Abstract
Bovine coronavirus (BCoV) is one of the agents causing bovine respiratory disease complex (BRDC), with single infection tending to be mild to moderate; the probability of developing pneumonia in BRDC may be affected by viral and bacterial combinations. Previously, we reported that bovine respiratory syncytial virus (BRSV) infection enhances adherence of Pasteurella multocida (PM) to cells derived from the bovine lower respiratory tract but that BRSV infection in cells derived from the upper respiratory tract reduces PM adherence. In this study, we sought to clarify whether the modulation of bacterial adherence to cells derived from the bovine upper and lower respiratory tract is shared by other BRDC-related viruses by infecting bovine epithelial cells from the trachea, bronchus and lung with BCoV and/or PM. The results showed that cells derived from both the upper and lower respiratory tract were susceptible to BCoV infection. Furthermore, all cells infected with BCoV exhibited increased PM adherence via upregulation of two major bacterial adhesion molecules, intercellular adhesion molecule-1 (ICAM-1) and platelet-activating factor receptor (PAF-R), suggesting that compared with BRSV infection, BCoV infection differentially modulates bacterial adherence. In summary, we identified distinct interaction between bovine respiratory viruses and bacterial infections.
Keywords: Bacterial adhesion, Bovine coronavirus, Bovine respiratory disease complex, Intercellular adhesion molecule-1, Pasteurella multocida, Platelet-activating factor receptor
1. Introduction
Bovine coronavirus (BCoV) is a member of the family Coronavirus, genus Betacoronavirus, species Betacoronavirus 1. BCoV is a pneumo-enteric virus that infects the upper and lower respiratory tract as well as the intestine (Saif, 2010). Some studies have reported that BCoV is among the most frequently detected viruses in calves with respiratory symptoms (O’Neill et al., 2014; Castells et al., 2019) and multiple infection with Pasteurella multocida (PM) were found in farms with respiratory symptoms (Kishimoto et al., 2017). The pathogenicity of BCoV infection in the bovine respiratory tract is considered milder than that of other bovine respiratory disease complex (BRDC)-related viruses, such as bovine respiratory syncytial virus (BRSV) (Beaudeau et al., 2010).
As bacterial infection of the lower respiratory tract can lead to severe pneumonia (Bryson, 1985), clarifying the mechanism of bacterial invasion from the upper to lower respiratory tract is of great importance. Our previous study revealed that PM adherence to BRSV-infected bovine respiratory epithelial cells (BRECs) is dependent on the site of origin from the upper or lower part of the respiratory tract (Sudaryatma et al., 2019). Interestingly, the number of adhering bacteria decreased and increased in BRSV-infected cells derived from the upper and lower respiratory tract, respectively. These changes in the number of adhering bacteria were caused by down- and upregulation, respectively, of two bacterial adhesion molecules (Sudaryatma et al., 2020a, 2020b). Reduced and enhanced bacterial adherence to BRSV-infected upper and lower BRECs would permit bacteria to pass from the upper to lower respiratory tract, inducing severe pneumonia. The tendency to cause pneumonia differs markedly among BRDC-related viruses. Thus, differences in bacterial adherence to BRDC-related virus-infected upper and lower BRECs need to be clarified. In this study, we investigated bacterial adherence to BCoV-infected BRECs and analyzed expression of bacterial adhesion molecules. The results of this study will contribute to our understanding of the possible mechanism underlying differences in pathogenicity among bovine respiratory viruses.
2. Methods, techniques
2.1. Virus and bacterium
The BCoV used in this study was isolated from nasal swab of cattle showing mild respiratory symptoms. Swab samples were tested for BCoV by qRT-PCR, as described by our group (Mekata et al., 2020). The samples were inoculated onto a monolayer of human rectal tumor cells (HRT-18 G) in serum-free Opti-MEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (FUJIFILM Wako Pure Chemical, Osaka, Japan). After 60 min. of adsorption, the cells were washed, and fresh serum-free Opti-MEM with 0.5 μg/mL TPCK trypsin (Sigma-Aldrich, St. Louis, MO, USA) was added. The cultures were maintained for 5 days at 37 °C in 5% CO2. Three blind passages were conducted by collecting the supernatant via centrifugation at 3000 rpm for 10 min at 4 °C and inoculating onto fresh monolayer cells following the methods described above. Further passage for virus propagation was conducted using Dulbecco's Modified Eagle Medium (DMEM; Nacalai Tesque, Kyoto, Japan) with 10 % fetal bovine serum (FBS) without the addition of TPCK trypsin. Virus isolation was confirmed by qRT-PCR. The virus titer was measured based on the median tissue culture infectious dose (TCID50) on Madin-Darby Bovine Kidney (MDBK) cells for 10 days. The cells were stained with 0.2 % crystal violet in 4% paraformaldehyde. The propagated viruses were stored at −80 °C until use. PM strain 2368, capsular type B isolated from nasal swab of cattle was used as described previously (Sudaryatma et al., 2018). For infection, the virus and bacterium were diluted in antibiotic- and serum-free Dulbecco’s modified Eagle’s medium:Nutrient Mixture F-12 (DMEM/F12), GlutaMAX (Thermo Fisher Scientific) to achieve a 5000 TCID50/mL and a multiplicity of infection (MOI) of 100, respectively.
2.2. Culture of bovine respiratory epithelial cells
Bovine respiratory epithelial cells (BRECs) from the trachea (bTEC), bronchus (bBEC), and lung (bLEC) used in this study were previously established by our group (Sudaryatma et al., 2019). The cells were maintained by subculturing every 5–7 days in a T-75 flask coated with collagen coating type I (Sumilon, Sumitomo Bakelite, Tokyo, Japan) using a culture medium comprising DMEM/F12, GlutaMAX supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 1 μg/mL amphotericin-B, 10 ng/mL epidermal growth factor, 1% insulin-transferrin-selenium, 1% nonessential amino acid, and 2 mM l-glutamine (FUJIFILM Wako Pure Chemical). The culture medium was changed every 3 days until the cells reached 100 % confluence and were ready for subculture.
2.3. BCoV and/or PM infection of BRECs
Each type of BREC (5 × 105 cells/mL) was seeded and then inoculated with BCoV (50,000 TCID50); cells without the virus served as a negative control. The supernatant and cell lysate were collected at 1, 3, 6, and 12 h postinfection (hpi) and at 3, 5, and 7 days postinfection (dpi). BCoV-infected and uninfected cells were exposed to PM (MOI 100) for 1 h at 37 °C, and the number of adhering bacteria was measured in colony forming units (CFUs) as described previously (Sudaryatma et al., 2019). The number of adhering bacteria was also counted at 1, 2 and 3 dpi. For microscopic observation, PM was labeled with fluorescein isothiocyanate (FITC; Dojindo Laboratories, Kumamoto, Japan), as described previously (Sudaryatma et al., 2018). BCoV-infected and uninfected cells were exposed to FITC-labeled PM for 1 h and observed by fluorescence microscopy (DP-74, Olympus, Tokyo, Japan); cell nuclei were visualized by Hoechst 33342 staining (ab145597; Abcam, Cambridge, UK).
2.4. Real-time RT-PCR to quantify viral RNA and cytokine mRNA
Viral or cellular RNA was extracted from supernatants or cells using a NucleoSpin Virus (TaKaRa Bio, Kusatsu, Japan) or an RNeasy Mini Kit (Qiagen, Venlo, Netherlands), respectively. All qRT-PCR assays were performed in triplicate using a LightCycler 96 system (Roche Diagnostics, Rotkreuz, Switzerland) and a One Step PrimeScript RT-PCR Kit (TaKaRa Bio). Synthetic control DNA was designed based on the BCoV Kakegawa strain (Accession No. AB354579). The primers (forward: 5′-GGACCCAAGTAGCGATGAG-3′, reverse: 5′-GACCTTCCTGAGCCTTCAATA-3′), probe (5′-FAM/ATTCCGACT/Zen/AGGTTTCCGCCTGG/IBFQ-3′) and amplification conditions used in this study were previously reported (Kishimoto et al., 2017; Mekata et al., 2020). To analyze mRNA expression of bovine cytokines, qRT-PCR was performed using One Step TB Green PrimeScript RT-PCR Kit II (TaKaRa Bio). The primers used to quantify mRNA levels of bovine IL-1β, IL-6, TNF-α, ICAM-1 and GAPDH have been described previously (Sobotta et al., 2017; Sudaryatma et al., 2020b). The primers for PAF-R (Forward; 5′-CACGCAGCAGGTGCAAATAC-3′ and Reverse; 5′ TGGGGCACGAAACAGATGAT-3′) were designed based on the reference sequence in this study (Accession No. NM001040538). Relative mRNA expression between the infected sample and uninfected control was calculated using the 2−ΔΔCT method, and the result is expressed as the fold change.
2.5. Western blot analysis
BCoV-infected cells in a 6-well plate were lysed with 300 μl lysis buffer (150 mM NaCl, 50 mM Tris−HCl, pH 7.6, 1% NP-40, protease inhibitor, and phosphatase inhibitor) and sonicated twice for 10 s each. The protein concentrations of the samples were determined using a TaKaRa BCA Protein Assay Kit (TaKaRa Bio). Protein samples (10 μg per well) were loaded into each lane of a 4–15 % SDS polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA, USA), separated by electrophoresis, and transferred to a PVDF membrane (Bio-Rad Laboratories) using a Transblot semidry system (Bio-Rad Laboratories). The membranes were blocked overnight at 4 °C with blocking buffer (25 mM Tris, pH 8.0, 125 mM NaCl, 0.1 % Tween 20, and 3% bovine serum albumin) and then incubated with either mouse anti-ICAM-1 (ab2213; Abcam), rabbit anti-PAF-R (ab104162; Abcam) or mouse anti-GAPDH (ab9482; Abcam) primary antibodies at room temperature for 1 h. After vigorous washing, the membranes were incubated with a secondary goat anti-mouse IgG (ab205719; Abcam) or anti-rabbit IgG (ab205718; Abcam) antibody conjugated to horseradish peroxidase at room temperature for 1 h. The chemiluminescent signals were detected using the western blot hyper HRP substrate (TaKaRa Bio) and a ChemiDoc Touch Imaging System (Bio-Rad Laboratories).
2.6. Statistical analysis
Data are reported as the mean and standard error of the mean (SEM) of at least three independent experiments. Statistical analyses were performed using one-way analysis of variance (ANOVA) and Tukey’s Multiple Comparison Test. A p-value <0.05 was considered statistically significant. The data analysis was performed using RStudio software (RStudio, Boston, MA USA).
3. Results
3.1. BCoV isolation and the viral susceptibility of BRECs
BCoVs were isolated from 7 nasal swab samples of 32 qPCR-positive cattle. The seven virus isolates were named C13, C19, CS5, U39, S5, S13 and S15. Because the C13 strain produced the highest number of viral copies in the culture medium (data not shown), we used this strain for further experiments. Although viral copy numbers increased or remained stable during viral passages, the cytopathic effect (CPE) in BCoV-infected HRT-18 G cells was not clear. Thus, the virus titer was determined by using MDBK cells.
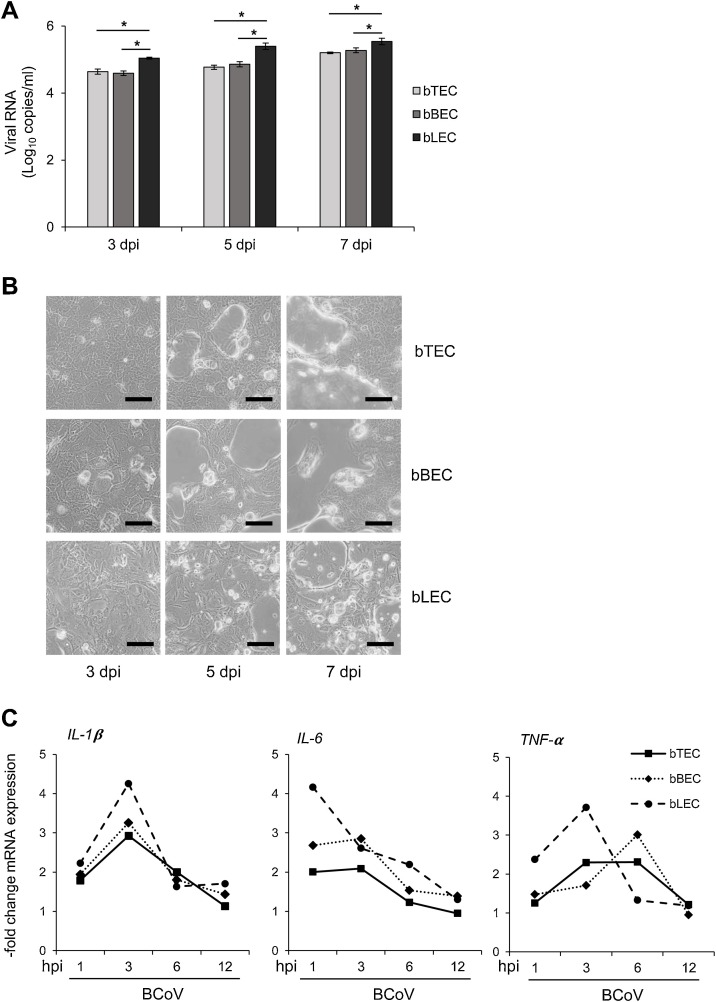
To assess the susceptibility of BRECs to BCoV, qRT-PCR was performed to measure the viral copy number in the culture medium at different time points. Viral RNA was detected from 3 to 7 dpi in bTECs, bBECs, and bLECs (Fig. 1 A), with significant differences in viral copy number among the three cell types (p < 0.05). bLECs produced the greatest amount of viral RNA compared with bTECs and bBECs. In addition, we observed a clear CPE in all BCoV-infected BRECs, including floating cells and circular cells (Fig. 1B). BCoV-infected BRECs expressed different levels of mRNA associated with inflammation (Fig. 1C). For example, the peak of IL-1® expression was observed at 3 hpi in all three cell types. However, the highest level of IL-6 expression was observed at 3 hpi in bTECs and bBECs and at 1 hpi in bLECs. TNF-α expression in bTECs and bLECs peaked at 3 hpi, but it reached a peak at 6 hpi in bBECs.
Fig. 1.
Bovine respiratory coronavirus (BCoV) infection in bovine respiratory epithelial cells (BRECs) and inflammatory responses.
BRECs from the trachea (bTEC), lung (bLEC), and bronchus (bBEC) were infected with 5000 TCID50 of BCoV. Culture medium and cell lysates were collected at 3, 5, and 7 days postinfection (dpi). (A) BCoV RNA in the culture medium was quantified by qRT-PCR. The graph shows the mean viral copy number (± SEM; n = 3; * p < 0.05). (B) The BCoV-induced cytopathic effect in BRECs at 3, 5 and 7 dpi (bar = 150 μm). (C) Dynamics of mRNAs encoding bovine IL-1β, IL-6, and TNF-α in BCoV-infected BRECs. Data represent the mean of relative mRNA expression between the infected samples and uninfected controls (n = 3).
3.2. BCoV infection of BRECs enhances PM adherence
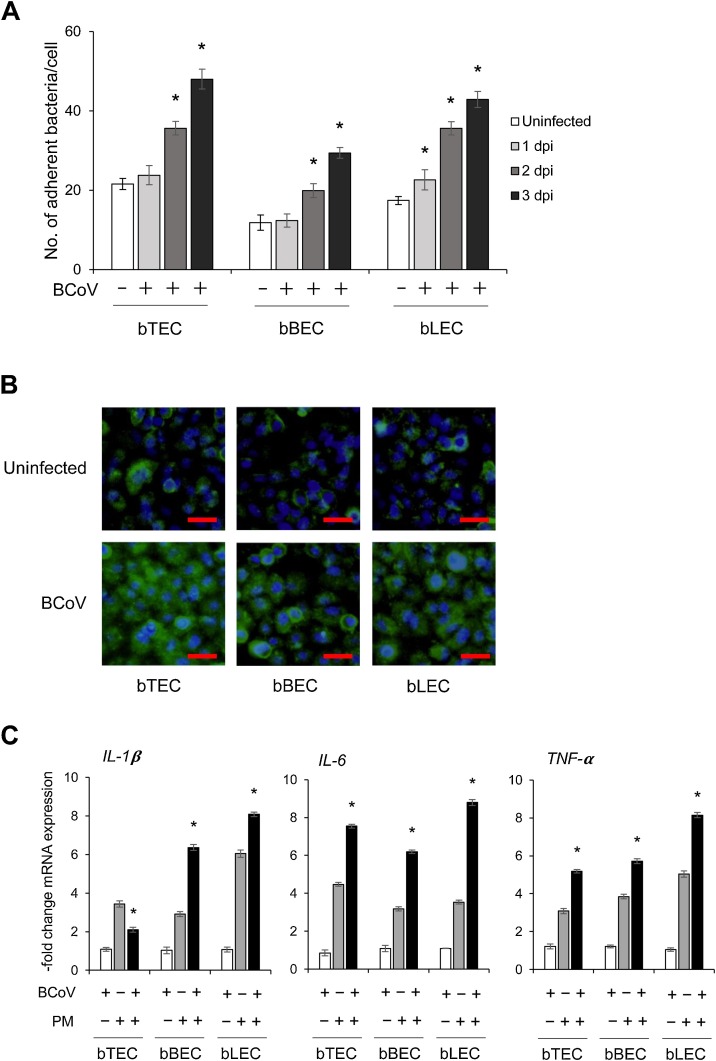
To examine whether bacterial adherence to BRECs can be enhanced by BCoV infection, cells were infected with BCoV for three days and then inoculated with PM. Compared to cells not infected with the virus, the number of adhering bacteria increased significantly for all BCoV-infected BRECs at 2–3 dpi (p < 0.05) (Fig. 2 A and B). Specifically, rates of increasing numbers of adhering bacteria were 1.7- and 2.2-fold in bTECs, 1.7- and 2.5-fold in bBECs, and 2.0 and 2.4-fold in bLECs at 2 and 3 dpi compared to virus-uninfected cells, respectively. Moreover, coinfection of BCoV with bacteria stimulated expression of inflammatory cytokines (Fig. 2C), and the levels of IL-6 and TNF-α mRNA expression in bTECs coinfected with BCoV and PM were significantly higher than those in cells infected with PM alone (p < 0.05). In contrast, expression of IL-1β in bTECs coinfected with BCoV and PM was lower than that in cells infected with only PM. Nevertheless, mRNA expression of IL-6, TNF-α and IL-1β in bBECs and bLECs coinfected with BCoV and PM was significantly higher than that in cells only infected with PM (p < 0.05).
Fig. 2.
Bovine respiratory coronavirus (BCoV) infection enhances the adherence of Pasteurella multocida (PM) to bovine respiratory epithelial cells (BRECs).
(A) BRECs preinfected with 5000 TCID50 of BCoV for 1, 2, and 3 days were exposed to PM (MOI = 100) for 1 h. The number of PM adhering to BRECs was counted on Brucella agar. Data are expressed as the mean (±SEM; n = 3; *p < 0.05 compared to uninfected cells). (B) FITC-labeled PM (green) adherence to BCoV-infected and -uninfected BRECs at 3 dpi was observed with a fluorescence microscope; nuclei were stained with Hoechst-33342 (blue) (scale bar: 30 μm). (C) Expression of bovine IL-1β, IL-6, and TNF-α mRNA in BRECs infected with BCoV alone at 3 dpi, PM alone at 1 hpi, or BCoV 3 dpi plus PM 1 hpi. Data represent the mean of relative mRNA expression between the virus and/or bacteria infected samples and uninfected controls (±SEM; n = 3; * p < 0.05 compared to PM alone).
3.3. BCoV infection upregulates ICAM-1 and PAF-R expression in BRECs
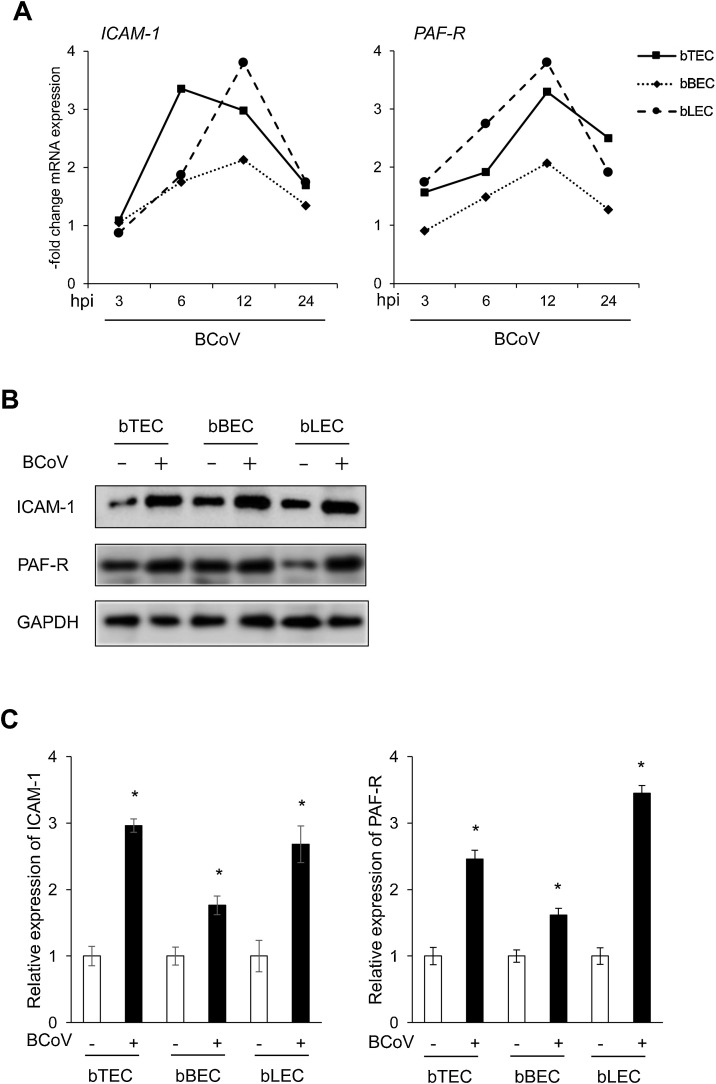
We recently demonstrated that expression of two cell surface receptors, platelet-activating factor receptor (PAF-R) and intercellular adhesion molecule-1 (ICAM-1), determines the adherence of PM to BRSV-infected BRECs (Sudaryatma et al., 2020a, 2020b). Based on this finding, we measured the expression levels of these receptors by qRT-PCR and western blotting. As depicted in Fig. 3 A, BCoV infection stimulated mRNA expression of PAF-R and ICAM-1 in BRECs. We observed a peak of PAF-R expression at 12 hpi in all cell types infected with BCoV (3.2-, 2.1- and 3.8-fold increase, respectively). In bTECs, ICAM-1 expression reached its peak (3.4-fold increase) at 6 h after BCoV infection, and the highest level of ICAM-1 expression in bBECs and bLECs was observed at 12 hpi (2.1- and 3.8-fold increase, respectively). Consistent with the results for mRNA expression, levels of PAF-R and ICAM-1 protein expression increased significantly after BCoV infection in all cells compared with uninfected cells (Fig. 3B and C), and this difference was statistically significant (p < 0.05).
Fig. 3.
Expression levels of surface receptors on bovine respiratory epithelial cells (BRECs) infected with bovine respiratory coronavirus (BCoV).
(A) mRNA expression of bovine ICAM-1 and PAF-R in BRECs infected with BCoV (5000 TCID50) at 3, 6, 12, and 24 h postinfection (hpi). Data represent the mean of relative mRNA expression between the infected samples and uninfected controls (n = 3). (B) Protein expression of bovine ICAM-1 and PAF-R in BCoV-infected and uninfected BRECs. (C) Relative values of ICAM-1 and PAF-R protein expression were calculated by normalizing the band density of GAPDH in the respective sample (lane); values were then compared to those in uninfected cells. Data represent the mean (±SEM; n = 3; *p < 0.05 compared to uninfected cells).
4. Discussion
Bovine respiratory coronavirus infection tends to induce milder respiratory symptoms than BRSV infection (Beaudeau et al., 2010; Gershwin et al., 2015; Oma et al., 2016). In general, understanding the mechanism of differences in pathogenicity between BCoV and BRSV is important to predict the prognosis of affected animals and minimize damage due to BRDC. In this study, we found that BCoV infection induced the adherence of PM to bovine upper and lower respiratory epithelial cells through upregulation of ICAM-1 and PAF-R. This result was in contrast to the outcome of BRSV infection, which reduced PM adherence and ICAM-1 expression in cells derived from the upper respiratory tract (Sudaryatma et al., 2019, 2020b). PM is one of the major bacteria in the upper respiratory tract and microbiota in the upper respiratory tract is an important defense mechanism to prevent invasion of bacteria into the lower respiratory tract (Allen et al., 1992; Man et al., 2017). Hence, this study provides a possible reason for the difference in pathogenicity between BRSV and BCoV infection in BRDC.
Here, we show that all BRECs from three respiratory tissues were susceptible to BCoV. Indeed, we observed time-dependent replication of BCoV in bTECs (representative of the upper respiratory tract), bBECs (representative of the upper part of the lower respiratory tract), and bLECs (representative of the lower part of the lower respiratory tract). Moreover, robust viral replication in bLECs correlated with a clear CPE and high mRNA expression of inflammatory cytokines (Fig. 1B and C). Although most of BCoV infection induce mild respiratory symptoms, invasion of BCoV into the lower respiratory tract appears to induce severe inflammation and pneumonia (Amoroso et al., 2020). In general, HRT-18 G cells are used for BCoV isolation due to their high susceptibility as well as the high productivity of the virus in these cells. BCoV-infected HRT-18 G cells exhibited a slight CPE in this study, though it was not a clear effect. Therefore, we used MDBK cells to determine the virus titer. Recent field isolates of BCoV in Japan have reportedly lost hemagglutination activity in chicken red blood cells (Kanno et al., 2018). We also confirmed the same feature in our isolates (data not shown). Accordingly, the characteristics of BCoVs circulating in Japan might differ from those of prior strains, explaining why the CPE in HRT-18 G cells was difficult to observe.
This study showed that PM adherence to upper and lower BRECs was significantly enhanced by preinfection with BCoV. Furthermore, we confirmed upregulation of ICAM-1 and PAF-R in BCoV-infected bovine upper and lower respiratory epithelial cells. A similar tendency has been observed in human respiratory cells infected with mildly pathogenic viruses such as rhinovirus and coronavirus NL63 (Ishizuka et al., 2003; Golda et al., 2011). Infection with these viruses enhanced expression of cellular receptors in respiratory epithelial cells and bacterial adherence. Upregulation of bacterial adhesion molecules in the upper respiratory tract during viral infection can be read in two ways. One is associated with more severe damage in the upper respiratory tract. Another is an important defense system for the host to prevent infection in the lower respiratory tract. BRSV infection downregulates expression of these molecules in the upper respiratory tract and would enhance bacterial invasion into the lower respiratory tract (Sudaryatma et al., 2019, 2020b). We do not know why BCoV and BRSV infection induce the different type of cell response. BRSV NS1 and NS2 proteins function to inhibit the interferon responses and the virus evades the host immunity (Schlender et al., 2000; Bossert et al., 2003). These kinds of protein might affect to the expression of bacteria adhesion molecules and induce bacterial invasion into the lower respiratory tract. Thus, it might be important to clear the virus-mediated modulation of bacterial adhesion molecule expression to identify the pathogenicity of BRDC-related virus.
Under our conditions, BCoV infection enhanced adherence of bacteria to BRECs, inducing overproduction of inflammatory cytokines. Although mRNA expression levels of IL-6 and TNF-α were increased in coinfected upper respiratory cells (bTECs), IL-1β was decreased. Regardless, all three inflammatory cytokines were increased in coinfected lower respiratory cells (bBECs and bLECs). IL-1β is responsible for neutrophil recruitment and inflammation in the lungs (Ichinohe et al., 2009). However, an excessive production of this cytokine has been associated with several autoinflammatory diseases (McDermott and Tschopp, 2007). Thus, control of IL-1β expression in upper respiratory epithelial cells coinfected with BCoV and PM might be important to maintain a homeostasis in the upper respiratory tract.
In conclusion, our data demonstrate that BCoV infection enhances PM adherence and triggers inflammatory responses in cells derived from the upper and lower respiratory tracts. BCoV infection upregulated both ICAM-1 and PAF-R expression, enhancing bacterial adherence to BCoV-infected cells. The study presents evidence for a distinct mechanism of BRDC-related infections in response to viral infections in respiratory epithelial cells. Understanding this complex interplay will help in the development of new therapeutic strategies for BRDC.
Funding
This work was supported by a grant (grant No. JPMJSA1908) from the Japan Science and Technology Agency (JST) for Science and Technology Research Partnership for Sustainable Development (SATREPS). The first author (W.F.) was supported by the training program from Japan Veterinary Medical Association (JVMA) for Training Program for Asian Veterinarians II for fiscal year 2019 (TP-FAVII).
Role of the funding source
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author, H.M., upon reasonable request.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We kindly thank many clinical veterinarians and owners for the collection of bovine nasal swab samples.
References
- Allen J.W., Viel L., Bateman K.G., Rosendal S. Changes in the bacterial flora of the upper and lower respiratory tracts and bronchoalveolar lavage differential cell counts in feedlot calves treated for respiratory diseases. Can. Vet. J. 1992;56:177–183. [PMC free article] [PubMed] [Google Scholar]
- Amoroso M.G., Lucifora G., Degli Uberti B., Serra F., De Luca G., Borriello G., De Domenico A., Brandi S., Cuomo M.C., Bove F., Riccardi M.G., Galiero G., Fusco G. Fatal interstitial pneumonia associated with bovine coronavirus in cows from Southern Italy. Viruses. 2020;12:1331. doi: 10.3390/v12111331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudeau F., Ohlson A., Emanuelson U. Associations between bovine coronavirus and bovine respiratory syncytial virus infections and animal performance in Swedish dairy herds. J. Dairy Sci. 2010;93:1523–1533. doi: 10.3168/jds.2009-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert B., Marozin S., Conzelmann K.K. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 2003;77:8661–8668. doi: 10.1128/JVI.77.16.8661-8668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson D.G. Calf pneumonia. Vet. Clin. North Am. Food Anim. Pract. 1985;1:237–257. doi: 10.1016/S0749-0720(15)31326-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castells M., Giannitti F., Caffarena R.D., Casaux M.L., Schild C., Castells D., Riet-Correa F., Victoria M., Parreño V., Colina R. Bovine coronavirus in Uruguay: genetic diversity, risk factors and transboundary introductions from neighboring countries. Arch. Virol. 2019;164:2715–2724. doi: 10.1007/s00705-019-04384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershwin L.J., Van Eenennaam A.L., Anderson M.L., McEligot H.A., Shao M.X., Toaff-Rosenstein R., Taylor J.F., Neibergs H.L., Womack J., Bovine Respiratory Disease Complex Coordinated Agricultural Project Research T. Single pathogen challenge with agents of the bovine respiratory disease complex. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golda A., Malek N., Dudek B., Zeglen S., Wojarski J., Ochman M., Kucewicz E., Zembala M., Potempa J., Pyrc K. Infection with human coronavirus NL63 enhances streptococcal adherence to epithelial cells. J. Gen. Virol. 2011;92:1358–1368. doi: 10.1099/vir.0.028381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T., Lee H.K., Ogura Y., Flavell R., Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka S., Yamaya M., Suzuki T., Takahashi H., Ida S., Sasaki T., Inoue D., Sekizawa K., Nishimura H., Sasaki H. Effects of rhinovirus infection on the adherence ofstreptococcus pneumoniaeto cultured human airway epithelial cells. J. Infect. Dis. 2003;188:1928–1939. doi: 10.1086/379833. [DOI] [PubMed] [Google Scholar]
- Kanno T., Ishihara R., Hatama S., Uchida I. A long-term animal experiment indicating persistent infection of bovine coronavirus in cattle. J. Vet. Med. Sci. 2018;80:1134–1137. doi: 10.1292/jvms.18-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto M., Tsuchiaka S., Rahpaya S.S., Hasebe A., Otsu K., Sugimura S., Kobayashi S., Komatsu N., Nagai M., Omatsu T., Naoi Y., Sano K., Okazaki-Terashima S., Oba M., Katayama Y., Sato R., Asai T., Mizutani T. Development of a one-run real-time PCR detection system for pathogens associated with bovine respiratory disease complex. J. Vet. Med. Sci. 2017;79:517–523. doi: 10.1292/jvms.16-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W.H., De Steenhuijsen Piters W.A.A., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott M.F., Tschopp J. From inflammasomes to fevers, crystals and hypertension: how basic research explains inflammatory diseases. Trends Mol. Med. 2007;13:381–388. doi: 10.1016/j.molmed.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Mekata H., Hamabe S., Sudaryatma P.E., Kobayashi I., Kanno T., Okabayashi T. Molecular epidemiological survey and phylogenetic analysis of bovine respiratory coronavirus in Japan from 2016 to 2018. J. Vet. Med. Sci. 2020;82:726–730. doi: 10.1292/jvms.19-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill R., Mooney J., Connaghan E., Furphy C., Graham D.A. Patterns of detection of respiratory viruses in nasal swabs from calves in Ireland: a retrospective study. Vet. Rec. 2014;175:351. doi: 10.1136/vr.102574. [DOI] [PubMed] [Google Scholar]
- Oma V.S., Tråvén M., Alenius S., Myrmel M., Stokstad M. Bovine coronavirus in naturally and experimentally exposed calves; viral shedding and the potential for transmission. Virol. J. 2016;13:100. doi: 10.1186/s12985-016-0555-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J. Bovine respiratory coronavirus. Vet. Clin. N. Am. Food Anim. Pract. 2010;26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlender J., Bossert B., Buchholz U., Conzelmann K.K. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 2000;74:8234–8242. doi: 10.1128/jvi.74.18.8234-8242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotta K., Bonkowski K., Liebler-Tenorio E., Germon P., Rainard P., Hambruch N., Pfarrer C., Jacobsen I.D., Menge C. Permissiveness of bovine epithelial cells from lung, intestine, placenta and udder for infection with Coxiella burnetii. Vet. Res. 2017;48:23. doi: 10.1186/s13567-017-0430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudaryatma P.E., Nakamura K., Mekata H., Sekiguchi S., Kubo M., Kobayashi I., Subangkit M., Goto Y., Okabayashi T. Bovine respiratory syncytial virus infection enhances Pasteurella multocida adherence on respiratory epithelial cells. Vet. Microbiol. 2018;220:33–38. doi: 10.1016/j.vetmic.2018.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudaryatma P.E., Mekata H., Kubo M., Subangkit M., Goto Y., Okabayashi T. Co-infection of epithelial cells established from the upper and lower bovine respiratory tract with bovine respiratory syncytial virus and bacteria. Vet. Microbiol. 2019;235:80–85. doi: 10.1016/j.vetmic.2019.06.010. [DOI] [PubMed] [Google Scholar]
- Sudaryatma P.E., Saito A., Mekata H., Kubo M., Fahkrajang W., Mazimpaka E., Okabayashi T. Bovine respiratory syncytial virus enhances the adherence of Pasteurella multocida to bovine lower respiratory tract epithelial cells by upregulating the platelet-activating factor receptor. Front. Microbiol. 2020;11:1676. doi: 10.3389/fmicb.2020.01676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudaryatma P.E., Saito A., Mekata H., Kubo M., Fahkrajang W., Okabayashi T. Bovine respiratory syncytial virus decreased Pasteurella multocida adherence by downregulating the expression of intercellular adhesion molecule-1 on the surface of upper respiratory epithelial cells. Vet. Microbiol. 2020;246:108748. doi: 10.1016/j.vetmic.2020.108748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, H.M., upon reasonable request.