Abstract
Children infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) develop less severe coronavirus disease 2019 (COVID-19) than adults. The mechanisms for the age-specific differences and the implications for infection-induced immunity are beginning to be uncovered. We show by longitudinal multimodal analysis that SARS-CoV-2 leaves a small footprint in the circulating T cell compartment in children with mild/asymptomatic COVID-19 compared to adult household contacts with the same disease severity who had more evidence of systemic T cell interferon activation, cytotoxicity and exhaustion. Children harbored diverse polyclonal SARS-CoV-2-specific naïve T cells whereas adults harbored clonally expanded SARS-CoV-2-specific memory T cells. A novel population of naïve interferon-activated T cells is expanded in acute COVID-19 and is recruited into the memory compartment during convalescence in adults but not children. This was associated with the development of robust CD4+ memory T cell responses in adults but not children. These data suggest that rapid clearance of SARS-CoV-2 in children may compromise their cellular immunity and ability to resist reinfection.
Keywords: COVID-19, SARS-CoV-2, Memory T cells, Interferon-activated naive T cells, Single cell transcriptomics, T cell receptor repertoire, Clonal dynamics
Graphical abstract
1. Introduction
Infection with the respiratory pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the pandemic coronavirus disease 2019 (COVID-19) [1,2]. Disease severity varies widely from asymptomatic infection in the majority of individuals, to severe life-threatening disease in a minority of patients [[3], [4], [5]]. Older age, male sex and comorbidities such as hypertension, cardiovascular disease and diabetes have been identified as independent risk factors for severe disease and death [6]. There is now consistent evidence across multiple different settings and locations that the severity of COVID-19 infection increases substantially with age [7].
Recently, a number of investigators have examined the local and systemic immune responses of children to SARS-CoV-2 to determine the potential mechanisms for these age-specific differences in susceptibility to infection and disease. Most notably, it has been shown that children have an enhanced antiviral sensing and stronger antiviral interferon response in the upper airways due to higher basal expression of the MDA5 and RIG-1 viral pattern recognition receptors and interferon gene signatures in nasal epithelial cells, macrophages and dendritic cells [8,9]. This pre-activated innate immune system may be more efficient at clearing SARS-CoV-2 infection. Indeed, analysis of three child household contacts of adults with PCR-confirmed symptomatic COVID-19 suggested that children may mount an effective early antiviral immune response that eliminates the virus without any detected PCR evidence of SARS-CoV-2 infection [10]. These differences in the innate immune response of children may also be detectable in the immunophenotype of circulating neutrophils, dendritic cells, monocytes and natural killer (NK) cells in the peripheral blood [11].
The adaptive immune response to SARS-CoV-2, particularly the humoral component provided by antibodies and memory B cells, can be either protective or pathogenic in COVID-19 [12,13]. There is evidence that antibodies against endemic human coronaviruses (hCoV) cross-react with SARS-CoV-2 and these may be back-boosted upon infection [[14], [15], [16]]. However, this pre-existing cross-reactive humoral immunity does not appear to provide protection, and may potentially be harmful by locking in the memory B cell responses and preventing the emergence of de novo naïve B cell responses to novel SARS-CoV-2 antigens, a phenomenon known as original antigenic sin [15,[17], [18], [19]]. Analysis of pre-pandemic serum has also shown that healthy elderly individuals have higher immunoglobulin class-switched IgA and IgG antibodies that cross-react with SARS-CoV-2, whereas children have elevated cross-reactive SARS-CoV-2 IgM antibodies [20], suggesting that they have less exposure to hCoV and are less antigen-experienced but more polyreactive. Furthermore, it has been suggested that pathogenic responses associated with severe COVID-19 are linked to SARS-CoV-2 IgA and neutrophil hyperactivation [13] and that the antibodies produced by children differ from adults in their Fc-dependent antibody effector functions such as antibody-dependent cellular cytotoxicity, phagocytosis and complement activation [13,20].
There is emerging evidence that cellular immunity against SARS-CoV-2 provided by T cells may be similarly impacted by prior exposure to hCoV. Using overlapping peptide pools for in vitro T cell stimulation assays, investigators have detected CD4+ and CD8+ T cells cross-reactive against SARS-CoV-2 spike (S), membrane (M), nucleocapsid (N) and open reading frames (ORF) in pre-pandemic blood samples from unexposed individuals [[21], [22], [23], [24], [25]]. Consistent with this, SARS-CoV-2 T cell responses have been shown to be lower in children and increase with age and time after infection [26].
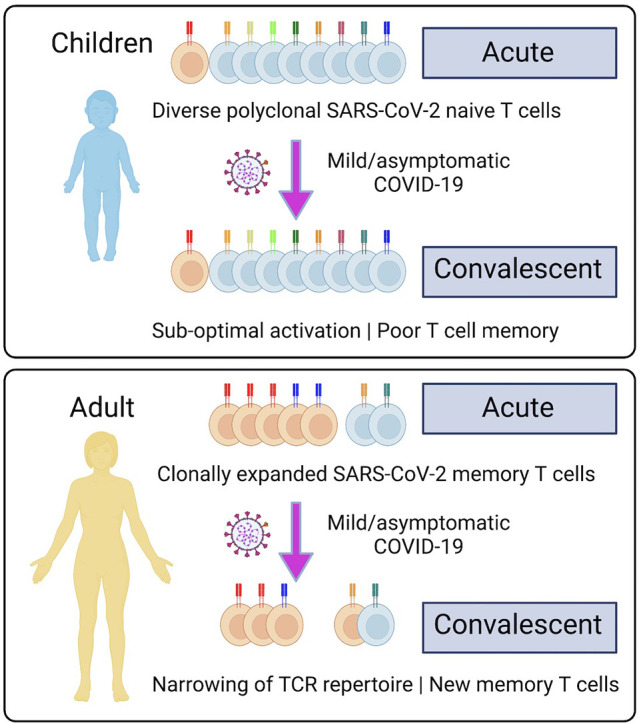
We performed longitudinal analysis of the immune response of seven children and five adults from the same household with mild/asymptomatic COVID-19 in the community confirmed by reverse transcription polymerase chain reaction (RT-PCR) and an additional two unrelated adults who were ventilated in the intensive care unit with severe life-threatening COVID-19. We analyzed the cellular phenotype, serum antibody response to SARS-CoV-2, cytokine profile, in vitro memory T cell responses to recombinant S and RBD proteins, and simultaneous single cell transcriptome and TCR and B cell receptor (BCR) repertoire sequencing of 433,301 single cells obtained from acute and convalescent blood samples. This multimodal analysis includes the TCR repertoire of the 158,975 CD4+ T cells (of which 746 were SARS-CoV-2-specific) and 105,273 CD8+ T cells (of which 798 were SARS-CoV-2-specific). This identified novel subpopulations of naïve T cells, including acutely expanded clusters of interferon-activated naïve T cells which differentiate into memory T cells in convalescence. We show that mild/asymptomatic COVID-19 results in systemic activation of both the innate and adaptive immune compartments in adults. In contrast, children had less activation of the circulating T and B cells. Children have more SARS-CoV-2-specific T cells which predominantly have a naïve phenotype and diverse TCR repertoire. Adults have fewer SARS-CoV-2-specific T cells which are more antigen-experienced and often harbor clonally expanded exhausted memory T cells. Circulating T cells in children retain their naïve state and did not generate many antigen-specific memory T cells despite infection with the virus. In contrast, adults generated more memory T cells from the naïve interferon-activated and this was associated with the development robust SARS-CoV-2-specific memory T cell responses in adults, but not children. This failure of infection-induced immunity places children at the risk of recurrent infection and progressive restriction of their T cell repertoire and responses as they grow older.
2. Materials and methods
2.1. Subjects
2.1.1. Mild/asymptomatic COVID-19 children and adult family members
Participants were recruited under the Immunophenotyping of COVID-19 and other Severe Acute Respiratory Infections Study (2020/PID00920) approved by the Sydney Children's Hospital Network (SCHN) Human Research Ethics Committee (2020/ETH00837). Potential child participants were identified through admission to the Children's Hospital at Westmead following a positive SARS-CoV-2 reverse transcriptase polymerase chain reaction (RT-PCR). Families were approached and both children and positive household adults were consented to longitudinal blood sampling. Blood used in these analyses were collected in lithium heparin and serum-separation tubes (Becton Dickinson, USA), and collected both acutely (within 1–10 days of first positive RT-PCR) and approximately one-month post infection. All patients were ambulant with either mild symptoms, consisting of a cough or sneeze, or asymptomatic (WHO Clinical Progress Scale of 1 out of 10). All consented samples used in these analyses were collected between 23rd July 2020 and 24th October 2020.
2.1.2. Severe COVID-19 patients
Participants were recruited under the South-East Queensland COVID Consortium study approved by the Gold Coast Hospital and Health Service Human Research Ethics Committee (HREC/2020/QGC/63082). Patient recruitment was undertaken by research team members at the Gold Coast University Hospital. A waiver of informed consent, with the opportunity to opt-out was used for this study. Both patients were elderly males with severe COVID-19 who were intubated and ventilated ICU (WHO Clinical Progress Scale of 7 out of 10). Acute samples were taken on the 14th April 2020. A13 had second sample 8 days later on the 22nd April 2020 and A14 had convalescent sample taken on the 10th September 2020.
2.2. PBMC processing and cryopreservation
PBMCs were diluted 1:1 with PBS and the mix overlaid in a 2:1 ratio with Ficoll-Paque in 50 ml Falcon tube. Cells were centrifuged at 1700 rpm for 30 min with brake off at room temperature (18–21 °C). Mononuclear cells at the interface were collected and transferred to a new Falcon tube and washed ×2 with ice-cold PBS/0.5% FBS by centrifugation at 1300 rpm for 8 min with brake on. Cells were resuspended in 5 ml culture media (RPMI/1% HEPES) for counting with a haemocytometer and Trypan blue. Cells were then resuspended in 500 μl culture media at a concentration of 2 × 10 [7]/ml and 500 μl freezing media (200 ml culture media, 200 ml FBS and 100 ml DMSO) and cryovials transferred to the −80 °C freezer in a Styrofoam box for 24–48 h before storage in liquid nitrogen. Frozen PBMCs were thawed and centrifuged at 400 g for 5 min. Samples were washed with twice with 4% FBS/PBS and stained with DAPI (0.1 μg/ml) for 5 min at room temperature prior sorting of live DAPI— cells on the AriaIII (BD Biosciences).
2.3. Serum antibody testing
Serum was prepared from clotted tubes by centrifugation at 1000 g for 10 min. IgG titres to the SARS-CoV-2 Spike protein were measured by our previously published high sensitivity flow cytometry cell-based assay [27] which has been modeled on autoantibody detection test used in clinical diagnostic testing of neuroimmunological disorders [28,29]. HEK293 cells were transfected to express early-clade SARS-CoV-2 Spike antigens. Diluted serum (1:80) was added to live Spike-expressing cells. Codon-optimised, wild-type SARS-CoV-2 strain Wuhan Spike protein ORF with 18 amino acids deleted from the cytoplasmic tail was cloned within the MCS of a lentiviral expression vector, pLVX-IRES-ZsGreen1, using EcoRI and XbaI restriction sites, resulting in pSpike-IRES-ZsGreen vector. All synthetic gene fragments were ordered through IDT. Cells were then incubated with Alexa Fluor 647-conjugated anti-human IgG (H + L) (ThermoFisher Scientific). Cell events were acquired on LSRII flow cytometer (BD Biosciences, USA), and median fluorescence intensity (MFI), a proxy of antibody titres was analyzed. The threshold for a positive result was determined if the delta MFI (ΔMFI = MFI transfected cells – MFI untransfected cells) was above the positive threshold (mean ΔMFI + 4SD of 24 pre-pandemic age-matched controls) in at least two of three quality-controlled experiments. The sensitivity of the assay was superior to several commercial assays at 98% (95% CI: 92–99%) [27]. Data were analyzed using FlowJo 10.4.1 (TreeStar, USA), Excel (Microsoft, USA) and GraphPad Prism (GraphPad Software, USA).
2.4. Serum cytokine bead array
We used the BD Cytometric Bead Array (CBA) kit to measure serum cytokines. Cytokine standards were made up in assay diluent as per the manufacturer's instructions. Capture bead mix was made up in Capture Bead Diluent for Serum/Plasma to a final concentration of 25 μL/test. Serum samples were diluted 1:1 in assay diluent. To each well 25 μL of beads was added, followed by 25 μL of standard or 25 μL of diluted serum sample. Samples were incubated with beads in the dark for 1 h at room temp. Detection master mix was prepared as per the manufacturer's instructions and 25 μL added to each sample and then samples incubated for a further 2 h in the dark at room temperature. Beads were then washed twice in wash buffer and then acquired on BD FACS Canto II flow cytometer (BD Pharmingen). Analysis was performed on FCAP array software v 3.0 (BD Biosciences). Cytokine concentrations were imported into R (version 4.1.2), log transformed, and visualized with ggplot2 (version 3.3.5).
2.5. Fluorescence activated cell sorting (FACS)
Thawed cells were resuspended in 2% FBS/PBS and plated in a 96-well V-bottom plate. Antibody cocktails were prepared in FACS buffer (0.1% BSA/0.1% sodium azide/PBS). Cells were pelleted by centrifugation at 490 g for 5 min at 4 °C. Cells were then stained with 50 μL of Zombie UV Fixable viability dye (diluted 1/500 in PBS) for 20 min on ice in the dark. Cells were washed 3 times with FACS buffer. Cells were then incubated with 50 μL of blocking agents (normal mouse serum 1/20, Fc block 1/10) for 15 min on ice. Antibody cocktails were prepared in FACS Buffer and 50 μL added to each sample and then incubated for 30 min on ice in the dark. Cells were washed 3 times in FACS buffer and then fixed by resuspending in 150 μL of 1% formaldehyde for 20 min at room temperature. Cells were then washed and resuspended in FACS buffer and run on FACSymphony (BD Pharmingen). Samples were analyzed using FlowJo software (Tree Star).
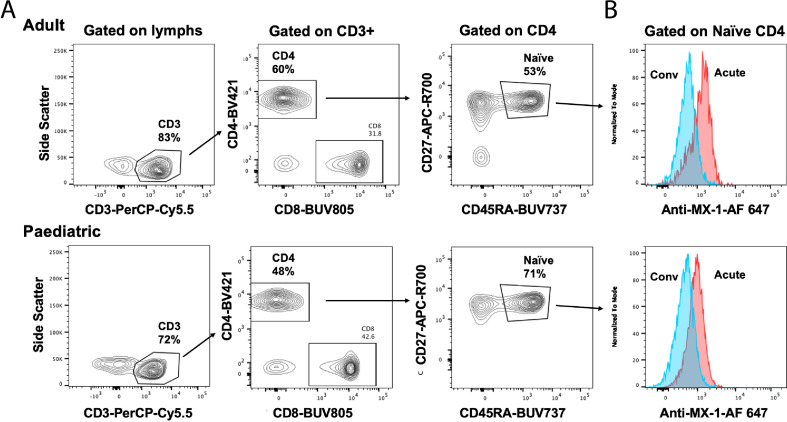
Intracellular staining for MX-1 was performed for 1 × 105 thawed PBMC using the Transcription Factor Buffer Set (BD Biosciences) according to the manufacturer's directions. Permeabilized cells were stained with CD3-PerCP-Cy5.5, CD4-BUV395, CD8-BUV805, CD45RA-BUV737, CD27-APC-R700 (BD Biosciences) and 1 μg MX-1-AF647 (Abcam) according to manufacturer's directions and analyzed on a 5-laser Fortessa X20 (BD Biosciences) as previously described [30].
2.6. Recombinant RBD and S protein
Expression plasmids encoding His-tagged SARS-CoV-2 RBD (residues 319 to 541 of SARS-CoV-2 S protein) or S protein (with a C-terminal trimerization domain) were cloned into pCEP4 vector and transfected into Expi293F (ThermoFisher Scientific) and the proteins expressed for 7 days at 37 °C [31]. The proteins were captured from the clarified cell culture using TALON resin (ThermoFisher Scientific) and eluted with imidazole. The full trimeric S protein was further purified by size exclusion chromatography (Superose 6 resin) to remove dissociated S1 and S2 domains. The protein purity was assessed by visualization on SDS-PAGE gel.
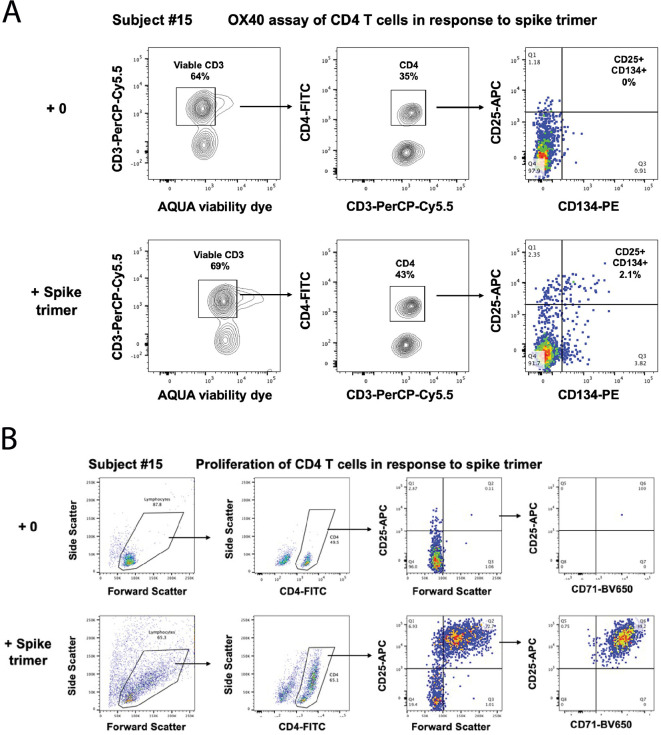
2.7. OX40 antigen-specific memory T cell assay
Antigen-specific CD4 T-cells responding to recall antigens were measured in cultures of 300,000 PBMC in 200 μl/well of a 96-well plate, in Iscove's Modified Dulbecco's Medium (IMDM; Thermofisher, Waltham, MA, USA) containing 10% human serum (Wayne Dyer, Australian Red Cross Lifeblood, Sydney, Australia), and incubated for 44–48 h incubation, in a 5% CO2 incubator, as previously described [32]. Separate cultures were incubated with different antigens including: (i) culture medium only negative control well; (ii) anti-CD3/anti-CD28/anti-CD2 T cell activator (1/100 dilution) polyclonal positive control well; (iii) 5 μg/ml recombinant SARS-CoV-2 S trimer; and (iii) 5 μg/ml recombinant SARS-CoV-2 RBD. 100 μl of PBMC from the respective cultures were stained with CD3-PerCP-Cy5.5, CD4-FITC, CD25-APC, and CD134-PE (BD Biosciences, San Jose, CA, USA), and live/dead fixable NIR dead cell stain kit reagent according to manufacturer's directions and analyzed on a 5-laser Fortessa X20 (BD Biosciences) as previously described [30]. Antigen-specific CD4+ T cells were gated and expressed as CD25+CD134+ % of CD3+CD4+ live T cells as previously described [32]. Cultures were classified as positive for antigen-specific CD4+ T cells if the CD25+CD134+ % of CD4+ CD3+ T cells was ≥0.2% [33].
2.8. Antigen-specific T cell proliferation and RNA extraction
Antigen-specific T cell proliferation was measured in cultures of PBMC incubated with different controls and antigens in separate wells as above for the OX40 assays, except cells were incubated for 7 days. 100 μl of PBMC were then stained with CD3-PerCP-Cy5.5, CD4-FITC, CD25-APC, and CD71-BV650 according to manufacturer's directions and analyzed on a 5-laser Fortessa X20 (BD Biosciences) as previously described [30]. Antigen-specific proliferating CD4+ T cells were gated as % of Forward Scatter high and CD25+CD71+CD4+CD3+ T cells. Proliferating cells remaining in the cultures were further expanded by incubating for a further 7 days with 20 IU/mL IL-2 (Roche Life Science Products). After expansion, cells from each well were used for RNA extraction, using the Maxwell RSC SimplyRNA Tissue kit and the Maxwell RSC automated extraction system (Promega, Madison, WI) as previously described [34].
2.9. Single cell RNA transcriptome and TCR repertoire sequencing
Single cell transcriptomic libraries were generated using the 5’v2 Gene expression and immune profiling kit (10× Genomics). Viable PBMCs were sorted into 2% FBS/PBS and cell counts were performed using a haemocytometer. Up to 40,000 cells were loaded into each lane of Chromium Next GEM Chip K Single Cell Kit (10× Genomics) to achieve a recovery cell number of approximately 20,000 cells. Subsequent cDNA and TCR libraries were generated according to manufacturer's instructions. Generated libraries were sequenced on the NovaSeq S4 flow cell (Illumina) at Read 1 = 28, i7 index = 10, i5 index = 10 and Read 2: 90 cycles according to manufacturer's instructions.
2.10. Transcriptomic analysis
2.10.1. Pre-processing of raw sequencing files
Single-cell sequencing data was demultiplexed, aligned and quantified using Cell Ranger (10× Genomics) against the human reference genome (10× Genomics, July 7, 2020 release) with default parameters. Filtering and quality control was performed using Seurat [35] on raw data containing 522,926 cells where 433,301 cells were retained satisfying thresholds of both <10% mitochondria content and number of genes between 200 and 5000. ‘SCTransform’ was used for normalization with regression of batch, gender and cell mitochrondria content covariates [36].
2.10.2. Annotation of cell identities
Cell annotation was performed using ‘reference-based mapping’ pipeline implemented in Azimuth algorithm in Seurat [37]. Azimuth annotated T cells were re-clustered and T cell sub-populations were manually annotated based on UMAP clustering and markers defined by ‘FindAllMarkers’ function in Seurat.
2.10.3. Differential gene expression analysis
Raw counts from defined cell populations were normalized using scran / scater [38,39] and differential gene expression analysis was performed using Limma voom [40] with regression of Batch and Gender covariates. DGE analysis was not performed on dendritic cells (all sub populations) and CD8 IFN activated Naïve cell populations due to low cell numbers (<100 cells) sampled in the dataset.
2.10.4. Gene signature scores
Gene signature scores was generated using ‘AddModuleScore’ function in Seurat. Cell sub-populations with <5 cells within sample groups were excluded from the analysis. To compare the gene signatures across different sub-populations within sample groups, gene scores were weighted based on the proportion of positive expressing cells within the sub-population. The interferon gene signature was generated from aggregating unique interferon response genes sourced from [[41], [42], [43], [44]]. T cell exhaustion signature was sourced from [45]. CD8+ cytotoxic T cell signature was sourced from [44].
2.11. Analysis of the OneK1K cohort
OneK1K Cohort Study [46] was established to investigate the effects of genetic variation on gene expression at single cell resolution. Original cohort includes >1000 individuals recruited from the Royal Hobart Hospital, Hobart Eye Surgeons as well as from the retirement villages within Hobart, Australia prior to the COVID-19 pandemic. We have selected 26 age- and sex- matched individuals from this cohort to compare cell type proportions with the COVID-19 patients. The study was approved by the Tasmanian Health and Medical Human Research Ethics Committee (H0012902). Informed consent was obtained from all participants.
Peripheral blood samples were collected into vacutainer tubes containing either FICOLL™ and sodium heparin (8 mL CPT™; BD Australia, North Ryde, NSW; 362,753) or K2EDTA (10 mL; BD Australia, North Ryde, NSW; Catalogue: 366643). During single cell library preparation equal numbers of live cells were combined for 12–14 samples per pool. Pooled single cell suspensions partitioned and barcoded using the 10× Genomics Chromium Controller and the Single Cell 3’ Library and Gel Bead Kit version 2 (PN-120237). The pooled cells were super-loaded onto the Chromium Single Cell Chip A (PN-120236) to target 20,000 cells per pool. Libraries for all samples were multiplexed and sequenced across five 2 × 150 cycle flow cells on an Illumina NovaSeq 6000. The Cell Ranger Single Cell Software Suite (version 2.2.0) was used to process data produced by the Illumina NovaSeq 6000 sequencer into transcript count tables. Raw base calls from multiple flow cells were demultiplexed into separate pools of samples. Reads from each pool were then mapped to the GRCh38 genome using STAR [47]. Cells for each individual were identified using the Demuxlet computational tool [48]. The most likely individual for each droplet was determined using the genotype posterior probability estimate from imputation of 265,053 exonic SNPs (R2 > 0.3 and MAF > 0.05). In all approaches, α was set to 0.5, assuming a 50/50 ratio and other parameters were kept as default. Droplets which were identified as doublets by both Demuxlet and Scrublet [49] were removed from the dataset.
We used our COVID-19 dataset as a reference to guide the cell type classification of the OneK1K cohort using the Symphony approach [50]. First, we selected the top 5000 highly variable genes conditioned by batch information and normalized the data using factor normalization and logarithmic transformation as implemented in Seurat. Next, we centered and standardized the normalized gene expression data for the highly variable genes and stored the means and standard deviations for each gene across all cells. We performed singular value decomposition on the scaled data. We applied harmony to align the gene expression embeddings by batch using a theta value of 2 and performing 100 clustering iterations and a maximum of 20 rounds of harmony clustering and correction. We applied the same normalization strategy for the OneK1K data and projected the data onto the reference by scaling the OneK1K data using the reference means and standard deviation and aligning the data using Symphony. Finally, we assign the cell type labels to the OneK1K cells using a lazy K-nearest neighbour classifier with k = 5.
2.12. TCR repertoire analysis
Following processing with 10× Genomics cellranger vdj (v6.1.2) using the human reference the resulting VDJ contigs were post-processed using stand-alone IgBLAST (v1.14) [51] to generate further alignment details. Where a single barcode was associated with more than one chain for either the TRB or TRA loci the VDJ with the highest UMI count was retained. Clonal lineages were defined by IgBLAST called V, J and CDR3 amino acid sequences for both TRA and TRB, if available, or by a single chain if paired chains were not available. Expanded clonotypes were defined within each sample (subject and time point) as those observed across 2 or more cells, while longitudinal clonotypes were those from a subject that were observed at both the acute and convalescent timepoints.
TRBs of reported specificities were collected from immuneCODE Multiplex Identification of T cell Receptor Antigen specificity (MIRA) release 002.2 [52], and VDJdb v2021-09-05 [53]. Additional TRBs were added from [54], [55] and [56]. TRBs were formatted to consistent format, where ambiguous TRBVs reported as a separate entry were created for each TRB.
To account for the private SARS-CoV-2 responses that may not be captured in the public databases, bulk TRB sequencing was undertaken following proliferation of the outputs of the OX40 assays. The bulk sequencing assay was adapted from [57]. RNA was reverse transcribed to cDNA that incorporated a 10 bp universal molecular identifier using a modification of the SmartSeq2 protocol [58] described in (Massey et al., 2020).
TRBs of reported or inferred specificity were mapped to 10× VDJs by matching of TRB clonotype labels. Where the same TRB was reported to bind multiple epitopes, all epitopes were associated with the TRB clonotype. SARS-CoV-2-annotated clonotypes were defined as any clonotype matching a SARS-CoV-2 reported VDJ regardless of poly-specificity or those observed in the bulk repertoire sequencing from the proliferation assay at an enrichment of at least 64-fold above baseline.
TCR clonotype and annotation data were merged with 10× GEX via cell barcodes. Repertoire metrics were summarised in RStudio (v1.4.1106, RStudio Team (2021). RStudio: Integrated Development Environment for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com) using tidyverse package [59]. Shannon entropy was calculated for CD4+ and CD8+ T cell compartments for each subject to explore the diversity [60]. Clonotype distribution across cell types and time points was explored using Upset plots [61] as implemented by the ComplexHeatmap package [62].
2.13. Statistics
Statistical analysis was performed using Prism software (GraphPad) or in R. We used unpaired Student's t-test to compare between 2 groups and paired Student's t-tests to compare longitudinal differences within the same individuals. We used the one-way ANOVA with Tukey's correction for comparisons between multiple groups. We used Fisher's exact test for 2 × 2 contingency tables and Chi-square for 2 × 3 contingency tables. Correlation between variables was measured by Pearson's correlation coefficient with a one-sided Student's t-test.
3. Results
3.1. Longitudinal tracking of the immune response to SARS-CoV-2 in children and adults
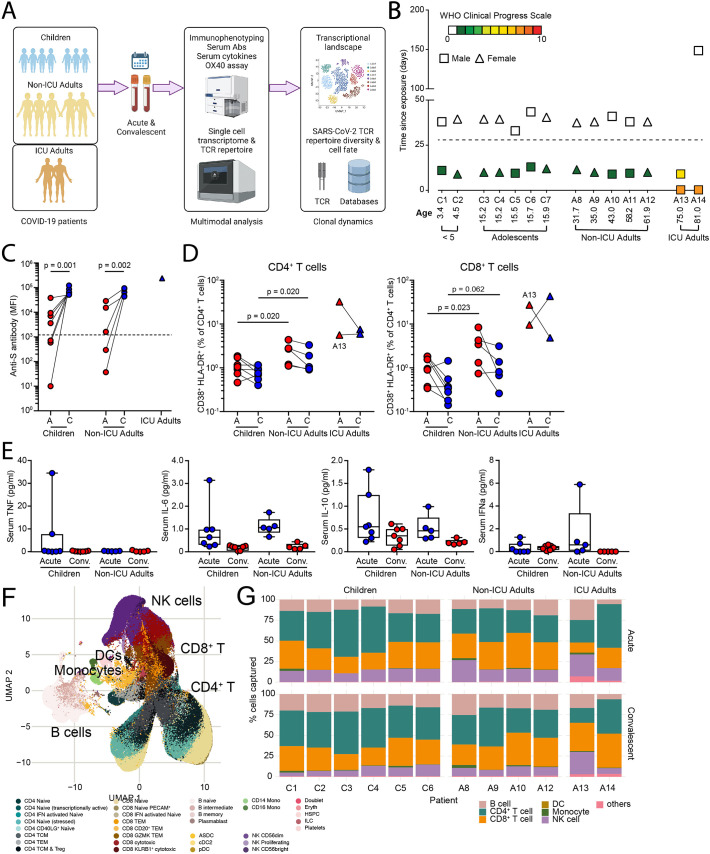
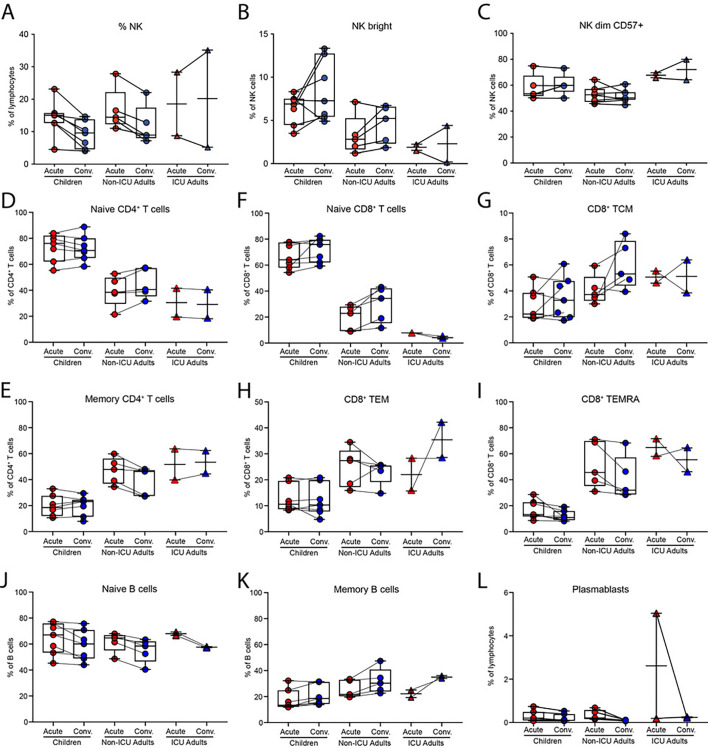
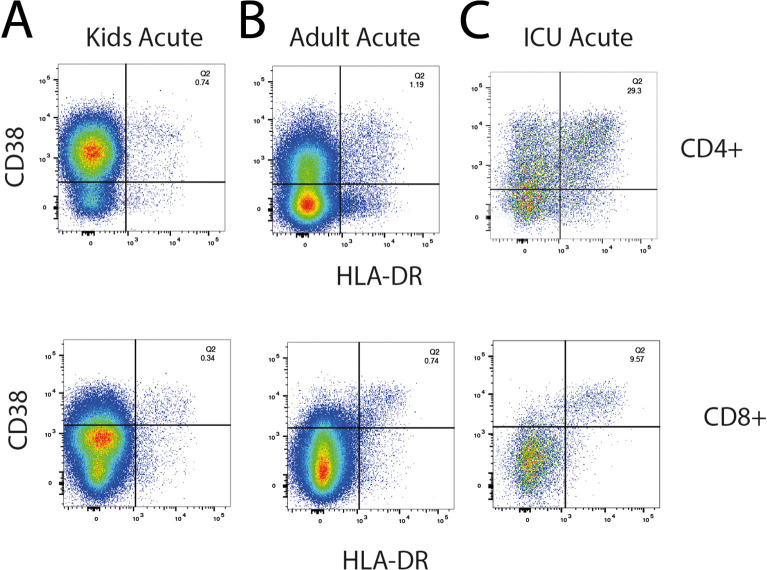
To compare the immune response of children and adults with the same disease severity, we collected acute and convalescent blood samples from seven children (<16 years of age) and five adults (>30 years of age) from the same household with mild/asymptomatic disease (WHO Clinical Progress Scale of 0 or 1 out 10); in addition, two adults who were intubated and ventilated in the intensive care unit (ICU) with severe disease (WHO Clinical Progress Scale of 7 out of 10) were analyzed (Fig. 1A, B). Two children were < 5 years of age and 5 were adolescents (Fig. 1B). Two of the children (C3 and C4) were identical twins. The second blood sample from one ICU adult (A13) was collected when the patient was still acutely ill and does not represent a true ‘convalescent’ sample. Both children and adults developed antibodies to S protein in the convalescent phase, with the highest antibody titres detected in the ICU adults (Fig. 1C). High dimensional flow cytometry showed consistent differences in the distribution of circulating natural killer (NK) cells, naïve and memory B and T cells that reflected age-specific differences between children and adults (Fig. S1). Nevertheless, by fluorescence activated cell sorting (FACS) analysis, there was evidence of increased T cell activation in the acute stage with 2-fold expansion of activated CD38+HLA-DR+ CD4+ T cells and 3-fold expansion of CD38+HLA-DR+ CD8+ T cells in non-ICU adults compared to children (Fig. 1D and Fig. S2). ICU adults had the highest frequency of CD38+HLA-DR+ T cells. Serum cytokine analysis detected 50- to 100-fold elevation of interleukin-6 (IL-6) in the ICU adults compared to acute children, convalescent children, acute non-ICU adult and convalescent non-ICU adult (p = 0.001, one-way ANOVA with Tukey's corrections) (Fig. 1E). Levels of TNF, IL-10 and interferon-α were not statistically different.
Fig. 1.
Multimodal analysis of children and adults with mild COVID-19.
(A) Study overview.
(B) Patient demographic and clinical severity score. For A13 the second blood sample was collected on day 8 during the acute phase.
(C) Serum anti-S protein antibodies.
(D) CD38+HLA-DR+ activated CD4+ (left) and CD8+ (right) T cells in children, non-ICU, and ICU adults during Acute and Convalescent phases.
(E) Serum TNF, IL-6, IL-10, and interferon-α levels.
(F) UMAP showing 433,301 single cells from children, non-ICU, and ICU adults during Acute and Convalescent phases.
(G) Stacked barplot showing cellular composition of PBMCs in children, non-ICU, and ICU adults during Acute (top) and Convalescent (bottom) phases from the single cell RNA sequencing.
We next generated single cell transcriptomes from acute and convalescent samples from these 14 COVID-19 patients. In total, we sequenced 522,926 cells and excluded samples from two patients (C7 and A11 from the same household) due to batch effects. The remaining 433,301 cells were visualized in 2D Euclidean space by uniform manifold approximation and projection (UMAP) (Fig. 1F). We initially annotated clusters of cells which consisted of populations of 69,074 B lineage cells, 171,393 CD4+ T cells, 127,069 CD8+ T cells, 376 dendritic cells (DCs), 4135 monocytes, 56,807 NK cells, and 4447 other cell types (including HSCs, platelets, erythrocytes, and doublets) (Fig. 1G). Analysis of the cellular composition identified similar age-specific changes to the circulating NK, B and T cell populations as observed by the FACS (Fig. 1G and Table S1). Taken together, these data show that, in addition to age-specific differences in the circulating immune compartments in children and adults, there is evidence for greater systemic T cell activation in adults.
3.2. Deconvolution of the circulating immune compartment in children and adults with COVID-19
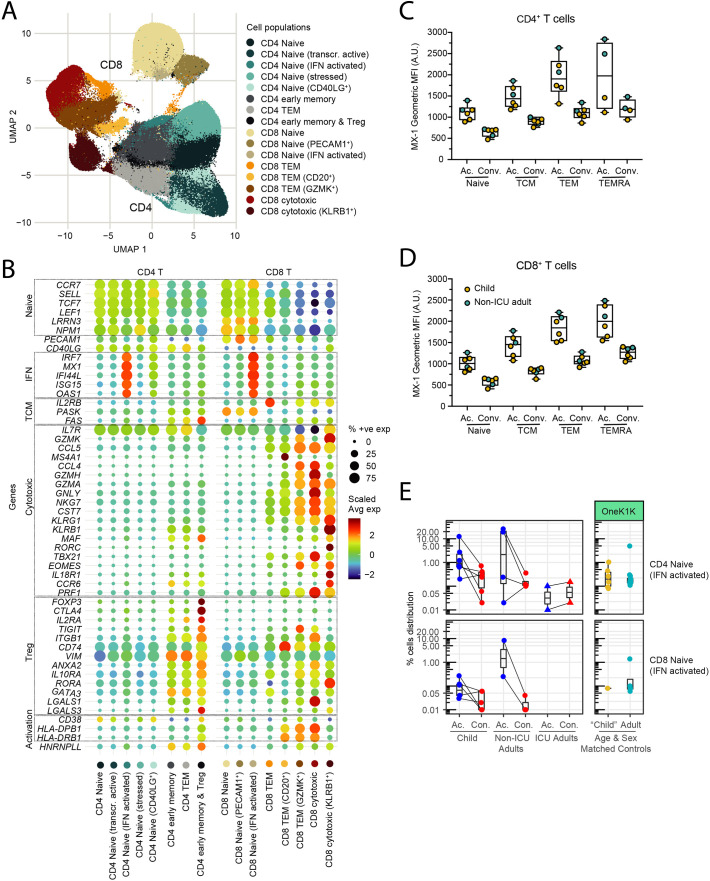
We resolved the circulating innate and adaptive immune compartments in children and adults into populations of DCs (plasmacytoid dendritic cells, pDC; AXL+SIGLEC6+ dendritic cells, ASDCs; type 2 conventional dendritic cell, cDC2), NK cells (proliferating; CD56dim; CD56bright), monocytes (CD14+; CD16+), other cells (platelets; erythroblasts; HSPCs; innate lymphoid cells, ILCs; doublets), B lineage cells (naïve, memory, intermediate and plasmablasts), CD4+ T cells (naïve; stressed naïve; transcriptionally active naïve; interferon-activated naïve; CD40LG+ naïve; early memory; effector memory (TEM); early memory and regulatory T cells (Treg), and CD8+ T cells (naïve; PECAM-1+ naïve; interferon-activated naïve; TEM; GZMK+ TEM; CD20+ TEM; cytotoxic; KLRB1+ cytotoxic T cells) (Fig. 2A). Semi-automated annotation of the T cell compartment based on the expression of canonical genes revealed marked heterogeneity, particularly in the naïve T cell compartment with several previously unappreciated subpopulations of naïve CD4+ and CD8+ T cells that showed evidence of TCR-independent bystander activation by upregulation of CD40LG, stress-induced genes, transcriptional activity, and interferon-induced genes (Fig. 2B). Analysis of the genes expressed by the different T cell clusters revealed interferon-activated naïve CD4+ and CD8+ T cells express many interferon-induced genes including IFI44L, MX1, ISG15, IRF7 and OAS1 (Fig. 2B and Table S2). We confirmed the increased protein expression of MX-1 by FACS (Fig. S3). Similarly, we identified a number of memory and effector subpopulations including CD8+ GZMK+ TEM and KLRB1+ cytotoxic cells based on their transcriptional profile and TCR receptor usage. CD8+ GZMK+ TEM are a newly described subset of exhausted-like memory T cells expressing TOX, TIGIT, FGFBP2, TBX21, SLAMF7, ZEB2, NKG7, GZMH and PRF1 (Table S2) that give rise to dysfunctional, exhausted effector T cells [63]. KLRB1+ cytotoxic T cells include MR1-restricted mucosal-associated invariant T (MAIT) cells expressing TRAV1/TRAJ33 and IL-17-producing cytotoxic T cells (Tc17) expressing MAF, RORC, TBX21, EOMES, IL18R1, CCR6, PRF1, GZMK, GZMA, NKG7, CST7 and GNLY (Table S2). KLRB1+ cytotoxic T cells are reported to be tissue-homing IL-17A producing cells [64]; however, we did not detect upregulated expression of IL17A or related IL-17 family genes.
Fig. 2.
Decomposition of the T cell compartment in children and adults with COVID-19.
(A) UMAP showing 171,393 CD4+ T cells and 127,069 CD8+ T cells from children, non-ICU, and ICU adults during Acute and Convalescent phases.
(B) Expression of genes associated with the naïve, interferon response (IFN), T central memory (TCM), T cell cytotoxicity, regulatory T cell (Treg) and activation states by different subclusters of CD4+ and CD8+ T cells shown in the UMAP.
(C) Detection of interferon-induced MX-1 protein in subpopulations of CD4+ T cells during Acute and Convalescence. Gating are as follows for Naïve T cells = CD45RA+CD45RO−; TCM = CD45RA−CD45RO+CCR7+CD62L+; TEM = CD45RA−CD45RO+CCR7−CD62L−; TEMRA = CD45RA+CD45RO−CCR7−CD62L−.
(D) Detection of interferon-induced MX-1 protein in subpopulations of CD8+ T cells during Acute and Convalescence. T cell markers are as in (C).
(E) Detection of interferon-activated naïve CD4+ T cells (top) and CD8+ T cells (bottom) in children, non-ICU, and ICU adults during Acute and Convalescent phases (left) and in healthy age and sex-matched donors in the OneK1K cohort (right).
Intracellular flow cytometry confirmed the upregulated expression of the interferon-inducible MX-1 protein in several cell types, including naïve CD4+ (Fig. 2C) and CD8+ T cells (Fig. 2D), particularly in the acute phase. To determine if these subpopulations of naïve T cells were unique to COVID-19 or were also present in individuals who had not been exposed to SARS-CoV-2, we examined the distribution of T cells in age-matched PBMCs from the OneK1K cohort [46] which was collected pre-2019 (Fig. 2E). This analysis showed that these novel interferon-activated naïve CD4+ and CD8+ T cells are also present in healthy adults who have not been infected with SARS-CoV-2. Thus, the circulating T cell compartment in children and adults is heterogeneous with multiple previously unrecognized subpopulations of naïve and activated T cells, including interferon-activated naïve CD4+ and CD8+ T cells.
3.3. Differential gene expression between children and adults infected with SARS-CoV-2
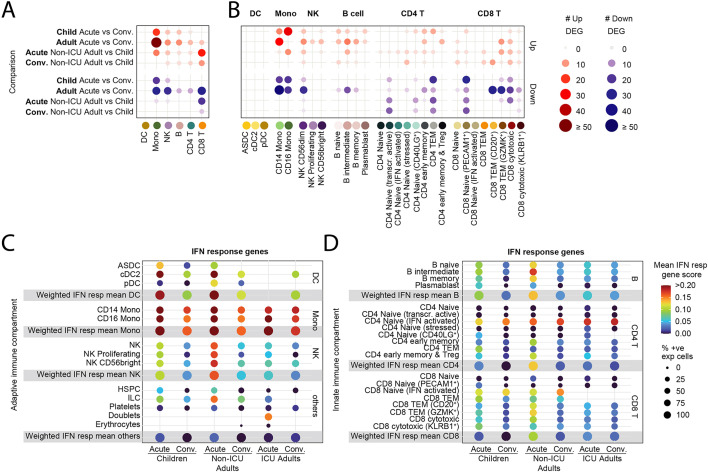
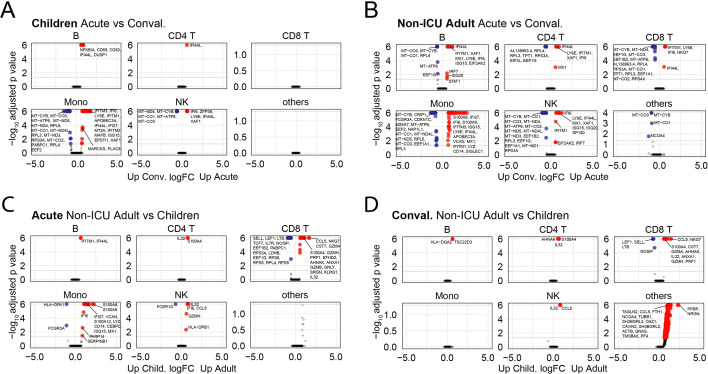
We analyzed for differentially expressed genes (DEGs) between each subpopulation to determine the differences between children and non-ICU adults who both had mild/asymptomatic COVID-19. We detected very few DEGs in the adaptive B and T cell compartments in children compared to adults as they transitioned from acute to convalescence (6 upregulated, no downregulated genes in children; 22 upregulated, 43 downregulated genes in non-ICU adults; p = 0.003, Fisher's exact test) (Fig. 3A). In contrast, there were many DEGs in the innate NK cell and monocyte compartment in both groups (25 upregulated, 24 downregulated genes in children; 67 upregulated, 50 downregulated genes in non-ICU adults; p = 0.285, Fisher's exact test). Notably, there was upregulation of interferon-induced genes (e.g., IFI66, IFI44L and XAF1) during acute infection and mitochondrial oxidative phosphorylation (OXPHOS) genes (e.g., MT-ATP6, MT-CYB, MT-ND4 and MT-CO1) upon recovery (Fig. S4). In contrast, there were many differentially expressed genes in both the innate and adaptive immune compartments in non-ICU adults (Fig. 3A). Notably, there was significant upregulation of interferon-induced genes in the B, CD4+ and CD8+ T cell compartments, as well as the NK cell and monocyte compartments, during acute infection, and upregulation of OXPHOS genes upon recovery (Fig. S2). Direct comparison of acute children and acute non-ICU adult PBMCs revealed the largest differences were in the monocyte and CD8+ T cell compartment (Fig. 3A and Fig. S4). There was upregulation of naïve/T memory stem cell genes (e.g., IL7R, LEF1, NOSIP, SELL and TCF7) in CD8+ T cells in children and upregulation of cytotoxicity genes (e.g., CST7, GNLY, GZMA, GZMB, GZMH, NKG7 and PRF1) in non-ICU adults. These acute gene expression differences between children and non-ICU adults in the CD8+ T cell compartment were less prominent but nevertheless persisted into convalescence (Fig. S4). We next determined which subpopulation of cells was responsible for the observed differences by analyzing the DEGs for each subcluster (Fig. 3B). There was a significant number of differentially expressed genes for all cell clusters except for the DC subsets and the interferon-activated naïve CD8+ T cells. Importantly, greater gene expression differences were observed in non-ICU adults than in children, particularly in the NK, B and CD8+ T cell subpopulations (Fig. 3B and Fig. S4).
Fig. 3.
Transcriptomic differences between children and adults with mild COVID-19.
(A) Dotplot showing the number of differentially expressed genes (DEGs) in the innate (DC, monocyte, and NK cell) and adaptive (B, CD4+ T and CD8+ T cell) compartments between children and non-ICU adults during Acute and Convalescent phases. Upregulated genes are shown in red; downregulated genes are shown in blue.
(B) Dotplot showing the number of DEGs in the innate and adaptive immune cell subclusters between children and non-ICU adults during Acute and Convalescent phases. Upregulated genes are shown in red; downregulated genes are shown in blue.
(C) Dotplot showing expression of interferon response genes by innate immune cells from children, non-ICU, and ICU adults during Acute and Convalescence.
(D) Dotplot showing expression of interferon response genes by adaptive immune cells from children, non-ICU, and ICU adults during Acute and Convalescence. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
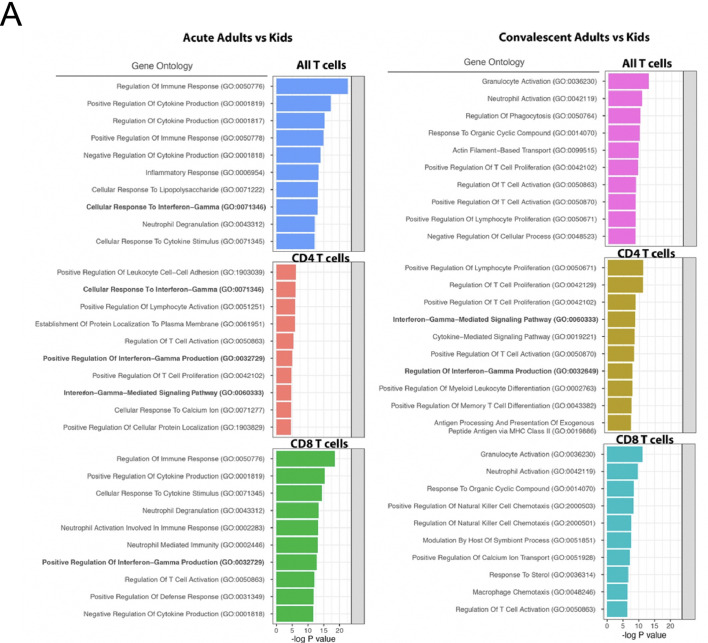
The most consistently upregulated genes were interferon-induced genes. We therefore generated an interferon gene signature (Table S3), and scored each cell subpopulation in children, non-ICU adults and ICU adults (Fig. 3C and D). These data confirmed the high expression of interferon-induced genes, particularly in the monocyte subpopulations and novel interferon-activated naïve CD4+ and CD8+ T cells. Interferon gene signatures were elevated in the acute phase, persisted in the convalescent phase and was higher in non-ICU adults than children. We next performed pseudobulk DEG analysis and GO analysis to confirm that differences in the expression of interferon-regulated genes between children and adults were statistically significant (Fig. S5). During the acute stage, there was statistically significant upregulation of biological processes involved in the production of interferon-γ (in CD4+ and CD8+ T cells), interferon-γ mediated signaling (in CD4+ T cells) and cellular response to interferon-γ (in all T cells and CD4+ T cells). Persistent upregulation of interferon-γ mediated signaling and production was observed in convalescent phase only in CD4+ T cells. These data show that SARS-CoV-2 infection leaves a more profound immunological footprint, especially in the adaptive B and T cell compartments, in adults compared to children with the same disease severity.
3.4. Differences between SARS-CoV-2-specific T cells in children and adults
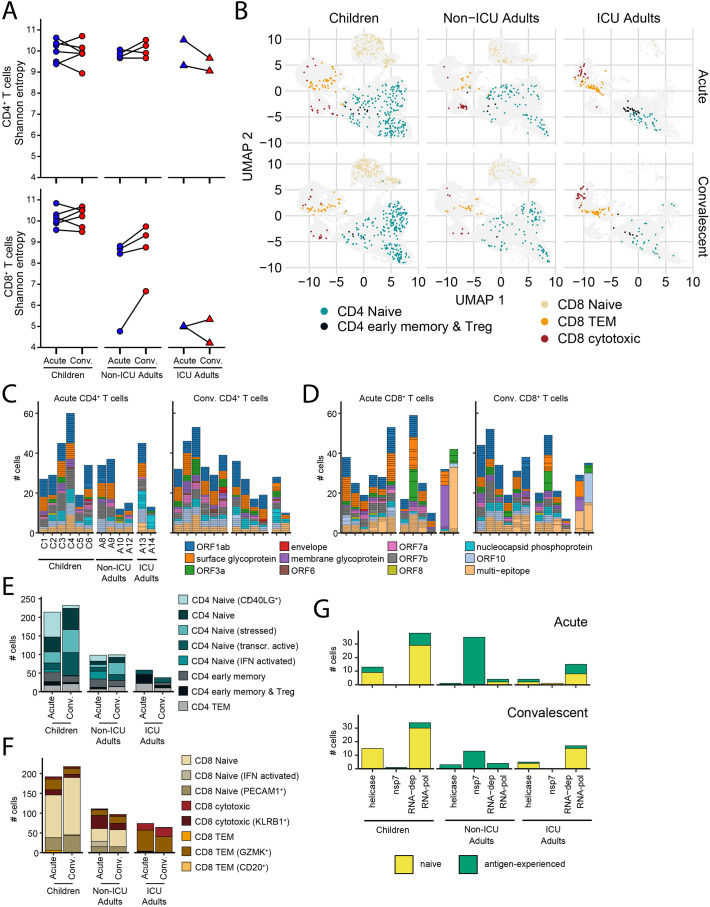
We next analyzed the 158,975 CD4+ T cells and 105,273 CD8+ T cells where we were able to sequence and reconstruct the TCR. There was similar clonal diversity in the CD4+ T cell compartment in children and adults as measured by Shannon Entropy index (Fig. 4A). However, there were significant differences in the CD8+ T cell compartment where children had the most diverse TCRs followed by non-ICU adults and then ICU adults (Fig. 4A). Within each group the diversity did not differ between acute and convalescent timepoints. TCR sequences were annotated using ImmuneCODE [52] and VDJdb [53], large-scale databases of TCR sequences and binding associations with the addition of SARS-CoV-2 TCR sequences reported in the literature and the SARA-CoV-2-annotated cells mapped on to the T cell clusters in the UMAP for children, non-ICU adults and ICU adults (Fig. 4B). The ImmuneCODE database aggregates >135,000 TCR sequences that have been shown with high confidence to react against SARS-CoV-2, while VDJdb collects and curates TCRs of diverse specificity from direct submissions and mining of the literature. These databases do not represent the full landscape of SARS-CoV-2 specificity as they are highly dependent on the peptide pools used to stimulate the T cells. We therefore sought to explore additional SARS-CoV-2 responding T cells within our donors by bulk TRB sequencing pools of proliferated T cells that had been stimulated with recombinant S and RBD proteins. Clonotypes with enriched frequencies after in vitro antigen-specific expansion were considered to have putative SARS-CoV-2 specificity (Table S3). In total, we annotated significantly more SARS-CoV-2-specific T cells in children (431 of 81,686 in acute and 474 of 84,567 cells in convalescence) than non-ICU (209 of 46,216 in acute and 196 of 49,620 in convalescence) and ICU adults (132 of 17,081 in acute and 102 of 19,292 in convalescence) (p < 0.0001, Fisher's exact test). We matched 746 CD4+ and 798 CD8+ T cells in our dataset that were potentially reactive against the envelope, surface glycoprotein, membrane glycoprotein, nucleocapsid phosphoprotein, ORF1ab, ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10 of SARS-CoV-2 (Fig. 4C and D). Children harbored more SARS-CoV-2-annotated CD4+ T cells (mean 76 cells, 74 clones) than non-ICU (mean 49 cells, 49 clones) and ICU adults (48 cells, 40 clones); however, these T cells had similarly diverse TCR repertoires capable of recognizing multiple components of the virus (Fig. 4C). Notably, both ICU adults had clonally expanded SARS-CoV-2-annotated CD4+ T cells. The differences were more apparent in the CD8+ T cell compartment where children had more SARS-CoV-2-specific CD8+ T cells (mean of 75 cells, 66 clones in children compared to mean of 41 clones in non-ICU adults and 12.5 in ICU adults) and these included a small number of expanded clonotypes in four of six children (Fig. 4D). SARS-CoV-2-annotated CD8+ T cell clonal expansions were more marked in several adults, particularly ICU adults where the SARS-CoV-2 annotated repertoire in both acute and convalescent phase was dominated by a few clones.
Fig. 4.
Clonal analysis of SARS-CoV-2-specific T cells.
(A) Shannon entropy score for CD4+ (top) and CD8+ (bottom) T cells in children, non-ICU, and ICU adults in Acute and Convalescent phases. Higher scores indicate higher diversity of the TCR repertoire.
(B) Transcriptional state of T cells annotated as SARS-CoV-2-specific in children, non-ICU, and ICU adults in Acute (top) and Convalescent (bottom) phases. Children have more naïve CD4+ and CD8+ SARS-CoV-2-specific T cells than non-ICU adults.
(C) Epitope specificity of CD4+ T cells for different components of SARS-CoV-2 virus in children, non-ICU, and ICU adults in Acute (left) and Convalescent (right) phases. Each stack in the stacked barplot represents a single clone. Children have more diverse, polyclonal SARS-CoV-2-specific CD4+ T cells than non-ICU adults.
(D) Epitope specificity of CD8+ T cells for different components of SARS-CoV-2 virus in children, non-ICU, and ICU adults in Acute (left) and Convalescent (right) phases. Each stack in the stacked barplot represents a single clone. Children have more diverse, polyclonal SARS-CoV-2-specific CD8+ T cells than non-ICU adults.
(E) Transcriptional state of SARS-CoV-2-annotated CD4+ T cells in children, non-ICU, and ICU adults in Acute and Convalescent phases.
(F) Transcriptional state of SARS-CoV-2-annotated CD8+ T cells in children, non-ICU, and ICU adults in Acute and Convalescent phases.
(G) Number of RTC-specific T cells in children, non-ICU, and ICU adults in Acute (top) and Convalescent (bottom) phases. Naïve CD4+ and CD8+ T cells are yellow and antigen-experienced T cells are green. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We aggregated the SARS-CoV-2-annotated CD4+ T cells and noted that this compartment in children comprised predominantly naïve cell clusters (naïve, CD40LG+, stressed, transcriptionally active and interferon-activated) at both the acute and convalescent timepoints (Fig. 4E). Adults, whose repertoire was dominated by virus-specific TEM, early memory and Tregs, had significantly less naïve and more antigen-experienced SARS-CoV-2-annotated CD4+ T cells compared to children (351 naïve, 102 antigen-experienced in children; 133 naïve, 64 antigen-experienced in non-ICU; 25 naïve, 71 antigen-experienced in ICU adults; p < 0.001, Chi-square). These differences were more pronounced in the CD8+ T cell repertoire where children had more naïve cells than adults (329 naïve, 123 antigen-experienced in children; 118 naïve, 90 antigen-experienced in non-ICU; 4 naïve, 134 antigen-experienced in ICU adults; p < 0.001, Chi-square) (Fig. 4F).
We also analyzed for the expression of TCRs directed against conserved early components of the SARS-CoV-2 replication-transcription complex (RTC) encoded within ORF1ab, including RNA-polymerase cofactor non-structural protein 7 (NSP7), RNA-dependent RNA polymerase (NSP12) and RNA helicase (NSP13), that have been proposed to cross-react with hCoV (Swadling et al., 2021). Children had more RTC-specific T cells, and these were predominantly naïve compared to non-ICU adults where they were almost exclusively antigen-experienced and ICU adults where they were a mix of naïve and experienced T cells (Fig. 4G). Interestingly, the transition from acute to convalescence was associated with attrition of RTC-specific antigen-experienced T cells in both children and adults. Taken together, these data reveal differences in the SARS-CoV-2-specific T cell compartment in children and adults that may reflect prior antigen exposure to cross-reactive hCoV and virus-induced changes to T cell composition and repertoire.
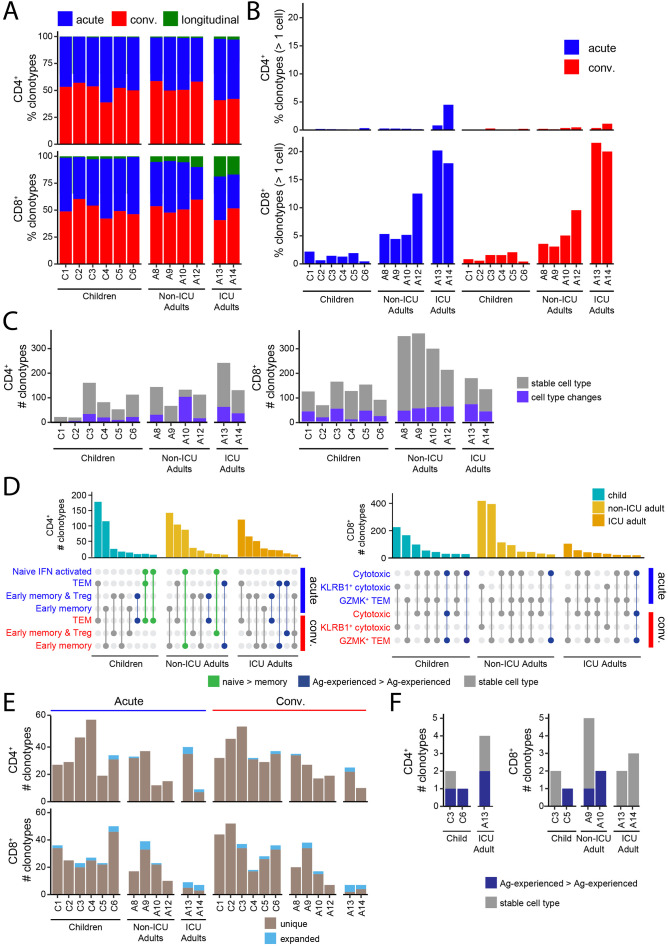
3.5. Annotation and tracking of T cell clonotypes in children and adults
Analysis of the annotated clonotypes revealed that most T cells were unique and only could be detected at either the acute or convalescent timepoints, but rarely both (Fig. 5A). T cell clonotypes that could be tracked longitudinally were more prevalent in the CD8+ T cell compartment in ICU adults. The CD4+ compartment was almost entirely comprised of unique clonotypes (single cells) with a higher proportion of clonotypes being expanded (> 1 cell) within the CD8+ T cell compartment for both the children and adults (Fig. 5B). Notably, adults had a higher frequency of expanded clonotypes compared to children, especially the ICU adults. Clonally expanded T cells detected at both acute and convalescent timepoints were considered longitudinal clones.
Fig. 5.
Clonal dynamics of SARS-CoV-2-specific T cells.
(A) Stacked barplot showing the percentage of SARS-CoV-2-specific CD4+ (upper) and CD8+ (lower) clonotypes that are unique at the Acute (red) or Convalescent (blue) phase of infection or present in both (green) for each subject. Longitudinal clonotypes that are present in both acute and convalescent phases were more common in non-ICU adults than children.
(B) Graph showing the percentage of CD4+ (upper) and CD8+ (lower) longitudinal clonotypes that are detected at both the Acute and Convalescent phase of infection. Longitudinal clonotypes were more common in CD8+ than CD4+ T cells.
(C) Counts for CD4+ (left) and CD8+ (right) clonotypes coloured by whether their cell state remains the same (grey) or changes to a different cell type (purple) between the Acute and Convalescent phases.
(D) Top 8 cell type distributions for CD4+ (left) and CD8+ (right) longitudinal clonotypes for children, non-ICU adults, and ICU adults. The barplots indicate the number of clonotypes with the cell type distribution pattern depicted below each bar where a filled circle indicates that the cell type on the y-axis is present. Distributions are coloured to indicate whether they represent transitions from naïve to antigen-experienced (green), transitions between antigen-experienced compartments (blue) or are the same cell type across the two timepoints (grey).
(E) Clonotype counts for SARS-CoV-2-annotated clonotypes for all donors for the CD4+ (upper) and CD8+ (lower) compartments coloured by whether the clonotype was unique (light brown) or expanded (light blue) at either the Acute (left) or Convalescent (right) phase.
(F) Clonotype counts for longitudinal SARS-CoV-2-annotated CD4+ (left) and CD8+ (right) T cells for the subset of donors that harbor them. Clonotypes are grouped and coloured by whether they have the same cell type at both timepoints (grey) or altered their cell types between Acute and Convalescent phases (dark blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A small number of CD4+ and CD8+ T cell clones were detected in both the acute and convalescent phase in both children and adults making it possible to track their trajectories (Fig. 5C). The CD4+ T cells tended to be antigen-experienced and unique at the acute timepoint. In contrast, the CD8+ T cells were almost exclusively made up of antigen-experienced cells which were a mix of both unique and clonally expanded cells in the acute phase. We tracked the transcriptional state of these longitudinal clones to determine if they had undergone cellular differentiation and found that most clonotypes were stable with few transitions from one T cell state to another (Fig. 5C). We identified 27 out of 38,051 (0.07%) naïve CD4+ T cells in children that transitioned to TEM and 97 out of 15,022 (0.65%) in non-ICU adults that transitioned to early memory T cells (p < 0.0001, Fisher's exact test) (Fig. 5D). Interestingly, child TEM and non-ICU adult CD4+ early memory T cells all originated from the novel interferon-activated naïve CD4+ T cell pool. These cells were not annotated as SARS-CoV-2-specific in the ImmuneCODE and VDJdb databases but, given their differentiation trajectory, may represent clonotypes responding to the virus. No transitions from naïve to memory were identified in the CD8+ T cell clonotypes. Thus, naïve CD4+ T cells were significantly more likely to differentiate into memory T cells in adults than children with the same disease severity.
3.6. Longitudinal tracking of SARS-CoV-2-annotated T cells in children and adults
We next examined the trajectories of SARS-CoV-2-annotated T cells. This revealed that in children they were rarely clonally expanded, consisting of 1.39% of acute and 1.30% of convalescent CD4+ T clones, and 6.52%, of acute and 4.19% of convalescent CD8+ T clones (Fig. 5E). Even among the non-ICU adult CD8+ compartment, clonal expansions of the SARS-CoV-2-annotated cells were rare (acute 7.87%, convalescent 7.32%) (Fig. 5E). The most evidence for SARS-CoV-2-specific T cell clonal expansion was in the ICU adult CD8+ compartment where 50% and 57% of the SARS-CoV-2-annotated clonotypes were expanded at both sampling timepoints, respectively. Only 6 or 7 clonotypes, equating to 0.71% and 1.95% in kids and non-ICU adults, respectively, were found at both timepoints (Fig. 5F). Of the expanded clones, 25% of the CD8+ clonotypes from the ICU adults were longitudinal cloned present at both timepoints. For ICU adult A13, the 7 days between sampling may make repeated detection on the same clonotypes more likely, but this was also observed for ICU adult A14, who was sampled 149 days apart. These CD8+ SARS-CoV-2-specific T cells showed no evidence of phenotypic differentiation (Fig. 5F). Taken together, these data suggest that children have a large pool of diverse, polyclonal naïve virus-specific T cells that largely remain intact, whereas the adult repertoire include clonally expanded memory T cells that undergo activation and attrition, possibly via terminal differentiation.
3.7. Functional state of SARS-CoV-2-specific T cells in children and adults
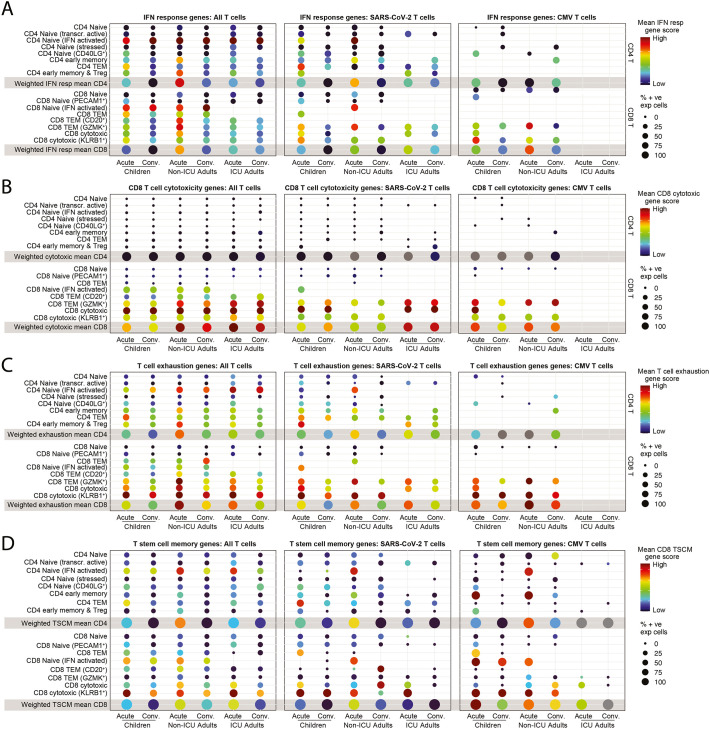
The TCR annotations enabled further analysis of the interferon response gene signature that was differentially expressed at the global level between T cells from children and non-ICU adults (Fig. 3D). The interferon response gene signature (Table S3) was enriched in SARS-CoV-2-specific T cells and we were able to detect high scores in cells, such as subpopulations of CD8+ T cells, that were not evident at the global level (Fig. 6A). Interferon-activated naive CD4+ T cells and CD20+ CD8+ T cells from non-ICU adults had the highest interferon response scores. We were also able to annotate several TCR specificities against cytomegalovirus (CMV), Epstein-Barr virus (EBV) and influenza. There were sufficient numbers of CMV-specific T cells (138 CD4+ and 271 CD8+ clones), but not enough EBV- or influenza-specific T cells, to enable their analysis. No CMV-specific T cell clones were detected in ICU adults. Interestingly, we detected interferon-response gene activation during the acute phase in a small number of CMV-specific cytotoxic KLRB1+ cytotoxic and GZMK+ TEM (Fig. 6A). This suggests that there may be bystander T cell activation, particularly in acute non-ICU adults where the score was higher.
Fig. 6.
T cell interferon activation, cytotoxicity, and exhaustion states.
(A) Interferon response gene scores for all T cells (left), SARS-CoV-2-specific T cells (middle) and CMV-specific T cells (right) in children, non-ICU, and ICU adults in Acute and Convalescent phases.
(B) CD8+ T cell cytotoxicity gene scores for all T cells (left), SARS-CoV-2-specific T cells (middle) and CMV-specific T cells (right) in children, non-ICU, and ICU adults in Acute and Convalescent phases.
(C) T cell exhaustion gene scores for all T cells (left), SARS-CoV-2-specific T cells (middle) and CMV-specific T cells (right) in children, non-ICU, and ICU adults in Acute and Convalescent phases.
(D) T stem cell memory (TSCM) gene scores for all T cells (left), SARS-CoV-2-specific T cells (middle) and CMV-specific T cells (right) in children, non-ICU, and ICU adults in Acute and Convalescent phases.
To further investigate the functional state of SARS-CoV-2-specific T cells, we generated a cytotoxicity gene signature (Table S3). Globally, this signature was strongest in CD8+ cytotoxic T cells, and stronger in adults, particularly ICU adults, than children, and absent in the CD8+ naïve T cell and CD4+ T cell populations (Fig. 6B). Interestingly, KLRB1+ cytotoxic T cells, despite their expression of cytotoxicity genes such as GZMK, GZMA, NKG7, CST7 and GNLY (Table S2), did not have a high cytotoxicity score. CD8+ GZMK+ TEM cells in adults expressed a higher cytotoxicity score than children. Overall, there was evidence for increased cytotoxicity in adults compared to children, and this was most noticeable during acute infection. We also detected upregulation of cytotoxic genes in CMV-specific effector cells in the acute stage in children and in both stages in non-ICU adults.
We next generated a T cell exhaustion signature (Table S3). T cell exhaustion scores were significantly higher in CD8+ than CD4+ T cells where it was predominantly expressed by CD4+ interferon-activated naïve T cells (Fig. 6C). Expression was highest in ICU adults, followed by non-ICU adults and then children. Expression was higher during acute infection than in convalescence. In the CD8+ T cell compartment there was evidence for exhaustion of the GZMK+ TEM exhausted memory precursors and cytotoxic T cell subpopulations, particularly the KLRB1+ cytotoxic T cells. Exhaustion scores were higher during acute infection than convalescence and higher in non-ICU adults than children. This pattern was also evident in the SARS-CoV-2- and CMV-annotated T cells, but the smaller number of cells in these groups meant that several cell types were missing.
Finally, we generated a CD8 T stem cell memory (TSCM) signature (Table S3). TSCM scores were most highly enriched in the KLRB1+ cytotoxic T cells. Interestingly, TSCM genes were also expressed by interferon-activated naïve CD8+ and CD4+ T cells, particularly in the non-ICU adult setting (Fig. 6D). Taken together, these data suggest that SARS-CoV-2-specific T cells in children are less activated, less cytotoxic, and less exhausted than their adult counterparts.
3.8. Memory T cell responses to SARS-CoV-2 in children and adults
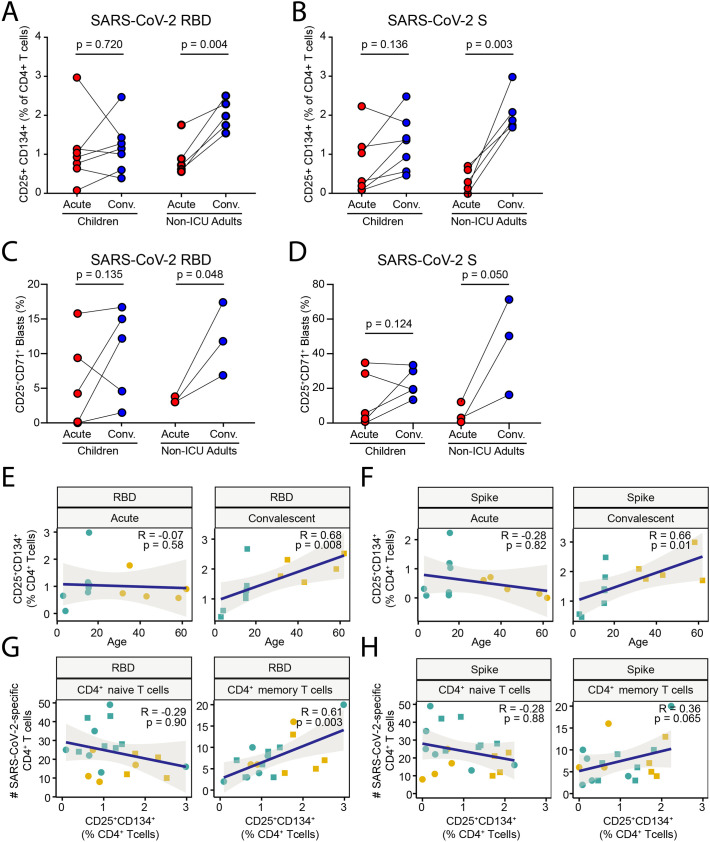
To determine the functional consequences of these age-specific differences in T cell composition and transcriptional state we performed in vitro stimulation with recombinant RBD and S protein to detect antigen-induced upregulation of CD25 and CD134 (OX40) in CD4+ T cells [32]. This assay specifically detects CD45RO+ memory T cells that are activated after secondary stimulation and not naïve T cells that have not encountered antigen before [65]. This analysis showed that during acute infection, children had variable responses to RBD, which did not significantly change upon recovery (Fig. 7A). In contrast, paired samples from non-ICU adults showed a consistent increase in the memory CD4+ T cell response to RBD in all patients. A similar pattern was observed in the CD4+ T cell response to S protein, with no significant increase in children in contrast to the uniform increase in paired samples from all non-ICU adults (Fig. 7B and Fig. S6). These differences were reflected in the T cell proliferation assay, which showed significant increase in proliferative responses to RBD (Fig. 7C) and S protein (Fig. 7D and Fig. S6) in adults but not children. It should be noted that this was an unconventional 14-day proliferation assay due to the low cell numbers because of the limitations on the amount of blood that can be ethically drawn from children for research. There was a moderately positive correlation between the acquisition of memory CD4+ T cell responses in the convalescent phase and age to RBD (Fig. 7E) and S antigen (Fig. 7F). There was also a moderately positive correlation between responses to RBD and the number of SARS-CoV-2-specific memory CD4+ T cells (Fig. 7G) and a trend towards correlation for responses to S protein (Fig. 7H). These data are consistent with the single cell transcriptomic and TCR repertoire analysis and show that natural infection with SARS-CoV-2 induces T cell memory in adults more efficiently than in children.
Fig. 7.
Memory T cell responses to SARS-CoV-2 in children and adults.
(A) Frequency of CD25+CD134+CD4+ T cells in cultures of PBMCs stimulated with recombinant SARS-CoV-2 RBD protein from children and non-ICU adults in Acute and Convalescent phases.
(B) Frequency of CD25+CD134+CD4+ T cells in cultures of PBMCs stimulated with recombinant SARS-CoV-2 Spike (S) protein from children and non-ICU adults in Acute and Convalescent phases.
(C) CD4+ T cell proliferative response in 14-day cultures of PBMCs stimulated with recombinant SARS-CoV-2 RBD protein from children and non-ICU adults in Acute and Convalescent phases.
(D) CD4+ T cell proliferative response in cultures of PBMCs stimulated with recombinant SARS-CoV-2 S protein from children and non-ICU adults in Acute and Convalescent phases.
(E) Linear regression of CD25+CD134+ response to RBD protein by CD4+ T cells with age in Acute (left) and Convalescent (right) phases. The magnitude of the memory response to RBD scales linearly with age in convalescent samples.
(F) Linear regression of CD25+CD134+ response to S protein by CD4+ T cells with age in Acute (left) and Convalescent (right) phases. The magnitude of the memory response to RBD scales linearly with age in convalescent samples.
(G) Linear regression of proliferative response to RBD protein by CD4+ T cells with number of SARS-CoV-2-specific naïve (left) and memory (right) T cells. The magnitude of the proliferative memory response to RBD scales linearly with age in convalescent samples.
(H) Linear regression of proliferative response to S protein by CD4+ T cells with number of SARS-CoV-2-specific naïve (left) and memory (right) T cells.
4. Discussion
The immune response to SARS-CoV-2 and the immunopathogenesis of severe life-threatening COVID-19 has been the focus of intense investigation since the first cluster of pneumonia cases were reported in Wuhan, China on the 31st of December 2019. Important insights have derived from the application of innovative single cell technologies and tissue sampling techniques to deconvolute the local and systemic immune response [66]. While initial studies have examined adults across the disease severity spectrum, it is only recently that efforts have been directed more towards understanding the immune response of children exposed to SARS-CoV-2. These studies have contributed to a detailed picture in which children are able to rapidly eliminate the virus due to their higher steady state expression of interferon genes and pre-activated innate immune system, especially in the upper respiratory tract [8,9]. These landmark studies by Loske et al. and Yoshida et al. were designed to detect high level differences in the innate and adaptive immune of children and adults across the COVID-19 disease severity spectrum. For example, only 9 of the 19 children and 5 of the 18 adults in the Yoshida et al. study was mild/asymptomatic [9]. However, Loske et al. [8] did not analyze the TCR repertoire and Yoshida et al. [9] did not annotate the TCRs for SARS-CoV-2 specificity and neither tracked T cell clonal dynamics longitudinally. Nevertheless, similar local innate immune defense mechanisms may operate to protect children from SARS and Middle Eastern Respiratory Syndrome (MERS) to which they are also less susceptible [67,68]. However, this innate resistance to SARS-CoV-2 infection may come at a cost and it is still unclear how the rapid clearance of viral antigens impacts on the adaptive immune response and the generation of immunological memory in children. This is particularly relevant as there is emerging public health concerns over the relative merits and risks of infection- versus vaccine-induced immunity in children. Here, we have concentrated on the systemic immune response in children and adults with the same mild/asymptomatic disease and tracked responses during acute infection and in the convalescent recovery phase to ascertain the factors that may contribute to age-specific differences in COVID-19 severity and its consequences for SARS-CoV-2 immunity.
There are several strengths and weaknesses to our study design. Unlike other cohorts which compared children and adults across the spectrum of COVID-19 disease severity, we chose to focus on children and adults with the same asymptomatic/mild disease to ensure that we were comparing like with like.
This resolves any confounding factors relating to disease heterogeneity and the impact of variables such as medical interventions and treatments. Furthermore, we compared children with adult family members cohabiting in the same household. This removes any confounding impact of host genetics [69] and environmental exposures [70,71] which may impact on the composition and state of immune system activation [72]. These factors make our study unique, and we were able to detect statistically significant differences despite the seemingly small patient sample size.
Our multimodal analysis of the acute and convalescent immune response revealed that COVID-19 leaves a deeper immunological footprint in the adaptive immune system in adults than children. While both children and adults make similar antibody responses against S protein, adults had more circulating activated CD38+ HLA-DR+ T cells. CD38 and HLA-DR are classical markers of viral infection which may also be induced by bystander activation [73,74] and trogocytosis [74]. The ICU adults with the highest frequency of activated CD38+ HLA-DR+ T cells also had elevated serum IL-6 levels. Deconvolution of the circulating immune compartment by high dimensional flow cytometry and single cell RNA sequencing revealed age-specific differences in cellular composition of the NK, B and T cell compartments. However, we also detected widespread upregulation of interferon-induced genes in both the innate (monocytes and NK cells) and adaptive compartment (B, CD4+ and CD8+ T cells) in adults during acute infection, but this signature was largely limited to the innate compartment in children. This may reflect differences in timing with early production of interferon in children and late interferon production in adults. The importance of interferons in antiviral immunity and resistance to SARS-CoV-2 infection has been well recognized [75]. What was surprising was the fact that interferon gene signatures were largely restricted to the innate immune compartment, suggesting that SARS-CoV-2 infection left only a small immunological footprint in the circulating B and T cells in children. These data are consistent with evidence for reduced breadth of SARS-CoV-2 antibodies in children [76].
Our analysis also revealed novel subpopulations of naïve T cells, including interferon-activated naïve CD4+ and CD8+ T cells which were expanded during acute infection and declined in the convalescent recovery phase in children, but which nevertheless were also detectable in healthy adults. Interestingly, naïve T cells exposed to interferon or interferon-induced cytokines, such as IL-15, exhibit signs of activation but do not undergo cell proliferation [77]. In this regard, it is notable that persistent secretion of type I and type III interferon and activated naïve T cells have been reported in patients with post-COVID-19 syndrome [78] and it will be interesting to determine if such patients have persistent expansion of interferon-activated naïve CD4+ and CD8+ T cells. Interferon activation of naïve CD8+ T cells have been postulated to enhance their homing, survival, differentiation, antiviral and antibacterial effector functions [79,80]. Importantly, clonal tracking revealed that interferon-activated CD4+ T cells were the precursors of TEM in children and CD4+ early memory T cells in adults. These transitioning expanded T cell clones may represent unannotated SARS-CoV-2-specific T cells. In this regard, the expression of TSCM signature genes by non-ICU adult interferon-activated naïve CD4+ and CD8+ T cells is intriguing. TSCM cells are a rare subset of long-lived memory T cells with the stem cell-like self-renewal and multipotent capacity to generate effector and memory T cells [81]. These data suggests that interferon-activated naïve T cells may be precursors to TSCM. Thus, interferon-activated naïve T cells are a novel cell population and add to the growing recognition from single cell analyses that seemingly homogeneous cell populations, in this case the naïve T cell pool, may be more heterogeneous than previously recognized [66,82]. Furthermore, this suggests that traditional nomenclature based on cell surface markers may be inadequate.
We simultaneously sequenced the transcriptome and TCR repertoire of circulating T cells to characterize the SARS-CoV-2-reactive T cell compartment and the impact of COVID-19 on T cell fate and antiviral memory. Children had more diverse TCR repertoires than adults, consistent with their immunological age and previous reports [9,83]. Interestingly, the SARS-CoV-2-specific T cell compartment in children consisted predominantly of naïve T cells with a diverse repertoire capable of recognizing multiple T cell epitopes, including T cells that recognize components of the RTC. RTC-specific memory T cells have recently been proposed to cross-react with hCoV and mediate immunity during abortive SARS-CoV-2 infection in adult health care workers [84]. These data suggest that cross-reactive antigen-specific T cells generated by VDJ recombination during ontogeny may still be naïve and not yet selected by antigen experience into the memory pool, particularly in young children who have not been repeatedly exposed to cross-reactive hCoV [[85], [86], [87]]. In contrast, adults harbored fewer SARS-CoV-2-specific T cells, and these included a large number of clonally expanded memory T cells, including to the RTC, that may have been selected by prior infection with hCoV. Such pre-existing cross-reactive T cell memory has been implicated as a risk factor for severe COVID-19 in the elderly [23]. On the other hand, recent infection with endemic hCoV have also been associated with less severe COVID-19, possibly by “back-boosting” pre-existing immunity [88].
In addition to the differences in cellular composition, we also detected differences in the transcriptional state of the T cells in children and adults. It has been reported that patients admitted to ICU with severe COVID-19 may have impaired cytotoxicity compared to non-ICU patients as measured by reduced secretion of IL-2 and interferon-γ following in vitro polyclonal stimulation [89]. However, our analysis showed that ICU adults had enhanced cytotoxicity scores (particularly in SARS-CoV-2-specific CD8+ T cells) compared to non-ICU adults, whereas children had the lowest cytotoxicity scores. This cytotoxicity score encompasses 67 genes that includes, but are not limited to, IL-2 and interferon-γ. Increased T cell cytotoxicity in severe COVID-19 has also been reported by other investigators [90]. Intriguingly, it has also been reported that severely ill COVID-19 patients show features of impaired T cell exhaustion from the single cell RNA sequencing of expanded T cells generated by in vitro stimulation with SARS-CoV-2 peptide pools [91]. In contrast, our transcriptomic analysis of unstimulated T cells shows a clear hierarchy of exhaustion with children having the least exhausted CD8+ T cells followed by non-ICU and then ICU adults. This is consistent with a number of studies showing evidence of T cell exhaustion in patients with severe COVID-19 [[92], [93], [94], [95], [96]], although it may be difficult to distinguish exhausted from activated cell states [97,98]. The exhaustion in both non-ICU and ICU adult T cells was accompanied by mitochondrial dysfunction and increased expression of OXPHOS genes, particularly in the two ICU adults. Metabolic dysregulation in patients with severe COVID-19 has been previously described [99,100]. Taken together, these data suggest that SARS-CoV-2-specific T cells from adult were more activated and terminally differentiated than children despite having the same disease severity.
One puzzling aspect of our data is the fact that many naïve SARS-CoV-2-specific T cells remained suggesting that they either failed to be activated or were transiently activated and returned to a naïve state. When naïve longitudinal clones were detected, we only observed recruitment into the memory pool of small numbers of interferon-activated naïve T cells, and this was largely in non-ICU adults. This may reflect sampling differences between timepoints and biological differences between children and adults. In adults, there is evidence for activation, cytotoxicity, and exhaustion of circulating SARS-CoV-2-specific T cells. Under these circumstances, it is possible that hCoV cross-reactive memory and cytotoxic T cells are preferentially recruited at the expense of naïve T cells into the SARS-CoV-2 response where they undergo terminal differentiation leading to clonal attrition and contraction of the SARS-CoV-2-specific memory and effector T cell pool in convalescence. This exclusion of adult naïve T cells from the response by pre-existing cross-reactive memory T cells is consistent with T cell original antigenic sin [101]. Such imprinting may bias T cell responses and promote the immune escape of SARS-CoV-2 and its variants. The description of pre-existing RTC-specific T cells that expand in adult health care workers with abortive SARS-CoV-2 infection [84] argues against this notion. However, we found antigen-experienced RTC-specific T cells decreased in number in both adults and children, consistent with their clonal attrition. In children, we have shown that, apart from the interferon-activated naïve T cells, there is little evidence for activation of circulating T cells. Therefore, it is possible that naïve SARS-CoV-2-specific T cells are sub-optimally activated in children due to the rapid clearance of viral antigens [[8], [9], [10]]. Rapid viral clearance may also shut down the secretion of interferon and interferon-induced IL-15, cytokines that have been shown to be needed for the generation of CD8+ memory T cells [102,103]. This has important implications for the development of T cell memory and the protection afforded by infection-induced immunity in children.
Longitudinal analysis detected the generation of robust CD4+ T cell memory responses to RBD and S protein in adults but not children by the OX40 assay, which detects memory and not naïve CD4+ T cell responses [32,65]. These data are consistent with murine studies showing that CD8+ T cells of adult origin were more likely to develop into memory T cells than those of fetal origin [104]. In children, this result agrees with the single cell transcriptomic and TCR data showing little evidence for activation of circulating SARS-CoV-2-specific T cells and recruitment of naïve T cells into the memory pool. Adults also did not show evidence of de novo naïve T activation or recruitment but, in contrast to the children, had an expanded SARS-CoV-2-specific memory T cell pool. We noted that OX40 responses were positively correlated with age and the number of SARS-CoV-2-specific memory CD4+ T cells. Interestingly, single cell analysis of in vitro stimulated PBMCs suggest that severe COVID-19 is associated with increased cytotoxic CD4+ and decreased regulatory T cell activity [90]. We note that early memory T cells and Tregs dominated the SARS-CoV-2-specific CD4+ T cell compartment in the ICU adults. There was also more evidence of acute systemic T cell activation, cytotoxicity and exhaustion in both SARS-CoV-2-specific and non-specific adult T cells which declined in convalescence. Therefore, the development of RBD and S protein memory T cell responses in adults may not only be due to the emergence of new memory T cells from interferon-activated naïve T cells but also recovery of pre-existing memory T cells from exhaustion or active suppression by regulatory T cells.
Several studies from Hong Kong, Australia and the United States have shown impaired humoral and cellular immune responses in children exposed to SARS-CoV-2 [10,26,76,105]. This agrees with our data showing impaired generation of T cell memory responses in children compared to adults. It is possible that the rapid efficient elimination of virus by the innate immune system reduces the antigen availability and prolonged cytokine exposure needed to generate long-lived cellular immunity. Under this scenario, since responses are short-lived, children are dependent on their primed innate immune system for continuing protection from reinfection. These data collectively contradict a recent study showing that children may actually generate more robust adaptive immune responses to SARS-CoV-2 than adults [106]. In a large cohort of convalescent primary school children and their teachers from England, it was shown that children make higher titres of cross-reactive antibodies and T cell memory responses than adults. However, this involved a heterogeneous study group with an uneven mix of male and female patients who are not hospitalized but may otherwise suffer from varying degrees of disease severity. Another point of difference is that our study examines the response of a naïve patient group to the second wave of COVID-19 in Sydney which occurred after a period of a time when there were no SARS-CoV-2 cases in the community. The study from England began recruiting after the lockdown was lifted in June 2020 in the setting of widespread community transmission and may include children and adults who have been multiply exposed to SARS-CoV-2.
A critical question arising from all these studies is the risk of reinfection following COVID-19 in children. Our data supports other studies that would suggest that potent innate immunity undermines the adaptive immune response. If correct this suggests that vaccination, for example with of the BNT162b2 Covid-19 vaccine [107], will be required to bypass this immune bottleneck in the upper airway in children and allow them to generate long-lasting immunity. Future studies involving more innovative technologies with smaller sample requirements, including nasal sampling of local immune responses, may provide a more complete picture of the dynamic clonal landscape of both local and systemic T cell responses to SARS-CoV-2 and hCOV.
4.1. Limitations of the study
A major limitation of our study is that sample size of our cohort is relatively small. In addition, only two of the children were < 5 years of age and 5 were adolescent. However, we had carefully designed the study to longitudinally track the clonal dynamics of SARS-CoV-2-specific T cells in children and adults with the same asymptomatic/mild disease severity. This avoids the problem of comparing heterogeneous patient populations across the spectrum of COVID-19 disease severity and the confounding effects of disease treatments. Moreover, we studied children and adults from the same households to eliminate the confounding effects of host genetics and environmental factors that contribute to variation in the human immune system [72]. We complemented our cohort with an additional two ICU patients with severe COVID-19 and the findings relating to these may be regarded as circumstantial. In total, we analyzed 433,301 single cells directly ex vivo, including the TCR specificity of 158,975 CD4+ T cells and 105,273 CD8+ T cells, making it one of the largest single cell datasets to be made available.
Another limitation of our study is the short duration to follow up, and it will be interesting to see the impact of immunity on intercurrent hCoV and SARS-CoV-2 reinfection rates with longer follow-up. Accordingly, large-scale prospective studies involving longitudinal study of homogeneous patient groups with the same disease severity will be needed to determine the relative merits and risks of infection- vs vaccine-induced immunity against SARS-CoV-2. Due to technical reasons and the volume of blood sample that can be ethically drawn from children, we were only able to test memory CD4+ T cell responses to RBD and S protein. In addition, the identification of SARS-CoV-2-specific T cells in our study was dependent on the annotation of their TCR in ImmuneCODE and VDJdb databases, which are almost certainly incomplete. Another limitation of our study is the absence of pre-exposure blood samples and longitudinal viral loads to determine the kinetics of viral clearance, interferon response and baseline cross-reactive immunity to hCoV. Such data would provide direct evidence that viral persistence is a driver of T cell activation, exhaustion, and memory formation in adults compared to children.
The following are the supplementary data related to this article.
Supplementary Fig. S1.
Representative FACS plots showing the gating for CD38+HLA-DR+ CD4+ T cells (top) and CD8+ T cells (bottom)
(A) Child in Acute Phase
(B) Non-ICU adult in Acute Phase
(C) ICU adult in Acute Phase.
Supplementary Fig. S2.
Flow cytometric analysis of PBMCs in children and adults with COVID-19. Gates for each population are as follows:
(A) NK cells. CD3-CD56+
(B) NK bright cells. CD3-CD56bright NK
(C) NK dim cells. CD3-CD56dim NK
(D) Naïve CD4+ T cells. abTCR+gdTCR-CD4+ CD45RA + CCR7+
(E) Memory CD4+ T cells. abTCR+gdTCR-CD4+ CXCR5-CD45RA-
(F) Naïve CD8+ T cells. – abTCR+gdTCR-CD8+ CD45RA + CCRA7+
(G) CD8+ central memory T cells (TCM). abTCR+gdTCR-CD8 + CD45RA- CCR7+
(H) CD8+ effector memory T cells (TEM). abTCR+gdTCR-CD8 + CD45RA-CCR7-
(I) CD8+ terminally differentiated effector memory T cells (TEMRA). abTCR+gdTCR-CD8 + CD45RA + CCR7-
(J) Naïve B cells. CD3-CD20 + CD21 + CD27-CD10-
(K) Memory B cells. CD3-CD20 + CD21 + CD27+
(L) Plasmablasts. CD56-CD3-CD4-CD8-CD19 + CD38hiCD27+.
Supplementary Fig. S3.
Representative FACS plot showing expression of MX-1 in naïve CD4+ T cells.
(A) FACS gating strategy for adult (top) and child (bottom).
(B) Histogram showing overlay of MX-1 staining in Acute (red) and Convalescent phases (cyan) for adult (top) and child (bottom).
Supplementary Fig. S4.
Volcano plots showing top 50 differentially expressed genes. Significantly upregulated genes are shown in red and downregulated genes in blue.
(A) Between acute and convalescent samples in children.
(B) Between acute and convalescent samples in non-ICU adults.
(C) Between non-ICU adults and children in the acute phase.
(D) Between non-ICU adults and children in the convalescent phase.
Supplementary Fig. S5.
Pseudobulk differential gene expression analysis showing differences in Gene Ontology (GO) terms between Acute Adults vs Kids (left) and Convalescent Adult vs Kids (right).
Supplementary Fig. S6.
FACS gating strategy for OX40 assay and T cell proliferation assay. (A) Representative FACS plot showing gating strategy for detection of CD25+CD134+CD4+ T cells upon stimulation with nil (top panels) and Spike trimer (bottom panels).(B) Representative FACS plot showing gating strategy for proliferation of CD4+ T cells upon stimulation with nil (top panels), and Spike trimer (bottom panels).
Tally of the number of cells in each cluster for each patient group.
Expression of marker genes in each T cell cluster.
List of genes used to generate the interferon, T cytotoxic, T exhausted and T memory stem cell signatures.
Funding
P.I.C. and T.G.P. are supported by Mrs. Janice Gibson and the Ernest Heine Family Foundation. T.G.P., P.N.B. and J.E.P. are supported by National Health and Medical Research Council (NHMRC) Fellowships APP1155678, APP1145817 and APP1107599, respectively. W.H.K. is supported by the UNSW Cellular Genomics Futures Institute. RD is supported by a UNSW Scientia PhD Scholarship. WK is supported by a Research Training Program scholarship. This work is supported by the Garvan Institute COVID Catalytic Grant, UNSW COVID-19 Rapid Response Research Initiative, National Institutes of Health Centers of Excellence for Influenza Research and Response (CEIRR) COVID-19, Snow Medical Foundation BEAT COVID-19 and Griffith University funding.
Author contributions
P.N.B. is the coordinating principal investigator for clinical site. R.N., P.S.H., P.N.B. and T.G.P. conceived and designed the study. R.N., P.S.H., P.N.B., A.H.-J., A.B., B.T., N.W. and D.C. recruited patients and collected patient data. D.R.C. leads the biospecimen research services. L.Z., A.Y., C.L.L., T.V. and R.B. collected and processed patient samples. R.B. managed the project. W.H.K. and T.G.P. designed experiments. W.H.K. performed single cell transcriptome and repertoire sequencing. M.S. performed bulk TCR sequencing. W.H.K., K.J., J.A.-H., S.Y., J.E.P., W.K. and T.G.P. analyzed the sequencing data. K.J. analyzed the TCR repertoire. C.P. and J.J.Z. performed in vitro T cell stimulation. S.R.-D. and E.K.D. performed and analyzed the flow cytometry and cytokine bead array. F.L., V.M., F.X.Z.L and F.B. measured anti-S antibodies. R.R. and D.C. generated the recombinant RBD and S proteins. P.I.C., A.K.D., C.G.G., J.E.P., R.N., P.S.H., P.N.B. and T.G.P. provided supervision. W.H.K., K.J., J.D.S., P.I.C., A.K.D., R.N., P.S.H., E.K.D., P.N.B. and T.G.P. wrote the manuscript.
Declaration of Competing Interest
The authors declare no competing financial interests.
Acknowledgments
We thank the patients and their families. We thank Miles Davenport, Tony Basten and Robert Brink for critical discussions and comments on the manuscript. We thank Eric Lim, Hira Saeed, and staff in the Garvan-Weizmann Centre for Cellular Genomics for technical support. Drawings were created with BioRender.com.
Data availability
Sequencing data for this study has been deposited in the Gene Expression Omnibus with the accession number GSE196456.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S., China Medical Treatment Expert Group for, C Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen T., Wu D., Chen H., Yan W., Yang D., Chen G., Ma K., Xu D., Yu H., Wang H., Wang T., Guo W., Chen J., Ding C., Zhang X., Huang J., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368 doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan R.E., Adab P., Cheng K.K. Covid-19: risk factors for severe disease and death. BMJ. 2020;368 doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 7.O'Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., Fontanet A., Cauchemez S., Salje H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590:140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 8.Loske J., Rohmel J., Lukassen S., Stricker S., Magalhaes V.G., Liebig J., Chua R.L., Thurmann L., Messingschlager M., Seegebarth A., Timmermann B., Klages S., Ralser M., Sawitzki B., Sander L.E., Corman V.M., Conrad C., Laudi S., Binder M., Trump S., Eils R., Mall M.A., Lehmann I. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol. 2021;40:319–324. doi: 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida M., Worlock K.B., Huang N., Lindeboom R.G.H., Butler C.R., Kumasaka N., Conde C.D., Mamanova L., Bolt L., Richardson L., Polanski K., Madissoon E., Barnes J.L., Allen-Hyttinen J., Kilich E., Jones B.C., de Wilton A., Wilbrey-Clark A., Sungnak W., Pett J.P., Weller J., Prigmore E., Yung H., Mehta P., Saleh A., Saigal A., Chu V., Cohen J.M., Cane C., Iordanidou A., Shibuya S., Reuschl A.K., Herczeg I.T., Argento A.C., Wunderink R.G., Smith S.B., Poor T.A., Gao C.A., Dematte J.E., Investigators N.S.S., Reynolds G., Haniffa M., Bowyer G.S., Coates M., Clatworthy M.R., Calero-Nieto F.J., Gottgens B., O’Callaghan C., Sebire N.J., Jolly C., de Coppi P., Smith C.M., Misharin A.V., Janes S.M., Teichmann S.A., Nikolic M.Z., Meyer K.B. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature. 2022;602:321–327. doi: 10.1038/s41586-021-04345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tosif S., Neeland M.R., Sutton P., Licciardi P.V., Sarkar S., Selva K.J., Do L.A.H., Donato C., Quan Toh Z., Higgins R., Van de Sandt C., Lemke M.M., Lee C.Y., Shoffner S.K., Flanagan K.L., Arnold K.B., Mordant F.L., Mulholland K., Bines J., Dohle K., Pellicci D.G., Curtis N., McNab S., Steer A., Saffery R., Subbarao K., Chung A.W., Kedzierska K., Burgner D.P., Crawford N.W. Immune responses to SARS-CoV-2 in three children of parents with symptomatic COVID-19. Nat. Commun. 2020;11:5703. doi: 10.1038/s41467-020-19545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neeland M.R., Bannister S., Clifford V., Dohle K., Mulholland K., Sutton P., Curtis N., Steer A.C., Burgner D.P., Crawford N.W., Tosif S., Saffery R. Innate cell profiles during the acute and convalescent phase of SARS-CoV-2 infection in children. Nat. Commun. 2021;12:1084. doi: 10.1038/s41467-021-21414-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zohar T., Alter G. Dissecting antibody-mediated protection against SARS-CoV-2. Nat. Rev. Immunol. 2020;20:392–394. doi: 10.1038/s41577-020-0359-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartsch Y.C., Wang C., Zohar T., Fischinger S., Atyeo C., Burke J.S., Kang J., Edlow A.G., Fasano A., Baden L.R., Nilles E.J., Woolley A.E., Karlson E.W., Hopke A.R., Irimia D., Fischer E.S., Ryan E.T., Charles R.C., Julg B.D., Lauffenburger D.A., Yonker L.M., Alter G. Humoral signatures of protective and pathological SARS-CoV-2 infection in children. Nat. Med. 2021;27:454–462. doi: 10.1038/s41591-021-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., Hicks P., Manzoni T.B., Oniyide O., Ramage H., Mathew D., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., D’Andrea K., Kuthuru O., Dougherty J., Pattekar A., Kim J., Han N., Apostolidis S.A., Huang A.C., Vella L.A., Kuri-Cervantes L., Pampena M.B., Unit U.P.C.P., Betts M.R., Wherry E.J., Meyer N.J., Cherry S., Bates P., Rader D.J., Hensley S.E. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864. doi: 10.1016/j.cell.2021.02.010. (e1810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., Garcia-Sastre A. Immunological imprinting of the antibody response in COVID-19 patients. Nat. Commun. 2021;12:3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., Roustan C., Bolland W., Thompson R., Agua-Doce A., Hobson P., Heaney J., Rickman H., Paraskevopoulou S., Houlihan C.F., Thomson K., Sanchez E., Shin G.Y., Spyer M.J., Joshi D., O'Reilly N., Walker P.A., Kjaer S., Riddell A., Moore C., Jebson B.R., Wilkinson M., Marshall L.R., Rosser E.C., Radziszewska A., Peckham H., Ciurtin C., Wedderburn L.R., Beale R., Swanton C., Gandhi S., Stockinger B., McCauley J., Gamblin S.J., McCoy L.E., Cherepanov P., Nastouli E., Kassiotis G. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis T. On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 1960;104:572–578. [Google Scholar]
- 18.Dhenni R., Phan T.G. The geography of memory B cell reactivation in vaccine-induced immunity and in autoimmune disease relapses. Immunol. Rev. 2020;296:62–86. doi: 10.1111/imr.12862. [DOI] [PubMed] [Google Scholar]
- 19.Zhang A., Stacey H.D., Mullarkey C.E., Miller M.S. Original antigenic sin: how first exposure shapes lifelong anti-influenza virus immune responses. J. Immunol. 2019;202:335–340. doi: 10.4049/jimmunol.1801149. [DOI] [PubMed] [Google Scholar]
- 20.Selva K.J., van de Sandt C.E., Lemke M.M., Lee C.Y., Shoffner S.K., Chua B.Y., Davis S.K., Nguyen T.H.O., Rowntree L.C., Hensen L., Koutsakos M., Wong C.Y., Mordant F., Jackson D.C., Flanagan K.L., Crowe J., Tosif S., Neeland M.R., Sutton P., Licciardi P.V., Crawford N.W., Cheng A.C., Doolan D.L., Amanat F., Krammer F., Chappell K., Modhiran N., Watterson D., Young P., Lee W.S., Wines B.D., Mark Hogarth P., Esterbauer R., Kelly H.G., Tan H.X., Juno J.A., Wheatley A.K., Kent S.J., Arnold K.B., Kedzierska K., Chung A.W. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat. Commun. 2021;12:2037. doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501. doi: 10.1016/j.cell.2020.05.015. (e1415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., Mallal S., Lammers M., Rubiro P., Quiambao L., Sutherland A., Yu E.D., da Silva Antunes R., Greenbaum J., Frazier A., Markmann A.J., Premkumar L., de Silva A., Peters B., Crotty S., Sette A., Weiskopf D. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacher P., Rosati E., Esser D., Martini G.R., Saggau C., Schiminsky E., Dargvainiene J., Schroder I., Wieters I., Khodamoradi Y., Eberhardt F., Vehreschild M., Neb H., Sonntagbauer M., Conrad C., Tran F., Rosenstiel P., Markewitz R., Wandinger K.P., Augustin M., Rybniker J., Kochanek M., Leypoldt F., Cornely O.A., Koehler P., Franke A., Scheffold A. Low-avidity CD4(+) T cell responses to SARS-CoV-2 in unexposed individuals and humans with severe COVID-19. Immunity. 2020;53:1258–1271. doi: 10.1016/j.immuni.2020.11.016. (e1255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I., Wang L.F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G., Tan Y.J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 25.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Rohmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Muller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 26.Cohen C.A., Li A.P.Y., Hachim A., Hui D.S.C., Kwan M.Y.W., Tsang O.T.Y., Chiu S.S., Chan W.H., Yau Y.S., Kavian N., Ma F.N.L., Lau E.H.Y., Cheng S.M.S., Poon L.L.M., Peiris M., Valkenburg S.A. SARS-CoV-2 specific T cell responses are lower in children and increase with age and time after infection. Nat. Commun. 2021;12:4678. doi: 10.1038/s41467-021-24938-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tea F., Ospina Stella A., Aggarwal A., Ross Darley D., Pilli D., Vitale D., Merheb V., Lee F.X.Z., Cunningham P., Walker G.J., Fichter C., Brown D.A., Rawlinson W.D., Isaacs S.R., Mathivanan V., Hoffmann M., Pohlman S., Mazigi O., Christ D., Dwyer D.E., Rockett R.J., Sintchenko V., Hoad V.C., Irving D.O., Dore G.J., Gosbell I.B., Kelleher A.D., Matthews G.V., Brilot F., Turville S.G. SARS-CoV-2 neutralizing antibodies: Longevity, breadth, and evasion by emerging viral variants. PLoS Med. 2021;18 doi: 10.1371/journal.pmed.1003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tea F., Lopez J.A., Ramanathan S., Merheb V., Lee F.X.Z., Zou A., Pilli D., Patrick E., van der Walt A., Monif M., Tantsis E.M., Yiu E.M., Vucic S., Henderson A.P.D., Fok A., Fraser C.L., Lechner-Scott J., Reddel S.W., Broadley S., Barnett M.H., Brown D.A., Lunemann J.D., Dale R.C., Brilot F., Australasian & New Zealand, M.O.G.S.G Characterization of the human myelin oligodendrocyte glycoprotein antibody response in demyelination. Acta Neuropathol. Commun. 2019;7:145. doi: 10.1186/s40478-019-0786-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez J.A., Houston S.D., Tea F., Merheb V., Lee F.X.Z., Smith S., McDonald D., Zou A., Liyanage G., Pilli D., Denkova M., Lechner-Scott J., van der Walt A., Barnett M.H., Reddel S.W., Broadley S., Ramanathan S., Dale R.C., Brown D.A., Brilot F. Validation of a flow cytometry live cell-based assay to detect myelin oligodendrocyte glycoprotein antibodies for clinical diagnostics. J. Appl. Lab. Med. 2022;7:12–25. doi: 10.1093/jalm/jfab101. [DOI] [PubMed] [Google Scholar]
- 30.Zaunders J., Munier C.M.L., McGuire H.M., Law H., Howe A., Xu Y., de St Groth B.F., Schofield P., Christ D., Milner B., Obeid S., Dyer W.B., Saksena N.K., Kelleher A.D. Mapping the extent of heterogeneity of human CCR5+ CD4+ T cells in peripheral blood and lymph nodes. Aids. 2020;34:833–848. doi: 10.1097/QAD.0000000000002503. [DOI] [PubMed] [Google Scholar]
- 31.Rouet R., Mazigi O., Walker G.J., Langley D.B., Sobti M., Schofield P., Lenthall H., Jackson J., Ubiparipovic S., Henry J.Y., Abayasingam A., Burnett D., Kelleher A., Brink R., Bull R.A., Turville S., Stewart A.G., Goodnow C.C., Rawlinson W.D., Christ D. Potent SARS-CoV-2 binding and neutralization through maturation of iconic SARS-CoV-1 antibodies. MAbs. 2021;13:1922134. doi: 10.1080/19420862.2021.1922134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaunders J.J., Munier M.L., Seddiki N., Pett S., Ip S., Bailey M., Xu Y., Brown K., Dyer W.B., Kim M., de Rose R., Kent S.J., Jiang L., Breit S.N., Emery S., Cunningham A.L., Cooper D.A., Kelleher A.D. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40) J. Immunol. 2009;183:2827–2836. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
- 33.Hsu D.C., Zaunders J.J., Plit M., Leeman C., Ip S., Iampornsin T., Pett S.L., Bailey M., Amin J., Ubolyam S., Avihingsanon A., Ananworanich J., Ruxrungtham K., Cooper D.A., Kelleher A.D. A novel assay detecting recall response to mycobacterium tuberculosis: comparison with existing assays. Tuberculosis (Edinb) 2012;92:321–327. doi: 10.1016/j.tube.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki K., Levert A., Yeung J., Starr M., Cameron J., Williams R., Rismanto N., Stark T., Druery D., Prasad S., Ferrarini C., Hanafi I., McNally L.P., Cunningham P., Liu Z., Ishida T., Huang C.S., Oswald V., Evans L., Symonds G., Brew B.J., Zaunders J. HIV-1 viral blips are associated with repeated and increasingly high levels of cell-associated HIV-1 RNA transcriptional activity. Aids. 2021;35:2095–2103. doi: 10.1097/QAD.0000000000003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. (e1821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fleming L.M., Yeung B., Rogers A.J., McElrath J.M., Blish C.A., Gottardo R., Smibert P., Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587. doi: 10.1016/j.cell.2021.04.048. (e3529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy D.J., Campbell K.R., Lun A.T., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Law C.W., Chen Y., Shi W., Smyth G.K. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kerneis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S., Jeong S.J., Lee H.K., Park S.H., Park S.H., Choi J.Y., Kim S.H., Jung I., Shin E.C. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M.H., Salloum S., Wang J.Y., Wong L.P., Regan J., Lefteri K., Manickas-Hill Z., Gao C., Li J.Z., Sadreyev R.I., Yu X.G., Chung R.T., Collection, M.C, Processing, T Type I, II, and III interferon signatures correspond to coronavirus disease 2019 severity. J. Infect. Dis. 2021;224:777–782. doi: 10.1093/infdis/jiab288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P., Farber D.L., Sims P.A. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Utzschneider D.T., Gabriel S.S., Chisanga D., Gloury R., Gubser P.M., Vasanthakumar A., Shi W., Kallies A. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat. Immunol. 2020;21:1256–1266. doi: 10.1038/s41590-020-0760-z. [DOI] [PubMed] [Google Scholar]
- 46.Yazar S., Alquicira-Hernandez J., Wing K., Senabouth A., Gordon M.G., Andersen S., Lu Q., Rowson A., Taylor T.R.P., Clarke L., Maccora K., Chen C., Cook A.L., Ye C.J., Fairfax K.A., Hewitt A.W., Powell J.E. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science. 2022;376 doi: 10.1126/science.abf3041. [DOI] [PubMed] [Google Scholar]
- 47.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang H.M., Subramaniam M., Targ S., Nguyen M., Maliskova L., McCarthy E., Wan E., Wong S., Byrnes L., Lanata C.M., Gate R.E., Mostafavi S., Marson A., Zaitlen N., Criswell L.A., Ye C.J. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2018;36:89–94. doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291. doi: 10.1016/j.cels.2018.11.005. (e289) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang J.B., Nathan A., Weinand K., Zhang F., Millard N., Rumker L., Moody D.B., Korsunsky I., Raychaudhuri S. Efficient and precise single-cell reference atlas mapping with symphony. Nat. Commun. 2021;12:5890. doi: 10.1038/s41467-021-25957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nolan S., Vignali M., Klinger M., Dines J.N., Kaplan I.M., Svejnoha E., Craft T., Boland K., Pesesky M., Gittelman R.M., Snyder T.M., Gooley C.J., Semprini S., Cerchione C., Mazza M., Delmonte O.M., Dobbs K., Carreno-Tarragona G., Barrio S., Sambri V., Martinelli G., Goldman J.D., Heath J.R., Notarangelo L.D., Carlson J.M., Martinez-Lopez J., Robins H.S. A large-scale database of T-cell receptor beta (TCRbeta) sequences and binding associations from natural and synthetic exposure to SARS-CoV-2. Res Sq. 2020:rs.3.rs–51964. [Google Scholar]
- 53.Bagaev D.V., Vroomans R.M.A., Samir J., Stervbo U., Rius C., Dolton G., Greenshields-Watson A., Attaf M., Egorov E.S., Zvyagin I.V., Babel N., Cole D.K., Godkin A.J., Sewell A.K., Kesmir C., Chudakov D.M., Luciani F., Shugay M. VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. 2020;48:D1057–D1062. doi: 10.1093/nar/gkz874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Low J.S., Vaqueirinho D., Mele F., Foglierini M., Jerak J., Perotti M., Jarrossay D., Jovic S., Perez L., Cacciatore R., Terrot T., Pellanda A.F., Biggiogero M., Garzoni C., Ferrari P., Ceschi A., Lanzavecchia A., Sallusto F., Cassotta A. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science. 2021;372:1336–1341. doi: 10.1126/science.abg8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lineburg K.E., Grant E.J., Swaminathan S., Chatzileontiadou D.S.M., Szeto C., Sloane H., Panikkar A., Raju J., Crooks P., Rehan S., Nguyen A.T., Lekieffre L., Neller M.A., Tong Z.W.M., Jayasinghe D., Chew K.Y., Lobos C.A., Halim H., Burrows J.M., Riboldi-Tunnicliffe A., Chen W., D'Orsogna L., Khanna R., Short K.R., Smith C., Gras S. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065. doi: 10.1016/j.immuni.2021.04.006. (e1055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Francis J.M., Leistritz-Edwards D., Dunn A., Tarr C., Lehman J., Dempsey C., Hamel A., Rayon V., Liu G., Wang Y., Wille M., Durkin M., Hadley K., Sheena A., Roscoe B., Ng M., Rockwell G., Manto M., Gienger E., Nickerson J., Collection, M.C, Processing, T, Moarefi A., Noble M., Malia T., Bardwell P.D., Gordon W., Swain J., Skoberne M., Sauer K., Harris T., Goldrath A.W., Shalek A.K., Coyle A.J., Benoist C., Pregibon D.C. Allelic variation in class I HLA determines CD8(+) T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abk3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shugay M., Britanova O.V., Merzlyak E.M., Turchaninova M.A., Mamedov I.Z., Tuganbaev T.R., Bolotin D.A., Staroverov D.B., Putintseva E.V., Plevova K., Linnemann C., Shagin D., Pospisilova S., Lukyanov S., Schumacher T.N., Chudakov D.M. Towards error-free profiling of immune repertoires. Nat. Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 58.Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 59.Wickham H., Averick M., Bryan J., Chang W., McGowan L.D.A., François R., Grolemund G., Hayes A., Henry L., Hester J. Welcome to the Tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- 60.Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;7:379–423. [Google Scholar]
- 61.Lex A., Gehlenborg N., Strobelt H., Vuillemot R., Pfister H. UpSet: visualization of intersecting sets. IEEE Trans. Vis. Comput. Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 63.Galletti G., De Simone G., Mazza E.M.C., Puccio S., Mezzanotte C., Bi T.M., Davydov A.N., Metsger M., Scamardella E., Alvisi G., De Paoli F., Zanon V., Scarpa A., Camisa B., Colombo F.S., Anselmo A., Peano C., Polletti S., Mavilio D., Gattinoni L., Boi S.K., Youngblood B.A., Jones R.E., Baird D.M., Gostick E., Llewellyn-Lacey S., Ladell K., Price D.A., Chudakov D.M., Newell E.W., Casucci M., Lugli E. Two subsets of stem-like CD8(+) memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 2020;21:1552–1562. doi: 10.1038/s41590-020-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Billerbeck E., Kang Y.H., Walker L., Lockstone H., Grafmueller S., Fleming V., Flint J., Willberg C.B., Bengsch B., Seigel B., Ramamurthy N., Zitzmann N., Barnes E.J., Thevanayagam J., Bhagwanani A., Leslie A., Oo Y.H., Kollnberger S., Bowness P., Drognitz O., Adams D.H., Blum H.E., Thimme R., Klenerman P. Analysis of CD161 expression on human CD8+ T cells defines a distinct functional subset with tissue-homing properties. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3006–3011. doi: 10.1073/pnas.0914839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phetsouphanh C., Xu Y., Bailey M., Pett S., Zaunders J., Seddiki N., Kelleher A.D. Ratios of effector to central memory antigen-specific CD4(+) T cells vary with antigen exposure in HIV+ patients. Immunol. Cell Biol. 2014;92:384–388. doi: 10.1038/icb.2013.101. [DOI] [PubMed] [Google Scholar]
- 66.Tian Y., Carpp L.N., Miller H.E.R., Zager M., Newell E.W., Gottardo R. Single-cell immunology of SARS-CoV-2 infection. Nat. Biotechnol. 2022;40:30–41. doi: 10.1038/s41587-021-01131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rajapakse N., Dixit D. Human and novel coronavirus infections in children: a review. Paediatr. Int. Child Health. 2021;41:36–55. doi: 10.1080/20469047.2020.1781356. [DOI] [PubMed] [Google Scholar]
- 68.Zimmermann P., Curtis N. Coronavirus infections in children including COVID-19: an overview of the epidemiology, clinical features, diagnosis, treatment and prevention options in children. Pediatr. Infect. Dis. J. 2020;39:355–368. doi: 10.1097/INF.0000000000002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orru V., Steri M., Sole G., Sidore C., Virdis F., Dei M., Lai S., Zoledziewska M., Busonero F., Mulas A., Floris M., Mentzen W.I., Urru S.A., Olla S., Marongiu M., Piras M.G., Lobina M., Maschio A., Pitzalis M., Urru M.F., Marcelli M., Cusano R., Deidda F., Serra V., Oppo M., Pilu R., Reinier F., Berutti R., Pireddu L., Zara I., Porcu E., Kwong A., Brennan C., Tarrier B., Lyons R., Kang H.M., Uzzau S., Atzeni R., Valentini M., Firinu D., Leoni L., Rotta G., Naitza S., Angius A., Congia M., Whalen M.B., Jones C.M., Schlessinger D., Abecasis G.R., Fiorillo E., Sanna S., Cucca F. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J., Maecker H.T., Davis M.M. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carr E.J., Dooley J., Garcia-Perez J.E., Lagou V., Lee J.C., Wouters C., Meyts I., Goris A., Boeckxstaens G., Linterman M.A., Liston A. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liston A., Carr E.J., Linterman M.A. Shaping variation in the human immune system. Trends Immunol. 2016;37:637–646. doi: 10.1016/j.it.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 73.Kim T.S., Shin E.C. The activation of bystander CD8(+) T cells and their roles in viral infection. Exp. Mol. Med. 2019;51:1–9. doi: 10.1038/s12276-019-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia X., Chua B.Y., Loh L., Koutsakos M., Kedzierski L., Olshansky M., Heath W.R., Chang S.Y., Xu J., Wang Z., Kedzierska K. High expression of CD38 and MHC class II on CD8(+) T cells during severe influenza disease reflects bystander activation and trogocytosis. Clin. Transl. Immunol. 2021;10 doi: 10.1002/cti2.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J.S., Shin E.C. The type I interferon response in COVID-19: implications for treatment. Nat. Rev. Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., Idzikowski E., Stelitano D., Bovier F.T., Davis-Porada J., Matsumoto R., Poon M.M.L., Chait M., Mathieu C., Horvat B., Decimo D., Hudson K.E., Zotti F.D., Bitan Z.C., La Carpia F., Ferrara S.A., Mace E., Milner J., Moscona A., Hod E., Porotto M., Farber D.L. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tough D.F., Sun S., Zhang X., Sprent J. Stimulation of naive and memory T cells by cytokines. Immunol. Rev. 1999;170:39–47. doi: 10.1111/j.1600-065x.1999.tb01327.x. [DOI] [PubMed] [Google Scholar]
- 78.Phetsouphanh C., Darley D.R., Wilson D.B., Howe A., Munier C.M.L., Patel S.K., Juno J.A., Burrell L.M., Kent S.J., Dore G.J., Kelleher A.D., Matthews G.V. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022;23:210–216. doi: 10.1038/s41590-021-01113-x. [DOI] [PubMed] [Google Scholar]
- 79.Urban S.L., Berg L.J., Welsh R.M. Type 1 interferon licenses naive CD8 T cells to mediate anti-viral cytotoxicity. Virology. 2016;493:52–59. doi: 10.1016/j.virol.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jergovic M., Coplen C.P., Uhrlaub J.L., Besselsen D.G., Cheng S., Smithey M.J., Nikolich-Zugich J. Infection-induced type I interferons critically modulate the homeostasis and function of CD8(+) naive T cells. Nat. Commun. 2021;12:5303. doi: 10.1038/s41467-021-25645-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gattinoni L., Speiser D.E., Lichterfeld M., Bonini C. T memory stem cells in health and disease. Nat. Med. 2017;23:18–27. doi: 10.1038/nm.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen A., Khoo W.H., Moran I., Croucher P.I., Phan T.G. Single cell RNA sequencing of rare immune cell populations. Front. Immunol. 2018;9:1553. doi: 10.3389/fimmu.2018.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Naylor K., Li G., Vallejo A.N., Lee W.W., Koetz K., Bryl E., Witkowski J., Fulbright J., Weyand C.M., Goronzy J.J. The influence of age on T cell generation and TCR diversity. J. Immunol. 2005;174:7446–7452. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- 84.L. Swadling, M.O. Diniz, N.M. Schmidt, O.E. Amin, A. Chandran, E. Shaw, C. Pade, J.M. Gibbons, N. Le Bert, A.T. Tan, A. Jeffery-Smith, C.C.S. Tan, C.Y.L. Tham, S. Kucykowicz, G. Aidoo-Micah, J. Rosenheim, J. Davies, M. Johnson, M.P. Jensen, G. Joy, L.E. McCoy, A.M. Valdes, B.M. Chain, D. Goldblatt, D.M. Altmann, R.J. Boyton, C. Manisty, T.A. Treibel, J.C. Moon, C.O. Investigators, L. van Dorp, F. Balloux, A. McKnight, M. Noursadeghi, A. Bertoletti, M.K. Maini, Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2, Nature 601 (2022) 110-117. [DOI] [PMC free article] [PubMed]
- 85.Pierce C.A., Preston-Hurlburt P., Dai Y., Aschner C.B., Cheshenko N., Galen B., Garforth S.J., Herrera N.G., Jangra R.K., Morano N.C., Orner E., Sy S., Chandran K., Dziura J., Almo S.C., Ring A., Keller M.J., Herold K.C., Herold B.C. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abd5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gorse G.J., Donovan M.M., Patel G.B. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus-associated illnesses. J. Med. Virol. 2020;92:512–517. doi: 10.1002/jmv.25715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.To, K.K, Cheng V.C., Cai J.P., Chan K.H., Chen L.L., Wong L.H., Choi C.Y., Fong C.H., Ng A.C., Lu L., Luo C.T., Situ J., Chung T.W., Wong S.C., Kwan G.S., Sridhar S., Chan J.F., Fan C.Y., Chuang V.W.M., Kok K.H., Hung I.F., Yuen K.Y. Seroprevalence of SARS-CoV-2 in Hong Kong and in residents evacuated from Hubei province, China: a multicohort study. Lancet Microbe. 2020;1:e111–e118. doi: 10.1016/S2666-5247(20)30053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sagar M., Reifler K., Rossi M., Miller N.S., Sinha P., White L.F., Mizgerd J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Invest. 2021;131 doi: 10.1172/JCI143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazzoni A., Salvati L., Maggi L., Capone M., Vanni A., Spinicci M., Mencarini J., Caporale R., Peruzzi B., Antonelli A., Trotta M., Zammarchi L., Ciani L., Gori L., Lazzeri C., Matucci A., Vultaggio A., Rossi O., Almerigogna F., Parronchi P., Fontanari P., Lavorini F., Peris A., Rossolini G.M., Bartoloni A., Romagnani S., Liotta F., Annunziato F., Cosmi L. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Invest. 2020;130:4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meckiff B.J., Ramirez-Suastegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., Sette A., Ay F., Seumois G., Ottensmeier C.H., Vijayanand P. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183:1340–1353. doi: 10.1016/j.cell.2020.10.001. (e1316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kusnadi A., Ramirez-Suastegui C., Fajardo V., Chee S.J., Meckiff B.J., Simon H., Pelosi E., Seumois G., Ay F., Vijayanand P., Ottensmeier C.H. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Biasi S., Meschiari M., Gibellini L., Bellinazzi C., Borella R., Fidanza L., Gozzi L., Iannone A., Lo Tartaro D., Mattioli M., Paolini A., Menozzi M., Milic J., Franceschi G., Fantini R., Tonelli R., Sita M., Sarti M., Trenti T., Brugioni L., Cicchetti L., Facchinetti F., Pietrangelo A., Clini E., Girardis M., Guaraldi G., Mussini C., Cossarizza A. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat. Commun. 2020;11:3434. doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laing A.G., Lorenc A., Del Molino Del Barrio I., Das A., Fish M., Monin L., Munoz-Ruiz M., McKenzie D.R., Hayday T.S., Francos-Quijorna I., Kamdar S., Joseph M., Davies D., Davis R., Jennings A., Zlatareva I., Vantourout P., Wu Y., Sofra V., Cano F., Greco M., Theodoridis E., Freedman J.D., Gee S., Chan J.N.E., Ryan S., Bugallo-Blanco E., Peterson P., Kisand K., Haljasmagi L., Chadli L., Moingeon P., Martinez L., Merrick B., Bisnauthsing K., Brooks K., Ibrahim M.A.A., Mason J., Lopez Gomez F., Babalola K., Abdul-Jawad S., Cason J., Mant C., Seow J., Graham C., Doores K.J., Di Rosa F., Edgeworth J., Shankar-Hari M., Hayday A.C. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 97.Rha M.S., Shin E.C. Activation or exhaustion of CD8(+) T cells in patients with COVID-19. Cell. Mol. Immunol. 2021;18:2325–2333. doi: 10.1038/s41423-021-00750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wherry E.J., Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Siska P.J., Decking S.M., Babl N., Matos C., Bruss C., Singer K., Klitzke J., Schon M., Simeth J., Kostler J., Siegmund H., Ugele I., Paulus M., Dietl A., Kolodova K., Steines L., Freitag K., Peuker A., Schonhammer G., Raithel J., Graf B., Geismann F., Lubnow M., Mack M., Hau P., Bohr C., Burkhardt R., Gessner A., Salzberger B., Wagner R., Hanses F., Hitzenbichler F., Heudobler D., Luke F., Pukrop T., Herr W., Wolff D., Spang R., Poeck H., Hoffmann P., Jantsch J., Brochhausen C., Lunz D., Rehli M., Kreutz M., Renner K. Metabolic imbalance of T cells in COVID-19 is hallmarked by basigin and mitigated by dexamethasone. J. Clin. Invest. 2021;131 doi: 10.1172/JCI148225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thompson E.A., Cascino K., Ordonez A.A., Zhou W., Vaghasia A., Hamacher-Brady A., Brady N.R., Sun I.H., Wang R., Rosenberg A.Z., Delannoy M., Rothman R., Fenstermacher K., Sauer L., Shaw-Saliba K., Bloch E.M., Redd A.D., Tobian A.A.R., Horton M., Smith K., Pekosz A., D’Alessio F.R., Yegnasubramanian S., Ji H., Cox A.L., Powell J.D. Metabolic programs define dysfunctional immune responses in severe COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Klenerman P., Zinkernagel R.M. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–485. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 102.Kolumam G.A., Thomas S., Thompson L.J., Sprent J., Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schluns K.S., Lefrancois L. Cytokine control of memory T-cell development and survival. Nat. Rev. Immunol. 2003;3:269–279. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 104.Smith N.L., Patel R.K., Reynaldi A., Grenier J.K., Wang J., Watson N.B., Nzingha K., Yee Mon K.J., Peng S.A., Grimson A., Davenport M.P., Rudd B.D. Developmental origin governs CD8(+) T cell fate decisions during infection. Cell. 2018;174:117–130 e114. doi: 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 105.Toh Z.Q., Anderson J., Mazarakis N., Neeland M., Higgins R.A., Rautenbacher K., Dohle K., Nguyen J., Overmars I., Donato C., Sarkar S., Clifford V., Daley A., Nicholson S., Mordant F.L., Subbarao K., Burgner D.P., Curtis N., Bines J.E., McNab S., Steer A.C., Mulholland K., Tosif S., Crawford N.W., Pellicci D.G., Do L.A.H., Licciardi P.V. Comparison of Seroconversion in Children and Adults With Mild COVID-19. JAMA Netw Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dowell A.C., Butler M.S., Jinks E., Tut G., Lancaster T., Sylla P., Begum J., Bruton R., Pearce H., Verma K., Logan N., Tyson G., Spalkova E., Margielewska-Davies S., Taylor G.S., Syrimi E., Baawuah F., Beckmann J., Okike I.O., Ahmad S., Garstang J., Brent A.J., Brent B., Ireland G., Aiano F., Amin-Chowdhury Z., Jones S., Borrow R., Linley E., Wright J., Azad R., Waiblinger D., Davis C., Thomson E.C., Palmarini M., Willett B.J., Barclay W.S., Poh J., Amirthalingam G., Brown K.E., Ramsay M.E., Zuo J., Moss P., Ladhani S. Children develop robust and sustained cross-reactive spike-specific immune responses to SARS-CoV-2 infection. Nat. Immunol. 2022;23:40–49. doi: 10.1038/s41590-021-01089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.E.B. WalterK.R. TalaatC. SabharwalA. GurtmanS. LockhartG.C. PaulsenE.D. BarnettF.M. MunozY. MaldonadoB.A. PahudJ.B. DomachowskeE.A.F. SimoesU.N. SarwarN. KitchinL. CunliffeP. RojoE. KucharM. RametI. MunjalJ.L. PerezR.W. Frenck Jr.E. LagkadinouK.A. SwansonH. MaX. XuK. KouryS. MatherT.J. BelangerD. CooperO. TureciP.R. DormitzerU. SahinK.U. JansenW.C. GruberGroup, C.C.T. Evaluation of the BNT162b2 Covid-19 vaccine in children 5 to 11 years of ageN. Engl. J. Med.38620223546.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tally of the number of cells in each cluster for each patient group.
Expression of marker genes in each T cell cluster.
List of genes used to generate the interferon, T cytotoxic, T exhausted and T memory stem cell signatures.
Data Availability Statement
Sequencing data for this study has been deposited in the Gene Expression Omnibus with the accession number GSE196456.