Abstract
RBP16 is a guide RNA (gRNA)-binding protein that was shown through immunoprecipitation experiments to interact with ∼30% of total gRNAs in Trypanosoma brucei mitochondria. To gain insight into the biochemical function of RBP16, we used affinity chromatography and immunoprecipitation to identify RBP16 protein binding partners. By these methods, RBP16 does not appear to stably interact with the core editing machinery. However, fractionation of mitochondrial extracts on MBP–RBP16 affinity columns consistently isolated proteins of 12, 16, 18 and 22 kDa that were absent from MBP control columns. We describe here our analysis of one RBP16-associated protein, p22. The predicted p22 protein has significant sequence similarity to a family of multimeric, acidic proteins that includes human p32 and Saccharomyces cerevisiae mam33p. Glutaraldehyde crosslinking of recombinant p22 identified homo-multimeric forms of the protein, further substantiating its homology to p32. We confirmed the p22–RBP16 interaction and demonstrated that the two proteins bind each other directly by ELISA utilizing recombinant p22 and RBP16. p32 family members have been reported to modulate viral and cellular pre-mRNA splicing, in some cases by perturbing interaction of their binding partners with RNA. To determine whether p22 similarly affects the gRNA binding properties of RBP16, we titrated recombinant p22 into UV crosslinking assays. These experiments revealed that p22 significantly stimulates the gRNA binding capacity of RBP16. Thus, p22 has the potential to be a regulatory factor in T.brucei mitochondrial gene expression by modulating the RNA binding properties of RBP16.
INTRODUCTION
The expression of most mitochondrial genes in Trypanosoma brucei requires a remarkable process termed RNA editing (reviewed in 1,2). Editing involves the remodeling of pre-mRNAs through the site-specific insertion and deletion of uridine residues to create mature mRNAs. This process is catalyzed by a series of enzymes that are associated in a ribonucleoprotein complex (3,4). Within the editing complex, the genetic information for uridine insertion and deletion is transferred from small trans-acting RNAs called guide RNAs (gRNAs) to mRNAs through base pairing interactions (5). Thus, the efficiency of editing relies on the ordered assembly of pre-mRNA, gRNA and protein molecules involved in catalysis and/or regulation of the process. By analogy to other RNA processing events, it is also presumed that accessory factors that are not components of the core editing complex regulate the specificity, accuracy and/or efficiency of the process. One mechanism by which proteins are likely to regulate RNA editing is through associations with gRNA, as gRNA is an essential element in the reaction. For example, accessory factors may promote association of gRNAs with the editing machinery by positioning, unwinding, annealing or stabilizing gRNAs or gRNA–mRNA interactions. It is also probable that gRNA-binding proteins regulate gRNA usage or stability. Furthermore, these processes are likely to be modulated in response to physiological conditions and developmental stage (6,7).
We have previously described a gRNA-binding protein from T.brucei mitochondria that we called RBP16 (for RNA-binding protein of 16 kDa) (8). RBP16 was isolated based upon its affinity for poly(U) and its ability to be UV crosslinked to synthetic gRNA in vitro. UV crosslinking competition assays demonstrated that RBP16 can bind to different gRNA molecules. RBP16 gRNA binding is mediated largely through the oligo(U) tail, a non-encoded element at the gRNA 3′-end (9), although contacts between RBP16 and the encoded portion of the gRNA function to stabilize the interaction (10). We demonstrated the native interaction between RBP16 and non-encoded (U) extensions on gRNA and rRNA 3′-ends by immunoprecipitation (8) and in organello labeling experiments (11). The RBP16 protein contains a cold shock domain (CSD) at its N-terminus, placing RBP16 as a member of the eukaryotic Y-box protein family (12). The C-terminus of RBP16 is rich in arginine and glycine, reminiscent of the RGG RNA-binding motif (13). Taken together, the presence of the CSD and the RNA binding properties of RBP16 suggest a role for the protein in gene expression, and likely in RNA editing. However, the precise function(s) of RBP16 in T.brucei mitochondria and the significance of RBP16 gRNA binding have not been determined. Thus, the identification of RBP16-associated polypeptides and enzyme activities may direct future studies regarding the role of RBP16 in trypanosome biology. In the studies presented here, RBP16 affinity chromatography and immunoprecipitation experiments were used to investigate a potential physical interaction between RBP16 and editing-related proteins. In addition, novel RBP16-associated proteins were isolated from mitochondrial lysates via interaction with a maltose-binding protein (MBP)–RBP16 fusion protein covalently bound to an agarose matrix. We describe here the isolation and characterization of a 22 kDa RBP16-associated protein (p22). p22 displays significant homology to a family of proteins, including human p32, which was first identified as a component of the ASF/SF2 pre-mRNA splicing factor (14). We show that p22 significantly stimulates the RBP16–gRNA interaction. Thus, p22 has the potential to be a regulatory factor in T.brucei mitochondrial gene expression through modulation of the RNA binding properties of RBP16.
MATERIALS AND METHODS
Cell culture, mitochondrial vesicle isolation and mitochondrial extract preparation
Procyclic form T.brucei brucei clone IsTaR1 stock EATRO 164 was grown as described (15). Bloodstream form T.brucei strain 427 (provided by George Cross) was cultured in HMI-9 medium according to a published protocol (16). Mitochondrial vesicles from procyclic form parasites were isolated and stored according to the method of Harris et al. (17).
Cleared mitochondrial extracts used for affinity chromatography were prepared as follows. Isolated mitochondria (0.5–1.0 × 1011 cell equivalents) were centrifuged for 15 min at 4°C and resuspended in 900 µl/1010 cell equivalents of sonication buffer (25 mM HEPES pH 8.0, 10 mM MgOAc, 50 mM KCl, 1 mM CHAPS, 0.5 mM DTT). Mitochondrial vesicles were sonicated three times for 30 s, supplemented with 10% glycerol and centrifuged at 100 000 g for 1 h at 4°C. The resulting supernatant was used for affinity chromatography. The protein concentration of supernatants, as determined using the Bio-Rad protein assay with BSA as the standard, was typically in the range 2.0–2.5 mg/ml.
Nucleic acid preparation
Constructs encoding the gRNAs gA6[14] and gCYb[558], with 17 and 15 nt oligo(U) tails, respectively, were previously described (18). RNAs were synthesized in vitro using an Ambion T7 Maxiscript kit and purified by gel electrophoresis on 6% acrylamide–7 M urea.
Total cellular RNA was isolated from procyclic and bloodstream form cells using the method of Chomszyski and Sacchi (19). Genomic DNA was isolated from procyclic form cells as described (20).
Southern blot, northern blot and semi-quantitative PCR
For Southern analysis T.brucei genomic DNA (7 µg) was digested with either EcoRI, BamHI, SalI or KpnI. Digested DNA was electrophoresed on a 0.7% agarose gel and transferred to nylon membrane. A radioactive 552 nt DNA probe was generated by PCR amplification of the 5′-portion of the p22 cDNA sequence using primers E-SL22 (5′-GCGAATTCGCTATTATTAGAACAGTTTCTG-3′) and p22-a (5′-CGGGATCCCGCATTGTTGATCACAAACTCGCC-3′). Hybridization conditions were as described (21).
For northern analysis, 10 µg total procyclic form RNA was electrophoresed on a 1.5% formaldehyde–agarose gel and transferred to nylon membrane. The same DNA probe as used for Southern analysis was hybridized to p22 RNA using previously described conditions (21).
For semi-quantitative PCR (22), cDNA was synthesized from 2.6 µg total procyclic or bloodstream form RNA using primer RXS-dT (5′-GAGAATTCTCGAGGTCGACTTTTTTTTTTTTTTTTT-3′). Each cDNA sample was then serially diluted 1:10, 1:100 and 1:1000. For PCR amplification of a portion of the p22 cDNA, 1 µl of undiluted or diluted cDNA was used as template with primers p22-Nco (5′-CATGCCATGGTATCGGACCAACGACTTTC-3′) and p22-F (5′-CGGAATTCCTTACGAAACAAATTTGTTAATGCTGCTC-3′). As a positive control for a developmentally regulated RNA, ATPase subunit 9 cDNA (23) was amplified using primers ATP9rev (5′-ATAGGCCGATAGCTTCCGTG-3′) and E-SL22 (see above). Amplified products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining. For figure preparation, photographs of ethidium bromide stained gels were analyzed on a BioRad GS-700 Imaging Densitometer using Molecular Analyst v.1.5 software (Bio-Rad). The contrast on scanned images was inverted.
Protein expression and purification
A MBP–RBP16 fusion was produced as previously described (8). A MBP control protein was also expressed from the pMal-C2 plasmid (New England Biolabs) and purified by amylose affinity chromatography. His-tagged RBP16 was produced as follows. The mature full-length RBP16 open reading frame (ORF) was PCR amplified from a pBluescript plasmid containing the entire ORF using primers Tb16K5′exp3 (5′-GCGAATTCCATATGAACAAGGGTAAGGTGATATCG-3′) and RBP16-3′exphis1 (5′-CCGCTCGAGAAAGTCATCGCTGAAGCTCTG-3′). The reaction products were digested and ligated into the NdeI/XhoI site of pET-21a (Novagen) and transformed into Escherichia coli strain BL21(DE3)pLysS: S (Novagen). Expression in bacteria generated a fusion protein with a C-terminal 6×His tag sequence. Cells were grown at 37°C to an optical density of 0.6 at 600 nm and induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were collected by centrifugation for 10 min at 5000 g 2.5 h after addition of IPTG. Cells were weighed and resuspended in 3 ml of lysis buffer (10 mM Tris pH 6.8, 300 mM NaCl, 10 mM imidazole, 10% glycerol, 0.1% Na deoxycholate, 0.01% NP40, 10 mM MgCl2, 1 mM PMSF, 10 µM leupeptin, 0.06 mg/ml lysozyme and 0.06 mg/ml DNase) per gram cells and sonicated. The supernatant fraction was collected after centrifugation at 12 000 g for 20 min at 4°C. A final concentration of 0.05% polyethylenimine was added and the supernatant was rocked for 20 min at 4°C. The suspension was then centrifuged at 12 000 g for 10 min at 4°C. A one-twentieth volume of ProBond resin (Invitrogen), pre-equilibrated with wash buffer (10 mM Tris pH 6.8, 300 mM NaCl, 30 mM imidazole), was added and the mixture was rocked for 1 h at 4°C. The resin was poured into a column and washed with at least 20 bed vol wash buffer. The recombinant His-tagged protein was eluted from the column in buffer containing 250 mM imidazole, 10 mM Tris pH 6.8, and 300 mM NaCl.
For expression of p22 as a MBP–p22 fusion, the mature p22 ORF was PCR amplified from genomic DNA using p22-Nde (5′-GGAATTCCATATGGTATCGGACCAACGACTTTCTGAAGCC-3′) and p22-F (see above). The resulting product was digested with NdeI and EcoRI and ligated into pMalC2. Expression and purification of MBP–p22 was done using a standard protocol (24). The amylose affinity purified fusion protein was treated with 2% Factor Xa (New England Biolabs) for 2 h at room temperature in amylose column buffer (24) containing 1 mM CaCl2 to remove the MBP moiety. The released p22 was further purified by anion exchange chromatography as follows. Factor Xa-cleaved MBP–p22 was dialyzed overnight into buffer A (20 mM Tris pH 8.0, 25 mM KCl) and loaded onto a Q-Sepharose (Amersham Pharmacia) column equilibrated in buffer A. The column was washed with 10 bed vol buffer A containing 100 mM KCl and bound proteins were eluted with a step gradient of 300–500 mM KCl in buffer A.
The purity and integrity of expressed proteins was examined by Coomassie staining of 12.5% SDS–PAGE gels. Protein concentrations were determined using the Bio-Rad protein assay with BSA standards.
RBP16 affinity chromatography
Purified MBP–RBP16 and MBP control proteins were dialyzed against 30 mM HEPES pH 8.0. For each experiment, 4–6 mg protein was coupled to 0.5 ml of Affigel-10 matrix (Bio-Rad) by the following protocol. Affigel-10 (0.5 ml) was washed five times with 10 ml of cold distilled water in each of two 15 ml conical tubes. MBP or MBP–RBP16 ligands were added to the gel matrix in 30 mM HEPES pH 8.0, at a concentration of 1 mg/ml. Coupling was performed overnight at 4°C with gentle rocking. Supernatants were removed and unbound protein was measured using the Bio-Rad protein assay. Coupling efficiencies were generally between 80 and 90%. Unreacted groups on the Affigel-10 matrix were blocked by incubation in 100 mM ethanolamine pH 8.0, for 1 h at 4°C. Coupled matrices were washed twice with 30 mM HEPES pH 8.0, once with elution buffer (25 mM HEPES pH 8.0, 10 mM MgOAc, 10% glycerol, 1 mM CHAPS, 0.5 mM DTT) containing 1 M KCl and twice with elution buffer containing 200 mM KCl. All washes were performed with 10 ml of buffer. Cleared mitochondrial extracts were incubated with MBP-coupled Affigel-10 matrix for 1 h at 4°C with rocking. The mixture was poured into a 1 ml column and the flow-through collected. Flow-through from the MBP column was then incubated with the MBP–RBP16 affinity column. Both MBP and MBP–RBP16 columns were washed with 10 ml of elution buffer containing 200 mM KCl, and bound proteins were sequentially eluted with 1 ml each of elution buffers containing 500 mM KCl and 1% SDS. In some experiments, bound proteins were released with elution buffer containing 300 mM KCl prior to the 500 mM KCl elution. After each elution, columns were washed with an additional 10 ml of buffer. All solutions were stored at 4°C prior to use and all steps were performed at 4°C. Protein profiles were analyzed by silver staining of SDS–PAGE gels. In some cases, proteins were concentrated using Centricon-10 (Amicon) prior to SDS–PAGE analysis.
Antibodies and immunoprecipitation experiments
Antibodies directed against MBP–RBP16 and MBP–p22 were produced and affinity purified at Bethyl Laboratories (Montgomery, TX). Pre-immune IgG was purified from pre-immune serum using protein A–Sepharose. Affinity purified anti-RBP16 antibodies and pre-immune IgG were crosslinked with dimethylpimelimidate to protein A–Sepharose and immunoprecipitations performed using a standard protocol (25). Immunoprecipitates were electrophoresed on 12.5% SDS–PAGE gels, transferred to Nytran (Schleicher & Schuell) and analyzed by western blotting with antibodies directed against gBP21 (provided by Uli Göringer) (26), TBRGG1 (provided by Ettiene Pays) (27) and REAP-1 (provided by Steve Hajduk) (28). Total mitochondrial extracts were analyzed as a positive control.
In a separate set of experiments, immunoprecipitates were analyzed for the presence of gRNA-binding proteins by UV crosslinking and for the presence of terminal uridylyl transferase (TUTase) activity as described below. These experiments were performed using anti-MBP–RBP16 antibodies prepared by the SUNY at Buffalo Monoclonal Antibody Center (8).
Enzyme activity and UV crosslinking assays
Adenylation, TUTase and RNase assays were performed essentially as described (3). UV crosslinking of RBP16 affinity column fractions or immunoprecipitates to radiolabeled gCYb[558] was done as described (18).
In experiments designed to determine the effect of p22 on RBP16–gRNA interactions, RBP16 was UV crosslinked to in vitro transcribed, [32P]UTP-labeled gA6[14] gRNA as described (18), using 1 µM His-RBP16. Reactions were performed either in the absence of additional proteins or in the presence of increasing amounts of p22 or the similarly charged protein, glucose oxidase (GOd; Sigma) (29). Assuming that p22 is trimeric (see below), 0.3-, 1-, 2-, 3- and 4-fold molar excesses of p22 or GOd were used. Crosslinked proteins were resolved on 15% tricine gels, dried and autoradiographed. Non-saturated autoradiographs were analyzed by densitometry using Multianalyst v.1.1 (Bio-Rad).
Cloning the p22 gene
The p22 protein was eluted from an MBP–RBP16 affinity column with 300 and 500 mM KCl. The fractions were combined, concentrated using a Centricon-10 microconcentrator and separated on a 12.5% SDS–PAGE gel. Proteins were transferred to a Pro-Blot membrane (Applied Biosystems) and the N-terminal sequence obtained from ProSeq Inc. (Boxford, MA). For the internal peptide sequence, p22 was excised directly from a 12.5% SDS–PAGE gel. Internal peptide sequences were obtained through the Harvard Microchemistry Facility by microcapillary reverse phase HPLC nano-electrospray tandem mass spectrometry on a Finnigan LCQ quadrupole ion trap mass spectrometer. Several overlapping p22 peptides corresponded to the predicted translation of a sheared T.brucei (strain GUTat 10.1) genomic DNA fragment present in a library constructed at The Institute for Genomic Research (TIGR) (accession no. AQ649494). The genomic DNA fragment contained approximately the 3′-half of the p22 gene, including the putative stop codon. The 5′-portion of p22 was amplified from oligo(dT)-primed procyclic cDNA in the following manner. Oligonucleotides p22-b (5′-CGGGATCCCGGCTAGTTTTCCGTCATGCCGG-3′) and p22-a (see above) were prepared corresponding to the p22 genomic DNA fragment and used in nested PCR in conjunction with an oligonucleotide corresponding to the splice leader sequence (E-SL22, see above). The product amplified with p22-b and E-SL22 was reamplified with E-SL22 and p22-a. The resulting PCR products were inserted into the EcoRI/BamHI site of pBluescriptII SK– (Stratagene) and used to transform DH5α competent cells (Life Technologies). Two ampr colonies were selected, the plasmids isolated and nucleotide sequences obtained for both directions by automated DNA sequencing. Both 5′-p22 clones contained identical nucleotide sequences. The 5′-p22 nucleotide sequence was subsequently used for construction of PCR primers p22-Nde and p22-F (see above). The PCR product amplified with p22-Nde and p22-F from genomic DNA represented the exact p22 ORF. The PCR product was digested with NdeI and BamHI, filled in with T4 polymerase (Life Technologies) and phosphorylated with T4 kinase (Life Technologies). The PCR product was ligated into the EcoRV site of pBluescriptII SK– subsequent to dephosphorylation of the digested plasmid with calf intestinal alkaline phosphatase (Life Technologies). Five resulting clones were sequenced in both directions to ascertain the complete p22 coding sequence. p22 homlogs were identified using the Basic Local Alignment Search Tool (BLAST) to compare the predicted p22 amino acid sequence to protein sequences in several protein databases. Global protein sequence alignments were generated using Clustal W (30).
Glutaraldehyde crosslinking assay
Recombinant p22 and His-tagged RBP16 used in glutaraldehyde crosslinking assays were expressed and purified as described above. Recombinant human replication protein A (RPA) (31) was a generous gift from Jen-Sing Liu and Tom Melendy. Proteins of interest were incubated for 20 min at room temperature. Glutaraldehyde was then added to a final concentration of 0.2% and reactions were incubated for an additional 10 min at room temperature. SDS–PAGE sample buffer was added to stop the reactions. Samples were analyzed by SDS–PAGE on a 12.5% polyacrylamide gel and proteins were visualized by silver staining.
ELISAs
ELISAs were performed in triplicate and carried out in 96-well vinyl plates at room temperature. To prepare the immobilized substrate, wells were coated for 2 h with purified protein (His-RBP16 or BSA; Sigma) in 50 µl of TBS (50 mM Tris pH 8.0, 100 mM NaCl). The wells were then washed three times with TBS-T (TBS with 0.05% v/v Triton X-100) and blocked with 5% (w/v) dry milk and 2% (v/v) fetal bovine serum (FBS) in TBS-T for 60 min. After washing three times with TBS-T, increasing amounts of p22 were added to the wells in 50 µl of TBS-T supplemented with 1 mM MgCl2, 1 mM CaCl2 and 40 U/ml micrococcal nuclease and incubated for 30 min. After washing three times with TBS-T, the plates were incubated with affinity purified rabbit polyclonal antibody against p22 diluted 1:4000 in TBS-T with 0.5% (w/v) dry milk and 1% (v/v) FBS for 60 min. The plates were then washed five times with TBS-T and incubated with horseradish peroxidase-conjugated goat anti-rabbit antibody (1:10 000 in TBS-T) with 0.5% (w/v) dry milk and 1% (v/v) FBS for 60 min. After washing four times with TBS-T and three times with TBS, the plate wells were incubated with 50 µl of visualization buffer (110 mM sodium acetate pH 5.5) containing the chromogenic substrate 3,3′,5,5′-tetramethylbenzidine (0.1 mg/ml) and hydrogen peroxide (0.0075% v/v). After 10 min, the reaction was stopped by addition of 50 µl of 2 M sulfuric acid. The assays were quantified spectrophotometrically by measuring absorbance at 450 nm. A negative control well containing only 300 ng BSA was quantified and subtracted from each value.
RESULTS
RBP16 affinity chromatography
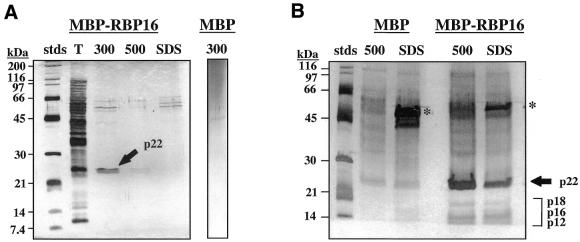
One approach to discerning RBP16 function is the identification of its protein binding partners. To isolate polypeptides that associate with RBP16, MBP–RBP16 was covalently bound to agarose beads using the Affigel-10 matrix. Mitochondrial extracts were initially applied to a MBP column both to visualize non-specific protein binding and to pre-clear mitochondrial extracts of non-specifically binding proteins. The flow-through from the MBP column was subsequently loaded onto a MBP–RBP16 column. Interacting proteins were eluted from both the MBP and MBP–RBP16 columns with increasing concentrations of KCl and, finally, with 1% SDS. The proteins were separated by SDS–PAGE and protein profiles were analyzed by silver staining. Representative experiments are shown in Figure 1. A protein with an apparent molecular weight of 22 kDa, that we termed p22, was consistently present in the 300 and 500 mM KCl fractions of the MBP–RBP16 column (Fig. 1A). This protein was absent from MBP control column fractions. The interaction between RBP16 and p22 was not dependent upon RNA, as micrococcal nuclease treatment of mitochondrial extracts did not abrogate p22 binding (not shown). Upon concentration of affinity column eluates, substantial non-specific protein binding to the MBP–RBP16 column was observed in the higher molecular mass ranges, as determined by comparison to eluates from the MBP control column (Fig. 1B). This precluded identification of specific RBP16-interacting proteins in this region of the gel. However, three additional, less abundant proteins of ∼12, ∼16 and ∼18 kDa (bracket in Fig. 1B) that specifically bound to the RBP16 column were detected.
Figure 1.
Isolation of RBP16-associated proteins by affinity chromatography. (A) S100 extract of T.brucei mitochondria was first incubated with MBP–Affigel-10 matrix. The flow-through from this column was then incubated with MBP–RBP16 bound to Affigel-10 beads. After washing, bound proteins were eluted from both columns with 300 and 500 mM KCl and, finally, 1% SDS. Eluted fractions were separated by SDS–PAGE and visualized by silver staining. Stds, molecular mass markers; T, total mitochondrial S100 extract; 300, 300 mM KCl eluate; 500, 500 mM KCl eluate; SDS, 1% SDS eluate. The major RBP16-interacting protein, p22, is indicated by an arrow. The less abundant bands between 50 and 60 kDa represent MBP–RBP16 that leached out of the column. (B) Affinity chromatography was performed as described in (A), except that the 300 mM KCl elution was omitted and samples were concentrated using a Centricon-10 prior to electrophoresis and silver staining. Asterisks indicate MBP and MBP–RBP16. p22 is indicated by an arrow. The less abundant RBP16-interacting proteins p12, p16 and p18 are indicated with a bracket.
Analysis of affinity column eluates for editing-associated activities and proteins
To assess whether RBP16 forms a stable association with the editing machinery, we analyzed MBP–RBP16 affinity column eluates for the presence of enzymes that co-purify with RNA editing activity (3,4). TUTase, protein adenylation (as an indicator of RNA ligase) and RNase activity were present only in the flow-through fractions, demonstrating that these factors do not physically associate with RBP16 by this method and that p22 does not possess any of these activities. The lack of an interaction between RBP16 and TUTase was verified by immunoprecipitation experiments (data not shown). Furthermore, the titration of anti-RBP16 antibodies into unfractionated mitochondrial extracts had no effect on TUTase activity (data not shown). RBP16 immunoprecipitates and affinity column fractions were also analyzed by western blotting for the presence of the gRNA-binding proteins gBP21 (26) and TBRGG1 (27), as well as the RNA editing-associated protein REAP-1 (28), using the respective antibodies donated generously by other laboratories. None of these proteins were detected in either affinity column eluates or RBP16 immunoprecipitates. In addition, no gRNA-binding proteins were detected in MBP–RBP16 affinity column fractions or in RBP16 immunoprecipitates by UV crosslinking to radiolabeled in vitro transcribed gRNA. Taken together, these data do not support a stable physical interaction between RBP16 and the RNA editing machinery.
Cloning and analysis of the p22 gene
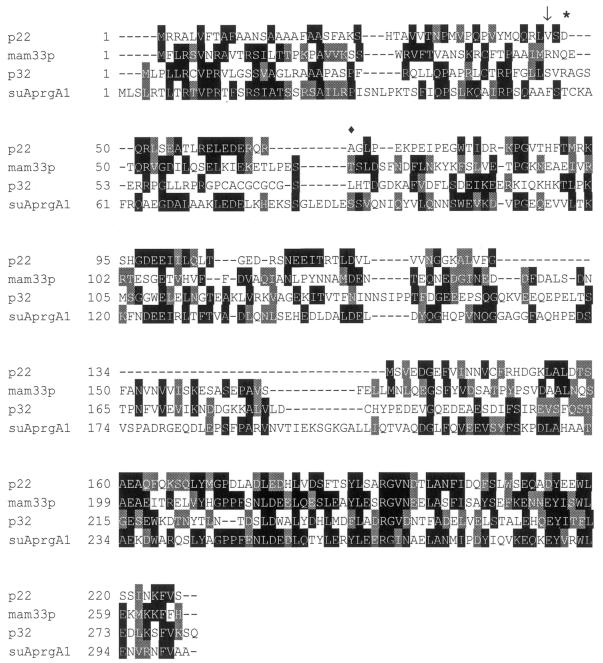
The major RBP16-interacting protein, p22, was purified by MBP–RBP16 affinity chromatography and N-terminal and internal peptide sequences were obtained. The N-terminal sequence begins with a valine (Fig. 2), suggesting that p22 is transported into the mitochondria via a cleaved signal peptide. The internal peptide sequence was also obtained and several overlapping internal peptides matched the predicted translation of a T.brucei genomic fragment from a library constructed at TIGR (AQ649494). The genomic fragment contained approximately the 3′-half of the p22 ORF, including the predicted stop codon. The 5′-segment was amplified from total oligo(dT)-primed procyclic form cDNA using oligonucleotide primers constructed from the genomic sequence in conjunction with a primer corresponding to the 5′ spliced leader sequence. Subsequently, the entire p22 ORF was amplified from genomic DNA using primers corresponding to the extreme ends of the ORF. The p22 ORF contains three methionine codons upstream of the N-terminal valine of the mature protein. Translation initiation at the first methionine residue would result in a cleaved mitochondrial import sequence of 46 amino acids. The N-terminal sequence of the predicted p22 pre-protein exhibits significant homology to the mitochondrial import sequence of several other kinetoplastid proteins (32) and the p22 protein is predicted by the MitoProt II algorithm (33) to be mitochondrially localized. The p22 cDNA possesses a 66 nt 5′-untranslated region (UTR), excluding the spliced leader sequence. The 3′-UTR sequence was not determined. The p22 ORF contains 681 nt and encodes a 227 amino acid pre-protein. The mature p22 protein beginning with Val47 contains 181 amino acids and has a calculated molecular weight of 20.5 kDa and a pI of 4.5. The p22 nucleotide and amino acid sequences have been deposited in the GenBank database under accession no. AF376060.
Figure 2.
p22 is homologous to the p32 family of proteins. Clustal W was used to align the pre-protein sequences of T.brucei p22 (accession no. AF376060), S.cerevisiae mam33p (accession no. P40513), human p32 (accession no. Q07021) and A.nidulans suAprgA1 (accession no. CAB62571). Black boxes indicate residues identical to those in p22; gray boxes indicate conserved residues. N-terminal amino acids of the mature proteins are indicated by an arrow (p22 V47), an asterisk (mam33p Q48) and a diamond (p32 L74). The mature N-terminus of suAprgA1 has not been determined.
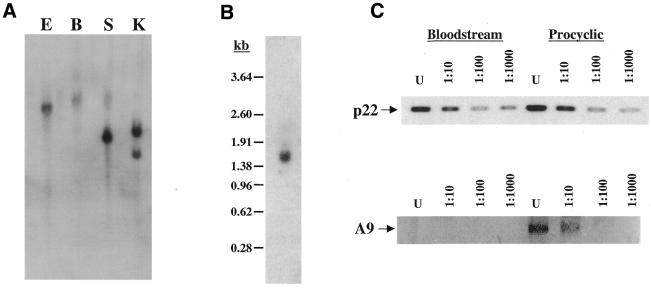
To determine p22 gene copy number, T.brucei genomic DNA was digested with several restriction enzymes and Southern blot analysis was performed (Fig. 3A). A single hybridizing band was detected in samples digested with either EcoRI, BamHI or SalI, which do not cut within the p22 sequence. Two bands were detected when DNA was digested with KpnI, for which one site exists within the p22 ORF. These results indicate that p22 is present in the genome as a single copy. Northern blot analysis of T.brucei procyclic form RNA revealed one transcript of ∼1500 nt (Fig. 3B). Since the cDNA sequence containing the spliced leader, 5′-UTR and ORF spans 789 nt, this indicates that the p22 transcript possesses a 3′-UTR of ∼700 nt, which is similar in length to 3′-UTRs of other reported T.brucei RNAs. To determine whether p22 RNA levels are developmentally regulated during the trypanosome life cycle, we performed semi-quantitative RT–PCR using RNA from both procyclic and bloodstream form parasites (22). As shown in Figure 3C, RNA levels appear similar in the two life cycle stages, suggesting that p22 is constitutively expressed. A control PCR reaction amplifying a portion of the ATPase subunit 9 cDNA (Fig. 3C) demonstrates that the conditions were sufficient to detect developmental regulation of RNA levels.
Figure 3.
Analysis of p22 DNA and RNA. (A) Trypanosoma brucei genomic DNA was digested with EcoRI (E), BamHI (B), SalI (S) or KpnI (K), electrophoresed on a 0.7% agarose gel and transferred to nylon membrane. The blot was probed with a 32P-labeled PCR product spanning 552 bp of the p22 gene. KpnI cuts once within the p22 sequence; no sites for the other enzymes are present within the p22 gene. The faint upper band in the SalI lane is presumably the result of incomplete digestion by this enzyme. (B) Total procyclic form RNA was analyzed by northern blotting using the probe described in (A). Size markers are indicated in kb on the left. (C) p22 RNA levels were compared in bloodstream and procyclic form cells by semi-quantitative PCR using serial dilutions of oligo(dT)-primed cDNA. Semi-quantitative PCR of the ATPase subunit 9 cDNA was included as a positive control to demonstrate that conditions were sufficient for detection of developmental regulation of RNA levels. U, undiluted cDNA; 1:10, 1:100 and 1:1000 indicate the respective cDNA dilutions.
p22 sequence analysis and identification of p22 homologs
A BLAST search of several databases with the predicted p22 amino acid sequence identified a family of proteins with similarity to p22. The first described member of this family was human p32 protein, which was identified by its co-purification with the SR-type splicing factor ASF/SF2 (14). Since the identification of human p32, homologs of p32 have been identified in various organisms, including turkey (34), chicken (35), mouse (36), yeast (37) and the filamentous fungus Aspergillus nidulans (38). Human p32 protein has been reported to regulate in vitro pre-mRNA splicing through modulation of both the RNA binding activity and phosphorylation status of ASF/SF2 (29). There is also evidence for involvement of this protein in transcription, nuclear import and facilitation of protein–protein interactions (see Discussion). Interestingly, while p32 is clearly present and functional in both the nucleus and cytosol (29,35,39,40), the bulk of the protein appears to be mitochondrially localized (37,39,41). The role of p32 homologs in the mitochondrion has not been well established, however. A global alignment of the T.brucei p22 pre-protein and three p32 homologs from human (14), Saccharomyces cerevisiae (37) and A.nidulans (38) is shown in Figure 2. Overall, T.brucei p22 is 23% identical and 41% similar to human p32 and 24% identical and 37% similar to S.cerevisiae mam33p. All p32 homologs, including T.brucei p22, share the greatest homology at their C-termini. Indeed, amino acids 88–181 of the mature T.brucei p22 protein display 35% identity and 61% similarity to the C-terminus of yeast mam33p. The various homologs of p32 all have pI values in the range 4–4.5, suggesting that they have similar biochemical properties.
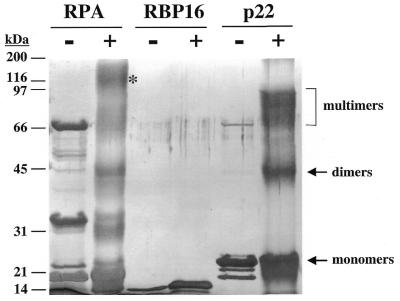
A crystal structure has recently been solved for human p32 revealing seven adjacent antiparallel β-strands followed by two C-terminal α-helices (42). The region of highest primary sequence conservation approximately corresponds to the two C-terminal α-helices. In addition, the crystal structure reveals a homo-trimeric structure for p32, and biochemical studies have observed multimeric forms of the protein (43). Gel filtration of yeast mam33p indicated that this protein forms homo-oligomeric complexes comprised of three or four subunits (37). To further verify that T.brucei p22 is a homolog of human p32, we analyzed the subunit structure of p22. An MBP–p22 fusion protein was expressed in E.coli, purified by amylose affinity chromatography and cleaved with Factor Xa. The released p22 protein was purified by anion exchange chromatography on Q-Sepharose and dialyzed into low salt buffer. Glutaraldehyde was added to a portion of the p22 sample to a final concentration of 0.2% to facilitate protein–protein crosslinking. The hetero-trimer human RPA (31) was included as a positive control and RBP16 was included as a negative control. Untreated and crosslinked proteins were analyzed by SDS–PAGE as shown in Figure 4. While the untreated p22 sample revealed only monomeric p22, the glutaraldehyde crosslinked sample contained p22 molecules of molecular masses corresponding to monomers, dimers and trimers. The largest crosslinked band was somewhat indistinct, suggesting that even higher order multimers may be present. The propensity of the T.brucei p22 protein to form homo-oligomers provides further evidence that this protein is a homolog of human p32 protein.
Figure 4.
p22 forms homo-oligomers. Bacterially expressed p22 was incubated either in the absence (–) or presence (+) of 0.2% glutaraldehyde prior to SDS–PAGE and silver staining. Human RPA is a hetero-trimeric protein comprised of 14, 32 and 70 kDa subunits, and was included as a positive control. RBP16 was included as a negative control. Molecular masses are indicated on the left in kilodaltons. The asterisk denotes hetero-trimeric RPA. Arrows and the bracket indicate p22 monomers, dimers and multimers. The faint band at 66 kDa in the p22 lane minus glutaraldehyde is uncleaved MBP–p22.
Direct interaction of p22 and RBP16
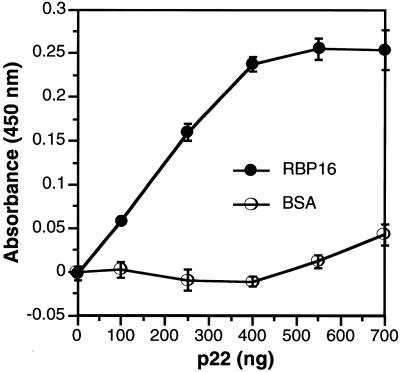
Since p22 was isolated by RBP16 affinity chromatography from mitochondrial extract, the possibility remained that the interaction between RBP16 and p22 was indirect. While we ruled out RNA bridging by micrococcal nuclease treatment of the mitochondrial extract (see above), we could not exclude the possibility that the RBP16–p22 interaction was mediated by a third protein. To determine whether p22 and RBP16 interact directly, we examined binding of recombinant p22 and recombinant RBP16 by ELISA (44; Fig. 5). His-RBP16 or BSA was used to coat the wells of an ELISA plate and the immobilized proteins were challenged with increasing amounts of recombinant p22. The wells were extensively washed and p22 binding was detected with affinity purified anti-p22 antibody followed by horseradish peroxidase-conjugated goat anti-rabbit antibody. After incubation with the chromogenic substrate 3,3′,5,5′-tetramethylbenzidine and hydrogen peroxide, p22 binding was quantified spectrophotometrically by measuring absorbance at 450 nm. As shown in Figure 5, binding of p22 to the negative control (BSA) remained at essentially background levels, whereas binding to RBP16 increased as a function of p22 concentration. These results confirm the RBP16–p22 interaction that was detected by affinity chromatography and demonstrate that the p22 and RBP16 proteins interact directly.
Figure 5.
p22 interacts directly with RBP16. Three hundred nanograms of either His-RBP16 or BSA was immobilized in triplicate in a 96-well ELISA plate and challenged with increasing amounts of recombinant p22 for 30 min as described in Materials and Methods. Interactions were detected with affinity purified rabbit polyclonal antibody against p22 followed by horseradish peroxidase-conjugated anti-rabbit antibody. The wells were incubated with a chromogenic substrate and absorbance was measured at 450 nm. A negative control well containing only 300 ng BSA was quantified and subtracted from each value.
Stimulation of RBP16 gRNA binding activity by p22
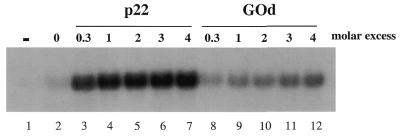
To assess the functional significance of the RBP16–p22 interaction, we analyzed whether p22 can modulate the gRNA binding properties of RBP16. Increasing amounts of bacterially expressed p22 were titrated into a UV crosslinking assay containing gA6[14] and His-RBP16 (Fig. 6). p22 significantly stimulated RBP16–gRNA binding under all conditions tested. The similarly charged protein GOd had only a slight effect on the RBP16–gRNA interaction. At a 0.3-fold molar concentration of p22 compared to RBP16, p22 stimulated gRNA binding 7.5-fold after correction for the non-specific increase observed with GOd (Fig. 6, compare lanes 3 and 8). Addition of p22 at between 1- and 4-fold molar excess over RBP16 stimulated gRNA binding by 9- to 10-fold over the control (Fig. 6, compare lanes 4–7 to 9–12). These molar values were calculated based on the assumption that p22 is primarily trimeric. However, even assuming monomeric p22, addition of p22 leads to a 7- to 8-fold increase in RBP16–gRNA crosslinking after correction for the GOd control (Fig. 6, compare lane 3 to 9 and lane 4 to 11). Thus, in contrast to what has been observed for the human p22 homolog p32, which abrogates the RNA interactions of its binding partner (29), we find that p22 significantly increases the RNA binding capacity of its protein binding partner, RBP16.
Figure 6.
p22 stimulates RBP16–gRNA binding. One micromolar His-RBP16 was incubated with in vitro transcribed gA6[14] gRNA either in the absence of p22 (lane 2) or presence of increasing molar excesses of p22 (lanes 3–7). Glucose oxidase (GOd), which possesses a similar charge to p22, was tested as a control (lanes 8–12). The p22 molar excesses were calculated based on a p22 trimer. Assuming a p22 monomer, the molar excesses of p22 in lanes 3–7 would be 1-, 3-, 6-, 9- and 12-fold. –, gRNA in the absence of any protein.
DISCUSSION
The identification of protein–protein interactions can provide valuable information relating to the biological function(s) of the respective proteins. We report here experiments aimed at identifying protein–protein interactions involving the mitochondrial gRNA-binding protein RBP16. Affinity chromatography of mitochondrial extracts on an MBP–RBP16 matrix identified four RBP16-interacting proteins of 12, 16, 18 and 22 kDa. We isolated the major RBP16 protein binding partner, which we term p22, and showed that p22 can dramatically stimulate the gRNA binding properties of RBP16.
Sequence analysis indicates that p22 is a homolog of the human p32 (14) and yeast mam33p (37) proteins. p32 family proteins are acidic, homo-oligomeric proteins, and these properties are shared by T.brucei p22. Mature p22 contains >20% glutamic and aspartic acid residues and has an acidic pI of 4.5. The acidic nature of p22 suggests that it may interact with the basic RGG domain of RBP16 (8). Similarly, human p32 binds arginine and glycine-rich motifs of Epstein–Barr virus (EBV) nuclear antigen-1 (45), and this interaction is specific in that p32 does not bind other arginine-rich proteins (46). Experiments to delineate the regions of RBP16 that interact with p22 are underway in our laboratory. We isolated the trypanosome p22 protein from mitochondria and N-terminal sequencing indicates the presence of a cleaved mitochondrial import sequence. While it is clear that p32 and mam33p are primarily mitochondrially localized, their mitochondrial function is unknown. One study (41) indicated a defect in oxidative phosphorylation in yeast lacking mam33p, however, another study could not reproduce this finding (37). Several studies have also reported the presence of p32 in extramitochondrial locations (35,40,43,47). Thus, it is plausible that the intracellular localization of p32 family proteins, including p22, may vary depending on the physiological state of the cell and external cellular stimuli. Indeed, the location of p32 was shown to shift from the mitochondria to the nucleus upon adenovirus infection of HeLa cells (47). It will be of interest to determine whether T.brucei p22 is present in subcellular locations other than in mitochondria.
Human p32 is the best-studied member of the protein family to which p22 belongs, and multiple functions have been ascribed to p32. Many of these functions involve interaction with viral proteins. For example, p32 cooperates with the human immunodeficiency virus-1 (HIV-1) Tat protein and EBV EBNA-1 to activate transcription of reporter constructs containing the appropriate viral sequences (46,48,49). A role for p32 in EBV DNA replication has also been described (46). Most intriguing from the standpoint of RBP16–p22 interaction are the reported functions of p32 in the regulation of RNA processing. p32 was originally identified by co-purification with the SR family splicing factor ASF/SF2 (14) and recent studies have indicated a role for p32 in pre-mRNA splicing regulation (29,36,50). For example, both p32 and its murine homolog (YL2) have been shown to modulate the splicing inhibitory effect of the HIV-1 Rev protein on HIV-1 pre-mRNA (36,50). Moreover, in both in vitro and in vivo studies, p32 inhibited the splicing enhancer and splicing repressor functions of ASF/SF2 by preventing stable association of ASF/SF2 with cellular pre-mRNA. p32 also inhibits the phosphorylation of ASF/SF2 which, in turn, is expected to perturb spliceosome formation (29). Functions not involving association with nucleic acids have also been attributed to p32. For example, p32 has been implicated as a compartment-specific regulator of signal transduction, since it directly inhibits substrate phosphorylation by the mitochondrially localized fraction of protein kinase C µ (51). Finally, the avian p32 homolog co-localizes with myosin and other cytoskeletal elements and promotes the assembly of myosin filaments (35).
The multifunctional nature of p32 suggests several possible implications of the p22–RBP16 interaction in trypanosome RNA editing. The multimeric structure of p32 and its ability to bind to a diverse group of molecules suggest that p22 may function as a protein assembly factor. More specifically, p22 could promote association of RBP16 with kRNA editing factors and/or other RNA processing proteins by bringing them into proximity via trimer formation. Similarly, p22 could promote RNA–RNA interactions during editing by bringing together multiple RNA-bound RBP16 molecules or by facilitating interaction of gRNA-bound RBP16 with pre-mRNA-binding proteins. p32 has been shown to regulate the phosphorylation status of cellular proteins through both substrate and kinase binding (29,51). While there is no evidence that RBP16 is phosphorylated, we recently demonstrated that RBP16 is mono-, di- and trimethylated on several arginine residues (52). MALDI-TOF analysis of native RBP16 indicates that some of these methylation events are mutually exclusive, suggesting a regulatory role for RBP16 methylation. By analogy to the role of p32 in regulating protein phosphorylation, T.brucei p22 could potentially modulate the methylation status of RBP16.
Finally, the ability of p32 to regulate the RNA binding properties of ASF/SF2 (29) suggested to us that p22 could similarly regulate the RNA binding affinity and/or specificity of RBP16. Indeed, we have demonstrated in these studies that the in vitro association of RBP16 with gRNA is dramatically stimulated by p22. It will be important for functional studies on RBP16 to determine what fraction of RBP16 is associated with p22 in vivo and whether the majority of gRNAs associate with this fraction. It will also be of interest in the future to determine whether p22 modulates the RNA binding specificity of RBP16. The N-terminal cold shock and C-terminal RGG domains of RBP16 preferentially bind poly(U) and poly(G), respectively (M.Miller and L.Read, unpublished results). Thus, by specifically binding one of the two RBP16 domains, p22 could abrogate the RNA binding capacity of that domain and thereby change the overall RNA binding specificity of RBP16. Trypanosoma brucei cells disrupted in their expression of p22 are being generated in our laboratory. This will allow us to test the in vivo effect of p22 on RBP16–RNA interactions and, ultimately, on mitochondrial gene expression.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs Uli Göringer, Steve Hajduk and Ettiene Pays for providing antibodies and to Dr George Cross for providing culture-adapted bloodstream form T.brucei cells. We thank Drs Jen-Sing Liu and Tom Melendy for providing recombinant human RPA and for advice on glutaraldehyde crosslinking. We also thank Dr Kevin Militello for critical reading of the manuscript. This work was supported by NIH grants GM53502 and AI47329 to L.K.R.
DDBJ/EMBL/GenBank accession no. AF376060
REFERENCES
- 1.Estevez A,M. and Simpson,L. (1999) Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene, 240, 247–260. [DOI] [PubMed] [Google Scholar]
- 2.Stuart K., Allen,T.E., Heidmann,S. and Seiwert,S.D. (1997) RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev., 61, 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rusché L.N., Cruz-Reyes,J., Piller,K.J. and Sollner-Webb,B. (1997) Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J., 16, 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panigrahi A.K., Gygi,S.P., Ernst,N.L., Igo,R.P., Palazzo,S.S., Schnaufer,A., Weston,D.S., Carmean,N., Salavati,R., Aebersold,R. and Stuart,K.D. (2001) Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol., 21, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seiwert S.D. and Stuart,K. (1994) RNA editing: transfer of genetic information from gRNA to precursor mRNA in vitro. Science, 266, 114–117. [DOI] [PubMed] [Google Scholar]
- 6.Koslowsky D.J., Riley,G.R., Feagin,J.E. and Stuart,K. (1992) Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol. Cell. Biol., 12, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley G.R., Myler,P.J. and Stuart,K. (1995) Quantitation of RNA editing substrates, products and potential intermediates: implications for developmental regulation. Nucleic Acids Res., 23, 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayman M.L. and Read,L.K. (1999) Trypanosoma brucei RBP6 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem., 274, 12067–12074. [DOI] [PubMed] [Google Scholar]
- 9.Blum B. and Simpson,L. (1990) Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell, 62, 391–397. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier M., Miller,M.M. and Read,L.K. (2000) RNA-binding properties of the mitochondrial Y-box protein, RBP16. Nucleic Acids Res., 28, 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Militello K.T., Hayman,M.L. and Read,L.K. (2000) Transcriptional and post-transcriptional in organello labelling of Trypanosoma brucei mitochondrial RNA. Int.J. Parasitol., 30, 643–647. [DOI] [PubMed] [Google Scholar]
- 12.Graumann P.L. and Marahiel.M.A. (1998) A superfamily of proteins that contain the cold shock domain. Trends Biochem. Sci., 23, 286–290. [DOI] [PubMed] [Google Scholar]
- 13.Burd C.G. and Dreyfuss,G. (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science, 265, 615–621. [DOI] [PubMed] [Google Scholar]
- 14.Krainer A.R., Mayeda,A., Kozak,D. and Binns,G. (1991) Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K and Drosophila splicing regulators. Cell, 66, 383–394. [DOI] [PubMed] [Google Scholar]
- 15.Brun R. and Schönenberger,V. (1979) Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop., 36, 289–292. [PubMed] [Google Scholar]
- 16.Hirumi H. and Hirumi,K. (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol., 75, 985–989. [PubMed] [Google Scholar]
- 17.Harris M.E., Moore,D.R. and Hajduk,S.L. (1990) Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem., 265, 11368–11376. [PubMed] [Google Scholar]
- 18.Read .L.K., Göringer,H.U. and Stuart,K. (1994) Assembly of mitochondrial ribonucleoprotein complexes involves specific guide RNA (gRNA)-binding proteins but does not require preedited mRNA. Mol. Cell. Biol., 14, 2629–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 20.Carrington M. (1993) Preparation of DNA and RNA from Trypanosoma brucei. In Hyde,J.E. (ed.), Protocols in Molecular Parasitology. Humana Press, Totowa, NJ, pp. 89–100. [DOI] [PubMed]
- 21.Read L.K., Myler,P.J. and Stuart,K. (1992) Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J. Biol. Chem., 267, 1123–1128. [PubMed] [Google Scholar]
- 22.Zhang J., Ruyechan,W. and Williams,N. (1998) Developmental regulation of two nuclear RNA binding proteins, p34 and p37, from Trypanosoma brucei.Mol. Biochem. Parasitol., 92, 79–88. [DOI] [PubMed] [Google Scholar]
- 23.Chi T.B., Brown,S.V. and Williams,N. (1998) Subunit 9 of the mitochondrial ATP synthase of Trypanosoma brucei is nuclearly encoded and developmentally regulated. Mol. Biochem. Parasitol., 92, 29–38. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.R., Seidman,J.G., Smith,J.A. and Struhl,K. (1998) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 25.Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Köller J., Müller,U.F., Schmid,B., Missel,A., Kruft,V., Stuart,K. and Göringer,H.U. (1997) Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem., 272, 3749–3757. [DOI] [PubMed] [Google Scholar]
- 27.Vanhamme L., Perez-Morga,D., Marchal,C., Speijer,D., Lambert,L. Geuskens,M., Alexandre,S., Ismaili,N., Göringer,H.U., Benne,R. and Pays,E. (1998) Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem., 273, 21825–21833. [DOI] [PubMed] [Google Scholar]
- 28.Madison-Antenucci S., Sabatini,R.S., Pollard,V.W. and Hajduk,S.L. (1998) Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J., 17, 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen-Mahrt S.K., Estmer,C., Ohrmalm,C., Matthews,D.A., Russell,W.C. and Akusjarvi,G. (1999) The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J., 18, 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henricksen L.A. and Wold,M.S. (1994) Replication protein A mutants lacking phosphorylation sites for p34cdc2 kinase support DNA replication. J. Biol. Chem., 269, 24203–24208. [PubMed] [Google Scholar]
- 32.Häusler T., Stierhof,Y.-D., Blattner,J. and Clayton,C. (1997) Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur. J. Cell Biol., 73, 240–251. [PubMed] [Google Scholar]
- 33.Claros M.G. and Vincens,P. (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem., 241, 779–786. [DOI] [PubMed] [Google Scholar]
- 34.Simos G. and Georgatos,S.D. (1994) The lamin B receptor-associated protein p34 shares sequence homology and antigenic determinants with the splicing factor 2-associated protein, p32. FEBS Lett., 346, 225–228. [DOI] [PubMed] [Google Scholar]
- 35.Okagaki T., Nakamura,A., Suzuki,T., Ohmi,K. and Kohama,K. (2000) Assembly of smooth muscle myosin by the 38k protein, a homologue of a subunit of pre-mRNA splicing factor-2. J. Cell Biol., 148, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo Y., Yu,H. and Peterlin,B.M. (1994) Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J. Virol., 68, 3850–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seytter T., Lottspeich,F., Neupert,W. and Schwartz,E. (1998) Mam33p, an oligomeric, acidic protein in the mitochondrial matrix of Saccharomyces cerevisiae is related to the human complement receptor gC1q-R. Yeast, 14, 303–310. [DOI] [PubMed] [Google Scholar]
- 38. Van den Brulle J., Steidl,S. and Brakhage,A.A. (1999) Cloning and characterization of an Aspergillus nidulans gene involved in the regulation of penicillin biosynthesis. Appl. Environ. Microbiol., 65, 5222–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltys B.J., Kang,D. and Gupta,R.S. (2000) Localization of P32 protein (gC1q-R) in mitochondria and at specific extramitochondrial locations in normal tissues. Histochem. Cell Biol., 114, 245–255. [DOI] [PubMed] [Google Scholar]
- 40.Brokstad K.A., Kalland,K.-H., Russell,W.C. and Matthews,D.A. (2001) Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun., 281, 1161–1169. [DOI] [PubMed] [Google Scholar]
- 41.Muta T., Kang,D., Kitajima,S., Fujiwara,T. and Naotaka,H. (1997) p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem., 272, 24363–24370. [DOI] [PubMed] [Google Scholar]
- 42.Jiang J., Zhang,Y., Drainer,A.R. and Xu,R.M. (1999) Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl Acad. Sci. USA, 96, 3572–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghebrehiwet B., Lim,B.L., Peerschke,E.I., Willis,A.C. and Reid,K.B. (1994) Isolation, cDNA cloning and overexpression of a 33-kD cell surface glycoprotein that binds to the globular “heads” of C1q. J. Exp. Med., 179, 1809–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Y., Loo,Y.-M., Militello,K.T. and Melendy,T.E. (1999) Interactions of the papovavirus DNA replication proteins, bovine papilloma virus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol., 73, 4899–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Finan,J.E., Middeldorp,J.M. and Hayward,S.D. (1997) P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology, 236, 18–29. [DOI] [PubMed] [Google Scholar]
- 46. Van Scoy S., Watakabe,I., Krainer,A.R. and Hearing,J. (2000) Human p32: a coactivator for Epstein-Barr virus nuclear antingen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology, 275, 145–157. [DOI] [PubMed] [Google Scholar]
- 47.Matthews D.A. and Russell,W.C. (1998) Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and nucleus. J. Gen. Virol., 79, 1677–1685. [DOI] [PubMed] [Google Scholar]
- 48.Yu L., Shang,Z., Loewenstein,P.M., Desai,K., Tang,Q., Mao,D., Symington,J.S. and Green,M. (1995) Molecular cloning and characterization of a cellular protein that interacts with the human immunodeficiency virus type 1 tat transactivator and encodes a strong transcriptional activation domain. J. Virol., 69, 3007–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu L., Loewenstein,P.M., Zhang,Z. and Green,M. (1995) In vitro interaction of the human immunodeficiency virus type 1 tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J. Virol., 69, 3017–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tange T.O., Jensen,T.H. and Kjems,J. (1996) In vitro interaction between human immunodeficiency virus type 1 rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem., 271, 10066–10072. [DOI] [PubMed] [Google Scholar]
- 51.Storz P., Hausser,A., Link,G., Dedio,J., Ghebrehiwet,B., Pfizenmaier,K. and Johannes,F.-J. (2000) Protein kinase C µ is regulated by the multifunctional chaperone protein p32. J. Biol. Chem., 275, 24601–24607. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier M., Xu,Y., Wang,X., Zahariev,S., Pongor,S., Aletta,J.M. and Read,L.K. (2001) Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei.Mol. Biochem. Parasitol., 118, 49–59. [DOI] [PubMed] [Google Scholar]