Abstract
Isothermal amplification methods are a promising trend in virus detection because of their superiority in rapidity and sensitivity. However, the generation of false positives and limited multiplexity are major bottlenecks that must be addressed. In this study, we developed a multiplex Argonaute (Ago)-based nucleic acid detection system (MULAN) that integrates rapid isothermal amplification with the multiplex inclusiveness of a single Ago for simultaneous detection of multiple targets such as SARS-CoV-2 and influenza viruses. Owing to its high specificity, MULAN can distinguish targets at a single-base resolution for mutant genotyping. Moreover, MULAN also supports portable and visible devices with a limit of detection of five copies per reaction. Validated by SARS-CoV-2 pseudoviruses and clinical samples of influenza viruses, MULAN showed 100% agreement with quantitative reverse-transcription PCR. These results demonstrated that MULAN has great potential to facilitate reliable, easy, and quick point-of-care diagnosis for promoting the control of infectious diseases.
Keywords: Argonaute, Multiplex detection, SARS-CoV-2 detection, Point-of-care diagnosis, Isothermal amplification
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread worldwide, creating a major public health concern. The early clinical features of SARS-CoV-2 infection are similar to those of common respiratory viruses, such as influenza viruses, but with higher fatality (Standl et al., 2021), and evolved single-base mutants are more infectious and deadlier than wild-type (WT) SARS-CoV-2 (Ali et al., 2021; Korber et al., 2020; Wang et al., 2021). Therefore, multiplex detection and mutation genotyping have become increasingly important for evaluating the transmissibility and pathogenicity of the virus (Volz et al., 2021).
Quantitative reverse-transcription polymerase chain reaction (RT-qPCR) is well recognized as the gold standard detection technique for viruses including SARS-CoV-2 (Oliveira et al., 2020); however, it has certain limitations, such as bulky equipment and long readout time (Esbin et al., 2020). Emerging isothermal methods, such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) are promising alternative tools for rapid and sensitive detection (El Wahed et al., 2021; Yan et al., 2020). However, these isothermal methods face the challenge of nonspecific amplification, which may produce false results (Hardinge and Murray, 2019; Schneider et al., 2019; Wang et al., 2019), as observed in the detection of SARS-CoV-2 (de Oliveira et al., 2021; Pang et al., 2020). In addition, multiplex detection derived from isothermal amplification is challenging owing to the nonspecific direct readout, which is usually generated from a fluorescent dye.
In recent years, clustered regularly interspaced short palindromic repeat (CRISPR)-based diagnostic methods have been established in combination with RT-LAMP or RT-RPA isothermal amplification methods (Chen et al., 2018; Kellner et al., 2019; Li et al., 2019). The multiplexed nucleic acid detection platform was developed by screening and characterizing additional CRISPR-associated protein (Cas) orthologs to allow for the detection of up to four targets; a single Cas enzyme is responsible only for a single target with the corresponding probe (Gootenberg et al., 2018). Nevertheless, the complexity of multiplex detection, high cost, and instability caused by guide RNA utilization constrain the diagnostic application of CRISPR methods.
Thermophilic Argonaute (Ago)-based nucleic acid detection is an alternative method for recognizing target nucleic acids mediated by guide DNAs (gDNAs) without any restriction of the target sequence (Jolly et al., 2020; Liu et al., 2018). Profiting from the stability of gDNA, the flexibility of design, and the high specificity of endonuclease activity, Ago-based methods have been developed for the detection of tumor and pathogen biomarkers (He et al., 2019; Liu et al., 2021; Song et al., 2020; Xun et al., 2021b). In particular, the Ago enzymes of the hyperthermophilic Pyrococcus furiosus (PfAgo) and Thermus thermophilus (TtAgo) have been applied to achieve efficient multiplex detection of rare mutations (Liu et al., 2021; Song et al., 2020). For application in virus detection, Ago's stepwise cleavage has been explored to enable the sequence-specific detection of human papillomavirus (He et al., 2019; Xun et al., 2021b). However, these methods are still combined with PCR in a two-step protocol and require bulky instruments and a lid-opening operation, along with a risk of contamination. The recently reported one-pot PfAgo-based detection for COVID-19 diagnosis integrates rapid amplification and PfAgo cleavage, but it requires prefabricated capillaries and a specific device that is expensive and unavailable for common instruments, and only supplies single-plex detection (Xun et al., 2021a).
In this study, we established an Ago-integrated RT-LAMP for one-pot multiplex detection, named the multiplex Ago-based nucleic acid detection system (MULAN). We first integrated this new platform in a one-pot system via a special sealed tube and detected the virus using a portable device. With an elaborate design of gDNAs and reporters, we also applied this new method to simultaneously detect SARS-CoV-2 WT and its D614G mutant. Moreover, triplex MULAN detection of SARS-CoV-2 and influenza A and B viruses in a single reaction was achieved via clinical validation. In short, this platform provides a rapid, sensitive, and easy-to-implement solution for a range of medical diagnoses.
2. Results and discussion
2.1. Design and optimization of MULAN
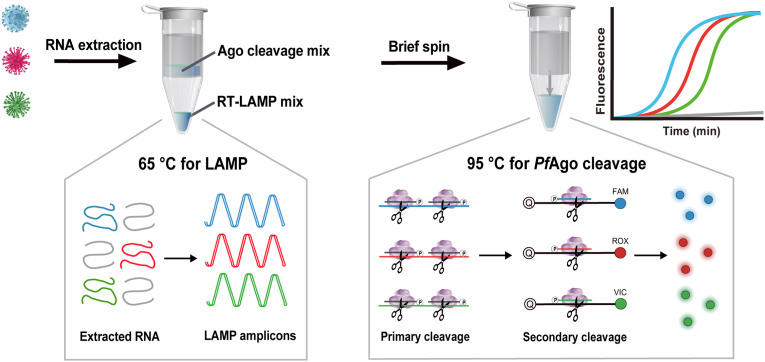
We designed MULAN to detect multiple targets in a one-pot reaction, as shown in Scheme 1 .
Scheme. 1.
Schematic illustration of the PfAgo-based MULAN for simultaneous detection of three targets.
Isothermal LAMP is difficult to use for multiplex detection because of its low specificity due to non-specific amplification. The conventional readouts of LAMP, for example, SYBR Green dye, cannot specifically recognize the target product. MULAN takes advantage of specific gDNA-directed PfAgo cleavage to precisely recognize the LAMP products at a constant temperature of 95 °C. Briefly, two responding gDNAs complementary to one strand of concatenated amplicons were designed for each target. Each target amplicon corresponded to two specific gDNAs and one fluorescence-modified reporter. The gDNA triggered the primary cleavage of one strand of target LAMP amplicons to generate renewed gDNA, which then directed the secondary specific cleavage of reporters with different fluorescence modifications. Then, the results were determined by the fluorescent readout of different channels corresponding to the different targets. Consequently, the fluorescence intensity was observed in the presence of the input target, whereas nonspecific amplicons could not trigger the cleavage of the reporter to generate fluorescence. We anticipated that MULAN would integrate rapid amplification and PfAgo's cleavage abiding by sequence base-pairing, providing high sensitivity and orthogonal specificity for multiple targets. Importantly, in order to avoid contamination, the MULAN was conducted in a one-pot system via a divided chamber, which allows the amplification products to mix with PfAgo's cleavage without lid opening.
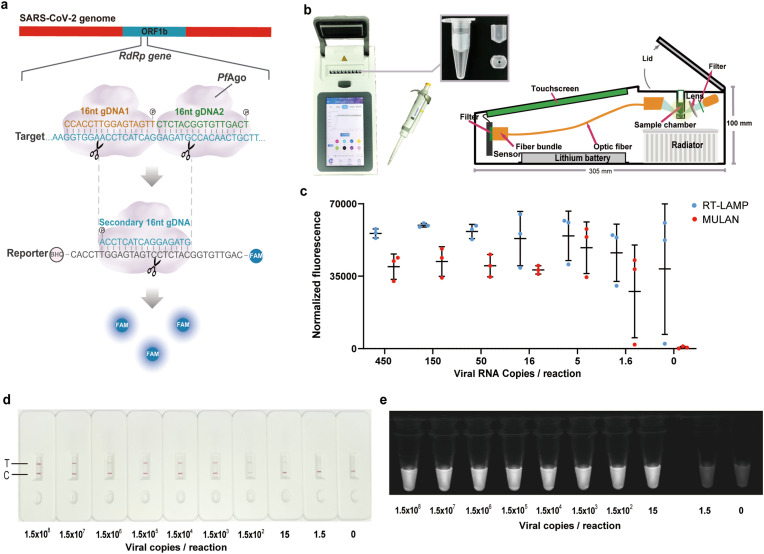
Initially, we established the system individually for each selected target independently, including SARS-CoV-2 and influenza viruses A and B. For SARS-CoV-2, for example, we designed and screened a set of primers for RT-LAMP to target the RNA-dependent RNA polymerase (RdRp) gene of the SARS-CoV-2 genome, which is conserved in SARS-CoV-2. The concatenated amplicons were generated with samples at concentrations as low as 1.5 copies per reaction, and even in the absence of template (Fig. S1). Next, we designed two 16-nt gDNAs complementary to one strand of the RdRp amplicon and a fluorescent reporter with a sequence complementary to the cleavage product (Fig. 1 a). To improve the efficiency of PfAgo catalytic cleavage, we systematically optimized the concentrations of PfAgo, gDNA, and metal ions (Fig. S2). In addition, we optimized the time required for PfAgo cleavage and found that 15 min was adequate for analysis (Fig. S3). Furthermore, we validated that MULAN detected two out of three replicates at 1.6 copies of SARS-CoV-2 RNA, while the qRT-PCR detected two replicates at only five copies (Fig. S4). We further evaluated the specificity of MULAN using total nucleic acids extracted from 21 species of in vitro-cultured respiratory pathogens that exhibited similar symptoms of respiratory infection, and no cross-reactivity was observed (Fig. S5). Similarly, MULAN was applied to detect influenza A and B viruses, and it was demonstrated that MULAN can detect two out of three replicates at 1.5 copies per reaction for each virus (Fig. S6).
Fig. 1.
Portable and visible detection based on MULAN. (a) Design of gDNAs and reporter for SARS-CoV-2 detection. (b) Device and consumables needed to perform the portable MULAN assay. (c) One-pot portable detection using diluted SARS-CoV-2 RNA. Error bars indicate the standard deviation of three replicates. (d) Lateral flow dipstick detection of SARS-CoV-2 RNA with decreasing input concentrations. (e) Blue-light irradiation results for detection of SARS-CoV-2 RNA with decreasing input concentrations. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. One-pot portable MULAN assays
By designing a mini tube affiliated with a routine 0.2 mL reaction tube, we explored MULAN as a one-pot system without a lid-opening operation. RT-LAMP amplicons in the lower chamber were mixed with the upper components of PfAgo via brief centrifugation. To allow a Point-of-Care Testing (POCT) solution, we developed a mini isothermal fluorescence detector (MIFD), which is a lightweight, stand-alone, real-time device (Fig. 1b). We determined the detection time to be 35 min for RT-LAMP and 15 min for PfAgo cleavage, which was sufficient to detect low-load viral RNA (Fig. S7). However, we observed fluorescence signals even from the non-template controls in RT-LAMP alone (Figs. S7a–b). When the entire process of MULAN was carried out, a strong fluorescence signal at the end time was observed for the sample with the target input; in contrast, no signals from the non-template controls were observed (Figs. S7c–d). These results demonstrate that the MULAN assay accurately eliminates the false-positive effect of nonspecific amplification caused by the LAMP reaction. We next verified that the limit of detection (LoD) of one-pot MULAN was five copies per reaction (Fig. 1c), and two out of three replicates at 1.6 copies per reaction were detected. This limit of detection was close to the that of the two-step MULAN, and even superior to that of the two-step detection system of Cas13a-based Specific High-sensitivity Enzymatic Reporter unlocking (SHERLOCK; LoD of 42 copies per reaction within 60 min) (Patchsung et al., 2020). As reported in CRISPR-based detection, an attempt was made to combine two steps into a one-pot assay; however, this approach has been proven to be much less sensitive than the two-step approach, as well as more challenging for experimental optimization (Fozouni et al., 2021; Joung et al., 2020). The detection sensitivity of one-pot MULAN is also higher than that of RT-qPCR detection systems such as RealStar® SARS-CoV-2 in Europe and CDC COVID-19 in the United States (LoD of 10 copies per reaction in 100 min) (Uhteg et al., 2020).
For visualization and instrument-free implementation, we also developed a paper-based detection method to obtain readouts that are visible to the naked eye. We designed a reporter with a biotin modification at the 3′-terminus, which binds a colloidal gold-labeled anti-biotin antibody (Fig. S8a). To avoid interference from the hook effect (Fig. S8b), we found the final optimal concentration of reporter loaded on the lateral flow dipstick pad to be 13.3 nM (Fig. S8c). The detection limitwas 15 copies/reaction (Fig. 1d), and the interpretation of the results is shown in Fig. S8d. By observing the products of MULAN under blue light irradiation (wavelength: 450 nm), an LoD of 15 copies per reaction was obtained (Fig. 1e). In conclusion, MULAN exhibits important application potential for on-site testing, which is crucial for outpatients and communities.
2.3. Genotyping of the SARS-CoV-2 and its mutant
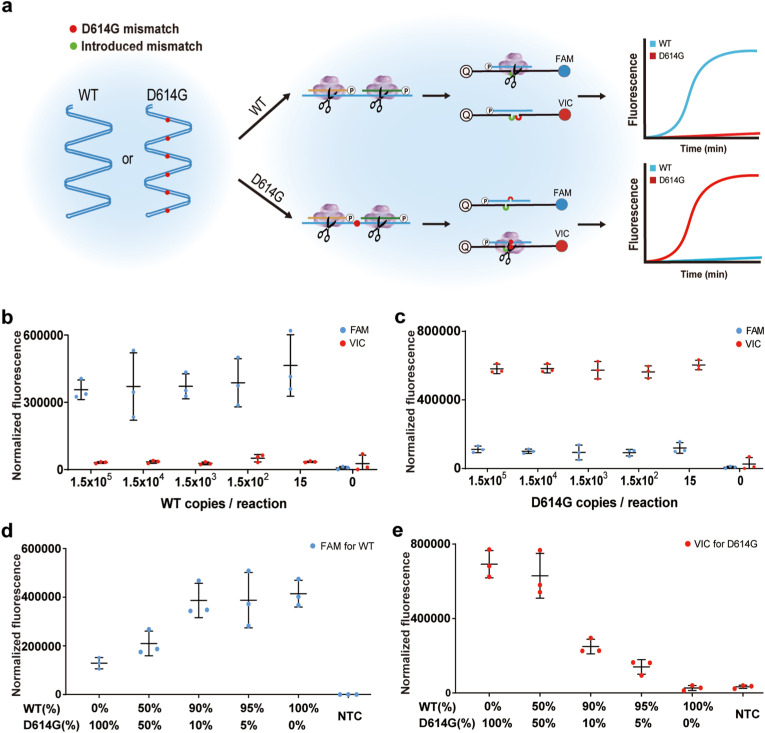
Based on the highly specific recognition of the target sequence by PfAgo, we developed MULAN to discriminate between WT and the single-base mutant (Fig. 2 a). We used a set of universal primers to obtain amplicons and the same primary gDNAs for specific cleavage of either WT or the mutant. Consequently, the different secondary gDNAs corresponding to WT and the mutant possessed a single-nucleotide difference. To ensure the precise detection of WT or the mutant in the samples, an extra mismatch was introduced via the fluorescent reporters, which was inspired by our previous investigation on Ago-based detection for single nucleotide variant diagnosis (Liu et al., 2021). Using the D614G mutant as an example, we designed two types of 30-nt reporters with one introduced mismatch against the 11th position of the secondary gDNA (Fig. S9a). To achieve the optimal discrimination effect, we screened the reporter sequences by introducing three different types of mismatch nucleotides. One pair of reporters was obtained to generate a high fluorescence signal for a specific target and a low fluorescence signal for another nonspecific target in a single reaction (Fig. S9b). We again screened four sets of LAMP primers in combination with the selected reporters to discriminate the amplicons effectively (Fig. S10).
Fig. 2.
MULAN detection for WT and the D614G mutant of SARS-CoV-2. (a) Schematics of MULAN detection for WT and the mutant in a one-pot reaction. The bubbles between the secondary gDNA and reporter represent the mismatches on designed gDNAs. LoD assay for the detection using template of the WT plasmid (b) and the D614G plasmid (c). Fluorescence detection of WT (d) and the D614G (e) mutant with different mutation frequencies. Error bars indicate the standard deviation of three replicates. “NTC” refers to non-template control.
Next, we evaluated the LoD of this discrimination MULAN using serial dilutions of plasmids containing either WT or D614G sequences. These samples were detected with the expected difference between FAM and VIC fluorescence signals, demonstrating an LoD of 15 copies per reaction (Fig. 2b and c). To validate whether this genotyping can be applied to detect a mutation carried at different frequencies, we used mixed templates of WT and the mutant to obtain mutant frequencies of 0%, 5%, 10%, 50%, and 100% (Fig. 2d and e). The results show that VIC fluorescence, representing D614G, can be detected using templates with a 5% mutation frequency. This finding suggests that MULAN, which can distinguish mutants with low mutation frequencies, is a powerful tool for the investigation of infectious viral variants. Compared with the mainstream sequencing methods for mutant detection, MULAN is faster, more economical and does not require bulky equipment. Considering the easy-to-implement design, MULAN can be quickly deployed in the screening of emerging viral mutations, such as the recent outbreak of the Omicron variant (B.1.1.529) in some resource-limited regions.
2.4. Triplex detection of SARS-CoV-2 and influenza viruses
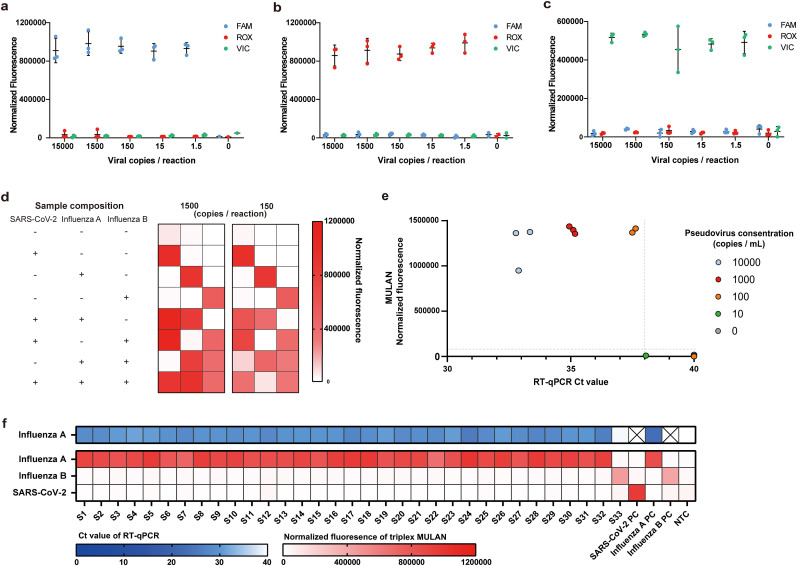
As influenza viruses and SARS-CoV-2 lead to similar clinical symptoms, simultaneous discrimination of the three pathogens would help inform diagnosis and guide treatment. We further conducted triplex detection to target SARS-CoV-2, as well as influenza A and B viruses. The reaction that was introduced to the one-pot detection system consisted of all three sets of primers, gDNAs, and reporters. The presence of each target RNA could be determined by the corresponding fluorescence readout from the MULAN reaction, such as FAM fluorescence for SARS-CoV-2, VIC fluorescence for influenza A virus, and ROX fluorescence for influenza B virus. Considering that the triplex system is more complicated than the singleplex system, we first evaluated the LoD of triplex MULAN to detect each individual virus using titrated plasmids. The presence of 1.5 copies per reaction of each target was precisely detected by the corresponding fluorescence pattern (Fig. 3 a–c). Across various concentrations of each target, MULAN can specifically produce fluorescence that responds only to the respective target without observed cross-activity. In addition, as there were some clinical cases of two-virus coinfection among these three viruses (Azekawa et al., 2020, Cuadrado-Payan et al., 2020, Huang et al., 2021), we validated triplex MULAN in the detection of mimic samples co-infected with two or three viruses.
Fig. 3.
Triplex MULAN detection of SARS-CoV-2, influenza A, and influenza B. LoD assay of triplex MULAN detection using diluted SARS-CoV-2 (a), influenza A (b), and influenza B (c) templates, respectively. Error bars indicate the standard deviation of three replicates. (d) Triplex MULAN detection with mixed samples of three target plasmids. The depth of red in the heat map represents the average fluorescence intensity of three replicates. (e) Triplex MULAN detection and commercial RT-qPCR test of SARS-CoV-2 pseudoviruses. The vertical dotted line represents the threshold of RT-PCR (Ct value < 38 was determined to be positive), and the horizontal dotted line represents the threshold of MULAN (the mean of the normalized fluorescence from samples with water input for three replicates). (f) Triplex MULAN detection of clinical samples with influenza viruses compared to commercial RT-qPCR test. S1–S33 refer to clinical samples. “PC” and “NTC” refer to the positive control and non-template control, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Because the amplification efficiency of each primer set was different, the generation of amplicons for each target varied and affected the sensing capability. We found that, when using an equal ratio of three primer sets, the fluorescence intensity of influenza A was much stronger than those of SARS-CoV-2 and influenza B. To achieve balanced amplification efficiency for the three targets, we adjusted the primer ratio of SARS-CoV-2, influenza A, and influenza B to 4:2:4 (Fig. S11). Using this optimized primer ratio, the triplex MULAN orthogonally detected 150 or 1500 copies per reaction of double coinfection samples or triple coinfection samples (Figs. 3d and S12). Notably, no cross-activity was observed for the absent virus. These results imply that MULAN endows LAMP to achieve accurate triplex detection assisted by a single endonuclease. Although CRISPR detection has also successfully detected different targets in a single reaction, each target requires an individual Cas protein or engineered Cas protein, increasing the complexity and cost of the system (Gootenberg et al., 2018; Myhrvold et al., 2018; Ooi et al., 2021).
2.5. Validation with pseudoviruses and clinical samples
Finally, we performed clinical validation of triplex MULAN from pseudoviruses and clinical samples. Pseudoviruses have been used to validate the detection of SARS-CoV-2, since clinical samples from COVID-19 patients were extremely limited. We also adapted a commercial SARS-CoV-2 RT-PCR test kit (Wondfo Biotech Technologies) for parallel comparison. The results demonstrate that MULAN shares 100% agreement with RT-PCR results. Samples with three replicates at 1000 copies/mL all obtained positive readouts, and two replicates at 100 copies/mL were detected (Fig. 3e). The triplex MULAN assay was then validated using nasopharyngeal swabs from patients with confirmed influenza viruses, in comparison with a commercial RT-PCR kit for influenza A (BioGerm Medical Technology). The RT-PCR results identified 32 positives (Ct values from 27 to 35) and one negative for influenza A (Table S1). We found that the MULAN assay showed 100% (32 out of 32) sensitivity to the RT-PCR results, and no cross-fluorescence readout was observed (Fig. 3f). These demonstrated that the multiplex MULAN was able to discriminate the species of diverse viruses existing in clinical samples.
3. Conclusions
In summary, we have developed MULAN based on Ago's precise and orthogonal cleavage for the detection of SARS-CoV-2 as well as influenza A and B viruses. MULAN provides a promising solution for diagnosing multiple pathogens and has clear advantages, including fast reaction, high sensitivity, genotyping capability, and POCT potential compared with existing diagnostic techniques (Table S2). MULAN not only solves the problem of low specificity in isothermal amplification, but is also simple in terms of design and implementation for multiplex detection compared with currently advanced CRISPR-based tests that require multiple enzymes. Moreover, MULAN integrates two continuous reactions in designed chambers to allow the reaction to occur within a single tube with no lid-opening operation, thus preventing contamination risk. The portable application of MULAN provides economical and easy-to-operate detection of multiple pathogens, which is in high demand for managing infectious diseases.
CRediT authorship contribution statement
Xingyu Ye: Methodology, Conceptualization, Investigation, Writing – original draft, Writing – review & editing, Visualization. Haiwei Zhou: Methodology, Validation. Xiang Guo: Methodology, Visualization, Formal analysis. Donglai Liu: Visualization, Investigation, Validation. Zhonglei Li: Methodology, Funding acquisition. Junwei Sun: Methodology, Investigation, Visualization. Jun Huang: Methodology, Funding acquisition. Tao Liu: Investigation. Pengshu Zhao: Investigation. Heshan Xu: Investigation. Kai Li: Writing – review & editing. Hanming Wang: Validation. Jihua Wang: Methodology. Li Wang: Investigation. Weili Zhao: Investigation. Qian Liu: Writing – review & editing, Visualization, Funding acquisition, Supervision. Sihong Xu: Supervision, Writing – review & editing, Investigation, Validation. Yan Feng: Supervision, Conceptualization, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Ministry of Science and Technology (2020YFA0907700), National Natural Science Foundation of China (31770078), Cross-Institute Research Fund of Shanghai Jiao Tong University (20X990010002), and Zhejiang University Special Scientific Research Fund for COVID-19 Prevention and Control (2020XGZX022).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114169.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ali F., Kasry A., Amin M. Med. Drug. Discov. 2021;10:100086. doi: 10.1016/j.medidd.2021.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azekawa S., Namkoong H., Mitamura K., Kawaoka Y., Saito F. IDCases. 2020;20 doi: 10.1016/j.idcr.2020.e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado-Payan E., Montagud-Marrahi E., Torres-Elorza M., Bodro M., Blasco M., Poch E., Soriano A., Pineiro G.J. Lancet. 2020;395(10236):e84. doi: 10.1016/S0140-6736(20)31052-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira K.G., Estrela P.F.N., Mendes G.M., Dos Santos C.A., Silveira-Lacerda E.P., Duarte G.R.M. Analyst. 2021;146(4):1178–1187. doi: 10.1039/d0an02066d. [DOI] [PubMed] [Google Scholar]
- El Wahed A.A., Patel P., Maier M., Pietsch C., Ruster D., Bohlken-Fascher S., Kissenkotter J., Behrmann O., Frimpong M., Diagne M.M., Faye M., Dia N., Shalaby M.A., Amer H., Elgamal M., Zaki A., Ismail G., Kaiser M., Corman V.M., Niedrig M., Landt O., Faye O., Sall A.A., Hufert F.T., Truyen U., Liebert U.G., Weidmann M. Anal. Chem. 2021;93(4):2627–2634. doi: 10.1021/acs.analchem.0c04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. RNA. 2020;26(7):771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P., Son S., Diaz de Leon Derby M., Knott G.J., Gray C.N., D'Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardinge P., Murray J.A.H. Sci. Rep. 2019;9(1):7400. doi: 10.1038/s41598-019-43817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R.Y., Wang L.Y., Wang F., Li W.Q., Liu Y., Li A.T., Wang Y., Mao W.X., Zhai C., Ma L.X. Chem. Commun. 2019;55(88):13219–13222. doi: 10.1039/c9cc07339f. [DOI] [PubMed] [Google Scholar]
- Huang B.R., Lin Y.L., Wan C.K., Wu J.T., Hsu C.Y., Chiu M.H., Huang C.H. J. Microbiol. Immunol. Infect. 2021;54(2):336–338. doi: 10.1016/j.jmii.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly S.M., Gainetdinov I., Jouravleva K., Zhang H., Strittmatter L., Bailey S.M., Hendricks G.M., Dhabaria A., Ueberheide B., Zamore P.D. Cell. 2020;182(6):1545–1559 e1518. doi: 10.1016/j.cell.2020.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.G., Woolley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Cell. 2020;182(4):812–827 e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. ACS Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Liu Q., Guo X., Xun G., Li Z., Chong Y., Yang L., Wang H., Zhang F., Luo S., Cui L., Zhao P., Ye X., Xu H., Lu H., Li X., Deng Z., Li K., Feng Y. Nucleic Acids Res. 2021;49(13):e75. doi: 10.1093/nar/gkab274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yu Z., Zhu J., Wang S., Xu D., Han W. Front. Chem. 2018;6:223. doi: 10.3389/fchem.2018.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira B.A., Oliveira L.C., Sabino E.C., Okay T.S. Rev. Inst. Med. Trop. Sao Paulo. 2020;62:e44. doi: 10.1590/S1678-9946202062044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi K.H., Liu M.M., Tay J.W.D., Teo S.Y., Kaewsapsak P., Jin S., Lee C.K., Hou J., Maurer-Stroh S., Lin W., Yan B., Yan G., Gao Y.G., Tan M.H. Nat. Commun. 2021;12(1):1739. doi: 10.1038/s41467-021-21996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., Xu J.Y., Liu Y.M., Peng H.Y., Feng W., Cao Y.R., Wu J.J., Xiao H.Y., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H.Q., Le X.C. Anal. Chem. 2020;92(24):16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J.S., Abudayyeh O.O., Zhang F., Horthongkham N., Uttamapinant C. Nat. Biomed. Eng. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Schneider L., Blakely H., Tripathi A. Electrophoresis. 2019;40(20):2706–2717. doi: 10.1002/elps.201900167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Hegge J.W., Mauk M.G., Chen J., Till J.E., Bhagwat N., Azink L.T., Peng J., Sen M., Mays J., Carpenter E.L., van der Oost J., Bau H.H. Nucleic Acids Res. 2020;48(4):e19. doi: 10.1093/nar/gkz1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standl F., Jockel K.H., Brune B., Schmidt B., Stang A. Lancet Infect. Dis. 2021;21(4):e77. doi: 10.1016/S1473-3099(20)30648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhteg K., Jarrett J., Richards M., Howard C., Morehead E., Geahr M., Gluck L., Hanlon A., Ellis B., Kaur H., Simner P., Carroll K.C., Mostafa H.H. J. Clin. Virol. 2020;127:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole A., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Consortium C.-U., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Cell. 2021;184(1):64–75 e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhao P., Si X., Li J., Dai X., Zhang K., Gao S., Dong J. Front. Microbiol. 2019;10:2959. doi: 10.3389/fmicb.2019.02959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Chen J., Gao K., Wei G.W. Genomics. 2021;113(4):2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun G., Lane S.T., Petrov V.A., Pepa B.E., Zhao H. Nat. Commun. 2021;12(1):2905. doi: 10.1038/s41467-021-23185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun G., Liu Q., Chong Y., Guo X., Li Z., Li Y., Fei H., Li K., Feng Y. Bioresour. Bioprocess. 2021;8(1):46. doi: 10.1186/s40643-021-00401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Clin. Microbiol. Infect. 2020;26(6):773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.