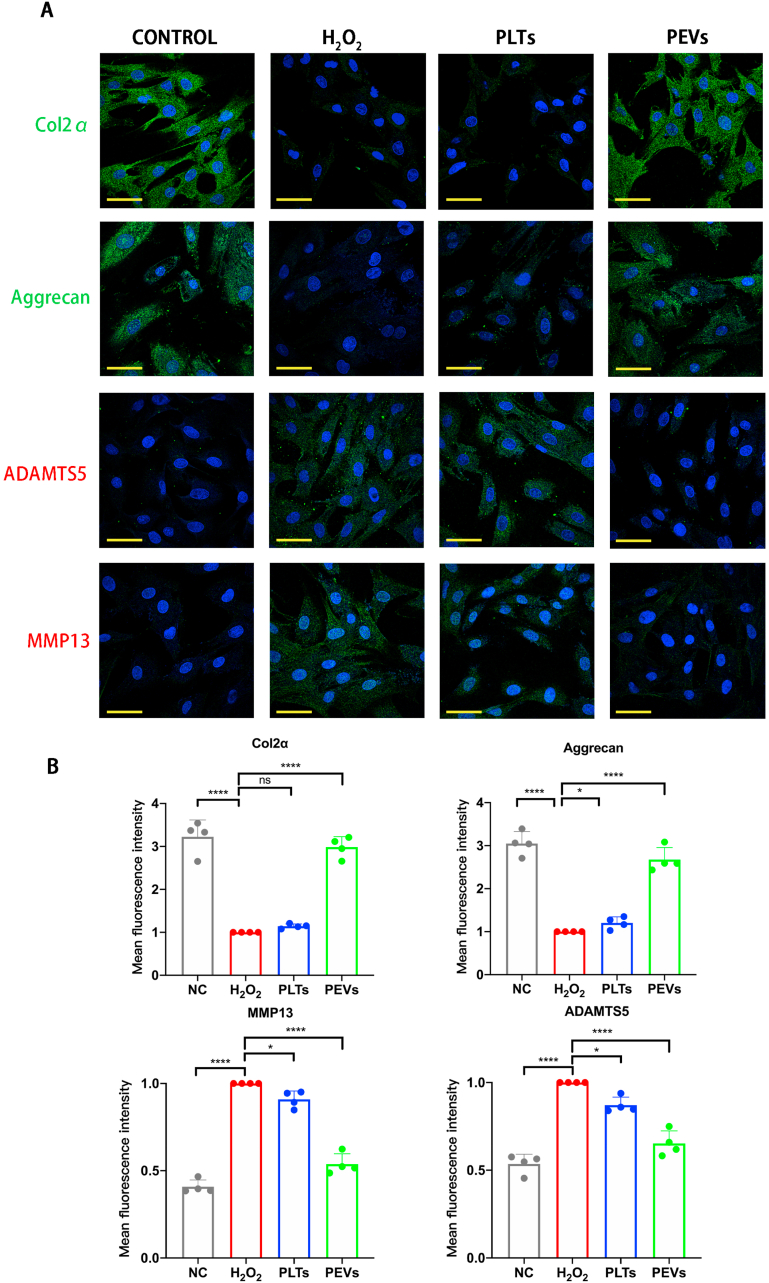
We examined the expression of genes related to ECM synthesis and degradation using qPCR to study the effect of PEVs on the metabolism of NP cells. The results illustrated that H2O2 stimulation increased the expression of catabolic genes (Mmp3, Mmp13, and Adamts5) and decreased the expression of anabolic genes (Col2α, Sox9, and Acan). After PEVs treatment, the expression of catabolic genes in the NP cells is downregulated and that of anabolic genes is upregulated (Fig. 4A and B). Then, we examined the protein levels involved in cell anabolism and catabolism using western blotting. Considering the mRNA expression data, the PEVs increased anabolic protein (Col2α, Sox9, and Aggrecan) levels and reduced catabolic protein (MMP3, MMP13, and ADAMTS5) levels in H2O2-stimulated NP cells (Fig. 4C and D). According to the result and reference [91], we draw a schematic of the effect of PEVs in NP cells by repairing damaged mitochondria and regulating ECM-degrading enzymes (Fig. 4E). The immunofluorescence confirmed that PEVs enhanced the fluorescence intensities of Col2α and Aggrecan and attenuated the fluorescence intensities of ADAMTS5 and MMP13 in H2O2-stimulated NP cells (Fig. 5A and B). The results indicate that PEVs can enhance cellular anabolism and attenuate cellular catabolism in pathological NP cells.
Fig. 4.
PEVs regulate metabolism proteins in degenerative NP cells. (A–B) The effects of PLTs and PEVs on mRNA expression levels of Col2α, Sox9, Acan, Mmp3, Mmp13, and Adamts5 with 200 μM H2O2 stimulation. (C) Western blot analysis showing Col2α, Sox9, Aggrecan, MMP3, MMP13, and ADAMTS5 protein levels after treatment with 100 μg/mL PEVs (with 200 μM H2O2 stimulation) and 100 μg/mL PLTs. βactin was used as a loading control. Uncropped gel is in SI Fig. 4B. (D) Quantitative analysis of metabolism protein expression differences in each group. (E) The schematic of PEVs regulating metabolism proteins in degenerative NP cells by repairing damaged mitochondria and regulating ECM-degrading enzymes. Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 3). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
Fig. 5.
Effects of PEVs on metabolic proteins of NP cells. (A) Immunofluorescence assay was performed to assess expression of Col2α, Aggrecan, ADAMTS5, and MMP13. Scale bars = 50 μm. (B) Quantification of specific immunofluorescence intensities according to (A). Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 4). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
3.5. PEVs uptake and mitochondrial colocalization
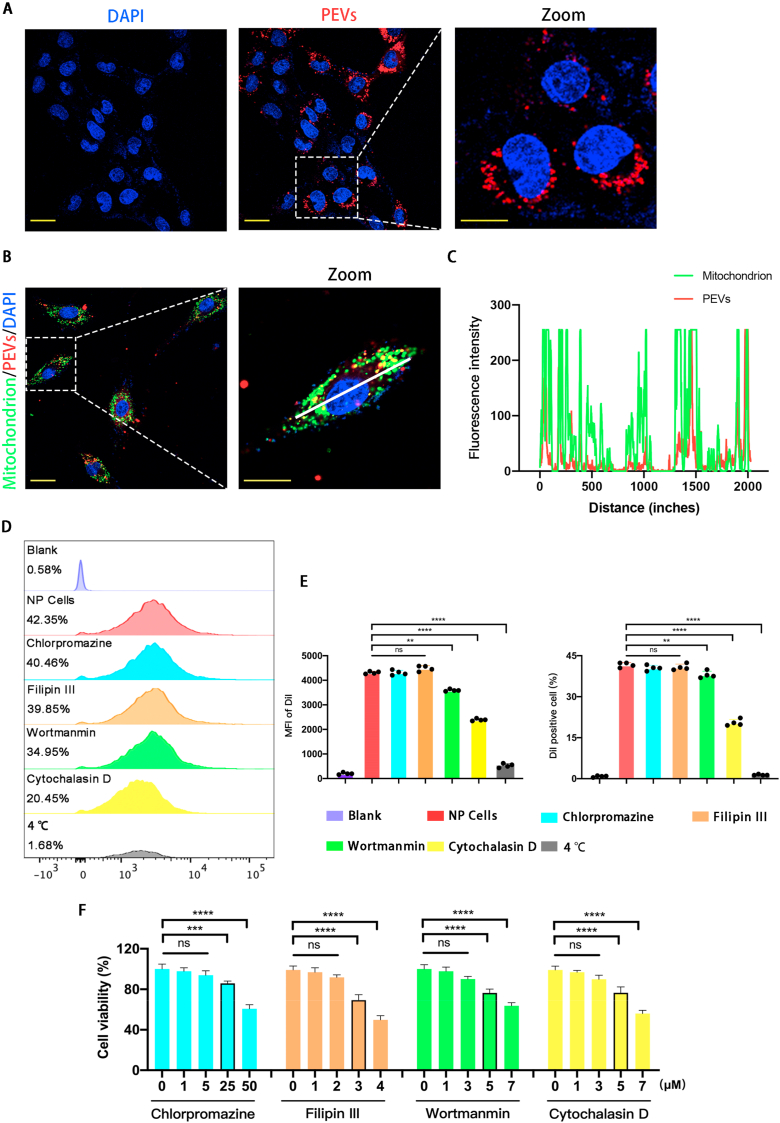
We first labeled the mitochondria of NP cells with a MitoTracker Green probe to test the ability of NP cells to take up PEVs. Then, we labeled the PEVs with a DiI membrane probe. Finally, we incubated the labeled NP cells with PEVs (Fig. 6A). The confocal microscopy pictures a partial overlap of DiI and MitoTracker Green fluorescence, and this indicates that PEVs can co-localize with the mitochondria of NP cells after being endocytosed by NP cells (Fig. 6B). Subsequently, we drew a figure of distribution of PEVs (Red) and NP cells (Green) using Image J software. Fig. 6C illustrated PEVs colocalized with mitochondria. Then, we validated the endocytic pathway of PEVs in NP cells using chlorpromazine, filipin III, wortmannin, and cytochalasin D to inhibit clathrin-mediated endocytosis, caveolae-mediated endocytosis, micropinocytosis, and actin-mediated endocytosis, respectively. We used DiI to label PEVs and DiO to label NP cells. According to flow cytometry, the percentage of DiI/DiO-positive NP cells is 38.20%, 36.40%, 35.05%, 16.32%, 30.95%, and 1.02% in the control, chlorpromazine, filipin III, cytochalasin D, wortmannin, and 4 °C groups, respectively (Fig. 6D and E). The results showed that the cellular uptake of PEVs was suppressed at 4 °C by cytochalasin D or by wortmannin, but not by Filipin III or chlorpromazine. Therefore, the uptake of PEVs with diameters of 100–200 nm by NP cells is mediated by actin-mediated endocytosis. Further, a small number of PEVs are endocytosed by micropinocytosis because of their larger size. The previous study demonstrated that chlorpromazine, filipin III, wortmannin, and cytochalasin D are toxic [92]. To obtain CCK-8 results, the chosen concentrations were 25, 3, 5, and 5 μM for chlorpromazine, filipin III, wortmannin, and cytochalasin D, respectively.
Fig. 6.
PEVs uptake and mitochondrial colocalization. (A) Fluorescence confocal microscopy image of PEVs uptake. Scale bar = 50 μm. (B–C) Fluorescence confocal microscopy images and line-scanning profiles of NP cells mitochondria colocalized with PEVs. Red: PEVs, Green: mitochondria, Blue: DAPI-labeled nuclei. Scale bar = 50 μm. (D) NP cells measured by flow cytometry after cells were pretreated with each inhibitor (chlorpromazine, filipin III, wortmannin, and cytochalasin D) for 0.5 h and cultured with PEVs for 24 h at 37 °C or at 4 °C. (E) Quantification of specific Immunofluorescence intensities (MFI) and the percentage of Dil positive cells by flow cytometry. (F) The toxicity of different concentration of each inhibitor (chlorpromazine, filipin III, wortmannin, and cytochalasin D) in NP cells by CCK-8 kit assay. Significance between six groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 4). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
3.6. Effects of PEVs on mitochondrial function of cells in vitro
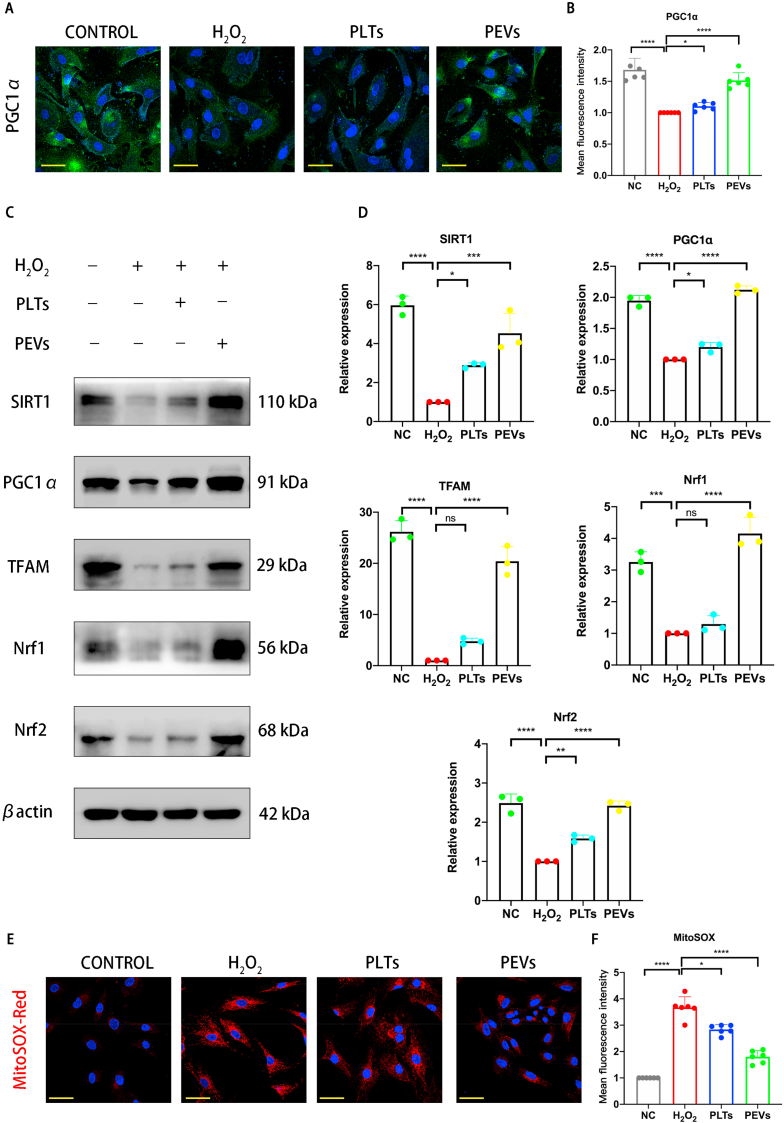
We detected the abundance of PGC1α (a regulator of mitochondrial genesis) in NP cells using immunofluorescence staining to confirm that PEVs could restore mitochondrial function in degenerated NP cells. The results illustrated that the H2O2 stimulation significantly downregulated the abundance of PGC1α, whereas the treatment of PEVs restored the abundance of PGC1α (Fig. 7A and B). Treatment with PLTs resulted in only slight upregulation of PGC1α. Studies have suggested that the activation of AMPK can promote the expression of SIRT1, PGC1α, and TFAM, which can help maintain mitochondrial function in cells [58]. Then, we examined the levels of these mitochondria-related proteins (SIRT1, PGC1α, TFAM, Nrf1, and Nrf2) by western blotting. The results elucidated that these proteins were downregulated following H2O2 stimulation (Fig. 7C and D). In contrast, PLTs and PEVs increased the abundance of these proteins, and the PEV treatment was more effective than the PLTs treatment. Next, we used MitoSOX Red staining to observe the effect of PEVs on mitochondrial ROS levels. MitoSOX Red staining results showed low ROS level in the mitochondria of the NC group and significantly increased levels in the H2O2 group (Fig. 7E and F). The ROS level of PEVs group was significantly lower than those in the H2O2 and PLTs groups. These data indicate that PEVs can reduce mitochondrial ROS levels in NP cells.
Fig. 7.
Effects of PEVs on mitochondrial functional proteins. (A) Fluorescence expression of mitochondrial functional protein PGC1α. Scale bar = 50 μm. (B) Quantification of specific immunofluorescence intensities according to (A). (C) Western blot analysis showing SIRT1, PGC1α, TFAM, Nrf1, and Nrf2 protein levels after treatment with 100 μg/mL PEVs (with 200 μM H2O2 stimulation) and 100 μg/mL PLTs. βactin was used as a loading control. Uncropped gel is in SI Fig. 4C. (D) Quantitative analysis of mitochondrial functional protein expression differences in each group. (E–F) Fluorescence intensity and quantitative analysis of mitochondria using MitoSOX Red staining. Scale bar = 50 μm. Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 3). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
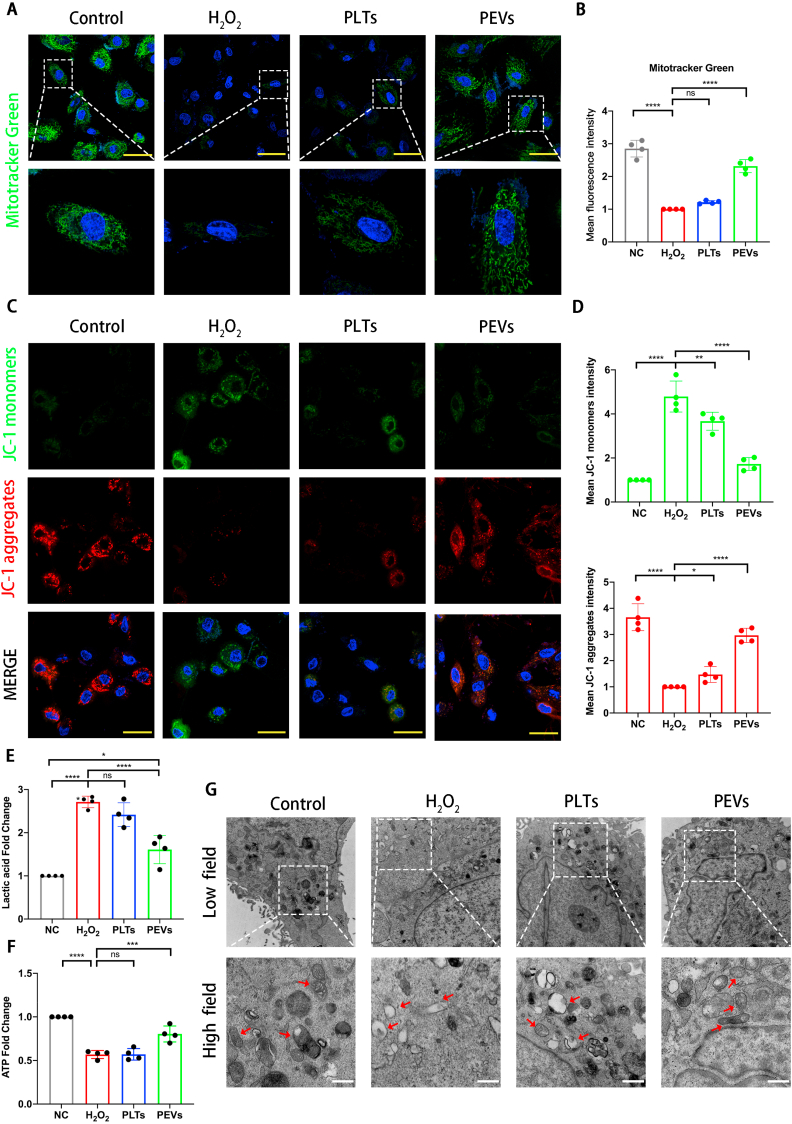
We evaluated the mitochondrial mass and mitochondrial membrane potential (ΔΨm) of NP cells after PEVs treatment to further investigate the effect of PEVs on mitochondrial function. Compared with the H2O2 group, PEV-treated NP cells showed enhanced mitochondrial mass, as depicted by the increased MitoTracker Green staining (Fig. 8A and B). Furthermore, decreased ΔΨm is a hallmark event of cellular oxidative stress in early apoptosis, and it can be easily detected by the transformation of JC-1 from red to green fluorescence [78]. Normal NP cells emit strong red and weak green fluorescence using the JC-1 probe and indicate a higher ΔΨm (Fig. 8C and D). After H2O2 stimulation, the green fluorescence started to increase, and the red fluorescence diminished, which indicate a lower ΔΨm. We found a significant increase in red fluorescence and a decrease in green fluorescence in the PEVs group, which indicates an increased ΔΨm. These results suggest that PEVs can attenuate oxidative stress-induced mitochondrial dysfunction in NP cells.
Fig. 8.
PEVS restored mitochondrial membrane potential homeostasis and mitochondrial function. Scale bar = 20 μm. (A) Protective effect of PEVs was evaluated by MitoTracker Green. (B) Quantification of fluorescence intensities according to (A). (C) Detection of mitochondrial membrane potentials in four groups of cells by JC-1 fluorescence probe. Scale bar = 50 μm. (D) Quantification of fluorescence intensities according to (C). (E) Detection of lactic acid content in supernatant of NP cells culture medium. (F) ATP levels of NP cells was detected. (G) Transmission electron microscopy (TEM) images of mitochondria in NP cells. Scale bar = 0.5 μm. Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 4). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
We further detected the content of lactic acid and ATP produced by NP cells under H2O2 stimulation to further observe mitochondrial function of cells. The increase in the lactic acid induced by H2O2 stimulation reflected an elevated level of glycolysis as the main product of the glycolysis; this was attenuated by PEVs treatment (Fig. 8E). In addition, the ATP concentration of NP cells decreased after H2O2 stimulation. Interestingly, this concentration increases in PEVs group (Fig. 8F). In conclusion, PEVs can correct the mitochondrial dysfunction in H2O2-treated NP cells.
Then, we observed the mitochondrial morphology of NP cells by TEM and quantified the percentage of mitochondrial vacuoles (Fig. 8G). The TEM results revealed that the percentage of mitochondrial vacuoles/mitochondria in the PEVs group is markedly lower than those in the H2O2 and PLTs groups. These data suggested that PEVs can restore mitochondrial function in NP cells. Therefore, we considered the supplement of mitochondrial related proteins by PEVs results in restoring the mitochondrial function of NP cells.
3.7. Proteomic analysis
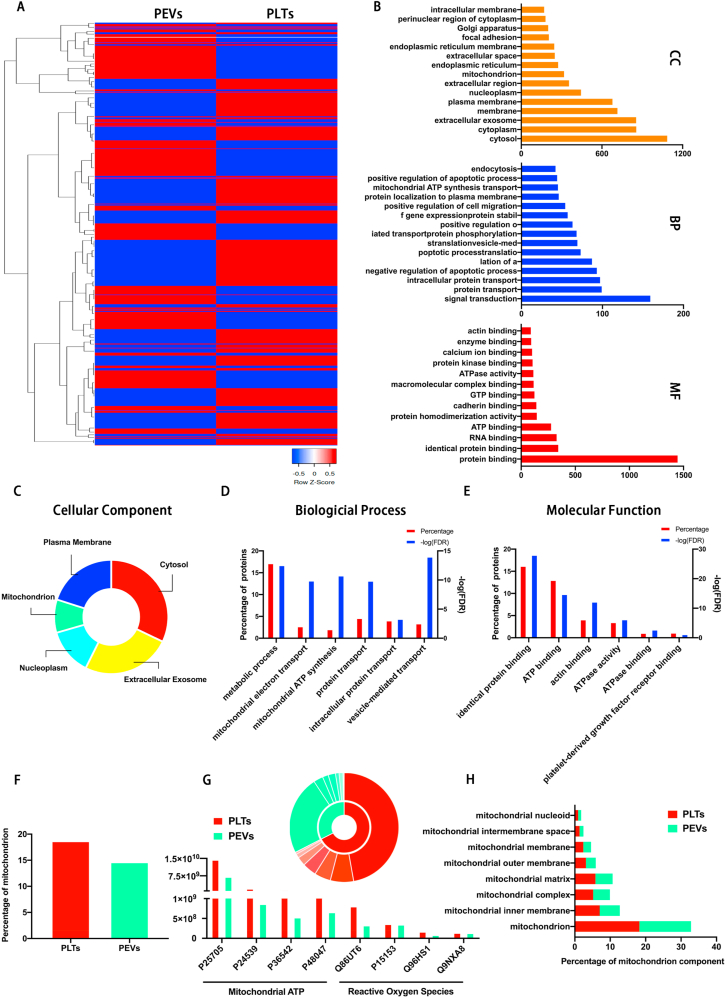
We evaluated PEVs and PLTs using proteomics to further clarify the reasons for the therapeutic effects of PEVs. The results showed that the PEVs contained 1732 PLTs parental proteins. Compared with PLTs, there are 790 upregulated proteins and 942 downregulated proteins (Fig. 9A). Through GO enrichment analysis, we classify these parental proteins into biological processes, molecular functions, and cellular components (Fig. 9B). The proteomic analysis elucidated that the proteins in PEVs are derived from the cytosol (1085), extracellular exosome (854), plasma membrane (678), nucleoplasm (444), and mitochondrion (318) (Fig. 9C). The GO biological process analysis revealed that 17.26% of PEVs proteins are involved in metabolic processes, 3.47% in vesicle-mediated transport, 2.8% in mitochondrial electron transport, and 2.14% in mitochondrial ATP synthesis biological processes (Fig. 9D). The GO molecular functional analysis revealed that these components are involved in the binding of specific proteins, ATP, ATPase, actin, and platelet-derived growth factor receptors, and this can contribute to the phagocytosis and localization of PEVs by NP cells (Fig. 9E). Next, the mitochondrial protein contents of PEVs are similar with PLTs (Fig. 9F), which suggests that PEVs retain the mitochondrial components of platelets. In addition, PEVs and PLTs contained SIRT1 proteins (P48047), which can play an essential role in the protection of mitochondrial function both in vitro and in vivo. Compared with PLTs, PEVs retain partial mitochondrial ATP and ROS proteins (Fig. 9G). Moreover, enrichment analysis illustrated that different parts of mitochondria were enriched in PEVs, including mitochondria, mitochondrial inner membrane, mitochondrial complex, mitochondrial matrix, mitochondrial intermembrane space, mitochondrial outer membrane, and mitochondrial nucleoid (Fig. 9H). These results suggest that PEVs may repair impaired mitochondria by replenishing mitochondria-related proteins after endocytosis by NP cells, which can restore the mitochondrial function of NP cells. The data helped our follow-up focus on investigating the role of SIRT1 in NP cells, as further demonstrated by PEVs restoring mitochondrial function in NP cells.
Fig. 9.
Proteomic analysis of PEVs and PLTs. (A) Heat map of 1732 parental proteins in PEVs and PLTs. (B) GO classification of parental proteins in biological process, molecular function, and cellular component. (C–E) GO classification of PEVs proteins in cellular component, biological process, and molecular function. (F) Comparison of mitochondrial contents in PLTs and PEVs. (G) The content and categories of proteins in PLTs and PEVs involved in mitochondrial ATP and reactive oxygen species (ROS). (H) Summary of significantly enriched GO terms in mitochondrial content of PLTs and PEVs.
3.8. Effects of inhibitor and activator on pathological NP cells
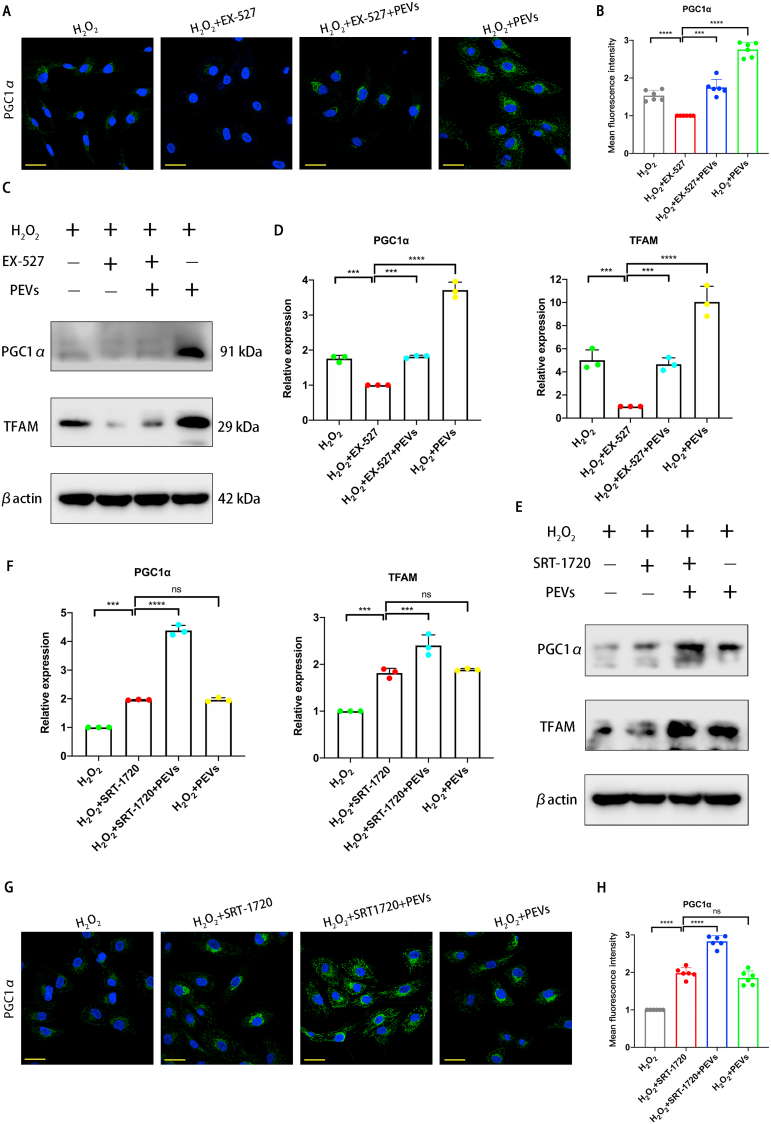
According to the result of immunofluorescence staining, PGC1α was present at a low protein level in pathological NP cells (H2O2 group), and its fluorescence intensity almost disappeared after the addition of the selective SIRT1 inhibitor (H2O2+EX-527 group). Compared with the H2O2+EX-527 group, the H2O2+EX-527+PEVs group exhibited elevated protein levels of PGC1α, while the H2O2+PEVs group showed a significant increase in PGC1α protein abundance (Fig. 10A and B). The Western blot analysis results indicated that the mitochondria-related protein abundances of PGC1α and TFAM were lower in the H2O2+EX-527 group compared with those in the H2O2 group (Fig. 10C and D). Compared with those in the H2O2 +EX-527 group, the proteins in the H2O2+PEVs group were effectively increased. Then, we examined the abundances of the mitochondria-related proteins PGC1α and TFAM with the addition of the SIRT1 activator (Fig. 10E and F). The results indicate that these proteins are upregulated following SRT-1720 stimulation (H2O2+SRT-1720 group). Moreover, the mitochondria-related protein abundances were increased significantly in the H2O2+SRT-1720+PEVs group. The result of PGC1α immunofluorescence staining illustrated that SRT-1720 stimulation significantly upregulated the abundance of PGC1α, whereas the effect of treatment of PEVs was same as that of SRT-1720 (Fig. 10G and H). Treatment with SRT-1720 and PEVs (H2O2+SRT-1720+PEVs group) resulted in a significant upregulation of PGC1α. Taken together, the data demonstrate that PEVs restoring mitochondrial function in pathological NP cells by activating the SIRT1–PGC1α–TFAM pathway via SIRT1 supplementation.
Fig. 10.
Regulation of mitochondrial function by PEVs via SIRT1 in pathological NP cells. (A) Effects of EX-527 (the selective SIRT1 inhibitor) on the fluorescence expression of mitochondrial functional protein PGC1α in each group. Scale bar = 50 μm. (B) Quantification of specific immunofluorescence intensities according to (A). (C) Western blot analysis showing the effects of EX-527 on PGC1α and TFAM protein levels in pathological NP cells. βactin was used as loading control. Uncropped gel is in SI Fig. 4D. (D) Quantitative analysis of mitochondrial functional protein levels in each group. (E) Western blot analysis showing the effects of SRT-1720 (the selective SIRT1 activator) on PGC1α and TFAM protein levels in pathological NP cells. βactin was used as loading control. Uncropped gel is in SI Fig. 4E. (F) Quantitative analysis of mitochondrial functional protein levels in each group. (G) Effects of SRT-1720 on the fluorescence expression of mitochondrial functional protein PGC1α in each group. Scale bar = 50 μm. (H) Quantification of specific immunofluorescence intensities according to (G). Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 3). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
3.9. Effects of rotenone on PEVs-treated pathological NP cells
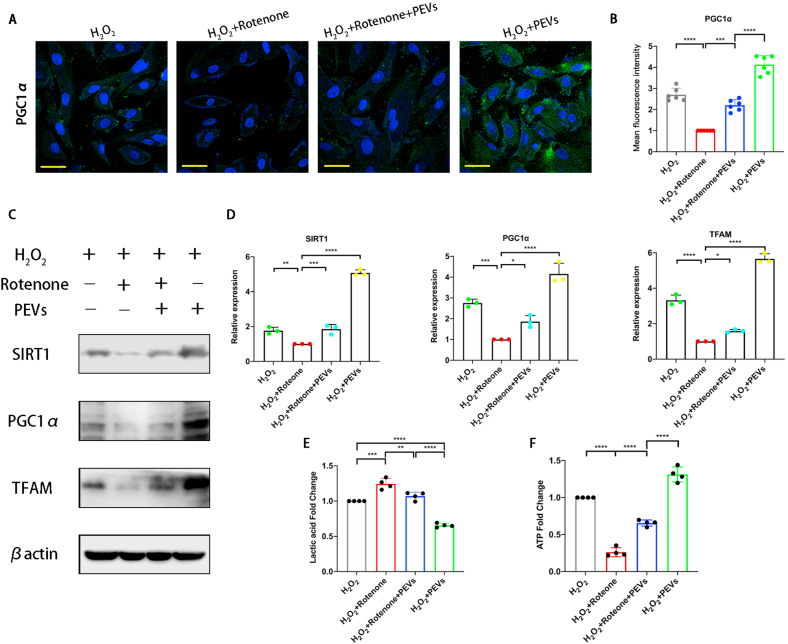
We used rotenone to suppress electron transport in the mitochondria to verify that PEVs exerted a therapeutic effect on cells by repairing mitochondria. The protein level of PGC1α was low in pathological NP cells (H2O2 group), and the fluorescence intensity of PGC1α diminished to almost zero after the addition of rotenone (H2O2+rotenone group). Although the H2O2+PEVs group exhibit the elevated protein levels of PGC1α compared with the H2O2 group, the H2O2+PEVs + rotenone group showed a significant decrease in PGC1α protein abundance compared with the H2O2+PEVs group (Fig. 11A and B). The results of western blotting revealed that the mitochondria-related protein abundances of SIRT1, PGC1α, and TFAM were decreased in the H2O2+rotenone group compared with those in the H2O2 group (Fig. 11C and D). In contrast, these proteins were effectively increased in the H2O2+PEVs group compared to the H2O2 group. However, rotenone significantly inhibited the elevating effect of PEVs on these proteins. Further, the effect of PEVs on the mitochondrial function (lactic acid and ATP) of pathological NP cells was tested; the concentration of lactic acid decreased in H2O2+PEVs group compared with H2O2+PEVs + Rotenone group (Fig. 11E), and the ATP concentration increased in H2O2+PEVs group compared with H2O2+PEVs + Rotenone group (Fig. 11F). These data indicated that H2O2-induced cellular was mitochondrial function inhibited by rotenone, and the therapeutic effect of PEVs protected mitochondrion of NP cells by alleviating the oxidative stress damage.
Fig. 11.
Effect of rotenone on PEVs treated pathological NP cells. (A) Effects of the inhibitor rotenone on the fluorescence expression of mitochondrial functional protein PGC1α in each group. Scale bar = 50 μm. (B) Quantification of specific immunofluorescence intensities according to (A). (C) Western blot analysis showing the effects of rotenone on SIRT1, PGC1α, and TFAM protein levels in pathological NP cells. βactin was used as loading control. Uncropped gel is in SI Fig. 4F. (D) Quantitative analysis of mitochondrial functional protein levels in each group. (E) Detection of lactic acid concentration in supernatant of pathological NP cells culture medium. (F) ATP levels of pathological NP cells was detected. Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 3). (Scale bars are listed above; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
3.10. PEVs retard disc degeneration in the rat IVDD model
We hypothesized that PEVs could be an effective strategy to treat IVDD because PEVs could restore the mitochondrial function of NP cells in vitro. To this end, we established a rat IVDD model by needle puncture of the rat caudal IVD. First, we confirmed the selected caudal intervertebral plane for puncture using manual palpation and X-ray fluoroscopy (Fig. 12A). Second, we injected the PEVs and PLTs into the IVD. Finally, the IVD disc height and index of the CT images are measured and calculated, respectively (Fig. 12B). Repair of the nucleus pulposus (NP) was observed by magnetic resonance imaging (MRI). At 4 and 8 weeks, the NC group showed a high T2-weighted signal, which indicated high water content in the NP by measuring the signal intensity of NP (Fig. 12C). The vehicle group showed a progressively lower T2-weighted signal. Compared to the vehicle group, the PEVs group showed a relatively stable T2-weighted signal at 4 and 8 weeks after injection, whereas the PLTs group showed a significant lower in the signal. Statistical analysis results indicated significant difference in T2-weighted signal between the PEVs and vehicle groups at 4 and 8 weeks (Fig. 12D); meanwhile, no significant difference was observed between the signals of the PLTs and vehicle groups. These results demonstrated that PEVs can alleviate the inflammatory microenvironment to retard IVDD and restore the water content of NP, whereas PLTs do not exert a significant effect. Subsequently, we used microcomputed tomography (micro-CT) and disc height index (DHI) analyses to assess disc space height (Fig. 12E). Throughout the time course, no significant change in the intervertebral space height was observed in the NC group. At 4 and 8 weeks, the IVD height of rats in the vehicle and PLTs groups started to decrease, whereas this phenomenon was not apparent in the PEV group. The statistical results of micro-CT were similar to those of MRI (Fig. 12F), and these data indicated that PEVs can maintain the stability of the IVD and retard the development of IVDD.
Fig. 12.
Radiological data of IVDD rat experiments by the imaging system. (A) Overview of animal experiments and x-ray view of the puncture modeling in the IVDD rat. (B) Measurement and calculation formula of intervertebral disc height index in rat. (C) Representative MRI images of rat caudal vertebrae at 4 and 8 weeks. (The arrow represents the nucleus pulposus (NP)). (D) Quantitative analysis of T2-weighted signal intensity of the NP in each group. (E) Representative Micro-CT images of the caudal vertebrae of rats at 4 and 8 weeks. (F) Change in DHI% in each group. Significance between four groups was calculated using two-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 9). (∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
Then we stain IVD from 4- and 8-weeks rats with H&E, A&B, and S&O (Fig. 13A, B, C). The NC group exhibited a normal gel-like appearance of the NP, normal morphology of the fibrocartilage, no AF breakage, no serpentine appearance within the AF, and a clear border between the NP and the AF. However, the Vehicle and PLTs groups showed gradual NP atrophy and AF breakage at 4 and 8 weeks. The structure and NP content of the IVD in the PEVs group were significantly repaired compared to the Vehicle and PLTs groups. S&R staining revealed that the IVD height was obviously reduced in the Vehicle group at 4–8 weeks; the AF was severely broken, and the NP was replaced by AF (Fig. 13D). In contrast, the PEVs group showed a normal appearance of AF and a clear boundary between the NP and AF regions. The histological grade results suggest that intact NP tissue is observed in the NC group at 4 and 8 weeks (Fig. 13E and F). The Vehicle group exhibited severely degenerated NP tissue with a narrowing of the IVD height. The PLTs group showed moderately degenerated NP tissue; the PEVs group promoted NP tissue repair and IVD height restoration. These data indicate that PEVs can retard IVDD in vivo.
Fig. 13.
Histological evaluation of the IVDD rat experiment. (A) The H&E staining images of intervertebral discs of rats at 4 and 8 weeks. (B) Alcian blue staining images. (C) Safranin O-Fast Green staining images. (D) Sirius Red staining images. (E–F) Changes of histological grades evaluation at 4 and 8 weeks in each group. Significance between four groups was calculated using one-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 6). (Scale bars = 400 μm; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001).
3.11. Effects of PEVs on the mitochondrial function of IVD in vivo
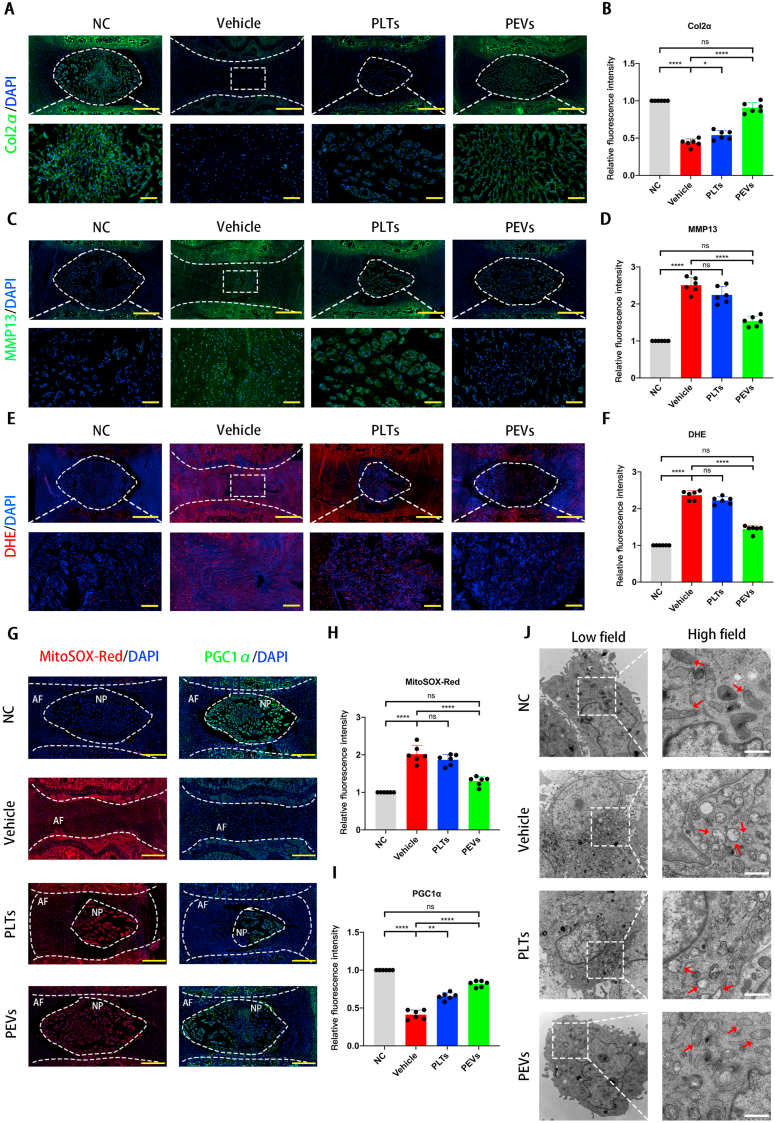
The IVDD tissues are characterized by a loss of ECM and the high expression of matrix-degrading enzymes such as matrix metalloproteinases (MMPs). We used the immunofluorescence to detect Col2α and MMP13 expression in the different groups. Compared to the NC group, the Vehicle and PLTs groups showed significantly lower Col2α levels (Fig. 14A and B). The PEVs treatment significantly elevated Col2α levels compared to the Vehicle and PLTs groups. Meanwhile, the fluorescence intensity of MMP13 was significantly higher in the Vehicle and PLTs groups compared with that for the NC group (Fig. 14C and D). In contrast, PEVs dramatically reduced the fluorescence intensity of MMP13 compared to that in the PLTs group. Overall, these data suggested that PEVs can suppress matrix degradation in vivo.
Fig. 14.
Effects of PEVs on the mitochondrial function of IVD in vivo. (A) Immunofluorescence staining images of Col2α of intervertebral discs (IVD). (B) Quantification of specific immunofluorescence intensities according to Col2α. (C) Immunofluorescence staining images of MMP13 of IVD. (D) Quantification of specific immunofluorescence intensities according to MMP13. (E) Immunofluorescence staining images of DHE on ROS levels in nucleus pulposus in rat IVD. (F) Quantification of specific immunofluorescence intensities according to DHE. (G) Fluorescent staining images of MitoSox Red and PGC1α in rats IVD. (H) Quantification of intensities according to MitoSox Red. (I) Quantification of specific immunofluorescence intensities according to PGC1α. (J) Transmission electron microscopy (TEM) images of mitochondrial morphology in NP cells in rats IVD. Scale bar = 0.5 μm. Significance between four groups was calculated using two-way ANOVA with Tukey's post-hoc test. Data are presented as mean ± SD (n = 6). (Scale bars = 400 μm; ∗: P < 0.05, ∗∗: P < 0.01, ∗∗∗: P < 0.005, and ∗∗∗∗: P < 0.001). Complete slides (A, C, E, G, and H) are in SI Fig. 5.
Then we used DHE fluorescent probe to evaluate the effect of PEVs on ROS levels of NP tissue in rat IVD (Fig. 14E and F). The ROS level of NP tissue, which is indicated by the red fluorescence intensity, was low in the NC group and significantly increased after the puncture. The PEVs group showed a significantly downregulated ROS level compared with the PLTs group although the PLTs group exhibited a slight effect in reducing the ROS level. The result suggested that PEVs can attenuate ROS levels of NP tissue in vivo.
Next, we performed frozen histological section experiments to observe ROS levels of mitochondria in rat IVD. We utilized MitoSOX Red staining to detect the effect of PEVs on the ROS levels of mitochondria of rats IVD (Fig. 14G and H). The ROS level of mitochondria is low in the NC group and gradually increased in the vehicle group. Moreover, ROS production indicated by MitoSOX Red staining in the PEVs group was significantly lower than that in degenerated IVD group (vehicle group) and PLTs group. These results indicate that PEVs can reduce ROS levels of mitochondria in rats IVD.
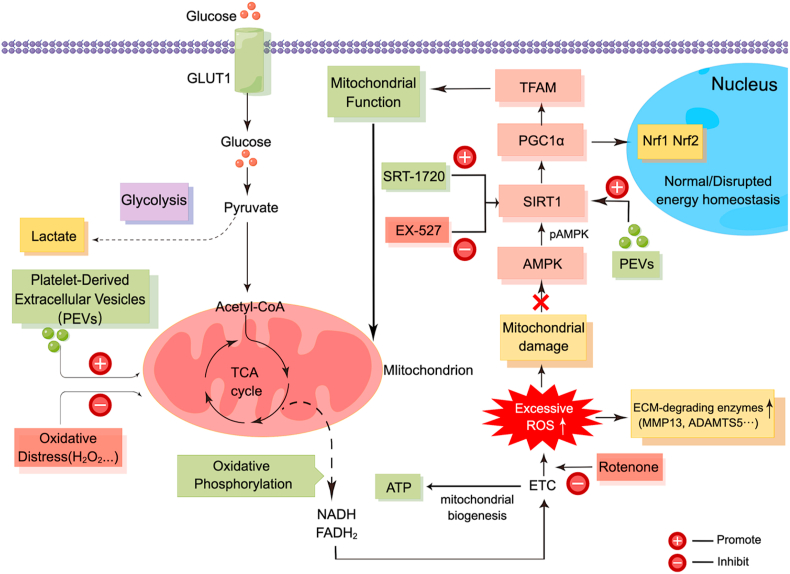
PGC1α is the major regulator of mitochondrial biogenesis in the AMPK–SIRT1–PGC1α–TFAM pathway, and it is the most representative mitochondrial functional protein [93]. Then, we detected PGC1α abundance in the rat IVD to determine the repairing effect of PEVs on the mitochondria in vivo. Compared to the NC group, which showed higher levels of PGC1α, the vehicle and PLTs groups showed a significant reduction in PGC1α levels (Fig. 14G, I). However, the PEVs group showed significantly higher PGC1α fluorescence intensity than the Vehicle and PLTs groups. These results suggest that PEVs promote the restoration of the mitochondrial function to maintain the homeostasis of IVD tissues. We also evaluated mitochondrial morphology on NP tissue after 8 weeks by TEM and quantified the percentage of mitochondrial vacuoles (Fig. 14J). The results showed that the percentage of mitochondrial vacuoles/mitochondria in PEVs group was markedly lower than in the vehicle and PLTs groups. In other words, the data suggest that PEVs can reduce ROS level and restore mitochondrial function in vivo. A schematic of PEVs regulating mitochondrial pathway to promote the repair of damaged mitochondria (Fig. 15).
Fig. 15.
The schematic of PEVs regulating mitochondrial function to promote the repair of impaired mitochondria. Glycolysis, oxidative phosphorylation, and the AMPK–SIRT1–PGC1α–TFAM pathway helps to maintain an optimal level of mitochondrial function and intracellular homeostasis in healthy NP cells. In degenerated NP cells, oxidative distress damage and reactive oxygen species (ROS) accumulate, causing damage to the mitochondria, thereby inhibiting AMPK signaling and activity, downregulating SIRT1, decreasing levels of PGC1α and TFAM, and further impairing the mitochondrial function in NP cells. GLUT1, glucose transporter type 1; TCA, tricarboxylic acid; ETC, electron transport chain; ROS, reactive oxygen species; ATP, adenosine-5′-triphosphate; AMPK, AMP-activated protein kinase; SIRT1, NAD-dependent protein deacetylase sirtuin-1; PGC1α, peroxisome proliferator-activated receptor γ co-activator 1α; TFAM, Mt transcription factor A; Nrf1, nuclear respiratory factor 1; Nrf2, nuclear respiratory factor 2; PEVs, platelet-derived extracellular vesicles; SRT-1720, the selective SIRT1 activator; EX-527, the selective SIRT1 inhibitor.
4. Discussion
In this study, we developed an efficient strategy for IVDD treatment by injecting PEVs. The three major findings are as follows: 1. “Artificial EVs” prepared by membrane extrusion are good substitutes for cell-derived natural extracellular vesicles. 2. PEVs can alleviate oxidative stress damage and promote the repair of damaged mitochondria in NP cells by activating the SIRT1–PGC1α–TFAM pathway in vitro. 3. Currently, no related research on PEVs in treating rat IVDD exists. We injected PEVs into IVD of rats and validated the retardation effect of PEVs on the degeneration of NP in rats. These findings suggest that the injection of PEVs may be a promising strategy for the treatment of IVDD.
Recent studies have found that PEVs possess molecular markers on their surface such as CD9, CD81, and CD63, which promote intercellular and intracellular signaling and mediate the targeting of PEVs to sites of inflammation, thrombosis, and tumors [94]. Further, studies have reported that PEVs can regulate cell function by delivering proteins, lipids, nucleic acids, and other biologically active molecules to target cells for intercellular communication [45]. Moreover, the function of PEVs in the regulation of tissue repair has been evaluated [95]. The PEVs can cross tissue barriers in the human body, such as the synovium, lymph, and bone marrow because of their 100–200 nm size and phospholipid bilayer structure, and therefore, they are easily absorbed by tissues or cells [37]. Our study found that PEVs can be internalized by NP cells through the actin-mediated endocytosis pathway and localized to the mitochondria.
During bone metabolism, the NP cells utilize glucose and other metabolites as energy sources. These cells generate energy through glycolysis and oxidative phosphorylation of glucose to maintain optimal mitochondrial function [91]. We found that PEVs regulated cellular metabolism by activating the AMPK–SIRT1–PGC1α–TFAM pathway to maintain mitochondrial homeostasis. The mitochondria are the main producers of ROS in the cells [64]. The accumulation of intracellular ROS can regulate cellular metabolism, ATP production, and synthesis of nuclear and mitochondrial DNA-encoded genes [96]. However, excessive ROS levels can lead to oxidative stress damage, which further results in mitochondrial dysfunction [97]. Anabolic proteins are upregulated, and catabolic proteins are downregulated when intracellular ROS levels are elevated [98,99]. Depolarized mitochondria are characterized by an imbalance in ΔΨm, and this is an early event in apoptosis [100]. Moreover, aging-related diseases are caused by dysfunctional mitochondria that fail to provide cellular energy [101]. In this study, we verified that H2O2 stimulation causes ROS accumulation in NP cells. Moreover, H2O2 induced the upregulation of Mmp3, Mmp13, and Adamts5 protein levels and downregulated Col2α, Aggrecan, and Sox9 protein levels. Further, the levels of mitochondria-related proteins (SIRT1, PGC1α, TFAM, Nrf1, and Nrf2) were reduced in NP cells after H2O2 stimulation. H2O2 could impair cellular metabolism by disrupting mitochondrial protein levels, which leads to the degeneration of the ECM and eventually IVDD.
Based on previous studies and references [71,91], AMPK, SIRT1, PGC1α, and TFAM were defined as mitochondrial-related proteins. These proteins are key regulators of metabolism in the mitochondrion of NP cells. The key protein to regulate mitochondrial function of cells is AMPK, which regulates energy metabolism and intracellular homeostasis through the downstream mediators, SIRT1, PGC1α, which is the expert regulator of mitochondrial biogenesis and TFAM [102]. The mitochondrial dysfunction in cells is linked to the decreased expression of SIRT1, PGC1α, TFAM, Nrf1, and Nrf2, respectively. The activation of the AMPK–SIRT1–PGC1α–TFAM pathway promotes the homeostasis of energy metabolism and mitochondrial function in cells, further limiting the progression of degenerative diseases [71,103,104]. Moreover, a study has demonstrated that SIRT1 can play an essential role in the protection of mitochondrial function both in vitro and in vivo [105]. The increased expression of SIRT1 might be a new therapeutic approach for degenerative diseases [[106], [107], [108]].
After a series of in vitro and in vivo studies, we found that PEVs could modulate the SIRT1–PGC1α–TFAM pathway, which helps maintain the homeostasis of the IVD. We showed that PEVs could attenuate oxidative stress and inflammation in NP cells. Further, we showed that PEVs decreased the levels of catabolic proteins and elevated those of anabolic proteins. In addition, PEVs can retain some growth factors and mitochondrial proteins from PLTs [30]. We found that their mitochondrial proteins were similar by comparing the proteomic analyses of PEVs and PLTs. Therefore, we tested the effect of PEVs on the expression of mitochondria-related proteins in NP cells, and the results showed that PEVs promoted the abundance of these mitochondrial proteins. In rat IVDD models, we verified that PEVs could retard IVDD, and these results were consistent with those of cellular experiments.
Over the past few years, studies have consistently concluded that PEVs can be inflammation-targeted and play an important role in intercellular communication [37,45]. However, the current therapeutic strategies for PEVs continue to face a few challenges in basic research. The cellular origin of PEVs is critical; we must identify the safest cell source for PEVs to avoid immunogenicity issues and ensure that PEVs do not produce toxic components. In addition, we must minimize off-target effects by balancing the natural processing and degradation processes of PEVs when designing therapeutic strategies for specific targets. Appropriate preclinical models must be established, and specific biological processes, cellular components, and molecular functions must be selected to address these issues. The current study demonstrated that PEVs can be phagocytosed and targeted to the mitochondria in NP cells, which helps provide mitochondria-associated proteins to restore their function. During this process, the PEVs alleviate oxidative stress, inhibit cellular inflammation, and restore mitochondrial function to maintain homeostasis in the IVDD. This was similar to previous studies showing that PEVs could enhance wound or tissue healing by improving mitochondrial function and promoting musculoskeletal or nerve regeneration by inducing cell differentiation [44,109,110]. In the emerging field of nanomaterial therapy research, PEVs provide pivotal insights into degenerative diseases associated with the homeostasis of mitochondrial function.
5. Conclusion
We demonstrated that PEVs with high antioxidant activity and inflammatory targeting could retard the progression of IVDD by reducing oxidative stress and inflammatory response. Moreover, PEVs could activate the SIRT1–PGC1α–TFAM pathway to maintain mitochondrial function. Our results showed that the injection of PEVs can be a promising strategy for treating patients with IVDD.
Credit author statement
Zhanqiu Dai and Chen Xia: Conceptualization, Methodology, Data Curation, and Writing-Original Draft; Tingxiao Zhao: Methodology and Data Curation; Haoli Wang and Hongsen Tian: Formal analysis and Data Curation; Ouyuan Xu: Methodology and Data Curation; Pengfei Chen, Jun Zhang and Xunbin Zhu: Supervision, Project administration, Funding acquisition, and Writing -Review & Editing.
Funding
The study was supported by the National Nature Science Fund of China (Grant No. 82072414 and U21A20351), the National Key R&D Program of China (Grant No. 2020YFC1107104), the Natural Science Fund of Zhejiang Province (Grant No. LY21H060002), the Innovative Talent Support Program Project of Zhejiang Provincial Health Commission (Grant No. 2021433298), the Public Projects of Zhejiang Province (LGF19H060013 and LGD22H060004), the Key research and development plan in Zhejiang Province (Grant No. 2020C03043), and the Zhejiang Provincial Program for the cultivation of high-level innovative health talents. We thank Wu Lingyun and Yang Ping in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for their technical assistance on cryo-TEM.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Graphical Abstract and Figure 1 were created using Figdraw (www.figdraw.com). Figures 2A, 4E, 15, 12A, and SI Figure 1 were created with biorender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100512.
Contributor Information
Xunbin Zhu, Email: zhuxb22@163.com.
Jun Zhang, Email: spinezhangjun@aliyun.com.
Pengfei Chen, Email: pengfei_chen@zju.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Knezevic N.N., et al. Low back pain. Lancet. 2021;398(10294):78–92. doi: 10.1016/S0140-6736(21)00733-9. [DOI] [PubMed] [Google Scholar]
- 2.Hartvigsen J., et al. What low back pain is and why we need to pay attention. Lancet. 2018;391(10137):2356–2367. doi: 10.1016/S0140-6736(18)30480-X. [DOI] [PubMed] [Google Scholar]
- 3.Vos T., et al. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanayama M., et al. Cross-sectional magnetic resonance imaging study of lumbar disc degeneration in 200 healthy individuals. J. Neurosurg. Spine. 2009;11(4):501–507. doi: 10.3171/2009.5.SPINE08675. [DOI] [PubMed] [Google Scholar]
- 5.Cheung K.M., et al. Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty-three individuals. Spine. 2009;34(9):934–940. doi: 10.1097/BRS.0b013e3181a01b3f. [DOI] [PubMed] [Google Scholar]
- 6.Roberts S., et al. Histology and pathology of the human intervertebral disc. J Bone Joint Surg Am. 2006;88(Suppl 2):10–14. doi: 10.2106/JBJS.F.00019. [DOI] [PubMed] [Google Scholar]
- 7.Chen B.L., et al. Surgical versus non-operative treatment for lumbar disc herniation: a systematic review and meta-analysis. Clin. Rehabil. 2018;32(2):146–160. doi: 10.1177/0269215517719952. [DOI] [PubMed] [Google Scholar]
- 8.Yavin D., et al. Lumbar fusion for degenerative disease: a systematic review and meta-analysis. Neurosurgery. 2017;80(5):701–715. doi: 10.1093/neuros/nyw162. [DOI] [PubMed] [Google Scholar]
- 9.Pan A., et al. Adjacent segment degeneration after lumbar spinal fusion compared with motion-preservation procedures: a meta-analysis. Eur. Spine J. 2016;25(5):1522–1532. doi: 10.1007/s00586-016-4415-6. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto K., et al. Adjacent segment degeneration after fusion spinal surgery-a systematic review. Int. Orthop. 2019;43(4):987–993. doi: 10.1007/s00264-018-4241-z. [DOI] [PubMed] [Google Scholar]
- 11.Rogerson A., Aidlen J., Jenis L.G. Persistent radiculopathy after surgical treatment for lumbar disc herniation: causes and treatment options. Int. Orthop. 2019;43(4):969–973. doi: 10.1007/s00264-018-4246-7. [DOI] [PubMed] [Google Scholar]
- 12.Bowles R.D., Setton L.A. Biomaterials for intervertebral disc regeneration and repair. Biomaterials. 2017;129:54–67. doi: 10.1016/j.biomaterials.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukomska B., et al. Challenges and controversies in human mesenchymal stem cell therapy. Stem Cell. Int. 2019;2019 doi: 10.1155/2019/9628536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia C., et al. Mesenchymal stem cell-derived exosomes ameliorate intervertebral disc degeneration via anti-oxidant and anti-inflammatory effects. Free Radic. Biol. Med. 2019;143:1–15. doi: 10.1016/j.freeradbiomed.2019.07.026. [DOI] [PubMed] [Google Scholar]
- 15.Liao Z., et al. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics. 2019;9(14):4084–4100. doi: 10.7150/thno.33638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing H., et al. Injectable exosome-functionalized extracellular matrix hydrogel for metabolism balance and pyroptosis regulation in intervertebral disc degeneration. J. Nanobiotechnol. 2021;19(1):264. doi: 10.1186/s12951-021-00991-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C., et al. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8(1):237–255. doi: 10.7150/thno.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everts P., et al. Platelet-rich plasma: new performance understandings and therapeutic considerations in 2020. Int. J. Mol. Sci. 2020;21(20) doi: 10.3390/ijms21207794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akeda K., et al. Platelet-rich plasma in the management of chronic low back pain: a critical review. J. Pain Res. 2019;12:753–767. doi: 10.2147/JPR.S153085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y., et al. Effect of platelet-rich plasma on intervertebral disc degeneration in vivo and in vitro: a critical review. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/8893819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oneto P., Etulain J. PRP in wound healing applications. Platelets. 2021;32(2):189–199. doi: 10.1080/09537104.2020.1849605. [DOI] [PubMed] [Google Scholar]
- 22.Everts P.A., et al. Platelet rich plasma in orthopedic surgical medicine. Platelets. 2021;32(2):163–174. doi: 10.1080/09537104.2020.1869717. [DOI] [PubMed] [Google Scholar]
- 23.Gupta S., Paliczak A., Delgado D. Evidence-based indications of platelet-rich plasma therapy. Expet Rev. Hematol. 2021;14(1):97–108. doi: 10.1080/17474086.2021.1860002. [DOI] [PubMed] [Google Scholar]
- 24.Beitzel K., et al. US definitions, current use, and FDA stance on use of platelet-rich plasma in sports medicine. J. Knee Surg. 2015;28(1):29–34. doi: 10.1055/s-0034-1390030. [DOI] [PubMed] [Google Scholar]
- 25.Zallio F., et al. A single-center pilot prospective study of topical application of platelet-derived eye drops for patients with ocular chronic graft-versus-host disease. Biol. Blood Marrow Transplant. 2016;22(9):1664–1670. doi: 10.1016/j.bbmt.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 26.Everts P.A., et al. Differences in platelet growth factor release and leucocyte kinetics during autologous platelet gel formation. Transfus. Med. 2006;16(5):363–368. doi: 10.1111/j.1365-3148.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 27.Machlus K.R., Italiano J.E., Jr. The incredible journey: from megakaryocyte development to platelet formation. J. Cell Biol. 2013;201(6):785–796. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holinstat M. Normal platelet function. Cancer Metastasis Rev. 2017;36(2):195–198. doi: 10.1007/s10555-017-9677-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gremmel T., Frelinger A.L., 3rd, Michelson A.D. Platelet physiology. Semin. Thromb. Hemost. 2016;42(3):191–204. doi: 10.1055/s-0035-1564835. [DOI] [PubMed] [Google Scholar]
- 30.Levoux J., et al. Platelets facilitate the wound-healing capability of mesenchymal stem cells by mitochondrial transfer and metabolic reprogramming. Cell Metabol. 2021;33(2):283–299 e9. doi: 10.1016/j.cmet.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Chen E., et al. Platelet-derived respiratory-competent mitochondria transfer to mesenchymal stem cells to promote wound healing via metabolic reprogramming. Platelets. 2022;33(2):171–173. doi: 10.1080/09537104.2021.1961717. [DOI] [PubMed] [Google Scholar]
- 32.Xiang Y.Z., et al. Platelet activation, and antiplatelet targets and agents: current and novel strategies. Drugs. 2008;68(12):1647–1664. doi: 10.2165/00003495-200868120-00004. [DOI] [PubMed] [Google Scholar]
- 33.van der Meijden P.E.J., Heemskerk J.W.M. Platelet biology and functions: new concepts and clinical perspectives. Nat. Rev. Cardiol. 2019;16(3):166–179. doi: 10.1038/s41569-018-0110-0. [DOI] [PubMed] [Google Scholar]
- 34.Koupenova M., et al. Circulating platelets as mediators of immunity, inflammation, and thrombosis. Circ. Res. 2018;122(2):337–351. doi: 10.1161/CIRCRESAHA.117.310795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H.S., Chang H.H. Platelets in inflammation and immune modulations: functions beyond hemostasis. Arch. Immunol. Ther. Exp. 2012;60(6):443–451. doi: 10.1007/s00005-012-0193-y. [DOI] [PubMed] [Google Scholar]
- 36.Nurden A.T. The biology of the platelet with special reference to inflammation, wound healing and immunity. Front Biosci (Landmark Ed) 2018;23(4):726–751. doi: 10.2741/4613. [DOI] [PubMed] [Google Scholar]
- 37.Puhm F., Boilard E., Machlus K.R. Platelet extracellular vesicles: beyond the blood. Arterioscler. Thromb. Vasc. Biol. 2021;41(1):87–96. doi: 10.1161/ATVBAHA.120.314644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Estevez B., Du X. New concepts and mechanisms of platelet activation signaling. Physiology. 2017;32(2):162–177. doi: 10.1152/physiol.00020.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung H., Kang Y.Y., Mok H. Platelet-derived nanovesicles for hemostasis without release of pro-inflammatory cytokines. Biomater. Sci. 2019;7(3):856–859. doi: 10.1039/c8bm01480a. [DOI] [PubMed] [Google Scholar]
- 40.Liao W., et al. Exosomes: the next generation of endogenous nanomaterials for advanced drug delivery and therapy. Acta Biomater. 2019;86:1–14. doi: 10.1016/j.actbio.2018.12.045. [DOI] [PubMed] [Google Scholar]
- 41.Melki I., et al. Platelet microvesicles in health and disease. Platelets. 2017;28(3):214–221. doi: 10.1080/09537104.2016.1265924. [DOI] [PubMed] [Google Scholar]
- 42.Boilard E. Extracellular vesicles and their content in bioactive lipid mediators: more than a sack of microRNA. J. Lipid Res. 2018;59(11):2037–2046. doi: 10.1194/jlr.R084640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burnouf T., et al. Platelet microparticles: detection and assessment of their paradoxical functional roles in disease and regenerative medicine. Blood Rev. 2014;28(4):155–166. doi: 10.1016/j.blre.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Tao S.C., Guo S.C., Zhang C.Q. Platelet-derived extracellular vesicles: an emerging therapeutic approach. Int. J. Biol. Sci. 2017;13(7):828–834. doi: 10.7150/ijbs.19776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson J., et al. Prospective therapeutic applications of platelet extracellular vesicles. Trends Biotechnol. 2021;39(6):598–612. doi: 10.1016/j.tibtech.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Dyer M.R., et al. Platelet-derived extracellular vesicles released after trauma promote hemostasis and contribute to DVT in mice. J. Thromb. Haemostasis. 2019;17(10):1733–1745. doi: 10.1111/jth.14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcoux G., et al. Platelet-derived extracellular vesicles convey mitochondrial DAMPs in platelet concentrates and their levels are associated with adverse reactions. Transfusion. 2019;59(7):2403–2414. doi: 10.1111/trf.15300. [DOI] [PubMed] [Google Scholar]
- 48.Boilard E., Duchez A.C., Brisson A. The diversity of platelet microparticles. Curr. Opin. Hematol. 2015;22(5):437–444. doi: 10.1097/MOH.0000000000000166. [DOI] [PubMed] [Google Scholar]
- 49.Xie J., et al. Nanomaterial-based blood-brain-barrier (BBB) crossing strategies. Biomaterials. 2019;224 doi: 10.1016/j.biomaterials.2019.119491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franco A.T., Corken A., Ware J. Platelets at the interface of thrombosis, inflammation, and cancer. Blood. 2015;126(5):582–588. doi: 10.1182/blood-2014-08-531582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez E., et al. Platelet-derived- extracellular vesicles promote hemostasis and prevent the development of hemorrhagic shock. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-53724-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chimen M., et al. Appropriation of GPIbalpha from platelet-derived extracellular vesicles supports monocyte recruitment in systemic inflammation. Haematologica. 2020;105(5):1248–1261. doi: 10.3324/haematol.2018.215145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daneshmandi L., et al. Emergence of the stem cell secretome in regenerative engineering. Trends Biotechnol. 2020;38(12):1373–1384. doi: 10.1016/j.tibtech.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang B., et al. Mesenchymal stem cell-derived extracellular vesicles in tissue regeneration. Cell Transplant. 2020;29 doi: 10.1177/0963689720908500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng C., et al. vol. 2017. Oxid Med Cell Longev; 2017. (ROS: Crucial Intermediators in the Pathogenesis of Intervertebral Disc Degeneration). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shadel G.S., Horvath T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163(3):560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitaker R.M., et al. Mitochondrial biogenesis as a pharmacological target: a new approach to acute and chronic diseases. Annu. Rev. Pharmacol. Toxicol. 2016;56:229–249. doi: 10.1146/annurev-pharmtox-010715-103155. [DOI] [PubMed] [Google Scholar]
- 58.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505(7483):335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallace D.C., Fan W., Procaccio V. Mitochondrial energetics and therapeutics. Annu. Rev. Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finkel T. The metabolic regulation of aging. Nat. Med. 2015;21(12):1416–1423. doi: 10.1038/nm.3998. [DOI] [PubMed] [Google Scholar]
- 61.Picard M. Mitochondrial synapses: intracellular communication and signal integration. Trends Neurosci. 2015;38(8):468–474. doi: 10.1016/j.tins.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 62.Koch R.E., Josefson C.C., Hill G.E. Mitochondrial function, ornamentation, and immunocompetence. Biol. Rev. Camb. Phil. Soc. 2017;92(3):1459–1474. doi: 10.1111/brv.12291. [DOI] [PubMed] [Google Scholar]
- 63.Hickman T.T., Rathan-Kumar S., Peck S.H. Development, pathogenesis, and regeneration of the intervertebral disc: current and future insights spanning traditional to omics methods. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.841831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dan Dunn J., et al. Reactive oxygen species and mitochondria: a nexus of cellular homeostasis. Redox Biol. 2015;6:472–485. doi: 10.1016/j.redox.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graca A.L., et al. Therapeutic effects of platelet-derived extracellular vesicles in a bioengineered tendon disease model. Int. J. Mol. Sci. 2022;23(6) doi: 10.3390/ijms23062948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu L., Deng Q.J. Role of platelet-derived extracellular vesicles in traumatic brain injury-induced coagulopathy and inflammation. Neural Regen Res. 2022;17(10):2102–2107. doi: 10.4103/1673-5374.335825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rannou F., et al. Intervertebral disc degeneration: the role of the mitochondrial pathway in annulus fibrosus cell apoptosis induced by overload. Am. J. Pathol. 2004;164(3):915–924. doi: 10.1016/S0002-9440(10)63179-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steinberg G.R., Kemp B.E. AMPK in health and disease. Physiol. Rev. 2009;89(3):1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 69.Tang B.L. Sirt1 and the mitochondria. Mol. Cell. 2016;39(2):87–95. doi: 10.14348/molcells.2016.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang X., et al. The diabetes medication canagliflozin promotes mitochondrial remodelling of adipocyte via the AMPK-Sirt1-Pgc-1alpha signalling pathway. Adipocyte. 2020;9(1):484–494. doi: 10.1080/21623945.2020.1807850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y., et al. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor gamma coactivator 1alpha. Arthritis Rheumatol. 2015;67(8):2141–2153. doi: 10.1002/art.39182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang H., Ward W.F. PGC-1alpha: a key regulator of energy metabolism. Adv. Physiol. Educ. 2006;30(4):145–151. doi: 10.1152/advan.00052.2006. [DOI] [PubMed] [Google Scholar]
- 73.Kunkel G.H., Chaturvedi P., Tyagi S.C. Mitochondrial pathways to cardiac recovery: TFAM. Heart Fail. Rev. 2016;21(5):499–517. doi: 10.1007/s10741-016-9561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin W., et al. Advantages of pure platelet-rich plasma compared with leukocyte- and platelet-rich plasma in promoting repair of bone defects. J. Transl. Med. 2016;14:73. doi: 10.1186/s12967-016-0825-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y., et al. Exosomes derived from platelet-rich plasma administration in site mediate cartilage protection in subtalar osteoarthritis. J. Nanobiotechnol. 2022;20(1):56. doi: 10.1186/s12951-022-01245-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaib S., et al. Role of mitochondrial membrane potential and lactate dehydrogenase A in apoptosis. Anti Cancer Agents Med. Chem. 2022;22(11):2048–2062. doi: 10.2174/1871520621666211126090906. [DOI] [PubMed] [Google Scholar]
- 77.Kalpage H.A., et al. Tissue-specific regulation of cytochrome c by post-translational modifications: respiration, the mitochondrial membrane potential, ROS, and apoptosis. FASEB J. 2019;33(2):1540–1553. doi: 10.1096/fj.201801417R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elefantova K., et al. Detection of the mitochondrial membrane potential by the cationic dye JC-1 in L1210 cells with massive overexpression of the plasma membrane ABCB1 drug transporter. Int. J. Mol. Sci. 2018;19(7) doi: 10.3390/ijms19071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perelman A., et al. JC-1: alternative excitation wavelengths facilitate mitochondrial membrane potential cytometry. Cell Death Dis. 2012;3:e430. doi: 10.1038/cddis.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wan Y., et al. Aptamer-conjugated extracellular nanovesicles for targeted drug delivery. Cancer Res. 2018;78(3):798–808. doi: 10.1158/0008-5472.CAN-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim H.S., et al. Cell-membrane-derived nanoparticles with notch-1 suppressor delivery promote hypoxic cell-cell packing and inhibit angiogenesis acting as a two-edged sword. Adv. Mater. 2021;33(40):e2101558. doi: 10.1002/adma.202101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Geeurickx E., et al. The generation and use of recombinant extracellular vesicles as biological reference material. Nat. Commun. 2019;10(1):3288. doi: 10.1038/s41467-019-11182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luan X., et al. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol. Sin. 2017;38(6):754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jhan Y.Y., et al. Engineered extracellular vesicles with synthetic lipids via membrane fusion to establish efficient gene delivery. Int. J. Pharm. 2020;573 doi: 10.1016/j.ijpharm.2019.118802. [DOI] [PubMed] [Google Scholar]
- 85.Grangier A., et al. Technological advances towards extracellular vesicles mass production. Adv. Drug Deliv. Rev. 2021;176 doi: 10.1016/j.addr.2021.113843. [DOI] [PubMed] [Google Scholar]
- 86.Pokrovskaya I.D., et al. 3D ultrastructural analysis of alpha-granule, dense granule, mitochondria, and canalicular system arrangement in resting human platelets. Res Pract Thromb Haemost. 2020;4(1):72–85. doi: 10.1002/rth2.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miwa S., et al. Mitochondrial dysfunction in cell senescence and aging. J. Clin. Invest. 2022;132(13) doi: 10.1172/JCI158447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Habiballa L., Salmonowicz H., Passos J.F. Mitochondria and cellular senescence: implications for musculoskeletal ageing. Free Radic. Biol. Med. 2019;132:3–10. doi: 10.1016/j.freeradbiomed.2018.10.417. [DOI] [PubMed] [Google Scholar]
- 89.Zhang M., et al. Conscription of immune cells by light-activatable silencing NK-derived exosome (LASNEO) for synergetic tumor eradication. Adv. Sci. 2022;9(22):e2201135. doi: 10.1002/advs.202201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong H., et al. SERPINA1 is a hub gene associated with intervertebral disc degeneration grade and affects the nucleus pulposus cell phenotype through the ADIRF-AS1/miR-214-3p axis. Transl. Res. 2022;245:99–116. doi: 10.1016/j.trsl.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Mobasheri A., et al. The role of metabolism in the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2017;13(5):302–311. doi: 10.1038/nrrheum.2017.50. [DOI] [PubMed] [Google Scholar]
- 92.Liu X., et al. Fusogenic reactive oxygen species triggered charge-reversal vector for effective gene delivery. Adv. Mater. 2016;28(9):1743–1752. doi: 10.1002/adma.201504288. [DOI] [PubMed] [Google Scholar]
- 93.Canto C., Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 2009;20(2):98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gasecka A., et al. Platelet extracellular vesicles as biomarkers for arterial thrombosis. Platelets. 2017;28(3):228–234. doi: 10.1080/09537104.2016.1254174. [DOI] [PubMed] [Google Scholar]
- 95.Wu J., et al. Platelet-rich plasma-derived extracellular vesicles: a superior alternative in regenerative medicine? Cell Prolif. 2021;54(12) doi: 10.1111/cpr.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li H., Slone J., Huang T. The role of mitochondrial-related nuclear genes in age-related common disease. Mitochondrion. 2020;53:38–47. doi: 10.1016/j.mito.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 97.Kang L., et al. Restoration of autophagic flux rescues oxidative damage and mitochondrial dysfunction to protect against intervertebral disc degeneration. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/7810320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khan N.M., et al. Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human Osteoarthritis chondrocytes. Free Radic. Biol. Med. 2017;106:288–301. doi: 10.1016/j.freeradbiomed.2017.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liebel F., et al. Irradiation of skin with visible light induces reactive oxygen species and matrix-degrading enzymes. J. Invest. Dermatol. 2012;132(7):1901–1907. doi: 10.1038/jid.2011.476. [DOI] [PubMed] [Google Scholar]
- 100.Fearon U., et al. Hypoxia, mitochondrial dysfunction and synovial invasiveness in rheumatoid arthritis. Nat. Rev. Rheumatol. 2016;12(7):385–397. doi: 10.1038/nrrheum.2016.69. [DOI] [PubMed] [Google Scholar]
- 101.Karbowski M., Neutzner A. Neurodegeneration as a consequence of failed mitochondrial maintenance. Acta Neuropathol. 2012;123(2):157–171. doi: 10.1007/s00401-011-0921-0. [DOI] [PubMed] [Google Scholar]
- 102.Liu-Bryan R., Terkeltaub R. Emerging regulators of the inflammatory process in osteoarthritis. Nat. Rev. Rheumatol. 2015;11(1):35–44. doi: 10.1038/nrrheum.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang C., et al. Allicin regulates energy homeostasis through Brown adipose tissue. iScience. 2020;23(5) doi: 10.1016/j.isci.2020.101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao X., et al. Peroxisome proliferator-activated receptor gamma coactivator 1alpha and FoxO3A mediate chondroprotection by AMP-activated protein kinase. Arthritis Rheumatol. 2014;66(11):3073–3082. doi: 10.1002/art.38791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Price N.L., et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metabol. 2012;15(5):675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matsushita T., et al. The overexpression of SIRT1 inhibited osteoarthritic gene expression changes induced by interleukin-1beta in human chondrocytes. J. Orthop. Res. 2013;31(4):531–537. doi: 10.1002/jor.22268. [DOI] [PubMed] [Google Scholar]
- 107.Moon M.H., et al. SIRT1, a class III histone deacetylase, regulates TNF-alpha-induced inflammation in human chondrocytes. Osteoarthritis Cartilage. 2013;21(3):470–480. doi: 10.1016/j.joca.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 108.Zheng Z., et al. Ginsenoside Rb1 reduces H2O2induced HUVEC dysfunction by stimulating the sirtuin1/AMPactivated protein kinase pathway. Mol. Med. Rep. 2020;22(1):247–256. doi: 10.3892/mmr.2020.11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Antich-Rossello M., et al. Platelet-derived extracellular vesicles for regenerative medicine. Int. J. Mol. Sci. 2021;22(16) doi: 10.3390/ijms22168580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gomes F.G., et al. Synergy of human platelet-derived extracellular vesicles with secretome proteins promotes regenerative functions. Biomedicines. 2022;10(2) doi: 10.3390/biomedicines10020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.