Abstract
Employing the biparental exogenous plasmid isolation method, conjugative plasmids conferring mercury resistance were isolated from the microbial community of the rhizosphere of field grown alfalfa plants. Five different plasmids were identified, designated pSB101–pSB105. One of the plasmids, pSB102, displayed broad host range (bhr) properties for plasmid replication and transfer unrelated to the known incompatibility (Inc) groups of bhr plasmids IncP-1, IncW, IncN and IncA/C. Nucleotide sequence analysis of plasmid pSB102 revealed a size of 55 578 bp. The transfer region of pSB102 was predicted on the basis of sequence similarity to those of other plasmids and included a putative mating pair formation apparatus most closely related to the type IV secretion system encoded on the chromosome of the mammalian pathogen Brucella sp. The region encoding replication and maintenance functions comprised genes exhibiting different degrees of similarity to RepA, KorA, IncC and KorB of bhr plasmids pSa (IncW), pM3 (IncP-9), R751 (IncP-1β) and RK2 (IncP-1α), respectively. The mercury resistance determinants were located on a transposable element of the Tn5053 family designated Tn5718. No putative functions could be assigned to a quarter of the coding capacity of pSB102 on the basis of comparisons with database entries. The genetic organization of the pSB102 transfer region revealed striking similarities to plasmid pXF51 of the plant pathogen Xylella fastidiosa.
INTRODUCTION
The deliberate release of genetically engineered microorganisms (GEMs) into the environment has raised concern about the potential negative ecological impact of the organisms and the nucleic acid they carry. One concern is related to the genetic stability of released GEMs. The acquisition of genes from the indigenous bacterial community by released GEMs could alter their properties and may modulate the potential ecological effects exerted by GEMs. To assess the nature of such events the acquisition of mercury resistance by deliberately released strains has been studied.
Genetic determinants conferring resistance to mercury (HgR) are ubiquitous in bacterial species of various natural environments (1). Mercury resistance genes (mer) are often located on mobile genetic elements such as transposons or conjugative plasmids. Self-transmissible plasmids, some of which have broad host range (bhr) properties, have been isolated from bulk soil, rhizosphere soils and the phyllosphere of plants employing Pseudomonas spp. as recipients (2–4). During a deliberate release experiment with a Pseudomonas strain tagged with the lacZY genes, acquisition of mercury resistance plasmids in the phyllosphere of sugar beet was readily observed (5). The acquisition of such plasmids was correlated with a specific stage in sugar beet development and was shown to temporarily increase the ecological fitness of the GEM compared to its plasmid-free parent strain (6).
In the context of a joint project the first deliberate release of GEMs was performed in Germany (7). The GEMs were derivatives of Sinorhizobium meliloti strain 2011 genetically tagged with the firefly luciferase gene (luc) mediating bioluminescence (8). These bacteria were released in 1995 at field plots of the Federal Agricultural Research Center (FAL, Braunschweig), mainly for monitoring purposes. The question arose whether self-transmissible plasmids conferring mercury resistance present at the release site were able to transfer to and be maintained in S.meliloti and whether these were related to plasmids already chracterized.
Thus the main objective of this work was the identification and characterization of conjugative mercury resistance plasmids residing in the microbial population of the alfalfa rhizosphere. Self-transmissible plasmids conferring HgR were exogenously isolated employing an S.meliloti recipient. One environmental mercury resistance plasmid, designated pSB102, which exhibited bhr properties was selected for sequence analysis. The molecular structure of this plasmid pSB102 as well as its relatedness to other plasmids and chromosomal DNA fragments are reported.
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains and Pseudomonas sp. B131 and its derivatives were grown in Luria–Bertani (LB) medium at 37°C and 30°C, respectively. Sinorhizobium meliloti strains were grown at 30°C in Tryptone yeast medium. The final concentrations of antibiotics for selective growth were 200 µg ml–1 streptomycin and 100 µg ml–1 rifampicin. Bacterial strains were stored at –20°C in 50% glycerol.
Table 1. Bacterial strains and plasmids used.
Rif, rifampicin; Ap, ampicillin; Km, kanamycin; Gm, gentamicin; Hg, mercury.
Construction of the S.meliloti strain FP2 tagged with green fluorescent protein (GFP)
The gfp marker gene was introduced into S.meliloti strain M4 using suicide plasmid pAG308-1 (12). Plasmid pAG308-1 contains a Tn5-derived mini transposon, carrying a promoterless gfp gene and a kanamycin resistance gene. pAG308-1 was mobilized from E.coli strain S17-1 8pir to S.meliloti M4 in filter matings. Neomycin-resistant transconjugants were selected on TY agar containing 50 µg ml–1 neomycin and 7 µg ml–1 nalidixic acid. Transconjugants arose at a frequency of ∼4.4 × 10–8 per recipient cell. A selected colony designated FP2 exhibited strong fluorescence, indicating that the promoterless gfp gene was under the control of a strong indigenous S.meliloti promoter (data not shown).
Exogenous plasmid isolation from the bacterial community of the alfalfa rhizosphere
Conjugative plasmids of the indigenous bacterial community inhabiting the root surface of field grown alfalfa plants were isolated using gfp-tagged S.meliloti strain FP2 as recipient in filter crosses. For each mating 20 g root material from two different field plots at the FAL in Braunschweig was collected. The roots were cut into pieces and transferred to 200 ml Erlenmeyer flasks. An aliquot of 100 ml of PBS buffer (2) was added and the flasks were shaken for 20 min at 250 r.p.m. The supernatant was centrifuged for 5 min at 1000 g to remove soil particles. Bacterial cells were pelleted by 10 min centrifugation of the supernatant at 8000 g. The pellet was resuspended in 2 ml of PBS. In the first mating 0.6 ml of rhizosphere suspension, corresponding to 3.3 g root material, was mixed o/n (overnight) with 0.3 ml of cultures of S.meliloti FP2. In the second mating 1 ml of rhizosphere suspension, corresponding to 5 g root material, was mixed o/n with 0.5 ml of culture of S.meliloti FP2. The mating mixture as well as the appropriate controls of 1 ml of rhizosphere suspension and 1 ml of recipient culture were concentrated by 30 s centrifugation at 14 000 g and were spotted on nitrocellulose filters (Sartorius AG, Göttingen, Germany). Filters were incubated at 30°C for 24 h on R2A agar plates (Difco Laboratories, Detroit, MI) supplemented with 100 µg ml–1 cycloheximide. The titer of rhizosphere bacteria was determined on R2A supplemented with 100 µg ml–1 cycloheximide. Mercury resistance served as the selection marker for plasmid transfer and streptomycin was used to counterselect against the bacterial community. Transconjugants were selected on TY medium containing 100 µg ml–1 cycloheximide, 200 µg ml–1 streptomycin and 20 µg ml–1 mercury(II) chloride (HgCl2). Sinorhizobium meliloti transconjugants were identified on the basis of their green fluorescence phenotype under UV irradiation.
Plasmid conjugation tests by filter matings
An aliquot of 1 ml of a donor culture was mixed o/n with 0.5 ml of a culture of the recipient strain and concentrated by 30 s centrifugation at 14 000 g. Matings were performed as described above, using filters placed on TY or LB medium. Serial dilutions of mating mixtures were plated onto TY or LB plates supplemented with the appropriate antibiotics and heavy metals.
Analysis of plasmids for antibiotic and heavy metal resistances
Five milliliters of TY medium containing various concentrations of the selective agent under investigation were inoculated with 50 µl of a late logarithmic culture of each plasmid-harboring strain. After incubation o/n the OD580 values of the cultures were determined and compared with OD580 values of the control cultures of S.meliloti FP2 lacking the corresponding plasmid. The following compounds were used in resistance tests: 0–10 µg ml–1 tetracycline, 0–36 µg ml–1 amikacin, 0–15 µg ml–1 gentamicin, 0–70 µg ml–1 spectinomycin, 0–120 µg ml–1 chloramphenicol, 0–150 µg ml–1 novobiocin, 0–10 µg ml–1 nurseothricin, 0–5 µg ml–1 norfloxacin, 0–30 µg ml–1 rifampicin, 0–40 µg ml–1 HgCl2, 0–20 µg ml–1 phenylmercuric acetate (PMA, C6H5HgOCOCH3), 0–0.5 mM cadmium sulfate (CdSO4), 0–3 mM cobalt chloride (CoCl2), 0–10 mM copper sulfate (CuSO4), 0–20 mM ferric chloride (FeCl3), 0–3 mM nickel sulfate (NiSO4) and 0–5 mM zinc sulfate (ZnSO4).
Subcloning, sequencing and annotation of plasmid pSB102
To determine the DNA sequence of pSB102, plasmid DNA was digested with EcoRI and ligated with EcoRI-linearized pBluescript II KS(+) vector. Plasmid DNA was isolated using a modified HB lysis method (13). The protocol was modified to include purification by CsCl–ethidium bromide buoyant density centrifugation (14). The preparation of competent E.coli cells and the transformation procedures were performed according to Sambrook et al. (15). Ten EcoRI fragments ranging in size from 0.5 to 11 kb were cloned in vector pBluescript II KS(+). The ends of the EcoRI fragments were sequenced with M13 standard forward and reverse primers. Using this sequence information, two types of specific walking primers were designed. One type was used for walking on the cloned fragments. The second type, outwardly directed from the EcoRI fragments, was employed for walking on pSB102 DNA to connect the fragments. For sequencing purposes plasmid DNA was isolated using the Nucleobond AX 100 kit (Macherey-Nagel, Düren, Germany) according to the protocol provided by the manufacturer. DNA was sequenced by the dideoxy chain termination method (16) using PCR-based sequencing with a Taq Dye Deoxy Terminator Cycle-Sequencing Kit on an ABI Prism 377 DNA Sequencer (Applied Biosystems, Perkin Elmer) by IIT Biotech/Bioservice GmbH (Bielefeld, Germany). Base calling was done using PHRED. Walking primers were designed with PRIDE from the Staden package. Sequence assembly was done with the GAP4 sequence assembly tool from the Staden package (17). Annotation was performed using the automated genome interpretation system GenDB (Center for Genome Research, Bielefeld, Germany). Open reading frames (ORFs) were predicted by GLIMMER 2.0 (18). Each identified ORF was the subject of BLAST database similarity searches (19). The complete sequence was searched for σ70-dependent promoters using Neural Network Promoter Prediction (NNPP) (20). Repeat regions within the pSB102 sequence were analyzed with the computer program REPuter (21). Prediction of transmembrane helices was performed by the dense alignment surface (DAS) method using the DAS transmembrane prediction server (22). Amino acid sequence similarities were calculated with the ALIGN computer program (23). Prediction of signal peptides was performed with the neural network systems SignalP (24). The sequence of plasmid pSB102 is deposited in the EMBL nucleotide sequence database under accession no. AJ304453.
Classification of plasmids by PCR analyses
Template DNA was obtained from single colonies of S.meliloti. For this purpose bacterial colonies were suspended in 50 µl of lysis buffer (0.5 M NaOH, 0.25% SDS) and boiled for 5 min. An aliquot of 1.5 µl of a 10-fold dilution of the boiled sample was used in PCR. PCR was carried out in a volume of 25 µl, containing 1.6 µM both forward and reverse primers, 0.2 mM each dNTP, 1 U Taq polymerase (Qiagen, Hilden, Germany) and 1× PCR buffer (Qiagen). After an initial step of 5 min at 94°C, PCR amplification involved 30 cycles of 1 min at 94°C for denaturation, 1 min at the annealing temperature recommended for the respective primer system and 1 min polymerisation at 72°C. The final primer extension reaction was done for 10 min at 72°C. PCR products were analyzed in 1.5% agarose gels with TA buffer (40 mM Tris, 10 mM sodium acetate, 1 mM EDTA). Primers employed for PCR-based classification were specific for incompatibility (Inc) groups A/C (25), N, P and W (26). PCR analyses were performed using plasmids RA1, RN3, R702 and pSa (Table 1) as positive controls.
RESULTS AND DISCUSSION
Isolation and characterization of conjugative mercury resistance plasmids from the microbial community of the alfalfa rhizosphere
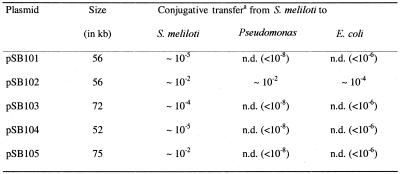
To assess the presence of conjugative mercury resistance plasmids at a field site where S.meliloti GEMs tagged with the firefly luc gene had been released, the exogenous plasmid isolation method was employed. Using the alfalfa bacterial rhizosphere communities, sampled twice during November 1998 and July 1999 from two different field plots, as donors in filter matings with gfp-tagged S.meliloti strain FP2 as recipient, mercury-resistant colonies exhibiting the GFP phenotype were readily recovered. According to their EcoRI restriction pattern a total of five different plasmids designated pSB101–pSB105 were identified (Table 2). During the first mating in November 1998 two plasmids were isolated, which differed in their EcoRI restriction fragment patterns. Transconjugants obtained from the plot I sample carried plasmid pSB101 and transconjugants obtained from the plot II sample carried plasmid pSB102. In the second mating performed in July 1999 five distinct plasmids were recovered. Transconjugants obtained from the plot I sample carried plasmids pSB102 and pSB104 and transconjugants obtained from the plot II sample carried plasmids pSB101, pSB103 and pSB105. The fact that plasmids pSB101 and pSB102 were repeatedly recovered over time at a spatial distance of at least 3 m may indicate that these two plasmids were abundant in bacterial communities of the field site.
Table 2. Characterization of selected properties of exogenously isolated mercury resistance plasmids.
aThe frequencies of conjugative transfers are given per recipient cell. Sinorhizobium meliloti strain FP2 served as the donor and S.meliloti strain L331, Pseudomonas sp. strain B131 and E.coli strain HBR101 served as recipients in the respective mating experiments. Transconjugants were selected on LB or TY medium containing 20 µg ml–1 HgCl2 and 100 µg ml–1 rifampicin. The values given in parentheses indicate the limit of detection. n.d., not detected.
As a first step in the characterization of the newly identified plasmids, their sizes were determined on the basis of their restriction fragment patterns in agarose gels. These analyses revealed sizes ranging from 52 to 75 kb (Table 2). To address the question of whether additional resistances to antibiotics or heavy metals were located on the various plasmids, resistance tests were performed using the antibiotics and heavy metals listed in Materials and Methods. All plasmids conferred resistance to inorganic (HgCl2) and organic (PMA) mercury compounds. Plasmids pSB103 and pSB104 were resistant to 10 µg ml–1 HgCl2 but were unable to grow at 20 µg ml–1 HgCl2, whereas all other plasmids conferred resistance to 20 µg ml–1 HgCl2. Plasmids pSB101 and pSB102 showed resistance to 10 µg ml–1 PMA, while all the other plasmids were only able to grow at 4 µg ml–1 PMA. Plasmid-free S.meliloti FP2 was unable to grow in liquid medium at 5 µg ml–1 HgCl2 or 2 µg ml–1 PMA. None of these five plasmids displayed resistance against the antibiotics tetracycline, amikacin, gentamicin, spectinomycin, chloramphenicol, novobiocin, norfloxacin, nurseothricin and rifampicin or heavy metal ions of cadmium, cobalt, copper, iron, nickel and zinc.
To characterize the host range of the newly isolated plasmids, mating experiments were performed. Sinorhizobium meliloti FP2 served as the donor strain and three different bacterial strains, S.meliloti L331 belonging to the α-Proteobacteria and Pseudomonas sp. B131 and E.coli HBR101 belonging to the γ-Proteobacteria, were used as recipients. As expected, all plasmids were self-transmissible. The transfer frequencies were in the range 10–2–10–5 (Table 2). Plasmid pSB102 was transferred to all recipients used and thus displayed bhr properties, whereas the host range of all other plasmids was restricted to S.meliloti (Table 2). Employing a Tn5-tagged derivative of plasmid pSB102, transfer from E.coli to Ralstonia eutropha strain 11098, which belongs to the β-Proteobacteria, was also observed (unpublished results). PCR replicon typing employing primers specific for Inc groups IncP-1, IncW, IncN and IncA/C, respectively, yielded no PCR products, suggesting that plasmid pSB102 belongs to an as yet undescribed group of bhr plasmids.
Sequence analysis of bhr mercury resistance plasmid pSB102 employing an approach which minimizes the number of sequencing reactions
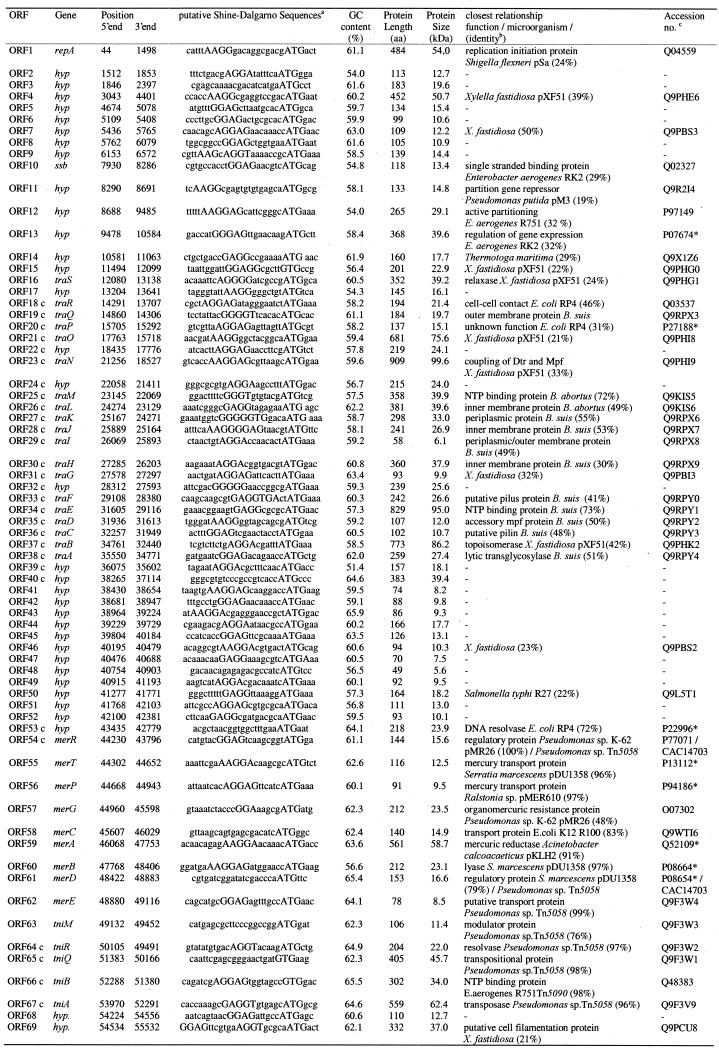
The 55 578 bp sequence of plasmid pSB102 was determined by primer walking. Employing this strategy the entire double-stranded DNA sequence was established with a minimal number of 260 sequencing reactions, leading to a 2.5-fold coverage of the genome. The average GC content of the plasmid is 59.8%. No obvious significant deviations in the GC content were observed. Potential ORFs were identified with an interpolated Markov model using GLIMMER2.0 (18). GLIMMER2.0 predicted a total of 101 frequently overlapping ORFs. The annotation of plasmid pSB102 was performed using the automated genome interpretation system GenDB. Taking the results of database searches and the existence of putative Shine–Dalgarno sequences (Table 3) into account, the number of ORFs was reduced from 101 to 66. BLASTX searches with short sequence segments of pSB102 revealed the existence of three further ORFs (ssb, traC and traD), which were not predicted by GLIMMER2.0.
Table 3. Location of the coding regions of pSB102 and the closest relatives of the deduced proteins.
c, complementary DNA strand.
aThe putative Shine–Dalgarno (SD) sequences of plasmid pSB102 are indicated by bold capital letters. Start codons are given in capitals. Overlaps of start codons with previous stop codons are underlined.
bIdentities were overall identities at the amino acid sequence level.
cAccession numbers are given in SPTREMBL format or, as indicated by *, in SWISSPROT format.
Significant similarities to database entries for 46 of a total of 69 hypothetical ORF-encoded proteins were identified. Putative functions could be assigned to 39 ORFs of pSB102. These genetic functions comprise genes for: (i) mating pair formation (Mpf); (ii) DNA transfer and replication (Dtr); (iii) plasmid replication (Rep); (iv) plasmid maintenance (Mai); and (v) a mercury resistance transposon. The relevant information about putative proteins of pSB102, such as GC content of coding regions, number of amino acid residues and molecular weight, as well as the closest relatives among protein sequences stored in the databases, are summarized in Table 3. The genetic map of pSB102 is shown in Figure 1. The complete sequence was searched for σ70-dependent promoters using the NNPP program (20). The NNPP program predicted a total of 110 putative promoters using a threshold score of 0.8, which recognizes 60% of all promoters, with a rate of false positive predictions of 0.4%. Finally, 14 putative pSB102 promoters were selected from the NNPP predicted promoters by the logic of gene organization (Table 4 and Fig. 1). One promoter, PmerR, was chosen due to conformity with published promoter/operator sequences of mercury resistance operons.
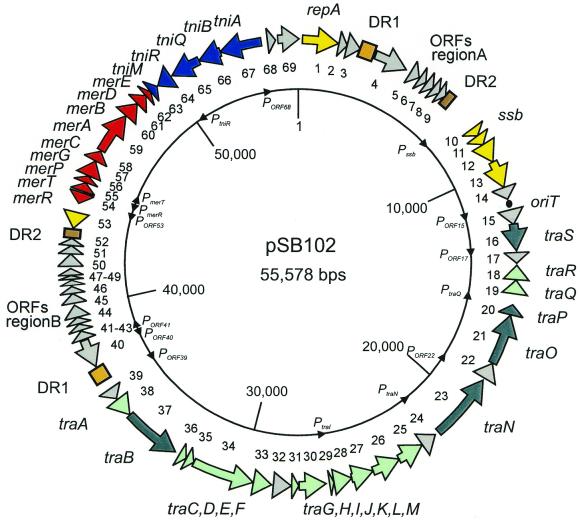
Figure 1.
Genetic map of plasmid pSB102. The 69 ORFs identified in the nucleotide sequence of plasmid pSB102 (Table 3) are located on a circular map. ORFs are shown as arrows. The numbers of the ORFs are given as the inner circle, the names of the putative genes are given as the outer circle. Putative promoters are indicated as black triangles on the inner circle. Different colors of the arrows indicate transfer genes with mating pair formation (Mpf) and DNA processing/transfer (Dtr) functions (light and deep green, respectively), genes encoding plasmid replication and maintenance (Rep and Mai) (yellow), Tn5718 encoded genes involved in mercury resistance and transposition (red and blue, respectively) as well as genes to which no similarities to database entries were detected (gray). Long DRs are indicated as light and dark brown rectangles, respectively. The putative origin of transfer (oriT) is shown as a filled circle.
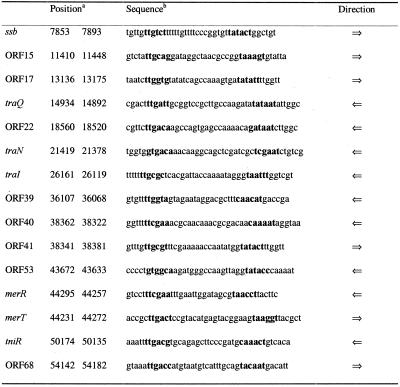
Table 4. Putative promoters identified in the pSB102 sequence.
aThe numbers indicate the position of the first and the last nucleotide in the pSB102 sequence, respectively.
bPutative –10 and –35 promoter sequences, predicted using the Neural Network Promoter Prediction program, are printed in bold.
Putative mating pair formation genes of plasmid pSB102 show highest similarity to Brucella sp. virB genes encoding a type IV secretion system
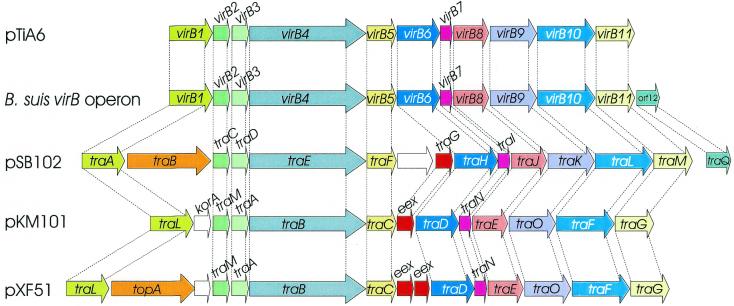
Transfer of conjugative plasmids requires the interaction of a number of cell surface components, to form a mating bridge between donor and recipient strains. The putative Mpf genes of pSB102, designated traA to traM, show highest similarity to the virB gene cluster of Brucella suis (27) and Brucella abortus (28; Fig. 2). Moreover, significant similarity was noted to the tra genes of plasmid pXF51 (29), transfer (tra) genes of bhr plasmid pKM101 and the virB gene cluster of the Agrobacterium tumefaciens plasmid pTiA6.
Figure 2.
Alignment of various gene clusters encoding type IV secretion systems. The VirB gene clusters of A.tumefaciens plasmid pTiA6, of B.suis/abortus chromosomal locus virB and the Mpf genes of plasmid pSB102, of the IncN plasmid pKM101 and of the X.fastidiosa plasmid pXF51 were aligned. Coding regions are shown as arrows. Genes encoding similar products are displayed in the same color. GenBank accession numbers of the respective nucleotide sequences are as follows: A.tumefaciens pTiA6, J03216; B.suis, AF141604; X.fastidiosa plasmid pXF51, AE003851; IncN plasmid pKM101, U09868.
traA, traC. The predicted pSB102 TraA protein contains the amino acid motifs 22AVVHVES28, 69GVAQVN74 and 127YYSG130 related to those of blocks A, B and C associated with the active site of the E.coli soluble lytic transglycosylase (30). The core region of propilin-like proteins, such as TrbC of IncP-1 plasmids, was found to be highly conserved, including a LepB cleavage site of E.coli leader peptidase I and two transmembrane helices in the central part (31). The pSB102 TraC protein contains an N-terminal peptidase cleavage site (31AFA↓QG35) at positions 33–34 and two predicted transmembrane helices in the remaining part of the protein (positions 49–66 and 84–91). Hence, pSB102 TraC may be post-translationally processed at the N-terminus, by removal of a 33 amino acid leader peptide and cyclization. IncP-1-like C-terminal maturation of the pSB102 propilin by a TraF-like peptidase might occur, because a cyclization recognition site (94AELA97) is conserved near the C-terminus (31).
traE, traF, traH. The pSB102 TraE protein contains a Walker A nucleoside triphosphate (NTP)-binding domain 457GQSGAGKTV465 (32) which is conserved in VirB4 homologs. The Agrobacterium VirB4 protein is tightly associated with the cytoplasmic membrane and is proposed to form dimers (33). The traF encoded protein of pSB102 displays a putative signal peptide cleavage site at residues 20–21 (18ANA↓QI22), indicating that it may be exported. The TraH protein of plasmid pSB102 contains five putative transmembrane domains as predicted with the DAS transmembrane prediction server (22).
traI. TraI of pSB102 contains a lipoprotein consensus (14LAGC17) sequence, suggesting that, like Agrobacterium VirB7, it is anchored to the outer membrane. In Agrobacterium the periplasmic VirB9 protein interacts via disulfide bridges with the VirB7 lipoprotein. The pSB102 TraI protein contains a single cysteine residue which is located in the putative signal sequence, whereas the plasmid pSB102 VirB9-like protein, TraK, lacks any cysteine residues. Therefore, if TraI and TraK of pSB102 form a complex, this interaction is not mediated by a disulfide bond. Using a yeast two-hybrid system circumstantial evidence has been presented that Agrobacterium VirB7 and VirB9 could also form a heterodimer without disulfide bond formation (34).
traM. Key players of the type IV secretion system are energy generators, probably in the form of ATPases. The pSB102 TraM protein contains four highly conserved motifs (35), which are common among the members of the VirB11 family: a nucleotide-binding site type A (Walker A Box 183GETGSGKTT191), an aspartate box (204QRIITIEDVPEL215), a nucleotide-binding site type B (Walker B Box 250LRMKPTRILLAELRG264) and a histidine box (276SGHGGSITSLHAG288). Using the GeNomenclature classification system for hexameric NTPases (36; http://cpmcnet.columbia.edu/dept/figurski/genomenclature/), which allows the online submission of VirB11 homologous proteins, the pSB102 TraM protein grouped to the VirB11 subfamily of NTPases. Interestingly, the VirB11 subfamily exclusively comprises NTPases implicated in the delivery of virulence factors, such as pertussis toxin, or the A.tumefaciens-mediated delivery of oncogenic T-DNA to plant cells.
traQ, traR. Two accessory genes, traQ and traR, related to ORF12 of the B.suis and B.abortus virB cluster and to trbM of IncP-1α plasmid RP4 and IncP-1β plasmid R751, respectively, complete the pSB102 putative Mpf genes. ORF12 of B.suis was first discovered as an addition to the virB operon. No similar ORFs were present in either the Agrobacterium pTi plasmid or in IncP-1α or IncW plasmids. IncP-1β plasmid R751 of Enterobacter aerogenes, however, encodes an outer membrane protein specified by the upf30.5 gene (P71192), which shows similarity to TraQ of pSB102. The putative pSB102 TraQ protein has a lipoprotein consensus sequence (13LAAC16) and a predicted signal sequence cleavage site at positions 14–15 within this motif and one predicted transmembrane helix (positions 99–105). TraR shows homology to IncP-1α plasmid RP4 (Q03537) and IncP-1β plasmid R751 (P71188) encoded protein TrbM. pSB102 TraR contains an export signal sequence cleavage site between amino acids 22 and 23 (20AQA↓ED24), indicating that the TraR protein may be exported. Downstream of trbM a putative ρ-independent transcription terminator is located (positions 13660–13685).
traB. In contrast to the virB gene cluster of B.suis, the 11 virB homologous transfer genes of pSB102 are intermixed with virB unrelated genes (Fig. 2). TraB shows similarity to type I and type III topoisomerases of various organisms, such as plasmid pXF51 encoded putative TopA (Table 3), TopA of E.coli K12 (P06612*), VC1730 of Vibrio cholerae (Q9KRB2*), To of Pseudomonas aeruginosa (AAG06399) and TopA of Zymomonas mobilis (Q9X3X7).
traG, ORF32. The traG gene encoding a putative entry exclusion function of pSB102 was found to be related to the Xylella fastidiosa plasmid pXF51 (29) encoded putative protein XFa0038 (Q9P9Q7), a hypothetical 8.5 kDa protein of IncW plasmid R388 (O50332), lipoprotein MagB05 (AAG24432) of Actinobacillus actinomycetemcomitans plasmid pVT745 (37) and the IncN plasmid pKM101 entry exclusion protein (Q46700) (38). The predicted pSB102 TraG protein contains a putative conserved lipoprotein modification/processing site (17LSG↓C20). The hypothetical ORF32 encoded protein contains four putative transmembrane domains and a putative signal peptide cleavage site (57LLA↓AR61) between amino acids 59 and 60. The ORF32 encoded protein may therefore function in the transmembrane transport machinery.
Putative genes for the processing of DNA for transfer and establishment of the plasmid in the recipient cell are scattered over plasmid pSB102
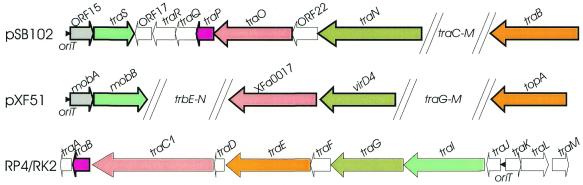
The Dtr genes of plasmid pSB102 (traB, traN, traO, traP and traS) were identified by comparison with the putative Dtr genes of plasmids pXF51 (29) and RK2/RP4 (39). They were found to be scattered over one half of plasmid pSB102 and are intermixed with Mpf genes (Figs 1 and 3). This arrangement contrasts with other conjugative bhr plasmids, such as IncP-1 or IncN, where Dtr genes are clustered in a single region (40).
Figure 3.
Alignment of the oriT and the Dtr gene region of plasmid pSB102, the X.fastidiosa plasmid pXF51 and IncP-1α plasmid RK2/RP4. The Dtr genes are shown as arrows. Genes encoding similar products are displayed in the same color. ORFs of unknown function or genes without a similar product in the pSB102 Dtr gene region are shown as white arrows. Genes displaying highest identities of their deduced amino acid sequences are indicated by bold lines. GenBank accession numbers of the respective nucleotide sequences are as follows: X.fastidiosa plasmid pXF51, AE003851; IncP-1α plasmid RK2/RP4, L27758.
oriT, ORF15, traS. The pSB102 encoded putative relaxase TraS is most closely related to the X.fastidiosa plasmid pXF51 encoded relaxase MobB (Table 3). Next nearest relatives are the A.actinomycetemcomitans plasmid pVT745 (AAG24403) encoded relaxase, TaxC of IncX plasmid R6K (P17232) and Agrobacterium encoded relaxase VirD2 (P06668*), which are all grouped into the IncP-1-type family of DNA relaxases. The pSB102 TraS protein possesses three conserved motifs of IncP-1-type relaxases (41). Upstream of the relaxase gene traS pSB102 contains a conserved oriT nic site (11424TATCCTG↓C11417) in the complementary DNA strand, identical to the RP4 and the A.tumefaciens pTiC58 oriT core sequences (40). Between pSB102 oriT and traS is located putative ORF15, which codes for a protein that shows similarity to the putative MobA protein of X.fastidiosa plasmid pXF51 (Table 3). Interestingly, mobA is preceeded by the same oriT core sequence 39556TATCCTG↓C39563 (AE003851) and followed by the relaxase gene mobB.
traN. The pSB102 TraN protein contains two conserved motifs of the TraG family of coupling proteins (42), which are responsible for recruiting the relaxosome DNA–protein complex and coupling it with the membrane-located Mpf functions (40). The first motif is a potential NTP-binding Walker motif A (199GNPLHGKT206), while the second motif includes a potential NTP-binding Walker motif B (484LMLDDE489).
traO, traP. In some conjugation systems, the establishment of transferred DNA in the recipient cell is aided by plasmid-encoded enzymes, which are transferred from the donor to the recipient cell. Classic examples are plasmid DNA primases, such as the Sog polypeptide of plasmid ColIb-P9 (43). Plasmid pSB102 TraO and TraP proteins show similarities to proteins of the primase gene region of IncP-1 plasmids (Fig. 3). Putative TraP protein of pSB102 is most closely related to hypothetical protein XFa0017 of X.fastidiosa plasmid pXF51 (Table 3). Both proteins display similarity to the C-terminal part of plasmid-encoded primases, such as PriL (Q9X6J9) of IncM plasmid pACM1 (44), but not to the N-terminal part containing the common primase sequence motif EGYATA (45).
The putative replication gene of plasmid pSB102 is related to the replication gene of the broad host range plasmid pSa
repA. The pSB102 encoded RepA protein shows highest similarity to RepA of IncW plasmid pSa (Table 3). The pSB102 RepA protein is unusually large (484 amino acids), compared to the pSa RepA protein (333 amino acids). An alignment of these two sequences indicated an addition at the N-terminus of the pSB102 encoded RepA. The C-terminal 302 amino acids of pSB102 RepA show 35% identity to RepA of pSa. The oriV of IncW plasmid pSa contains six 17 bp direct repeats, termed iterons (46). Plasmid pSB102 lacks iterons, in contrast to other plasmids encoding IncW-related RepA proteins.
ORF2, ORF3, ORF68, ORF69. Hypothetical ORF2 and ORF3 are located 13 bp downstream of the pSB102 repA gene. ORF2 overlaps the start of ORF3 by 8 bp (ATGCCTAA) (Table 3). The putative proteins encoded by ORF2 and ORF3 do not display any significant similarity to database entries. Hypothetical ORF68 and ORF69, preceeded by putative promoter PORF68, are located upstream of the pSB102 repA gene.
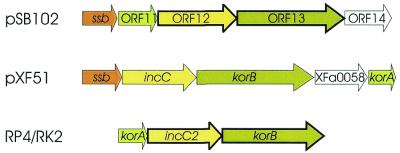
The putative maintenance region of plasmid pSB102 shows similarities to the central control region of IncP-1 plasmids
Putative pSB102 Mai genes are packed close together and are preceeded upstream by putative promoter Pssb (Table 4). ORF11, ORF12 and ORF13 are arranged in the same order in pSB102 as the related genes korA, incC and korB in the central control region of IncP-1α plasmids RK2 and RP4 (Fig. 4).
Figure 4.
Alignment of genes in the maintenance (Mai) region of plasmids pSB102, pXF51 and RK2/RP4. Putative Mai genes are shown as arrows. Genes encoding similar products are displayed in the same color. ORFs of unknown function are shown as white arrows. Genes displaying highest identities of their deduced amino acid sequences are indicated by bold lines. GenBank accession numbers of the repective nucleotide sequences are as follows: X.fastidiosa plasmid pXF51, AE003851; IncP-1α plasmid RK2/RP4, L27758.
ssb. The first gene in the pSB102 Mai region, the ssb gene, encodes a single-stranded DNA-binding protein. Nearest relatives of the putative pSB102 SSB protein are the SSB proteins of RK2 (Q02327) and R751 (Q56467). Plasmid-encoded SSB proteins presumably reduce SSB starvation in cells receiving single-stranded DNA during conjugation (40).
ORF11, ORF12, ORF13. The ORF11 encoded protein shows weak similarity to the KorA protein of Pseudomonas putida IncP-9 plasmid pM3, whereas the ORF12 encoded protein is most closely related to KorB of IncP-1α plasmid RK2 (Table 3). The ORF13 encoded protein carries three conserved ATP-binding motifs, residues 10KGGVGKSA17, 29LRNKRVLVIDF39 and 115FDFCIIDTNP124, related to the respective motifs of short IncC proteins of RK2/RP4 and R751. Both plasmids encode two in-frame IncC products which differ in their length. The short IncC protein of IncP-1 plasmids was shown to support partitioning of plasmids (47).
ORF53. Plasmid pSB102 ORF53 encodes another plasmid-stabilizing protein, a DNA resolvase, which may be involved in multimer resolution. The pSB102 ORF53 encoded protein shows 72% identity to ParA of RP4 (Table 3), which is part of the parABCDE multimer resolution system. Plasmid pSB102 specifies a distinct res site to RP4, as the deduced consensus sequence of the RP4 res site (48) was not found upstream of the pSB102 parA gene. A mercury resistance encoding transposon grouping to the Tn5053/Tn402 family, whose members are known as res site hunters (49), is located 265 bp upstream of ORF53.
The mercury resistance transposon Tn5718 belongs to the Tn5053 family of transposons
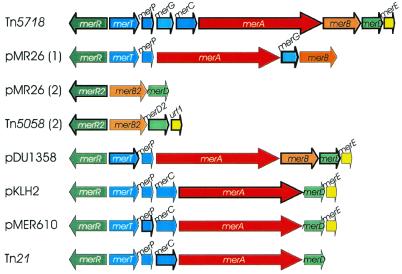
Plasmid pSB102 carries a mercury resistance transposon, designated Tn5718, which is 10 414 bp in size. It is delimited by 25 bp inverted repeats (IRs) and flanked by 5 bp direct repeats (DRs). Tn5718 contains 14 putative ORFs, nine of which were identified as mer genes and five of these genes, named tniA, tniB, tniQ, tniR and tniM (Fig. 1), are putatively involved in Tn5718 transposition. The 25 bp IRs of Tn5718 show significant similarity to integron repeats (50), which are also found as IRs at the ends of mercury resistance transposon Tn5053 (51) and integron-carrying transposon Tn5090/Tn402 (52). Transposons Tn5053 and Tn5090 have been shown to display an unusual strict insertion preference for the non-transcribed spacer of the par locus of IncP-1 plasmid RP1 (49). A 38 nt Tn21-like IR element is fused to merR in the same way as the mer-proximal IR in Tn501 and Tn21. The fusion of a Tn21-like relic IR was reported for other mercury resistance operons (51), such as Tn5053, Tn5041, pKLH2 and pMR26 (53).
The Tn5718 mercury resistance gene cluster contains many of the known mer genes identified to date. Compared to the arrangement of genes in representative mer operons, the structure of the Tn5718 mer genes is one of the most complex reported so far. Figure 5 shows an alignment of the mer operons of Pseudomonas sp. K62 plasmid pMR26, Pseudomonas sp. ED23-33 transposon Tn5058 (GenBank accession no. Y17897), Serratia marcescens plasmid pDU1358, Acinetobacter calcoaceticus plasmid pKLH2, Ralstonia sp. plasmid pMER610 and Shigella flexneri plasmid R100 transposon Tn21, which are closely related to individually encoded Tn5718 mer genes.
Figure 5.
Alignment of various mer operons mediating resistance to inorganic mercury and organomercurials located on plasmids and transposons. Genes displaying highest identities of their deduced amino acid sequences are indicated by bold arrows. Regulatory genes are shown in green, transfer genes in blue, enzyme genes in red and genes of unknown function in yellow. GenBank accession numbers of the respective nucleotide sequences are as follows: Pseudomonas sp. K62 pMR26 mer operon 1, D83080; pMR26 mer operon 2, AB013925; Pseudomonas sp. ED23-33 mer operon 2, Y17897; Serratia marcescens plasmid pDU1358, M24940 and M15049; Acinetobacter calcoaceticus plasmid pKLH2, L04303; Ralstonia sp. plasmid pMER610, Y08993; Tn21, AF071413.
merR. The deduced protein encoded by the merR gene is 100% identical to MerR2 of the organomercurial resistance operon 2 from plasmid pMR26 and Pseudomonas sp. ED23-33 Tn5058 (Table 3). The merR gene and the merTPGCABDE genes are transcribed divergently from a compact promoter/operator region, in which their potential –10 RNA polymerase recognition hexamers overlap by 4 bp (PmerT 44261TAAGGT44266 and PmerR 44268cTAACCT44263c).
merT, merP, merC, merG. Peculiar to Tn5718, the gene merG, which is supposed to reduce cellular permeability to phenylmercury (54), is inserted between merP and merC. The MerT and MerC proteins are integral membrane proteins (55). Their postulated function is the transport of Hg2+ across the cytoplasmic membrane from the periplasmic Hg2+-binding protein MerP to cytoplasmic mercuric reductase MerA. MerP of Tn5718 contains a consensus heavy metal-binding motif 30GMXCXXC36, which has been correlated with the specific binding of Hg2+ (56). merT, merP and merC genes are frequently found in various mer operons related to Tn21, whereas the merG gene is only found in the broad spectrum mer operon of plasmid pMR26 of Pseudomonas strain sp. K-62 (53). The deduced Tn5718 MerG protein carries a signal peptide cleavage site (25ALA↓YD29), like MerG from pMR26. Deletion of merG in pMR26 had no effect on inorganic mercury resistance, but rendered the bacterium more sensitive to organic mercury (54).
merA, merB. MerA and MerB are the major two enzymes involved in mercury detoxification. The Tn5718 merA gene encodes a cytoplasmic redox enzyme of 561 amino acids belonging to the class I pyridine nucleotide-disulfide oxidoreductases. The gene merB is located 14 bp downstream of merA in Tn5718. The merB encoded enzyme, organomercurial lyase, cleaves the covalent C–Hg bond of certain organomercurials.
merD, merE. MerD, which is co-transcribed with the mer structural genes, was described as a second regulatory protein. In pDU1358 MerD down-regulates expression of the mer operon by specifically binding to the mer promoter/operator sequence (57). The function of the most promoter-distal gene, merE, which is frequently found in other mer operons, is still unknown. Tn5718 encoded merE overlaps the 3′-end of merD by 4 bp at a 5′-ATGA-3′ motif (Table 3), indicating translational coupling as a regulatory mechanism. The putative Tn5718 MerE protein contains a characteristic cysteine pair (Cys28 and Cys30) and adjacent residues (LTCPCHL) which are conserved in all MerE polypeptides, therefore MerE may play a role in mercury transport (58).
The transposition module of Tn5718 contains five putative transposition genes, designated tniA, tniB, tniQ, tniR and tniM. With the exception of tniB, the deduced amino acid sequences of the four remaining transposition genes show highest similarity to TniA, TniQ, TniR and Urf2 of Pseudomonas sp. ED23-33 mercury resistance transposon Tn5058 (Table 3).
tniA, tniB, tniQ. The Tn5718 encoded TniA protein contains the D,D(35)E motif, which is characteristic of transposases of members of the IS3/retroposon superfamily (59). The predicted TniB protein of Tn5718 contains two sites, 68GPTNNGKS75 and 148MLVID152, which are potentially involved in binding and hydrolysis of NTPs (52). The GTG start codon of the following gene tniQ overlaps the TGA stop codon of tniB (Table 3), suggesting that translational coupling may be involved in tniQ expression. Although the function of TniQ in transposition is unknown, deletion mutation analysis with tniA, tniB and tniQ of Tn5053 have shown that these genes were essential for transposition (60).
tniR, tniM. The fourth gene of Tn5718, tniR, encodes a recombinase of the resolvase/invertase family. The potential resolvase (res) region of Tn5718 is located upstream of the tniR gene in the center of a pair of 14 bp imperfect inverted repeats (IR1, bp 50113–50126; IR2, bp 50142–50129). The sequence of the Tn5718 res region is identical to the res region of Pseudomonas sp. ED23-33 transposon Tn5058 and Xylella campestris transposon Tn5053 (60). Thus, Tn5718 may transpose via a two-step replicative pathway including co-integrate formation and resolution at the res region. The fifth gene of the transposition module, designated tniM, shows highest smilarity to Urf2 of Pseudomonas sp. ED23-33 mercury resistance transposon Tn5058 (Table 3), which is closely related to modulator proteins ORF2 (Q48362) of Tn501 and TnpM (P04162*) of Tn21. TnpM has been proposed to enhance transposition and to modulate co-integrate resolution in Tn21 (61).
Two clusters of hypothetical ORFs of unknown function exist on plasmid pSB102
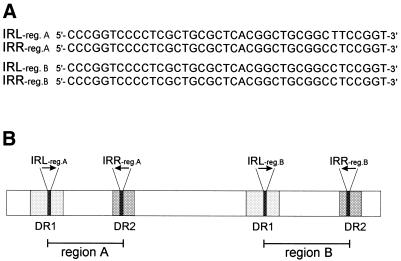
Plasmid pSB102 contains two regions, designated regions A and B, where hypothetical ORFs of unknown function are clustered. These regions are flanked by two nearly identical pairs of 39 bp IRs (Fig. 6A). The first 39 bp IR (bp 2903–2941 and 6690–6728) is imperfect (one mismatch), whereas the second 39 bp IR (bp 36998–37036 and 42533–42571) is perfectly conserved. These two 39 bp IRs are part of two different pairs of long DRs, which are 460 and 301 bp in size, respectively (Fig. 6B). The imperfect 460 bp DR (DR1) contains 24 mismatches, while the 301 bp DR (DR2) is perfectly conserved (Fig. 6B). The function of these direct and inverted repeats remains an enigma.
Figure 6.
Long DRs, containing IRs, flank regions A and B of plasmid pSB102. (A) The nucleotide sequences of the two 39 bp IRs (IRreg.A and IRreg.B) are given in forward and reverse direction for left and right hand repeats, repectively. (B) Schematic representation of 460 and 301 bp DRs, which contain the left and right hand arms of the 39 bp IRs, respectively. The two pairs of 39 bp IRs are indicated by black arrows. The 460 bp imperfect DR (DR1) is shown as light grey rectangles, the 301 bp DR (DR2) is shown as dark grey rectangles. Region A encompasses nt 3492–7317 of plasmid pSB102 and region B encompasses nt 37587–43160.
Hypothetical ORFs in region A. Region A contains six putative ORFs of unknown function. The first of these hypothetical ORFs, ORF4, encodes a putative protein of 452 amino acids. The C-terminal part of the ORF4 protein is related to X.fastidiosa plasmid pXF51 encoded hypothetical protein XFa0062 (Q9PHE6), with a size of 284 amino acids, whereas residues 78–135 of ORF4 are similar to residues 73–130 of the 297 amino acid anti-restriction protein ArdC (Q9R2K1) of IncW plasmid pSa. The downstream hypothetical ORFs, ORF5–ORF9, encode small proteins, with sizes ranging from 99 to 138 amino acids. ORF5, ORF6, ORF8 and ORF9 show no similarities to any database entries. ORF7 displays similarity to hypothetical proteins XF2067 (Table 3) and XFa0020 (Q9PHI5) encoded by X.fastidiosa genomic DNA and plasmid pXF51, respectively.
Hypothetical ORFs in region B. Region B is 5224 bp in size and contains 13 hypothetical ORFs of unknown function. The first ORF of this region, ORF40 (383 amino acids), is located on the complementary strand, whereas the remaining 12 ORFs are divergently orientated and encode proteins of rather small sizes, ranging from 49 to 166 amino acids. The deduced proteins of 11 of these ORFs, ORF40–ORF45, ORF47–ORF49, ORF51 and ORF52 did not reveal any significant similarities to database entries.
The striking clustering of genes in regions A and B encoding proteins of unknown function which display weak or no similarities to database entries rises two questions: (i) what functions are encoded by these genes; and (ii) why do the genes accumulate in these two regions of the plasmid? At present, transposon mutagenesis is under way to assess whether some of these putative proteins are involved in plasmid stability, replication or transfer.
Plasmid pSB102 defines a new family of environmental bhr plasmids
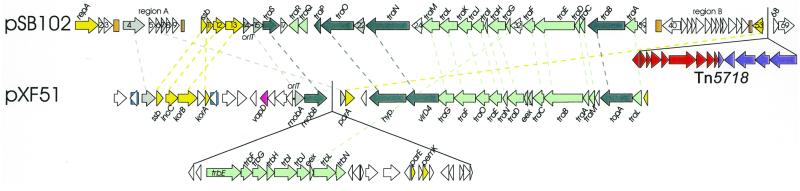
As shown in the previous sections, plasmid pSB102 is closely related to X.fastidiosa plasmid pXF51 (29), whose sequence was established in the course of the X.fastidiosa genome sequencing project (62). With the exception of the replication region, both plasmids share structural characteristics (Fig. 7). The common features are as follows. (i) The presence of a putative topoisomerase encoding gene between Mpf genes coding for a putative lytic transglycosylase and putative propilin, respectively. This arrangement contrasts with the location of the topoisomerase encoding gene traE in IncP-1α plasmid RP4, where traE is located between traF and traD in the Dtr gene region (see Fig. 3). (ii) The orientation of the relaxase gene is unique in that transcription is oriented towards the other Dtr genes. In those bhr plasmids characterized at the nucleotide sequence level to date, the relaxase genes are orientated in the same direction as the other Dtr genes (Fig. 3). (iii) The organization of the Mai region is nearly the same on both plasmids. Homologs of the IncP-1-related korA, incC and korB genes (Fig. 4) are also encoded by pXF51. Interestingly, this common set of survival genes is preceeded by the gene ssb on both plasmids. This finding may be indicative of the importance of single-stranded DNA-binding proteins in the maintenance of or establishment of plasmids in their bacterial hosts. (iv) Both plasmids carry at least one pair of unrelated DRs spanning several hundred base pairs. Whether these repeated sequences are structural components of plasmids involved in transfer, replication or maintenance or whether they confer some mobility to the ORFs they border remains an open question.
Figure 7.
Genetic organization of plasmid pSB102 and pXF51 encoded genes involved in plasmid transfer and maintenance. ORFs are shown as arrows. Related ORFs are connected by dashed lines. The colors of the arrows and dashed lines indicate different postulated functions of the encoded proteins: light green is reserved for mating pair formation (Mpf) functions, dark green for DNA transfer processing functions (Dtr), yellow for replication and maintenance. ORFs encoding proteins of unknown function are shown in grey, if they are related, or without color, if they are unrelated. The virulence-associated gene vapD of pXF51 is shown in pink. Direct repeats are indicated as rectangles with different shades of brown and blue for the different pairs of repeats in pSB102 and pXF51, respectively. GenBank accession numbers of the respective nucleotide sequences are as follows: pSB102, AJ304453; X.fastidiosa plasmid pXF51, AE003851.
In conclusion, plasmid pSB102, together with pXF51, is representative of a new class of environmental plasmids. Surprisingly, both plasmids reside in plant-associated bacteria, either in bacterial communities inhabiting the plant rhizosphere (pSB102) or in a plant pathogenic organism (pXF51). This may indicate that both plasmids are variants of an archetypical plasmid class associated with phytosphere bacteria. Therefore, such plasmids may well contribute to the genomic plasticity of plant-associated bacteria.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the BMBF (BEO 0311202 and BEO 0311952) and EU BIO4 CT98-0483 (‘TRAFFIC’ project). Support of the EU for EU-BIOTECH Concerted Action BIO4-CT-099 ‘MECBAD’, which provided opportunities for constructive discussions, is gratefully acknowledged.
DDBJ/EMBL/GenBank accession no. AJ304453
REFERENCES
- 1.Osborn A.M., Bruce,K.D., Strike,P. and Ritchie,D.A. (1997) Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev., 19, 239–262. [DOI] [PubMed] [Google Scholar]
- 2.Drønen A.K., Torsvik,V., Goksøyr,J. and Top,E.M. (1998) Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol. Ecol., 27, 381–394. [Google Scholar]
- 3.Lilley A.K., Bailey,M.J., Day,M.J. and Fry,J.C. (1996) Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol. Ecol., 20, 211–227. [Google Scholar]
- 4.Smit E., Wolters,A. and van Elsas,J.D. (1998) Self-transmissible mercury resistance plasmids with gene-mobilizing capacity in soil bacterial populations: influence of wheat roots and mercury addition. Appl. Environ. Microbiol., 64, 1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilley A.K. and Bailey,M.J. (1997) The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl. Environ. Microbiol., 63, 1577–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lilley A.K. and Bailey,M.J. (1997) Impact of plasmid pQBR103 acquisition and carriage on the phytosphere fitness of Pseudomonas fluorescens SBW25: burden and benefit. Appl. Environ. Microbiol., 63, 1584–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwieger F. and Tebbe,C.C. (2000) Effect of field inoculation with Sinorhizobium meliloti L33 on the composition of bacterial communities in rhizospheres of a target plant (Medicago sativa) and a non-target plant (Chenopodium album)—linking of 16S rRNA gene-based single-strand conformation polymorphism community profiles to the diversity of cultivated bacteria. Appl. Environ. Microbiol., 66, 3556–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selbitschka W., Dresing,U., Hagen,M., Niemann,S. and Pühler,A. (1995) A biological containment system for Rhizobium meliloti based on the use of recombination-deficient (recA–) strains. FEMS Microbiol. Ecol., 16, 223–232. [Google Scholar]
- 9.Grant S.G.N., Jessee,J., Bloom,F.R. and Hanahan,D. (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl Acad. Sci. USA, 87, 4645–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer H.W. and Roulland-Dussoix,D. (1969) A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol., 41, 459–472. [DOI] [PubMed] [Google Scholar]
- 11. de Lorenzo V., Eltis,L., Kessler,B. and Timmis,K.N. (1993) Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene, 123, 17–24. [DOI] [PubMed] [Google Scholar]
- 12.Dröge M., Pühler,A. and Selbitschka,W. (2000) Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge.Mol. Gen. Genet., 263, 471–482. [DOI] [PubMed] [Google Scholar]
- 13.Horowicz D.I. and Burke,J.F. (1981) Rapid and efficient cosmid cloning. Nucleic Acids Res., 9, 2989–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spitzbarth M., Pühler,A. and Heumann,W. (1979) Characterization of plasmids isolated from Rhizobium meliloti. Arch. Microbiol., 121, 1–7. [Google Scholar]
- 15.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staden R., Beal,K.F. and Bonfield,J.K. (2000) The Staden package, 1998. Methods Mol. Biol., 132, 215–130. [DOI] [PubMed] [Google Scholar]
- 18.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reese M.G., Harris,N.L. and Eeckman,F.H. (1996) Large scale sequencing specific neural networks for promoter and splice site recognition. In Hunter,L. and Klein,T.E. (eds), Biocomputing: Proceedings of the 1996 Pacific Symposium. World Scientific Publishing Co, Singapore.
- 21.Kurtz S. and Schleiermacher,C. (1999) REPuter: fast computation of maximal repeats in complete genomes. Bioinformatics, 15, 426–427. [DOI] [PubMed] [Google Scholar]
- 22.Cserzö M., Wallin,E., Simon,I., von Heijne,G. and Elofsson,A. (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng., 10, 673–676. [DOI] [PubMed] [Google Scholar]
- 23.Myers E.W. and Miller,W. (1988) Optimal alignments in linear space. Comput. Appl. Biosci., 4, 11–17. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 25.Llanes C., Gabant,P., Couturier,M., Bayer,L. and Plesiat,P. (1996) Molecular analysis of the replication elements of the broad-host-range RepA/C replicon. Plasmid, 36, 26–35. [DOI] [PubMed] [Google Scholar]
- 26.Götz A., Pukall,R., Smit,E., Tietze,E., Prager,R., Tschäpe,H., van Elsas,J.D. and Smalla,K. (1996) Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol., 62, 2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O‘Callaghan D., Cazevieille,C., Allardet-Servent,A., Boschiroli,M.L., Bourg,G., Foulongne,V., Frutos,P., Kulakov,Y. and Ramuz,M. (1999) A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol., 33, 1210–1220. [DOI] [PubMed] [Google Scholar]
- 28.Sieira R., Comerci,D.J., Sánchez,D.O. and Ugalde,R.A. (2000) A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol., 182, 4849–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marques M.V., da Silva,A.M. and Gomes,S.L. (2001) Genetic organization of plasmid pXF51 from the plant pathogen Xylella fastidiosa.Plasmid, 45, 184–199. [DOI] [PubMed] [Google Scholar]
- 30.Mushegian A.R., Fullner,K.J., Koonin,E.V. and Nester,E.W. (1996) A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc. Natl Acad. Sci. USA, 93, 7321–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenbrandt R., Kalkum,M., Lurz,R. and Lanka,E. (2000) Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J. Bacteriol., 182, 6751–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker J.E., Saraste,M., Runswick,M.J. and Gay,N.J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J., 1, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang T.A., Zhou,X.R., Graf,B. and Christie,P.J. (1999) Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol., 32, 1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baron C., Thorstenson,Y.R. and Zambryski,P.C. (1997) The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J. Bacteriol., 179, 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krause S., Bárcena,M., Pansegrau,W., Lurz,R., Carazo,J.M. and Lanka,E. (2000) Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl Acad. Sci. USA, 97, 3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planet P.J., Kachlany,S.C., DeSalle,R. and Figurski,D. (2000) Phylogeny of genes for secretion of the widepsread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl Acad. Sci. USA, 98, 2503–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galli D.M., Chen,J., Novak,K.F. and Leblanc,D.J. (2001) Nucleotide sequence and analysis of conjugative plasmid pVT745. J. Bacteriol., 183, 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pohlmann R.F., Genetti,H.D. and Winans,S.C. (1994) Entry exclusion of IncN plasmid pKM101 is mediated by a single hydrophilic protein containing a lipid attachment motif. Plasmid, 31, 158–165. [DOI] [PubMed] [Google Scholar]
- 39.Pansegrau W., Lanka,E., Barth,P.T., Figurski,D.H., Guiney,D.G., Haas,D., Helinski,D.R., Schwab,H., Stanisich,V.A. and Thomas,C.M. (1994) Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J. Mol. Biol., 239, 623–663. [DOI] [PubMed] [Google Scholar]
- 40.Zechner E.L., de la Cruz,F., Eisenbrandt,R., Grahn,A.M., Koraimann,G., Lanka,E., Muth,G., Pansegrau,W., Thomas,C.M., Wilkins,B.M. and Zatyka,M. (2000) Conjugative-DNA transfer processes. In Thomas,C.M. (ed.), The Horizontal Gene Pool - Bacterial Plasmids and Gene Spread. Harwood Academic, Amsterdam, The Netherlands, pp. 87–174.
- 41.Pansegrau W., Schröder,W. and Lanka,E. (1994) Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem., 28, 2782–2789. [PubMed] [Google Scholar]
- 42.Lessl M., Pansegrau,W. and Lanka,E. (1992) Relationship of DNA-transfer-systems: essential transfer factors of plasmids RP4, Ti and F share common sequences. Nucleic Acids Res., 25, 6099–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilkins B.M. and Thomas,A.T. (2000) DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol. Microbiol., 38, 650–657. [DOI] [PubMed] [Google Scholar]
- 44.Preston K.E., Radomski,C.C.A. and Venezia,R.A. (2000) Nucleotide sequence of a 7-kb fragment of pACM1 encoding an IncM DNA primase and other putative proteins associated with conjugation. Plasmid, 44, 12–23. [DOI] [PubMed] [Google Scholar]
- 45.Strack B., Lessl,M., Calendar,R. and Lanka,E. (1992) A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J. Biol. Chem., 267, 13062–13072. [PubMed] [Google Scholar]
- 46.Okumura M.S. and Kado,C.I. (1992) The region essential for efficient autonomous replication of pSa in Escherichia coli. Mol. Gen. Genet., 235, 55–63. [DOI] [PubMed] [Google Scholar]
- 47.Macartney D.P., Williams,D.R., Stafford,T. and Thomas,C.M. (1997) Divergence and conservation of the partitioning and global regulation functions in the central control region of the IncP plasmids RK2 and R751. Microbiology, 143, 2167–2177. [DOI] [PubMed] [Google Scholar]
- 48.Eberl L., Kristensen,C.S., Givskov,M., Grohmann,E., Gerlitz,M. and Schwab,H. (1994) Analysis of the multimer resolution system encoded by the parCBA operon of broad-host-range plasmid RP4. Mol. Microbiol., 12, 131–141. [DOI] [PubMed] [Google Scholar]
- 49.Minakhina S., Kholodii,G., Mindlin,S., Yurieva,O. and Nikiforov,V. (1999) Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol., 33, 1059–1068. [DOI] [PubMed] [Google Scholar]
- 50.Brown H.J., Stokes,H.W. and Hall,R.M. (1996) The integrons In0, In2 and In5 are defective transposon derivatives. J. Bacteriol., 178, 4429–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kholodii G.Ya., Yurieva,O.V., Lomovskaya,O.L., Gorlenko,Z.M., Mindlin,S.Z. and Nikiforov,V.G. (1993) Tn5053, a mercury resistance transposon with integron’s ends. J. Mol. Biol., 230, 1103–1107. [DOI] [PubMed] [Google Scholar]
- 52.Rådström P., Sköld,O., Swedberg,G., Flensburg,J., Roy,P.H. and Sundström,L. (1994) Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu and the retroelements. J. Bacteriol., 176, 3257–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kiyono M., Omura,T., Inuzuka,M., Fujimori,H. and Pan-Hou,H. (1997) Nucleotide sequence and expression of the organomercurial-resistance determinants from a Pseudomonas K-62 plasmid pMR26. Gene, 189, 151–157. [DOI] [PubMed] [Google Scholar]
- 54.Kiyono M. and Pan-Hou,H. (1999) The merG gene product is involved in phenylmercury resistance in Pseudomonas strain K-62. J. Bacteriol., 181, 726–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misra T.K. (1992) Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid, 27, 4–16. [DOI] [PubMed] [Google Scholar]
- 56.Sahlman L., Wong,W. and Powlowski,J. (1997) A mercuric ion uptake role for the integral inner membrane protein, MerC, involved in bacterial mercuric ion resistance. J. Biol. Chem., 272, 29518–29526. [DOI] [PubMed] [Google Scholar]
- 57.Mukhopadhyay D., Yu,H.R., Nucifora,G. and Misra,T.K. (1991) Purification and functional characterization of MerD. A coregulator of the mercury resistance operon in gram-negative bacteria. J. Biol. Chem., 266, 18538–18542. [PubMed] [Google Scholar]
- 58.Liebert C.A., Hall,R.M. and Summers,A.O. (1999) Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev., 63, 507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mahillon J. and Chandler,M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kholodii G.Ya., Mindlin,S.Z., Bass,I.A., Yurieva,O.V., Minakhina,S.V. and Nikiforov,V.G. (1995) Four genes, two ends and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol. Microbiol., 17, 1189–1200. [DOI] [PubMed] [Google Scholar]
- 61.Hyde D.R. and Tu,C.P.D. (1985) tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell, 42, 629–638. [DOI] [PubMed] [Google Scholar]
- 62.Simpson A.J., Reinach,F.C., Arruda,P., Abreu,F.A., Acencio,M., Alvarenga,R., Alves,L.M., Araya,J.E., Baia,G.S., Baptista,C.S. et al. (2000) The genome sequence of the plant pathogen Xylella fastidiosa. Nature, 406, 151–157. [DOI] [PubMed] [Google Scholar]