Abstract
Type 1 fimbriae of Salmonella enterica serovar Typhimurium are surface appendages that carry adhesins specific for mannosylated host glycoconjugates. Regulation of the major fimbrial subunit is thought to be controlled by a number of ancillary fim genes, including fimZ, fimY, fimW, and fimU. Previous studies using a FimZ mutant have indicated that this protein is necessary for fimA expression, and in vitro DNA binding assays determined that FimZ is a transcriptional activator that binds directly to the fimA promoter. To determine the role of FimY as a potential regulator of fimbrial expression, a fimY mutant of serovar Typhimurium was generated by allelic exchange. This mutant was found to be phenotypically nonfimbriate. No transcription from the fimA promoter was detected in a fimY mutant containing a fimA-lacZ reporter construct located on the chromosome. In addition, transcription from the cloned fimY promoter was not detected in Escherichia coli unless both FimZ and FimY were present, indicating that these proteins also act as coactivators of fimY expression. Consistent with these results, there is no transcription from a fimY-lacZ reporter construct within a serovar Typhimurium fimY or fimZ mutant. Studies using the fimY-lacZ construct reveal that expression of this gene varies with environmental conditions in a manner similar to fimA expression. Extensive in vitro DNA binding assays using extracts from E. coli that overexpress FimY, as well as partially purified FimY, were unable to identify a specific interaction between FimY and the fimA or fimY promoter. The results indicate that FimY is a positive regulator of fimbrial expression and that this protein acts in cooperation with FimZ to regulate the expression of Salmonella type 1 fimbrial appendages.
Type 1 fimbriae are bacterial adhesins characterized by their ability to mediate mannose-sensitive binding to eukaryotic cells in vitro. These fimbriae are common adherence factors expressed by both Escherichia coli and Salmonella and have been detected on many other members of the family Enterobacteriaceae (7, 16, 35). Numerous studies of E. coli have established the importance of type I fimbriae as virulence factors during urinary tract infections (2, 13, 30, 40). In Salmonella, these appendages have been implicated in initiating intestinal colonization, and they may contribute to tissue tropism by adhering to specific mannosylated host proteins (3, 20, 37, 51). In addition, type 1 fimbriae of Salmonella are known to mediate binding to a number of human epithelial cell lines in vitro (4, 19, 29, 32, 51). The phenotypic expression of type 1 fimbriae is phase variable, allowing a transition between fimbriate and nonfimbriate phenotypes (1, 17, 21). Serial subculturing of bacteria in static liquid broth has been reported to select for highly fimbriate bacteria, while growth on solid media selects for poorly fimbriate bacteria (15, 42). Fimbrial phase variation in E. coli is due, in part, to inversion of a 314-bp DNA element found upstream of the gene encoding the major fimbrial subunit, fimA (1). This inversion event requires the action of two site-specific recombinases, fimB and fimE, located upstream of the fim structural genes (22, 33).
In Salmonella enterica serovar Typhimurium, variation of type 1 fimbrial expression appears to occur through a mechanism distinct from that described in E. coli (10). Regardless of the fimbrial phenotype, the fimA promoter region was found to be oriented in the direction that would promote fimA transcription (10). In addition, fimbriate E. coli strains lysogenized with a serovar Typhimurium λfimA lacZ fusion produce no detectable β-galactosidase activity, indicating that E. coli Fim proteins do not activate serovar Typhimurium fimA expression (47). Four genes, located within the serovar Typhimurium fim gene cluster, have been implicated as regulators of fimA expression (47). The gene products from two of these genes, fimZ and fimW, exhibit a relatedness to prokaryotic transcriptional regulators, and one, fimU, encodes an arginine tRNA molecule that is known to effect both serovar Typhimurium and Salmonella enterica serovar Enteritidis type 1 fimbrial expression (12, 50). There are no apparent homologues to the E. coli recombinases FimB and FimE within the serovar Typhimurium gene cluster, and no genes for regulatory proteins related to FimZ, -Y, or -W have been found within the E. coli fim gene cluster (48).
A serovar Typhimurium fimZ mutant was constructed previously and found to be phenotypically nonfimbriate. In addition, this mutant demonstrated significantly decreased levels of fimA expression when compared to the parental strain (54). The FimZ protein was partially purified and found to bind to the promoter region of fimA, approximately 100 bp upstream of the transcription initiation site. Amino acid sequence analysis revealed that FimZ is related to a number of transcriptional activators, including BvgA, a response regulator of a two-component system in Bordetella pertussis that activates several virulence factors in that organism (14). Similar to FimZ, the C-terminal amino acid sequence of FimY appears to contain a helix-turn-helix DNA binding motif, yet examination of the entire FimY sequence identifies very limited homology to known prokaryotic proteins. Previously we have demonstrated that both FimZ and FimY are necessary for fimA expression in a recombinant E. coli host (54). Here we report the construction and characterization of a fimY mutant in serovar Typhimurium. The nonfimbriate phenotype of this mutant and the location of the FimY gene within the fim gene cluster on the chromosome, as well as the requirement for a functional FimY to mediate fimA expression, imply the involvement of this protein in fimbrial regulation. To further define the role of FimY, a fimY-lacZ reporter was constructed. Similar to expression of fimA, fimY expression requires the presence of both FimY and FimZ. In addition, expression of fimY is increased under environmental conditions that also promote fimA expression. Attempts to identify a FimY binding site on the fimA promoter region using in vitro DNA binding assays were unsuccessful, suggesting that other Salmonella proteins may be necessary for the action of FimY. The results reported here support the model in which serovar Typhimurium fimA expression requires both FimY and FimZ and these proteins are essential components of the regulatory cascade involved in fimbrial production.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
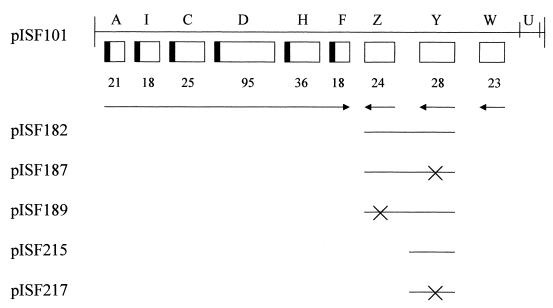
The strains and recombinant molecules used in this study are shown in Table 1. The fimbriate strain serovar Typhimurium LB5010 (8) was used to construct the fimY mutant, LBY100. The mutation was subsequently introduced into the strongly fimbriate and invasive serovar Typhimurium strain SL1344 by P22 transduction using lysates of serovar Typhimurium LBY100 (44). Serovar Typhimurium IS145 is a λfimA lacZ lysogen used as a single-copy reporter of fimA expression, and its construction has been described previously (47). Construction and characterization of the fimZ mutant LBZ100 has been reported previously (54). The fimZ mutation was also transduced into serovar Typhimurium SL1344 to generate the strain SL1344JTZ. All strains were cultured on Luria-Bertani (LB) medium and incubated at 37°C, or 30°C for lysogens, for 24 or 48 h. Plasmids were prepared by standard techniques, and manipulation of recombinant DNA was performed using conventional procedures (44). All plasmids used in this study are derivatives of pISF101 carrying the serovar Typhimurium fim gene cluster cloned into pACYC184 (New England Biolabs, Beverly, Mass.), as shown in Fig. 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant features | Reference or source |
|---|---|---|
| Serovar Typhimurium | ||
| LB5010 | Wild type; fimbriate with complete fim gene cluster | 8 |
| LBY100 | LB5010 fimY::kan Kanr | This study |
| ISF145 | LB5010 λfimA lacZ lysogen | 47 |
| ISF145Y | LBY100 λfimA lacZ lysogen | This study |
| SL1344 | Wild type; fimbriate with complete fim gene cluster | 28 |
| SL1344JTY | SL1344 fimY::kan Kanr | This study |
| SL1344JTZ | SL1344 fimZ::kan Kanr | This study |
| E. coli | ||
| SY327 | Host for suicide vector pGP704 | 39 |
| JM109 | Host for fimY lacZ reporter plasmids and FimY expression plasmid; Δ(lacZ) | 53 |
| Plasmids | ||
| pISF101 | fimAICDHFZYWU | 49 |
| pISF145 | fimA-lacZ reporter fusion | 47 |
| pISF182 | fimZ and fimY cloned into pACYC184; FimZ+ FimY+ | 54 |
| pISF187 | pISF182 with translation terminator inserted into fimY; FimZ+ FimY− | 54 |
| pISF189 | pISF182 with translation terminator inserted into fimZ; FimZ− FimY+ | 54 |
| pISF215 | fimY cloned into pACYC184; FimY+ | This study |
| pISF217 | pISF215 with translation terminator inserted into fimY; FimY− | This study |
| pISF234 | fimY-lacZ reporter fusion | This study |
| pISF237 | Single-copy fimY-lacZ reporter fusion | This study |
| pISF241 | fimY-malE fusion for FimY purification | This study |
FIG. 1.
Genetic organization of the Salmonella fim gene cluster. The sizes of the polypeptides encoded by the genes are shown below the boxes. fimA (A) is the gene encoding the major fimbrial subunit, whereas fimZ (Z) and fimY (Y) are those described in the text. The arrows indicate the direction of transcription as determined by S1 nuclease mapping for fimA, primer extension analysis for fimZ and fimW (W), and sequence analysis for fimY. The derivatives of pISF101 utilized in this study are indicated below the map, with solid lines representing the DNA retained by each derivative. For pISF187, -189, and -217, the crosses indicate the locations of the inserted translation terminators.
The construction of plasmids pISF182, pISF187, and pISF189 has been described previously (54). The plasmid pISF215 possesses only the fimY gene of the fim gene cluster and was constructed following digestion of pISF101 with BamHI and BglII and religation to remove all fim genes except fimY. The plasmid pISF217 was constructed by insertion of a universal translation terminator into a unique EcoRV site within fimY on pISF215. The fimA-lacZ (pISF145) and fimY-lacZ (pISF234) multicopy reporter constructs were generated by ligating a PCR product of the respective promoter regions into the promoterless lacZ vector, pMC1403 (9). Single-copy lacZ reporters were constructed using an ampicillin-resistant derivative of the single-copy pDF41 plasmid ligated to the promoterless lacZ, -Y, and -A genes from pMC1403 (25). This plasmid, designated pGS375 (kindly supplied by George Stauffer, University of Iowa) was digested with EcoRI and BamHI and ligated to a PCR product of the fimY promoter region to generate a single-copy fimY-lacZ reporter (pISF237). All plasmids were sequenced through the fusion to confirm the fidelity of the construct.
Detection of type 1 fimbriae.
Bacteria were serially subcultured in 10 ml of LB broth and incubated without shaking for 48 h to select for highly fimbriate cultures. The cells were harvested by centrifugation and gently resuspended in the residual fluid as described previously (15, 41). Subsequently, 50 μl of bacterial suspension was mixed with 50 μl of a 3% (vol/vol) suspension of guinea pig erythrocytes in phosphate-buffered saline. Mannose-sensitive hemagglutination was determined by incubation of the bacterial suspension with cells resuspended in phosphate-buffered saline containing 3% (wt/vol) α-methyl-d-mannoside. The mannose-sensitive adhesin was considered to be present if the red blood cells agglutinated only in the absence of mannose within 1 min. Fimbrial antigens were detected using monospecific serovar Typhimurium antifimbrial serum as described previously (26). The titers of the hemagglutination and antibody agglutination reactions were determined as the reciprocal of the highest bacterial or serum dilution resulting in hemagglutination or bacterial agglutination, respectively, and they are described in detail elsewhere (11). For transmission electron microscopy, aliquots of 48-h bacterial suspensions were placed on carbon-coated grids and stained for 1 min with phosphotungstic acid before visualization at 50,000× or 20,000× with a Hitachi H-600 electron microscope (24).
Construction of the serovar Typhimurium fimY mutant.
The plasmid pIS182, which possesses an intact fimY gene (Fig. 1), was linearized at a unique EcoRV site within fimY. A HincII digest of a DNA cassette containing a kanamycin resistance determinant, isolated from the plasmid pUC4K (Pharmacia, Piscataway, N.J.), was prepared and subsequently ligated into the fimY gene at the EcoRV site. Following isolation of kanamycin-resistant E. coli HB101 (6) transformants, the plasmid carrying the insertionally inactive fimY gene was isolated by standard techniques (44). The disrupted fimY determinant was then cloned into the suicide vector pGP704 (kindly supplied by John Mekalanos, Harvard Medical School) and maintained in the permissive E. coli host, SY327 (39). Recombinant DNA was prepared from kanamycin- and ampicillin-resistant transformants and analyzed by restriction digestions. The appropriate construct was then introduced into serovar Typhimurium LB5010, and kanamycin-resistant but ampicillin-sensitive transformants were selected. Further analysis of putative fimY mutants was completed by Southern hybridization using random-primed dUTP-labeled DNA probes (Genius kit; Boehringer Mannheim, Indianapolis, Ind.) specific for the fimY gene or the kanamycin-resistant determinant. Chromosomal DNA was digested to completion with BglII and transferred to nitrocellulose. All hybridizations were performed under high-stringency conditions as described elsewhere (24).
β-Galactosidase assays.
Assays for β-galactosidase were performed in triplicate by the method of Miller (38), using the chloroform-sodium dodecyl sulfate lysis procedure, and λfimA lacZ lysogens or fimA lacZ and fimY lacZ plasmid transformants. The strains were grown on LB agar for 24 h or in static liquid LB broth for 48 h before analysis. Subcultures were performed by transferring one loopful of cells to a second 10-ml broth culture or picking one colony and replating. All assays were performed independently at least twice with less than 20% variability.
Partial purification of the FimY–maltose-binding protein fusion and gel mobility shift assays.
The plasmid pISF241 was used to purify a FimY–maltose-binding protein fusion. pISF241 was constructed from the vector pMal-c2 (New England Biolabs), which contains the β-galactosidase coding region fused to the maltose-binding protein of E. coli. The β-galactosidase gene was removed by digestion with BamHI and PstI, and the remaining vector was ligated to a PCR product of the fimY coding region digested with BamHI and PstI. The resulting construct was confirmed by sequencing it through the junction. pISF241 was introduced into E. coli JM109 and grown at room temperature to an optical density at 600 nm of ∼0.5 before induction with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The culture was allowed to grow for an additional 12 h at room temperature before the cells were collected and harvested by sonication and resuspended in column buffer (20 mM Tris-HCl, 200 mM NaCl, 1 mM EDTA). The FimY–maltose-binding protein fusion was separated from the crude extract by binding to an amylose-agarose bead resin and eluted from the resin by washing with column buffer plus 10 mM maltose, according to the manufacturer's instructions.
Gel mobility shift assays were performed with various concentrations of the above-described FimY preparation as well as various concentrations of crude extracts from E. coli JM109 transformed with pISF215 or pISF217 (described above). The preparation of the 452-bp fimA promoter region used as target DNA has been described elsewhere (49). A 564-bp DNA fragment containing the fimY promoter was generated by PCR. End labeling was performed by removing the 5′ phosphate from the promoter fragments with calf intestine alkaline phosphatase and then incubating the fragments with T4 polynucleotide kinase and [γ-32P]ATP. Assays were performed by standard techniques (23), except that 0.25 μg of unlabeled single-stranded sperm carrier DNA was added to each incubation mixture and no bovine serum albumin was added. The DNA was subsequently mixed with appropriate twofold dilutions (up to 5 μg) of FimY protein extracts, and all volumes were adjusted with sterile distilled water. The samples were loaded onto a nondenaturing polyacrylamide gel and electrophoresed at 200 V. The mobilities of the DNA fragments were analyzed by autoradiography. In all experiments, the concentration of protein was determined by the use of a commercially available Bradford protein assay kit (Pierce, Rockford, Ill.).
RESULTS
Construction of the fimY mutant of serovar Typhimurium LB5010.
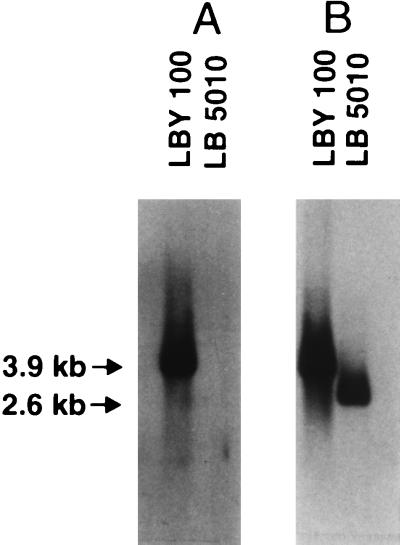
The fimY mutant, serovar Typhimurium LBY100, was constructed following transformation of serovar Typhimurium LB5010 with the suicide vector, pGP704, carrying an insertionally inactive fimY gene. Kanamycin-resistant and ampicillin-sensitive bacteria that had retained the inactivated gene but lost the plasmid vector were isolated and further analyzed. Genomic DNA was prepared from both the parental and the mutant strains and used in Southern hybridization analysis to confirm the location of the mutated allele (Fig. 2). Genomic preparations were restricted with BglII and hybridized to a 1,300-bp DNA probe possessing the kanamycin resistance gene. In addition, the restricted DNA was probed with a 470-bp DNA fragment comprising nucleotides of the fimY gene itself. The probe possessing the resistance determinant hybridized to a 3.9-kb DNA fragment found only in serovar Typhimurium LBY100, and no sequences homologous to the probe were detected in the parental strain. The size of this fragment is consistent with replacement of the parental allele with the fimY mutation. The fimY DNA probe hybridized to a 4.0-kb BglII DNA fragment from serovar Typhimurium LBY100 and a 2.7-kb fragment from serovar Typhimurium LB5010. The sizes of these fragments are consistent with insertion of the 1.3-kb kanamycin resistance cassette, which lacks a BglII restriction site, into the chromosome of serovar Typhimurium LBY100 and replacement of the intact fimY gene by allelic exchange. Confirmation of the location of the mutant allele was performed by additional restriction analysis using several endonucleases.
FIG. 2.
Southern hybridization profiles of genomic DNA isolated from serovar Typhimurium LB5010 (wild type) and LBY100 (fimY). (A) DNA digested with BglII and probed with sequences from the Kanr cassette. (B) DNA digested with BglII and probed with a fimY gene probe. The sizes of the DNA fragments are as shown.
Characterization of the fimY mutant of serovar Typhimurium LB5010.
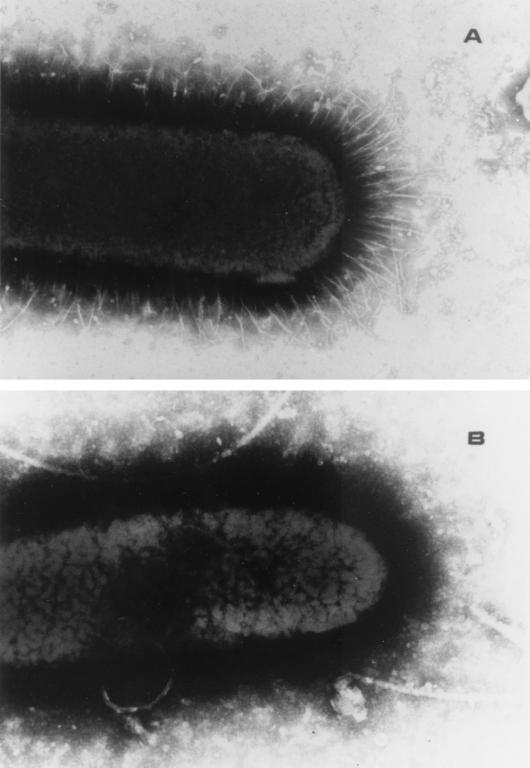
The ability of the fimY mutant to mediate mannose-sensitive hemagglutination of guinea pig erythrocytes was investigated. Serovar Typhimurium LBY100 was grown under optimal conditions for the expression of type I fimbriae, and unlike the parental strain, the bacteria were unable to mediate hemagglutination even after multiple subcultures in static liquid broth. In addition, the strain was examined for its reactivity with a fimbria-specific antiserum and observed under the transmission electron microscope. These results are summarized in Table 2, and they demonstrate that serovar Typhimurium LBY100 does not express surface-associated type 1 fimbriae under conditions that normally promote the expression of these appendages. Serovar Typhimurium LBY100 was never observed to express type 1 fimbriae on its surface regardless of culture conditions. Figure 3 shows a transmission electron micrograph of the nonfimbriate LBY100 fimY mutant and the fimbriate LB5010 parental strain after 48 h of growth in static liquid broth.
TABLE 2.
Phenotypic expression of type 1 fimbriae by serovar Typhimurium
| Strain | Plasmid (relevant genotype) | Serum titera | Slide agglutinationb | Hemagglutination titerc | Presence of fimbriae on bacteriad |
|---|---|---|---|---|---|
| LB5010 | None | 80 | + | 8 | + |
| LBY100 | None | <20 | − | <2 | − |
| pISF215 (fimY+) | 640 | + | 32 | ND | |
| pISF217 (fimY) | <20 | − | <2 | ND | |
| SL1344 | None | 2,560 | + | 16 | + |
| SL1344JTY | None | 20 | − | <2 | − |
| pISF215 (fimY+) | 10,240 | + | 32 | ND |
Reciprocal of the highest serum dilution causing bacterial agglutination.
Agglutination by fimbria-specific polyclonal antisera observed after 60 s. +, present; −, absent.
Reciprocal of the highest bacterial dilution causing hemagglutination.
Fimbriae observed by transmission electron microscopy. +, present; −, absent ND, not done.
FIG. 3.
Transmission electron micrographs of the fimbriate serovar Typhimurium LB5010 parental strain (A) and the nonfimbriate serovar Typhimurium LBY100 fimY mutant (B). Magnification, ×32,000.
To insure that insertion of the kanamycin cassette on the chromosome did not result in abrogation of fimZ expression, serovar Typhimurium LBY100 was transformed with a plasmid carrying the fimY gene alone (pISF215) as well as a plasmid carrying a mutation in the fimY gene (pISF217). The pISF215 plasmid was able to restore type 1 fimbrial expression in the fimY mutant, as evidenced by hemagglutination and reactivity with fimbria-specific antiserum. Unlike the parental strain, expression of fimbriae in the pISF215 transformant was constitutive and occurred under all conditions, most likely due to the high level of FimY produced by the gene carried on the multicopy plasmid. The gene carried on pISF217 was not able to restore the fimbrial phenotype to serovar Typhimurium LBY100, as summarized in Table 2. The plasmid pISF215 could not restore fimbriation to a previously characterized strain, serovar Typhimurium LBZ100 (54), that carries a fimZ mutation on its chromosome.
The fimY mutation of serovar Typhimurium LBY100 was introduced, by P22 phage transduction, into a second strain of serovar Typhimurium, SL1344. This strain, designated SL1344JTY, was also found to be nonfimbriate even after serial subcultures in static broth and could be complemented by transformation with a functional fimY gene (Table 2).
Expression of β-galactosidase by a fimA-lacZ reporter in serovar Typhimurium LBY100.
The serovar Typhimurium λfimA lacZ lysogen, which has been described previously (47), was used as a source of recombinant phage to generate a λfimA lacZ lysogen of the LBY100 mutant. Table 3 shows the results of β-galactosidase expression by the serovar Typhimurium LBY100 lysogen grown under conditions normally favoring optimal fimbrial expression or on solid medium, which is known to select for poorly fimbriate bacteria. There was no detectable β-galactosidase activity by the fimY mutant regardless of the conditions of growth. In contrast, the parental strain of serovar Typhimurium exhibited previously reported levels of enzyme expression consistent with a fimbriate strain when grown in broth, compared to lower but detectable levels of expression when grown on agar (47). Transformation of the serovar Typhimurium LBY100 lysogen with pISF215, carrying a functional copy of fimY, resulted in constitutively high levels of fimA expression regardless of the conditions of growth. Transformation of the lysogen with plasmids carrying a nonfunctional fimY gene (pISF217) resulted in strains that did not produce detectable β-galactosidase activity. Even the multicopy plasmid carrying the fimA-lacZ fusion (47) did not express detectable levels of β-galactosidase when introduced into serovar Typhimurium LBY100, regardless of the conditions of culture (data not shown).
TABLE 3.
Expression of β-galactosidase by λfimA-lacZ reporter fusions
| Strain | Plasmid (relevant genotype) | β-galactosidase production by bacteria growna:
|
|
|---|---|---|---|
| On agar | In broth | ||
| Serovar Typhimurium | |||
| ISF145 | None | 12.1 ± 3.5 | 40.5 ± 1.3 |
| ISF145Y | None | 0 | 0 |
| pISF215 (fimY+) | 448.0 ± 10.5 | 483.3 ± 28.0 | |
| pISF217 (fimY) | 0 | 0 | |
| E. coli λfimA lacZ | pISF187 (fimZ+) | ND | 0 |
| pISF241 (MBP-fimY)b | ND | 0 | |
| pISF187 + pISF241 | ND | 145 ± 6.0 | |
β-Galactosidase activity is reported in Miller (38) units, and the data represent the mean ± standard deviation for one culture assayed in triplicate. All assays were performed independently at least twice with <20% variability. ND, not done.
pISF241 encodes the maltose-binding protein (MBP)–FimY fusion.
Expression of β-galactosidase from the fimYlacZ reporter in E. coli and serovar Typhimurium.
To investigate the level of fimY expression, a 564-bp PCR fragment containing the fimY promoter region was fused to a promoterless lacZ gene carried on a single-copy replicon. This plasmid, designated pISF237, was used to investigate fimY expression in an E. coli host lacking genes that affect Salmonella fim expression, as shown in Table 4. No expression was observed when this fusion was introduced by itself into E. coli JM109, indicating that no E. coli proteins are able to independently activate expression of the serovar Typhimurium fimY gene. The addition of plasmids carrying a functional fimZ or fimY gene (pISF187 or pISF189, respectively) into E. coli carrying the fimY-lacZ fusion did not significantly increase fimY expression. However, pISF182, carrying both fimZ and fimY, resulted in a 70-fold increase in fimY expression. We have previously reported that fimA expression is dependent on the presence of both FimZ and FimY in a similar manner (54). Similar results were obtained when a multicopy fimY-lacZ fusion was introduced into E. coli and compared to strains transformed with pISF187, pISF189, and pISF182 (data not shown).
TABLE 4.
Expression of β-galactosidase by fimY-lacZ reporter constructs in E. coli and serovar Typhimurium
| Strain | Plasmid (relevant genotype) | β-Galactosidase expressiona |
|---|---|---|
| E. coli JM109 | pISF237 (fimY lacZ) | 0 |
| + pISF187 (fimZ+ fimY)b | 0 | |
| +pISF189 (fimZ fimY+) | 0 | |
| + pISF182 (fimZ+ fimY) | 69.4 ± 1.1 | |
| Serovar Typhimurium | ||
| SL1344 | pISF237 | 52.1 ± 2.3 |
| SL1344JTY | pISF237 | 0 |
| SL1344JTZ | pISF237 | 0 |
β-Galactosidase activity is reported in Miller (38) units, and the data represent the mean ± standard deviation for one culture assayed in triplicate. All experiments were performed independently at least three times with ≤20% variation.
pISF237 plus pISF187.
The requirement for the presence of both FimY and FimZ to activate fimY expression was confirmed by analysis using the fimY-lacZ reporter in serovar Typhimurium SL1344 and the SL1344 fimY and fimZ mutants. As shown in Table 4, detectable levels of β-galactosidase are expressed when serovar Typhimurium SL1344, with the fimY-lacZ reporter, is cultured in broth for 48 h. In contrast, there is no expression from the fimY promoter in a fimY or fimZ background. These strains were also transformed with the multicopy fimY-lacZ fusion, and a consistent pattern of expression was observed (data not shown).
Analysis of fimA and fimY expression after multiple serial subcultures.
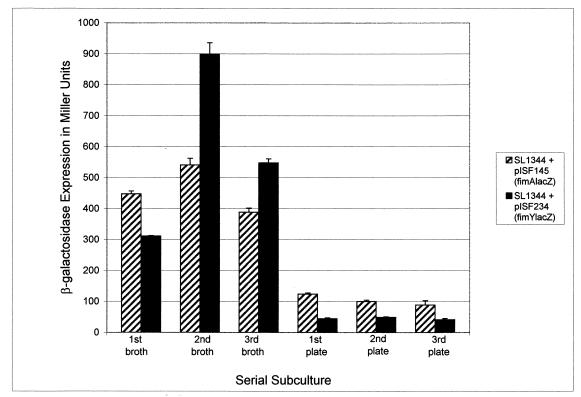
To determine if high levels of fimY expression correlate with high levels of fimA expression under conditions that select for strongly fimbriate bacteria, serial liquid subcultures of serovar Typhimurium SL1344, containing the fimA-lacZ or fimY-lacZ fusion, were analyzed. As shown in Fig. 4, three successive 48-h subcultures in broth resulted in an increase in fimY and fimA expression, as detected by the β-galactosidase assay. On the third subculture, the strains were transferred from broth onto agar plates and assayed after three successive 24-h subcultures. Both fimA and fimY expression dropped sharply after plating onto solid medium. This experiment was performed multiple times with a consistent decrease (≥3-fold) in gene expression from both fusions when the strains were transferred to solid medium. The difference between expression levels in broth and those observed for agar-grown bacteria was always greater for fimY, suggesting that this gene may be more responsive to environmental signals. These results are consistent with the function of FimY as an activator of fimA expression. In addition, the ability of these cultures to decrease expression of fimA following overnight growth on agar correlates with the change in fimbrial phenotype and indicates that this change occurs, at least in part, as a result of differential transcription of fimY and fimA.
FIG. 4.
fimA-lacZ (pISF145) and fimY-lacZ (pISF234) reporter plasmids were transformed into serovar Typhimurium SL1344 and assayed for β-galactosidase activity after multiple serial subcultures. The results represent the mean + standard deviation for one series assayed in triplicate.
Partial purification of FimY for use in in vitro DNA binding assays.
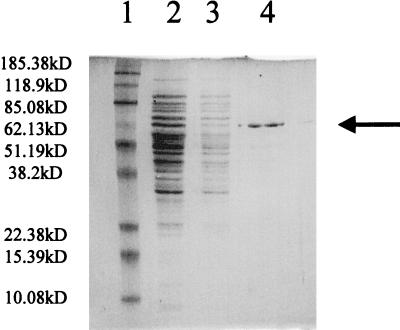
FimY was partially purified by construction of a fusion with the E. coli maltose-binding protein and separation of this fusion protein from crude extracts on an amylose-agarose bead resin. Figure 5 shows the sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the resulting protein extract after elution from the resin. The protein eluted from the column was approximately 70.5 kDa, consistent with fusion of the 27.8-kDa FimY protein and the 42.7 kDa maltose-binding protein. Up to 5 μg of this extract was combined with radiolabeled fimA or fimY containing promoter DNA fragments in gel mobility shift assays, and no altered mobility was observed compared to controls. However, the plasmid (pISF241) used to express the FimY fusion protein was introduced into serovar Typhimurium LBY100 and restored the ability of the mutant to mediate hemagglutination and express fimbriae. Also, in the presence of FimZ encoded by pISF187, the FimY fusion could activate expression of the fimA reporter constructs (Table 3). These results indicated that the N-terminal fusion with the maltose-binding protein did not severely alter the functional activity of FimY in vivo. Additional DNA binding assays were performed using crude extracts generated from E. coli transformed with pISF215(fimY+) or pISF217(fimY). These assays were also unable to detect a specific interaction between the fim promoters and FimY.
FIG. 5.
SDS-PAGE of partially purified FimY–maltose-binding protein fusion. Lane 1, molecular mass standards; lane 2, amylose resin column flowthrough; lane 3, amylose resin wash; lane 4, 5 μg of FimY fusion eluate (≈70.5 kDa). Arrow indicates FimY–maltose-binding protein.
DISCUSSION
Previous studies using recombinant E. coli strains have indicated that the presence of two regulatory proteins, FimZ and FimY, is necessary for activation of the serovar Typhimurium fimA gene (54). These studies also reported the construction of a serovar Typhimurium fimZ mutant and determined that this strain was phenotypically nonfimbriate even under conditions favoring fimbriation. To confirm the role of fimY as a positive regulator of fimbrial expression, a fimY mutant was constructed and characterized. Similar to the fimZ mutant, the fimY mutant was found to be nonfimbriate and expressed significantly reduced levels of fimA expression under all conditions studies. The lack of production of FimA subunits by this mutant was not due to a polar effect on fimZ expression, since a positive fimbrial phenotype, and concomitant fimA expression, could be restored following transformation of the fimY mutant with plasmids carrying only the fimY gene. The fimY mutation was introduced, following transduction, into serovar Typhimurium SL1344. Serovar Typhimurium SL1344 is invasive, strongly fimbriate, and, unlike LT2 strains of serovar Typhimurium, has been used extensively to investigate virulence. The serovar Typhimurium SL1344 fimY mutant was also observed to be nonfimbriate under all conditions, confirming that the FimY polypeptide plays a crucial role in fimbrial expression in two independent isolates.
Expression of fimY itself was analyzed by construction of a fimY-lacZ reporter. The results shown in Table 4 indicate that FimY is an autoregulatory protein but that this activation is only achievable when FimZ is present. Thus expression of fimY, similar to expression of fimA, was determined to be dependent upon the presence of both fimY and fimZ gene products, supporting the roles of these two proteins as coregulators of fimbrial production. Expression of fimY was also investigated following growth under conditions favoring fimbriation and was found to respond to these environmental conditions in a manner similar to fimA expression. In contrast to fimA, however, greater differences in fimY expression were observed when bacteria were grown in liquid media compared to growth on solid media. These results are consistent with a model in which the expression of fimY is influenced by environmental conditions in a regulatory cascade upstream of fimA. In addition, the overproduction of FimY results in constitutive expression of type 1 fimbriae, and similar observations have been made following overexpression of FimZ (54). These studies indicate that the concentrations of both FimY and FimZ in vivo may be critical for fimA regulation and fimbrial expression by the bacteria.
Our observations support the role of FimY as an activator of fimA expression. However, extensive in vitro DNA binding studies, under conditions in which FimZ has previously been shown to bind to the fimA promoter, were unable to establish a specific interaction between FimY and the fimA or fimY promoter regions. The inability to bind FimY to these DNA fragments suggests that other Salmonella factors are necessary for the transcriptional activity of this protein, and one or more of these factors may not be available in the binding assays used in these studies. For example, FimZ may be essential, in vivo, for the binding of FimY to a specific region of DNA. Alternatively, FimY may not be a DNA binding protein at all, but it may instead interact with FimZ in a manner that activates FimZ for binding to the promoter region of fimA. Studies analyzing FimZ-FimY protein interactions are currently under way in our laboratory.
Previously, we have reported that FimZ is a positive activator of fimA expression and that this activation is mediated by FimZ binding to the promoter region of fimA (54). The precise binding site of FimZ has been shown to extend from 47 to 100 bp upstream of the transcription initiation site of fimA (unpublished data). Consistent with the observed binding, in vitro, of purified FimZ to the fimA promoter region is the amino acid sequence relatedness of FimZ to BvgA, a transcriptional regulator of virulence gene expression in B. pertussis (14, 46). BvgA is a sensory response regulator that, along with the sensor kinase BvgS, makes up a two-component regulatory system in B. pertussis. Frequently, both components are encoded by contiguous genes on the bacterial chromosome (27). However, a complete two-component system is unlikely to be found within the fim gene cluster, since fimZ is flanked by fimF, a gene encoding a polypeptide required for fimbrial assembly (34, 36), and fimY. Examination of the amino acid sequence of the entire FimY polypeptide indicates that this protein has limited homology to prokaryotic transcriptional regulators and no apparent homology to sensory regulators of two-component systems. Closer examination of the C-terminal region of FimY reveals the presence of conserved kinase phosphorylation sites, suggesting that the action of this protein could depend upon phosphorylation. The inability to identify a specific binding site on the fimA promoter, even in the presence of phosphorylating agents, such as acetyl phosphate, may indicate that FimY acts within a unique phosphorelay system (31, 43, 52). However, it is unlikely that fimY encodes a traditional sensor kinase component, since it is uncharacteristically small and contains no apparent transmembrane domains.
The overexpression of FimY in Salmonella results in constitutive production of fimbriae on the surfaces of bacteria, since transformants of serovar Typhimurium LBY100 expressing fimY on a multicopy plasmid are fimbriate regardless of culture conditions. These transformants have lost the ability to vary phenotypic expression of type 1 fimbriae, and expression of fimA is constitutively high in this strain, similar to what has been detected in serovar Typhimurium producing large amounts of FimZ (54). Consequently, fimbrial phase variation may be modulated, at least in part, by the relative intracellular concentrations of regulators such as FimZ and FimY. However, additional fim genes are known to affect the ability of serovar Typhimurium to produce fimbriae (47), and the regulation of fimA expression is also influenced by the activity of the fimW and fimU genes specifically. In addition, global regulators, such as LRP, IHF, and HN-S, have been shown to affect fimA expression in E. coli (5, 18, 45), whereas little is known about the ability of this group of molecules to control fimA expression in Salmonella. In order to fully understand the molecular mechanisms of serovar Typhimurium fimA expression, each component of the fimA regulon will have to be examined individually. To date, our investigations have indicated that FimZ and FimY are positive coactivators of fimA that are necessary for the formation of fimbrial appendages on the surfaces of the bacteria and act at the level of fimA expression. One of these proteins, FimZ, has been established as a DNA binding protein (54), whereas FimY demonstrates no specific binding to the fimA promoter region. Nonetheless, both proteins appear to be critical for fimA expression and thus fimbrial formation in serovar Typhimurium.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Research Initiative of the USDA (97-35204-4616) and a predoctoral fellowship to J.K.T. from a National Institutes of Health Parasitism Training Grant (TE AI07511).
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA that controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham S N, Babu J P, Giampapa C S, Hasty D L, Simpson W A, Beachey E H. Protection against Escherichia coli-induced urinary tract infections with hybridoma antibodies directed against type 1 fimbriae or complementary d-mannose receptors. Infect Immun. 1985;48:625–628. doi: 10.1128/iai.48.3.625-628.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslanzadeh J, Paulissen L J. Role of type 1 and type 3 fimbriae on the adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol Immunol. 1992;36:351–359. doi: 10.1111/j.1348-0421.1992.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 4.Baumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomfield I C, Calie P J, Eberhardt K J, McClain M S, Eisenstein B I. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J Bacteriol. 1993;175:27–36. doi: 10.1128/jb.175.1.27-36.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix S. A complementary analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan K, Falkow S, Hull R A, Hull S I. Frequency among Enterobacteriaceae of the DNA sequences encoding type 1 pili. J Bacteriol. 1985;162:799–803. doi: 10.1128/jb.162.2.799-803.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullas L R, Ryu J. Salmonella typhimurium LT2 strains are r− m+ for all three chromosomally located systems of DNA restriction and modification. J Bacteriol. 1983;156:471–474. doi: 10.1128/jb.156.1.471-474.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban M J, Chou J, Cohen S. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clegg S, Hancox L S, Yeh K-S. Salmonella typhimurium phase variation and FimA expression. J Bacteriol. 1996;178:542–545. doi: 10.1128/jb.178.2.542-545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clegg S, Purcell B K, Pruckler J. Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect Immun. 1987;55:281–287. doi: 10.1128/iai.55.2.281-287.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clouthier S C, Collinson S K, White A P, Banser P A, Kay W W. tRNA(Arg) (fimU) and expression of SEF 14 and SEF 21 in Salmonella enteritidis. J Bacteriol. 1998;180:840–845. doi: 10.1128/jb.180.4.840-845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connel H, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotter P A, Miller J F. BvgAS-mediated signal transduction: analysis of phase locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect Immun. 1994;62:3381–3390. doi: 10.1128/iai.62.8.3381-3390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duguid J P, Anderson E S, Campbell I. Fimbriae and adhesive properties in Salmonellae. J Pathol Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- 16.Duguid J P, Gillies R R. Fimbriae and haemagglutinating activity in Salmonella, Klebsiella, Proteus and Chromobacterium. J Pathol Bacteriol. 1958;75:517–519. [Google Scholar]
- 17.Eisenstein B I. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science. 1981;214:337–338. doi: 10.1126/science.6116279. [DOI] [PubMed] [Google Scholar]
- 18.Eisenstein B L, Sweet D S, Vaughn V, Friedman D I. Integration host factor is required for the inversion that controls phase variation in Escherichia coli. Proc Natl Acad Sci USA. 1987;84:6506–6510. doi: 10.1073/pnas.84.18.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst R K, Dombroski D M, Merrick J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of Hep-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewen S W, Naughton P J, Grant G, Sojka M, Allen-Vercoe E, Bardoz S, Thorns C J, Pusztaj A. Salmonella enterica var. Typhimurium and Salmonella enterica var. Enteritidis express type 1 fimbriae in the rat in vivo. FEMS Immunol Med Microbiol. 1997;18:185–192. doi: 10.1111/j.1574-695X.1997.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 21.Freitag C S, Abraham J M, Clements J R, Eisenstein B I. Genetic analysis of the phase variation control of expression of type 1 fimbriae in Escherichia coli. J Bacteriol. 1985;162:668–675. doi: 10.1128/jb.162.2.668-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in E. coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 23.Garner M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the E. coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlach G-F, Clegg S, Ness N J, Swenson D L, Allen B L, Nichols W A. Expression of type 1 fimbriae and mannose-sensitive hemagglutinin by recombinant plasmids. Infect Immun. 1989;57:764–770. doi: 10.1128/iai.57.3.764-770.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghrist A C, Stauffer G V. Characterization of the Escherichia coli gcvR gene encoding a negative regulator of gcv expression. J Bacteriol. 1995;177:4980–4984. doi: 10.1128/jb.177.17.4980-4984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hancox L S, Yeh K-S, Clegg S. Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol Med Microbiol. 1998;19:289–296. doi: 10.1111/j.1574-695X.1997.tb01099.x. [DOI] [PubMed] [Google Scholar]
- 27.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 28.Hoiseth S K, Stocker B A D. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 29.Horiuchi S, Inagaki Y, Okamura N, Nakaya R, Yamamoto N. Type 1 pili enhance the invasion of Salmonella braenderup and Salmonella typhimurium to HeLa cells. Microbiol Immunol. 1992;36:593–602. doi: 10.1111/j.1348-0421.1992.tb02059.x. [DOI] [PubMed] [Google Scholar]
- 30.Hultgren S J, Schwan W R, Schaeffer A J, Duncan J L. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect Immun. 1985;50:370–377. doi: 10.1128/iai.50.2.370-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishige K, Nagasawa S, Tokishita S, Mizuno T. A novel device of bacterial signal transducers. EMBO J. 1994;13:5195–5202. doi: 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones G W, Richardson L A. The attachment to and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose resistant haemagglutinating activities. J Gen Microbiol. 1981;127:361–370. doi: 10.1099/00221287-127-2-361. [DOI] [PubMed] [Google Scholar]
- 33.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in E. coli. EMBO J. 1986;5:1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 35.Klemm P, Krogfelt K A. Type 1 fimbriae of Escherichia coli. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 9–26. [Google Scholar]
- 36.Krogfelt K A, Klemm P. Investigation of minor components of Escherichia coli type 1 fimbriae: protein, chemical, and immunological aspects. Microb Pathog. 1988;4:231–238. doi: 10.1016/0882-4010(88)90073-3. [DOI] [PubMed] [Google Scholar]
- 37.Lindquist B L, Lebenthal E, Lee P C, Stinson M W, Merrick J M. Adherence of Salmonella typhimurium to the small intestine of the rat. Infect Immun. 1987;55:3044–3050. doi: 10.1128/iai.55.12.3044-3050.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 39.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Induction and evasion of host defenses by type 1 piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 41.Old D C. Inhibition of the interaction between fimbrial haemagglutinins and erythrocytes by d-mannose and other carbohydrates. J Gen Microbiol. 1972;71:149–157. doi: 10.1099/00221287-71-1-149. [DOI] [PubMed] [Google Scholar]
- 42.Old D C, Duguid J P. Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol. 1970;103:447–456. doi: 10.1128/jb.103.2.447-456.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perraud A L, Wiess V, Gross R. Signalling pathways in two-component phosphorelay systems. Trends Microbiol. 1999;7:115–120. doi: 10.1016/s0966-842x(99)01458-4. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Schembri M A, Olsen P B, Klemm P. Orientation-dependent enhancement by H-NS of the activity of the type 1 fimbrial phase switch promoter in Escherichia coli. Mol Gen Genet. 1998;259:336–344. doi: 10.1007/s004380050820. [DOI] [PubMed] [Google Scholar]
- 46.Stibitz S. Mutations in the bvgA gene of Bordetella pertussis that differentially affect regulation of virulence determinants. J Bacteriol. 1994;176:5615–5621. doi: 10.1128/jb.176.18.5615-5621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swenson D L, Clegg S. Identification of ancillary fim genes affecting fimA expression in Salmonella typhimurium. J Bacteriol. 1992;174:7697–7704. doi: 10.1128/jb.174.23.7697-7704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swenson D L, Clegg S. Salmonella fimbriae. In: Klemm P, editor. Fimbriae: adhesion, genetics, biogenesis and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 105–113. [Google Scholar]
- 49.Swenson D L, Clegg S, Old D C. The frequency of fim genes among Salmonella serovars. Microb Pathog. 1991;10:487–492. doi: 10.1016/0882-4010(91)90115-q. [DOI] [PubMed] [Google Scholar]
- 50.Swenson D L, Kim K-J, Six E W, Clegg S. The gene fimU affects expression of Salmonella type 1 fimbriae and is related to the E. coli tRNA gene argU. Mol Gen Genet. 1994;244:216–218. doi: 10.1007/BF00283525. [DOI] [PubMed] [Google Scholar]
- 51.Tavendale A, Jardine C K H, Old D C, Duguid J P. Haemagglutinins and adhesion of Salmonella typhimurium to Hep-2 and Hela cells. J Med Microbiol. 1983;16:371–380. doi: 10.1099/00222615-16-3-371. [DOI] [PubMed] [Google Scholar]
- 52.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 54.Yeh K-S, Hancox L S, Clegg S. Construction and characterization of a fimZ mutant of Salmonella typhimurium. J Bacteriol. 1995;177:6861–6865. doi: 10.1128/jb.177.23.6861-6865.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]