Abstract
Detection of nucleic acids is crucial in many medical applications, and in particular for monitoring infectious diseases, as it has become perfectly clear after the pandemic infection of COVID-19. In this context, the development of innovative detection methods based on signal-amplification rather than analyte-amplification represents a significant breakthrough compared to existing PCR-based methodologies, allowing the development of new nucleic acid detection technologies suitable to be integrated in portable and low-cost sensor devices while keeping high sensitivities, thus enabling massive diagnostic screening. In this work, we present a novel molecular sensor for the ultrasensitive PCR-free detection of Hepatitis B Virus (HBV) based on electrochemiluminescence (ECL). Thanks to the combination of surface cooperative hybridization scheme with ECL detection strategy, our novel DNA sensor is able to detect HBV genome – both synthetic and extracted – with the unprecedented limit of detection (LoD) of 0.05 cps μL−1 for extracted sample, that is even lower than the typical LoD of PCR methodologies. The detection concept presented here for HBV detection is very versatile and can be extended to other pathogens, paving the way for future development of rapid molecular test for infectious diseases, both viral and bacterial, in Point-of-Care (PoC) format.
Keywords: Electrochemiluminescence, DNA sensor, Nucleic acid detection, Point-of-Care sensors, Virus
1. Introduction
The detection of nucleic acids is nowadays of extreme importance in many medical fields for early and accurate diagnosis, personalized therapy, and preventive screening. In this context, DNA identification and quantification through molecular methods provides relevant clinical advantages with respect to the traditional laboratory methods, such as bacterial cultures or antibody detection, being much faster (hours versus days for bacterial cultures), specific (allowing the detection of genotypes), sensitive (few copies of pathogens in a sample) and accurate (able to detect different microorganisms through the specific molecular markers, but also their vitality through the mRNA monitoring). However, molecular methods typically need an amplification step achieved through the well-known Polymerase Chain Reaction (PCR) that is intrinsically quite laborious because it involves a multi-step and expensive process (about $15–80 per sample) thus limiting its utilization to specialized laboratories (Espy et al., 2006). Therefore, although current PCR-based methods are well-established and consolidated, they are not suitable to be used by unskilled personnel near the patient at competitive costs. This represents a strong limitation for its massive use, limiting the potential of genome analysis for human health. This is proved by the diffused difficulty to perform massive and real time diagnosis of coronavirus disease 2019 (COVID-19) for prompt infection management and prevention which is stimulating new approaches and methodologies for molecular detection (Cheong et al., 2020; Fan et al., 2021; Nunez-bajo et al., 2020). In addition, it has become evident that COVID-19 antigen rapid diagnostic tests, although being fast and relatively cheap, yield a high percentage of false negative results.
Therefore, the development of new molecular methods allowing rapid, sensitive, and simple detection of pathogens would be a significant breakthrough in molecular diagnostics opening new technological frontiers in the genome detection, as represented by the Genetic Point of Care (G-PoC) testing (Petralia and Conoci, 2017; Wang et al., 2021). G-PoC sensors indeed would present some unique advantages, such as a simple operation (performable by untrained personnel), rapid results, low power supply and use of reagents, and convenience of use specifically in areas where resources are limited (Arduini et al., 2016; Chan et al., 2017).
In this scenario, PCR-free approaches are extremely appealing because they do not require any analyte-amplification reactions significantly simplifying the instrumentation needed and thus lowering costs. However, these approaches are extremely challenging since detection of whole genome sequences has to be achieved at lower concentrations. This is particularly true in the case of infectious diseases where the concentration of bacterial or viral genome can be few copies in few microliters of sample. For these reasons, only very few examples of PCR-free methods have been reported in the literature so far and, consequently, a limited number of devices has been proposed (Kerr et al., 2021). Furthermore, most of them use indirect strategies that target analytes different from the chased viral or bacterial genomes (Wen et al., 2012) and, in case of pathogens, they do not fulfil the Limit of Detection (LoD) requirements (10 copies of target-genome/reaction) (Petralia et al., 2017) or use complex instrumentation (Leonardi et al., 2018). Additionally, all the reported methods need pre-treatment or labelling of the sample, adding a further degree of complexity (M. Chen et al., 2020). Therefore, a fundamental prerequisite to achieve an effective PCR-free detection of the nucleic acid of pathogens is the design of a radically new biochemical strategy for molecular recognition combined with a highly efficient transduction method.
Among infections, Hepatitis B Virus (HBV) is one of the major worldwide infective agents (Schweitzer et al., 2015). The World Health Organization (WHO) estimates that in 2015 257 million people were living with HBV infection and that 887.000 deaths yearly (World Health Organization (WHO, 2016) are caused by the subsequent occurrence of hepatocellular carcinoma (HCC). From a genetic standpoint, HBV is featured by an incomplete circular partially double-stranded (ds)-DNA genome that can be present in a wide range of concentrations (from few copies to 109 copies μL−1) in several biological fluids such as serum and dried blood spots (Brechot et al., 1981; Gupta et al., 1992) The molecular diagnosis of HBV is carried out using standard real-time PCR methods. To overcome the above reported limitations of this methodology, several types of alternative biosensors have been proposed including immunobiosensors (Chen et al., 2014; Kannan et al., 2019), paper-based electrochemical sensors (Srisomwat et al., 2020) and label-free sensors based on electrochemiluminescence (ECL) transduction (Babamiri et al., 2018).
ECL can in fact provide a very high sensitivity with the use of very simple instrumentation (Kannan et al., 2019; Pang et al., 2017; Xiang et al., 2018) being very appealing for the development of cheap G-PoC (Delaney et al., 2011). ECL is based on a luminescent phenomenon induced by electrochemical stimulus on a specific molecular system (Qi and Zhang, 2020; Sojic, 2020). Thanks to the combination between electrochemical and spectroscopic methods, ECL possesses several advantages over chemiluminescence and photoluminescence, such as (i) superior temporal and spatial control on light emission, (ii) intrinsically very low background and high sensitivity (pmol L−1) due to the absence of excitation light, and (iii) broad dynamic range (i.e., more than six orders of magnitude) and rapid measurement (i.e., few seconds) with low volumes (≤1 mL). Finally, the electrical trigger allows both simplified optical architectures (light detection without any interference of light excitation) and the integration in miniaturized microelectronic systems. In particular, in the ECL luminophore/coreactant mechanism, used in most of analytical applications and instruments (Ma et al., 2019; Zanut et al., 2020a), the excited state of the luminophore is generated through its reaction with reactive radicals produced by the electrochemical oxidation or reduction of the coreactant (Fantacci et al., 2005; Hu et al., 2009; Yuan et al., 2012). Tris(2,2’-bipyridine)ruthenium(II) ([Ru(bpy)3]2+)/tri-n-propylamine (TPrA) (A. Fiorani et al., 2018) is the most efficient ECL-coreactant system widely exploited both in commercial assays (more than 2 billion tests per year) and in innovative biochemical sensors (Sciuto et al., 2015, 2019; Zanut et al., 2019, 2020a, 2020b). An alternative approach is represented by the so called “reductive oxidation” coreactant mechanism using peroxydisulfate (S2O8 2−) as coreactant, with negative potentials (Bolletta, 1982; Fiorani et al., 2018; White and Bard, 1982; Zhang et al., 2020). This has been proved to enhance the ECL intensity of about 450-fold compared with other alternative cathodic coreactants (H2O2, Na2C2O4) (Russell et al., 2016).
In this work, a novel molecular biosensor for ultrasensitive detection and quantification of HBV whole genome without target amplification is proposed. The method is based on the combination of surface cooperative hybridization of HBV genome and ECL transduction using the [Ru(phen)2dppz]2+ complex (phen = 1,10-phenanthroline; dppz = quinoxalino[2,3-f][1,10]phenanthroline) as ECL-active dye. (“Patent pending – Italian submission N. 102021000018422,” 2021) The analytical performances of the PCR-free biosensor using both synthetic HBV genome (SG ds-HBV) and samples extracted from real samples (EG ds-HBV) are presented and discussed.
2. Materials and methods
2.1. Chemicals and materials
Analytical grade reagents were used in this work. Phosphate buffered Saline (PBS) at different concentrations (0.1 M and 0.01 M) and pH (5.5, 6.5, 7.4 and 8.5), potassium persulfate (K2S2O8) > 99%, sulfuric acid (H2SO4) 98%, Potassium hexacyanoferrate (III) (K3[Fe(CN)6]) and Ethanol (C2H6O) 100% were purchased from Sigma-Aldrich, Fetal Bovine Serum (FBS) produced by Gibco® 100% was purchased from ThermoFisher Scientific. [Ru(phen)2dppz]2+ and [Os(bpy)2dppz]Cl2 were prepared as previously described (Li et al., 2016) (Deféver et al., 2011). 6-Mercapto-1-hexanol (C6H14OS) > 97% was purchased from Fluka.
Hepatitis B virus (HBV) clone complete genome (analytical synthetic genome - SG ds-HBV) was purchased from Clonit (ref 05960467) and consists of the HBV ds-DNA genome (3.2 kbps) inserted into plasmid PBR322 vector (3.8 kbps). It is provided in a TE (Tris 10 mM, EDTA 1 mM, pH = 8) solution. Extracted HBV real genome (EG ds-HBV) were provided by Clonit (extraction were carried out from human blood using Qiagen QIAamp DNA Mini Kit "ref. 51306", following the instructions for use) at concentration of 12 × 103 IU/mL (quantified by real time PCR).
Mycobacterium Tuberculosis clone complete genome (MTB) was obtained from Clonit (ref 05960564) provided in a TE (Tris 10 mM, EDTA 1 mM, pH = 8) solution to test the selectivity of the system.
Complementary P1 probe (HS-(CH2)6-GGTGAGTGATTGGAGGTT) and P2 Probe (HS-(CH2)6-CACATCAGGATTCCTAGG) were purchased from MWG (Germany).
2.2. Apparatus and measurements
All Electrochemiluminescent (ECL) and electrochemical (EC) measurements were conducted in static environment with a potentiostat (PGSTAT302N, Metrohm) using a single-compartment three-electrode Teflon cell in static condition with Au electrode (CHI101) as the working electrode (WE), a Pt wire as the counter electrode (CE), and an Ag/AgCl (saturated KCl) electrode as the reference electrode (RE). The ECL signal was measured with a photomultiplier tube (PMT, Hamamatsu R928) placed at a fixed height from the electrochemical cell, inside a dark box. A high-voltage power supply socket assembly with a transimpedance amplifier (Hamamatsu C6271) was used to supply 750 V to the PMT, using an external trigger connection to the potentiostatic DAC module. Light/current/voltage curves were recorded by collecting the amplified PMT output signal with the ADC module of the potentiostat. ECL spectra were collected by a SEC2000 Spectra system UV–visible spectrophotometer (ALS Co. Ltd., Japan). As EC technique it was used the cyclic voltammetry (CV) applying negative potential from 0 to −2 V, with a scan rate of 0.3 V s−1, in a buffer solution PBS 0.1M pH 5.5; using a transparent electrochemical cell. For the Electrochemical Impedance Spectroscopy (EIS) a buffer solution of PBS 0.1M, pH 5.5 containing [Fe (CN)6]−3/−4 5.0 mM was used. All EIS measurements have been done in the range of frequency 0.1 MHz−1 Hz, thus the current range has been kept equivalent at 1 mA. Finally, the value of open circuit potential (Eocp) was fixed at 0.228V for the entire measurement.
2.3. Electrode preparation
Gold electrodes were polished with 0.3 and 0.05 μm alumina slurry in order to obtain a mirror surface and then washed by sonication in a 50:50 ethanol and deionized water for 5 min. Furthermore, electrodes were cleaned electrochemically through cyclic voltammetry in 0.5 M H2SO4 scanning from −1 to 1 V (10 scans and 0.1 V s−1). Finally, they were dried with Ar gas. Next, P1 and P2 thiol 5’-terminated probes were dissolved in PBS 0.01 M (pH 7.4) buffer at a concentration of 10 μM and immobilized on the surface according to the following procedure: electrodes were kept at "Room Temperature(RT)" for 4 h immersed in the solution containing the thiol-terminated probes. Afterwards, they were gently washed with PBS 0.01 M (pH 7.4) followed by an overnight blocking procedure with 10 μΜ of C6H14OS dissolved in PBS 0.01 M (pH 7.4) to stabilize the assembled layer of P1 and P2 specific probes (Fig. 1 a).
Fig. 1.
Scheme of ECL-PCR-free strategy: (a) Immobilization of specific probes P1 and P2 and of (b) 6-Mercapto-1-hexanol on the surface of a gold electrode; (c) recognition of ds-HBV by surface cooperative hybridization and (d) intercalation of [Ru(phen)2dppz]2+ and HBV whole genome detection.
2.4. Analytical test procedure
Synthetic HBV genome (SG ds-HBV) samples (1, 5, 10, 100, 1000, 10000 cps μL−1) were prepared by diluting the starting clone solution (106 cps μL−1) in PBS 0.01 M at pH 5.5. P1/P2 modified electrodes were immersed in 1 mL of SG ds-HBV solutions at 50 °C in a chamber for 3 h. To intercalate [Ru(phen)2dppz]2+ in the anchored HBV genome, electrodes were immersed in a solution containing 14 μM of [Ru(phen)2dppz]2+ in PBS 0.01M at pH 5.5 and incubated for 2 h until the intercalation was completed. The Au electrode was then ready for the ECL procedure. The electrochemical cell was fixed in front of a PMT tube in order to always keep a 0.5 cm space between the cell and the PMT input window. The transparent nature of the cell allows the photocurrent emission, produced by [Ru(phen)2dppz]2+ complex, to be detected and amplified by the PMT tube. For real sample analysis, 0–0.75 copies μL−1 of EG ds-HBV (extracted from real samples) diluted in PBS pH 5.5 from the starting solution (12 × 103 IU/mL) were used. For the biological fluid simulation testing, SG ds-HBV (106 cps μL−1) has been diluted into 1 mL Fetal Bovine Serum (FBS) to a final concentration of 100, 1000 and 10000 cps μL−1, then the modified P1/P2 electrodes were immersed in these solutions and we repeated the same procedure as it is described above. Standard deviations were obtained by repeating the experiment with 5 independent samples. For details on the EC apparatus see the paragraph 2.2.
3. Results and discussion
To detect the whole double-stranded DNA genome of HBV (ds-HBV DNA) we developed an innovative strategy combining the surface cooperative hybridization of the ds-HBV whole genome with ECL transduction mechanism, as depicted in Fig. 1.
More specifically, the typical surface hybridization implies specific interactions, leading to a thermodynamic equilibrium between hybridized and unhybridized DNA, of immobilized single strain DNA probe with the target DNA. In the case of cooperative hybridization, instead, two NA probes (P1 and P2) – designed to recognize specific and different sequences on both parallel and anti-parallel strands of the ds-HBV – were immobilized on a gold electrode surface (Fig. 1a). The ds-HBV DNA is recognized by the two surface probes that independently hybridize two complementary filaments of the ds-HBV DNA and anchor it on the electrode surface (Fig. 1c and fully characterize by EIS see Fig. S1). This leads to an effective DNA recognition reaction endowed with higher affinity, as confirmed by the comparison of the two individual probes performances (Fig. S2) and by our previous molecular dynamic investigation showing higher binding enthalpy for ds-HBV DNA – P1/P2 interaction (−67.3 kJ mol−1) than the sum of the individual single probes interactions (−47.0 kJ mol−1) (Petralia et al., 2020). A sequence gap in the target of 138 bps between the two regions recognized by the two probes (Fig. 1d) was proved to be effective in minimising the steric hindrance (Petralia et al., 2020). To detect the whole genome recognition event occurring at the electrode surface, an ECL-active complex – [Ru(phen)2dppz]2+ – was used as efficient intercalating dye (“Patent pending – Italian submission N. 102021000018422,” 2021) in the final step of our PCR-free detection strategy (Fig. 1d). Intercalation methods are appealing since they do not require any extra chemical labelling step of target or probes (He et al., 2015; Lin et al., 2016); in addition, since hundreds of complexes can intercalate in the whole viral genome, a strong signal amplification can be obtained. Moreover, [Ru(phen)2dppz]2+ shows an intense luminescence at 600–650 nm when intercalated with a double-strand DNA genome (Chambron and Sauvage, 1991; Friedman et al., 1990; Nair et al., 1997), while in aqueous solution the emission is dramatically quenched, increasing even more the signal-to-noise ratio. This behaviour results from a differential population of two close-lying dppz-localized excited states, the non-emissive state being favoured in polar and H-bonding solvents (Hiort et al., 1993; Tuite et al., 1997). Therefore, once [Ru(phen)2dppz]2+ was intercalated to the HBV-SG anchored at the electrode surface, light emission at 600–650 nm was recorded at −0.8 V (Fig. S4) (vs Ag/AgCl reference electrode), i.e. upon the reduction of the S2O8 2− coreactant which ignites the ECL process.
The effectiveness of the molecular recognition mechanism and the analytical performances of the above detection strategy were firstly tested at different pH values (from 5.5 to 7.4), using the SG ds-HBV concentration of 100 cps μL−1 (Fig. S3). In fact, the use of acid hybridization buffer (pH = 5.5) may lead to protonation of the -G and –C groups of DNA which promotes a stronger bonding with probes (Dai et al., 2015; Fiorani et al., 2020; Petralia et al., 2020). However, at pH < 7.4 unwanted reactions involving electrochemically generated SO4 •- take place leading to a decreasing of the ECL signal (Huang et al., 2010). At pH = 7.4 we observed the highest ECL value intensity in agreement with previous results (Ismail et al., 2017). Finally, at pH > 7.4 the ECL intensity is strongly reduced due to the decreasing of the SG ds-HBV recognition efficiency by the P1 and P2 probes at the surface.
After the optimization of the ECL detection conditions, we tested our system with both the SG ds-HBV analytical synthetic genome and the EG ds-HBV extracted genome from real samples. Measurements on the synthetic genome have been performed in order to optimize the conditions and to investigate in details the mechanisms of the signal generation, while the analytical characterization has been performed on the genome extracted from real samples.
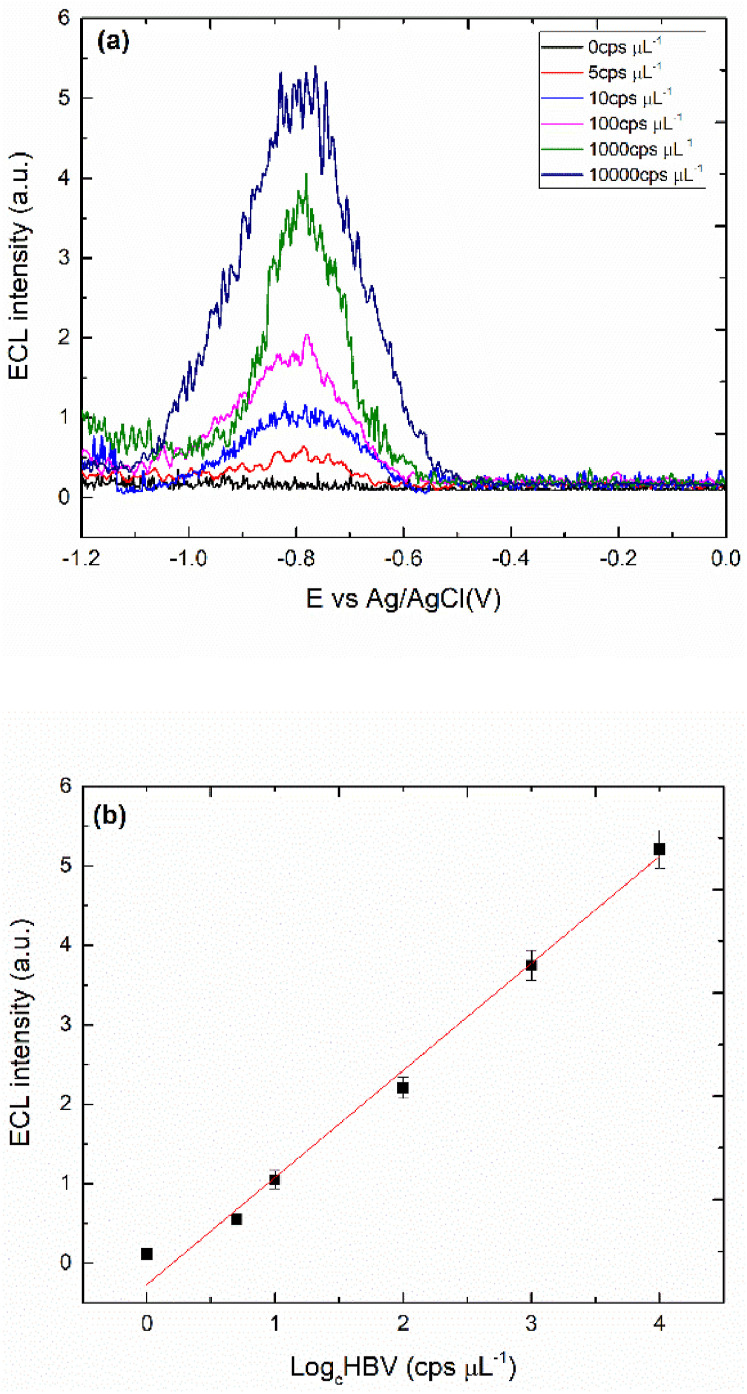
Fig. 2a reports the ECL intensities vs electrode potential measured in SG ds-HBV solutions at different concentrations (0–10000 cps μL−1). Fig. 2a suggests a logarithmic dependence of ECL intensities on HBV genome concentration and a recordable signal is still detected with few copies of HBV genome (5 cps μL−1), noteworthy without any analyte-amplification process. To assess the analytical performance of the method, ECL intensity vs the logarithm of the ds-HBV concentration was plotted. Fig. 2b displays a good linear behaviour in the 5 and 10000 cps μL−1 range with a correlation coefficient of 0.987 with a good reproducibility; thus, the increasement of the cathodic current held at −0.8V with and without the presence of [Ru(phen)2dppz]2+ (see Figs. S4 and S5). The calculated LoD (S/N = 3.3) was 2.4 cps μL−1 (i.e. 4.5 aM) with a sensitivity (slope of the curve) of 1.35 a.u./decade.
Fig. 2.
(a) ECL intensity vs potential for different SG ds-HBV concentrations (0–10000 cps μL−1), (b) Calibration plot analysis and dependence of the response of the electrode on concentration of SG ds-HBV with standard deviation of ±0.08–0.32, in 50 mM K2S2O8 in PBS 0.1 M. Working Au electrode with P1 and P2 formation vs Ag/AgCl reference electrode with scan rate 0.3 V s−1. PMT bias of 750 V. The error bar shows the standard deviation (n = 5).
At the heart of the transduction mechanism, the processes leading to light emission are described by equations (1), (2), (3), (4), (5)) (Reshetnyak and Koval’chuk, 1997; Sojic, 2020): by applying −0.80 V, both the reduction of peroxydisulfate (Eq. (1)) and [Ru(phen)2dppz]2+ (Eq. (2) see Fig. S6) (Liang et al., 2019; Wang et al., 2018; Zhou et al., 2020) take place, thus making the sequence of processes outlined by Eqs (3), (4), (5) possible. In particular, peroxydisulfate (S2O8 2−) is reduced to generate a sulphate anion (SO4 2−) and a sulphate radical anion (SO4 •-) (Eq. (1)),(Y. Chen et al., 2020) the latter also being indirectly obtained by the mediation of [Ru(phen)2dppz]+ (Eq. (3)). The reaction between [Ru(phen)2dppz]+ and the sulphate anion radical generates the [Ru(phen)2dppz]2+ excited state (Eq. (4)) which emits light (Eq. (5)).
| S2O82− + e−→SO4•- + SO42− | (1) |
| [Ru(phen)2dppz]2+ +e−→[Ru(phen)2dppz]+ | (2) |
| [Ru(phen)2dppz]++SO42−→[Ru(phen)2dppz]2++SO4•- | (3) |
| [Ru(phen)2dppz]++SO4•-→[Ru(phen)2dppz]2+*+SO42− | (4) |
| [Ru(phen)2dppz]2+*→[Ru(phen)2dppz]2++hν | (5) |
In order to evaluate its selectivity, the method was tested towards an unspecific target. Mycobacterium tuberculosis (MTB) synthetic clone at three different concentrations (10-100-1000 cps μL−1) was tested resulting in the absence of any ECL emission (Fig. S8 a-c), evidencing the high selectivity of P1 and P2 probes for SG ds-HBV.
To further validate the high sensitivity of the method, EG ds-HBV extracted from real samples was used. Noteworthy, much lower concentrations than those tested for SG ds-HBV could be detected. Fig. 3 a reports the ECL curves obtained for EG ds-HBV at concentrations below 0.75 cps μL−1 and a signal was still detected at 0.06 cps μL−1. The calibration curve shows a good linear correlation (Fig. 3b) with a LoD (S/N = 3.3) of 0.06 cps μL−1 (i.e. 0.1 aM).
Fig. 3.
(a) ECL intensity vs potential for different concentration of EG ds-HBV (0–0.75 cps μL−1); (b) calibration curve of ECL intensity vs LogC HBV concentration with standard deviation of ±0.011–0.017. In PBS 0.1 M, 14 μM intercalated [Ru(phen)2dppz]2+ and 50 mM K2S2O8. Working Au electrode with P1 and P2 formation vs Ag/AgCl reference electrode with scan rate 0.3 V s−1. PMT bias of 750V. The error bar shows the standard deviation (n = 5).
The different sensitivity between the synthetic SG-dsHBV (1.35 a.u./LogC) and the extracted real EG-ds HBV genomes (0.18 a.u./LogC) can be imputed to the different structure of the two macromolecules. Actually, the former consists of the HBV genome (3.2 kbps) inserted into plasmid PBR322 vector of 3.8 kbps leading to a circular genome of about 7.0 kbps (Petralia et al., 2017). On the contrary, the latter is a real extracted HBV genome consisting in an unusual genome partially dsDNA of about 3.2 kbps. Such a difference may be responsible for a change in the genome surface conformation and distribution of the ECL luminophore which in turn would affect the efficiency of the ECL generating mechanism (Zanut et al., 2021). The above data confirm the higher sensitivity of our method compared to standard Real time-PCR (RT-PCR) also for the real sample composed by extracted genome (10 cps μL−1, slope 0.2 – See Fig. S7) (Allice et al., 2007; Petralia et al., 2017).
An important factor in the electrically stimulated sensors is the applicability in biological fluids that can interfere with the electrical signal. The effect of biological matrixes on the modified gold electrodes was tested according to the experimental conditions described in section 2. To simulate the biological matrix, PBS was replaced by Fetal Bovin Serum (FBS) in the last step of triplex formation of our experimental procedure, and three different SG ds-HBV concentrations (100, 1000, and 10000 cps μL−1) were examined. ECL intensities measured in FBS are consistent with the one observed in PBS solution (Fig. S9). The matrix effect was evaluated by calculating the Real Standard Deviation (RSD) between FBS and PBS (Table S2), showing a satisfactory detection of SG ds-HBV in the range of 83–104%, thus confirming the efficient applicability of the proposed electrochemical DNA sensor in complex fluids without detectable interferences. These results are very promising for future application in real human complex matrixes such as plasma or blood.
Table 1 gathers the analytical performances of different methodologies for the detection of HBV DNA reported in the literature, compared with the ones obtained with our proposed PCR-free sensors. It can be noticed that our method represents an important step forward regarding sensitivity with respect to those already described. Stimulated by these results, we are currently applying this method to SARS-CoV2 detection for the development of molecular point-of-care test for low-cost massive screening.
Table 1.
Comparison of the analytical performance with different PCR-free strategies and techniques for the determination of HBV genome.
| Strategies | Techniques | Linear range | Sensitivity | Reference |
|---|---|---|---|---|
| HBV DNA sandwich assay | Anodic Stripping Voltammetry (ASV) | 0–500 × 108 cps μL−1 | 511 × 105 cps μL−1 | Li et al. (2015) |
| Label-free HBV DNA | Differential Pulse Voltammetry (DPV) | 50 × 108–100 × 1011 cps μL−1 | 8.71 × 105 cps μL−1 | Srisomwat et al., (2020) |
| PCR-free HBV genome detection | Square Wave Voltammetry (SWV) | 0.1–100 cps μL−1 | 1 cps μL−1a | Petralia et al., (2017) |
| Multicolors QDs based sensor | Electrochemiluminescence (ECL) | 0.0005–0.5 × 1011 cps μL−1 | 0.49 × 105 cps μL−1 | Liu et al. (2016) |
| Carbon sphere/Polyaniline | Differential Pulse Voltammetry (DPV) | 0.01 × 108–1 × 1011 cps μL−1 | 3.62 × 105 cps μL−1 | Salimian et al. (2019) |
| Label-free platform using graphene QDs | Differential Pulse Voltammetry (DPV) | 1012 - 5 × 1013 cps μL−1 | 1011 cps μL−1 | Xiang et al., (2018) |
| PCR-free EG ds-HBV | Electrochemiluminescence (ECL) | EG ds-HBV 0–0.75 ± 0.011–0.017 cps μL−1 |
EG ds-HBV 0.06 ± 0.011 cps μL−1 |
This work |
This LoD it was obtained using SG ds-DNA.
4. Conclusions
In summary, we developed a novel molecular sensor for ultrasensitive and PCR-free detection of HBV whole genome. Our method exploits a surface cooperative hybridization at miniaturized gold electrode with ECL transduction through the intercalation of [Ru(phen)2dppz]2+ complex, using potassium persulfate (K2S2O8) as coreactant. By combining the superior analytical performances of ECL and the signal amplification offered by the intercalation of a high number of ECL-active Ru complexes, we were able to detect HBV genome from real sample without any amplification step, reaching a LoD of 0.06 cps μL−1 (i.e. 0.1 aM). This approach may be performed with extremely simple and portable instrumentation and ongoing research in our laboratories is dedicate to the detection of viral genomes using the same strategy. The present results represent indeed an important step towards the development of rapid, sensitive and amplification free molecular Point-of-Care (PoC) test for monitoring infectious diseases.
CRediT authorship contribution statement
Pavlos Nikolaou: performed and analyzed the ECL experiments. Emanuele Luigi Sciuto: optimized the protocol for the DNA recognition and performed the RT-PCR analysis. Alessandra Zanut: performed and analyzed the ECL experiments. Salvatore Petralia: optimized the protocol for the DNA recognition and performed the RT-PCR analysis. Giovanni Valenti: planned and supervised the research and co-wrote the paper with contributions from other authors. Francesco Paolucci: planned and supervised the research and co-wrote the paper with contributions from other authors. Luca Prodi: planned and supervised the research and co-wrote the paper with contributions from other authors. Sabrina Conoci: planned and supervised the research and co-wrote the paper with contributions from other authors.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
The authors have a potential competing interest having filed the following Patent Application: Italian submission N. 102021000018422, 2021
Acknowledgements
This work is supported by the Italian Ministero dell'Istruzione, Università e Ricerca (PRIN 2020CBEYHC_002 project), and University of Bologna.
Authors thanks Dr. Carlo Roccio and Dr. Dario Russo from Clonit for the support in the molecular design and for providing extracted samples.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2022.114165.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Allice T., Cerutti F., Pittaluga F., Varetto S., Gabella S., Marzano A., Franchello A., Colucci G., Ghisetti V. COBAS AmpliPrep-COBAS TaqMan hepatitis B virus (HBV) test: a novel automated real-time PCR assay for quantification of HBV DNA in plasma. J. Clin. Microbiol. 2007;45:828–834. doi: 10.1128/JCM.00914-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arduini F., Micheli L., Moscone D., Palleschi G., Piermarini S., Ricci F., Volpe G. Electrochemical biosensors based on nanomodified screen-printed electrodes: recent applications in clinical analysis. Trends Anal. Chem. 2016;79:114–126. [Google Scholar]
- Babamiri B., Hallaj R., Salimi A. Ultrasensitive electrochemiluminescence immunosensor for determination of hepatitis B virus surface antigen using CdTe@CdS-PAMAM dendrimer as luminescent labels and Fe3O4 nanoparticles as magnetic beads. Sensor. Actuator. B Chem. 2018;254:551–560. [Google Scholar]
- Bolletta F. Polypyridine transition metal complexes as light emission sensitizers in the electrochemical reduction of the persulfate ion. Inorg. Chim. Acta. 1982;62:207–213. [Google Scholar]
- Brechot C., Scotto J., Charnay P., Hadchouel M., Degos F., Trepo C., Tiollais P. Detection of Hepatitis B virus DNA in liver and serum: a direct appraisal of the chronic carrier state. Lancet. 1981;318:765–768. doi: 10.1016/s0140-6736(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Chambron J., Sauvage J. Ru (bipy)2dppz2+ : a highly sensitive luminescent probe for micellar sodium dodecyl sulfate solutions. Chem. Phys. Lett. 1991;182:603–607. [Google Scholar]
- Chan H.N., Tan M.J.A., Wu H. Point-of-care testing: applications of 3D printing. Lab Chip. 2017;17:2713–2739. doi: 10.1039/c7lc00397h. [DOI] [PubMed] [Google Scholar]
- Chen M., Ning Z., Chen K., Zhang Y., Shen Y. Recent advances of electrochemiluminescent system in bioassay. J. Anal. Test. 2020;4:57–75. [Google Scholar]
- Chen Y., Qian C., Liu C., Shen H., Wang Z., Ping J., Wu J., Chen H. Biosensors and Bioelectronics Nucleic acid amplification free biosensors for pathogen detection. Biosens. Bioelectron. 2020;153:112049. doi: 10.1016/j.bios.2020.112049. [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang J., Liu Z., Wu G. Determination of hepatitis B virus surface antigen in serum with a sandwich immunoassay and capillary electrophoresis–electrochemical detection. Anal. Methods. 2014;6:2484–2489. [Google Scholar]
- Cheong J., Yu H., Lee C.Y., Lee Jung-uk, Choi H., Lee Jae-hyun, Lee H., Cheon J. Fast detection of SARS-CoV-2 RNA via the integration of plasmonic thermocycling and fluorescence detection in a portable device. Nat. Biomed. Eng. 2020;4:1159–1167. doi: 10.1038/s41551-020-00654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P., Yu T., Shi H., Xu J., Chen H. General strategy for enhancing electrochemiluminescence of semiconductor nanocrystals by hydrogen peroxide and potassium persulfate as dual coreactants. Anal. Chem. 2015;87:12372–12379. doi: 10.1021/acs.analchem.5b03890. [DOI] [PubMed] [Google Scholar]
- Deféver T., Druet M., Evrard D., Marchal D., Limoges B. Real-time electrochemical PCR with a DNA intercalating redox probe. Anal. Chem. 2011;83:1815–1821. doi: 10.1021/ac1033374. [DOI] [PubMed] [Google Scholar]
- Delaney J.L., Hogan C.F., Tian J., Shen W. Electrogenerated chemiluminescence detection in paper-based microfluidic sensors. Anal. Chem. 2011;83:1300–1306. doi: 10.1021/ac102392t. [DOI] [PubMed] [Google Scholar]
- Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A., Yao J.D.C., Wengenack N.L., Rosenblatt J.E., Cockerill F.R., Smith T.F. Real-time PCR in clinical microbiology : applications for routine laboratory testing. Clin. Microbiol. Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Yao B., Ding Y., Zhao J., Xie M., Zhang K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021;178:113015. doi: 10.1016/j.bios.2021.113015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantacci S., De Angelis F., Sgamellotti A., Marrone A., Re N. Photophysical properties of [Ru(phen)2(dppz)]2+ intercalated into DNA: an integrated Car-Parrinello and TDDFT study. J. Am. Chem. Soc. 2005;127:14144–14145. doi: 10.1021/ja054368d. [DOI] [PubMed] [Google Scholar]
- Fiorani A., Han D., Jiang D., Fang D., Paolucci F., Sojic N., Valenti G. Spatially resolved electrochemiluminescence through a chemical lens. Chem. Sci. 2020;11:10496–10500. doi: 10.1039/d0sc04210b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani, Andrea, Irkham, Valenti G., Paolucci F., Einaga Y. Electrogenerated chemiluminescence with peroxydisulfate as a coreactant using boron doped diamond electrodes. Anal. Chem. 2018;90:12959–12963. doi: 10.1021/acs.analchem.8b03622. [DOI] [PubMed] [Google Scholar]
- Fiorani A., Valenti G., Iurlo M., Marcaccio M., Paolucci F. Electrogenerated chemiluminescence: a molecular electrochemistry point of view. Curr. Opin. Electrochem. 2018;8:31–38. [Google Scholar]
- Friedman A.E., Chambron J.C., Sauvage J.P., Turro N.J., Barton J.K. A molecular light switch for DNA: Ru(bpy)2(dppz)2+ J. Am. Chem. Soc. 1990;112:4960–4962. [Google Scholar]
- Gupta B.P., Jayasuryan N., Jameel S. Direct detection of hepatitis B virus from dried blood spots by polymerase chain reaction amplification. J. Clin. Microbiol. 1992;30:1913–1916. doi: 10.1128/jcm.30.8.1913-1916.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Liu Y. Reusable and dual-potential responses electrogenerated chemiluminescence biosensor for synchronously cytosensing and dynamic cell surface N-glycan evaluation. Anal. Chem. 2015;87:9777–9785. doi: 10.1021/acs.analchem.5b02048. [DOI] [PubMed] [Google Scholar]
- Hiort C., Lincoln P., Nordén B. DNA binding of .DELTA.- and .LAMBDA.-[Ru(phen)2DPPZ]2+ J. Am. Chem. Soc. 1993;115:3448–3454. [Google Scholar]
- Hu L., Bian Z., Li H., Han S., Yuan Y., Gao L., Xu G. [Ru(bpy)2dppz]2+ Electrochemiluminescence switch and its applications for DNA interaction study and label-free ATP aptasensor. Anal. Chem. 2009;81:9807–9811. doi: 10.1021/ac901925x. [DOI] [PubMed] [Google Scholar]
- Huang H., Tan Y., Shi J., Liang G., Zhu J.J. DNA aptasensor for the detection of ATP based on quantum dots electrochemiluminescence. Nanoscale. 2010;2:606–612. doi: 10.1039/b9nr00393b. [DOI] [PubMed] [Google Scholar]
- Ismail L., Ferronato C., Fine L., Jaber F., Chovelon J. Elimination of sulfaclozine from water with SO4 •− radicals: evaluation of different persulfate activation methods. Appl. Catal. B Environ. 2017;201:573–581. [Google Scholar]
- Kannan P., Subramanian P., Maiyalagan T., Jiang Z. Cobalt oxide porous nanocubes-based electrochemical immunobiosensing of hepatitis B virus DNA in blood serum and urine samples. Anal. Chem. 2019;91:5824–5833. doi: 10.1021/acs.analchem.9b00153. [DOI] [PubMed] [Google Scholar]
- Kerr E., Farr R., Doeven E.H., Nai Y.H., Alexander R., Guijt R.M., Prieto-Simon B., Francis P.S., Dearnley M., Hayne D.J., Henderson L.C., Voelcker N.H. Amplification-free electrochemiluminescence molecular beacon-based microRNA sensing using a mobile phone for detection. Sensors Actuators, B Chem. 2021;330:129261. [Google Scholar]
- Leonardi A.A., Jose M., Faro L., Petralia S., Fazio B., Musumeci P., Conoci S., Irrera A., Priolo F. Ultrasensitive label- and PCR-free genome detection based on cooperative hybridization of silicon nanowires optical biosensors. ACS Sens. 2018;3:1690–1697. doi: 10.1021/acssensors.8b00422. [DOI] [PubMed] [Google Scholar]
- Li X., Scida K., Crooks Richard M. Detection of hepatitis B Virus DNA with a paper electrochemical sensor. Anal. Chem. 2015;87(17):9009–9015. doi: 10.1021/acs.analchem.5b02210. [DOI] [PubMed] [Google Scholar]
- Li G., Sun L., Ji L., Chao H. Ruthenium(II) complexes with dppz: from molecular photoswitch to biological applications. Dalton Trans. 2016;45:13261–13276. doi: 10.1039/c6dt01624c. [DOI] [PubMed] [Google Scholar]
- Liang X.X., Qian L., Huang R.F. Label-free and ultrasensitive electrochemiluminescence detection of oxidative DNA damage using DNA repair enzyme. Electrochim. Acta. 2019;312:313–319. [Google Scholar]
- Lin Y., Yang L., Yue G., Chen L., Qiu B., Guo L., Lin Z., Chen G. Label-free electrochemiluminescence biosensor for ultrasensitive detection of telomerase activity in HeLa cells based on extension reaction and intercalation of Ru(phen)3 2+ Anal. Bioanal. Chem. 2016;408:7105–7111. doi: 10.1007/s00216-016-9561-5. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang X., Ma Q., Lin Z., Chen S., Li Y., Lu L., Qu H., Su X. Multiplex electrochemiluminescence DNA sensor for determination of hepatitis B virus and hepatitis C virus based on multicolor quantum dots and Au nanoparticles. Anal. Chim. Acta. 2016;916:92–101. doi: 10.1016/j.aca.2016.02.024. [DOI] [PubMed] [Google Scholar]
- Ma C., Cao Y., Gou X., Zhu J.-J. Recent progress in electrochemiluminescence sensing and imaging. Anal. Chem. 2019;92:431–454. doi: 10.1021/acs.analchem.9b04947. [DOI] [PubMed] [Google Scholar]
- Nair R.B., Cullum B.M., Murphy C.J. Optical properties of [Ru(phen)2dppz]2+ as a function of nonaqueous environment. Inorg. Chem. 1997;36:962–965. doi: 10.1021/ic960862u. [DOI] [PubMed] [Google Scholar]
- Nunez-bajo E., Silva A., Collins P., Kasimatis M., Cotur Y., Asfour T., Tanriverdi U., Grell M., Kaisti M., Senesi G., Stevenson K., Güder F. Disposable silicon-based all-in-one micro-qPCR for rapid on-site detection of pathogens. Nat. Commun. 2020;11:6176. doi: 10.1038/s41467-020-19911-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X., Bian H., Wang W., Liu C., Khan M.S., Wang Q., Qi J., Wei Q., Du B. A bio-chemical application of N-GQDs and g-C3N4 QDs sensitized TiO2 nanopillars for the quantitative detection of pcDNA3-HBV. Biosens. Bioelectron. 2017;91:456–464. doi: 10.1016/j.bios.2016.12.059. [DOI] [PubMed] [Google Scholar]
- Patent pending . 2021. Italian Submission N. 102021000018422. [Google Scholar]
- Petralia S., Conoci S. PCR technologies for point of care testing: progress and perspectives. ACS Sens. 2017;2:876–891. doi: 10.1021/acssensors.7b00299. [DOI] [PubMed] [Google Scholar]
- Petralia S., Forte G., Zimbone M., Conoci S. The cooperative interaction of triplex forming oligonucleotides on DNA- triplex formation at electrode surface: molecular dynamics studies and experimental evidences. Colloids Surf. B Biointerfaces. 2020;187:110648. doi: 10.1016/j.colsurfb.2019.110648. [DOI] [PubMed] [Google Scholar]
- Petralia S., Sciuto E.L., Di Pietro M.L., Zimbone M., Grimaldi M.G., Conoci S. An innovative chemical strategy for PCR-free genetic detection of pathogens by an integrated electrochemical biosensor. Analyst. 2017;142:2090. doi: 10.1039/c7an00202e. [DOI] [PubMed] [Google Scholar]
- Qi H., Zhang C. Electrogenerated chemiluminescence biosensing. Anal. Chem. 2020;92:524–534. doi: 10.1021/acs.analchem.9b03425. [DOI] [PubMed] [Google Scholar]
- Reshetnyak O.V., Koval’chuk E.P. A possible scheme of electrochemiluminescence generation on platinum cathodes in aqueous solutions of peroxydisulfate. Electrochim. Acta. 1997;43:465–469. [Google Scholar]
- Russell R., Stewart A.J., Dennany L. Optimising electrogenerated chemiluminescence of quantum dots via co-reactant selection. Anal. Bioanal. Chem. 2016;408:7129–7136. doi: 10.1007/s00216-016-9557-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimian R., Shahrokhian S., Panahi S. Enhanced electrochemical activity of a hollow carbon sphere/polyaniline-based electrochemical biosensor for HBV DNA marker detection. ACS Biomater. Sci. Eng. 2019;5(5):2587–2594. doi: 10.1021/acsbiomaterials.8b01520. [DOI] [PubMed] [Google Scholar]
- Schweitzer A., Horn J., Mikolajczyk R.T., Krause G., Ott J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection : a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- Sciuto E.L., Santangelo M.F., Villaggio G., Sinatra F., Bongiorno C., Nicotra G., Libertino S. Photo-physical characterization of fluorophore Ru(bpy)32+ for optical biosensing applications. Sens. Bio-Sensing Res. 2015;6:67–71. [Google Scholar]
- Sciuto E.L., Villaggio G., Santangelo M.F., Laudani S., Federico C., Saccone S., Sinatra F., Libertino S. Study of a miniaturizable system for optical sensing application to human cells. Appl. Sci. 2019;9 [Google Scholar]
- Sojic N. Royal Society Of Chemistry (RSC); 2020. Analytical Electrogenerated Chemiluminescence: from Fundamentals to Bioassays. [Google Scholar]
- Srisomwat C., Teengam P., Chuaypen N., Tangkijvanich P., Vilaivan T., Chailapakul O. Chemical Pop-up paper electrochemical device for label-free hepatitis B virus DNA detection. Sensor. Actuator. B Chem. 2020;316:128077. [Google Scholar]
- Tuite E., Lincoln P., Norden B. Photophysical evidence that Δ- and Λ-[Ru(phen)2(dppz)]2+ intercalate DNA from the minor groove. J. Am. Chem. Soc. 1997;119:239–240. [Google Scholar]
- Wang C., Liu M., Wang Z., Li S., Deng Y., He N. Point-of-care diagnostics for infectious diseases: from methods to devices. Nano Today. 2021;37:101092. doi: 10.1016/j.nantod.2021.101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gao H., Qi H., Gao Q., Zhang C. Proximity hybridization-regulated immunoassay for cell surface protein and protein-overexpressing cancer cells via electrochemiluminescence. Anal. Chem. 2018;90:3013–3018. doi: 10.1021/acs.analchem.7b04359. [DOI] [PubMed] [Google Scholar]
- Wen Y., Pei H., Shen Y., Xi J., Lin M., Lu N., Shen X., Li J., Fan C. DNA Nanostructure-based Interfacial electrochemical analysis of microRNA. Sci. Rep. 2012;84:14–18. doi: 10.1038/srep00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White H.S., Bard A.J. Electrogenerated chemiluminescence. 41. Electrogenerated chemiluminescence and chemiluminescence of the Ru(2,21 - bpy)32+-S2O82- system in acetonitrile-water solutions. J. Am. Chem. Soc. 1982;104:6891–6895. [Google Scholar]
- World Health Organization (WHO) 2016. Global Health Sector Strategy on Viral Hepatitis. [Google Scholar]
- Xiang Q., Huang J., Huang H., Mao W., Ye Z. A label-free electrochemical platform for the highly sensitive detection of hepatitis B virus DNA using graphene quantum dots. RSC Adv. 2018;8:1820–1825. doi: 10.1039/c7ra11945c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Han S., Hu L., Parveen S., Xu G. Coreactants of tris(2,2′-bipyridyl)ruthenium(II) electrogenerated chemiluminescence. Electrochim. Acta. 2012;82:484–492. [Google Scholar]
- Zanut A., Fiorani A., Rebeccani S., Kesarkar S., Valenti G. Electrochemiluminescence as emerging microscopy techniques. Anal. Bioanal. Chem. 2019;411:4375–4382. doi: 10.1007/s00216-019-01761-x. [DOI] [PubMed] [Google Scholar]
- Zanut A., Fiorani A., Saito T., Ziebart N., Rapino S., Rebeccani S., Barbon A., Irie T., Josel H., Negri F., Marcaccio M., Windfuhr M., Imai K., Valenti G., Paolucci F. Insights into the mechanism of coreactant electrochemiluminescence facilitating enhanced bioanalytical performance. Nat. Commun. 2020;11:2668–2676. doi: 10.1038/s41467-020-16476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanut A., Palomba F., Rossi Scota M., Rebeccani S., Marcaccio M., Genovese D., Rampazzo E., Valenti G., Paolucci F., Prodi L. Dye-doped silica nanoparticles for enhanced ECL-based immunoassay analytical performance. Angew. Chem. Int. Ed. 2020;59:21858–21863. doi: 10.1002/anie.202009544. [DOI] [PubMed] [Google Scholar]
- Zanut A., Rossetti M., Marcaccio M., Ricci F., Paolucci F., Porchetta A., Valenti G. DNA-based nanoswitches: insights into electrochemiluminescence signal enhancement. Anal. Chem. 2021;93:10397–10402. doi: 10.1021/acs.analchem.1c01683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kerr E., Usman K.A.S., Doeven E.H., Francis P.S., Henderson L.C., Razal J.M. Cathodic electrogenerated chemiluminescence of tris(2,20-bipyridine)ruthenium(II) and peroxydisulfate at pure Ti3C2Tx MXene electrodes. Chem. Commun. 2020;56:10022–10025. doi: 10.1039/d0cc02993a. [DOI] [PubMed] [Google Scholar]
- Zhou T., Huang R., Huang M., Shen J., Shan Y., Xing D. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific MiRNA detection. Adv. Sci. 2020;7:1–10. doi: 10.1002/advs.201903661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.