Abstract
Retinoic acid (RA) binds and activates retinoid X receptor (RXR)/retinoic acid receptor (RAR) heterodimers, which regulate the transcription of genes that have retinoic acid response elements (RARE). The RAR isotypes (α, β and γ) are comprised of six regions designated A–F. Two isoforms of RARα, 1 and 2, have been identified in humans, which have different A regions generated by differential promoter usage and alternative splicing. We have isolated two new splice variants of RARα1 from human B lymphocytes. In one of these variants, exon 2 is juxtaposed to exon 5, resulting in an altered reading frame and a stop codon. This variant, designated RARα1ΔB, does not code for a functional receptor. In the second variant, exon 2 is juxtaposed to exon 6, maintaining the reading frame. This isoform, designated RARα1ΔBC, retains most of the functional domains of RARα1, but omits the transactivation domain AF-1 and the DNA-binding domain. Consequently, it does not bind nor transactivate RARE on its own. Nevertheless, RARα1ΔBC interacts with RXRα and, as an RXRα/RARα1ΔBC heterodimer, transactivates the DR5 RARE upon all-trans-RA binding. The use of RAR- and RXR-specific ligands shows that, whereas transactivation of the DR5 RARE through the RXRα/RARα1 heterodimer is mediated only by RAR ligands, transactivation through the RXRα/RARα1ΔBC heterodimer is mediated by RAR and RXR ligands. Whilst RARα1 has a broad tissue distribution, RARα1ΔBC has a more heterogeneous distribution, but with significant expression in myeloid cells. RARα1ΔBC is an infrequent example of a functional nuclear receptor which deletes the DNA-binding domain.
INTRODUCTION
Retinoic acid (RA) regulates the growth and differentiation of a wide variety of embryonic and adult cell types (1, and references therein). Two classes of receptors bind RA, the retinoic acid receptors (RAR) and the retinoic X receptors (RXR). They belong to the superfamily of steroid-thyroid nuclear hormone receptors (2). The known ligands for the RARs are all-trans-RA (ATRA) and 9-cis-RA and for the RXRs 9-cis-RA only (3–5). Each class of receptor is composed of three genes, named α, β and γ (6–8). Based on sequence homology, the nuclear receptors are structured in modules. The RARs are composed of six regions, A–F (Fig. 1A and C), and the RXRs of five regions, A–E. The A and B regions possess a promoter-specific, ligand-independent transcription activating function (AF-1) (9). The C region constitutes the DNA-binding domain, through which the RARs bind to retinoic acid response elements (RARE), which are specific DNA sequences generally located in the vicinity of target genes. RAREs consist of direct repeats of the consensus sequence (A,G)G(T,G)TCA separated by 1–5 nt (DR1-5) (10–13). RARs bind to RAREs as heterodimers with RXRs. The E region contains the ligand-binding domain, a dimerization interface, the ligand-dependent transcription activating function (AF-2) and the corepressor binding and the coactivator association domains (9,14,15).
Figure 1.
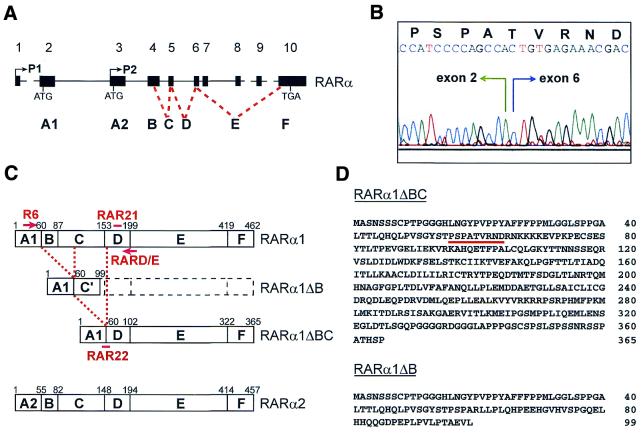
Isolation of novel RARα isoforms. (A) Schematic representation of the genomic structure of the human RARα gene (adapted from refs 24–26 and our unpublished observations). Exons, numbered 1–10, are represented as black boxes. Exons encoding the different domains, A–F, are indicated. Two promoters, P1 and P2, located in front of exons 1 and 3, give rise to the alternative domains A1 and A2, present in the isoforms RARα1 and RARα2 [detailed in (C)]. (B) Trace from the sequencing of the junction between exons 2 and 6 of the RARα1ΔBC isoform. The predicted amino acid sequence of the junction following the open reading frame is indicated. (C) Schematic representation of the human RARα isoforms. The predicted sequence of RARα1ΔB has an altered reading frame from R60 until the stop codon at position 100, designated the C′ domain. Oligonucleotides used as RT–PCR primers, R6 and RARD/E, or as Southern blotting probes, RAR21 and RAR22, are drawn at their approximated positions. (D) Predicted amino acid sequence of novel isoforms RARα1ΔBC and RARα1ΔB. Amino acids of the junction [detailed in (B)] are underlined.
The physiological effects of RA are mediated through activated RXR/RAR heterodimers that stimulate gene transcription (16). In the absence of RA, RXR/RAR binds the nuclear receptor corepressor (N-CoR) or its homolog SMRT, which recruit histone deacetylase leading to transcriptional repression of target genes (reviewed in 15,17). RA binding to RXR/RAR induces dissociation of corepressors, enabling the heterodimer to associate with nuclear receptor coactivator complexes, which include various histone acetyltransferases (15,17,18). These data indicate that RARE-bound, RA-activated RXR/RAR heterodimers recruit the transcriptional apparatus to RARE-containing genes.
Seven isoforms of RARα (RARα1–7), four of RARβ (RARβ1–4) and seven of RARγ (RARγ1–7) have been identified in mice (19–23). The isoforms differ in their 5′-untranslated region (5′-UTR) and their A region, which is either encoded by different exons or deleted. The most abundant isoforms of RARα in mice, RARα1 and RARα2, have also been cloned in human. The human RARα gene, located on chromosome 17q21, consists of 10 exons (Fig. 1A; adapted from refs 24–26 and our unpublished observations). Two promoters, located in front of exons 1 and 3, control expression of RARα1 and RARα2. Start codons of RARα1 and RARα2 lie in exons 2 and 3, respectively. RARα1 is expressed in a wide variety of tissues at similar levels (27). In contrast, RARα2 is expressed in a tissue-specific manner (21) and is up-regulated upon RA- or granulocyte colony-stimulating factor-induced differentiation (28,29). The specific function of each isoform is unknown. Targeted disruptions of single RAR isotypes show normal embryonic development and adult phenotypes (reviewed in 30). However, compound null mutants for RARα1 and total RARβ, for RARα1 and total RARγ or for other RAR and/or RXR isoforms exhibit malformations and are short lived (30). These data suggest that a degree of functional redundancy exists, but certain combinations of isoforms are irreplaceable, underlining the complexity of RA signaling.
In this paper, we report the identification of two new RARα isoforms which result from usage of the A1 region and alternative splicing of other regions. One of these isoforms, designated RARα1ΔB, splices out the B region and, as a consequence of an altered reading frame and a premature termination codon, lacks the rest of the functional domains. The second isoform, designated RARα1ΔBC, splices out the B and C regions, which comprise the AF-1- and DNA-binding domains, while the remaining functional domains are intact. RARα1ΔBC represents an infrequent example of a functional nuclear receptor that deletes the DNA-binding domain.
MATERIALS AND METHODS
Fresh cells and cell lines
Peripheral blood (PB) or bone marrow (BM) mononuclear cells (PBMC and BMMC, respectively) of normal donors and B chronic lymphocytic leukemia (B-CLL) patients were isolated by gradient centrifugation. PB CD3+ T cells, CD19+ B cells and CD56+ natural killer (NK) cells from normal donors were separated using immunomagnetic particles and processed for RNA purification as previously described (31). CD34+ hematopoietic progenitor cells from the BM of normal individuals were purified using the Ceprate LC kit (CellPro, Bothell, WA). Myeloid cell lines NB4 (PML-RARα-expressing acute promyelocytic leukemia), HL60 and U937, T cell acute leukemia cell line Jurkat, Epstein–Barr virus (EBV)-negative Burkitt lymphoma (BL) cell line DG75 and an EBV-transformed B lymphoblastoid cell line derived from normal B cells (B-LCL) were cultured in RPMI-1640 medium, supplemented with 10% fetal calf serum (FCS). Epithelial cell line MCF-7 and COS monkey fibroblasts were grown in DMEM with 10% FCS.
Cloning and sequencing
A λDR2 cDNA expression library (Clontech, Palo Alto, CA) made from total RNA obtained from malignant cells of a B-CLL patient (1) was screened by hybridization as previously described (32), using an RARα probe (1.9 kb EcoRI fragment containing the entire coding region of human RARα1 cDNA; 6). A positive clone was plaque purified, excised and circularized into the recombinant plasmid pDR2 as described by the manufacturer. The cloned insert was sequenced using an ABI automatic sequencer (Perkin Elmer, Branchburg, NJ), appearing to be a splice variant of RARα1. The plasmid was called pDR2-RARα1ΔBC (see Results). RARα cosmids 121 and 124 were gifts of E. Solomon (London, UK). Amplified RT–PCR products (described below) from two B-CLL patients (1 and 2) and from a normal individual (CD19+ 1) were cloned, using the pCR-Trap cloning vector primer kit (Genehunter, Nashville, TN), and sequenced. The DNAstar software was used for database searches and molecular biology programs (Madison, WI).
Semi-quantitative competitive RT–PCR
Total RNA was obtained from human or mouse tissues or obtained from the fresh cells and cell lines described above. Part of the RNA panel of human tissues was purchased (Clontech). RNA (1, 0.3 or 0.1 µg) was reverse transcribed using random primers for 15 min at 42°C, followed by 25 cycles of PCR consisting of 25 s at 95°C, 1 min at 59°C and 3 min at 72°C, in a Perkin Elmer thermal cycler. The upstream primer R6 (5′-GGTGCCTCCCTACGCCTTCT-3′) was located within exon 2 of the RARα gene. The downstream primer RARD/E (5′-AGAGGGCAGGGAAGGTTTCC-3′) was located within exon 6. PCR products were electrophoresed and blotted onto Hybond N+ nylon membranes (Amersham, Little Chalfont, UK). Filters were hybridized with [γ-32P]ATP-labeled oligonucleotide probes RAR21 (5′-GAGCTCCCCCACCTCCGGCGT-3′), upstream to RARD/E within exon 6, or RAR22 (5′-TCCCCAGCCACTGTGAGAAAC-3′), comprising the junction between exons 2 and 6, and autoradiographed. Mouse primers and probes were homologs of the human set: R6M (5′-AGTACCCCCCTACGCCTTCT-3′); RARD/EM (5′-AGAGGGCCGGGAAGGTCTCC-3′); RAR21M (5′-GAGCTCGCCCACCTCAGGAGT-3′); RAR22M (5′-TCCCCAGCCACGGTGCGAAAC-3′).
Constructions
For transient transfections, a pSG5-RARα1ΔBC construct was generated by excising a 2.2 kb BamHI fragment from pDR2-RARα1ΔBC and subcloning into the BamHI site of pSG5.The pSG5-hRARα1 (6) and pSG5-mRXRα (4) expression vectors and the RARE3-tk-luc luciferase reporter (33) were also used. The RSV-β plasmid (Promega, Madison, WI) was used as a control for transfection efficiency. To produce GST fusion recombinant protein, a pGEX-RARα1ΔBC construct was generated by PCR (20 cycles under the conditions described above), using pDR2-RARα1ΔBC as PCR template and primers carrying EcoRI and XhoI restriction sites at the 5′-end (DBC-Eco, 5′-TCTGAATTCATGGCCAGCAACAGCAGCTC-3′; DBC-Xho, 5′-AATCTCGAGTGTGTCCATGTGGCGTGGGC-3′). The PCR product was digested with EcoRI and XhoI and cloned into the EcoRI and XhoI sites of pGEX-4T-1 (Pharmacia, Uppsala, Sweden). The pGEX-RARα1 and pGEX-RXRα (34) constructs were also used.
Antibodies
A rabbit polyclonal antibody directed against the F region of RARα [RPα(F)] (35) was used in western blot analysis. A rabbit antiserum directed against the predicted A1–D domain junction of RARα1ΔBC was made by sequential intradermal injections using the synthetic peptide NH2-YSTPSPATVRNDRNKC-CONH2 (Syntem, Nîmes, France). This antibody, designated RPα1Δ, was used in immunofluorescence studies. Mouse monoclonal antibodies (mAb) directed against the F region of RARα [Ab9α(F)] (35) and against the D–E region of RXRα (4RX3A2) (36) were also used in immunofluorescence as well as in electrophoretic mobility shift assay (EMSA) experiments.
Western blot analysis
The immunoblotting procedures have been previously described (31,37). Briefly, 10 µg whole cell protein extracts or 1 µg purified recombinant protein was fractionated on 10% Tris–glycine/SDS/polyacrylamide gels and electrotransferred onto Hybond-ECL nitrocellulose membranes (Amersham). Filters were blocked for 3 h in 5% non-fat milk in PBS. After overnight incubation in PBS with RPα(F) diluted at 1:1000 or RPα1Δ diluted at 1:100, the filters were washed five times for 10 min, blocked for 10 min in 2.5% non-fat milk and incubated for 30 min with protein A linked to horseradish peroxidase (Amersham) diluted at 1:10 000. After five additional washes, the proteins were visualized using ECL chemiluminescent reagents.
Immunofluorescence
In order to localize RARα1ΔBC in the cell, immunofluorescence was undertaken on transiently transfected COS cells as previously described (37). Briefly, cells were fixed in 4% formaldehyde and incubated overnight at 4°C with RPα1Δ or Ab9α antibodies at 1:100 dilution. Cells were visualized using fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody at 1:100 dilution or cyanin 5 (cy5)-conjugated anti-mouse antibody at 1:50 dilution (Caltag, San Francisco, CA). Nuclei were stained with Hoechst 33258. Cells were analyzed by confocal fluorescence microscopy. As negative controls, non-transfected cells were stained in parallel using the antibodies.
Electrophoretic mobility shift assays
The procedures were similar to those previously described (37). RARα1, RXRα and RARα1ΔBC recombinant proteins were produced and purified from bacterial lysates and incubated, alone or in combinations, with double-stranded [γ-32P]ATP-labeled probes made using the following oligonucleotides and their complements: DR5 RARE (5′-GATCAGGGTTCACCGAAAGTTCACTCGCATATATTA-G-3′) and DR1 RXRE (5′-GATCAGGTCACAGGTCACAGGTCACAGTTCA-3′). For supershifts, 1 µg Ab9α(F) or 4RX3A2 mAb was added 10 min before the recombinant protein. Binding reactions were electrophoresed in 10% polyacrylamide gels for 1 h. Gels were dried and autoradiographed.
In vitro interaction assay
GST ‘pull-down’ assays were performed as previously described (37). Briefly, bacterial lysates containing GST–RXRα or GST protein were bound for 2 h on glutathione–Sepharose beads. After four washes, beads were incubated for 1 h at 4°C with whole cell protein extracts of COS cells transfected with pSG5-RARα1ΔBC or pSG5-RARα. After four washes, SDS loading buffer was added. Proteins were denatured for 10 min at 100°C, loaded onto SDS–PAGE gels and processed for immunoblotting using the RPα(F) antibody as described above.
Transfections and transactivation assays
COS cells were transfected by the calcium phosphate precipitation method and B-LCL cells by electroporation, as described previously (31). Briefly, 3 × 105 COS cells/35 mm dish were plated the day before transfection, then transfected with 0.1 µg each receptor plasmid following different combinations and 1 µg reporter constructs. B-LCL cells (20 × 106) were electroporated in the presence of 5 µg each expression construct, 7.5 µg RARE3-tk-luc and 2.5 µg RSV-β. The quantities of DNA in each experiment were equalized with the pSG5 vector. Cells were grown in carbon-treated FCS (Gemini, Calabasas, CA) in the presence or absence of 10–6 M ATRA (Hoffman-La-Roche, Basel, Switzerland) or synthetic RAR agonist CD336 or RXR agonist CD2809 (CIRD-Galderma, Sophia Antipolis, France). A reporter lysis buffer was added to cells 24 h after transfection and protein was extracted according to the manufacturer’s instructions (Promega). Standard assays were performed to measure luciferase (Promega) and β-galactosidase activities (Boehringer-Mannheim, Mannheim, Germany) using a Berthold luminometer. Luciferase activity was normalized to β-galactosidase activity. Each experiment was done in triplicate at least twice. Results are expressed as fold induction of luciferase activity induced by transfected receptors relative to the pSG5 vector.
RESULTS
Isolation of novel RARα splice variants
Exons 1 and 2 of the RARα gene encode the 5′-UTR and A1 region of the RARα1 isoform, exon 3 encodes the 5′-UTR and A2 region of the RARα2 isoform and exons 4 and 5 encompass regions B, C and the first 3 amino acids of D. The B–F regions are common to both isoforms (Fig. 1A and C; 21,26). Screening a λDR2 cDNA library from a B-CLL patient (1) with a full-length RARα1 cDNA probe revealed a clone which, upon sequencing, lacked exons 3–5 of the RARα gene. As the A1 region is retained in this clone and the major deleted regions are B and C, this variant receptor was designated RARα1ΔBC. The junction between exons 2 and 6 maintains the reading frame of the D region (Fig. 1B). As a consequence, RARα1ΔBC represents a short form of the RARα1 isoform where the A1 region is juxtaposed to the D–E–F regions (Fig. 1C and D). Using primers flanking the A1–D junction (R6 and RARD/E, Fig. 1C), an RT–PCR technique was set up to detect RARα1ΔBC in total RNA samples. By the use of such primers the RARα1 isoform was co-amplified in the same tube. Southern blot analysis and hybridization with the RAR21 oligonucleotide probe, which comprises sequences of exon 6 upstream of RARD/E (Fig. 1C), allowed specific and simultaneous detection of both RARα1 and RARα1ΔBC amplified fragments (Fig. 2A, left). Hybridization with the RAR22 oligonucleotide probe, which comprises the A1–D junction (Fig. 1C), allowed specific detection of the RARα1ΔBC fragment (Fig. 2A, right). The RARα1ΔBC fragment was subsequently cloned from three independent RT–PCR products, including one from the original patient (1), one from another B-CLL patient (2) and one from a normal individual (CD19+ 1). Sequencing of these clones confirmed the A1–D junction observed in the original RARα1ΔBC cDNA clone. The splice donor and acceptor sites were sequenced using RARα cosmids and primers R6 and RARD/E (data not shown). Sequences previously reported of the exon 2/intron 2 and intron 5/exon 6 boundaries were confirmed (24,26).
Figure 2.
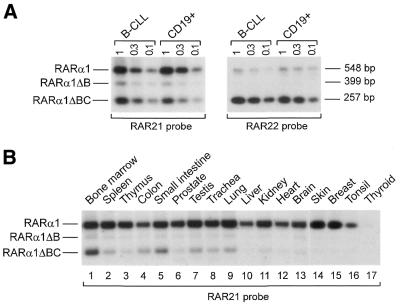
RARα1ΔBC mRNA is expressed at the highest levels in fresh hematopoietic cells. (A) Southern blot of semi-quantitative competitive RT–PCR products using different quantities of total RNA (1–0.1 µg, indicated at the top of the blots) from one B-CLL patient (no. 1) and one normal purified CD19+ B cell sample (no. 1) using primers R6 and RARD/E. Specific bands corresponding to RARα1, RARα1ΔB and RARα1ΔBC were detected using the RAR21 probe (left). The same blot was reprobed with RAR22 to the A1–D junction (right), which is specific for the RARα1ΔBC isoform. The size of the PCR products is indicated on the right. (B) As (A) with 1 µg RNA from different human tissues using the RAR21 probe.
The RT–PCR method generated, in addition to the fragments corresponding to the RARα1 and RARα1ΔBC mRNAs, another product of intermediate size detected with the RAR21 probe (Fig. 2A, left). Cloning and sequencing of this product from the B-CLL patient 1 and normal CD19+ 1 samples showed that it consisted of sequences of exon 2 juxtaposed to exon 5. Therefore, exon 4, which encodes the B and part of the C regions of RARα, was deleted. Thus, this fragment may represent a novel RARα isoform, which was designated RARα1ΔB (Fig. 1C). The junction between exons 2 and 5 resulted in an altered reading frame and a stop codon in the C region at position 100 (Fig. 1D). Therefore, RARα1ΔB represents a truncated form of RARα1 where the A region is fused to a short amino acid stretch derived from exon 5 (C region) and which does not code for a functional receptor. In conclusion, we have isolated two new RARα isoforms, RARα1ΔBC and RARα1ΔB, with the exceptional feature that other functional domains apart from the A region are spliced out.
Transcript and protein tissue expression and nuclear localization of RARα1ΔBC
The competitive RT–PCR technique described above was used to detect expression of RARα1ΔBC and RARα1ΔB mRNAs in different tissues and cell lines. As the RARα1 mRNA is expressed at similar levels in a broad spectrum of tissues, including hematopoietic cells (27), it provides a competitor template which is appropriate as an RT–PCR internal control. The RT–PCR conditions were set up to end the reaction in the exponential phase of amplification of RARα1, RARα1ΔB and RARα1ΔBC (data not shown). Therefore, Southern blot hybridization of the RT–PCRs with the RAR21 probe provided a semi-quantitative analysis of the expression of the novel isoforms. From a panel of human tissues, RARα1ΔBC mRNA was expressed in mononuclear cells from bone marrow (Fig. 2B, lane 1) and from peripheral blood (not shown). As previously shown (Fig. 2A), RARα1ΔBC mRNA was also expressed in normal CD19+ B lymphocytes and malignant B-CLL cells. In addition, it was expressed in CD3+ T lymphocytes and CD56+ NK cells (not shown). RARα1ΔBC mRNA was also detected as a less intense band in spleen, colon, small intestine, testis, trachea and lung (Fig. 2B, lanes 2, 4, 5 and 7–9, respectively). Lower levels or absence of RARα1ΔBC mRNA expression was observed in tissues such as thymus, prostate, liver, kidney, heart, brain, skin, breast, tonsil and thyroid (Fig. 2B, lanes 3, 6 and 10–17, respectively). RARα1ΔBC mRNA was not detected in a panel of mouse tissues which included brain, heart, kidney, liver, lung, skeletal muscles, spleen, lymph node and bone marrow in the presence of a positive signal for RARα1 (data not shown).
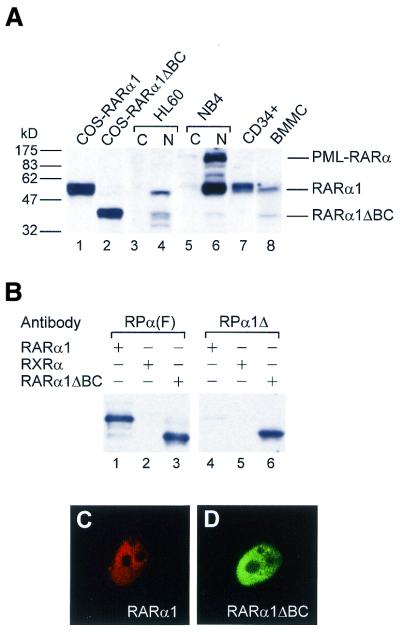
Western blot analysis of transiently transfected COS cells with a polyclonal antibody against the F region of RARα [RPα(F)] showed that, as expected from the nucleotide sequence of its cDNA, RARα1ΔBC is expressed as a 40 kDa protein (Fig. 3A, lane 2), smaller than wild-type RARα1 (50 kDa) (Fig. 3A, lane 1). Native RARα1ΔBC protein of the same size was detected in nuclear extracts of HL60 and NB4 myeloid cell lines and total extracts of BMMC (Fig. 3A, lanes 4, 6 and 8, respectively). However, it was not detected in total extracts of BM CD34+ hematopoietic progenitor cells (lane 7), PBMC (not shown) or in nuclear extracts of monocytic U937, B lymphoid DG75, T lymphoid Jurkat or epithelial MCF-7 cell lines (not shown), in the presence of the signal for the RARα1 isoform, which served as a control. Thus, from the panel of tissues and cell lines analyzed it can be concluded that RARα1ΔBC has a more restricted and weaker expression than RARα1. Furthermore, as it is not expressed in mouse, RARα1ΔBC may be a human-specific RARα isoform.
Figure 3.
The RARα1ΔBC protein is expressed in hematopoietic myeloid cells and is nuclear. (A) Western blot analysis using 10 µg whole cell extracts from COS cells transfected with pSG5-RARα1 (lane 1) or pSG5-RARα1ΔBC (lane 2), BM purified CD34+ cells (lane 7), BMMC (lane 8) and cytoplasmic and nuclear extracts from HL60 (lanes 3 and 4, respectively) and NB4 (lanes 5 and 6, respectively) cells, using the RARα antibody RPα(F). (B) Western blot analysis of purified recombinant proteins (RARα1, RXRα and RARα1ΔBC) with RARα(F) (lanes 1–3) or RPα1Δ made against the A1–D junction (lanes 4–6), showing that this antibody is specific for the RARα1ΔBC isoform. (C) Immunofluorescence analysis of COS cells transfected with RARα1, labeled with a mAb against RARα (Ab9α) and visualized with cyanin 5 (cy5)-conjugated anti-mouse antibody. (D) As (C) except that COS cells were transfected with RARα1ΔBC, labeled with RPα1Δ and visualized with a FITC-conjugated anti-rabbit antibody, showing that RARα1ΔBC, like RARα1, is located in the nucleus.
The presence of RARα1ΔBC in nuclear extracts is consistent with retention of the nuclear localization signal (NLS) of the RARα main isoforms, located in the D region (38). Nevertheless, experiments were performed to determine whether absence of the C domain, which is involved in DNA binding, affected subcellular localization of the receptor. A polyclonal antibody generated against the A1–D junction (RPα1Δ), which recognized RARα1ΔBC efficiently but RARα1 only marginally, as shown by immunoblotting using recombinant proteins (Fig. 3B, lane 6), was used. Immunofluorescence analysis of RARα1ΔBC-transfected COS cells labeled with the RPα1Δ antibody showed that the RARα1ΔBC protein was located in the nucleus (Fig. 3D), with a diffuse pattern similar to that of RARα1, as shown in RARα1-transfected COS cells labeled with a mAb against the F region [Ab9α(F)] (Fig. 3C). Staining of non-transfected control cells using the antibodies did not produce significant background (data not shown). Thus, absence of the DNA-binding domain did not affect the subcellular distribution of RARα1ΔBC.
RARα1ΔBC is found in a complex which binds the RARE and RXRE, through interaction with RXRα
In RARα, the B region contains the AF-1 activity and the C region contains the DNA-binding domain. To determine whether absence of the B and C regions might affect the DNA-binding capacity of RARα1ΔBC, EMSA analysis was performed using a DR5 RARE and the recombinant receptors. It was first observed that, on its own, RARα1ΔBC did not bind to a DR5 response element, unlike RARα1 or RXRα (Fig. 4A, compare lane 2 to lanes 3 and 4). However, when RARα1ΔBC was tested in the presence of RXRα, a retarded complex was observed (compare lanes 4 and 5). This complex was shifted with antibodies against either RARα (lane 6) or RXRα (lane 7). The intensity of the complexes was less than that observed with the complex between RARα1 and RXRα (lanes 8–10), suggesting that RXRα/RARα1 dimers bind the RARE more efficiently than RXRα/RARα1ΔBC dimers. Similar results were obtained with a DR1 RXRE (data not shown). Altogether, these results indicate that, though unable to bind DNA due to absence of the C region, RARα1ΔBC protein is still found in the complex which binds the RARE, probably through heterodimerization with RXRα.
Figure 4.
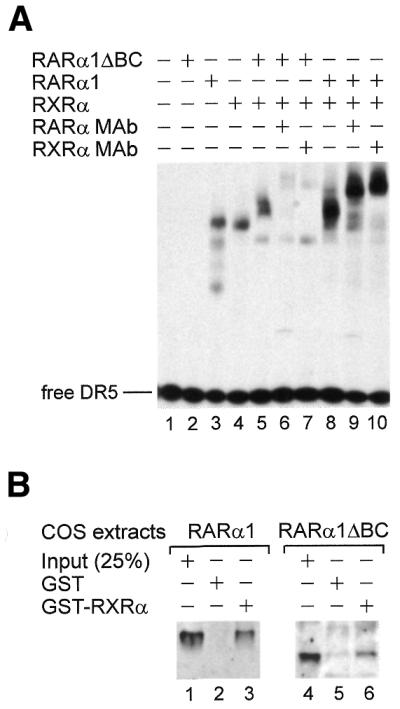
RARα1ΔBC alone does not bind a DR5 response element, but can bind RXRα and be found in the complex that binds DR5. (A) EMSA using purified recombinant proteins (RARα1, RXRα and RARα1ΔBC) shows that RARα1ΔBC does not bind the RARE DR5 (lane 2), unlike RARα1 and RXRα (lanes 3 and 4, respectively). When RARα1ΔBC and RXRα are incubated together (lane 5), a retarded complex is observed; this complex was shifted with a RARα mAb (Ab9α, lane 6) and a RXRα mAb (4RX3A2, lane 7). A complex is also observed when RARα1 and RXRα are co-incubated (lane 8), which shifted with the same antibodies (lanes 9 and 10). (B) GST ‘pull down’ experiments using immobilized GST–RXRα or GST proteins and whole cell extracts from COS cells transfected with RARα1 (lanes 1–3) or RARα1ΔBC (lanes 4–6). Like RARα1, RARα1ΔBC directly interacts with RXRα in vitro.
To obtain additional evidence that RARα1ΔBC may interact with RXRα, GST ‘pull down’ experiments were performed using a GST–RXRα fusion protein bound to glutathione–Sepharose beads. After immunoblotting, RARα1ΔBC was found to directly interact with RXRα in the absence of RA (Fig. 4B, lane 6), as was RARα1 (Fig. 4B, lane 3).
RARα1ΔBC and RXRα contribute to the transactivation mediated by the RARα1ΔBC/RXRα heterodimer on DR5 response elements
The ability of RARα1ΔBC to bind RXRα raises the question of a potential dominant negative function of RARα1ΔBC on RARα1. To test this hypothesis, the transactivation capacities of RARα1ΔBC were analyzed by luciferase reporter assays after transient co-transfection. Experiments performed in the presence of ATRA in COS cells (Fig. 5, white bars) or in B-LCL cells (not shown) showed that, as expected, RARα1ΔBC alone could not significantly induce the activity of a luciferase reporter under the control of DR5 response elements (compare white bars 1 and 4), whereas expression of RARα or RXRα induced 3- and 2.3-fold increases above empty vector (white bars 2 and 3, respectively). However, when co-expressed with RXRα, RARα1ΔBC significantly increased transactivation of the promoter, as did RARα1, though to a lesser extent (3.2- and 4.6-fold, white bars 7 and 5, respectively), showing that in vitro RARα1ΔBC is a functional receptor when heterodimerized with RXRα and does not exert a dominant negative effect. These conclusions were further supported when RARα1, RXRα and RARα1ΔBC were co-transfected (4.7-fold, white bar 8).
Figure 5.
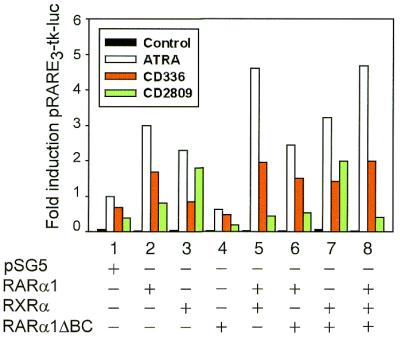
Transactivation of a RARE through combinations of the RXRα, RARα1 and RARα1ΔBC receptors upon binding to the retinoids ATRA (white bars), RAR agonist CD336 (red bars) or RXR agonist CD2809 (green bars). RARα1ΔBC alone does not produce significant activity of the RARE3-tk-luc reporter in COS cells (lane 4); the RXRα/RARα1ΔBC heterodimers transactivate a RARE upon binding to ATRA, CD336 or CD2809 (lane 7); RXRα/RARα1 transduces the transactivating signal mediated by ATRA and CD336 (lane 5); RARα1 fixation inhibits the transactivation mediated by CD2809 binding to RXRα (lanes 5 and 3, green bars, respectively). Luciferase activities were normalized to β-galactosidase activities. A representative experiment, done in triplicate, is shown. All the retinoids were used at 10–6 M. The results are expressed as fold induction produced by the transfected receptors related to pSG5 empty vector in the presence of ATRA.
As ATRA isomerizes in vivo into 9-cis-RA, the ligand of RXR, specific retinoids were used to dissect the contribution of the receptors to the transactivating capacity of the RXRα/RARα1ΔBC heterodimers. As shown in Figure 5 (red bars), transactivation of the DR5 RARE induced by the RAR agonist CD336 through the RXRα/RARα1ΔBC heterodimer was dependent on binding of the ligand to RARα1ΔBC [compare red bars 7 and 3, representing 2.1- and 1.2-fold increases over empty vector (red bar 1), respectively]. When the RXR agonist CD2809 was used (Fig. 5, green bars), transactivation mediated through RXRα (green bar 3) was inhibited by RARα1 fixation (green bar 5). Interestingly, CD2809 induced transactivation through the RXRα/RARα1ΔBC heterodimer (green bar 7). These findings suggest that transactivation of the DR5 RARE by the RXRα/RARα1 heterodimer is mediated only by RAR ligands, whilst both RAR and RXR ligands transactivate the DR5 RARE through binding to the RXRα/RARα1ΔBC heterodimer.
DISCUSSION
A novel isoform of the RARα gene derived from usage of promoter P1 and alternative splicing of the B and C regions was identified. This isoform, designated RARα1ΔBC, lacks the DNA-binding domain and part of the AF-1 domain, whereas the ligand-binding domain, AF-2 domain and other functional domains remain intact. A second isoform was detected, derived from usage of promoter P1 and alternative splicing of the B region. This isoform, designated RARα1ΔB, generates a premature termination codon and lacks most of the RAR functional domains, making it unlikely that it codes for a functional receptor. In addition, its mRNA expression appears to be minor, and we have not studied its significance further. In contrast, the RARα1ΔBC protein is expressed in hematopoietic cells, particularly in primary myeloid cells and cell lines. Whilst it was originally cloned from B-CLL cells, RARα1ΔBC mRNA was expressed at apparently equal levels in normal B lymphocytes and B-CLL cells, as observed by a sensitive RT–PCR assay, ruling out the hypothesis of an association of RARα1ΔBC expression with malignant B lymphocytes.
The different spatial and temporal expression of the RARα, β and γ isoforms suggests that they have specific activities. RARα1ΔBC tissue expression was found to be more restricted and weaker than that of RARα1, but with significant expression in myeloid cells. This suggests that this variant receptor may have distinct functions from RARα1. Both receptors were localized in the nucleus, as expected from retention of the nuclear translocation signal in the D domain (38), and were found in the same nuclear compartment. As for the rest of the RAR isoforms, the exact role of RARα1ΔBC remains to be elucidated. The RARs are phosphoproteins and phosphorylation of specific serine residues influence their functional properties. Two Ser-Pro motifs within the B region of RARα, S74 and S77, which are targets for CDK7 within TFIIH, are critical for AF-1 activity and transcription of RA-inducible genes (39–41). Another serine of RARα1, S157, can be phosphorylated in vitro by protein kinase C (42). Although the exact role of these post-translational modifications has not yet been defined, the absence of these residues in RARα1ΔBC further suggests a divergent role from RARα1.
The transcriptional properties of RARα1ΔBC and RARα1 were compared. Absence of the DNA-binding domain would explain why RARα1ΔBC bound to neither a DR5 RARE nor to a DR1 RXRE. Nevertheless, our results show that it can interact with RXRα in the absence of ligand, consistent with presence of the heterodimerization interface. Moreover, these heterodimers bind to RARE and RXRE, likely through the RXRα half-site, although apparently with less affinity than RXRα/RARα1 heterodimers. Despite this, the RXRα/RARα1ΔBC heterodimers are functional in transactivating a DR5 RARE in transfected COS cells in the presence of ATRA or an RAR-specific retinoid. Thus, our results rule out that RARα1ΔBC could exert a dominant negative effect and suggest that binding of the RXRα/RARα1ΔBC heterodimer through the RXR half-site of the RARE would be sufficient to transduce the activating signal triggered by RA. Furthermore, the results of our transactivation experiments using RAR- and RXR-specific retinoids allow us to conclude that whereas transactivation through RXRα/RARα1 is mediated preferentially or exclusively by RAR ligands (reviewed in 17), transactivation through RXRα/RARα1ΔBC may be mediated by both RAR and RXR ligands. An interesting question which remains to be elucidated is whether the response element repertoire of the RXRα/RARα1ΔBC heterodimer differs from that of the regular RXRα/RARα heterodimer in a target gene promoter context. Although further studies are needed to investigate their specific function, RARα1ΔB and RARα1ΔBC represent a new class of receptor variants which may provide another tier of control in RA signaling.
Acknowledgments
ACKNOWLEDGEMENTS
We thank P. Chambon for the RARα1 and RXRα plasmids and antibodies, H. de Thé for the RARα, PML-RARα and RARE3-tk-luc plasmids and useful discussions, E. Solomon for the RARα cosmids and Fabien Zassadowski, Nicole Balitrand, Michel Schmid and Christelle Dolliger for technical assistance. This work was supported by grants from the Leukaemia Research Fund of Great Britain, the Welsh Bone Marrow Transplant Research Fund, the Ligue Nationale Contre le Cancer of France, the Association pour la Recherche contre le Cancer and Subprograma para el Perfeccionamiento de Doctores y Tecnólogos del Ministerio de Educación y Ciencia of Spain.
REFERENCES
- 1.Wendling O., Ghyselinck,N.B., Chambon,P. and Mark,M. (2001) Roles of retinoic acid receptors in early embryonic morphogenesis and hindbrain patterning. Development, 128, 2031–2038. [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf D.J., Thummel,C., Beato,M., Herrlich,P., Schutz,G., Umesono,K., Blumberg,B., Kastner,P., Mark,M., Chambon,P. and Evans,R.M. (1995) The nuclear receptor family: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petkovich M., Brand,N.J., Krust,A. and Chambon,P. (1987) A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature, 330, 444–450. [DOI] [PubMed] [Google Scholar]
- 4.Leid M., Kastner,P., Lyons,R., Nakshatri,H., Saunders,M., Zacharewski,T., Chen,J.Y., Staub,A., Garnier,J.M., Mader,S. and Chambon,P. (1992) Purification, cloning and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell, 68, 377–395. [DOI] [PubMed] [Google Scholar]
- 5.Heyman R.A., Mangelsdorf,D.J., Dyck,J.A., Stein,R.B., Eichele,G., Evans,R.M. and Thaller,C. (1992) 9-cis Retinoic acid is a high affinity ligand for the retinoid X receptor. Cell, 68, 397–406. [DOI] [PubMed] [Google Scholar]
- 6.Brand N., Petkovich,M., Krust,A., Chambon,P., de The,H., Marchio,A., Tiollais,P. and Dejean,A. (1988) Identification of a second human retinoic acid receptor. Nature, 332, 850–853. [DOI] [PubMed] [Google Scholar]
- 7.Krust A., Kastner,P., Petkovich,M., Zelent,A. and Chambon,P. (1989) A third human retinoic acid receptor, hRAR-γ. Proc. Natl Acad. Sci. USA, 86, 5310–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangelsdorf D.J., Borgmeyer,U., Heyman,R.A., Zhou,J.Y., Ong,E.S., Oro,A.E., Kakizuka,A. and Evans,R.M. (1992) Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev., 6, 329–344. [DOI] [PubMed] [Google Scholar]
- 9.Nagpal S., Friant,S., Nakshatri,H. and Chambon,P. (1993) RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J., 12, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de The H., Vivanco-Ruiz,M.M., Tiollais,P., Stunnenberg,H. and Dejean,A. (1990) Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature, 343, 177–180. [DOI] [PubMed] [Google Scholar]
- 11.Leroy P., Nakshatri,H. and Chambon,P. (1991) Mouse retinoic acid receptor alpha 2 isoform is transcribed from a promoter that contains a retinoic acid response element. Proc. Natl Acad. Sci. USA, 88, 10138–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Umesono K., Murakami,K.K., Thompson,C.C. and Evans,R.M. (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid and vitamin D3 receptors. Cell, 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naar A.M., Boutin,J.M., Lipkin,S.M., Yu,V.C., Holloway,J.M., Glass,C.K. and Rosenfeld,M.G. (1991) The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell, 65, 1267–1279. [DOI] [PubMed] [Google Scholar]
- 14.Nagpal S., Saunders,M., Kastner,P., Durand,B., Nakshatri,H. and Chambon,P. (1992) Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell, 70, 1007–1019. [DOI] [PubMed] [Google Scholar]
- 15.Xu L., Glass,C.K. and Rosenfeld,M.G. (1999) Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev., 9, 140–147. [DOI] [PubMed] [Google Scholar]
- 16.Kastner P., Mark,M., Ghyselinck,N., Krezel,W., Dupe,V., Grondona,J.M. and Chambon,P. (1997) Genetic evidence that the retinoid signal is transduced by heterodimeric RXR/RAR functional units during mouse development. Development, 124, 313–326. [DOI] [PubMed] [Google Scholar]
- 17.Moras D. and Gronemeyer,H. (1998) The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol., 10, 384–391. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 19.Giguere V., Shago,M., Zirngibl,R., Tate,P., Rossant,J. and Varmuza,S. (1990) Identification of a novel isoform of the retinoic acid receptor gamma expressed in the mouse embryo. Mol. Cell. Biol., 10, 2335–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kastner P., Krust,A., Mendelsohn,C., Garnier,J.M., Zelent,A., Leroy,P., Staub,A. and Chambon,P. (1990) Murine isoforms of retinoic acid receptor gamma with specific patterns of expression. Proc. Natl Acad. Sci. USA, 87, 2700–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leroy P., Krust,A., Zelent,A., Mendelsohn,C., Garnier,J.M., Kastner,P., Dierich,A. and Chambon,P. (1991) Multiple isoforms of the mouse retinoic acid receptor alpha are generated by alternative splicing and differential induction by retinoic acid. EMBO J., 10, 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelent A., Mendelsohn,C., Kastner,P., Krust,A., Garnier,J.M., Ruffenach,F., Leroy,P. and Chambon,P. (1991) Differentially expressed isoforms of the mouse retinoic acid receptor beta generated by usage of two promoters and alternative splicing. EMBO J., 10, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagpal S., Zelent,A. and Chambon,P. (1992) RAR-beta 4, a retinoic acid receptor isoform is generated from RAR-beta 2 by alternative splicing and usage of a CUG initiator codon. Proc. Natl Acad. Sci. USA, 89, 2718–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand N.J., Petkovich,M. and Chambon,P. (1990) Characterization of a functional promoter for the human retinoic acid receptor-alpha (hRAR-alpha). Nucleic Acids Res., 18, 6799–6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandolfi P.P., Alcalay,M., Fagioli,M., Zangrilli,D., Mencarelli,A., Diverio,D., Biondi,A., Lo Coco,F., Rambaldi,A., Grignani,F., Rochette-Egly,C., Gaube,M.P., Chambon,P. and Pelicci,P.G. (1992) Genomic variability and alternative splicing generate multiple PML/RAR alpha transcripts that encode aberrant PML proteins and PML/RAR alpha isoforms in acute promyelocytic leukaemia. EMBO J., 11, 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hjalt T.A.H. and Murray,J.C. (1999) Genomic structure of the human retinoic acid receptor-alpha1 gene. Mamm. Genome, 10, 528–529. [DOI] [PubMed] [Google Scholar]
- 27. de The H., Marchio,A., Tiollais,P. and Dejean,A. (1989) Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J., 8, 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zelent A., Enver,T., Gallagher,R. and Waxman,S. (1996) RARα2 isoform in normal granulopoiesis and leukemia. Blood, 88 (suppl. 1), 52 (abstract). [Google Scholar]
- 29.Zelent A., Zhu,J., Lanotte,M., Gallagher,R., Waxman,S., Heyworth,C.M. and Enver,T. (1997) Differential expression of retinoic receptors during multilineage differentiation of haemopoietic progenitor cells—role of the RARα2 isoform in normal granulopoiesis and leukaemia. Blood, 90 (suppl. 1), 44–45 (abstract). [Google Scholar]
- 30.Mark M., Ghyselinck,N.B., Wendling,O., Dupe,V., Mascrez,B., Kastner,P. and Chambon,P. (1999) A genetic dissection of the retinoid signalling pathway in the mouse. Proc. Nutr. Soc., 58, 609–613. [DOI] [PubMed] [Google Scholar]
- 31.Parrado A., Noguera,M.E., Delmer,A., McKenna,S., Davies,J., Le Gall,I., Bentley,P., Whittaker,J., Sigaux,F., Chomienne,C. and Padua,R.A. (2000) Deregulated expression of leukemia translocation protein PLZF in B-cell chronic lymphocytic leukemias (B-CLL) does not affect cyclin A expression. Hematol. J., 1, 15–27. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 33. de The H., Lavau,C., Marchio,A., Chomienne,C., Degos,L. and Dejean,A. (1991) The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell, 66, 675–684. [DOI] [PubMed] [Google Scholar]
- 34. Le Douarin B., Zechel,C., Garnier,J.M., Lutz,Y., Tora,L., Pierrat,P., Heery,D., Gronemeyer,H., Chambon,P. and Losson,R. (1995) The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J., 14, 2020–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaub M.P., Rochette-Egly,C., Lutz,Y., Ali,S., Matthes,H., Scheuer,I. and Chambon,P. (1992) Immunodetection of multiple species of retinoic acid receptor alpha: evidence for phosphorylation. Exp. Cell Res., 201, 335–346. [DOI] [PubMed] [Google Scholar]
- 36.Rochette-Egly C., Lutz,Y., Pfister,V., Heyberger,S., Scheuer,I., Chambon,P. and Gaub,M.P. (1994) Detection of retinoid X receptors using specific monoclonal and polyclonal antibodies. Biochem. Biophys. Res. Commun., 204, 525–536. [DOI] [PubMed] [Google Scholar]
- 37.Delva L., Bastie,J.N., Rochette-Egly,C., Kraïba,R., Balitrand,N., Despouy,G., Chambon,P. and Chomienne,C. (1999) Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol. Cell. Biol., 19, 7158–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambon P., Zelent,A., Petkovich,M., Mendelsohn,C., Leroy,P., Krust,A., Kastner,P. and Brand,N. (1991) The family of retinoic acid nuclear receptors. In Saurat,J.H. (ed.), Retinoids: 10 Years On. Karger, Basel, Switzerland, pp. 10–27.
- 39.Rochette-Egly C., Oulad-Abdelghani,M., Staub,A., Pfister,V., Scheuer,I., Chambon,P. and Gaub,M.P. (1995) Phosphorylation of the retinoic acid receptor-alpha by protein kinase A. Mol. Endocrinol., 9, 860–871. [DOI] [PubMed] [Google Scholar]
- 40.Rochette-Egly C., Adam,S., Rossignol,M., Egly,J.M. and Chambon,P. (1997) Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell, 90, 97–107. [DOI] [PubMed] [Google Scholar]
- 41.Taneja R., Rochette-Egly,C., Plassat,J.L., Penna,L., Gaub,M.P. and Chambon,P. (1997) Phosphorylation of activation functions AF-1 and AF-2 of RAR alpha and RAR gamma is indispensable for differentiation of F9 cells upon retinoic acid and cAMP treatment. EMBO J., 16, 6452–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delmotte M.H., Tahayato,A., Formstecher,P. and Lefebvre,P. (1999) Serine 157, a retinoic acid receptor alpha residue phosphorylated by protein kinase C in vitro, is involved in RXR.RARalpha heterodimerization and transcriptional activity. J. Biol. Chem., 274, 38225–38231. [DOI] [PubMed] [Google Scholar]