Abstract
Genome instability is a primary factor leading to the activation of the p53 tumor suppressor protein. Telomeric repeat (TR) sequences are also responsible for genome integrity. By capping the termini of the chromosomes, TRs prevent them undergoing nucleolytic degradation, ligation or chromosome fusion. Interestingly, telomere shortening was suggested to activate p53, which in turn may cause primary cells to senesce. In order to elucidate the nature of a possible cross talk between the two, we introduced into cells TRs of defined length and investigated their effect on p53 activation and subsequent cellular response. We found that the introduction of a TR into cells leads to stabilization of the p53 protein. This stabilization was specific to TRs and was not observed in response to exposure of cells to plasmids containing non-TR sequences. p53 stabilization requires the presence of an intact p53 oligomerization domain. TR-activated p53 exhibited enhanced transcriptional activity. Eventually, TRs induced p53-dependent growth suppression, measured as a reduction in colony formation.
INTRODUCTION
Both p53 protein and telomeres play a pivotal role in the maintenance of genome stability. Each one of them was shown to be associated with DNA repair pathways and genomic integrity checkpoints; therefore, both could be regarded as guardians of the genome (1). Telomeres are nucleoprotein structures found at the ends of eukaryotic chromosomes. In humans, the telomeric DNA consists of 5–15 kb of (TTAGGG)n repeats (2). Telomere structure and function are regulated by a number of telomeric DNA binding proteins; the main ones are telomeric repeat binding factors (TRF)-1 and TRF-2 (3). Both telomeric DNA and trans-acting protein factors evolve in solving the end replication problem (4–6), protection of chromosome ends from recombination, fusion and degradation (7), and finally, prevent the chromosome ends from being recognized as damaged DNA. Each replication cycle of human primary somatic cells is associated with the loss of 50–150 bp of telomeric repeat (TR) DNA (8–10) and thus in the absence of specific repair mechanism, primary somatic cells stop proliferating when critical telomere length is reached (5,11). This irreversible cell-cycle arrest, termed replicative senescence, is mainly attributed to wild type p53 tumor suppressor activation (12–16). Exposure of chromosome ends and telomeric single-strand damage were recently shown to induce p53-dependent apoptosis or growth arrest, respectively (17). However, the molecular mechanism that underlies p53 activation under these circumstances is still unknown.
The p53 tumor suppressor is a multifunctional protein, which by exerting a variety of activities in the cell has a critical role in protection from cancer development (18–20). The protein, which is a transcription factor, binds the consensus DNA sequences of p53-specific target genes through the core domain and activates transcription through its N-terminus (21). The C-terminus of the protein is considered to be a regulatory domain, which contains several sites for post-translational modifications (22,23). It is well accepted that the main function of the p53 protein is to respond to damaged DNA through several pathways (24). Transcription activation of p21Waf1 and GADD45 leads to G1 cell-cycle arrest and DNA repair (25,26), whereas transcriptional activation of BAX and several other pro-apoptotic genes, may lead to p53-dependent apoptosis (27–32). In addition to sequence-specific DNA binding, p53 was also shown to interact with different types of damaged DNA. The C-terminus domain recognizes double-stranded and single-stranded (ss) DNA ends, internal deletion loops in DNA and helps catalyze the annealing of ssDNA to double strands (33–37). This recognition of damaged DNA may facilitate the activation of p53 sequence-specific DNA binding. A signaling cascade of p53 activation in response to DNA damage involves multiple post-translational modifications of the molecule. These include phosphorylations that are mediated by ataxia telangiectasia mutated (ATM) (38) and/or DNA-phosphatidylinositol 3 kinase (PK) (39) and p300-mediated acetylations (23,40,41). Those modifications are believed to relieve Mdm-2-mediated degradation (39), which allows effective accumulation of a transcriptionally active p53.
Inactivation of the p53 pathway was suggested to account for the escape from replicative senescence that is associated with critical telomere length (12–16). In contrast to normal cells, immortalized cells maintain their telomeres in order to keep a certain level of genomic stability by two means. One pathway that is predominantly used by immortal and cancer cells engages the action of the reverse transcriptase enzyme telomerase (42–45). The second pathway, alternative lengthening of telomeres (ALT), is a less frequently used mechanism based on recombination (46,47).
Our present study was addressed at the elucidation of the relationship between p53 and TRs. To that end, we have exposed the p53 protein to telomeric sequences either by co-transfection of the wild type p53 expression vectors and TR-containing plasmids into p53 null cells or by transfection of TR-containing plasmids into cells expressing endogenous wild type p53 protein. We found that TRs induced the stabilization of both exogenous and endogenous p53 proteins as compared with controls. Furthermore, the extreme C-terminus of p53, which was shown to be the non-specific DNA-binding domain, was found to be dispensable for stabilization induced by TRs, whereas the oligomerization domain was essential. The TR-stabilized p53 exhibited an enhanced transcriptional activity. Finally, activation of p53 by a TR-containing plasmid was associated with significant suppression of colony formation of a cell line containing functional p53. Inactivation of p53 by E6 oncoprotein abrogated this growth suppression effect. This suggests that TR sequences are important constituents of a novel pathway leading to p53 protein activation.
MATERIALS AND METHODS
Cell lines
H1299 cell line (ATCC number: CRL-5803) is a human non-small cell lung cancer, lacking p53 expression due to a 5′-intragenic deletion (48). The cell line was maintained in RPMI medium supplemented with 10% fetal calf serum (FCS). MCF-7 breast cancer cell line (ATCC number: HTB-22) and Hep G2 hepatoblastoma cells (ATCC number: HB-8065) were grown in DMEM supplemented with 10% FCS, 1.0 mM sodium pyruvate and 2 mM l-glutamine. WI-38 mortal cell line (ATCC number: CCL75) derived from a human embryonic lung was grown in MEM supplemented with 10% FCS, 1.0 mM sodium pyruvate and 2 mM l-glutamine. HCT116 colon cancer cells were the kind gift of Dr B. Vogelstein (The Johns Hopkins Oncology Center) and were grown in Mc Coy medium supplemented with 10% FCS and 2 mM l-glutamine.
All cell lines were grown at 37°C in a humidified atmosphere of 5% CO2 in air.
Plasmids
Expression plasmids for p53 were pCMV-Neo-Bam-p53 (provided by Dr B. Vogelstein, Johns Hopkins Oncology Center), p53-360-del and 342-stop were described previously (49).
The reporter plasmids used were Waf-1-Luc, Mdm-2-Luc, Bax-Luc (provided by Dr M. Oren, Weizmann Institute of Science).
pBlueScript-TEL was generated by the subcloning of telomere repeats (TTAGGG)40 insert (240 bp) from pHuR93 plasmid (50) into the PstI site of the pBlueScript. Plasmids containing T3AG3, T2AG2C and T2AG5 repeats were cloned into pSX-neo vector and were kindly provided by Dr T. de Lange (The Rockefeller University).
E6 expression plasmid was kindly provided by Dr L. Sherman (Tel-Aviv University).
All plasmids were purified by Quigen Plasmid Purification kit using the manufacturer’s provided protocol.
DNA transfections
Cells were grown in a complete medium and replated 16–24 h before transfection. For reporter gene assay and protein levels, study cells were transfected by FuGENE 6 Transfection Reagent (Boehringer Mannheim). DNA precipitates were left on the cells for 24 h, after which fresh complete medium was added for the periods indicated. HCT116 cells were transfected by Lipofectamine reagent (Life Technologies). Precipitates were left on the cells for 6 h, after which fresh complete medium was added.
Colony formation assay
HCT116 cells were replated 16–24 h before transfection in 10-cm dishes. The cells were transfected by Lipofectamine (26 µl) with pBlueScript, pBlueScript-TEL or their mixture as indicated. Two micrograms per plate of E6 expression plasmid (when indicated) and 1 µg/plate of pBabe-puro were included. A constant amount of DNA (13 µg/transfection) was maintained. Transfections were performed in a total volume of 6 ml serum-free DMEM medium. Six hours later, the serum-free medium was replaced by DMEM medium supplemented with 10% FCS. Forty-eight hours after transfection, cells were trypsinized and 1 × 105 cells plated out into medium containing puromycin (1 µg/ml). Sixteen days later, plates were fixed, stained with Crystal Violet and colonies were counted.
Western blot analysis
Nuclear extracts were prepared by a modified protocol according to that described before (51). Briefly, 106–107 cells were washed twice with cold phosphate-buffered saline (PBS), cell pellets were resuspended in 400 µl buffer A [10 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 10 µg/ml leupeptin, 1 µg/ml pepstatin and 10 µg/ml aprotinin] by gentle pipetting. After 15 min incubation on ice, 25 µl of 10% NP-40 was added and vortexed vigorously for 10 min. After centrifugation, cell pellets were resuspended in 150–300 µl of buffer C (20 mM HEPES–KOH pH 7.9, 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF, 10 µg/ml leupeptin, 1 µg/ml pepstatin and 10 µg/ml aprotinin). Tubes were transferred to a rotating platform for 15 min at 4°C. After centrifugation the supernatant was kept at –70°C. Extracts were analyzed for protein concentration (Bradford assay).
For total cellular extract, 106 cells were lyzed in sample buffer (140 mM Tris pH 6.8, 22.4% glycerol, 6% SDS, 10% β-mercaptoethanol and 0.02% bromophenol blue) boiled and loaded on 10% polyacrylamide gels containing SDS. Proteins were transferred to nitrocellulose membranes. The p53 protein was detected using p53 monoclonal antibodies: DO-1, PAb-1801 and PAb-421. Anti-p21 antibody (C-19) was purchased from Santa Cruz. The protein–antibody complexes were detected using a horseradish peroxidase (HRP)-conjugated secondary antibody using the super-signal enhanced chemiluminescene system (Pierce).
Luciferase assay
For luciferase assays, MCF-7 and H1299 cells were seeded in 12-well plates or WI-38 cells were seeded in 6-well plates and transfected with 200 ng of different p53-responsive luciferase constructs, 5 ng of p53 expression vector, 200 ng of cytomegalovirus (CMV) β-galactosidase expression vector and increasing amounts of pBlueScript-TEL. The total amount of DNA in each transfection was kept constant by complementing with pBlueScript vector control DNA. After 24–48 h the cells were rinsed twice with cold PBS and lyzed in the wells by cell culture lysis reagent (Promega) for 15 min at room temperature. A sample (100 µl) of lysate was quantified for luciferase activity in the presence of luciferin (Promega) and ATP using a Turner Design Model 20 Luminometer. Transfections were done in triplicate and normalized to β-galactosidase activity as an internal transfection control.
β-Galactosidase enzyme assay
A sample (10–15 µl) of cell lysate was mixed with 100 µl of Lac Z buffer (60 mM Na2CO3, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) and 20 µl of substrate [2 mg/ml o-nitrophenol-β-d-galactopyranoside (ONPG) in Na2CO3, 40 mM NaH2PO4]. The reaction was incubated at 37°C until a yellow color was visible and stopped by the addition of 1 M Na2CO3. The results were read at OD420 using a multi-well ELISA reader.
RESULTS
We were interested in studying the effect of a defined telomeric DNA sequence on the p53 protein, which potentially can recognize DNA both in a sequence-specific and non-sequence-specific manner (22). In designing our experiments we considered the observations that the treatment of cells with oligonucleotides, linearized plasmid DNA, circular DNA with a large gap or single-stranded circular phagemid, can all activate p53 and induce a p53-dependent growth arrest. Supercoiled and nicked plasmids that lack free ends of DNA were ineffective (52). In the above experiments, p53 activation was induced in a DNA sequence-independent manner. Introduction of exogenous DNA fragments containing the TR at the terminus (the ‘telomere seed’) was shown to induce chromosome breakage and de novo telomere formation at the site of the break (telomere seeding) (53–55). As in the telomere seeding experiments, cell lines with inactivated p53 were used, it was possible to follow the de novo telomere formation phenomenon, but not to investigate the effect of TR sequences on the wild type p53 protein.
Our experimental model consisted of a transient expression system in which p53-coding plasmids were transfected in conjunction with TR-containing plasmids and a luciferase reporter construct containing p53-responsive elements. To avoid p53 activation by free DNA ends in a sequence non-specific manner we used two double-stranded circular plasmids: (i) pBlueScript-TEL, which contains the 240 bp of TTAGGG repeats and (ii) an empty construct, pBlueScript. The 240 bp insert of TR (TTAGGG)40 that was originally cloned from the human genomic DNA (55) was subcloned by us into pBlueScript.
TR sequences induce p53 protein stabilization
An important manifestation of the activation of p53 is an increase in protein level. In unstressed cells, the p53 protein is barely detectable and has a very short half-life. This is due to rapid degradation via the Mdm-2/ubiquitin–proteosome pathway (56,57). However, in response to genotoxic stress or hypoxia p53 rapidly accumulates. This stabilization is mostly attributed to the disruption of the p53–Mdm-2 interaction that leads to attenuation in the degradation process (49).
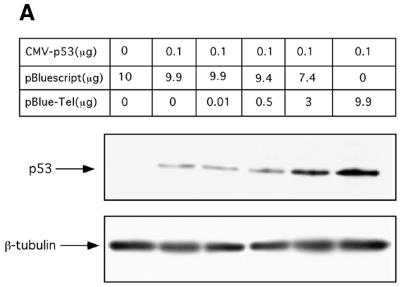
To examine the possibility that TRs induced p53 protein stabilization, H1299 p53 null cells were transiently co-transfected with p53-encoding construct and a mixture of pBlueScript/pBlueScript-TEL plasmids at various ratios. The amount of transfected DNA was kept constant in all transfections. Twenty-four hours later, total cellular extracts were prepared and the levels of p53 protein were examined by western blot analysis using DO-1 anti-p53 monoclonal antibody. As shown in Figure 1A, p53 protein levels were significantly augmented in a dose-dependent manner following the addition of increasing amounts of pBlueScript-TEL plasmid. These observations suggest that in response to the presence of TR sequences, the exogenously expressed p53 protein undergoes stabilization.
Figure 1.
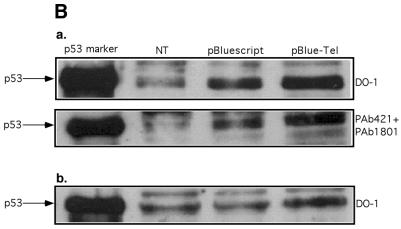
Stabilization of the exogenously and endogenously expressed p53 by TR sequences. (A) H1299 cells were transiently co-transfected with p53 expressing plasmid in the presence of pBlueScript, pBlueScript-TEL or their mixture as indicated in the table. Constant amount of DNA (10 µg/transfection) was maintained. Twenty-four hours later, the p53 protein level was determined by western blotting using DO-1 monoclonal antibody. Comparable loading is demonstrated by β-tubulin blotting. (B) Stabilization of endogenously expressed p53 by TR sequences. HepG2 (a, upper panels) or MCF-7 (b, lower panel) cells were transiently transfected with 20 µg of either pBlueScript or pBlueScript-TEL. Forty-eight hours later, p53 levels were determined by western blotting with the indicated anti-p53 monoclonal antibodies. NT, non-treated cells.
To further investigate the effect of TTAGGG repeat sequences on the stabilization of endogenously expressed wild type p53, we analyzed two tumor-derived human cell lines, MCF-7 and HepG2 that express endogenous wild type p53. HepG2 and MCF-7 cells were transiently transfected with the same amount of pBlueScript or pBlueScript-TEL plasmids. Twenty-four hours later, nuclear extracts from the cells were prepared and immunoblotted with DO-1. As can be seen in Figure 1B, a significant increase in the level of p53 protein was evident in the presence of pBlueScript-TEL as compared with that of the control vector. Immunoblotting with a mixture of other p53-specific monoclonal antibodies, PAb-1801 (N-terminus specific) and PAb-421 (C-terminus specific), revealed a similar pattern of significant accumulation of the endogenous p53 in the presence of pBlueScript-TEL. This phenomenon was not transient and could still be observed 48 h after the transfection (data not shown). Previously, it was shown that in certain MCF-7 cell line strains, p53 is localized mainly at the cytoplasmic compartment (58). However, our MCF-7 cell line strain expresses well detectable levels of nuclear p53 even in non-treated cells as was detected by DO-1 in the nuclear enriched fraction (Fig. 1B, lower panel).
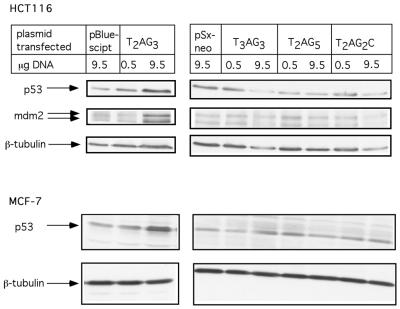
To address the question of the specificity of p53 activation in response to TTAGGG repeats, we performed a comparative analysis of p53 stabilization following the transfection of cells with plasmids containing repeats that are similar but not identical to TR. As demonstrated in Figure 2, as expected, transfection of HCT116 cells with T2AG3 (TR)-containing plasmid induced an increase in p53 protein levels. No changes in protein levels were evident when HCT116 cells were transfected with control plasmids (pBlueScript and pSx-neo) or with plasmids containing repeats such as, T3AG3, T2AG2C, T2AG5. T2AG3 induced p53 protein exhibited an increased transactivation potential as measured by increased expression of its target gene MDM-2. A similar pattern of preferential induction of p53 protein in response to TR transfection as compared with the non-TR sequences was also observed in the MCF-7 cell line (Fig. 2, bottom panel).
Figure 2.
p53 stabilization occurs preferentially by telomere repeats. HCT116 (upper panel) or MCF-7 (lower panel) cells were transfected with indicated amounts of double-stranded circular DNA plasmids. pBlueScript and pSx-neo are plasmids containing T2AG3 and T3AG3, T2AG5, T2AG2C inserts respectively. Forty-eight hours later, p53 levels, MDM-2 and β-tubulin levels were determined by western blotting.
p53 stabilization in response to TRs requires an intact oligomerization domain
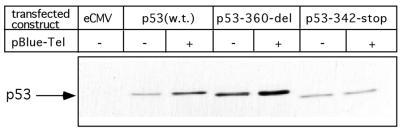
Stabilization of p53 by various stimuli may involve different mechanisms. We reasoned that identification of the specific p53 domains that are responsible for the stabilization of the molecule in response to TR-containing plasmid would provide us with additional clue(s) concerning the p53-telomere pathway. To this end, we transfected H1299 cells with wild type p53 as well as with two C-terminal deletion constructs together with TR-containing or control plasmids. The p53-360-del construct codes for a p53 protein without the 30 C-terminal amino acids. The p53-342-stop mutant codes for a p53 protein that has a stop codon mutation at the oligomerization domain (59). As seen in Figure 3, pBlueScript-TEL induced the stabilization of the full length as well as that of p53-360-del encoded p53 protein. In contrast, the level of the oligomerization-defective p53 protein, encoded by p53-342-stop, was unchanged.
Figure 3.
Identification of the p53 protein domain required for TR induced stabilization. H1299 cells were transiently co-transfected with different p53 expressing plasmids in the presence of pBlueScript, pBlueScript-TEL as indicated in the table. Constant amount of DNA (10 µg/transfection) was maintained. Twenty-four hours later, the p53 protein level was determined by western blotting using DO-1 monoclonal antibody. Comparable loading is demonstrated by β-tubulin blotting.
These results point to the possibility that an intact oligomerization domain of p53 is necessary for this mechanism of p53 stabilization.
TR sequences enhance p53-dependent transactivation
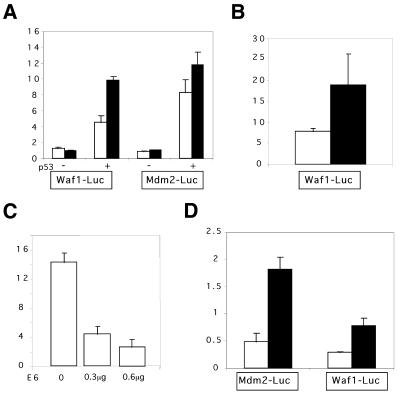
In the following experiments we examined whether TR-stabilized p53 protein also facilitates p53 transcriptional activity. To that end, we co-transfected H1299 p53 null cells with the p53 expression plasmids and the Waf-1-Luc or the Mdm-2-Luc in conjunction with the TR-containing plasmids. As seen in Figure 4A, co-transfection of p53 in conjunction with pBlueScript-TEL into H1299 cells significantly enhanced luciferase activity mediated by Waf-1-Luc reporter as compared with cells co-transfected with the control (pBlueScript) plasmid. It should be noted that no effect of either pBlueScript-TEL or pBlueScript was scored when Waf-1-Luc activity was assayed in the absence of p53. Similar patterns of enhancement in the transcriptional activity of p53 by pBlueScript-TEL were also observed with Mdm-2-Luc construct (Fig. 4A) and Bax-Luc (data not shown).
Figure 4.
TR sequences enhance the transcriptional activity of wild type p53. (A) H1299 cells were seeded (45 000 cells/well) in a 12 well plate and transiently transfected 16–24 h later with Waf-1-Luc or Mdm-2-Luc (0.2 µg/well), pBlueScript (white bars) or pBlueScript-TEL (black bars) (2.8 µg/well), and pCMV-p53 (1 ng/well when indicated). pCMVβ-GAL (0.2 µg/well) was included in each transfection as an internal control. Cells were assayed for luciferase and β-galactosidase activity 24–36 h after transfection. Relative luciferase activity was calculated by dividing the luciferase activity by the β-galactosidase activity of the same sample. Histograms show the mean of typical experiment performed in triplicate. Bars indicate the standard deviation of the mean. (B) TR sequences enhance the transcriptional activity of wild-type p53 in MCF-7 cells. MCF-7 cells were seeded (140 000 cells/well) in a 12 well plate and transiently transfected 16–24 h later with Waf-1-Luc in the presence of 2.8 µg/well pBlueScript (white bars) or pBlueScript-TEL (black bars). Luciferase and β-galactosidase assays were carried out as described above. (C) E6 inhibition of wild type p53 induced Waf-1-Luc in MCF-7 cells. Indicated amounts of pSG5-E6 were co-transfected with Waf-1-Luc (0.2 µg/well). pCMVβ-GAL (0.2 µg) was included in each transfection as an internal control. Luciferase and β-galactosidase assays were carried out as described before. (D) TR sequences enhance the transcriptional activity of wild type p53 in normal human fibroblasts. WI-38 cells, (passage 21–23) were seeded (100 000 cells/well) in a 6 well plate and transiently transfected 16–24 h later with Mdm-2-Luc or Waf-1-Luc (0.5 µg/well), 4.5 µg/well of pBlueScript (white bars) or pBlueScript-TEL (black bars). pCMVβ-GAL (0.2 µg/well) was included in each transfection as an internal control. Luciferase and β-galactosidase assays were carried out as described above.
To exclude the possibility that the enhanced transactivation potential of p53 by TRs may be the case only for exogenously expressed p53, we also examined other cell lines derived from different tissues, which express endogenous wild type p53. To this end, we used the breast cancer-derived MCF-7 epithelial cell line and the human diploid fibroblast cell strain WI-38. When we transfected MCF-7 cells with the reporter construct Waf-1-Luc, there was a significant enhancement in the p53-mediated luciferase expression in the presence of telomere-containing plasmid as compared with the empty one (Fig. 4B). The p53-dependency of this enhanced luciferase expression was further confirmed by including in the transfection experiments a construct encoding for the E6 oncoprotein. E6 induces rapid degradation of p53 via the ubiquitin–proteosome pathway (60). Figure 4C shows that luciferase activity in the MCF-7 cell line was strongly reduced as a function of E6 amount. This suggests that the measured luciferase activity is wild type p53 dependent. Similar results were obtained when WI-38 primary human fibroblasts were used instead. As seen in Figure 4D, transcriptional activation of the Mdm-2 and Waf-1 reporter genes was two to three times higher in the presence of the pBlueScript-TEL as compared with that in the presence of pBlueScript.
This suggests that the presence of extra-chromosomal TRs specifically enhances the transactivation potential of the p53 protein. This was the case both for exogenous and endogenous expressed wild type p53. Moreover, the effect does not seem to be cell type-specific. Indeed, enhancement of the p53 transcriptional activity, mediated by telomeric sequences, was equally evident in epithelial and in fibroblast cells and observed in primary (WI-38) as well as in transformed (H1299, MCF-7) cellular contexts.
Our experiments show that exposure of cells to pBlueScript-TEL plasmid induces an accumulation of both exogenously and endogenously expressed wild type p53 proteins. This stabilization was accompanied by significant up-regulation of the p53 transactivation activity. Collectively, the data suggest that the TRs would enhance activation of p53 probably by inducing protein stabilization, which in turn may facilitate transcriptional activity.
TR sequences induce growth suppression in the p53-dependent manner
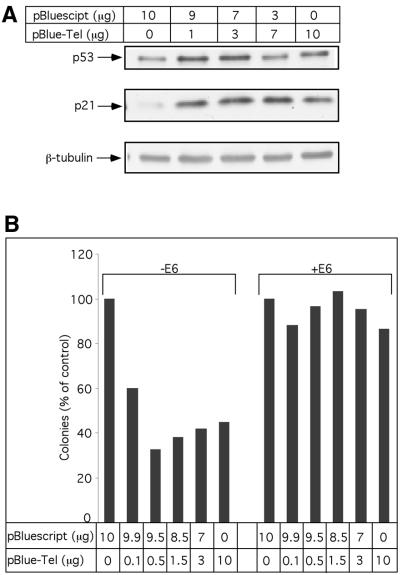
The above observations showing that plasmid containing TRs induces p53 protein stabilization and enhances p53-dependent transcription (including the transcription of growth suppressive p21 and pro-apoptotic BAX genes) prompted us to test the effect of telomeric sequence on cell growth and proliferation. For this purpose, we used the HCT116 colon cancer-derived cell line containing wild type p53 as well as functional DNA damage checkpoints (61,62). HCT116 cells were transfected with a mixture of pBlueScript/pBlueScript-TEL plasmids at various ratios. The total amount of transfected DNA was kept constant in all transfections. In agreement with the above observation with other cell lines, increasing amounts of TR-containing plasmid induces p53 protein accumulation, as compared with transfection with a control plasmid. Moreover, this p53 induction was accompanied by significant up-regulation in p21Waf–1 protein expression (Fig. 5A).
Figure 5.
p53-dependent suppression of colony formation in response to TR transfection. (A) Stabilization of endogenously expressed p53 by TR sequences. HCT116 cells were transiently transfected with pBlueScript, pBlueScript-TEL or their mixture as indicated. Twenty-four hours later, the levels of p53, p21 were determined by western blotting analysis. (B) HCT116 cells were transfected by Lipofectamine with pBlueScript, pBlueScript-TEL or their mixture as indicated. E6 expression plasmid (2 µg/plate; when indicated) and 1 µg/plate of pBabe-puro were included. Constant amount of DNA (13 µg/transfection) was maintained. Forty-eight hours later, selection with puromycin (1 µg/ml) was initiated. Sixteen days later, plates were fixed, stained with Crystal Violet and colonies were counted. The experiment was performed three times and representative results are shown.
In order to monitor the effect of extra-cellular TRs on cell growth in a long-term colony formation assay, we co-transfected puromycin resistance plasmid along with the test plasmids. Following selection the colonies were counted. As seen in Figure 5B, a significant reduction in the number of colonies which correlated with increasing amount of telomeric plasmid DNA was observed. To confirm that the reduction observed in the colony number is indeed a result of p53 activation, we also co-transfected E6 expression plasmid along with the increasing amount of telomeric plasmid. The addition of E6 completely alleviated the TR induced growth suppression effect (Fig. 5B).
The observed suppression in colony formation following transfection of TR sequences is most likely due to the increase in the p53 transcriptional activity observed in the presence of telomeric sequences. Of particular relevance is our above observation where the transactivation potential of p53 toward its direct target p21Waf–1 was significantly increased (Figs 1 and 5A) following telomeric plasmid transfection. This is in agreement with previous studies demonstrating that induction of p21Waf–1 cyclin-dependent kinase inhibitor by p53 accounts for the p53-mediated growth arrest (61,63,64). This may suggest that p53-dependent growth suppression in response to telomeric sequences could be a barrier to the genomic rearrangements induced by telomeres and/or prevent the immortalization mediated by ALT.
DISCUSSION
Both p53 and TR sequences were shown to play a pivotal role in the maintenance of genome stability (1,65). An association between telomere dysfunction and p53 activation has been established over the last few years. For example, it was shown by Chin et al. (66) that growth arrest and apoptosis observed in the late generations of mTR knockout mice were due to p53 activation. This points to the importance of p53 in a pathway that underlies the response to telomere dysfunction. Likewise, it was suggested that the critical shortening of telomeres or aberrations in their structure might be recognized as DNA damage by the p53-dependent cell-cycle check point system (17,67,68). However, the nature of the interaction between TR sequences and p53 has not been elucidated yet.
Here we report on the observation that TR sequences can induce p53 activation. We found that transfection of an intact plasmid containing a 240 bp TR insert induced the activation of p53 in several cell type systems. Both endogenously and exogenously expressed wild type p53 were stabilized in the presence of the pBlueScript-TEL plasmid but not in the presence of the pBlueScript control plasmid or the plasmids containing repeats with non-telomeric sequences. TR-activated p53 exhibited enhanced transcriptional activity and exerted a growth suppression activity that was manifested by a reduction in the number of colonies scored in the long-term colony formation assay.
As it is accepted that broken DNA serves as a DNA insult that induces stabilization of the p53 protein, we have used in our experiments circular plasmids that contain the telomeric repeat sequences. In agreement with others (52) we found that exposure of cells to telomeric oligonucleotides induced p53 stabilization, as did non-TR sequences. However, we assumed that probability of random break generated in cells following DNA transfection is similar with all constructs used. Thus, the specific response observed in the case of TR (T2AG3)-containing plasmid is probably not a result of exposure of cells to broken DNA.
Based on data presented here and that accumulated by others (1,17,68), we would like to propose several models describing a plausible cross talk between TR sequences and the p53 tumor suppressor protein that may in turn lead to the activation of the latter. One model would suggest a direct recognition of TR DNA sequence by p53. Another one may engage the existence of mediator protein(s) or enzymatic activity that anchor p53 onto the telomeres or transmit the signal. A third model, which may induce p53 activation, consists of a squelching mechanism by which exogenously introduced TRs compete out the endogenous telomeres for TRFs that otherwise suppress p53 activation.
p53 was shown to interact with DNA both through its ‘core’ domain (100–300 amino acids) and its C-terminus. The core domain of wild type p53 was shown to interact with a specific ‘consensus’ sequence found in p53 target genes that will lead to the specific transactivation of the latter (19). The C-terminus was suggested to bind damaged DNA in a non-sequence-specific manner. Several mechanisms by which alterations at the C-terminus affect the capacity of the p53 ‘core’ domain to interact with the p53-specific target genes were proposed. One of them involves a conformational change of the p53 molecule driven by its C-terminus domain. Phosphorylations and acetylations at the C-terminus allow allosteric change of the whole molecule that enhances specific DNA binding by the core domain (69). A similar effect can be achieved by the interaction of the C-terminus with damaged DNA (37), PAb-421 antibody (70) or C-terminus-derived peptides (71). An intriguing possibility, pertaining to our present study, is the observation that enhancement in the p53 transactivation potential by the presence of TRs is a result of a direct recognition of the (TTAGGG)n DNA motif by the p53 C-terminus domain. This interaction was suggested to facilitate the conformation conversion of latent p53 molecules into activated ones (72). Furthermore, the ability of the G-rich telomeric sequences to create quaternary structures and loops under physiological conditions (2) could provide an additional target sequence for the p53 C-terminus domain that is known to interact with loops resulting from insertion–deletion mismatching (35). The fact that in our experiments p53 protein deleted of its C-terminus is still activated by the TR suggests that this domain is not the single direct structural target for telomeric induced p53 activation. It is worth mentioning that in the present experiments we have used a circular intact plasmid that is most likely not to be regarded by the cell as damaged DNA. It is therefore not surprising that the extreme C-terminus of the p53 protein, which is referred to as the molecular domain that senses damaged DNA (22,35), is dispensable in the telomeric-induced p53-activation pathway. TR-activated p53 is probably not a mere result of the interaction of p53 with aberrant DNA sequences.
Another possibility for a direct interaction between p53 and telomeric DNA may involve the p53 ‘core’ domain. This assumption is supported by the existence of partial sequence similarity between the consensus p53 binding sequence (5′-PuPuPuC(A/T)(T/A)GPyPyPy-3′) that is recognized by the ‘core’ domain (21) and the (TTAGGGTTAGGG)n telomeric motif. Such a homology may permit the direct interaction of the p53 ‘core’ domain and the TR motif. Under such circumstances it is possible that either the C-terminus and/or the core domain of p53, even at low binding affinity, may activate sequence-specific DNA recognition and enhance p53 transcriptional activity. Although there are no reports claiming that interaction of the p53 ‘core’ domain with DNA leads to p53 stabilization, the possibility that following interaction with telomeric DNA, the p53 molecule undergoes a specific conformational change that renders it resistant to Mdm-2-mediated degradation cannot be excluded.
Participation of several DNA damage inducible and DNA repair proteins such as ATM, DNA-PK, PARP, Ku, Blm and Wrn both in the telomere integrity sensing and p53 activity modulation provides strong evidence towards a possible way by which telomere signals are transmitted to p53 (23,41). Indeed, stabilization of p53 following transfection of cells with plasmids containing TRs strongly referred to the involvement of post-translational modifications in the activation process. Introduction of extra-chromosomal DNA containing TRs into cells may initiate a DNA damage checkpoint. Activation of p53 by extra-chromosomal TRs could be described by competition with native telomeres for telomere binding proteins such as TRF-1 and TRF-2. It was shown that exposure of native chromosomal ends caused by squelching out of TRF-2 triggers p53 activation through ATM (17).
Our observation of a p53-dependent reduction in colony formation coupled with the induction of WAF-1 expression observed as a result of extra-chromosomal TR transfection, points to the possibility that enhancement in transactivation potential and p53 protein stabilization are specific stages in the signaling cascade culminated in this physiological response of growth suppression. This p53-dependent growth suppression could be the result of p53-mediated growth arrest and/or apoptosis. In addition, this finding of p53 signaling cells to undergo growth arrest may be the result of telomeric shortening occurring with aging and thus could potentially be a potent anti-cancer protection mechanism.
Previously, it was shown that transfection of cells with linearized exogenous DNA containing TRs at one terminus induces chromosome breakage and de novo telomere formation at the site of the break (53–55). The free DNA ends and double-stranded chromosomal breakage generated were suggested to induce a p53 response (24,73). The mechanism, by which circular plasmid-containing TRs cause in our hands a reduction in colony formation assayed by the long-term growth assay, is still to be determined.
In conclusion, we would like to put forward the hypothesis that p53-dependent growth inhibition induced by the presence of extra-chromosomal telomeric sequences could explain the low frequency of the ALT mechanism. The precise mechanism of ALT is not known, but it could include homologous recombination between endogenous telomeres and/or extra-chromosomal DNA containing telomeric sequences (74–77). Indeed, the immortalized cell lines that use the ALT pathway have either p53 deletion or inactivation of p53 by oncogenes (48,51,78–81).
As telomeres, telomerase and p53 are believed to be the target for future therapeutics in aging-associated and malignant disorders, understanding of their interrelationship is expected to have a significance impact on basic research and its clinical implications.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by grants from the Israel–USA Binational Science Foundation (BSF) and the Deutsch–Israelische Projektkooperation (DIP) and the Kadoori Foundation. V.R. is the incumbent of the Norman and Helen Asher Professorial Chair in Cancer Research at the Weizmann Institute.
REFERENCES
- 1. de Lange T. and Jacks,T. (1999) For better or worse? Telomerase inhibition and cancer. Cell, 98, 273–275. [DOI] [PubMed] [Google Scholar]
- 2.Wellinger R.J. and Sen,D. (1997) The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer, 33, 735–749. [DOI] [PubMed] [Google Scholar]
- 3.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Mammalian telomeres end in a large duplex loop. Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 4.Allsopp R.C., Vaziri,H., Patterson,C., Goldstein,S., Younglai,E.V., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA, 89, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu ,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Linchtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 6.Harley C.B. (1991) Telomere loss: mitotic clock or genetic time bomb? Mutat. Res., 256, 271–282. [DOI] [PubMed] [Google Scholar]
- 7. van Steensel B., Smogorzewska,A. and de Lange,T. (1998) TRF2 protects human telomeres from end-to-end fusions. Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 8.Harley C.B., Futcher,A.B. and Greider,C.W. (1990) Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. [DOI] [PubMed] [Google Scholar]
- 9.Hastie N.D., Dempster,M., Dunlop,M.G., Thompson,A.M., Green,D.K. and Allshire,R.C. (1990) Telomere reduction in human colorectal carcinoma and with ageing. Nature, 346, 866–868. [DOI] [PubMed] [Google Scholar]
- 10.Lindsey J., McGill,N.I., Lindsey,L.A., Green,D.K. and Cooke,H.J. (1991) In vivo loss of telomeric repeats with age in humans. Mutat. Res., 256, 45–48. [DOI] [PubMed] [Google Scholar]
- 11.Vaziri H. and Benchimol,S. (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr. Biol., 8, 279–282. [DOI] [PubMed] [Google Scholar]
- 12.Bond J.A., Wyllie,F.S. and Wynfordthomas,D. (1994) Escape from senescence in human diploid fibroblasts induced directly by mutant p53. Oncogene, 9, 1885–1889. [PubMed] [Google Scholar]
- 13.Bond J., Haughton,M., Blaydes,J., Gire,V., Wynford-Thomas,D. and Wyllie,F. (1996) Evidence that transcriptional activation by p53 plays a direct role in the induction of cellular senescence. Oncogene, 13, 2097–2104. [PubMed] [Google Scholar]
- 14.Shay J.W., Pereira-Smith,O.M. and Wright,W.E. (1991) A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res., 196, 33–39. [DOI] [PubMed] [Google Scholar]
- 15.Wynford-Thomas D. (1999) Cellular senescence and cancer. J. Pathol., 187, 100–111. [DOI] [PubMed] [Google Scholar]
- 16.Wynford-Thomas D. (1996) Telomeres, p53 and cellular senescence. Oncol. Res., 8, 387–398. [PubMed] [Google Scholar]
- 17.Karlseder J., Broccoli,D., Dai,Y., Hardy,S. and de Lange,T. (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science, 283, 1321–1325. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz D. and Rotter,V. (1998) p53-dependent cell cycle control: response to genotoxic stress. Semin. Cancer Biol., 8, 325–336. [DOI] [PubMed] [Google Scholar]
- 19.Levine J.A. (1997) p53, the cellular gatekeeper for growth and division. Cell, 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 20.Ryan K.M., Phillips,A.C. and Vousden,K.H. (2001) Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol., 13, 332–337. [DOI] [PubMed] [Google Scholar]
- 21.Kern S.E., Kinzler,K.W., Bruskin,A., Jarosz,D., Friedman,P., Prives,C. and Vogelstein,B. (1991) Identification of p53 as a sequence-specific DNA-binding protein. Science, 252, 1708–1711. [DOI] [PubMed] [Google Scholar]
- 22.Wolkowicz R. and Rotter,V. (1997) The DNA binding regulatory domain of p53: see the C. Pathol. Biol. (Paris), 45, 785–796. [PubMed] [Google Scholar]
- 23.Appella E. and Anderson,C.W. (2000) Signaling to p53: breaking the posttranslational modification code. Pathol. Biol. (Paris), 48, 227–245. [PubMed] [Google Scholar]
- 24.Giaccia A.J. and Kastan,M.B. (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev., 12, 2973–2983. [DOI] [PubMed] [Google Scholar]
- 25.Canman C.E., Wolff,A.C., Chen,C.Y., Fornace,A.J.,Jr and Kastan,M.B. (1994) The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res., 54, 5054–5058. [PubMed] [Google Scholar]
- 26.Kastan M.B., Zhan,Q., el-Deiry,W.S., Carrier,F., Jacks,T., Walsh,W.V., Plunkett,B.S., Vogelstein,B. and Fornace,A.J.,Jr (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 27.Miyashita T., Krajewski,S., Krajewska,M., Wang,H.G., Lin,H.K. and Liebermann,D.A. (1994) Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene, 9, 1799–1805. [PubMed] [Google Scholar]
- 28.Miyashita T. and Reed,J.C. (1995) Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell, 80, 293–299. [DOI] [PubMed] [Google Scholar]
- 29.Polyak K., Xia,Y., Zweier,J.L., Kinzler,K.W. and Vogelstein,B. (1997) A model for p53-induced apoptosis. Nature, 389, 300–305. [DOI] [PubMed] [Google Scholar]
- 30.Oda K., Arakawa,H., Tanaka,T., Matsuda,K., Tanikawa,C., Mori,T., Nishimori,H., Tamai,K., Tokino,T., Nakamura,Y. and Taya,Y. (2000) p53AIP1, a potential mediator of p53-dependent apoptosis and its regulation by Ser-46-phosphorylated p53. Cell, 102, 849–862. [DOI] [PubMed] [Google Scholar]
- 31.Vogelstein B., Lane,D. and Levine,A.J. (2000) Surfing the p53 network. Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 32.Oda E., Ohki,R., Murasawa,H., Nemoto,J., Shibue,T., Yamashita,T., Tokino,T., Taniguchi,T. and Tanaka,N. (2000) Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science, 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- 33.Bakalkin G., Yakovleva,T., Selivanova,G., Magnusson,K.P., Szekely,L., Kiseleva,E., Klein,G., Terenius,L. and Wiman,K.G. (1994) p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc. Natl Acad. Sci. USA, 91, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S., Elenbaas,B., Levine,A. and Griffith,J. (1995) p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell, 81, 1013–1020. [DOI] [PubMed] [Google Scholar]
- 35.Szak S.T. and Pietenpol,J.A. (1999) High affinity insertion/deletion lesion binding by p53. Evidence for a role of the p53 central domain. J. Biol. Chem., 274, 3904–3909. [DOI] [PubMed] [Google Scholar]
- 36.Wu L., Bayle,H., Elenbaas,B., Pavletich,N.P. and Levine,A.J. (1995) Alternatively spliced forms in the carboxy-terminal domain of the p53 protein regulate its ablity to promote annealing of complementary single strands of nucleic acids. Mol. Cell. Biol., 15, 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayaraman J. and Prives,C. (1995) Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell, 81, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 38.Banin S., Moyal,L., Shieh,S., Taya,Y., Anderson,C.W., Chessa,L., Smorodinsky,N.I., Prives,C., Reiss,Y., Shiloh,Y. and Ziv,Y. (1998) Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science, 281, 1674–1677. [DOI] [PubMed] [Google Scholar]
- 39.Shieh S.Y., Ikeda,M., Taya,Y. and Prives,C. (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell, 91, 325–334. [DOI] [PubMed] [Google Scholar]
- 40.Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 41.Jayaraman L. and Prives,C. (1999) Covalent and noncovalent modifiers of the p53 protein. Cell. Mol. Life Sci., 55, 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shay J.W. (1997) Telomerase in human development and cancer. J. Cell Physiol., 173, 266–270. [DOI] [PubMed] [Google Scholar]
- 43.Counter C.M., Avilion,A.A., LeFeuvre,C.E., Stewart,N.G., Greider,C.W., Harley,C.B. and Bacchetti,S. (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J., 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Counter C.M., Botelho,F.M., Wang,P., Harley,C.B. and Bacchetti,S. (1994) Stabilization of short telomeres and telomerase activity accompany immortalization of Epstein–Barr virus-transformed human B lymphocytes. J. Virol., 68, 3410–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 46.Bryan T.M., Englezou,A., Gupta,J., Bacchetti,S. and Reddel,R.R. (1995) Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J., 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryan T.M., Englezou,A., Dalla-Pozza,L., Dunham,M.A. and Reddel,R.R. (1997) Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nature Med., 3, 1271–1274. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi T., Carbone,D., Nau,M.M., Hida,T., Linnoila,I., Ueda,R. and Minna,J.D. (1992) Wild-type but not mutant p53 suppresses the growth of human lung cancer cells bearing multiple genetic lesions. Cancer Res., 52, 2340–2343. [PubMed] [Google Scholar]
- 49.Ashcroft M. and Vousden,K.H. (1999) Regulation of p53 stability. Oncogene, 18, 7637–7643. [DOI] [PubMed] [Google Scholar]
- 50.Moyzis R.K., Buckingham,J.M., Cram,L.S., Dani,M., Deaven,L.L., Jones,M.D., Meyne,J., Ratliff,R.L. and Wu,J.R. (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA, 85, 6622–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1474–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang L.C., Clarkin,K.C. and Wahl,G.M. (1996) Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest. Proc. Natl Acad. Sci. USA, 93, 4827–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farr C., Fantes,J., Goodfellow,P. and Cooke,H. (1991) Functional reintroduction of human telomeres into mammalian cells. Proc. Natl Acad. Sci. USA, 88, 7006–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanish J.P., Yanowitz,J.L. and de Lange,T. (1994) Stringent sequence requirements for the formation of human telomeres. Proc. Natl Acad. Sci. USA, 91, 8861–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barnett M.A., Buckle,V.J., Evans,E.P., Porter,A.C., Rout,D., Smith,A.G. and Brown,W.R. (1993) Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res., 21, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haupt Y., Maya,R., Kazak,A. and Oren,M. (1997) Mdm2 promotes the rapid degradation of p53. Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- 57.Kubbutat M.H., Jones,S.N. and Vousden,K.H. (1997) Regulation of p53 stability by Mdm2. Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- 58.Moll U., Riou,G. and Levine,A.J. (1992) Two distinct mechanisms alter p53 in breast cancer: mutation and nuclear exclusion. Proc. Natl Acad. Sci. USA, 89, 7262–7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X., Wang,X.W., Xu,L., Hagiwara,K., Nagashima,M., Wolkowicz,R., Zurer,I., Rotter,V. and Harris,C.C. (1999) COOH-terminal domain of p53 modulates p53-mediated transcriptional transactivation, cell growth and apoptosis. Cancer Res., 59, 843–848. [PubMed] [Google Scholar]
- 60.Scheffner M., Werness,B.A., Huibregtse,J.M., Levine,A.J. and Howley,P.M. (1990) The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell, 63, 1129–1136. [DOI] [PubMed] [Google Scholar]
- 61.Waldman T., Kinzler,K.W. and Vogelstein,B. (1995) p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res., 55, 5187–5190. [PubMed] [Google Scholar]
- 62.Bunz F., Hwang,P.M., Torrance,C., Waldman,T., Zhang,Y., Dillehay,L., Williams,J., Lengauer,C., Kinzler,K.W. and Vogelstein,B. (1999) Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest., 104, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunz F., Dutriaux,A., Lengauer,C., Waldman,T., Zhou,S., Brown,J.P., Sedivy,J.M., Kinzler,K.W. and Vogelstein,B. (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science, 282, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 64.el-Deiry W.S. (1998) p21/p53, cellular growth control and genomic integrity. Curr. Top. Microbiol. Immunol., 227, 121–137. [DOI] [PubMed] [Google Scholar]
- 65.Blackburn E.H. (2000) Telomere states and cell fates. Nature, 408, 53–56. [DOI] [PubMed] [Google Scholar]
- 66.Chin L., Artandi,S.E., Shen,Q., Tam,A., Lee,S.L., Gottlieb,G.J., Greider,C.W. and DePinho,R.A. (1999) p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell, 97, 527–538. [DOI] [PubMed] [Google Scholar]
- 67.Vaziri H. and Benchimol,S. (1996) From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: the telomer loss/DNA damage model of cell aging. Exp. Gerontol., 31, 295–301. [DOI] [PubMed] [Google Scholar]
- 68.Vaziri H., West,M.D., Allsopp,R.C., Davison,T.S., Wu,Y.S., Arrowsmith,C.H., Poirier,G.G. and Benchimol,S. (1997) ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly (ADP-ribose) polymerase. EMBO J., 16, 6018–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hupp T.R. (1999) Regulation of p53 protein function through alterations in protein-folding pathways. Cell. Mol. Life Sci., 55, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hupp T.R., Meek,D.W., Midgley,C.A. and Lane,D.P. (1992) Regulation of the specific DNA binding function of p53. Cell, 71, 875–886. [DOI] [PubMed] [Google Scholar]
- 71.Hupp T.R., Sparks,A. and Lane,D.P. (1995) Small peptides activate the latent sequence-specific DNA binding function of p53. Cell, 83, 237–245. [DOI] [PubMed] [Google Scholar]
- 72.Hupp T.R. and Lane,D.P. (1994) Allosteric activation of latent p53 tetramers. Curr. Biol., 4, 865–875. [DOI] [PubMed] [Google Scholar]
- 73.Nelson W.G. and Kastan,M.B. (1994) DNA strand breaks - the DNA template alteration that triggers p53 dependent DNA damage response pathways. Mol. Cell. Biol., 14, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith S. (2000) Recombination: a means to an end in human cells. Nature Genet., 26, 388–389. [DOI] [PubMed] [Google Scholar]
- 75.Kass-Eisler A. and Greider,C.W. (2000) Recombination in telomere-length maintenance. Trends Biochem. Sci., 25, 200–204. [DOI] [PubMed] [Google Scholar]
- 76.Regev A., Cohen,S., Cohen,E., Bar-Am,I. and Lavi,S. (1998) Telomeric repeats on small polydisperse circular DNA (spcDNA) and genomic instability. Oncogene, 17, 3455–3461. [DOI] [PubMed] [Google Scholar]
- 77.Tokutake Y., Matsumoto,T., Watanabe,T., Maeda,S., Tahara,H., Sakamoto,S., Niida,H., Sugimoto,M., Ide,T. and Furuichi,Y. (1998) Extra-chromosomal telomere repeat DNA in telomerase-negative immortalized cell lines. Biochem. Biophys. Res. Commun., 247, 765–772. [DOI] [PubMed] [Google Scholar]
- 78.Yeager T.R., Neumann,A.A., Englezou,A., Huschtscha,L.I., Noble,J.R. and Reddel,R.R. (1999) Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res., 59, 4175–4179. [PubMed] [Google Scholar]
- 79.Abdul-Ghani R., Ohana,P., Matouk,I., Ayesh,S., Ayesh,B., Laster,M., Bibi,O., Giladi,H., Molnar-Kimber,K., Sughayer,M.A., de Groot,N. and Hochberg,A. (2000) Use of transcriptional regulatory sequences of telomerase (hTER and hTERT) for selective killing of cancer cells. Mol. Ther., 2, 539–544. [DOI] [PubMed] [Google Scholar]
- 80.Wen J., Cong,Y.S. and Bacchetti,S. (1998) Reconstitution of wild-type or mutant telomerase activity in telomerase-negative immortal human cells. Hum. Mol. Genet., 7, 1137–1141. [DOI] [PubMed] [Google Scholar]
- 81.Bryan T.M., Marusic,L., Bacchetti,S., Namba,M. and Reddel,R.R. (1997) The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum. Mol. Genet., 6, 921–926. [DOI] [PubMed] [Google Scholar]