Abstract
This review reports the recent advances in surface-enhanced Raman scattering (SERS)-based lateral flow assay (LFA) platforms for the diagnosis of infectious diseases. As observed through the recent infection outbreaks of COVID-19 worldwide, a timely diagnosis of the disease is critical for preventing the spread of a disease and to ensure epidemic preparedness. In this regard, an innovative point-of-care diagnostic method is essential. Recently, SERS-based assay platforms have received increasing attention in medical communities owing to their high sensitivity and multiplex detection capability. In contrast, LFAs provide a user-friendly and easily accessible sensing platform. Thus, the combination of LFAs with a SERS detection system provides a new diagnostic modality for accurate and rapid diagnoses of infectious diseases. In this context, we briefly discuss the recent application of LFA platforms for the POC diagnosis of SARS-CoV-2. Thereafter, we focus on the recent advances in SERS-based LFA platforms for the early diagnosis of infectious diseases and their applicability for the rapid diagnosis of SARS-CoV-2. Finally, the key issues that need to be addressed to accelerate the clinical translation of SERS-based LFA platforms from the research laboratory to the bedside are discussed.
Abbreviations: LFA, lateral flow assay; SERS, surface-enhanced Raman scattering; POC, point-of-care; AuNPs, gold nanoparticles; NC, nitrocellulose; SARS-CoV-2, severe acute respiratory syndrome-coronavirus-2; HIV, human immunodeficiency virus; RT-PCR, real-time polymerase chain reaction; IgG, immunoglobulin G; IgM, immunoglobulin M; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeat; crRNAs, CRISPR RNAs; SEB, staphylococcal enterotoxin; NS1, nonstructural protein 1; IFA, indirect immunofluorescence assay; Si-AuNPs, silica-encapsulated AuNPs; KSHV, Kaposi’s sarcoma herpes virus; BA, bacillary angiomatosis; PRV, pseudorabies virus
Keywords: Surface-enhanced Raman scattering (SERS), Lateral flow assay (LFA), Point-of-care (POC), Infectious disease, SARS-CoV-2, In vitro diagnostics (IVD)
1. Introduction
Paper-based lateral flow assay (LFA) strips are simple tools intended to detect the presence of a specific target analyte in a sample. The LFA approach is considered a convenient point-of-care (POC) diagnostic tool because its operation does not require any expertise [[1], [2], [3], [4], [5], [6], [7]]. According to World Health Organization (WHO) guidelines, LFA meets the ASSURED criteria (affordable, sensitive, specific, user-friendly, rapid/robust, equipment-free, and deliverable for end users). Consequently, LFA strips have been extensively used for testing a wide range of analytes, including proteins [[8], [9], [10], [11], [12], [13]], nucleic acids [[14], [15], [16]], infectious viruses [[17], [18], [19]], and bacterial pathogens [[20], [21], [22], [23], [24]]. Antibody-conjugated gold nanoparticles (AuNPs) are the most commonly used detection probes in conventional colorimetric LFAs, because their color changes to red under the localized surface plasmon effects when they accumulate in the test line on a strip.
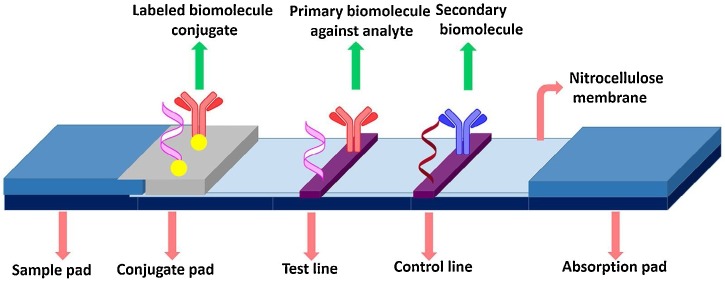
A commercially available LFA strip is composed of four components: a nitrocellulose (NC) membrane, a sample pad, a conjugation pad, and an absorption pad. Here, the NC membrane is used as a substrate for the immobilization of antibodies because the nitrate group in the NC membrane interacts with the peptide bonds in the antibodies through an electrostatic interaction. Therefore, capture and control antibodies are immobilized on the test and control lines of the NC membrane using an antibody dispenser [5,6]. The operating principle of LFA strips is straightforward, as shown in Fig. 1 . When a liquid sample containing the target antigens is loaded onto the LFA strip, the solution flows toward the absorption pad through capillary force. Thereafter, the target antigens in the solution interact with the detection antibody-conjugated AuNPs in the conjugation pad. Subsequently, the antigen–antibody-conjugated AuNP complexes flow toward the test line and are captured by the antibodies immobilized on the test line. Thus, sandwich immunocomplexes labeled with AuNPs are formed on the test line, and the AuNP accumulation induces a color change in the test line. The excess antibody-conjugated AuNPs continue to travel along the strip and are captured by the antibodies immobilized on the control line. The color change on the control line represents whether the LFA strip is working normally. The remaining residue flows to the absorption pad, which is used as a liquid sink. As a result, two red lines appear on completion of the assay in the presence of the target analyte, but only one red line appears on the control line in its absence. The most well-known commercial LFA strip is a pregnancy test kit. The increase in the concentration of human chronic gonadotropin hormone during pregnancy is an effective biomarker for the test. Most pregnant individuals have a positive test result one week after the first day of a missed menstrual period. However, such a kit is only used in qualitative tests (“yes” or “no”) for determining pregnancy. This detection strategy does not require any reading device because it only relies on visual detection; however, it suffers from limitations in terms of its “limit of quantitative analysis” and “poor sensitivity.”
Fig. 1.
Basic structure of LFA. Reprinted with permission from Ref. [7]. Copyright 2016 Elsevier Science.
To date, many researchers have attempted to address these problems in conventional LFA strips. To improve the quantification capability and detection sensitivity of analytes, various optical strip readers and corresponding detection labels have been employed. Several optical readers for reading fluorescence [[25], [26], [27], [28], [29]], chemiluminescence [[30], [31], [32], [33], [34], [35]], electrochemical [[36], [37], [38], [39], [40]], or surface plasmon enhancement [[41], [42], [43], [44], [45]] signals have been introduced for the detection of signals radiated from the test and control lines of an LFA strip. Subsequently, corresponding optical labels for optical readers, such as fluorescence microspheres [27], quantum dots [[46], [47], [48]], europium nanoparticles [49,50], carbon nanoparticles [51,52], up-conversion nanoparticles [53,54], and plasmonic nanoparticles [55,56], have also been developed. Among them, the fluorescence detection method has been most extensively used in reading optical signals from LFA strips; however, it suffers from poor detection sensitivity and low precision in many cases. Recently, SERS detection techniques have attracted considerable attention owing to their high sensitivity [57,58]. Electromagnetic enhancement takes place by increasing the electric field intensity by the molecule. Its mechanism can be explained by a phenomenon called “surface plasmon resonance”. Conduction band electrons easily produce a large oscillation at the surface plasmon resonance frequency in the local electric field, and it strongly depends on the shape and size of particles, the dielectric properties of the metal and the wavelength of incident light. When SERS nanotags (i.e., functional Au nanoparticles) are exposed to laser radiation, the incident field is substantially enhanced at the active sites under localized surface plasmon effects. This electromagnetic enhancement overcomes the low sensitivity problem inherent in fluorescence detection [59,60]. In particular, this SERS-based assay platform has recently exhibited broad applicability in the diagnosis of infectious diseases [61,62]. The measurement principle in a SERS-based LFA strip is the same as that in a conventional colorimetric LFA strip, except for the use of Raman reporter-labeled SERS nanotags. Using this SERS nanotag, the presence of target biomarkers can be identified through a color change on the test and control lines, similar to a colorimetric LFA strip. Additionally, the quantitative evaluation of target biomarkers with high sensitivity is possible by monitoring the intensity of a characteristic Raman peak of reporter molecules. Raman equipment can be conveniently miniaturized and combined with the SERS-LFA strip for on-site, POC diagnostics of infectious diseases [63,64]. Therefore, it is important to investigate the current status, challenges, and applications associated with SERS-based LFA platforms. It is also important for researchers to become more knowledgeable about the current status of SERS-based LFA platforms for detecting infectious diseases. In this context, we particularly focus on the recent advances in LFA platforms for the POC diagnosis of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) and the applicability of SERS-based LFA techniques for the diagnosis of viral or bacterial pathogens.
2. Recent application of LFA platforms for the diagnosis of infectious diseases
Infectious diseases, which are caused by pathogens such as bacteria and viruses, are responsible for many deaths worldwide. Respiratory infectious diseases, such as influenza, tuberculosis, and pneumonia, lead to 5 million deaths every year [[65], [66], [67]]. Moreover, the deaths caused by other infectious diseases, such as human immunodeficiency virus (HIV), malaria, and diarrhea, collectively comprise a further 6 million deaths annually [68]. Recently, the advent of a new SARS-CoV-2 has resulted in an ongoing global pandemic. Many pneumonia cases caused by SARS-CoV-2 have been reported in China [[69], [70], [71]]. Since then, the outbreak has rapidly spread across the globe and affected 213 countries and territories, leading to an unanticipated public health crisis worldwide. This disastrous pandemic demands a rapid and sensitive diagnostic tool to fight against the novel pathogen. Presently, real-time polymerase chain reaction (RT-PCR) is considered the gold standard method for diagnosing SARS-CoV-2 [72,73], but it requires centralized diagnostic services and highly skilled personnel. Diagnosis using RT-PCR requires a sequential process, including RNA extraction from the virus, reverse transcription, and repetitive thermocycling steps for DNA amplification. It takes approximately four hours for detecting SARS-CoV-2 using RT-PCR, but the turnaround time for diagnosing patients takes over 24 h because of the time required to deliver samples to the central clinical laboratory. Therefore, it is important to reduce the detection time and develop a POC sensing system for use outside of the clinical laboratory. In recent years, several LFA platforms for the POC detection of viral RNAs, immunoglobulin G (IgG)/ immunoglobulin M (IgM) antibodies, and antigens associated with SARS-CoV-2 have been reported [[74], [75], [76]]. In the case of LFA strips for detecting viral RNAs, the significant detection time for RT-PCR is the primary issue. In contrast, immunoassay LFA strips essentially require improvements in accuracy and sensitivity. In this section, we briefly introduce the current developments in LFA strips for the POC diagnosis of SARS-CoV-2.
2.1. POC LFA strips for detecting viral RNAs in SARS-CoV-2
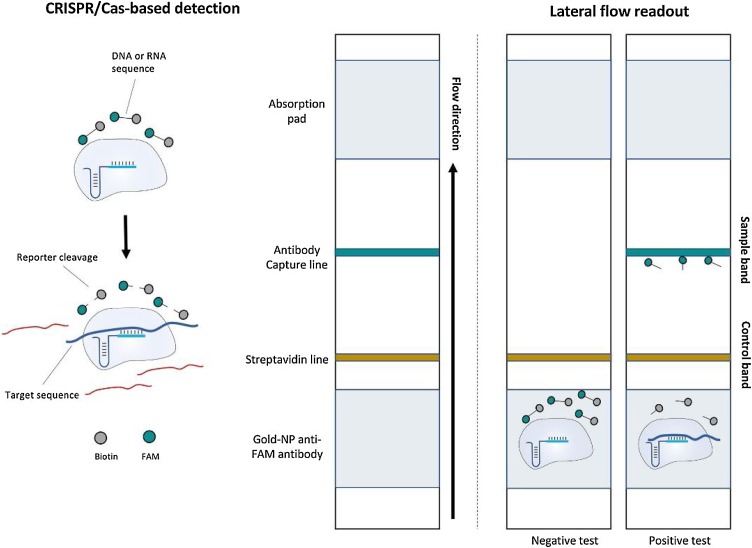
To reduce the time required for the detection of SARS-CoV-2 nucleic acids, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas-based LFA platforms have been recently reported. CRISPR-Cas is an RNA-mediated adaptive defense system in prokaryotic organisms, which protect against invading viruses [77]. This immune system is based on small RNAs for the specific detection and cleavage of foreign nucleic acids, such as viral RNA sequences [78]. Due to various CRISPR-Cas enzymes, which can be programmed by different CRISPR RNAs (crRNAs), CRISPR-Cas technology is amenable to the detection of different nucleic acids from virus targets [79]. To make this technique affordable for on-site diagnosis, an LFA strip has been designed and combined with the CRISPR-Cas system [80]. An RNA or DNA reporter (based on Cas enzymes) that has been labeled with fluorescein at one end and biotin at the other terminus is used in the lateral flow strip. At one end, the biotin moiety binds to streptavidin and the other fluorescein binds to AuNPs. If the trans-cleavage activity of the Cas enzyme is triggered, the RNA/DNA reporter will be cleaved and AuNP-labeled antibodies will flow to the test line, where a secondary antibody exists, and form a colored product, representing the presence of the target, as shown in Fig. 2 [81].
Fig. 2.
Schematic of lateral flow detection of CRISPR-based biosensing. The CRISPR-based lateral flow detection relies on the cleavage of a FAM-biotin reporter by the collateral activity of the Cas enzymes upon target recognition, allowing for detection on commercial lateral flow strips. The reporter accumulates anti-FAM antibody–AuNP conjugates at the first line on the strip (brown), preventing binding of the antibody–gold conjugates to protein A on the second line (green); the cleavage of the reporter would reduce accumulation at the first line and result in a signal at the second line. Reprinted with permission from Ref. [81]. Copyright 2020 American Chemical Society.
Sherlock Biosciences reported a protocol using the combination of the CRISPR-based Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) technique with a lateral flow strip for detecting SARS-CoV-2 [82]. Viral RNA sequences were detected in a range of 10–100 copies per microliter of input using synthetic coronavirus RNA fragments. The test was performed for RNA extracted from patient respiratory samples, and corresponding signals were read out using an LFA in less than one hour, without requiring any elaborate equipment.
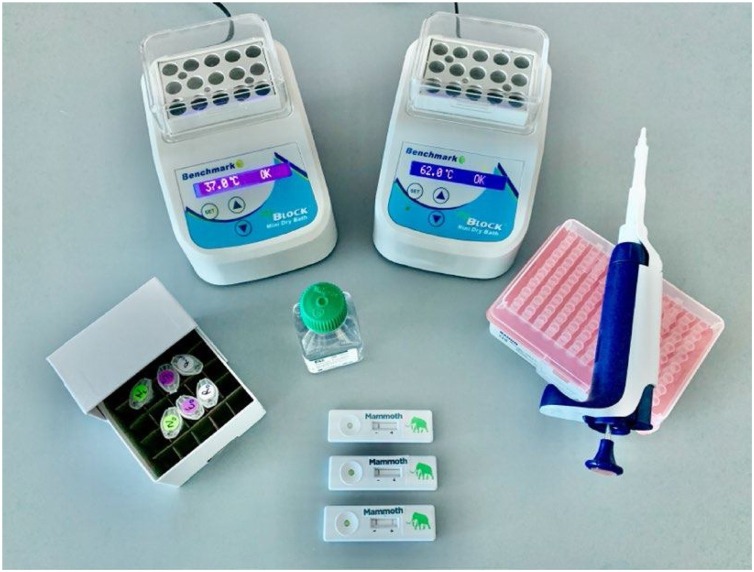
A CRISPR-Cas12-based LFA for detecting SARS-CoV-2 from respiratory RNA extracts, called SARS-CoV-2 DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR), was recently reported by Broughton et al. [83] in Mammoth Biosciences Company (the other leading company in the field of CRISPR diagnostics). In this LFA platform, reverse transcription and isothermal amplification using loop-mediated amplification were simultaneously performed for rapid diagnosis. The estimated LOD for the DETECTR assay was 10 copies per microliter reaction, and the accuracy of the DETECTR assay was comparable to that of RT-PCR (Fig. 3 ). The DETECTR offers some advantages over RT-PCR, such as isothermal signal amplification, averting the need for thermocycling, fast response (<40 min), high specificity, integration with a convenient readout format (i.e., lateral flow), and no requirement for advanced laboratory services.
Fig. 3.
Minimum equipment required to run the protocol. With appropriate biosafety level 2 requirements, the minimum equipment required to run the protocol following RNA extraction includes Eppendorf tubes with reagents, heat blocks or water bath (37 °C and 62 °C), nuclease-free water, pipettes and tips, and lateral flow strips. Reprinted with permission from Ref. [83]. Copyright 2020 Nature Publishing Group.
2.2. POC LFA strips for serological testing of SARS-CoV-2
Although molecular diagnostic methods, which are mostly based on PCR techniques, have been extensively applied in SARS-CoV-2 diagnosis, they cannot be used for monitoring disease progression. The presence of viral protein antibodies, which are generated against SARS-CoV-2 infection, can be used for extensive identification of past infection and immunity. SARS-CoV-2 possesses four main structural proteins: nucleocapsid protein (N), spike surface glycoprotein (S), envelope protein (E), and matrix protein (M) [84]. Thus, the immune system produces immunoglobulins (Ig) to fight against the pathogen.
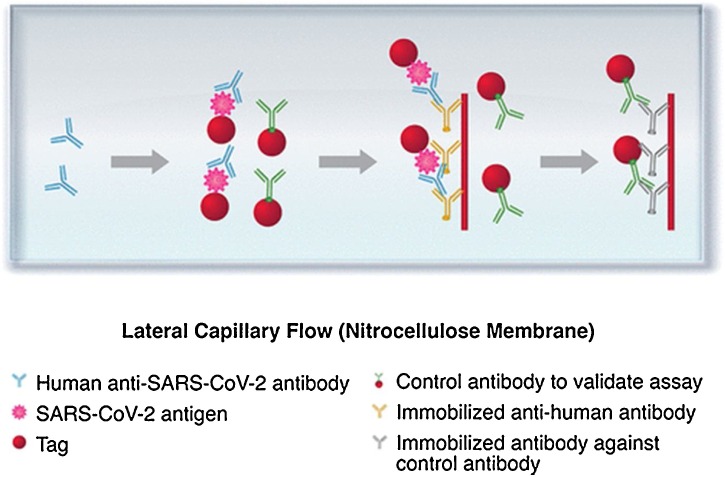
Serological testing is used to analyze the blood serum or plasma and can be expanded to test saliva, sputum, and other biological fluids to check the presence of IgM and IgG. The test plays a key role in monitoring disease progression, providing an evaluation of short- and long-term antibody responses as well as the abundance of the antibody. IgM can be detected in the serum after a few days and remains up to a few weeks after the infection. Therefore, IgM can be considered a biomarker for early-stage infection diagnoses. In contrast, IgG represents the prior or current situation of infection. Accordingly, IgG can indicate post-infection and potential immunity. Considering the above information, serological tests can be applied for detecting the current and past exposure to coronavirus [81], as shown in Fig. 4 . Herein, we focus on serological tests based on LFA for detecting SARS-CoV-2.
Fig. 4.
Lateral flow immunoassay for detecting anti-SARS-CoV-2 antibodies. Samples move via capillary flow on the NC membrane. When anti-SARS-CoV-2 antibodies are present, they bind to the labeled antigen and continue to move until they are captured by the immobilized antihuman antibodies. The presence of the captured antibody–antigen complex is visualized as a colored test band. The labeled control antibodies comigrate until they are captured at the control band. Reprinted with permission from Ref. [81]. Copyright 2020 American Chemical Society.
Chen et al. reported a lateral flow immunoassay for detecting anti-SARV-CoV-2 IgG in human serum [85]. The assay results were obtained within 10 min and revealed a potential application for clinical diagnostics. The assay can also be used to monitor the progression of SARS-CoV-2 as well as patients’ response to the treatment. In another study, Black et al. reported a lateral flow immunoassay for detecting anti-SARS-CoV-2 IgM and IgG in serum, plasma, and whole blood [86]. The samples were examined with the lateral flow immunoassay, and the results were compared with those obtained by ELISA. The results were found to be in good agreement, and no significant difference was observed when detecting IgM and IgG with LFI and ELISA at D0 and D7 (p = 1.00). The clinical sensitivity of the developed assay was 92 % 7 days after the PCR diagnosis of coronavirus on venous blood samples; the specificity for IgM and IgG was 92 % and 100 %, respectively. This assay serves as a rapid test that can be used anywhere and enable POC testing to detect the presence of anti-SARS-CoV-2 antibodies among different sample types.
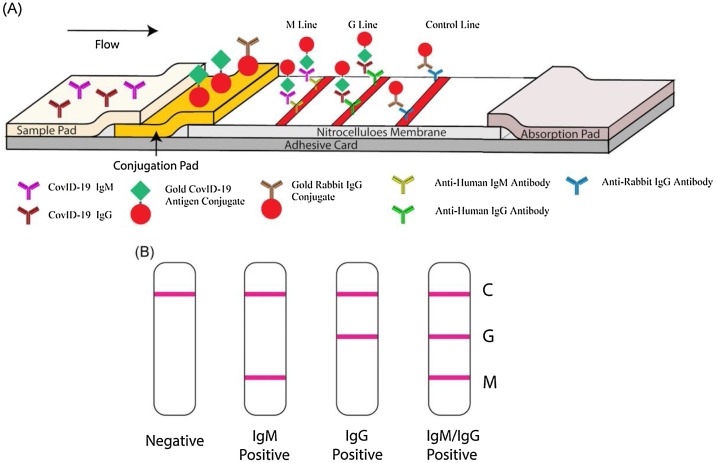
Li et al. [87] reported a lateral flow immunoassay that could simultaneously detect IgM and IgG antibodies against coronavirus in human blood within 15 min, as shown in Fig. 5 . This assay can successfully diagnose patients at different infection stages. The sensitivity and specificity of the assay were evaluated using blood samples collected from 397 PCR-confirmed SARS-CoV-2 patients and 128 negative patients; the sensitivity and specificity for this assay were 88.7 % and 90.6 %, respectively. Moreover, various blood samples, including venous and fingerstick blood, were assessed using the developed test. The test results obtained using fingertip blood were in good agreement with those obtained using venous blood, indicating that the test is applicable for rapid detection. Owing to the capability of detecting IgM and IgG simultaneously, the test can be used for early diagnoses and for monitoring disease progression during treatment.
Fig. 5.
Schematic of rapid SARS‐CoV‐2 IgM–IgG combined antibody test. A, Schematic of the detection device; B, illustration of different testing results; C, control line; G, IgG line; M, IgM line. IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome-coronavirus 2. Reprinted with permission from Ref. [87]. Copyright 2020 Wiley Interscience.
A patient cohort study was conducted by Lou et al. [88] to test total antibody (Ab), IgM antibody, and IgG antibody against SARS-CoV-2 in plasma samples through three different LFAs. The antigen used to develop the Ab and IgM assays was the receptor-binding domain, while the antigen of the recombinant nucleoprotein of SARS-CoV-2 was used for the IgG assay. For patients at the early stages of the disease (i.e., 0–7 days post exposure), the Ab assay showed the highest sensitivity (64.1 %) compared to the IgM and IgG assays (33.3 % for both). Two weeks later, the detection sensitivity for all Ab, IgM, and IgG assays increased to 100 %, 96.7 %, and 93.3 %, respectively. Furthermore, the assays showed excellent specificity ranging from 95 % to 100 %. This study demonstrated that serological testing is an important complement to molecular testing (i.e., RNA testing) for pathogen-specific diagnosis and provides important information for the assessment of immunity status of patients.
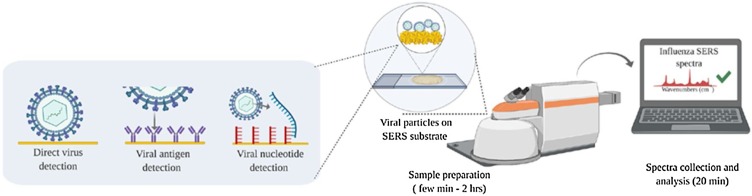
The diagnosis of an infectious disease such as SARS-CoV-2 does not require screening of various pathogens; instead, the presence or absence of a particular pathogen is considered. As the amount of pathogen loaded from patient samples depends on the infection stage, it is important to detect low concentrations of a pathogen for early diagnoses. For SARS-CoV-2 pathogens, the relation between viral amount and disease severity has not yet been fully elucidated. However, it is certain that a diagnostic approach to detect lower concentrations of its pathogens is very important. From this viewpoint, the SERS-based viral diagnostic technique has a strong potential to satisfy the requirements for highly sensitive detection (Fig. 6 ) [89]. In the following section, recent advances in SERS-based LFA techniques for early diagnosis of infectious diseases are introduced, although their application to SARS-CoV-2 diagnosis has not been reported thus far.
Fig. 6.
Summary of standard clinical and SERS-based viral diagnostics. Versatile SERS-based viral diagnostic approaches targeting the whole virus, surface markers, or viral nucleic acids. Reprinted with permission from Ref. [89]. Copyright 2020 American Institute of Physics.
3. SERS-based LFAs for the diagnosis of infectious diseases
Unlike fluorescence-based LFA strips, the detection principle of SERS-based LFA strips is identical to that of LFA strips that can be assessed by the naked eye because receptor-conjugated AuNPs are used as the detection probes. In SERS-based LFA strips, however, Raman reporter molecules are additionally adsorbed on the AuNP surface. Using these SERS nanotags, the presence of target species on the test line can be identified through color changes. Additionally, a quantitative analysis of a target species with high sensitivity is possible by monitoring the characteristic Raman peak intensity of Raman reporter molecules [90,91]. The SERS-based assay platform is considered a next-generation assay format because of its high sensitivity and multiplex detection capability [[92], [93], [94], [95]]. When SERS nanoparticles are used as detection probes, Raman signals are substantially improved at the hotspots by electromagnetic enhancements. Such enhancement overcomes the sensitivity problem inherent in fluorescence detection. SERS is capable of sensitive detection, and LFA provides a rapid and user-friendly sensing platform. Thus, the integration of SERS and LFA provides a robust and reliable platform for the POC diagnosis of various infectious diseases. Recently, SERS-based LFAs have been extensively used for detecting various target analytes, including proteins, nucleic acids, and infectious pathogens. Different elements, such as antibodies, aptamers, and DNA oligonucleotides, are also used as recognition receptors.
3.1. Immunoassays using SERS-LFA
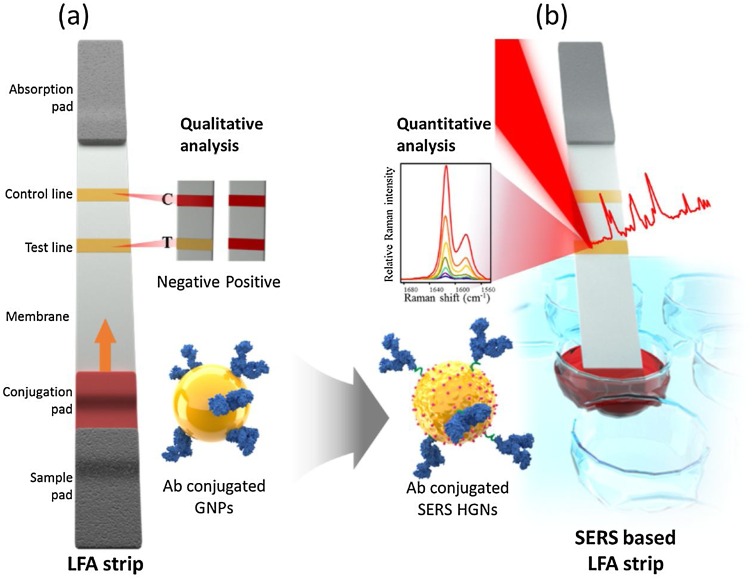
Hwang et al. [96] introduced a SERS-based LFA platform for the first time to detect staphylococcal enterotoxin (SEB) on an LFA strip. Here, Raman reporter-labeled hollow gold nanospheres were used as detection labels instead of the AuNP labels in conventional naked-eye-based LFA strips. This SERS-based LFA platform enables the rapid identification of SEB antigens through a color change. Additionally, its quantitative evaluation is possible by measuring the Raman signal of the test line, as shown in Fig. 7 . Using this SERS-based LFA platform, quantities as low as 0.001 ng/mL of SEB could be detected, making this approach approximately three orders of magnitude more sensitive than the ELSA-based assay method for SEB. This study demonstrated that SERS-based LFA can be utilized to overcome the inherent limitations of conventional LFAs and achieve more sensitive assay results without deviating from the original LFA strip design.
Fig. 7.
Schematic of (a) a conventional LFA strip and (b) the SERS-based LFA strip. Only one red band is observed in the control zone in the absence of the target antigen (negative), whereas two red bands appear in the presence of the target antigen (positive). Using the SERS-based LFA strip, highly sensitive quantification of target analytes is possible by monitoring the SERS peak intensity. Reprinted with permission from Ref. [96]. Copyright 2016 Royal Society of Chemistry.
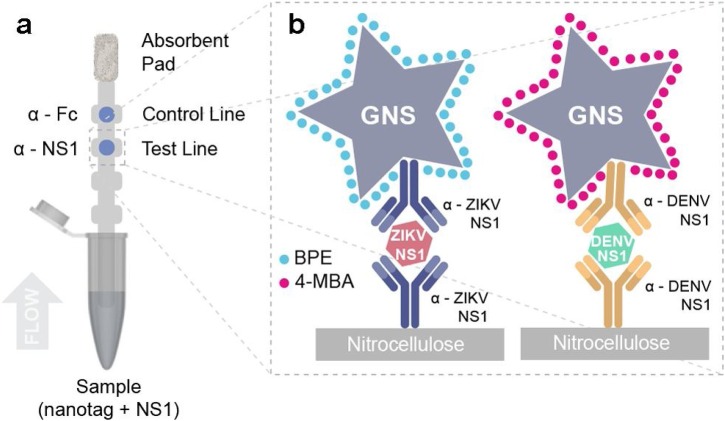
Sanchez-Purra et al. [97] developed a SERS-based duplex assay platform that could distinguish between Zika and dengue nonstructural protein 1 (NS1) biomarkers (Fig. 8 ). Zika and dengue viruses are mosquito-borne infectious diseases, both of which cause similar initial symptoms but possess different potential complications. Therefore, it has been of interest to develop a POC assay platform capable of differentiating between the two viral diseases. It is known that NS1 is present at high levels (>15 μg/mL) two days post infection for acute dengue, but the NS1 level of Zika virus is significantly lower. Thus, an increase in detection sensitivity is crucial for detecting Zika and would also be helpful in the early detection of both pathogens. In this study, the authors used highly sensitive SERS-encoded Au nanostars as SWERS nanotags that could distinguish between the different concentrations of NS1 for Zika and dengue viruses. Compared to LFA strips, the LODs were improved by 15-fold for Zika and 7.2-fold for dengue.
Fig. 8.
Sandwich immunoassay. (a) Schematic of dipstick sandwich immunoassay. (b) Sandwiches formed by each antibody pair, NS1 and GNS − Ab conjugate, for both ZIKV and DENV NS1, at the test line. Reprinted with permission from Ref. [97]. Copyright 2017 American Chemical Society.
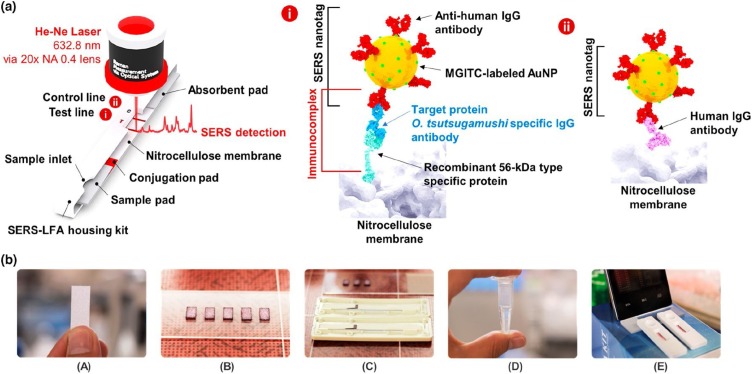
Lee et al. [98] developed a SERS-LFA technique for the accurate diagnosis of scrub typhus. It is an infectious disease caused by the intracellular bacterium Orientia tsutsugamushi (O. tsutsugamushi). Indirect immunofluorescence assay (IFA) is a standard serological diagnostic method for its diagnosis. Herein, target human antibodies, IgG and IgM created by O. tsutsugamushi, were detected by observing the increase in the titer concentration during the acute phase of the disease, as shown in Fig. 9 . In this IFA test, a fluorescence microscope is required because the titer values for O. tsutsugamushi-specific IgG and IgM are determined by the fluorescence titer images of a clinical sample. However, this fluorescence image-based diagnosis often suffers from the subjective determination of end points and the requirement of high-cost fluorescence microscopy. To address the current problems in IFA assays, patient-friendly SERS-LFA platforms have been developed for accurate POC diagnosis of scrub typhus. The feasibility of the proposed diagnostic technique was verified by testing 40 clinical samples and comparing the assay results with those obtained by IFA methods.
Fig. 9.
Schematic of the SERS-LFA process. (a) 56-kDa recombinant proteins immobilized on the test line capture the O. tsutsugamushi-specific IgG antibodies in the human serum. Sandwich immunocomplexes are formed when the antihuman IgG antibody-conjugated SERS nanotags bind with the target antibodies. Conversely, the human IgG antibodies are immobilized on the control line and directly bind with the antibody-conjugated SERS nanotags. (b) Fabrication procedure of SERS-LFA strips. Reprinted with permission from Ref. [98]. Copyright 2019 American Chemical Society.
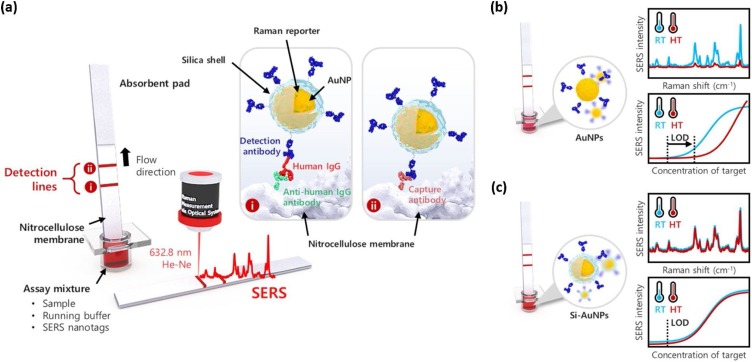
More recently, Jeon et al. [99] used silica-encapsulated AuNPs (Si-AuNPs) in LFAs to maintain thermal stability and reproducibility for use in high-temperature regions. For the diagnosis of dengue and Zika viruses, which are endemic in tropical regions, thermally stable SERS nanotags are essential. To address this issue, SERS-based immunoassays using Si-AuNPs in an LFA strip were performed, and the results were compared with those obtained using AuNPs. In case of LFA strips using AuNPs, the sensitivity worsened at 45 °C but LFA strips using Si-AuNPs maintained similar LOD values regardless of the increase in temperature, as shown in Fig. 10 . Therefore, it was concluded that this SERS-LFA strip using Si-AuNPs can be used as a stable diagnostic platform for tropical infectious diseases such as Zika or dengue viruses.
Fig. 10.
(a) Schematic of SERS-based LFA for human IgG using thermally stable Si-AuNPs. Antihuman IgG antibodies on the test line capture the Si-AuNPs; the excess SERS nanotags were captured by capture antibodies on the control line. (b) SERS spectra of the AuNP-loaded LFA strips and the corresponding calibration curves at 25 °C and 45 °C. (c) SERS spectra of the Si-AuNP-loaded LFA strips and the corresponding calibration curves at 25 °C and 45 °C. Reprinted with permission from Ref. [99]. Copyright 2020 Elsevier Science.
Besides the biomarkers of infectious diseases described above, various protein biomarkers, including infection markers [100], cytokines (IL-1β and IFN-γ) [101], prostate cancer antigen 3 [102], cytokine [103], mycovirus protein A [104], biomarkers of acute myocardial infarction (troponin I, isoenzyme MB of creatine kinase and myoglobin) [[105], [106], [107]], bisphenol A [108], and thyroid-stimulating hormone [109], have been successfully investigated using this SERS-LFA technique.
3.2. DNA detection using SERS-LFA
SERS-LFA platforms can also be used for detecting nucleic acid targets. An RT-PCR using fluorescence DNA TaqMan probes has been considered as a standard detection method, but it requires special equipment for nucleic acid amplification as well as trained personnel. Additionally, a long assay time is required because a low copy number of nucleic acids needs to be amplified through repetitive thermocycling steps. If a highly sensitive detection method is introduced for a low concentration of target genes, the assay time can be substantially reduced and a thermal amplification process is not required. The highly sensitive SERS detection method is a strong potential candidate for overcoming the low sensitivity problem inherent in conventional detection methods.
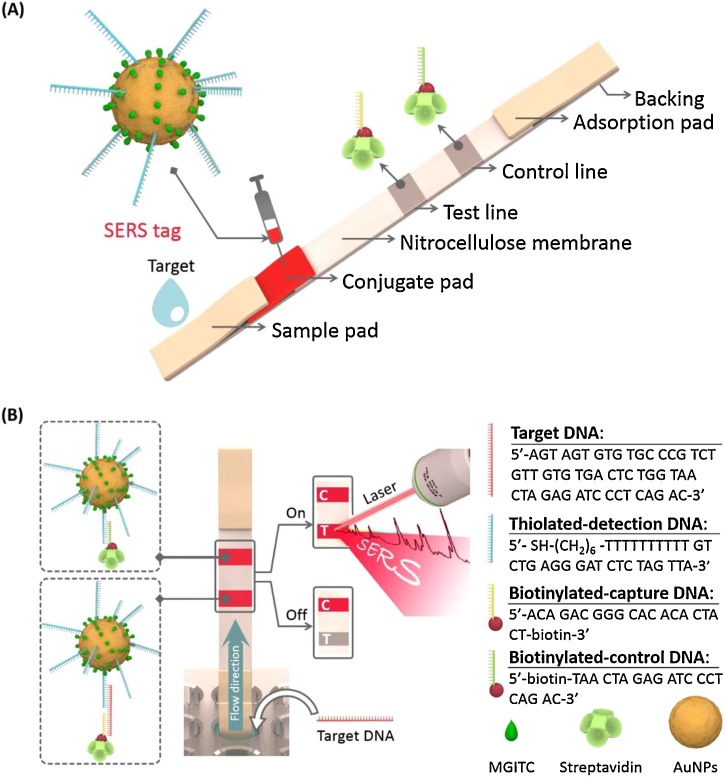
Fu et al. [110] developed a SERS-LFA platform for the sensitive detection of human immunodeficiency virus type 1 (HIV-1) DNA. HIV-1 is a retrovirus that makes the human immune system unable to resist disease infections. In the proposed SERS-LFA strip for HIV-1 DNA detection, Raman reporters and oligonucleotide-functionalized AuNPs were used as SERS nanotags. In contrast, capture DNA, which is complementary to an HIV-1 target DNA, and control DNA, which is complementary to the detection DNA on AuNPs, were immobilized on the test and control lines of the LFA strip, respectively (Fig. 11 ). A good linear relation between HIV-1 concentration and characteristic Raman intensity was obtained with a detection limit of 0.24 pg/mL.
Fig. 11.
(A) Schematic of the configuration and (B) measurement principle of SERS-based LFA for the quantification of HIV-1 DNA. (C is the control line and T is the test line). Reprinted with permission from Ref. [110]. Copyright 2016 Elsevier Science.
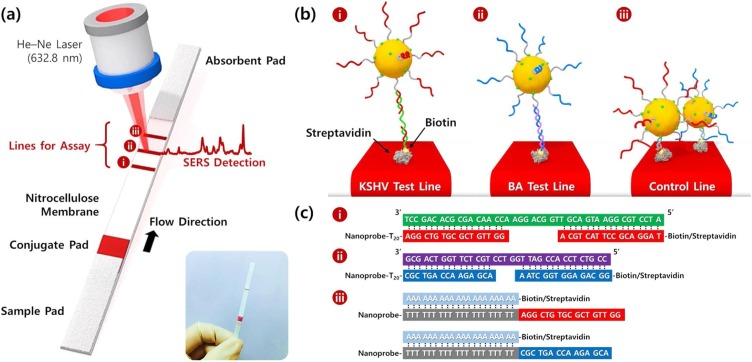
Inspired by the SERS-LFA strip for single DNA detection, Wang et al. [111] reported the application of a SERS-LFA strip for the detection of dual DNA markers. This strip enables the simultaneous detection of dual DNA markers in a single assay. Two target DNAs, associated with Kaposi’s sarcoma herpes virus (KSHV) and bacillary angiomatosis (BA), were used to evaluate the detection capability of the proposed SERS-DNA strip. A 1:1 M mixture of KSHV and BA detection DNA-labeled SERS nanotags was prepared, and the nanotags were captured by corresponding KSHV and BA capture DNAs immobilized on the first and second test lines of the LFA strip, respectively (Fig. 12 ). The LODs of KSHV and BA DNAs were determined to be 0.043 and 0.074 pM, respectively, which indicates a sensitivity 10,000 times better than that of colorimetric LFA strips, as shown in Fig. 12.
Fig. 12.
(a) Schematic of the LFA biosensor for simultaneous detection of two nucleic acids. The strip is composed of two test lines and one control line. (b) (i) KSHV DNA-AuNP complexes were captured by the probe KSHV DNAs on the first test line, (ii) BA DNA-AuNPs complexes were captured by the probe BA DNAs on the second test line, and (iii) excess KSHV and BA detection DNAs attached to AuNPs were captured by control DNAs through T20-A20 hybridization on the third control line. (c) Corresponding DNA hybridizations for two test lines (i and ii) and one control line (iii). Reprinted with permission from Ref. [111]. Copyright 2017 American Chemical Society.
3.3. Detection of viral and bacterial pathogens using SERS-LFAs
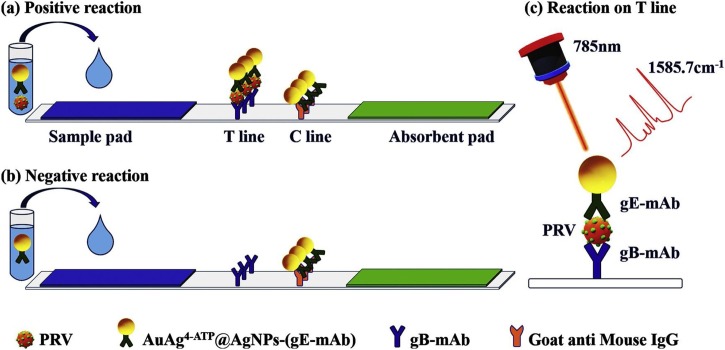
Several studies regarding the application of SERS-LFAs for the detection of viruses and bacterial pathogens have been reported. Recently, Shen et al. [112] reported a SERS-LFA strip for the detection of pseudorabies virus (PRV), which causes a serious infectious disease in wild animals. As shown in Fig. 13 , AuAg@Ag SERS nanotags were captured at both T and C lines for a sample containing wild-type PRV, but SERS nanotags were only captured at the C line for a sample without the target virus. The characteristic Raman peak intensity of the Raman reporter molecule (4-aminothiophenol) at 1586 cm−1 at the T line increased concomitantly with the PRV concentration. In this study, the linear detection range for wild-type PRV was 41–650 ng/mL, and the LOD was estimated to be 5 ng/mL.
Fig. 13.
Simple illustration of SERS-LFS. (a) Positive reaction: the AuAg4−ATP@AgNPs-(gE-mAb) is captured at both the T line and C line. (b) Negative reaction: the AuAg4−ATP@AgNPs-(gE-mAb) is only captured at the C line. (c) Principle illustration of positive reaction at T line. Reprinted with permission from Ref. [112]. Copyright 2019 Elsevier Science.
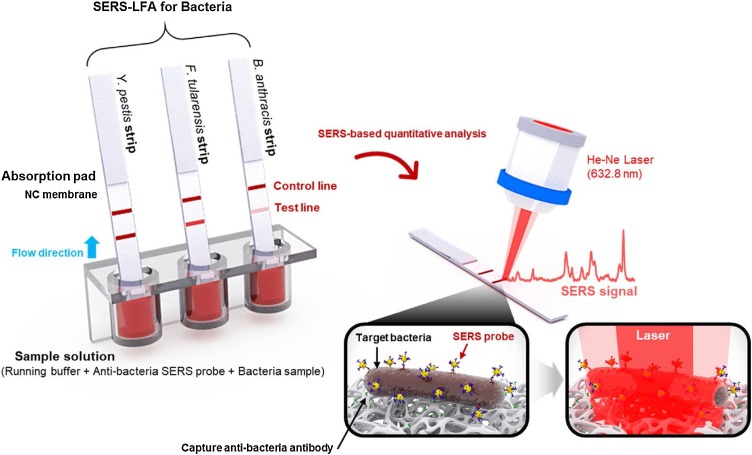
Wang et al. [113] developed SERS-LFA strips for the simultaneous detection of three high-risk bacterial pathogens categorized as lethal infectious agents: Yersinia pestis (Y. pestis), Francisella tularensis (F. tularensis), and Bacillus anthracis (B. anthracis) (Fig. 14 ). In this study, highly accurate quantitative analyses of three different pathogenic bacteria were possible by monitoring the characteristic Raman peak intensities of the Raman reporter-labeled SERS tags accumulated at the T and C lines. The LODs of the three bacterial pathogens––Y. pestis, F. tularensis, and B. anthracis––measured using this SERS-LFA strips were estimated as 43.4, 45.8, and 357 CFU/mL, respectively, which were approximately 2–4 orders of magnitude more sensitive than those of commercial colorimetric LFA kits. Moreover, this approach consumes small volumes of the sample (40 μL) and features a short assay time (15 min). Thus, this SERS-LFA platform has significant potential for the early detection of bacterial pathogens in the field.
Fig. 14.
Operating principle of the SERS-based LFA sensing platform. LFA strips for Y. pestis, F. tularensis, and B. anthracis were dipped into wells of a 96-well ELISA plate containing mixtures of SERS nanotags and different concentrations of bacteria in the buffer solution. The formed immunocomplexes migrated through the capillary action toward to the test line, where their Raman signals were measured and analyzed. Reprinted with permission from Ref. [113]. Copyright 2018 Elsevier Science.
4. Conclusion and future perspectives
Herein, we review recent advances in various LFA techniques for the diagnosis of infectious diseases. Particularly, we focused on the POC diagnosis of SARS-CoV-2 and SERS-based LFA techniques, which has a strong potential for overcoming the limit inherent in conventional colorimetric and fluorescence LFA techniques. As described above, LFAs provide a user-friendly and easily accessible device platform, and SERS detection methods serve as a highly sensitive sensing tool. Therefore, the combination of LFAs with SERS detection tools provides a conceptually new diagnostic modality for rapid and accurate diagnoses of infectious diseases. From this viewpoint, it is important to be knowledgeable regarding the current status of SERS-based LFA platforms. To accelerate the clinical translation of SERS-based LFAs to the area of in vitro diagnostics, however, the following issues need to be addressed.
First, more stable and reproducible SERS nanotags should be developed. Conventional LFA strips using AuNPs are known to be thermally unstable in high-temperature regions. Consequently, they are restricted for use in the diagnosis of tropical infectious diseases, such as dengue fever and Zika virus infection. High temperatures in tropical regions induce the desorption of Raman reporter molecules from the AuNP surface and impair their performance as SERS nanotags [99,114]. Various silica- or polymer-encapsulated SERS nanotags have been recently developed to resolve this problem [115,116]. Another important issue of SERS nanotags is their signal reproducibility. As the reproducibility of SERS active sites on the test line is difficult to achieve, the application of SERS-LFAs in quantitative analysis has been hindered. To date, various hybrid SERS nanotags, such as core-shell nanoparticles [[117], [118], [119], [120]], nanorods [[121], [122], [123]], and nanostars [[124], [125], [126], [127]], have been developed to resolve this problem. More reproducible and stable SERS nanotags should be continuously developed to make SERS-LFAs robust for in vitro diagnostic platforms.
Second, advanced data treatment technologies, including neural networks and machine learning algorithms, are required for efficient analyses of information-rich Raman spectral data [[128], [129], [130], [131], [132], [133], [134]]. These software algorithms will be helpful for the high-throughput analysis of clinical data as well as for rapid and efficient data analysis of multiple biomarkers of infectious diseases. Therefore, the above-described software algorithms should be embedded into a computer system connected with the SERS-LFA platforms to perform more reliable clinical analysis by avoiding the misinterpretation of complicated Raman data.
Third, the combination of SERS-based LFA strips with a portable Raman system is essential to realize on-site diagnostic platforms for infectious diseases [63,135,136]. Fortunately, it is convenient to miniaturize a Raman system, and various miniaturized Raman systems are commercially available. Nonetheless, there is still a need for a robust, user-friendly system to collect data with limited training. An easily accessible data library for clinical Raman spectral data is also required to extend the use of the system for multiple disease targets.
Finally, a closer collaboration between clinical doctors and laboratory researchers is critical for the fast translation from basic sciences to clinical applications. For the development of a new in vitro diagnostic modality in clinical diagnostics, the collection and analysis of patient samples are essential for approved medical research. Thus, the linkage between clinicians and laboratory researchers facilitates clinical research.
Considering the recent, ongoing COVID-19 pandemic, the development of a rapid and sensitive diagnostic tool is essential for preventing the spread of the disease. Presently, RT-PCR is considered the gold standard method for diagnosing SARS-CoV-2 but it is essential to reduce the detection time less than 30 min for its rapid POC diagnosis. Several research groups are trying to reduce the time required for the sample pre-treatment, such as RNA extraction from the virus and reverse transcription steps, using microfluidic devices [137,138]. Additionally, our research group recently reported that it is possible to significantly reduce the number of thermocycles in PCR process using the highly sensitive SERS-PCR technique [93]. Therefore, it will be possible to diagnose SARS-CoV-2 less than 1 h by combining SERS-PCR technique with a microfluidic platform. In the case of SERS-LFA platform, the key issue is a critical improvement of its accuracy using appropriate antibodies (antigen kit) or recombinant proteins (antibody kit) associated with SARS-CoV-2. We believe that the SERS-based assay system has a strong potential to serve as a new platform for the early diagnoses of various infectious viral and bacterial pathogens.
Funding
This work was supported by the National Research Foundation of Korea (grant numbers 2019R1A2C3004375 and 2020R1A5A1018052).
CRediT authorship contribution statement
Kihyun Kim: Conceptualization, Validation, Investigation. Leila Kashefi-Kheyrabadi: Methodology, Investigation, Writing - review & editing. Younju Joung: Conceptualization, Investigation, Visualization. Kyeongnyeon Kim: Formal analysis, Investigation, Visualization. Hajun Dang: Investigation, Visualization. Sachin Ganpat Chavan: Validation, Formal analysis. Min-Ho Lee: Conceptualization, Methodology, Writing - review & editing. Jaebum Choo: Funding acquisition, Conceptualization, Methodology, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Biographies
Kihyun Kim: Received his BS degree from Department of Bionano Engineering at Hanyang University, South Korea in 2017. He is currently studying for his PhD degree in Department of Chemistry at Chung-Ang University.
Leila Kashefi-Kheyrabadi: Obtained her Ph.D. in Analytical Chemistry from Isfahan University, Iran in 2013. She is currently working as a senior researcher in the School of Integrative Engineering at Chung-Ang University, South Korea. Her research interest is the development of highly sensitive and easy-to-use biosensors for diagnostics applications.
Younju Joung: Received her BS degree from Department of Bionano Engineering at Hanyang University, South Korea in 2019. She is currently studying for her PhD degree in Department of Chemistry at Chung-Ang University.
Kyeongnyeon Kim: Received his BS degree from Department of Molecular Life Science at Hanyang University, South Korea in 2019. He is currently studying for his MS degree in Department of Chemistry at Chung-Ang University.
Hajun Dang: Received his BS degree from Department of Bionano Engineering at Hanyang University, South Korea in 2017. He is currently studying for his PhD degree in Department of Chemistry at Chung-Ang University.
Sachin Ganpat Chavan: Received his MS degree in Microbiology from S.R.T.M. University, Nanded, India in 2013. He is currently studying for his PhD degree in the Integrative Engineering at Chung-Ang University in South Korea.
Min-Ho Lee: Obtained his PhD in Biophotonics from Texas Rice University in 2005. He is currently an Associate Professor in the Integrative Engineering at Chung-Ang University in South Korea. His works are focused on development of the highly sensitive biosensor platforms.
Jaebum Choo: Obtained his PhD in laser-induced spectroscopy from Texas A&M University in 1994. Now, he is a Distinguished Professor in Department of Chemistry at Chung-Ang University in South Korea. He served as a President of Korea Biochip Society in 2015. His current research programs are centered on the development of ultra-sensitive optical sensors for rapid and accurate in vitro diagnostics. He has given 120 invited lectures in the USA, Europe and Asia, and has published over 280 research papers in refereed journals and 8 book chapters.
References
- 1.Lia F., You M., Li S., Hue J., Liu C., Gong Y., Yang H., Xua F. Paper-based point-of-care immunoassays: recent advances and emerging trends. Biotechnol. Adv. 2020;39:107442. doi: 10.1016/j.biotechadv.2019.107442. [DOI] [PubMed] [Google Scholar]
- 2.Mahmoudi T., de la Guardia M., Shirdel B., Mokhtarzadeh A., Baradaran B. Recent advancement in structural improvements of lateral flow assays toward point-of-care testing. Trends Analyt. Chem. 2019;116:13–30. [Google Scholar]
- 3.Nguyen V.-T., Song S., Park S., Joo C. Recent advances in high-sensitivity detection methods for paper-based lateral-flow assay. Biosens. Bioelectron. 2020;15:112015. doi: 10.1016/j.bios.2020.112015. [DOI] [PubMed] [Google Scholar]
- 4.Mahmoudi T., de la Guardia M., Baradaran B. Lateral flow assays towards point-of-care cancer detection: a review of current progress and future trends. Trends Analyt. Chem. 2020;125:115842. [Google Scholar]
- 5.Huang X., Aguilar Z.P., Xu H., Lai W., Xiong Y. Membrane-based lateral flow immunochromatographic strip with nanoparticles as reporters for detection: a review. Biosens. Bioelectron. 2016;75:166–180. doi: 10.1016/j.bios.2015.08.032. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee R., Jaiswa A. Recent advances in nanoparticle-based lateral flow immunoassay as a point-of-care diagnostic tool for infectious agents and diseases. Analyst. 2018;143:1970–1996. doi: 10.1039/c8an00307f. [DOI] [PubMed] [Google Scholar]
- 7.Bahadir E.B., Sezgintürk M.K. Lateral flow assays: principles, designs and labels. Trends Analyt. Chem. 2016;82:286–306. [Google Scholar]
- 8.Shen M., Li N., Lu Y., Cheng J., Xu Y. An enhanced centrifugation-assisted lateral flow immunoassay for the point-of-care detection of protein biomarkers. Lab Chip. 2020;20:2626–2634. doi: 10.1039/d0lc00518e. [DOI] [PubMed] [Google Scholar]
- 9.Lee K.W., Kim K.R., Chun H.J., Jeong K.Y., Hong D.-K., Lee K.-N., Yoon H.C. Time-resolved fluorescence resonance energy transfer-based lateral flow immunoassay using a raspberry-type europium particle and a single membrane for the detection of cardiac troponin I. Biosens. Bioelectron. 2020;163:112284. doi: 10.1016/j.bios.2020.112284. [DOI] [PubMed] [Google Scholar]
- 10.Grant B.D., Anderson C.E., Williford J.R., Alonzo L.F., Glukhova V.A., Boyle D.S., Weigl B.H., Nichols K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020;92:11305–11309. doi: 10.1021/acs.analchem.0c01975. [DOI] [PubMed] [Google Scholar]
- 11.Brazaca L.C., Moreto J.R., Martin A., Tehrani F., Wang J., Zucolotto V. Colorimetric paper-based immunosensor for simultaneous determination of fetuin B and clusterin toward early alzheimer’s diagnosis. ACS Nano. 2019;13:13325–13332. doi: 10.1021/acsnano.9b06571. [DOI] [PubMed] [Google Scholar]
- 12.Li T., Jalbani Y.M., Zhang G., Zhao Z., Wang Z., Zhao Y., Zhao X., Chen A. Rapid authentication of mutton products by recombinase polymerase amplification coupled with lateral flow dipsticks. Sens. Actuators B. 2019;290:242–248. [Google Scholar]
- 13.Ren W., Mohammed S.I., Wereley S., Irudayaraj J. Magnetic focus lateral flow sensor for detection of cervical Cancer biomarkers. Anal. Chem. 2019;91:2876–2884. doi: 10.1021/acs.analchem.8b04848. [DOI] [PubMed] [Google Scholar]
- 14.Mukama O., Wu J., Li Z., Liang Q., Yi Z., Lu X., et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020;159:112143. doi: 10.1016/j.bios.2020.112143. [DOI] [PubMed] [Google Scholar]
- 15.He X., Liu Z., Yang Y., Li L., Wang L., Li A., Qu Z., Xu F. Sensitivity enhancement of nucleic acid lateral flow assays through a physical–chemical coupling method: dissoluble saline barriers. ACS Sens. 2019;4:1691–1700. doi: 10.1021/acssensors.9b00594. [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Xiong E., Tian T., Cheng M., Lin W., Wang H., Zhang G., Sun J., Zhou X. Clustered regularly interspaced short palindromic repeats/cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14:2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 17.Ivanov A.V., Safenkova I.V., Zherdev A.V., Dzantiev B.B. Nucleic acid lateral flow assay with recombinase polymerase amplification: solutions for highly sensitive detection of RNA virus. Talanta. 2020;210:120616. doi: 10.1016/j.talanta.2019.120616. [DOI] [PubMed] [Google Scholar]
- 18.Le T.T., Chang P., Benton D.J., McCauley J.W., Iqbal M., Cass A.E.G. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017;89:6781–6786. doi: 10.1021/acs.analchem.7b01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan J.C., Pettitt J., George J.S., Fakoli L.S., Taweh F.M., Bateman S.L., Bennett R.S., Norris S.L., Spinnler D.A. Lateral flow immunoassays for Ebola virus disease detection in Liberia. J. Infect. Dis. 2016;214:S222–S228. doi: 10.1093/infdis/jiw251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scharinger E.J., Dietrich R., Wittwer T., Märtlbauer E., Schauer K. Multiplexed lateral flow test for detection and differentiation of Cronobacter sakazakii serotypes O1 and O2. Front. Microbiol. 2017;8:1826. doi: 10.3389/fmicb.2017.01826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y., Wang H., Zhang P., Sun C., Wang X., Wang X., Yang R., Wang C., Zhou L. Rapid multiplex detection of 10 foodborne pathogens with an up-converting phosphor technology-based 10-channel lateral flow assay. Sci. Rep. 2016;6:21342. doi: 10.1038/srep21342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Xu C.-q., Guo T., Hong L. An automated bacterial concentration and recovery system for pre-enrichment required in rapid Escherichia coli detection. Sci. Rep. 2018;8:17808. doi: 10.1038/s41598-018-35970-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh J., Sharma S., Nara S. Evaluation of gold nanoparticle based lateral flow assays for diagnosis of enterobacteriaceae members in food and water. Food Chem. 2015;170:470–483. doi: 10.1016/j.foodchem.2014.08.092. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z.-K., Zhang Q.-Y., Yang N.-N., Xu M.-G., Xu J.-F., Jing M.-L., Wu W.-X., Lu Y.-D., Shi F., Chen C.-F. Rapid and sensitive detection of Salmonella in chickens using loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Microbiol. Biotechnol. 2019;29:454–464. doi: 10.4014/jmb.1712.12010. [DOI] [PubMed] [Google Scholar]
- 25.Hemmig E., Temiz Y., Gökçe O., Lovchik R.D., Delamarche E. Transposing lateral flow immunoassays to capillary-driven microfluidics using self-coalescence modules and capillary-assembled receptor carriers. Anal. Chem. 2020;92:940–946. doi: 10.1021/acs.analchem.9b03792. [DOI] [PubMed] [Google Scholar]
- 26.Hu J., Zhang Z.L., Wen C.Y., Tang M., Wu L.L., Liu C., Zhu L., Pang D.W. Sensitive and quantitative detection of C-reaction protein based on immunofluorescent nanospheres coupled with lateral flow test strip. Anal. Chem. 2016;88:6577–6584. doi: 10.1021/acs.analchem.6b01427. [DOI] [PubMed] [Google Scholar]
- 27.Xie Q.Y., Wu Y.H., Xiong Q.R., Xu H.Y., Xiong Y.H., Liu K., Jin Y., Lai W.H. Advantages of fluorescent microspheres compared with colloidal gold as a label in immunochromatographic lateral flow assays. Biosens. Bioelectron. 2014;54:262–265. doi: 10.1016/j.bios.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Berlina A.N., Taranova N.A., Zherdev A.V., Vengerov Y.Y., Dzantiev B.B. Quantum dot-based lateral flow immunoassay for detection of chloramphenicol in milk. Anal. Bioanal. Chem. 2013;405:4997–5000. doi: 10.1007/s00216-013-6876-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang Z., Li H., Li C., Yu Q., Shen J., de Saeger S. Development and application of a quantitative fluorescence-based immunochromatographic assay for fumonisin B1 in maize. J. Agr. Food Chem. 2014;62:6294–6298. doi: 10.1021/jf5017219. [DOI] [PubMed] [Google Scholar]
- 30.Wang S., Ge L., Song X., Yu J., Ge S., Huang J., Zeng F. Paper-based chemiluminescence ELISA: lab-on-paper based on chitosan modified paper device and wax-screen-printing. Biosens. Bioelectron. 2012;31:212–218. doi: 10.1016/j.bios.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 31.Zhao M., Li H., Liu W., Guo Y., Chu W. Plasma treatment of paper for protein immobilization on paper-based chemiluminescence immunodevice. Biosens. Bioelectron. 2016;79:581–588. doi: 10.1016/j.bios.2015.12.099. [DOI] [PubMed] [Google Scholar]
- 32.Liu W., Cassano C.L., Xu X., Fan Z.H. Laminated paper-based analytical devices (LPAD) with origami-enabled chemiluminescence immunoassay for cotinine detection in mouse serum. Anal. Chem. 2013;85:10270–10276. doi: 10.1021/ac402055n. [DOI] [PubMed] [Google Scholar]
- 33.Zangheri M., Cevenini L., Anfossi L., Baggiani C., Simoni P., Di Nardo F., Roda A. A simple and compact smartphone accessory for quantitative chemiluminescence-based lateral flow immunoassay for salivary cortisol detection. Biosens. Bioelectron. 2015;64:63–68. doi: 10.1016/j.bios.2014.08.048. [DOI] [PubMed] [Google Scholar]
- 34.Wang S., Ge L., Song X., Yan M., Ge S., Yu J., Zeng F. Simple and covalent fabrication of a paper device and its application in sensitive chemiluminescence immunoassay. Analyst. 2012;137:3821–3827. doi: 10.1039/c2an35266d. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y., Chu W., Liu W., Guo X., Jin Y., Li B. Paper-based chemiluminescence immunodevice for the carcinoembryonic antigen by employing multi-enzyme carbon nanosphere signal enhancement. Microchim. Acta. 2018;185:187. doi: 10.1007/s00604-018-2726-5. [DOI] [PubMed] [Google Scholar]
- 36.Ma C., Li W., Kong Q., Yang H., Bian Z., Song X., Yu J., Yan M. 3D origami electrochemical immunodevice for sensitive point-of-care testing based on dual signal amplification strategy. Biosens. Bioelectron. 2015;63:7–13. doi: 10.1016/j.bios.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz-Vega G., Kitsara M., Pellitero M.A., Baldrich E., del Campo F.J. Electrochemical lateral flow devices: towards rapid immunomagnetic assays. ChemElectroChem. 2017;4:880–889. [Google Scholar]
- 38.Fan Y., Shi S., Ma J., Guo Y. A paper-based electrochemical immunosensor with reduced graphene oxide/thionine/gold nanoparticles nanocomposites modification for the detection of cancer antigen 125. Biosens. Bioelectron. 2019;135:1–7. doi: 10.1016/j.bios.2019.03.063. [DOI] [PubMed] [Google Scholar]
- 39.Min J., Nothing M., Coble B., Zheng H., Park J., Im H., Weber G.F., Castro C.M., Swirski F.K., Weissleder R., Lee H. Integrated biosensor for rapid and pointof-care sepsis diagnosis. ACS Nano. 2018;4:3378–3384. doi: 10.1021/acsnano.7b08965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Channon R.B., Yang Y., Feibelman K.M., Geiss B.J., Dandy D.S., Henry C.S. Development of an electrochemical paper-Based analytical device for trace detection of virus particles. Anal. Chem. 2018;90:7777–7783. doi: 10.1021/acs.analchem.8b02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco-Covian L., Montes-Garcia V., Girard A., Fernandez-Abedul M.T., Perez-Juste J., Pastoriza-Santos I., Faulds K., Graham D., Blanco-Lopez M.C. Au@Ag SERRS tags coupled to a lateral flow immunoassay for the sensitive detection of pneumolysin. Nanoscale. 2017;9:2051–2058. doi: 10.1039/c6nr08432j. [DOI] [PubMed] [Google Scholar]
- 42.Fernanda Cardinal M., Rodríguez-González B., Alvarez-Puebla R.A., Pérez-Juste J., LizMarzán L.M. Modulation of localized surface plasmons and SERS response in gold dumbbells through silver coating. J. Phys. Chem. C. 2010;114:10417–10423. [Google Scholar]
- 43.Zhang D., Huang L., Liu B., Ni H., Sun L., Su E., Chen H., Gu Z., Zhao X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers in lateral flow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018;106:204–211. doi: 10.1016/j.bios.2018.01.062. [DOI] [PubMed] [Google Scholar]
- 44.Zhang D., Huang L., Liu B., Su E., Chen H.-Y., Gu Z., Zhao X. Quantitative detection of multiplex cardiac biomarkers with encoded SERS nanotags on a single T line in lateral flow assay. Sens. Actuators B. 2018;277:502–509. doi: 10.1016/j.bios.2018.01.062. [DOI] [PubMed] [Google Scholar]
- 45.Tian L., Morrissey J.J., Kattumenu R., Gandra N., Kharasch E.D., Singamaneni S. Bioplasmonic paper as a platform for detection of kidney cancer biomarkers. Anal. Chem. 2012;84:9928–9934. doi: 10.1021/ac302332g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taranova N.A., Berlina A.N., Zherdev A.V., Dzantiev B.B. ’Traffic light’ immunochromatographic test based on multicolor quantum dots for the simultaneous detection of several antibiotics in milk. Biosens. Bioelectron. 2015;63:255–261. doi: 10.1016/j.bios.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 47.Berlina A., Taranova N., Zherdev A., Vengerov Y., Dzantiev B. Quantum dot-based lateral flow immunoassay for detection of chloramphenicol in milk. Anal. Bioanal. Chem. 2013;405:4997–5000. doi: 10.1007/s00216-013-6876-3. [DOI] [PubMed] [Google Scholar]
- 48.Li X., Lu D., Sheng Z., Chen K., Guo X., Jin M., Han H. A fast and sensitive immunoassay of avian influenza virus based on label-free quantum dot probe and lateral flow test strip. Talanta. 2012;100:1–6. doi: 10.1016/j.talanta.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 49.Juntunen E., Myyryläinen T., Salminen T., Soukka T., Pettersson K. Performance of fluorescent europium(III) nanoparticles and colloidal gold reporters in lateral flow bioaffinity assay. Anal. Biochem. 2012;428:31–38. doi: 10.1016/j.ab.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Rundström G., Jonsson A., Mårtensson O., Mendel-Hartvig I., Venge P. Lateral flow immunoassay using europium (III) chelate microparticles and time-resolved fluorescence for eosinophils and Neutrophils in whole blood. Clin. Chem. 2007;53:342–348. doi: 10.1373/clinchem.2006.074021. [DOI] [PubMed] [Google Scholar]
- 51.Blažková M., Mičková-Holubová B., Rauch P., Fukal L. Immunochromatographic colloidal carbon-based assay for detection of methiocarb in surface water. Biosens. Bioelectron. 2009;25:753–758. doi: 10.1016/j.bios.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 52.Posthuma-Trumpie G., Wichers J., Koets M., Berendsen L.J.M. A. Van Amerongen, Carbon nanoparticles in lateral flow methods to detect genes encoding virulence factors of Shiga toxin-producing Escherichia coli. Anal. Bioanal. Chem. 2012;402:593–600. doi: 10.1007/s00216-010-4334-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bobosha K., Tjon Kon Fat E.M., van den Eeden S.J.F., Bekele Y., van der Ploeg-van Schip J.J., de Dood C.J., Dijkman K., Franken K.L.M.C., Wilson L., Aseffa A., Spencer J.S., Ottenhoff T.H.M., Corstjens P.L.A.M., Geluk A. Field-evaluation of a new lateral flow assay for detection of cellular and humoral immunity against Mycobacterium leprae. PLoS Neglected Trop. Dis. 2014;8:e2845. doi: 10.1371/journal.pntd.0002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan Z., Zhou L., Zhao Y., Wang J., Huang L., Hu K., Liu H., Wang H., Guo Z., Song Y., Huang H., Yang R. Rapid quantitative detection of Yersinia pestis by lateral-flow immunoassay and up-converting phosphor technology-based biosensor. Sens. Actuators B Chem. 2006;119:656–663. doi: 10.1016/j.snb.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talley C.E., Jackson J.B., Oubre C., Grady N.K., Hollars C.W., Lane S.M., Huser T.R., Nordlander P., Halas N.J. Surface-enhanced Raman scattering from individual Au nanoparticles and nanoparticle dimer substrates. Nano Lett. 2005;5:1569–1574. doi: 10.1021/nl050928v. [DOI] [PubMed] [Google Scholar]
- 56.Schwartzberg A.M., Oshiro T.Y., Zhang J.Z., Huser T., Talley C.E. Improving nanoprobes using surface-enhanced Raman scattering from 30-nm hollow gold particles. Anal. Chem. 2006;78:4732–4736. doi: 10.1021/ac060220g. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y., Jin Y., Xiao X., Zhang T., Yang H., Zhao Y., Wang J., Jiang K., Fan S., Li Q. Flexible, transparent and highly sensitive SERS substrates with cross-nanoporous structures for fast on-site detection. Nanoscale. 2018;10:15195–15204. doi: 10.1039/c8nr01628c. [DOI] [PubMed] [Google Scholar]
- 58.Chen H., Park S.-G., Choi N., Moon J.-I., Dang H., Das A., Lee S., Kim D.-G., Chen L., Choo J. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020;167:112496. doi: 10.1016/j.bios.2020.112496. [DOI] [PubMed] [Google Scholar]
- 59.Lee S., Chon H., Yoon S.-Y., Lee E.K., Chang S.-I., Lim D.W., Choo J. Fabrication of SERS-fluorescence dual modal nanoprobes and application to multiplex cancer cell imaging. Nanoscale. 2012;4:124–129. doi: 10.1039/c1nr11243k. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y., Zong S., Li N., Wang Z., Chen B., Cui Y. SERS-based dynamic monitoring of minimal residual disease markers with high sensitivity for clinical applications. Nanoscale. 2019;11:2460–2467. doi: 10.1039/c8nr06929h. [DOI] [PubMed] [Google Scholar]
- 61.Hamm L., Gee A., de Silva Indrasekara A.S. Recent advancement in the surface-enhanced Raman spectroscopy-based biosensors for infectious disease diagnosis. Appl. Sci. (Basel) 2019;9:1448. [Google Scholar]
- 62.Choi N., Lee J., Ko J., Jeon J.H., Rhie G.-e., deMello A.J., Choo J. Integrated SERS-based microdroplet platform for the automated immunoassay of F1 antigens in Yersinia pestis. Anal. Chem. 2017;89:8413–8420. doi: 10.1021/acs.analchem.7b01822. [DOI] [PubMed] [Google Scholar]
- 63.Quang L.X., Lim C., Seong G.H., Choo J., Do K.J., Yoo S.-K. A portable surface-enhanced Raman scattering sensor integrated with a lab-on-a-chip for field analysis. Lab Chip. 2008;8:2214–2219. doi: 10.1039/b808835g. [DOI] [PubMed] [Google Scholar]
- 64.Xu K., Zhou R., Takei K., Hong M. Toward flexible surface‐enhanced Raman scattering (SERS) sensors for point‐of‐care Diagnostics. Adv. Sci. 2019;6:1900925. doi: 10.1002/advs.201900925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murray C.J., Lopez A.D., Chin B., Feehan D., Hill K.H. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: a quantitative analysis. Lancet. 2006;368:2211–2218. doi: 10.1016/S0140-6736(06)69895-4. [DOI] [PubMed] [Google Scholar]
- 66.Oxford J.S. Influenza A pandemics of the 20th century with special reference to 1918: virology, pathology and epidemiology. Rev. Med. Virol. 2000;10:119–133. doi: 10.1002/(sici)1099-1654(200003/04)10:2<119::aid-rmv272>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 67.Taubenberger J.K., Morens D.M. 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 2006;12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pashchenko O., Shelby T., Banerjee T., Santra S. A comparison of optical, electrochemical, magnetic, and colorimetric point-of-care biosensors for infectious disease diagnosis. ACS Infect. Dis. 2018;4:1162–1178. doi: 10.1021/acsinfecdis.8b00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobayashi T., Jung S.-M., Linton N.M., Kinoshita R., Hayashi K., Miyama T., Anzai A., Yang Y., Yuan B., Akhmetzhanov A.R. Communicating the risk of death from novel coronavirus disease (COVID-19) J. Clin. Med. 2020;9:580. doi: 10.3390/jcm9020580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT-PCR: pitfalls and potential. BioTechniques. 1999;26:124–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 73.Kageyama T., Kojima S., Shinohara M., Uchida K., Fukushi S., Hoshino F.B., Takeda N., Katayama K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003;41:1548–1557. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma H., Zeng W., He H., Zhao D., Jiang D., Zhou P., Cheng L., Li Y., Ma X., Jin T., IgA Serum. IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 76.Hoffman T., Nissen K., Krambrich J., Rönnberg B., Akaberia D., Esmaeilzadeha M., Salaneck E., Lindahl J., Lundkvist Å. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect. Ecol. Epidemiol. 2020;10:1754538. doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chertow D.S. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 79.Li Y., Li S., Wang J., Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37:730–743. doi: 10.1016/j.tibtech.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 80.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aman R., Mahas A., Mahfouz M. Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth. Biol. 2020;9:1226–1233. doi: 10.1021/acssynbio.9b00507. [DOI] [PubMed] [Google Scholar]
- 82.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotech. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Espejo A.P., Akgun Y., Al Mana A.F., Tjendra Y., Millan N.C., Gomez-Fernandez C., Cray C. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020;154:293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, using lnthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 86.Black M.A., Shen G., Feng X., Garcia Beltran W., Feng Y., Vasudevaraja V., Allison D., Lin L.H., Gindin T., Astudillo M., Yang D., Murali M., Iafrate A.J., Jour G., Cotzia P., Snuderl M. Analytical performance of lateral flow immunoassay for SARS-CoV-2 exposure screening on venous and capillary blood samples. MedRxiv. 2020 doi: 10.1101/2020.05.13.20098426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., Zhang Y., Wang J., Huang B., Lin Y., Yang J., Cai W., Wang X., Cheng J., Chen Z., Sun K., Pan W., Zhan Z., Chen L., Ye F. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lou B., Li T., Zheng S., Su Y., Li Z., Liu W., Yu F., Ge S.-X., Zou Q.-D., Yuan Q., Lin S., Hong C.-M., Yao X.-Y., Zhang X.-J., Wu D.-H., Zhou G.-L., Hou W.-H., Li T.-T., Zhang Y.-L., Zhang S.-Y., Fan J., Zhang J., Xia N.-S., Chen Y. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. Eur. Respir. J. 2020 doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tadesse L.F., Safir F., Ho C.-S., Hasbach X., Khuri-Yakub B., Jeffrey S.S., Saleh A.A.E., Dionne J. Toward rapid infectious disease diagnosis with advances in surface-enhanced Raman spectroscopy. J. Chem. Phys. 2020;152:240902. doi: 10.1063/1.5142767. [DOI] [PubMed] [Google Scholar]
- 90.Zhang D., Huang L., Liu B., Ge Q., Dong J., Zhao X. Rapid and ultrasensitive quantification of multiplex respitory tract infection pathogen via lateral flow microarray based on SERS nanotags. Theranostics. 2019;9:4849–4859. doi: 10.7150/thno.35824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z., Leustean L., Inci F., Zheng M., Demirci U., Wang S. Plasmonic-based platforms for diagnosis of infectious diseases at the point-of-care. Biotechnol. Adv. 2019;37:107440. doi: 10.1016/j.biotechadv.2019.107440. [DOI] [PubMed] [Google Scholar]
- 92.Chisanga M., Muhamadali H., Ellis D.I., Goodacre R. Enhancing disease diagnosis: biomedical applications of surface-enhanced Raman scattering. Appl. Sci. 2019;9:1163. [Google Scholar]
- 93.Wu Y., Choi N., Chen H., Dang H., Chen L., Choo J. Performance evaluation of surface-enhanced Raman scattering-polymerase chain reaction sensirs for future use in sensitive genetic assays. Anal. Chem. 2020;92:2628–2634. doi: 10.1021/acs.analchem.9b04522. [DOI] [PubMed] [Google Scholar]
- 94.Hunter R., Sohi A.N., Ali N., Khatoon Z., Berthiaume V.R., Alarcon E.I., Godin M., Anis H. Optofluidic label-free SERS platform for rapid bacteria detection in serum. Sens. Actuators B. 2019;300:126907. [Google Scholar]
- 95.Kelly J., Patrick R., Patrick S., Bell S.E.J. Surface-enhanced Raman spectroscopy for the detection of a metabolic product in the headspace above live bacterial cultures. Angew. Chem. Int. Ed. 2018;57:15686–15690. doi: 10.1002/anie.201808185. [DOI] [PubMed] [Google Scholar]
- 96.Hwang J., Lee S., Choo J. Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B. nanoscale. 2016;8:11418–11425. doi: 10.1039/c5nr07243c. [DOI] [PubMed] [Google Scholar]
- 97.Sanchez-Purra M., Carre-Camps M., de Puig H., Bosch I., Gehrke L., Hamad-Schifferli K. Surface-enhanced Raman spectroscopy-based sandwich immunoassays for multiplexed detection of Zika and Dengue viral biomarkers. ACS Infect. Dis. 2017;3:767–776. doi: 10.1021/acsinfecdis.7b00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee S.H., Hwang J., Kim K., Jeon J., Lee S., Ko J., Lee J., Kang M., Chung D.R., Choo J. Quantitative serodiagnosis of scrub typhus using surface-enhanced Raman scattering-based lateral flow assay platforms. Anal. Chem. 2019;91:12275–12282. doi: 10.1021/acs.analchem.9b02363. [DOI] [PubMed] [Google Scholar]
- 99.Jeon J., Lee S.H., Joung Y., Kim K., Choi N., Choo J. Improvement of reproducibility and thermal stability of surface-enhanced Raman scattering-based lateral flow assay strips using silica-encapsulated gold nanoparticles. Sens. Actuators B. 2020;321:128521. [Google Scholar]
- 100.Liu X., Yang X., Li K., Liu H., Xiao R., Wang W., Wang C., Wang S. Fe3O4@Au SERS tags-based lateral flow assay for simultaneous detection of serum amyloid A and C-reactive protein in unprocessed blood sample. Sens. Actuators B. 2020;320:128350. [Google Scholar]
- 101.Keller T., Brem S., Tran V., Sritharan O., Schaefer D., Schluecker S. Rational design of thiolated polyenes as trifunctional Raman reporter molecules in surface-enhanced Raman scattering nanotags for cytokine detection in a lateral flow assay. J. Biophotonics. 2020;13:e201960126. doi: 10.1002/jbio.201960126. [DOI] [PubMed] [Google Scholar]
- 102.Fu X., Wen J., Li J., Lin H., Liu Y., Zhuang X., Tian C., Chen L. Highly sensitive detection of prostate cancer specific PCA3 mimic DNA using SERS-based competitive lateral flow assay. Nanoscale. 2019;11:15530–15536. doi: 10.1039/c9nr04864b. [DOI] [PubMed] [Google Scholar]
- 103.Wang Y., Sun Y., Hou Y., Zhang C., Li D., Li H., Yang M., Fan C., Sun B. A SERS-based lateral flow assay biosensor for quantitative and ultrasensitive detection of interleukin-6 in unprocessed whole blood. Biosens. Bioelectron. 2019;141:111432. doi: 10.1016/j.bios.2019.111432. [DOI] [PubMed] [Google Scholar]
- 104.Russo L., Sanchez-Purra M., Rodriguez-Quijada C., Leonardo B.M., Puntes V., Hamad-Schifferli K. Detection of resistance protein A (MxA) in paper-based immunoassays with surface enhanced Raman spectroscopy with AuAg nanoshells. Nanoscale. 2019;11:10819–10827. doi: 10.1039/c9nr02397f. [DOI] [PubMed] [Google Scholar]
- 105.Zhang D., Huang L., Liu B., Su E., Chen H.-Y., Gu Z., Zhao X. Quantitative detection of multiplex cardiac biomarkers with encoded SERS nanotags on a single T line in lateral flow assay. Sens. Actuators B. 2018;277:502–509. doi: 10.1016/j.bios.2018.01.062. [DOI] [PubMed] [Google Scholar]
- 106.Bai T., Wang M., Cao M., Zhang J., Zhang K., Zhou P., Liu Z., Liu Y., Guo Z., Lu X. Functionalized Au@Ag-Au nanoparticles as an optical and SERS dual probe for lateral flow sensing. Anal. Bioanal. Chem. 2018;410:2291–2303. doi: 10.1007/s00216-018-0850-z. [DOI] [PubMed] [Google Scholar]
- 107.Zhang D., Huang L., Liu B., Ni H., Sun L., Su E., Chen H., Gu Z., Zhao X. Quantitative and ultrasensitive detection of multiplex cardiac biomarkers inlateralflow assay with core-shell SERS nanotags. Biosens. Bioelectron. 2018;106:204–211. doi: 10.1016/j.bios.2018.01.062. [DOI] [PubMed] [Google Scholar]
- 108.Lin L.-K., Stanciu L.A. Bisphenol A detection using gold nanostars in a SERS improved lateral flow immunochromatographic assay. Sens. Actuators B. 2018;276:222–229. [Google Scholar]
- 109.Choi S., Hwang J., Lee S., Lim D.W., Joo H., Choo J. Quantitative analysis of thyroid-stimulating hormone (TSH) using SERS-based lateral flow immunoassay. Sens. Actuators B. 2017;240:358–364. [Google Scholar]
- 110.Fu X., Cheng Z., Yu J., Choo P., Chen L., Choo J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016;78:530–537. doi: 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]
- 111.Wang X., Choi N., Cheng Z., Ko J., Chen L., Choo J. Simultaneous detection of dual nucleic acids using a SERS-based lateral flow assay biosensor. Anal. Chem. 2017;89:1163–1169. doi: 10.1021/acs.analchem.6b03536. [DOI] [PubMed] [Google Scholar]
- 112.Shen H., Xie K., Huang L., Wang L., Ye J., Xiao M., Ma L., Jia A., Tang Y. A novel SERS-based lateral flow assay for differential diagnosis of wild-type pseudorabies virus and gE-deleted vaccine. Sens. Actuators B. 2019;282:152–157. [Google Scholar]
- 113.Wang R., Kim K., Choi N., Wang X., Lee J., Jeon J.H., Rhie G.-E., Choo J. Highly sensitive detection of high-risk bacterial pathogens using SERS-based lateral flow assay strips. Sens. Actuators B. 2018;270:72–79. [Google Scholar]
- 114.Borzenkov M., Chirico G., D’Alfonso L., Sironi L., Collini M., Cabrini E., Dacarro G., Milanese C., Pallavicini P., Taglietti A., Bernhard C., Denat F. Thermal and chemical stability of thiol bonding on gold nanostars. Langmuir. 2015;31:8081–8091. doi: 10.1021/acs.langmuir.5b01473. [DOI] [PubMed] [Google Scholar]
- 115.Huang J., Kim K.H., Choi N., Chon H., Lee S., Choo J. Preparation of silica-encapsulated hollow gold nanosphere tags using layer-by-layer method for multiplex surface-enhanced Raman scattering detection. Langmuir. 2011;27:10228–10233. doi: 10.1021/la201739n. [DOI] [PubMed] [Google Scholar]
- 116.Fernández-López C., Mateo-Mateo C., Álvarez-Puebla R.A., Pérez-Juste J., Pastoriza-Santos I., Liz-Marzán L.M. Highly controlled silica coating of PEG-capped metal nanoparticles and preparation of SERS-encoded particles. Langmuir. 2009;25:13894–13899. doi: 10.1021/la9016454. [DOI] [PubMed] [Google Scholar]
- 117.Choi N., Dang H., Das A., Sim M.S., Chung I.Y., Choo J. SERS biosensors for ultrasensitive detection of multiple biomarkers expressed in cancer cells. Biosens. Bioelectron. 2016;164:112326. doi: 10.1016/j.bios.2020.112326. [DOI] [PubMed] [Google Scholar]
- 118.Yu H., Xiao M., Lai W., Alam M.F., Zhang W., Pei H., Wan Y., Li L. A Self-calibrating surface-enhanced Raman scattering-active system for bacterial phenotype detection. Anal. Chem. 2020;92:4491–4497. doi: 10.1021/acs.analchem.9b05614. [DOI] [PubMed] [Google Scholar]
- 119.Li M., Wang J.-Y., Chen Q.-Q., Lin L.-H., Radjenovic P., Zhang H., Luo S.-Y., Tian Z.-Q., Li J.-F. Background-free quantitative surface enhanced Raman spectroscopy analysis using core-shell nanoparticles with an inherent internal standard. Anal. Chem. 2019;91:15025–15031. doi: 10.1021/acs.analchem.9b03703. [DOI] [PubMed] [Google Scholar]
- 120.Yang H., Li B.Q., Jiang X., Shao J. Hybrid nanostructure of SiO2@Si with Au-nanoparticles for surface enhanced Raman spectroscopy. Nanoscale. 2019;11:13484–13493. doi: 10.1039/c9nr03813b. [DOI] [PubMed] [Google Scholar]
- 121.Wei W., Wang Y., Ji J., Zuo S., Li W., Bai F., Fan H. Fabrication of large-area arrays of vertically aligned gold nanorods. Nano Lett. 2018;18:4467–4472. doi: 10.1021/acs.nanolett.8b01584. [DOI] [PubMed] [Google Scholar]
- 122.Tian L., Su M., Yu F., Xu Y., Li X., Li L., Liu H., Tan W. Liquid-state quantitative SERS analyzer on self-ordered metal liquid-like plasmonic arrays. Nat. Commun. 2018;9:1–12. doi: 10.1038/s41467-018-05920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Botta R., Eiamchai P., Horprathum M., Limwichean S., Chananonnawathorn C., Patthanasettakul V., Nuntawong N. Investigation of silver nanorods as reusable SERS-active substrates for trace level detection of 2-MIB volatile organic compound. Sens. Actuators B. 2018;271:122–127. [Google Scholar]
- 124.Tanwar S., Haldar K.K., Sen T. DNA origami directed Au nanostar dimers for single-molecule surface-enhanced Raman scattering. J. Am. Chem. Soc. 2017;139:17639–17648. doi: 10.1021/jacs.7b10410. [DOI] [PubMed] [Google Scholar]
- 125.Jimenez de Aberasturi D., Serrano-Montes A.B., Langer J., Henriksen-Lacey M., Parak W., Liz-Marzan L.M. Surface enhanced Raman scattering encoded gold nanostars for multiplexed cell discrimination. Chem. Mat. 2016;28:6779–6790. [Google Scholar]
- 126.Niu W., Chua Y.A.A., Zhang W., Huang H., Lu X. Highly symmetric gold nanostars: crystallographic control and surface-enhanced Raman scattering property. J. Am. Chem. Soc. 2015;137:10460–10463. doi: 10.1021/jacs.5b05321. [DOI] [PubMed] [Google Scholar]
- 127.Indrasekara A.S.D.S., Meyers S., Shubeita S., Feldman L.C., Gustafsson T., Fabris L. Gold nanostar substrates for SERS-based chemical sensing in the femtomolar regime. Nanoscale. 2014;6:8891–8899. doi: 10.1039/c4nr02513j. [DOI] [PubMed] [Google Scholar]
- 128.Lussier F., Thibault V., Charron B., Wallace G.Q., Masson J.-F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. Trends Analyt. Chem. 2020;124:115796. [Google Scholar]
- 129.Ho C.-S., Jean N., Hogan C.A., Blackmon L., Jeffrey S.S., Holodniy M., Banaei N., Saleh A.A.E., Ermon S., Dionne J. Rapid identification of pathogenic bacteria using Raman spectroscopy and deep learning. Nat. Commun. 2019;10:4927. doi: 10.1038/s41467-019-12898-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kloss S., Kampe B., Sachse S., Rösch P., Straube E., Pfister W., Kiehntopf M., Popp J. Culture independent Raman spectroscopic identification of urinary tract infection pathogens: a proof of principle study. Anal. Chem. 2013;85:9610. doi: 10.1021/ac401806f. [DOI] [PubMed] [Google Scholar]
- 131.Jiang J., Sell D., Hoyer S., Hickey J., Yang J., Fan J.A. Free-form diffractive metagrating design based on generative adversarial networks. ACS Nano. 2019;13:8872–8878. doi: 10.1021/acsnano.9b02371. [DOI] [PubMed] [Google Scholar]
- 132.He H., Xu M., Zong C., Zheng P., Luo L., Wang L., Ren B. Speeding pp the line-scan Raman imaging of living cells by deep convolutional neural network. Anal. Chem. 2019;91:7070–7077. doi: 10.1021/acs.analchem.8b05962. [DOI] [PubMed] [Google Scholar]
- 133.Shi H., Wang H., Meng X., Chen R., Zhang Y., Su Y., He Y. Setting up a surface-enhanced Raman scattering database for artificial intelligence-based label-free discrimination of tumor suppressor genes. Anal. Chem. 2018;90:14216–14221. doi: 10.1021/acs.analchem.8b03080. [DOI] [PubMed] [Google Scholar]
- 134.Weng S., Xu X., Li J., Wong S.T.C. Combining deep learning and coherent anti-Stokes Raman scattering imaging for automated differential diagnosis of lung cancer. J. Biomed. Opt. 2017;22:1–10. doi: 10.1117/1.JBO.22.10.106017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Huang Z., Siddhanta S., Zheng G., Kickler T., Barman I., Ishan Rapid. Label-free optical spectroscopy platform for diagnosis of heparin-induced thrombocytopenia. Angew. Chem. Int. Ed. 2020;59:5972–5978. doi: 10.1002/anie.201913970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kim S., Kim T.G., Lee S.H., Kim W., Bang A., Moon S.W., Song J., Shin J.-H., Yu J.S., Choi S. Label-free surface-enhanced Raman spectroscopy biosensor for on-site breast cancer detection using human tears. ACS Appl. Mater. Interfaces. 2020;12:7897–7904. doi: 10.1021/acsami.9b19421. [DOI] [PubMed] [Google Scholar]
- 137.Bhalla N., Payam A.F., Pan Y., Yang Z. Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Microfluidic immunoassays for sensitive and simultaneous detection of IgG/IgM/antigen of SARS-CoV-2 within 15 min. Anal. Chem. 2020;92:9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]